- Key Laboratory of Animal Epidemiology and Zoonosis, Ministry of Agriculture, National Animal Protozoa Laboratory, College of Veterinary Medicine, China Agricultural University, Beijing, China
Neospora caninum, of the Apicomplexa phylum, is a common cause of abortions in cattle and nervous system dysfunction in dogs. Rhoptry proteins of Apicomplexa play an important role in virulence. The objectives of this study were to study functions of NcROP5 in N. caninum by deleting the NcROP5 gene from the wild Nc-1 strain. We selected NcROP5 in ToxoDB and successfully constructed an NcROP5 gene-deleted vector, pTCR-NcROP5-CD KO. Then we screened the NcROP5 knockout strains (ΔNcROP5) at the gene, protein and transcription levels. Plaque assay, host cell invasion assay and intracellular proliferation test showed that the ΔNcROP5 strain had less plaque space, weakened invasion capacity and slower intracellular growth. Animal testing showed significantly lower cerebral load of ΔNcROP5 than the load of the Nc-1 strain, as well as a loss of virulence for the ΔNcROP5 strains. Phenotypic analyses using the label-free LC-MS/MS assay-based proteomic method and KEGG pathway enrichment analysis showed a reduction of NcGRA7 transcription and altered expression of multiple proteins including the apicomplexan family of binding proteins. The present study indicated that ROP5 is a key virulence factor in N. caninum in mice. The proteomic profiling of Nc-1 and ΔNcROP5 provided some data on differential proteins. These data provide a foundation for future research of protein functions in N. caninum.
Introduction
Neospora caninum is a common cause of abortions in cattle and nervous system dysfunction in dogs (Hall et al., 2005; Lyon, 2010). The parasite has become an international concern due to the connection of the parasite infection to abortions in dairy and beef cattle worldwide (Dubey, 1999). Antibodies to N. caninum have been detected in humans in Brazil, Korea, Northern Ireland, and the United States, although viable parasites have not been isolated (Dubey et al., 2007). Investigations are ongoing to determine whether the pathogen is a potential threat to human health.
Neospora caninum and Toxoplasmosis gondii belong to the same Apicomplexa family and share similar morphology, host range and clinical symptoms, although they appear to be divergent in their pathogenicity in mice (Lyon, 2010). A tachyzoite of the T. gondii RH strain can lead to death in a mouse (Saeij et al., 2006; Taylor et al., 2006), whereas the same outcome requires more than 106 tachyzoites of the N. caninum (Tao et al., 2014; Arranz-Solís et al., 2015). Rhoptry proteins (ROPs), including TgROP5, TgROP16, TgROP17, and TgROP18 are impotant virulence factors of T. gondii (TgROPs) (Yamamoto et al., 2009; Etheridge et al., 2014; Behnke et al., 2015; Shwab et al., 2015). Deletion of the TgROP5 gene, which forms a complex with TgROP18, results in a complete loss of virulence (Reese and Boothroyd, 2011). We hypothesized that the above genes had similar functions in N. caninum. Unexpectedly, we found that ROP18 in N. caninum was a pseudogene (Tao et al., 2014). We suspected that another rhoptry gene of N. caninum might play a crucial role in virulence. Transgenic strains of T. gondii and Plasmodium falciparum of Apicomplexa parasites have been widely used to study the functions of parasite proteins (Ghorbal et al., 2014; Hui et al., 2014; Shen et al., 2014; Hammoudi et al., 2015; Williams et al., 2015; Li et al., 2016). However, no laboratory strains and genome-editing techniques have been developed and no transgenic strains constructed for the study of protein functions of N. caninum. We sought to construct a ROP5-knockout strain of N. caninum and determine the function of ROP5 in this parasite.
Firstly, we constructed a recombinant plasmid containing the untranslated region of the NcROP5 gene and obtained a NcROP5-deficienct strain (ΔNcROP5). The NcROP5-deficienct parasite exhibited, weakened host invasion, proliferation and virulence, compared with the wild parent strain. The technique we developed and the new transgenic strain laid ground work for future research of this highly pathogenic parasite.
Materials and Methods
Ethics Statement
The experiments were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals, Ministry of Science and Technology, China. All experimental proceduresw ere approved by the Institutional Animal Care and Use Committee of China Agricultural University (under the certificate of Beijing Laboratory Animal employee ID: 18049). The mice were humanely euthanized by cervical dislocation after anesthetization by subcutaneous injection of atropine (0.02 mg/kg) when they were unable to reach food or water for more than 24 h and lost 20% body weight.
Cell Culture
Vero cells and human foreskin fibroblast (HFF) cells were purchased from the ATCC (Manassas, VA, USA) and serially passaged in our laboratories as previously described (Gaskell et al., 2009). The cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, MACGENE, Beijing, China) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco, USA) in a humidified incubator containing 5% CO2 at 37°C. The cells were passaged every 3–6 days by trypsinization.
Parasite Culture and Preparation
The N. caninum Nc-1 wild-type strain and the newly constructed NcROP5-deficient strain (described below) were cultured as tachyzoites by serial passages in HFF or vero cells as previously described (Gaskell et al., 2009). Briefly, the parasites were cultured in DMEM (pH 7.4) supplemented with L-glutamine, 2% heat-inactivated FBS, penicillin (100 U/mL) and streptomycin (100 μg/mL) (Sigma–Aldrich, St. Louis, MO, USA) at 37°C in an atmosphere with 5% CO2. The parasites were harvested by filtering through a 5.0 μm pore filter (Millipore, MA, USA), centrifuged, washed twice with cold phosphate-buffered saline (PBS), and centrifuged (2,000 rpm for 10 min each time) (Zhang et al., 2007).
Construction of the NcROP5-Deficient N. caninum Strain
The pTCR-CD plasmid was used at the Key Laboratory of Animal Parasitology (Beijing City, China) as previously described (Li et al., 2016). The plasmid contains the chloramphenicol resistance gene (CmR), red fluorescence protein gene (RFP), 5-fluorine cytosine gene (CodA), and ampicillin resistance gene (Amp). Nctublin promoter was used to drive the CmR-RFP fusion gene and CodA gene expression. The fragments of the NcROP5 5′ (2826 bp) and 3′ UTR regions (2297 bp) were amplified using the primer pairs F1/R1 and F2/R2 (Supplementary Table S1), which were designed based on the N. caninum Liverpool protein coding gene sequence1 (FR823393, Chromosome: XII). Primers F1, R1, F2, and R2 introduced KpnI, XhoI, XmaI, and SpeI sites, respectively (Supplementary Table S1). After amplification, the fragments were cloned into a pEASY-T-Blunt vector (TransGen Biotech Co., Ltd, Beijing). Then the DNA insert was sequenced (Beijing Sunbiotech Co. Ltd., China) and analyzed by blast in ToxoDB1 to confirm the authenticity of the cloned sequence. All alignments were performed using DNAMAN version v5.2.2 and the correct plasmids were double digested with KpnI and XhoI for the 5′ UTR and XmaI and SpeI for the 3′ UTR (NEB, USA). Then, the fragments were ligated to the pTCR-CD vector to produce the NcROP5 gene deleted vector, pTCR-NcROP5-CD KO.
All constructs were verified by sequencing (Beijing Sunbiotech Co. Ltd, China). After digestion with NotI (NEB, Beijing), the linearized pTCR-NcROP5-CD KO plasmids were purified using ethanol precipitation and then resuspended using cytomix (Hui et al., 2014). Nc-1 tachyzoites (1 × 107), mixed with 50 μg of the above plasmids, were transferred to a 0.4 mm gap cuvette and electroporated with 2 kV at 25 μFd and 50 Ω with the Gene Pulser Xcell electroporation system (Bio-Rad, USA). Then the parasites cultured in an atmosphere containing 5% CO2 at 37°C for 24 h prior to the addition of chloramphenicol (20 μM) and 5-fluorine cytosine (40 μM). The parasites were cultured consecutively to the 8th generation and then screened to confirm the purity of the selected strains.
For complementation analysis, pDMG plasmid (Hui et al., 2014; Tao et al., 2014; Li et al., 2016) was used to express NcROP5 in the ΔNcROP5 strain. The complete coding sequence of NcROP5 was amplified using primers F3/R3 (Supplementary Table S1) that appended the flanking EcoRV and AvrII restriction endonuclease sites. The amplification products were introduced into the same sites of a modified pDMG plasmid with GFP replaced by HA (Li et al., 2016). The pDMG-NcROP5-HA vector was electroporated into the ΔNcROP5 strain. The transgenic parasites were grown under pyrimethamine selection pressure. The selected NcROP5 complementary strain, named as iΔNcROP5, was identified by western blotting and immunofluorescence assays (IFAs).
Validation of the ΔNcROP5 Strain and the Complemented Strain
PCR
To screen the NcROP5 knockout parasites (ΔNcROP5), we designed primers targeting the NcROP5 fragment (ToxoDB: NCLIV_060730, 800 bp) to detect if the gene could be amplified from different clones. Polymerase chain reaction (PCR) was conducted using the T100TM Thermal Cycler (Bio-Rad, USA) with PCR SuperMix (TransGen, China) following manufacturer’s instructions. The primer sequences were F4/R4 (Supplementary Table S1). The Nc5 gene (336 bp) served as the internal reference with the primer pair F5/R5 (Supplementary Table S1). The PCR conditions were as follows: 95°C for 5 min, 30 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 1 min, and 72°C for 10 min. The PCR product was identified using electrophoresis.
Western Blot
Purified parasites (Nc-1, three ΔNcROP5 clones and one iΔNcROP5) were lysed using RIPA buffer (Beyotime, Beijing) supplemented with a cocktail of protease inhibitors (Sigma, USA). The lysates were resolved on a 10% (w/v) SDS-PAGE gel. After electrophoresis, separated strips were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, MA, USA) together with a visible pre-stained protein marker (TransGen Biotech Co., Ltd, Beijing). The membranes were blocked with 5% (w/v) skim milk (BD Difco, USA) in PBS for 1 h at 37°C and then incubated with the mouse anti-rNcROP5 antibody (Tao et al., 2014) (National Animal Protozoa Laboratory in China Agricultural University, 1:500) for ΔNcROP5 and the mouse anti-HA antibody (Sigma–Aldrich, 1:500) for iΔNcROP5. The N. caninum F-actin subunit beta (NcActin, National Animal Protozoa Laboratory in China Agricultural University) was used as a loading control and was incubated with the rabbit anti-rNcActin antibody (1:2000). After five washes with PBST (1% Tween-20), the membranes were incubated for 1 h with a horseradish peroxidase (HRP)-labeled goat anti-mouse IgG (H+L) secondary antibody (Sigma, USA) diluted 1:5000 and an HRP-labeled goat anti-mouse IgG secondary antibody (Sigma, USA) diluted 1:10,000 in PBS with 5% BSA. Finally, the reactive bands were visualized using enhanced chemiluminescence reagents (CoWin Biotech Co., Ltd, Beijing).
Immunofluorescence Assay (IFA)
Immunofluorescence Assay was used to detect the NcROP5 subcellular localization as previously described (Li et al., 2016). Appropriate numbers of parasites were coated on glass coverslips in 12-well plates and then fixed in 4% paraformaldehyde, permeabilized with 0.25% Triton X-100 and blocked with 3% BSA. Subsequently, the cells were incubated with a mouse anti-HA monoclonal antibody, mouse anti-NcROP5 and rabbit anti-NcSRS2 antibody (National Animal Protozoa Laboratory in China Agricultural University) followed by FITC-conjugated goat-anti mouse IgG (H+L), Cy3-conjugated goat-anti mouse IgG (H+L), and Cy3-conjugated goat-anti rabbit IgG (H+L) (Sigma, USA). The nuclear DNA was stained with Hoechst33258 (Sigma, USA). Extracellular iΔNcROP5 parasites were identified by IFA. The images were obtained using a Leica confocal microscope system (Leica, TCS SP52, Germany).
Invasion Assays
Cell invasion assays were performed using the following protocol (Tao et al., 2014). Freshly egressed parasites (Nc-1, ΔNcROP5 and iΔNcROP5 strains) were allowed to settle onto the HFF monolayers on 12-well plates for 30 min at 37°C prior to incubation. Extracellular parasites were removed by washing three times with PBS. The cells were incubated for 20 h prior to IFA using anti-NcSRS2 primary and FITC-conjugated secondary antibodies (1:100 dilution). The number of vacuoles representing successful invasion parasites was counted under a fluorescence microscope. A total of three replicates (each clone in duplicate) were performed.
Plaque Assay
Plaque assays were set up immediately following the initiation of the invasion assays using the same parasite suspensions. Nc-GFP strain (kindly provided by Professor Xuenan Xuan, Obihiro University of Agriculture and Veterinary Medicine, Japan), as a null vector constructing group, which is a transgenic parasite by transfecting Nc-1 wild-type strain with the pDMG plasmid, was propagated as tachyzoites by serial passages in HFF cells as previously described (Gaskell et al., 2009). The plaque assay was performed as previously described (Hui et al., 2014). Five hundred parasites were added to each well of a 6-well plate containing confluent host cell HFF monolayers. The plate was incubated for 7 days in a 37°C incubator with 5% CO2, fixed with 4% paraformaldehyde for 15 min and stained with 2% crystal violet for 15 min at room temperature. The stained wells were washed with deionized water, air dried and visualized using a fluorescence microscope (IX71, Olympus, Japan). The plaque area measurement was performed as previously described (Roos et al., 1994).
Replication Assay
The Nc-1, ΔNcROP5 and iΔNcROP5 parasites were harvested and counted. Five-hundred parasites were added to confiuent vero and HFF cells on 25 mm coverslips. The coverslips were allowed to sit at 37°C for 30 min in DMEM with 20% FBS and then washed three times with PBS. The coverslips were incubated at 37°C for 24 h in DMEM with 2% FBS and then fixed with 4% paraformaldehyde for 15 min. IFAs were performed using anti-NcSRS2 primary (Tao et al., 2014) and FITC-conjugated secondary antibodies (Proteintech, USA). The number of parasites per vacuole was counted under a fluorescence microscope. The experiments were performed in triplicates and repeated three times, and 200 vacuoles were counted per coverslip.
Egress Assay
Neospora caninum tachyzoites, as well as T. gondii, must form parasitophorous vacuole (PV) after invading the host cell (Tilley et al., 1997). They proliferate in the PV and then egress out of PV. Then, the tachyzoites invades other cells again (Soldati and Meissner, 2004). In our experiments, parasites were inoculated in 12-well plates containing prepared HFF cells for 30 h. Egress was stimulated with 10 μM of the Ca2+ ionophore A23187 from Streptomyces chartreusensis (Sigma, USA) for 2 min at 37°C prior to fixation with paraformaldehyde (Williams et al., 2015). The IFA was performed as described above using the mouse anti-NcSRS2 primary and FITC-conjugated secondary molecules. The average number of ruptured vacuoles was determined by counting a minimum of 100 vacuoles per slide and three slides for each experiment. Three independent experiments were performed.
Gene Expression Assay
Total RNA was extracted from 1 × 107 tachyzoites of the Nc-1 wild-type strain and three ΔNcROP5 clones with TRIzol reagent (Invitrogen, USA). cDNA was synthesized using the EasyScript First-Strand cDNA Synthesis SuperMix kit (TransGen, China). Specific primers were designed using Primer Premier 5.0 (Hui et al., 2014), including primers for rhoptry necks (RON2 and RON4), rhoptrys (ROP4, ROP5, ROP7, ROP16, and ROP17), dense granules (GRA2, GRA6, and GRA7) and the endogenous reference gene NcActin (Supplementary Table S1). The specificity of these primers was evaluated using conventional quantitative real-time PCR (qRT-PCR). The qRT-PCR was conducted using the ABI Prism 7500 System (Biosystems Inc., USA) with SYBR Green II (Takara Biotechnology, Dalian, Co., Ltd) following manufacturer’s instructions. The resulting RNA concentrations were normalized using Ncactin (Wang et al., 2017), and the relative expression levels of the target genes were analyzed using the ABI Prism 7500 software v2.0.5 (Biosystems Inc., USA). The RT-PCR conditions were as follows: 94°C for 5 s, 40 cycles of 94°C for 5 s and 60°C for 30 s. The relative expression of genes was calculated using the 2-ΔΔCt method and standard deviation was calculated from three replicates (Cárdenas-Mondragón et al., 2016; Gagnaire et al., 2016).
Label-Free LC-MS/MS Assay
The N. caninum Nc-1 and ΔNcROP5 strains were cultured as tachyzoites in HFF cells as previously described (Gaskell et al., 2009) for 96 h. For each parasite, a total of 5 × 107 tachyzoites were harvested and isolated by washing in cold PBS, centrifugation, resuspension in cold PBS, syringing three times through a 27-gaure needle, filtering through a 5.0 μm pore filter (Millipore, MA, USA), washing three times with PBS, and finally centrifugation at 1800 rpm for 10 min (Tao et al., 2014). Then the parasites were ultrasonically lysed in protein lysis buffer (8.0 M urea, 100 mM pH 8.0 Tris-HCl, 1×cocktail) using an ultrasonic liquid processor (Sonic & Materials INC., USA). The lysates were sonicated five times for 2 s each at 30% amplitude. Then the sonicated lysates were centrifuged for 30 min at 4°C at 12000 g and obtained the supernatants (Deutsch et al., 2014). After quantitative assay by the Bradford method (Bradford Protein Assay kit) (Deutsch et al., 2014), 60 μg protein was reduced, alkylated and trypsin digested. Each sample was separated by a high performance liquid chromatography (HPLC) system (Ultimate 3000, Thermo Scientific), and analyzed by mass spectrometry (Q-Exactive HF, Thermo Scientific). The spectra were identified and analyzed by the Mascot software (Thermo Scientific, USA) and Proteome discoverer 2.1 (Thermo Scientific, USA) on the basis of database listed in Supplementary Table S2 (Sandberg et al., 2012).
Mouse Virulence Assay
To compare the virulence of the ΔNcROP5 strain with the parental Nc-1 strain and complemented iΔNcROP5 strain, tachyzoites (3 × 106) were injected i.p. into 6-week-old female BALB/c mice (five mice per strain), which were purchased from the Laboratory Animal Center of Academy of Military Medical Sciences (Beijing, China). Survival was monitored for 30 days. To examine the cerebral load of parasites, tachyzoites were injected i.p. into 6-week-old female BALB/c mice at a dose of 1 × 106 (five mice). The mouse brains were examined for parasites on the 30th day (Pan et al., 2014).
Statistical Analysis
The statistical analysis of all of the data was performed using GraphPad Prism 5 v. 5.01 (San Diego, CA, USA). The results were expressed as mean ± SD and evaluated by non-parametric tests. Values of P < 0.05 and P < 0.01 were considered statistically significant and very significant, respectively.
Results
Successful Construction of the NcROP5 Knockout Strain
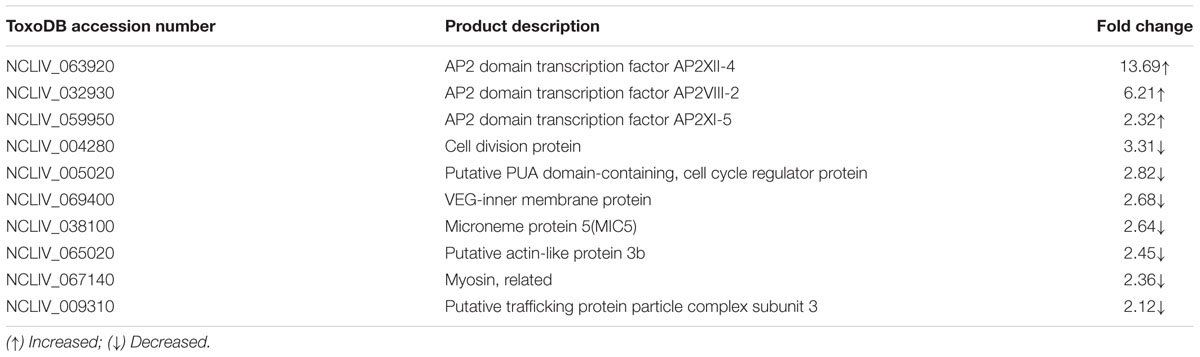
Using the knockout plasmid, pTCR-CD, we successfully constructed the NcROP5 knockout strain. To use the plasmid for deleting a gene, we ligated the 5′ UTR and 3′ UTR regions of the target gene. Then, we analyzed the genome of N. caninum Liverpool strain in ToxoDB2 and identified the sequences with high identity to TgROP5, which were NcLIV_060730, NcLIV_060740, and NcLIV_060741. Compared to the tandem cluster of polymorphic alleles of TgROP5 (TgROP5A, TgROP5B, and TgROP5C) (Behnke et al., 2011), we believed that NcROP5 would contain NcROP5A, NcROP5B, and NcROP5C, coded by the above three genes, respectively. Homology of nucleotide and protein sequence between NcLIV_060730 and TgROP5A were 52.72 and 76.76%, respectively. Since TgROP5 is an important virulence factor of T. gondii (Behnke et al., 2012), we suspected that NcROP5 might play an active role in N. caninum virulence. To examine the physiological role of ROP5 in the Nc-1 strain, NcROP5 KO parasites were generated by targeted gene disruption. The non-coding region (UTR) sequences of NcROP5, NcROP5 5′ UTR and 3′ UTR, were cloned (Supplementary Figure S1A) and ligated to the pTCR-CD vector, forming pTCR-NcROP5-CD KO (Supplementary Figure S1B). The plasmid was identified by restriction enzyme digestion (Supplementary Figure S1A) and sequenced and then transfected into Nc-1. Theoretically, the entire ROP5 coding region, containing NcROP5A, NcROP5B, and NcROP5C, could be replaced with the CmR and RFP genes (Supplementary Figure S1C). RFP was expressed at normal levels in the suspected ROP5 mutants (Figure 1A). Following CmR selection of the RFP and 5-fluorine cytosine-negative parasites, we isolated three mutants out of 218 clones.
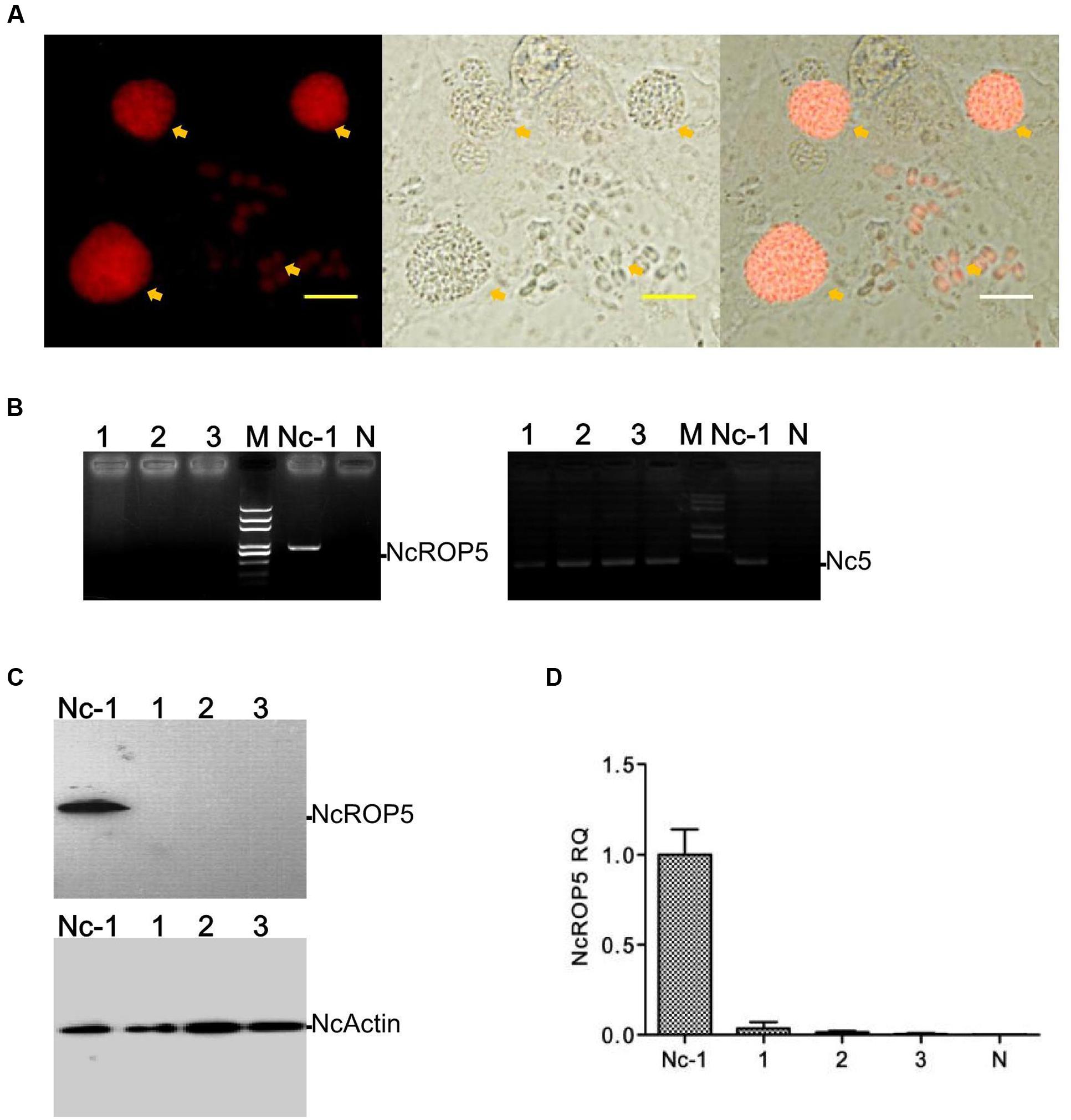
FIGURE 1. Identification of the ΔNcROP5 strains. (A) The NcROP5-disrupted (ΔNcROP5) strain was identified using a fluorescent inverted microscope. The positive parasites showed red fluorescent (Yellow arrow). (B) ORF-specific PCR confirming the deletion of the NcROP5 gene locus. An expected 800 bp band of the ROP5 gene from the parental Nc-1 strain was amplified compared to the absence of this band in the three ΔNcROP5 clones (1, 2, and 3) and in the plasmid DNA (negative control, N). Nc5 gene served as a Neospora caninum specific gene. (C) Western blots with NcROP5 anti-mouse antibodies with NcROP5 of 61 kDa in the parental Nc-1 strain. No NcROP5 protein was detected in the ΔNcROP5 monoclonal strains (1, 2, and 3). N. caninum F-actin subunit beta (NcActin) was used as a loading control. (D) Quantitative RT-PCR was used to analyze the transcription levels of the NcROP5 gene in the ΔNcROP5 clones (1, 2, and 3) and compared with the parental strain (Nc-1). Each bar indicates the relative quantity (RQ) ±SD. Scale bar, 10 μm.
To generate the ROP5 gene-deficient Nc-1 strain, the NcROP5 gene was targeted and replaced by CmR-RFP. PCR was used to confirm the deletion of the NcROP5 gene. NcROP5 gene amplification was negative in the parasite genomes (Figure 1B). NcROP5 deficiency was confirmed using western blotting (Figure 1C) and RT-PCR (Figure 1D). We also successfully generated a strain with complete knockout of the NcROP5 gene. These results suggested that NcROP5 was not an essential, surviving gene in the parasites.
Successful Construction of the NcROP5 Complementary Strain
To verify the biological role of NcROP5, we obtained the complete coding sequence of NcROP5A without the termination codon TAA (NcLIV_060730, 1647 bp, Supplementary Figure S2A) and generated the complementary plasmid pDMG-NcROP5-HA. The expression of NcROP5 with an HA-tag and stop codon TAA at the C-terminus was driven under the control of the NcGRA1 promoter (Supplementary Figure S2B). We correctly identified the plasmid by restriction enzyme digestion and DNA sequencing (Supplementary Figure S2A), and then electroporated it into one of the ΔNcROP5 strains. After repeated passage, we selected strains that stably expressed NcROP5 (named as iΔNcROP5). NcROP5 expression in the complementary strain based on WB and RT-PCR was comparable to the expression of the wild strain (Figures 2A,B). IFA was used to analyze the subcellular localization of NcROP5 in the iΔNcROP5 strains using a mouse anti-HA monoclonal antibody. NcROP5 was co-localized with HA at the anterior end of the extracellular parasites (Figure 2C).
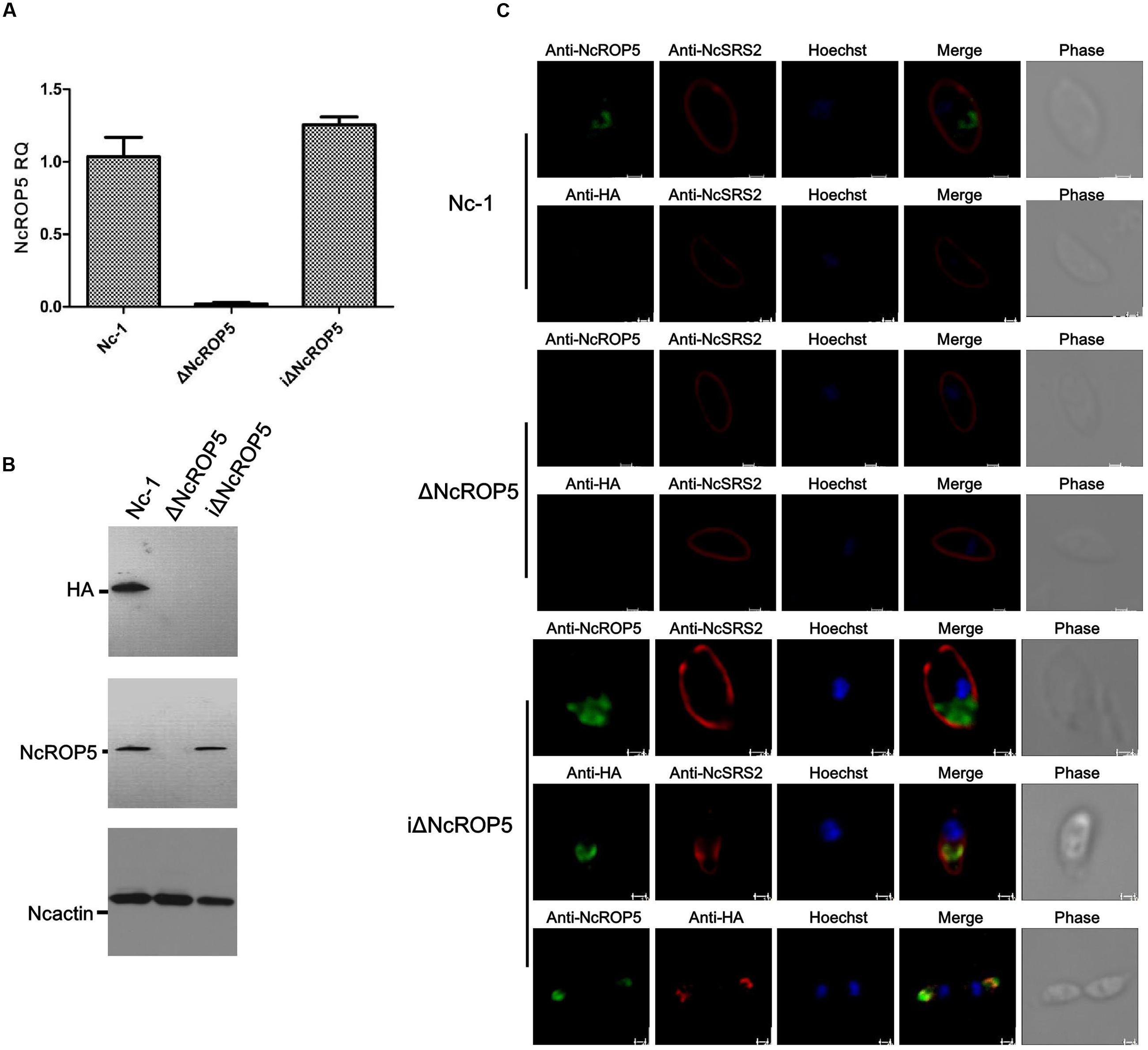
FIGURE 2. Identification of ΔNcROP5 complementary strain, iΔNcROP5. (A) RT-PCR analysis of NcROP5 gene transcription. (B) Western blotting analysis of Nc-1, ΔNcROP5 and iΔNcROP5 tachyzoites. The expected band was detected by the anti-HA monoclonal antibody and anti-rNcROP5 polyclonal antibody. NcActin was used as the control. (C) IFA analysis of iΔNcROP5 tachyzoites. The HA-tagged ROP5 and ROP5 proteins (green) were localized at the anterior end of the parasites, and HA co-localized with ROP5. The shapes of parasites were stained with anti-NcSRS2 (red), and the nuclear DNA was stained with Hoechst (blue). Scale bar, 1.0 μm.
The NcROP5 Gene Is Critical in the Invasion and Proliferation of N. caninum
Plaque formation measures the viability of parasites in cell culture and reflects parasite motility on the surface of the host cell layer, invasion, intracellular growth and egress (Plattner et al., 2008). We assessed the ΔNcROP5 strain using the plaque assay. The ΔNcROP5 strain formed noticeably smaller and considerably fewer plaques than the parental strain and the complementary strain (Figures 3A,B). The ΔNcROP5 strain showed significant weakening of host cell invasion (Figure 3C). Intracellular replication assays were performed to investigate if deleting ROP5 in the Nc-1 strain weakened the ability of the parasite to proliferate in host cells. Parasites that invaded host cells 24 h after inoculation were analyzed for intracellular growth by counting the number of parasites per vacuole. Due to the lack of synchronization of host cell invasion, the intracellular vacuoles contained 1, 2, 4, or 8 parasites. However, intracellular replication of ΔNcROP5 in Vero and HFF cells was lower than that of the wild type parasite (Nc-1). As shown in Figure 3D, the distribution of the eight parasites per vacuole was significantly lower for the ROP5 knockout strain than that for Nc-1, Nc-GFP, and iΔNcROP5, indicating that the intracellular replication of the ΔNcROP5 strain was slower than that of the other three strains. However, there was no significant difference in egress between the four strains (Figure 3E). These findings indicated that NcROP5 expression in N. caninum is a critical determinant of parasite phenotype, including host cell invasion and proliferation.
FIGURE 3. Deletion of the NcROP5 gene affects the Nc-1 strain phenotype. (A,B) Plaque assays showed very low plaque formation of the ΔNcROP5 strain compared with the strains harboring NcROP5. The assay was performed three times independently, and the plaque area was quantified from three independent experiments. At least 10 fields were quantified per strain in every experiment. Each bar indicates mean ± SD; “Pi” indicates pixels of each plaque. (C) The ΔNcROP5 strain possess significantly reduced cell invasion compared with the Nc-1, Nc-GFP, and iΔNcROP5 strains. The number of parasite vacuoles was counted after immunofluorescent staining with anti-NcSRS2 antibodies in the cell invasion assay. (D) Intracellular replication assay showed that ΔNcROP5 strain proliferated more slowly than the other three strains. The IFA was performed with anti-NcSRS2 antibodies. At least 100 vacuoles were examined for each per strain. (E) Egress assay showed no difference between strains indicating that NcROP5 is not involved in parasite egress. ∗P < 0.01 and ∗∗P < 0.001 (Non-parametric tests). Scale bar, 10 μm.
Identification of Peptides and Proteins in Nc-1 and ΔNcROP5
To analyze the cause of the ΔNcROP5 phenotype changes, we compared protein expression levels in Nc-1 and ΔNcROP5. Nc-1 and ΔNcROP5 tachyzoites were analyzed by label-free LC-MS/MS for quantification of peptides. A total of 18,809 peptides with FDR ≤ 0.01 were identified, and a total of 1149 proteins with expression difference ≥ 1.2-fold between the two strains were functionally annotated according to the N. caninum genome annotation in the ToxoDB database (Supplementary Figure S3A and Table S3). The three main annotation types were biological process, cellular component and molecular function (Supplementary Figures S3B–D). The differentially expressed proteins were classified into 31 functional groups (Supplementary Figure S3E), with biological process possessing 12 GO (gene ontology annotation) terms, cellular component 10 GO terms, and molecular function 9 GO terms. Cellular process and metabolic process were major biological processes (32 and 31%, respectively), cell part and cell are major cellular components (25% each), and binding and catalytic activities were major molecular functions (53 and 37%, respectively).
Molecularly Enriched Pathways Associated with Differentially Expressed Proteins in Nc-1 and ΔNcROP5
To further identify biological pathways of the differentially expressed proteins, the number of proteins expressed in Nc-1 and ΔNcROP5 and differences between the two strains are shown in Supplementary Figure S4A. Demographic analysis demonstrated that 76 proteins were increased and 89 proteins were decreased with a 2.0-fold change; 198 proteins were increased and 252 proteins were decreased with a 1.5-fold change; and 477 proteins were increased and 671 proteins were decreased with a 1.2-fold change. We predicted that 477 increased proteins participated in 92 biological pathways and 671 decreased proteins participated in 88 biological pathways by KEGG pathway analysis3. The differentially expressed proteins were matched with the proteins annotated in the KEGG pathway database, and they were compared to determine their involvement in the KEGG pathway. We found that five KEGG pathways were clearly affected (P ≤ 0.05) (Supplementary Figure S4B and Table S4), followed by mismatch repair, SNARE interactions in vesicular transport, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, ABC transporters and RNA degradation.
To better comprehend the causes of phenotypic differences, we analyzed representative differentially expressed proteins and uncommented proteins by gene and sequence alignment. Blast analysis (ToxoDB-28_Neospora caninum_Annotated Proteins. fasta) showed that increased proteins included apetela 2 (AP2) family proteins (NCLIV_063920, NCLIV_032930, and NCLIV_059950) and dense granule protein (NCLIV_004260), and decreased proteins included cell division and proliferation proteins (NCLIV_004280, NCLIV_005020, NCLIV_069400, NCLIV_065020, NCLIV_067140, and NCLIV_009310), and secreted proteins (NCLIV_038100) (Table 1).
Rhoptry neck proteins (RONs), ROPs and dense granule proteins (GRAs) were related to invasion, proliferation, and formation of PV (Sinai and Joiner, 2001; Dubremetz and Lebrun, 2012; Nolan et al., 2015). To detect whether these genes were affected by NcROP5, we selected RON2, RON4, ROP4, ROP7, ROP16, ROP17, GRA2, GRA6, and GRA7 genes to evaluate their mRNA expression. Quantitative RT-PCR (qRT-PCR) analysis of 1 × 107 Nc-1, ΔNcROP5 and iΔNcROP5 tachyzoites was conducted to determine the potential involvement of NcROP5 in transcriptional regulation of other related genes. The qRT-PCR analysis indicated that RON2 (R2 = 0.9838, slope = -3.392), RON4 (R2 = 0.9849, slope = -3.425), ROP4 (R2 = 0.9961, slope = -3.381), ROP7 (R2 = 0.9918, slope = -3.466), ROP16 (R2 = 0.9907, slope = -3.372), ROP17 (R2 = 0.9899, slope = -3.373), GRA2 (R2 = 0.9911, slope = -3.407), and GRA6 (R2 = 0.9951, slope = -3.475) were not altered at the transcription level in ΔNcROP5. Notably, NcGRA7 (R2 = 0.9879, slope = -3.331) expression was significantly down-regulated by approximately twofold in ΔNcROP5 (Figure 4). All genes have similar amplification efficiency to the reference gene Ncactin (R2 = 0.9912, slope = -3.392). The results suggested that NcROP5 expression might affect the expression of other genes, including NcGRA7.
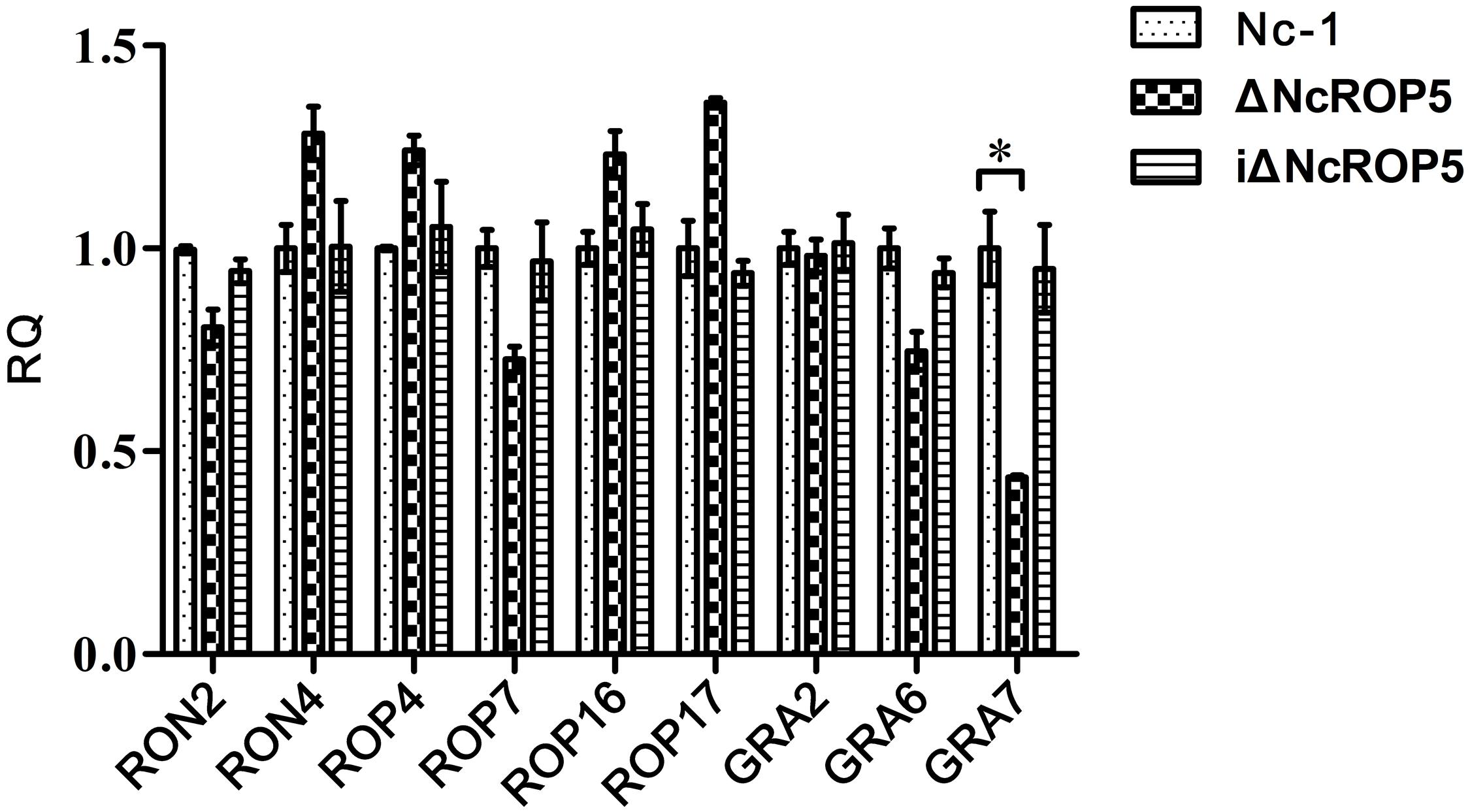
FIGURE 4. The transcription of differential genes in Nc-1, ΔNcROP5 and iΔNcROP5. Quantitative RT-PCR analyses of the transcription levels of the indicated genes in ΔNcROP5 parasites compared with the parental strain (Nc-1) and the complementary strain (iΔNcROP5). Each bar indicates the relative quantity (RQ) ±SD. The RQs of the ΔNcROP5 and iΔNcROP5 parasites were expressed as fold of the expression by Nc-1, which was artificially set as 1. The data presented are representative of three independent experiments each performed in duplicate. ∗P < 0.05.
Knockout of NcROP5 Reduced Parasite Virulence
To investigate if NcROP5 is necessary for Nc-1 virulence, mice were infected intraperitoneally with 1 × 106 Nc-1, Nc-GFP, ΔNcROP5, or iΔNcROP5 tachyzoites, followed by observation of parasite load in the brain and survival rate. Significant differences were observed between the ΔNcROP5 group and the other groups (Nc-1, Nc-GFP and iΔNcROP5). Signs of illness such as rough coats, inactivity, or nervous system signs (hind limb weakness, head tilting, or walking in circles) were observed 2 or 5 days before death. In addition, rough hair coats and reduced activity were observed in 2 Nc-1-infected mice from day 28 post-infection, 1 Nc-GFP-infected mouse from day 29 post-infection and 1 iΔNcROP5-infected animal from day 26 post-infection. There were no clinical signs in ΔNcROP5-infected mice during the 30 days observation period. The cerebral parasite load of the ΔNcROP5 group was approximately one-fourth of the load in the Nc-1 and Nc-GFP groups. The iΔNcROP5 group had the most parasites in the brain (Figure 5A). Consistent with the low parasite load in the brain, the survival rate of the ΔNcROP5-infected animals was higher than that of the other groups. Actually, all ΔNcROP5-infected mice survived, compared with the survival rate of 40–60% for the other groups (Figure 5B). These results demonstrate a requirement of NcROP5 for Nc-1 virulence.
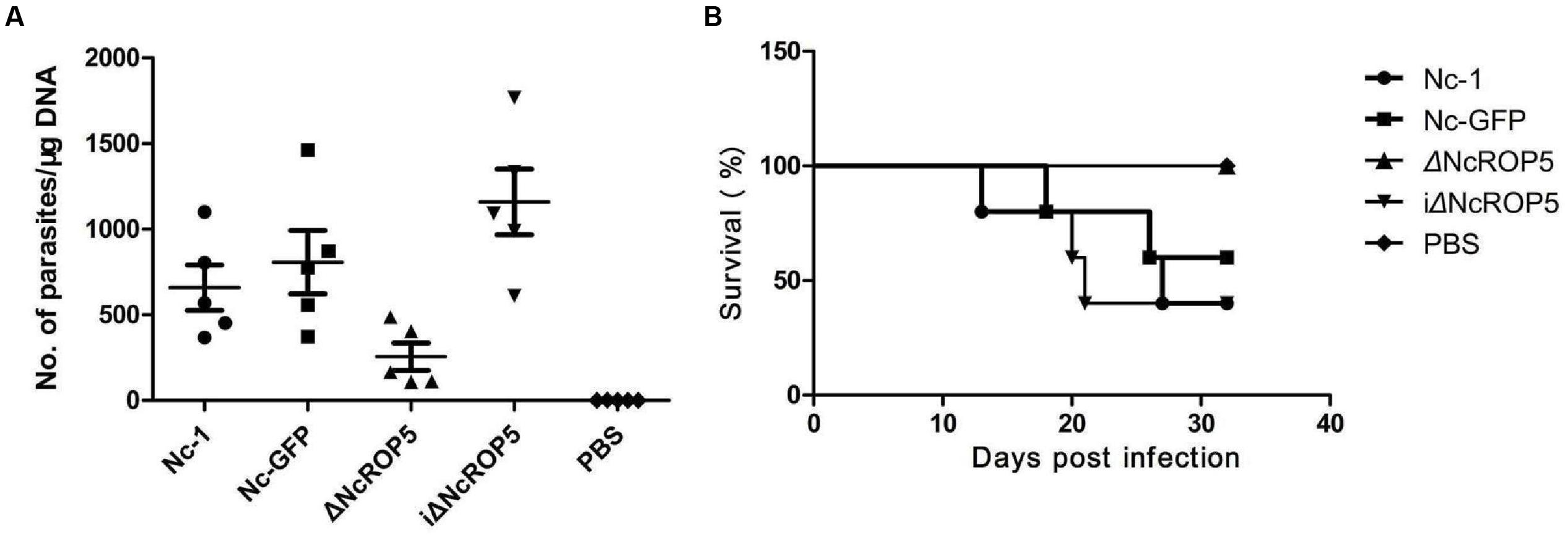
FIGURE 5. The ROP5 gene is critical in N. caninum virulence. (A) Brain parasite load of ΔNcROP5 was significantly reduced compared to Nc-1, Nc-GFP, and iΔNcROP5. BALB/c mice (n = 5) were injected i.p. with 1 × 106 tachyzoites and the number of parasites in the brain was determined 30 days after parasite inoculation. (B) All ΔNcROP5-inoculated mice survived compared with 40–60% survival of Nc-1, Nc-GFP and iΔNcROP5-inoculated animals. BALB/c mice (n = 5) were infected with 3 × 106 tachyzoites and monitored for 30 days. Two independent experiments were performed.
Discussion
Neospora caninum and T. gondii are obligate intracellular parasites that belong to the phylum Apicomplexa, which includes Cryptosporidium, Eimeria, Plasmodium, and Theileria (Hajj et al., 2007; Plattner et al., 2008). Toxoplasma is a widely used model organism for studying protein functions in Apicomplexa (Shen et al., 2014). In T. gondii, the methods of gene deletion and protein expression regulation have been developed to study protein functions and provided valuable information (Payne et al., 2011). However, related research reports about functions of proteins in N. caninum are limited to epidemiological studies, the expression and localization of cloned genes, and immunogenicity (Liu et al., 2013; Li et al., 2014; Hecker et al., 2015; Pastor-Fernández et al., 2016). The overexpression of some genes and addition of labeling with biotin in N. caninum have been reported (Tao et al., 2014; Mota et al., 2016). Few reports on N. caninum have investigated the loss of one or more genes and their effects on gene functions (Mota et al., 2016).
The genome-editing technique is very important and has advanced researches on the roles of gene functions in some organisms. We constructed a pTCR-CD knockout plasmid and used the plasmid to edit genes in N. caninum successfully with the example of the NcROP5 gene. This step was a very positive development for research on N. caninum protein functions. The roles of NcROP5 were partially deduced using the strain lacking the NcROP5 gene based on the PCR assay results, gene transcription levels and protein expression levels. The study provides a reference method and lays the foundation for further studies on NcROP5 functions and other tachyzoite proteins. We selected three ΔNcROP5 monoclonal strains and observed their phenotypes. The phenotypes of the ΔNcROP5 strains were similar, with a weakened invasion capacity, slower intracellular proliferation rate and smaller sized plaques. However, there were significant differences compared to the Nc-1, Nc-GFP, and ΔNcROP5 complement strains. The slight phenotypic differences between iΔNcROP5 and the reference strains might be due to the efficient promoter NcGRA1 of pDMG-NcROP5-HA, while ROP5 is encoded in a multicopy gene in the reference strain Nc-1. The results indicated that the NcROP5 gene played an important role in N. caninum virulence. The phenotypic changes in the ΔNcROP5 strains were probably caused by the abnormal expression of other genes associated with NcROP5.
Proteomics analysis was performed to explore the proteins expression characteristics of Nc-1 and ΔNcROP5, but regrettably there were few interesting proteins secreted by microneme, rhoptry, or dense granule. GRA7 transcription level was reduced in ΔNcROP5 parasites, and for the reason that qRT-PCR was more sensitive than label-free analysis. In this part, we noticed some mysterious proteins, AP2 protein family. The apicomplexan AP2 (ApiAP2) family of binding proteins might be a major class of transcriptional regulators in Apicomplexan (Radke et al., 2013). There are 68 ApiAP2-encoding genes predicted in the Toxoplasma genome (Altschul et al., 2010). We crudely inferred the functions of the genes according to other Apicomplexan parasites, because they have not been fully studied in N. caninum. In T. gondii, AP2XI-5 targets more than 300 gene promoters and actively controls the transcription of these genes. AP2XI-5 was reported to be enriched at numerous active promoters, especially crucial virulence factors such as the rhoptry and microneme proteins (Walker et al., 2013b). In addition, TgAP2XI-4 regulates bradyzoite gene expression during parasite differentiation and cyst formation (Walker et al., 2013a), and TgAP2XI-3 dampens proliferation of parasites (Lai et al., 2012). In our study, the expression levels of AP2VIII-2, AP2XII-4, and AP2XII-5 were increased and AP2VIII-3 was not detected in the ΔNcROP5 group. These findings might be related to the loss of NcROP5 or abnormal expression of virulence factors NcROPs, MICs, GRAs and some metabolic enzymes as result of NcROP5 deletion. Meanwhile expression of some proteins, such as actin, myosin, and cell division protein, was decreased in the ΔNcROP5 strain, consistent with the slow proliferation of ΔNcROP5. The lower expression of microneme protein 5 (MIC5) was one possible reason for the weaker invasion capacity of ΔNcROP5, because MIC5 as a secreted protein modulates parasite adhesion and invasion (Brydges et al., 2008; Gaji et al., 2011).
Toxoplasmosis gondii strains are primarily divided into three types based on genomic analysis (type I, type II, and type III) (Grigg et al., 2001; Su et al., 2003). Previous studies identified a closely related ROP5 gene of T. gondii that was absolutely critical for pathogenesis in mice; the deletion of this gene resulted in a complete loss of virulence in mice even following infection with 106 parasites (Reese and Boothroyd, 2011). For N. caninum, various authors have described evidence of different toxicity among isolates. They demonstrated Nc-liv and Nc-Spain 7 had a higher virulence than Nc-1 (Regidorcerrillo et al., 2010; Caspe et al., 2012), and Nc-Spain 2H, Nc-Spain 3H, Nc-Spain 5H, Nc-Spain 6, Nc-Spain 7, and Nc-Spain 9 isolates showed different parasite burdens and brain lesions during the late phase of infection (Pereira et al., 2010). Although among N. caninum isolates, Nc-1 is less virulent in cattle than other isolates, it still leads to nervous system deficiency and deaths (Pan et al., 2014). In our study, the pathogenicity of the ΔNcROP5 strain from Nc-1 was greatly decreased, similar to TgROP5. The phylogenetic tree was showed that NcROP5 is closely related to TgROP5 (Ghorbal et al., 2014). ROP5 likely has similar functions in Apicomplexa and mediates parasite virulence (Reese and Boothroyd, 2011; Walzer et al., 2013). In Toxoplasma, ROP5, ROP18, and GRA7 form a complex that plays critical roles in parasite virulence (Hermanns et al., 2016). ROP5 and ROP17 were also reported to control T. gondii RH strain virulence through the complex (Etheridge et al., 2014). However, ROP18 has been proven to be a pseudogene and not a virulence factor in N. caninum (Reid et al., 2012). Surprisingly, the loss of NcROP5 led to a reduction in NcGRA7 transcription in our study. In contrast, TgGRA7 bound the host cell immunity-related GTPase A6 and enhanced its polymerization, rapid turnover, and eventual disassembly (Alaganan et al., 2014). TgGRA7 was considered a virulence factor, which was in contrast to the NcGRA7 immunogenicity analysis (Vemulapalli et al., 2007; Alaganan et al., 2014). Therefore, further research is needed to clarify the association between NcROP5, NcROP17, and NcGRA7, although we have defined NcROP5 as a factor that affects N. caninum virulence in mice.
Conclusion
Our findings here highlight the role of ROP5 as an important virulence factor in N. caninum and may contribute to a better understanding of the possible mechanisms underlying the interactions between parasites and their hosts.
Author Contributions
Conceived and designed the experiments: LM, JL, and QL. Performed the experiments: LM, ML, and YF. Analyzed the data: LM, XZ, and QL. Contributed reagents/materials/analysis tools: QL and JL. Wrote the paper: LM and QL.
Funding
This study was supported by the National Key Basic Research Program (973 program) of China (Grant No. 2015CB150300), Beijing Municipal Natural Science Foundation (Grant No. 6131001) and Earmarked fund for Modern Agro-industry Technology Research System (CARS-37).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Professor Silvia N. J. Moreno (University of Georgia, USA) for kindly providing the pTCY vector. We thank Professor Xuenan Xuan (Obihiro University of Agriculture and Veterinary Medicine, Japan) for kindly providing the Nc-GFP strain. We are grateful to Dr. Jin Zhu (Therapeutic Goods Administration, Australia) for his assistance in the preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00370/full#supplementary-material
FIGURE S1 | Construction of pTCR-NcROP5-CD KO. (A) Products of NcROP5 5′ UTR and 3′ UTR by PCR and identification of pTCR-NcROP5-CD KO by enzyme digestion. 1: NcROP5 5′ UTR; 2: NcROP5 3′ UTR; 3, 5: The knockout plasmid pTCR-NcROP5-CD KO; 4: Double enzyme restriction of pTCR-NcROP5-CD KO for NcROP5 5′ UTR; 6: Double enzyme restriction of pTCR-NcROP5-CD KO for NcROP5 3′ UTR; M1: Trans2K plus DNA marker; M2: Trans15k DNA marker. (B,C) Schemes of the targeted disruption of the NcROP5 gene by double homologous recombination. The pTCR-NcROP5-CD KO plasmid contains the 5′ UTR and 3′ UTR sequences flanking the CmR and RFP marker, which confers resistance to chloramphenicol.
FIGURE S2 | Construction of ΔNcROP5 complementary strain, iΔNcROP5. (A) Products of NcROP5A by PCR and identification of the pDMG-NcROP5-HA plasmid by enzyme digestion. 1: NcROP5A coding gene. 2: Double enzyme restricted pDMG-NcROP5-HA. M1: DNA 2000 plus M2: Trans 15K. (B) Diagram of the pDMG-NcROP5-HA construct.
FIGURE S3 | Gene ontology analyses of differential proteins in Nc-1 and ΔNcROP5. (A) Differential analysis between Nc-1 and ΔNcROP5. (B) Biological process.  Cellular process (31.88%),
Cellular process (31.88%),  Metabolic process (31.38%),
Metabolic process (31.38%),  Single-Organism Process (16.86%),
Single-Organism Process (16.86%),  Localization (7.80%),
Localization (7.80%),  Biological regulation (4.36%),
Biological regulation (4.36%),  Cellular component organization or biogenesis (3.27%),
Cellular component organization or biogenesis (3.27%),  Response to stimulus (2.68%),
Response to stimulus (2.68%),  Signaling (1.26%),
Signaling (1.26%),  Multicellular organismal process (0.25%),
Multicellular organismal process (0.25%),  Multi-Organism process (0.08%),
Multi-Organism process (0.08%),  Detoxofication (0.08%),
Detoxofication (0.08%),  Developmental process (0.08%); (C) Cellular component.
Developmental process (0.08%); (C) Cellular component.  Cell part (24.92%),
Cell part (24.92%),  Cell (24.92%),
Cell (24.92%),  Organelle (13.85%),
Organelle (13.85%),  Membrane (11.73%),
Membrane (11.73%),  Macromolecular complex (11.62%),
Macromolecular complex (11.62%),  Membrane part (6.15%),
Membrane part (6.15%),  Organelle part (5.25%),
Organelle part (5.25%),  Membrane-enclosed lumen (1.01%),
Membrane-enclosed lumen (1.01%),  Extracellular region (0.45%),
Extracellular region (0.45%),  Supramolecular fiber (0.11%); (D) Molecular function.
Supramolecular fiber (0.11%); (D) Molecular function.  Binding (52.84%),
Binding (52.84%),  Catalytic activity (37.34%),
Catalytic activity (37.34%),  Structural molecule activity (3.67%),
Structural molecule activity (3.67%),  Transporter activity (2.66%),
Transporter activity (2.66%),  Nucleic acid binding transcription factor activity (1.10%),
Nucleic acid binding transcription factor activity (1.10%),  Molecular function regulator (0.92%),
Molecular function regulator (0.92%),  Electron carrier activity (0.92%),
Electron carrier activity (0.92%),  Antioxidant activity (0.46%),
Antioxidant activity (0.46%),  Metallochaperone activity (0.09%); (E) Number of proteins involved in biological process, cellular component and molecular function.
Metallochaperone activity (0.09%); (E) Number of proteins involved in biological process, cellular component and molecular function.
FIGURE S4 | The number of differential proteins and classification of enriched pathways in Nc-1 and ΔNcROP5. (A) Increased and decreased proteins are divided into three groups: differentially expressed proteins with a 1.2-, 1.5-, and 2.0-fold difference. (B) Proteins with 1.5-fold difference are categorized by enrichment analysis to 39 pathways.
Footnotes
References
Alaganan, A., Fentress, S. J., Tang, K., Wang, Q., and Sibley, L. D. (2014). Toxoplasma GRA7 effector increases turnover of immunity-related GTPases and contributes to acute virulence in the mouse. Proc. Natl. Acad. Sci. U.S.A. 111, 1126–1131. doi: 10.1073/pnas.1313501111
Altschul, S. F., Wootton, J. C., Zaslavsky, E., and Yu, Y.-K. (2010). The construction and use of log-odds substitution scores for multiple sequence alignment. PLoS Comput. Biol. 6:e1000852. doi: 10.1371/journal.pcbi.1000852
Arranz-Solís, D., Aguado-Martíne, A., Müller, J., Regidor-Cerrillo, J., Ortega-Mora, L. M., and Hemphill, A. (2015). Dose-dependent effects of experimental infection with the virulent Neospora caninum Nc-Spain7 isolate in a pregnant mouse model. Vet. Parasitol. 211, 133–140. doi: 10.1016/j.vetpar.2015.05.021
Behnke, M. S., Fentress, S. J., Mashayekhi, M., Li, L. X., Taylor, G. A., and Sibley, L. D. (2012). The polymorphic pseudokinase ROP5 controls virulence in Toxoplasma gondii by regulating the active kinase ROP18. PLoS Pathog. 8:e1002992. doi: 10.1371/journal.ppat.1002992
Behnke, M. S., Khan, A., Lauron, E. J., Jimah, J. R., Wang, Q., Tolia, N. H., et al. (2015). Rhoptry proteins ROP5 and ROP18 are major murine virulence factors in genetically divergent south american strains of Toxoplasma gondii. PLoS Genet. 11:e1005434. doi: 10.1371/journal.pgen.1005434
Behnke, M. S., Khan, A., Wootton, J. C., Dubey, J. P., Tang, K., and Sibley, L. D. (2011). Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc. Natl. Acad. Sci. U.S.A. 108, 9631–9636. doi: 10.1073/pnas.1015338108
Brydges, S. D., Harper, J. M., Parussini, F., Coppens, I., and Carruthers, V. B. (2008). A transient forward-targeting element for microneme-regulated secretion in Toxoplasma gondii. Biol. Cell 100, 253–264. doi: 10.1042/BC20070076
Cárdenas-Mondragón, M. G., Ares, M. A., Panunzi, L. G., Pacheco, S., Camorlinga-Ponce, M., Girón, J. A., et al. (2016). Transcriptional profiling of Type II toxin–antitoxin genes of Helicobacter pylori under different environmental conditions: identification of HP0967–HP0968 system. Front. Microbiol. 7:1872. doi: 10.3389/fmicb.2016.01872
Caspe, S. G., Moore, D. P., Leunda, M. R., Cano, D. B., Lischinsky, L., Regidor-Cerrillo, J., et al. (2012). The Neospora caninum–spain 7 isolate induces placental damage, fetal death and abortion in cattle when inoculated in early gestation. Vet. Parasitol. 189, 171–181. doi: 10.1016/j.vetpar.2012.04.034
Deutsch, D. R., Fröhlich, T., Otte, K. A., Beck, A., Habermann, F. A., Wolf, E., et al. (2014). Stage-specific proteome signatures in early bovine embryo development. J. Proteome Res. 13, 4363–4376. doi: 10.1021/pr500550t
Dubey, J. P. (1999). Recent advances in Neospora and neosporosis. Vet. Parasitol. 84, 349–367. doi: 10.1016/S0304-4017(99)00044-8
Dubey, J. P., Schares, G., and Ortega-Mora, L. M. (2007). Epidemiology and control of Neosporosis and Neospora caninum. Clin. Microbiol. Rev. 20, 323–367. doi: 10.1128/CMR.00031-06
Dubremetz, J. F., and Lebrun, M. (2012). Virulence factors of Toxoplasma gondii. Microbes Infect. 14, 1403–1410. doi: 10.1016/j.micinf.2012.09.005
Etheridge, R., Alaganan, A., Tang, K., Lou, H. J., Turk, B., and Sibley, L. D. (2014). The Toxoplasma pseudokinase ROP5 forms complexes with ROP18 and ROP17 kinases that synergize to control acute virulence in mice. Cell Host Microbe 15, 537–550. doi: 10.1016/j.chom.2014.04.002
Gagnaire, A., Gorvel, L., Papadopoulos, A., Von Bargen, K., Mège, J., and Gorvel, J. (2016). COX-2 inhibition reduces Brucella bacterial burden in draining lymph nodes. Front. Microbiol. 7:1987. doi: 10.3389/fmicb.2016.01987
Gaji, R. Y., Flammer, H. P., and Carruthers, V. B. (2011). Forward targeting of Toxoplasma gondii proproteins to the micronemes involves conserved aliphatic amino acids. Traffic 7, 840–853. doi: 10.1111/j.1600-0854.2011.01192.x
Gaskell, E. A., Judith, E. S., John, W. P., Dave, R. W., and Glenn, A. M. (2009). A unique dual activity amino acid hydroxylase in Toxoplasma gondii. PLoS ONE 4:e4801. doi: 10.1371/journal.pone.0004801
Ghorbal, M., Gorman, M., Macpherson, C. R., Martins, R. M., Scherf, A., and Lopezrubio, J. J. (2014). Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 32, 819–821. doi: 10.1038/nbt.2925
Grigg, M. E., Bonnefoy, S., Hehl, A. B., Suzuki, Y., and Boothroyd, J. C. (2001). Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294, 161–165. doi: 10.1126/science.1061888
Hajj, H. E., Maryse, L., Stefan, T. A., Henri, V., Gilles, L., and Jean, F. O. D. (2007). ROP18 Is a rhoptry kinase controlling the intracellular proliferation of Toxoplasma gondii. PLoS Pathog. 3:e14. doi: 10.1371/journal.ppat.0030014
Hall, C. A., Reichel, M. P., and Ellis, J. T. (2005). Neospora abortions in dairy cattle: diagnosis, mode of transmission and control. Vet. Parasitol. 128, 231–241. doi: 10.1016/j.vetpar.2004.12.012
Hammoudi, P., Jacot, D., Mueller, C., Di Cristina, M., Dogga, S. K., Marq, J., et al. (2015). Fundamental roles of the golgi-associated toxoplasma aspartyl protease, ASP5, at the host-parasite interface. PLoS Pathog. 11:e1005211. doi: 10.1371/journal.ppat.1005211
Hecker, Y. P., Cantón, G., Regidor-Cerrillo, J., Chianini, F., Morrell, E., Lischinsky, L., et al. (2015). Cell mediated immune responses in the placenta following challenge of vaccinated pregnant heifers with Neospora caninum. Vet. Parasitol. 214, 247–254. doi: 10.1016/j.vetpar.2015.10.015
Hermanns, T., Müller, U. B., Könen-Waisman, S., Howard, J. C., and Steinfeldt, T. (2016). The Toxoplasma gondii rhoptry protein ROP18 is an Irga6-specific kinase and regulated by the dense granule protein GRA7. Cell. Microbiol. 18, 244–259. doi: 10.1111/cmi.12499
Hui, W., Tao, L., Jing, L., Muzi, L., Huizhu, N., and Qun, L. (2014). A nuclear factor of high mobility group box protein in Toxoplasma gondii. PLoS ONE 9:e111993. doi: 10.1371/journal.pone.0111993
Lai, B. S., Witola, W. H., El Bissati, K., Zhou, Y., Mui, E., Fomovska, A., et al. (2012). Molecular target validation, antimicrobial delivery, and potential treatment of Toxoplasma gondii infections. Proc. Natl. Acad. Sci. U.S.A. 109, 14182–14187. doi: 10.1073/pnas.1208775109
Li, J., He, P., Yu, Y., Du, L., Gong, P., Zhang, G., et al. (2014). Detection of Neospora caninum-DNA in feces collected from dogs in Shenyang (China) and ITS1 phylogenetic analysis. Vet. Parasitol. 205, 361–364. doi: 10.1016/j.vetpar.2014.06.036
Li, M., Wang, H., Liu, J., Hao, P., Ma, L., and Liu, Q. (2016). The apoptotic role of metacaspase in Toxoplasma gondii. Front. Microbiol. 6:1560. doi: 10.3389/fmicb.2015.01560
Liu, G., Cui, X., Hao, P., Yang, D., Liu, J., and Liu, Q. (2013). GRA 14, a novel dense granule protein from Neospora caninum. Acta Biochim. Biophys. Sin. 45, 607–609. doi: 10.1093/abbs/gmt036
Lyon, C. (2010). Update on the diagnosis and management of Neospora caninum infections in dogs. Top. Companion Anim. Med. 25, 170–175. doi: 10.1053/j.tcam.2010.07.005
Mota, C. M., Oliveira, A. C. M., Davoli-Ferreira, M., Silva, M. V., Santiago, F. M., Nadipuram, S. M., et al. (2016). Neospora caninum activates p38 MAPK as an evasion mechanism against innate immunity. Front. Microbiol. 7:1456. doi: 10.3389/fmicb.2016.01456
Nolan, S. J., Romano, J. D., Luechtefeld, T., and Coppens, I. (2015). Neospora caninum recruits host cell structures to its parasitophorous vacuole and salvages lipids from organelles. Eukaryot. Cell 14, 454–473. doi: 10.1128/EC.00262-14
Pan, H., Na, Y., Xia, C., Jing, L., Daoyu, Y., and Qun, L. (2014). First isolation of Neospora caninum from blood of a naturally infected adult dairy cow in Beijing, China. J. Parasitol. 100, 812–816. doi: 10.1645/14-498.1
Pastor-Fernández, I., Regidor-Cerrillo, J., Jiménez-Ruiz, E., Álvarez-García, G., Marugán-Hernández, V., Hemphill, A., et al. (2016). Characterization of the Neospora caninum NcROP40 and NcROP2Fam-1 rhoptry proteins during the tachyzoite lytic cycle. Parasitology 143, 97–113. doi: 10.1017/S0031182015001511
Payne, T. M., Payne, A. J., and Knoll, L. J. (2011). A Toxoplasma gondii mutant highlights the importance of translational regulation in the apicoplast during animal infection. Mol. Microbiol. 82, 1204–1216. doi: 10.1111/j.1365-2958.2011.07879.x
Pereira, G. D., Regidorcerrillo, J., Collantesfernández, E., Aguadomartínez, A., Del, P. I., Minguijón, E., et al. (2010). Pathogenic characterization in mice of Neospora caninum isolates obtained from asymptomatic calves. Parasitology 137, 1057–1068. doi: 10.1017/S0031182009991855
Plattner, F., Yarovinsky, F., Romero, S., Didry, D., Carlier, M. F., Sher, A., et al. (2008). Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe 3, 77–87. doi: 10.1016/j.chom.2008.01.001
Radke, J. B., Lucas, O., De Silva, E. K., Ma, Y., Sullivan, W. J., Weiss, L. M., et al. (2013). ApiAP2 transcription factor restricts development of the Toxoplasma tissue cyst. Proc. Natl. Acad. Sci. U.S.A. 110, 6871–6876. doi: 10.1073/pnas.1300059110
Reese, M. L., and Boothroyd, J. C. (2011). A conserved non-canonical motif in the pseudoactive site of the ROP5 pseudokinase domain mediates its effect on Toxoplasma virulence. J. Biol. Chem. 286, 29366–29375. doi: 10.1074/jbc.M111.253435
Regidorcerrillo, J., Gómezbautista, M., Del, P. I., Jiménezruiz, E., Aduriz, G., and Ortegamora, L. M. (2010). Influence of Neospora caninum intra-specific variability in the outcome of infection in a pregnant BALB/c mouse model. Vet. Res. 41, 472–476. doi: 10.1051/vetres/2010024
Reid, A. J., Vermont, S. J., Cotton, J. A., Harris, D., Hill-Cawthorne, G. A., Knen-Waisman, S., et al. (2012). Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: coccidia differing in host range and transmission strategy. PLoS Pathog. 8:e1002567. doi: 10.1371/journal.ppat.1002567
Roos, D. S., Donald, R. G., Morrissette, N. S., and Moulton, A. L. (1994). Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Method Cell Biol. 45, 27–63. doi: 10.1016/S0091-679X(08)61845-2
Saeij, J. P. J., Boyle, J. P., Coller, S., Taylor, S., Sibley, L. D., Brooke-Powell, E. T., et al. (2006). Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314, 1780–1783. doi: 10.1126/science.1133690
Sandberg, A., Lindell, G., Källström, B. N., Branca, R. M., Danielsson, K. G., Dahlberg, M., et al. (2012). Tumor proteomics by multivariate analysis on individual pathway data for characterization of vulvar cancer phenotypes. Mol. Cell. Proteom. 7, M112.016998. doi: 10.1074/mcp.M112.016998
Shen, B., Brown, K. M., Lee, T. D., and Sibley, L. D. (2014). Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio 5:e1114-14. doi: 10.1128/mBio.01114-14
Shwab, E. K., Jiang, T., Pena, H. F. J., Gennari, S. M., Dubey, J. P., and Su, C. (2015). The ROP18 and ROP5 gene allele types are highly predictive of virulence in mice across globally distributed strains of Toxoplasma gondii. Int. J. Parasitol. 46, 141–146. doi: 10.1016/j.ijpara.2015.10.005
Sinai, A. P., and Joiner, K. A. (2001). The Toxoplasma gondii protein ROP2 mediates host organelle association with the parasitophorous vacuole membrane. J. Cell Biol. 154, 95–108. doi: 10.1083/jcb.200101073
Soldati, D., and Meissner, M. (2004). Toxoplasma as a novel system for motility. Curr. Opin. Cell Biol. 16, 32–40. doi: 10.1016/j.ceb.2003.11.013
Su, C., Evans, D., Cole, R. H., Kissinger, J. C., Ajioka, J. W., and Sibley, L. D. (2003). Recent expansion of Toxoplasma through enhanced oral transmission. Science 299, 414–416. doi: 10.1126/science.1078035
Tao, L., Hui, W., Jing, L., Huizhu, N., and Qun, L. (2014). ROP18 is a key factor responsible for virulence difference between Toxoplasma gondii and Neospora caninum. PLoS ONE 9:e99744. doi: 10.1371/journal.pone.0099744
Taylor, S., Barragan, A., Su, C., Fux, B., Fentress, S. J., Tang, K., et al. (2006). A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 314, 1776–1780. doi: 10.1126/science.1133643
Tilley, M., Fichera, M. E., Jerome, M. E., Roos, D. S., and White, M. W. (1997). Toxoplasma gondii sporozoites form a transient parasitophorous vacuole that is impermeable and contains only a subset of dense-granule proteins. Infect. Immun. 65, 4598–4605.
Vemulapalli, R., Sanakkayala, N., Gulani, J., Schurig, G. G., Boyle, S. M., Lindsay, D. S., et al. (2007). Reduced cerebral infection of Neospora caninum in BALB/c mice vaccinated with recombinant Brucella abortus RB51 strains expressing N. caninum SRS2 and GRA7 proteins. Vet. Parasitol. 148, 219–230. doi: 10.1016/j.vetpar.2007.06.029
Walker, R., Gissot, M., Croken, M. M., Huot, L., Hot, D., Kim, K., et al. (2013a). TheToxoplasma nuclear factor TgAP2XI-4 controls bradyzoite gene expression and cyst formation. Mol. Microbiol. 87, 641–655. doi: 10.1111/mmi.12121
Walker, R., Gissot, M., Huot, L., Alayi, T. D., Hot, D., Marot, G., et al. (2013b). Toxoplasma transcription factor TgAP2XI-5 regulates the expression of genes involved in parasite virulence and host invasion. J. Biol. Chem. 43, 31127–31138. doi: 10.1074/jbc.M113.486589
Walzer, K. A., Adomako-Ankomah, Y., Dam, R. A., Herrmann, D. C., Schares, G., Dubey, J. P., et al. (2013). Hammondia hammondi, an avirulent relative of Toxoplasma gondii, has functional orthologs of known T. gondii virulence genes. Proc. Natl. Acad. Sci. U.S.A. 110, 7446–7451. doi: 10.1073/pnas.1304322110
Wang, J., Tang, D., Li, W., Xu, J., Liu, Q., and Liu, J. (2017). A new microneme protein of Neospora caninum, NcMIC8 is involved in host cell invasion. Exp. Parasitol. 175, 21–27. doi: 10.1016/j.exppara.2017.01.004
Williams, M. J., Alonso, H., Enciso, M., Egarter, S., Sheiner, L., Meissner, M., et al. (2015). Two essential light chains regulate the MyoA lever arm to promote Toxoplasma gliding motility. mBio 6:e00845–15. doi: 10.1128/mBio.00845-15
Yamamoto, M., Standley, D. M., Takashima, S., Saiga, H., Okuyama, M., Kayama, H., et al. (2009). A single polymorphic amino acid on Toxoplasma gondii kinase ROP16 determines the direct and strain-specific activation of Stat3. J. Exp. Med. 206, 2747–2760. doi: 10.1084/jem.20091703
Zhang, G., Sugimoto, C., and Fujisaki, K. (2007). Apical membrane antigen 1 is a cross-reactive antigen between Neospora caninum and Toxoplasma gondii, and the anti-NcAMA1 antibody inhibits host cell invasion by both parasites. Mol. Biochem. Parasitol. 151, 205–212. doi: 10.1016/j.molbiopara.2006.11.005
Keywords: Neospora caninum, gene editing, rhoptry protein 5, virulence factor, protozoan parasites
Citation: Ma L, Liu J, Li M, Fu Y, Zhang X and Liu Q (2017) Rhoptry protein 5 (ROP5) Is a Key Virulence Factor in Neospora caninum. Front. Microbiol. 8:370. doi: 10.3389/fmicb.2017.00370
Received: 18 October 2016; Accepted: 22 February 2017;
Published: 07 March 2017.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Tiago W. P. Mineo, Federal University of Uberlandia, BrazilShivangi Agarwal, Northwestern University, USA
Javier Regidor Cerrillo, Complutense University of Madrid, Spain
Copyright © 2017 Ma, Liu, Li, Fu, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qun Liu, qunliu@cau.edu.cn
 Lei Ma
Lei Ma Qun Liu
Qun Liu