- 1Department of Life Sciences, Yeungnam University, Gyeongsan, South Korea
- 2Department of Biology, The Research Institute of Natural Science, Gyeongsang National University, Jinju, South Korea
Previous studies of Veronica and related genera were weakly supported by molecular and paraphyletic taxa. Here, we report the complete chloroplast genome sequence of Veronica nakaiana and the related species Veronica persica and Veronicastrum sibiricum. The chloroplast genome length of V. nakaiana, V. persica, and V. sibiricum ranged from 150,198 bp to 152,930 bp. A total of 112 genes comprising 79 protein coding genes, 29 tRNA genes, and 4 rRNA genes were observed in three chloroplast genomes. The total number of SSRs was 48, 51, and 53 in V. nakaiana, V. persica, and V. sibiricum, respectively. Two SSRs (10 bp of AT and 12 bp of AATA) were observed in the same regions (rpoC2 and ndhD) in three chloroplast genomes. A comparison of coding genes and non-coding regions between V. nakaiana and V. persica revealed divergent sites, with the greatest variation occurring petD-rpoA region. The complete chloroplast genome sequence information regarding the three Veroniceae will be helpful for elucidating Veroniceae phylogenetic relationships.
Introduction
Chloroplast (cp) are photosynthetic organelles that provide energy to green plants (Douglas, 1990). Chloroplast genomes are valuable sources of phylogenetic information because of their relatively stable genome structure and higher evolutionary rate relative to mitochondrial genomes. Chloroplast genomes consist of a large inverted repeat (IR) separated by a large single-copy (LSC) region and a small single-copy (SSC) region. Approximately 100–130 genes are located along the circular genome structure of chloroplasts. These genes exhibit a highly conserved gene order and contests (Wicke et al., 2011) and typically encode ~79 proteins, ~30 transfer RNAs and four ribosomal RNAs. However, some parasitic plants contain fewer genes than photosynthetic plants (Funk et al., 2007; Wicke et al., 2013). Cp genome sequences are useful genetic markers for DNA barcoding, transplastomic studies and evolutionary studies from the population level, as well as for phylogenetic relationships (Bock and Khan, 2004; Jansen et al., 2007).
The genus Veronica is a member of the family Plantaginaceae in the order Lamiales, which consists of 24 families and ~23,000 species (Angiosperm Phylogeny Group, 2009). Almost all families of Lamiales are included in Scrophulariaceae. However, molecular analysis of Scrophulariaceae was only recently conducted and reclassification (Olmstead and Reeves, 1995; Albach et al., 2005). Veronica were traditionally circumscribed to Scrophulariaceae. However, molecular studies conducted by Olmstead et al. (2001) separated several families, including Veroniceae (Plantaginaceae). The tribe Veroniceae was first established in 1828 by Duby to include the genera Bseeya, Botryopleuron, Calorhabdos, Hebe, Lagotis, Paederota, Picrorhiza, Synthris, Wulfenia, Veronicastrum, and Veronica. Subsequent molecular studies discussed the tribe Veroniceae and proposed a revised classification for the genus Veronica. Albach and Chase (2001) and Albach and Meudt (2010) included Veronica, Hebe, Beccabunga, Synthyris, Cochlidiosperma, Pellidosperma, Chamaedrys, Stenocarpon, Pocilla, Pentasepalae, and Pseudolysimachium into a genus Veronica.
DNA barcoding can be used to elucidate plant relationships at the species level. Ribosomal ITS sequences (Alvarez and Wendel, 2003) and non-coding region trnL intron (Taberlet et al., 2006) and psbA-trnH regions (Kress et al., 2005) are useful markers for identifying plant specimens and relationships. Previous phylogenetic analyses of Veroniceae used markers of the nuclear DNA ITS and CYC2 and plastid DNA rbcL, psbA-trnH, trnL-trnL-trnF, rps16 introns, and rpoB-trnC (Albach and Chase, 2001; Wagstaff et al., 2002; Albach et al., 2004a,b, 2005; Albach and Meudt, 2010). However, the relationship among the subgenus in Veronica (including subgenus Pseudolysimachium) was paraphyletic and not well supported (Albach et al., 2004a,b; Albach and Meudt, 2010).
Here, we report the first chloroplast genome of three Veroniceae (Veronica nakaiana, subgenus Pseudolysimachium; Veronica persica, subgenus Pocilla; Veronicastrim sibiricum). The specific goals of the present study were to: (1) present the complete chloroplast genome sequence of three Veroniceae, (2) evaluate the phylogenetic position of 78 coding genes in three Veroniceae plants, (3) compare the coding and non-coding regions of three Veroniceae plants and suggest effective regions of the chloroplast genome for phylogenic analyses in Veroniceae.
Materials and Methods
Plant Samples and Sequencing
This research was approved by the Korean National Arboretum. V. nakaiana, V. persica, and V. sibiricum were obtained from the living collections at the greenhouse of Yeungnam University. Total DNA was extracted using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA) and quantified using a HiGenTM Gel and PCR Purification System (Biofact Inc., Daejeon, Korea). Genomic DNA was sequenced using Illumina Miseq (Illumina Inc., San Diego, CA). V. nakaiana, V. persica, and V. sibiricum were sequenced to produce 3,205,654–4,475,346 raw reads and 301 bp were obtained from them. These paired-end reads were aligned with Sesamum indicum (JN637766.2), and 207,913 to 1,450,476 reads were mapped to the reference genomes. The fold coverage of V. nakaiana, V. persica, and V. sibiricum was 1223, 1911, and 404, respectively. The genome coverage was estimated using the CLC Genomics Workbench v7.0.4 software (CLC-bio, Aarhus, Denmark).
Annotation, Genome Mapping, and Sequence Analysis
DOGMA (Wyman et al., 2004) was used to annotate the V. nakaiana, V. persica, and V. sibiricum. Initial annotation, putative starts, stops, and intron positions were determined by comparison with homologous genes in other cp genomes. tRNA genes were annotated using DOGMA and tRNAscan-SE (Schattner et al., 2005). To compare the structure and gene content within the three Veroniceae, plants were aligned using MAFFT (Katoh et al., 2002). A circular cp genome map was drawn using the OGDRAW program (Lohse et al., 2007).
Repeat Structure
REPuter was used to visualize both forward, palindrome, reverse and complement sequences, with a minimum repeat size of 30 bp and a sequence identity greater than 90% (Kurtz and Schleiermacher, 1999). The simple sequence repeats (SSRs) in the three Veroniceae plant cp genomes were detected using Phobos v. 3.3.12 (http://www.ruhr-uni-bochum.de/ecoevo/cm/cm_phobos.htm). Repeats were ≥10 sequence length and three repeat units for mono-, di-, tri-, tetra-, penta-, and hexanucleotides.
Phylogenetic Analysis
A total of 78 coding genes of 18 Lamiles plants were compiled into a single file of 71,314 bp and aligned with MAFFT (Katoh et al., 2002). Seventeen Lamiales (including V. nakaiana, V. persica, and V. sibiricum) were selected as the in-groups and Buxus was included as the outgroup (Supplementary Table 1). Maximum likelihood (ML) analyses were performed using RAxML v7.4.2 with 1000 bootstrap replicates and the GTR+I+G model (Stamatakis, 2006).
Divergence Hotspot Identification
The three completed chloroplast genome sequences (V. nakaiana, V. persica, and V. sibiricum) were aligned using MAFFT (Katoh et al., 2002). To analyze nucleotide diversity (Pi), the total number of mutations (Eta) and average number of nucleotide differences (K) were determined using DnaSP (Librado and Rozas, 2009).
Result
Genome Organization of Three Veroniceae
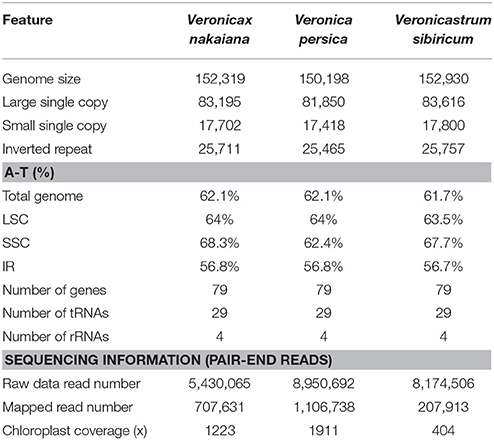
The total genome size of the Veroniceae (Figure 1) is 152,319 bp in V. nakaiana (Genebank accession no. KT633216), 150,198 bp in V. persica (Genebank accession no. KT724052), and 152,930 bp in V. sibiricum (GeneBank accession no. KT724053). Chloroplast genomes displayed a typical quadripartite structure, consisting of a pair of IRs (25,465–25,757 bp) separated by the LSC (81,850–83,616 bp) and SSC (17,418–17,800 bp) regions (Figure 1, Table 1). Among the tree species, the cp genomes (LSC, SSC, and IR regions) of V. sibiricum are larger than that of the other species, and V. persica is smallest (Table 1). Both contain 112 different genes, including 79 proteins (nine large ribosomal subunits, 12 small ribosomal subunits, four DNA-dependent RNA polymerases, one translation initiation factor, 45 genes encoding photosynthesis-related proteins and eight genes encoding other proteins), 29 tRNA genes and 4 rRNA genes were annotated (Figure 1, Table 1) in three Veroniceae chloroplast genomes.
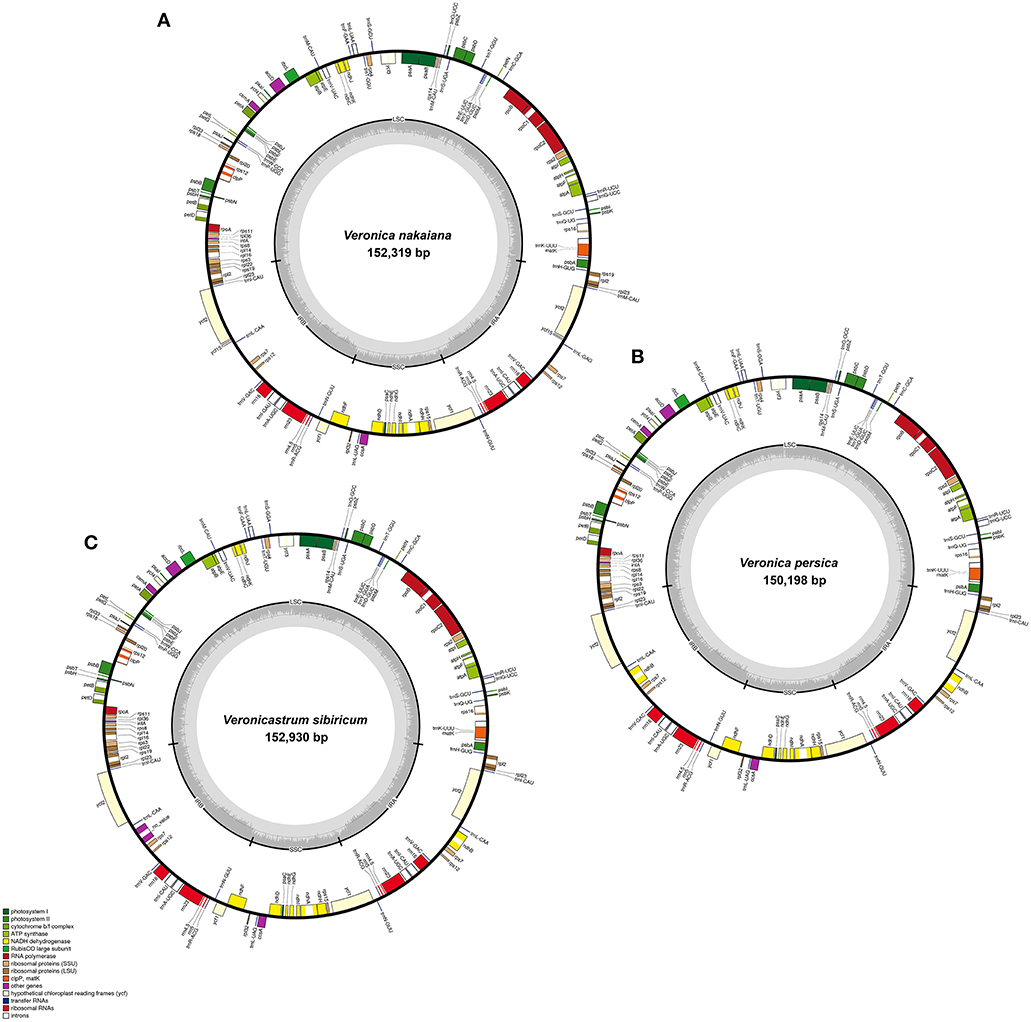
Figure 1. Chloroplast genome of Veronica nakaiana, Veronica persica, and Veronicastrum sibiricum. Genes inside the circle are transcribed clockwise, gene outside are transcribed counter-clockwise. The dark gray inner circle corresponds to the GC content, the light-gray to the AT content. (A) Veronica nakaiana chloroplast genome, (B) Veronica persica chloroplast genome, (C) Veronicastrum sibiricum chloroplast genome.
The overall AT content was 61.7–62.1%, indicating nearly identical levels among the three complete Veroniceae cp genomes. The AT content was 63.5–64, 62.4–68.3, and 56–56.8% of the genome for the LSC, SSC, and IR regions, respectively (Table 1).
IR Expansion and Contraction
The IR/LSC and IR/SSC borders of the Veroniceae cp genomes were compared (Figure 2). In the V. nakaiana chloroplast genome, the IR extended into the rps19 gene, resulting in a short rps19 pseudogene of 3 bp at the IR/LSC border. However, the rps19 gene of V. persica and V. sibiricum was separated from the LSC/IRb border by 3–47 bp and not duplicated in the IRa/LSC border. The position of ycf1 in the IR regions varied from 1255 to 1323 bp. The ndhF gene and ycf1 gene overlapped in the IRb/SSC border (72 bp in V. nakaiana, 72 bp in V. persica, and 108 bp in V. sibiricum). The trnH gene of the three Veroniceae species was located in the LSC region (separated from the IR/LSC border by 0–7 bp).
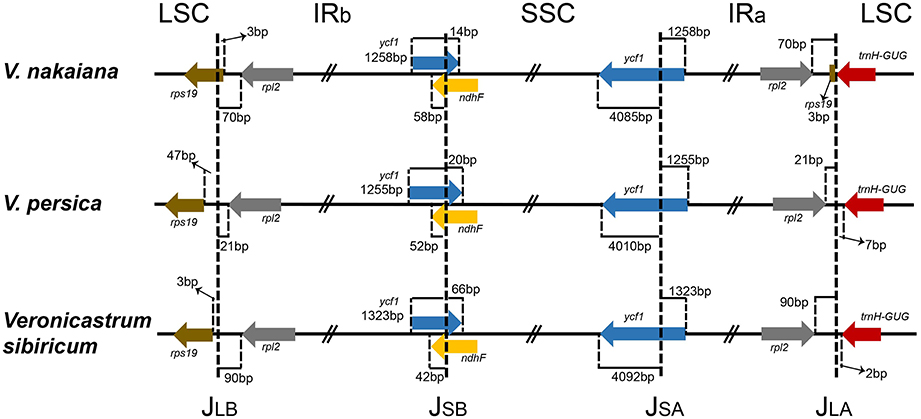
Figure 2. Comparison of the LSC, IR, and SSC junction positions among three Veroniceae chloroplast genomes.
SSR and Repeat Analysis
We used REPuter to analyze the repeat sequence of three Veroniceae cp genomes and found forward repeats, palindrome repeats and reverse repeats of at least 30 bp long per repeat unit with a sequence identity of ≥90% (Figure 3). V. nakaiana contained 15 forward repeats, 18 palindrome repeats, and 2 reverse repeats (Supplementary Table 2). Overall, 33 repeats were 30–40 bp long, with two repeats 41 long. V. persica contained 16 forward repeats and 19 palindrome repeats. Of these, 32 repeats were 30–40 bp long, two repeats were 41 bp long, and one repeat was 42 bp long. V. sibiricum contained 22 forward repeats and 21 palindrome repeats. Thirty-two repeats were 30–40 bp long, seven repeats were 40–50 bp long, and four repeats were 56 bp long (Supplementary Table 2). Most of these repeats had lengths between 30 and 41 bp. A total of 113 repeats were detected in these cp genomes, 46% (53 repeats) of which were direct repeats, 51% (58 repeats) were palindromic repeats, and 3% (2 repeats) reverse repeats. The five repeats shared among V. nakaiana, V. persica, and V. sibiricum were as follows: a 30-bp sequence in the rrn4.5-rrn5 spacer, psaB, psaA, ycf2, and a 41 bp repeat that occurred in the rps12-trnV-GAC intergenic spacer. Four repeats were only shared between V. nakaiana and V. sibiricum: a 30 bp direct repeat in the psbI-trnS-GCU intergenic spacer and trnS-GGA tRNA, and a 30 bp palindrome repeat in the accD-psaI intergenic spacer and rpl16 intron.
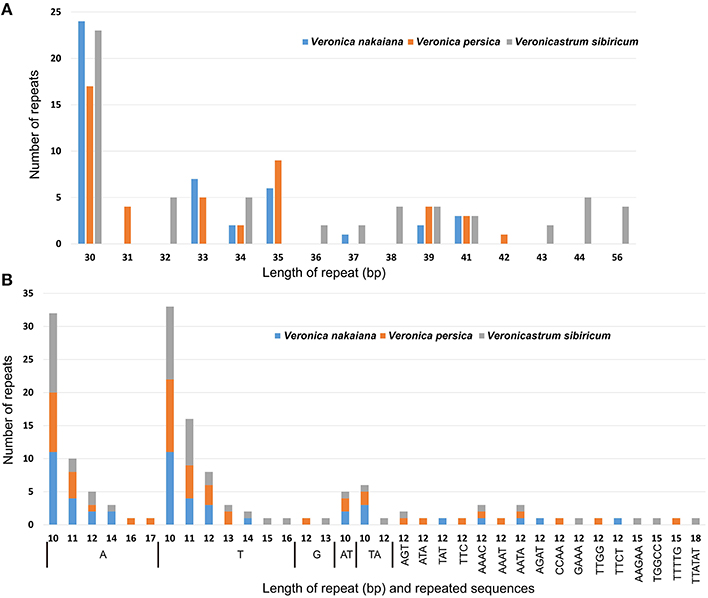
Figure 3. Analyses of repeated sequences in the three Veroniceae chloroplast genomes. (A) Frequency of repeat sequences determined by REPuter. (B) Frequency of simple sequence repeats (SSRs) by Phobos.
SSRs are repeated DNA sequences consisting of tandem repeats 1–10 bp in length per unit distributed throughout the genome (Figure 3B). SSRs are highly polymorphic and therefore useful as molecular markers and for population genetics (Powell et al., 1995; Zhang et al., 2012). We detected SSRs longer than 10 bp in three chloroplast genomes. Additionally, the total number of SSRs was 48 in V. nakaiana, 51 in V. persica, and 53 in V. sibiricum (Supplementary Table 3). The majority of SSRs in all species are A/T mononucleotides. Chloroplast genome SSRs were composed of adenine or thymine repeats, and rarely contained tandem guanine (G) or cytosine (C) repeats (Kuamg et al., 2011; Qian et al., 2013). Most SSRs were located in intergenic regions (69%), whereas 14 and 17% were located in introns and genes, respectively. Overall, 25 SSRs were located in the genes in three Veroniceae. Among these, three (rpoA, rpoC2, ndhD) were shared by all three Veroniceae species.
Phylogenetic Analyses of 78 Coding Genes in Chloroplast Genome
Phylogenetic analysis of coding genes was not resolved in previous studies of Lamiales plants. Previous phylogenetic studies of Veroniceae have focused on non-coding regions such as rapidly evolving chloroplast DNA (Rahmanzadeh et al., 2005; Schäferhoff et al., 2010). Our analyses of the extended data with much wider coding genes in Lamiales support a monophyletic family. The monophyly of the Lamiales families, its circumscription is clearly revealed. The position of Veroniceae within the Plantaginaceae corresponded with that reported by Schäferhoff et al. (2010).
In this study, maximum likelihood phylogenetic analysis of 78 protein coding genes was conducted and 18 taxa were aligned (Figure 4). This alignment was used for analyses of 71,314 bp. The Asterids clade was monophyletic and well supported by the bootstrap value (100%). Within the order, two well-supported clades, Lamiales and Solanales, were distinguished. Veroniceae was highly supported (100%) and a sister to Scrophulariaceae, Pedaliaceae, and Lamiaceae.
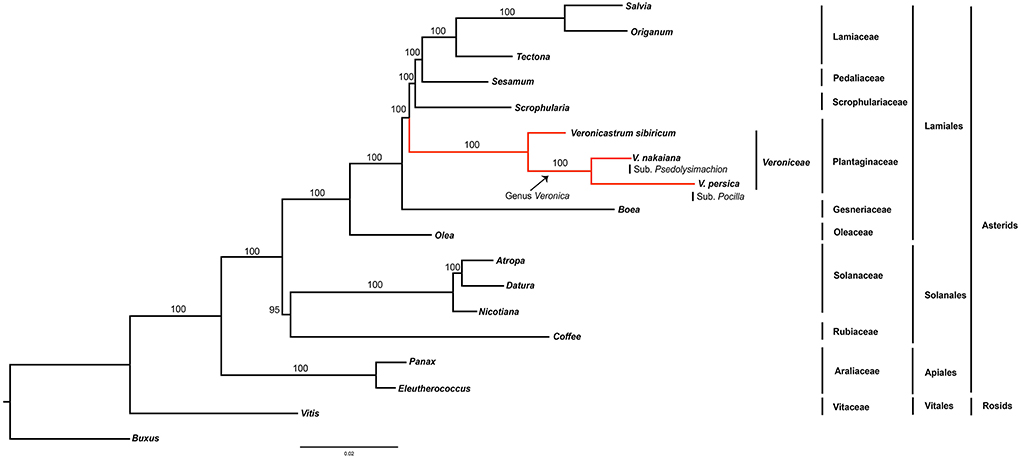
Figure 4. Phylogenetic tree reconstruction of 18 taxa using maximum likelihood based on concatenated sequences of 78 protein coding genes. Bootstrap support values >50% are given at the nodes.
Divergence Sequence Hotspots in Veroniceae
The coding genes, non-coding regions and intron regions were compared among the three Veroniceae species divergence hotspots. We generated more 200 bp of 114 regions (coding genes, non-coding regions and intron regions) from three Veroniceaea species and the nucleotide variability (Pi) values calculated with the DnaSP 5.0 software.
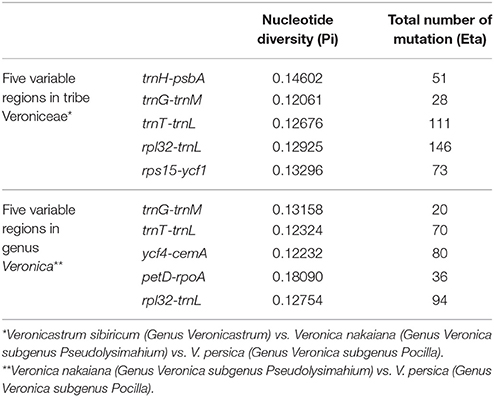
Among Veroniceae three species (V. nakaiana, V. persica, and V. sibiricum) values ranged from 0.00403 (rpl2 gene) to 0.14602 (trnH-psbA region). The IR region is much more conserved than the LSC and SSC regions. Five of these, trnH-psbA, trnG-trnM, trnT-trnL, rpl32-trnL, and rps15-ycf1, showed high levels of variation (Table 2). Three of these (trnH-psabA, trnG-trnM, trnT-trnL) were located in the LSC and two region (rpl32-trnL and rps15-ycf1) were in the SSC region (Figure 5A).
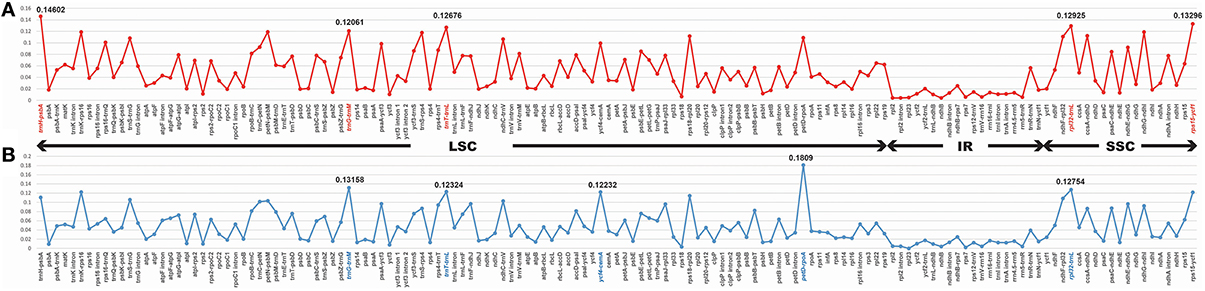
Figure 5. Comparison of the nucleotide variability (Pi) values in Veroniceae. (A) Veronica nakaiana vs. V. persica vs. Veronicastrum sibiricum, (B) Veronica nakaiana vs. V. persica.
The sequence divergence between subgenus Pseudolysimachium and subgenus Pocilla varied from 0 (rpl23 gene) to 0.1809 (petD-rpoA region), and most of the variable loci were in the LSC regions (four in the LSC region and one in the SSC region). Five regions (trnG-trnM, trnT-trnL, ycf4-cemA, petD-rpoA, and rpl32-trnL) were highly informative phylogenetic markers (Figure 5B, Table 2).
Discussion
The Chloroplast Genome of Veroniceae
Recently, some taxonomic studies have used the chloroplast genome to evaluate the relationship of related species. For example, the chloroplast genome of two species of Machilus (Song et al., 2015) and four species of Tilia (Cai et al., 2015) were utilized to evaluate the phylogenomics and relationship within the family. Comparative analyses of three members of the Veroniceae chloroplast genome also showed highly conserved structures and genes. The size of V. nakaiana, V. persica, and V. sibiricum ranged from 150,198 to 152,930 bp. Three Veroniceae contained the same coding genes, tRNAs and rRNAs. However, the start codon of the rps19 gene in V. sibiricum was changed to GTG. Similar findings have been detected in Cymbidium (Yang et al., 2013).
Land plants have a highly conserved chloroplast genome, but four junctions with changed genome sizes (Plunkett and Downie, 2000; Hansen et al., 2007; Qian et al., 2013). For example, Geraniaceae (Blazier et al., 2011) was found to lack a copy of the IR region, Tetracentron (Sun et al., 2013) showed expansion/contraction of the IR region, subfamily Apioideae (Plunkett and Downie, 2000) showed variations in JLB (LSC/IRb region) and Elaeagnaceae contained trnH gene duplication in the IR region (Choi et al., 2015).
We compared three Veroniceae species, among which V. nakaiana was unique in that its rps19 gene was duplicated and JLB (LSC/IRb) and JLA (LSC/IRa) differences occurred (Figure 2). The rpl2 genes of V. nakaiana and V. sibiricum were separated by >50 bp at the JLB, whereas that in V. persica was separated by 21 bp. Three types of JLB based on expansion/contraction have been characterized in Veroniceae.
Divergence Region of Veroniceae Plastid Genome
The ITS region and cpDNA non-coding regions have been widely used to investigate taxonomy and molecular phylogeny at the interspecific level (Taberlet et al., 1991; Baldwin, 1992). Recently, studies of the chloroplast genome showed divergent sequences in plants (Parks et al., 2009; Yang et al., 2013). In Veroniceae, Albach (Albach and Chase, 2001, 2004; Albach et al., 2004a,b,c; Albach and Meudt, 2010) tested the ITS and CYC2 of nuclear ribosomal DNA and some chloroplast DNA (trnL-F, trnL, rps16, rpoB-trnC, and trnH-psbA). However, the relationships among the 12 genera in Veronica are not well supported.
Based on our study, the largest sequence divergence regions were trnH-psbA, trnG-trnM, trnT-trnL, rpl23-trnL, and rps15-ycf1 in Veroniceae plants (V. nakaiana, V. persica, and V. sibiricum). In Veronica, trnG-trnM, trnT-trnL, ycf4-cemA, petD-rpoA, and rpl2-trnL was a highly variable locus (Figure 5B). The total sequence divergence hotspot regions in Veroniceae and Veronica were trnH-psbA, trnG-trnM, trnT-trnL, rpl32-trnL, rps15-ycf1, ycf4-cemA, petD-rpoA, and rpl32-trnL (Table 2).
Here, we showed the eight DNA variable regions that could be used as molecular markers in Veroniceae and Veronica (Figure 5, Table 2 and Supplementary Table 4). The results presented here will be helpful to systematics of Veroniceae and evaluating the relationships among the 12 subgenera of Veronica.
Author Contributions
Conceived and designed the experiments: SP, KC, MC. Performed the experiments: KC, SP. Analyzed the data: KC, MC. Wrote the paper: KC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work supported by grants from Scientific Research (KNA1-2-13, 14-2) of the Korea National Arboretum.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00355
References
Albach, D. C., and Chase, M. W. (2004). Incongruence in Veroniceae (Plantaginaceae): evidence from two plastid and nuclear ribosomal DNA region. Mol. Phylogenet. Evol. 32, 183–197. doi: 10.1016/j.ympev.2003.12.001
Albach, D. C., and Chase, M. W. (2001). Paraphyly of Veronica (Veroniceae: Scrophulariaceae): evidence from the internal transcribed spacer (ITS) sequences of nuclear ribosomal DNA. J. Plant Res. 114, 9–18. doi: 10.1007/PL00013971
Albach, D. C., Martinez-Ortega, M. M., and Chase, M. A. (2004b). Veronica: parallel morphological evolution and phylogeography in the Mediterranean. Plant Syst. Evol. 246, 177–194. doi: 10.1007/s00606-004-0148-9
Albach, D. C., Martinez-Ortega, M. M., Fischer, M. A., and Chase, M. W. (2004a). Evolution of Veroniceae: a phylogenetic perspective. Ann. Mo. Bot. Gard. 91, 275–302.
Albach, D. C., Martinez-Ortega, M. M., Fischer, M. A., and Chase, M. W. (2004c). A new classification of the tribe Veroniceae-problems and a possible solution. Taxon 53, 429–452. doi: 10.2307/4135620
Albach, D. C., and Meudt, H. M. (2010). Phylogeny of Veronica in the Southern and Northern hemispheres based on plastid, nuclear ribosomal and nuclear low-copy DNA. Mol. Phygenet. Evol. 54, 457–471. doi: 10.1016/j.ympev.2009.09.030
Albach, D. C., Meudt, H. M., and Oxelman, B. (2005). Piecing together the “new” Plantaginaceae. Am. J. Bot. 92, 297–315. doi: 10.3732/ajb.92.2.297
Alvarez, I., and Wendel, J. F. (2003). Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29, 417–434. doi: 10.1016/S1055-7903(03)00208-2
Angiosperm Phylogeny Group (2009). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161, 105–121. doi: 10.1111/j.1095-8339.2009.00996.x
Baldwin, B. G. (1992). Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Mol. Phylogenet. Evol. 1, 3–16. doi: 10.1016/1055-7903(92)90030-K
Blazier, J. C., Guisinger, M. M., and Jansen, R. K. (2011). Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol. Biol. 76, 263–272. doi: 10.1007/s11103-011-9753-5
Bock, R., and Khan, M. S. (2004). Taming plastids for a green future. Trends Biotechnol. 22, 311–318. doi: 10.1016/j.tibtech.2004.03.005
Cai, J., Ma, P.-F., Li, H.-T., and Li, D.-Z. (2015). Complete plastid genome sequencing of four Tilia species (Malvaceae): a comparative analysis and phylogenetic implications. 10:e0142705. doi: 10.1371/journal.pone.0142705
Choi, K. S., Son, O., and Park, S. (2015). The chloroplast genome of Elaeagnus macrophylla and trnH duplication event in Elaeagnaceae. PLoS ONE 10:e0138727. doi: 10.1371/journal.pone.0138727
Douglas, S. E. (1990). Plastid evolution: origins, diversity, trends. Curr. Opin. Genet. Dev. 8, 655–661. doi: 10.1016/S0959-437X(98)80033-6
Funk, H. T., Berg, S., Krupinska, K., Maier, U. G., and Krause, K. (2007). Complete DNA sequence of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 7:45. doi: 10.1186/1471-2229-7-45
Hansen, D. R., Dastidar, S. G., Cai, Z., Penaflor, C., Kuehl, J. V., Boore, J. L., et al. (2007). Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxs (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol. Phylogen. Evol. 45, 547–563. doi: 10.1016/j.ympev.2007.06.004
Jansen, R. K., Cai, Z., Raubeson, L. A., Danuell, H., de Pamphiliis, C. W., Leebens-Mack, J., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A. 104, 19369–19374. doi: 10.1073/pnas.0709121104
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A., and Janzen, D. H. (2005). Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. U.S.A. 102, 8369–8374. doi: 10.1073/pnas.0503123102
Kuamg, D. Y., Wu, H., Wang, Y. L., Gao, L. M., Zhang, S. Z., and Lu, L. (2011). Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome 54, 663–773. doi: 10.1139/g11-026
Kurtz, S., and Schleiermacher, C. (1999). REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics 15, 426–427. doi: 10.1093/bioinformatics/15.5.426
Librado, P., and Rozas, J. (2009). DnaSP v5: software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lohse, M., Dechsel, O., and Bock, R. (2007). OrganellarGenomeDRAW (ORDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274. doi: 10.1007/s00294-007-0161-y
Olmstead, R. G., and Reeves, P. A. (1995). Evidence for the polyphyly of the Scrophulariaceae based on chloroplast rbcL and ndhF sequences. Ann. Mo. Bot. Gard. 82, 176–193. doi: 10.2307/2399876
Olmstead, R. G., de Pamphilis, C. W., Wolfe, A. D., Young, N. D., Elisons, W. J., and Reeves, P. A. (2001). Disintegration of the Scrophulariacae. Am. J. Bot. 88, 348–361. doi: 10.2307/2657024
Parks, M., Cronn, R., and Liston, A. (2009). Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7:84. doi: 10.1186/1741-7007-7-84
Plunkett, M. G., and Downie, S. R. (2000). Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Syst. Bot. 25, 648–667. doi: 10.2307/2666726
Powell, W., Morgante, M., McDevitt, R., Vendramin, G. G., and Rafalski, J. A. (1995). Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc. Natl. Acad. Sci. U.S.A. 92, 7759–7763. doi: 10.1073/pnas.92.17.7759
Qian, J., Song, J., Gao, H., Zhu, Y., Xu, J., Pang, X., et al. (2013). The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE 8:e57607. doi: 10.1371/journal.pone.0057607
Rahmanzadeh, R., Müller, K., Fischer, E., Bartels, D., and Borsch, T. (2005). The Linderniaceae and Gratiolaceae are further lineages distinct from the Scrophulariaceae (Lamiales). Plant Biol. 7, 67–78. doi: 10.1055/s-2004-830444
Schäferhoff, B., Fleischmann, A., Fischer, E., Albach, D. C., Borsh, T., Heubl, G., et al. (2010). Towards resolving Lamiales relationships: insights from rapidly evolving chloroplast sequences. BMC Evol. Biol. 10:352. doi: 10.1186/1471-2148-10-352
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. doi: 10.1093/nar/gki366
Song, Y., Dong, W., Liu, B., Xu, C., Yao, X., Gao, J., et al. (2015). Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus ynnanensis and Machilus balansae in the family Lauraceae. Front. Plant Sci. 6:662. doi: 10.3389/fpls.2015.00662
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Sun, Y.-X., Moore, M. J., Meng, A.-P., Soltis, P. S., Soltis, D. E., Li, J.-Q., et al. (2013). Complete plastid genome sequencing of Trochodendraceae reveals a significant expansion of the inverted repeat and suggests a paleogene divergence between the two extant species. PLoS ONE 8:e60429. doi: 10.1371/journal.pone.0060429
Taberlet, P., Coissac, E., Pompanon, F., Gielly, L., Miquel, C., Valentini, A., et al. (2006). Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14. doi: 10.1093/nar/gkl938
Taberlet, P., Gielly, L., Pautou, G., and Bouver, J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105–1109. doi: 10.1007/BF00037152
Wagstaff, S. J., Bayly, M. J., Garnock-Jones, P. J., and Albach, D. C. (2002). Classification origin and diversification of the New Zealand Hebes (Scrophulariaceae). Ann. Mo. Bot. 36, 425–437. doi: 10.2307/3298656
Wicke, S., Müller, K. F., de Pamphilis, C. W., Quandt, D., Wickett, N. J., Zhang, Y., et al. (2013). Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25, 3711–3725. doi: 10.1105/tpc.113.113373
Wicke, S., Schneeweiss, G. M., de Pamphilis, C. W., Müller, K. F., and Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 51, 273–297. doi: 10.1007/s11103-011-9762-4
Wyman, S. K., Jansen, R. K., and Boore, J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3244. doi: 10.1093/bioinformatics/bth352
Yang, J.-B., Tang, M., Li, H.-T., and Zhang, D.-Z. (2013). Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 13:84. doi: 10.1186/1471-2148-13-84
Keywords: veroniceae, veronica, chloroplast genome, phylogenetic tree, divergent regions
Citation: Choi KS, Chung MG and Park SJ (2016) The Complete Chloroplast Genome Sequences of Three Veroniceae Species (Plantaginaceae): Comparative Analysis and Highly Divergent Regions. Front. Plant Sci. 7:355. doi: 10.3389/fpls.2016.00355
Received: 22 December 2015; Accepted: 07 March 2016;
Published: 23 March 2016.
Edited by:
Manuel Acosta, Universidad de Murcia, SpainCopyright © 2016 Choi, Chung and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: SeonJoo Park, sjpark01@ynu.ac.kr
 Kyoung Su Choi
Kyoung Su Choi Myong Gi Chung2
Myong Gi Chung2 SeonJoo Park
SeonJoo Park