- State Key Laboratory of Agrobiotechnology and MOA Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing, China
Alternative σ factors in bacteria redirect RNA polymerase to recognize alternative promoters, thereby facilitating coordinated gene expression necessary for adaptive responses. The gene sig8 (sav_741) in Streptomyces avermitilis encodes an alternative σ factor, σ8, highly homologous to σB in Streptomyces coelicolor. Studies reported here demonstrate that σ8 is an important regulator of both avermectin production and stress responses in S. avermitilis. σ8 inhibited avermectin production by indirectly repressing expression of cluster-situated activator gene aveR, and by directly initiating transcription of its downstream gene sav_742, which encodes a direct repressor of ave structural genes. σ8 had no effect on cell growth or morphological differentiation under normal growth conditions. Growth of a sig8-deletion mutant was less than that of wild-type strain on YMS plates following treatment with heat, H2O2, diamide, NaCl, or KCl. sig8 transcription was strongly induced by these environmental stresses, indicating response by σ8 itself. A series of σ8-dependent genes responsive to heat, oxidative and osmotic stress were identified by EMSAs, qRT-PCR and in vitro transcription experiments. These findings indicate that σ8 plays an important role in mediating protective responses to various stress conditions by activating transcription of its target genes. Six σ8-binding promoter sequences were determined and consensus binding sequence BGVNVH-N15-GSNNHH (B: C, T or G, V: A, C or G, S: C or G, H: A, C or T, N: any nucleotide) was identified, leading to prediction of the σ8 regulon. The list consists of 940 putative σ8 target genes, assignable to 17 functional groups, suggesting the wide range of cellular functions controlled by σ8 in S. avermitilis.
Introduction
Soil-inhabiting filamentous Streptomyces are characterized by the ability to produce valuable secondary metabolites having antimicrobial, anticancer, anthelmintic, immunosuppressive, or other biological activities (Challis and Hopwood, 2003). Biosynthesis of secondary metabolites is usually accompanied by initiation of morphological differentiation, and precisely controlled by complex regulatory networks involving cluster-situated regulators (CSRs) and higher-level global/pleiotropic regulators in response to various environmental and endogenous signals (van Wezel and McDowall, 2011; Liu et al., 2013; Niu et al., 2016; Urem et al., 2016). Elucidation of these regulatory networks is essential for strain improvement by metabolic engineering approaches.
Streptomyces avermitilis is an important industrial microorganism used for production of avermectins, effective anthelmintic agents widely applied in agricultural and medical fields (Burg et al., 1979; Egerton et al., 1979). aveR, located at the left edge of the ave gene cluster, encodes a LuxR-family cluster-situated activator (Kitani et al., 2009; Guo et al., 2010). The aveR promoter is recognized by a housekeeping σ factor, σhrdB (Zhuo et al., 2010). We showed that two extracytoplasmic function (ECF) σ factors, σ6 (SAV663) and σ25 (SAV3351), inhibit avermectin production by indirectly affecting aveR transcription (Jiang et al., 2011; Luo et al., 2014). The other 57 σ factors in S. avermitilis remain to be characterized. Elucidation of their functions will help to clarify the regulatory networks involved in avermectin biosynthesis.
With very few exceptions, bacterial σ factors belong to the σ70 family, which is divided into four groups (Groups 1–4) based on differential possession of four conserved domains (σ1, σ2, σ3, and σ4), phylogenic relationships and physical functions (Osterberg et al., 2011). Group 1 housekeeping sigma factors possess all four domains and are necessary for growth. Group 2 sigma factors also contain four domains, but are dispensable for growth. Group 3 alternative σ factors lack a σ1 domain and are involved mainly in stress response and differentiation processes. Group 4 ECF sigma factors contain only σ2 and σ4 domains and usually respond to extracytoplasmic stimuli. The first identified alternative σ factor is σB in Bacillus subtilis (Haldenwang and Losick, 1980), which functions as a master regulator that controls >200 genes in response to a wide variety of stress/starvation stimuli including glucose, phosphate, or oxygen starvation, heat or cold shock, ethanol, acid, or osmotic stress, nitric oxide, and antibiotic-induced cell wall damage (Lee et al., 2005; Hecker et al., 2007). sigB and its homologes are widespread among Gram-positive bacteria, and have diverse functions (Hecker et al., 2007). SCO0600, the σB homolog in Streptomyces coelicolor, responds to osmotic stress, but not to heat, ethanol, or H2O2 stress (Cho et al., 2001; Lee et al., 2004a, 2005). σB is also involved in secondary metabolism and morphological differentiation in S. coelicolor. The sigB deletion mutant of this species displayed overproduction of actinorhodin (ACT), reduced production of undecylprodigosin (RED) and lack of aerial mycelium formation on R2YE or NA plates (Cho et al., 2001). In Streptomyces hygroscopicus 5008, sigB transcription was enhanced by heat stress or treatment with a reactive oxygen species (ROS) inhibitor, suggesting involvement of σB in response to changes in temperature or ROS level (Wei et al., 2012).
We demonstrated recently that SAV742, a novel AraC-family transcriptional regulator in S. avermitilis, directly represses ave structural genes and controls cell growth and morphological differentiation (Sun et al., 2016). The gene adjacent to sav_742, sav_741 (sig8), encodes an alternative σ factor, σ8, homologous to σB in S. coelicolor. We describe here characterization of σ8 as an important regulator that controls avermectin biosynthesis and multiple stress responses in S. avermitilis. σ8 represses avermectin biosynthesis in part through direct initiation of sav_742 transcription. Unlike its S. coelicolor homolog σB, σ8 responds to heat, osmotic and oxidative stress by directly regulating expression of its own gene and certain other stress protection genes, but is not involved in morphological differentiation or cell growth. Moreover, we predicted the σ8 regulon based on the consensus σ8-binding promoter sequence.
Materials and Methods
Primers, Plasmids, Strains, and Growth Conditions
The strains and plasmids used or constructed in this study are listed in Table 1, and the primers in Supplementary Table S1. Culture conditions for Escherichia coli and S. avermitilis strains were as described previously (Liu W. et al., 2015). MM, R2YE (Kieser et al., 2000) and YMS (Ikeda et al., 1988) plates were used for phenotypic observation of S. avermitilis strains. Insoluble fermentation medium FM-I (Chen et al., 2007) was used for routine avermectin production. Soluble fermentation medium FM-II (Guo et al., 2010) was used to grow mycelia for biomass analysis, and for RNA isolation following stress treatment.
Gene Deletion, Complementation, and Overexpression
A sig8 in-frame gene deletion mutant was constructed using a homologous recombination strategy. Two homologous fragments flanking sig8 were amplified by PCR from wild-type (WT) genomic DNA. A 529-bp 5′-flanking region (positions -337 to +192 relative to the sig8 start codon) was amplified with primers SD2A/SD2B, and a 541-bp 3′-flanking region (positions +835 to +1375) was amplified with primers SD3A/SD3B. The two fragments were cut with HindIII/BamHI and BamHI/EcoRI, respectively, and then simultaneously cloned into HindIII/EcoRI-digested pKC1139 to generate sig8 deletion vector pKCDsig8. Non-methylated pKCDsig8 was transformed into WT protoplasts, and double-crossover recombinant strains were isolated as reported previously (Zhao et al., 2007). Resulting sig8 deletion mutants were verified by colony PCR using primers SD23A/SD23B (flanking the exchange regions) and SD39A/SD39B (located within the deletion region of sig8) (Supplementary Figure S1), followed by DNA sequencing. When primers SD23A/SD23B were used, a 1.2-kb band appeared, whereas a 1.9-kb band was detected in WT genomic DNA. When primers SD39A/SD39B were used, only WT DNA generated a 390-bp band. We thus obtained sig8 gene deletion mutant Dsig8, in which 840-bp sig8 ORF was mostly deleted (from positions +193 to +834 relative to the start codon) (Supplementary Figure S1). The deletion part of sig8 covered coding region for all three functional domains (σ2, σ3, and σ4); Thus, the remaining fragment was unlikely to be functional.
To construct a sig8sav_742 double deletion mutant, the pKCDsig8 vector was transformed into D742 protoplasts (Sun et al., 2016). The expected mutant, Dsig8–742, was isolated by selection of the Dsig8 mutant.
For complementation of Dsig8, a 1618-bp PCR fragment containing the sig8 promoter and open reading frame (ORF) was amplified with primers SD1B/SD1D, and inserted into the integrative vector pSET152 to give sig8-complemented vector pSET152-sig8, which was then transformed into Dsig8 to obtain complemented strain Csig8.
For overexpression of sig8, a 987-bp fragment carrying the sig8 ORF was amplified using primers SD1B/SD1C, and inserted into pJL117 to generate pJL117-sig8, in which sig8 was controlled by ermE∗p (Streptomyces strong constitutive promoter). The 1.3-kb EcoRI/HindIII fragment containing ermE∗p and sig8 ORF from pJL117-sig8 was ligated into pKC1139 to generate sig8 overexpression vector pKC1139-erm-sig8, which was transformed into WT protoplast to construct sig8 overexpression strain Osig8.
Production and Analysis of Avermectins
Fermentation of S. avermitilis strains and HPLC (high-pressure liquid chromatography) analysis of avermectin yield in fermentation broth were performed as described previously (Chen et al., 2007).
Quantitative Real-Time RT-PCR (qRT-PCR)
S. avermitilis mycelia grown in FM-I or FM-II were collected at various time points for RNA isolation. Triturated samples were suspended in TRIzol reagent (Tiangen, China) for RNA isolation. Genomic DNA contamination was removed by treatment of RNA samples with RNase-free DNase I (TaKaRa, China). 4 μg total RNA was used for cDNA synthesis. qRT-PCR was performed using primers listed in Supplementary Table S1 to analyze transcription levels of the tested genes as described previously (Luo et al., 2014), with expression level of housekeeping gene 16S rRNA as internal control. Each experiment was repeated three times.
Overexpression and Renaturation of His6-σ8
The 953-bp sig8 coding region (279 amino acids) was amplified from WT genomic DNA using primers SD1A/SD1B. The obtained PCR product was inserted into pET-28a (+) to generate pET-sig8. After confirmation by DNA sequencing, pET-sig8 was transformed into E. coli BL21 (DE3) for His6-σ8 (N-terminal His6-tagged σ8 recombinant protein) overexpression. Following IPTG induction, cells containing His6-σ8 were harvested, washed, resuspended in a lysis buffer (Luo et al., 2014), and disrupted by sonication on ice. His6-σ8 present as inclusion body in E. coli was purified, solubilized, and renatured as described previously (Luo et al., 2014). Purified renatured soluble His6-σ8 was quantified and stored at -80°C for use in vitro transcription experiments and electrophoretic mobility shift assays (EMSAs).
Electrophoretic Mobility Shift Assays (EMSAs)
EMSA probes carrying respective tested promoter regions were obtained by PCR using primers listed in Supplementary Table S1, and labeled at their 3′ ends with digoxigenin (DIG). Conditions for binding reaction and detection were as described previously (Sun et al., 2016). To confirm binding specificity between His6-σ8 and DNA probes, a ∼300-fold excess of each specific or non-specific (hrdB) unlabeled probe was added to the reaction mixture before incubation.
In Vitro Transcription Assay
DNA templates containing respective promoter and partial coding region were amplified by PCR using primers listed in Supplementary Table S1. Conditions for transcription assays and detection of transcripts were as described previously (Luo et al., 2014). Reaction mixtures contained 0.4 pmol DNA template and various amounts of renatured His6-σ8. Transcription was initiated by addition of 3.7 pmol E. coli core RNA polymerase (core-RNAP) (New England Biolabs, USA).
5′ Rapid Amplification of cDNA Ends (5′ RACE)
The transcriptional start site (TSS) of selected genes was identified using a 5′/3′ RACE kit (Roche, 2nd Generation). S. avermitilis strains were cultured in FM-II at 28°C for 2 days, and then subjected to various stress conditions. Mycelia were harvested after various durations of stress treatment and used for RNA isolation. 4 μg total RNA was reverse transcribed with 40 pmol gene-specific primer SP1. Purified cDNAs were added to oligo-dA tails at the 3′ end by TdT (terminal deoxynucleotidyl transferase, TaKaRa) treatment at 37°C for 30 min. The tailed cDNA was used as template for first-round PCR using second inner gene-specific primer SP2 and oligo-dT anchor primer. The resulting PCR product was diluted to appropriate concentration, and used as template for second-round PCR with nested primer SP3 and an anchor primer. The purified final PCR product was sequenced (Invitrogen Biotechnology Corporation, China). TSS was determined as the first nucleotide following the oligo-dT sequence.
Results
σ8 Regulates Avermectin Production by Repressing ave Gene Expression during the Late Fermentation Stage
The sig8 (sav_741) gene in the S. avermitilis chromosome contains 840 nucleotides (nt) and encodes a 279-amino-acid σ70-family alternative σ factor, σ8. The downstream convergently transcribed gene sav_742, located 377 nt from sig8 (Figure 1A), encodes an AraC-family transcriptional regulator that was recently characterized as a global regulator of avermectin biosynthesis, cell growth, and morphological development (Sun et al., 2016). The upstream convergently transcribed gene sav_740, located 215 nt from sig8, encodes a hypothetical protein. BLAST analysis revealed that σ8 displays high amino acid sequence identities with S. coelicolor σB (77.22%) and S. hygroscopicus σB (78.29%).
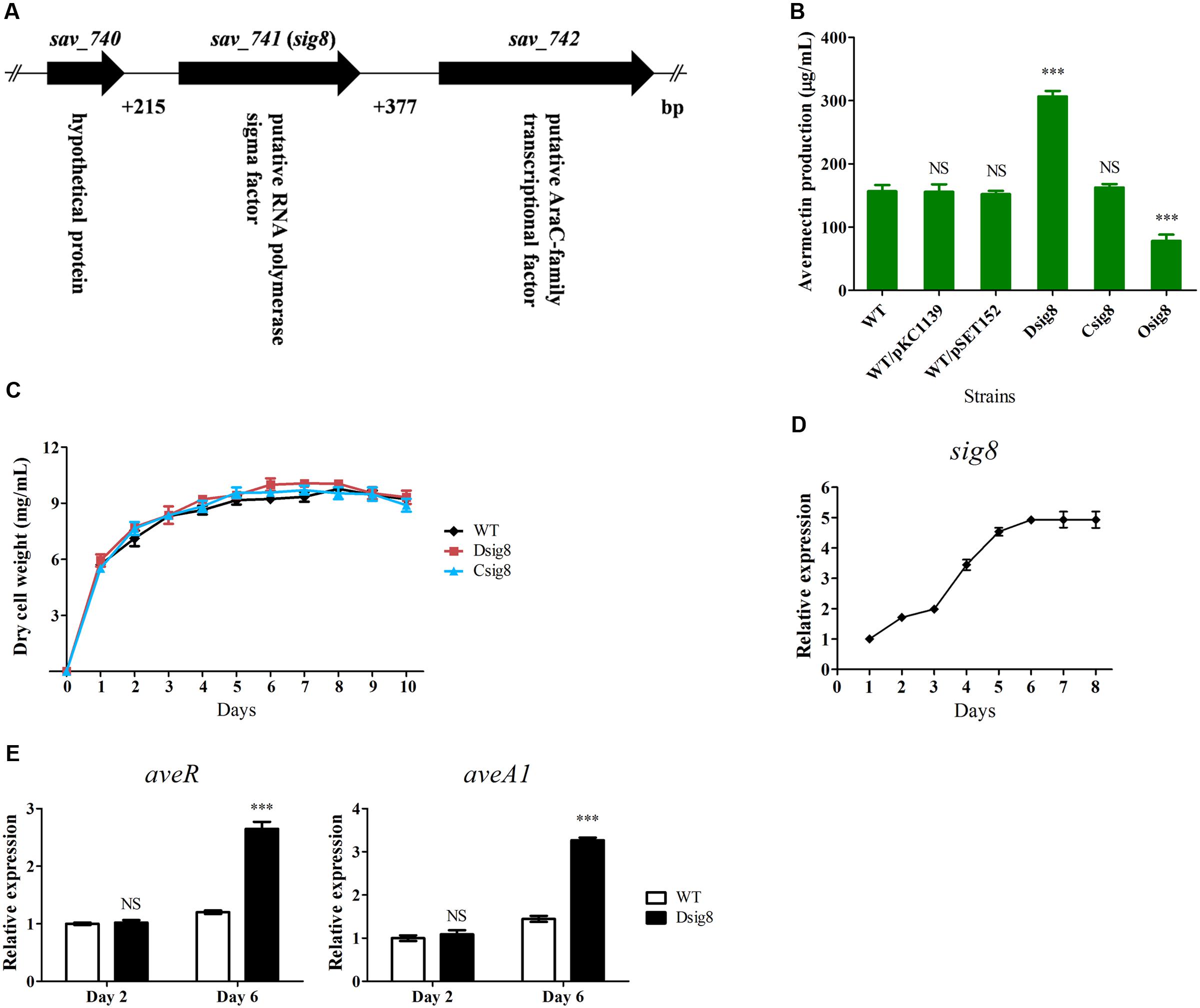
FIGURE 1. Effects of σ8 on avermectin production, cell growth and ave gene transcription of S. avermitilis. (A) Schematic diagram of sig8 and its neighboring genes. (B) Comparative avermectin yield in WT, sig8 deletion mutant (Dsig8), complemented strain (Csig8) and overexpression strain (Osig8) grown in FM-I for 10 days. WT/pKC1139 and WT/pSET152: vector control strains constructed by transformation of plasmids pKC1139 and pSET152 into WT, respectively. Error bar: SD from three independent experiments. NS, not significant; ∗∗∗, P < 0.001 for comparison of values for mutant versus WT strains (Student’s t-test). (C) Growth curves of WT, Dsig8 and Csig8 in soluble FM-II. (D) Transcriptional profile of sig8 in WT grown in FM-I. Values were calculated by normalization against internal control gene 16S rRNA at specific time points. The relative value of sig8 at day 1 was assigned as 1. (E) qRT-PCR analysis of aveR and aveA1 from WT and Dsig8 grown in FM-I at days 2 and 6. Transcription level of each gene was expressed relative to WT value at day 2, defined as 1. NS, not significant; ∗∗∗, P < 0.001 (Student’s t-test).
To investigate the role of σ8 in avermectin biosynthesis, we constructed sig8 deletion mutant Dsig8, complemented strain Csig8 and overexpression strain Osig8, and compared their avermectin yields with that of WT ATCC31267 cultured in FM-I for 10 days. Relative to WT yield, that of Dsig8 was ∼96% higher, that of Osig8 was ∼50% lower, and that of Csig8 was similar (Figure 1B). Yields of two vector control strains, WT/pSET152 and WT/pKC1139, were almost the same as that of WT (Figure 1B). These findings indicate that σ8 has a negative effect on avermectin production.
To assess the effect of σ8 on S. avermitilis growth, we measured biomasses of WT, Dsig8 and Csig8 cultured in soluble FM-II. Biomass of Dsig8 and Csig8 was similar to that of WT (Figure 1C), indicating that σ8 has no effect on cell growth, and that avermectin overproduction in Dsig8 does not result from alteration of growth. Dsig8 and Osig8 grew normally on YMS, R2YE, and MM plates (Supplementary Figure S2), indicating that σ8 also has no effect on morphological differentiation.
To investigate the relationship between σ8 and avermectin production, we examined the sig8 transcription profile by qRT-PCR using RNAs isolated from WT in FM-I. sig8 transcript was detected throughout the fermentation process. Its level increased gradually from day 1, reached a maximum on day 6, and remained high thereafter, suggesting that σ8 plays its regulatory role mainly during the late stage of fermentation (Figure 1D).
The effect of σ8 on expression of ave genes was assessed by qRT-PCR analysis of transcription levels of aveR (CSR gene) and aveA1 (structural gene encoding AVES1 polyketide synthase) in WT and Dsig8 grown in FM-I for 2 days (early exponential phase) or 6 days (stationary phase). Transcription levels of aveR and aveA1 in Dsig8 did not differ from those in WT on day 2, but were notably higher on day 6 (Figure 1E), consistent with the observed avermectin overproduction in Dsig8. These findings indicate that σ8 controls avermectin production at the transcription level by repressing ave genes, primarily in the late fermentation stage.
σ8 Directly Activates Expression of Its Own Gene and sav_742, Indirectly Regulates ave Genes
σ factors function as initiators of transcription. The negative regulatory effects of σ8 on aveR and aveA1 are therefore likely to be indirect. To test this idea, we performed EMSAs using refolded soluble recombinant His6-σ8 and probes aveRp and aveA1p prepared by labeling promoter regions of aveR and aveA1. His6-σ8 did not bind to the probes even at high concentration (1 μM) (Figure 2A), confirming that σ8 indirectly represses ave genes.
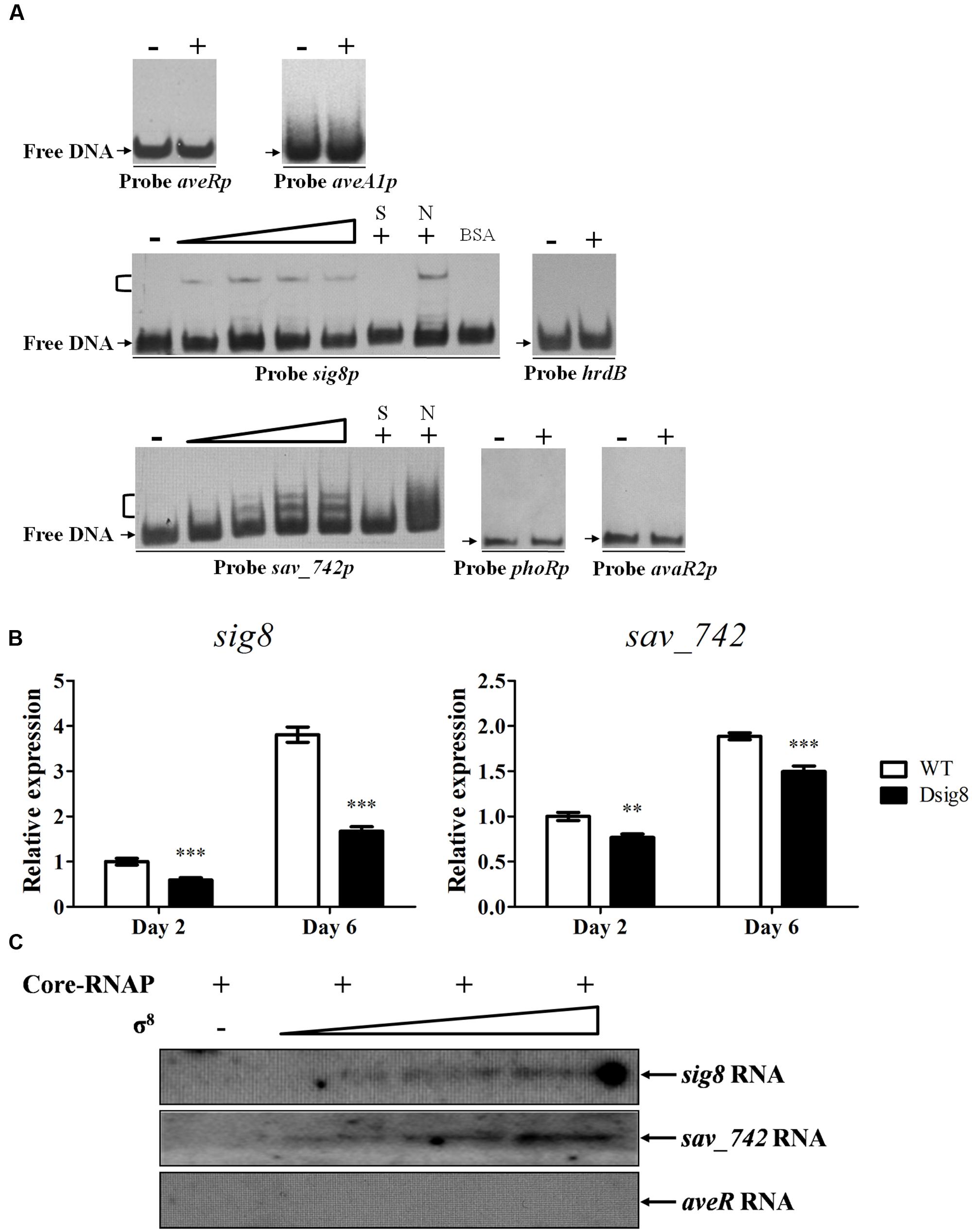
FIGURE 2. Initiation of sig8 and sav_742 transcription by σ8. (A) EMSAs of His6-σ8 interaction with probes aveRp, aveA1p, sig8p, sav_742p, phoRp, and avaR2p. Each lane contained 0.15 nM labeled probe. For competition assays, ∼300-fold excess of unlabeled specific (lanes S) or non-specific (lanes N) competitor DNA was used. Lanes –: EMSAs without His6-σ8. Lanes 2 to 5, for probes sig8p and sav_742p, contained respectively 0.25, 0.5, 0.75, and 1 μM renatured His6-σ8. 1 μM His6-σ8 was used for competition assays and probes aveRp, aveA1p, phoRp, avaR2p, and hrdB (lanes +). Negative probe: labeled non-specific probe hrdB. Negative protein control: 0.1% BSA. Arrows: free probes. Brackets: σ8-DNA complexes. (B) qRT-PCR analysis of sig8 and sav_742 in WT and Dsig8 grown in FM-I. Transcription level of each gene is expressed relative to WT value at day 2, defined as 1. ∗∗, P < 0.01; ∗∗∗, P < 0.001 (t-test). (C) In vitro transcription assay of sig8, sav_742 and aveR promoters by RNA polymerase containing σ8 (Eσ8). Each reaction mixture contained 0.4 pmol DNA template. Eσ8 holoenzyme was reconstituted by mixing 3.7 pmol Escherichia coli core-RNAP with various amounts (30, 70, 110 pmol) of renatured His6-σ8.
σB homologes typically initiate their own transcription (Hecker et al., 2007). The finding that σ8 indirectly represses ave genes suggested that σ8 initiates transcription of direct repressor(s) of aveR or aveA1. We recently characterized PhoP (Yang et al., 2015) and AvaR2 (Zhu et al., 2016) as direct repressors of aveR. We also found that SAV742 directly represses transcription of several ave structural genes including aveA1, but not aveR (Sun et al., 2016). To test the hypothesis that σ8 directly controls sig8 and regulatory genes phoP, avaR2 and sav_742, we performed EMSAs, qRT-PCR analysis and in vitro transcription assays.
EMSA results revealed that His6-σ8 formed complexes with promoter regions of sig8 (probe sig8p) and sav_742 (probe sav_742p), but not with those of avaR2 (probe avaR2p) or phoR-phoP operon (probe phoRp) (Figure 2A). No shifted band was observed for negative probe control hrdB or protein control BSA. Binding specificity was confirmed by competitive assays using excess unlabeled specific probe (lanes S) and non-specific probe hrdB (lanes N). qRT-PCR analysis showed that transcription levels of sig8 and sav_742 on days 2 and 6 were lower in Dsig8 than in WT (Figure 2B), indicating that σ8 positively regulates expression of its own gene and adjacent gene sav_742. The lower sav_742 transcription level and higher avermectin yield in Dsig8 are consistent with SAV742’s function as a repressor of avermectin production (Sun et al., 2016).
In vitro transcription experiments were performed to determine whether σ8 initiates transcription of sig8 and sav_742. Linear DNA fragments harboring sig8, sav_742, and aveR promoter regions were used as templates, and various amounts of renatured His6-σ8 were mixed with sufficient E. coli core-RNAP to reconstitute RNAP holoenzyme (Eσ8). Core-RNAP alone did not initiate transcription at the sig8 or sav_742 promoter, whereas the Eσ8 holoenzyme did (Figure 2C). When core-RNAP concentration was kept constant and σ8 concentration was increased, transcript levels of sig8 and sav_742 increased, but no aveR transcript was detected. These findings indicate that σ8 initiates transcription of its own gene and sav_742, but not that of aveR. The presence of sig8 and sav_742 transcripts in Dsig8 suggests that additional σ factors may recognize sig8 and sav_742 promoters and initiate their transcription, but with lower transcription efficiency.
Avermectin production in Dsig8 was very close to that in sig8sav_742 double deletion mutant Dsig8–742 (Supplementary Figure S3), but was higher than that in sav_742 deletion mutant D742 (Sun et al., 2016), suggesting that other σ8 targets may also affect avermectin production in Dsig8.
σ8 Responds to Various Environmental Stresses
Alternative σ factors are usually involved in modulation of stress responses (Osterberg et al., 2011). To determine which type(s) of stress σ8 responds to, we performed stress tests on YMS plates. Relative to growth of WT, that of sig8 deletion mutant Dsig8 was more sensitive to NaCl, KCl, H2O2, diamide, and heat (42°C) stresses, but similarly sensitive to tert-butyl hydroperoxide (TBHP) and sucrose stresses (Figure 3A). These findings indicate that σ8 is required for WT levels of resistance to multiple stresses, e.g., heat stress, salt stress, and oxidative stress (from H2O2 or diamide).
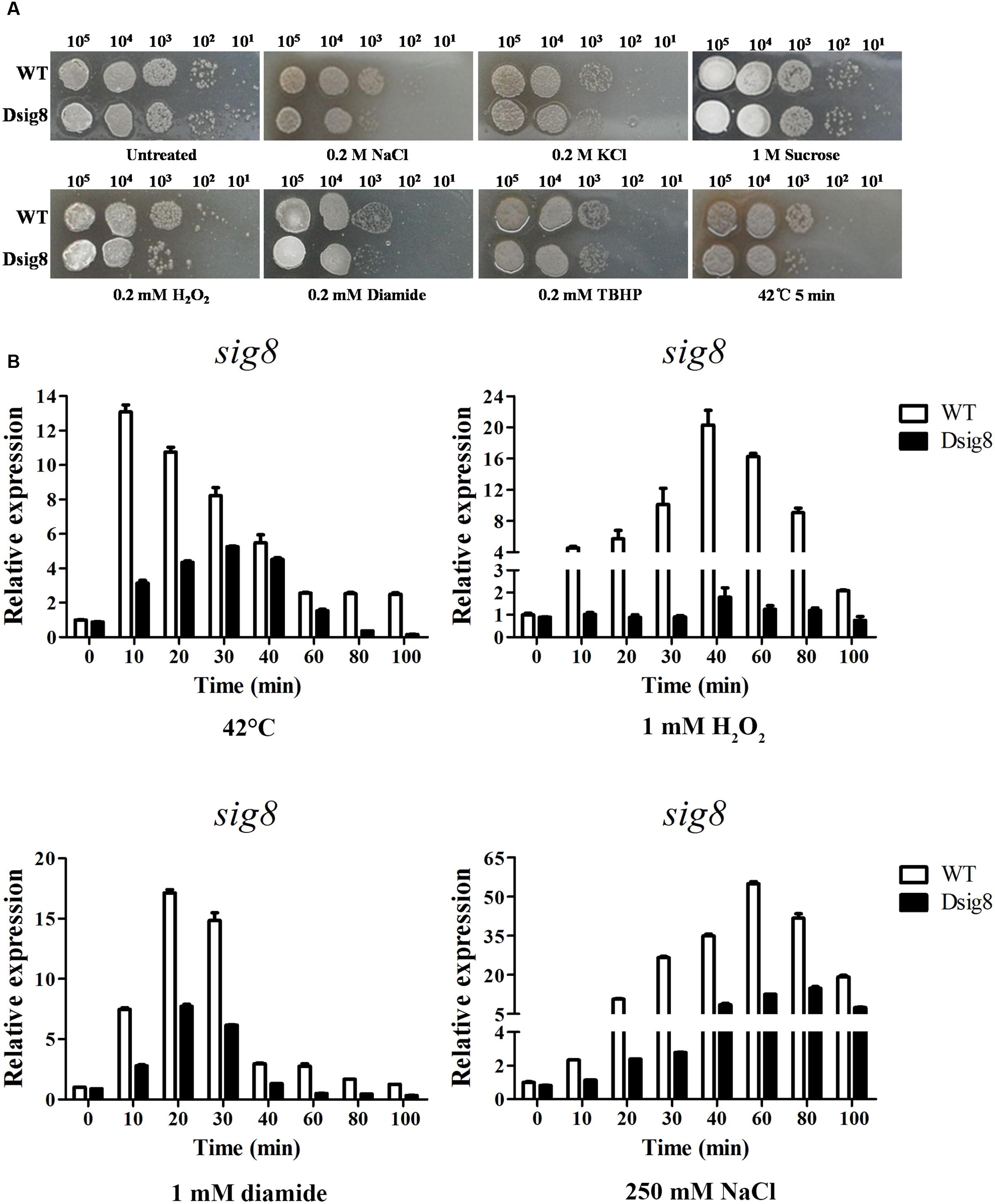
FIGURE 3. Role of σ8 in responses to osmotic, oxidative and heat stresses. (A) Sensitivity of WT and Dsig8 to stress conditions. For osmotic and oxidative stress assays, serial dilutions of spores were spotted onto YMS plates containing NaCl (0.2 M), KCl (0.2 M), sucrose (1 M), H2O2 (0.2 mM), diamide (0.2 mM), or TBHP (0.2 mM), and incubated at 28°C for 3 days. For heat stress assay, spore suspensions were treated at 42°C for 5 min and then spotted onto YMS plates. Each stress assay was repeated three times on YMS plates with consistent results. (B) Induction of sig8 transcription in WT and Dsig8 by stresses. RNAs used for qRT-PCR analysis were prepared from cells grown in FM-II for 2 days followed by treatment with 42°C, 1 mM H2O2, 1 mM diamide, or 250 mM NaCl for the indicated times. Relative values are shown as fold change relative to sig8 transcription level at the first time point (0 min) in WT, which was assigned as 1.
The observations that sig8 mutant Dsig8 was more sensitive to various environmental stresses, and that σ8 was autoregulated, suggested that sig8 itself may be induced by these environmental stresses in a σ8-dependent manner. We tested this possibility by qRT-PCR comparison of sig8 transcription levels in WT and Dsig8 under stress conditions. Cells were cultured in soluble FM-II for 2 days, and then treated with heat (42°C), H2O2, diamide, or NaCl. RNA samples were isolated from cells before (0 min) and after treatment (10, 20, 30, 40, 60, 80, and 100 min). For WT, sig8 transcription level increased to a maximal value within 10–60 min for each stress type (∼13-fold for heat; ∼20-fold for H2O2, ∼17-fold for diamide; ∼54-fold for NaCl). For Dsig8, maximal induction was sharply decreased under each stress type (∼6-fold for heat; ∼2-fold for H2O2, ∼8-fold for diamide; ∼17-fold for NaCl) and delayed for heat (from 10 to 30 min) and salt (from 60 to 80 min) (Figure 3B). These findings indicate that σ8 itself responds to various environmental stresses at the transcription level in either a σ8-dependent or σ8-independent manner. The rapid and robust induction of sig8 in WT may be due to σ8-dependent control, whereas the decreased and/or delayed induction in Dsig8 may be due to σ8-independent control.
Identification of σ8 Target Genes Related to Heat Stress
σ8 was strongly induced in response to a variety of stresses, suggesting that it mediates defensive systems against stresses. To identify σ8 target genes that respond to heat stress, we initially performed a series of EMSAs using His6-σ8 and potential promoter probes of heat shock genes, including dnaK1p for dnaK1-grpE1-dnaJ1-hspR operon, dnaK2p for dnaK2-grpE2-dnaJ2 operon, groES1p for groES1-groEL1 operon, groEL2p, htpGp, hsp18_1p and hsp18_2p. His6-σ8 bound specifically to dnaK1p, but not to other probes tested (Figure 4A). In vitro transcription analysis confirmed that dnaK1 transcription was initiated by σ8 (Figure 4B). dnaK1 transcription level in Dsig8 was downregulated at two time points (Figure 4C). These findings demonstrate that σ8 is a direct activator of dnaK1.
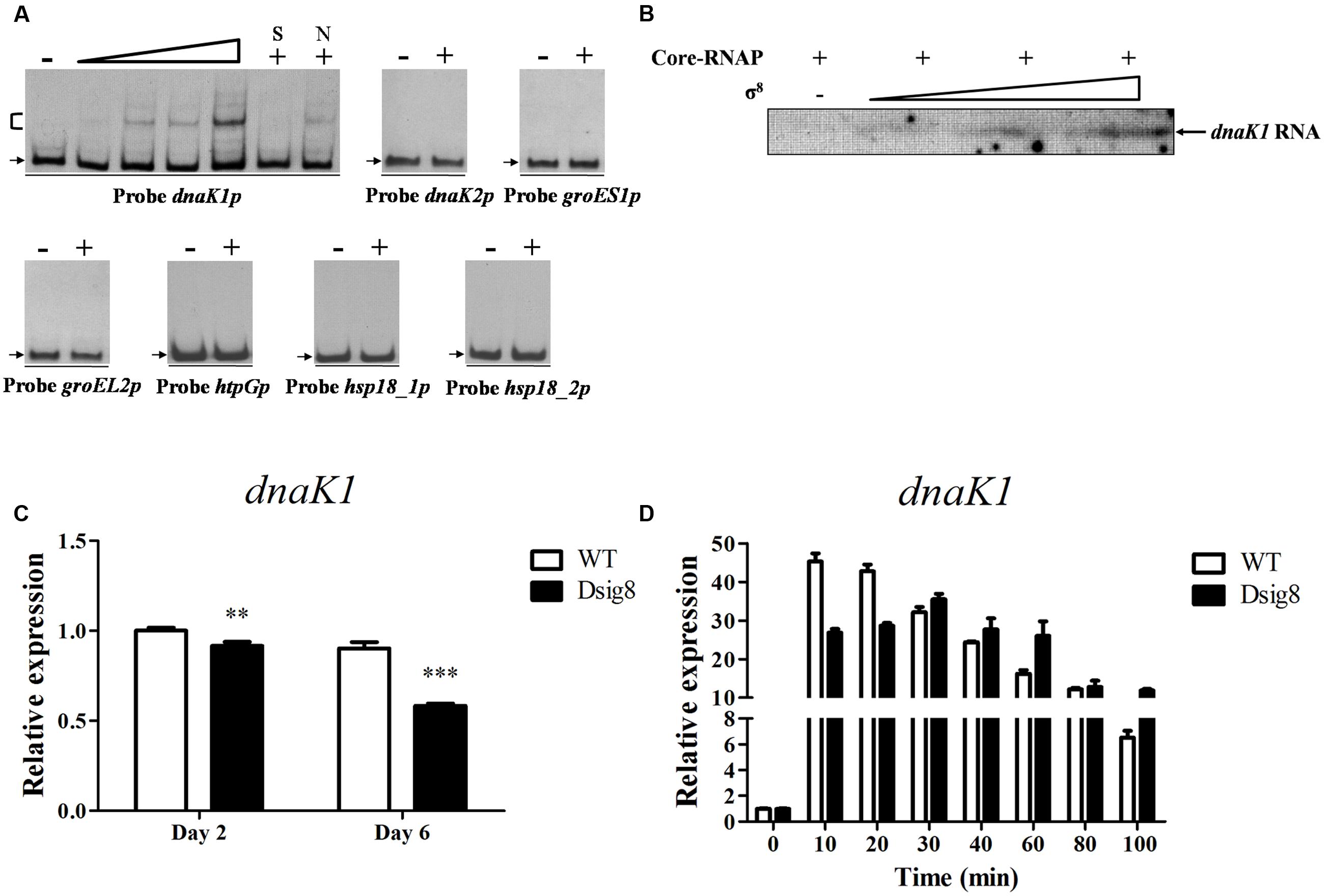
FIGURE 4. σ8-dependent transcription of dnaK1. (A) EMSAs of His6-σ8 with dnaK1, dnaK2, groES1, groEL2, htpG, hsp18_1, and hsp18_2 promoter regions under conditions as described for Figure 2A. (B) In vitro transcription assay of the dnaK1 promoter by Eσ8. (C) qRT-PCR analysis of dnaK1 in WT and Dsig8 grown in FM-I. The WT value at day 2 was defined as 1. ∗∗, P < 0.01; ∗∗∗, P < 0.001 (t-test). (D) Induction of dnaK1 by heat (42°C) in WT and Dsig8 grown in FM-II. dnaK1 transcription level before temperature rise (0 min) in WT was assigned as 1.
grpE1, dnaJ1, and hspR are cotranscribed with dnaK1, and therefore are also σ8 targets. dnaK1, grpE1, and dnaJ1 encode molecular chaperones for interaction with denatured proteins and facilitate refolding to the native state following heat stress, and hspR encodes a putative transcriptional repressor of its operon. Transcription levels of dnaK1 in WT and Dsig8 under heat stress were recorded to determine whether dnaK1p for its operon was induced in a σ8-dependent manner. In WT, dnaK1 transcription level increased rapidly to ∼45-fold after 10 min exposure to 42°C, and then gradually declined. In Dsig8, maximal induction was reduced to ∼35-fold and delayed after 30 min (Figure 4D). These findings indicate that σ8 facilitates rapid adaption to heat stress mainly by direct regulation of the dnaK1-grpE1-dnaJ1-hspR operon. The slower, lower-degree induction of dnaK1p in Dsig8 presumably depends on other factors not addressed here.
Identification of σ8-dependent Genes Involved in Responses to Oxidative Stress
σ8 responds to H2O2 and diamide, which respectively cause peroxidative and thiol-oxidative stress. Bacteria often respond to H2O2 stress by producing catalases and peroxidases that degrade H2O2. We showed that responses to H2O2 stress in S. avermitilis involve three catalase genes (katA1, katA2, katA3), the ahpCD operon (encoding alkyl hydroperoxide reductase and alkylhydroperoxidase), and two peroxide-sensing transcriptional factor genes (oxyR, catR) (Liu et al., 2016). In S. coelicolor, ECF-σR plays a key role in response to thiol-oxidative stress, and major σR targets include trx genes (encoding the thioredoxin system) and msh genes (for biosynthesis of mycothiol, the major thiol buffer) (Kallifidas et al., 2010). S. avermitilis contains one sigR homologous gene (sig22), six thioredoxin genes (trxA1, trxA2, trxA3, trxA4, trxA5, trxA6), two thioredoxin reductase genes (trxB1, trxB2), and four msh genes (mshA, mshB, mshC, mshD). Possible interactions of σ8 with these oxidative stress-related genes were investigated by EMSAs. His6-σ8 bound specifically to the bidirectional promoter probes catR_katA1_int and oxyR_ahpCD_int, and to promoter probes trxA3p, trxB2p, mshAp, mshCp, mshDp and sig22p, but not to probes katA2p, katA3p, trxA1p, trxA2p, trxA4p, sav_2810p for sav_2810-2809-trxA5-2807-cyp12 operon, trxA6p, trxB1p, or mshBp (Figure 5A). In vitro transcription analysis showed that σ8 initiated transcription of catR, oxyR, trxA3, trxB2, mshA, mshC, mshD and sig22, but not that of katA1 or ahpC (Figure 5B). Transcription levels of catR, oxyR, trxA3, trxB2, mshA, mshC, mshD, and sig22 were all reduced in Dsig8 relative to WT on day 6, or days 2 and 6 (Figure 5C). These findings indicate that catR, oxyR, trxA3, trxB2, mshA, mshC, mshD, and sig22 are directly activated by σ8.
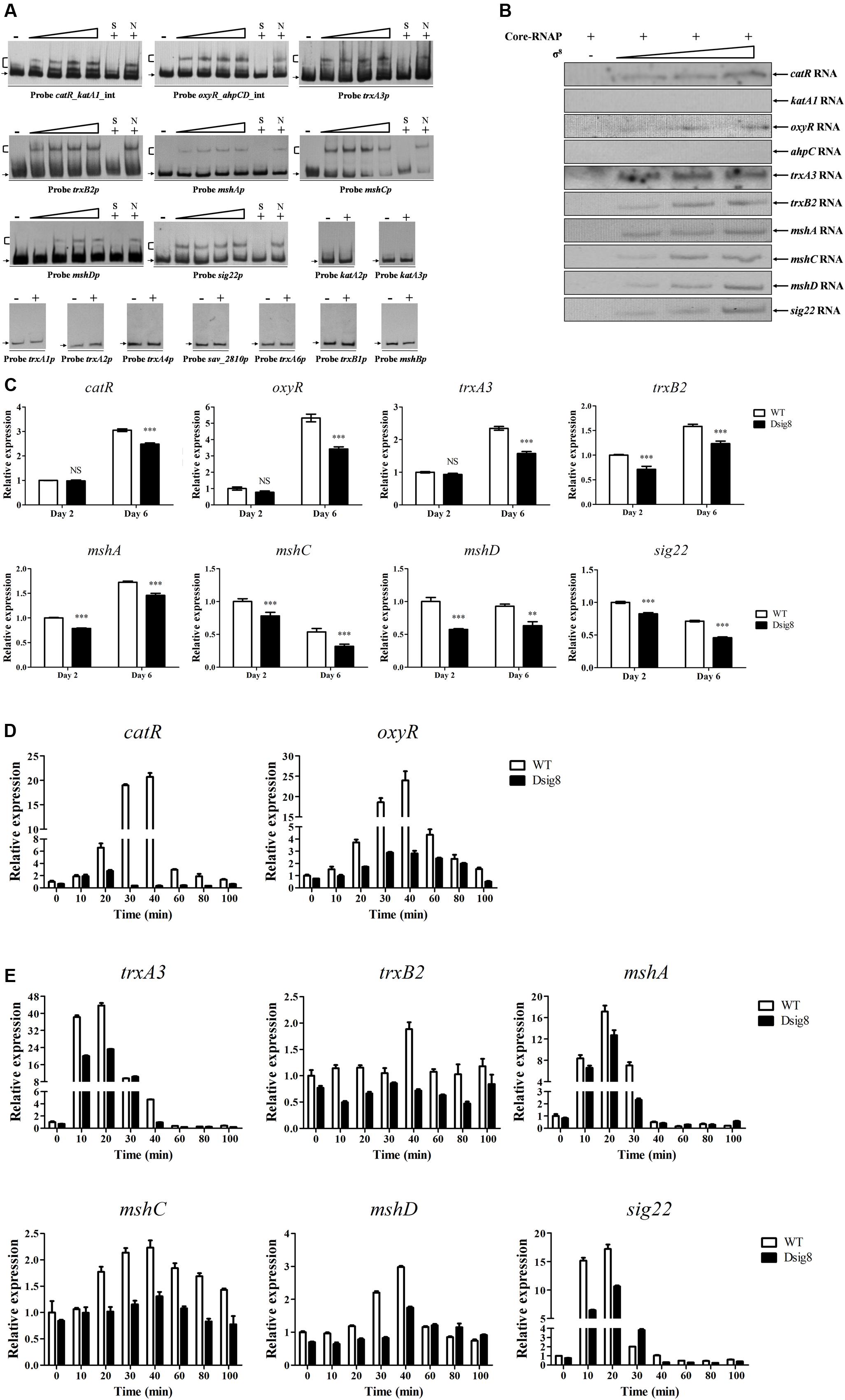
FIGURE 5. Identification of σ8 target genes involved in responses to oxidative stress. (A) EMSAs of His6-σ8 with promoters of genes involved in oxidative stress responses. (B) In vitro transcription analysis of catR, katA1, oxyR, ahpC, trxA3, trxB2, mshA, mshC, mshD, and sig22 promoters by Eσ8. (C) qRT-PCR analysis of eight newly identified σ8 target genes in WT and Dsig8 grown in FM-I. Transcription level of each gene is expressed relative to WT value at day 2, defined as 1. NS, not significant; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (t-test). (D) Induction of catR and oxyR by 1 mM H2O2 in WT and Dsig8 grown in FM-II. Relative value of each gene in WT before treatment (0 min) was assigned as 1. (E) Induction of trxA3, trxB2, mshA, mshC, mshD, and sig22 by 1 mM diamide in WT and Dsig8 grown in FM-II.
For analysis of oxidative stress responses, WT and Dsig8 were treated with 1 mM H2O2 or diamide for various durations. In WT, catR and oxyR were induced to maximal transcription level (∼20- and ∼23-fold, respectively) by H2O2 within 40 min (Figure 5D). In Dsig8, H2O2 treatment caused only a slight increase of transcription level (∼4-fold induction) of these two genes. These findings indicate that σ8 mediates H2O2 induction of the peroxide-sensing regulators CatR and OxyR. Diamide treatment of WT caused notable induction of trxA3 (∼43-fold), mshA (∼17-fold) and sig22 (∼17-fold) within 20 min, and slight induction of mshC (∼2.2-fold), mshD (∼3-fold) and trxB2 (∼1.8-fold) within 40 min, whereas diamide treatment of Dsig8 had much lower (for trxA3, mshA, mshC, mshD, and sig22) or no effect (for trxB2) on expression of these genes (Figure 5E). These findings indicate that σ8 helps S. avermitilis to cope with thiol-oxidative stress by activating transcription of its target genes trxA3, trxB2, mshA, mshC, mshD, and sig22.
Identification of σ8-dependent Genes Involved in Responses to Osmotic Stress
The σ8 homolog, S. coelicolor σB, responds to osmotic stress caused by NaCl and KCl. To investigate σ8 target genes involved in osmoprotective functions, a series of candidates were selected for EMSA analysis: ect genes (encoding enzymes for biosynthesis of ectoine) (Bursy et al., 2008), opu genes (encoding ABC transporters for uptake of osmoprotectants) (Horn et al., 2005), osmC (encoding a putative osmotically inducible protein) (Gutierrez and Devedjian, 1991), osaAB (encoding a two-component system) and katB (encoding a catalase). osaAB and catB (katB homolog) were reported to be essential for osmoadaptation in S. coelicolor (Cho et al., 2000; Bishop et al., 2004). EMSAs revealed specific binding of His6-σ8 to the promoter probes ectAp for ectA-ectB-ectC-ectD operon, opuBA1p, opuBC1p, sav_5148p for sav_5148-opuBB2-opuBA2-opuBB1-opuBC2-sav_5143 operon, osaAp, osaBp, and katBp, but not to those of opuAAp for opuAA-opuAB operon or osmCp (Figure 6A). In vitro transcription assay confirmed initiation of transcription of ectA, opuBA1, opuBC1, sav_5148, osaA, osaB, and katB by σ8 (Figure 6B). Transcription levels of seven newly identified σ8 target genes were all lower in Dsig8 than in WT (Figure 6C), demonstrating the role of σ8 as an activator of these genes. ectB, ectC, and ectD are cotranscribed with ectA, and opuBB2, opuBA2, opuBB1, and opuBC2 are cotranscribed with sav_5148; therefore, these are also target genes of σ8.
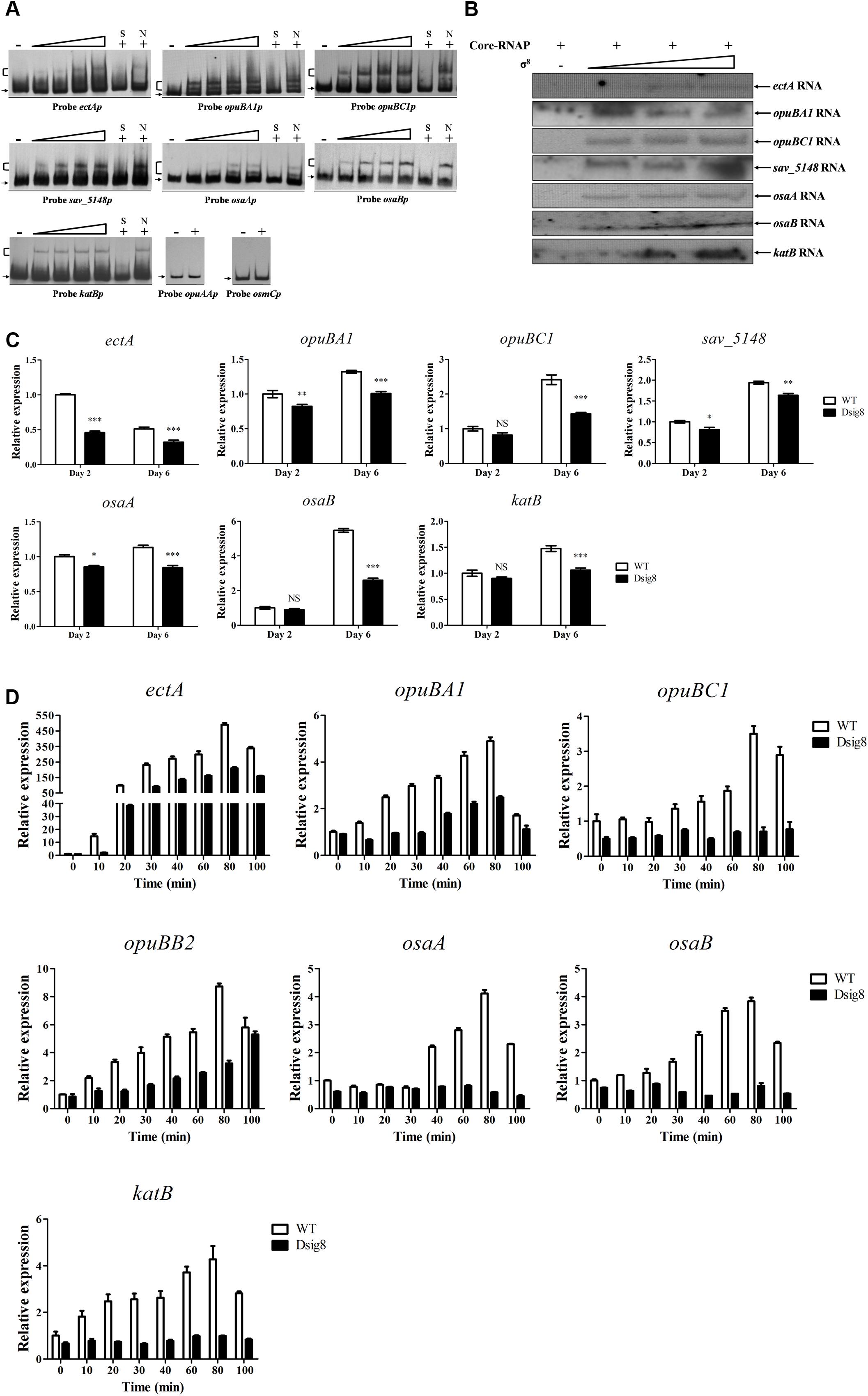
FIGURE 6. Identification of σ8 target genes involved in osmoprotection. (A) EMSAs of His6-σ8 with promoters of genes involved in osmoprotection. (B) In vitro transcription analysis of ectA, opuBA1, opuBC1, sav_5148, osaA, osaB, and katB promoters by Eσ8. (C) qRT-PCR analysis of ectA, opuBA1, opuBC1, opuBB2, osaA, osaB, and katB in WT and Dsig8 grown in FM-I. Transcription level of each gene is expressed relative to WT value at day 2, defined as 1. NS, not significant; ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (t-test). (D) Induction of seven newly identified σ8 target genes by 250 mM NaCl in WT and Dsig8 grown in FM-II. Relative value of each gene in WT before NaCl treatment (0 min) was assigned as 1.
σ8 may also affect induction of its target osmoprotection-related genes by salt stress in S. avermitilis. ectA transcription in WT was sharply increased to ∼490-fold by 80 min of NaCl treatment (Figure 6D), suggesting that ectoine plays as a key osmoprotective role. Maximal induction by NaCl treatment in WT was ∼3.5-fold to 8.7-fold for opuBA1, opuBC1, opuBB2 (selected for qRT-PCR analysis for its operon), osaA, osaB, and katB (Figure 6D). In Dsig8, NaCl treatment had no effect on transcription of opuBC1, osaA, osaB, or katB, indicating that salt stress induces these genes in a σ8-dependent manner. Maximal induction by NaCl treatment on ectA, opuBA1, and opuBB2 transcription was lower in Dsig8 than in WT, suggesting that these three genes are induced by salt stress in both σ8-dependent and σ8-independent manners.
Prediction of the σ8 Regulon
σ8 in S. avermitilis evidently plays a pleiotropic role in control of avermectin production and in protection against a variety of stresses. To clarify broader roles of σ8 in this species, more σ8 target genes need to be identified. The recognition and binding sites of σ70-family factors are the -35 and -10 hexamers of its target promoters. We conducted 5′-RACE assays to determine promoter structures of several σ8 targets and identified a consensus σ8-binding sequence. TSSs of sig8, dnaK1, oxyR, trxA3, sig22, and opuBC1 were determined by 5′-RACE analysis of WT gene transcripts under stress conditions (Supplementary Figure S4), leading to the putative -35 and -10 promoter sequences shown in Figure 7. Analysis of these promoter sequences using the PREDetector web-based program1 revealed a consensus sequence BGVNVH-N15-GSNNHH (Figure 7), which resembles that of σB-specific promoters of S. coelicolor (GNNTN-N14-16-GGGTAY) (Y: C or T) (Lee et al., 2004b).
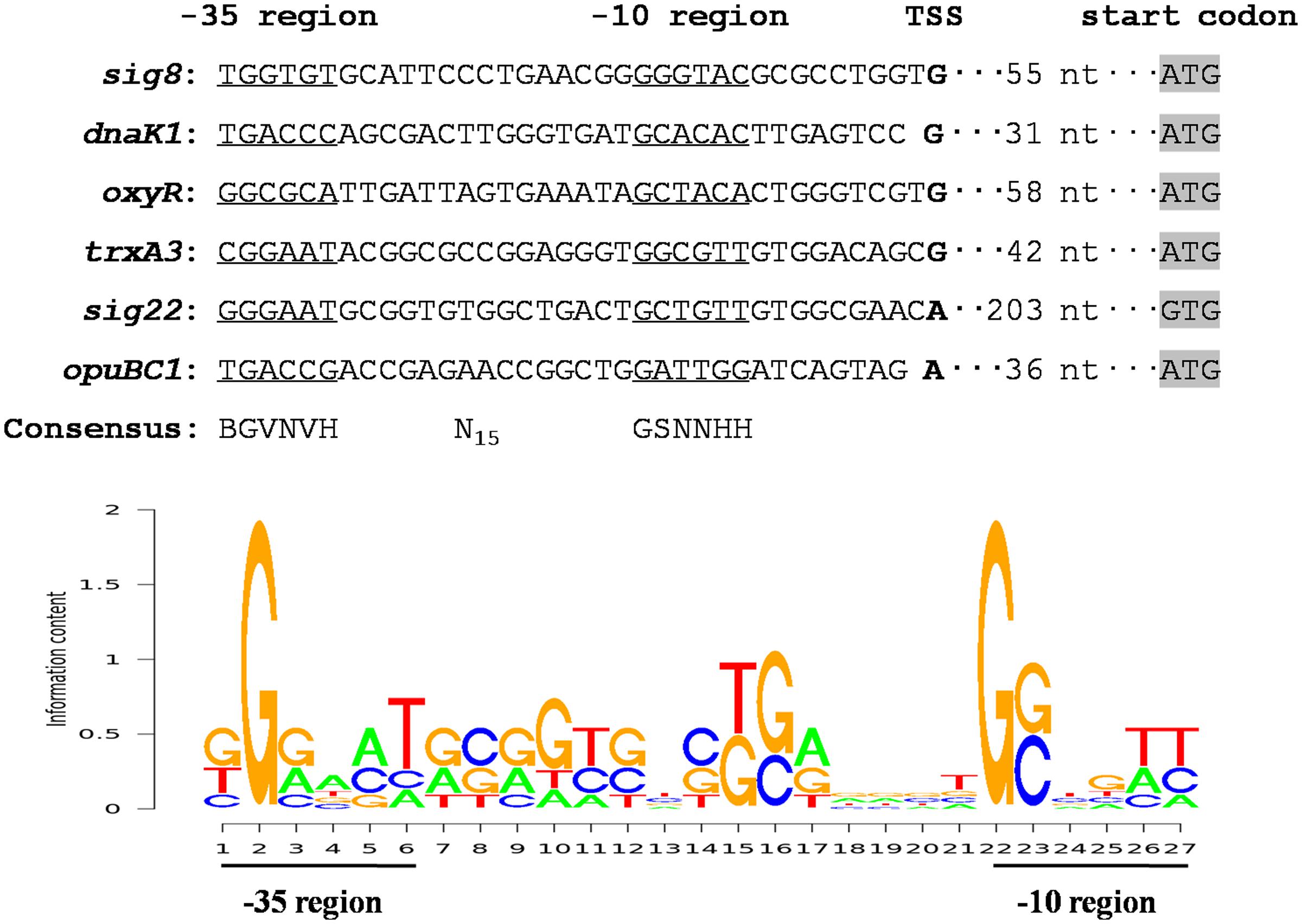
FIGURE 7. Analysis of consensus σ8-binding promoter sequence using the PREDetector program. Putative -35 and -10 regions are underlined. Shading: translational start codons. Boldface: TSSs. In the sequence logo of σ8-binding consensus, appearance frequency of each base is proportional to the height of the corresponding letter.
RNA polymerase is not sensitive to variations in spacer length between the -35 and -10 regions, and the consensus σB-binding sequence is GNNTN-N14-16-GGGTAY. We therefore employed the sequence pattern BGVNVH-N14-16-GSNNHH to scan the S. avermitilis genome using the PREDetector program to predict candidate members of the σ8 regulon, and selected putative promoters that were located within 300 nt upstream of the translational start codons. Using a cut-off score of ≥6.5, we identified 940 putative σ8 target genes, of which 453 have unknown function or are unclassified (Supplementary Table S2). The remaining 487 genes were assigned to 17 functional groups. Among these 487 genes, 179 are associated with regulatory functions, according to the KEGG pathways database for S. avermitilis2. These findings suggest the extent biological significance of σ8 in S. avermitilis.
Discussion
Important roles of alternative σ factors in Streptomyces species in regulation of secondary metabolism are suggested by several previous studies. In S. coelicolor, σB (Cho et al., 2001; Lee et al., 2004b) and σK (Mao et al., 2009) are involved in regulation of ACT and RED biosynthesis, although the regulatory mechanisms are unclear. In Streptomyces chattanoogensis L10, alternative σ factor WhiGch promotes natamycin biosynthesis by directly activating two structural genes, scnC and scnD (Liu S. P. et al., 2015). In the present study, we characterized alternative σ factor σ8 in S. avermitilis, and demonstrated that it indirectly represses avermectin production through its effects on expression of CSR gene aveR and structural gene aveA1. σ8 is the first among the 10 alternative σ factors in S. avermitilis to be characterized. Our findings augment the limited knowledge of regulation of secondary metabolism by alternative σ factors in Streptomyces.
AveR is an essential activator for transcription of the ave cluster (Guo et al., 2010). We showed that σ8 initiates transcription of SAV742, which directly represses expression of ave structural genes aveA1, aveD, aveF, and aveA4 (Sun et al., 2016), but not that of aveR. Thus, σ8 controls avermectin production through at least two pathways: (i) directly initiating transcription of sav_742; (ii) indirectly repressing expression of aveR. The observation that σ8 has an indirect effect on aveR expression suggests that it regulates aveR through a “cascade” mechanism. However, our search for aveR upstream regulator(s) that mediate the repression of σ8 on aveR showed that the two known aveR direct repressor genes, phoP (Yang et al., 2015) and avaR2 (Zhu et al., 2016), are not targets of σ8. Future studies will identify such direct regulator(s) of aveR and clarify the mechanisms underlying σ8 function in avermectin production.
Alternative σ factors help to control morphological differentiation and stress responses, in addition to secondary metabolism. In S. coelicolor, σB is required for aerial mycelium formation and osmoprotection (Cho et al., 2001); σB-dependent σL and σM are involved in sporulation; σH is activated under heat stress and osmotic stress, and plays a crucial role in morphological development (Sevcikova et al., 2001); σI responds to osmotic stress, but has no effect on differentiation (Homerova et al., 2012); σF and σWhiG affect spore formation (Chater et al., 1989; Potuckova et al., 1995; Lee et al., 2005). In S. hygroscopicus, sigB is induced by heat stress and ROS inhibitor (Wei et al., 2012). The present study showed that σ8 is involved in protection against heat, osmotic and oxidative stresses, but has no effect on morphological differentiation. The differing functions of σB homologes in various Streptomyces species presumably reflect differences in regulatory mechanisms for σB activity/expression, and in σB-mediated regulatory pathways for adaptation to various environmental stresses.
In many Gram-positive bacteria, activity of σB homologs is modulated by their cognate anti-σ factors (Hecker et al., 2007), which inhibit transcription activity of σB by binding to it. Under stress conditions, σB is released free of its anti-σ factor, and subsequently initiates transcription of its target genes related to stress protection. The anti-σ factor gene is typically located adjacent to the sigB locus. In S. coelicolor, the anti-σB factor gene rsbA (sco0599) is upstream of sigB (sco0600) (Lee et al., 2004a). In S. avermitilis, the rsbA-homolog gene prsR (sav_7158) is not located near sig8 (sav_741), and sig8-adjacent genes have no similarity to rsbA, suggesting that the regulatory mechanism of σ8 activity is different from that of σB. Further studies to identify regulators associated with σ8, based on co-immunoprecipitation assays within S. avermitilis cells, are in progress.
σ8 responds rapidly to heat stress by directly activating transcription of the dnaK1-grpE1-dnaJ1-hspR operon. Heat shock proteins include not only chaperones for refolding denatured proteins, but also proteases for degrading more denatured proteins. S. avermitilis has three putative heat shock protease genes lonA, htpX1, and htpX2, and whether they are under direct control of σ8 needs to be investigated. Under H2O2 stress, σ8 directly activates transcription of H2O2-sensing regulator genes catR and oxyR, but not of kat or ahpCD, suggesting that CatR and OxyR mediate H2O2 induction of peroxide-scavenging enzymes. We showed that OxyR in S. avermitilis directly activates expression of antioxidant enzyme genes ahpCD, katA1, katA2, and katA3 in response to H2O2 stress (Liu et al., 2016). The targets of CatR remain to be characterized. σ8 thus exerts its protective effect against H2O2 damage in S. avermitilis mainly through a cascade regulatory mechanism involving control of OxyR. σR homologes and their predicted targets, e.g., trx and msh genes are well-conserved in Streptomyces (Kim et al., 2012). Two trx genes, three msh genes and sig22 (sigR homolog) in S. avermitilis are directly controlled by σ8 in response to diamide, indicating that σ8 facilitates a rapid response to thiol-oxidative stress through both direct and cascade regulatory mechanisms. σ8 targets involved in osmoprotection, identified in this study, are ect genes, opuB genes, osaAB and katB. The specific substrate of OpuB transporter in S. avermitilis remains unknown. Our findings are consistent with the observations that ectoine is osmoprotectant, and osaAB and katB are required for σB-dependent osmoprotection in S. coelicolor (Cho et al., 2000, 2001; Bursy et al., 2008; Fernández Martínez et al., 2009), indicating a conserved role of ectoine, osaAB and katB in osmoadaptation in Streptomyces.
Our present findings, taken together, demonstrate clearly that σ8 is a pleiotropic regulator of avermectin production and responses to heat, osmotic and oxidative stresses in S. avermitilis. A proposed model of the σ8-mediated regulatory network is presented in Figure 8. σ8 is a good example of a regulator that links stress responses to antibiotic production. Regulatory pathways of specific stress responses are potential targets for genetic manipulation to increase antibiotic yields. For example, disruption of osaB led to a 37% increase in avermectin yield (Godinez et al., 2015), and deletion of sig8 increased avermectin yield ∼96%. Continued elucidation of such regulatory mechanisms will contribute to improvements in antibiotic production.
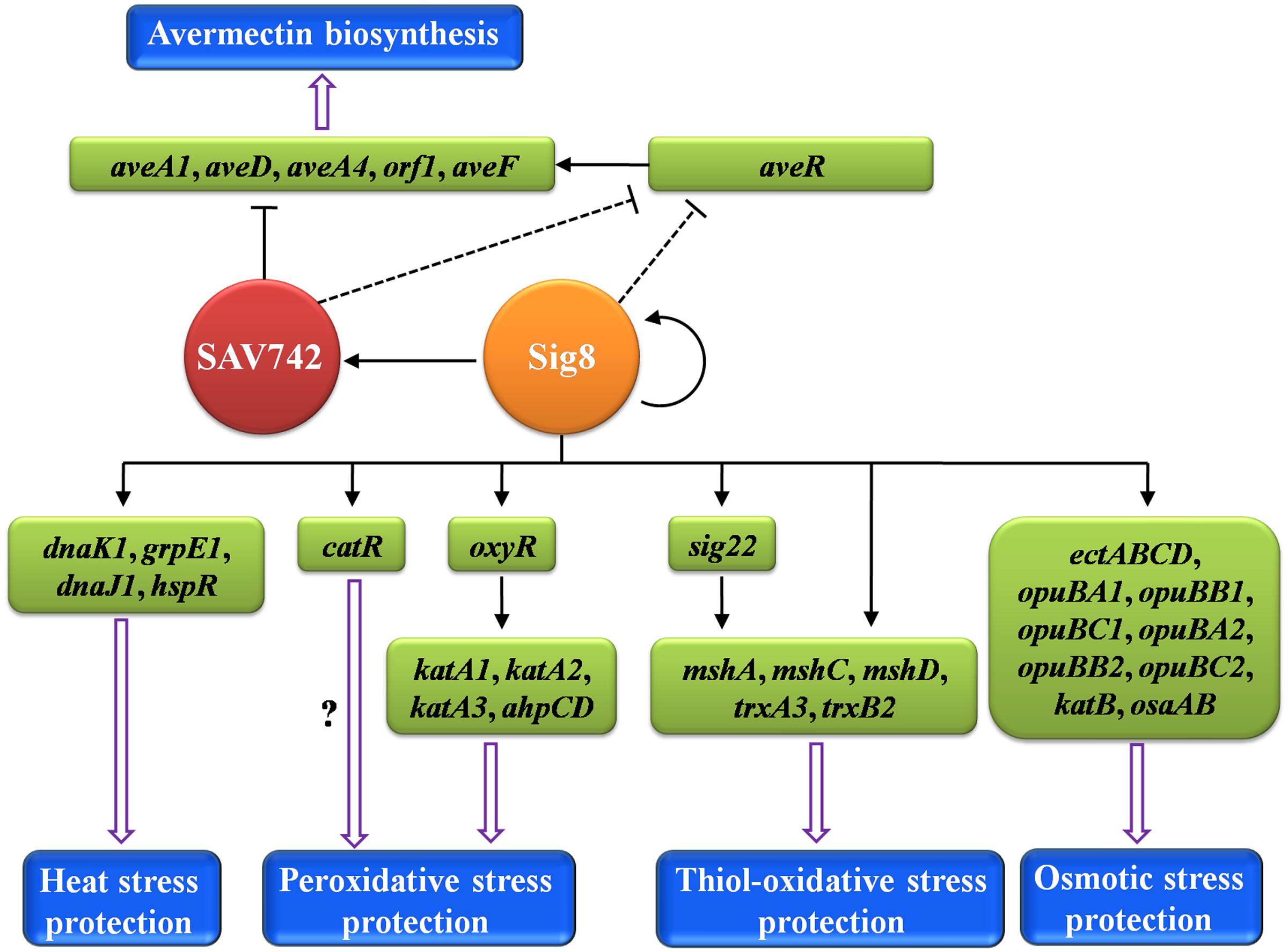
FIGURE 8. Proposed model of σ8-mediated regulatory network in S. avermitilis. σ8 exerts pleiotropic regulatory effects on avermectin biosynthesis and responses to four types of stress. Solid arrows: direct activation. Solid bars: direct repression. Dashed bars: indirect repression. Hollow arrows: avermectin biosynthesis or response to stress.
Using the identified σ8 consensus binding sequence, we predicted 940 putative σ8 target genes. It is unlikely that σ8 binds to all the predicted targets, more knowledge of the promoter sequence recognized by σ8 is necessary for precise prediction of σ8 regulon. Studies using high-throughput technologies (e.g., ChIP-seq) are in progress to experimentally verify additional σ8 targets in S. avermitilis and thereby elucidate the wide range of cellular functions controlled by this important σ factor.
Author Contributions
YW and DS: designed the research, DS: performed experiments, QW, ZC, and JL: contributed study materials, YW and DS: wrote the manuscript.
Funding
This study was supported by National Natural Science Foundation of China (grant no. 31170045).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We are grateful to Dr. S. Anderson for English editing of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00736/full#supplementary-material
Footnotes
References
Bierman, M., Logan, R., O’Brien, K., Seno, E. T., Rao, R. N., and Schoner, B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49. doi: 10.1016/0378-1119(92)90627-2
Bishop, A., Fielding, S., Dyson, P., and Herron, P. (2004). Systematic insertional mutagenesis of a streptomycete genome: a link between osmoadaptation and antibiotic production. Genome Res. 14, 893–900. doi: 10.1101/gr.1710304
Burg, R. W., Miller, B. M., Baker, E. E., Birnbaum, J., Currie, S. A., Hartman, R., et al. (1979). Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15, 361–367. doi: 10.1128/AAC.15.3.361
Bursy, J., Kuhlmann, A. U., Pittelkow, M., Hartmann, H., Jebbar, M., Pierik, A. J., et al. (2008). Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol. 74, 7286–7296. doi: 10.1128/AEM.00768-08
Challis, G. L., and Hopwood, D. A. (2003). Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. U.S.A. 100(Suppl. 2), 14555–14561. doi: 10.1073/pnas.1934677100
Chater, K. F., Bruton, C. J., Plaskitt, K. A., Buttner, M. J., Mendez, C., and Helmann, J. D. (1989). The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell 59, 133–143. doi: 10.1016/0092-8674(89)90876-3
Chen, Z., Wen, J., Song, Y., Wen, Y., and Li, J. (2007). Enhancement and selective production of avermectin B by recombinants of Streptomyces avermitilis via intraspecific protoplast fusion. Chin. Sci. Bull. 52, 616–622. doi: 10.1007/s11434-007-0119-y
Cho, Y. H., Lee, E. J., Ahn, B. E., and Roe, J. H. (2001). SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol. Microbiol. 42, 205–214. doi: 10.1046/j.1365-2958.2001.02622.x
Cho, Y. H., Lee, E. J., and Roe, J. H. (2000). A developmentally regulated catalase required for proper differentiation and osmoprotection of Streptomyces coelicolor. Mol. Microbiol. 35, 150–160. doi: 10.1046/j.1365-2958.2000.01685.x
Egerton, J. R., Ostlind, D. A., Blair, L. S., Eary, C. H., Suhayda, D., Cifelli, S., et al. (1979). Avermectins, new family of potent anthelmintic agents: efficacy of the B1a component. Antimicrob. Agents Chemother. 15, 372–378. doi: 10.1128/AAC.15.3.372
Fernández Martínez, L., Bishop, A., Parkes, L., Del Sol, R., Salerno, P., Sevcikova, B., et al. (2009). Osmoregulation in Streptomyces coelicolor: modulation of SigB activity by OsaC. Mol. Microbiol. 71, 1250–1262. doi: 10.1111/j.1365-2958.2009.06599.x
Godinez, O., Dyson, P., del Sol, R., Barrios-Gonzalez, J., Millan-Pacheco, C., and Mejia, A. (2015). Targeting the osmotic stress response for strain improvement of an industrial producer of secondary metabolites. J. Microbiol. Biotechnol. 25, 1787–1795. doi: 10.4014/jmb.1503.03042
Guo, J., Zhao, J., Li, L., Chen, Z., Wen, Y., and Li, J. (2010). The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol. Genet. Genomics 283, 123–133. doi: 10.1007/s00438-009-0502-2
Gutierrez, C., and Devedjian, J. C. (1991). Osmotic induction of gene osmC expression in Escherichia coli K12. J. Mol. Biol. 220, 959–973. doi: 10.1016/0022-2836(91)90366-E
Haldenwang, W. G., and Losick, R. (1980). Novel RNA polymerase sigma factor from Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 77, 7000–7004. doi: 10.1073/pnas.77.12.7000
Hecker, M., Pane-Farre, J., and Volker, U. (2007). SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61, 215–236. doi: 10.1146/annurev.micro.61.080706.093445
Homerova, D., Sevcikova, B., Rezuchova, B., and Kormanec, J. (2012). Regulation of an alternative sigma factor σI by a partner switching mechanism with an anti-sigma factor PrsI and an anti-anti-sigma factor ArsI in Streptomyces coelicolor A3(2). Gene 492, 71–80. doi: 10.1016/j.gene.2011.11.011
Horn, C., Bremer, E., and Schmitt, L. (2005). Functional overexpression and in vitro re-association of OpuA, an osmotically regulated ABC-transport complex from Bacillus subtilis. FEBS Lett. 579, 5765–5768. doi: 10.1016/j.febslet.2005.09.063
Ikeda, H., Kotaki, H., Tanaka, H., and Omura, S. (1988). Involvement of glucose catabolism in avermectin production by Streptomyces avermitilis. Antimicrob. Agents Chemother. 32, 282–284. doi: 10.1128/AAC.32.2.282
Jiang, L., Liu, Y., Wang, P., Wen, Y., Song, Y., Chen, Z., et al. (2011). Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis. Biotechnol. Lett. 33, 1955–1961. doi: 10.1007/s10529-011-0673-x
Kallifidas, D., Thomas, D., Doughty, P., and Paget, M. S. (2010). The σR regulon of Streptomyces coelicolor A3(2) reveals a key role in protein quality control during disulphide stress. Microbiology 156, 1661–1672. doi: 10.1099/mic.0.037804-0
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics: A Laboratory Manual. Norwich: John Innes Foundation.
Kim, M. S., Dufour, Y. S., Yoo, J. S., Cho, Y. B., Park, J. H., Nam, G. B., et al. (2012). Conservation of thiol-oxidative stress responses regulated by SigR orthologues in actinomycetes. Mol. Microbiol. 85, 326–344. doi: 10.1111/j.1365-2958.2012.08115.x
Kitani, S., Ikeda, H., Sakamoto, T., Noguchi, S., and Nihira, T. (2009). Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 82, 1089–1096. doi: 10.1007/s00253-008-1850-2
Lee, E. J., Cho, Y. H., Kim, H. S., Ahn, B. E., and Roe, J. H. (2004a). Regulation of σB by an anti- and an anti-anti-sigma factor in Streptomyces coelicolor in response to osmotic stress. J. Bacteriol. 186, 8490–8498. doi: 10.1128/JB.186.24.8490-8498.2004
Lee, E. J., Cho, Y. H., Kim, H. S., and Roe, J. H. (2004b). Identification of σB-dependent promoters using consensus-directed search of Streptomyces coelicolor genome. J. Microbiol. 42, 147–151.
Lee, E. J., Karoonuthaisiri, N., Kim, H. S., Park, J. H., Cha, C. J., Kao, C. M., et al. (2005). A master regulator σB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 57, 1252–1264. doi: 10.1111/j.1365-2958.2005.04761.x
Li, L., Guo, J., Wen, Y., Chen, Z., Song, Y., and Li, J. (2010). Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J. Ind. Microbiol. Biotechnol. 37, 673–679. doi: 10.1007/s10295-010-0710-0
Liu, G., Chater, K. F., Chandra, G., Niu, G., and Tan, H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/MMBR.00054-12
Liu, S. P., Yu, P., Yuan, P. H., Zhou, Z. X., Bu, Q. T., Mao, X. M., et al. (2015). Sigma factor WhiGch positively regulates natamycin production in Streptomyces chattanoogensis L10. Appl. Microbiol. Biotechnol. 99, 2715–2726. doi: 10.1007/s00253-014-6307-1
Liu, W., Zhang, Q., Guo, J., Chen, Z., Li, J., and Wen, Y. (2015). Increasing avermectin production in Streptomyces avermitilis by manipulating the expression of a novel TetR-family regulator and its target gene product. Appl. Environ. Microbiol. 81, 5157–5173. doi: 10.1128/AEM.00868-15
Liu, X., Sun, M., Cheng, Y., Yang, R., Wen, Y., Chen, Z., et al. (2016). OxyR is a key regulator in response to oxidative stress in Streptomyces avermitilis. Microbiology doi: 10.1099/mic.0.000251 [Epub ahead of print].
Luo, S., Sun, D., Zhu, J., Chen, Z., Wen, Y., and Li, J. (2014). An extracytoplasmic function sigma factor, σ 25, differentially regulates avermectin and oligomycin biosynthesis in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 98, 7097–7112. doi: 10.1007/s00253-014-5759-7
Macneil, D. J., and Klapko, L. M. (1987). Transformation of Streptomyces avermitilis by plasmid DNA. J. Ind. Microbiol. 2, 209–218. doi: 10.1007/BF01569542
Mao, X. M., Zhou, Z., Hou, X. P., Guan, W. J., and Li, Y. Q. (2009). Reciprocal regulation between SigK and differentiation programs in Streptomyces coelicolor. J. Bacteriol. 191, 6473–6481. doi: 10.1128/JB.00875-09
Niu, G., Chater, K. F., Tian, Y., Zhang, J., and Tan, H. (2016). Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40, 554–573. doi: 10.1093/femsre/fuw012
Osterberg, S., del Peso-Santos, T., and Shingler, V. (2011). Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65, 37–55. doi: 10.1146/annurev.micro.112408.134219
Potuckova, L., Kelemen, G. H., Findlay, K. C., Lonetto, M. A., Buttner, M. J., and Kormanec, J. (1995). A new RNA polymerase sigma factor, σF, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 17, 37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x
Sevcikova, B., Benada, O., Kofronova, O., and Kormanec, J. (2001). Stress-response sigma factor σH is essential for morphological differentiation of Streptomyces coelicolor A3(2). Arch. Microbiol. 177, 98–106. doi: 10.1007/s00203-001-0367-1
Sun, D., Zhu, J., Chen, Z., Li, J., and Wen, Y. (2016). SAV742, a novel AraC-family regulator from Streptomyces avermitilis, controls avermectin biosynthesis, cell growth and development. Sci. Rep. 6:36915. doi: 10.1038/srep36915
Urem, M., Swiatek-Polatynska, M. A., Rigali, S., and van Wezel, G. P. (2016). Intertwining nutrient-sensory networks and the control of antibiotic production in Streptomyces. Mol. Microbiol. 102, 183–195. doi: 10.1111/mmi.13464
van Wezel, G. P., and McDowall, K. J. (2011). The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat. Prod. Rep. 28, 1311–1333. doi: 10.1039/c1np00003a
Wei, Z. H., Wu, H., Bai, L., Deng, Z., and Zhong, J. J. (2012). Temperature shift-induced reactive oxygen species enhanced validamycin A production in fermentation of Streptomyces hygroscopicus 5008. Bioprocess Biosyst. Eng. 35, 1309–1316. doi: 10.1007/s00449-012-0718-0
Yang, R., Liu, X., Wen, Y., Song, Y., Chen, Z., and Li, J. (2015). The PhoP transcription factor negatively regulates avermectin biosynthesis in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 99, 10547–10557. doi: 10.1007/s00253-015-6921-6
Zhao, J., Wen, Y., Chen, Z., Song, Y., and Li, J. (2007). An adpA homologue in Streptomyces avermitilis is involved in regulation of morphogenesis and melanogenesis. Chin. Sci. Bull. 52, 623–630. doi: 10.1007/s11434-007-0105-4
Zhu, J., Sun, D., Liu, W., Chen, Z., Li, J., and Wen, Y. (2016). AvaR2, a pseudo gamma-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol. Microbiol. 102, 562–578. doi: 10.1111/mmi.13479
Keywords: Streptomyces avermitilis, avermectins, alternative σ factor, σ8, stress response
Citation: Sun D, Wang Q, Chen Z, Li J and Wen Y (2017) An Alternative σ Factor, σ8, Controls Avermectin Production and Multiple Stress Responses in Streptomyces avermitilis. Front. Microbiol. 8:736. doi: 10.3389/fmicb.2017.00736
Received: 18 February 2017; Accepted: 10 April 2017;
Published: 24 April 2017.
Edited by:
Michael Sauer, University of Natural Resources and Life Sciences, Vienna, AustriaReviewed by:
Paloma Liras, Universidad de León, SpainDavid Salvador Zamorano Sanchez, University of California, Santa Cruz, USA
Copyright © 2017 Sun, Wang, Chen, Li and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Wen, wen@cau.edu.cn
 Di Sun
Di Sun Ying Wen
Ying Wen