- 1Guangdong Institute of Microbiology, State Key Laboratory of Applied Microbiology Southern China, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Open Laboratory of Applied Microbiology, Guangzhou, China
- 2School of Bioscience and Bioengineering, South China University of Technology, Guangzhou, China
- 3College of Food Science, South China Agricultural University, Guangzhou, China
- 4Department of Food Science and Technology, Jinan University, Guangzhou, China
Staphylococcus aureus is one of the most important food-borne pathogens globally. It produces various toxins and invasive enzymes and can be found in numerous food products. Milk is an important source of staphylococcal food poisoning. After pasteurization, this microorganism or its enterotoxins might still remain in pasteurized milk. Therefore, this study was to investigate the contamination of S. aureus in 258 pasteurized milk from 39 cities of China. The prevalence and levels of S. aureus in these samples as well as antibiotic susceptibility profiles, virulence genes, biofilm formation, and biofilm related genes, spa typing and MLST were used to determine the characterization among the isolates. It was found 3.9% of samples were detected S. aureus in 8 of 39 cities in China. The contaminated level were not very excessive which showed the MPN values of the most positive samples (9/10) were less than 1 MPN/g. All pasteurized milk-related S. aureus isolates have ability to produce biofilm and harbored icaA, icaD, eno, clfA, clfB, fnbA, fnbB, fib genes, other biofilm related genes icaC were showed in 91.7% of isolates and cna gene were showed in 50%, except bap gene which were free in all isolates. The antibiotic susceptibility test showed that all isolates were resistant or intermediate-resistant to different concentrations of the antibiotics. Furthermore, 75.0% of the isolates were resistant to three or more antibiotic classes, which indicated multidrug resistance. The isolates had virulence potential, which showed 66.7% (8/12) of the isolates carried one or more virulence-associated genes. Molecular typing by MLST and spa typing enabled classification of these isolates into a total of 11 sequence types (STs) and spa types, which indicated high genetic diversity. Most of these types were related to various clinical S. aureus infections. Thus, the findings of this study reflect the potential risk of S. aureus infection in China. Our study also provides comprehensive analysis of the prevalence of S. aureus in pasteurized milk and helps ensure more accurate treatment of human infection with effective antibiotics.
Introduction
Staphylococcus aureus is a pathogen associated with serious community and hospital-acquired diseases. It has low nutritional requirements and widely exists in nature. In China, microbial food poisoning accounted for 53.7% of food poisoning emergencies in 2015. Of which, S. aureus was an important pathogenic factor in these cases (Wu et al., 2018a). In the United States, S. aureus causes approximately 241,000 cases of food poisoning each year (Scallan et al., 2011; Kadariya et al., 2014). This foodborne pathogen is considered as one of the world’s leading causes of disease outbreaks related to food consumption, being responsible for a variety of manifestations and diseases (Jamali et al., 2015).
Staphylococcus aureus produces a variety of toxins and invasive enzymes such as staphylococcal enterotoxins (SEs), hemolysins, Panton-Valentine leukocidin (PVL), toxic shock syndrome toxin-1 (TSST-1), plasma coagulase, and deoxyribonuclease (Spanu et al., 2012). Differentiation between virulent and non-virulent strains is significant for evaluating the potential implications of the presence of this microorganism for food safety and public health. In which, SEs are active at concentrations ranging from high nanogram to low microgram quantities (Larkin et al., 2009) and are resistant to conditions (heat treatment, low pH) that retaining their activity in the digestive tract after ingestion (Evenson et al., 1988; Argudín et al., 2010). In addition, TSST-1 is a superantigenic exotoxin that causes toxic shock syndrome and PVL is a bacteriophage-encoded bicomponent leukotoxin that is in some strains of S. aureus and plays a key role in leukocytolysis and tissue necrosis (Shallcross et al., 2013). S. aureus often develops antibiotic resistance. Isolation of single- or multiple-drug resistant S. aureus (MRSA) strains from food, the environment, and clinics has been constantly reported (Gould et al., 2012; Rasigade et al., 2014). The ability of S. aureus to form biofilms helps the bacterium to survive in hostile environments within the host and is considered to be responsible for chronic or persistent infections (Costerton et al., 1999). The ability of some strains to synthesize biofilms could increase their pathogenicity since established biofilms can tolerate antimicrobial agents, thus making the bacterium extremely difficult to eradicate (Zmantar et al., 2010).
Nowadays, molecular typing methods are crucial in epidemiological investigations of food processing and enhance the resolution of surveillance (Wu et al., 2015). Of various subtyping approaches, multilocus sequence typing (MLST) is a widely accepted method of DNA sequence based typing that based on analysis of relatively conserved genes that encode essential proteins. For S. aureus, the level of discrimination provided by MLST is sufficient to provide a relatively detailed picture of the global dissemination of the organism (Saunders and Holmes, 2007). spa typing is another efficient typing method for S. aureus, it based on sequencing of the polymorphic X region of the protein A gene (spa) (Hallin et al., 2009). Similar to MLST, it suggests that spa typing is suitable for epidemiology and evolutionary investigations based on studies of European and international isolates (Wu et al., 2018b).
Milk is an important source of staphylococcal food poisoning. There are several foodborne outbreaks of S. aureus intoxications have been documented to be associated with consumption of contaminated milk (De Buyser et al., 1985; Miwa et al., 2001; Fetsch et al., 2014). In addition, raw milk and raw milk products are frequently contaminated with different types of S. aureus around the world (Tham et al., 1990; Rosengren et al., 2010; Yu et al., 2010; Jamali et al., 2015; Riva et al., 2015). Milk is a good substrate for S. aureus growth and enterotoxin production. Enterotoxins can retain their biological activity after pasteurization (Asao et al., 2003; Rall et al., 2008). In China, studies have reported that some S. aureus strains persist in powdered infant formula (Wang et al., 2012). Currently, only a few full-scale and systematic studies have been performed on S. aureus prevalence and contamination levels in pasteurized milk in China. This study aimed to investigate S. aureus contamination in pasteurized milk obtained from different Chinese cities. For this purpose, we analyzed the S. aureus prevalence and contamination levels, antibiotic susceptibility profiles, virulence genes, biofilm formation, biofilm-related genes, spa typing results, and multilocus sequence typing (MLST) results to characterize the isolates.
Materials and Methods
Sampling
From July 2011 to June 2016, a total of 258 pasteurized milk samples were collected from supermarkets, fairs, and farmers’ markets (Supplementary Table 1). The samples were obtained from 39 cities located in a total of 29 provinces and 2 directly controlled municipalities in China, which covered most of the provincial capitals of China (Supplementary Figure 1). The samples were placed in a cold box at approximately 4°C, tightly sealed with sterile plastic wrap, transported to an accredited laboratory, and subjected to microbiological analysis within 24 h.
Isolation and Detection
The samples were qualitatively and quantitatively analyzed to detect S. aureus. For qualitative analysis, samples were examined according to GB 4789.30-2010 (National Food Safety Standards of China) with slight modification. The most probable number (MPN) method was used for quantitative analysis. Approximately 25 g of the food sample was homogenized in 225 mL tryptic soy broth with 10% sodium chloride (Huankai, Guangzhou, China). Subsequently, 1-mL, 0.1-mL, and 0.01-mL aliquots were transferred to tubes containing 9, 10, and 10 mL in triplicate with trypticase soy broth (Huankai) supplemented with 10% NaCl. The tubes were then incubated at 37°C for 48 h.
A loopful of enrichment broth culture (10 μL) was streaked onto chromogenic S. aureus agar plates (Huankai) and incubated at 37°C for 24 h. Of the colonies obtained, 1–4 pink colonies were purified on nutrient agar medium. The purified colonies were analyzed via the coagulase activity test involving freeze-dried rabbit plasma (Huankai), and the API STAPH test strips (bioMérieux, Marcy-l’Étoile, France) were then used. The MPN value was determined on the basis of the number of positive tube(s) in each of the three sets using the MPN table.
Antimicrobial Susceptibility Testing
The Kirby–Bauer disk diffusion method was used to test antibiotic susceptibility, and diameter interpretations were based on the protocol specified in the guidelines of Clinical and Laboratory Standards Institute [CLSI] (2015). S. aureus ATCC 25923 and Escherichia coli ATCC 25922 were used as quality control organisms. All isolates were assessed for antimicrobial susceptibility to 24 antibiotics (Oxoid, United Kingdom): amoxicillin/clavulanic acid, ampicillin, cefepime, cefoxitin, penicillin G, ceftazidime, amikacin, gentamicin, kanamycin, streptomycin, chloramphenicol, clindamycin, erythromycin, telithromycin, ciprofloxacin, norfloxacin, tetracycline, linezolid, trimethoprim/sulfamethoxazole (1:19), rifampicin, quinupristin/dalfopristin, teicoplanin, nitrofurantoin, and fusidic acid. The CLSI zone diameter breakpoints were used to interpret the antimicrobial susceptibilities of the analyzed strains.
In vivo Biofilm Formation
Biofilm production was quantified using a microtiter plate assay (MPA) described previously (Vasudevan et al., 2003), with slight modifications. S. aureus strains were individually grown at 37°C overnight in brain heart infusion broth (BHI). The overnight culture was diluted 1:100 in fresh BHI, and 200-μL aliquots of each prepared suspension were transferred into three wells of 96-well tissue culture treated polystyrene microplates (CELLSTAR® Cell Culture Microplates, Greiner Bio-one, Frickenhausen, Germany). After cultivation at 37°C for 48 h, the wells were washed three times with 200 mL sterile phosphate-buffered saline (PBS, pH 7.4) and dried at room temperature. The adherent bacterial cells were fixed with 200 μL methanol for 15 min, and the plates were emptied and left to dry overnight. The adherent cells were then stained with 1% crystal violet for 10 min and washed twice with water. The dye bound to the adherent cells was dissolved with 150 mL 95% ethanol and optical density (OD) was measured at 590 nm with a spectrophotometer (SpectroStar Nano, BMG Labtech). Uninoculated wells containing BHI served as blanks. Blank-corrected absorbance values of S. aureus strains were used for reporting biofilm production. Isolates were considered biofilm producers when their OD values were three times greater than the standard deviation of the mean Dc. Additionally, isolates showing biofilm-producing ability were classified as weak (Dc < OD ≤ 2 × Dc), moderate (2 × Dc < OD ≤ 4 × Dc), or strong (OD > 4 × Dc) biofilm producers.
DNA Extraction and PCR
After the isolates were cultivated for at 37°C 24 h in BHI, genomic DNA was extracted using a Genomic DNA Extraction kit (Magen Biotech, Guangzhou, China) according to the manufacturer’s instructions. The genomic DNA concentration was determined at 260 nm by using a NanoDrop-ND-1000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, MA, United States). The mecA/mecC gene, which confers methicillin resistance to S. aureus, was detected by PCR performed using primers described previously (Perez-Roth et al., 2001; Stegger et al., 2012). PCR was also used to detect the presence of virulence genes. The detection of 18 SE genes (sea, seb, sec, sed, see, seg, seh, sei, sej, sek, sel, sem, sen, seo, sep, seq, ser, and seu), the tsst gene and the presence of lukSF-PV (pvl) genes was performed as previously described (Jarraud et al., 2002; Varshney et al., 2009). For adhesion genes (clfA, clfB, fnbA, fnbB, fib, and cna) and biofilm-producing genes (icaA, icaD, icaC, bap, and eno), PCR was performed according to previous studies (Zmantar et al., 2010; Artini et al., 2013; Salaberry et al., 2015; Pereyra et al., 2016). The primer sequences used in the PCR assays, expected amplified product sizes, and references are listed in Supplementary Table 2. The amplicons were stained with Goldview, loaded onto 1.5% agarose gels, electrophoresed at 120 V for 0.5 h, and visualized by a UV transilluminator gel imaging system (GE Healthcare, WI, United States). The images were saved as TIFF files for analysis.
Molecular Typing
All the isolates were characterized by MLST and spa typing. The MLST analysis was based on DNA sequencing of the seven housekeeping genes arcC, aroE, glpF, gmk, pta, tpi, and yqil, as described previously (Enright et al., 2000). The primers spa-1113f (5′-TAA AGA CGA TCC TTC GGT GAG C-3′) and spa-1514r (5′-CAG CAG TAG TGC CGT TTG CTT-3′) were used for spa amplification (Shopsin et al., 1999). The PCR products were purified with a PCR purification kit (Qiagen, Genmany) and were sequenced in both directions with Big Dye fluorescent terminators on an ABI 3730XL sequencer (Applied BioSystems). For each MLST locus, an allele number was assigned to each distinct sequence variant, and a distinct sequence type (ST) number was attributed to each distinct combination of alleles for the seven genes. STs were determined using the S. aureus MLST database.1 Clonal complex (CC) analysis was performed using eBURST v.3 as previously described (Feil et al., 2004). The spa types were randomly assigned using the SpaServer website.2 Sequence Type Analysis and Recombinational Tests software (S.T.A.R.T. ver. 2; https://pubmlst.org/software/analysis/start2) was used to analyze the MLST data.
Results
Isolation and Identification of S. aureus From Pasteurized Milk
Of the 258 pasteurized milk samples, 10 samples were contaminated with coagulase-positive staphylococci, and 12 isolates obtained from these positive samples were confirmed as S. aureus by the API STAPH test. The qualitative and quantitative results for these positive samples are shown in Table 1. Overall, the mean prevalence of S. aureus was 3.9%, and the MPN values for the samples showing the highest levels (9/10) were less than 1 MPN/g. Positive samples were obtained from 8 of the 39 Chinese cities included (Supplementary Figure 1): 33.3% of the positive samples were from Zhanjiang and Xining cities, and 16.7% were obtained from the other six cities, respectively. Besides, among the different types of sampling source, the positive samples isolated from fairs was 4.2% (6/144) followed by farmer’s market (3.8%, 1/26) and supermarkets (3.4%, 3/88).
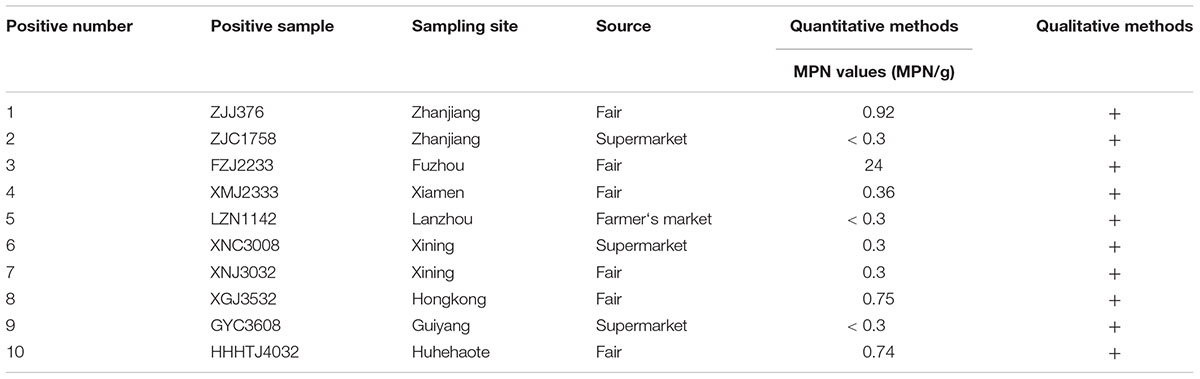
Table 1. Prevalence and contamination levels of Staphylococcus aureus in pasteurized milk samples from China.
Antibiotic Resistance
Most S. aureus isolates showed resistance and intermediate resistance to different antibiotics concentrations. The resistance patterns of S. aureus to the tested antimicrobial agents are presented in Table 2. The phenotypic resistance profiles of the S. aureus isolates are as follows: ampicillin, 97.7%; penicillin G, 83.3%; erythromycin, 50.0%; kanamycin, 41.7%; telithromycin, 33.3%; clindamycin, 33.3%; gentamicin, 33.3%; tetracycline, 33.3%; norfloxacin, 25.0%; ciprofloxacin, 16.7%; amikacin, 16.7%; rifampicin, 16.7%; chloramphenicol, 8.3%; fusidic acid, 16.7%; ceftazidime, 8.3%; streptomycin, 8.3%; amoxicillin/clavulanic acid, 8.3%; trimethoprim/sulfamethoxazole (1:19), 8.3% and quinupristin/dalfopristin, 8.3%. All the isolates were susceptible to cefepime, cefoxitin, linezolid, and nitrofurantoin. The mecA/mecC gene was free in all S. aureus isolates. Combining with antibiotic resistance test, there was none of MRSA isolates identified. However, only one isolate was susceptible to all the antimicrobial agents tested, whereas two isolates were resistant to two antibiotics and 75.0% of isolates (9/12) were resistant to three or more classes of antibiotics which showed multi-drug resistance including one strain Sta1758 were resistant to more than 10 antibiotics.
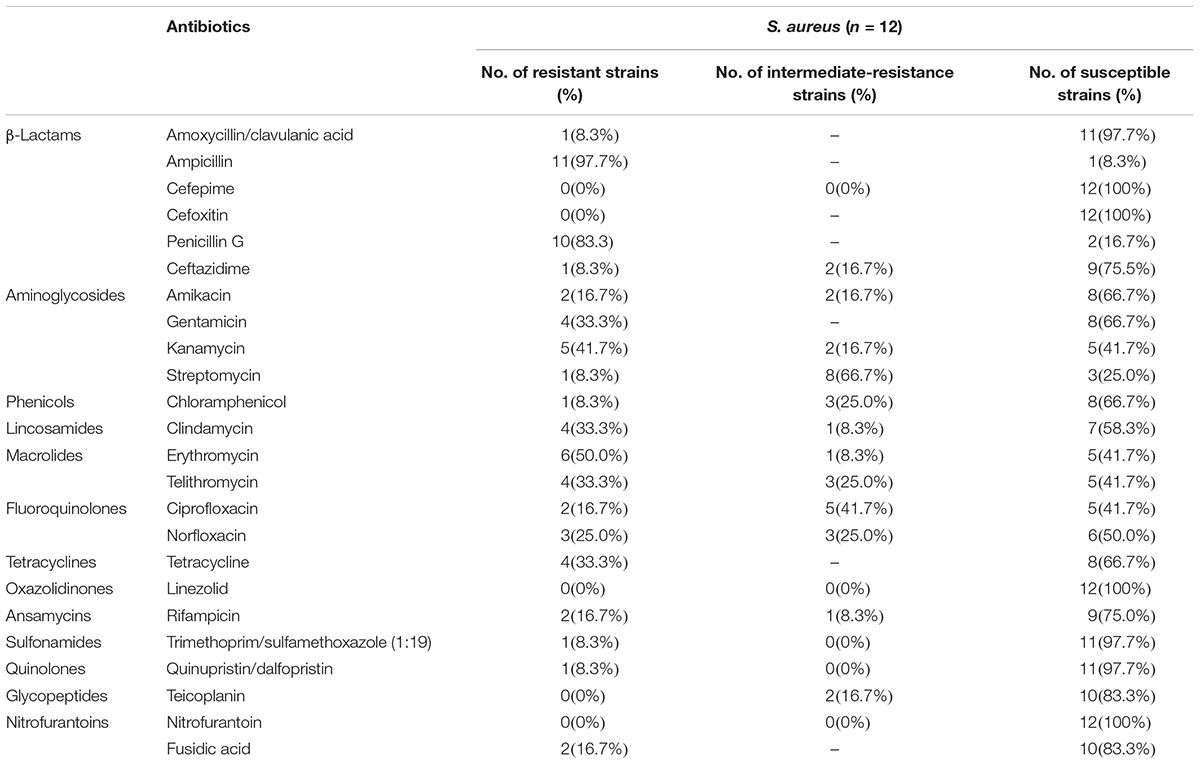
Table 2. Antimicrobial susceptibility tests for Staphylococcus aureus isolates obtained from pasteurized milk samples from China.
In vitro Biofilm Production and Presence of Biofilm-Related Genes
The MPA results showed that all the S. aureus strains could produce biofilms, including four isolates (33.3%, 4/12) showing moderate and eight isolates (76.7%, 8/12) showing strong biofilm formation (Table 3). Table 3 also provides the results for the presence of S. aureus adhesion and biofilm-associated genes. Overall, icaA, icaD, eno, clfA, clfB, fnbA, fnbB, and fib were found in all 12 isolates, whereas the bap gene was free in all isolates. In addition, icaC was present in 91.7% (11/12) of the isolates and the cna gene in 50% (6/12). Thus, most of the biofilm-producing genes and adhesion genes were present in these S. aureus strains isolated from the pasteurized milk samples.
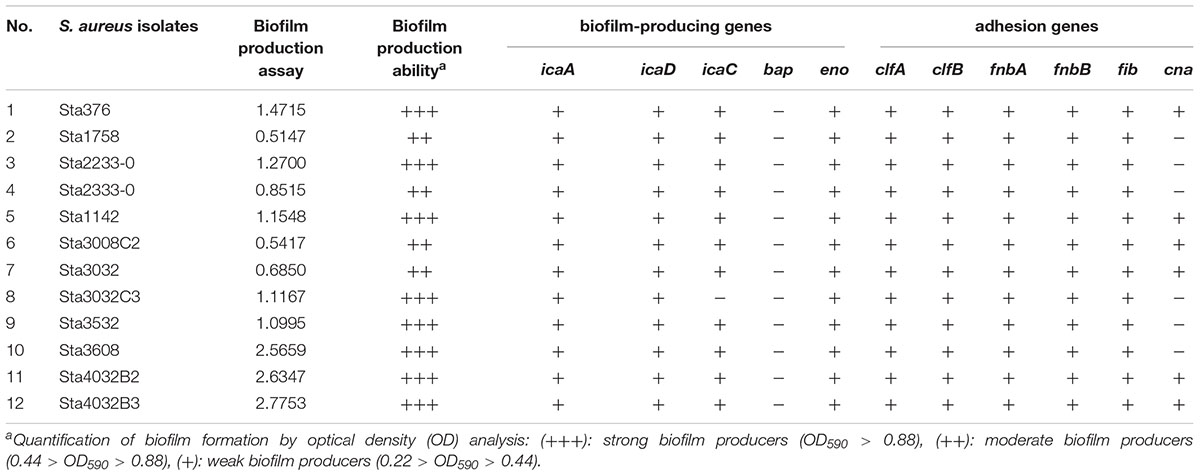
Table 3. Biofilm-formation ability and biofilm-associated genes in Staphylococcus aureus isolates obtained from pasteurized milk samples.
Prevalence of Virulence-Associated Genes
The prevalence of virulence-associated genes is shown in Figure 1. Of the S. aureus isolates analyzed, 66.7% (8/12) carried one or more virulence-associated genes. Of the 20 virulence genes investigated, seg, sei, and sem were most frequently detected which showed 41.7% of prevalence, respectively. sea, seb, sed, see, seu, seq, sej, ser, sek, and pvl were not detected in the S. aureus isolates. The prevalence of the other virulence genes is as follows: sen (25%), sec (16.7%), sel (16.7%), seo (8.3%), sep (8.3%), seh (8.3%), and tsst (8.3%). Staphylococcal enterotoxin genes can be divided into classic SE genes (sea, seb, sec, sed, and see), egc cluster genes (seg, sei, sem, sen, seo, and seu), and other novel SE genes (sep, seq, she, sej, sel, ser, and sek). From the classic SE genes, only two isolates showed the sec gene, that is 7.7% (2/26) prevalence in the detected genes. The egc cluster accounted for 73.1% (19/26) of the detected genes, and other novel SE genes accounted for 15.4%.
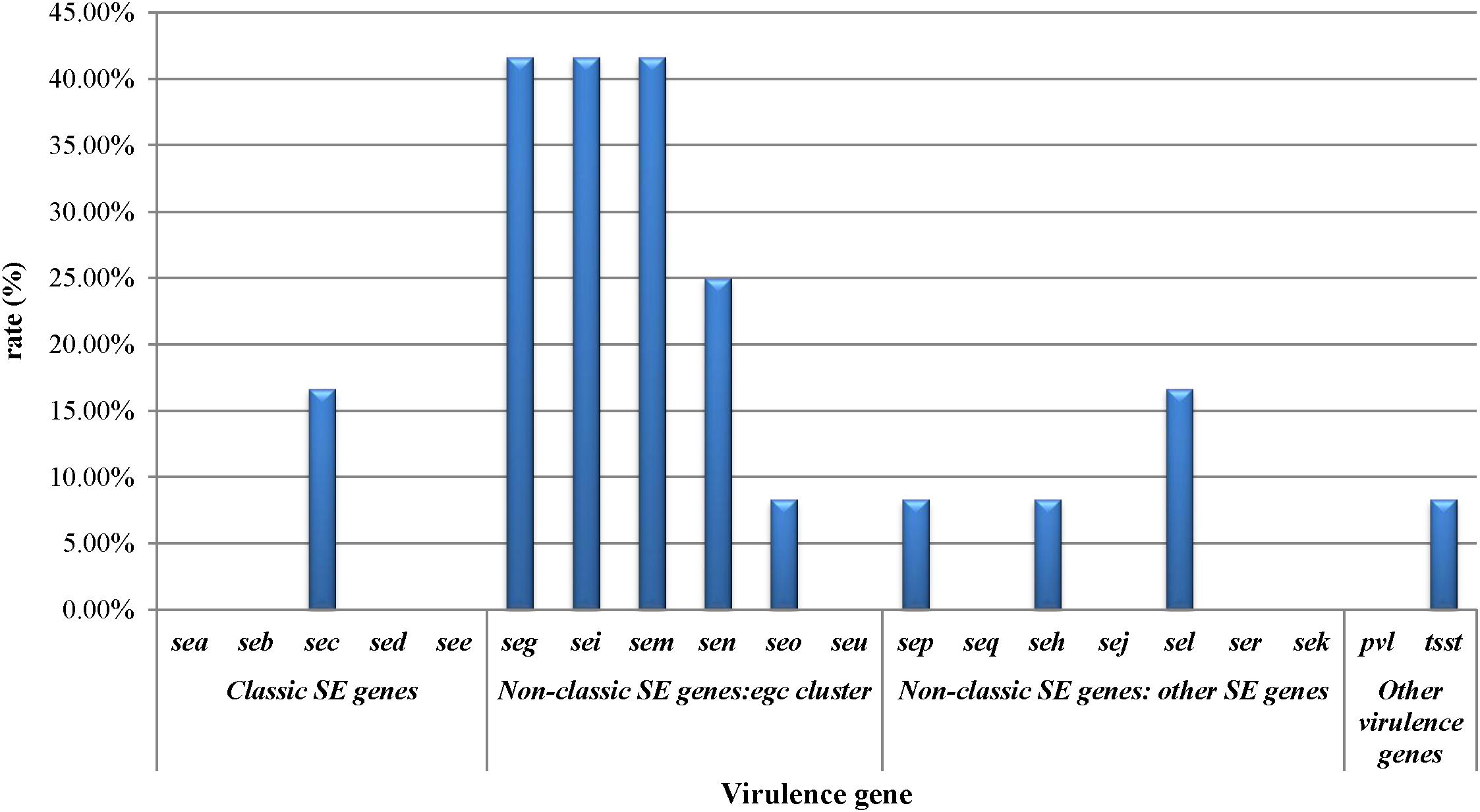
Figure 1. Staphylococcal toxin gene profiles of Staphylococcus aureus isolates obtained from pasteurized milk.
Molecular Characterization of S. aureus Isolates
MLST analysis of the 12 isolates yielded 11 STs showing high genetic diversity. Except ST188 was found in two strains, the other STs were present in single strains which included ST1, ST5, ST7, ST9, ST12, ST15, ST25, ST72, ST398, and ST4069. Of these, ST4069 was a newly assigned ST, which found a single-locus variant (SLV) on the aroE gene (assigned as aroE-472). spa typing showed results similar to those of MLST. Eleven spa types were detected: t189 was found in two strains, and t177, t899, t213, t3092, t148, t091, t002, t085, t078, and t034 were found in single isolates. The phylogenetic tree based on the seven concatenated MLST sequences in Figure 2 shows the relatedness between these isolates. ST188 correlated well with t189, which was also found in city of Xining.
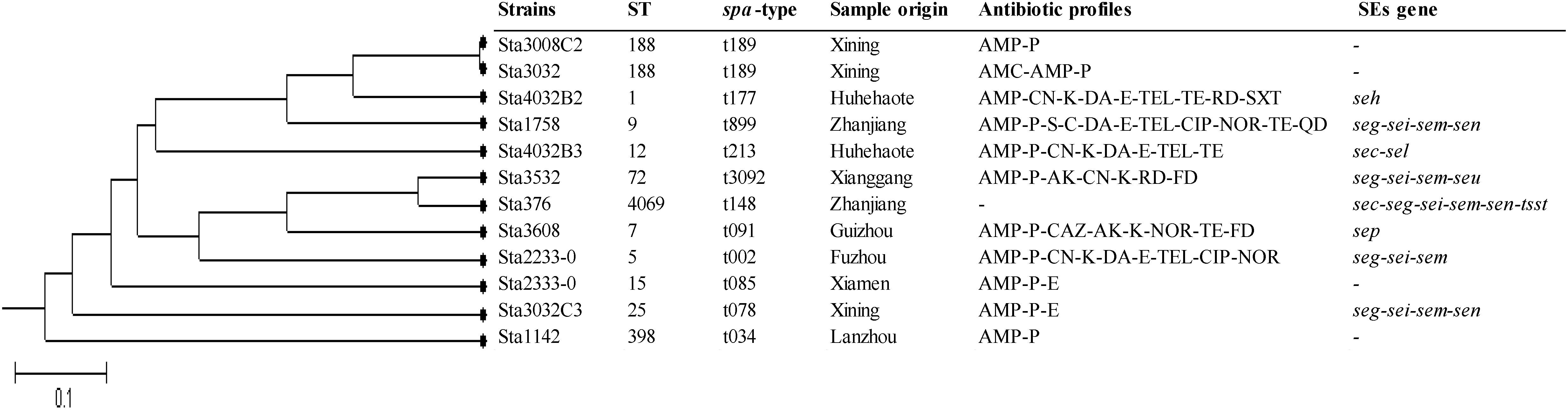
Figure 2. Unweighted pair group method with arithmetic mean (UPGMA) tree of the 7 multilocus sequence typing loci of S. aureus isolates obtained from pasteurized milk. This tree was generated using S.T.A.R.T. (version 2).
Discussion
Pasteurization, or heat treatment, of milk is an important milestone in for public health and has contributed to a dramatic decline in many infectious diseases. However, outbreaks associated with pasteurized milk continue to occur (Ryan et al., 1987; Dalton et al., 1997; Ackers et al., 2000). In China, milk has become an important component of a balanced diet, and the annual per capita milk consumption increased from 9.23 kg in 1992 to 24.87 kg in 2007 (Qian et al., 2011; Liu and Wang, 2013). SFP outbreaks associated with milk have occurred occasionally (Dai et al., 2009). There were limited researches on the prevalence of S. aureus from pasteurized milk in China. In the current study, 3.9% of the samples showed the presence of S. aureus. Compare with other countries’ studies, the prevalence of S. aureus from pasteurized milk in our study is lower. In Brazil, Rall et al. (2008) collected 162 raw and pasteurized milk samples and found 20.4% of the pasteurized milk samples were positive for S. aureus. In 2006, Gündoğan et al. (2006) analyzed 180 raw milk, pasteurized milk, and ice cream samples; 56.6% of the pasteurized milk samples were positive for S. aureus. However, it is maybe attribute to the sample sizes, sample source or geographic locations, which showed most of these S. aureus were isolated from farm, whereas our samples were collected from supermarkets, fairs or farmer’s markets. Quantitative analysis by MPN showed that only one S. aureus–positive sample reached a level of 10 MPN/g. In China, a limit of 100 CFU/g (mL) has been proposed for S. aureus in milk and the highest fraction of samples with values that exceeded this limit was observed at the end of food shelf-life (Helwigh and Korsgaard, 2007). Thus, the currently available data, along with our study, indicate that levels of S. aureus in pasteurized milk in China are not very high.
Staphylococcal enterotoxins are responsible for most SFP outbreaks (Dinges et al., 2000; Loir et al., 2003). This could account for retention of biological activity after pasteurization. This study was investigated the 18 SE genes among pasteurized milk related S. aureus isolates and found that 66.7% of the isolates carried one or more SE genes, suggesting the risk of S. aureus contamination in pasteurized milk in China. This rate is higher than that reported in a previous study (Chao et al., 2015) in which the prevalence of 18 SE genes was studied in S. aureus isolates obtained from different origins in China, showing that 54.4% of those isolates harbored SE genes. Generally, the sea gene has been the most common gene in the SFP outbreak isolates tested, followed by the sed gene from classic SE genes (Kérouanton et al., 2007), but they were not detected in the current study. sec was the only classical SE gene detected in our study, which is the most commonly occurring SE in milk (Lindqvist et al., 2002; Scherrer et al., 2004; Aragon-Alegro et al., 2007). In 2002, an outbreak of community-acquired foodborne illness caused by MRSA was attributed to SEC-containing coleslaw, which was produced in the United States (Jones et al., 2002). In addition, we observed higher prevalence of seg-sei-sem among the isolates in our study, these SE genes belonged to the egc cluster, which is widely distributed in clinical isolates and acts as a putative nursery of enterotoxin genes (Jarraud et al., 2001; Wu et al., 2018b). Thus, the hazard posed by these isolates harboring SE genes should not be ignored.
In the current study, all S. aureus strains obtained from pasteurized milk samples could produce biofilms and showed moderate or strong biofilm production capability. In addition, various genes have been reported to be involve in biofilm formation, of these genes the ica group genes especially icaA and icaD (Arciola et al., 2001) and the other group such as fnbA, fnbB, cna and eno genes that encode MSCRAMMs proteins (Atshan et al., 2012) are noticeable. Most of these genes were present in the S. aureus strains isolated from our samples. This result were similar with the research by Atshan et al. (2012) who detected 9 MSCRAMM and 4 biofilm related genes in the MSSA and MRSA isolates from clinically patients by PCR method and found fnbA, eno, ebpS, clfA, clfB, icaA, icaD, icaB, and icaC genes were detected in 100% of isolates. Furthermore, fib and cna were detected in 90% and 46.6% MSSA isolates, respectively, which is in agreement with our result. In our study, the bap gene was free in all S. aureus isolates. Many studies have reported that the bap gene was not detected or very low rates in the solutes studied (Melchior et al., 2007; Szweda et al., 2012; Khoramrooz et al., 2016; Pereyra et al., 2016; Felipe et al., 2017). Although there was inconclusive evidence for the correlation between phenotypic tests and the detection of specific genes associated with adhesion and biofilm formation, the presence of icaA, icaD, icaC, fnbA, clfA, clfB, fnbA, fnb, and fib genes indicated a significant association with biofilm formation in this study. The biofilm production in isolates with negative results for any of the genes studied may be attributable to other biofilm-related genes.
Antimicrobial resistance was noted in pasteurized milk related S. aureus isolates. Most of the isolates were resistant to at least one antimicrobial agent. However, resistance to ampicillin and penicillin was higher than that reported in many previous studies involving S. aureus isolates from dairy samples (Rosengren et al., 2010; Jamali et al., 2015; Riva et al., 2015). We also observed that 25.0% of the isolates were resistant to multiple antibiotics such as ampicillin, gentamicin, kanamycin, clindamycin, erythromycin, and telithromycin. These were similar to the study by Jamali et al.’ investigation, which showed the resistance profiles of S. aureus isolates from raw milk and dairy products for tetracycline, penicillin, clindamycin, erythromycin, streptomycin, kanamycin, chloramphenicol, and gentamicin (Jamali et al., 2015). Furthermore, many isolates were found intermediate-resistant to the selected antibiotics in this study. The reduced antibiotic susceptibility of these antibiotics suggests that these bacteria may be exposed to such antimicrobials for a long time (Howden et al., 2010). Thus, it should draw public attention and controlled use of antimicrobials would limit the emergence of drug-resistant bacteria.
MLST and spa genotyping have shown a highly clonal population structure for S. aureus in many studies (Kanika et al., 2011; Fetsch et al., 2014; Basanisi et al., 2017). The S. aureus MLST database3 currently has 5024 different STs. However, some STs are specific to a particular source. For example, ST398 occurs in livestock-associated S. aureus; ST5, ST45, and ST239 have been detected in hospital-associated S. aureus infections; and ST1, ST8, ST30, and ST59 are often related to community-associated S. aureus infections (Bens et al., 2006; Cui and Li, 2009; Neela et al., 2009; Skov, 2009; Deleo et al., 2010). Generally, spa typing showed highly concordant with MLST. In the current study, MLST and spa typing enabled classification of pasteurized milk related S. aureus isolates into 11 types (ST188-t189, ST1-t177, ST9-899, ST12-t213, ST72-t3092, ST7-t091, ST5-t002, ST15-t085, ST25-t078, ST398-t034, and ST4069-t148). Most of these STs were detected in various of samples and relevant to a variety of clinical S. aureus infections (Krziwanek et al., 2009; Lee et al., 2011; Yu et al., 2012; Schaumburg et al., 2015). Therefore, the hazards of these strains for consumers cannot be ignored.
Conclusion
To conclude, our results provide information regarding the genetic background of S. aureus strains obtained from pasteurized milk samples in China. To our knowledge, our study is the first systematic investigation of prevalence and contamination levels for S. aureus isolated from pasteurized milk samples in China. Different source strains showed different types in this study. These isolates had virulence potential, which half of them carried enterotoxin genes and infection-related molecular type. The resistance patterns can help determine more appropriate treatments for human infections in China. All the isolates had moderate or strong biofilm-production capability, and most of them harbored biofilm-related genes, indicating that these potentially virulent bacteria could persist in the food-manufacturing environment.
Author Contributions
QW, JZ, JD, SW, and JH conceived and designed the experiments. JD, JH, and FZ performed the experiments. JD, SW, HW, and TL analyzed the data. XY, LX, YD, SZ, and JW contributed reagents, materials, and analysis tools. SW, JD, and QW contributed to the writing of the manuscript.
Funding
We would like to acknowledge the financial support of National Natural Science Foundation of China (No. 31801657), China Postdoctoral Science Foundation (2017M612623), and GDAS’ Special Project of Science and Technology Development (2017GDASCX-0201, 2017GDASCX-0817).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00641/full#supplementary-material
Footnotes
- ^https://pubmlst.org/saureus/
- ^https://spaserver2.ridom.de
- ^https://pubmlst.org/bigsdb?db=pubmlst_saureus_isolates&page=profiles
References
Ackers, M. L., Schoenfeld, S., Markman, J., Smith, M. G., Nicholson, M. A., Dewitt, W., et al. (2000). An outbreak of Yersinia enterocolitica O:8 infections associated with pasteurized milk. J. Infect. Dis. 181, 1834–1837. doi: 10.1086/315436
Aragon-Alegro, L. C., Konta, E. M., Suzuki, K., Silva, M. G., Júnior, A. F., Rall, R., et al. (2007). Occurrence of coagulase-positive Staphylococcus in various food products commercialized in Botucatu, SP, Brazil and detection of toxins from food and isolated strains. Food Control 18, 630–634. doi: 10.1016/j.foodcont.2006.02.010
Arciola, C. R., Baldassarri, L., and Montanaro, L. (2001). Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 39, 2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001
Argudín, M. Á, Mendoza, M. C., and Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2, 1751–1773. doi: 10.3390/toxins2071751
Artini, M., Papa, R., Scoarughi, G. L., Galano, E., Barbato, G., Pucci, P., et al. (2013). Comparison of the action of different proteases on virulence properties related to the staphylococcal surface. J. Appl. Microbiol. 114, 266–277. doi: 10.1111/jam.12038
Asao, T., Kumeda, Y., Kawai, T., Shibata, T., Oda, H., Haruki, K., et al. (2003). An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol. Infect. 130, 33–40. doi: 10.1017/S0950268802007951
Atshan, S. S., Shamsudin, M. N., Sekawi, Z., Lung, L. T. T., Hamat, R. A., Karunanidhi, A., et al. (2012). Prevalence of adhesion and regulation of biofilm-related genes in different clones of Staphylococcus aureus. J. Biomed. Biotechnol. 2012:976972. doi: 10.1155/2012/976972
Basanisi, M. G., Bella, G. L., Nobili, G., Franconieri, I., and Salandra, G. L. (2017). Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol. 62:141. doi: 10.1016/j.fm.2016.10.020
Bens, C. C., Voss, A., and Klaassen, C. H. (2006). Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 44, 1875–1876. doi: 10.1128/JCM.44.5.1875-1876.2006
Chao, G., Bao, G., Cao, Y., Yan, W., Yan, W., Zhang, X., et al. (2015). Prevalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int. J. Food Microbiol. 211:142. doi: 10.1016/j.ijfoodmicro.2015.07.018
Clinical and Laboratory Standards Institute [CLSI] (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Approved Standard-M02-A12. Wayne, PA: The Clinical and Laboratory Standards Institute.
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Cui, S., and Li, J. C. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemoth. 64, 680–683. doi: 10.1093/jac/dkp275
Dai, C. F., Wen, J., Lu, X. U., Huang, G. X., Sun, H., Hu, Z. K., et al. (2009). Investigation of a case on milk product poisoning caused by Staphylococcus aureus. Chin. J. Food Hyg. 03, 262–264.
Dalton, C. B., Austin, C. C., Sobel, J., Hayes, P. S., Bibb, W. F., Graves, L. M., et al. (1997). An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 336, 100–105. doi: 10.1056/NEJM199701093360204
De Buyser, M., Janin, F., and Dilasser, F. (1985). Contamination of ewe cheese with Staphylococcus aureus: study of an outbreak of food poisoning. Zentralblatt fur Bakteriologie Mikrobiologie Hygiene 14, 677–678.
Deleo, F. R., Otto, M., Kreiswirth, B. N., and Chambers, H. F. (2010). Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375, 767–768. doi: 10.1016/S0140-6736(10)61370-0
Dinges, M. M., Orwin, P. M., and Schlievert, P. M. (2000). Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13, 16–34. doi: 10.1128/CMR.13.1.16
Enright, M. C., Day, N. P. J., Davies, C. E., Peacock, S. J., and Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015.
Evenson, M. L., Hinds, M. W., Bernstein, R. S., and Bergdoll, M. S. (1988). Estimation of human dose of staphylococcal enterotoxin A from a large outbreak of staphylococcal food poisoning involving chocolate milk. Int. J. Food Microbiol. 7, 311–316. doi: 10.1016/0168-1605(88)90057-8
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Felipe, V., Morgante, C. A., Somale, P. S., Varroni, F., Zingaretti, M. L., Bachetti, R. A., et al. (2017). Evaluation of the biofilm forming ability and its associated genes in Staphylococcus species isolates from bovine mastitis in Argentinean dairy farms. Microb. Pathog. 104, 278–286. doi: 10.1016/j.micpath.2017.01.047
Fetsch, A., Contzen, M., Hartelt, K., Kleiser, A., Maassen, S., Rau, J., et al. (2014). Staphylococcus aureus food-poisoning outbreak associated with the consumption of ice-cream. Int. J. Food Microbiol. 187:1. doi: 10.1016/j.ijfoodmicro.2014.06.017
Gould, I. M., David, M. Z., Esposito, S., Garau, J., Lina, G., Mazzei, T., et al. (2012). New insights into meticillin-resistant Staphylococcus aureus (MRSA) pathogenesis, treatment and resistance. Int. J. Antimicrob. Ageing 39:96. doi: 10.1016/j.ijantimicag.2011.09.028
Gündoğan, N., Citak, S., and Turan, E. (2006). Slime production, DNase activity and antibiotic resistance of Staphylococcus aureus isolated from raw milk, pasteurised milk and ice cream samples. Food Control 17, 389–392. doi: 10.1016/j.foodcont.2005.01.006
Hallin, M., Friedrich, A. W., and Struelens, M. J. (2009). spa typing for epidemiological surveillance of Staphylococcus aureus. Methods Mol. Biol. 551:189. doi: 10.1007/978-1-60327-999-4_15
Helwigh, B., and Korsgaard, H. (2007). The Community Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents, Antimicrobial Resistance and Foodborne Outbreaks in the European Union in 2006. Parma: European Food Safety Authority.
Howden, B. P., Davies, J. K., Johnson, P. D. R., Stinear, T. P., and Grayson, M. L. (2010). Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23, 99–139. doi: 10.1128/CMR.00042-09
Jamali, H., Paydar, M., Radmehr, B., and Ismail, S. (2015). Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control 54, 383–388. doi: 10.1016/j.foodcont.2015.02.013
Jarraud, S., Mougel, C., Thioulouse, J., Lina, G., Meugnier, H., Forey, F., et al. (2002). Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (Alleles), and human disease. Infect. Immun. 70:631. doi: 10.1128/IAI.70.2.631-641.2002
Jarraud, S., Peyrat, M. A., Lim, A., Tristan, A., Bes, M., Mougel, C., et al. (2001). egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immun. 166:669.
Jones, T. F., Kellum, M. E., Porter, S. S., Michael, B., and William, S. (2002). An outbreak of community-acquired foodborne illness caused by methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 8, 82–84. doi: 10.3201/eid0801.010174
Kadariya, J., Smith, T. C., and Thapaliya, D. (2014). Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. BioMed. Res. Int. 2014:9. doi: 10.1155/2014/827965
Kanika, B., Wang, X., Susan, D., Marcus, Z., Da, R. L., and Zhang, Y. (2011). Methicillin-resistant Staphylococcus aureus in retail meat, Detroit, Michigan, USA. Emerg. Infect. Dis. 17:1135. doi: 10.3201/eid1706.101905
Krouanton, A., Hennekinne, J. A., Letertre, C., Petit, L., Chesneau, O., Brisabois, A., et al. (2007). Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int. J. Food Microbiol. 115, 369–375. doi: 10.1016/j.ijfoodmicro.2006.10.050
Khoramrooz, S. S., Mansouri, F., Marashifard, M., Malek Hosseini, S. A., Akbarian, C.-O. F., Ganavehei, B., et al. (2016). Detection of biofilm related genes, classical enterotoxin genes and agr typing among Staphylococcus aureus isolated from bovine with subclinical mastitis in southwest of Iran. Microb. Pathog. 97, 45–51. doi: 10.1016/j.micpath.2016.05.022
Krziwanek, K., Metzgercek, S., and Mittermayer, H. (2009). Methicillin-resistant Staphylococcus aureus ST398 from human patients, Upper Austria. Emerg. Infect. Dis. 15, 766–769. doi: 10.3201/eid1505.080326
Larkin, E. A., Carman, R. J., Krakauer, T., and Stiles, B. G. (2009). Staphylococcus aureus: the toxic presence of a pathogen extraordinaire. Curr. Med. Chem. 16, 4003–4019. doi: 10.2174/092986709789352321
Lee, Y. T., Lin, D. B., Wang, W. Y., Tsao, S. M., Yu, S. F., Wei, M. J., et al. (2011). First identification of methicillin-resistant Staphylococcus aureus MLST types ST5 and ST45 and SCCmec types IV and Vt by multiplex PCR during an outbreak in a respiratory care ward in central Taiwan. Diagn. Microb. Infect. Dis. 70, 175–182. doi: 10.1016/j.diagmicrobio.2010.12.021
Lindqvist, R., Sylvén, S., and Vågsholm, I. (2002). Quantitative microbial risk assessment exemplified by Staphylococcus aureus in unripened cheese made from raw milk. Int. J. Food Microbiol. 78, 155–170. doi: 10.1016/S0168-1605(02)00237-4
Liu, R., and Wang, L. (2013). Study on China’s dairy consumption and influencing factors. Agric. Outlook 3, 71–75.
Loir, Y. L., Baron, F., and Gautier, M. I. (2003). Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2, 63–76.
Melchior, M. B., Fink-Gremmels, J., and Gaastra, W. (2007). Extended antimicrobial susceptibility assay for Staphylococcus aureus isolates from bovine mastitis growing in biofilms. Vet. Microbiol. 125, 141–149. doi: 10.1016/j.vetmic.2007.05.019
Miwa, N., Kawamura, A., Masuda, T., and Akiyama, M. (2001). An outbreak of food poisoning due to egg yolk reaction-negative Staphylococcus aureus. Int. J. Food Microbiol. 64, 361–366. doi: 10.1016/S0168-1605(00)00446-3
Neela, V., Zafrul, A. M., Mariana, N. S., Belkum, A. V., Yun, K. L., and Rad, E. G. (2009). Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J. Clin. Microbiol. 47:4138. doi: 10.1128/JCM.01363-09
Pereyra, E. A. L., Picech, F., Renna, M. S., Baravalle, C., Andreotti, C. S., Russi, R., et al. (2016). Detection of Staphylococcus aureus adhesion and biofilm-producing genes and their expression during internalization in bovine mammary epithelial cells. Vet. Microbiol. 183, 69–77. doi: 10.1016/j.vetmic.2015.12.002
Perez-Roth, E., Claverie-Martın, F., Villar, J., and Mendez-Alvarez, S. (2001). Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J. Clin. Microbiol. 39,4037–4041. doi: 10.1128/JCM.39.11.4037-4041.2001
Qian, G., Guo, X., Guo, J., and Wu, J. (2011). China’s dairy crisis: impacts, causes and policy implications for a sustainable dairy industry. Int. J. Sust. Dev. World 18, 434–441. doi: 10.1080/13504509.2011.581710
Rall, V. L., Vieira, F. P., Rall, R., Vieitis, R. L., Fernandes, A. Jr., Candeias, J. M., et al. (2008). PCR detection of staphylococcal enterotoxin genes in Staphylococcus aureus strains isolated from raw and pasteurized milk. Vet. Microbiol. 132, 408–413. doi: 10.1016/j.vetmic.2008.05.011
Rasigade, J. P., Dumitrescu, O., and Lina, G. (2014). New epidemiology of Staphylococcus aureus infections. Clin. Microbiol. Infet. 20, 587–588. doi: 10.1111/1469-0691.12718
Riva, A., Borghi, E., Cirasola, D., Colmegna, S., Borgo, F., Amato, E., et al. (2015). Methicillin-resistant Staphylococcus aureus in raw milk: prevalence, SCCmec typing, enterotoxin characterization, and antimicrobial resistance patterns. J. Food Protect. 78, 1142. doi: 10.4315/0362-028X.JFP-14-531
Rosengren, A., Fabricius, A., Guss, B., Sylvén, S., and Lindqvist, R. (2010). Occurrence of foodborne pathogens and characterization of Staphylococcus aureus in cheese produced on farm-dairies. Int. J. Food Microbiol. 144, 263–269. doi: 10.1016/j.ijfoodmicro.2010.10.004
Ryan, C. A., Nickels, M. K., Hargrettbean, N. T., Potter, M. E., Endo, T., Mayer, L., et al. (1987). Massive outbreak of antimicrobial-resistant salmonellosis traced to pasteurized milk. JAMA 258:3269. doi: 10.1001/jama.1987.03400220069039
Salaberry, S. R. S., Saidenberg, A. B. S., Zuniga, E., Melville, P. A., Santos, F. G. B., Guimarães, E. C., et al. (2015). Virulence factors genes of Staphylococcus spp. isolated from caprine subclinical mastitis. Microb. Pathog. 85, 35–39. doi: 10.1016/j.micpath.2015.05.007
Saunders, N. A., and Holmes, A. (2007). Multilocus sequence typing (MLST) of Staphylococcus aureus. Methods Mol. Biol. 391:71. doi: 10.1007/978-1-59745-468-1_6
Scallan, E., Hoekstra, R. M., Angulo, F. J., Robert, V. T., Marc-Alain, W., Sharon, L. R., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Schaumburg, F., Pauly, M., Anoh, E., Mossoun, A., Wiersma, L., Schubert, G., et al. (2015). Staphylococcus aureus complex from animals and humans in three remote African regions. Clin. Microbiol. Infect. 21, 345.e1–345.e8. doi: 10.1016/j.cmi.2014.12.001
Scherrer, D., Corti, S., Muehlherr, J. E., Zweifel, C., and Stephan, R. (2004). Phenotypic and genotypic characteristics of Staphylococcus aureus isolates from raw bulk-tank milk samples of goats and sheep. Vet. Microbiol. 101, 101–107. doi: 10.1016/j.vetmic.2004.03.016
Shallcross, L. J., Ellen, F., Johnson, A. M., and Hayward, A. C. (2013). The role of the Panton-valentine leucocidin toxin in staphylococcal disease: a systematic review and meta-analysis. Lancet Infect. Dis. 13, 43–54. doi: 10.1016/S1473-3099(12)70238-4
Shopsin, B., Gomez, M., Montgomery, S. O., Smith, D. H., Waddington, M., Dodge, D. E., et al. (1999). Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556.
Skov, R. L. (2009). Community-associated meticillin-resistant Staphylococcus aureus as a cause of hospital-acquired infections. J. Hosp. Infect. 73, 364–370. doi: 10.1016/j.jhin.2009.07.004
Spanu, V., Spanu, C., Virdis, S., Cossu, F., Scarano, C., and De Santis, E. P. (2012). Virulence factors and genetic variability of Staphylococcus aureus strains isolated from raw sheep’s milk cheese. Int. J. Food Microbiol. 153, 53–57. doi: 10.1016/j.ijfoodmicro.2011.10.015
Stegger, M., Andersen, P. S., Kearns, A., Pichon, B., Holmes, M. A., Edwards, G., et al. (2012). Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA LGA251. Clin. Microbiol. Infect. Dis. 18, 395–400. doi: 10.1111/j.1469-0691.2011.03715.x
Szweda, P., Schielmann, M., Milewski, S., Frankowska, A., and Jakubczak, A. (2012). Biofilm production and presence of ica and bap genes in Staphylococcus aureus strains isolated from cows with mastitis in the eastern Poland. Polish J. Microbiol. 61, 65–69.
Tham, W. A., Hajdu, L. J., and Danielssontham, M. L. (1990). Bacteriological quality of on-farm manufactured goat cheese. Epidemiol. Infect. 104, 87–100. doi: 10.1017/S095026880005456X
Varshney, A. K., Mediavilla, J. R., Robiou, N., Guh, A., Wang, X., Gialanella, P., et al. (2009). Diverse enterotoxin gene profiles among clonal complexes of Staphylococcus aureus isolates from the Bronx, New York. Appl. Environ. Microbiol. 75:6839. doi: 10.1128/AEM.00272-09
Vasudevan, P., Nair, M. K., Annamalai, T., and Venkitanarayanan, K. S. (2003). Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet. Microbiol. 92:179. doi: 10.1016/S0378-1135(02)00360-7
Wang, X., Meng, J., Zhang, J., Zhou, T., Zhang, Y., Yang, B., et al. (2012). Characterization of Staphylococcus aureus isolated from powdered infant formula milk and infant rice cereal in China. Int. J. Food Microbiol. 153, 142–147. doi: 10.1016/j.ijfoodmicro.2011.10.030
Wu, S., Huang, J., Wu, Q., Zhang, F., Zhang, J., Lei, T., et al. (2018a). Prevalence and characterization of Staphylococcus aureus isolated from retail vegetables in China. Front. Microbiol. 9:1263. doi: 10.3389/fmicb.2018.01263
Wu, S., Huang, J., Wu, Q., Zhang, J., Zhang, F., Yang, X., et al. (2018b). Staphylococcus aureus isolated from retail meat and meat products in China: incidence, antibiotic resistance and genetic diversity. Front. Microbiol. 9:2767. doi: 10.3389/fmicb.2018.02767
Wu, S., Wu, Q., Zhang, J., Chen, M., and Yan, Z. (2015). Prevalence, antibiotic resistance and genetic diversity of Listeria monocytogenes isolated from retail ready-to-eat foods in China. Food Control 47, 340–347. doi: 10.1016/j.foodcont.2014.07.028
Yu, F., Li, T., Huang, X., Xie, J., Xu, Y., Tu, J., et al. (2012). Virulence gene profiling and molecular characterization of hospital-acquired Staphylococcus aureus isolates associated with bloodstream infection. Diagn. Microbiol. Infect. Dis. 74, 363–368. doi: 10.1016/j.diagmicrobio.2012.08.015
Yu, X. J., Yan, J., Zhang, J. F., Xue, C. G., Dong, R., Xie, P. H., et al. (2010). Risk assessment of Staphylococcus aureus in raw milk and establishment of prevention and control measures. China Dairy Industry 38, 53–58.
Keywords: Staphylococcus aureus, pasteurized milk, risk assessment, virulence genes, antibiotic resistance, biofilm formation, MLST, spa typing
Citation: Dai J, Wu S, Huang J, Wu Q, Zhang F, Zhang J, Wang J, Ding Y, Zhang S, Yang X, Lei T, Xue L and Wu H (2019) Prevalence and Characterization of Staphylococcus aureus Isolated From Pasteurized Milk in China. Front. Microbiol. 10:641. doi: 10.3389/fmicb.2019.00641
Received: 27 November 2018; Accepted: 13 March 2019;
Published: 02 April 2019.
Edited by:
Julio Parra-Flores, University of the Bío Bío, ChileReviewed by:
Yajaira Esquivel Hernández, Universidad Autónoma de Querétaro, MexicoJianfeng Wang, Jilin University, China
Copyright © 2019 Dai, Wu, Huang, Wu, Zhang, Zhang, Wang, Ding, Zhang, Yang, Lei, Xue and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, wuqp203@163.com
†These authors have contributed equally to this work
 Jingsha Dai1†
Jingsha Dai1† Shi Wu
Shi Wu Qingping Wu
Qingping Wu Jumei Zhang
Jumei Zhang Juan Wang
Juan Wang Yu Ding
Yu Ding Liang Xue
Liang Xue Haoming Wu
Haoming Wu