- 1State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin, China
- 2Department of Plant Nutrition, College of Resources and Environmental Sciences, China Agricultural University, Beijing, China
- 3School of Life Science, Northeast Forestry University, Harbin, China
Ammonium transporters (AMTs) are plasma membrane proteins that exclusively transport ammonium/ammonia. These proteins are encoded by an ancient gene family with many members. The molecular characteristics and evolutionary history of AMTs in woody plants are still poorly understood. We comprehensively evaluated the AMT gene family in the latest release of the Populus trichocarpa genome (version 3.0; Phytozome 9.0), and identified 16 AMT genes. These genes formed four clusters; AMT1 (7 genes), AMT2 (2 genes), AMT3 (2 genes), and AMT4 (5 genes). Evolutionary analyses suggested that the Populus AMT gene family has expanded via whole-genome duplication events. Among the 16 AMT genes, 15 genes are located on 11 chromosomes of Populus. Expression analyses showed that 14 AMT genes were vegetative organs expressed; AMT1;1/1;3/1;6/3;2 and AMT1;1/1;2/2;2/3;1 had high transcript accumulation level in the leaves and roots, respectively and strongly changes under the nitrogen-dependent experiments. The results imply the functional roles of AMT genes in ammonium absorption in poplar.
Introduction
For most higher plant species, the main sources of nitrogen are ammonium (NH+4), nitrate (NO−3), and amino acids, which are present in the soil as organic and inorganic complexes and compounds (Williams and Miller, 2001). The ammonium transporter (AMT) is responsible for transporting ammonium/ammonia from extracellular into intracellular locations. In plant, once ammonium is uptaked into root cells by AMTs, it is ultimately directed into glutamine via glutamine synthase (GS). Less energy is required for uptake and assimilation of NH+4 than that of NO−3 (Bloom et al., 1992). However, a high concentration of NH+4 can be toxic to plants as indexed by an inhibitory growth (Britto and Kronzucker, 2002).
Recently, some AMT genes have been identified and cloned from diverse plant species. Previous studies on phylogenetic analyses of the AMT gene family revealed two distinct subfamilies: the AMT1 subfamily (AMT1 cluster) and the AMT2 subfamily (AMT2/3/4 cluster) (Loqué and von Wirén, 2004; Koegel et al., 2013). The biochemical properties of proteins encoded by AMT1 cluster genes, and the related regulation mechanisms were reported in the model plant Arabidopsis thaliana (Loqué et al., 2007; Yuan et al., 2007, 2009, 2013; Lanquar et al., 2009). The proteins encoded by AMT1 cluster genes have a high-affinity NH+4-transport function. For example, both AtAMT1;1 and AtAMT1;3 account for 30–35% of the capacity for NH+4 uptake in nitrogen-deficient roots, and AtAMT1;2 for 18–26% (Loqué et al., 2006; Yuan et al., 2007). AtAMT1;4, which is pollen-specific expressed, contributes to nitrogen nutrition of the pollen via NH+4 uptake or retrieval (Yuan et al., 2009).
Populus, a model system for trees and woody perennial plants, is widely distributed throughout the northern hemisphere. Members of the Populus genus are fast-growing trees that are capable of growing under low- or high-NH+4 and NO−3 conditions (Min et al., 1999). It is necessary to a better understanding on how the uptake and transport of NH+4 and NO−3 are regulated in this genus. In a previous study, 14 AMT genes were identified in the Populus trichocarpa genome version 1v1. PtaAMT1;2/1;5/1;6/2;1/2;2 were confirmed to have NH+4-transporter functions in yeast (Couturier et al., 2007). The expression of PtaAMT1;2 and PtaAMT3;1 were induced by ectomycorrhiza (Selle et al., 2005; Luo et al., 2009).
In this study, we investigated the evolution and transcription profiles of Populus AMT genes by describing the expanded AMT gene family consisting of 16 genes, which were dentified in the latest release of the P. trichocarpa genome (version 3.0; Phytozome 9.0), analyzing the phylogeny, gene structure, conserved domain, and genome location. Moreover, we comprehensively analyzed the tissue and nitrogen-dependent transcription profiles of AMT genes in Populus.
Materials and Methods
Plant Seedlings and Growth Conditions
Cuttings of P. simonii × P. nigra were pots-cultivated (organic substrate and vermiculite, 1:1 vol/vol) at Northeast Forestry University Forest Farm, Harbin, China for 3 months under the following conditions; photosynthetic photon flux density (PPFD) of 100 μmol·m-2·s-1, 16-h-light/8-h-dark photoperiod, and 22°C. The plantlets were harvested, and several whole plantlets were frozen in liquid nitrogen and stored at −80°C. New branches were cut into segments of equal length before transferring into modified Long-Ashton medium, pH 5.5 (Dluzniewska et al., 2007). The medium was replaced every 2 days. After 3 weeks, the plantlets were treated with nitrogen at various concentrations. For the nitrogen-free medium, 0.5 mM KNO3 and 0.5 mM NH4Cl were replaced with 0.5 mM KCl. To supply NH+4 or NO−3, the medium contained 2 mM (NH4)2SO4 and 0.5 mM KCl or 4 mM KNO3 and 2 mM MgSO4, respectively. After the treatments, whole plantlets were harvested, frozen in liquid nitrogen, and stored at −80°C until analysis.
Identification of AMT Gene Family Members in Populus
We downloaded the Hidden Markov Model (HMM) profile file (Ammonium_transp.hmm) of the Pfam AMT domain (PF00909) from the Pfam database (Finn et al., 2010). The protein sequences of P. trichocarpa were downloaded from Phytozome 9.0 (http://phytozome.jgi.doe.gov/pz/portal.html). We used the HMM modules of PF00909 with HMMER (v 3.0) software to search the proteome of P. trichocarpa (Eddy, 2009). Proteins with e-values of less than 5E-40 were included in further analyses. Various splicing variants of one gene or incomplete genes were discarded. We searched for the ammonium-domain in all of the collected proteins using Interproscan (http://www.ebi.ac.uk/Tools/pfa/iprscan/) and SMART software (Letunic et al., 2012).
For each putative protein, the grand average of hydropathicity (GRAVY) was calculated using ProtParam (http://web.expasy.org/protparam/). We used TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) to predict the transmembrane domains in each AMT protein.
Phylogenetic Analysis and Chromosomal Location
According to the method of Koegel et al. (2013), we aligned full-length amino acid sequences of AMTs with ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method and Poisson correction model with MEGA5 software (Tamura et al., 2011). To confirm the reliability of the phylogenetic tree, bootstrap resampling tests were carried out 1000 times.
Information on the chromosomal location of all of the AMT genes was downloaded from Phytozome 9.0, and duplicated regions among chromosomes were identified as described by Tuskan et al. (2006). The criterion for tandemly duplicated genes in Populus was the occurrence of five or fewer gene loci within a 100-kb region.
Gene Structure and Conserved Motifs
We used the Gene Structure Display Server (GSDS) program to illustrate the exon/intron organization of individual AMT genes (Guo et al., 2007). The Ka/Ks ratio was computed using KaKs_Calculator 2.0 (Wang et al., 2010).
RNA Isolation and Quantitative RT–PCR Analysis
Total RNA was extracted from leaf, stem, and root tissues using the CTAB method (Chang et al., 1993). The integrity of the extracted RNA was verified by 1.5% agar gel electrophoresis. Approximately 2 μg RNA was used to synthesize first-strand cDNA using the PrimerScript RT Reagent Kit, after removing genomic DNA with gDNAEraser (Takara Biotechnology, Dalian, China). Primer Premier 5.0 (Premier Biosoft, Palo Alto, CA, USA) software was used to design specific primers for semi-quantitative PCR analysis. The primer sequences are listed in Supplementary Table 1. A 7500 Real-Time PCR System (Applied Biosystems) was used to conduct a three-step PCR procedure. In the organ-dependent and nitrogen-dependent expression analyses, transcript levels were normalized to that of the PtrActin2 gene.
Results
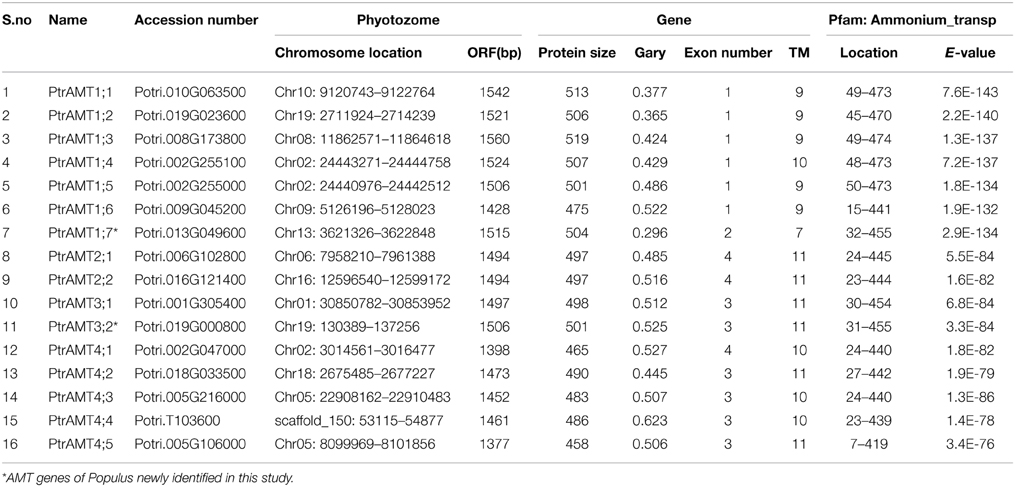
Identification of AMT Genes in Populus
By referring to the method of Wang et al. (2004) and Chai et al. (2012), the HMM profile “PF00909” was performed against the P. trichocarpa genome to identify AMT genes. We ultimately identified 16 putative AMT proteins and the related encoding genes from the P. trichocarpa genome. We assigned the names to the 2 AMT genes that are not described previously (Table 1). The length of encoded proteins ranged from 458 amino acids (a.a.) to 519 a.a., and their sequences had 7 to 11 trans-membrane domains (TMDs). All of the putative proteins had low GRAVY values (range: 0.369–0.623).
Phylogenetic and Gene Structural Analyses of AMT Genes
To evaluate the evolutionary relationships among orthologous AMT genes, we constructed a phylogenetic tree with the Neighbor-Joining (N-J) method using MEGA5 software with 8 different plant species (Figure 1). The results revealed two major clades and four clusters. Among the 16 AMT genes in Populus, 7 genes were in the AMT1 cluster, and the remaining AMT genes were in three other separate clusters (AMT2, AMT3, and AMT4).
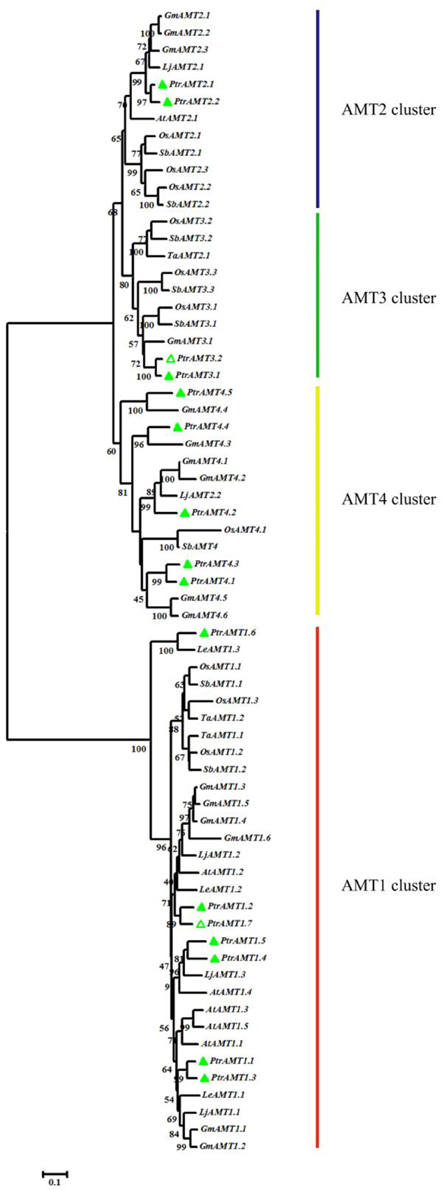
Figure 1. Phylogenetic tree of proteins encoded by AMT genes from Arabidopsis thaliana, Lycopersicon esculentum, Giycine max, Lotus japonicas, Sorghum bicolor, Oryza sativa, Triticum aestivum, and Populus trichocarpa. Protein sequences were aligned by Clustalw and tree was constructed by MEGA5 using N-J method, with 1000 bootstrap replicates. Green hollow triangle are the new AMTs in this study.
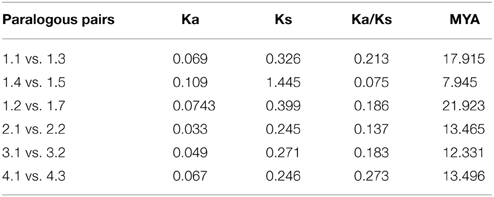
To investigate the divergence of paralogs and the evolutionary relationships among Populus AMT proteins, we aligned full-length sequences of the 16 proteins using ClustalW, and constructed a phylogenetic tree with the Neighbor-Joining method using MEGA5 software (Figure 2A). We identified 6 paralogous pairs, and then determined their substitution rate ratios (non-synonymous vs. synonymous mutations; Ka/Ks). All of 6 paralogous pairs had Ka/Ks ratios of less than 0.5. We deduced that the divergence time of these paralogous pairs ranged from 1.07 to 21.92 million years ago (Table 2). These results indicated that all of the 6 Populus AMT gene pairs evolved under the influence of purifying selection.
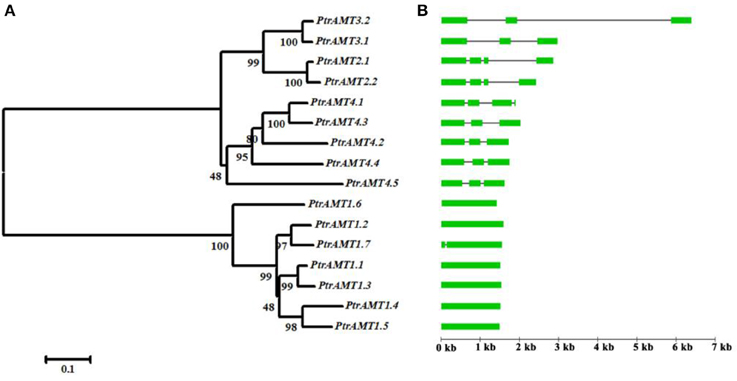
Figure 2. Paralog phylogenetic tree, gene structures, and motif distribution of Populus AMT gene family. (A) Multiple alignment of full-length AMT protein sequences was conducted using Clustalw. Phylogenetic tree was constructed using MEGA5 by the N-J method, with 1000 bootstrap replicates. (B) Gene structures of AMT genes. Green boxes show coding exons, black lines show introns.
The AMT genes in the same cluster had similar exon/intron structures and similar numbers of exons and introns (Figure 2B). Genes in the AMT1 cluster had 1 exon, except for PtrAMT1;7 who had 2 exons. And those in the AMT2 cluster had 4 exons. Genes in the AMT3 cluster had 3 exons, and those in the AMT4 cluster had 3 exons, except for PtrAMT4;1, which had 4 exons.
We further analyzed the exon/intron structure of the 6 paralogous pairs of Populus AMT genes. 4 of the 6 paralogous pairs were well conserved in terms of exon/intron structure, with similar numbers of introns and similar gene lengths. There were greater variations in gene structure among the other 2 paralogous pairs (PtrAMT4;1/4;3, and PtrAMT1;2/1;7). These differences were rooted in single- and double-intron loss or gain events during the structural evolution of AMT paralogs (Figure 3). As shown in Figure 2B, the size of exons was generally well conserved among most members of the 4 AMT clusters. Interestingly, Comparing with other members in AMT2 subfamily, PtrAMT3;2 had 2 long introns, but CDS sequence was similar to PtrAMT3;1. Therefore, the substantial differences in gene structure resulted from differences in the size of exons and introns among the various genes.
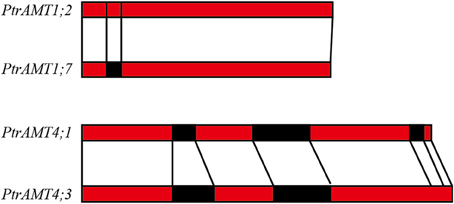
Figure 3. Duplicated genes in AMT gene family in Populus. Schematic diagram of gene structure is based on three duplicated paralogous pairs. Exons (red boxes) and introns (black boxes) are shown. Vertical lines show corresponding regions. Numbers of nucleotides are shown beside exons.
For finding distinctively domain of poplar AMTs, we aligning all the poplar AMT protein sequences with AtAMT1;1, AtAMT2;2 and EcAmtB which crystal structures was well characterized (Khademi et al., 2004; Pantoja, 2012). Comparing with AtAMT1;1, poplar AMT1 subfamily members also have conserved C-terminal domain and N-terminal domain (Supplementary Figure 1). While PtrAMT1;6 was similar to LeAMT1;3 who has a short N-terminal domain. In contrast to all of the TMDs present in EcAmtB, all of the poplar AMT gene family members have accordingly conserved TMDs. These results suggested that the AMT gene family members are well conserved both in terms of gene structure and specific domain of AMT proteins.
Chromosomal Location and Gene Duplication of the Populus AMT Gene Family
To explore the relationship between AMT genes and segmental duplications in the Populus genome, we analyzed the segmental and tandem duplication events in the AMT gene family in Populus. Based on the location information for AMTs in Phytozome 9.0, the genes were marked on the physical map of the Populus linkage groups (LG). The Populus AMT genes showed a heterogeneous distribution pattern among the chromosomes (Figure 4). We localized 15 of the 16 AMT genes on 11 of 19 LG of Populus. Only PtrAMT4;4 was located on unattributed scaffold fragments.
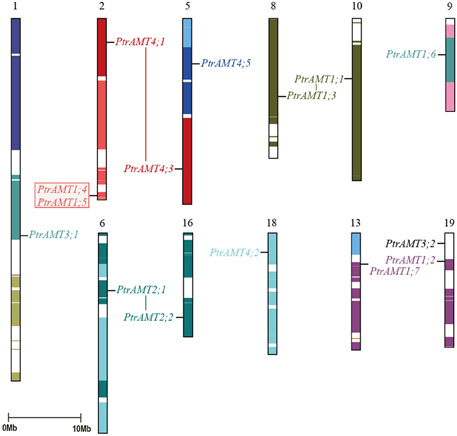
Figure 4. Chromosomal location and gene duplication of Populus AMT gene family. Same-colored boxes show segmental duplicated homologous regions. These regions were identified based on duplication coordinates from the Populus genome assembly 3.0. Duplicated paralogous pairs of AMT genes are connected by colored lines. Red box shows two tandemly duplicated gene pairs.
A previous study showed that paralogous segments of the Populus genome arose from whole-genome duplication during the salicoid duplication event (Tuskan et al., 2006). In the AMT gene family, 14 of the 15 mapped genes were located in duplicated blocks. 4 block pairs harbored 4 paralogous pairs of AMT genes (PtrAMT1;1/1;3, PtrAMT2;1/2;2, PtrAMT1.2/1.7 and PtrAMT4;1/4;3), which arose via a whole-genome duplication event. Paralogous pair PtrAMT1;4/1;5 were arranged in tandem repeats on LG 2 and LG 13, but both lacked corresponding duplicates. Out of 12 AMT genes, 2 genes (PtrAMT3;1, and PtrAMT4;5) also lacked corresponding duplicates. Only PtrAMT3;2 was not located in duplicated blocks. The corresponding homologs of these genes may have been lost after the duplication event, or genes may have arisen after the salicoid duplication event. In conclusion, duplication events and tandem repeats are expected to contribute to the expansion of the AMT gene family in the Populus genome.
Transcription Patterns of Populus AMT Genes in Various Tissues
To investigate the transcription patterns of Populus AMT genes during development, we used real-time quantitative RT-PCR to analyze AMT gene transcript levels in young leaves, mature leaves, old leaves, stems, and roots of P. simonii × P. nigra (Figure 5). Because of significantly difference of transcript accumulation of poplar AMT genes, we used square root value of relative transcript ratio of each gene for display express pattern, and raw date was show in Supplementary Table 4. Finally, we detected transcripts of 14 AMT genes: AMT1;1/1;2/1;3/1;4/1;5/1;6/2;1/2;2/3;1/3;2/4;1/4;3/4;4/4;5, but there were relatively low transcript levels of AMT1;4/1;5/3;1/4;1/4;3/4;4/4;5 in the 5 nutritive organs. We detected transcripts of AMT1;1/1;3/1;4/1;6/2;1/2;2/3;1/3;2/4;1 in all 5 tested tissues. AMT4;3 leaf-specific transcribed, and AMT4;4 stem-specific transcribed. There were high transcript levels of AMT1;3/1;6 in the leaves and AMT3;1 in the root. However, transcripts of AMT1;5/4;5 were not detected in the stem or root. A previous study showed that PtaAMT1;2 was specifically expressed in the root and PtaAMT1;6/2;1/3;1 in the shoot (Couturier et al., 2007). However under our experiment conditions, all the 4 genes mentioned above were detected in 5 tissues. Among them, AMT1;6 had high transcript accumulation in leaves, AMT1;2/3;1 had high transcript accumulation in roots, and AMT2;1 were expressed similarly in all the 5 tissues, except in young leaves. These differences in transcription patterns may be due to the highly heterozygous genetic background of P. simonii × P. nigra and/or differences in experimental conditions.
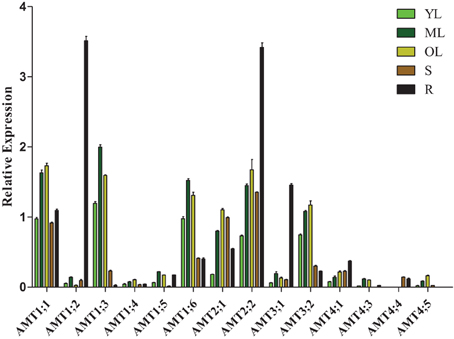
Figure 5. Relative transcript levels of AMT genes in different tissues of Populus. YL, young leaf; ML, mature leaf; OL, old leaf; R, root; S, stem. The average expression of each gene was calculated with square root of relative transcript ratio of each gene for display express pattern. Error bars indicate SE.
Populus AMT Transcription Patterns in Response to Different Nitrogen Concentrations
To better understand the function of AMT genes in Populus, we examined the transcription patterns of poplar AMT genes in P. simonii × P. nigra under nitrogen-dependent experiment. We selected 10 genes (AMT1;1/1;2/1;3/1;4/1;5/1;6/2;1/2;2/3;1/3;2) with high transcript accumulation in the leaf and root to evaluate transcription patterns.
In leaves of plantlets under nitrogen-starvation conditions, AMT1;1 was up-regulated, AMT1;3/3;2 were unchanged and down-regulated, respectively, at 4 h, and then up-regulated at 24 and 48 h. AMT1;4/1;6/2;1/3;1 were down-regulated, while AMT1;5 was down-regulated at 4 h, unchanged at 24 h, and further down-regulated at 48 h (Figure 6A, Supplementary Table 5A). In the roots of nitrogen-starved plantlets, AMT1;1/1;6/2;2/3;1/3;2 were up-regulated; AMT1;3 was down-regulated; AMT1;4 was unchanged at this condition. AMT1;2 was up-regulated at 4 and 48 h but down-regulated at 24 h. AMT1;2/1;5 was up-regulated at 4 and 48 h but down-regulated at 24 h. AMT2;1 was up-regulated at 4 h and but down-regulated at 24 and 48 h (Figure 6B, Supplementary Table 5B).
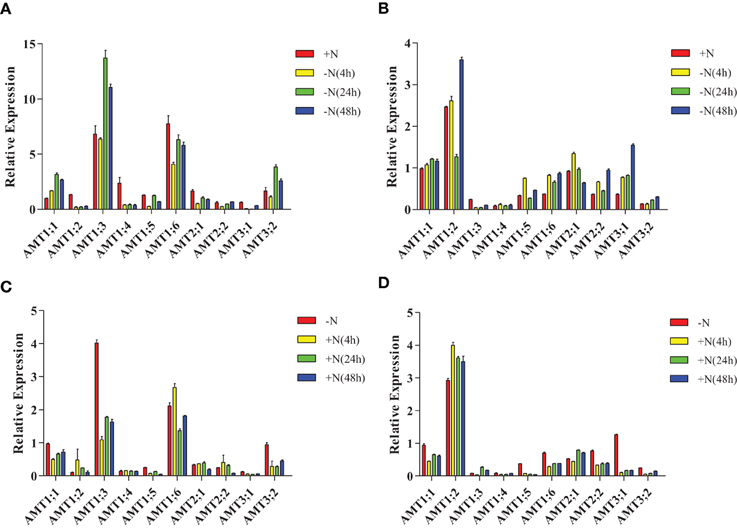
Figure 6. Expression patterns of Populus AMT genes under nitrogen-starvation and ammonium-resupply conditions. Plants were grown in modified Long–Ashton medium for 2 weeks, and then transferred to nitrogen-free medium (A,B) then, after 2 days, plantlets were transferred to medium with ammonium as the sole nitrogen source for 2 days (C,D). The average expression of each gene was calculated with square root of relative transcript ratio of each gene for display express pattern. Error bars indicate SE.
In the leaves of plantlets under NH+4-resupply conditions, AMT1;1/1;3/1;5/3;2 were down-regulated and AMT1;4 was unchanged. AMT1;6 was up-regulated at 4 h, but down-regulated at 24 h. AMT2;1/2;2 were up-regulated at 4 and 24 h, but down-regulated at 48 h (Figure 6C, Supplementary Table 5C). In the roots, AMT1;2 was up-regulated, AMT1;1/1;5/1;6/ 2;2/3;1/3;2 were down-regulated, and AMT1;4 was unchanged. AMT1;3/2;1 was down-regulated at 4 h but up-regulated at 24 h (Figure 6D, Supplementary Table 5D).
Interestingly, after plants were resupplied with different concentrations of NH+4, only AMT2;1 transcripts had high accumulation when the concentration of NH+4 was increased in roots (Figure 7, Supplementary Table 6), while the transcripts accumulation of AMT1;3/1;5/1;6/2;2/3;1 were reduced. The transcription of AMT1;4/3;2 was up-regulation under 0.1 mM NH+4 condition, but down-regulated under 0.4, 1, and 4 mM NH+4, respectively. AMT1;1 transcripts were up-regulated under 0.1, 0.4, and 1 mM NH+4conditions, but was down-regulated under 4 mM NH+4 condition. The expression level of AMT1;2 was strongly decreased under resupplied 0.1 and 0.4 mM NH+4 condition, while resupplied 1 and 4 mM NH+4 led to the transcripts of AMT1;2 was significantly accumulated.
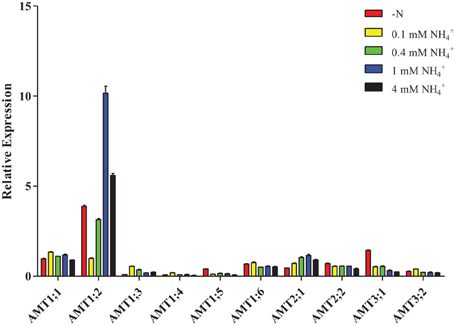
Figure 7. Relative transcript levels of AMT1;2 in roots of Populus in response to different concentrations of ammonium. Plantlets were grown in nitrogen-free medium for 2 days, and then transferred to medium containing indicated concentrations of ammonium for 1 day. The average expression of each gene was calculated with square root of relative transcript ratio of each gene for display express pattern. Error bars indicate SE.
Discussion
AMT Gene Family in Populus
We retrieved a total 16 AMT genes from the recently released Populus genome (Phytozome 9.0, Populus trichocarpa 3.0) with improved annotation. Couturier et al. (2007) analyzed an earlier version of the Populus genome (1v1) and found 14 AMT genes; 6 in the AMT1 cluster, 2 in the AMT2 cluster, 1 in the AMT3 cluster, and 5 in the AMT4 cluster. In the present study, we found 2 new AMT genes in Populus (PtrAMT1;7/3;2). All of these genes have completely ammonium transport region in their protein sequence.
The evolution of the AMT/MEP/Rh superfamily of integral membrane proteins is extremely complex. Within each of these families, various cases, including duplication and expansion events, gene losses, and horizontal gene transfer events may occur (Couturier et al., 2007; McDonald et al., 2010). In Populus, expansion of the AMT gene family can be ascribed to duplication events and tandem repeats. In this study, the phylogenetic analysis and chromosomal location information revealed that duplication events, tandem events, and the loss of duplicates after duplication events occurred in the Populus AMT gene family. A previous study revealed that the Populus genome has undergone two whole-genome duplication events that significantly contributed to the amplification of many multigene families. One of the whole-genome duplication events was the salicoid duplication event that occurred 65 million years ago (Tuskan et al., 2006). Many previous studies have provided evidence for gene duplication in several gene families, including the GS gene family and the NRT gene family (Castro-Rodríguez et al., 2011; Bai et al., 2013). The ratio of putative Populus NRT homologs to corresponding genes in Arabidopsis was reported to be 1.4–1.6 (Bai et al., 2013), compared with a ratio of 3.5 for the AMT gene family. This result supports the hypothesis that plant species from different environments organize NH+4 transport with different numbers of NH+4 transporters (Loqué and von Wirén, 2004).
In the evolutionary history of Populus, members of the AMT gene family have undergone rigorous selection. The structure of Populus AMT genes is well conserved and these genes have different numbers of exons. A previous study reported that most genes in the AMT1 cluster have one exon and no introns, except for LjAMT1;1, which has an intron in its open reading frame (ORF) (Salvemini et al., 2001). In Populus, PtrAMT1;7 also has an intron in ORF, but could not detected it in all the nutritive organ, it may express in specific tissue.
In AMT gene family, function of extracellular N-terminus play a role for oligomer stability. In Lycopersicon esculentum, LeAMT1;1/1;2 were detected as a trimeric complex in planta, but in the paraloge LeAMT1;3 who had a short N-terminus, trimeric complexes were not detected (Graff et al., 2011). This may indicate that PtrAMT1;6 is similar to LeAMT1;3 who maintain dimer and monomer complexes on plasma membrane. Previous studies on AtAMT1;1 showed that protein activity could be controlled by phosphorylation site T460, which was localized in C-terminus conserved domain (Loqué et al., 2007; Lanquar et al., 2009). When compared with AtAMT1;1, all the members of poplar AMT1 subfamily members have the conserved phosphorylation site T, except PtrAMT1;6 whose site was replaced by S; but this site was not conserved in AMT2 subfamily members (Supplementary Figure 1). These results may indicate phosphorylation at specific site of poplar AMT1 may be equally important for regulate ammonium up-take under various external environment conditions, and there were possible different regulation mechanisms between AMT1 and AMT2 subfamily members.
Transcript Profiles of AMT Genes in Populus
In the Arabidopsis roots, the genes in the AMT1 cluster encode proteins responsible for NH+4 uptake (Yuan et al., 2007). AtAMT1;1/1;2/1;3/2;1 account for 90% of high-affinity NH+4-uptake capacity in the root, while AtAMT1;4 is responsible for the high-affinity NH+4-uptake capacity of pollen (Loqué et al., 2006; Yuan et al., 2007, 2009). However, in poplar, the physiological function of AMTs is still not well known. When comparing with Arabidopsis, there are more AMT gene family members in poplar than in Arabidopsis. These may indicate function redundancy of AMTs in poplar; or execute special function depend on differential tissue expression, like AtAMT1;3 who can mediate lateral root branching (Lima et al., 2010).
In this study, transcripts of 14 AMT genes were detected in nutritive organs. There were relatively high transcript levels of AMT1;1/1;3/1;6/2;1/2;2/3;2 in the leaves, AMT1;1/2;1/2;2 in the stems, and AMT1;1/1;2/2;2/3;1 in the roots. These results indicate that these genes may play different physiological functions in ammonium utilization. Based on our observations, we propose that AMT1;1/1;2/2;2/3;1 may be suspected to be responsible for ammonium uptake from the soil; and the others may be involved in ammonium redistribution, for example, AMT1;1/2;1/2;2 may play key roles in the ammonium transport from roots to shoots, AMT1;6 may participate in the retrieval and import from apoplast of leaves (von Wirén et al., 2000), and AMT1;1/1;3/1;6/2;1/2;2/3;2 may be in charge of ammonium retrieval from old leaves to young leaves (Couturier et al., 2007). Noteworthy, paralogous pairs PtrAMT3;1/3;2 had different intron length and express pattern and in roots and leaves, these results indicate that these two genes may have different transcriptional regulation mechanism and/or different function in specially tissue or cell.
A comprehensive analysis of RNA-seq data and Microarray data (Yang et al., 2008) from popgenie v3 (http://www.popgenie.org/) confirms that AMT1;2 prefers to be expressed in roots, but less in the leaves and stems, while AMT1;6 prefer to be expressed in leaves, but had low expression level in the stems and roots (Supplementary Tables 2, 3).
In this study, we propose that the ammonium-dependent expression of some PtrAMTs may be controlled by a local ammonium signal in roots or a systematic N signal in leaves, respectively. The expression pattern of AMT1;1/2;2/3;1 have acutely change under nitrogen starvation and ammonium supply in roots, these results are similar to ZmAMT1;1a/1;3 who could be controlled by a local ammonium signal (Gu et al., 2013), and this expression pattern may improve NH+4 up-take efficiency. Although AMT1;2 showed up-regulated transcription under nitrogen starvation and NH+4-resupply conditions, but the external ammonium concentration can affect AMT1;2 transcript levels in roots, this result suggests that AMT1;2 may play a housekeeping role in root who is always ready to transport ammonium in the roots. In addition, AMT1;1/1;2/2;2/3;1 had a high expression level in roots, they may make up the loss of ammonium transport in the roots of some down-regulated AMT genes under nitrogen-dependent experiment. Under nitrogen starvation and ammonium supply condition, transcripts accumulation of AMT1;1/1;3/3;2 may regulate by the whole-plant N status, who had high transcripts accumulation after nitrogen starvation for 24 h. While AMT1;6 had oppositely expression pattern under nitrogen starvation, these results may indicate that expression of AMT1;6 was controlled by ammonium concentration in the apoplast of leaves.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was financially supported by the Innovation Project of the State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University) (2013B03), the National High Technology Research and Development Program of China (2013AA102702), and by Fundamental Research Funds for the Central Universities (DL13EA03-01).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00337/abstract
References
Bai, H., Euring, D., Volmer, K., Janz, D., and Polle, A. (2013). The nitrate transporter (NRT) gene family in poplar. PLoS ONE 8:e72126. doi: 10.1371/journal.pone.0072126
Bloom, A. J., Sukrapanna, S. S., and Warner, R. L. (1992). Root respiration associated with ammonium and nitrate absorption and assimilation by barley. Plant Physiol. 99, 1294–1301. doi: 10.1104/pp.99.4.1294
Britto, D. T., and Kronzucker, H. J. (2002). NH+4 toxicity in higher plants: a critical review. J. Plant Physiol. 159, 567–584. doi: 10.1078/0176-1617-0774
Castro-Rodríguez, V., García-Gutiérrez, A., Canales, J., Avila, C., Kirby, E. G., and Cánovas, F. M. (2011). The glutamine synthetase gene family in Populus. BMC Plant Biol. 11:119. doi: 10.1186/1471-2229-11-119
Chai, G., Hu, R. B., Zhang, D. Y., Qi, G., Cao, Y. P., Chen, P., et al. (2012). Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genomics 13:253. doi: 10.1186/1471-2164-13-253
Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. doi: 10.1007/BF02670468
Couturier, J., Montanini, B., Martin, F., Brun, A., Blaudez, D., and Chalot, M. (2007). The expanded family of ammonium transporters in the perennial poplar plant. New Phytol. 174, 137–150. doi: 10.1111/j.1469-8137.2007.01992.x
Dluzniewska, P., Gessler, A., Dietrich, H., Schnitzler, J. P., Teuber, M., and Rennenberg, H. (2007). Nitrogen uptake and metabolism in Populus× canescens as affected by salinity. New Phytol. 173, 279–293. doi: 10.1111/j.1469-8137.2006.01908.x
Eddy, S. R. (2009). A new generation of homology search tools based on probabilistic inference. Genome Inform. 23, 205–211. doi: 10.1142/9781848165632_0019
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. doi: 10.1093/nar/gkp985
Graff, L., Obrdlik, P., Yuan, L., Loque, D., Frommer, W. B., and von Wirén, N. (2011). N-terminal cysteines affect oligomer stability of the allosterically regulated ammonium transporter LeAMT1;1. J. Exp. Bot. 62, 1361–1373. doi: 10.1093/jxb/erq379
Gu, R., Duan, F., An, X., Zhang, F., von Wirén, N., and Yuan, L. (2013). Characterization of AMT-Mediated High-Affinity Ammonium Uptake in Roots of Maize (Zea mays L.). Plant Cell Physiol. 54, 1515–1524. doi: 10.1093/pcp/pct099
Guo, A. Y., Zhu, Q. H., Chen, X., and Luo, J. C. (2007). GSDS: a gene structure display server. Yi Chuan 29, 1023–1026. doi: 10.1360/yc-007-1023
Khademi, S., O'Connell, J. R. D., Remis, J., Robles-Colmenares, Y., Miercke, L. J., and Stroud, R. M. (2004). Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science 305, 1587–1594. doi: 10.1126/science.1101952
Koegel, S., Ait Lahmidi, N., Arnould, C., Chatagnier, O., Walder, F., Ineichen, K., et al. (2013). The family of ammonium transporters (AMT) in Sorghum bicolor: two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 198, 853–865. doi: 10.1111/nph.12199
Lanquar, V., Loque, D., Hormann, F., Yuan, L., Bohner, A., Engelsberger, W. R., et al. (2009). Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell Online 21, 3610–3622. doi: 10.1105/tpc.109.068593
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Lima, J. E., Kojima, S., Takahashi, H., and von Wirén, N. (2010). Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-Dependent manner. Plant Cell. 22, 3621–3633. doi: 10.1105/tpc.110.076216
Loqué, D., Lalonde, S., Looger, L. L., Von Wirén, N., and Frommer, W. B. (2007). A cytosolic trans-activation domain essential for ammonium uptake. Nature 446, 195–198. doi: 10.1038/nature05579
Loqué, D., and von Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305. doi: 10.1093/jxb/erh147
Loqué, D., Yuan, L., Kojima, S., Gojon, A., Wirth, J., Gazzarrini, S., et al. (2006). Additive contribution of AMT1; 1 and AMT1; 3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 48, 522–534. doi: 10.1111/j.1365-313X.2006.02887.x
Luo, Z. B., Janz, D., Jiang, X., Göbel, C., Wildhagen, H., Tan, Y., et al. (2009). Upgrading root physiology for stress tolerance by ectomycorrhizas: insights from metabolite and transcriptional profiling into reprogramming for stress anticipation. Plant Physiol. 151, 1902–1917. doi: 10.1104/pp.109.143735
McDonald, S. M., Plant, J. N., and Worden, A. Z. (2010). The mixed lineage nature of nitrogen transport and assimilation in marine eukaryotic phytoplankton: a case study of Micromonas. Mol. Biol. Evol. 27, 2268–2283. doi: 10.1093/molbev/msq113
Min, X., Yaeesh Siddiqi, M., Guy, R. D., Glass, A. D. M., and Kronzucker, H. J. (1999). A comparative study of fluxes and compartmentation of nitrate and ammonium in early-successional tree species. Plant Cell Environ. 22, 821–830. doi: 10.1046/j.1365-3040.1999.00450.x
Pantoja, O. (2012). High affinity ammonium transporters: molecular mechanism of action. Front. Plant Sci. 3:34. doi: 10.3389/fpls.2012.00034
Salvemini, F., Marini, A., Riccio, A., Patriarca, E. J., and Chiurazzi, M. (2001). Functional characterization of an ammonium transporter gene from Lotus japonicus. Gene 270, 237–243. doi: 10.1016/S0378-1119(01)00470-X
Selle, A., Willmann, M., Grunze, N., Gessler, A., Weiss, M., Nehls, U. (2005). The high-affinity poplar ammonium importer PttAMT1. 2 and its role in ectomycorrhizal symbiosis. New Phytol. 168, 697–706. doi: 10.1111/j.1469-8137.2005.01535.x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
von Wirén, N., Lauter, F. R., Ninnemann, O., Gillissen, B., Walch-Liu, P., Engels, C., et al. (2000). Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J. 21, 167–175. doi: 10.1046/j.1365-313x.2000.00665.x
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, G., Kong, H. Z., Sun, Y. J., Zhang, X. H., Zhang, W., Altman, N., et al. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-Like proteins. Plant Physiol. 135, 1084–1099. doi: 10.1104/pp.104.040436
Williams, L. E., and Miller, A. J. (2001). Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Biol. 52, 659–688. doi: 10.1146/annurev.arplant.52.1.659
Yang, X., Kalluri, U. C., Jawdy, S., Gunter, L. E., Yin, T., Tschaplinski, T. J., et al. (2008). The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol. 148, 1189–1200. doi: 10.1104/pp.108.121921
Yuan, L., Graff, L., Loqué, D., Kojima, S., Tsuchiya, Y. N., Takahashi, H., et al. (2009). AtAMT1; 4, a pollen-specific high-affinity ammonium transporter of the plasma membrane in Arabidopsis. Plant Cell Physiol. 50, 13–25. doi: 10.1093/pcp/pcn186
Yuan, L., Gu, R., Xuan, Y., Smith-Valle, E., Loqué, D., Frommer, W. B., et al. (2013). Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell Online 25, 974–984. doi: 10.1105/tpc.112.108027
Yuan, L., Loqué, D., Kojima, S., Rauch, S., Ishiyama, K., Inoue, E., et al. (2007). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell Online 19, 2636–2652. doi: 10.1105/tpc.107.052134
Keywords: ammonium transporter, poplar, genome-wide analysis, evolutionary mechanism, expression profile, ammonium deficiency
Citation: Wu X, Yang H, Qu C, Xu Z, Li W, Hao B, Yang C, Sun G and Liu G (2015) Sequence and expression analysis of the AMT gene family in poplar. Front. Plant Sci. 6:337. doi: 10.3389/fpls.2015.00337
Received: 19 December 2014; Accepted: 29 April 2015;
Published: 21 May 2015.
Edited by:
Jirong Huang, University of Tokyo, JapanReviewed by:
Omar Pantoja, Universidad Nacional Autónoma de México, MexicoJianyong Li, Cornell University, USA
Copyright © 2015 Wu, Yang, Qu, Xu, Li, Hao, Yang, Sun and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangyu Sun, School of Life Science, Northeast Forestry University, 26 Hexing Road, HarBin 150040, China, sungy@nefu.edu.cn;
Guanjun Liu, State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, 26 Hexing Road, HarBin 150040, China, liuguanjun2013@nefu.edu.cn
†These authors have contributed equally to this work.
 Xiangyu Wu
Xiangyu Wu Han Yang
Han Yang Chunpu Qu
Chunpu Qu Zhiru Xu
Zhiru Xu Wei Li1
Wei Li1 Guangyu Sun
Guangyu Sun Guanjun Liu
Guanjun Liu