- 1Department of Biological Sciences, College of Life Science, Shanxi University, Taiyuan, China
- 2Shanxi Key Laboratory of Crop Genetics and Molecular Improvement, Institute of Crop Science, Shanxi Academy of Agricultural Sciences, Taiyuan, China
- 3Biotechnology Research Insititute, Chinese Academy of Agricultural Sciences, Beijing, China
- 4National Key Facility for Gene Resources and Gene Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 5Triticeae Research Institute, Sichuan Agricultural University, Chengdu, China
- 6Department of Biotechnology, College of Agriculture, Sichuan Agricultural University, Chengdu, China
- 7Beijing Anzhen Hospital Affiliated to the Capital Medical University/Beijing Institute of Heart Lung and Blood Vessel Diseases, Beijing, China
The Auxin/indole-3-acetic acid (Aux/IAA) gene family plays key roles in the primary auxin-response process and controls a number of important traits in plants. However, the characteristics of the Aux/IAA gene family in hexaploid bread wheat (Triticum aestivum L.) have long been unknown. In this study, a comprehensive identification of the Aux/IAA gene family was performed using the latest draft genome sequence of the bread wheat “Chinese Spring.” Thirty-four Aux/IAA genes were identified, 30 of which have duplicated genes on the A, B or D sub-genome, with a total of 84 Aux/IAA sequences. These predicted Aux/IAA genes were non-randomly distributed in all the wheat chromosomes except for chromosome 2D. The information of wheat Aux/IAA proteins is also described. Based on an analysis of phylogeny, expression and adaptive evolution, we prove that the Aux/IAA family in wheat has been replicated twice in the two allopolyploidization events of bread wheat, when the tandem duplication also occurred. The duplicated genes have undergone an evolutionary process of purifying selection, resulting in the high conservation of copy genes among sub-genomes and functional redundancy among several members of the TaIAA family. However, functional divergence probably existed in most TaIAA members due to the diversity of the functional domain and expression pattern. Our research provides useful information for further research into the function of Aux/IAA genes in wheat.
Introduction
Auxin, the first phytohormone discovered, controls many aspects of plant physiology and morphology including embryogenesis, lateral root initiation, leaf expansion, inflorescence and fruit set (Vanneste and Friml, 2009), and is involved in gene stimulation and regulating the transcription of multiple genes on the molecular level. Several primer auxin-responsive genes have been identified containing the Aux/IAA, GRETCHEN HAGEN 3 (GH3), and SMALL AUXIN-UP RNA (SAUR) gene families (Abel and Theologis, 1996). With a key role in the auxin signaling pathway, the Aux/IAA gene is well known as the transcriptional repressor of the Auxin Response Factor (ARF) gene family to regulate downstream auxin-regulated genes (Rogg et al., 2001). Moreover, Aux/IAAs can mediate the pathway interaction between auxin and light signaling (Halliday et al., 2009) or other hormone signaling such as brassinosteroids (Song et al., 2009), jasmonic acid (Kazan and Manners, 2009), and ethylene (Strader et al., 2010).
The Aux/IAA family members encode short-lived nuclear proteins ranging from 18 to 36 kD (Paul et al., 2005). Canonical proteins of the Aux/IAA family share four conserved motifs known as domains I–IV. Domain I at the N-terminus contains a leucine repeat (LxLxLx) motif (Tiwari et al., 2001). Domain II is involved in the instability of proteins (Kepinski and Leyser, 2004). Domains III and IV mediate homo-dimerization and hetero-dimerization between the Aux/IAA and ARF proteins via C-terminal dimerization binding sites (Hagen and Guilfoyle, 2002; Tiwari et al., 2004).
Since the initial isolation of Aux/IAA family genes in soybean (Glycine max) (Walker and Key, 1982), 29 members in Arabidopsis thaliana (Paul et al., 2005), 35 members in Populus trichocarpa (Kalluri et al., 2007), 26 members in tomato (Wu et al., 2012), sorghum (Wang et al., 2010a), and grape (Birsen et al., 2013), and 31 members in rice (Jain et al., 2006), and maize (Wang et al., 2010b) have been identified. Several Aux/IAA family genes control a number of important plant traits. OsIAA2 enhances the resistance of rice to pathogens (Chen et al., 2009); OsIAA5 (Peleg et al., 2011), and OsIAA6 (Jung et al., 2015) are involved in drought tolerance; SbIAA1 relates to stress response (Wang et al., 2010a); and VvIAA4 (Birsen et al., 2013), VvIAA9 (Jung et al., 2014), SlIAA9 (Wang et al., 2005a; Mazzucato et al., 2015), SlIAA15 (Deng et al., 2012), and SlIAA17 (Su et al., 2014) are key regulators of the fruit set process.
Despite extensive studies of Aux/IAA in many other plants, little is known about this gene family in bread wheat (Triticum aestivum L.), one of the most widely grown crops in the world. Until now, only one Aux/IAA gene, TaIAA1, was reported to be regulated by epibrassinolide and light (Singla et al., 2006). Bread wheat (AABBDD; 2n = 6x = 42) is a result of hybridization between T. turgidum (AABB; 2n = 4x = 28), an allotetraploid originating from a cross of T. urartu (AA; 2n = 14) and Aegilops speltoides (SS; 2n = 14), and A. tauschii (DD; 2n = 14) which occurred approximately 0.43 MYA (million years ago). It is problematic to isolate Aux/IAA family genes in bread wheat due to its huge genome that comprises a high scale (>80%) of repetitive sequences (Wicker et al., 2011).
In 2012, the draft genome of bread wheat “Chinese Spring” (“CS”) reading by whole-genome shotgun sequencing was published (Brenchley et al., 2012). Soon afterwards, the sequencing data of T. urartu (Ling et al., 2013) and A. tauschii (Jia et al., 2013), two progenitors of bread wheat, was released in 2013. These resources have provided a wealth of information about the coding genes of wheat (Saintenac et al., 2013). However, little is known about the distribution and position of these genes on each wheat chromosome and their evolution during the two polyploidization events. Recently, a 17-gigabase draft genome of the bread wheat “CS” produced through sequencing isolated chromosome arms was published (Mayer et al., 2014), making possible the isolation and analysis of gene families on a genomic scale.
In this study, a genome-wide isolation of Aux/IAA domains in bread wheat was performed using sequence resources. Eighty-four sequences were isolated and divided into 34 groups based on homology among the A, B and D sub-genomes. Detailed information about wheat Aux/IAA genes (TaIAAs) was acquired. In addition, the expansion pattern and selection pressure analysis of the Aux/IAA family in wheat was deduced.
Materials and Methods
Searching for the Aux/IAA Family in Wheat, T. urartu and A. tauschii
The whole-genome sequences of T. aestivum was downloaded from the wheat genome URGI database (http://wheat-urgi.versailles.inra.fr/) and Ensembl database (http://plants.ensembl.org). Based on these sequences, a local nucleotide and protein database was established by Basic Local Alignment Search Tool (BLAST, ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/). The hidden Markov model (HMM) profile of the Aux/IAA family (PF02309) was extracted from the Pfam database (http://pfam.sanger.ac.uk) and the Aux/IAA HMM profile was used to search the local protein database for target hits with the Aux/IAA-domain by HMMER3.0 (http://hmmer.janelia.org/). All non-redundant sequences with expected values lower than 1E-5 were selected and received a conserved domain check using the Pfam tool (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/) web server. According to the sequence ID of the Aux/IAA protein in wheat, the coding sequences and genome sequences were isolated from the local nucleotide database. Using the same method, the Aux/IAA genes of T. urartu and A. tauschii were defined from the T. urartu genomic database (http://gigadb.org/dataset/100050) and A. tauschii genomic database (http://gigadb.org/dataset/100054), respectively.
Phylogenetic Relationships, Gene Structure, and Chromosome Location of Wheat Aux/IAA Genes
The exon/intron organization of each Aux/IAA gene was illustrated in the Gene Structure Display Server program (Hu et al., 2015) by comparing their coding sequences with genomic sequences. The position of each Aux/IAA gene in the wheat chromosomes and the genome sequences of the corresponding wheat chromosomes were determined by BLAST, and the results were displayed using the MapInspect tool (http://mapinspect.software.informer.com/). An unrooted phylogenetic tree for wheat Aux/IAA genes was constructed using MEGA 6.0 (Tamura et al., 2013) via the Neighbor-Joining (NJ) method. Duplicated genes in the branch ends of each group belonging to the A, B or D sub-genomes of wheat were regarded as the homologous copies of one Aux/IAA gene. All of the TaIAAs were named according to their chromosome position and homology among the three wheat genomes.
Physical and Chemical Properties, Secondary Structure Prediction, Motif Display, and Phylogenetic Analysis for the Wheat Aux/IAA Proteins
The basic physical and chemical parameters of TaIAA proteins were predicted by the ProtParam tool (http://www.expasy.org/tools/protparam.html). The secondary structures of TaIAAs were analyzed by the net service NPS (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html). The conserved domains were investigated using Clustal W (Larkin et al., 2007) by multiple alignment analyses and visualized in Jalview (Waterhouse et al., 2009). Motifs of the TaIAA proteins were displayed with the MEME (Bailey et al., 2009) online tool. MEME found four motifs and the other parameters were defaulted. The phylogenetic analysis of Aux/IAA proteins was performed using the NJ method of MEGA 6.0. The protein sequences of other species such as OsIAAs, AtIAAs, SlIAAs, and VvIAAs were download from the NCBI database (http://www.ncbi.nlm.nih.gov/) according to the Genbank number.
Expression Analysis for TaIAAs
The coding sequences of the TaIAAs were submitted to the Plex database (http://www.plexdb.org/) to search for corresponding probes. These probes were then used to query the GeneVestigator database (https://genevestigator.com/gv/) to obtain the expression data of TaIAA genes in 19 organs of wheat. All transcript data was transformed by log2 and the heat map was viewed in the MeV tool (http://www.tm4.org/mev.html).
Expansion Pattern Analysis for TaIAAs
Segmental replication and tandem duplication are the main repeat methods of the gene family among the lineage-specific expansion. In this study, 84 genomic sequences of TaIAAs were investigated for segmental replication and tandem duplication events according to the method established in previous studies (Guo and Qiu, 2013). Paralogs from different sub-genomes of wheat were regarded as segmental replication, while two or more TaIAAs situated in the same location of the chromosome were defined as tandem duplication, based on the wheat genome annotation results in the URGI database.
Collinearity Analysis for the Aux/IAA Family
Phylogenetic trees for the wheat-T. urartu and wheat-A. tauschii Aux/IAA genes were constructed in MEGA 6.0. Two genes from different species situated in the same branch of the phylogenetic tree were designated orthologs (Koonin, 2005). Based on these orthologous Aux/IAA genes, collinearity maps of the Tu-wheat A genome and the Aet-wheat D genome were output by the genome visualization tool CIRCOS (Krzywinski et al., 2009).
Ka and Ks Calculations
The rate of Ka (non-synonymous substitution rate)/Ks (synonymous substitution rate) was applied to compare the rates of codon evolution in the sub-genome of wheat using barley as an outgroup (Akhunov et al., 2013). The orthologous gene pairs between barley and wheat were used to calculate Ka and Ks in the PAL2NAL server (Suyama et al., 2006) using the codeml program of phylogenetic analysis by maximum likelihood (PAML; Yang, 1997). The barley Aux/IAA sequences were downloaded from the PlantTF database (http://planttfdb_v1.cbi.pku.edu.cn:9010/).
Results
Searching for Aux/IAA Genes in Wheat
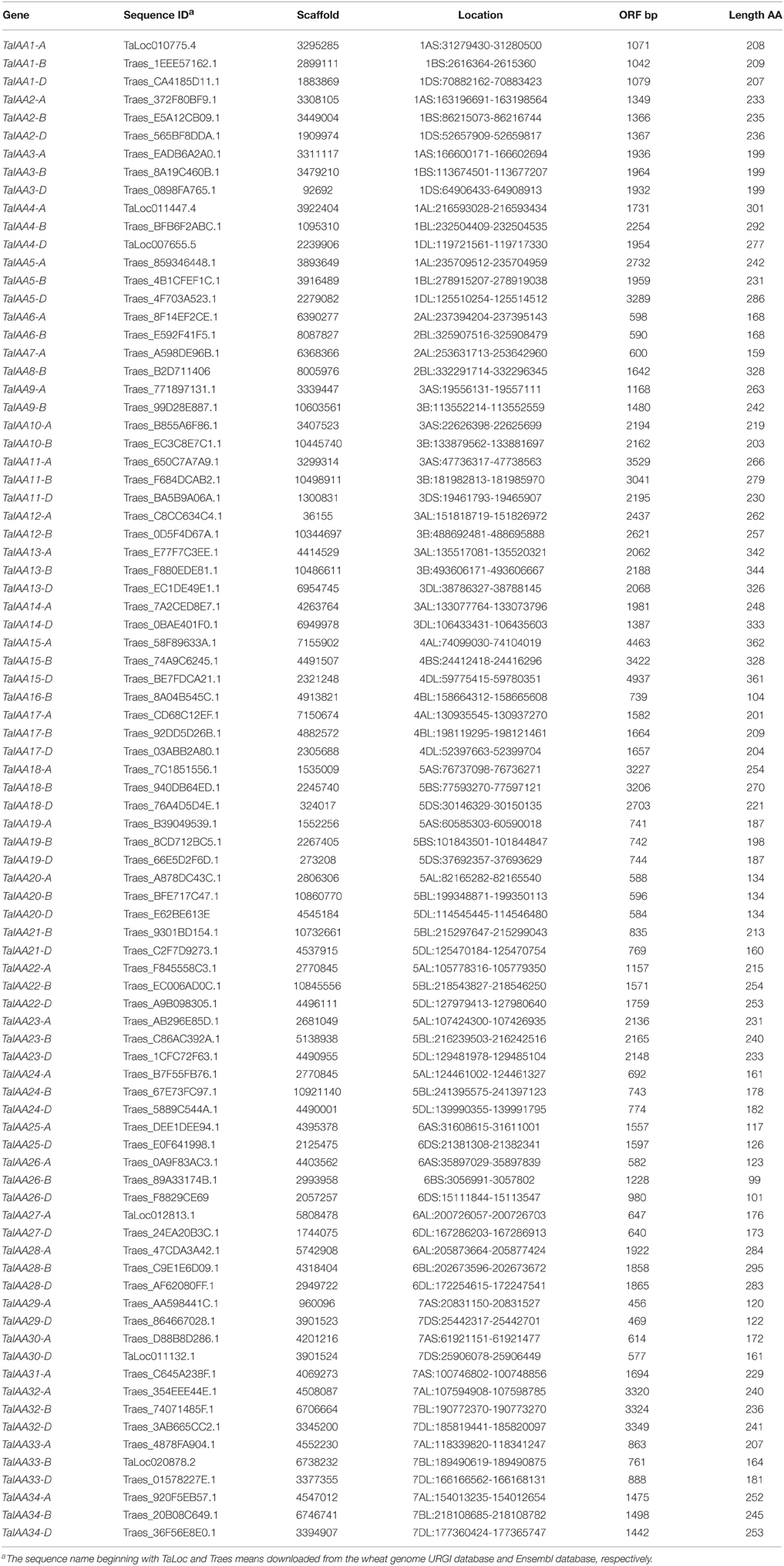
A 22.8 GB local database of wheat nucleotide and protein sequences was built. By retrieving wheat protein sequence databases using the Aux/IAA HMM file, 127 non-redundant sequences were obtained. Next, 84 full-length protein sequences were detected that contained conserved Aux/IAA domains. In addition, corresponding coding sequences and genome sequences were isolated from the local nucleotide database and each Aux/IAA gene was located by searching the wheat chromosome genomic sequences using BLASTn (Table 1). An un-rooted tree of the 84 Aux/IAA full-length coding sequences was constructed (Figure 1A) to reveal the phylogenetic relationship and all the sequences were divided into 34 groups. Among them, 30 groups have two or three genes from different wheat sub-genomes, and these were regarded as different copies of one member of the Aux/IAA gene family.
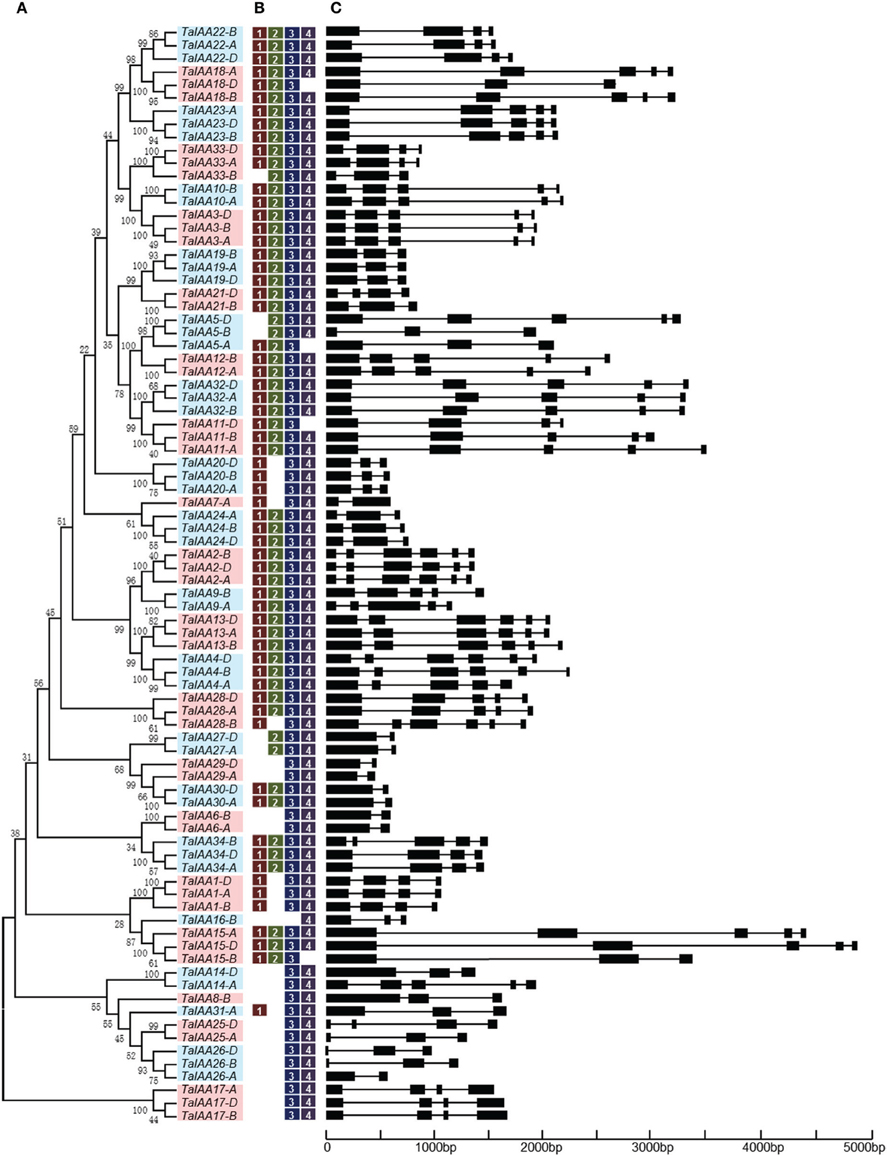
Figure 1. Phylogenetic relationship, motif and gene structure of wheat Aux/IAA genes. (A) The phylogenetic tree of TaIAAs constructed from a complete alignment of 84 wheat Aux/IAA genes using MEGA 6.0 by the neighbor-joining method with 1000 bootstrap replicates. Bootstrap scores are indicated on the nodes and the 34 members of TaIAA, most of which contain duplicated genes, are indicated by blue or pink block. (B) The conserved motifs of TaIAAs. Motifs were identified by MEME software using the deduced amino-acid sequences of the TaIAAs. The relative position of each identified motif in all TaIAA proteins is shown as 1, 2, 3, and 4, which represented the four conserved domains of IAA. (C) Exon/intron structures of TaIAA genes. Exons are represented by black boxes and introns by black lines. The sizes of exons and introns can be estimated using the scale below.
Genome Distribution of Wheat Aux/IAA Genes
The genome distribution of 84 wheat Aux/IAA genes is shown in Figure 2. Respectively, 31, 27, and 26 Aux/IAA genes are non-randomly distributed in the three wheat sub-genomes. According to chromosome position and genomic homology, all these wheat Aux/IAA genes (TaIAAs) were named TaIAA1-A~TaIAA34-D and distributed on every wheat chromosome except for chromosome 2D (Figure 2). Each of the 20 TaIAA genes (TaIAA1, 2, 3, 4, 5, 11, 13, 15, 17, 18, 19, 20, 22, 23, 24, 26, 28, 32, 33, and 34) contains three copies in chromosome A, B and D; 10 TaIAAs have two copies each including TaIAA-A/-B (TaIAA6, 9, 10, and 12), TaIAA-B/-D (TaIAA21), and TaIAA-A/-D (TaIAA14, 25, 27, 29 and 30); and TaIAA7, 8, 16, and 31 have just one copy in wheat chromosomes.
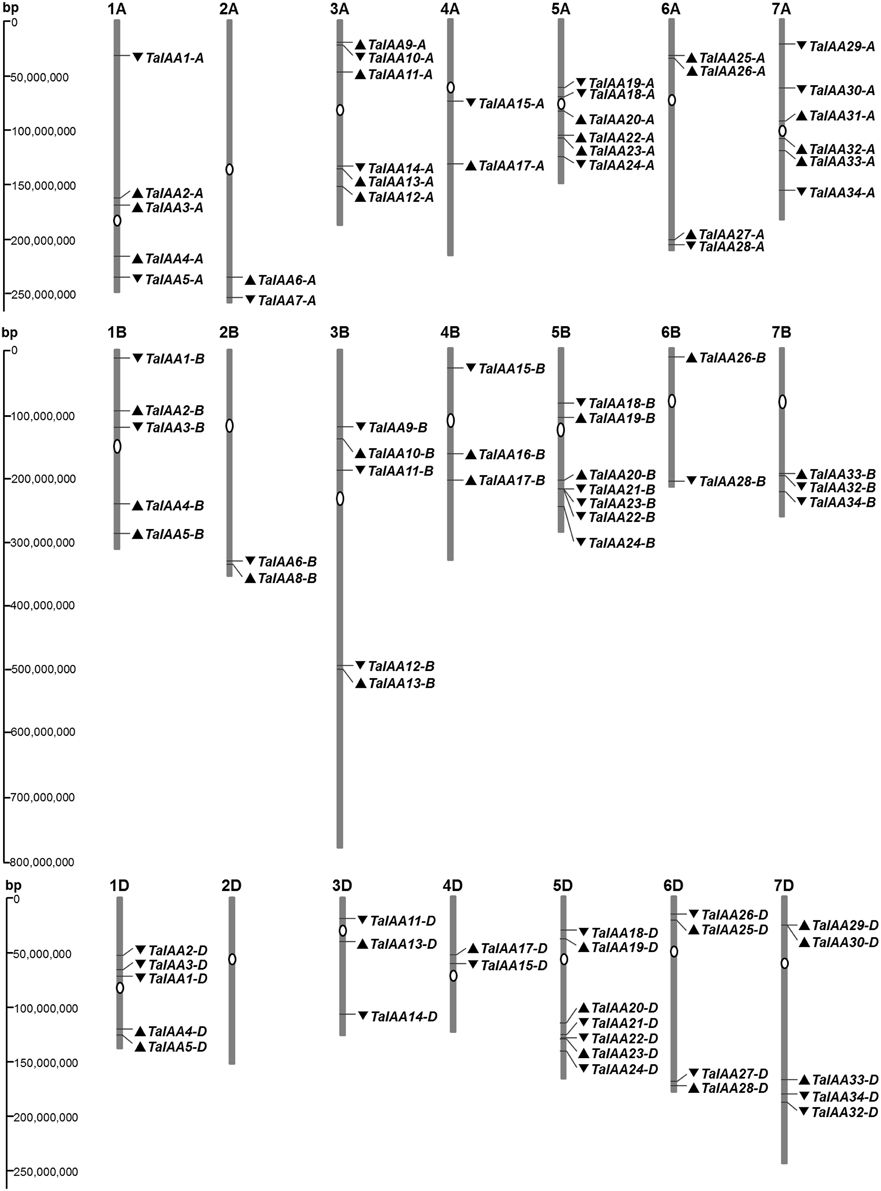
Figure 2. Chromosome distribution of TaIAA family in wheat. White ovals on the chromosomes (vertical bar) indicate the position of centromeres. The arrows next to gene names show the direction of transcription. The position of each gene can be estimated using the left scale. The chromosome numbers are indicated at the top of each bar.
There is a high homology among the scaffolds of one TaIAA member which belongs to the A, B or D sub-genome (Figure 3), proving that the TaIAA genes experienced two segmental replication events in wheat except for the four single-copy TaIAA genes. However, it could also be the case that the segmental replication genes of the four TaIAAs were lost after the expansion event. Furthermore, three genomic loci containing two TaIAA genes each were defined in the A sub-genome, and the B or D sub-genomes have four such loci (Supplementary Table 1), implying that tandem duplication is also an expansion pattern of the TaIAA gene family.
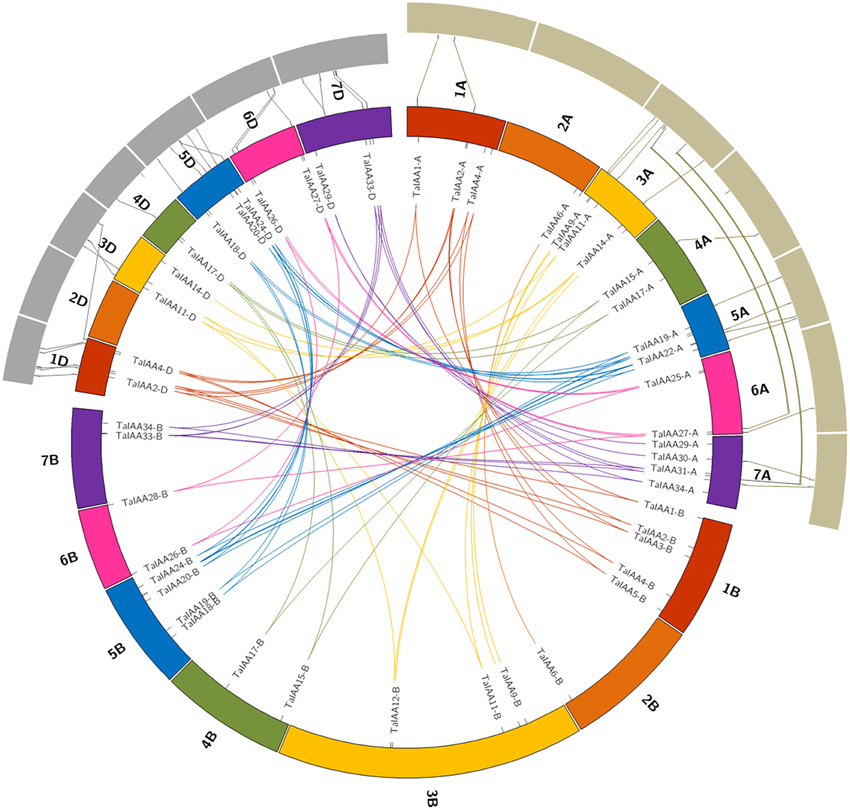
Figure 3. Duplicated TaIAA genes of wheat homologous groups and the collinearity among TaIAAs, TuIAAs, and AetIAAs. Seven homologous groups of wheat chromosomes are displayed in different colors. Duplicated genes of each homo-group are linked using lines with corresponding color. The gray annulus on the periphery represents chromosomes of T. urartu and A. tauschii. The collinearity among TaIAAs, TuIAAs, and AetIAAs were signified by the lines between the orthologous gene-pairs.
Gene Structure of Wheat Aux/IAA Genes
Schematics of gene structures generated by the GSDS utility are shown in Figure 1C. In the TaIAA gene family, the number of introns ranges from 1 to 5. Among the 30 TaIAAs which contains two or three copies, 17 TaIAA genes (TaIAA1, 2, 3, 6, 10, 12, 13, 17, 19, 20, 22, 23, 24, 27, 29, 30, and 32) have the same gene structures, and the remaining 13 have only one or two differences in the number of introns in each group. Overall, a highly similar gene structure is exhibited in the same phylogenetic cluster of the TaIAA genes, suggesting that duplicated genes may have the same function.
Characteristics of TaIAA Protein Sequences
Some physical and chemical properties of wheat Aux/IAA proteins are shown in Supplementary Table 2. The predicted molecular mass varies from 21.9 kD for TaIAA1-A to 37.9 kD for TaIAA15-A amongst the Aux/IAA proteins. The negative grand average of the hydropathicity (GRAVY) index showed that all wheat Aux/IAA polypeptides are hydrophilic except TaIAA26-A, suggesting that they are more likely nucleoproteins than membrane proteins. In addition, 69 out of 84 (82.1%) wheat Aux/IAA proteins possess a stability index of more than 40 and might be unstable in vitro.
The results of multiple alignment and motif distribution analyses of TaIAA proteins showed that 48 out of 84 (57.1%) wheat Aux/IAA proteins contain the same domains, known as domains I, II, III and IV (Supplementary Figure 1). Motifs 1, 2, 3, and 4 are located in these four domains, respectively (Figures 1B, 4). Eighteen (21.4%) wheat Aux/IAA proteins missed one domain (I, II, or IV), while 17 (20.2%) proteins missed domains I & II; just one (1.2%) protein, TaIAA16-B, lacked three domains (I & II & III). Generally, TaIAA proteins within the same phylogenetic group have similar domains and motifs. Domain I (Motif 1) contains the LxLxLx motif, a typical leucine-rich region that was shown in most TaIAA proteins, which has been shown to act as a strong transcriptional repressor (Tiwari et al., 2004). Domain II contains VGWPP, the core sequence of the target site for wheat Aux/IAA protein degradation. Dominant mutation in this region causes Aux/IAA proteins to fail to resolve via the ubiquitin pathway (Kepinski and Leyser, 2005). Domains III and IV are comparatively more conserved. A βαα structure existing in domain III (Motif 3) appeared among all the TaIAA proteins except TaIAA16-B (Figure 1B). It was found that this fold plays an important role in the dimerization of Aux/IAA proteins. Most of the wheat Aux/IAA proteins include two hypothetical nuclear localization signals (NLS). The first bipartite NLS is comprised of two elements: one between domains I and II, and the other in domain II (Supplementary Figure 1). The second element, the SV40-type NLS, is located in domain IV (Supplementary Figure 1). The possible function of these NLSs may be to transfer TaIAA proteins into the nucleus. Interestingly, the conserved residue in the bipartite NLS is KP in TaIAA proteins, while it is KR in rice Aux/IAA proteins (Jain et al., 2006). Therefore, further study into the bipartite NLS in wheat is required. In addition, the existence of phosphorylation sites in several TaIAA proteins partly indicates that these proteins can be extrapolated as short-lived proteins (Supplementary Figure 1).
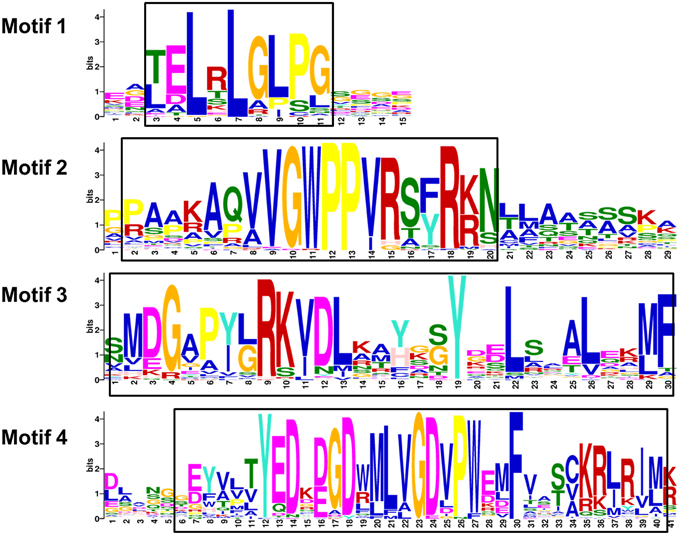
Figure 4. Motifs of wheat Aux/IAA proteins. The sequence logos are based on multiple alignment analysis of 84 wheat Aux/IAA proteins performed with Clustal W. The bit score indicates the information content for each position in the sequence. Positions of conserved domains are boxed.
Phylogenetic Analysis of TaIAA Proteins
The phylogenetic tree was built with the sequences of 159 Aux/IAA proteins, including 84 TaIAAs, 31 OsIAAs, 29 AtIAAs, and 15 Aux/IAA proteins with known functions from other species. The information of all the above sequences is listed in Supplementary Table 3.
The phylogenetic tree shows that TaIAA proteins can be classified into two major groups: A and B (Figure 5). Based on the clustering tree of OsIAAs (Jain et al., 2006), groups A and B can be further separated into several subgroups: A1, A3, and A4 contain monocotyledon IAA proteins; A5 contains dicotyledon proteins; and A2, B1, B2, B3, and B4 are all types of IAA protein. Moreover, monocot and dicot IAA proteins are not gathered for one class in every subgroup, while the paralogous proteins of each TaIAA are clustered to the same branch, similar to the clustering result in Figure 1A. Orthologous genes usually have similar biological functions (Li et al., 2005). In group A1, OsIAA5 was induced by drought (Peleg et al., 2011), suggesting that TaIAA12 may be related to drought resistance. In addition, OsIAA2 and OsIAA6 in group B1 were induced by pathogens (Chen et al., 2009), OsIAA1 in group A2 regulates plant type (Song et al., 2009), SbIAA1 in group A3 may be related to stress response (Wang et al., 2010a), and OsIAA4 in group B2 regulates plant tiller (Song and Xu, 2013), implying that their orthologs, such as TaIAA1, 9, 10, 13, and 24, may have similar functions in wheat.
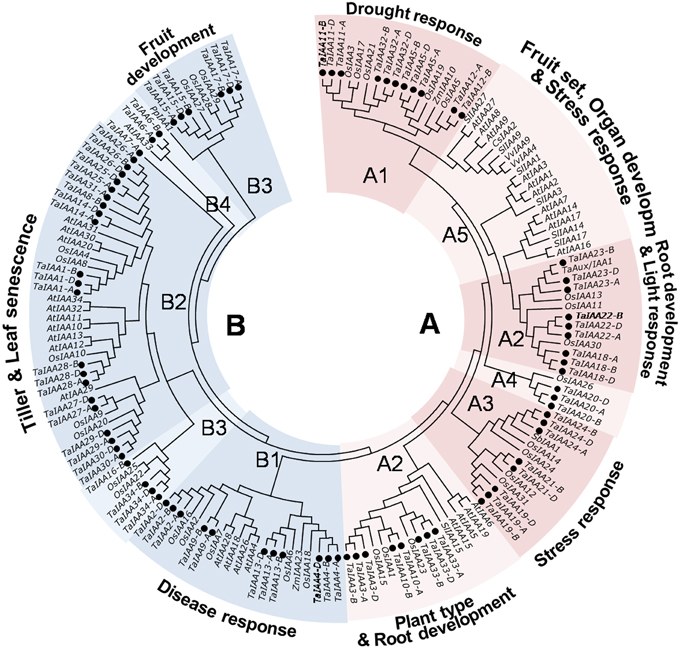
Figure 5. Phylogenetic relationship of Aux/IAA proteins among wheat and another species. The full-length amino-acid sequences of 85 wheat, 31 rice, 29 Arabidopsis, 7 tomato, 2 maize, 2 grape, 1 sorghum, and 1 pear genes were aligned by Clustal W and the phylogenetic tree was constructed using MEGA 6.0 by the neighbor-joining method with 1000 bootstrap replicates. Each TaIAA protein is indicated by a black dot. Two major groups, group A and B, are represented by the red and blue. The functions of some clades were annotated.
Expression Pattern of TaIAAs in Wheat
The coding sequences of 34 TaIAAs were used to search the Plex database for corresponding probes. All gene copies of each TaIAA gene share one probe. The probes of TaIAA20 and TaIAA34 were not found. Several genes have the same probe, including TaIAA2, 9; TaIAA18, 22, 33; and TaIAA29, 30. Finally, 28 probes for 32 TaIAAs were obtained from the GeneVestigator database (Supplementary Table 4). The expression profile of 32 TaIAAs in 19 wheat organs covering the seedling to adult stage is shown as a heat map (Figure 6). TaIAAs were divided into eight classes according to the subgroups of phylogenetic analysis. In general, the majority of genes in groups A3, B1, B2, and B3 are expressed in vegetative organs of wheat, and genes in group A3 are also expressed in the pistil and embryo. Moreover, all genes in A1, A2 and TaIAA14 of B2 have high expression levels in the vast majority of wheat organs throughout the entire growing stage. In addition, TaIAA6 and TaIAA27 are specifically expressed in the roots, while TaIAA16, TaIAA28, and TaIAA31 are specifically expressed in inflorescence, flag leaf and seed, respectively. TaIAA1, 7, 10, 17, 29, and 30 showed low expression in wheat.
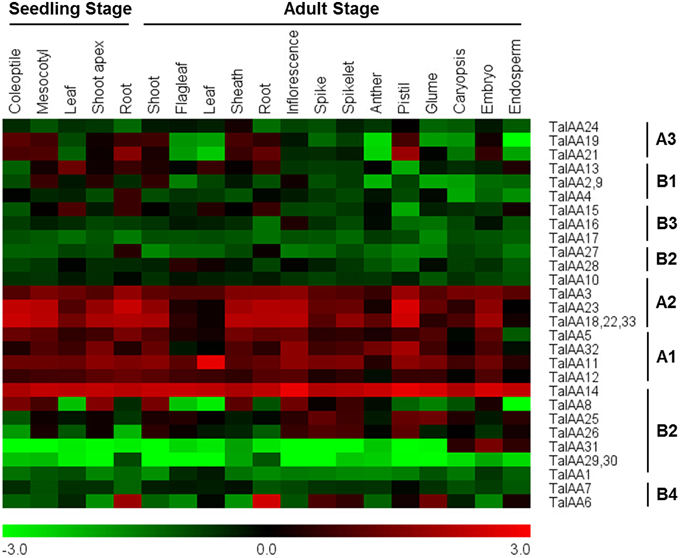
Figure 6. Heatmap of expression profiles for TaIAA genes across different organs of seedling and adult stages. The expression data were generated from GeneVestigator database and viewed in MeV software. The relative expression level of a particular gene in each row was normalized against the mean value by log2 transformation. The color scale below represents expression values, green indicating low levels and red indicating high levels of transcript abundance.
Synchronic Analyses of Aux/IAA Families among T. urartu, A. tauschii, and Wheat
Twenty-seven TuIAA, 31 TaIAA-A, 28 AetIAA, and 26 TaIAA-D sequences were used to construct the phylogenetic tree, and pairs of 22 and 21 Ta-A/Tu and Ta-D/Aet orthologs were identified (Supplementary Figure 2). The synchronic analysis results of 16 pairs (just 16 of the 27 TuIAA sequences have chromosome location information (Supplementary Table 5) of TuIAA and TaIAA-A showed that there are 14 (87.5%) homologous genes located on the same chromosome, including 1A, 3A, 4A, 5A, and 6A, while 20 of the 21 pairs (95.2%) between AetIAA and TaIAA-D are located on 1D, 3D, 4D, 5D, 6D, and 7D (Figure 3). In general, there is a good collinearity in Aux/IAA families among T. urartu, A. tauschii, and wheat, which suggests that the evolution of the Aux/IAA family has been conservative following the formation of hexaploid wheat. However, TaIAA27-A and TaIAA32-A on wheat chromosomes 6AL and 7AL, respectively correspond to chromosome 3A of T. urartu, and TaIAA5-D on wheat chromosome 1DL corresponds to chromosome 3DL in A. tauschii. Furthermore, a difference in chromosome location caused by pericentric also exists in several orthologous gene pairs, indicating that chromosomal inversion and crossover have occurred in the evolutionary process of the Aux/IAA family in wheat.
Adaptive Evolution Analysis of the TaIAA Family
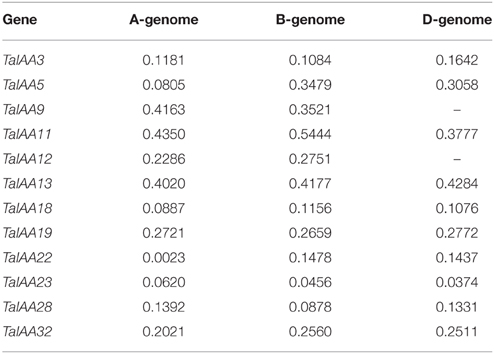
To determine which type of Darwinian selection decided the process of gene divergence after duplication, the Ka/Ks substitution ratio was applied to the coding sequences of 12 pairs of orthologs between barley and wheat Aux/IAA family (Supplementary Figure 3). In general, Ka/Ks ratio > 1 means positive selection, ratio < 1 means purifying selection and ratio = 1 means neutral evolution (Akhunov et al., 2013). The Ka/Ks ratios were always less than 1 for TaIAAs and ranged from 0.0023 to 0.5444 (Table 2), suggesting that the Aux/IAA family has undergone purifying selection in wheat.
Discussion
Genome-wide Isolation of Gene Families in Hexaploid Bread Wheat
Sequencing projects provide an opportunity for the isolation of gene families in a genome-wide scan. However, it is a great challenge for analysis of polyploid genomes because the relatedness of homeologous sub-genome sequences makes it difficult to assign isolated sequences to the specific chromosome from which they are derived. Until now, some gene families, such as WRKY genes (Okay et al., 2014) and nucleotide-binding site (NBS) domain-containing genes (Bouktila et al., 2015), were isolated in wheat. Yet the position of these family genes on wheat genomes and their homologous relationship was still unknown, which leads to the studies on functional divergence and redundancy of duplicated genes could not be proceeded. So compared with rice or maize, the genomic research into hexaploid wheat has been slow for a long time in crops. With the benefit of the new bread wheat draft genome recently through sequencing isolated chromosome arms, we isolated the Aux/IAA gene family in wheat. Unlike previous studies, all the TaIAA genes were mapped in homoeologous chromosome groups and sequences in the same group of the phylogenetic tree were considered the duplicated genes of one TaIAA member. Based on this perspective, the following analysis reflects some special duplicable and functional characteristics of the Aux/IAA gene family in wheat.
Expansion and the Fate of Duplicated Aux/IAA Family Genes in Wheat
In this study, we isolated 27 and 28 Aux/IAA genes from the genomes of T. urartu and A. tauschii, respectively, which are similar in their number of Aux/IAA family genes with tomato (26), sorghum (26), A. thaliana (29), rice (31), and maize (31). We deduce that the allopolyploidization event that crossed T. urartu and A. speltoides resulted in the Aux/IAA family being replicated in T. turgidum. This tetraploid emmer wheat subsequently hybridized with A. tauschii, resulting in the second replication of the Aux/IAA family in hexaploid bread wheat. The tandem duplication also occurred via this process, eventually forming the 84 wheat Aux/IAA sequences including 31, 27, and 26 genes from the A, B, and D sub-genomes of wheat, respectively. A good collinearity of Aux/IAAs was shown between wheat and the A and D genome donors T. urartu and A. tauschii (Figure 3).
After the expansion of Aux/IAAs in wheat, the duplicated genes underwent an evolutionary process of purifying selection that can be inferred by their Ka/Ks ratios (Table 2). Therefore, the duplicated genes from the A, B, or D sub-genomes of wheat have a high conservation and share one probe in the expression database (Figure 6). However, the inversion and crossover also happened in wheat chromosomes during the evolutionary process, causing the different fates of several duplicated Aux/IAA genes. One such fate was the loss of duplicated genes such as TaIAA14-B or TaIAA17-B (Figure 1). Moreover, the positions of some duplicated genes changed, involving not only the shift of linear array location and transcription direction (Figure 2), but also the exchange of position between two chromosomes. Based on the collinearity of Aux/IAA genes between T. urartu and wheat, we infer that TaIAA27 and TaIAA32 were originally on chromosome 3A and then transferred to chromosome 6A and 7A, respectively by interchromosomal translocation (Figure 3). The translocation phenomenon has appeared in previous research (Salse et al., 2008; Ma et al., 2013). In addition, the other important peculiar feature of duplicated Aux/IAA genes is the altering of the conserved motif, including degenetation and neofunctionalization according to Yang's view (Yang et al., 2006). For example, TaIAA11-A, TaIAA11-B, the orthologs of the D subgenome and AEGTA27928 all contain the four motifs of the Aux/IAA family, while TaIAA11-D lacks motif 4. Other duplicated genes obtained a new motif. Compared with TaIAA5-A, the motifs of TaIAA5-B and TaIAA5-D transform Motif 1 + 2 + 3 to Motif 2 + 3 + 4, which may bring out a novel function.
Functional Divergence of TaIAAs
Based on the phylogenetic tree of Aux/IAA proteins (Figure 5), we infer that the Aux/IAA genes divided before the split of monocots and dicots (90 MYA). Later, the second differentiation happened in a lineage-specific manner in the rice lineage and Triticeae before the divergence of Triticum and Aegilops (3~40 MYA). Therefore, the wheat Aux/IAA family has experienced at least two changes.
Despite several genes, including TaIAA29 and TaIAA30, that may have a functional redundancy because of their identical expression patterns (Figure 6), the functions of most TaIAAs are probably diverse due to the diversity of the functional domain and expression pattern.
In this study, TaIAA27 did not contain domain I and the protein may function differently to the classical IAA repressors. Reference on the research in Arabidopsis (Dreher et al., 2006), absent of Domain II in TaIAA proteins may lead to enhanced auxin responses by blocking the degradation of IAA proteins, such as TaIAA14. However, TaIAA1 and TaIAA7 present a very low expression level in various organs throughout the growing period of wheat, suggesting that the degradation of these proteins were independent on domain II- mediate auxin responses. Moreover, TaIAA20, which also lacks domain II, has not had any expression data detected at all, implying that TaIAA20 might be a pseudogene like some AtIAAs (Reed, 2001). In addition, TaIAA34 similarly has no expression data but it does have all four domains, which implies that TaIAA34 is expressed through a particular pattern according to similar research into OsIAAs (Jain et al., 2006) and ZmIAAs (Wang et al., 2010b).
The genes in groups A1 and A2 of the phylogenetic tree exercise an important function in terms of root development and responding to the environment. Some TaIAAs in these two groups are highly expressed in various organs and involved in almost the entire growth process of wheat. Among them, TaIAA11 from group A1 is highly expressed in leaves, as well as its orthologous OsIAA3 (Nakamura et al., 2006), which shows that TaIAA11 play a key role in leaf development. TaIAA23 from group A2 shows higher expression levels in leaves and roots than in other wheat organs, and TaIAA23-B shows a sequence similarity as high as 95.3% with TaAux/IAA1 (AJ575098) which is sensitive to light and induced by auxin and brassinosteroids (Singla et al., 2006). In addition, TaIAA27 and TaIAA31 exhibit tissue-specific expression in roots and seeds, respectively.
Conclusion
Bread wheat is an important worldwide crop with a huge genome and highly repetitive transposable elements. In summary, 34 Aux/IAA genes including 84 duplicated genes in total were isolated from the wheat genome and located in 41 wheat chromosomes (except chromosome 2D). The TaIAA family has been replicated twice in the two allopolyploidization events of bread wheat, when the tandem duplication also occurred. The duplicated genes have undergone an evolutionary process of purifying selection, resulting in the high conservation of copy genes among the sub-genomes and functional redundancy among several members of the TaIAA family. However, functional divergence probably existed in most TaIAA members due to the diversity of the functional domain and expression pattern.
Author Contributions
Conceived and designed the experiments: XyL, ZC, LQ. Performed the experiments: LQ, XZ, HZ, XL, WZ, LC, PL. Analyzed the data: LQ. Contributed reagents/materials/analysis tools: XyL, LQ, JM, XH. Wrote the manuscript: LQ, LZ, ZC.
Funding
This study is funded by National Natural Science Foundation of China (31171839), Shanxi Province Science Foundation for Youths (2015021145), Technologies R & D Program of the Shanxi Academy of Agricultural Sciences (15YGG01), The Key Program of the Shanxi Academy of Agricultural Sciences (YZD1501), Shanxi Province Technologies R & D Progaram (20150311001-1), Shanxi Province S & T Infrastructure Development Program (2015012001-13), and Shanxi Province finance supported agricultural projects (2014ZYFZ-03).
Conflict of Interest Statement
The Associate editor, Dr Tiegang Lu, declares that despite being affiliated with the same institution as the co-author Dr Xiao Han, the review process was handled objectively. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00770
References
Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. doi: 10.1104/pp.111.1.9
Akhunov, E. D., Sehgal, S., Liang, H., Wang, S., Akhunova, A. R., Kaur, G., et al. (2013). Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol. 161, 252–265. doi: 10.1104/pp.112.205161
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Birsen, C., Ozan, K., and Ahmet, C. O. (2013). Genome-wide analysis of Aux/IAA genes in Vitis vinifera: cloning and expression profiling of a grape Aux/IAA gene in response to phytohormone and abiotic stresses. Acta. Physiol. Plant. 35, 365–377. doi: 10.1007/s11738-012-1079-7
Bouktila, D., Khalfallah, Y., Habachi-Houimli, Y., Mezghani-Khemakhem, M., Makni, M., and Makni, H. (2015). Full-genome identification and characterization of NBS-encoding disease resistance genes in wheat. Mol. Genet. Genomics 290, 257–271. doi: 10.1007/s00438-014-0909-2
Brenchley, R., Spannagl, M., Pfeifer, M., Barker, G. L., D'Amore, R., Allen, A. M., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710. doi: 10.1038/nature11650
Chen, S. Y., Li, X. Q., Zhao, A. G., Wang, L. J., Li, X. F., Shi, Q., et al. (2009). Genes and pathways induced in early response to defoliation in rice seedlings. Curr. Issues Mol. Biol. 11, 81–100.
Deng, W., Yang, Y., Ren, Z., Audran-Delalande, C., Mila, I., Wang, X., et al. (2012). The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytol. 194, 379–390. doi: 10.1111/j.1469-8137.2012.04053.x
Dreher, K. A., Brown, J., Saw, R. E., and Callis, J. (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18, 699–714. doi: 10.1105/tpc.105.039172
Guo, Y., and Qiu, L. J. (2013). Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 8:e76809. doi: 10.1371/journal.pone.0076809
Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. doi: 10.1023/A:1015207114117
Halliday, K. J., Martínez-García, J. F., and Josse, E. M. (2009). Integration of light and auxin signaling. Cold Spring Harb. Perspect Biol. 1:a001586. doi: 10.1101/cshperspect.a001586
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jain, M., Kaur, N., Garg, R., Thakur, J. K., Tyagi, A. K., and Khurana, J. P. (2006). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 6, 47–59. doi: 10.1007/s10142-005-0005-0
Jia, J. Z., Zhao, S. C., Kong, X. Y., Li, Y. R., Zhao, G. Y., He, W., et al. (2013). Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496, 91–95. doi: 10.1038/nature12028
Jung, C. J., Hur, Y. Y., Yu, H. J., Noh, J. H., Park, K. S., and Lee, H. J. (2014). Gibberellin application at pre-bloom in grapevines down-regulates the expressions of VvIAA9 and VvARF7, negative regulators of fruit set initiation, during parthenocarpic fruit development. PLoS ONE 9:e95634. doi: 10.1371/journal.pone.0095634
Jung, H., Lee, D. K., Choi, Y. D., and Kim, J. K. (2015). OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 236, 304–312. doi: 10.1016/j.plantsci.2015.04.018
Kalluri, U. C., Difazio, S. P., Brunner, A. M., and Tuskan, G. A. (2007). Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 7:59. doi: 10.1186/1471-2229-7-59
Kazan, K., and Manners, J. M. (2009). Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 14, 373–382. doi: 10.1016/j.tplants.2009.04.005
Kepinski, S., and Leyser, O. (2004). Auxin-induced SCF-TIR1-Aux/IAA interaction involves stable modification of the SCF/TIR1 complex. Proc. Natl. Acad. Sci. U.S.A. 101, 12381–12386. doi: 10.1073/pnas.0402868101
Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. doi: 10.1038/nature03542
Koonin, E. V. (2005). Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39, 309–338. doi: 10.1146/annurev.genet.39.073003.114725
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, W. H., Yang, J., and Gu, X. (2005). Expression divergence between duplicate genes. Trends Genet. 21, 602–607. doi: 10.1016/j.tig.2005.08.006
Ling, H. Q., Zhao, S., Liu, D., Wang, J., Sun, H., Zhang, C., et al. (2013). Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496, 87–90. doi: 10.1038/nature11997
Ma, J., Stiller, J., Berkman, P. J., Wei, Y., Rogers, J., Feuillet, C., et al. (2013). Sequence-based analysis of translocations and inversions in bread wheat (Triticum aestivum L.). PLoS ONE 8:e79329. doi: 10.1371/journal.pone.0079329
Mayer, K. F. X., Rogers, J., Doležel, J., Pozniak, C., Eversole, K., Feuillet, C., et al. (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788. doi: 10.1126/science.1251788
Mazzucato, A., Cellini, F., Bouzayen, M., Zouine, M., Mila, I., Minoia, S., et al. (2015). A TILLING allele of the tomato Aux/IAA9 gene offers new insights into fruit set mechanisms and perspectives for breeding seedless tomatoes. Mol. Breeding 35:22. doi: 10.1007/s11032-015-0222-8
Nakamura, A., Umemura, I., Gomi, K., Hasegawa, Y., Kitano, H., Sazuka, T., et al. (2006). Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 46, 297–306. doi: 10.1111/j.1365-313X.2006.02693.x
Okay, S., Derelli, E., and Unver, T. (2014). Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol. Genet. Genomics 289, 765–281. doi: 10.1007/s00438-014-0849-x
Paul, J. O., Yoko, O., José, M. A., April, C., Chang, C., Ecker, J. R., et al. (2005). Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17, 3282–3300. doi: 10.1105/tpc.105.036723
Peleg, Z., Reguera, M., Tumimbang, E., Walia, H., and Blumwald, E. (2011). Cytokinin-mediated source /sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol. J. 9, 747–758. doi: 10.1111/j.1467-7652.2010.00584.x
Reed, J. W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. doi: 10.1016/S1360-1385(01)02042-8
Rogg, L. E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. doi: 10.1105/tpc.13.3.465
Saintenac, C., Jiang, D., Wang, S., and Akhunov, E. (2013). Sequence-based mapping of the polyploid wheat genome. G3 3, 1105–1114. doi: 10.1534/g3.113.005819
Salse, J., Bolot, S., Throude, M., Jouffe, V., Piegu, B., Quraishi, U. M., et al. (2008). Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20, 11–24. doi: 10.1105/tpc.107.056309
Singla, B., Chugh, A., Khurana, J. P., and Khurana, P. (2006). An early auxin-responsive Aux/IAA gene from wheat (Triticum aestivum) is induced by epibrassinolide and differentially regulated by light and calcium. J. Exp. Bot. 57, 4059–4070. doi: 10.1093/jxb/erl182
Song, Y., and Xu, Z. F. (2013). Ectopic overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) Gene OsIAA4 in rice induces morphological changes and reduces responsiveness to auxin. Int. J. Mol. Sci. 14, 13645–13656. doi: 10.3390/ijms140713645
Song, Y., You, J., and Xiong, L. (2009). Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinos-teroid hormone responses and plant morphogenesis. Plant Mol. Biol. 70, 297–309. doi: 10.1007/s11103-009-9474-1
Strader, L. C., Chen, G. L., and Bartel, B. (2010). Ethylene directs auxin to control root cell expansion. Plant J. 64, 874–884. doi: 10.1111/j.1365-313X.2010.04373.x
Su, L., Bassa, C., Audran, C., Mila, I., Cheniclet, C., Chevalier, C., et al. (2014). The auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expansion. Plant Cell Physiol. 55, 1969–1976. doi: 10.1093/pcp/pcu124
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, 609–612. doi: 10.1093/nar/gkl315
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tiwari, S. B., Hagen, G., and Guilfoyle, T. J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543. doi: 10.1105/tpc.017384
Tiwari, S. B., Wang, X. J., Hagen, G., and Guilfoyle, T. J. (2001). Aux/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822. doi: 10.1105/tpc.010289
Vanneste, S., and Friml, J. (2009). Auxin: a trigger for change in plant development. Cell 136, 1005–1016. doi: 10.1016/j.cell.2009.03.001
Walker, J. C., and Key, J. L. (1982). Isolation of cloned cDNAs to auxin-responsive polyA RNAs of elongating soybean hypocotyl. Proc. Natl. Acad. Sci. U.S.A. 79, 7185–7189. doi: 10.1073/pnas.79.23.7185
Wang, H., Jones, B., Li, Z. G., Frasse, P., Delalande, C., Regad, F., et al. (2005a). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17, 2676–2692. doi: 10.1105/tpc.105.033415
Wang, S. K., Bai, Y. H., Shen, C. J., Wu, Y. R., Zhang, S. N., Jiang, D., et al. (2010a). Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genomics 10, 533–546. doi: 10.1007/s10142-010-0174-3
Wang, Y., Deng, D., Bian, Y., Lv, Y., and Xie, Q. (2010b). Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays. L.). Mol. Biol. Rep. 37, 3991–4001. doi: 10.1007/s11033-010-0058-6
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wicker, T., Mayer, K. F., Gundlach, H., Martis, M., Steuernagel, B., Scholz, U., et al. (2011). Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell 23, 1706–1718. doi: 10.1105/tpc.111.086629
Wu, J., Peng, Z., Liu, S., He, Y., Cheng, L., Kong, F., et al. (2012). Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol. Genet. Genomics 287, 295–311. doi: 10.1007/s00438-012-0675-y
Yang, X., Tuskan, G. A., and Cheng, M. Z. (2006). Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 142, 820–830. doi: 10.1104/pp.106.083642
Keywords: bread wheat genome, Aux/IAA family, chromosome location, expansion pattern, function prediction
Citation: Qiao L, Zhang X, Han X, Zhang L, Li X, Zhan H, Ma J, Luo P, Zhang W, Cui L, Li X and Chang Z (2015) A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.). Front. Plant Sci. 6:770. doi: 10.3389/fpls.2015.00770
Received: 19 June 2015; Accepted: 07 September 2015;
Published: 30 September 2015.
Edited by:
Tiegang Lu, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Shichen Wang, Kansas State University, USAYongqing Li, South China Botanical Garden, Chinese Academy of Sciences, China
Copyright © 2015 Qiao, Zhang, Han, Zhang, Li, Zhan, Ma, Luo, Zhang, Cui, Li and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyan Li, Beijing Anzhen Hospital Affiliated to the Capital Medical University/Beijing Institute of Heart Lung and Blood Vessel Diseases, No. 2 Anzhen Road, Beijing 100029, China, xiaoyanli82@163.com;
Zhijian Chang, Institute of Crop Science, Shanxi Academy of Agricultural Sciences, No. 81 Longcheng Street, Taiyuan 030031, Shanxi, China, czjsxaas@126.com
 Linyi Qiao
Linyi Qiao Xiaojun Zhang2
Xiaojun Zhang2 Xiao Han
Xiao Han Lei Zhang
Lei Zhang Zhijian Chang
Zhijian Chang