- National Institute of Plant Genome Research, New Delhi, India
Small heat shock proteins (sHSPs) are a diverse group of proteins and are highly abundant in plant species. Although majority of these sHSPs were shown to express specifically in seed, their potential function in seed physiology remains to be fully explored. Our proteomic analysis revealed that OsHSP18.2, a class II cytosolic HSP is an aging responsive protein as its abundance significantly increased after artificial aging in rice seeds. OsHSP18.2 transcript was found to markedly increase at the late maturation stage being highly abundant in dry seeds and sharply decreased after germination. Our biochemical study clearly demonstrated that OsHSP18.2 forms homooligomeric complex and is dodecameric in nature and functions as a molecular chaperone. OsHSP18.2 displayed chaperone activity as it was effective in preventing thermal inactivation of Citrate Synthase. Further, to analyze the function of this protein in seed physiology, seed specific Arabidopsis overexpression lines for OsHSP18.2 were generated. Our subsequent functional analysis clearly demonstrated that OsHSP18.2 has ability to improve seed vigor and longevity by reducing deleterious ROS accumulation in seeds. In addition, transformed Arabidopsis seeds also displayed better performance in germination and cotyledon emergence under adverse conditions. Collectively, our work demonstrates that OsHSP18.2 is an aging responsive protein which functions as a molecular chaperone and possibly protect and stabilize the cellular proteins from irreversible damage particularly during maturation drying, desiccation and aging in seeds by restricting ROS accumulation and thereby improves seed vigor, longevity and seedling establishment.
Introduction
Seeds, during development acquire remarkable protective mechanisms that allow them to survive desiccation to extremely low water content and to maintain their germinability even after many years of storage. Current evidence indicate that acquisition of desiccation tolerance and seed longevity is a multifunctional trait and diverse mechanisms have been proposed to be involved in acquiring such traits in seeds. As for example, presence and abundance of specific mono, di and oligosaccharides, soluble sugars and specific proteins along with the mechanisms related to ROS removal were shown to play important roles in the acquisition of desiccation tolerance and seed longevity in orthodox seeds (Blackman et al., 1995; Wehmeyer and Vierling, 2000). Previous studies also suggest that orthodox seeds essentially acquire desiccation tolerance during seed development particularly during maturation drying when most of these protective molecules are highly abundant (Roberts, 1973). Among these protective molecules, sHSPs have recently been suggested to play an important role in seed desiccation tolerance and longevity (Wehmeyer and Vierling, 2000; Sun et al., 2002; Tsvetkova et al., 2002; He and Yang, 2013). sHSPs are known to function as molecular chaperones that facilitate protein folding and prevent irreversible protein aggregation during developmental and adverse environmental conditions. Plants synthesize at least 21 different types of sHSPs and are grouped into six different classes on the basis of their subcellular localization and sequence alignments. This sHSP family has members with molecular size ranging from 16 to 42 kDa and invariably contains a conserved ACD in the C-terminal. These sHSPs are believed to play diverse role in plant biology (de Jong et al., 1998). Even though, sHSPs are known more for their extensive role in plant defense against abiotic stresses, their role in seed physiology has recently came into light (Wehmeyer and Vierling, 2000; Sun et al., 2001; Guo et al., 2007; Perez et al., 2009). Certain sHSPs have been reported as preferentially expressed in seeds particularly during development, maturation, and germination, highlighting their role in desiccation tolerance and longevity of seeds (Wehmeyer and Vierling, 2000; Scharf et al., 2001; Sun et al., 2002; Tsvetkova et al., 2002; Neta-Sharir et al., 2005; Volkov et al., 2005; Sarkar et al., 2009; Zhou et al., 2012). A heat shock transcription factor from sunflower (HaHSFA9) overexpressed in tobacco shows an increase in the accumulation of certain sHSPs and consequent improvement of seed vigor and longevity (Prieto-Dapena et al., 2006). Recently, a sHSP from Nelumbo nucifera was shown to increase seed germination vigor and seedling thermo tolerance in transgenic Arabidopsis (Zhou et al., 2012). In rice, 23 sHSP genes have been identified and majority of these genes are expressed in seed indicating their potential role in seed physiology. However, the role of sHSPs particularly in seed desiccation tolerance and longevity has not been well elucidated (Sarkar et al., 2009).
Rice (Oryza sativa) is a monocot model crop which feeds nearly half of the world population. Most of the cultivated rice varieties have a low or null dormancy and variable periods of storage life. Even under best storage conditions, rice seeds lose their viability and germination vigor (He and Yang, 2013). Therefore, a focused effort is required to elucidate and identify the proteins and underlying mechanisms implicated in seed vigor, viability, and longevity. More so, since the production of high quality seeds for food and germplasm preservation is also dependent on seed vigor and longevity. To identify such aging responsive proteins in rice seeds, a proteomic approach has been adopted in this study and eventually a small HSP (OsHSP18.2, a class II heat shock protein, LOC_Os01g08860) has been identified. Subsequently, the OsHSP18.2 cDNA has been cloned from rice. Further, to get detailed insight of the involvement and participation of this sHSP protein in seed desiccation tolerance and longevity, transcript accumulation was analyzed in different organs, during seed development, aging, germination, and stress conditions. Further, OsHSP18.2 was bacterially expressed, purified, and biochemically characterized. Finally the ability of this gene to enhance seed vigor and longevity has been analyzed through seed specific overexpression in Arabidopsis.
Materials and Methods
Plant Materials and Stress Treatments
Rice seeds (O. sativa indica cv. PB-1) were imbibed overnight in dark and sown in glass bottles over moist cotton bed and a layer of germination paper. The bottles were then kept for normal growth in growth room 28 ± 1°C and 65% humidity levels. For different stress treatments, 7 day-old seedlings were subjected to various stresses as described by Liu et al. (2009). For time course heat stress, 10 day-old seedlings were transferred to 42°C for 15 min, 30 min, 1, 2, 3, 5, and 24 h.
Controlled Deterioration Treatment
Seeds harvested on the same day from plants grown under identical conditions were used for all comparisons. Rice and Arabidopsis seeds were imbibed for 1 h and then blot dried. The seeds were treated with a combination of high temperature (45°C) and high humidity (100% Relative humidity) for 6 days (for rice) and 4 days (for Arabidopsis) to induce artificial aging (Delouche and Baskin, 1973; Verma et al., 2013). CDT treated rice seeds were used for protein extraction for 2 D electrophoresis. Transgenic Arabidopsis seeds subjected to CDT were used to score seed viability via germination percentage, tetrazolium assay and H2O2 accumulation via DAB staining.
Protein Extraction, 2D Electrophoresis, and Protein Identification
Artificially aged rice seeds (250 mg) were ground to fine powder and homogenized with 2 ml of extraction buffer (20 mM Tris-HCl, pH 7.5, 250 mM sucrose, 10 mM EGTA, 1 mM PMSF, 1 mM DTT, and 1% Triton X-100). Homogenate was centrifuged at 15000 g for 15 min at 4°C and supernatant was collected as total soluble protein. Protein (500 μg) was precipitated with ice cold 100% acetone and pellets were solubilised in 250 μl of DeStreak Rehydration solution (GE) with 2% IPG buffer pH 3-10 NL. The protein sample was used to rehydrate IPG strips 13 cm, pH 3-10NL overnight. Isoelectric focussing (IEF) was performed with rehydrated strips on Ettan IPGphor 3 (GE) at 100V–2 h; 500V–1 h; 1000V–1 h; 8000V–30 min and finally at 8000V–3 h. After IEF, the strips were equilibrated first with 1% (w/v) DTT in equilibration buffer (10 ml of 50 mM Tris-Cl (pH 8.8), 2% SDS, 6 M urea and 30% glycerol) followed by 4% (w/v) iodoacetamide in equilibration buffer. The strips were then loaded on 12.5% polyacrylamide gels for SDS-PAGE. Gels were stained with PlusOne Silver Staining Kit (GE) and analyzed with Image Master Platinum seven software (GE). The protein spots were excised manually and in-gel digestion was performed using trypsin (Sigma). Peptide analysis was carried out on 4800 MALDI-TOF/TOF (Applied Biosystems/MDS SCIEX) and peptides were identified using Mascot search engine run on GPS explorer version 3.6 (Applied Biosystems) using the following parameters: 800–4000 m/z interval MS peak filtering, monoisotopic, MSDB version 20060831 (3239079 sequences; 1079594700 residues), enzyme trypsin with maximum allowance of one missed cleavage, taxonomy Viridiplantae (Green Plants; 247347 sequences), ±100 ppm peptide tolerance, ±0.3 Da fragment mass tolerance, with fixed modification as carbamidomethyl (C) and variable modification as oxidation (M). Protein samples with probability score above default threshold level (p < 0.05) as determined by Mascot were considered for further analysis.
Quantitative Real-time PCR
For real time quantification of the transcripts, total RNA was isolated from rice seeds and seedlings using modified guanidine hydrochloride protocol and TRI reagent, respectively (Singh et al., 2003). Total RNA was treated with DNaseI from Ambion. First-strand cDNA was synthesized from the Dnase treated RNA using the Verso cDNA Synthesis Kit (Thermo). cDNA was then quantified by Nanodrop and dilutions were adjusted for 50 ng/μl. Real time quantification was performed as described in Kaur et al. (2008). Primers used for real time are described in Supplementary Table S1.
Cloning and Transformation of OsHSP18.2
Full length CDS and 5′ upstream sequence of OsHSP18.2 (LOC_Os01g08860) was obtained from Ensembl plant O. sativa indica group database available at http://ensembl.gramene.org and GenomeIndia’s manually curated database of rice proteins respectively (Gour et al., 2014). Complete ORF was amplified based on the primers designed from these sequences (Supplementary Table S1) from O. sativa indica var. PB-1. The coding region was cloned into pJET1.2 vector and sequence was confirmed. Finally the coding region was subcloned into modified pCAMBIA2301 vector under the control of napin promoter for seed specific expression. The construct was then transferred to Agrobacterium strain GV3101 and finally Arabidopsis plants were transformed by floral dipping (Clough and Bent, 1998). The 5′ upstream promoter sequence of OsHSP18.2 was scanned for the presence of cis–acting regulatory elements with online available softwares PLACE (Higo et al., 1999) and PlantCARE database (Lescot et al., 2002).
Bacterial Overexpression and Purification of Recombinant OsHSP18.2
CDS of OsHSP18.2 was subcloned into bacterial expression vector pET23b (Novagen) and transformed into Escherichia coli host strain BL21DE3. Transformed cells were grown in LB medium with appropriate antibiotic till A600 reached 0.5 and then induced by adding 0.5 mM IPTG. After 6 h growth at 37°C, cells were harvested. Finally, 6X His tagged recombinant HSPs were purified from soluble fraction using nickel-charged affinity columns (GE) following the manufacturer’s protocol.
Thermal Inactivation Assay
Chaperone activity of OsHSP18.2 was assayed by the method of Lee et al. (1995). CS and purified OsHSP18.2 were combined in equimolar concentration (150 nM) in 50 mM HEPES-KOH buffer, pH-8 with total reaction volume of 500 μl. Controls were made without HSP and with Lysozyme as the control protein. The tubes were incubated in a water bath set at 38°C for 60 min after which the tubes were shifted to 22°C to allow refolding of CS for another 60 min. Twenty micro liter aliquots were removed and CS activity was assayed every 20 min starting at zero minute.
Citrate Synthase Assay
Citrate synthase activity was measured according to Eigentler et al. (2012). Reaction was set up in a cuvette, 25 μl each of 12.2 mM acetyl coA and 10% triton X-100 was added with 100 μl of 1.01 mM DTNB solution (prepared in 1 M Tris-Cl, pH-8.1). Twenty micro liter of sample was added from the refolding reaction and the final volume was brought to 1 ml. Finally 50 μl of 10 mM Oxaloacetate was added to start the reaction. The absorbance was recorded in a Biorad spectrophotometer set at 412 nm for 2 min at an interval of 20 s. The initial rate of reaction was compared with non-denatured CS and the data was presented as percentage reactivation relative to this activity.
Tetrazolium Staining
Seeds were stained with 1% solution of 2,3,5-triphenyltetrazolium chloride (Sigma) to differentiate between viable and non-viable seeds according to Verma et al. (2013). Seeds were photographed using Zeiss SteREO Discovery V12 microscope fitted with Axiocam ICc 3 camera.
DAB Staining
Seeds treated with or without CDT were stained with 3,3′- DAB according to Yi et al. (2010). Seeds were incubated overnight in DAB staining solution (0.1 mg/ml DAB in 50 mM tris actetate buffer, pH 5.0) at 25°C in dark. Next day seeds were bleached in 80% ethanol for 10 min at 70°C. Seeds were photographed using Zeiss SteREO Discovery V12 microscope fitted with Axiocam ICc 3 camera.
Germination Assay Under Stress Treatments
Seeds from WT, VC, and transgenic lines were harvested at the same time and stored at room temperature till further use. Germination assays were carried out as described in Saxena et al. (2013). Three biological replicates with n = 50 seeds were used. Seeds were surface sterilized and plated on to either ½ MS or ½ MS supplemented with NaCl (150 mM), PEG (-0.4 MPa). For heat stress, sterilized seeds were incubated at 45°C for 1 h and then plated on to ½ MS. Seeds were considered germinated when radicle protruded beyond the testa and seedling establishment was considered positive with the emergence of green cotyledonary leaves (Silva-Correia et al., 2014).
Statistical Analysis
Statistical analysis was performed with one way ANOVA using Duncan’s multiple comparison test to determine significant differences among samples. Differences were taken as significant when P < 0.01.
Results
OsHSP18.2 is an Aging and Seed Vigor Associated Protein
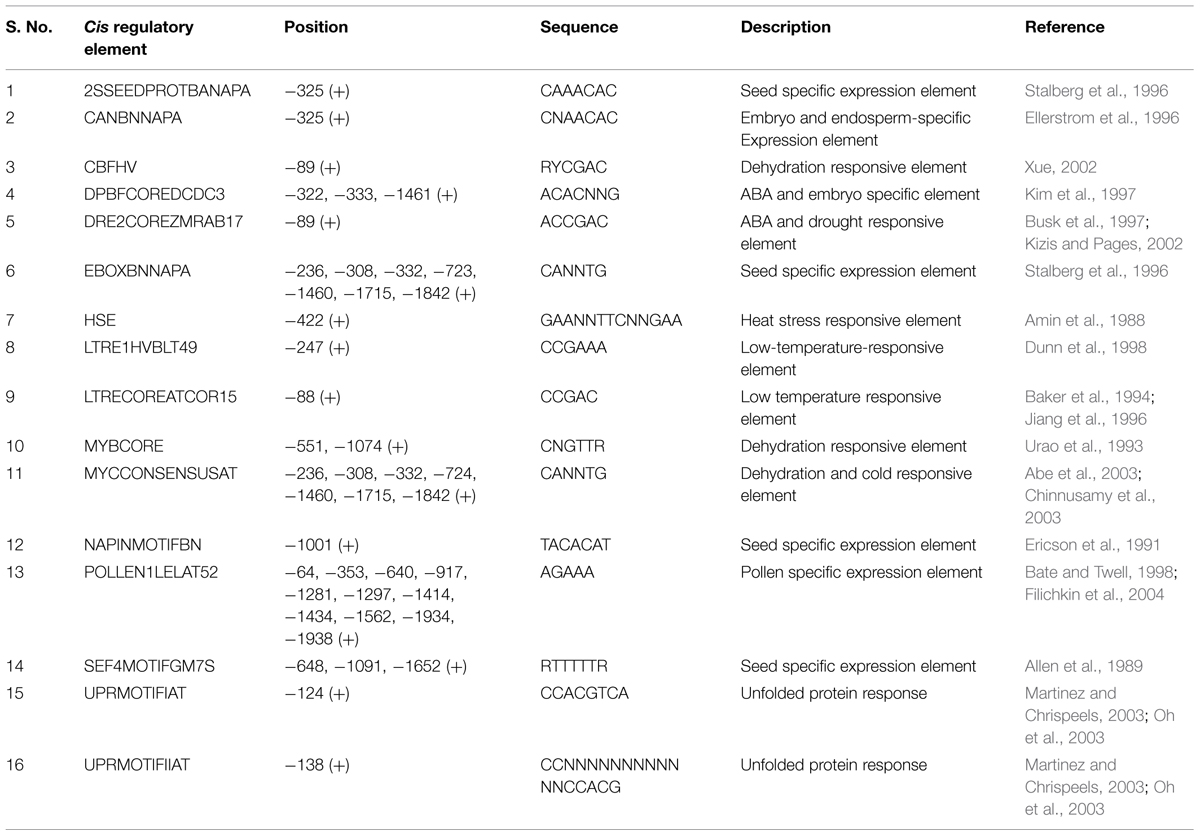
Controlled deterioration treatment has been used widely for quick evaluation of seed vigor, longevity and also to artificially induce aging related changes in seeds (Delouche and Baskin, 1973; TeKrony, 1995; Lanteri et al., 1996; Prieto-Dapena et al., 2006; Oge et al., 2008). Therefore, to identify the seed vigor and aging responsive proteins in rice seed, changes in protein expression after CDT were analyzed using proteomic approach. For CDT, dry seeds were subjected to a combination of high temperature and humidity as described in Section “Materials and Methods.” Subsequently total soluble proteins were extracted from control and deteriorated seeds and were separated by two dimensional polyacrylamide gel electrophoresis (2 D-PAGE). Following silver staining, protein spot showing apparent variation indicated in Figure 1 was excised from the gel. Excised spot was subjected to trypsin digestion and peptides were extracted and analyzed by MALDI-TOF/TOF. Subsequently peptides were identified using Mascot search engine run on GPS explorer 3.6. Only the best matches with high confidence level were selected. The protein was identified as OsHSP18.2 (LOC_Os01g08860). OsHSP18.2 is an intronless gene which encodes a protein of 166 aa with an isoelectric point of 5.61 and a predicted molecular mass of 18.2 kDa. Sequence analysis revealed a conserved 90 amino acid alpha crystalline domain, a characteristic feature of sHSP family (Supplementary Figure S1). Pairwise alignment of OsHSP18.2 with its Arabidopsis homolog, AtHSP17.0 (AT5G12020.1) shows 59% sequence identity between them (Supplementary Figure S2).
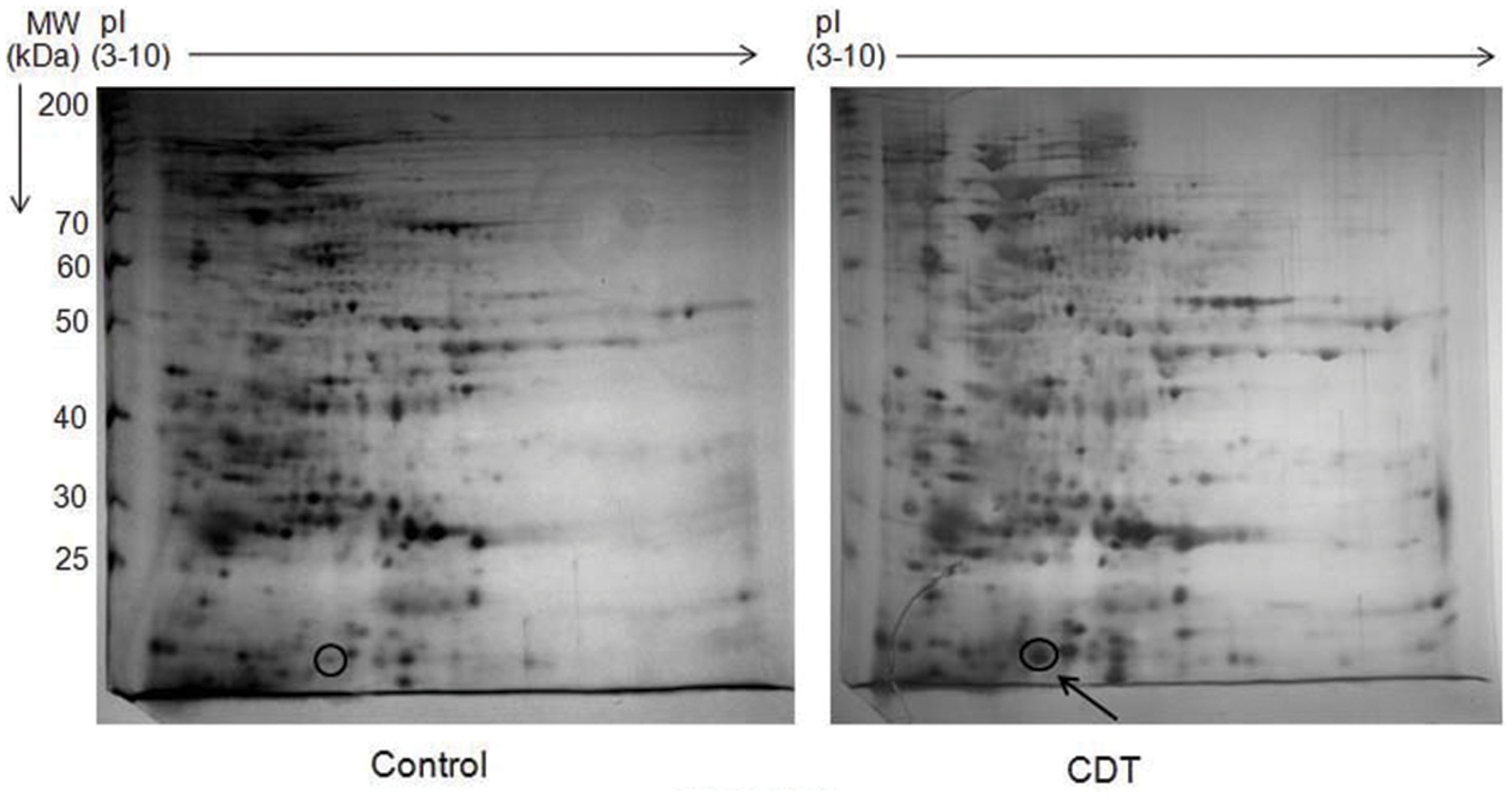
FIGURE 1. 2D profile of rice seed proteins under control and CDT. Total 500 μg of protein from Control and CDT treated seeds (6 days) was used for IEF using a non-linear IPG strip of pH 3–10. Second dimension was run on 12.5% SDS-PAGE and gels were stained with silver stain. Gel images were analyzed with Imagemaster 2D Platinum 7.0 (GE Healthcare) and differentially expressed spots were identified. Encircled spot was eluted and sequenced using MALDI-TOF/TOF.
OsHSP18.2 Upregulates in Seed, During Seed Maturation, and Upon Accelerated Aging
To elucidate the function and mechanism of OsHSP18.2 in seed maturation and desiccation tolerance, we initially investigated the transcript accumulation of OsHSP18.2 in seed along with other organs. As shown in Figure 2A, OsHSP18.2 transcript was highly abundant in seed and more specifically in embryo than endosperm. Subsequently, to get the refined view of transcript accumulation during seed development, flowers were tagged according to the day after pollination (DAP) as described by Agarwal et al. (2011). Accumulation of OsHSP18.2 transcript was found to be relatively low during initial stages of development, i.e., S1 till S4 stage (0–20 DAP) then strikingly increased in the late maturation stage S5 (21–29 DAP) and reached highest levels in dry mature seed (Figure 2B). This data essentially revealed that OsHSP18.2 markedly increased at the later stages of seed development consistent with the time when seed actually acquires desiccation tolerance and longevity.
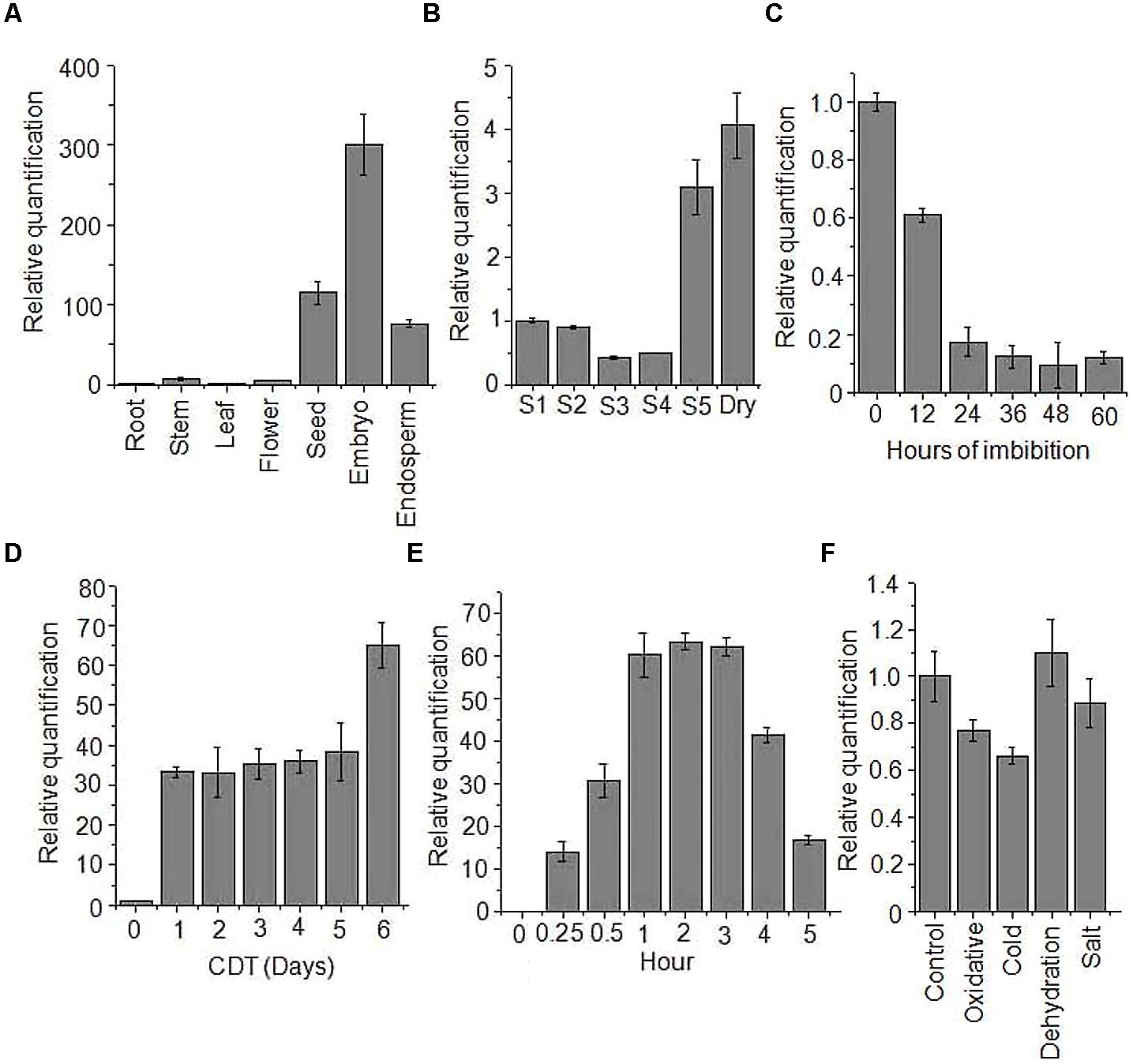
FIGURE 2. Expression profiling of OsHSP18.2. Transcript accumulation was analyzed by quantitative real time PCR (A) in different organs, (B) during seed development, (C) during seed germination, (D) under CDT, (E) under heat stress, and (F) under various abiotic stresses. Actin was used as an endogenous control. The relative expression value of each gene was normalized to an endogenous control and calculated using the ΔΔCT method (Applied Biosystems). Expression values are the mean ± SD of three independent biological measurements.
Transcript accumulation was also studied during germination and data showed a sharp decline in transcript level as germination proceeds (Figure 2C). Next, we investigated the correlation of transcript accumulation of OsHSP18.2 and aging in rice seeds. For this, rice seeds were subjected to CDT treatment for 6 days and transcript accumulation pattern was studied. Results clearly revealed a significant increase in transcript accumulation due to artificial aging even after 1 day of CDT which was further increased to more than 60-fold after 6 days of CDT (Figure 2D). To examine the possible involvement of OsHSP18.2 in abiotic stress tolerance, transcript accumulation was checked in rice seedlings challenged with various stresses (Figures 2E,F). As shown in Figure 2E, heat stress triggered maximum induction of OsHSP18.2 transcript as expected while other abiotic stresses had minor changes in transcript levels (Figure 2F). This analysis clearly revealed that OsHSP18.2 is upregulated during seed maturation and upon aging and indicates its participation in maturation drying and seed longevity. Transcriptional induction of OsHSP18.2 in rice seedlings challenged with heat stress also indicates its participation in thermal stress tolerance. Investigation of the 5′ upstream sequence of OsHSP18.2 revealed the presence of many interesting cis-acting regulatory elements which are possibly responsible for abiotic stress responsiveness and seed and embryo specific expression. A list of some of these cis-acting elements is shown in Table 1.
Purified Recombinant OsHSP18.2 Forms Higher Order Oligomers
sHSPs are known to form multimers with varying number of subunits, most commonly between 2 and 48 subunits (Basha et al., 2012). In order to explore this, we isolated and cloned OsHSP18.2 coding region into bacterial expression vector pET23b and transformed into E. coli expression host BL21DE3. The recombinant protein was induced by IPTG and was found to be expressed in soluble phase (Figure 3A). Subsequently, 6X His tagged OsHSP18.2 protein was purified using nickel charged affinity chromatography. Size exclusion chromatography was used to check the oligomeric state of OsHSP18.2 and the purified protein was shown to elute as an oligomer of ∼202 kDa thus having approximately 12 subunits which agrees to the dodecameric nature of wheat sHSP16.9 (van Montfort et al., 2001; Figure 3B). The gel exclusion fractions were run on SDS-PAGE and revealed the presence of a single band in the 202 kDa peak fraction corresponding to the single subunit size of 18.2 kDa (Figure 3C). In order to confirm this oligomeric association of OsHSP18.2, the purified protein was run on native PAGE (Figure 3D) which revealed a band across 240 kDa marker band thus endorsing the gel filtration results.
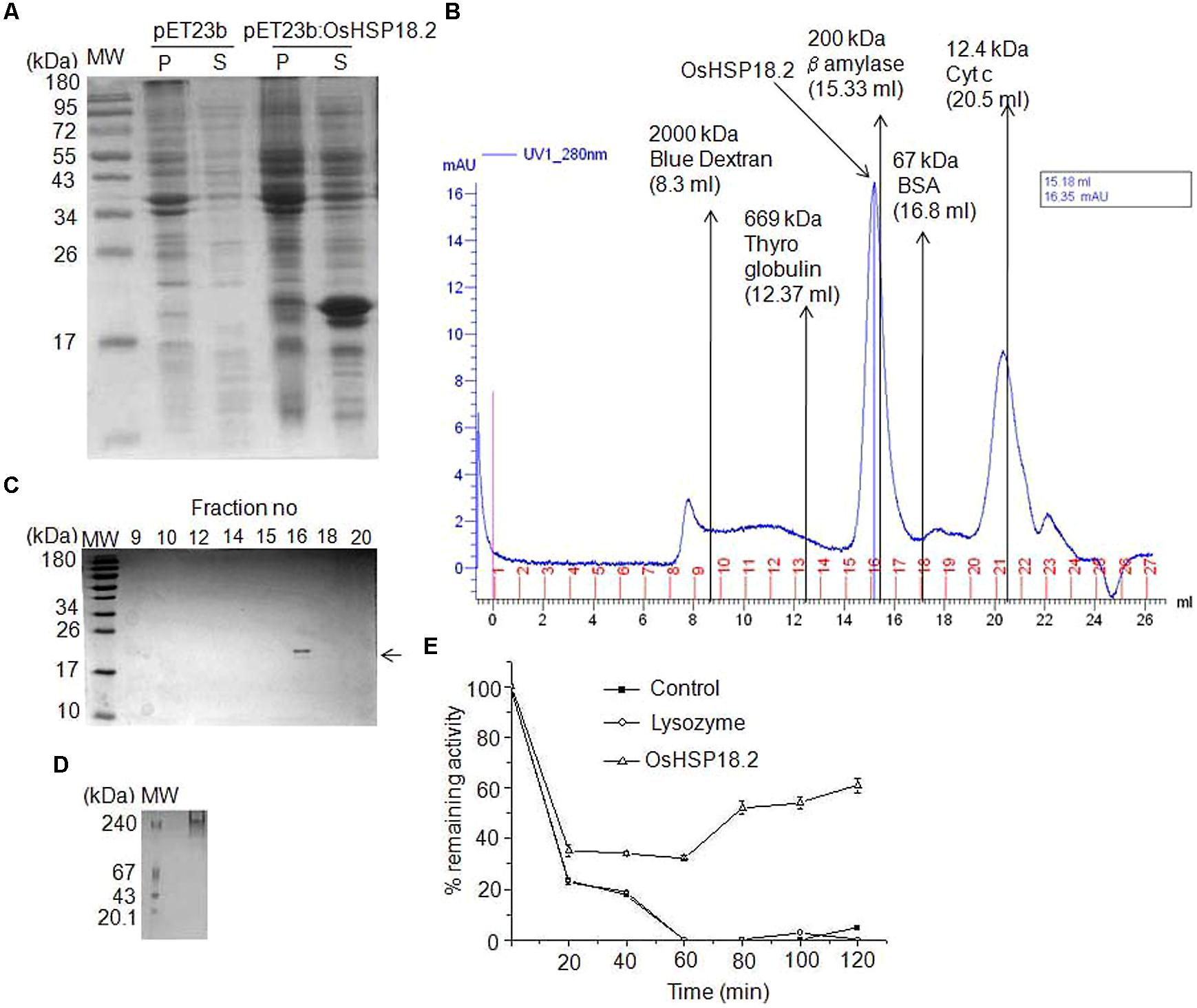
FIGURE 3. Characterization of recombinant OsHSP18.2. (A) Bacterial overexpression of OsHSP18.2 in Escherichia coli. Proteins were separated on a 15% SDS-PAGE gel. pET23b empty vector transformed cells were used as a control. Pellet (P) and Supernatant (S) were run for control and OsHSP18.2 transformed cells along with the molecular weight marker. (B) Size exclusion chromatography on Superose 6 column revealed the homooligomeric nature of OsHSP18.2. Purified protein and appropriate molecular weight markers were run and their elution time (Ve) to void volume (Vo) ratio (Ve/Vo) was plotted against their molecular weight in log scale and fitted in an exponential function where y = 512304e-4.306x. Approximate molecular weight of OsHSP18.2 was calculated from the above equation as ∼202 kDa which reflects dodecameric conformation. (C) Gel filtration fractions as run on 15% SDS-PAGE show a single band of OsHSP18.2 in fraction number 16 corresponding to the gel filtration peak. (D) Five percent native polyacrylamide gel shows a protein band of OsHSP18.2 across the 240 kDa marker band. (E) OsHSP18.2 prevents irreversible thermal inactivation of CS at 38°C. CS was incubated without HSP (—■—), with lysozyme (—○—) and with OsHSP18.2 (—Δ—) at 38°C for 60 min and then shifted to 22°C for refolding. CS enzyme activity was assayed at the mentioned time points. Values are the mean ± SE of three independent experiments.
OsHSP18.2 is an Active Chaperone and Prevents Thermal Denaturation of CS
Many small HSPs are known to have in vitro chaperone activity. Therefore, to assess the chaperone activity of OsHSP18.2, we used CS as a substrate for refolding experiments (Collada et al., 1997; Lee et al., 1997). CS monomers (150 nM) were incubated with or without HSP and with lysozyme as control at 38°C for 60 min and a quick decrease in CS activity was observed (Figure 3E). Control without HSP and with lysozyme showed 20% remaining activity after just 20 min of heat stress which came down to almost zero after 60 min of heat stress whereas CS with OsHSP18.2 retained 30% activity even after 60 min of heat. After 60 min, all the combinations were shifted to 22°C, temperature permissive for refolding. Even at 22°C, control lacking HSP and lysozyme control did not display any regain in CS activity but CS with OsHSP18.2 showed a regain of 60% of native CS activity. This result clearly demonstrated that OsHSP18.2 functions as a molecular chaperone that facilitates protein folding and prevents thermal denaturation.
Seed Specific Overexpression of OsHSP18.2 in Arabidopsis thaliana Improves Seed Vigor and Longevity
To examine the functional implication of sHSP in seed vigor and longevity, OsHSP18.2 was overexpressed in seeds in Arabidopsis thaliana using the napin promoter. Seed specific OsHSP18.2 transcript accumulation was examined through quantitative RT PCR and significant levels of transcripts were observed in seeds of transgenic lines. Based on this analysis, three independent homozygous T3 lines were selected and subsequently used to assess their germination vigor and longevity. To evaluate seed vigor and longevity, seeds were subjected to CDT and germination performance was analyzed. Under normal conditions, transformed and control seeds (empty vector transformed or WT) exhibited 100% germination (Figure 4A), however, after CDT, control seeds showed <20% germination as opposed to OsHSP18.2 transformed seeds where remarkably >50% germination was observed in each line (Figure 4B). In addition, TZ staining was carried out to examine the potential viability of these seeds (Berridge et al., 1996). TZ precipitates to red colored 2,3,5 triphenyl formazan by the activity of dehydrogenases present in the living cells thus staining them red. As expected under normal condition, OsHSP18.2 expressing and control seeds were stained dark red indicating their viability while after CDT, only OsHSP18.2 transformed seeds exhibited dark red staining (i.e., viable) in contrast to control seeds which remain unstained or were stained pale red (non-viable; Figure 4C). Aging in seeds is also accompanied with ROS accumulation (Bailly, 2004). ROS adversely affects cellular proteins and enzymes and renders them inactive. Therefore, we wanted to check if the overexpression of OsHSP18.2 protects the seed against ROS mediated damage. DAB staining was performed for this purpose as it stains the areas of H2O2 production (Mao and Sun, 2015). Results clearly demonstrated that WT and VC seeds accumulated more H2O2 after CDT in comparison to OsHSP18.2 overexpressing lines (Figure 4D). These data clearly indicate that WT or empty vector transformed seeds subjected to controlled deterioration exhibited higher mortality, reduced germination and more H2O2 accumulation while seeds from transgenic lines overexpressing OsHSP18.2 exhibited less mortality, better germination and less H2O2, thus demonstrating the role of OsHSP18.2 toward maintaining seed viability and longevity during aging.
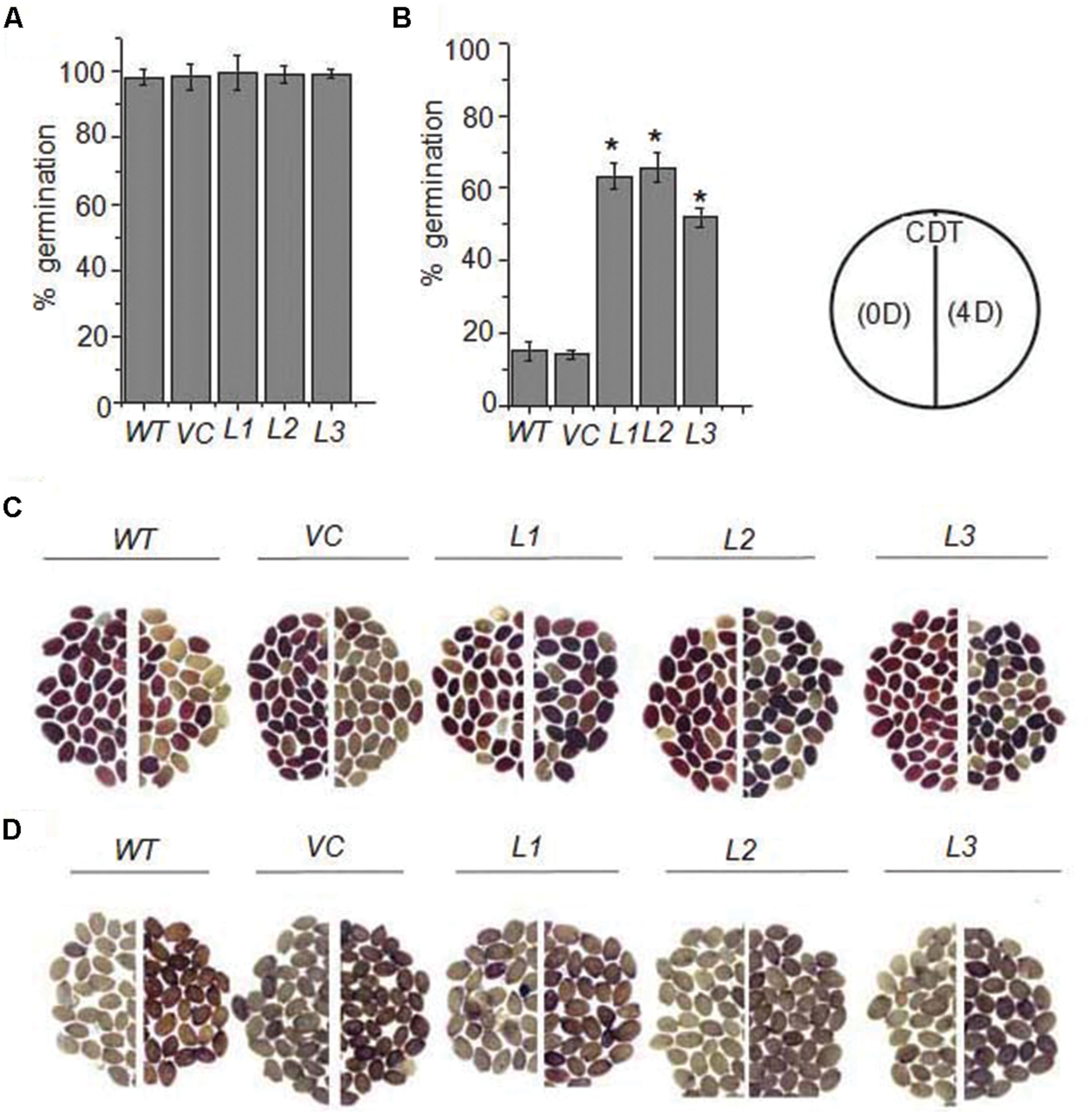
FIGURE 4. Germination and viability of OsHSP18.2 overexpressing seeds after CDT. Six week-old seeds were subjected to 4 days of CDT as described in Materials and Methods. Germination analysis of WT, empty vector VC, and three OsHSP18.2 transgenic lines (L1, L2, and L3) before (A) and after (B) 4 days of CDT. (C) Tetrazolium assay for seed viability and (D) DAB staining for H2O2 production of WT, empty vector (VC), and three OsHSP18.2 transgenic lines (L1, L2, and L3) before (left) and after 4 days of CDT (right). Age matched seeds were used for all germination experiments. In graph the values are mean ± SE of three independent sets (n = 50). Asterisk indicate significant difference between WT and test genotypes (∗P < 0.01).
OsHSP18.2 Transformed Arabidopsis Seeds Display Improved Seed Germination and Seedling Establishment Under Abiotic Stress Conditions
Germination performance of these transgenic seeds under various stress situations were also evaluated, since seed vigor also implies the ability to complete germination under widely variable environmental conditions. Germination of WT, VC, and transgenic seeds was assessed under heat, dehydration, and salt stress and was considered complete when radicle emerged beyond testa. While moderate level of these stresses (i.e., 37°C heat, 100 mM NaCl and -0.25 MPa PEG) did not much affect the germination (data not shown) but slightly elevated stresses revealed a significant difference in germination pattern among control and overexpressing seeds (Figure 5). In normal conditions, OsHSP18.2 transformed and control seeds showed 100% germination. For heat stress, seeds were treated at 45°C for 1 h and then plated on ½ MS, results revealed that >90% of transgenic seeds could complete germination in all lines by the end of 5 days whereas only 70% seeds of WT and VC could germinate by this time (Figure 5C). At 150 mM NaCl concentration, >90% seeds of transgenic lines could germinate compared to 80% of WT and VC (Figure 5E). Dehydration stress was provided by -0.4 MPa PEG and as expected overexpression lines performed better than control lines (Figure 5G). All seeds were monitored for 7 days and seedling establishment was checked which was taken as the emergence of green cotyledons over this period as indicated in previous study (Silva-Correia et al., 2014). Results suggest that in addition to seed germination a significant difference was also observed for seedling establishment among control and OsHSP18.2 overexpressing seeds in adverse conditions (Figure 5).
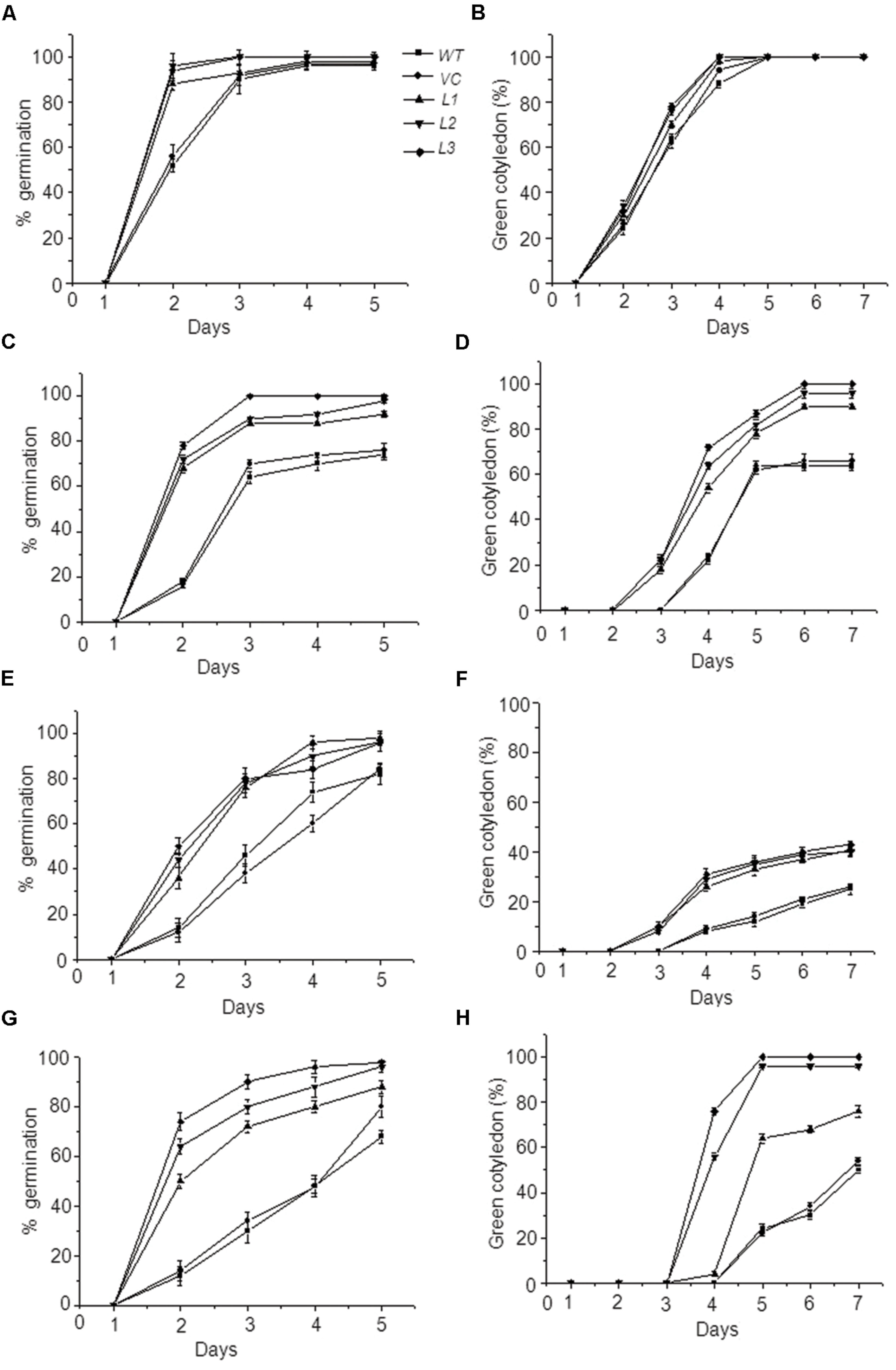
FIGURE 5. Germination and seedling establishment of OsHSP18.2 overexpressing seeds under abiotic stresses. Germination percentage (A,C,E,G) and green cotyledon emergence (B,D,F,H) of WT, VC, and transgenic seeds overexpressing OsHSP18.2 scored under (A,B) Control conditions, (C,D) Heat (45°C), (E,F) Salt (150 mM NaCl), and (G,H) Dehydration (-0.4 MPa PEG). Age matched seeds were surface sterilized and plated on either ½ MS or ½ MS supplemented for various stresses. Plates were stratified at 4°C for 3 days and transferred to growth chamber at 22 ± 2°C. Values are mean ± SE of three independent sets (n = 50).
Overall results conclude with strong evidence that OsHSP18.2 is an aging responsive protein which plays an important role in maintaining seed vigor and longevity as well as seedling emergence by protecting structural damage to proteins and restricting ROS accumulation during prolonged storage.
Discussion
Research on small HSPs has been more emphasized toward plant stress tolerance, however, their role in seed physiology has not been elucidated properly, particularly in rice where majority of the sHSPs are specifically expressed in seed (Sarkar et al., 2009). In this study, our proteomic analysis identified OsHSP18.2 as an aging responsive protein whose abundance significantly increases after artificial aging, indicating its involvement in rice seed vigor and longevity. Previous studies clearly revealed that similar molecular events accompany both in artificial and natural seed aging and thus artificial aging induced protein is also likely to be induced by natural aging (Catusse et al., 2008; Rajjou et al., 2008). Subsequently, we showed that OsHSP18.2 transcript is markedly increased at the late maturation stage and is highly abundant in dry seeds and sharply decreases after germination. This expression pattern of OsHSP18.2 also supports its involvement in desiccation tolerance and longevity as other protective molecules that are generally associated with seed desiccation tolerance and longevity in orthodox seeds are also accumulated in maturation phase and are highly abundant in dry seeds. High transcript accumulations of various sHSPs during seed maturation and in dry seed were also reported in several plant species like pea, sunflower, Arabidopsis and rice (Coca et al., 1994; DeRocher and Vierling, 1994; Wehmeyer et al., 1996; Sarkar et al., 2009; Omar et al., 2011; Zhou et al., 2012). Accelerated aging induced increase in OsHSP18.2 transcript level in rice seed also strengthens our hypothesis that OsHSP18.2 indeed participates in seed vigor and longevity. Notably, a high abundance of sHSPs was reported in beech seeds stored for 8 years (Kalemba and Pukacka, 2008). In addition, studies also suggest that induction of certain sHSPs in embryo is a routine part of seed development and seed desiccation tolerance program (DeRocher and Vierling, 1994).
Moreover, analysis of OsHSP18.2 promoter also revealed motifs like 2SSEEDPROTBANAPA, CANBNNAPA, DPBFCOREDCDC3, EBOXBNNAPA, NAPINMOTIFBN, and SEF4MOTIFGM7S which are responsible for seed and embryo specific expression (Table 1). In addition to this there are many abiotic stress and hormone responsive elements as well which include the heat stress response element HSE (GAANNTTCNNGAA) and well known dehydration responsive element CBFHV (RYCGAC), MYBCORE and MYCCONSENSUSAT thus explaining a higher expression of the transcript during heat and dehydration stress. Surprisingly, few low temperature responsive sequences were also present but our real time experiments revealed not much induction of OsHSP18.2 transcript during cold stress. DPBFCOREDCDC3 (ACACNNG) and DRE2COREZMRAB17 (ACCGAC) responsible for ABA inducible expression are also present. Expression data available on GENEVESTIGATOR also reveals slightly up regulation in response to ABA and Salicylic acid treatment (Hruz et al., 2008). Multiple repeats of pollen specific expression element POLLEN1LELAT52 (AGAAA) present in the promoter suggests that developmentally regulated expression of sHSPs during desiccation associated plant stages such as pollen formation, sporulation, and seed development emphasizes their crucial role during the acquisition of desiccation tolerance (DeRocher and Vierling, 1994). Interestingly, some highly conserved sequence motifs like UPRMOTIFIAT (CCACGTCA) and UPRMOTIFIIAT (CCNNNNNNNNNNNNCCACG) for unfolded protein response are also present as these elements have been also shown to occur in the promoters of other heat shock protein (HSP90) in Arabidopsis. Thus, these sequences might drive the expression of OsHSP18.2 gene during the accumulation of unfolded polypeptides in the cell.
Subsequently, our biochemical study clearly demonstrated that OsHSP18.2 forms oligomeric complex, is dodecameric in nature and function as a molecular chaperone (Figure 3). Studies on wheat sHSP16.9 also demonstrated a dodecameric structure of this heat shock protein although homologs from other species have been shown to have structures with subunits ranging between 2 and >48 subunits (van Montfort et al., 2001; Basha et al., 2012). This is well known that sHSPs function in plant stress adaptation by binding to vulnerable cellular proteins during stress conditions, prevent their aggregation and thus hold them in a competent state for refolding by other chaperones (Ehrnsperger et al., 1997; Veinger et al., 1998; Murakami et al., 2004). Our data clearly establishes that OsHSP18.2 works as a chaperone at high temperature and prevents irreversible thermal inactivation of CS. Comparative studies have shown that addition of pea HSP18.1 significantly increased the refolding of Luciferase even though DnaK system was alone sufficient for stabilization and refolding thus emphasizing its role in restricting denaturation and assisting refolding (Lee and Vierling, 2000). Taken together the increased accumulation of OsHSP18.2 in later stages of seed maturation and aging and its function as a molecular chaperones, suggests that OsHSP18.2 might protect vulnerable cellular proteins during maturation drying, desiccation and aging in seeds. This hypothesis fits well as during seed maturation, desiccation, and storage, seed, particularly embryo faces severe dehydration and oxidative stress that can potentially damage seed proteins (Tamarit et al., 1998; McDonald, 1999; Ingrosso et al., 2000; Rajjou and Debeaujon, 2008). In addition, OsHSP18.2 was shown to be highly induced in rice seedlings exposed to thermal stress, indicating its participation also in thermal stress tolerance.
Our subsequent functional studies essentially demonstrated that OsHSP18.2 has the ability to improve seed vigor and longevity (Figure 4). This improved seed vigor and longevity correlates well with the reduced deleterious ROS accumulation in transformed seeds. Similar to our observation, heterologous expression of a sHSP from sacred lotus in Arabidopsis also lead to enhanced germination of transgenic seeds after accelerated aging treatment (CDT; Zhou et al., 2012). In addition, certain members of the sHSP family have been known to be involved in scavenging of these reactive oxygen species (Härndahl et al., 1999; Fedoroff, 2006). Our results also demonstrated faster and better germination and seedling establishment under adverse situations in OsHSP18.2 overexpressing lines. Collectively, our findings clearly establish that OsHSP18.2 is seed vigor and aging responsive protein which functions as a molecular chaperone in stabilizing the cellular proteins from irreversible damage and containing ROS accumulation during seed development particularly during seed maturation, drying and storage. Finally, our results also propose that this gene can be a good candidate to improve seed vigor and longevity in terms of better seed germination and seedling establishment in wide environmental stress conditions.
Funding
This work was supported by the institute core grant from NIPGR, Department of Biotechnology, Government of India.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
HK, BP, NK, VR, PS and SG thank NIPGR, Council of Scientific and Industrial Research and University Grant Commission, Government of India, for research fellowships. Authors acknowledge the proteomics facility and central instrumentation facility, NIPGR, India.
Abbreviations
ABA, abscisic acid; ACD, α-crystallin domain; CDT, controlled deterioration treatment; CS, citrate synthase; DAB, 3,3′-diaminobenzidine tetrahydrochloride; DTNB, 5,5′-dithiobis (2-nitrobenzoic acid); PEG, poly ethylene glycol; sHSP, small heat shock Protein; TZ, 2,3,5-triphenyltetrazolium chloride; VC, vector control; WT, wild type.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00713
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signalling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Agarwal, P., Kapoor, S., and Tyagi, A. K. (2011). Transcription factors regulating the progression of monocot and dicot seed development. Bioessays 33, 189–202. doi: 10.1002/bies.201000107
Allen, R. D., Bernier, F., Lessard, P. A., and Beachy, R. N. (1989). Nuclear factors interact with a soybean beta-conglycinin enhancer. Plant Cell 1, 623–631. doi: 10.1105/tpc.1.6.623
Amin, J., Ananthan, J., and Voellmy, R. (1988). Key features of heat shock regulatory elements. Mol. Cell. Biol. 8, 3761–3769.
Bailly, C. (2004). Active oxygen species and antioxidants in seed biology. Seed. Sci. Res. 14, 93–108. doi: 10.1079/SSR2004159
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Basha, E., O’Neill, H. O., and Vierling, E. (2012). Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem. Sci. 37, 106–117. doi: 10.1016/j.tibs.2011.11.005
Bate, N., and Twell, D. (1998). Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 37, 859–869. doi: 10.1023/A:1006095023050
Berridge, M. V., Tan, A. S., McCoy, K. D., and Wang, R. (1996). The biochemical and cellular basis of cell proliferation assays that use tetrazolium salts. Biochemica 4, 15–19.
Blackman, S. A., Obendorf, R. L., and Leopold, A. C. (1995). Desiccation tolerance in developing soybean seeds: the role of stress proteins. Physiol. Plantarum 93, 630–638. doi: 10.1111/j.1399-3054.1995.tb05110.x
Busk, P. K., Jensen, A. B., and Pages, M. (1997). Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J. 11, 1285–1295. doi: 10.1046/j.1365-313X.1997.11061285.x
Catusse, J., Strub, J. M., Job, C., Van Dorsselaer, A., and Job, D. (2008). Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc. Natl. Acad. Sci. U.S.A. 105, 10262–10267. doi: 10.1073/pnas.0800585105
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coca, M. A., Almoguera, C., and Jordano, J. (1994). Expression of sunflower low-molecular-weight heat-shock proteins during embryogenesis and persistence after germination localization and possible functional implications. Plant Mol. Biol. 25, 479–492. doi: 10.1007/BF00043876
Collada, C., Gomez, L., Casado, R., and Aragoncillo, C. (1997). Purification and in vitro chaperone activity of a class I small heat-shock protein abundant in recalcitrant chestnut seeds. Plant Physiol. 115, 71–77. doi: 10.1104/pp.115.1.71
de Jong, W. W., Caspers, G. J., and Leunissen, J. A. M. (1998). Genealogy of the α-crystallin—small heat-shock protein superfamily. Int. J. Biol. Macromol. 22, 151–162. doi: 10.1016/S0141-8130(98)00013-0
Delouche, J. C., and Baskin, C. C. (1973). Accelerated ageing techniques for predicting the relative storability of seed lots. Seed Sci. Technol. 1, 427–452.
DeRocher, A. E., and Vierling, E. (1994). Developmental control of small heat shock protein expression during pea seed maturation. Plant J. 5, 93–102. doi: 10.1046/j.1365-313X.1994.5010093.x
Dunn, M. A., White, A. J., Vural, S., and Hughes, M. A. (1998). Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.) Plant Mol. Biol. 38, 551–564. doi: 10.1023/A:1006098132352
Ehrnsperger, M., Gräber, S., Gaestel, M., and Buchner, J. (1997). Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 16, 221–229. doi: 10.1093/emboj/16.2.221
Eigentler, A., Draxl, A., Wiethüchter, A., Kuznetsov, A. V., Lassing, B., and Gnaiger, E. (2012). Laboratory protocol: citrate synthase a mitochondrial marker enzyme. Mitochondrial. Physiol. Netw. 17.04, 1–11.
Ellerstrom, M., Stalberg, K., Ezcurra, I., and Rask, L. (1996). Functional dissection of a napin gene promoter: identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol. Biol. 32, 1019–1027. doi: 10.1007/BF00041385
Ericson, M. L., Muren, E., Gustavsson, H.-O., Josefsson, L.-G., and Rask, L. (1991). Analysis of the promoter region of napin genes from Brassica napus demonstrates binding of nuclear protein in vitro to a conserved sequence motif. Eur. J. Biochem. 197, 741–746. doi: 10.1111/j.1432-1033.1991.tb15966.x
Fedoroff, N. (2006). Redox regulatory mechanisms in cellular stress responses. Ann. Bot. 98, 289–300. doi: 10.1093/aob/mcl128
Filichkin, S. A., Leonard, J. M., Monteros, A., Liu, P. P., and Nonogaki, H. (2004). A novel endo-beta-mannanase gene in tomato LeMAN5 is associated with anther and pollen development. Plant Physiol. 134, 1080–1087. doi: 10.1104/pp.103.035998
Gour, P., Garg, P., Jain, R., Joseph, S. V., Tyagi, A. K., and Raghuvanshi, S. (2014). Manually curated database of rice proteins. Nucleic Acids Res. 42, D1214–D1221. doi: 10.1093/nar/gkt1072
Guo, S. J., Zhou, H. Y., Zhang, X. S., Li, X. G., and Meng, Q. W. (2007). Overexpression of CaHSP26 in transgenic tobacco alleviates photoinhibition of PSII and PSI during chilling stress under low irradiance. J. Plant. Physiol. 164, 126–136. doi: 10.1016/j.jplph.2006.01.004
Härndahl, U., Hall, R. B., Osteryoung, K. W., Vierling, E., Bornman, J. F., and Sundby, C. (1999). The chloroplast small heat shock protein undergoes oxidation dependent conformational changes and may protect plants from oxidative stress. Cell Stress Chaperon. 4, 129–138. doi: 10.1379/1466-1268(1999)004<0129:TCSHSP>2.3.CO;2
He, D., and Yang, P. (2013). Proteomics of rice seed germination. Front. Plant Sci. 4:246. doi: 10.3389/fpls.2013.00246
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., et al. (2008). GENEVESTIGATOR V3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 420747. doi: 10.1155/2008/420747
Ingrosso, D., D’Angelo, S., di Carlo, E., Perna, A. F., Zappia, V., and Galletti, P. (2000). Increased methyl esterification of altered aspartyl residues in erythrocyte membrane proteins in response to oxidative stress. Eur. J. Biochem. 267, 4397–4405. doi: 10.1046/j.1432-1327.2000.01485.x
Jiang, C., Iu, B., and Singh, J. (1996). Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 30, 679–684. doi: 10.1007/BF00049344
Kalemba, E. M., and Pukacka, S. (2008). Changes in late embryogenesis abundant proteins and a small heat shock protein during storage of beech (Fagus sylvatica L.) seeds. Environ. Exp. Bot. 63, 274–280. doi: 10.1016/j.envexpbot.2007.12.011
Kaur, H., Shukla, R. K., Yadav, G., Chattopadhyay, D., and Majee, M. (2008). Two divergent genes encoding L-myo-inositol 1 -phosphate synthase1 (CaMIPS1) and 2 (CaMIPS2) are differentially expressed in chickpea. Plant Cell Environ. 31, 1701–1716. doi: 10.1111/j.1365-3040.2008.01877.x
Kim, S. Y., Chung, H. J., and Thomas, T. L. (1997). Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 11, 1237–1251. doi: 10.1046/j.1365-313X.1997.11061237.x
Kizis, D., and Pages, M. (2002). Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J. 30, 679–689. doi: 10.1046/j.1365-313X.2002.01325.x
Lanteri, S., Nada, E., Belletti, P., Quagliotti, L., and Bino, R. J. (1996). Effects of controlled deterioration and osmoconditioning on germination and nuclear replication in seeds of pepper (Capsicum annuum L.). Ann. Bot. 77, 591–597. doi: 10.1093/aob/77.6.591
Lee, G. J., Pokala, N., and Vierling, E. (1995). Structural and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J. Biol. Chem. 270, 10432–10438. doi: 10.1074/jbc.270.18.10432
Lee, G. J., Roseman, A. M., Saibil, H. R., and Vierling, E. (1997). A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16, 659–671. doi: 10.1093/emboj/16.3.659
Lee, G. J., and Vierling, E. (2000). A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 122, 189–197. doi: 10.1104/pp.122.1.189
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van der Peer, Y. R., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal of tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liu, J. G., Qin, Q. L., Zhang, Z., Peng, R. H., Xiong, A. S., Chen, J. M., et al. (2009). OsHSF7 gene in rice, Oryza sativa L., encodes a transcription factor that functions as a high temperature receptive and responsive factor. BMB Rep. 42, 16–21. doi: 10.5483/BMBRep.2009.42.1.016
Mao, Z., and Sun, W. (2015). Arabidopsis seed-specific vacuolar aquaporins are involved in maintaining seed longevity under the control of ABSCISIC ACID INSENSITIVE 3. J. Exp. Bot. 66, 4781–4794. doi: 10.1093/jxb/erv244
Martinez, I. M., and Chrispeels, M. J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15, 561–576. doi: 10.1105/tpc.007609
McDonald, M. B. (1999). Seed deterioration: Physiology, repair and assessment. Seed Sci. Technol. 27, 177–237.
Murakami, T., Matsuba, S., Funatsuki, H., Kawaguchi, K., Saruyama, H., Tanida, M., et al. (2004). Over-expression of a small heat shock protein, sHSP17.7, confers both heat tolerant and UV-B resistant to rice plants. Mol. Breed. 13, 165–175. doi: 10.1023/B:MOLB.0000018764.30795.c1
Neta-Sharir, I., Isaacson, T., Lurie, S., and Weiss, D. (2005). Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. Plant Cell 17, 1829–1838. doi: 10.1105/tpc.105.031914
Oge, L., Bourdais, G., Bove, J., Collet, B., Godin, B., Granier, F., et al. (2008). Protein repair L-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 20, 3022–3037. doi: 10.1105/tpc.108.058479
Oh, D. H., Kwon, C. S., Sano, H., Chung, W. I., and Koizumi, N. (2003). Conservation between animals and plants of the cis-acting element involved in the unfolded protein response. Biochem. Biophys. Res. Commun. 301, 225–230. doi: 10.1016/S0006-291X(02)03019-X
Omar, S. A., Fu, Q. T., Chen, M. S., Wang, G. J., Song, S. Q., and Elsheery, N. I. (2011). Identification and expression analysis of two small heat shock protein cDNAs from developing seeds of biodiesel feedstock plant Jatropha curcas. Plant Sci. 181, 632–637. doi: 10.1016/j.plantsci.2011.03.004
Perez, D. E., Hoyer, J. S., Johnson, A. I., Moody, Z. R., Lopez, J., and Kaplinsky, N. J. (2009). BOBBER1 is a noncanonical Arabidopsis small heat shock protein required for both development and thermotolerance. Plant Physiol. 151, 241–252. doi: 10.1104/pp.109.142125
Prieto-Dapena, P., Castano, R., Almoguera, C., and Jordano, J. (2006). Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol. 142, 1102–1112. doi: 10.1104/pp.106.087817
Rajjou, L., and Debeaujon, I. (2008). Seed longevity: Survival and maintenance of high germination ability of dry seeds. C. R. Biol. 331, 796–805. doi: 10.1016/j.crvi.2008.07.021
Rajjou, L., Lovigny, Y., Groot, S. P. C., Belghazi, M., Job, C., and Job, D. (2008). Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol. 148, 620–641. doi: 10.1104/pp.108.123141
Sarkar, N. K., Kim, Y. K., and Grover, A. (2009). Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10:393. doi: 10.1186/1471-2164-10-393
Saxena, S. C., Salvi, P., Kaur, H., Verma, P., Petla, B. P., Rao, V., et al. (2013). Differentially expressed myo-inositol monophosphatase gene (CaIMP). in chickpea (Cicer arietinum L.). encodes a lithium-sensitive phosphatase enzyme with broad substrate specificity and improves seed germination and seedling growth under abiotic stresses. J. Exp. Bot. 64, 5623–5639. doi: 10.1093/jxb/ert336
Scharf, K. D., Siddique, M., and Vierling, E. (2001). The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperon. 6, 225–237. doi: 10.1379/1466-1268(2001)006<0225:TEFOAT>2.0.CO;2
Silva-Correia, J., Freitas, S., Tavares, R. M., Lino-Neto, T., and Azevedo, H. (2014). Phenotypic analysis of the Arabidopsis heat stress response during germination and early seedling development. Plant Methods 10, 7. doi: 10.1186/1746-4811-10-7
Singh, G., Kumar, S., and Singh, P. (2003). A quick method to isolate RNA from wheat and other carbohydrate-rich seeds. Plant Mol. Biol. Rep. 21, 93a. doi: 10.1007/BF02773401
Stalberg, K., Ellerstom, M., Ezcurra, I., Ablov, S., and Rask, L. (1996). Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 199, 515–519. doi: 10.1007/BF00195181
Sun, W., Bernard, C., Cotte, B. V., Montagu, M. V., and Verbruggen, N. (2001). AtHSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J. 27, 407–415. doi: 10.1046/j.1365-313X.2001.01107.x
Sun, W. N., Montagu, M. V., and Verbruggen, N. (2002). Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta 1577, 1–7. doi: 10.1016/S0167-4781(02)00417-7
Tamarit, J., Cabiscol, E., and Ros, J. (1998). Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J. Biol. Chem. 273, 3027–3032. doi: 10.1074/jbc.273.5.3027
TeKrony, D. M. (1995). “Accelerated aging,” in Seed Vigour Testing Seminar, ed. H. A. van de Venter (Zurich: International Seed Testing Association), 53–73.
Tsvetkova, N. M., Horváth, I., Török, Z., Wolkers, W. F., Balogi, Z., and Shigapova, N., (2002). Small heat-shock proteins regulate membrane lipid polymorphism. Proc. Natl. Acad. Sci. U.S.A. 99, 13504–13509. doi: 10.1073/pnas.192468399
Urao, T., Yamaguchi-Shinozaki, K., Urao, S., and Shinozaki, K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5, 1529–1539. doi: 10.1105/tpc.5.11.1529
van Montfort, R. L., Basha, E., Friedrich, K. L., Slingsby, C., and Vierling, E. (2001). Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 8, 1025–1030. doi: 10.1038/nsb722
Veinger, L., Diamant, L., Buchner, P., and Goloubinoff, P. (1998). The small heat-shock protein IbpB from Escherichia coli stabilizes stress denatured proteins for subsequent refolding by a multichaperone network. J. Biol. Chem. 273, 11032–11037. doi: 10.1074/jbc.273.18.11032
Verma, P., Kaur, H., Petla, B. P., Rao, V., Saxena, S. C., and Majee, M. (2013). PROTEIN L-ISOASPARTYL METHYLTRANSFERASE2 is differentially expressed in chickpea and enhances seed vigor and longevity by reducing abnormal isoaspartyl accumulation predominantly in seed nuclear proteins. Plant Physiol. 161, 1141–1157. doi: 10.1104/pp.112.206243
Volkov, R. A., Panchuk, I. I., and Schöffl, F. (2005). Small heat shock proteins are differentially regulated during pollen development and following heat stress in tobacco. Plant Mol. Biol. 57, 487–502. doi: 10.1007/s11103-005-0339-y
Wehmeyer, N., Hernandez, L. D., Finkelstein, R. R., and Vieling, E. (1996). Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiol. 112, 747–757. doi: 10.1104/pp.112.2.747
Wehmeyer, N., and Vierling, E. (2000). The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol. 122, 1099–1108. doi: 10.1104/pp.122.4.1099
Xue, G.-P. (2002). Characterisation of the DNA-binding profile of barley HvCBF1using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res. 30:e77. doi: 10.1093/nar/gnf076
Yi, S. Y., Lee, D. J., Yeom, S. I., Yoon, J., Kim, Y. H., Kwon, S. Y., et al. (2010). A novel pepper (Capsicum annuum) receptor-like kinase functions as a negative regulator of plant cell death via accumulation of superoxide anions. New Phytol. 185, 701–715. doi: 10.1111/j.1469-8137.2009.03095.x
Keywords: sHSP, chaperone, seed vigor, CDT, stress
Citation: Kaur H, Petla BP, Kamble NU, Singh A, Rao V, Salvi P, Ghosh S and Majee M (2015) Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress. Front. Plant Sci. 6:713. doi: 10.3389/fpls.2015.00713
Received: 09 July 2015; Accepted: 25 August 2015;
Published: 14 September 2015.
Edited by:
Girdhar Kumar Pandey, University of Delhi, IndiaReviewed by:
Sitakanta Pattanaik, University of Kentucky, USASoumendra K. Naik, Ravenshaw University, India
Copyright © 2015 Kaur, Petla, Kamble, Singh, Rao, Salvi, Ghosh and Majee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manoj Majee, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India, manojmajee@nipgr.ac.in
 Harmeet Kaur
Harmeet Kaur Bhanu P. Petla
Bhanu P. Petla Nitin U. Kamble
Nitin U. Kamble Ajeet Singh
Ajeet Singh Venkateswara Rao
Venkateswara Rao Prafull Salvi
Prafull Salvi Shraboni Ghosh
Shraboni Ghosh Manoj Majee
Manoj Majee