- 1Dairy Microbiology Division, Indian Council of Agricultural Research – National Dairy Research Institute, Karnal, India
- 2School of Food Science and Engineering, Hefei University of Technology, Hefei, China
With the aim to bioprospect potent riboflavin producing lactobacilli, the present study was carried out to evaluate the relative mRNA expression of riboflavin biosynthesis genes namely Rib 1, Rib 2, Rib 3, and Rib 4 from potent riboflavin producers obtained from our previous studies. All the four genes were successfully cloned and sequenced for further analysis by in silico procedures. As studied by non-denaturing Polyacrylamide gel electrophoresis, no difference in size of all the four genes among those of various lactobacilli was observed. The relative fold increase in mRNA expression in Rib 1, Rib 2, Rib 3, and Rib 4 genes has been observed to be 10-, 1-, 0.7-, and 8.5-fold, respectively. Due to increase in relative mRNA expression for all the Rib genes as well as phenotypic production attribute, KTLF1 strain was used further for expression studies in milk and whey. The fold increase in mRNA expression for all the four Rib genes was higher at 12 and 18 h in milk and whey respectively. After exposure to roseoflavin, resistant variant of KTLF1 showed considerable increase in expression of all the targets genes. This is the first ever study to compare the mRNA expression of riboflavin biosynthesis pathway genes in lactobacilli and it also under lines the effect of media and harvesting time which significantly affect the expression of rib genes. The use of roseoflavin-resistant strains capable of synthesizing riboflavin in milk and whey paves a way for an exciting and economically viable biotechnological approach to develop novel riboflavin bio-enriched functional foods.
Introduction
In the recent years, many researchers have shown burgeoning interest in riboflavin which has now regarded as an essential component of cellular biochemistry (Thakur et al., 2016a). Several bacteria have the trait to synthesize riboflavin and its pathway has been studied in bacteria whereas humans lack its biosynthesis ability (Perkins and Pero, 2002). The microorganisms harbor the genetic structure to synthesize B vitamins particularly riboflavin to obtain bio-enriched food (Capozzi et al., 2011). Due to adaptability to fermentation processes, lactic acid bacteria (LAB) act an ideal candidates for in situ riboflavin production in food (Arena et al., 2014). Though, ability for riboflavin biosynthesis is strain specific (Capozzi et al., 2012). An alternative RNA structure involving the RFN element serves a model for regulation of riboflavin biosynthesis (Gelfand et al., 1999; Vitreschak et al., 2002). Riboflavin metabolism and transport genes are regulated at transcription attenuation and translation initiation level in Gram-positive bacteria and Gram-negative bacteria respectively (Vitreschak et al., 2002). Four genes (rib1, rib2, rib3, and rib4) are required for biosynthesis of riboflavin from guanosine triphosphate (GTP) and ribulose-5- phosphate (Perkins et al., 1999). According to these authors, these genes are located in an operon and their order differs from that of enzymatic reactions (Richter et al., 1992, 1993, 1997). There are mainly two promoters responsible for transcription of riboflavin genes where all the four genes are controlled are primarily controlled by the ribP1 promoter (Perkins et al., 1999). The rib3 and rib4, are regulated from a second promoter (ribP2) and regulatory region RFN (Perkins et al., 1999).
There are number of reports where overexpression in riboflavin production was observed after exposure to range of roseoflavin (a chemical analog to riboflavin) (Burgess et al., 2004, 2006; del Valle et al., 2014). In these studies, riboflavin overproduction directly correlated with the spontaneous roseoflavin resistant strains (Burgess et al., 2006; Capozzi et al., 2011). The tolerance to the toxic roseoflavin signifies the mutations in the regulatory region of the rib operon which ultimately give rise to riboflavin over producing phenotype. Lately, in situ bacterial overproduction of the B group vitamins, including riboflavin is of significant interest (Burgess et al., 2009; Capozzi et al., 2012). In particular for riboflavin, promising results have been reported for the production of yogurt (Burgess et al., 2006) or pasta and bread (Capozzi et al., 2011; Arena et al., 2014) and Soymilk (del Valle et al., 2014). Many researchers (Jayashree et al., 2011; Guru and Viswanathan, 2013; del Valle et al., 2014; Thakur and Tomar, 2015a; Thakur et al., 2016c) have studied the riboflavin production in LAB in MRS, Riboflavin free media, milk and whey but no one has ever reported the expression levels of riboflavin biosynthesis genes. The Lactobacilli used for present study were previously isolated and identified from various niches (human feces, fermented bamboo shoots, and curd) (Thakur and Tomar, 2015a; Thakur et al., 2015a, 2016c). Among them Lactobacilli isolated from fermented bamboo shoots (Manipur, India) have shown highest riboflavin producing properties as well as displayed probiotic and appreciable techno-functional properties (Thakur et al., 2015a). In the continuance of our previous reports, the present study reveals the first ever profile of mRNA expression of four Rib genes (molecular determinants for riboflavin biosynthesis which form a complete functional rib operon) in four different media by harvesting the test isolates at different intervals of time. There are few reports where the regulatory mechanism of riboflavin biosynthesis has been studied in roseoflavin resistant variants in LAB. However, there exists per se no such report for Lactobacillus species till date.
Materials and Methods
Bacterial Strains and Growth Conditions
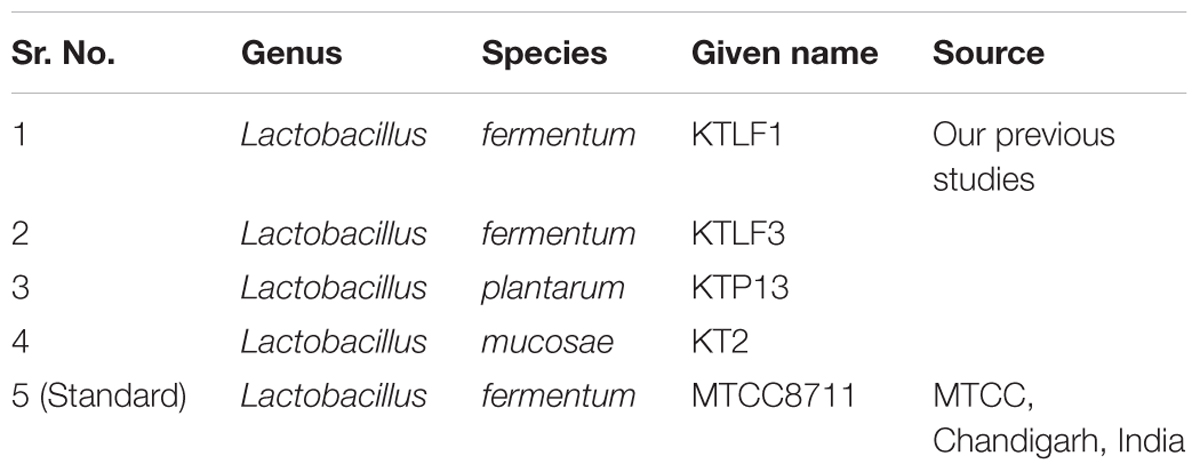
The Lactobacillus strains (Table 1) used in this work were confirmed for riboflavin production by an array of analytical methods viz. Polymerase chain reaction (PCR) based method (presence of riboflavin biosynthesis genes), Spectrophotometric method, Microbiological assay method, and High Performance Liquid Chromatography in our previous studies (Thakur and Tomar, 2015a; Thakur et al., 2016b). All the strains stored previously at -80°C in MRS supplemented with glycerol (20% v/v) were routinely cultured on de Man-Rogosa -Sharp (MRS) medium (Sigma- Aldrich, St. Louis, MO, USA) for this study.
Cloning, Transformation, and Sequencing
Purified PCR products (HiPuraATM purification kit, Himedia, India) were used for cloning of all the four genes. The cloning vector used in this study was PTZ57R/T clone vector amp (InstClone PCR cloning kit, Stratagene, USA). The clones were transformed into competent cells of Escherichia coli (E. coli) (XL1 blue). The successful clones were picked from Luria broth+ ampicillin plates and amplified for target genes by colony PCR method followed by plasmid isolation. The positive clones were identified by PCR analysis of plasmid DNA by using primers used in our previous study (Thakur et al., 2016b). The nucleotide sequencing was performed by sequencing services provided by Xcelris Labs, Ltd, Ahmedabad, India. The chromatograms of sequences obtained were analyzed and converted to Fasta using Bio-Edit Software. Nucleotide sequence similarity searches were performed for the obtained sequences by matching with previously published complete genome of Lactobacillus species of interest.
Size Variation in Rib Genes by Polyacrylamide Gel Electrophoresis (PAGE)
Non-denaturing PAGE was used to detect the difference is size of all the four Rib genes amplified in different lactobacilli. Silver staining was used to view the band pattern in the PAGE after the final gel run.
Growth in MRS, RAM, Milk and Whey Based Media
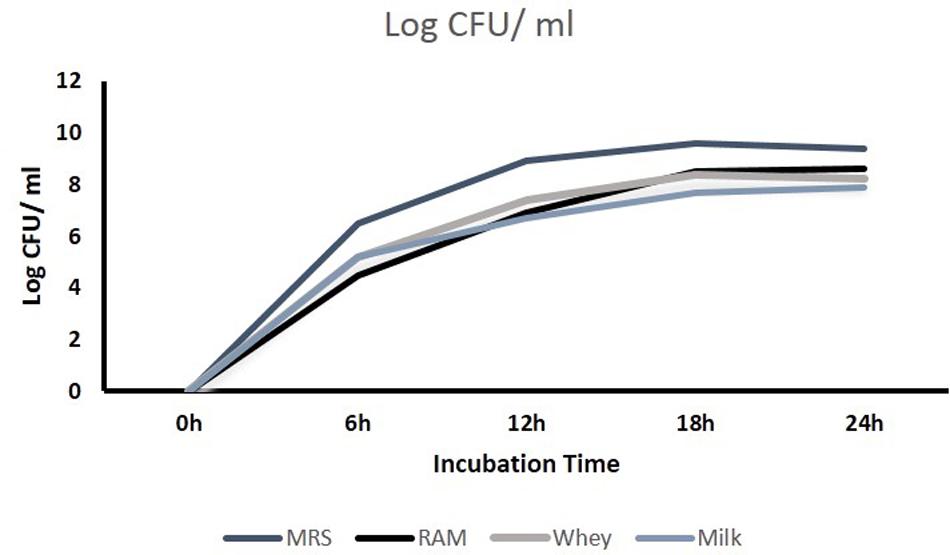
The test isolates were washed thrice with saline solution (0.85% m/v NaCl), resuspended in this solution and used to inoculate at 2% (v/v) riboflavin-free culture medium (Riboflavin Assay Medium, Difco, Becton, Dickinson, and Co., Sparks, MD, USA), reconstituted skim milk and whey based medium and then incubated without agitation at 37°C for 18 h. The optical density of selected isolates was observed before harvesting them for RNA isolation. The log count/ml was checked at lag, log, and stationary phases of growth.
Designing of Primers
The primers (Table 2) were designed by aligning sequences of riboflavin operon using CLUSTALW program. The Lactobacillus fermentum IFO3956 strain was considered for primer selection: GenBank accession number NC_010610. The house keeping genes for normalizing real time reaction were synthesized for REC gene essential for the repair and maintenance of DNA and TUF gene encoding elongation factor from the database of genome in NCBI.
RNA Extraction, cDNA Synthesis, and RT-PCR
Selected Lactobacilli were grown in 10 ml of MRS, Riboflavin Assay Medium (devoid of riboflavin) (RAM), Skim milk and Whey based medium. The RNA was extracted after 6, 12, 18, and 24 h of incubation using TRIzol reagent followed by cell lysis by lysozyme (10 mg/100 ml) (Sigma, USA) (Ramiah et al., 2007). The quality of isolated RNA was checked (Sambrook and Russell, 2001). RNA was quantified and its purity of RNA was judged and used for reverse transcription. The cDNA was prepared with cDNA kits (RevertAidTM First strand c-DNA synthesis kit, Fermentas, India), according to the manufacturer’s instructions. SYBR Green I Master mix (Roche) on 2 μL of diluted cDNA (1:1) using exon-spanning primers, 5 μL of SYBR green buffer 2X (Roche) and 2.5 pmol of each primer (Table 2) for a total volume reaction of 10 μL were used for qualitative PCR. The amplification was run in Lightcycler® 480 II, and the results were analyzed using Lightcycler® 480 II software release 1.50 SP3. The PCR conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles of amplification at 63°C for 30 s and 72°C for 30 s. At the end of the each run a melting curve was achieved from 70 to 95°C and continuous fluorescence measurement was taken. A melting curve analysis was performed in order to verify the specificity of real-time PCR (RT-PCR) and finally, a cooling step to 4°C was achieved. Fluorescence was measured once every cycle after the extension step using filters for SYBR Green I (excitation at 465 nm and emission at 510 nm). To calculate the threshold cycle value, the normalized fluorescence data was converted to a log scale and threshold value was determined. The quantitative data of RT-PCR is generated on the basis of number of cycles required for optimal amplification generated fluorescence to reach a specific threshold of detection (the quantification cycle:Cq values) (Bustin et al., 2009). The comparative critical threshold (ΔΔCT) method was used to calculate the relative expression ratios in which the amount of target RNA is adjusted to a reference (internal target RNA) (Livak and Schmittgen, 2001). The Prism 7.00 was used to analyze the RT-PCR data sets.
Isolation of Roseoflavin-Resistant Strains
The roseoflavin-resistant strains of KTLF1 was performed (according to Burgess et al., 2004, 2006) by exposing wild strain to increasing concentrations of roseoflavin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) in RAM. Further experiments were carried out by using subsequent inoculum in 1 mL of RAM supplemented with roseoflavin. From the culture grown at the maximum range of roseoflavin, 15 separated colonies were randomly isolated after spreading onto MRS agar plates, and those stocks were stored at -80°C in CDM roseoflavin-free supplemented with 20% of glycerol (Russo et al., 2014).
Principal Component Analysis
To discriminate the riboflavin producing isolates and riboflavin biosynthesis genes on the basis of mRNA expression levels in different media and at different time and principal component analysis (PCA) treatment, IBM SPSS Statistics 21.0 software program (IBM, Armonk, NY, USA) was used (ANOVA followed by a Tukey’s post hoc test). P < 0.05 was considered as statistically significant.
Results
Cloning Transformation and Sequencing
Purified amplicons were ligated into PTZ57R/T cloning vector. The ligates were transformed into competent cells of E. coli XL1 blue strain. The recombinant clones showed white (recombinant) colonies on LB agar plates supplemented with ampicillin (100 μg/ml). Ten randomly selected recombinant clones were analyzed by colony PCR for Rib genes. Consequently, positive clones were used for plasmid DNA analysis and isolated plasmids were further confirmed for their size by PCR and subsequently sequenced (Supplementary File). The sequences obtained from the isolates were compared by BLAST analysis for similarity check with three reference strains of Lactobacillus submitted to NCBI GenBank (Supplementary File). No size variation was found in the studied genes in different lactobacilli after staining the PAGE with silver staining (Supplementary File).
Growth of Isolates in MRS, RAM, Milk and Whey for RNA Isolation and Gene Expression
The CFU/ml was observed for KTLF1 at different growth phases (Figure 1) and the mRNA and the cDNA were prepared as described above. During the early growth, expression levels remain low and get elevated at later phase. The expression levels of Rib 1, Rib 4 were significantly higher in all the media used at four time intervals (Figure 2), whereas Rib2 and Rib 3 have shown almost constant regulation with different variables (Isolates, Media, and Time) (Figure 2). After incubation in different media at different time, the level of mRNAs expression was found to be changed in all the tested isolates. Particularly in KTLF1 strain, the mRNA expression level increased significantly in MRS at 12–18 h, in RAM at 6–12 h, in Milk at 6 h and in whey the upregulation was observed at 12 h (Figure 3). Among all the media used, RAM has shown increase in relative expression followed by MRS, Milk and Whey (Figure 4). Overall, there was no statistical difference in expression levels of Rib2 and Rib3 at different variables but a gradual decrease in the mean expression with time was recorded. Figure 5 revealed the marked difference of increase in the intensity of RT-PCR products at different variables in KTLF1 in MRS and RAM which correspond significantly to change in mRNA expression profile. In order to study the effect of different parameters mentioned on riboflavin production, relative expression levels of the Rib 1, Rib 2, Rib 3, and Rib 4 genes were calculated in comparison with lowest riboflavin producing strain obtained from our previous study. The gene expression profiles for each strain grown under these conditions were different (Figure 2). Among the five tested strains, KT2 has shown significant variation in gene expression profile across the different incubation time intervals. The Rib 3 and Rib4 genes had a steady transcript level (fold change equal to one) in all the tested strains except in KT2 strain. The most significant difference was found at 6 and 24 h. This showed that expression of these genes was not much affected with the strain in two different media. Whereas, in KTLF1 (Figure 3), effect of media on different genes was significant in Milk and whey however, the relative expression levels of these genes was steady in MRS and RAM across different incubation. In particular, the expression levels of these genes were found to be lowered with increased incubation time in all the four media. In KTLF1, all the genes have shown significant variation in their expression in milk, Whey and MRS (Figure 4). In milk and whey, the fold increase was found in descending order with the increased incubation times, whereas, in whey media, except Rib 1, remaining three genes have shown steady expression over different incubation periods. In MRS and RAM, except Rib 1 and Rib 4, remaining two genes have shown significant change in expression profile.
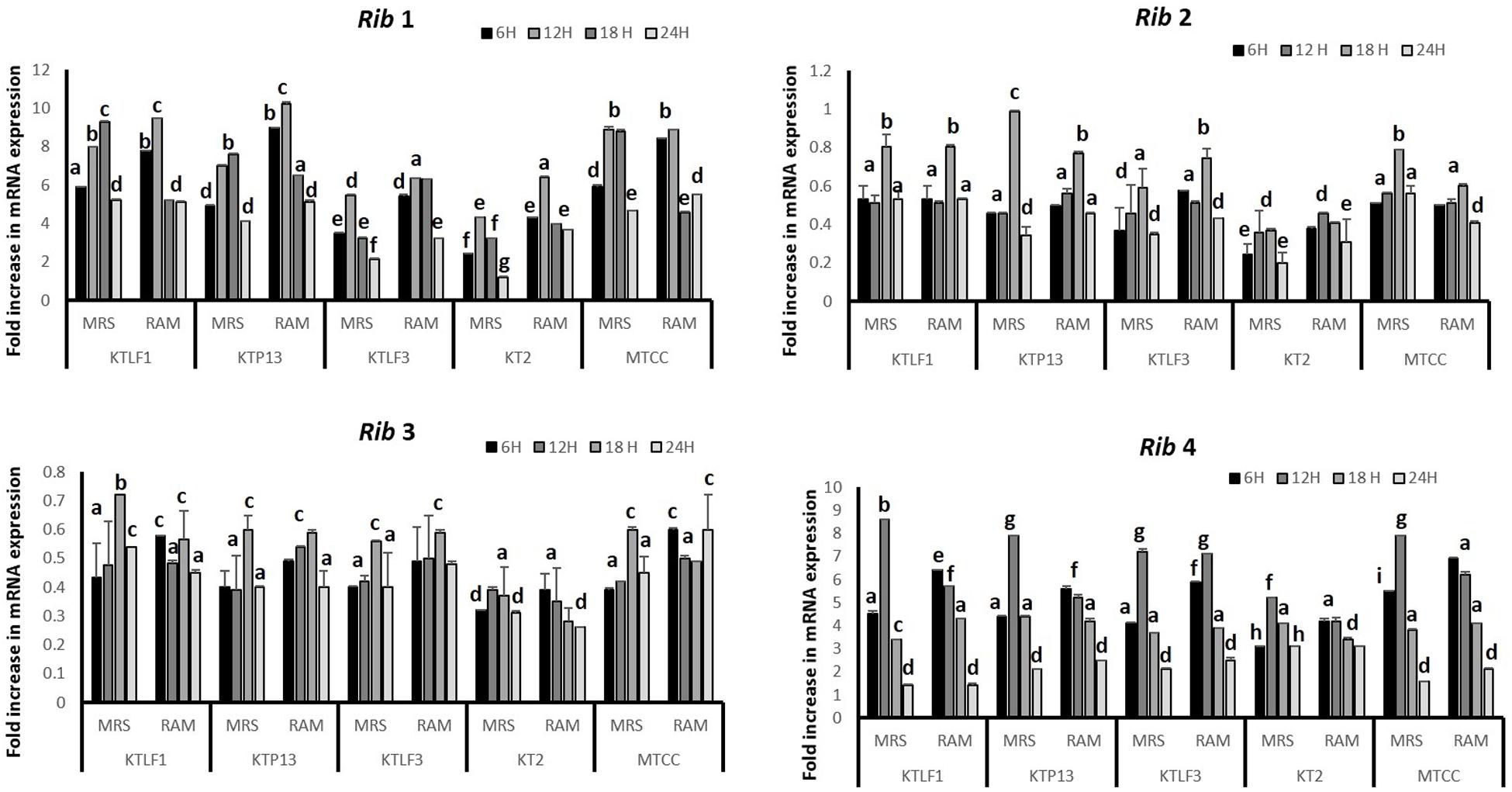
FIGURE 2. Comparative mRNA expression profile (fold increase) of four test isolates with respect to reference culture (relative expression is considered as 1) in MRS and RAM. Values are mean ± SD (n = 3) and different letters denote the significant difference at P < 0.05 (abcd refer to statistical differences with respect to MRS and RAM. Strain MTCC8711 is taken as control.
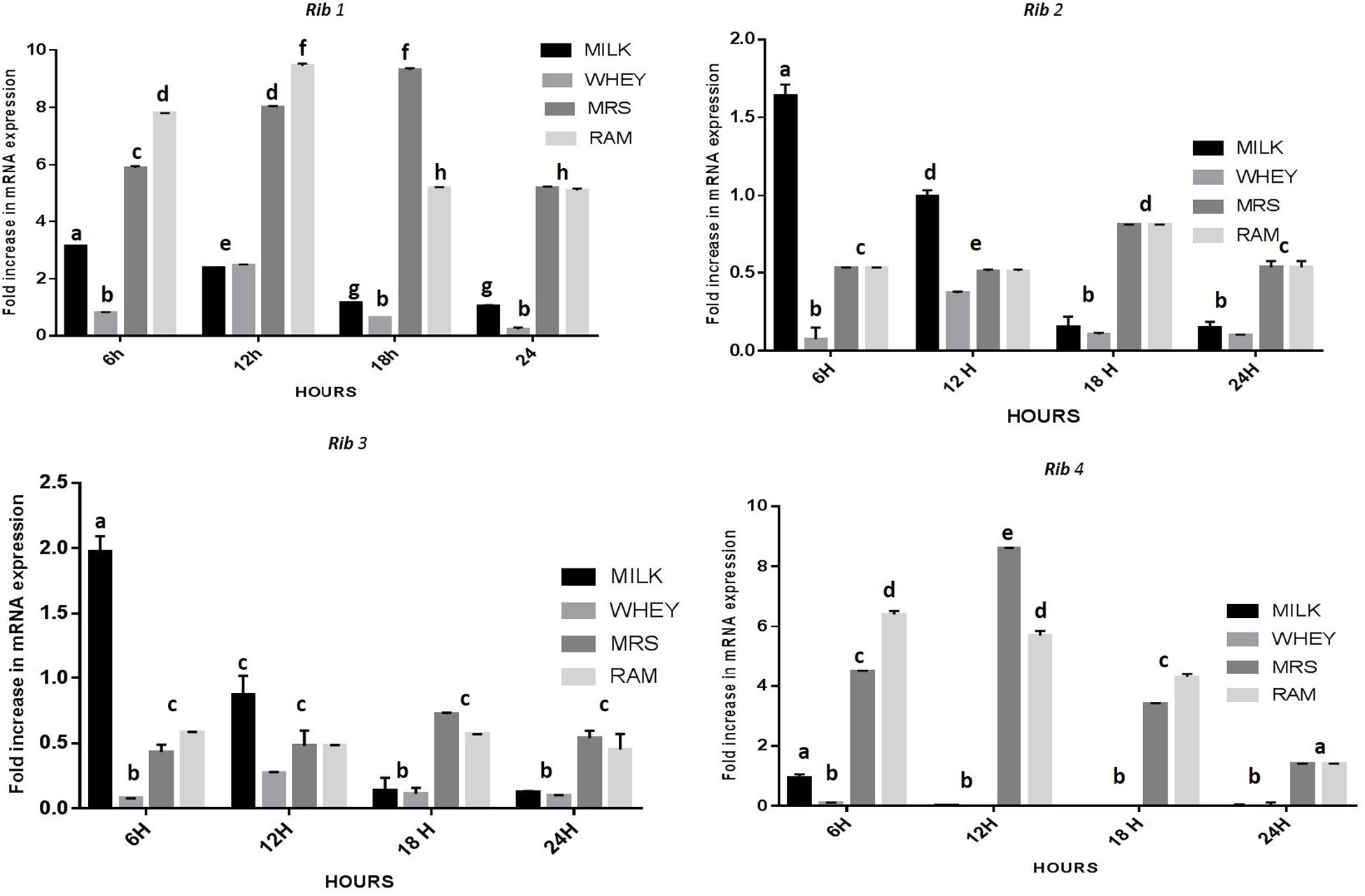
FIGURE 3. Comparative mRNA expression profile (fold increase) of KTLF1 in MRS, RAM, Whey and Milk. Values are mean ± SD (n = 3) and different letters denote show the significant difference at P < 0.05.
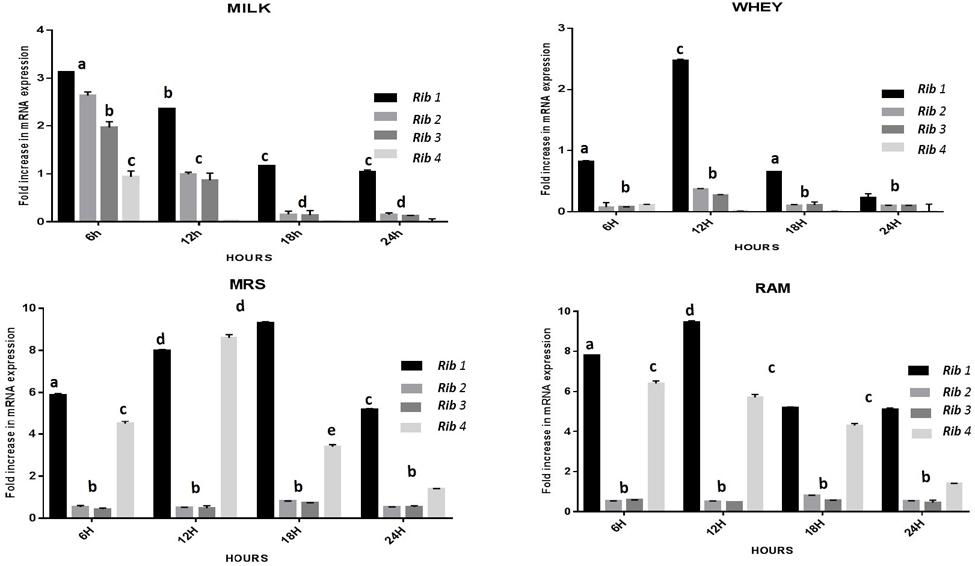
FIGURE 4. Overall fold increase in relative mRNA expression of riboflavin structural genes in KTLF1 with respect to media and time intervals over control (MTCC8711). Values are mean ± SD (n = 3) and different letters denote show the significant difference at P < 0.05.
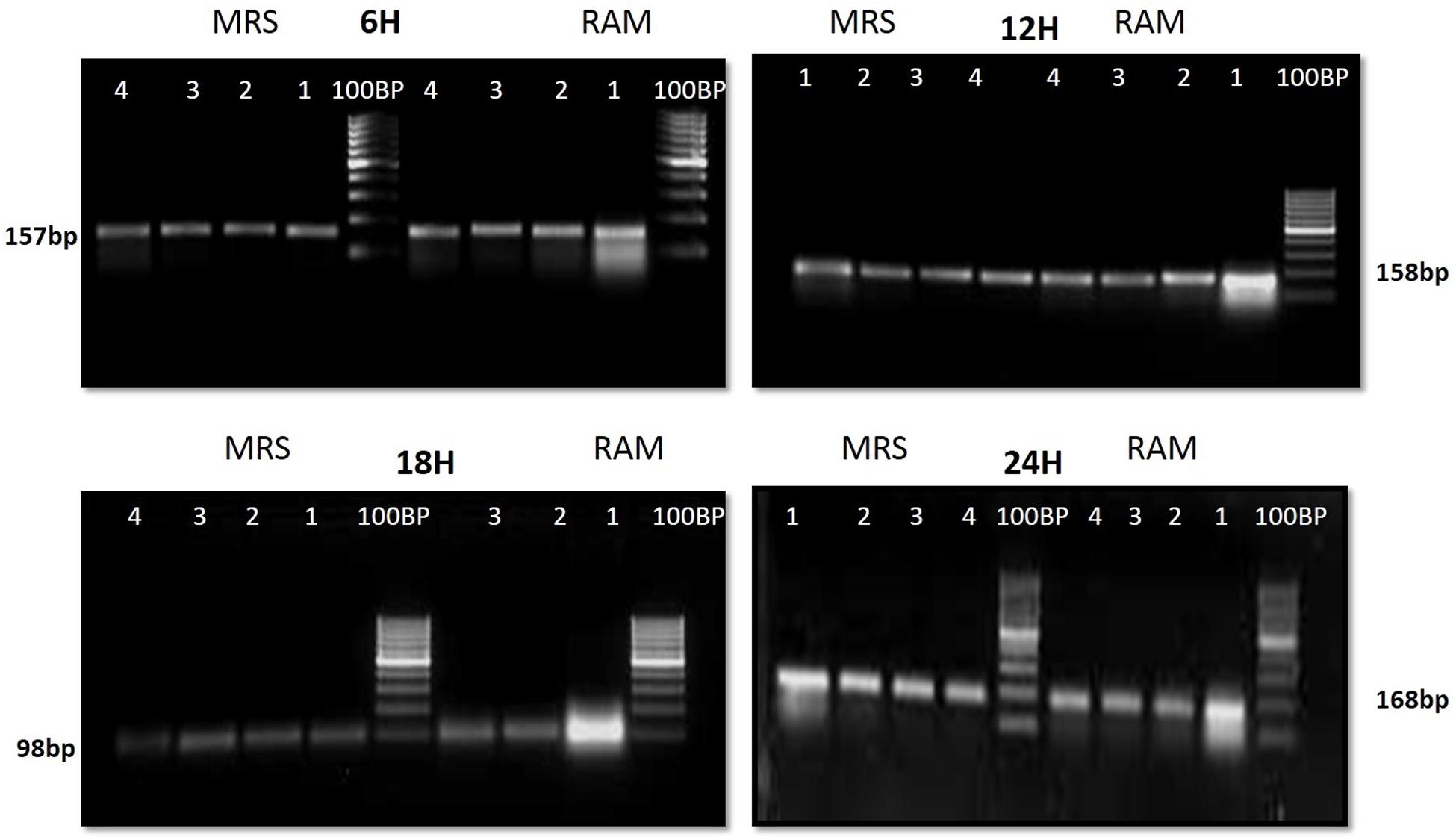
FIGURE 5. 1.5% agrose gel electrophoresis to discriminate the band intensity after RT-PCR in KTLF1 where 1, 2, 3, and 4 denote rib 1, rib 2, rib 3, and rib 4 genes.
Principal Component Analysis
The multivariate analysis was used for comparison of experimental data obtained for five lactobacilli for mRNA expression of four Rib genes (PCA). Two dimensional plots are drawn in Figure 6. In plot 1 (Figure 6A), contribution of the media with respect to variance is shown. The first two components present the total variance of 53.7%. The discrimination of isolates along PC1 is mainly due to RAM and Whey. Whereas along PC2, the isolate KT2 is discriminated vis-à-vis other isolates. In plot 2 (Figure 6B), the first two components present the total variance of 73.7%. The discrimination along samples along PC1 is mainly due to Rib 1 and Rib 4, whereas Rib 2 and Rib 3 are responsible for discrimination along PC2.
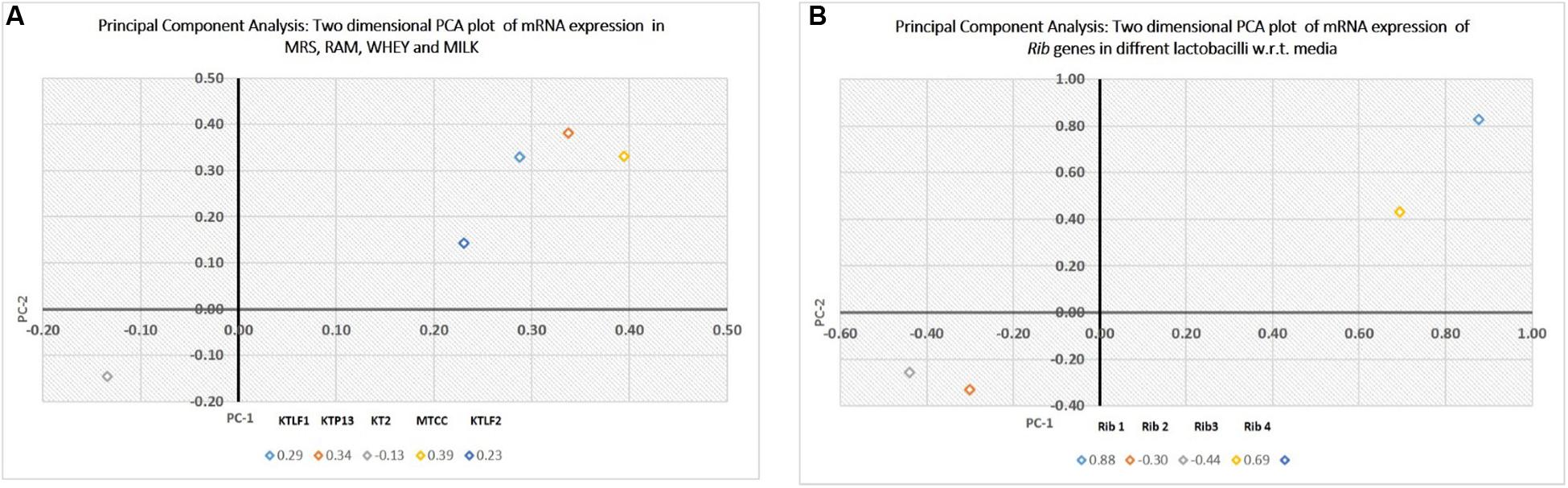
FIGURE 6. (A) Principal component analysis (PCA), expressed as two dimensional plot with respect to media as variables for the first two principal components. (B) PCA, expressed as two dimensional plot with respect to Rib genes for the first two principal components.
mRNA Expression of Rib Genes in Roseflavin Resistant Variant KTLF1 (4) and Riboflavin Overproduction
By following the procedure described by Burgess et al. (2006), KTLF1 was exposed to roseoflavin, a structural analog of riboflavin, which induces mutations in riboflavin-producing strains leading to a novel producer phenotype of the vitamin. Roseoflavin resistant variants were isolated and inoculated in the riboflavin-free medium and incubated for 16 h at 30°C. Both wild type and variant strains were re-inoculated in MRS, RAM, Milk and Whey based media at 37°C, 24 h and harvested for RNA isolation and riboflavin production. From five isolated variants, only KTLF1 variant [KTLF1 (4)] was able to up regulate mRNA expression of Rib genes and the increase in riboflavin production was more than 3.5-fold (4.5 mg/L) as compared with wild type strain in a culture medium without riboflavin (Figure 7A). The spike in mRNA expression level was observed after the exposure of roseoflavin in all the four genes across the media at different times of incubation (Figure 7B). The maximum up regulation was observed in Rib1 followed by Rib4. The fold increase in expression was lower as to that of other wild type strains (Figure 7B).
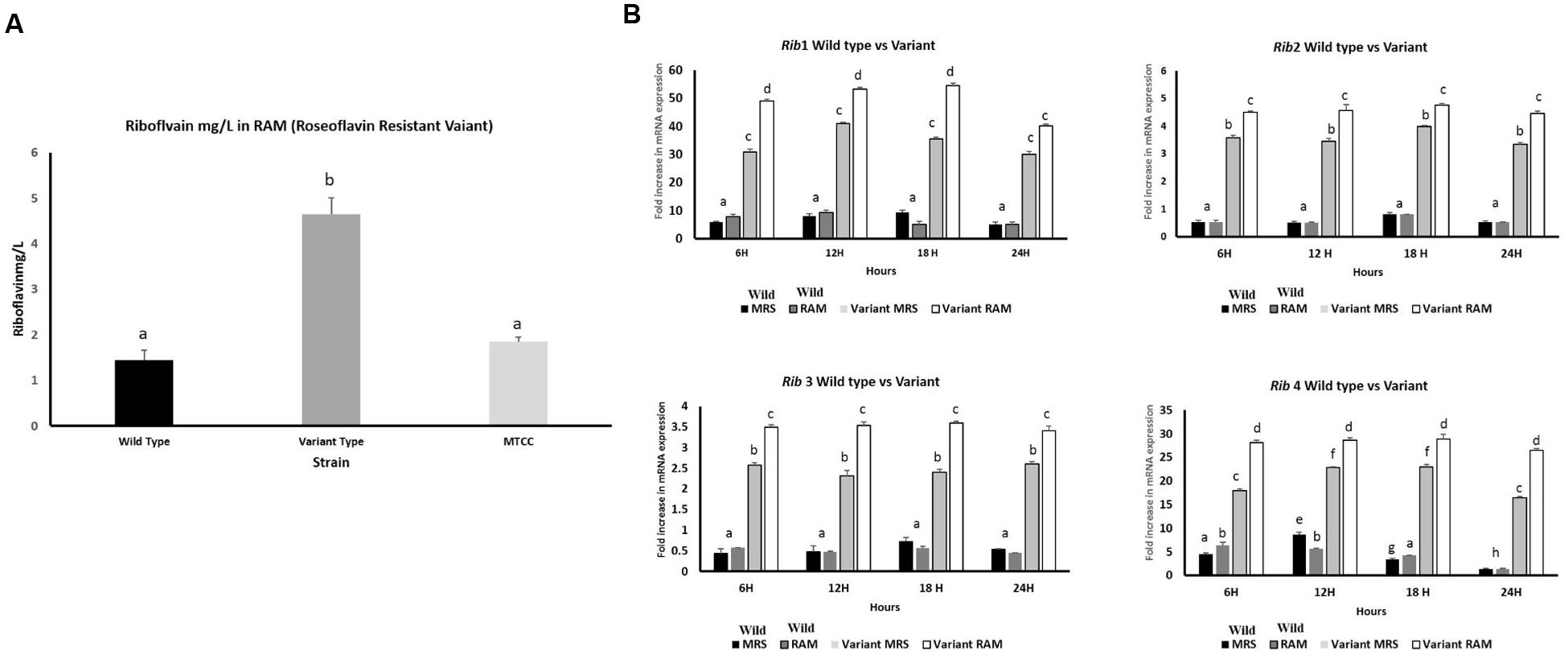
FIGURE 7. (A) Riboflavin production by roseoflavin-resistant KTLF (4) and wild strain in RAM. The line above the bar represents the SD of the mean and different letters denote show the significant difference at P < 0.05. (B) Over expression of Rib genes after exposure to roseoflavin in KTLF (4). Values are mean ± SD (n = 3) and different letters denote show the significant difference at P < 0.05.
Discussion
Consumers are increasingly becoming conscious for their nutritional requirements, thus, vitamins produced in situ by microbes may suit their needs and expectations (LeBlanc et al., 2013; Thakur et al., 2015b). Since little information is available on what factors affect riboflavin production in LAB, the aim of this study was to investigate the influence of incubation time and difference strains on the expression of the rib genes by a wild type strains. Also, the vitamin production by the roseoflavin resistant strain under these conditions was also evaluated in mutant strain. As it is known that riboflavin operon is inducible, it is essential to evaluate the mRNA levels rib genes in different strains in different media (MRS, RAM, Milk and Whey) at different interval of growth time (6, 12, 18, and 24 h). The isolates used in this study are prospected from dairy, non- dairy sources (fermented bamboo shoots, human feces) (Thakur and Tomar, 2015a). The isolate KTLF3 was isolated from fermented bamboo shoots collected from Manipur, North East Region (Ethnic) of India. The riboflavin producing isolates used in this study are well-characterized by in vitro methods for their functional probiotic (Thakur and Tomar, 2015b) as well as technological properties (Thakur et al., 2016c). Arena et al. (2014) have also reported the probiotic lactobacilli for riboflavin production as well as its overproduction by using roseoflavin. In our study, riboflavin-producing strains were selected on the basis of mRNA expression of riboflavin biosynthesis genes. Out of four strains, three strains were able to show change in relative expression in the targeted genes with different incubation and media variables. Thus, KTLF1 was confirmed as prolific riboflavin producer and hence was selected to screen the mRNA profile of these genes in milk and whey based media. Milk and whey enriched in riboflavin have shown the increase in relative expression after the riboflavin in the media was utilized by the bacteria for its growth. The riboflavin may be required by bacteria in small amounts, but it constitutes a vital growth factor for Enterococcus faecalis, Streptococcus pyogenes, Listeria monocytogenes, and some lactobacilli (Koser, 1968). The biosynthetic deficiency correlates well with the absence of riboflavin in the growth media as the riboflavin operon is an inducible one where the quantity of riboflavin inhibits its production by bacteria. Unlike, other media, the RAM has shown higher expression levels which is in accordance with the aforementioned hypothesis.
KTLF1 was further selected to observe the overexpression of Rib genes after exposure to certain levels of roseoflavin. These results clearly indicated the roseoflavin exposure led to 3.5-fold increase in riboflavin production besides increase in the expression of all the rib genes by the mutant strain compared with those obtained with wild type strains, being more marked this difference at Figures 7A,B. The tolerance to the toxic roseoflavin signifies the mutations in the regulatory region of the rib operon which ultimately give rise to riboflavin overproducing phenotype. Till date, riboflavin overproduction is led either by employing genetic engineering (Burgess et al., 2004, 2006). The increase in expression levels of Rib1 and Rib4 followed by Rib2 and Rib3 is due to increased transcription of riboflavin biosynthesis genes because riboflavin metabolism and transport genes are being regulated at transcription attenuation. Further, threefold (4.5 mg/L) increase in riboflavin production in a culture medium without riboflavin was in agreement with the all these reports. Overexpression of all the four genes contributes to enhanced riboflavin production (Burgess et al., 2004) which was also observed in our study. Roseoflavin-resistant strains of Leu. mesenteroides over produced up to 0.5 mg l-1 of riboflavin, whereas riboflavin-overproducing Lactobacillus plantarum and Propionibacterium freudenreichii were able to synthesize up to around 0.6 and 3 mg l-1 respectively (Burgess et al., 2006). According to del Valle et al. (2014) roseoflavin resistant strains increased six times (1860 ± 20 ng/mL) the initial riboflavin levels of soy milk.
The PCA plots have well-discriminated the isolates on the basis of their levels of mRNA expression. The Rib genes are also put into two different groups due to their high and low expression with respect to variables. Overall, this study reveals that isolates showed variations for expressing their Rib genes which qualifies riboflavin production as strain specific attribute.
Conclusion
The expression profile as well as phenotypic production of riboflavin have revealed that both the genotypic and phenotypic traits are dependent on riboflavin in media used for growth. Though the genotypic as well as phenotypic expression of riboflavin in milk and whey is lower as compared to riboflavin free media but it is better to use riboflavin producing bacteria than riboflavin consuming ones. From this study, it is clear that milk and whey can be used for development of riboflavin enriched fermented products. To the best of our knowledge, the present study reports for the first time the mRNA expression profile of riboflavin biosynthesis genes in lactobacilli. Furthermore, exposure of roseoflavin led to over expression of Rib genes in the variant of KTLF1 as compared to wild strains facilitates the enhanced riboflavin content in the final product. Lactobacilli isolated from fermented bamboo shoots have shown highest riboflavin producing properties as well as displayed probiotic and appreciable techno-functional properties which can be further explored to develop functional bamboo shoot foods. The riboflavin enriched products could be introduced by using these isolates as starters to prevent or treat riboflavin deficiencies which are still to be addressed.
Author Contributions
KT is the first author and has carried out the research work as a part of her Ph.D. program. The data analysis and manuscript writing was done by KT. ST has corrected the manuscript and helped in experimental work. Z-JW has helped revising the manuscript.
Funding
The authors thankfully acknowledge the support extended by the Indian Council of Agricultural Research (ICAR), New Delhi, India, and the Director of the National Dairy Research Institute (NDRI), Karnal, India. We would also acknowledge the major projects of science and technology in Anhui Province (15czz03115).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The help of Dr. Biswajit Brahma throughout the expression study is duly acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00427/full#supplementary-material
References
Arena, M. P., Russo, P., Capozzi, V., Lopez, P., Fiocco, D., and Spano, G. (2014). Probiotic abilities of riboflavinover producing Lactobacillus strains: a novel promising application of probiotics. Appl. Microbiol. Biotechnol. 98, 7569–7581. doi: 10.1007/s00253-014-5837-x
Burgess, C., O’Connell-Motherway, M., Sybesma, W., Hugenholtz, J., and van Sinderen, D. (2004). Riboflavinproduction in Lactococcus lactis: potential for in situ production of vitamin-enriched foods. Appl. Environ. Microbiol. 70, 5769–5777. doi: 10.1128/AEM.70.10.5769-5777.2004
Burgess, C. M., Smid, E. J., Rutten, G., and van Sinderen, D. (2006). A general method for selection of riboflavinoverproducing food grade micro-organisms. Microb. Cell Fact. 5:24. doi: 10.1186/1475-2859-5-24
Burgess, C. M., Smid, E. J., and van Sinderen, D. (2009). Bacterial vitamin B2, B11 and B12overproduction: an overview. Int. J. Food Microbiol. 133, 1–7. doi: 10.1016/j.ijfoodmicro.2009.04.012
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines:minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Capozzi, V., Menga, V., Digesu, A. M., De Vita, P., van Sinderen, D., Cattivelli, L., et al. (2011). Biotechnological production of vitamin B2-enriched bread and pasta. J. Agric. Food Chem. 59, 8013–8020. doi: 10.1021/jf201519h
Capozzi, V., Russo, P., Duenas, M. T., Lopez, P., and Spano, G. (2012). Lactic acid bacteria producing B-group vitamins:a great potential for functional cereals products. Appl. Microbiol. Biotechnol. 96, 1383–1394. doi: 10.1007/s00253-012-4440-2
del Valle, J. M., Lainoa, J. E., Savoy de Giori, G., and LeBlanc, J. G. (2014). Riboflavin producing lactic acid bacteria as abiotechnological strategy to obtain bio-enriched soymilk. Food Res. Int. 62, 1015–1019. doi: 10.1016/j.foodres.2014.05.029
Gelfand, M. S., Mironov, A. A., Jomantas, J., Kozlov, Y., and Perumov, D. A. (1999). A conserved RNA structure elementinvolved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 15, 439–442. doi: 10.1016/S0168-9525(99)01856-9
Guru, V., and Viswanathan, K. (2013). Riboflavin production in milk whey using probiotic bacteria – Lactobacillus acidophilus and Lactococcus lactis. Ind. J. Fund. Appl. Life Sci. 3, 169–176.
Jayashree, S., Rajendhran, J., Jayaraman, K., Kalaichelvana, G., and Gunasekaran, P. (2011). Improvement of riboflavin production by Lactobacillus fermentum isolated from yogurt. Food Biotechnol. 25, 240–251. doi: 10.1080/08905436.2011.590769
Koser, S. A. (1968). Vitamin Requirements of Bacteria and Yeasts. Springfield, IL: Charles C. Thomas.
LeBlanc, J. G., Milani, C., de Giori, G. S., Sesma, F., van Sinderen, D., and Ventura, M. (2013). Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 24, 160–168. doi: 10.1016/j.copbio.2012.08.005
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data usingreal-time quantitative PCR and the 2-[Delta][Delta] CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Perkins, J., and Pero, J. (2002). “Biosynthesis of riboflavin, biotin, folic acid, and cobalamin,” in Bacillus subtilis and Its Closest Relatives: From Genes to Cells, eds A. Sonenshine, J. Hoch, and R. Losick (Washington, DC: ASM Press), 271–276. doi: 10.1128/9781555817992.ch20
Perkins, J. B., Sloma, A., Hermann, T., Theriault, K., Zachgo, E., Erdenberger, T., et al. (1999). Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J. Ind. Microbiol. Biotechnol. 22, 8–18. doi: 10.1038/sj.jim.2900587
Ramiah, K., Van Reenen, C. A., and Dicks, L. M. T. (2007). Expression of the mucusadhesion genes Mub and MapA, adhesion-like factor EF-Tu and bacteriocin gene plaA of Lactobacillus plantarum 423, monitored with real-time PCR. Int. J. Food Microbiol. 116, 405–409. doi: 10.1016/j.ijfoodmicro.2007.02.011
Richter, G., Fischer, M., Krieger, C., Eberhardt, S., Lüttgen, H., and Gerstenschläger, I. (1997). Biosynthesis of riboflavin: characterization of the bifunctionaldeaminase-reductase of Escherichia coli and Bacillus subtilis. J. Bacteriol. 179, 2022–2028. doi: 10.1128/jb.179.6.2022-2028.1997
Richter, G., Ritz, H., Katzenmeier, G., Volk, R., Kohnle, A., Lottspeich, F., et al. (1993). Biosynthesis of riboflavin: cloning, sequencing, and mapping, and expression of the gene coding for GTP cyclohydrolase II of Escherichia coli. J. Bacteriol. 175, 4045–4051. doi: 10.1128/jb.175.13.4045-4051.1993
Richter, G., Volk, R., Krieger, C., Lahm, H. W., Rothlisberger, U., and Bacher, A. (1992). Biosynthesis of riboflavin:cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate of Escherichia coli. J. Bacteriol. 174, 4050–4056. doi: 10.1128/jb.174.12.4050-4056.1992
Russo, P., Capozzi, V., Arena, M. P., Spadaccino, G., Duenas, M. T., Lopez, P., et al. (2014). Riboflavin-overproducingstrains of Lactobacillus fermentum for riboflavin-enriched bread. Appl. Microbiol. Biotechnol. 98, 3691–3700. doi: 10.1007/s00253-013-5484-7
Sambrook, J. F., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Thakur, K., Lule, V. K., Rajni, C. S., and Kumar, N. (2016b). Riboflavin producing probiotic Lactobacilli as a biotechnological strategy to obtain riboflavin-enriched fermented foods. J. Pure Appl. Microbiol. 10, 161–166.
Thakur, K., Nanda, D. K., Kumar, N., and Tomar, S. K. (2015a). Phenotypic and genotypic characterization of indigenous Lactobacillus species from diverse niches of India. Curr. Trends Biotechnol. Pharm. 9, 222–227.
Thakur, K., and Tomar, S. K. (2015a). Exploring indigenous Lactobacillus species from diverse niches for riboflavin production. J. Young Pharm. 7, 122–127. doi: 10.5530/jyp.2015.2.11
Thakur, K., and Tomar, S. K. (2015b). Invitro study of Riboflavin producing lactobacilli as potential probiotic. LWT Food Sci. Technol. 68, 570–578. doi: 10.1016/j.lwt.2015.12.059
Thakur, K., Tomar, S. K., Brahma, B., and De, S. (2016c). Screening of riboflavin producing lactobacilli by PCR based approach and microbiological assay. J. Agric. Food Chem. 64, 1950–1956. doi: 10.1021/acs.jafc.5b06165
Thakur, K., Tomar, S. K., and De, S. (2015b). Lactic acid bacteria as a cell factory for riboflavin production. Microbiol. Biotechnol. 9, 441–451. doi: 10.1111/1751-7915.12335
Thakur, K., Tomar, S. K., Singh, A. K., Mandal, S., and Arora, S. (2016a). Riboflavin and health: a review of recent human research. Crit. Rev. Food Sci. Nutr. doi: 10.1080/10408398.2016.1145104 [Epub ahead of print].
Keywords: riboflavin, lactobacilli, fermented bamboo shoots, milk, whey, mRNA, roseoflavin
Citation: Thakur K, Tomar SK and Wei Z-J (2017) Comparative mRNA Expression Profiles of Riboflavin Biosynthesis Genes in Lactobacilli Isolated from Human Feces and Fermented Bamboo Shoots. Front. Microbiol. 8:427. doi: 10.3389/fmicb.2017.00427
Received: 31 May 2016; Accepted: 28 February 2017;
Published: 17 March 2017.
Edited by:
Jyoti Prakash Tamang, Sikkim University, IndiaReviewed by:
Prakash M. Halami, Central Food Technological Research Institute (CSIR), IndiaKeshab Chandra Mondal, Vidyasagar University, India
Copyright © 2017 Thakur, Tomar and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sudhir K. Tomar, sudhirndri@gmail.com
 Kiran Thakur1,2
Kiran Thakur1,2 Sudhir K. Tomar
Sudhir K. Tomar Zhao-Jun Wei
Zhao-Jun Wei