- 1Chair of Microbial Ecology, Technische Universität München, Freising, Germany
- 2ZIEL—Institute for Food & Health, Technische Universität München, Freising, Germany
- 3Department of Genome-Oriented Bioinformatics, Technische Universität München, Freising, Germany
The aim of this study was to analyze the adaptation of the environmental Listeria weihenstephanensis DSM 24698 to anaerobiosis. The complete circular genome sequence of this species is reported and the adaptation of L. weihenstephanensis DSM 24698 to oxygen availability was investigated by global transcriptional analyses via RNAseq at 18 and 34°C. A list of operons was created based on the transcriptional data. Forty-two genes were upregulated anaerobically and 62 genes were downregulated anaerobically. The oxygen dependent gene expression of selected genes was further validated via qPCR. Many of the differentially regulated genes encode metabolic enzymes indicating broad metabolic adaptations with respect to oxygen availability. Genes showing the strongest oxygen-dependent adaption encoded nitrate (narGHJI) and nitrite (nirBD) reductases. Together with the observation that nitrate supported anaerobic growth, these data indicate that L. weihenstephanensis DSM 24698 performs anaerobic nitrate respiration. The wide overlap between the oxygen-dependent transcriptional regulation at 18 and 34°C suggest that temperature does not play a key role in the oxygen-dependent transcriptional regulation of L. weihenstephanensis DSM 24698.
Introduction
Listeria are Gram-positive, facultative anaerobic, non-spore-forming, rod-shaped bacteria, 0.5 μm in width and 1.0–1.5 μm in length, with a low G+C content (McLauchlin and Rees, 2009), which can be isolated from a variety of environments, including soil, water, wastewater, sludge, feces, silage, food, and food processing environments (Paillard et al., 2005; Vilar et al., 2007; O'Connor et al., 2010; Linke et al., 2014). The genus Listeria currently contains 17 species which are, based on comparative genomics, divided in the Listeria sensu stricto and the Listeria sensu lato group (Chiara et al., 2015). The Listeria sensu stricto clade includes Listeria monocytogenes (Pirie, 1940), Listeria innocua (Seeliger, 1981), Listeria seeligeri (Rocourt and Grimont, 1983), Listeria welshimeri (Rocourt and Grimont, 1983), Listeria ivanovii (Seeliger et al., 1984), comprising the subspecies L. ivanovii subsp. ivanovii and L. ivanovii subsp. londoniensis (Boerlin et al., 1992) and Listeria marthii (Graves et al., 2010). The Listeria sensu lato group is further divided into three subclades (Chiara et al., 2015), for which recently even the implementation of three new genera was proposed (Orsi and Wiedmann, 2016). One group consists of only one species, Listeria grayi (Errebo Larsen and Seeliger, 1966), one includes Listeria fleischmanii (Bertsch et al., 2013), comprising the subspecies L. fleischmannii subsp. coloradonensis and L. fleischmannii subsp. fleischmannii (den Bakker et al., 2013), Listeria floridensis, and Listeria aquatica (den Bakker et al., 2014), and one is composed of Listeria rocourtiae (Leclercq et al., 2010), L. weihenstephanensis (Lang-Halter et al., 2013), Listeria cornellensis, Listeria riparia, Listeria grandensis (den Bakker et al., 2014), Listeria newyorkensis, and Listeria booriae (Weller et al., 2015).
The Listeria senu stricto group includes two pathogenic species, L. monocytogenes and L. ivanovii. L. monocytogenes infects both humans and animals and is known as an important opportunistic human foodborne pathogen, which causes, after ingestion of contaminated food, human listeriosis, a rare but severe disease (reviewed in Allerberger and Wagner, 2010). L. ivanovii is an animal pathogen (Gill et al., 1997; Chand and Sadana, 1999).
Whereas, especially the two pathogens L. monocytogenes and L. ivanovii which belong to the Listeria sensu stricto group are well-investigated, almost nothing is known about the members from the Listeria sensu lato group, which appear to be environmental species that have not yet been associated with human or animal diseases. In this study we investigated the adaptation of L. weihenstephanensis DSM 24698 to oxygen restriction. L. weihenstephanensis DSM 24698 was isolated from the water plant Lemna trisulca from a fresh water pond in Bavaria, Germany. The isolate is non-haemolytic and shows optimal growth at pH 7–8 and a temperature of 28–34°C (Lang-Halter et al., 2013).
The adaptation to oxygen restriction is of special interest, not only because the bacterium might encounter microaerophilic of anaerobic conditions in certain environmental niches, but also because the pathogens L. monocytogenes and L. ivanovii have to deal with oxygen restriction in the gastrointestinal tract of the host.
In this study, the adaptation of L. weihenstephanensis DSM 24698 to oxygen restriction was analyzed at two different temperatures. It has been reported earlier that in L. monocytogenes massive temperature dependent transcriptional changes can be observed. Best studied examples are the upregulated expression of flagellar genes at environmental temperature (Williams et al., 2005) and the increased expression of the classical L. monocytogenes virulence genes important for adhesion to and invasion into host cells and cell to cell spread at host temperature (reviewed in Vázquez-Boland et al., 2001). The temperature dependent regulation of many virulence genes is mediated by the transcriptional activator PrfA, whose activity is activated via a RNA thermosensor (Johansson et al., 2002). Furthermore, it has been shown recently that L. monocytogenes also regulates the expression of genes involved in central metabolic pathways like nitrogen metabolism in a temperature dependent manner (Kaspar et al., 2014), also suggesting a niche dependent metabolic adaptation.
To investigate whether the oxygen dependent metabolic adaptation in the environmental bacterium L. weihenstephanensis DSM 24698 is influenced by temperature, the transcriptional response to anaerobiosis was analyzed at 18°C, a potential environmental temperature, and 34°C.
Materials and Methods
Strain and Culture Conditions
The strain used in this study was L. weihenstephanensis DSM 24689 (= WS 4560; Lang-Halter et al., 2013).
For cultivation on solid media, bacteria from the glycerol stock were cultivated on BHI agar and incubated for 1–2 days at 24°C and then stored at 4°C. For overnight liquid cultures, a single bacterial colony was inoculated in 50 ml BHI medium (Brain Heart Infusion, Merck, Darmstadt, Germany) overnight (17 h) at 24°C under continuous shaking (150 rpm). Overnight cultures were diluted 1:100 in fresh BHI medium in a 51-ml total volume for growth analysis. For aerobic growth the cultures were grown in 200-ml flasks, for anaerobic growth cultures were grown in 50-ml sealed CELLSTAR® tubes (Greiner Bio-One, Frickenhausen, Germany). Oxygen availability was analyzed in parallel cultures to which the redox indicator resazurin (0.0001% w/v, 7-Hydroxy-3H-phenoxazin-3-one-10-oxide; Sigma-Aldrich, Taufkirchen, Germany) was added. When oxygen in the medium became limited, resazurin changed color from blue to pink and further to colorless when the culture became anaerobic. All cultures were incubated at the specific experimental temperature under continuous shaking (150 rpm). The optical density at 600 nm (OD600) was measured using the spectrophotometer Lambda Bio+ (PerkinElmer, Rodgau, Germany) every hour until the stationary phase was reached. One additional measurement was taken after 24 h. Three biologically independent experiments were performed for each growth condition. The mean values of the growth analysis were used for the calculation of the doubling time and the maximum OD600 (OD600 max) with the GrowthRates v2.0 calculator (Hall et al., 2014).
Small Scale In vitro Growth Analyses
For in vitro growth analysis in a 200-μl micro-volume a Bioscreen C growth curve reader was used (Oy Growth Curves Ab Ltd., Helsinki, Finland). Overnight cultures were diluted 1:200 in a modified minimal medium supplied with 0 or 10 mM sodium nitrate (NaNO3). The composition of the medium with glutamine as nitrogen source was described previously (Kaspar et al., 2014). As carbon source 1 g L−1 D (+)-glucose monohydrate (Fluka, Darmstadt, Germany) was used. Furthermore, 5 g L−1 yeast extract (Oxoid, Wesel, Germany) were added. Cultures were incubated at 34°C with continuous medium shaking (shaking step 60). For anaerobic growth analyses, cultures were overlaid with 200 μl sterile paraffin oil (Roth, Karlsruhe, Germany). The OD600 of each well was automatically recorded every hour over a period of 24 h. Mean values and standard deviation were calculated from at least four independent biological experiments each including technical duplicates. A student's t-test (Two-Sample Assuming Unequal Variance, Microsoft Excel) was performed for statistical analysis.
Genome Sequencing Using Illumina
The L. weihenstephanensis DSM 24698 genome was sequenced on the MiSeq platform (Illumina, San Diego, USA) using v2 chemistry (400 cycles; Illumina, München, Germany). Genomic DNA was isolated from a 3-ml overnight culture grown at 30°C. The isolated DNA was diluted to 20 ng μl−1. Fifty-five microliters were transferred to a Snap Cap microTUBE (Covaris, Brighton, United Kingdom) and the DNA was fragmented in the Covaris sonnicator S220 (45 s, 175 watt, 10% duty factor, 200 cycles per burst). The sample was stored at −20°C until further processing. The library for sequencing was prepared with a modified version of the standard PCR-free TruSeq DNA sample preparation protocol (Huptas et al., 2016).
Genome Sequencing Using PacBio
For DNA sequencing using the Pacific Biosciences PacBio RS II platform, an overnight culture was diluted 1:100 in BHI medium and grown aerobically at 18°C under constant shaking (150 rpm) to an OD600 = 0.86. The cells were pelleted by centrifugation (18°C, 8 min, 4,186 × g). The DNA isolation and sequencing run were performed by GATC Biotech (Konstanz, Germany).
Genome Assembly and Annotation
The raw PacBio sequence data were assembled with the hierarchical genome-assembly process (Chin et al., 2013) v3 workflow of the SMRT Analysis software, v2.3. Then, Pilon (Walker et al., 2014) was used to improve the assembly further by incorporating the Illumina sequence data. The resulting contig was circularized manually. An unusual high coverage region was further analyzed. This region contained a prophage sequence. The attachment sites of this prophage were determined and the sequence was extracted from the chromosomal sequence and circularized. Afterwards, reads were mapped against the bacterial chromosomal integration site and the circularized phage chromosome to verify the correct breakpoint sequences.
Both sequences were submitted to GenBank and structurally annotated by the NCBI Prokaryotic Annotation Pipeline (PGAP; Tatusova et al., 2016). Functional annotation was performed by the RAST server (Overbeek et al., 2014) and the PEDANT system (Walter et al., 2009) to get additional functional annotation like Clusters of Orthologous Groups (COGs; Galperin et al., 2015) assignment or annotation of genes not characterized by PGAP.
The sequencing project was registered in the NCBI BioProject database with accession number PRJNA275474. Genomic raw sequence data were submitted to the sequence read archive (SRA) with accession number SRS845980. The GenBank accession number for the chromosome is CP011102 and for the phage genome CP011103.
Transcriptional Profiling
Cell Harvesting and RNA Extraction
L. weihenstephanensis DSM 24698 cultures (51 ml) were grown aerobically or anaerobically in BHI medium at the specific temperature to an OD600 = 0.85–0.95. The RNA was isolated following the protocol previously published (Müller-Herbst et al., 2014), but performing the disruption by bead-beating (FastPrep-24, MP Biomedicals, USA) four times for 45 s at 6.5 m s−1. After the first DNAse I (RQ1 RNase-Free DNase Promega, Mannheim, Germany) treatment, RNA was purified from proteins by chloroform extraction (100 μl). Total RNA from the aqueous phase was further purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's RNA cleanup protocol. An additional on-column DNA digest (Qiagen RNase-free DNase, 2 U μl−1) was performed according to the manufacturer's instructions before RNA was eluted in nuclease-free water (30-μl final volume).
Library Preparation and RNA Sequencing
The RNAs were isolated as described above, but, after the second purification step with chloroform, only 60 μg RNA from the aqueous phase were further purified with the RNeasy Mini Kit following the RNA cleanup protocol including the optional on-column DNAse I treatment. Five micrograms of purified RNA was then mRNA-enriched by 16S and 23S rRNA depletion (MICROBExpress Kit, Ambion, Life Technologies, Darmstadt, Germany), following the manufacturers‘ instructions. The samples were then quantified using the 2.0 Qubit Fluorometer (Life Technologies, Darmstadt, Germany) and qualitatively analyzed by the Agilent 2100 Bioanalyzer (RNA 6000 Nano Kit; Agilent Technologies, Waldbronn, Germany). For library preparation, 300 ng of each total RNA were fragmented in 50 μl nuclease-free water (S220 sonicator, Covaris, 180 s, 175 watt, 10% duty factor and 200 cycles per burst) to obtain fragments with a normal distribution around 150 bp. The dephosphorylation of the RNA fragments was carried out using antarctic phosphatase (15 U), 1X antarctic phosphatase buffer (New England BioLabs, Frankfurt, Germany) with the addition of RNase inhibitor SUPERase In (65 units, Ambion, Life Technologies, Darmstadt, Germany) in a 65.5-μl final volume for 1 h at 37°C. Seven hundred microliters of QIAzol lysis reagent were added to the RNA sample. The RNAs were then purified with the miRNeasy Mini Kit following the protocol for purification of total RNA from animal cells omitting the cell homogenization steps (Qiagen) and eluted in a nuclease-free water (35.5-μl final volume). For phosphorylation, T4 Polynucleotide Kinase, 1X T4 Polynucleotide Kinase Buffer (New England BioLabs), ATP (1 mM, New England BioLabs), and RNase inhibitor SUPERase In (50 U) were added to the RNA (50-μl final volume) and the sample was incubated for 1 h at 37°C. Afterwards, RNA was purified again (miRNeasy Mini Kit) with a final elution in 30 μl nuclease free water. After further quality and quantity control by Bioanalyzer and Qubit 100 ng of the samples were finally concentrated to 20 ng/μl by a Concentrator 5301 (Eppendorf, Hamburg, Germany). Ligation of 3′- and 5′-adapters, reverse transcription and PCR amplification were performed following the instructions from the TruSeq Small RNA Sample Preparation Guide (Illumina, February 2013). The cDNAs derived from the aerobic and anaerobic culture were mixed, before the size separation of the amplified cDNA fragments on a ready to use 6% Novex TBE polyacrylamide gel (Life Technologies, Darmstadt, Germany) was performed. The fragments were size-selected between 200 and 350 bp and purified by filtration (0.22 μm Spin-X filter, Corning, USA) and precipitation. The indexed libraries were diluted to a 2-nM concentration in Buffer EB (Qiagen). For denaturation, 10 μl of 0.1 M NaOH (1 M NaOH [pH 13.5] was diluted with buffer EB to a 0.1 M NaOH solution) were added to 10 μl of the 2-nM library and the sample was incubated for 5 min a RT. Finally, the library was diluted with pre-chilled HT1 buffer (Illumina) to a concentration of 10 pM in 0.5 mM NaOH. Finally, the libraries were sequenced on the MiSeq sequencer (Illumina) using a MiSeq Reagent Kit v2 (50 cycles, Illumina) resulting in 50 bp single-end reads.
Bioinformatics
The Illumina RNA-Seq reads were mapped to the chromosomal sequence of L. weihenstephanensis DSM 24698 using Bowtie for Illumina implemented in Galaxy (Blankenberg et al., 2010; Goecks et al., 2010) with commonly used settings (seed length 28 nt, maximum number of mismatches in the seed is 2). Data processing steps to receive SAM and BAM files were performed as described previously (Landstorfer et al., 2014). The number of reads mapping on each gene were visualized and calculated in Artemis v. 15.0.0 (Rutherford et al., 2000; Carver et al., 2012), based on the GenBank file of the L. weihenstephanensis DSM 24698 reference genome. The number of reads for all conditions were normalized to the total number of reads of the smallest library (L. weihenstephanensis DSM 24698 18°C anaerobic) for a more accurate comparison between temperatures.
A second transcript analysis tool (Rockhopper, McClure et al., 2013) was used for verification and to determine non-coding RNAs (ncRNAs) as well as to infer and annotate operon structures (Fortino et al., 2014).
Differential Expression Analysis for the RNA-Seq Data
Differential gene expression was analyzed according to Mühlig et al. (2014), using the Bioconductor package edgeR (Robinson et al., 2010). Genes with ≤1 cpm (counts per million) in total under both the aerobic and anaerobic condition were filtered and not used in the analysis. The transcription of genes under aerobic conditions served as reference. Upregulated genes are expected to show a stronger transcription anaerobically, while downregulated genes are expected to be less transcribed upon oxygen-limitation. Genes showing a log2fold change in relative transcription (log2FC) ≥ 1 or ≤ −1, a false discovery rate (FDR) < 0.3 and a BH-corrected p < 0.05 were considered as being significantly up- or downregulated. The data discussed in this paper have been deposited in NCBI′s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE94124 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94124).
Quantitative Real Time PCR (qPCR)
To validate the RNA-Seq data, qPCR analyses were performed as previously described (Müller-Herbst et al., 2014). The qPCR reactions were carried out in duplicates for three biologically independent repetitions, which were also independent of the samples used for RNA-Seq, in an ICycler (Bio-Rad Laboratories GmbH, München, Germany) or in a SmartCycler® System (Cepheid, CA, USA). Oligonucleotides used for qPCR analyses are summarized in Table 1. Data were analyzed using the 2−ΔΔCt method (Schmittgen and Livak, 2008). Transcription of the housekeeping gene encoding ClpX, the ATP-binding subunit of ATP-dependent Clp protease, was used for normalization.
Results
Complete Genome Sequence of L. weihenstephanensis DSM 24698
A key prerequisite for the analyses of the transcriptional adaption of L. weihenstephanensis DSM 24698 to the oxygen availability via RNA NGS was a high quality genome sequence. Up till now, only a draft genome sequence (den Bakker et al., 2014), finally resulting in 71 contigs (GenBank accession number AODJ00000000.1) was available. In this study, the complete genome of L. weihenstephanensis DSM 24698 was analyzed via in-house genome sequencing (MiSeq) and additional automated sequencing (PacBio RS II). The assembly of 1,789,120 high quality reads with a mean length of 189 bp from the MiSeq sequencing reaction resulted in 23 contigs with a total assembly size of 3,380,080 bp. The assembly of the PacBio RS II sequencing reaction of 78,900 high quality reads with a mean read length of 6,091 bp resulted, after incorporation of the MiSeq data, in one single circular chromosome with a ~50- to 180-fold coverage (Figure S1).
Worth mentioning is that after the assembly a ~40 kb region with an unusual high coverage pattern followed by a region with below average coverage was observed (Figure S1). This region was further analyzed using PHAST (Zhou et al., 2011) and revealed to encode phage genes. The observed abnormal coverage pattern would be explained if it is assumed that in the culture used for sequencing a lysogenic phage switched to the lytic cycle. The circularized phage chromosome of L. weihenstephanensis DSM 24698 phage 01 (LWP01) has a size of 41,687 bp and contains 57 coding regions. Blast analysis revealed that the protein sequence of the phage DNA polymerase (UE_5225) and a putative phage tail tape measure protein (UE_5365) matched specifically with proteins encoded in the genome of L. monocytogenes and L. ivanovii strains with a 79–82% and 79–81% identity, indicating its putative relationship to other Listeria phages.
The complete genome sequence of L. weihenstephanensis DSM 24698 including the prophage sequence has a size of 3,406,292 bp and a GC-content of 41.51%. Three thousand two hundred twenty-nine genes were identified via the automated annotation from NCBI, including 3,008 coding genes, 131 pseudogenes, 19 rRNAs (5S, 16S, 23S), 70 tRNAs and 1 non-coding RNA. One thousand nine hundred thirty-five genes were assigned to one or more COG categories (Table S1). Based on transcriptional data (see below) 647 putative operons which comprised 1,951 genes (Table S2) and 210 putative non-coding RNAs, among them 10 long non-coding RNAs (>200 bp; Table S3), were identified.
Interestingly, the chromosome of L. weihenstephanensis DSM 24698 harbors genes encoding a nitrate (UE46_11980-UE46_11975-UE46_11970-UE46_11965, narGHJI) and a nitrite (UE46_01300-UE46_01305, nirBD) reductase. These genes are absent in L. monocytogenes and all other members of the Listeria sensu stricto phylogenetic clade, but are present, at least partially, in all recently described species of the Listeria sensu lato clade, except for L. floridensis.
Aerobic and Anaerobic Growth of L. weihenstephanensis DSM 24698
It has already been described that L. weihenstephanensis DSM 24698 is able to grow anaerobically on solid medium (Lang-Halter et al., 2013). To further investigate the oxygen dependent growth of L. weihenstephanensis DSM 24698, the bacterium was grown aerobically and anaerobically at 18 and 34°C in liquid culture. L. weihenstephanensis DSM 24698 grew aerobically and anaerobically at both temperatures (Figure 1). At both temperatures the OD600 max was higher for aerobic compared to anaerobic growth (18°C: aerobic OD600 max = 3.697, anaerobic OD600 max = 1.590; 34°C: aerobic OD600 max = 2.980, anaerobic OD600 max = 1.380). Although, the OD600 max for aerobic, respectively anaerobic growth was higher at 18°C than at 34°C, the doubling time at 18°C was higher than at 34°C (aerobic: doubling time at 18°C = 123.7 ± 1.8 min, doubling time at 34°C = 59.3 ± 2.6 min; anaerobic: doubling time at 18°C = 148.3 ± 5.7 min, doubling time at 34°C = 73.1 ± 5.0 min). At both temperatures the doubling time for anaerobic growth was higher than that for anaerobic growth.
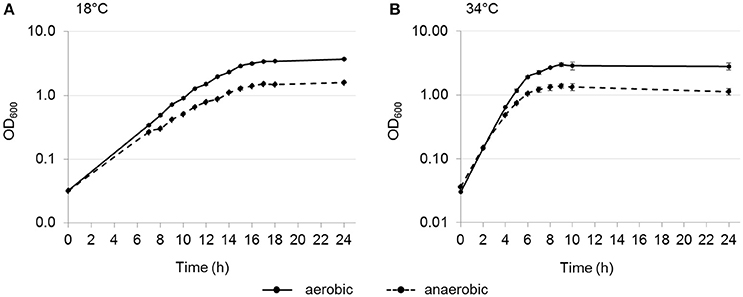
Figure 1. Oxygen-dependent growth of L. weihenstephanensis DSM 24698. Growth curves for L. weihenstephanensis DSM 24698 grown at 18°C (A) and 34°C (B) aerobically (solid line) and anaerobically (dotted line) are shown. OD600 was measured every hour. All data points represent means ± standard error from three independent biological replicates.
Oxygen Dependent Transcriptional Adaptation of L. weihenstephanensis DSM 24698 at 18 and 34°C
To analyze the oxygen dependent transcription of L. weihenstephanensis DSM 24698, bacteria were cultivated aerobically and anaerobically at 18 and 34°C to an OD600 of 0.85–0.95 before cells were harvested for RNA isolation. At this OD600 resazurin controls indicated high or low oxygen availability.
In total, 104 genes showed an oxygen dependent change in transcription. The anaerobically up- and down-regulated genes are listed in the Supplementary Material [Table S4a (upregulated genes) and Table S4b (downregulated genes)]. Forty-two genes were upregulated anaerobically and 62 genes were downregulated anaerobically. Most of the genes that were identified as being regulated depending on the oxygen availability only at one of the investigated temperatures showed the same tendency of regulation also at the other temperature, albeit not fulfilling all the filtering criteria applied during data analysis. For a better overview, the regulated genes were functionally classified according to their COGs (Figure 2).
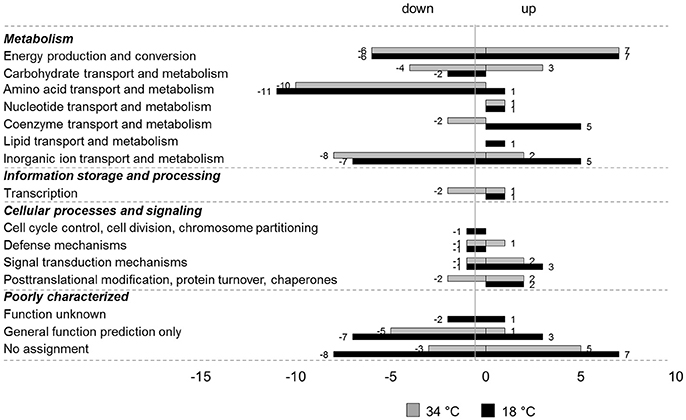
Figure 2. COG classification of genes regulated by oxygen at 18 and 34°C in L. weihenstephanensis DSM 24698. Genes up- or downregulated anaerobically at 18°C (black bars) or 34°C (gray bars) in L. weihenstephanensis DSM 24698 were grouped according to the NCBI COGs. Bars represent the number of genes up- or down-regulated for each category. Since one gene can belong to several COG categories the total number of regulated genes given in this figure is higher than the absolute number of regulated genes.
Most of the anaerobically downregulated genes encode proteins involved in metabolism, in particular in energy production and conversion, amino acid transport and metabolism and inorganic ion processes. Some genes were involved in transcription and in cellular processes and signaling and many were poorly characterized with an unknown or only predicted function or not assigned to any COG category. Genes exhibiting a high negative log2FC at both the temperatures encoded for example catalase (UE46_00145, cat), pyruvate dehydrogenase (UE46_11900-UE46_11895-UE46_11890-UE46_11885, pdhABCD) and glutamate synthase (UE46_10735 and UE46_10730, gltBD; Table S4b).
Also, for the upregulated genes the majority of the regulated genes encoded proteins involved in metabolic pathways in particular in energy, carbohydrate, coenzyme, and inorganic ion processes. Again, many regulated genes were poorly characterized. A small number of the anaerobically upregulated genes were involved in transcription and in cellular processes and signaling. Among the anaerobically upregulated genes the genes showing the highest log2FC at both temperatures encoded nitrate (narGHJI) and nitrite (nirBD) reductase.
Validation of RNA Sequencing
To validate the transcriptional data, qPCR analyses of selected genes were performed. Among the anaerobically upregulated genes, the genes encoding subunits of the nitrate and nitrite reductases (narH, nirD), the molybdenum cofactor biosynthesis protein MoaC (UE46_11930, moaC), the catalytic subunit of class III anaerobic ribonucleoside-triphosphate reductase (UE46_00920, nrdD), a protein annotated as hypothetical by NCBI (NCBI_PGAP) but annotated as nitric oxide dioxygenase by RAST (UE46_14170), and a pyruvate formate lyase (formate acetyltransferase, UE46_08310, pflB) were selected (Figure 3A).
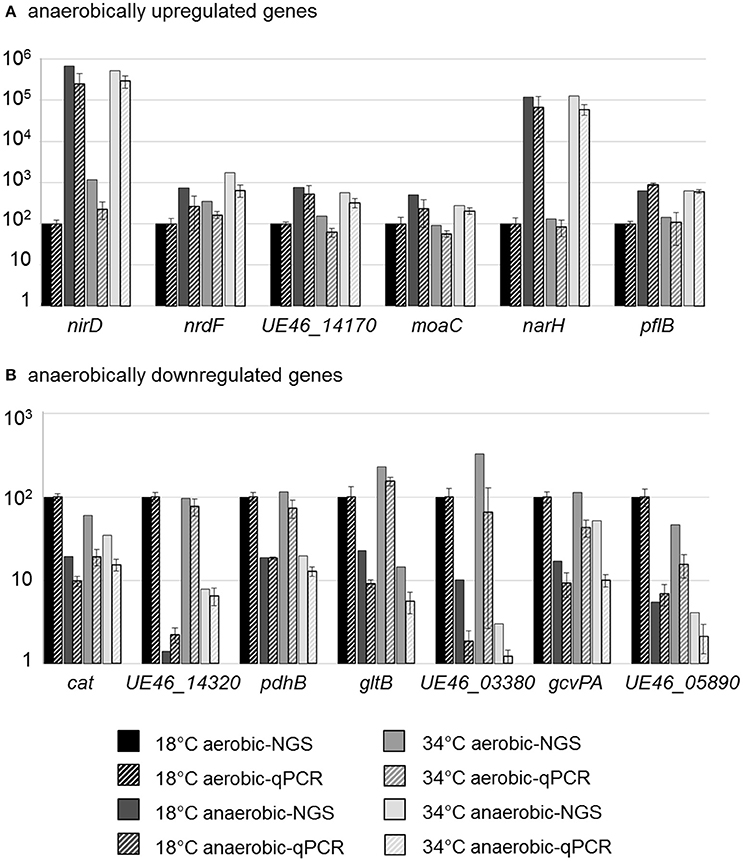
Figure 3. qPCR validation of RNA-NGS data for differently expressed genes. Bars represent the transcriptional level of selected upregulated (A) or downregulated (B) genes analyzed by NGS (solid bars) or qPCR (striped bars). The depicted data show the relative transcription in % compared to the reference condition (18°C aerobic) which was set to 100%. Bars for qPCR data represent mean values and standard errors from three biologically independent experiments, including technical duplicates.
Among the anaerobically downregulated genes, those encoding subunits of pyruvate dehydrogenase (pdhB), of glutamate synthase (gltB), of glycine dehydrogenase, which is part of the glycine cleavage system in L. monocytogenes (Mujahid et al., 2013; UE46_08610, gcvPA), catalase (cat), an ATP binding protein of a putative iron ABC transporter (UE46_03380), a putative L-cystine-binding protein (UE46_14320), and a hypothetical protein (UE46_05890) were investigated (Figure 3B).
Results from qPCR analysis confirmed the data from the RNA-Seq analysis, which therefore can be regarded as veritable.
Nitrate Supported Anaerobic growth of L. weihenstephanensis DSM 24698
The genome analysis revealed that nitrate and nitrite reductases are present in L. weihenstephanensis DSM 24698. The genes narGHJI and nirBD showed the strongest transcriptional induction during anaerobic growth, leading to the hypothesis that under anaerobic growth conditions L. weihenstephanensis DSM 24698 might switch from aerobic respiration to nitrate respiration.
To test the hypothesis that the presence of nitrate could support the anaerobic growth of L. weihenstephanensis DSM 24698, growth analyses were performed. Growth was analyzed (Bioscreen C) at 34°C in the presence and absence of sodium nitrate (10 vs. 0 mM) under both aerobic and anaerobic conditions. First experiments in BHI medium did not reveal a significant growth induction in the presence of sodium nitrate (data not shown). However, the same experiments performed in a modified minimal medium (Figure 4) showed that anaerobically the presence of sodium nitrate led to a statistically significant increase of growth which was characterized by gaining a higher OD600 before reaching the stationary phase. Also, in the aerobic culture a slight growth induction in the presence of sodium nitrate was observed, albeit this increase was not statistically significant. It might well be that that at high cell densities a low oxygen tension in the medium results in a switch to nitrate respiration in the “aerobic” culture.
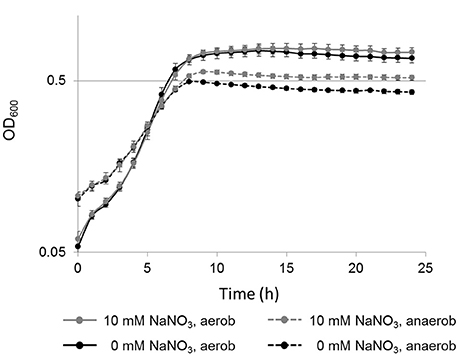
Figure 4. Nitrate-dependent growth of L. weihenstephanensis DSM 24698. Growth curves (y-axis logarithmic scale) for L. weihenstephanensis DSM 24698 grown at 34°C in a modified minimal medium aerobically (solid lines) and anaerobically (dotted lines), in the presence (gray lines) or absence (black lines) of nitrate (10 vs. 0 mM NaNO3) are shown. OD600 was measured every hour. All data points represent means ± standard error from seven independent biological replicates. Statistically significant differences in growth between cultures supplemented with NaNO3and those without NaNO3 were observed during anaerobic (student's t-test, p < 0.05 for time point 8 h, p < 0.005 for time points 9–24 h), but not during aerobic growth.
Discussion
Genome Sequence of L. weihenstephanensis DSM 24698
In this study the genome sequence of L. weihenstephanensis DSM 24698 was revealed, being the first complete circularized chromosome of a member of the Listeria sensu lato phylogenetic group (Chiara et al., 2015). With a size of 3.4 Mbp the genome of L. weihenstephanensis DSM 24698 is larger than the genome of the members of the Listeria sensu stricto group with a size of ~2.8–3.0 Mbp (Glaser et al., 2001; Hain et al., 2006; den Bakker et al., 2010; Steinweg et al., 2010; Buchrieser et al., 2011), but comparable to the genome size of L. rocourtiae, L. cornellensis, L. riparia, L. grandensis, L. newyorkensis, and L. booriae (3.2–3.5 Mbp; den Bakker et al., 2014; Weller et al., 2015), which belong to the same Listeria sensu lato phylogenetic subclade as L. weihenstephanasis (Chiara et al., 2015). The main virulence genes from the human pathogen L. monocytogenes which allow attachment to and invasion into host cells and cell to cell spread are missing in L. weihenstephanensis DSM 24698 as well as in all other members of the Listeria sensu lato group, which readily explains why none of these recently described species has been associated with human or animal disease.
Genes present in L. weihenstephanensis DSM 24698 and at least partially in all the other members of the Listeria sensu lato phylogenetic clade, except for L. floridensis, are the genes encoding nitrate and nitrite reductases. This goes in line with the observation that all the members of the Listeria sensu lato group except for L. floridensis have the capability to reduce nitrate (den Bakker et al., 2014).
Anaerobic Nitrate and Nitrite Reduction in L. weihenstephanensis DSM 24698
The genes encoding nitrate reductase (narGHJI) were induced during anaerobic growth of L. weihenstephanensis DSM 24698 in BHI medium which did not contain supplementary nitrate. This indicates an oxygen-dependent transcriptional regulation of the nar-genes, which has also been described for other bacteria like Staphylococcus aureus (Fuchs et al., 2007) and Bacillus subtilis (Ye et al., 2000). Interestingly, the addition of sodium nitrate did not support anaerobic growth of L. weihenstephanensis DSM 24698 in BHI medium, but in a modified minimal medium, suggesting that nitrate respiration causes a growth advantage only under nutrient limited conditions. Preliminary results indeed indicate an accumulation of nitrite in the supernatant of anaerobically grown L. weihenstephanensis DSM 24698 cultures in modified minimal medium to which sodium nitrate was added (Table S5). Interestingly, the amount of glucose that was added as carbon source to the minimal medium seamed to influence the reduction of nitrate to nitrite. The less glucose was present in the medium, the more nitrite was detected in the culture supernatant. It is therefore tempting to speculate, that in BHI, which contains glucose in a concentration of 2 g L−1, the presence of glucose inhibits nitrate reduction. This would explain the observation that in BHI the supplementation of nitrate did not improve anaerobic growth. Such an inhibitory effect of glucose on nitrate respiration has also been described for other bacteria like Lactobabacillus plantarum (Brooijmans et al., 2009).
Besides genes encoding nitrate reductase, a gene cluster, which is located next to the nitrate reductase operon and which encodes genes involved in the biosynthesis of the molybdenum cofactor (UE46_11960-UE46_11920) was anaerobically stronger transcribed than aerobically. The molybdenum cofactor is essential for the functionality of nitrate reductases (reviewed in Coelho and Romão, 2015). Nitrate respiration has neither been described yet for any Listeria spp. nor for any member of the genus Brochothrix, the other genus in the family Listeriaceae. However, other members of the order Bacillales like B. subtilis (Hoffmann et al., 1998) and Staphylococcus carnosus (Neubauer and Götz, 1996) are able to perform anaerobic nitrate respiration.
Both, B. subtilis and S. carnosus further reduce nitrite to ammonia (Neubauer and Götz, 1996; Hoffmann et al., 1998). Our data indicate that nitrite reduction to ammonia might also occur in L. weihenstephanensis DSM 24698. We observed a strong upregulation of genes encoding nitrite reductase (nirBD) under anaerobic conditions. A reduction of nitrite to ammonia under anaerobic conditions would supply ammonia which could be used for anabolic processes.
It has been shown recently that ammonium is a better nitrogen source for L. monocytogenes than glutamine (Kaspar et al., 2014). Under low ammonium concentrations like e.g., in the host cell cytosol, transcription of genes encoding glutamine synthase (glnA), and glutamate synthase (gltAB) was induced in L. monocytogenes. In this study, a stronger transcription of gltAB was observed during aerobic growth, a condition under which ammonium could not be generated from nitrate via nitrite by anaerobic respiration. Furthermore, the stronger transcription of genes encoding a putative glutamine ABC transporter (UE46_13335 and UE46_13340 [glnQ]) and an ammonia channel protein (UE46_09395) aerobically might reflect a lower nitrogen source supply under aerobic conditions.
Worth mentioning is also the anaerobic upregulation of a gene encoding a putative nitric oxide dioxygenase (UE46_14170). This enzyme might be involved in the detoxification of nitric oxide, which can be produced in the presence of nitrite via Nar-type nitrate reductases (Vine et al., 2011; Rowley et al., 2012).
Common Features and Differences in the Oxygen Dependent Adaptation of L. weihenstephanensis DSM 24698 and L. monocytogenes
All these adaptations of L. weihenstephanensis DSM 24698 to changes in oxygen supply described so far were not reported for L. monocytogenes, which might be due to the lack of nitrate and nitrite reductases in the human pathogen. However, some similarities between the oxygen dependent adaptations of these two species were also observed. Anaerobically, transcription of genes encoding an anaerobic ribonucleosid-triphosphat reductase system (nrdDG) was induced both in L. weihenstephanensis and in L. monocytogenes (Müller-Herbst et al., 2014).
In both organisms the gene encoding catalase (cat) was stronger transcribed aerobically. This protein is involved in the detoxification of hydrogen peroxide, a toxic byproduct of aerobic respiration. Furthermore, in both species there was a stronger aerobic transcription of genes encoding proteins of the glycine cleavage system [gcvT-gcvPA-gcvPB; lmo1348-lmo1350 in L. monocytogenes (Mujahid et al., 2013; Müller-Herbst et al., 2014), UE46_08615-UE46_08610-UE46_08615 in L. weihenstephanensis DSM 24698]. The net reaction catalyzed by the glycine cleavage system is the reversible conversion of Glycine + H4folate + NAD+ to 5,10-methylene-H4folate + CO2 + NH3 + NADH + H+ (reviewed in Kikuchi et al., 2008). Together with a serine hydroxymethyltransferase (UE46_02440), which was also stronger transcribed aerobically in L. weihenstephanensis DSM 24698, the overall reaction catalyzes the conversion of 2 glycine + NAD+ + H2O to serine + CO2 + NH3 + NADH + H+ (reviewed in Kikuchi et al., 2008). In both reactions NH3 and NADH/H+ could arise. Because of the need for balancing the NADH/H+ to NAD+ ratio to assure the catabolic capacity of the organism, it makes perfect sense for L. monocytogenes to reduce NADH/H+ generating reactions anaerobically, when NADH/H+ can only be converted to NAD+ via fermentative pathways and not via the respiratory chain. In L. weihenstephanensis DSM 24698, in which a respiratory chain is supposed to be active during anaerobic nitrate respiration, the main function of these systems might be the provision of NH3 as nitrogen source under aerobic growth conditions.
Also the genes encoding another NADH/H+ producing enzyme complex, the pyruvate dehydrogenase, were anaerobically downregulated in both L. weihenstephanensis and L. monocytogenes. Conversion of pyruvate to acetyl-CoA via pyruvate dehydrogenase is the major pathway to remove pyruvate from the metabolic pool during aerobic growth. However, also anaerobically pyruvate must be removed to allow carbon catabolism to continue. In L. monocytogenes pyruvate is mainly converted to lactate via lactate dehydrogenase anaerobically, thereby recycling NADH/H+ to NAD (Romick et al., 1996). In L. weihenstephanensis DSM 24698 the physiological role of the anaerobic downregulation of pyruvate dehydrogenase remains to be elucidated. However, downregulation of pyruvate dehydrogenase genes during nitrate respiration has also been described for B. subtilis (Ye et al., 2000).
In L. weihenstephanensis DSM 24698 an anaerobic upregulation of a gene cluster encoding a pyruvate formate lyase-activating protein (pflC) and a formate acetyltransferase (pflB) was observed, which together are responsible for the conversion of pyruvate to acetyl-CoA and formate. Formate can be used as electron donor in the nitrate respiratory chain (reviewed in Unden and Bongaerts, 1997) and could thereby compensate the lower production of NADH/H+.
In general, our data indicated that, albeit there are some similarities between the adaptations to oxygen concentration in L. monocytogenes and L. weihenstephanensis DSM 24698, the general adaptation to anaerobiosis differs in these two closely related species. In both organisms a gene encoding the subunit E1 of 2-oxoglutarate dehydrogenase is missing (Glaser et al., 2001, this study) causing a split of the citric acid cycle into an oxidative and a reductive branch, which has been shown via elegant physiological experiments for L. monocytogenes (Trivett and Meyer, 1971; Eisenreich et al., 2006). An incomplete citric acid cycle further implicates that pyruvate can't be oxidized completely to carbon dioxide. Even aerobically, fermentative pathways have to be used simultaneously to aerobic respiration, not to generate energy, but to get rid of pyruvate, the accumulation of which would inhibit glucose catabolism. Anaerobically, L. monocytogenes shifts its metabolism to fermentation only to ensure its energy production. L. weihenstephanensis DSM 24698 on the other site has the capacity to perform nitrate respiration for energy production during anaerobic growth. Nevertheless, both aerobic respiration and anaerobic nitrate respiration has to be accompanied by fermentative pathways to get rid of pyruvate.
Temperature-Independent Adaptation to Oxygen Availability
For L. monocytogenes it has been shown previously that temperature has a massive impact on transcriptional regulation. Best studied examples are the temperature dependent regulation of virulence genes via PrfA (Leimeister-Wächter et al., 1992; Johansson et al., 2002), of flagella genes (Williams et al., 2005) and metabolic adaptations, e.g., temperature dependent expression of genes encoding enzymes involved in nitrogen metabolism (Kaspar et al., 2014). These temperature-dependent adaptations are thought to allow a perfect adaptation to the respective ecological niche as saprophyte in the environment or as an intracellular pathogen in the human host (reviewed in Freitag et al., 2009). The study of the oxygen dependent adaptation of L. weihenstephanensis DSM 24698 at 18 and 34°C, performed in this study, did not reveal a massive impact of the temperature on transcription, at least not on the oxygen dependent adaptation. Differences that were observed between the oxygen dependent transcriptional profiles at 18 and 34°C are more likely due to stringent selection criteria applied for the determination of regulated genes, than to a real effect of temperature, as most of the genes showing a significant regulation at just one of the two investigated temperatures, showed the same tendency of regulation also at the other temperature. This underlines the hypothesis that L. weihenstephanensis is a true environmental species which has no need to adapt to a host.
Conclusion
This study is the first one that provided a complete genome, a transcriptome and an operon map of a member of the Listeria sensu lato phylogenetic clade. The capability of L. weihenstephanensis DSM 24698 to reduce nitrate and probably nitrite, which is shared, according to genome comparison, at least partially with all other members of the Listeria sensu lato group except for L. floridensis, clearly separates this phylogenic group from the members of the Listeria sensu stricto group. However, further studies need to be conducted before a reclassification of the Listeria sensu lato group in novel genera, which was suggested recently (Orsi and Wiedmann, 2016), should be considered.
Author Contributions
EF: Performed experiments, data analysis, wrote the manuscript. MW: Bioinformatics. CH: Performed together with EF, DNA sequencing, bioinformatics. SS: Supervised the project, discussions, corrected the manuscript. SM: Designed experiments, data analysis, wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the German Research Foundation (Deutsche Forschungsgemeinschaft) grant GRK 1482.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01672/full#supplementary-material
References
Allerberger, F., and Wagner, M. (2010). Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infect. 16, 16–23. doi: 10.1111/j.1469-0691.2009.03109.x
Bertsch, D., Rau, J., Eugster, M. R., Haug, M. C., Lawson, P. A., Lacroix, C., et al. (2013). Listeria fleischmannii sp. nov., isolated from cheese. Int. J. Syst. Evol. Microbiol. 63, 526–532. doi: 10.1099/ijs.0.036947-0
Blankenberg, D., Von Kuster, G., Coraor, N., Ananda, G., Lazarus, R., Mangan, M., et al. (2010). Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19, Unit 19.10, 11–21. doi: 10.1002/0471142727.mb1910s89
Boerlin, P., Rocourt, J., Grimont, F., Grimont, P. A. D., Jacquet, C., and Piffaretti, J. C. (1992). Listeria ivanovii subsp. londoniensis subsp. nov. Int. J. Syst. Bacteriol. 42, 69–73. doi: 10.1099/00207713-42-1-69
Brooijmans, R. J., de Vos, W. M., and Hugenholtz, J. (2009). Lactobacillus plantarum WCFS1 electron transport chains. Appl. Environ. Microbiol. 75, 3580–3585. doi: 10.1128/AEM.00147-09
Buchrieser, C., Rusniok, C., Garrido, P., Hain, T., Scortti, M., Lampidis, R., et al. (2011). Complete genome sequence of the animal pathogen Listeria ivanovii, which provides insights into host specificities and evolution of the genus Listeria. J. Bacteriol. 193, 6787–6788. doi: 10.1128/JB.06120-11
Carver, T., Harris, S. R., Berriman, M., Parkhill, J., and McQuillan, J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. doi: 10.1093/bioinformatics/btr703
Chand, P., and Sadana, J. R. (1999). Outbreak of Listeria ivanovii abortion in sheep in India. Vet. Rec. 145, 83–84. doi: 10.1136/vr.145.3.83
Chiara, M., Caruso, M., D'Erchia, A. M., Manzari, C., Fraccalvieri, R., Goffredo, E., et al. (2015). Comparative genomics of Listeria sensu lato: genus-wide differences in evolutionary dynamics and the progressive gain of complex, potentially pathogenicity-related traits through lateral gene transfer. Genome Biol. Evol. 7, 2154–2172. doi: 10.1093/gbe/evv131
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Coelho, C., and Romão, M. J. (2015). Structural and mechanistic insights on nitrate reductases. Protein Sci. 24, 1901–1911. doi: 10.1002/pro.2801
den Bakker, H. C., Cummings, C. A., Ferreira, V., Vatta, P., Orsi, R. H., Degoricija, L., et al. (2010). Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11:688. doi: 10.1186/1471-2164-11-688
den Bakker, H. C., Manuel, C. S., Fortes, E. D., Wiedmann, M., and Nightingale, K. K. (2013). Genome sequencing identifies Listeria fleischmannii subsp. coloradonensis subsp. nov., isolated from a ranch. Int. J. Syst. Evol. Microbiol. 63, 3257–3268. doi: 10.1099/ijs.0.048587-0
den Bakker, H. C., Warchocki, S., Wright, E. M., Allred, A. F., Ahlstrom, C., Manuel, C. S., et al. (2014). Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov. and Listeria grandensis sp. nov., from agricultural and natural environments. Int. J. Syst. Evol. Microbiol. 64, 1882–1889. doi: 10.1099/ijs.0.052720-0
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. doi: 10.1093/nar/30.1.207
Eisenreich, W., Slaghuis, J., Laupitz, R., Bussemer, J., Stritzker, J., Schwarz, C., et al. (2006). 13C isotopologue perturbation studies of Listeria monocytogenes carbon metabolism and its modulation by the virulence regulator PrfA. Proc. Natl. Acad. Sci. U.S.A. 103, 2040–2045. doi: 10.1073/pnas.0507580103
Errebo Larsen, H., and Seeliger, H. P. R. (1966). “A mannitol fermenting Listeria, Listeria grayi sp. n,” in Proceedings of the Third International Symposium on Listeriosis (Bilthoven), 35.
Fortino, V., Smolander, O. P., Auvinen, P., Tagliaferri, R., and Greco, D. (2014). Transcriptome dynamics-based operon prediction in prokaryotes. BMC Bioinformatics 15:145. doi: 10.1186/1471-2105-15-145
Freitag, N. E., Port, G. C., and Miner, M. D. (2009). Listeria monocytogenes- from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7, 623–628. doi: 10.1038/nrmicro2171
Fuchs, S., Pané-Farré, J., Kohler, C., Hecker, M., and Engelmann, S. (2007). Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189, 4275–4289. doi: 10.1128/JB.00081-07
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
Gill, P. A., Boulton, J. G., Fraser, G. C., Stevenson, A. E., and Reddacliff, L. A. (1997). Bovine abortion caused by Listeria ivanovii. Aust. Vet. J. 75:214. doi: 10.1111/j.1751-0813.1997.tb10069.x
Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., et al. (2001). Comparative genomics of Listeria species. Science 294, 849–852. doi: 10.1126/science.1063447
Goecks, J., Nekrutenko, A., Taylor, J. and Galaxy Team (2010). Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86. doi: 10.1186/gb-2010-11-8-r86
Graves, L. M., Helsel, L. O., Steigerwalt, A. G., Morey, R. E., Daneshvar, M. I., Roof, S. E., et al. (2010). Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 60, 1280–1288. doi: 10.1099/ijs.0.014118-0
Hain, T., Steinweg, C., Kuenne, C. T., Billion, A., Ghai, R., Chatterjee, S. S., et al. (2006). Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes. J. Bacteriol. 188, 7405–7415. doi: 10.1128/JB.00758-06
Hall, B. G., Acar, H., Nandipati, A., and Barlow, M. (2014). Growth rates made easy. Mol. Biol. Evol. 31, 232–238. doi: 10.1093/molbev/mst187
Hoffmann, T., Frankenberg, N., Marino, M., and Jahn, D. (1998). Ammonification in Bacillus subtilis utilizing dissimilatory nitrite reductase is dependent on resDE. J. Bacteriol. 180, 186–189.
Huptas, C., Scherer, S., and Wenning, M. (2016). Optimized Illumina PCR-free library preparation for bacterial whole genome sequencing and analysis of factors influencing de novo assembly. BMC Res. Notes 9:269. doi: 10.1186/s13104-016-2072-9
Johansson, J., Mandin, P., Renzoni, A., Chiaruttini, C., Springer, M., and Cossart, P. (2002). An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561. doi: 10.1016/S0092-8674(02)00905-4
Kaspar, D., Auer, F., Schardt, J., Schindele, F., Ospina, A., Held, C., et al. (2014). Temperature- and nitrogen source-dependent regulation of GlnR target genes in Listeria monocytogenes. FEMS Microbiol. Lett. 355, 131–141. doi: 10.1111/1574-6968.12458
Kikuchi, G., Motokawa, Y., Yoshida, T., and Hiraga, K. (2008). Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 84, 246–263. doi: 10.2183/pjab.84.246
Landstorfer, R., Simon, S., Schober, S., Keim, D., Scherer, S., and Neuhaus, K. (2014). Comparison of strand-specific transcriptomes of enterohemorrhagic Escherichia coli O157:H7 EDL933 (EHEC) under eleven different environmental conditions including radish sprouts and cattle feces. BMC Genomics 15:353. doi: 10.1186/1471-2164-15-353
Lang-Halter, E., Neuhaus, K., and Scherer, S. (2013). Listeria weihenstephanensis sp. nov., isolated from the water plant Lemna trisulca taken from a freshwater pond. Int. J. Syst. Evol. Microbiol. 63, 641–647. doi: 10.1099/ijs.0.036830-0
Leclercq, A., Clermont, D., Bizet, C., Grimont, P. A., Le Flèche-Matéos, A., Roche, S. M., et al. (2010). Listeria rocourtiae sp. nov. Int. J. Syst. Evol. Microbiol. 60, 2210–2214. doi: 10.1099/ijs.0.017376-0
Leimeister-Wächter, M., Domann, E., and Chakraborty, T. (1992). The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174, 947–952. doi: 10.1128/jb.174.3.947-952.1992
Linke, K., Rückerl, I., Brugger, K., Karpiskova, R., Walland, J., Muri-Klinger, S., et al. (2014). Reservoirs of Listeria species in three environmental ecosystems. Appl. Environ. Microbiol. 80, 5583–5592. doi: 10.1128/AEM.01018-14
McClure, R., Balasubramanian, D., Sun, Y., Bobrovskyy, M., Sumby, P., Genco, C. A., et al. (2013). Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41:e140. doi: 10.1093/nar/gkt444
McLauchlin, J., and Rees, C. (2009). Bergey's Manual of Systematic Bacteriology, 2nd Edn, Vol. 3, Dordrecht: Springer, 244–257.
Mühlig, A., Behr, J., Scherer, S., and Müller-Herbst, S. (2014). Stress response of Salmonella enterica serovar Typhimurium to acidified nitrite. Appl. Environ. Microbiol. 80, 6373–6382. doi: 10.1128/AEM.01696-14
Mujahid, S., Orsi, R. H., Vangay, P., Boor, K. J., and Wiedmann, M. (2013). Refinement of the Listeria monocytogenes σB regulon through quantitative proteomic analysis. Microbiology 159, 1109–1119. doi: 10.1099/mic.0.066001-0
Müller-Herbst, S., Wüstner, S., Mühlig, A., Eder, D., M Fuchs, T., Held, C., et al. (2014). Identification of genes essential for anaerobic growth of Listeria monocytogenes. Microbiology 160, 752–765. doi: 10.1099/mic.0.075242-0
Neubauer, H., and Götz, F. (1996). Physiology and interaction of nitrate and nitrite reduction in Staphylococcus carnosus. J. Bacteriol. 178, 2005–2009. doi: 10.1128/jb.178.7.2005-2009.1996
O'Connor, L., O'Leary, M., Leonard, N., Godinho, M., O'Reilly, C., Coffey, L., et al. (2010). The characterization of Listeria spp. isolated from food products and the food-processing environment. Lett. Appl. Microbiol. 51, 490–498. doi: 10.1111/j.1472-765X.2010.02928.x
Orsi, R. H., and Wiedmann, M. (2016). Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 100, 5273–5287. doi: 10.1007/s00253-016-7552-2
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Paillard, D., Dubois, V., Thiebaut, R., Nathier, F., Hoogland, E., Caumette, P., et al. (2005). Occurrence of Listeria spp. in effluents of French urban wastewater treatment plants. Appl. Environ. Microbiol. 71, 7562–7566. doi: 10.1128/AEM.71.11.7562-7566.2005
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rocourt, J., and Grimont, P. A. D. (1983). Notes: Listeria welshimeri sp. nov. and Listeria seeligeri sp. nov. Int. J. Syst. Bacteriol. 33, 866–869. doi: 10.1099/00207713-33-4-866
Romick, T. L., Fleming, H. P., and McFeeters, R. F. (1996). Aerobic and anaerobic metabolism of Listeria monocytogenes in defined glucose medium. Appl. Environ. Microbiol. 62, 304–307.
Rowley, G., Hensen, D., Felgate, H., Arkenberg, A., Appia-Ayme, C., Prior, K., et al. (2012). Resolving the contributions of the membrane-bound and periplasmic nitrate reductase systems to nitric oxide and nitrous oxide production in Salmonella enterica serovar Typhimurium. Biochem. J. 441, 755–762. doi: 10.1042/BJ20110971
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Seeliger, H. P. (1981). [Nonpathogenic listeriae: L. innocua sp. n. (Seeliger et Schoofs, 1977) (author's transl)]. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 249, 487–493.
Seeliger, H. P. R., Rocourt, J., Schrettenbrunner, A., Grimont, P. A. D., and Jones, A. N. D. D. (1984). Listeria ivanovii sp. nov. Int. J. Syst. Bacteriol. 34, 336–337. doi: 10.1099/00207713-34-3-336
Steinweg, C., Kuenne, C. T., Billion, A., Mraheil, M. A., Domann, E., Ghai, R., et al. (2010). Complete genome sequence of Listeria seeligeri, a nonpathogenic member of the genus Listeria. J. Bacteriol. 192, 1473–1474. doi: 10.1128/JB.01415-09
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016 44, 6614–6624. doi: 10.1093/nar/gkw569
Trivett, T. L., and Meyer, E. A. (1971). Citrate cycle and related metabolism of Listeria monocytogenes. J. Bacteriol. 107, 770–779.
Unden, G., and Bongaerts, J. (1997). Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320, 217–234. doi: 10.1016/S0005-2728(97)00034-0
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domínguez-Bernal, G., Goebel, W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640. doi: 10.1128/CMR.14.3.584-640.2001
Vilar, M. J., Yus, E., Sanjuán, M. L., Diéguez, F. J., and Rodríguez-Otero, J. L. (2007). Prevalence of and risk factors for Listeria species on dairy farms. J. Dairy Sci. 90, 5083–5088. doi: 10.3168/jds.2007-0213
Vine, C. E., Purewal, S. K., and Cole, J. A. (2011). NsrR-dependent method for detecting nitric oxide accumulation in the Escherichia coli cytoplasm and enzymes involved in NO production. FEMS Microbiol. Lett. 325, 108–114. doi: 10.1111/j.1574-6968.2011.02385.x
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9:e112963. doi: 10.1371/journal.pone.0112963
Walter, M. C., Rattei, T., Arnold, R., Güldener, U., Münsterkötter, M., Nenova, K., et al. (2009). PEDANT covers all complete RefSeq genomes. Nucleic Acids Res. 37, D408–D411. doi: 10.1093/nar/gkn749
Weller, D., Andrus, A., Wiedmann, M., and den Bakker, H. C. (2015). Listeria booriae sp. nov. and Listeria newyorkensis sp. nov., from food processing environments in the USA. Int. J. Syst. Evol. Microbiol. 65, 286–292. doi: 10.1099/ijs.0.070839-0
Williams, T., Joseph, B., Beier, D., Goebel, W., and Kuhn, M. (2005). Response regulator DegU of Listeria monocytogenes regulates the expression of flagella-specific genes. FEMS Microbiol. Lett. 252, 287–298. doi: 10.1016/j.femsle.2005.09.011
Ye, R. W., Tao, W., Bedzyk, L., Young, T., Chen, M., and Li, L. (2000). Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182, 4458–4465. doi: 10.1128/JB.182.16.4458-4465.2000
Keywords: Listeria weihenstephanensis, complete circular genome sequence, adaptation to anaerobiosis, nitrate respiration, transcriptome, temperature
Citation: Ferrari E, Walter MC, Huptas C, Scherer S and Müller-Herbst S (2017) Complete Circular Genome Sequence and Temperature Independent Adaptation to Anaerobiosis of Listeria weihenstephanensis DSM 24698. Front. Microbiol. 8:1672. doi: 10.3389/fmicb.2017.01672
Received: 05 May 2017; Accepted: 17 August 2017;
Published: 01 September 2017.
Edited by:
Michael Sauer, University of Natural Resources and Life Sciences, AustriaCopyright © 2017 Ferrari, Walter, Huptas, Scherer and Müller-Herbst. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siegfried Scherer, siegfried.scherer@wzw.tum.de
†Present Address: Elena Ferrari, Institut für Prophylaxe und Epidemiologie der Kreislaufkrankheiten, Ludwig-Maximilians Universität München, München, Germany
Mathias C. Walter, Bundeswehr Institute of Microbiology, München, Germany
 Elena Ferrari1,2†
Elena Ferrari1,2† Mathias C. Walter
Mathias C. Walter Christopher Huptas
Christopher Huptas Siegfried Scherer
Siegfried Scherer Stefanie Müller-Herbst
Stefanie Müller-Herbst