- Zhejiang Key Lab of Crop Germplasm, Department of Agronomy, Zhejiang University, Hangzhou, China
Low temperature is a major abiotic stress affecting crop growth and productivity. A better understanding of low temperature tolerance mechanisms is imperative for developing the crop cultivars with improved tolerance. We herein performed an Illumina RNA-sequencing experiment using two barley genotypes differing in freezing tolerance (Nure, tolerant and Tremois, sensitive), to determine the transcriptome profiling and genotypic difference under mild freezing shock treatment after a very short acclimation for gene induction. A total of 6474 differentially expressed genes, almost evenly distributed on the seven chromosomes, were identified. The key DEGs could be classified into six signaling pathways, i.e., Ca2+ signaling, PtdOH signaling, CBFs pathway, ABA pathway, jasmonate pathway, and amylohydrolysis pathway. Expression values of DEGs in multiple signaling pathways were analyzed and a hypothetical model of mild freezing shock tolerance mechanism was proposed. Expression and sequence profile of HvCBFs cluster within Frost resistance-H2, a major quantitative trait locus on 5H being closely related to low temperature tolerance in barley, were further illustrated, considering the crucial role of HvCBFs on freezing tolerance. It may be concluded that multiple signaling pathways are activated in concert when barley is exposed to mild freezing shock. The pathway network we presented may provide a platform for further exploring the functions of genes involved in low temperature tolerance in barley.
Introduction
Low temperature is one of the major adverse environmental factors that limit crop production. To cope with low temperature, plants have evolved various signal perception and response pathways, which subsequently lead to complicated biochemical and physiological changes, such as reprogramming of gene expression, modification of metabolic pathways, alteration in lipid composition, accumulation of osmoprotectants, and synthesis of antioxidants (Hughes and Dunn, 1996; Dai et al., 2009; Chinnusamy et al., 2010).
Low temperature perception is the first step for plants to develop the tolerance reactions, where a physical response is converted into complex biochemical and physiological processes (Ruelland and Zachowski, 2010). Temperature decrease can be sensed by plant cells through their membrane rigidification effect. Fatty acid desaturase (FAD) encode the enzymes that control the unsaturation of phospholipids and affect membrane fluidity (Miquel et al., 1993). In Arabidopsis, a fad2 mutant defective in oleate desaturase exhibits plasma membrane rigidification and low temperature sensitivity (Miquel et al., 1993; Vaultier et al., 2006). Membrane rigidification-activated mechano-sensitive or ligand-activated Ca2+ channels lead to Ca2+ influx into cytosol. This was thought to be an important initial event to temperature change in plant cells, as calcium chelators and calcium channel blockers could prevent cold-induced protein phosphorylation and gene expression (Monroy et al., 1997; Chinnusamy et al., 2006; Winfield et al., 2010). The spatial and temporal patterns of Ca2+ signals in plant cells are characteristic for particular stimuli and can be interpreted by Ca2+-binding proteins (CBPs) that act as Ca2+ sensors including calmodulins (CaMs), calmodulin-like proteins (CMLs), calcineurin B-like proteins (CBLs), and calcium-dependent protein kinases (CDPKs; McCormack et al., 2005; Kaplan et al., 2006). After binding Ca2+, CBPs undergo a conformational change that enables them to activate or inactivate target proteins (DeFalco et al., 2010). For instance, CaMs bind to calmodulin-binding transcription factor (CAMTA) and CBLs interact with CBL-interacting protein kinases (CIPKs; Doherty et al., 2009; Batistic et al., 2010; Thoday-Kennedy et al., 2015). Afterwards, these downstream effectors initiate a series of events, resulting in extensive reprogramming of gene expression in response to low temperature (Winfield et al., 2010). Ca2+ signal can also be amplified through phospholipids such as phosphatidic acids (PtdOH), which are lipid secondary messengers mediating low temperature signaling in plants (Vergnolle et al., 2005; Chinnusamy et al., 2006). During low temperature treatment, PtdOH could be synthesized by two pathways: directly by the action of a PLD or by the combined action of a PLC, which produces diacylglycerol (DAG), followed by the action of a diacylglycerol kinase (DGK; Ruelland et al., 2002). Low temperature induced activation of both phospholipase pathways mainly depends on Ca2+ entry into cells and PLC activity accounts for almost 80% of low temperature-induced PtdOH (Ruelland et al., 2002).
The C-repeat binding factors (CBFs)-dependent pathway, which plays a central role among multiple low temperature regulatory pathways has been extensively studied and well-characterized (Chinnusamy et al., 2007). As members of a large AP2/EREBP family of DNA-binding proteins, CBFs are rapidly induced by low temperature and can bind to the DRE/CRT cis-element in the promoter regions of many cold responsive (COR) genes, which encode cryoprotective proteins for protecting cells against low temperature induced damage (Chinnusamy et al., 2003; Campoli et al., 2009). A common feature of CBFs in grass genomes is that CBFs are organized in clusters of tandemly duplicated paralogs (Tondelli et al., 2011). In barley, a quantitative trait locus on 5H, Frost resistance-H2 (Fr-H2), contains a cluster of HvCBFs and largely contributes to low temperature tolerance (Francia et al., 2004; Knox et al., 2010). A recent study has identified the full-length sequences of HvCBFs in Fr-H2 using a high-resolution genetic map of Fr-H2 (Pasquariello et al., 2014).
Abscisic acid (ABA) accumulates when plants are exposed to low temperature, and is subsequently perceived by ABA receptors such as the proteins in the pyrabactin resistance/pyrabactin resistance 1-like/regulatory components of ABA receptor (PYR/PYL/RCAR) family. PYR/PYL/RCAR proteins can inhibit the activity of the clade A protein phosphatase type 2C (PP2C), relieving the repression of sucrose non-fermenting 1-related protein kinase 2 (SnRK2) activity by a clade A PP2C, which in turn phosphorylates abscisic acid responsive element (ABRE) binding protein/abscisic acid responsive element binding factor (AREB/ABF) to regulate the expression of low temperature responsive genes (Furihata et al., 2006; Ma et al., 2009; Park et al., 2009). A few of COR genes with ABRE in their promoters, such as COR47, can response to ABA and gain an increased expression level (Xiong et al., 2001). Moreover, ABA biosynthesis contributes to the maximum induction of low temperature responsive genes during the later stage of low temperature response (Zhang et al., 2014).
Blocking endogenous jasmonate biosynthesis and signaling renders plants hypersensitive to freezing stress, suggesting that jasmonate positively regulate freezing tolerance in plants (Hu et al., 2013). Jasmonate can be perceived by its receptor coronatine insensitive 1 (COI1) and subsequently facilitates the degradation of jasmonate-zim-domain proteins (JAZ), which act as repressors of jasmonate signaling via their physical interactions with a wide array of transcription factors (Thines et al., 2007; Yan et al., 2009).
Carbohydrate metabolism pathway was also altered in response to low temperature (Jiang et al., 2014). Starch degradation generates soluble sugars (such as sucrose, glucose, and trehalose), which can modulate cellular osmotic potential to protect plasma membrane from low temperature damage (Bogdanovic et al., 2008).
To better understanding the functional characterization of low temperature induced genes in cereals, more attention should be paid to choice of genotypes and experimental conditions, such as treatment and sampling time (Campoli et al., 2009). It has been reported that many COR genes show the maximal expression within 2 days after low temperature treatment (Ganeshan et al., 2008), therefore, sampling after short-term treatment could be ideal to capture the transcriptome profile of low temperature perception and signaling. Our previous study was focused on the change of chlorophyll fluorescence parameters during recovery after freezing shock in barley (Dai et al., 2007), however, no research has been done on the systematic transcriptome profiling of freezing shock response genes in barley. RNA-sequencing (RNA-Seq) is an efficient tool for gene expression profiling studies as well as simultaneous identification of mutations (Morozova et al., 2009), and it has been applied in low temperature tolerance research of many plant species, such as Solanum lycopersicoides (Chen et al., 2015), Chrysanthemum nankingense (Ren et al., 2014), and Camellia sinensis (Wang et al., 2013).
Therefore, we herein performed mild freezing shock treatment and RNA-Seq analysis using two barley cultivars, Nure (low temperature tolerant) and Tremois (low temperature sensitive; Francia et al., 2004; Rizza et al., 2011) to explore the perception and signaling pathways activated by short-term freezing treatment, in order to draw an outline of early regulatory gene network in response to freezing shock in barley.
Materials and Methods
Plant Materials and Mild Freezing Shock Treatment
Two barley (Hordeum vulgare L.) cultivars, Nure, and Tremois, were used in this study. Nure is a winter barley cultivar originated from Italy with superior low temperature tolerance, while Tremois is a spring barley cultivar originated from France with inferior low temperature tolerance (Francia et al., 2004). Compared with Tremois, Nure has higher winter survival rate, better frost tolerance, and more accumulation of COR proteins (Francia et al., 2004; Rizza et al., 2011).
Seeds of both cultivars, kindly provided by Dr. Luigi Cattivelli in CRA Agricultural Research Council, Italy, were sown in plastic pots (170 mm × 220 mm) filled with peat in November 24th 2012. Plants were grown in a glasshouse at Zhejiang University, Hangzhou, China. Temperature was set at 20/14°C (day/night), and relative humidity was 75%. Mild freezing shock treatment was conducted on three-leaf-stage seedlings under natural condition. As the freezing shock treatment, plants were moved to the open air in a fine day of winter from 5 p.m. to 8 a.m. of the next day, with the temperature fluctuation from -2 to 0°C. The plants kept in glasshouse were used as control. The plants of both control and treatment were sampled immediately after freezing shock treatment (at 8 a.m.) for analysis.
Sampling and RNA Isolation
The third fully expended leaves of 10 seedlings were collected, mixed and flash frozen in liquid nitrogen. Total RNA was extracted from an approximately 0.5 g frozen sample with TRIzol Reagent (Invitrogen, Carlsbad, CA), purified with RNeasy Mini Kit (Qiagen, Germantown, MD) and quality-checked with the Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA), and the RNA samples were frozen at −80°C until used.
Library Construction, Sequencing, and Data Processing
Library construction and sequencing were conducted as previously described in Dai et al. (2014). Briefly, the poly-A containing mRNA was enriched from total RNA by poly-T oligo-attached magnetic beads, and then fragmented into small pieces randomly. Using the fragments as templates, first-strand cDNA was synthesized with random hexamer-primers. Second-strand cDNA was synthesized using DNA polymerase I, dNTPs, and RNase H. The products were amplified by PCR and purified after end-repairing and adaptor ligations with the Illumina TruSeq™ RNA Sample Preparation Kit (Illumina, San Diego, CA). PCR products were loaded onto Illumina HiSeq2000 platform (Illumina, San Diego, CA) for 2 × 100 bp paired-ends sequencing. Raw reads were trimmed by removing all adaptor sequences, empty reads and low quality reads (Q < 30 and length < 50 bp) to obtain clean reads. The clean reads were mapped to the assembly genome of cv. Morex (Mayer et al., 2012) downloaded from Ensembl Genomes 2013 (http://plants.ensembl.org/index.html, Kersey et al., 2014) using Tophat v2.0.8b with default parameters (Trapnell et al., 2009). Cuffdiff of Cufflinks v2.1.1 was used to assemble the mapped reads, estimate the abundances, and analyze differentially expressed genes (DEGs) (Roberts et al., 2011).
Duplicated reads in the BAM files created by TopHat were removed by MarkDuplicates program of Picard v1.7 (http://picard.sourceforge.net). Uniquely mapped single-end and paired-end results were used for the SNVs (single nucleotide variants) and indel (insertion or deletion variant) calling. Raw SNVs and indel were called using SAM-tools mpileup and bcftools (Li and Durbin, 2009), and then were filtered with mapping quality score ≥ 50 and reads coverage > 4.
Identification of the DEGs and Quantitative RT-PCR Analysis
Transcripts abundance of each gene was normalized by using the fragments per kilobase per million map reads (FPKM) method (Li et al., 2014). The ratio of [FPKM (treatment)]/[FPKM (control)] or [FPKM (control)]/[FPKM (treatment)] was calculated as fold-change and the subtraction of |[FPKM (treatment)]—[FPKM (control)]|as number-change. According to previous reports, both fold-change and number-change should be taken into account (Zenoni et al., 2010; Li et al., 2014). In our study, DEGs were determined by the following criterion: “fold-change > 2 and number-change > 20” or “fold-change > 4 and number-change > 2.” This threshold took both highly expressed genes with relatively smaller fold change and lowly expressed genes with relatively greater fold change into consideration. An online software jvenn (http://bioinfo.genotoul.fr/jvenn/example.html) was used to generate Venn diagram (Bardou et al., 2014).
Expression of 10 randomly selected DEGs was determined by quantitative real-time PCR (qRT-PCR) to validate RNA-Seq data. All the primers are listed in Table S1. qRT-PCR was performed on a CFX96 system (Bio-Rad, USA) as described by Zeng et al. (2014) with some modifications. The actin gene was used as an internal control (Zheng et al., 2011).
Gene Annotation, GO Enrichment, and KEGG Analysis
Protein sequences of barley and Arabidopsis were downloaded from Ensembl Genomes 2013 (http://plants.ensembl.org/index.html), and Blastp of Blast v2.2.28 was performed with the parameters “-max_target_seqs 1 -evalue 1E-5” to annotate barley homologous genes to Arabidopsis (Altschul et al., 1990). Setting “max target seqs” as “1,” only the best match result was considered.
Using nucleotide sequences of DEGs as queries, Blastx of Blast2GO v3.0 was performed with default parameters to align against NCBI non-redundant (nr) protein database for homology search. GO annotation and KEGG pathways mapping of DEGs were performed using Blast2GO v3.0 (Conesa et al., 2005).
Presentation of Potential Mild Freezing Shock Signaling Pathways in Figure 1
Ca2+ signaling pathway was presented based on the reports of Batistic et al. (2010), Doherty et al. (2009), Hunt et al. (2004), Lecourieux et al. (2006), McCormack et al. (2005), and Townley and Knight (2002). PtdOH signaling was drawn as described by Arisz et al. (2013) with some modifications. CBFs-dependent low temperature response pathway was drawn referring to Chinnusamy et al. (2007) and Guan et al. (2013). ABA pathway was presented based on the reports of Acharya et al. (2013), Ding et al. (2015), Nambara and Marion-Poll (2005), Valdés et al. (2012), and Yoshida et al. (2015). Jasmonate pathway and amylohydrolysis pathway were presented mostly based on KEGG map00592 and map00500, respectively.
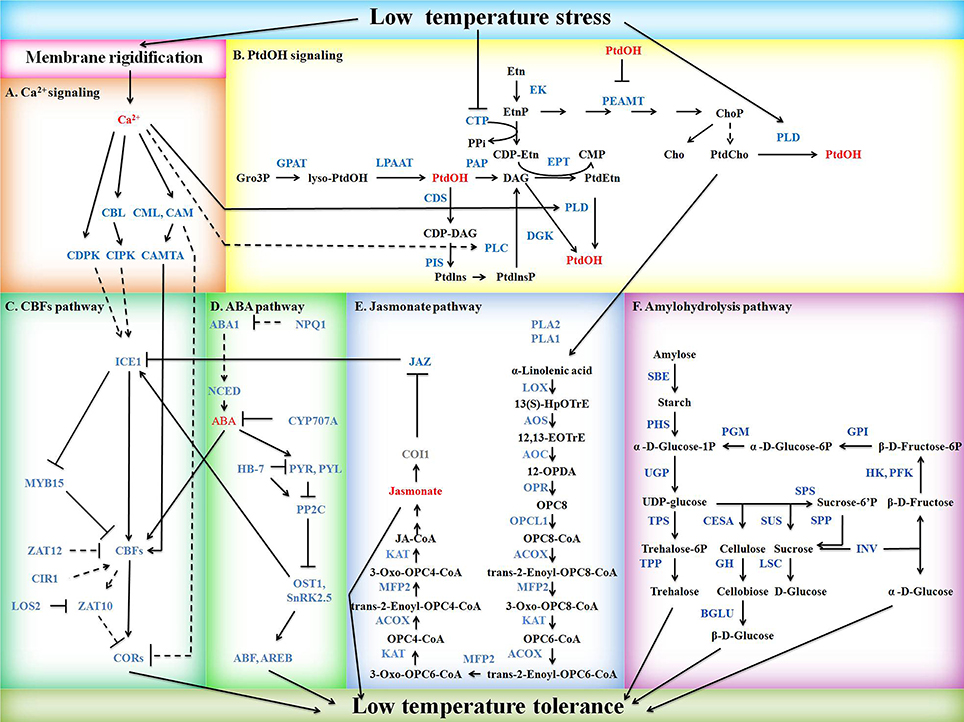
Figure 1. Schematic diagram of potential mild freezing shock signaling pathways in barley. Signaling molecules, DEGs, non-DEGs, and metabolites or others are shown in red, blue, gray, and black, respectively. Solid arrows indicate positive regulation whereas lines ending with a bar stand for negative regulation. Regulations are direct (solid line) or indirect (dash line). (A) Ca2+ signaling. CAM, calmodulin; CAMTA, calmodulin-binding transcription factor; CBL, calcineurin B-like protein; CDPK, calcium-dependent protein kinase. CIPK, CBL-interacting protein kinase; CML, calmodulin-like protein. (B) PtdOH signaling. CDS, cytidine diphosphate-diacylglycerol synthase; CTP, phosphorylcholine cytidylyltransferase; DGK, diacylglycerol kinase; EK, ethanolamine kinase; EPT, cytidine diphosphate-ethanolamine phosphotransferase; GPAT, glycerol 3-phosphate acyltransferase; LPAAT, lysophosphatidic acid acyltransferase; PAP, phosphatidic acid phosphatase; PEAMT, phosphoethanolaminemethyltranferase; PIS, phosphatidylinositol synthase; PLC, phospholipase C; PLD, phospholipase D; PtdOH, phosphatidic acid. (C) CBFs pathway. CBF, C-repeat binding factor; CIR1, circadian 1; COR, cold responsive genes; ICE1, inducer of CBF expression 1; LOS2, low expression of osmotically responsive genes 2; MYB15, Myb domain protein 15. (D) ABA pathway. ABA1, ABA deficient 1; ABF, ABA-responsive elements-binding factor; AREB, ABA-responsive element-binding protein; CYP707A, cytochrome P450-family 707-subfamily A; HB-7, homeobox-7; NCED, nine-cis-epoxycarotenoid dioxygenase; NPQ1, non-photochemical quenching 1; OST1, open stomata 1; PP2C, protein phosphatase 2C; PYL, pyrabactin resistance 1-like; PYR, pyrabactin resistance; SnRK2.5, sucrose non-fermenting 1-related protein kinase 2.5. (E) Jasmonate pathway. ACOX, acyl-CoA oxidase; AOC, allene-oxide cyclase; AOS, allene oxide synthase; COI1, coronatine insensitive 1; JAZ, jasmonate-zim-domain protein 11; KAT, ketoacyl-CoA acyltransferase; LOX, linoleate 13S-lipoxygenase; MFP2, enoyl-CoA hydratase; OPCL1, OPC-8:0 CoA ligase 1; OPR, 12-oxophytodienoate reductase; PLA1, phospholipase A1; PLA2, phospholipase A2. (F) Amylohydrolysis pathway. BGLU, beta-glucosidase; CESA, cellulose synthase; GH, 4-beta-D-glucan 4-glucanohydrolase; GPI, glucose-6-phosphate isomerase; HK, hexokinase; INV, invertase; LSC, levansucrase; PFK, phosphofructokinase; PGM, phosphoglucomutase. PHS, alpha-glucan phosphorylase; SBE, starch branching enzyme; SPP, sucrose-phosphate phosphatase; SPS, sucrose-phosphate synthase; SUS, sucrose synthase; TPP, trehalose-6-phosphate phosphatase; TPS, trehalose-6-phosphate synthase; UGP, UDP-glucose pyrophosphorylase.
Identification of DEGs Encoding HvCBFs within Fr-H2 Locus
Using nucleotide sequences of HvCBFs within Fr-H2 reported by Pasquariello et al. (2014) as a query, Blastn of Blast v2.2.28 was conducted with the parameters “-max_target_seqs 1 -evalue 1E-5” in order to identify the HvCBFs cluster in our DEGs (Altschul et al., 1990). Distributions of HvCBFs in FR-H2 locus shown in Figure 4 refer to Pasquariello et al. (2014).
Genetic Difference Analysis
In order to understand the genetic difference between Nure and Tremois, |log2[FPKM(Nure)/FPKM(Tremois)]|was calculated for both treatment and control. Here, the greater is the value, the greater is genotypic difference. The number of DEGs on a certain value of |log2[FPKM(Nure)/FPKM(Tremois)]|was counted to form two trendlines in Figure S1, which revealed changes of DEGs number as genetic difference become greater. Only |log2[FPKM(Nure)/FPKM(Tremois)]|not larger than 4 were presented.
Results
RNA-Seq Performance
After a very short acclimation for gene induction, over 113 million raw reads (10.52 Gb) for the four libraries of two barley genotypes in control and mild freezing shock treatment were yielded by the 100-bp paired-end transcriptome sequencing (Table S2). The sequences of raw data have been deposited in the National Center for Biotechnology Information Sequence Read Archive, www.ncbi.nlm.nih.gov (accession nos. SAMN03952824 and SAMN03952823). After removing adaptor sequences, empty reads, and poor quality reads, about 84.1% raw reads were determined as clean reads (Table S2). Approximately 88.0, 86.6, 86.6, and 85.8% of clean reads for Nure in control (Nure_C), Nure in treatment (Nure_T), Tremois in control (Tremois_C), and Tremois in treatment (Tremois_T) were mapped to the assembly genome of cv. Morex, and yielded 40,321, 37,714, 38,997, 39,065 transcripts, respectively (Table S2).
Overall Analysis of DEGs
Based on the criterion for screening DEGs, a total of 6474 genes were determined to be differentially expressed under low temperature treatment in comparison to the control (Table S3). Overall, these DEGs were almost evenly distributed on the seven chromosomes in barley (Table 1). Nure had less DEGs than Tremois, and both genotypes had a relatively higher proportion of up-regulated DEGs (Figure 2). There were 1303 up-regulated DEGs and 1051 down-regulated DEGs in both genotypes (Figure 2). However, 770 and 1474 DEGs were up-regulated only in Nure and Tremois, respectively (Figure 2). As many as 1266 DEGs were unchanged in Nure but down-regulated in Tremois (Figure 2). There were 3906 DEGs and 4465 DEGs in control and treatment showing genotypic difference between Nure and Tremois (Figure S1).
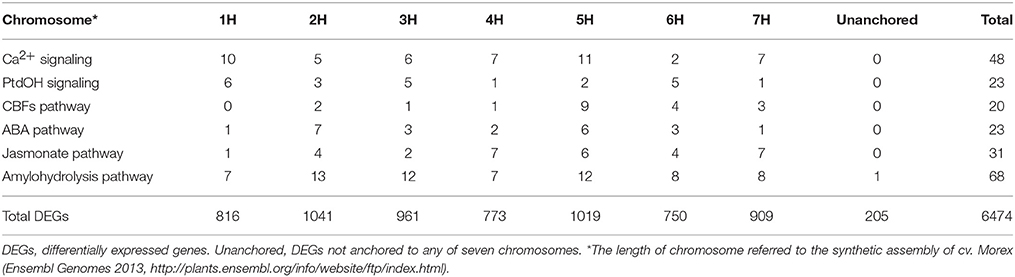
Table 1. Chromosome distribution of differentially expressed genes (DEGs) in low temperature signaling pathways.
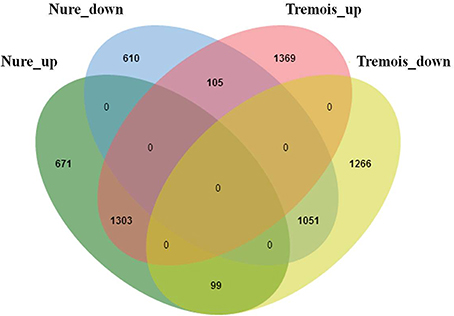
Figure 2. Overlapping of differentially expressed genes (DEGs) in Nure and Tremois after freezing shock. Nure_up, DEGs up-regulated in Nure. Nure_down, DEGs down-regulated in Nure. Tremois_up, DEGs up-regulated in Tremois. Tremois_down, DEGs down-regulated in Tremois.
GO (gene ontology) analysis of the whole 6474 DEGs was performed and the top five GO terms in each of the three major categories, molecular function, biological process and cellular component, were presented in Figure 2. In both genotypes, “ion binding,” “biological process,” and “plastid” were the most enriched terms in molecular function, biological process and cellular component, respectively, followed by “molecular function,” “biosynthetic process,” and “cellular component,” and then “kinase activity,” “cellular nitrogen compound metabolic process,” and “cytoplasmic membrane-bounded vesicle” (Figure 3). Then KEGG (Kyoto encyclopedia of genes and genomes) pathway enrichment was further investigated. A total of 1058 DEGs encoding 464 enzymes were assigned to 129 KEGG pathways, including lipid, carbohydrate, amino acid, energy, and other metabolic process (Table S4). DEGs encoding transferases made up the largest proportion, followed by hydrolases and oxidoreductases (Figure S2).
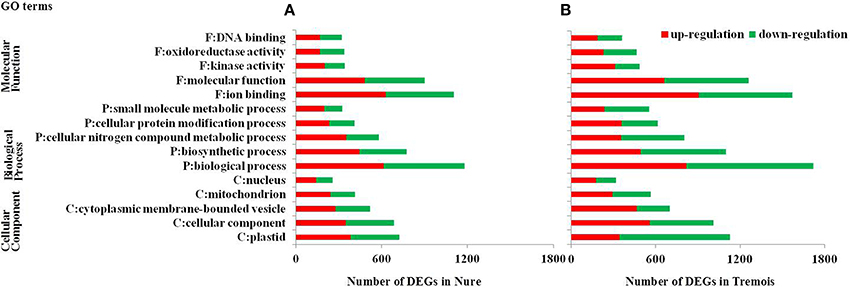
Figure 3. Gene Ontology (GO) analysis of differentially expressed genes (DEGs) in Nure (A) and Tremois (B). F, Molecular Function; P, Biological Process; C, Cellular Component; DEGs, differentially expressed genes. Only the top five GO terms in each of the three major categories, molecular function, biological process, and cellular component, are presented.
DEGs involved in low temperature perception, Ca2+ signaling, PtdOH signaling, CBFs pathway, ABA pathway, jasmonate pathway, and amylohydrolysis pathway were identified according to gene annotations and resulted in a hypothetical model of mild freezing shock tolerance mechanism in barley (Figure 1). The Ca2+, PtdOH, CBFs, and ABA pathways were mainly based on DEGs homologous to Arabidopsis identified by Blastp and relevant studies in Arabidopsis, while jasmonate and amylohydrolysis pathway were mainly based on KEGG maps. Annotations and FPKMs of DEGs in Figure 1 are listed in Table S5.
In order to assess the reliability of RNA-Seq data, we randomly selected 10 DEGs to investigate their expression in Nure and Tremois using qRT-PCR. There were highly significant correlations in both Nure and Tremois between RNA-Seq data and qRT-PCR results (Figure S3), indicating the high reliability of RNA-Seq data in the current study.
DEGs Involved in Ca2+ Signaling
A total of 48 DEGs involved in Ca2+ signaling were identified, i.e., 5 CaMs, 23 CMLs, 2 CBLs, 2 CDPKs, 2 CAMTAs, and 14 CIPKs (Figure 1A, Table S5). One CAM, one CDPK, three CIPKs and ten CMLs were up-regulated, while one CAM, one CML, and six CIPKs were down-regulated in both genotypes (Table S5). Two CAMs, two CMLs, two CBLs, and two CIPKs decreased in Nure but were unchanged in Tremois, while one CAM, one CIPK, one CDPK, two CAMTAs, and seven CMLs were unchanged in Nure but increased in Tremois after mild freezing shock (Table S5). CML4 (MLOC_71936) and CML6 (MLOC_62796) were up-regulated in Nure but unchanged in Tremois, while CML22 (MLOC_61579), CIPK26 (MLOC_63556), and CIPK11 (MLOC_12021) were unchanged in Nure but down-regulated in Tremois (Table S5). There was difference in the distribution of the 48 DEGs: 11 DEGs were anchored to chromosome 5H but only 2 DEGs were found in chromosome 6H (Table 1).
DEGs Involved in PtdOH Signaling
We identified 23 DEGs involved in PtdOH signaling (Figure 1B, Table S5). One PLD, one glycerol 3-phosphate acyltransferase (GPAT), one DGK, and two phosphatidic acid phosphatases (PAPs) increased, while one GPAT, one ethanolamine kinase (EK), one phosphorylcholine cytidylyltransferase (CTP), one cytidine diphosphate diacylglycerol synthase (CDS), and two phosphoethanolaminemethyltranferases (PEAMTs) decreased in both Nure and Tremois after mild freezing shock (Table S5). One PLD and one GPAT were down-regulated in Nure but unchanged in Tremois, while one PLD, one lysophosphatidic acid acyltransferase (LPAAT), one PAP, one EK, one phosphatidylinositol synthase (PIS), and one PLC were unchanged in Nure but up-regulated in Tremois (Table S5). The DEG encoding lipid phosphate phosphatase (LPP3, MLOC_72673) in Arabidopsis was down-regulated in Nure but up-regulated in Tremois (Table S5). LPAAT (MLOC_69398), cytidine diphosphate-ethanolamine phosphotransferase (EPT, MLOC_19179), and PLC (MLOC_58308) were unchanged in Nure but showed a 55.5, 66.1, and 51.0% decrease in Tremois after mild freezing shock (Table S5). As for the chromosome distribution, 1H harbors the most DEGs of PtdOH signaling pathway (Table 1).
DEGs Involved in CBFs Pathway
The 20 DEGs involved in CBFs pathway were interspersed on all seven chromosomes except 1H, where 5H having 9 DEGs (Table 1). Inducer of CBF expression 1 (ICE1), CBFs, circadian 1 (CIR1), low expression of osmotically responsive genes 2 (LOS2), and CORs were found to be positive regulators, while Myb domain protein 15 (MYB15), ZAT12, and ZAT10 were negative regulators of CBFs-dependent low temperature response pathway (Figure 1C).
DEG (MLOC_46407) homologous to ICE1 in Arabidopsis was identified to be unchanged in Nure but up-regulated in Tremois (Table S5). Four HvCBFs, e.g., HvCBF3 (MLOC_4956), HvCBF6 (MLOC_63682), HvCBF9 (MLOC_41994), and HvCBF14 (MLOC_59224) were up-regulated in both genotypes, while HvCBF1 (MLOC_75886) and HvCBF4B (MLOC_54227) were unchanged and down-regulated in Nure respectively, but up-regulated in Tremois. (Table S5). Two homologous genes of CIR1 in Arabidopsis were found out in our DEGs (MLOC_57684 and MLOC_62877) but their expression levels showed a dramatic difference. MLOC_62877 increased in both genotypes while MLOC_57684 was unchanged in Nure and decreased in Tremois by 53.6% after mild freezing shock (Table S5). MLOC_37167, a homologous DEG of LOS2 in Arabidopsis exhibited an 87.5% decrease in Nure after exposure to low temperature (Table S5). Two CORs (MLOC_14884 and MLOC_70331) were both up-regulated by low temperature with different expression patterns. MLOC_14884, had higher expression level than MLOC_70331 in all samples (Table S5). Both MLOC_14884 and MLOC_70331 showed lower expression values under control and higher expression values under treatment in Nure (Table S5).
Three DEGs (MLOC_34646, MLOC_43806 and MLOC_73037) homologous to MYB15 in Arabidopsis were collectively up-regulated in Nure but unchanged in Tremois (Table S5). Three ZAT10 homologs (MLOC_54674, MLOC_65033 and MLOC_70662) were up-regulated in all samples with an increase of at least 6.32 (Table S5). Three DEGs (MLOC_19822, MLOC_63032, and MLOC_76196) homologous to ZAT12 in Arabidopsis showed low expression levels under normal condition and were up-regulated by mild freezing shock in both cultivars (Table S5).
Among the DEGs, Five HvCBFs were identified within the Fr-H2 locus by Blastn. HvCBF9 is on the positive strand while HvCBF4B, HvCBF14, HvCBF6, and HvCBF3 are on the negative strand (Figure 4A). A startling up-regulation of HvCBF9 were indicated as its FPKMs were 5.2 and 2.8 under control while 296.3 and 201.4 after mild freezing shock to Nure and Tremois, respectively (Figure 4B). HvCBF14 also increased largely after mild freezing shock, with a 43.8- and 17.1-fold increase in Nure and Tremois, respectively (Figure 4B, Table S5). An SNV of HvCBF9 was identified as guanine (G) in Nure and cytosine (C) in Tremois at physical location 464369740 on 5H (Figure 4A). An insertion of “TC” within HvCBF14 in the physical location of 46435429 in Tremois compared with Nure was discovered (Figure 4A). A total of seven SNVs within HvCBF3 were identified, five of which were located in the coding domain sequence (CDS) area (Figure 4A). Only SNVs/indels causing sequence differences between Nure and Tremois were shown in Figure 4.
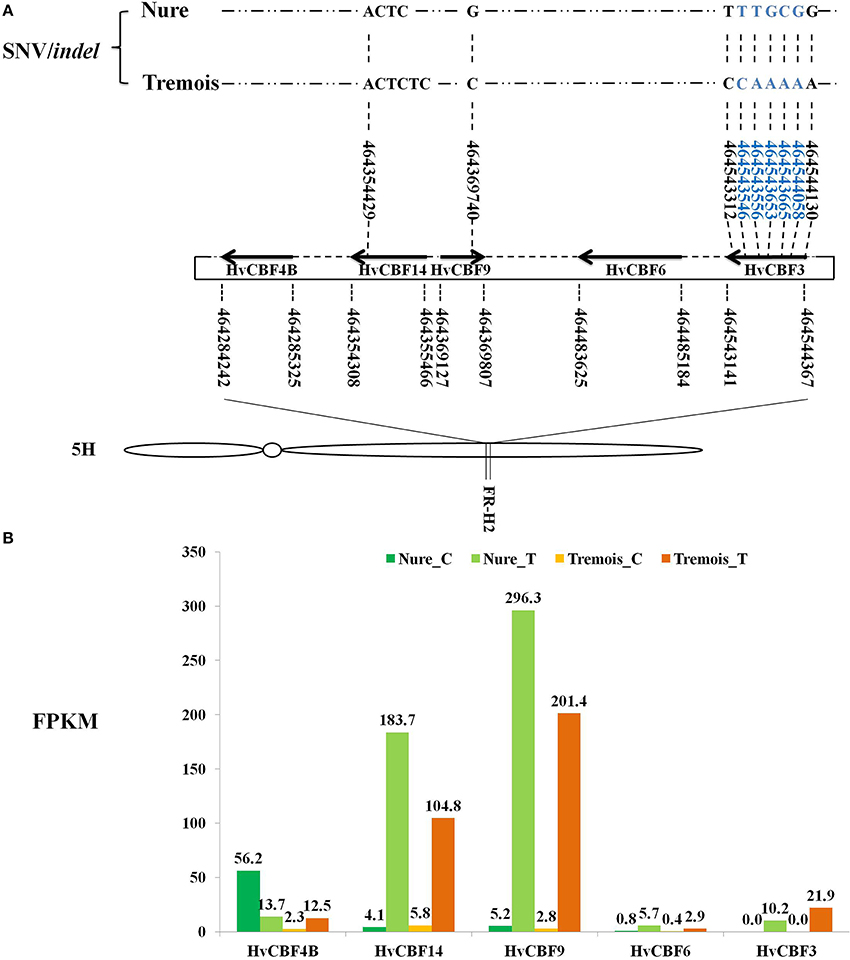
Figure 4. Feature of HvCBFs in FR-H2 locus of Chromosome 5H in barley. (A) Distribution and SNV/indel characteristics of HvCBFs in FR-H2 locus. Boundaries of HvCBFs in FR-H2 locus are illustrated by bold arrows. The numbers under and above gene names are physical locations of genes and SNV/indel, respectively. Right and left arrows show positive and negative strand of HvCBFs on the assembly genome. SNVs/indels sequence differed between Nure and Tremois are shown, and SNVs/indels in blue locate in the coding domain sequence (CDS) regionof HvCBF3. Indel, insertion or deletion variant. SNV, single nucleotide variant. Distributions of HvCBFs in FR-H2 locus refer to Pasquariello et al. (2014). (B) Expression values of HvCBFs in FR-H2 locus. Fragments per kilo base per million map reads (FPKMs) of samples are labeled above each bar. Nure_C, Nure in control. Nure_T, Nure in treatment. Tremois_C, Tremois in control. Tremois_T, Tremois in treatment.
DEGs Involved in ABA Pathway
Abscisic acid deficient 1 (ABA1), 9-cis-epoxycarotenoid dioxygenase (NCED), PYR, PYL, open stomata 1 (OST1), sucrose non-fermenting 1-related protein kinase 2.5 (SnRK2.5), ABF, and AREB are positive regulators while non-photochemical quenching 1 (NPQ1), cytochrome P450-family 707-subfamily A (CYP707A), homeobox-7 (HB-7), and protein phosphatase 2C (PP2C) are negative regulators in ABA biosynthesis and signaling (Figure 1D). Among positive regulators, PYL5 (MLOC_71349) and OST1 (MLOC_69212) were up-regulated in Nure but unchanged in Tremois, while ABF3 (MLOC_20326 and MLOC_66546) were unchanged in Nure but down-regulated in Tremois (Table S5). Among negative regulators, HB-7 (MLOC_69710) was down-regulated in Nure but unchanged in Tremois and HAI3 (MLOC_8131), which encoded a cluster A PP2C, being unchanged in Nure but up-regulated in Tremois (Table S5). Furthermore, according to the results of Blastn, the up-regulated HvCBF3 shared the similar nucleotide sequences with CBF4 in Arabidopsis, which positively regulated ABA signaling (Figure 1D). The 23 DEGs in ABA pathway were largely anchored to chromosomes 2H and 5H of assembly genome of Morex (Table 1).
DEGs Involved in Jasmonate Pathway
In total, 23 DEGs encoding 10 key enzymes during endogenous jasmonate biosynthesis from PtdCho and 8 DEGs encoding 4 JAZ proteins (negative regulators in jasmonate signaling) were identified (Figure 1E, Table S5). Among enzymes participating in jasmonate biosynthesis, phospholipase A (PLA2, MLOC_6428 and MLOC_24486) were up-regulated in Nure but unchanged in Tremois, while linoleate 13S-lipoxygenase (LOX, MLOC_64972) was up-regulated in Nure and down-regulated in Tremois (Table S5). Approximately 80% of DEGs encoding enzymes involved in jasmonate biosynthesis were up-regulated (Table S5). A DEG (MLOC_59388) encoding homolog of JAZ12 in Arabidopsis was unchanged in Nure but up-regulated in Tremois (Table S5).
DEGs Involved in Amylohydrolysis Pathway
A total of 68 DEGs encoding 17 enzymes involved in amylohydrolysis were identified. Starch branching enzyme (SBE, MLOC_63722), beta-glucosidase (BGLU, MLOC_79756), invertase (INV, MLOC_56803), phosphofructokinase (PFK, MLOC_15972), and glucose-6-phosphate isomerase (GPI, MLOC_1497) were up-regulated in Nure but unchanged in Tremois. However, cellulose synthases (CESAs, MLOC_7825, and MLOC_43749), BGLUs (MLOC_37976, MLOC_12571, and MLOC_37740), sucrose-phosphate synthase (SPS, MLOC_63746), sucrose-phosphate phosphatase (SPP, MLOC_6173), INVs (MLOC_60935, MLOC_72784 and MLOC_79590), and GPIs (MLOC_61974 and MLOC_40899) were unchanged in Nure but down-regulated in Tremois (Table S5). Regarding the chromosome distribution of DEGs of amylohydrolysis pathway, 2H had the largest number of DEGs, followed by 3H and 5H (Table 1).
Discussion
In the current study, we performed a RNA-Seq analysis to reveal the transcriptome reprogramming of barley plants under mild freezing shock after a very short acclimation for gene induction, which is different from the normal crop environmental conditions. We used such a treatment to mimic the natural condition with gradually change of temperature but non-lethal to winter barley, i.e., Nure, in order to potentially trigger as many genes as possible in response to low temperature. Moreover, this is not a study on the function of specific genes, so we pooled samples from 10 plants, but not single leaf from one for RNA-seq, which could average the differences between the responses of individual seedlings to the mild freezing treatment. And also, we only randomly selected some DEGs and conducted a “technical” validation of the fold-change ratios of the sequenced transcripts. This type of approach is widely used in many other studies, e.g., Yokosho et al. (2014).
An early response genes network of mild freezing shock was investigated by classifying DEGs into different freezing shock response pathways, named Ca2+ signaling, PtdOH signaling, CBFs pathway, ABA pathway, jasmonate pathway, and amylohydrolysis pathway (Figure 1). Membrane rigidification may be the first reaction to low temperature perception in plants, and reduced membrane fluidity is coupled with enhanced low temperature inducibility due to the action of a number of genes (Miquel et al., 1993). Four DEGs encoding homologs of FAD in Arabidopsis nearly collectively showed higher FPKMs under treatment in freezing tolerant cultivar Nure than freezing sensitive cultivar Tremois (Table S3), suggesting that higher content of unsaturated fatty acid after freezing shock may contribute to the superior freezing tolerance in Nure. This was consistent with previous study that plants preferentially accumulate polyunsaturated fatty acids under low temperature and genetically increasing of unsaturated fatty acids could enhance cold tolerance, and chilling tolerant rice cultivars contained greater proportion of unsaturated fatty acids than sensitive ones (Kodama et al., 1994; Ariizumi et al., 2002).
As a ubiquitous intracellular second messenger, Ca2+ participates in plant freezing response not only via Ca2+ signature but also by the cross-talk with other signaling pathways. Ca2+ may modulate PtdOH signaling by activating PLC and PLD, two pivotal enzymes in PtdOH synthesis (Hunt et al., 2004; Lecourieux et al., 2006). CAM is a potential negative regulator of COR, as its overexpression represses low temperature-induced COR expression in plants (Townley and Knight, 2002). CAM7 (MLOC_19924) and CAM9 (MLOC_65903) were down-regulated in freezing tolerant accession Nure and CAM7 (MLOC_36991) was up-regulated in freezing sensitive varieties Tremois, which consists with the higher COR expression in Nure, suggesting the potential negative regulation of CAM on freezing tolerance in barley (Table S5). The cross-talk between Ca2+ signaling and CBFs pathway is also evident by the positive regulation of CAMTA on freezing induced expression of CBF1 and CBF2 in Arabidopsis (Doherty et al., 2009). In the present study, CMTA4 (MLOC_44450 and MLOC_58709) were unchanged in Nure but up-regulated in Tremois, which may explain the higher expression value of HvCBF1 under treatment in Tremois.
PtdOH signaling is also involved in mild freezing shock response. Low temperature can inhibit the activity of CTP, resulting in a limited availability of cytidine diphosphate-ethanolamine (CDP-Etn) and an accumulation of DAG as precursor for PtdEtn synthesis, the latter of which could be an additional source for freezing induced PtdOH through DGK activity (Inatsugi et al., 2002; Arisz et al., 2013). Decreased CTP (MLOC_19066), EPT (MLOC_19179) and increased DGK (MLOC_58803) after freezing shock indicated the presence of a similar regulation mechanism in barley (Table S5). Besides, reduced CTP may cause the accumulation of EtnP, which can be repeatedly methylated by PEAMT to form ChoP and then PtdCho (Figure 1B). As the rate-limiting enzyme for PtdCho synthesis, PEAMT (MLOC_15214 and MLOC_66415) showed a 58.0 and 560% higher FPKM under treatment in Nure than in Tremois, respectively (Table S5). PtdCho can then be catalyzed by PLD to generate PtdOH (Figure 1B).
Chromosome 5H of barley is very important for cold-responsive CBFs pathway, as 9 out of the 20 DEGs involved in CBFs pathway were anchored to it (Table 1) and the HvCBFs are clustered within Fr-H2 locus on 5H. Among the five identified HvCBFs located in Fr-H2, HvCBF14 and HvCBF9 may be the major transcription factors for freezing tolerance. It was evident that FPKMs are increased sharply after freezing shock and the higher transcript level in freezing tolerant variety Nure (Figure 4). The expression difference of HvCBFs between Nure and Tremois may be partly related to the variation of gene copy number. For example, multiple copies of HvCBF4 were found in Nure as compared to a single copy in Tremois (Knox et al., 2010). Though an indel in HvCBF14 and a SNV in HvCBF9 led to sequence differences between Nure and Tremois, these sequence variations may not result in amino acid change, because both variable sites are located in the 3′ end of CDS rather than in the coding region (Figure 4). Furthermore, two subgroups of the five HvCBFs can be divided according to the phylogenetic analysis in previous reports (Skinner et al., 2005): HvCBF6 and HvCBF3 belong to HvCBF3-subgroup, while HvCBF4B, HvCBF14 and HvCBF9 are classified into HvCBF4-subgroup. We postulate that HvCBF4-subgroup possibly contributes more to freezing shock tolerance than HvCBF3- subgroup in barley based on their expression values (Figure 4). Moreover, it has been reported that HvCBFs in Fr-H2 could be affected by the major vernalisation gene (VRN-H1) on 5H in barley (Stockinger et al., 2007; Comadran et al., 2012). However, in the current study, none of the three genes determining growth habit in the vernalization pathway, HvVRN1, HvVRN2, and HvVRN3/HvFT1 (Comadran et al., 2012), belonged to the 6474 DEGs, which may due to the short-term freezing shock treatment, sampling tissues or other factors, such as photoperiod (Stockinger et al., 2007).
Although, ICE1 can bind to the promoter of CBF3 and enhance its expression in Arabidopsis, it is only slightly up-regulated by low temperature. It was reasoned that the freezing induced posttranslational modification of ICE1 such as phosphorylation is essential for its activation on CBFs (Chinnusamy et al., 2003). Furthermore, a well-known protein kinase in ABA signaling, SnRK2.6/OST1, was reported to be activated by low temperature in an ABA-independent mechanism and can phosphorylate and stabilize ICE1 to positively modulate CBFs pathway in Arabidopsis (Ding et al., 2015). DEGs (MLOC_69212, MLOC_11726, and MLOC_3013) homologous to OST1 in Arabidopsis were up-regulated by mild freezing shock and Nure always showed a greater FPKM (Table S5). It has been proposed that, low temperature may alter the conformation of a sensor protein, which could be one or more of the PYL proteins, to activate OST1 (Ding et al., 2015). It has been reported that PYL5 in Arabidopsis was able to modulate drought resistance by inhibiting clade A PP2Cs and the inhibition could be ABA-independent (Santiago et al., 2009; Hao et al., 2011). Coincidentally, DEGs (MLOC_71349 and MLOC_65591) homologous to PYL5 in Arabidopsis were up-regulated after freezing shock with larger increase in Nure (Table S5). More speculatively, we infer that, PYL5 (MLOC_71349 and MLOC_65591) in barley may be multiple sensory proteins that involves in the OST1-mediated CBFs dependent pathway in response to low temperature. This hypothesis may also be partly supported by the previous report that ABA receptors are candidates to improve abiotic stress tolerance of crops and constitutive overexpression of a cytosolic ABA receptor (OsPYL/RCAR5) in rice confers drought and salt tolerance (Kim et al., 2014).
As the pivot point of mild freezing response, CBFs pathway interacts with other signaling pathways to form an overall freezing tolerance in plants. Apart from the cross talk with PtdOH pathway and ABA pathway, CBFs pathway is also modulated by jasmonate. Three genes (MLOC_18524, MLOC_4800, and MLOC_80547) homologous to jasmonate receptor COI1 in Arabidopsis were detected to be uninfluenced by low temperature (data not shown as they were not DEGs). Among the 8 DEGs encoding homolog of JAZ in Arabidopsis, 7 DEGs showed higher FPKM in Nure under treatment (MLOC_61774, MLOC_10653, MLOC_45755, MLOC_11970, MLOC_59389, MLOC_9995, and MLOC_37630), 2 DEGs were up-regulated in Nure but unchanged in Tremois (MLOC_61774 and MLOC_10653) (Table S5). Given the inhibition of JAZ on ICE1 (Figures 3C,E; Table S5; Hu et al., 2013), our result may indicate that the expression profile of the DEG (MLOC_46407) homologous to ICE1 in Arabidopsis, which was unchanged in Nure but up-regulated in Tremois.
Soluble sugar, such as trehalose, act as osmoprotectant in freezing tolerance by lowering osmotic potential to protect membranes, enzymes, and other structures against irreversible damage and denaturation (Bartels and Sunkar, 2005). Previous study shows that overexpression of two crucial trehalose synthetases, trehalose 6-phosphate phosphatase (TPP) and trehalose 6-phosphate synthase (TPS), enhances freezing tolerance in plants (Pramanik and Imai, 2005; Miranda et al., 2007). We also found that TPS (MLOC_11773) and three out of the four TPPs (MLOC_11773, MLOC_49094, and MLOC_52798) in barley increased significantly after freezing shock (Table S5).
In conclusion, multifarious pathways are activated in concert to perceive and transduce the signal in barley exposed to mild freezing shock. Besides, cross-talk occurred among different signaling pathways in response to mild freezing shock. The pathway network we obtained may provide a platform for further exploring the functions of genes involved in low temperature tolerance in barley.
Author Contributions
FD conceived and designed this project. XW, GJ, and FD carried out the bioinformatics analysis. FD, XW, DW, QY, and JZ performed all the experimental parts. GZ and ZHC participated in its design and coordination. XW, FD, DW, ZC, and GZ wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Luigi Cattivelli (CRA Agricultural Research Council) for providing seeds of Nure and Tremois. This work was supported by the National Natural Science Foundation of China (31471480 and 31330055), Natural Science Foundation of Zhejiang Province (LR15C130001), the Fundamental Research Funds for the Central Universities and Jiangsu Collaborative Innovation Center for Modern Crop Production.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00106
Figure S1. Genotype difference analysis of differentially expressed genes (DEGs) in Nure and Tremois. FPKM: fragments per kilobase per million map reads. FPKM = 109 × total_ fragments_in_exons/total_fragments_counted/transcript_length.
Figure S2. Enzyme code distribution of differentially expressed genes (DEGs) in Nure and Tremois.
Figure S3. Correlation of gene expression ratio between RNA-Seq and qRT-PCR in Nure (A) and Tremois (B).
Table S1. Primers of differentially expressed genes (DEGs) selected for quantitative real-time PCR (qRT-PCR) expression analysis.
Table S2. Mapping of RNA-Seq data to barley cv. Morex WGS contigs.
Table S3. Annotations and expression values of the 6474 differentially expressed genes (DEGs).
Table S4. KEGG overview of differentially expressed genes (DEGs).
Table S5. Expression values of differentially expressed genes (DEGs) in low temperature signaling pathways.
Abbreviations
ABA1, abscisic acid deficient 1; ABRE, abscisic acid responsive element; AREB/ABF, abscisic acid responsive element binding protein/abscisic acid responsive element binding factor; CaM, calmodulin; CAMTA, calmodulin-binding transcription factor; CBFs, C-repeat binding factors; CBLs, calcineurin B-like proteins; CBPs, Ca2+-binding proteins; CDP-Etn, cytidine diphosphate-ethanolamine; CDPKs, calcium-dependent protein kinases; CDS, coding domain sequence; CIPKs, calcineurin B-like protein-interacting protein kinases; CIR1, circadian 1; CMLs, calmodulin-like proteins; COI1, coronatine insensitive 1; COR, cold responsive genes; CYP707A, cytochrome P450-family 707-subfamily A; DAG, diacylglycerol; DEGs, differentially expressed genes; DGK, diacylglycerol kinase; FAD, fatty acid desaturase; FPKM, fragments per kilobase per million map reads; Fr-H2, Frost resistance-H2; GO, gene ontology; HB-7, homeobox-7; ICE1, inducer of CBF expression 1; indel, insertion or deletion variant; JAZ, jasmonate-zim-domain protein; KEGG, Kyoto encyclopedia of genes and genomes; LOS2, low expression of osmotically responsive genes 2; MYB15, Myb domain protein 15; NCED, 9-cis-epoxycarotenoid dioxygenase; NPQ1, non-photochemical quenching 1; nr, non-redundant; Nure_C, Nure in control; Nure_T, Nure in treatment; OST1, open stomata 1; PLC, phospholipase C; PLD, phospholipase D; PP2C, protein phosphatase type 2C; PPIs, polyphosphoinositides; PtdCho, phosphatidylcholine; PtdEtn, phosphatidylethanolamine; PtdOH, phosphatidic acid; PYL, pyrabactin resistance PYR-like; PYR, pyrabactin resistance; RCAR, regulatory components of ABA receptor; RNA-Seq, RNA sequencing; SnRK2, sucrose non-fermenting 1-related protein kinase 2; SNVs, single nucleotide variants; TPP, trehalose 6-phosphate phosphatase; TPS, trehalose 6-phosphate synthase; Tremois_C, Tremois in control; Tremois_T, Tremois in treatment.
References
Acharya, B. R., Jeon, B. W., Zhang, W., and Assmann, S. M. (2013). Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol. 200, 1049–1063. doi: 10.1111/nph.12469
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ariizumi, T., Kishitani, S., Inatsugi, R., Nishida, I., Murata, N., and Toriyama, K. (2002). An increase in unsaturation of fatty acids in phosphatidylglycerol from leaves improves the rates of photosynthesis and growth at low temperatures in transgenic rice seedlings. Plant Cell Physiol. 43, 751–758. doi: 10.1093/pcp/pcf087
Arisz, S. A., van Wijk, R., Roels, W., Zhu, J. K., Haring, M. A., and Munnik, T. (2013). Rapid phosphatidic acid accumulation in response to low temperature stress in Arabidopsis is generated through diacylglycerol kinase. Front. Plant Sci. 4:1. doi: 10.3389/fpls.2013.00001
Bardou, P., Mariette, J., Escudié, F., Djemiel, C., and Klopp, C. (2014). jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15:293. doi: 10.1186/1471-2105-15-293
Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. doi: 10.1080/07352680590910410
Batistic, O., Waadt, R., Steinhorst, L., Held, K., and Kudla, J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61, 211–222. doi: 10.1111/j.1365-313X.2009.04045.x
Bogdanovic, J., Mojovic, M., Milosavic, N., Mitrovic, A., Vucinic, Z., and Spasojevic, I. (2008). Role of fructose in the adaptation of plants to cold-induced oxidative stress. Eur. Biophys. J. Biophy. 37, 1241–1246. doi: 10.1007/s00249-008-0260-9
Campoli, C., Matus-Cádiz, M. A., Pozniak, C. J., Cattivelli, L., and Fowler, D. B. (2009). Comparative expression of Cbf genes in the Triticeae under different acclimation induction temperatures. Mol. Genet. Genomics 282, 141–152. doi: 10.1007/s00438-009-0451-9
Chen, H., Chen, X., Chai, X., Qiu, Y., Gong, C., Zhang, Z., et al. (2015). Effects of low temperature on mRNA and small RNA transcriptomes in Solanum lycopersicoides leaf revealed by RNA-Seq. Biochem. Biophys. Res. Commun. 464, 768–773. doi: 10.1016/j.bbrc.2015.07.029
Chinnusamy, V., Zhu, J. K., and Sunkar, R. (2010). Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639, 39–55. doi: 10.1007/978-1-60761-702-0_3
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2006). Gene regulation during cold acclimation in plants. Physiol. Plant. 126, 52–61. doi: 10.1111/j.1399-3054.2006.00596.x
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Chinnusamy, V, Ohta, M., Kanrar, S., Lee, B. H., Hong, X. H., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Gene Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Comadran, J., Kilian, B., Russell, J., Ramsay, L., Stein, N., Ganal, M., et al. (2012). Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44, 1388–1392. doi: 10.1038/ng.2447
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Dai, F., Chen, Z. H., Wang, X., Li, Z., Jin, G., Wu, D., et al. (2014). Transcriptome profiling reveals mosaic genomic origins of modern cultivated barley. Proc. Natl. Acad. Sci. U.S.A. 111, 13403–13408. doi: 10.1073/pnas.1414335111
Dai, F., Huang, Y., Zhou, M., and Zhang, G. (2009). The influence of cold acclimation on antioxidative enzymes and antioxidants in sensitive and tolerant barley cultivars. Biol. Plant. 53, 257–262. doi: 10.1007/s10535-009-0048-5
Dai, F., Zhou, M., and Zhang, G. (2007). The change of chlorophyll fluorescence parameters in winter barley during recovery after freezing shock and as affected by cold acclimation and irradiance. Plant Physiol. Biochem. 45, 915–921. doi: 10.1016/j.plaphy.2007.09.006
DeFalco, T. A., Bender, K. W., and Snedden, W. A. (2010). Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 425, 27–40. doi: 10.1042/BJ20091147
Ding, Y. L., Li, H., Zhang, X. X., Xie, Q., Gong, Z. Z., and Yang, S. H. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32, 278–289. doi: 10.1016/j.devcel.2014.12.023
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Francia, E., Rizza, F., Cattivelli, L., Stanca, A. M., Galiba, G., Tóth, B., et al. (2004). Two loci on chromosome 5H determine low-temperature tolerance in a 'Nure' (winter) x 'Tremois' (spring) barley map. Theor. Appl. Genet. 108, 670–680. doi: 10.1007/s00122-003-1468-9
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993. doi: 10.1073/pnas.0505667103
Ganeshan, S., Vitamvas, P., Fowler, D. B., and Chibbar, R. N. (2008). Quantitative expression analysis of selected COR genes reveals their differential expression in leaf and crown tissues of wheat (Triticum aestivum L.) during an extended low temperature acclimation regimen. J. Exp. Bot. 59, 2393–2402. doi: 10.1093/jxb/ern112
Guan, Q., Wu, J., Zhang, Y., Jiang, C., Liu, R., Chai, C., et al. (2013). A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 25, 342–356. doi: 10.1105/tpc.112.108340
Hao, Q., Yin, P., Li, W., Wang, L., Yan, C., Lin, Z., et al. (2011). The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol. Cell 42, 662–672. doi: 10.1016/j.molcel.2011.05.011
Hu, Y., Jiang, L., Wang, F., and Yu, D. (2013). Jasmonate regulates the inducer of Cbf Expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
Hughes, M. A., and Dunn, M. A. (1996). The molecular biology of plant acclimation to low temperature. J. Exp. Bot. 47, 291–305. doi: 10.1093/jxb/47.3.291
Hunt, L., Otterhag, L., Lee, J. C., Lasheen, T., Hunt, J., Seki, M., et al. (2004). Gene-specific expression and calcium activation of Arabidopsis thaliana phospholipase C isoforms. New Phytol. 162, 643–654. doi: 10.1111/j.1469-8137.2004.01069.x
Inatsugi, R., Nakamura, M., and Nishida, I. (2002). Phosphatidylcholine biosynthesis at low temperature: Differential expression of CTP: phosphorylcholine cytidylyltransferase isogenes in Arabidopsis thaliana. Plant Cell Physiol. 43, 1342–1350. doi: 10.1093/pcp/pcf169
Jiang, H., Li, W., He, B., Gao, Y., and Lu, J. (2014). Sucrose metabolism in grape (Vitis vinifera L.) branches under low temperature during overwintering covered with soil. Plant Growth Regul. 72, 229–238. doi: 10.1007/s10725-013-9854-z
Kaplan, B., Davydov, O., Knight, H., Galon, Y., Knight, M. R., Fluhr, R., et al. (2006). Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18, 2733–2748. doi: 10.1105/tpc.106.042713
Kersey, P. J., Allen, J. E., Christensen, M., Davis, P., Falin, L. J., Grabmueller, C., et al. (2014). Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 42, D546–D552. doi: 10.1093/nar/gkt979
Kim, H., Lee, K., Hwang, H., Bhatnagar, N., Kim, D. Y., Yoon, I. S., et al. (2014). Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 65, 453–464. doi: 10.1093/jxb/ert397
Knox, A. K., Dhillon, T., Cheng, H., Tondelli, A., Pecchioni, N., and Stockinger, E. J. (2010). CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor. Appl. Genet. 121, 21–35. doi: 10.1007/s00122-010-1288-7
Kodama, H., Hamada, T., Horiguchi, G., Nishimura, M., and Iba, K. (1994). Genetic enhancement of cold tolerance by expression of a gene for chloroplast ω-3 fatty acid desaturase in transgenic tobacco. Plant Physiol. 105, 601–605.
Lecourieux, D., Raneva, R., and Pugin, A. (2006). Calcium in plant defence-signalling pathways. New Phytol. 171, 249–269. doi: 10.1111/j.1469-8137.2006.01777.x
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, S., Tighe, S. W., Nicolet, C. M., Grove, D., Levy, S., Farmerie, W., et al. (2014). Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study. Nat. Biotechnol. 32, 915–925. doi: 10.1038/nbt.2972
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Mayer, K. F., Waugh, R., Brown, J. W., Schulman, A., Langridge, P., Platzer, M., et al. (2012). International barley genome sequencing consortium. a physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716. doi: 10.1038/nature11543
McCormack, E., Tsai, Y. C., and Braam, J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Miquel, M., James, D. Jr., Dooner, H., and Browse, J. (1993). Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc. Natl. Acad. Sci. U.S.A. 90, 6208–6212. doi: 10.1073/pnas.90.13.6208
Miranda, J. A., Avonce, N., Suárez, R., Thevelein, J. M., Van Dijck, P., and Iturriaga, G. (2007). A bifunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis. Planta 226, 1411–1421. doi: 10.1007/s00425-007-0579-y
Monroy, A. F., Labbé, E., and Dhindsa, R. S. (1997). Low temperature perception in plants: effects of cold on protein phosphorylation in cell-free extracts. FEBS Lett. 410, 206–209. doi: 10.1016/S0014-5793(97)00589-9
Morozova, O., Hirst, M., and Marra, M. A. (2009). Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genomics Hum. Genet. 10, 135–151. doi: 10.1146/annurev-genom-082908-145957
Nambara, E., and Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Pasquariello, M., Barabaschi, D., Himmelbach, A., Steuernagel, B., Ariyadasa, R., Stein, N., et al. (2014). The barley Frost resistance-H2 locus. Funct. Integr. Genomics 14, 85–100. doi: 10.1007/s10142-014-0360-9
Pramanik, M. H., and Imai, R. (2005). Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol. Biol. 58, 751–762. doi: 10.1007/s11103-005-7404-4
Ren, L., Sun, J., Chen, S., Gao, J., Dong, B., Liu, Y., et al. (2014). A transcriptomic analysis of Chrysanthemum nankingense provides insights into the basis of low temperature tolerance. BMC Genomics 15:844. doi: 10.1186/1471-2164-15-844
Rizza, F., Pagani, D., Gut, M., Prasil, I. T., Lago, C., Tondelli, A., et al. (2011). Diversity in the response to low temperature in representative barley genotypes cultivated in Europe. Crop Sci. 51, 2759–2779. doi: 10.2135/cropsci2011.01.0005
Roberts, A., Pimentel, H., Trapnell, C., and Pachter, L. (2011). Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329. doi: 10.1093/bioinformatics/btr355
Ruelland, E., Cantrel, C., Gawer, M., Kader, J. C., and Zachowski, A. (2002). Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 130, 999–1007. doi: 10.1104/pp.006080
Ruelland, E., and Zachowski, A. (2010). How plants sense temperature. Environ. Exp. Bot. 69, 225–232. doi: 10.1016/j.envexpbot.2010.05.011
Santiago, J., Rodrigues, A., Saez, A., Rubio, S., Antoni, R., Dupeux, F., et al. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60, 575–588. doi: 10.1111/j.1365-313X.2009.03981.x
Skinner, J. S., von Zitzewitz, J., Szucs, P., Marquez-Cedillo, L., Filichkin, T., Amundsen, K., et al. (2005). Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol. 59, 533–551. doi: 10.1007/s11103-005-2498-2
Stockinger, E. J., Skinner, J. S., Gardner, K. G., Francia, E., and Pecchioni, N. (2007). Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J. 51, 308–321. doi: 10.1111/j.1365-313X.2007.0141.x
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. Nature 448, 661–662. doi: 10.1038/nature05960
Thoday-Kennedy, E. L., Jacobs, A. K., and Roy, S. J. (2015). The role of the CBL-CIPK calcium signalling network in regulating ion transport in response to abiotic stress. Plant Growth Regul. 76, 3–12. doi: 10.1007/s10725-015-0034-1
Tondelli, A., Francia, E., Barabaschi, D., Pasquariello, M., and Pecchioni, N. (2011). Inside the CBF locus in Poaceae. Plant Sci. 180, 39–45. doi: 10.1016/j.plantsci.2010.08.012
Townley, H. E., and Knight, M. R. (2002). Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 128, 1169–1172. doi: 10.1104/pp.010814
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Valdés, A. E., Overnäs, E., Johansson, H., Rada-Iglesias, A., and Engstrom, P. (2012). The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 80, 405–418. doi: 10.1007/s11103-012-9956-4
Vaultier, M. N., Cantrel, C., Vergnolle, C., Justin, A. M., Demandre, C., Benhassaine-Kesri, G., et al. (2006). Desaturase mutants reveal that membrane rigidification acts as a cold perception mechanism upstream of the diacylglycerol kinase pathway in Arabidopsis cells. Febs Lett. 580, 4218–4223. doi: 10.1016/j.febslet.2006.06.083
Vergnolle, C., Vaultier, M. N., Taconnat, L., Renou, J. P., Kader, J. C., Zachowski, A., et al. (2005). The cold-induced early activation of phospholipase C and D pathways determines the response of two distinct clusters of genes in Arabidopsis cell suspensions. Plant Physiol. 139, 1217–1233. doi: 10.1104/pp.105.068171
Wang, X. C., Zhao, Q. Y., Ma, C. L., Zhang, Z. H., Cao, H. L., Kong, Y. M., et al. (2013). Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 14:415. doi: 10.1186/1471-2164-14-415
Winfield, M. O., Lu, C., Wilson, I. D., Coghill, J. A., and Edwards, K. J. (2010). Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol. J. 8, 749–771. doi: 10.1111/j.1467-7652.2010.00536.x
Xiong, L., Ishitani, M., Lee, H., and Zhu, J. K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13, 2063–2083. doi: 10.1105/tpc.13.9.2063
Yan, J., Zhang, C., Gu, M., Bai, Z., Zhang, W., Qi, T., et al. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. doi: 10.1105/tpc.109.065730
Yokosho, K., Yamaji, N., and Ma, J. F. (2014). Global transcriptome analysis of Al-induced genes in an Al-cccumulating species, common buckwheat (Fagopyrum esculentum Moench). Plant Cell Physiol. 55, 2077–2091. doi: 10.1093/pcp/pcu135
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Zeng, J., He, X., Wu, D., Zhu, B., Cai, S., Nadira, U. A., et al. (2014). Comparative transcriptome profiling of two Tibetan wild barley genotypes in responses to low potassium. PLoS ONE 9:e100567. doi: 10.1371/journal.pone.0100567
Zenoni, S., Ferrarini, A., Giacomelli, E., Xumerle, L., Fasoli, M., Malerba, G., et al. (2010). Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol. 152, 1787–1795. doi: 10.1104/pp.109.149716
Zhang, D., Shi, Y., and Yang, S. (2014). “ABA regulation of the cold stress response in Plants,” in Abscisic Acid: Metabolism, Transport and Signaling, ed D. Zhang (Springer Netherlands), 337–363.
Keywords: barley (Hordeum vulgare L.), differentially expressed genes, freezing shock, RNA-sequencing, signaling pathway
Citation: Wang X, Wu D, Yang Q, Zeng J, Jin G, Chen Z-H, Zhang G and Dai F (2016) Identification of Mild Freezing Shock Response Pathways in Barley Based on Transcriptome Profiling. Front. Plant Sci. 7:106. doi: 10.3389/fpls.2016.00106
Received: 15 September 2015; Accepted: 20 January 2016;
Published: 08 February 2016.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Meixue Zhou, University of Tasmania, AustraliaNicola Pecchioni, Council for Agricultural Research and Economics, Italy
Copyright © 2016 Wang, Wu, Yang, Zeng, Jin, Chen, Zhang and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Dai, fdai@zju.edu.cn
 Xiaolei Wang
Xiaolei Wang Dezhi Wu
Dezhi Wu Zhong-Hua Chen
Zhong-Hua Chen Guoping Zhang
Guoping Zhang Fei Dai
Fei Dai