- 1School of Life Sciences, Chongqing University, Chongqing, China
- 2Institute of Cotton Research, Chinese Academy of Agricultural Sciences, the State Key Laboratory of Cotton Biology, Henan, China
Target of Rapamycin (TOR) is an eukaryotic protein kinase and evolutionally conserved from the last eukaryotic common ancestor (LECA) to humans. The growing evidences have shown that TOR signaling acts as a central controller of cell growth and development. The downstream effectors of TOR have been well-identified in yeast and animals by using the immunosuppression agent rapamycin. However, less is known about TOR in plants. This is largely due to the fact that plants are insensitive to rapamycin. In this study, AZD8055 (AZD), the novel ATP-competitive inhibitor of TOR, was employed to decipher the downstream effectors of TOR in Arabidopsis. One AZD insensitive mutant, TOR-inhibitor insensitive-1 (trin1), was screened from 10,000 EMS-induced mutation seeds. The cotyledons of trin1 can turn green when its seeds were germinated on ½ MS medium supplemented with 2 μM AZD, whereas the cotyledons greening of wild-type (WT) can be completely blocked at this concentration. Through genetic mapping, TRIN1 was mapped onto the long arm of chromosome 2, between markers SGCSNP26 and MI277. Positional cloning revealed that TRIN1 was an allele of ABI4, which encoded an ABA-regulated AP2 domain transcription factor. Plants containing P35S::TRIN1 or P35S::TRIN1-GUS were hypersensitive to AZD treatment and displayed the opposite phenotype observed in trin1. Importantly, GUS signaling was significantly enhanced in P35S::TRIN1-GUS transgenic plants in response to AZD treatment, indicating that suppression of TOR resulted in the accumulation of TRIN1. These observations revealed that TOR controlled seed-to-seedling transition by negatively regulating the stability of TRIN1 in Arabidopsis. For the first time, TRIN1, the downstream effector of TOR signaling, was identified through a chemical genetics approach.
Introduction
TOR, which belongs to the phosphoinositide 3-kinase (PI3K)-related kinase family, is an evolutionarily conserved Ser/Thr kinase in structure and function from the last eukaryotic common ancestor (LECA) to humans. TOR regulates cell growth, metabolism and development in response to dynamic and diverse environmental stresses and challenges (Horváth et al., 2006; Wullschleger et al., 2006; Laplante and Sabatini, 2012; Ren et al., 2012; Robaglia et al., 2012; Yuan et al., 2013). Two TOR genes were originally identified by genetic screening for rapamycin insensitive mutants in budding yeast (Saccharomyces cerevisiae) (Heitman et al., 1991b; Cafferkey et al., 1993; Kunz et al., 1993). Subsequently, only one TOR gene was identified in animals, humans and Arabidopsis (Sabatini et al., 1994; Sabers et al., 1995; Menand et al., 2002). TOR protein contains five conserved regions: HEAT repeats, FAT domain, FRB domain, kinase domain and FATC domain (Menand et al., 2002; Mahfouz et al., 2006). In yeast and mammals, TOR forms two structurally and functionally distinct protein complexes: TORC1 (TOR complex 1) and TORC2 (TOR complex 2; Loewith et al., 2002). TOR, regulatory-associated protein of mTOR (RAPTOR) and lethal with SEC13 protein 8 (LST8) constitute the core of TORC1, which regulates cell growth and metabolism in response to nutrients and energy requirements (Martin and Hall, 2005; Wang and Proud, 2009). Rapamycin can specifically bind to FK506 binding protein 12 (FKBP12), which interacts with the FRB domain of TOR and forms a rapamycin-FKBP12-TOR complex to inhibit the activity of TORC1 in yeast and animals (Heitman et al., 1991b; Zheng et al., 1995). On the other hand, TORC2 is insensitive to rapamycin and the core components include TOR, LST8 and rapamycin-insensitive companion of mTOR (RICTOR). TORC2 controls spatial cell growth by regulating cytoskeletal structure and polarity (Sarbassov et al., 2004; De Virgilio and Loewith, 2006; Wullschleger et al., 2006). It seems that the components of TORC2 are much less conserved across the eukaryotic species than that of TORC1, which suggests that the functions of TORC2 may likely vary across species. For example, RAPTOR, the core member of TORC1, has been identified in Arabidopsis, but the homologs of RICTOR, the key defining effector of TORC2, are missing in most examined plants (Xiong and Sheen, 2014; Rexin et al., 2015), suggesting that plants most likely have the distinct TORC2 throughout evolution.
A recent study showed that TOR could directly phosphorylate transcription factor E2Fa to activate S-phase genes in root meristems (Xiong and Sheen, 2013), and this finding revealed an important role of TOR in the regulation of cell cycle. TOR also played an essential role in the regulation of primary and secondary metabolism in plants. Disruption of TOR by reduction of TOR expression or kinase activity led to the accumulation of high levels of starch, triacylglycerides, amino acids, TCA intermediates and secondary metabolites (Deprost et al., 2007; Ren et al., 2012; Caldana et al., 2013). Genetic and physiological studies combining with large-scale transcript and metabolite profiling analyses have revealed that TOR regulates plant growth, development, flowering, senescence and life span by modulating transcription, translation, cell cycle, autophagy and metabolism (Deprost et al., 2007; Ahn et al., 2011; Ren et al., 2011, 2012; Moreau et al., 2012; Xiong and Sheen, 2012, 2014; Caldana et al., 2013).
The lesions of TOR result in lethality in yeast, animals and plants (Heitman et al., 1991a; Menand et al., 2002; Ren et al., 2011). This severely prevented people from identifying the downstream effectors of TOR signaling through classic genetic approaches. Significant discoveries on the functions of TORC1 did not occur until rapamycin was found and applied for the study of TOR in yeast and animals (Heitman et al., 1991a; Brown et al., 1994). Rapamycin is the first generation of TOR inhibitors, and it inhibits the activity of TORC1 only in the presence of FKBP12. Although FKBP12 is a non-essential protein for cell growth, it plays a key role in mediating the cytotoxicity of rapamycin on cell growth. Large amount of information about TORC1 and its downstream targets have been well documented in yeast and animals (Burnett et al., 1998; Nojima et al., 2003; Martin et al., 2004; Ahn et al., 2011). However, information on TOR signaling in plants is limited, which is mainly due to its insensitivity to rapamycin (Xu et al., 1998). Plants are anchored in soil and rapamycin is produced by the soil bacterium Streptomyces hygroscopicus. To escape from rapamycin inhibition, plants have adapted an evolutionary mutation in the FKBP12 gene that results in its loss of function to bind rapamycin and thus fail to mediate rapamycin to inhibit TOR activity (Xu et al., 1998; Sormani et al., 2007). To assess TOR signaling in plants by using rapamycin, Sormani et al. and Ren et al. independently generated rapamycin-hypersensitive plants by introducing yeast or human FKBP12 into Arabidopsis (Sormani et al., 2007; Ren et al., 2012). However, rapamycin inhibits the activity of TOR only in the presence of yeast or animals FKBP12 in plants (Sormani et al., 2007; Ren et al., 2012). This largely restricts the usage of rapamycin on various plants. In addition, previous studies have shown that rapamycin cannot fully inhibit the growth of plants harboring exogenous FKBP12 even at a concentration of 20 μM (Ren et al., 2012), which is 2000-fold greater than the dosage used in yeast and animals, suggesting that rapamycin partially inhibits TOR signaling and the broader functions of TOR cannot be deciphered by using rapamycin alone in plants.
To resolve this issue relating to rapamycin, active-site TOR inhibitors (asTORis), which are the second generation inhibitors of TOR, are well developed in mammalian cells. Unlike rapamycin, asTORis which include PP242, Torin1, Torin2, AZD, and KU63794 can directly and specifically target the ATP-binding pocket of the TOR kinase domain to suppress both the functions of TORC1 and TORC2 by competing with ATP in mammalian cells (Feldman et al., 2009; García-Martinez et al., 2009; Chresta et al., 2010; Liu et al., 2011). Recent studies have shown that AZD displayed highly selective and inhibitory effects on TOR activity in flowering plants, including A. thaliana, O. sativa, and L. japonicas (Calabrese and Blain, 2009; Montané and Menand, 2013). Importantly, the lesser copy numbers of TOR, the more inhibitory effects of AZD was observed in Arabidopsis (Calabrese and Blain, 2009; Montané and Menand, 2013). Based on these observations, in this study, we generated an EMS-induced mutation library and identified a trin1 mutant from this library through large-scale genetic screening and positional cloning. We found that TRIN1 acted as a negative effector of TOR signaling to modulate cotyledons greening in Arabidopsis. For the first time, the downstream target of TOR signaling was identified by using asTORis-based chemical genetics approaches.
Materials and Methods
Plant Materials and Growth Conditions
In this study, WT Arabidopsis Columbia (Columbia-0), Landsberg erecta (Ler) ecotype and abi4-1 were used. Arabidopsis seeds were surface sterilized by using liquid methods. The seeds first treated with 70% ethanol for 2 min and the supernatant was discarded; then, with 10% sodium hypochlorite containing 0.3% Tween-20 for 5 min, and the supernatant was discarded; followed by four or five rinses with sterile water, centrifugation for 2 min at 4000 g each time, and the supernatant was discarded. Finally, the seeds were suspended in 0.1% sterile agarose and kept at 4°C for 2 days. Sterilized seeds were plated on plates, and then grown in a controlled environment at 22°C under 16 h 60–80 μE·m−2 s−1 continuous light and 8 h darkness.
EMS Mutagenesis of Arabidopsis Col WT Seeds and Genetic Mapping
A total of ~10,000 seeds were placed in a 50-mL centrifuge tube and ultrapure water was added to about 1 cm above the seeds. The seeds were soaked at room temperature for 12 h. Water was decanted and 20 mL 0.3% ethyl methanesulfonate (EMS) (v/v) in water was added. The seeds were incubated for 15 h at room temperature (shock was introduced every hour), followed by decanting of the EMS and rinsing with 40 mL ultrapure water (5 times, 5 min each). The seeds were sown in soil and grown in the greenhouse until maturity.
For mapping, trin1 homozygous mutants were crossed with WT Landsberg erecta (Ler), and F2 seeds were obtained. Genomic DNA was isolated from the F2 plants that exhibited the AZD-insensitive phenotype, and the gene was mapped using simple sequence repeat (SSR) markers.
Generation of Overexpression Constructs and Transformation of Arabidopsis Plants
Total DNA was extracted from Arabidopsis Col WT seedlings using the DNAprep Pure Plant Kit (BioTeke, Beijing, China). The promoter of TRIN1 (2051 bp in length), full-length CDS of TRIN1 (987 bp), and GUS (1812 bp) were amplified by RT-PCR using the TransStar Taq Polymerase Mix kit (TRANSGEN, Beijing, China) following the manufacturer's instructions. The corresponding restriction enzyme sites were introduced into 5′ and 3′ end of the respective primer (Supplemental Table 1). The PCR products of the promoter of TRIN1 (PTRIN1) and CDS of TRIN1 and GUS were cloned into the TA cloning vector pEASY-T1 Simple (TRANSGEN, Beijing, China) and verified by DNA sequencing. Then the GUS coding sequence was subcloned downstream of the TRIN1 promoter or coding sequence (Supplemental Figure 1), and the TRIN1 coding sequence was subcloned downstream of the cauliflower mosaic virus 35S promoter in vector p8GWN to generate clones P35S::TRIN1-HA, P35S::TRIN1-GUS, P35S::GUS and PTRIN1::GUS (Supplemental Figure 1). A gateway system–based entry vector was generated by cloning the recombinant plasmids into p8GWN (Ren et al., 2011). These recombinant constructs were transformed into pEarleyGate303 (Earley et al., 2006) through LR recombination reactions (Supplemental Figure 1).
The constructs were introduced into Agrobacterium tumefaciens strain LBA4404 and used in the transformation of WT Col plants. Transgenic Arabidopsis lines were generated by Agrobacterium mediated transformation using the floral dip method (Zhang et al., 2006) for developmental and phenotypic analyses. Transgenic lines were selected on ½ MS medium containing 50 mg/L kanamycin. Plants were allowed to reproduce for two generations, and the T3 homozygous plants were used in the analysis.
Quantitative Real-time PCR
Total RNA of trin1, TRIN1-OE5, and WT seedlings that were treated for 36 h in mediums containing DMSO, AZD (5 μM), and Torin1 (10 μM) was isolated using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China). Total RNA was treated with RNase-free DNase (Promega). PrimeScript® RT reagent kit (Takara, Dalian, China) was used for reverse transcription, following the manufacturer's instructions. Relative transcript levels were assayed by one-step real-time PCR analysis using the CFX96 real-time PCR system (BIO-RAD, USA). Real-time primers were designed by Primer Premier 5.0 and the details are presented in Supplemental Table 2. AtACTIN2 was used as an internal control. Reactions were performed in a final volume of 20 μL containing 10 μL of 2 × Power Top Green qPCR SuperMix (TRANSGEN, Beijing, China), 50 ng of cDNA, and 500 nM of each of the forward and reverse primers. The following default program was used: 94°C for 5 min, followed by 40 cycles of 94°C for 15 s and 60°C for 30 s each, and a dissociation stage of 95°C for 15 s, 60°C for 30 s, and 95 °C for 15 s. RNA relative quantification analyses were performed with the Bio-Rad CFX Manager software. The data were expressed as the mean ± SD of three independent experiments. Each data point was determined in triplicate in each of the three biological replicates and expressed as the mean ± SD.
GUS Staining and Quantitative Determination of GUS Activity
Transgenic plants staining for GUS activity using X-Gluc were performed as previously described (Menand et al., 2002). The first true leaves of the P35S::TRIN1-GUS OE-2 plants were incubated with GUS staining solution [50 mM NaOH, 0.5 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 0.05% (v/v) Triton X-100 (pH 7.5)] for 8–12 h at 37°C, and a 70% ethanol wash was performed to remove chlorophyll from the leaves. Images were captured using a stereomicroscope (OLYMPUS MVX10, Japan). Each treatment was performed using three biological replicates.
Quantitative GUS assay was performed using the MarkerGene™ β-glucuronidase (GUS) reporter gene activity detection kit (Marker Gene Technologies, Inc., Eugene, OR, USA). Total proteins in extracts of the first true leaves were quantified using the Bradford assay (Bradford, 1976). Fluorometric quantification of GUS activity was performed using 4-methylumbelliferone (4-MU) substrate, and fluorescence was determined on a Tecan 200 fluorometer (Tecan, Durham, NC, USA) using 360 and 465 nm as excitation and emission wavelengths, respectively. GUS activity of the extracts was calculated in nmol 4-MU per minute per mg protein.
Measurement of Chlorophyll Content and Shoots
Chlorophyll content was measured after treatments using the specified agents for 0–6 days. Chlorophyll was extracted from the plant leaves and quantified (Weaver and Amasino, 2001). Arabidopsis seeds were plated on ½ MS culture medium containing the specified agents, and the length of emerging shoots were measured after 10 days. Three biological experiments, each consisting of 30 plants per treatment were measured.
Results
AZD is Able to Block Seed-to-seedling Transition in Arabidopsis
TOR protein is a highly conserved Ser/Thr kinase protein. Among all the (Arabidopsis thaliana TOR) AtTOR functional domains, including HEAT repeats, FAT, FRB, kinase, and FATC domains, the kinase domain of AtTOR showed the highest amino acid sequence identical to that of yeast and human (Supplemental Figure 2). AZD, a novel ATP competitive inhibitor of mTORC1 and mTORC2, can directly bind to the kinase domain of TOR to compete with ATP (Chresta et al., 2010). Montane and Menand showed that AZD could specifically inhibit the TOR activity in Arabidopsis (Supplemental Figure 3A), resulting in short roots and shoots, bleached cotyledons and a severe delay in seedling development (Montané and Menand, 2013). To identify the downstream components of the TOR signaling pathway in Arabidopsis, AZD was applied to screen AZD-insensitive mutant seeds derived from the EMS mutagenesis library in this study. To determine the optimal AZD concentration for mutants screening, WT seeds were treated with different concentrations of AZD dissolved in DMSO. Consisted with the findings of previous reports, AZD inhibited seedling growth and development in a dose-dependent manner. With increasing concentrations of AZD, the developing seedlings were subjected to different degrees of inhibition (Figure 1A). When applied concentration of AZD reached to 1 μM, the fresh weight of seedlings was only half of that treated with DMSO control (Figure 1A), suggesting that 1 μM AZD can be used as the 50% growth inhibitory dose (GI50) of AZD in Arabidopsis (Figure 1B). Additionally, the cotyledons did not turn green and the seedlings completely ceased to grow and develop with 2 μM AZD. Thus, 2 μM AZD was selected as the optimal concentration for screening AZD-insensitive mutant seeds.
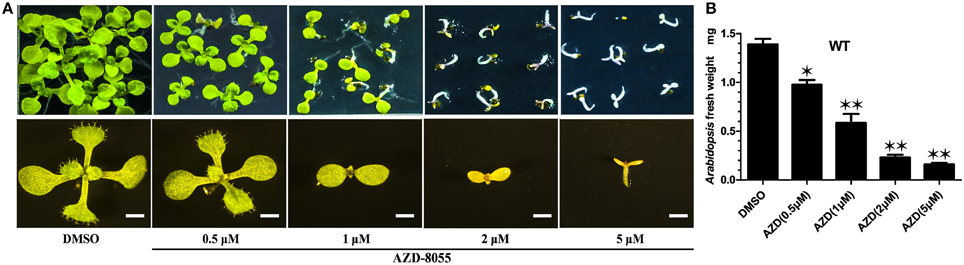
Figure 1. AZD arrests early seedlings establishment in Arabidopsis. (A) Representative images of WT plants growing on ½ MS medium containing increasing concentrations of AZD for 10 days (upper panel). The lower panel shows enlarged images of the individual seedling in the upper panel. Bar = 1 mm. (B) Fresh weight of WT seedlings growing on different AZD concentrations for 10 days. Each graph represents the average of 30 seedlings that were conducted in triplicate. Error bars indicate ±SD for triplicates. Asterisks denote Student's t-test significant difference compared with DMSO (*P < 0.05; **P < 0.01).
Screening, Genetic Analysis and Fine Mapping of trin1 Mutant
Approximately 100,000 EMS-induced M2 mature seeds were harvested and then cultured on ½ MS medium (Murashige and Skoog, 1962) containing 2 μM AZD. Because seed-to-seedling transition of WT plants can be blocked by using 2 μM AZD, the 15 days after germination greening seedlings grown on 2 μM AZD medium were selected as AZD-insensitive mutants for further study. Nine independent AZD-insensitive mutants in total were obtained from this screening and the first of these mutants was named as TOR-inhibitor insensitive-1 (trin1).
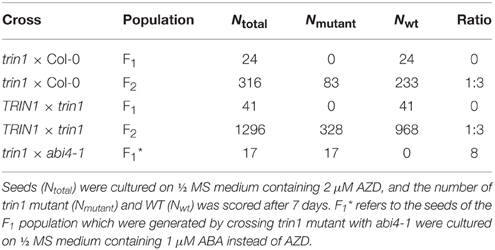
The trin1 mutant was backcrossed with Col-0 WT plants for three times. The results consistently showed that the AZD-insensitive phenotype was absent in every F1 generation of hybrid plants, and the F2 population segregated into WT and mutant at a ratio of 3:1 (Table 1), indicating that the AZD-insensitive phenotype was caused by a monogenic recessive mutation. The trin1 homozygous mutant plants were crossed with Arabidopsis Ler plants to generate the segregation population for fine mapping of the genetic loci of trin1.
Genomic DNA was isolated from 500 F2 plants that exhibited the AZD-insensitive phenotype, and the mutation was roughly mapped to chromosome 2 between SSR markers nga361 and nga168. More markers between nga361 and nga168 were designed. A candidate region with the mutation was mapped to between 63.27 and 67.39 cM. To identify point mutations, genomic DNA between 63.27 and 67.39 cM on chromosome 2 were sequenced. Analysis of sequencing results indicated that the trin1 mutant was allelic to abi4. To verify whether TRIN1 shared the same genetic locus with ABI4, trin1 mutant plants were crossed with abi4-1 which was insensitive to ABA (Finkelstein et al., 1998). All the F1 population seeds were insensitive to ABA (Table 1), thus confirming that the trin1 mutant was indeed allelic to abi4. Two changes were found in the coding sequence of trin1 based on the sequencing results, one was a 3-bp (AAC) deletion at positions 566–568; the other was a single nucleotide change, an A-to-G substitution at position 685 that led to a missense mutation of T to A (Figure 2).
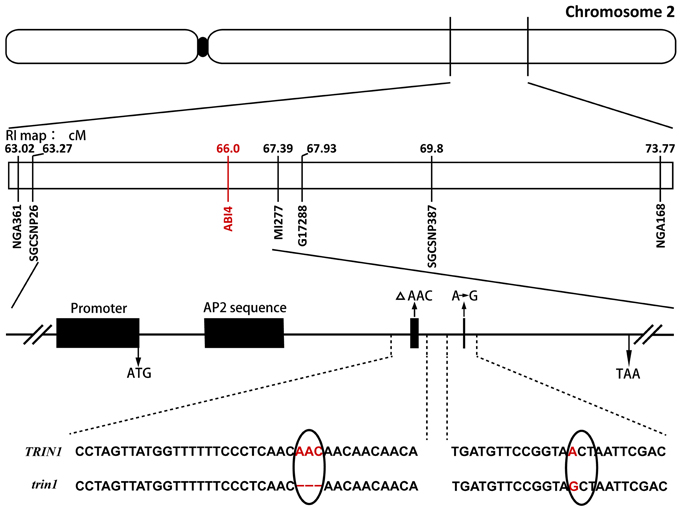
Figure 2. Fine mapping of TRIN1 on chromosome 2. Sequence analysis of trin1 shows a 3-bp (AAC) deletion (as indicated by a Δ) at positions 566–568 in the coding sequence that leads to deletion of the amino acid asparagine. Furthermore, a point mutation changing the first base of codon 229 of trin1 from A to G was identified (marked in red); it results in the substitution of threonine (ACT) with an alanine (GCT). The black solid oval represents the centromere.
Analysis of Phylogenetic Tree of TRIN1 Sequences and Phosphorylation Sites of trin1 Protein
TRIN1/ABI4 is one member of the AP2/ERF family, which can specifically bind to the CE1 element in the promoters of abiotic stress responsive genes and regulate their expression (Mizoi et al., 2012). Homolog of Arabidopsis TRIN1 has been reported in various plant species, such as Oryza sativa and Zea mays (Niu et al., 2002). Phylogenetic tree analysis showed that Arabidopsis TRIN1 was evolutionarily conserved across plant species, whereas no homologs were found in yeast and animals. The closest evolutionary relationship of TRIN1 was observed between Capsella rubella and Arabidopsis lyrata. On the other hand, the most distant phylogenetic relationship of TRIN1 was detected between Aquilegia coerulea and Arabidopsis thaliana (Supplemental Figure 4).
TOR is a well-known serine/threonine kinase. AZD-insensitivity of trin1 suggested that TRIN1 likely functioned as a downstream effector of TOR signaling. The phosphorylation of TRIN1 probably relays the TOR signaling cascade in plants. We next asked whether the threonine230 (ACT) replaced by an alanine230 in trin1 protein affected the phosphorylation status of TRIN1. Online tools were employed to predict the putative phosphorylation sites in TRIN1. Interestingly, threonine230 was one of five most possible targets of the upstream kinase (Supplemental Figure 3B). Once threonine230 was replaced by alanine230, only four putative phosphorylation sites existed in trin1 protein. These results demonstrated that threonine230 of TRIN1 likely played a crucial role in TOR signaling transduction in Arabidopsis.
TRIN1 Acts as a Key Player to Integrate ABA and TOR Signaling During Seed-to-seedling Transitional Stage
To further decipher the functions of TRIN1 in Arabidopsis, we generated TRIN1 overexpression plants by introducing P35S::TRIN1-HA into WT (Col) Arabidopsis. A total of 28 independent transgenic plants were obtained. 21 independent lines showed AZD hypersensitive phenotypes, suggesting that the AZD-sensitive phenotypes resulted from the overexpression of TRIN1 rather than the T-DNA insertion events. Torin1 has a different structure from that of AZD and is also a well-established TOR inhibitor in Arabidopsis (Montané and Menand, 2013; Schepetilnikov et al., 2013), but AZD is a more selective and potent agent against TOR than Torin1. To examine whether TRIN1 overexpression lines were sensitive to different TOR inhibitors, Torin1 was selected as a parallel control. Five out of twenty-one lines were selected to perform this examination. As expected, these lines showed hyper-sensitive to Torin1 as AZD (Supplemental Figure 5), indicating that different TOR inhibitors can co-target TOR kinase and generate a similar phenotype. TRIN1 transcriptional levels of these lines were measured. P35S::TRIN1-OE5 (overexpression line 5) showed the highest TRIN1 expression level, which was 109-fold higher than that of WT (Figure 3E). Meanwhile, the transcription level of TRIN1 had no significant difference among DMSO, ABA, AZD, and Torin1 treatments in P35S::TRIN1-OE5 plants. The lowest TRIN1 expression level was observed in P35S::TRIN1-OE1, which was 13-fold higher than that of WT. Importantly, the higher level of TRIN1 expressed, the more sensitive to AZD showed (Supplemental Figure 5), indicating that the amount of TRIN1 was tightly associated with the sensitivity of AZD in Arabidopsis.
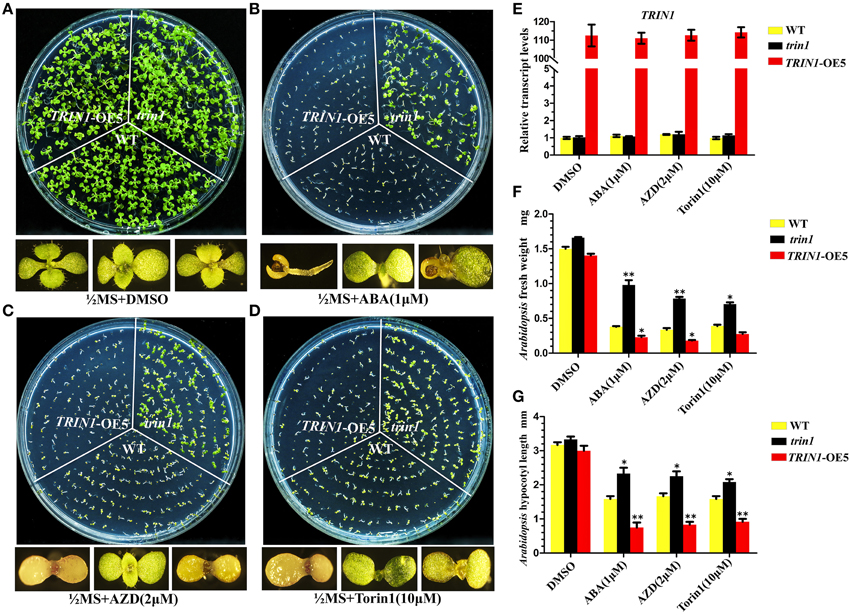
Figure 3. Seeds of trin1, TRIN1-OE5, and WT were treated with ABA and mTOR kinase inhibitors. (A–D) trin1, TRIN1-OE5, and WT seeds were cultured on different medium for 10 days; Enlarged pictures of TRIN1-OE5, trin1, and WT plants from left to right in the bottom. AZD, ABA, and Torin1 were dissolved in DMSO. (A) Using ½ MS containing 1 μg/mL DMSO as controls. (B–D) trin1 plants are insensitive to ABA, AZD and Torin1, whereas TRIN1-OE5 plants are sensitive. (E) qRT-PCR analysis of ABI4 transcripts from 10-day-old WT, trin1, and TRIN1-OE5 plants treated with DMSO, AZD (5 μM), and Torin1 (10 μM) for 36 h. The data represent the mean ±SE of n = 3 independent experiments. (F) Arabidopsis fresh weight of trin1, TRIN1-OE5, and WT plants treated as described in (A–D). The data represent the mean ±SE of n = 3 independent experiments each containing 30 plants per treatment. Asterisks denote Student's t-test significant difference compared with WT plants (**P < 0.01). (G) Arabidopsis shoots length of trin1, TRIN1-OE5, and WT plants treated as described in (A–D). The length from aboveground to rosette was measured. The data represents an average of 30 seedlings with three duplicates. Error bars indicate ±SD for triplicates. Asterisks denote Student's t-test significant difference compared with WT plants (*P < 0.05; **P < 0.01).
Since TRIN1 is allelic to ABI4, abi4 was insensitive to ABA (Finkelstein et al., 1998). To determine whether the trin1 and P35S::TRIN1-OE5 (TRIN1-OE5) seeds were insensitive or sensitive to ABA, the seeds of trin1, TRIN1-OE5, and WT were cultured on ½ MS medium supplemented with ABA, and ½ MS medium containing AZD and Torin1 were set as control (Figure 3). On ½ MS + DMSO medium, seedlings growth of trin1 and TRIN1-OE5 was quite similar to WT plants (Figure 3A). However, on ABA medium, trin1 plants were significantly resistant to ABA but TRIN1-OE5 plants were more sensitive to ABA than WT (Figure 3B). In addition, trin1 seeds could normally germinate and grow on ½ MS medium containing 1 μM ABA, 2 μM AZD, and 10 μM Torin1, whereas TRIN1-OE5 plants were overly sensitive to ABA, AZD, and Torin1 (Figures 3B–D). Importantly, the fresh weight of trin1 seedlings was significantly heavier than that of WT when they were grown on ½ MS medium containing ABA, AZD, or Torin1 (Figure 3F). The shoots length of trin1 was also significantly increased compared to that of the WT plants on ½ MS medium containing ABA, AZD, or Torin1 (Figure 3G). Together, these results indicated that TRIN1/ABI4 integrated TOR and ABA signaling in regulating plants growth and development in Arabidopsis.
TOR Regulates Chlorophyll Metabolism Via TRIN1/ABI4 at the Photoautotrophic Stage
Cotyledons greening is an extremely important transition from heterotrophism to autrophism in plants. As previously described, inhibition of TOR signaling suppresses the transition from heterotrophism to autrophism in plants (cotyledons cannot turn green). To determine the effect of TOR signaling pathway on chlorophyll metabolism at the photoautotrophic stage, Arabidopsis seedlings of trin1, TRIN1-OE5, and WT were treated with AZD. AZD interfered with the progress of chlorophyll metabolism, which in turn resulted in that leaves did not retain its green color in WT plants (Figure 4B). TRIN1-OE5 plants were more sensitive to AZD than WT plants (Figures 4C,D), whereas trin1 could rescue the phenotype of AZD inhibition (Figures 4A,D). Relative chlorophyll content of WT, trin1, and TRIN1-OE5 plant leaves were also measured by using 7-day-old seedlings treated with 5 μM AZD and 10 μM Torin1 for 0–6 days (Figures 4E,F). With time, the chlorophyll contents of WT, trin1, and TRIN1-OE5 were decreased, whereas trin1 showed a delayed reduction in chlorophyll content.
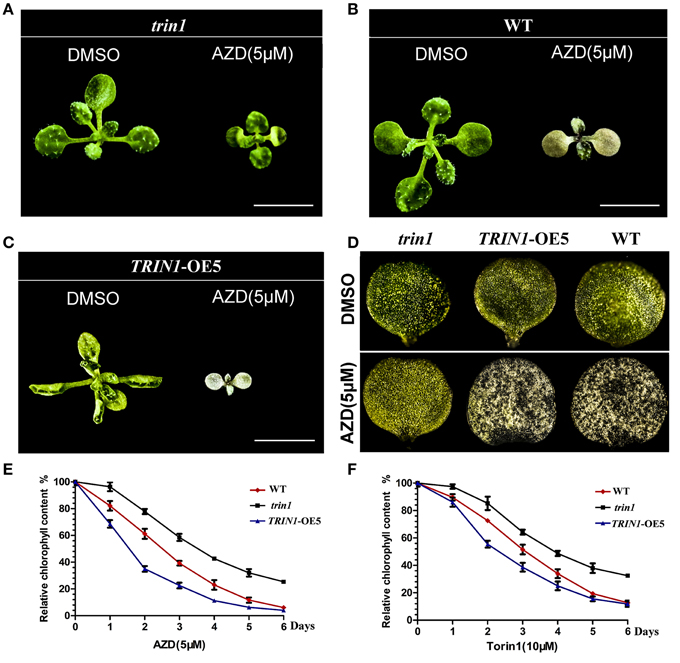
Figure 4. The effect of AZD on the phenotype of trin1 mutant and TRIN1-OE5 plants. (A–C) The phenotype of trin1, TRIN1-OE5, and WT plants, 10-day-old seedlings were transferred to ½ MS medium treated for 5 days with or without 5 μM AZD. Bar = 5 mm. (D) Images of the first cotyledon of trin1, TRIN1-OE5, and WT plants treated as described in (A–C) were captured under a stereomicroscope (2.5 × magnification). (E,F) Seeds of WT, trin1, and TRIN1-OE5 were cultured on ½ MS for 7 days. Then, the seedlings were treated for 0–6 days with 5 μM AZD or 10 μM Torin1, and the relative chlorophyll content of plant leaves were measured. The data represents the mean ± SD of n = 3 independent experiments.
To further examine the roles of TOR in chlorophyll synthesis and degradation, we analyzed the transcription levels of the key genes associated with chlorophyll synthesis and decomposition using qRT-PCR (Figure 5). The differential expressions of Pheide a oxygenase (PAO), Pheophytinase (PPH), chlorophyll a/b-binding protein 3 (CAB3), and HEMA1 were examined. PAO is a key regulator of chlorophyll catabolism(Pruzinská et al., 2003). PPH is a chloroplast-located hydrolase, which specifically dephytylates the Mg-free chlorophyll pigment and yield pheophorbide (Schelbert et al., 2009). PAO and PPH play important roles in the chlorophyll degradation pathway. The results showed that a significant upregulation of PAO mRNA expression in TRIN1-OE5 plants compared to that observed in the WT after AZD or Torin1 treatment. On the other hand, PAO showed a lower degree of induction after AZD treatment in the trin1 mutant when compared to WT (Figure 5A). The similar transcript changes of PPH were observed in WT, trin1, and TRIN1-OE5 with AZD or Torin1 treatment (Figure 5B), indicating that TOR and TRIN1 have important roles in the regulation of chlorophyll catabolism. CAB3, which is a member of chlorophyll a/b-binding protein family, encodes the most abundant Cab mRNA in developing embryos and young leaves (Leutwiler et al., 1986). HEMA1 gene encodes glutamyl-tRNA reductase (GluTR) which plays a vital role in chlorophyll biosynthesis in Arabidopsis thaliana, (Ilag et al., 1994). Interestingly, the transcript levels of CAB3 and HEMA1 were significantly up-regulated with AZD or Torin1 treatment in trin1 mutant compared to that observed with DMSO (Figures 5C,D). However, transcript levels of CAB3 and HEMA1 were down-regulated with AZD or Torin1 treatment in TRIN1-OE5 plants compared to that of DMSO (Figures 5C,D). These data indicated that the rate of decomposition of chlorophyll was higher than the synthesis rate, showing that TRIN1 reduced greening of cotyledons and induced chlorophyll degradation by AZD or Torin1 treatment. Therefore, TOR and TRIN1 play important roles in chlorophyll metabolism in Arabidopsis.
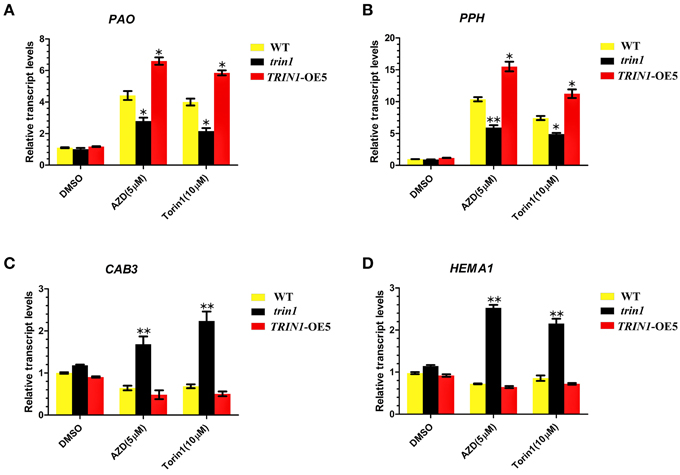
Figure 5. TOR regulates transcription of genes related to chlorophyll synthesis and decomposition. (A–D) qRT-PCR analysis of PAO, PPH, CAB3, and HEMA1 transcripts in 10- day-old WT, trin1, and TRIN1-OE5 plants treated with DMSO, AZD (5 μM), and Torin1 (10 μM) for 36 h. The data represents the mean ± SD of n = 3 independent experiments. Asterisks denote Student's t-test significant difference compared with WT plants (*P < 0.05; **P < 0.01).
TOR Signaling Regulates the Stability of TRIN1/ABI4 Protein
Early studies have shown that the degradation of ABI4 is crucial for its functions in plant growth and development (Gregorio et al., 2014). To determine whether TOR regulates the stability of TRIN1 protein in Arabidopsis, we fused TRIN1 with GUS and generated P35S::TRIN1-GUS transgenic plants. Seventeen independent transgenic plants were obtained. Nine lines showed AZD hypersensitive phenotype. Three out of nine lines were selected for assessment of GUS activity in leaves (Supplemental Figure 6). The most significant difference in GUS expression level in P35S::TRIN1-GUS OE-2 was observed between DMSO and AZD treatments. We respectively examined GUS activity of leaves and root tips of P35S::TRIN1-GUS OE-2 plants. High levels of GUS activity were observed in leaves of P35S::TRIN1-GUS OE-2 plants treated with TOR inhibitors (Figures 6A–C). Additionally, we quantified the GUS activity in response to DMSO, Torin1, and AZD treatments in the leaves. The quantitative results suggested that the GUS signals were significantly increased when TOR was inhibited by Torin1 or AZD (Figure 6H). Interestingly, GUS signaling was detectable in the root tips but not in the division and elongation regions in Arabidopsis root when P35S::TRIN1-GUS OE-2 plants growing on ½ MS medium in the absence of TOR inhibitors (Figure 6E), suggesting that that TRIN1 protein can be likely degraded in some plant tissues. Consistently, GUS activity in the roots of P35S::TRIN1-GUS OE-2 plants was significantly increased in response to TOR inhibitors treatment (Figures 6D–G). To further examine whether the increasing of GUS signals occurred at transcript level, GUS relative transcript levels were analyzed using qRT-PCR and RT-PCR (Figures 6I,J). The GUS transcript levels of the PTRIN1::GUS plants showed no significant differences among DMSO, Torin1, and AZD treatments, consistent with the previous study (Finkelstein et al., 2011). These results suggested that TOR could likely accelerate the degradation of TRIN1 protein to promote plant growth and development.
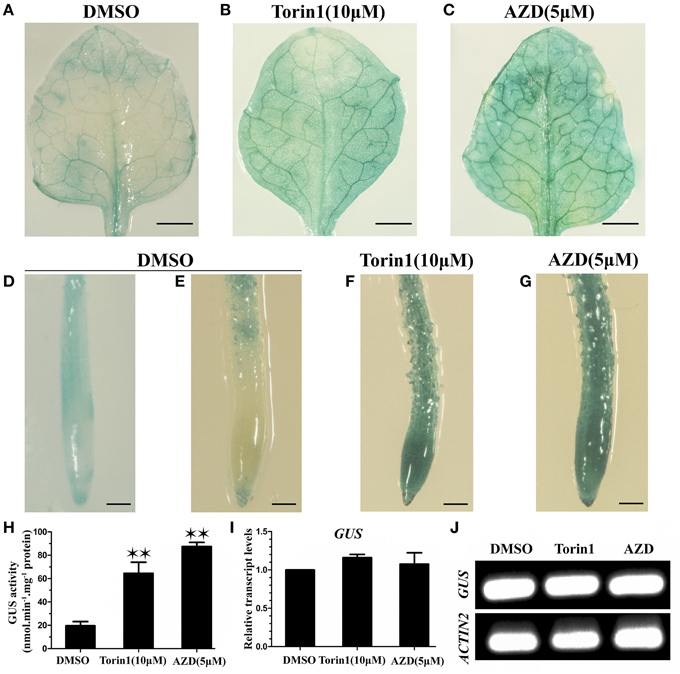
Figure 6. Suppression of TOR results in the accumulation of TRIN1 protein in Arabidopsis. (A–G) GUS staining of the first true leaves of P35S::TRIN1-GUS OE-2 plants. (A–C) GUS staining of leaves of 2-week-old P35S::TRIN1-GUS plants treated with DMSO, Torin1, and AZD for 48 h. Bar = 1 mm. (D) GUS staining of root tips of 2-week-old P35S::GUS plants treated with DMSO for 48 h. (E–G) GUS staining of root tips of 2-week-old P35S::TRIN1-GUS plants treated with DMSO, Torin1, and AZD for 48 h. Bar = 0.1 mm. (H) GUS activity of the first true leaves of 2-week-old P35S::TRIN1-GUS plants was measured by fluorometric quantification, plants were treated as described in (A–C). Asterisks denote Student's t-test significant difference compared with DMSO (*P < 0.05; **P < 0.01). (I) qRT-PCR analysis of GUS relative transcript level of 2-week-old PTRIN1::GUS plants treated with DMSO, AZD (5 μM) and Torin1 (10 μM) for 48 h. The data represent the mean ± SD of n = 3 independent experiments. (J) RT-PCR analysis of GUS transcript level of 2-week-old PTRIN1::GUS plants treated as described in (I).
Discussion
AZD is a Highly Selective Inhibitor of TOR
TOR is a highly conserved Ser/Thr kinase protein. Catalytic domain of Arabidopsis TOR shows more than 75% sequence similarity compared with that of ScTOR (yeast TOR) and HsTOR (human TOR; Supplemental Figure 2). Several studies have shown that AZD is able to inhibit TOR kinase activity across species (Calabrese and Blain, 2009; Chresta et al., 2010; Montané and Menand, 2013). The previous studies showed that AZD inhibited TOR kinase activity with a half maximal inhibitory concentration (IC50) of 0.8 nM in mammalian cells (Chresta et al., 2010). However, the IC50s of AZD for DNA-PK, PI3Kδ and PI3Kα are increased to 1370, 3200, and 3590 nM, respectively (Supplemental Table 3; Chresta et al., 2010). In addition, AZD showed undetectable inhibitory activity against more than 260 different protein kinases even at 10 μM (Chresta et al., 2010). These results demonstrated that AZD acted as a potent and selective mTOR kinase inhibitor with at least 1000-fold specificity over other PI3K or PIKK family members (Chresta et al., 2010). Since TOR belongs to the PI3K superfamily of kinases, it is possible that other PI3Ks are the off-targets of AZD in Arabidopsis. However, no homologs of PI3K were found in Arabidopsis (Supplemental Table 4), indicating that the off-target effects of AZD were irrelevant in Arabidopsis. Recent studies have further shown that AZD displayed potent inhibitory effects on various plants in a dose-dependent manner (Montané and Menand, 2013). Since Torin1 has been widely used as a TOR inhibitor in Arabidopsis (Montané and Menand, 2013; Schepetilnikov et al., 2013; Xiong et al., 2013), we therefore examined whether trin1 or TRIN1-OE plants were insensitive or sensitive to Torin1 as the action of AZD. The results showed that both trin1 and TRIN1-OE plants with Torin1 treatment exhibited the same growth pattern as that of AZD (Figure 3). However, Torin1 is the first generation of asTORis with 300-fold selectivity on TOR over other PI3Ks and PIKKs, whereas AZD belongs to the second generation of asTORis with improved pharmacodynamics (Thoreen et al., 2009; Chresta et al., 2010; Caldana et al., 2013; Montané and Menand, 2013; Supplemental Table 3). Our recent observation also showed that the transcriptional profile of AZD-treated Arabidopsis was highly overlapping with that of the previous TOR suppression lines generated by independent groups (Ren et al., 2012; Caldana et al., 2013; Xiong et al., 2013; Dong et al., 2015), AZD is therefore the priority selection to minimize the off-targets in this study.
TRIN1 is a Novel Effector Downstream of TOR Signaling
The previous studies showed that ABI4 played an important role in the initiation of plastid retrograde signaling (Acevedo-Hernández et al., 2005; Koussevitzky et al., 2007; León et al., 2012). ABI4 can bind to the promoter of a retrograde-regulated gene LHCB through a conserved motif (Koussevitzky et al., 2007). Chloroplast retrograde signaling is required for the initiation and balance Photosynthesis Associated Nuclear Gene (PhANG) expression. ABI4 is a repressor of the PhANG genes such as LHCB and RBCS in young seedlings and acts as a negative regulator of PhANGs (Acevedo-Hernández et al., 2005; Koussevitzky et al., 2007). Excessive accumulation of the ABI4 protein causes stunted growth in plants. Proteasome regulation of transcriptional regulators has been well-characterized for two ABA response factors: ABI3 and ABI5 (Zhang et al., 2005; Stone et al., 2006). Interestingly, although the ABI4 protein was regulated by post-transcription (Finkelstein et al., 2011; Ludwików, 2015), the degradation of ABI4 protein through ubiquitination remains to be studied. Here, we screened and identified a trin1 mutant which was insensitive to TOR kinase inhibitors by screening EMS mutagenesis library. Fine mapping and sequencing results showed that the trin1 mutant was allelic to abi4. A point mutation that changed the first base of codon 229 of trin1 from A to G was identified, resulting in a threonine (ACT) to be replaced by an alanine (GCT; Table 1 and Figure 2). Furthermore, a 3-bp (AAC) deletion at positions 566–568 was found in trin1, which resulted in deletion of an asparagine. Interestingly, there were five consecutive asparagines in the TRIN1 protein, but only four consecutive asparagines were existed in trin1 mutant. It remains to be dissected whether the asparagine deletion results in a change of protein structure or function. In this study we showed that TOR could play an important role in TRIN1/ABI4 degradation. The trin1 mutant was insensitive to mTOR kinase inhibitors and TRIN1 overexpression lines were sensitive to mTOR kinase inhibitors (Figures 3C,D), suggesting that TRIN1 maybe function as a regulator or component of the TOR signaling pathway in Arabidopsis.
TOR Regulates Cotyledons Greening Via TRIN1/ABI4
Based on the findings of this study, we present a potential working model highlighting the roles of TOR and TRIN1 in regulating the cotyledons greening in Arabidopsis (Figure 7). In this model, TOR functions as a master integrator for sensing and signaling the environmental stresses (water, nutrition and light energy stresses; Deprost et al., 2007; Ren et al., 2012; Robaglia et al., 2012; Rexin et al., 2015). Nutrition and energy can activate the activity of TOR while stresses and stress-induced ABA likely suppresses the functions of TOR (Deprost et al., 2007; Ren et al., 2012; Xiong et al., 2013; Rexin et al., 2015; Figure 7). TOR modulates cotyledons greening and hypocotyl elongation in a positive way, whereas TRIN1 acts as a downstream effector of TOR and can mediate TOR signaling to regulate cotyledons greening and hypocotyl elongation in a negative manner (Figure 7).
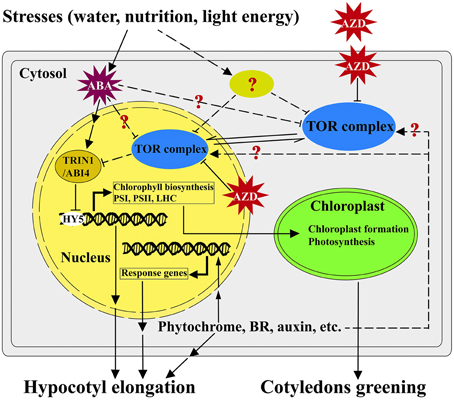
Figure 7. A working model of TOR-TRIN1 pathway for modulating cotyledons greening. A simplified model shows how TOR regulates cotyledons greening through TRIN1 in Arabidopsis. In this model, AZD is able to suppress the kinase activity of TOR. TOR negatively regulates the stability of the TRIN1/ABI4 protein in nucleus. TRIN1 functions as a negative regulator of chloroplast biogenesis and hypocotyl elongation through inhibiting the activity of Elongated Hypocotyl 5 (HY5). Hypocotyl elongation could be regulated by phytochrome, hormones and other signals might or might not mediate via TOR signaling. Arrows and T-bars represent enhancement and inhibition, respectively. The solid lines indicate the known confirmed or direct interactions. The dashed lines indicate experimentally unproven or indirect links.
It should be noted that the early studies showed that interference of TOR could significantly suppress hypocotyl elongation and chlorophyll biosynthesis in Arabidopsis (Deprost et al., 2007; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013), but little is known about the downstream effectors of TOR to mediate these phenotypes. This is largely due to the early embryo lethality of tor mutants which prevents people from screening downstream effectors via classic genetic approaches. In this model, AZD-TOR inducible inhibition system breaks the bottleneck and provides a platform to dissect TOR signaling pathway. For the first time, TRIN1 was identified as the downstream regulator of TOR by using AZD-TOR chemical genetics approach. Interestingly, TRIN1 is allelic to ABI4, which is a key regulator in abscisic acid (ABA) signaling cascade. It is well known that ABA plays pivotal roles in cotyledons greening through activating the expression of ABI4 (Chang and Walling, 1991; Acevedo-Hernández et al., 2005; Yamaguchi-Shinozaki and Shinozaki, 2006). The cotyledons greening regulated by TRIN1 in this study are highly consistent with the functions of ABI4 (Figure 7), indicating TRIN1/ABI4 mediates TOR signaling during cotyledons greening. Hypocotyl elongation is also a complex process which is influenced by phytochrome, Brassinosteriod (BR), auxin, ethylene and other signals (Figure 7). However, little is known about whether these signals modulate hypocotyl elongation via TOR signaling or not (Figure 7). Previous observations show that ABI4 is a negative inhibitor of Elongated Hypocotyl 5 (HY5) which is a key player in regulating the expression of PhANGs and hypocotyl elongation (Susek et al., 1993; McCormac and Terry, 2004; Acevedo-Hernández et al., 2005; Koussevitzky et al., 2007; Jarvis and López-Juez, 2013; Terry and Smith, 2013). Overexpression of ABI4 suppresses HY5 and thus inhibits hypocotyl elongation in Arabidopsis (Acevedo-Hernández et al., 2005; Koussevitzky et al., 2007; Finkelstein et al., 2011; Jarvis and López-Juez, 2013; Ludwików, 2015). Altogether, these results demonstrate that TOR regulates cotyledons greening and hypocotyl elongation through TRIN1/ABI4 in Arabidopsis (Figure 7). This study provides a platform to dissect functions of TOR signaling cascade in plants. Further dissection of TOR-TRIN1 signaling cascade will significantly advance our understanding of TOR signaling in plants.
Author Contributions
MR, LL, and YS designed the experiments. LL, YS, KW, XZ, PD, ZL, and MR performed the experiments. PD and FL analyzed the data. MR and LL wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Chongqing Frontier and Applied Basic Research (No. CSTC2014JCYJA80012), National Basic Research Program of China (No. 2013CB127100), the Open Project Funding of State Key Laboratory of Cotton Biology (No. CB2014A08 and CB2015A14), and Promote Scientific Research and Cooperation and High level Personnel Training Project in America and Oceania supported by Ministry of Education (No. 0903005109094/003).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00861
Supplemental Figure 1. The vectors of P35S::TRIN1-HA, P35S::TRIN1-GUS, P35S::GUS and PTRIN1::GUS.
Supplemental Figure 2. Amino acid sequence alignment of the catalytic domain of TOR in Saccharomyces cerevisiae, Arabidopsis thaliana and Homo sapiens (http://www.ncbi.nlm.nih.gov/protein/). The highly conserved amino acids are highlighted in the red box. The yellow alignment depicts amino acids with 75% similarity.
Supplemental Figure 3. AtTOR protein structure and predicted phosphorylation sites of TRIN1 and trin1 protein. (A) The interaction of AtTOR domains and RAPTOR, FKBP12, rapamycin (RAP), LST8, and TOR kianse inhibitors (Ku63794, Torin1, and AZD-8055). HEAT repeats: Huntingtin, Elongation factor 3, subunit of protein phosphatase 2A and TOR1; FAT: FRAP, ATM, and TRRAP domain; FRB: FKP12-rapamycin binding domain; FATC: Carboxy-terminal FAT domain; RAPTOR: Regulatory associate protein of TOR; LST8: Lethal with sec-13 protein 8 (Perry and Kleckner, 2003). (B) Predicted phosphorylation sites of TRIN1 and trin1 protein. The websites of predicting phosphorylation sites is: http://KinasePhos.mbc.nctu.edu.tw/; a point mutation that results in a change in the first base of codon 229 of trin1 from A to G was identified, which results in the substitution of threonine (ACT) with an alanine (GCT), we predicted the threonine sites of TRIN1 and trin1 protein. The different sites of TRIN1 and trin1 were marked with a red oval and rectangle, respectively. Predicted phosphorylation sites of the TRIN1 protein is presented at the left, and the right panel shows phosphorylation sites of the trin1 protein.
Supplemental Figure 4. Phylogenetic tree of the TRIN1 sequences of 30 taxa. TRIN1 sequences were collected from different source databases (Phytozome http://www.phytozome.net, version 8.0; Plaza http://bioinformatics.psb.ugent.be/plaza/ version 2.5 and NCBI Genbank http://www.ncbi.nlm.nih.gov/genbank/) based on sequence similarity with Arabidopsis thaliana TRIN1 gene (Wind et al., 2013). The evolutionary history was inferred using the UPGMA method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. The analysis involved 33 nucleotide sequences. Evolutionary analyses were conducted in MEGA5.
Supplemental Figure 5. P35S::TRIN1 overexpression lines were sensitive to AZD. (A–C) P35S::TRIN1 overexpression lines and WT seeds were cultured on ½ medium containing DMSO, 1 μM AZD and 10 μM Torin1 for 15 days. Images of representative seedlings were captured. (D) RT-PCR analysis of TRIN1 transcript level of 2-week-old P35S::TRIN1 overexpression lines. The name of lanes is the same as (A) from left to right.
Supplemental Figure 6. The GUS activity of P35S::TRIN1-GUS overexpression lines. GUS staining of the first true leaves of 2-week-old P35S::TRIN1-GUS lines treated with DMSO and 5 μM AZD for 48 h.
Supplemental Table 1. Primers for cloning promoter or full-length CDS.
Supplemental Table 2. Primers for quantitative real-time PCR.
Supplemental Table 3. The targets of AZD8055 and Torin1 and their IC50.
Supplemental Table 4. The comparison of the homologs of PI3Ks between animals and plants.
References
Acevedo-Hernández, G. J., León, P., and Herrera-Estrella, L. R. (2005). Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 43, 506–519. doi: 10.1111/j.1365-313X.2005.02468.x
Ahn, C. S., Han, J. A., Lee, H. S., Lee, S., and Pai, H. S. (2011). The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23, 185–209. doi: 10.1105/tpc.110.074005
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brown, E. J., Albers, M. W., Shin, T. B., Ichikawa, K., Keith, C. T., Lane, W. S., et al. (1994). A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369, 756–758. doi: 10.1038/369756a0
Burnett, P. E., Barrow, R. K., Cohen, N. A., Snyder, S. H., and Sabatini, D. M. (1998). RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. U.S.A. 95, 1432–1437. doi: 10.1073/pnas.95.4.1432
Cafferkey, R., Young, P. R., McLaughlin, M. M., Bergsma, D. J., Koltin, Y., Sathe, G. M., et al. (1993). Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13, 6012–6023.
Calabrese, E. J., and Blain, R. B. (2009). Hormesis and plant biology. Environ. Pollut. 157, 42–48. doi: 10.1016/j.envpol.2008.07.028
Caldana, C., Li, Y., Leisse, A., Zhang, Y., Bartholomaeus, L., Fernie, A. R., et al. (2013). Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 73, 897–909. doi: 10.1111/tpj.12080
Chresta, C. M., Davies, B. R., Hickson, I., Harding, T., Cosulich, S., Critchlow, S. E., et al. (2010). AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 70, 288–298. doi: 10.1158/0008-5472.CAN-09-1751
Deprost, D., Yao, L., Sormani, R., Moreau, M., Leterreux, G., Nicolaï, M., et al. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8, 864–870. doi: 10.1038/sj.embor.7401043
De Virgilio, C., and Loewith, R. (2006). Cell growth control: little eukaryotes make big contributions. Oncogene 25, 6392–6415. doi: 10.1038/sj.onc.1209884
Dong, P., Xiong, F., Que, Y., Wang, K., Yu, L., Li, Z., et al. (2015). Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front. Plant Sci. 6:677. doi: 10.3389/fpls.2015.00677
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Feldman, M. E., Apsel, B., Uotila, A., Loewith, R., Knight, Z. A., Ruggero, D., et al. (2009). Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7:e38. doi: 10.1371/journal.pbio.1000038
Finkelstein, R., Lynch, T., Reeves, W., Petitfils, M., and Mostachetti, M. (2011). Accumulation of the transcription factor ABA-insensitive (ABI)4 is tightly regulated post-transcriptionally. J. Exp. Bot. 62, 3971–3979. doi: 10.1093/jxb/err093
Finkelstein, R. R., Wang, M. L., Lynch, T. J., Rao, S., and Goodman, H. M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10, 1043–1054. doi: 10.1105/tpc.10.6.1043
García-Martinez, J. M., Moran, J., Clarke, R. G., Gray, A., Cosulich, S. C., Chresta, C. M., et al. (2009). Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem. J. 421, 29–42. doi: 10.1042/BJ20090489
Gregorio, J., Hernández-Bernal, A. F., Cordoba, E., and León, P. (2014). Characterization of evolutionarily conserved motifs involved in activity and regulation of the ABA-INSENSITIVE (ABI) 4 transcription factor. Mol. Plant 7, 422–436. doi: 10.1093/mp/sst132
Heitman, J., Movva, N. R., and Hall, M. N. (1991a). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909. doi: 10.1126/science.1715094
Heitman, J., Movva, N. R., Hiestand, P. C., and Hall, M. N. (1991b). FK-506-binding protein proline rotamase is a target for the immunosuppressive agent FK-506 in Saccharomyces-cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 88, 1948–1952. doi: 10.1073/pnas.88.5.1948
Horváth, B. M., Magyar, Z., Zhang, Y. X., Hamburger, A. W., Bakó, L., Visser, R. G., et al. (2006). EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 25, 4909–4920. doi: 10.1038/sj.emboj.7601362
Ilag, L. L., Kumar, A. M., and Söll, D. (1994). Light regulation of chlorophyll biosynthesis at the level of 5-aminolevulinate formation in Arabidopsis. Plant Cell 6, 265–275. doi: 10.1105/tpc.6.2.265
Jarvis, P., and López-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. doi: 10.1126/science.1140516
Kunz, J., Henriquez, R., Schneider, U., Deuter-Reinhard, M., Movva, N. R., and Hall, M. N. (1993). Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73, 585–596. doi: 10.1016/0092-8674(93)90144-F
Laplante, M., and Sabatini, D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274–293. doi: 10.1016/j.cell.2012.03.017
León, P., Gregorio, J., and Cordoba, E. (2012). ABI4 and its role in chloroplast retrograde communication. Front. Plant Sci. 3:304. doi: 10.3389/fpls.2012.00304
Leutwiler, L. S., Meyerowitz, E. M., and Tobin, E. M. (1986). Structure and expression of three light-harvesting chlorophyll a/b-binding protein genes in Arabidopsis thaliana. Nucleic Acids Res. 14, 4051–4064. doi: 10.1093/nar/14.10.4051
Liu, Q., Wang, J., Kang, S. A., Thoreen, C. C., Hur, W., Ahmed, T., et al. (2011). Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl) benzo[h][1,6]naphthyridin-2 (1H) -one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. J. Med. Chem. 54, 1473–1480. doi: 10.1021/jm101520v
Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J. L., Bonenfant, D., et al. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468. doi: 10.1016/S1097-2765(02)00636-6
Ludwików, A. (2015). Targeting proteins for proteasomal degradation-a new function of Arabidopsis ABI1 protein phosphatase 2C. Front. Plant Sci. 6:310. doi: 10.3389/fpls.2015.00310
Mahfouz, M. M., Kim, S., Delauney, A. J., and Verma, D. P. (2006). Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18, 477–490. doi: 10.1105/tpc.105.035931
Martin, D. E., and Hall, M. N. (2005). The expanding TOR signaling network. Curr. Opin. Cell Biol. 17, 158–166. doi: 10.1016/j.ceb.2005.02.008
Martin, D. E., Soulard, A., and Hall, M. N. (2004). TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119, 969–979. doi: 10.1016/j.cell.2004.11.047
McCormac, A. C., and Terry, M. J. (2004). The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signalling pathways during de-etiolation. Plant J. 40, 672–685. doi: 10.1111/j.1365-313X.2004.02243.x
Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C., et al. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. U.S.A. 99, 6422–6427. doi: 10.1073/pnas.092141899
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Montané, M. H., and Menand, B. (2013). ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 64, 4361–4374. doi: 10.1093/jxb/ert242
Moreau, M., Azzopardi, M., Clément, G., Dobrenel, T., Marchive, C., Renne, C., et al. (2012). Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24, 463–481. doi: 10.1105/tpc.111.091306
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Niu, X., Helentjaris, T., and Bate, N. J. (2002). Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. Plant Cell 14, 2565–2575. doi: 10.1105/tpc.003400
Nojima, H., Tokunaga, C., Eguchi, S., Oshiro, N., Hidayat, S., Yoshino, K., et al. (2003). The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278, 15461–15464. doi: 10.1074/jbc.C200665200
Perry, J., and Kleckner, N. (2003). The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112, 151–155. doi: 10.1016/S0092-8674(03)00033-3
Pruzinská, A., Tanner, G., Anders, I., Roca, M., and Hörtensteiner, S. (2003). Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. U.S.A. 100, 15259–15264. doi: 10.1073/pnas.2036571100
Ren, M., Qiu, S., Venglat, P., Xiang, D., Feng, L., Selvaraj, G., et al. (2011). Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 155, 1367–1382. doi: 10.1104/pp.110.169045
Ren, M., Venglat, P., Qiu, S., Feng, L., Cao, Y., Wang, E., et al. (2012). Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24, 4850–4874. doi: 10.1105/tpc.112.107144
Rexin, D., Meyer, C., Robaglia, C., and Veit, B. (2015). TOR signalling in plants. Biochem. J. 470, 1–14. doi: 10.1042/BJ20150505
Robaglia, C., Thomas, M., and Meyer, C. (2012). Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 15, 301–307. doi: 10.1016/j.pbi.2012.01.012
Sabatini, D. M., Erdjument-Bromage, H., Lui, M., Tempst, P., and Snyder, S. H. (1994). RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78, 35–43. doi: 10.1016/0092-8674(94)90570-3
Sabers, C. J., Martin, M. M., Brunn, G. J., Williams, J. M., Dumont, F. J., Wiederrecht, G., et al. (1995). Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 270, 815–822. doi: 10.1074/jbc.270.2.815
Sarbassov, D. D., Ali, S. M., Kim, D. H., Guertin, D. A., Latek, R. R., Erdjument-Bromage, H., et al. (2004). Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302. doi: 10.1016/j.cub.2004.06.054
Schelbert, S., Aubry, S., Burla, B., Agne, B., Kessler, F., Krupinska, K., et al. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21, 767–785. doi: 10.1105/tpc.108.064089
Schepetilnikov, M., Dimitrova, M., Mancera-Martínez, E., Geldreich, A., Keller, M., and Ryabova, L. A. (2013). TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 32, 1087–1102. doi: 10.1038/emboj.2013.61
Sormani, R., Yao, L., Menand, B., Ennar, N., Lecampion, C., Meyer, C., et al. (2007). Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 7:26. doi: 10.1186/1471-2229-7-26
Stone, S. L., Williams, L. A., Farmer, L. M., Vierstra, R. D., and Callis, J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428. doi: 10.1105/tpc.106.046532
Susek, R. E., Ausubel, F. M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. doi: 10.1016/0092-8674(93)90459-4
Terry, M. J., and Smith, A. G. (2013). A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front. Plant Sci. 4:14. doi: 10.3389/fpls.2013.00014
Thoreen, C. C., Kang, S. A., Chang, J. W., Liu, Q. S., Zhang, J. M., Gao, Y., et al. (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032. doi: 10.1074/jbc.M900301200
Wang, X., and Proud, C. G. (2009). Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol. 19, 260–267. doi: 10.1016/j.tcb.2009.03.005
Weaver, L. M., and Amasino, R. M. (2001). Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 127, 876–886. doi: 10.1104/pp.010312
Wind, J. J., Peviani, A., Snel, B., Hanson, J., and Smeekens, S. C. (2013). ABI4: versatile activator and repressor. Trends Plant Sci. 18, 125–132. doi: 10.1016/j.tplants.2012.10.004
Wullschleger, S., Loewith, R., and Hall, M. N. (2006). TOR signaling in growth and metabolism. Cell 124, 471–484. doi: 10.1016/j.cell.2006.01.016
Xiong, Y., McCormack, M., Li, L., Hall, Q., Xiang, C. B., and Sheen, J. (2013). Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186. doi: 10.1038/nature12030
Xiong, Y., and Sheen, J. (2012). Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 287, 2836–2842. doi: 10.1074/jbc.M111.300749
Xiong, Y., and Sheen, J. (2013). Moving beyond translation Glucose-TOR signaling in the transcriptional control of cell cycle. Cell Cycle 12, 1989–1990. doi: 10.4161/cc.25308
Xiong, Y., and Sheen, J. (2014). The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 164, 499–512. doi: 10.1104/pp.113.229948
Xu, Q., Liang, S., Kudla, J., and Luan, S. (1998). Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J. 15, 511–519. doi: 10.1046/j.1365-313X.1998.00232.x
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Chang, Y. C., and Walling, L. L. (1991). Abscisic acid negatively regulates expression of chlorophyll a/b binding protein genes during soybean embryogeny. Plant Physiol. 97, 1260–1264. doi: 10.1104/pp.97.3.1260
Yuan, H. X., Xiong, Y., and Guan, K. L. (2013). Nutrient sensing, metabolism, and cell growth control. Mol. Cell 49, 379–387. doi: 10.1016/j.molcel.2013.01.019
Zhang, X., Garreton, V., and Chua, N. H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543. doi: 10.1101/gad.1318705
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Keywords: TRIN1, TOR, TOR kinase inhibitors, cotyledons greening, Arabidopsis
Citation: Li L, Song Y, Wang K, Dong P, Zhang X, Li F, Li Z and Ren M (2015) TOR-inhibitor insensitive-1 (TRIN1) regulates cotyledons greening in Arabidopsis. Front. Plant Sci. 6:861. doi: 10.3389/fpls.2015.00861
Received: 23 May 2015; Accepted: 29 September 2015;
Published: 19 October 2015.
Edited by:
Hartmut Stützel, Leibniz Universität Hannover, GermanyReviewed by:
Rita Maria Zrenner, Leibniz-Institute of Vegetable and Ornamental Crops (IGZ), GermanyTsu-Wei Chen, Leibniz Universität Hannover, Germany
Copyright © 2015 Li, Song, Wang, Dong, Zhang, Li, Li and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maozhi Ren, renmaozhi@cqu.edu.cn
†These authors have contributed equally to this work.
 Linxuan Li1†
Linxuan Li1† Maozhi Ren
Maozhi Ren