- AgResearch Limited, Palmerston North, New Zealand
The synthesis of anthocyanin pigments and proanthocyanidins (condensed tannins) is regulated by MYB-bHLH-WDR (MBW) transcription factor complexes in all angiosperms studied to date. Tr-MYB133 and Tr-MYB134 were isolated from Trifolium repens and encode R2R3-MYBs that antagonize the activity of MBW activation complexes. These two genes are conserved in other legume species, and form two sub-clades within the larger anthocyanin/proanthocyanidin clade of MYB repressors. However, unlike petunia and Arabidopsis, these R2R3-MYB repressors do not prevent ectopic accumulation of anthocyanins or proanthocyanidins. Instead, they are expressed when anthocyanins or proanthocyanidins are being synthesized, and provide feedback regulation to MBW complexes. This feedback occurs because Tr-MYB133 and Tr-MYB134 are themselves regulated by MBW complexes. Tr-MYB133 is regulated by MBW complexes containing anthocyanin-related R2R3-MYB proteins (Tr-RED LEAF), while Tr-MYB134 is regulated by complexes containing the proanthocyanidin R2R3-MYBs (Tr-MYB14). Other features of the MBW gene regulation networks are also conserved within legumes, including the ability for the anthocyanin MBW complexes to activate the expression of the AN1/TT8 clade bHLH factor. The regulation of Tr-MYB133 and Tr-MYB134 by distinct, pathway-specific MBW complexes has resulted in subspecialization for controlling anthocyanin or proanthocyanidin synthesis.
Introduction
Anthocyanins and proanthocyanidins (syn. condensed tannins) are related flavonoid compounds that are produced by plants throughout development, and in response to biotic and abiotic stresses (Dixon and Paiva, 1995). Anthocyanins are red/purple/blue pigments that provide color to flowers, fruits and vegetative tissues in plants, fulfilling a variety of physiological requirements. In flowers and fruits, anthocyanins provide visual cues to pollinators and seed distributers, respectively, and can form highly elaborate patterns (e.g., stripes, spots), especially when combined with other types of pigments (Davies et al., 2012). In vegetative tissues, anthocyanins are commonly produced in response to environmental conditions that may compromise photosynthetic activity (e.g., high light, cold, nutrient deficiency), which can result in photoinhibition and the production of reactive oxygen species (ROS; Gould, 2004). Anthocyanins ameliorate the effect of these stresses by screening underlying photosynthetic tissue from excess light (Neill and Gould, 2003; Hughes et al., 2005; Albert et al., 2009; Hatier et al., 2013), and may also act as antioxidants in planta to neutralize ROS (Gould et al., 2002; Landi et al., 2014). Proanthocyanidins are structurally related to anthocyanins, being composed of anthocyanidin-based subunits (flavan-3-ols) that are polymerized into proanthocyanidin molecules (Zhao et al., 2010). The accumulation and distribution of proanthocyanidins varies widely between plant species, being found in seed coats of most angiosperms, but also accumulating in flowers, fruits, and leaves in some taxa (Dixon et al., 2005). They act as feeding deterrents for chewing insects and herbivores because of their astringency properties, and may also have structural functions (Barbehenn and Constabel, 2011).
Anthocyanins and proanthocyanidins offer health benefits to humans and other animals. Dietary anthocyanins have been associated with improvements in a range of risk factors for chronic diseases including metabolic syndrome, cardiovascular disease, and certain cancers (Martin et al., 2011), and there is increasing interest in fortifying fruits and vegetables with increased quantities of anthocyanins (Butelli et al., 2008; Espley et al., 2013). In contrast, proanthocyanidins offer a range of health and production benefits to grazing ruminants (e.g., sheep, cattle), due to their protein-binding properties. Dietary protein from forage legumes is rapidly fermented in the rumen, producing methane gas within stable foams and resulting in the potentially lethal condition, pasture bloat. The inclusion of proanthocyanidins in the diet prevents pasture bloat, by protecting protein from premature fermentation. This increases animal weight and milk production because of improved amino acid absorption, and also reduces methane and nitrogen emissions (Douglas et al., 1999; Beauchemin et al., 2007). The major forage legumes grown for pastoral agriculture are alfalfa (Medicago sativa) and white clover (Trifolium repens), which lack significant quantities of proanthocyanidins in their leaves. Therefore, there is strong interest in developing high-proanthocyanidin forage legumes using genetic technologies (Dixon et al., 2013).
Anthocyanin and proanthocyanidin biosynthesis is regulated primarily at the transcriptional level, by MBW transcription factor complexes (Koes et al., 2005). Both anthocyanin and proanthocyanidin biosynthesis share the same WDR protein and bHLH2/AN1/TT8 clade bHLH factor, which form the MBW complex together with R2R3-MYB proteins (Zhang et al., 2003; Baudry et al., 2004; Davies et al., 2012). These R2R3-MYB proteins provide specificity to the complex and determine which pathways are regulated (anthocyanins vs. proanthocyanidins; Ramsay and Glover, 2005; Heppel et al., 2013). While all three components are necessary to activate anthocyanin synthesis, the R2R3-MYB genes largely determine the pigmentation patterning, as these are typically encoded by small gene families with diverse spatial expression patterns (Schwinn et al., 2006; Albert et al., 2011; Lowry et al., 2012). For proanthocyanidin regulation, multiple R2R3-MYB factors have been identified that act within MBW complexes. However, unlike the small gene families of anthocyanin-related R2R3-MYB genes, the proanthocyanidin-related MYBs studied to date appear to differ in their functions. For example, MYB14 from Trifolium arvense and Medicago truncatula activates the accumulation of proanthocyanidins when expressed in T. repens or Medicago (Hancock et al., 2012; Liu et al., 2014), while other proanthocyanidin-related MYBs are either less effective or are unable to activate proanthocyanidin synthesis when expressed alone (Sharma and Dixon, 2005; Verdier et al., 2012). It is not yet clear how these MYB genes act together, although it seems likely that they may act upon different subsets of target genes, to cooperatively activate proanthocyanidin synthesis (Deluc et al., 2008; Terrier et al., 2009; Liu et al., 2014).
The MBW complex operates within a gene regulation network that involves reinforcement and feedback repression. Current models suggest that the MBW complex activates the expression of the bHLH2/AN1/TT8 clade bHLH factor to provide reinforcement, and also activates the expression of R2R3-MYB and R3-MYB repressors to provide feedback repression (Albert et al., 2014a). In both petunia and Arabidopsis, the R2R3-MYB repressors (At-MYBL2 contains a partial R2 domain) are also expressed in leaves during non-stress conditions, preventing inappropriate synthesis of anthocyanins (Dubos et al., 2008; Albert et al., 2011). The R2R3-MYB repressors actively repress transcription through motifs (EAR, TLLLFR) in their C-terminal domains (Aharoni et al., 2001; Matsui et al., 2008; Albert et al., 2014a), while the small R3-MYB proteins act competitively to inhibit the formation of functional MBW complexes (Schellmann et al., 2002; Yuan et al., 2013; Albert et al., 2014a).
The R2R3-MYB repressors that regulate anthocyanin (Aharoni et al., 2001; Dubos et al., 2008; Matsui et al., 2008; Albert et al., 2011, 2014a; Salvatierra et al., 2013) and proanthocyanidin synthesis (Huang et al., 2014; Cavallini et al., 2015; Yoshida et al., 2015) act upon MBW complexes (Matsui et al., 2008; Albert et al., 2014a), which differs from the R2R3-MYB repressors that were first identified as regulators of cinnamic acid derivatives (e.g., At-MYB4; Tamagnone et al., 1998; Jin et al., 2000). Many features of the MBW gene regulation network are likely to be conserved in eudicots (Albert et al., 2014a), if not more widely across angiosperms (Albert et al., 2014b), but it is not yet known which features are conserved and which differ in legumes.
The synthesis of anthocyanins and proanthocyanidins in white clover (T. repens) is tightly regulated, forming intricate anthocyanin pigmentation patterns in leaves (Albert et al., 2015) and restricted accumulation of proanthocyanidins to specific organs and tissues (Abeynayake et al., 2012; Hancock et al., 2012). Improving the content and distribution of proanthocyanidins in white clover is highly desirable for agricultural purposes, but this first requires an understanding of how the gene regulation networks determine anthocyanin and proanthocyanidin synthesis. The roles of repressors for proanthocyanidin regulation in forage legumes are currently unknown, yet these have been proposed as interesting gene targets for increasing the anthocyanin or proanthocyanidin content through mutagenesis and/or breeding strategies (Albert et al., 2014c).
The aim of this research was to investigate how the anthocyanin and proanthocyanidin biosynthetic pathways are regulated in legumes, and determine if R2R3-MYB repressors perform similar roles for regulating flavonoid synthesis, as they do in other groups of angiosperms. In this study, two distinct R2R3-MYB repressors were identified and characterized in white clover for their roles for regulating anthocyanin and proanthocyanidin synthesis.
Materials and Methods
Plant Material and Growth Conditions
Trifolium repens cultivar ‘Sustain’ plants were germinated from seed while T. repens plants with the ‘red leaf’ or ‘red leaflet’ anthocyanin leaf markings were clonally propagated (four biological replicates). Plants were grown in pots within a greenhouse that was heated at 15°C and vented at 25°C with ambient lighting. Clonal copies of plants with the ‘red leaflet’ marking were also grown outside during winter [July 2013; mean monthly air temperature 6.1°C/9.7°C (min/max); mean min ground temperature 3.3°C; mean relative humidity 88.7%], to expose plants to cold temperatures that induce the anthocyanin leaf marking (Albert et al., 2015). Expanding leaves were sampled for RNA isolation early during the illuminated part of the day, while temperatures will still cool outside (>10°C). Inflorescences were sampled from the ‘red leaflet’ genotype grown in the greenhouse at two developmental stages; when the first flowers were beginning to open (Inf. A), and when half of the inflorescence had open flowers, but before any flowers senesced (Inf. B). Tissue was immediately frozen in liquid nitrogen upon collection. The presence of proanthocyanidins were detected by staining with p-dimethylaminocinnamaldehyde (DMACA; Li et al., 1996).
RNA Isolation
Total RNA was isolated from ∼100 mg leaf tissue with the ISOLATE II RNA Plant mini kit using lysis buffer APR (Bioline, Auckland, New Zealand). The presence of proanthocyanidins in inflorescences required modification of the cell lysis conditions, to prevent proanthocyanidins from binding nucleic acids and inhibiting RNA isolation and purification. Frozen tissue (∼100 mg) was immediately homogenized in lysis buffer BPR supplemented with 4% (w/v) cetyltrimethyl ammonium bromide and 4% (w/v) polyvinylpyrrolidone, and then processed as per manufacturer’s instructions.
Gene Isolation
First stand cDNA was synthesized from 4 μg total RNA (mixed tissues and developmental stages from leaves and inflorescences) using Tetro cDNA synthesis kit (Bioline), primed with NAg11 (Supplementary Table S1). PCR using degenerate oligonucleotides (NAg3/NAg4) was performed using 2 μL of first strand cDNA, 250 nM of each oligonucleotide with MyTaqTM Red polymerase mastermix (Bioline) in 50 μL reactions. Amplification was performed with the following cycling conditions: 94°C 2 min; (94°C 15 s, 58-1°C/cycle 30 s, 72°C 1 min) × 10; (94°C 15 s, 48°C 30 s, 72°C 1 min) × 30; 72°C 5 min, 12°C hold. PCR products were sequenced, and primers were designed to extend sequences by 3′ RACE. 3′ RACE PCR was performed upon cDNAs primed with NAg11, essentially as described in Albert et al. (2015); primers NAg34/NAg12, and NAg35/NAg13 were used for the primary and secondary PCR reactions, respectively, for Tr-MYB133; primers NAg72/NAg12, and NAg73/NAg13 were used for the primary and secondary PCR reactions, respectively, for Tr-MYB134. The sequences were extended further by 5′ RACE using the SMARTer® RACE cDNA amplification kit (Clontech), following the manufacturer’s instructions, with gene-specific primers: Tr-MYB133, NAg49 and NAg47; Tr-MYB134, NAg45, and NAg46. Full cDNAs were amplified with primers NAg100/NAg103 and NAg72/NAg96 for Tr-MYB133 and Tr-MYB134, respectively.
Phylogenetic Analysis
A maximum likelihood phylogenetic tree was generated, with 1000 bootstrap replicates, upon amino acid sequences for R2R3-MYB repressors which were aligned using MUSCLE in Geneious software (v7; Kearse et al., 2012) and manually adjusted.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
First strand cDNA was prepared from 1 μg DNAseI-treated total DNA using iScript reverse transcriptase mastermix (Biorad), and diluted 20-fold with water. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed essentially as described in Dorling et al. (2011) upon four biological replicates per treatment, with three technical replicates, using Kapa SYBR® FAST Universal qPCR reagents for Tr-MYB133, Tr-MYB134, Tr-AN1, DFR, GAPDH, and PP2 assays (KAPA Biosystems) or Kapa probe FAST Universal qPCR reagents for ACTIN, ANR, and Tr-MYB14 (Supplementary Table S1 for primer and probe sequences). Relative transcript abundance was determined relative to the geometric mean of ACTIN, GAPDH, and PP2 (Dorling et al., 2011).
Luciferase Assays
The ANR, Tr-MYB133, and Tr-MYB134 promoters from T. repens were amplified and cloned into a dual luciferase vector, pNWA62, as has previously been done for the DFR promoter (Albert et al., 2015). Effector constructs that constitutively express transcription factors from a CaMV35S promoter were generated by cloning the coding sequence into pENTR-D Topo, and LR-recombination (Life technologies) into the gateway adapted binary vector pRSH1. Binary vectors containing the effector and dual luciferase reporter constructs were transformed into Agrobacterium tumefaciens (GV3101). Agrobacterium strains were grown and cells were resuspended in 10 mM MgCl2 containing 250 μM acetosyringone and cultured for 4 h at room temperature, and cultures were infiltrated into Nicotiana benthamiana leaves. Dual luciferase assays were performed with six biological replicates, using DLAR-2B reagents (Targeting Systems) as described in Albert et al. (2014a). Data are expressed as ratios of firefly:renilla luciferase activity.
Statistical Analyses
One-way ANOVA was performed with post hoc Fishers LSD (5%) using Genstat software (version 15). Analyses were performed upon log10-transformed data to normalize variances.
Results
Two Distinct Clades of Flavonoid R2R3-MYB Repressors Exist in Legumes
A candidate gene approach was taken to identify R2R3-MYB repressor genes that may be associated with regulating anthocyanin and/or proanthocyanidin synthesis in white clover. Fa-MYB1 from strawberry (Aharoni et al., 2001), Ph-MYB27 from petunia (Albert et al., 2011, 2014a) and At-MYBL2 (a truncated R2R3-MYB repressor) from Arabidopsis (Dubos et al., 2008; Matsui et al., 2008) were used as queries to search sequence databases for Lotus japonicus, M. Truncatula, and Glycine max using BLASTp. Criteria used for screening putative repressors were the presence of the bHLH interaction motif ([D/E]Lx2[R/K]x3Lx6Lx3R; Zimmermann et al., 2004) and either an EAR motif (LxLxL, DLNxxP; Kagale et al., 2010) or similarity to the TLLLFR repression motif present in At-MYBL2 (Matsui et al., 2008). The putative repressors identified from Lotus (Lj-MYB133, Lj-MYB134), Medicago (Medtr4g85530, Medtr5g079670), and Glycine (Glyma20g01610, Glyma07g33960, Glyma02g41440) fall into two well-supported clades, represented by Lj-MYB133 and Lj-MYB134 from Lotus (Figure 1; Supplementary Figure S1). These sequences were aligned, and degenerate oligonucleotides were designed to the conserved DNA binding domain.
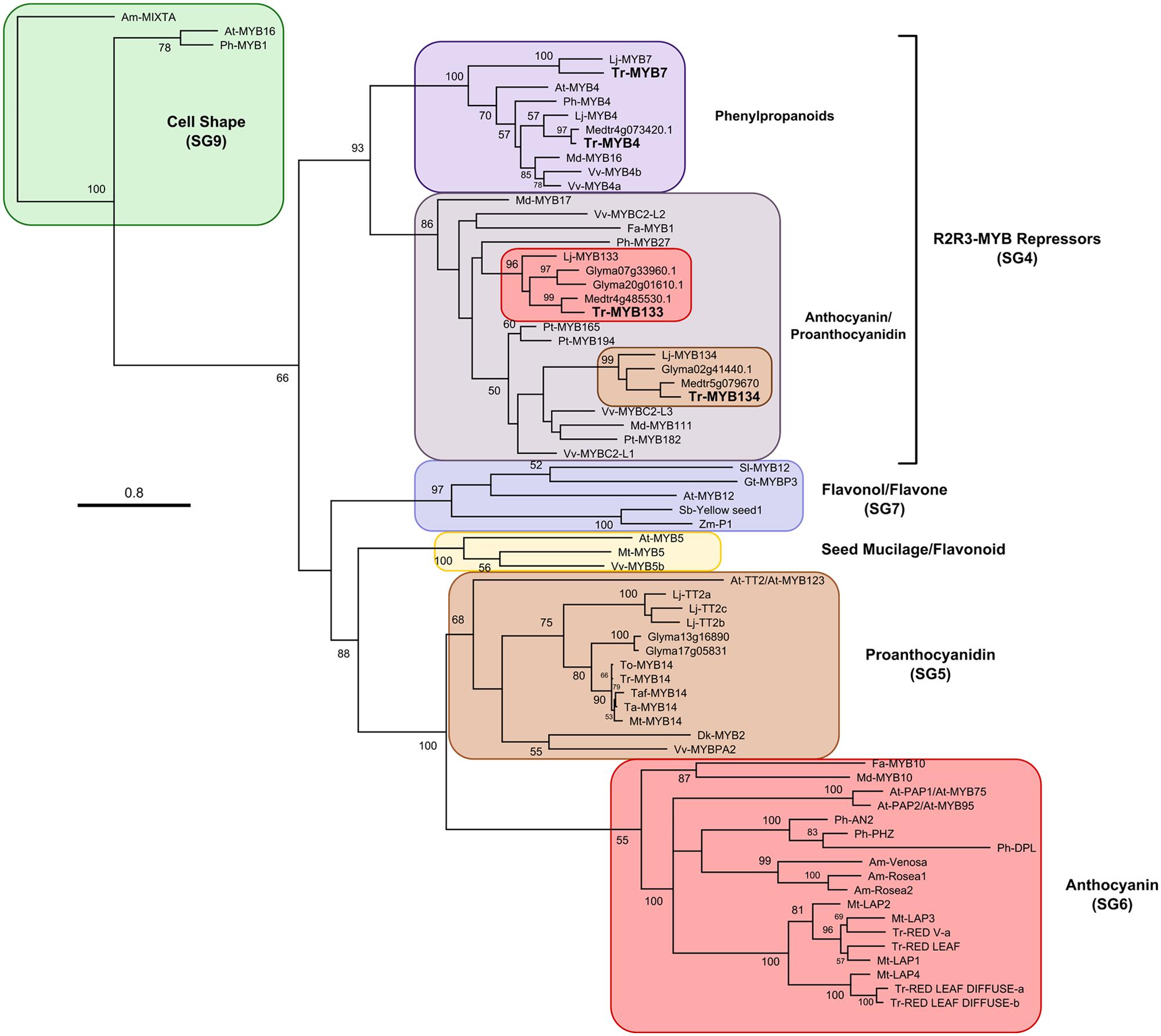
FIGURE 1. Maximum likelihood phylogenetic tree of R2R3-MYB proteins. Nodes with bootstrap support >50% from 1000 replicates are shown. Accession numbers for sequences used to construct this tree are provided in Supplementary Table S2. At, Arabidopsis thaliana; Am, Antirrhinum majus; Fa, Fragaria ananassa; Gt, Gentiana triflora; Gm, Glycine max; Lj, Lotus japonicus; Md, Malus domestica; Mt, Medicago truncatula; Ph, Petunia hybrida; Pt, Populus trichocarpa; Sl, Solanum lycopersicum; Ta, Trifolium arvense; Taf, T. affine; Tr, T. repens; To, T. occidentale; Vv, Vitis vinifera.
Two genes putatively encoding R2R3-MYB repressors were isolated from T. repens using a combination of PCR with degenerate oligonucleotides, and 3′ and 5′ rapid amplification of cDNA ends (RACE). The genes were named Tr-MYB133 and Tr-MYB134, based upon their sequence similarity to the corresponding homologs from L. japonicus (Shelton et al., 2012). Both Tr-MYB133 and Tr-MYB134 have the bHLH interaction motif present in the MYB DNA binding domain, and contain an LxLxL-type EAR repression motif in the C-terminus (Tr-MYB133 LDLNLELSL; Tr-MYB134 LNLEL).
Subsequent to isolating Tr-MYB133 and Tr-MYB134, an RNA-seq dataset became available from T. repens leaf tissue. Two additional R2R3-MYB repressors were identified, Tr-MYB4 and Tr-MYB7. These sequences belong to the MYB4-like clade of repressors, which are involved in regulating phenylpropanoid metabolism (e.g., cinnamic acid derivatives), rather than anthocyanins/proanthocyanidins directly, and therefore these were not pursued further.
The sequences of the putative R2R3-MYB repressors from T. repens were compared with other R2R3-MYB repressors and used to construct a phylogenetic tree (Figure 1). Tr-MYB133 and Tr-MYB134 fall within the well-supported clade of anthocyanin and proanthocyanidin repressors, which includes the functionally characterized anthocyanin repressors Fa-MYB1 and Ph-MYB27, and the proanthocyanidin repressors Vv-MYBC2-L1 and Pt-MYB165. The legume sequences form two distinct subclades, separating Tr-MYB133 and Tr-MYB134, raising the possibility that these two genes may have subspecialized, at least within legumes.
Tr-MYB133 and Tr-MYB134 Repress MBW Complex Activity
The activities of Tr-MYB133 and Tr-MYB134 were investigated by promoter activation/repression assays, using Agrobacterium-infiltrated N. benthamiana leaves. The promoters for DIHYDROFLAVONOL 4-REDUCTASE (DFR), a biosynthetic gene common to anthocyanin and proanthocyanidin pathways, and the proanthocyanidin biosynthetic gene ANTHOCYANIDIN REDUCTASE (ANR) were isolated from T. repens, and cloned into a dual luciferase reporter construct. Effector constructs expressing Tr-MYB133 or Tr-MYB134 were assayed with combinations of MBW constructs that have been characterized previously; the proanthocyanidin regulator Ta-MYB14 from Trifolium arvense (94% amino acid identity to Tr-MYB14; Hancock et al., 2012), the anthocyanin activator Tr-RED LEAF and bHLH factor Tr-AN1 (Albert et al., 2015).
The DFR promoter was activated when the R2R3-MYB anthocyanin regulator Tr-RED LEAF was co-infiltrated (Figure 2A), presumably forming MBW complexes with endogenous WDR and bHLH proteins (bHLH1 clade/JAF13) that are expressed in Nicotiana leaves (Albert et al., 2014a; Montefiori et al., 2015). Co-expression of Tr-AN1 (bHLH2 clade) enhanced the activation. However, co-transformation with Tr-MYB133 or Tr-MYB134 reduced the activity of the DFR promoter. Similarly, the R2R3-MYB proanthocyanidin regulator Ta-MYB14 activated the DFR promoter when the bHLH Tr-AN1 was coinfiltrated, but this was repressed by Tr-MYB133 and Tr-MYB134. The ANR promoter was activated when Ta-MYB14 was coinfiltrated with Tr-AN1, but this was repressed by Tr-MYB133 and Tr-MYB134 (Figure 2B). The anthocyanin regulator Tr-RED LEAF was unable to activate the ANR promoter, even when the bHLH Tr-AN1 was coinfiltrated.
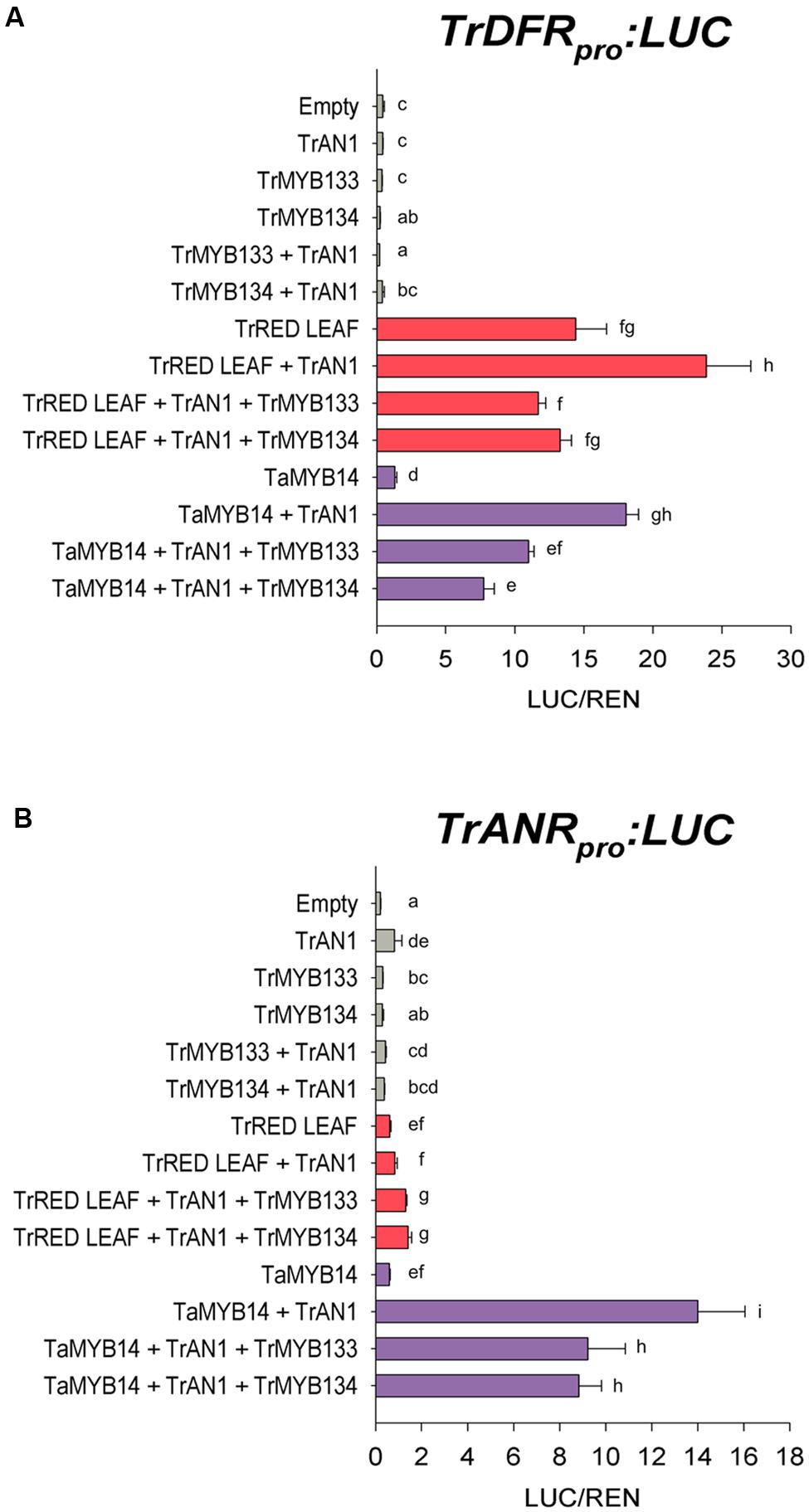
FIGURE 2. Tr-MYB133 and Tr-MYB134 repress flavonoid biosynthetic genes. Promoter activation/repression dual luciferase assays upon (A) the Tr-DFR (anthocyanin and proanthocyanidin biosynthetic gene) promoter or (B) the Tr-ANR promoter (proanthocyanin biosynthetic gene), using Agrobacterium-infiltrated Nicotiana benthamiana leaves. Combinations of activator R2R3-MYB genes Tr-RED LEAF (anthocyanin) or Ta-MYB14 (proanthocyanidin) were tested with or without the bHLH factor Tr-AN1, and the R2R3-MYB repressors Tr-MYB133 or Tr-MYB134. Firefly luciferase activity was normalized to Renilla luciferase activity (LUC/REN); mean ± SEM n = 5 biological replicates are shown. Mean that are significantly different are indicated by different letters, as determined by post hoc Fisher’s LSD (5%).
Tr-MYB133 and Tr-MYB134 Participate in Gene Regulation Networks
The expression patterns for Tr-MYB133, Tr-MYB134 and anthocyanin/proanthocyanidin biosynthesis and regulatory genes were examined to establish whether these genes are associated with the accumulation of these metabolites. This was conducted using T. repens germplasm and tissues that accumulate anthocyanins or proanthocyanidins, due to the expression of endogenous anthocyanin-related R2R3-MYB genes (e.g., Tr-RED LEAF), or the proanthocyanidin regulator Tr-MYB14, respectively.
Wild-type plants from cultivar ‘Sustain’ lack anthocyanin leaf markings (Figure 3A). In contrast, T. repens with ‘red leaf’ patterning have dark red leaves due to the accumulation of anthocyanins, which is regulated by the R2R3-MYB gene RED LEAFRL (Albert et al., 2015). Clonal copies of a T. repens genotype with the ‘red leaflet’ (distinct from ‘red leaf’) anthocyanin leaf marking were grown in a greenhouse (warm), or outside during winter (cold). Anthocyanin pigmentation associated with the ‘red leaflet’ locus (Vrl) only occurs in response to cold temperatures (Figure 3A). However, the R2R3-MYB activator responsible for ‘red leaflet’ patterning has not yet been conclusively identified or mapped to the Vrl locus (Albert et al., 2015). Developing inflorescences contain very little anthocyanin, and petals appear white or pale pink, although anthocyanin spots are present on the calyx.
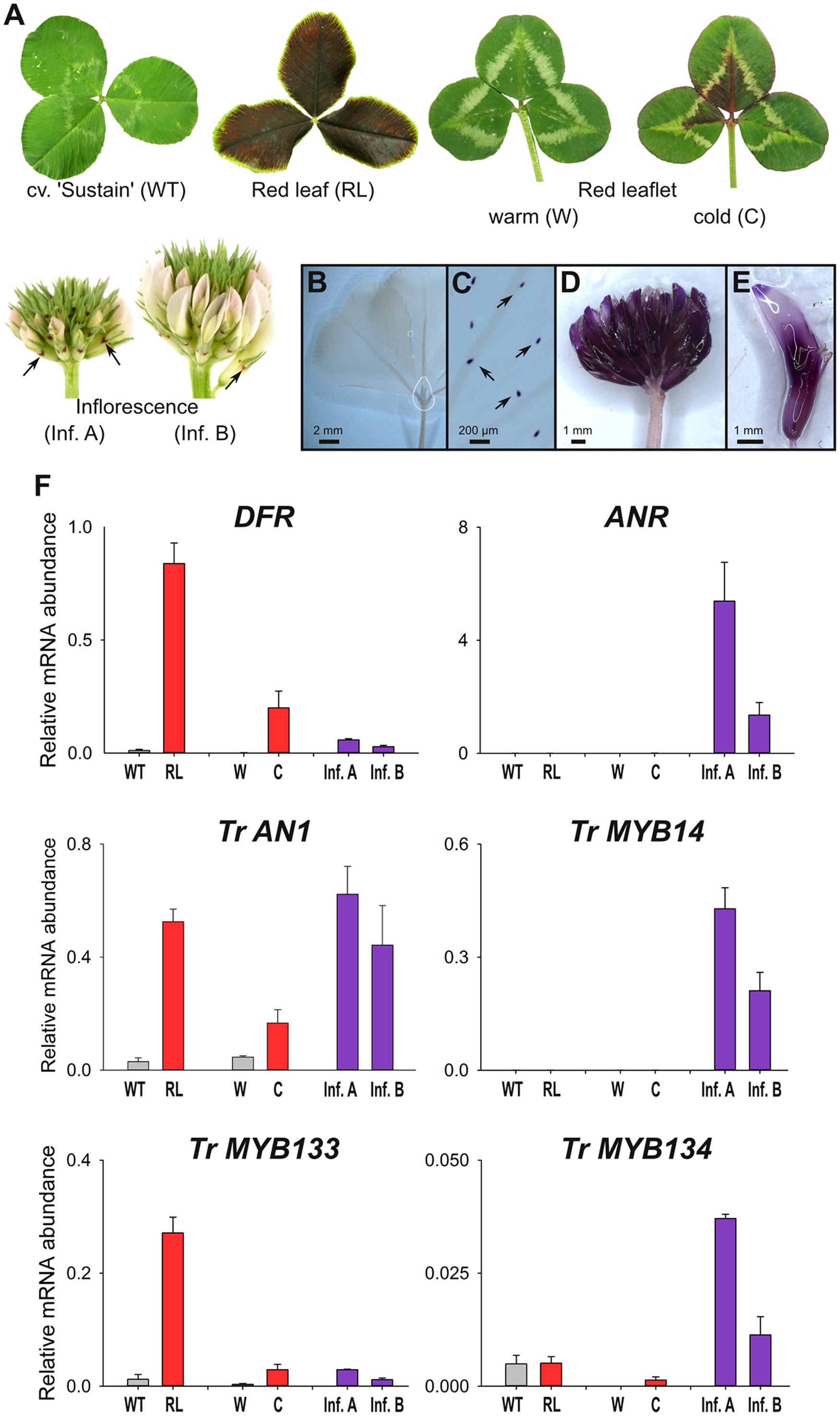
FIGURE 3. MYB133 and MYB134 are expressed in tissues accumulating anthocyanin and proanthocyanidins. (A) Anthocyanin pigmentation phenotypes of samples used for qRT-PCR. White clover ‘Sustain’ (WT) lacks anthocyanins in leaves, while a genotype with the ‘Red leaf’ (RL) anthocyanin mark accumulates anthocyanins throughout the lamina. A genotype with the ‘Red leaflet’ trait lack anthocyanins when grown under warm conditions (W), but are induced with cold temperature (C). Inflorescences have low quantities of anthocyanins, although anthocyanin spots (arrows) are present on the calyx. (B–E) Tissues stained with DMACA to detect proanthocyanidins (purple). (B) White clover leaves lack proanthocyanidins, except in their trichomes (C). (D) Proanthocyanidins accumulate in inflorescences and (E) within individual florets. (F) Relative transcript abundance of genes involved in anthocyanin and proanthocyanidin synthesis and regulation was determined by qRT-PCR. Mean ± SEM n = 4 biological replicates is shown.
The presence and spatial localization of proanthocyanidins was determined by staining tissues with DMACA, which reacts with polymerized flavanols to form an insoluble blue/purple precipitate. DMACA does not react with anthocyanin or other flavonoids (e.g., flavonols). Proanthocyanidins do not accumulate to high levels in white clover leaves (including high-anthocyanin genotypes; Figure 3B), although upon close examination DMACA staining is observed in trichomes (Figure 3C; see also Hancock et al., 2012). In contrast, inflorescences contain proanthocyanidins and stain strongly with DMACA (Figures 3D,E; see also Abeynayake et al., 2012).
Transcript abundance for genes involved in anthocyanin and proanthocyanidin biosynthesis and regulation was determined by quantitative RT-PCR (Figure 3F). Transcript abundance for DIHYDROFLAVONOL-4 REDUCTASE (DFR), a biosynthetic gene common to anthocyanin and proanthocyanidin pathways, strongly correlated with the presence of anthocyanins; the highest transcript levels were detected in the high-anthocyanin genotype (RL), and the ‘red leaflet’ plants exposed to cold (C) had ∼120-fold higher transcript levels for DFR than plants grown under glasshouse conditions (W). DFR transcripts were also detected in developing inflorescences at modest levels. By contrast, transcripts for the proanthocyanidin biosynthetic gene ANTHOCYANIDIN REDUCTASE (ANR) were detected in developing inflorescences, particularly in immature buds (Inf. A), but were only detected at trace levels in leaves (Figure 3F). Transcript abundance for the bHLH factor Tr-AN1 were elevated in leaf samples accumulating anthocyanin (RL, C) or in developing inflorescences, which accumulate proanthocyanidins. Transcripts for the proanthocyanidin regulator, Tr-MYB14, were abundant in developing inflorescences, particularly in immature buds (Inf. A), but were not detected in the leaf samples. Tr-MYB133 expression correlated with the accumulation of anthocyanins, with high transcript abundance detected in leaves of the high-anthocyanin genotype (RL), and ‘red leaflet’ plants exposed to cold (C) had ∼9-fold higher transcript levels for Tr-MYB133 than plants grown under glasshouse conditions (W). Tr-MYB133 transcripts were detected in inflorescences at low levels. By contrast, Tr-MYB134 expression was associated with proanthocyanidin accumulation, with highest transcript abundance detected in immature inflorescences. Trace levels of Tr-MYB134 were detected in leaf samples.
A transgenic white clover line expressing Ta-MYB14 from a CaMV35S promoter (Hancock et al., 2012) was also analyzed (Supplementary Figure S2). This line exhibits the ‘red midrib’ and ‘red V’ anthocyanin leaf markings, and thus anthocyanin-related R2R3-MYB genes are also expressed in leaves (Albert et al., 2015). Tr-MYB134 was highly expressed in leaves ectopically expressing Ta-MYB14, while transcript abundance of Tr-AN1 was not significantly altered compared to controls.
The expression patterns for Tr-MYB133 and Tr-MYB134 raised the possibility that these genes might be directly regulated by MBW complexes containing anthocyanin- or proanthocyanidin-specific R2R3-MYB activators, respectively. The ability for the anthocyanin R2R3-MYB regulator Tr-RED LEAF and the proanthocyanidin regulator Ta-MYB14 to regulate Tr-MYB133 and Tr-MYB134 was investigated by promoter activation assays. The promoters of Tr-MYB133 and Tr-MYB134 were isolated and cloned into a dual luciferase reporter construct. Tr-RED LEAF strongly activated the Tr-MYB133 promoter (acting with endogenous bHLH and WDR proteins), even without the addition of the bHLH partner Tr-AN1 (Figure 4A). Co-infiltration with Tr-MYB133 weakly repressed this activation. Ta-MYB14 only weakly activated the Tr-MYB133 promoter and required the co-infiltration of the bHLH Tr-AN1. Co-infiltration with either Tr-MYB133 or Tr-MYB134 strongly repressed the activation by Ta-MYB14.
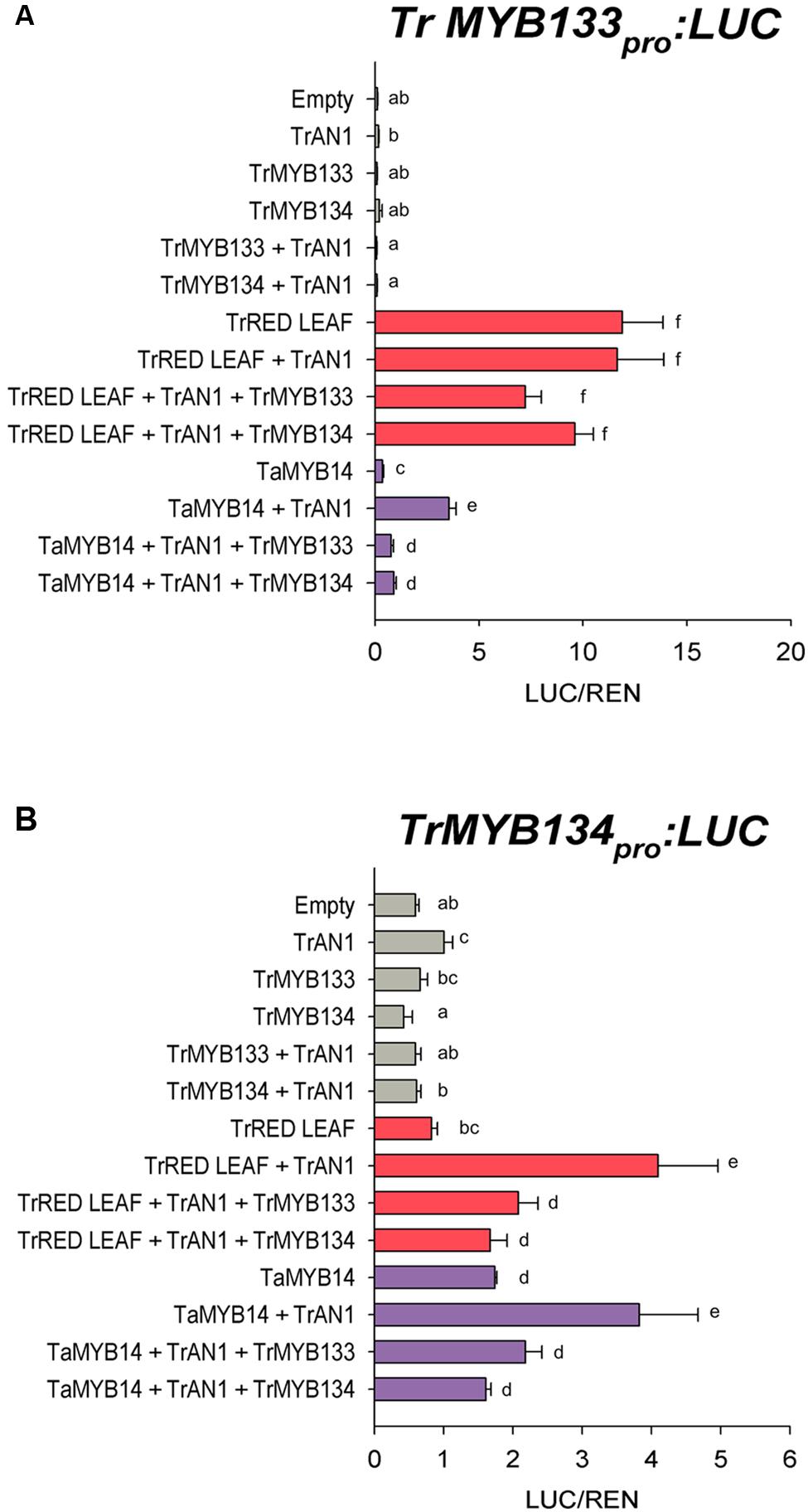
FIGURE 4. Tr-MYB133 and Tr-MYB134 are regulated by different MBW complexes. Promoter activation/repression dual luciferase assays upon (A) the Tr-MYB133 promoter or (B) the Tr-MYB134 promoter, using Agrobacteria-infiltrated N. benthamiana leaves. Combinations of activator R2R3-MYB genes Tr-RED LEAF (anthocyanin) or Ta-MYB14 (proanthocyanidin) were tested with or without the bHLH factor Tr-AN1, and the R2R3-MYB repressors Tr-MYB133 or Tr-MYB134. Firefly luciferase activity was normalized to Renilla luciferase activity (LUC/REN); mean ± SEM n = 5 biological replicates are shown. Means that are significantly different are indicated by different letters, as determined by post hoc Fisher’s LSD (5%).
The Tr-MYB134 promoter was responsive to the activity of Ta-MYB14, which was enhanced by the addition of Tr-AN1 (Figure 4B). Tr-RED LEAF did not activate the Tr-MYB134 promoter unless the bHLH factor Tr-AN1 was co-infiltrated, and this was repressed by either Tr-MYB133 or Tr-MYB134. However, Ta-MYB14 alone did activate the Tr-MYB134 promoter, and was enhanced with the addition of Tr-AN1. Co-infiltration with Tr-MYB133 or Tr-MYB134 repressed the activation by Ta-MYB14. The promoter activation assays agreed with the qPCR data, with the Tr-MYB133 promoter being more responsive to anthocyanin regulators, while the Tr-MYB134 promoter was more responsive to the proanthocyanidin regulator Ta-MYB14.
Discussion
In all angiosperm species studied to date, anthocyanin and proanthocyanidin biosynthesis is regulated by MBW complexes. The current understanding of how anthocyanins and proanthocyanidins are regulated has recently been extended to include the activities of repressive R2R3- and R3-MYB proteins, which operate within a conserved gene-regulation network (Dubos et al., 2008; Matsui et al., 2008; Albert et al., 2014a,b). In this study, the roles of R2R3-MYB repressors for regulating anthocyanin and proanthocyanidin biosynthesis were investigated in the forage legume, white clover. Two R2R3-MYB repressors, which are conserved in model legumes, were shown to repress the activity of anthocyanin and proanthocyanidin MBW complexes. These two repressors have subfunctionalized in terms of their regulation, providing a mechanism that allows anthocyanin and proanthocyanidin synthesis to be regulated separately.
Two Clades of Repressors in Legumes are Associated with Proanthocyanidin and Anthocyanin Regulation
Within the available sequence databases for M. truncatula, L. japonicus and G. max, two distinct R2R3-MYB genes exist that encode putative repressors of anthocyanin and/or proanthocyanidin biosynthesis in legumes. Using a candidate-gene approach, orthologs of these two genes were isolated from the forage legume, T. repens. These genes form two distinct clades when analyzed with other R2R3-MYB repressors, but share the bHLH interaction domain and EAR active repression motif, which are necessary for active R2R3-MYB repressors to assert their repressive activity (Aharoni et al., 2001; Albert et al., 2014a). These two genes are located on different linkage groups in Medicago and Glycine, and it is likely this will also be the case in Lotus and Trifolium, although data are currently not available for these genera [Lj-MYB133 is not present in the current genome assembly of L. japonicus (Shelton et al., 2012), and the T. repens genome is not available]. The conservation and retention of these two genes in legumes suggests that they may have non-redundant functions.
Tr-MYB133 and Tr-MYB134 have similar repressive activities, but based upon the transcript abundance patterns have different roles regulating flavonoid synthesis. In luciferase assays, Tr-MYB133 and Tr-MYB134 both repressed the promoters of the flavonoid genes DFR (common to anthocyanin and proanthocyanidin synthesis) and ANR (proanthocyanidin-specific; Figure 2). Interestingly, they did not exhibit specificity for the MBW complexes they acted upon, repressing complexes containing the R2R3-MYB activators Tr-RED LEAF (anthocyanin) and Ta-MYB14 (proanthocyanidin). While these two transcription factors have similar repressive activity, their expression patterns differ. The expression of Tr-MYB133 was strongly associated with the accumulation of anthocyanin pigments, while Tr-MYB134 expression was tightly associated with proanthocyanidin synthesis (Figure 3). This suggested that Tr-MYB133 and Tr-MYB134 have subspecialized to predominantly regulate anthocyanins and proanthocyanidins, respectively.
Subspecialization of R2R3-MYB repressors for anthocyanins vs. proanthocyanidins has yet to be demonstrated conclusively in other groups of plants, although several observations suggest this is likely to occur. In both grape (Vitis vinifera) and strawberry (Fragaria sp.), proanthocyanidins are synthesized at early stages of fruit development, while anthocyanins are produced during the late stages of ripening (Schaart et al., 2012; Huang et al., 2014). In grape, the R2R3-MYB repressor Vv-MYBC2-L1 has a bi-phasic expression pattern that correlates with the synthesis of both proanthocyanidins (early) and anthocyanins (late) during berry development (Huang et al., 2014). In contrast, Vv-MYBC2-L3 expression is associated with proanthocyanidin accumulation, and not anthocyanins (Cavallini et al., 2015). In strawberry, however, Fa-MYB1 expression is associated with anthocyanins and not proanthocyanidins (Aharoni et al., 2001; Lin-Wang et al., 2010; Schaart et al., 2012). Thus, it appears that R2R3-MYB repressor gene family members have subspecialized for proanthocyanidin or anthocyanin biosynthesis in grape and strawberry, and it is anticipated that this may also occur in more diverse species.
The association of Tr-MYB133 with anthocyanin synthesis, and Tr-MYB134 with proanthocyanidin synthesis may occur because these genes are themselves directly regulated by pathway-specific MBW complexes. In petunia, the R2R3-MYB repressor Ph-MYB27 is targeted by the MBW complexes that regulate anthocyanin synthesis to provide feedback repression, and it is proposed that this is conserved across eudicots (Albert et al., 2014a). This was examined by promoter activation assays upon the promoters of Tr-MYB133 and Tr-MYB134, using the anthocyanin R2R3-MYB Tr-RED LEAF (Albert et al., 2015) and the proanthocyanidin R2R3-MYB Ta-MYB14 (Hancock et al., 2012). These assays agreed with the gene expression data (Figures 3 and 4), demonstrating that the Tr-MYB133 promoter is more effectively activated by MBW complexes that contain an anthocyanin-related R2R3-MYB, compared to the proanthocyanidin regulator.
The Tr-MYB134 promoter was activated by MBW complexes containing either the anthocyanin- or proanthocyanidin-related R2R3-MYBs. Interestingly, Tr-RED LEAF was unable to activate the Tr-MYB134 promoter without the addition of Tr-AN1, which differs to the activation observed for the Tr-MYB133 promoter. The findings from the promoter activation assays appear to contrast slightly with the gene expression studies. White clover plants with the ‘red leaf’ anthocyanin leaf marking ectopically express the Tr-RED LEAF MYB gene (Albert et al., 2015), yet express Tr-MYB134 poorly (Figure 3). However, in tissues expressing Tr-MYB14 and accumulating proanthocyanidins, Tr-MYB134 was expressed. Similar observations have been made in Medicago, where ectopic expression of the proanthocyanidin regulators Mt-MYB5 and Mt-MYB14 resulted in 80- and 10-fold increases, respectively, in the expression of Medtr5g079670, the ortholog of Tr-MYB134 (Liu et al., 2014). This suggests that Tr-MYB134 is normally regulated by the proanthocyanidin-related R2R3-MYB factors, and this is conserved in other legumes.
MYB-bHLH-WDR Gene Regulation Networks Control Anthocyanin and Proanthocyanidin Synthesis in Legumes
Recently, a multi-species model was proposed for the MBW complex and the gene regulation networks that they operate within, integrating the activities of both activator and repressor transcription factors (Albert et al., 2014a). While this model was proposed for anthocyanin regulation, this also has implications for proanthocyanidin regulation, since anthocyanin and proanthocyanidin regulation occurs by a similar mechanism and shares MBW components. Key features of the models include (i) R2R3-MYB repressors are expressed in tissues to prevent ectopic synthesis of anthocyanins/proanthocyanidins; (ii) feedback repression occurs by both R2R3-MYB proteins containing active repression domains (EAR, TLLLFR), and by the mobile competitive R3-MYB proteins; (iii) MBW complexes can contain multiple MYB proteins (e.g., MYB activator + MYB repressor), bridged by dimerized bHLH proteins; (iv) hierarchical activation of the AN1-clade bHLH factor occurs to provide reinforcement.
Many commonalities exist between the proposed model (Albert et al., 2014a) and the findings from this study and with data from other legume species, although there are some interesting differences. While the core features of the MBW complex are highly conserved in legumes—including the requirement for the bHLH2/AN1/TT8 clade bHLH and WDR proteins for anthocyanin and proanthocyanidin regulation (Pang et al., 2009; Hellens et al., 2010; Verdier et al., 2012) and MBW complex assembly (Liu et al., 2014)—the involvement of MYB repressors differ. It was anticipated that Tr-MYB133 and Tr-MYB134 might be expressed highly in leaves to prevent ectopic accumulation of anthocyanins and proanthocyanidins. However, Tr-MYB133 and Tr-MYB134 were only expressed highly in tissues accumulating anthocyanins or proanthocyanidins, respectively, and do not appear to have roles preventing ectopic accumulation of these metabolites. This contrasts with petunia and Arabidopsis, where the R2R3-MYB repressor Ph-MYB27 and (truncated R2) R3-MYB repressor At-MYBL2 are expressed highly in leaves under non-stress conditions, while exposure to light-stress results in a dramatic reduction of expression (Dubos et al., 2008; Albert et al., 2011). This de-repression is likely to be important to allow plants to respond to changing environmental conditions, where the accumulation of anthocyanins is advantageous (e.g., to ameliorate light stress). Thus, the loss of the repressors Ph-MYB27 or At-MYBL2 results in enhanced accumulation of anthocyanins in petunia and Arabidopsis, respectively (Dubos et al., 2008; Albert et al., 2014a). It is unlikely that losing MYB133 or MYB134 activity in clover or other legumes (e.g., Medicago) will significantly alter the distribution of anthocyanins or proanthocyanidins, although it is anticipated the anthocyanin/proanthocyanidin content would increase in tissues that already accumulate these compounds. The difference in the regulation of MYB133 and MYB134 compared to repressors from other species raises questions about how legumes prevent ectopic accumulation of anthocyanins/proanthocyanidins and whether these activities are performed by additional repressive MYB family members, or by other transcription factors.
R2R3-MYB proteins provide feedback repression to MBW gene regulation networks in legumes (Figure 5). This feature of MBW gene regulation networks was observed in petunia and proposed to be conserved in eudicots. Such feedback regulation likely allows for fine-tuning of gene expression (Albert et al., 2014a; Cavallini et al., 2015). We have demonstrated that this also occurs for Tr-MYB133 and Tr-MYB134 in white clover, but these two genes have subspecialized to provide feedback repression for anthocyanin (Figure 5A) and proanthocyanidin biosynthesis (Figure 5B), respectively. These two repressors have the amino acid motif that is required to bind bHLH proteins, contain EAR repression motifs in their C-termini, and are from the Ph-MYB27/Fa-MYB1 clade of R2R3-MYB repressor (Figure 1). Thus, it is anticipated that Tr-MYB133 and Tr-MYB134 will be incorporated into MBW complexes, and recruited to promoters by activator R2R3-MYB proteins, as occurs for Ph-MYB27 (Albert et al., 2014a). It remains to be determined if competitive R3-MYB proteins also provide feedback repression upon anthocyanin and proanthocyanidin synthesis in legumes, as occurs in petunia and Arabidopsis (Zhu et al., 2009; Albert et al., 2014a).
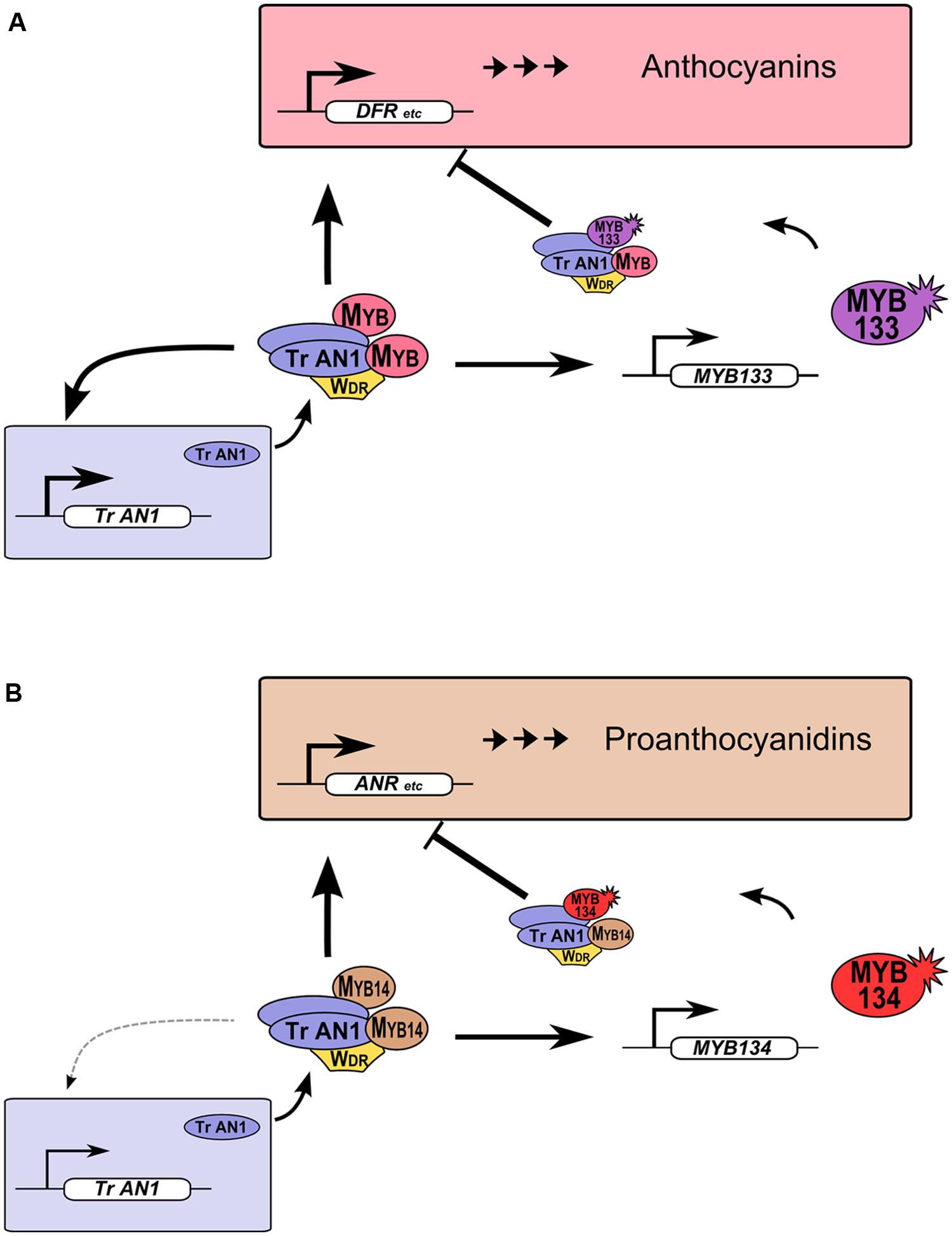
FIGURE 5. Tr-MYB133 and Tr-MYB134 participate in MBW regulatory networks. (A) Anthocyanin MBW gene regulation network. MBW complexes containing anthocyanin R2R3-MYB activate genes required for anthocyanin biosynthesis and transport, resulting in anthocyanin pigment accumulation. The AN1/TT8 clade bHLH (Tr-AN1) is also activated, to provide reinforcement, while Tr-MYB133 is activated to provide feedback repression upon the MBW complex. (B) The proanthocyanidin MBW gene regulation network activates genes required for proanthocyanidin biosynthesis and transport, and Tr-MYB134 to provide feedback repression. Proanthocyanidin MBW activation complexes may include other MYB activators, such as MYB5 or PAR (not shown). The AN1/TT8 clade bHLH gene does not appear to be a high affinity target of MYB14.
The activation of genes encoding R2R3-MYB repressors by MBW complexes appears to be widely conserved in eudicots. In petunia, ectopic expression of anthocyanin MYBs (Ph-DPL, Ph-PHZ) resulted in elevated expression of the repressor Ph-MYB27, and this was shown to occur by direct activation upon the Ph-MYB27 promoter by anthocyanin MBW complexes (Albert et al., 2014a). Similarly, ectopic expression of the anthocyanin MYB Vl-MYBA1 (Cutanda-Perez et al., 2009) or the proanthocyanidin MYBs Vv-MYBPA1 or Vv-MYBPA2 (Terrier et al., 2009) in grape cultures resulted in ectopic expression of Vv-MYBC2-L1. This agrees with the temporal expression pattern for Vv-MYBC2-L1, which correlates with both proanthocyanidin and anthocyanin synthesis during berry development (Huang et al., 2014). Interestingly, while the temporal expression of Vv-MYBC2-L3 suggests it may be associated with proanthocyanidin regulation (Cavallini et al., 2015), its expression was not elevated in grape cultures expressing Vv-MYBPA1/Vv-MYBPA2 (Terrier et al., 2009). It is not yet known if the apparent sub-specialization of particular R2R3-MYB repressor genes for anthocyanin or proanthocyanidin synthesis in grape (Vv-MYBC2-L3 – proanthocyanidins) or strawberry (Fa-MYB1 – anthocyanins; Aharoni et al., 2001; Schaart et al., 2012; ; Huang et al., 2014; Cavallini et al., 2015) occurs because they are regulated by distinct MBW complexes, or whether they regulated by developmental signals during fruit development and ripening.
The anthocyanin MBW complexes activate the expression of the Tr-AN1/Mt-TT8 bHLH genes. The ‘red leaf’ anthocyanin leaf marking in white clover occurs because the R2R3-MYB gene Tr-RED LEAF is ectopically expressed (Albert et al., 2015), which results in enhanced expression of Tr-AN1 (Figure 3). Similarly, Medicago plants expressing the anthocyanin R2R3-MYB gene Mt-LAP1 have enhanced expression of Mt-TT8 (Peel et al., 2009). Thus, the reinforcement of bHLH expression by MBW complexes containing anthocyanin R2R3-MYB genes also occurs in legumes, as it does in petunia and Arabidopsis (Baudry et al., 2006; Albert et al., 2014a). Interestingly, it is less convincing that the proanthocyanidin regulator MYB14 is a strong activator of the AN1/TT8 bHLH genes. Ectopic expression of Mt-MYB14 in Medicago only increased the expression of Mt-TT8 ∼2-fold (Liu et al., 2014), and overexpression of Ta-MYB14 in white clover did not significantly enhance Tr-AN1 expression (Supplementary Figure S2). Thus, Tr-AN1/Mt-TT8 may be a lower affinity target of MYB14, and these bHLH genes may be primarily regulated by developmental signals. Alternatively, they may be regulated by other proanthocyanidin-related R2R3-MYB genes that are present in legumes (e.g., Mt-MYB5, Mt-PAR), or by MBW complexes that contain more than one type of proanthocyanidin MYB (Verdier et al., 2012; Liu et al., 2014).
The models presented in this study (Figure 5) build upon the genetic and molecular data that supports the existence of MBW complexes for regulating both anthocyanins and proanthocyanidins in legumes (Pang et al., 2009; Peel et al., 2009; Hellens et al., 2010; Verdier et al., 2012; Liu et al., 2014; Albert et al., 2015), integrating the activities of R2R3-MYB repressors. The use of transient assays in N. benthamiana to investigate the activity of MBW genes, including MYB133 and MYB134, has been instrumental in overcoming the challenges posed by white clover – a species that is recalcitrant to molecular and genetic analysis (outcrossing, allotetraploid). The major limitation with such assays are that endogenous bHLH (bHLH1) and WDR proteins are expressed (Albert et al., 2014a; Montefiori et al., 2015), which can sometimes obscure the essential roles of the bHLH and WDR components. However, mutants for these MBW components in legumes completely lack anthocyanins and proanthocyanidins, such as Mendel’s classic ‘A’ (bHLH2) and ‘A2’ (WDR) genes in pea (Hellens et al., 2010) and Mt-WDR1 in Medicago (Pang et al., 2009), which highlights the essential role of these proteins for MBW complex activity. It is anticipated that the models presented in this study will be further strengthened by the analysis of mutants for MBW components and the MYB repressors in Medicago.
Conclusion
Two R2R3-MYB genes encoding flavonoid repressors are conserved in legumes, operating within the MBW gene regulation networks that control anthocyanin and proanthocyanidin synthesis. The MBW gene regulation network identified in white clover fulfills many of the key features proposed to be conserved in eudicots. This includes the involvement of R2R3-MYB repressors to provide feedback repression upon the MBW activation complexes and reinforcement in the expression of the bHLH factor. However, Tr-MYB133 and Tr-MYB134 are regulated by distinct MBW complexes, associated with anthocyanin and proanthocyanidin synthesis, respectively. This subspecialization provides a mechanism that allows for anthocyanin and proanthocyanidins to be regulated separately.
Author Contributions
NA designed the study, performed experimentation, analyses, and wrote the manuscript.
Funding
This work was supported with a Postdoctoral fellowship from AgResearch core funding, and with partial funding from Grasslanz Technology Limited.
Abbreviations
MBW, MYB-BHLH-WDR.
Footnotes: The nucleotide sequences reported in this manuscript have been submitted to NCBI with accession numbers KT699106, KT699107, KT699108 and KT699109.
Conflict of Interest Statement
AgResearch and Grasslanz Technology Limited hold a patent for the commercial use of Ta-MYB14 and related sequences.
The reviewer José Tomás Matus and handling Editor David Caparros-Ruiz declared their shared affiliation, and the handling Editor states that, nevertheless, the process met the standards of a fair and objective review.
Acknowledgments
I thank Anna Larking for greenhouse assistance, Pastoral Genomics for access to unpublished sequence data, and Andrew McLachlan for assistance with statistical analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01165
References
Abeynayake, S. W., Panter, S., Chapman, R., Webster, T., Rochfort, S., Mouradov, A., et al. (2012). Biosynthesis of proanthocyanidins in white clover flowers: cross talk within the flavonoid pathway. Plant Physiol. 158, 666–678. doi: 10.1104/pp.111.189258
Aharoni, A., De Vos, C. H. R., Wein, M., Sun, Z. K., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014a). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
Albert, N. W., Davies, K. M., and Schwinn, K. E. (2014b). Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signal. Behav. 9, e29526. doi: 10.4161/psb.29526
Albert, N. W., Davies, K. M., and Schwinn, K. E. (2014c). Repression – the dark side of anthocyanin regulation? Acta Hortic. 1048, 129–136. doi: 10.17660/ActaHortic.2014.1048.15
Albert, N. W., Griffiths, A. G., Cousins, G. R., Verry, I. M., and Williams, W. M. (2015). Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytol. 205, 882–893. doi: 10.1111/nph.13100
Albert, N. W., Lewis, D. H., Zhang, H., Irving, L. J., Jameson, P. E., and Davies, K. M. (2009). Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 60, 2191–2202. doi: 10.1093/jxb/erp097
Albert, N. W., Lewis, D. H., Zhang, H., Schwinn, K. E., Jameson, P. E., and Davies, K. M. (2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65, 771–784. doi: 10.1111/j.1365-313X.2010.04465.x
Barbehenn, R. V., and Constabel, C. P. (2011). Tannins in plant-herbivore interactions. Phytochemistry 72, 1551–1565. doi: 10.1016/j.phytochem.2011.01.040
Baudry, A., Caboche, M., and Lepiniec, L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46, 768–779. doi: 10.1111/j.1365-313X.2006.02733.x
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380.
Beauchemin, K. A., McGinn, S. M., Martinez, T. F., and McAllister, T. A. (2007). Use of condensed tannin extract from quebracho trees to reduce methane emissions from cattle. J. Anim. Sci. 85, 1990–1996. doi: 10.2527/jas.2006-686
Butelli, E., Titta, L., Giorgio, M., Mock, H.-P., Matros, A., Peterek, S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. doi: 10.1038/nbt.1506
Cavallini, E., Matus, J. T., Finezzo, L., Zenoni, S., Loyola, R., Guzzo, F., et al. (2015). The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470. doi: 10.1104/pp.114.256172
Cutanda-Perez, M. C., Ageorges, A., Gomez, C., Vialet, S., Terrier, N., Romieu, C., et al. (2009). Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 69, 633–648. doi: 10.1007/s11103-008-9446-x
Davies, K. M., Albert, N. W., and Schwinn, K. E. (2012). From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39, 619–638. doi: 10.1071/FP12195
Deluc, L., Bogs, J., Walker, A. R., Ferrier, T., Decendit, A., Merillon, J.-M., et al. (2008). The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 147, 2041–2053. doi: 10.1104/pp.108.118919
Dixon, R. A., Liu, C., and Jun, J. H. (2013). Metabolic engineering of anthocyanins and condensed tannins in plants. Curr. Opin. Biotechnol. 24, 329–335. doi: 10.1016/j.copbio.2012.07.004
Dixon, R. A., and Paiva, N. L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. doi: 10.2307/3870059
Dixon, R. A., Xie, D. Y., and Sharma, S. B. (2005). Proanthocyanidins – A final frontier in flavonoid research? New Phytol. 165, 9–28. doi: 10.1111/j.1469-8137.2004.01217.x
Dorling, S. J., Leung, S., Anderson, C. W. N., Albert, N. W., and McManus, M. T. (2011). Changes in 1-aminocyclopropane-1-carboxlate (ACC) oxidase expression and enzyme activity in response to excess manganese in white clover (Trifolium repens L.). Plant Physiol. Biochem. 49, 1013–1019. doi: 10.1016/j.plaphy.2011.04.007
Douglas, G. B., Stienezen, M., Waghorn, G. C., Foote, A. G., and Purchas, R. W. (1999). Effect of condensed tannins in birdsfoot trefoil (Lotus corniculatus) and sulla (Hedysarum coronarium) on body weight, carcass fat depth, and wool growth of lambs in New Zealand. N. Z. J. Agric. Res. 42, 55–64. doi: 10.1080/00288233.1999.9513353
Dubos, C., Gourrierec, J. L., Baudry, A., Huep, G., Lanet, E., Debeaujon, I., et al. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55, 940–953. doi: 10.1111/j.1365-313X.2008.03564.x
Espley, R. V., Bovy, A., Bava, C., Jaeger, S. R., Tomes, S., Norling, C., et al. (2013). Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol. J. 11, 408–419. doi: 10.1111/pbi.12017
Gould, K. S. (2004). Nature’s Swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004, 314–320. doi: 10.1155/S1110724304406147
Gould, K. S., McKelvie, J., and Markham, K. R. (2002). Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant Cell Environ. 25, 1261–1269. doi: 10.1046/j.1365-3040.2002.00905.x
Hancock, K. R., Collette, V., Fraser, K., Greig, M., Xue, H., Richardson, K., et al. (2012). Expression of the R2R3 MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes T. repens and Medicago sativa. Plant Physiol. 159, 1204–1220. doi: 10.1104/pp.112.195420
Hatier, J. H., Clearwater, M. J., and Gould, K. S. (2013). The functional significance of black-pigmented leaves: photosynthesis, photoprotection and productivity in Ophiopogon planiscapus ‘Nigrescens’. PLoS ONE 8:e67850. doi: 10.1371/journal.pone.0067850
Hellens, R. P., Moreau, C., Lin-Wang, K., Schwinn, K. E., Thomson, S. J., Fiers, M. W. E. J., et al. (2010). Identification of Mendel’s white flower character. PLoS ONE 5:e13230. doi: 10.1371/journal.pone.0013230
Heppel, S. C., Jaffe, F. W., Takos, A. M., Schellmann, S., Rausch, T., Walker, A. R., et al. (2013). Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 82, 457–471. doi: 10.1007/s11103-013-0074-8
Huang, Y. F., Vialet, S., Guiraud, J. L., Torregrosa, L., Bertrand, Y., Cheynier, V., et al. (2014). A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 201, 795–809. doi: 10.1111/nph.12557
Hughes, N. M., Neufeld, H. S., and Burkey, K. O. (2005). Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata. New Phytol. 168, 575–587. doi: 10.1111/j.1469-8137.2005.01546.x
Jin, H. L., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Kagale, S., Links, M. G., and Rozwadowski, K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152, 1109–1134. doi: 10.1104/pp.109.151704
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Koes, R., Verweij, W., and Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242. doi: 10.1016/j.tplants.2005.03.002
Landi, M., Guidi, L., Pardossi, A., Tattini, M., and Gould, K. S. (2014). Photoprotection by foliar anthocyanins mitigates effects of boron toxicity in sweet basil (Ocimum basilicum). Planta 240, 941–953. doi: 10.1007/s00425-014-2087-1
Li, Y.-G., Tanner, G., and Larkin, P. (1996). The DMACA–HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J. Sci. Food Agric. 70, 89–101. doi: 10.1002/(SICI)1097-0010(199601)70:1<89::AID-JSFA470>3.0.CO;2-N
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., Mcghie, T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10:50. doi: 10.1186/1471-2229-10-50
Liu, C., Jun, J. H., and Dixon, R. A. (2014). MYB5 and MYB14 Play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant Physiol. 165, 1424–1439. doi: 10.1104/pp.114.241877
Lowry, D. B., Sheng, C. C., Lasky, J. R., and Willis, J. H. (2012). Five anthocyanin polymorphisms are associated with an R2R3-MYB cluster in Mimulus guttatus (Phrymaceae). Am. J. Bot. 99, 82–91. doi: 10.3732/ajb.1100285
Martin, C., Butelli, E., Petroni, K., and Tonelli, C. (2011). How can research on plants contribute to promoting human health? Plant Cell 23, 1685–1699. doi: 10.1105/tpc.111.083279
Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55, 954–967. doi: 10.1111/j.1365-313X.2008.03565.x
Montefiori, M., Brendolise, C., Dare, A. P., Lin-Wang, K., Davies, K. M., Hellens, R. P., et al. (2015). In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 66, 1427–1436. doi: 10.1093/jxb/eru494
Neill, S. O., and Gould, K. S. (2003). Anthocyanins in leaves: light attenuators or antioxidants? Funct. Plant Biol. 30, 865–873. doi: 10.1071/FP03118
Pang, Y., Wenger, J. P., Saathoff, K., Peel, G. J., Wen, J., Huhman, D., et al. (2009). A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 151, 1114–1129. doi: 10.1104/pp.109.144022
Peel, G. J., Pang, Y., Modolo, L. V., and Dixon, R. A. (2009). The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J. 59, 136–149. doi: 10.1111/j.1365-313X.2009.03885.x
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Salvatierra, A., Pimentel, P., Moya-León, M. A., and Herrera, R. (2013). Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 90, 25–36. doi: 10.1016/j.phytochem.2013.02.016
Schaart, J. G., Dubos, C., Romero De La Fuente, I., Van Houwelingen, A. M. M. L., De Vos, R. C. H., Jonker, H. H., et al. (2012). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beerman, A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036–5046. doi: 10.1093/emboj/cdf524
Schwinn, K., Venail, J., Shang, Y., Mackay, S., Alm, V., Butelli, E., et al. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18, 831–851. doi: 10.1105/tpc.105.039255
Sharma, S. B., and Dixon, R. A. (2005). Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J. 44, 62–75. doi: 10.1111/j.1365-313X.2005.02510.x
Shelton, D., Stranne, M., Mikkelsen, L., Pakseresht, N., Welham, T., Hiraka, H., et al. (2012). Transcription factors of lotus: regulation of isoflavonoid biosynthesis requires coordinated changes in transcription factor activity. Plant Physiol. 159, 531–547. doi: 10.1104/pp.112.194753
Tamagnone, L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F. A., Roberts, K., et al. (1998). The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10, 135–154. doi: 10.1105/tpc.10.2.135
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verriès, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Verdier, J., Zhao, J., Torres-Jerez, I., Ge, S., Liu, C., He, X., et al. (2012). MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 109, 1766–1771. doi: 10.1073/pnas.1120916109
Yoshida, K., Ma, D., and Constabel, C. P. (2015). The MYB182 protein downregulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 167, 693–710. doi: 10.1104/pp.114.253674
Yuan, Y.-W., Sagawa, J. M., Young, R. C., Christensen, B. J., and Bradshaw, H. D. (2013). Genetic dissection of a major QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics 194, 255–263. doi: 10.1534/genetics.112.146852
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681
Zhao, J., Pang, Y., and Dixon, R. A. (2010). The mysteries of proanthocyanidin transport and polymerization. Plant Physiol. 153, 437–443. doi: 10.1104/pp.110.155432
Zhu, H.-F., Fitzsimmons, K., Khandelwal, A., and Kranz, R. G. (2009). CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2, 790–802. doi: 10.1093/mp/ssp030
Keywords: anthocyanin, condensed tannin, flavonoid, proanthocyanidin, repressor, transcription factor
Citation: Albert NW (2015) Subspecialization of R2R3-MYB Repressors for Anthocyanin and Proanthocyanidin Regulation in Forage Legumes. Front. Plant Sci. 6:1165. doi: 10.3389/fpls.2015.01165
Received: 06 October 2015; Accepted: 07 December 2015;
Published: 23 December 2015.
Edited by:
David Caparros-Ruiz, Centre for Research in Agricultural Genomics – CSIC–IRTA–UAB–UB, SpainReviewed by:
Ryo Nakabayashi, RIKEN Center for Sustainable Resource Science, JapanJosé Tomás Matus, Centre for Research in Agricultural Genomics, Spain
Copyright © 2015 Albert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nick W. Albert, nick.albert@plantandfood.co.nz
†Present address: Nick W. Albert, The New Zealand Institute for Plant and Food Research Limited, Private Bag 4442, Palmerston North, New Zealand
 Nick W. Albert
Nick W. Albert