Mitochondria and chloroplasts are two distinct organelles essential for plant viability. They evolved from prokaryotic endosymbionts and share a common ancestor with extant Gram-negative bacteria (Gray et al., 1999; Gould et al., 2008). Successful conversion of the free-living prokaryotes to the cytoplasmic organelles via endosymbiosis required conservation and adaptation of the outer membranes to the dramatic change of surroundings. In prokaryotes, the outer membrane serves as a physical barrier that protects cells from the extracellular environment and allows import of necessary nutrients, and also directly participates in interaction with other organisms (Nikaido, 2003). As part of the semi-autonomous organelles, by contrast, the outer membranes of mitochondria and chloroplasts have gained ability to participate in intracellular communication and organelle biogenesis, i.e., import and export of various ions and metabolites, import of nuclear-encoded proteins, various metabolic processes including the biosynthesis of membrane lipids, and division and movement of the organelles that require physical interaction with cytoplasmic components (Breuers et al., 2011; Inoue, 2011; Duncan et al., 2013). Our understanding of the organelle outer membranes have been advanced greatly in the last decade or so, and the last eight years have seen about a three-fold increase in the number of proteins identified or predicted to be in the chloroplast outer envelope of Arabidopsis thaliana (Arabidopsis) [total 117 proteins listed in Table 1; compare 34 proteins in Inoue (2007)]. This Research Topic is intended to provide snapshots of recent research on the organelle outer membranes. It collects seven original research, three review and two method articles, which can be divided into four groups according to the subjects – (1) outer membrane protein targeting, (2) functions, targeting and evolution of protein import components, (3) lipid metabolism, and (4) method development.
1. Protein Targeting to the Organelle Outer Membranes
All proteins identified so far in the organelle outer membranes are encoded in the nucleus (e.g., Table 1), and most of them use internal signals for targeting. This is distinct from the case for most nuclear-encoded proteins found inside the organelles: they are synthesized with N-terminal extensions, which are necessary and sufficient for proper targeting via the general pathway and cleaved upon import in the matrix (mitochondria) or stroma (chloroplasts). Lee et al. (2014) review the current knowledge of pathways and signals needed for targeting of three types of outer membrane proteins – signal-anchored (SA), tail-anchored (TA), and β-barrel proteins. SA and TA proteins are anchored to the membrane via a single transmembrane (TM) α-helix with either Nintermembrane space-Ccytosol (for SA) or Ncytosol-Cintermembrane space (for TA) orientation. β-Barrel proteins are integrated into the membrane via multiple TM-β-strands, whose formation appears to require evolutionarily conserved machinery in the membrane. Marty et al. (2014) have used a transient expression system with Nicotiana tabacum Bright Yellow-2 suspension cells to identify two types of targeting signals for mitochondria TA proteins. They have then performed database search, increasing the number of mitochondria TA proteins from 20 to 54. Interestingly, 16 of the mitochondria outer membrane proteins identified by the previous work (Duncan et al., 2013) and Marty et al. (2014) are also found in the chloroplast outer envelope membrane (Table 1). This may suggest the presence of targeting mechanisms and functions shared between the outer membranes of the two organelles.
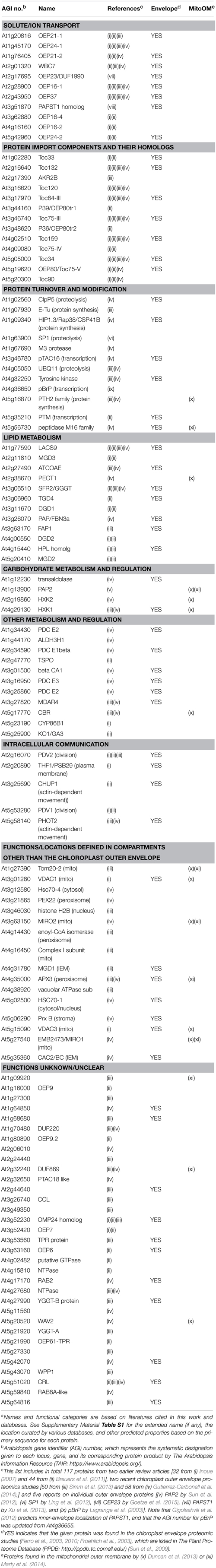
Table 1. One hundred and seventeen proteins identified or predicted to be in the outer membrane of the Arabidopsis chloroplast envelope.a
2. Functions, Targeting and Evolution of Protein Import Components
The most-studied chloroplast outer membrane proteins are subunits of the TOC (translocon at the outer-envelope-membrane of chloroplasts) machinery, which catalyzes the general pathway to import nuclear-encoded precursor proteins from the cytosol. Among the TOC components are homologous GTPases Toc159 and Toc34, which recognize the precursors and regulate their import, and Toc75, which forms a protein conducting channel. In Arabidopsis, there are four Toc159 isoforms which show substrate selectivity, two catalytically redundant Toc34 isoforms, and one functional Toc75 encoded on chromosome III (Table 1). Demarsy et al. (2014) review the current knowledge about how these subunits function and regulate protein import. Richardson et al. (2014) summarize available results and discuss functions, targeting and assembly of TOC subunits. Importantly, both review articles recognize outstanding questions about the TOC components, including the mechanisms of precursor recognition and their insertion into the membrane. By biochemical assays using chloroplasts isolated from pea seedlings, radiolabeled precursor proteins and recombinant proteins, Chang et al. (2014) demonstrate interaction of Toc159 isoforms called Toc132/Toc120 with a chloroplast superoxide dismutase (FSD1) that was predicted to comprise an exceptionally short import signal but has been shown otherwise, and also map the interaction domains beyond the N terminus. The interaction of FSD1 with Toc132, but not with Toc159, was also demonstrated by a split-ubiquitin yeast two-hybrid assay (Dutta et al., 2014). Grimmer et al. (2014) have used an in vivo approach, transiently producing GFP-tagged proteins in protoplasts of various Arabidopsis mutants and determining their N-terminal sequences by mass spectrometry analyses, and demonstrate that a plastid RNA binding protein is a substrate of Toc159. The Arabidopsis protoplast transient expression assay has also been used to define sequences required for targeting and membrane integration of a Toc159 ortholog (Lung et al., 2014). A previous genetic screening had demonstrated that Toc132 and Toc75 enhance root gravitropism signal transduction (Stanga et al., 2009). Strohm et al. (2014) now provide evidence supporting the involvement of plastids, instead of direct participation of TOC subunits, in the gravitropism signal transduction. Finally, Day et al. (2014) report phylogenetic relationships and in vitro targeting of the Toc75 homologs including the truncated forms of OEP80/Toc75-V, which are also known as P39 (Hsueh et al., 2014) and P36 (Nicolaisen et al., 2015) (Table 1).
3. Lipid Metabolism
Under phosphate starvation, phospholipids in the cell membranes, mainly those in extraplastidic compartments, are used as the source of free phosphates and substituted by galactolipids made in the chloroplast outer envelope. Murakawa et al. (2014) have used Arabidopsis mutants and feeding assays to show that the outer-envelope-dependent galactolipid synthesis is stimulated by sucrose supplementation and this stimulation in turn enhances utilization of the added sucrose for plant growth. This work nicely illustrates the physiological significance of the metabolic activity localized in the chloroplast outer envelope for plant growth and development.
4. Method Development
Hardre et al. (2014) report an attempt to apply biotin tagging and proteolysis to examine topology and membrane association of proteins in the spinach chloroplast. Although the work requires further refinement to achieve the desired specificity, the idea behind this approach is quite interesting. The toc159-null mutant is seedling-lethal thus has been examined as progenies of heterozygous parents. Tada et al. (2014) have established a method using Ziploc® container to grow the homozygous toc159 mutants on the sucrose-supplemented media to the point that viable seeds can be obtained. This cost-effective method should be useful to study not only the toc159-null plant but also other recessive lethal mutants of photosynthesis.
In summary, the collection highlights various questions about the organelle outer membranes and interdisciplinary approaches employed to address them. The future research should use these and other strategies to answer questions about the proteins of known functions, in particular those involved in protein homeostasis, as well as those of unknown functions (Table 1). The editor greatly acknowledges the excellent contributions of all the authors and constructive comments by expert reviewers to each of the articles.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Division of Molecular and Cellular Biosciences at the US National Science Foundation (Grant No. 1050602).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00278/full
Table S1. Extended names, curated locations and some other information of 117 proteins listed in Table 1.
References
Breuers, F. K., Brautigam, A., and Weber, A. P. (2011). The plastid outer envelope – a highly dynamic interface between plastid and cytoplasm. Front. Plant Sci. 2:97. doi: 10.3389/fpls.2011.00097
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chang, W., Soll, J., and Bolter, B. (2014). A new member of the psToc159 family contributes to distinct protein targeting pathways in pea chloroplasts. Front. Plant Sci. 5:239. doi: 10.3389/fpls.2014.00239
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Day, P. M., Potter, D., and Inoue, K. (2014). Evolution and targeting of Omp85 homologs in the chloroplast outer envelope membrane. Front. Plant Sci. 5:535. doi: 10.3389/fpls.2014.00535
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Demarsy, E., Lakshmanan, A. M., and Kessler, F. (2014). Border control: selectivity of chloroplast protein import and regulation at the TOC-complex. Front. Plant Sci. 5:483. doi: 10.3389/fpls.2014.00483
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Duncan, O., van der Merwe, M. J., Daley, D. O., and Whelan, J. (2013). The outer mitochondrial membrane in higher plants. Trends Plant Sci. 18, 207–217. doi: 10.1016/j.tplants.2012.12.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dutta, S., Teresinski, H. J., and Smith, M. D. (2014). A split-ubiquitin yeast two-hybrid screen to examine the substrate specificity of atToc159 and atToc132, two Arabidopsis chloroplast preprotein import receptors. PLoS ONE 9:e95026. doi: 10.1371/journal.pone.0095026
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ferro, M., Brugiere, S., Salvi, D., Seigneurin-Berny, D., Court, M., Moyet, L., et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9, 1063–1084. doi: 10.1074/mcp.M900325-MCP200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ferro, M., Salvi, D., Brugiere, S., Miras, S., Kowalski, S., Louwagie, M., et al. (2003). Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2, 325–345. doi: 10.1074/mcp.M300030-MCP200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Froehlich, J. E., Wilkerson, C. G., Ray, W. K., McAndrew, R. S., Osteryoung, K. W., Gage, D. A., et al. (2003). Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J. Proteome Res. 2, 413–425. doi: 10.1021/pr034025j
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gigolashvili, T., Geier, M., Ashykhmina, N., Frerigmann, H., Wulfert, S., Krueger, S., et al. (2012). The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5′-phosphosulfate to the cytosol. Plant Cell 24, 4187–4204. doi: 10.1105/tpc.112.101964
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Goetze, T. A., Patil, M., Jeshen, I., Bolter, B., Grahl, S., and Soll, J. (2015). Oep23 forms an ion channel in the chloroplast outer envelope. BMC Plant Biol. 15:47. doi: 10.1186/s12870-015-0445-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gould, S. B., Waller, R. F., and McFadden, G. I. (2008). Plastid evolution. Annu. Rev. Plant Biol. 59, 491–517. doi: 10.1146/annurev.arplant.59.032607.092915
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gray, M. W., Burger, G., and Lang, B. F. (1999). Mitochondrial evolution. Science 283, 1476–1481. doi: 10.1126/science.283.5407.1476
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Grimmer, J., Rodiger, A., Hoehenwarter, W., Helm, S., and Baginsky, S. (2014). The RNA-binding protein RNP29 is an unusual Toc159 transport substrate. Front. Plant Sci. 5:258. doi: 10.3389/fpls.2014.00258
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gutierrez-Carbonell, E., Takahashi, D., Lattanzio, G., Rodriguez-Celma, J., Kehr, J., Soll, J., et al. (2014). The distinct functional roles of the inner and outer chloroplast envelope of Pea (Pisum sativum) as revealed by proteomic approaches. J. Proteome Res. 13, 2941–2953. doi: 10.1021/pr500106s
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hardre, H., Kuhn, L., Albrieux, C., Jouhet, J., Michaud, M., Seigneurin-Berny, D., et al. (2014). The selective biotin tagging and thermolysin proteolysis of chloroplast outer envelope proteins reveals information on protein topology and association into complexes. Front. Plant Sci. 5:203. doi: 10.3389/fpls.2014.00203
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hsueh, Y. C., Flinner, N., Gross, L. E., Haarmann, R., Mirus, O., Sommer, M. S., et al. (2014). The chloroplast outer envelope protein P39 in Arabidopsis thaliana belongs to the Omp85 protein family. Proteins. doi: 10.1002/prot.24725. [Epub ahead of print].
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Inoue, K. (2007). The chloroplast outer envelope membrane: The edge of light and excitement. J. Integr. Plant Biol. 49, 1100–1111. doi: 10.1111/j.1672–9072.2007.00543.x
Inoue, K. (2011). Emerging roles of the chloroplast outer envelope membrane. Trends Plant Sci. 16, 550–557. doi: 10.1016/j.tplants.2011.06.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lagrange, T., Hakimi, M. A., Pontier, D., Courtois, F., Alcaraz, J. P., Grunwald, D., et al. (2003). Transcription factor IIB (TFIIB)-related protein (pBrp), a plant-specific member of the TFIIB-related protein family. Mol. Cell. Biol. 23, 3274–3286. doi: 10.1128/MCB.23.9.3274-3286.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lee, J., Kim, D. H., and Hwang, I. (2014). Specific targeting of proteins to outer envelope membranes of endosymbiotic organelles, chloroplasts, and mitochondria. Front. Plant Sci. 5:173. doi: 10.3389/fpls.2014.00173
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ling, Q., Huang, W., Baldwin, A., and Jarvis, P. (2012). Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338, 655–659. doi: 10.1126/science.1225053
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lung, S. C., Smith, M. D., Weston, J. K., Gwynne, W., Secord, N., and Chuong, S. D. (2014). The C-terminus of Bienertia sinuspersici Toc159 contains essential elements for its targeting and anchorage to the chloroplast outer membrane. Front. Plant Sci. 5:722. doi: 10.3389/fpls.2014.00722
Marty, N. J., Teresinski, H. J., Hwang, Y. T., Clendening, E. A., Gidda, S. K., Sliwinska, E., et al. (2014). New insights into the targeting of a subset of tail-anchored proteins to the outer mitochondrial membrane. Front. Plant Sci. 5, 426. doi: 10.3389/fpls.2014.00426
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Murakawa, M., Shimojima, M., Shimomura, Y., Kobayashi, K., Awai, K., and Ohta, H. (2014). Monogalactosyldiacylglycerol synthesis in the outer envelope membrane of chloroplasts is required for enhanced growth under sucrose supplementation. Front. Plant Sci. 5:280. doi: 10.3389/fpls.2014.00280
Nicolaisen, K., Missbach, S., Hsueh, Y. C., Ertel, F., Fulgosi, H., Sommer, M. S., et al. (2015). The Omp85-type outer membrane protein p36 of Arabidopsis thaliana evolved by recent gene duplication. J. Plant Res. 128, 317–325. doi: 10.1007/s10265-014-0693-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656. doi: 10.1128/MMBR.67.4.593-656.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Richardson, L. G., Paila, Y. D., Siman, S. R., Chen, Y., Smith, M. D., and Schnell, D. J. (2014). Targeting and assembly of components of the TOC protein import complex at the chloroplast outer envelope membrane. Front. Plant Sci. 5:269. doi: 10.3389/fpls.2014.00269
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Simm, S., Papasotiriou, D. G., Ibrahim, M., Leisegang, M. S., Muller, B., Schorge, T., et al. (2013). Defining the core proteome of the chloroplast envelope membranes. Front. Plant Sci. 4:11. doi: 10.3389/fpls.2013.00011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stanga, J. P., Boonsirichai, K., Sedbrook, J. C., Otegui, M. S., and Masson, P. H. (2009). A role for the TOC complex in Arabidopsis root gravitropism. Plant Physiol. 149, 1896–1905. doi: 10.1104/pp.109.135301
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Strohm, A. K., Barrett-Wilt, G. A., and Masson, P. H. (2014). A functional TOC complex contributes to gravity signal transduction in Arabidopsis. Front. Plant Sci. 5:148. doi: 10.3389/fpls.2014.00148
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sun, F., Carrie, C., Law, S., Murcha, M. W., Zhang, R., Law, Y. S., et al. (2012). AtPAP2 is a tail-anchored protein in the outer membrane of chloroplasts and mitochondria. Plant Signal. Behav. 7, 927–932. doi: 10.4161/psb.20769
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sun, Q., Zybailov, B., Majeran, W., Friso, G., Olinares, P. D., and van Wijk, K. J. (2009). PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37, D969–D974. doi: 10.1093/nar/gkn654
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tada, A., Adachi, F., Kakizaki, T., and Inaba, T. (2014). Production of viable seeds from the seedling lethal mutant ppi2-2 lacking the atToc159 chloroplast protein import receptor using plastic containers, and characterization of the homozygous mutant progeny. Front. Plant Sci. 5:243. doi: 10.3389/fpls.2014.00243
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xu, J., Yang, J., Wu, Z., Liu, H., Huang, F., Wu, Y., et al. (2013). Identification of a dual-targeted protein belonging to the mitochondrial carrier family that is required for early leaf development in rice. Plant Physiol. 161, 2036–2048. doi: 10.1104/pp.112.210831
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: Arabidopsis, chloroplast, membrane proteins, mitochondria, outer membrane
Citation: Inoue K (2015) Emerging knowledge of the organelle outer membranes – research snapshots and an updated list of the chloroplast outer envelope proteins. Front. Plant Sci. 6:278. doi: 10.3389/fpls.2015.00278
Received: 27 March 2015; Accepted: 07 April 2015;
Published: 30 April 2015.
Edited and reviewed by: Simon Gilroy, University of Wisconsin – Madison, USA
Copyright © 2015 Inoue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kentaro Inoue, kinoue@ucdavis.edu
 Kentaro Inoue
Kentaro Inoue