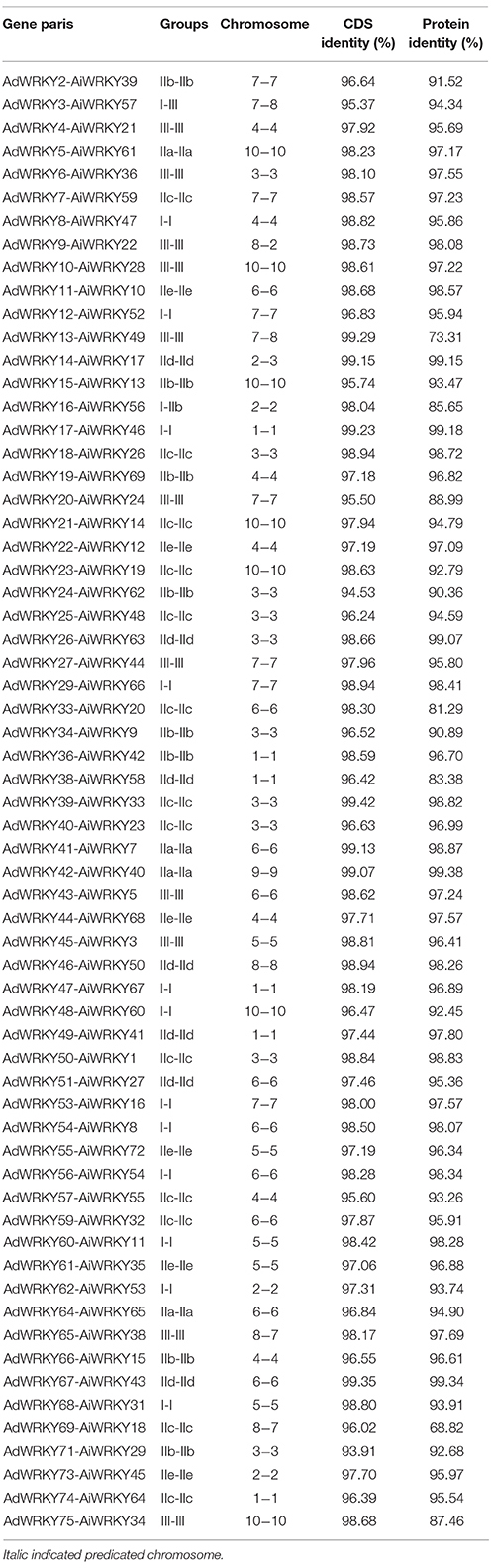
- 1Shandong Provincial Key Laboratory of Crop Genetic Improvement, Ecology and Physiology, Biotechnology Research Center, Shandong Academy of Agricultural Sciences, Jinan, China
- 2Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, Los Angeles, CA, USA
WRKY, an important transcription factor family, is widely distributed in the plant kingdom. Many reports focused on analysis of phylogenetic relationship and biological function of WRKY protein at the whole genome level in different plant species. However, little is known about WRKY proteins in the genome of Arachis species and their response to salicylic acid (SA) and jasmonic acid (JA) treatment. In this study, we identified 77 and 75 WRKY proteins from the two wild ancestral diploid genomes of cultivated tetraploid peanut, Arachis duranensis and Arachis ipaënsis, using bioinformatics approaches. Most peanut WRKY coding genes were located on A. duranensis chromosome A6 and A. ipaënsis chromosome B3, while the least number of WRKY genes was found in chromosome 9. The WRKY orthologous gene pairs in A. duranensis and A. ipaënsis chromosomes were highly syntenic. Our analysis indicated that segmental duplication events played a major role in AdWRKY and AiWRKY genes, and strong purifying selection was observed in gene duplication pairs. Furthermore, we translate the knowledge gained from the genome-wide analysis result of wild ancestral peanut to cultivated peanut to reveal that gene activities of specific cultivated peanut WRKY gene were changed due to SA and JA treatment. Peanut WRKY7, 8 and 13 genes were down-regulated, whereas WRKY1 and 12 genes were up-regulated with SA and JA treatment. These results could provide valuable information for peanut improvement.
Introduction
WRKY transcription factors, a large family of regulatory proteins, are widely distributed in plant and non-plant species (Eulgem et al., 2000; Riechmann et al., 2000; Zhang and Wang, 2005; Rushton et al., 2010; Rinerson et al., 2015). WRKY proteins are characterized by the WRKY domain, which includes about 60 amino acids with a conserved WRKYGQK heptapeptide (Eulgem et al., 2000; Rushton et al., 2010). WRKY proteins contain one or two WRKY domains and either one of two types of zinc finger motif at C-terminal (Eulgem et al., 2000; Rushton et al., 2010). WRKY proteins could be classified into three groups (I, II, and III) based on the number of WRKY domains and the type of the zinc finger motifs. Group I WRKY proteins include two WRKY domains and a zinc-finger motif (Eulgem et al., 2000; Rushton et al., 2010). Each group II and III WRKY proteins contain a single WRKY domain, and a CX4−5CX22−23HXH and CX7CX23HXC zinc-finger motifs in C-terminal region, respectively (Eulgem et al., 2000; Rushton et al., 2010). Group II is further subdivided into five subgroups (IIa-IIe) based on phylogenetic relationship (Eulgem et al., 2000).
By binding to W-box cis-element (C/TTGACT/C) in target gene promoter, WRKY proteins are involved in a variety of biological functions, including different plant developmental programs, as well as diverse abiotic and biotic stress response (Eulgem et al., 2000; Rushton et al., 2010). WRKY proteins were implicated to modulate plant development, such as, trichome morphogenesis (Johnson et al., 2002), flowering (Luo et al., 2013), seed development (Luo et al., 2005), dormancy and germination (Zhang et al., 2004; Zentella et al., 2007; Zou et al., 2008), and senescence (Robatzek and Somssich, 2002). Recent studies revealed that WRKY proteins were involved in response to abiotic stresses (Rushton et al., 2012), such as salt (Wu et al., 2009), drought (Ren et al., 2010; Jiang et al., 2012), cold (Zou et al., 2010), and wounding (Cheong et al., 2002). For instance, expression of AtWRKY46 gene could significantly induced by drought, H2O2 and salt stress, and wrky46 mutant in Arabidopsis was more sensitive to salt and osmotic stress compared to control (Ding et al., 2014). Expression of TaWRKY44 gene in tobacco could improve drought, salt and osmotic stress tolerance (Wang et al., 2015). Previous studies indicated that WRKY proteins play crucial roles in pathogen defense (Eulgem and Somssich, 2007; Rushton et al., 2010) and insect (Grunewald et al., 2008; Skibbe et al., 2008). Xu et al. (2006) found that expression of AtWRKY18 increased resistance to Pseudomonas syringae, but its co-expression with AtWRKY40 or AtWRKY60 made plants more susceptible to both P. syringae and Botrytis cinerea. Furthermore, increasing studies documented that WRKY proteins are involved in defense-relative hormone signal transduction, salicilic acid (SA) and jasmonic acid (JA)-mediated defenses, where SA triggered defenses against biotrophic pathogens and JA participates in the response to necrotrophic pathogens (Li et al., 2004; Schluttenhofer et al., 2014). For example, in Arabidopsis, AtWRKY50 and AtWRKY51 promoted SA biosynthesis (Gao et al., 2011); AtWRKY17 and AtWRKY33 genes were induced after JA treatment (Journot-Catalino et al., 2006). Overexpression of AtWRKY28 and AtWRKY46 genes could promote the expression of ICS1 and PBS3 genes through SA signaling pathway (van Verk et al., 2011). Additionally, recent study showed that the expression of 12 WRKY genes from Catharanthus roseus responded to JA (Schluttenhofer et al., 2014), and that the expression of 49 Salvia miltiorrhiza WRKY genes was signification up- or down-regulated by JA treatment (Li et al., 2015).
WRKY proteins were studied extensively in a variety of plant species (Eulgem et al., 2000; Wu et al., 2005; Wei et al., 2012a; Liu et al., 2014; Song and Nan, 2014; Song et al., 2014, 2016). However, current basic knowledge of WRKY proteins and the characterization of specific WRKY proteins involved in disease resistance from species in genus Arachis are still limited. Peanut (Arachis hypogaea L.) is an important oil crop grown throughout the tropics and subtropics regions. Especially in Asia, which accounts for 64% of the world yield, peanut provides a similar amount of calories from soybean (Bertioli et al., 2011). To date, 80 species in genus Arachis were identified and classified into nine taxonomic sections (Bertioli et al., 2011). Wild species are diploid, but cultivated peanut is allotetraploid (AABB). The wild ancestral species of cultivated peanut are generally considered to be Arachis duranensis and Arachis ipaënsis, which contributed the A and B sub-genomes, based on morphology, cytology, fertility of the interspecific hybrid and molecular studies (Kochert et al., 1996; Seijo et al., 2004, 2007; Ramos et al., 2006). Plant diseases have been a major reason for peanut yield losses. Interestingly, the disease resistant capacity of wild peanut was proved much higher than that of cultivated peanut (Herbert and Stalker, 1981; Pande and Narayana Rao, 2001; Simpson, 2001). Therefore, disease resistant genes from wild type species could be valuable resources for cultivated peanut improvement. Recently, the whole genome sequences of A. duranensis and A. ipaënsis have been released (Bertioli et al., 2016), which provided an important resource for genome wide analysis of the disease resistant genes. To fill the knowledge gap, we identified 75 AdWRKY and 77 AiWRKY proteins from A. duranensis (Aradu.V14167.a1.M1) and A. ipaënsis (Araip.K30076.a1.M1), respectively, by bioinformatics approaches, and then we studied the phylogenetic relationship, the genome-wide distribution pattern, gene duplication event, and selection pressure of WRKY genes in the two species. Additionally, we deduced potential disease-resistant Arachis WRKY proteins based on the functional study of Arabidopsis WRKY (AtWRKY) proteins and then we transferred the knowledge to cultivated peanut to determine the expression pattern of WRKY genes by quantitative real-time RT-PCR (qRT-PCR) in different tissues, and we monitored the WRKY transcriptional changes with SA and JA treatment to preliminarily validate the disease-resistant potential. Our results provide a comprehensive genome-wide knowledge of WRKY proteins in the two wild ancestral species of peanut and a preliminary knowledge of specific WRKY proteins potentially involved in disease resistance in peanut.
Materials and Methods
WRKY Proteins in A. duranensis and A. ipaënsis Genomes
The A. duranensis (Aradu.V14167.a1.M1) and A. ipaënsis (Araip.K30076.a1.M1) genome sequences were downloaded from http://peanutbase.org/ (Bertioli et al., 2016). The Hidden Markov Model (HMM) profile of the WRKY domain (PF03106) was downloaded from the pfam database (http://pfam.janelia.org). It was used to match each WRKY protein in genomes using HMMER program (Finn et al., 2011). To verify the reliability of results, all protein sequences were checked in the pfam database. The AtWRKY protein sequences were downloaded from Arabidopsis Information Resource website (TAIR, http://www.arabidopsis.org). WRKY sequences of Glycine max (GmWRKY), Lotus japonicus (LjWRKY) and Medicago truncatula (MtWRKY) were obtained from previous studies (Yin et al., 2013; Bencke-Malato et al., 2014; Song et al., 2014; Song and Nan, 2014).
Classification of A. duranensis and A. ipaënsis WRKY Proteins
The AtWRKY, AdWRKY, and AiWRKY domains were extracted based on pfam database. Multiple sequences alignment was executed by MAFFT 7.0 program (Katoh and Standley, 2013). AtWRKY domains were used as query to categorize the AdWRKY and AiWRKY proteins based on the phylogenetic tree described by previous study (Song et al., 2014). The phylogenetic trees were constructed using MEGA 6.0 (Tamura et al., 2013) using neighbor-joining model with 1000 replicates. Other phylogenetic trees were inferred according to the parameters.
The Genome-Wide Distribution Pattern, Gene Duplication Event and Selection Pressure of WRKY Genes
The chromosomal location information of AdWRKY and AiWRKY proteins was obtained from peanutbase website (http://peanutbase.org/). The map was generated using MapInspect software (http://mapinspect.software.informer.com/).
Different types of gene duplication existed in genome, while we focused only on tandem and segmental duplication events in this study. To identify gene duplication events in AdWRKY and AiWRKY genes, duplicated GmWRKY genes were used as query to construct phylogenetic trees of AdWRKY and AiWRKY genes, respectively. Colinearity between Arachis and Glycine is more conserved (Nagy et al., 2012). If GmWRKY and AdWRKY or AiWRKY genes were clustered in pairs in phylogenetic tree, the gene pairs were considered as orthologous genes (Dutilh et al., 2007; Altenhoff and Dessimoz, 2012). We classified gene duplication events of AdWRKY and AiWRKY based on the information from the duplicated GmWRKY (Yin et al., 2013). Moreover, we identified orthologous genes between AdWRKY and AiWRKY based on phylogenetic relationship using above method (Dutilh et al., 2007; Altenhoff and Dessimoz, 2012).
Non-synonymous (Ka) and synonymous (Ks) substitution of each duplicated AdWRKY and AiWRKY genes were calculated by PAL2NAL program (Suyama et al., 2006), which is based on codon model program in PAML (Yang, 2007). Generally, Ka/Ks (ω) = 1, >1, and < 1 indicated neutral, positive, and purifying selection, respectively. To detect whether the AdWRKY and AiWRKY genes underwent positive selection under site model and branch-site model, PAML program was applied in this study. In site model, M0 (one ratio), M1a (neutral), M2a (selection), M3 (discrete), M7 (beta), and M8 (beta + ω) were applied to selection pressure analysis. We detected absolute value in the ω ratio parameter among sites using likelihood ratio test (LRT) for M1a vs. M2a, M0 vs. M3 and M7 vs. M8. In Branch-site model, the ω ratio between clades was used for comparison. The phylogenetic trees were constructed using sequences from the amino acid by the MEGA 6.0 (Tamura et al., 2013). Posterior probabilities were estimated using the Bayes Empirical Bayes (BEB) method (Yang, 2007).
Plant Materials and Hormone Treatment
To analyze expression pattern of deduced potential SA- and JA- related WRKY genes in different tissues, root, stem, leaf, flower, and seed of cultivated peanut (Luhua 14) were harvested from experimental farm in September, 2015. To examine the expression of 13 deduced potential SA- and JA- related WRKY genes under SA and MeJA treatment, peanut seeds were germinated on humid filter paper in growth chamber at 28°C, and then growth for 4 weeks at room temperature (~32°C). SA and MeJA solution (0.1 mM) was applied to the leaves, respectively. Fresh leaves were harvested after 0, 6, 24, 36, and 48 h of treatments.
Gene Expression Analysis by qRT-PCR
Total RNA was extracted using CTAB method (Chang and Puryear, 1993). The first-strand cDNAs were obtained using 2 μg of DNA-free RNA using Reverse Transcriptase M-MLV System (Takara, Dalian, China).
Actin gene was used as a reference gene to quantitative the expression of AhWRKY genes (Xia et al., 2013). The reaction was carried out using fluorescent dye SYBR-Green (Takara, Dalian, China). qRT-PCR was carried out using Fast Start Universal SYBR Green Master (ROX) with a 7500 real-time PCR machine (ABI). The reaction was carried out as follows: 30 s at 95°C for denaturation, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. A melting curve analysis was performed at the end of the PCR run over a range of 55–99°C. Three biological replicates were used. The ΔΔCt method was used for quantification (Livak and Schmittgen, 2001). A pairwise student's t-test was performed to obtain the P values using JMP 9.0. If P < 0.05, we considered the WRKY genes as differential expressed genes.
Results and Discussion
WRKY Proteins in Two Wild Type Peanuts
A total of 75 and 77 WRKY proteins were identified from A. duranensis and A. ipaënsis, respectively, using bioinformatics approach (Tables S1, S2). They were named as AdWRKY1 to AdWRKY75, and AiWRKY1 to AiWRKY 77, respectively. Among 75 AdWRKY sequences, six were partially sequenced without complete sequence in the released genome draft, namely AdWRKY3, 11, 15, 24, 55, and 73. The length of other 69 full-length sequences ranged from 291 to 3477 bp (Table S1). AdWRKY8 and 35 sequences contained internal termination codon, indicating these two sequences were pseudogene or sequencing errors. In 77 AiWRKY sequences, nine were partially sequenced without complete sequence in the genome draft, including AiWRKY10, 24, 29, 30, 36, 40, 69, 70, and 71. The length of remaining 68 AiWRKY sequences ranged from 291 to 3564 bp (Table S2).
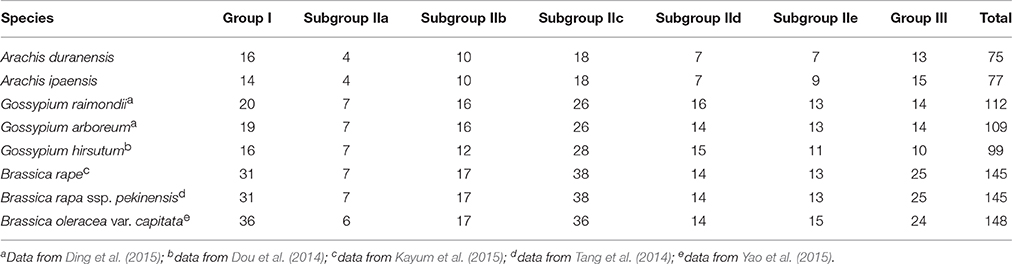
AdWRKY proteins could be classified into three groups, group I (16 sequences), group II (46 sequences) and group III (13 sequences). Similarly, AiWRKY proteins could be classified into group I (14 sequences), group II (48 sequences), and group III (15 sequences) based on the number of WRKY domain and the type of zinc finger motif (Table 1). Forty-six group II AdWRKY proteins could further be classified into five subgroups, IIa (4 sequences), IIb (10 sequences), IIc (18 sequences), IId (7 sequences), and IIe (7 sequences); The 48 group II AiWRKY could be classified into subgroup IIa (4 sequences), IIb (10 sequences), IIc (18 sequences), IId (7 sequences), and IIe (9 sequences) based on the phylogenetic trees (Table 1, Figures S1, S2). The number and type of AdWRKY and AiWRKY proteins in the corresponding subgroup was similar (Table 1). These results were consistent with the results described by Ding et al. (2015) that no significant gene domain or number difference was detected between two Gossypium species. Similar results were found in Brassica species (Table 1).
Previous studies found that WRKYGQK heptapeptide prone to mutate (Dou et al., 2014; Song and Nan, 2014; Song et al., 2014; Ding et al., 2015). WRKYGQK sequence is considered to be important for recognizing and binding to W-box elements. WRKYGKK sequences represented the major variant in AdWRKY and AiWRKY proteins. We found that WRKYGKK was also the most common variant in LjWRKY (Song et al., 2014) and GmWRKY (Song et al., 2016). The WRKYGKK sequence in tobacco WRKY12 bound specifically to WK-box (TTTTCCAC), which was significantly different from the consensus sequence of a W-box (C/TTGACT/C) (van Verk et al., 2008). WRKYGKK sequence was observed in AdWRKY21, 25, 26, 30, 59, 63, 69, and 72 proteins (Table S1). Among them, AdWRKY21, 25, 30, 59, 69, and 72 proteins belonged to subgroup IIc; AdWRKY26 belong to subgroup IId, and WRKYGKK sequence was located in the N-terminal WRKY domain sequence of AdWRKY63 protein, which belonged to group I (Table S1). Three WRKYGEK sequences were found in AdWRKY18 (subgroup IIc), AdWRKY20 (group III), and AdWRKY62 (group I). GRKYGEK, WRKYDEK, WRKYEEN, and WRKYGRK heptapeptides were detected in AdWRKY51 (subgroup IId), AdWRKY37 (group III), AdWRKY7 (subgroup IIc) and C-terminal WRKY domain sequence of AdWRKY63 (group I) (Table S1). In AiWRKY proteins, five WRKYGKK peptides were found in AiWRKY14, 18, 30, 32, and 48, which belonged to subgroup IIc (Table S2). Two WRKYGEK sequences were found in AiWRKY24 (group III) and AiWRKY26 (subgroup IIc) proteins, and two WRKYEEN sequences were identified in AiWRKY59 (subgroup IIc) and AiWRKY77 (subgroup IIc) proteins. Moreover, RKKYGQK, WCKYGEK, and WRKHGQK sequences were found in AiWRKY76 (group III), AiWRKY71 (subgroup IIc) and AiWRKY25 (group III) (Table S2). Group I AiWRKY contained one WRKYDKK and one WHKYGKK in WRKY N-terminal domain. These results showed that WRKYGQK sequence in subgroup IIc WRKY proteins prone to mutate. In soybean WRKY proteins we also found that some WRKY domains in subgroup IIc were likely to mutate (Song et al., 2016). These results suggested that subgroup IIc proteins might potentially carry out a variety of biological functions. It is noteworthy that mutation occurred in WRKYGQK sequence of AdWRKY63 and AiWRKY51, suggesting their possible involvement in multiple biological functions when bind to different W-box elements.
WRKY proteins contained two types of zinc-finger motif, CX4−5CX22−23HXH and CX7CX23HXC (Eulgem et al., 2000; Rushton et al., 2010). We found these two zinc-finger motifs were presented in peanut WRKY proteins. Furthermore, most of zinc-finger motifs in N-terminal of group I are CX4CX22HX1H but not CX4CX23HX1H. In other words, N-terminal zinc-finger motif contained one more amino acid residue between the second C and the first H in the zinc-finger structure than that in C-terminus (Tables S1, S2). On the other hand, CX5CX23HX1H always located in fabaceous group I WRKY proteins (Song and Nan, 2014; Song et al., 2014). However, we found AdWRKY3 (group I) contained CX7CX23HX1C zinc-finger motif. This type of zinc-finger structure was also found in Oryza sativa group I WRKY (Ross et al., 2007), indicating CX7CX23HX1C zinc-finger structure has appeared before the divergent of gramineous and leguminous species.
AdWRKY10, AdWRKY37, and AiWRKY28, belonged to group III, contained the nucleotide-binding site-leucine-rich repeat (NBS-LRR) domain. Previous studies showed that plants contained a larger number of NBS-LRR proteins to confer resistance to diverse pathogens (Jones and Dangl, 2006; McHale et al., 2006). It is known that some WRKY proteins contain NBS-LRR domain (Deslandes et al., 2003; Shen et al., 2007; Chang et al., 2009), and group III WRKY proteins are mainly involved in plant disease resistance (Kalde et al., 2003; Eulgem and Somssich, 2007). Our results suggested AdWRKY10, AdWRKY37, and AiWRKY28 were possibly involved in disease resistance.
Origin of WRKY Protein in Leguminous Plants Remain to be Solved
The phylogenetic relationship from WRKY domains of AtWRKY, MtWRKY, LjWRKY, GmWRKY, AdWRKY, and AiWRKY is consistent with the results of previous study (Zhang and Wang, 2005). The findings revealed that based on the phylogenetic trees WRKY sequences could be classified into eight subgroups: I-N, I-C, IIa, IIb, IIc, IId, IIe, and III (Zhang and Wang, 2005). However, phylogenetic relationship of MtWRKY and LjWRKY showed that some group II or III members were nested in subgroup I-N or subgroup I-C (Song and Nan, 2014; Song et al., 2014). As shown in Figure 1, subgroup I-C contained other group members, including AiWRKY56, MtWRKY4, GmWRKY130, GmWRKY131, GmWRKY183, and AtWRKY10. It indicated that the mixture was not only found in leguminous plants, but also in other dicotyledonous plants. Subgroup I-N contained MtWRKY2 from IIc, and AiWRKY6c from I-C. Group I proteins were found in other groups. For example, two WRKY domains of each AdWRKY63 and AiWRKY51 were found in subgroup IIc. Two WRKY domains of AdWRKY3 were located in subgroup III in the phylogenetic tree. These findings indicated that the origin of leguminous WRKY proteins is still to be clarified.
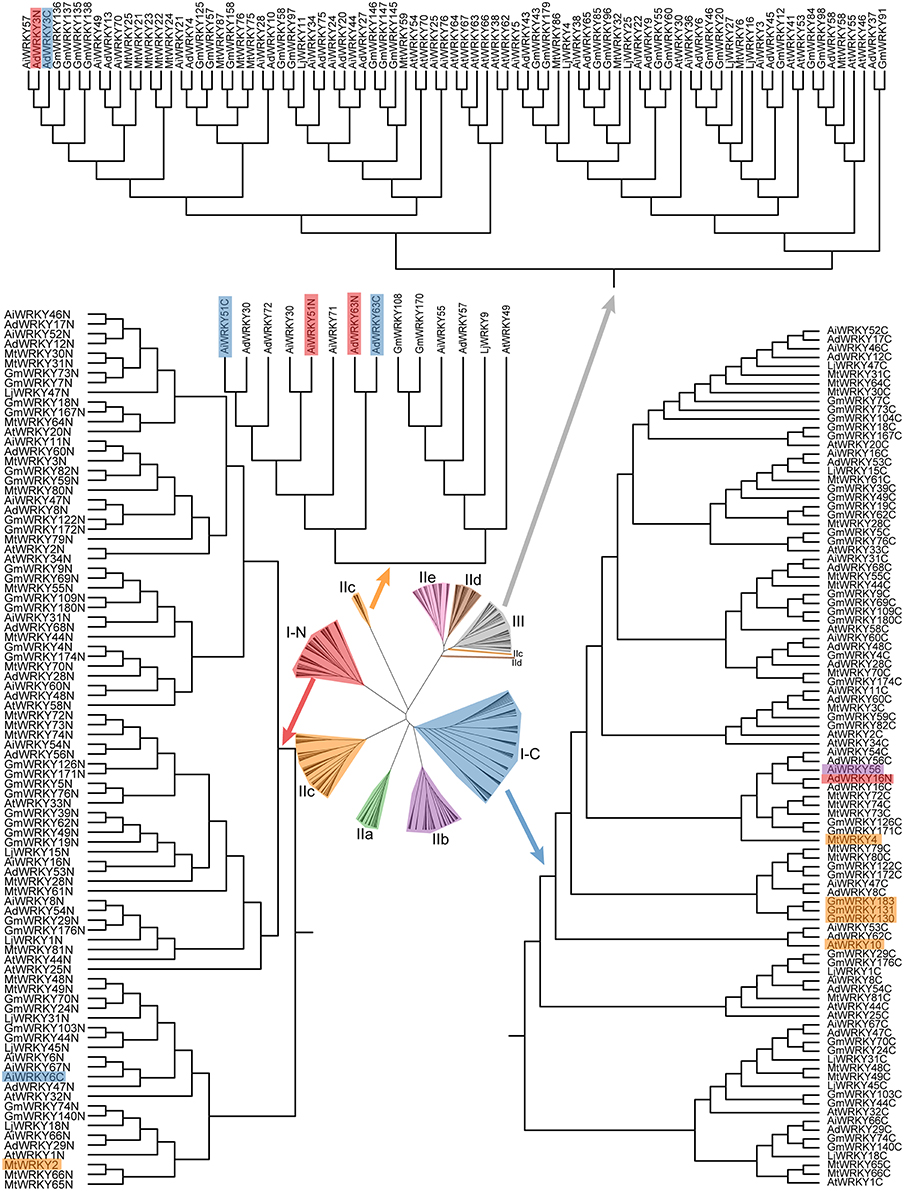
Figure 1. Phylogenetic tree of WRKY domains in six plants. The phylogenetic tree was constructed using MEGA 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
There are different opinions about origination of each type of WRKY proteins. In the beginning, researchers considered that the group II and III WRKY domains are the descendants originated from C-terminal WRKY domain of group I (Eulgem et al., 2000; Zhang and Wang, 2005). However, Zhu et al. (2013) found that Triticum aestivum IIc WRKY domains originated from the N-terminal WRKY domain of group I. Song et al. (2014) found that some L. japonicus and M. truncatula group II WRKY proteins derived from the N-terminal WRKY domain of group I. Wei et al. (2012a) demonstrated that group I WRKY protein firstly appeared in monocotyledons, followed by group III and II. In contrast, some researchers do not agree that group I WRKY protein is the ancestral member. Brand et al. (2013) reported that group I and other WRKY proteins likely originated from subgroup IIc. Recently, Rinerson et al. (2015) proposed two alternative hypotheses of WRKY protein evolution, “Group I Hypothesis” and “IIa + b Separate Hypothesis”. “Group I Hypothesis” considered all WRKY proteins evolving from C-terminal WRKY domains of group I proteins, whereas the “IIa + b Separate Hypothesis” considered groups IIa and IIb evolving directly from a single domain algal gene separated from group I-derived lineage (Rinerson et al., 2015).
WRKY Orthologous were Located in Syntenic Locus of Two Arachis Genomes
As shown in Figure 2, AdWRKY and AiWRKY genes were randomly distributed across 10 chromosomes. Chromosome A6 contained the largest number of WRKY genes (12), while chromosome A9 contained the least number of WRKY genes (1) in A. duranensis. In A. ipaënsis, 13 genes were distributed in chromosome B3, whereas only two WRKY genes were found in chromosome B9. The length of A. duranensis chromosome A7 (79.13 cM) and A8 (49.46 cM) is shorter than that of A. ipaënsis chromosome B7 (126.23 cM) and B8 (129.61 cM). However, chromosome 7 and 8 in these two species all contained 10 and 5 WRKY genes, respectively (Figure 2). There was no positive correlation between the chromosome length and the number of WRKY genes. In soybean, we found that most GmWRKY genes were located in the chromosome arm (Song et al., 2016). Same distribution pattern was observed in peanut. Previous study found more colinearity between Arachis and Glycine to compare with other leguminous plants (Nagy et al., 2012). The ends of chromosome exhibited stronger synteny than the central regions of chromosomes (Nagy et al., 2012).
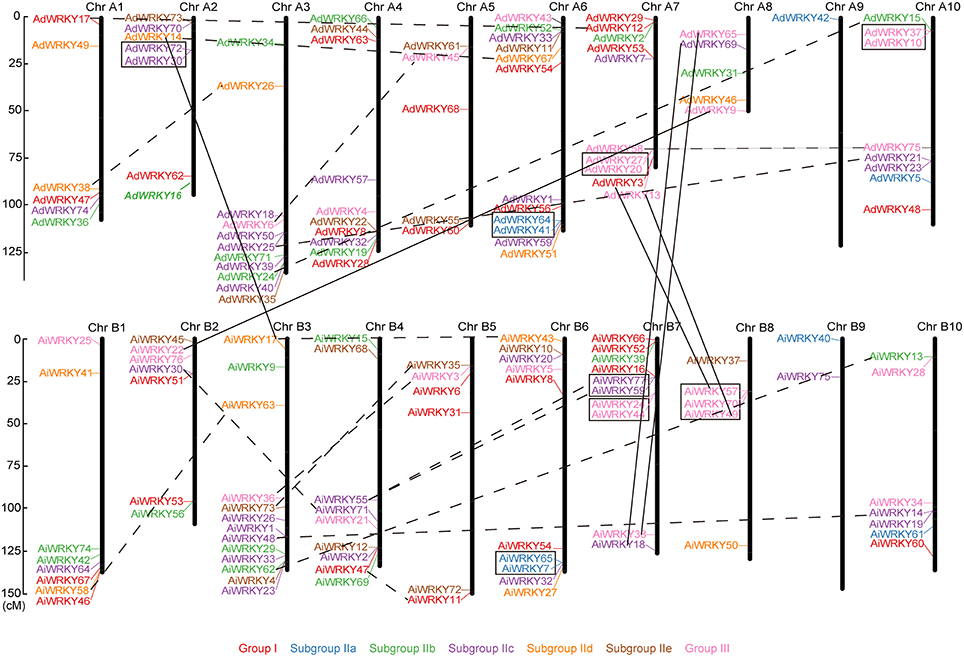
Figure 2. Chromosomal location of AdWRKY and AiWRKY genes. The chromosome numbers were shown at the top of each chromosome (black bars). The location of each WRKY gene was pointed out by a line. The solid lines indicated orthologous genes locations on the two peanut species' chromosomes. The dotted lines indicated the segmental duplicated genes. The black boxes indicated the tandem duplication genes. Italic indicated predicated chromosomal location in this study.
In this study, we detected 63 orthologous gene pairs according to the phylogenetic relationship of AdWRKY and AiWRKY genes (Table 2, Figure S3). Among which 56 orthologous gene pairs were found on the syntenic locus in A. duranensis and A. ipaënsis chromosome (Table 2, Tables S1, S2). However, the location of six AdWRKY genes (AdWRKY3, 9, 13, 14, 65, and 69) did not correspond to their orthologous gene in A. ipaënsis (AiWRKY57, 22, 49, 17, 38, and 18; Figure 2, Table 2). Physical mapping revealed that WRKY genes in Gossypium arboreum were not located in the corresponding chromosomes of G. raimondii, suggesting the occurrence of large chromosome rearrangement in the diploid cotton genomes (Ding et al., 2015).
AdWRKY16 gene has temporarily no precise location information in chromosome. Its orthologous gene, AiWRK56, was located in lower end of chromosome B2 (Figure 2). We speculated that AdWRKY16 gene might be distributed in the corresponding locus on chromosome A2 (Figure 2, italics bold).
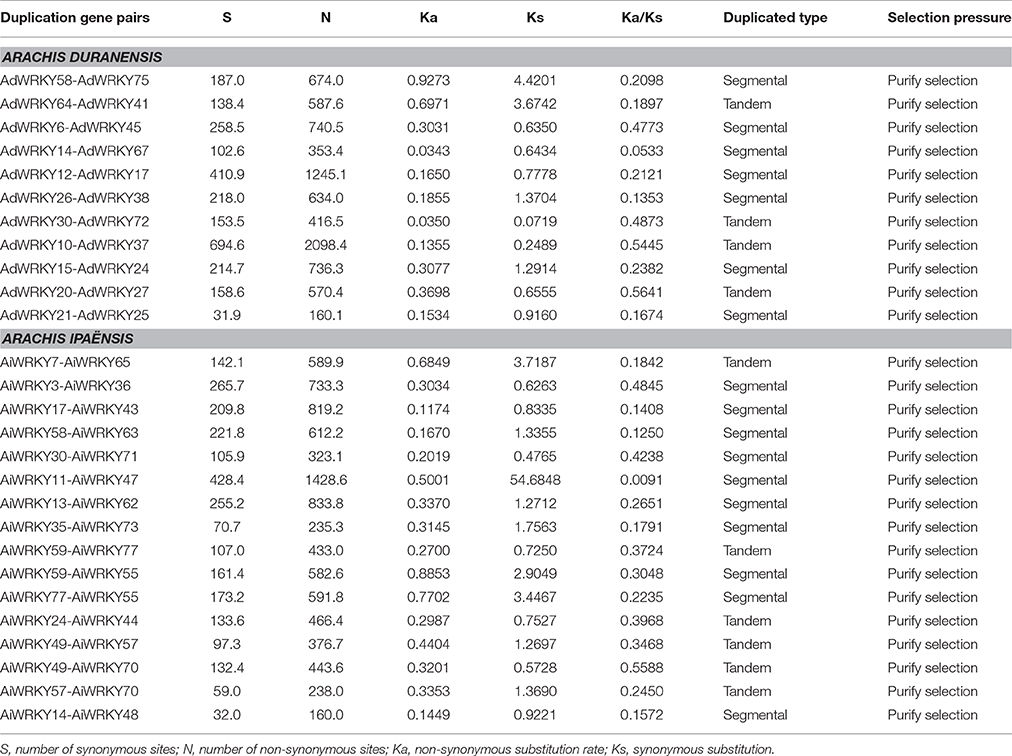
Segmental Duplication Events Played a Major Role in Arachis WRKY Gene Evolution
Gene duplication events occurred universally in WRKY genes (Cannon et al., 2004). Duplicated genes were considered to be the raw materials for the evolution of new biological functions and played crucial roles in adaption (Nei and Rooney, 2005). In this study, we employed duplicated GmWRKY genes (Yin et al., 2013) as query sequences to construct the molecular phylogenetic tree with AdWRKY and AiWRKY genes, respectively (Figures S4, S5). Results showed that gene duplication was detected for 22 AdWRKY and 26 AiWRKY genes (Table 3). Eight and 14 AdWRKY genes were experienced four tandem duplication and seven segmental duplication events, respectively (Figure 2, Table 3). Among the AiWRKY genes, four tandem duplication events with nine genes and 10 segmental duplication events with 17 genes were observed (Figure 2, Table 3). These results indicated that segmental duplication events played a major driving force for AdWRKY and AiWRKY evolution. This result is consistent with that most WRKY genes in soybean were derived from segmental duplication (Yin et al., 2013). However, the birth of WRKY gene in cotton and cocao genomes were considered through segmental duplication followed by tandem duplication (Dou et al., 2014; Ding et al., 2015). We calculated the Ks and Ka values and found that all duplicated AdWRKY and AiWRKY gene pairs with a ω value of < 1. This result indicated that purifying selection occurred on these duplicated gene pairs, which was agreed to the results of Brassica rapa (Tang et al., 2014).
Different Selection Pressure in Two Wild Peanut Species
Site model and branch-site model were used to estimate the selection pressure of AdWRKY and AiWRKY proteins. The results of site model showed that AdWRKY and AiWRKY proteins underwent purifying pressure during evolution (Table S3). It was showed that cotton WRKY proteins within each group are under strong purifying pressure (Ding et al., 2015). Li et al. (2015) found a strong acting of purifying selection in the evolution of Salvia miltiorrhiza WRKY proteins. Similarly, Group III WRKY proteins from L. japonicus (Song et al., 2014), M. truncatula (Song and Nan, 2014) and C. sativus (Ling et al., 2011) were considered to be under purifying selection. Although, AdWRKY and AiWRKY proteins underwent purifying selection, positive selected sites in group I (1 site), subgroup IIc (4 sites), IIe (5 sites), and group III (7 sites) AdWRKY proteins were detected using branch-site model (Table S4). In AiWRKY proteins, positive selected sites were discovered in group I (1 site), subgroup IIe (3 sites), and group III (1 site) (Table S5). More positive selected sites were detected in AdWRKY proteins than in AiWRKY proteins, indicating the existence of different degrees of positive selective pressure in AdWRKY and AiWRKY proteins. Purifying selection may generate genes with conserved functions or pseudogenization, but hard for neofunctionalization or subfunctionalization (Zhang, 2003). Based on this consideration, we tentatively suggested that AdWRKY group III and AiWRKY sungroup IIe genes may have various biological functions, instead, AdWRKY subgroup IIa, IIb and IId and AiWRKY subgroup IIa, IIb, IIc, and IId genes possibly have conserved biological functions.
Positively-selected sites were found using branch-site model. In cotton, group IIa and IId WRKY proteins were found many positive selection sites, while group I and subgroup IIc proteins had the lowest and no positive selection sites was found in cotton WRKY group III (Ding et al., 2015). GmWRKY group I, IIc, IIe, and III WRKY proteins had positive selection sites (Yin et al., 2013). The group IIc and III WRKY proteins from eggplant (Solanum melongena) and turkey berry (Solanum torvum) detected positive selection (Yang et al., 2015). Li et al. (2015) found that positive selection sites were presented in SmWRKY subgroup IIb, IIc, IId, IIe, and group III genes, while no positive selection site was detected in group I and subgroup IIa.
Arachis WRKY Gene Activities Respond to SA and MeJA
We intended to translate the knowledge from above wild ancestral peanut study to benefit disease resistance research in economic-important cultivated peanut. To obtain specific AdWRKY and AiWRKY that potentially involved in both SA and JA signaling pathways, we constructed phylogenetic tree using AdWRKY, AiWRKY, and AtWRKY proteins, respectively (Figures S6, S7), and then deduced SA- and JA-relative AdWRKY and AiWRKY proteins were determined if proteins were classified in the same clade of specific AtWRKY proteins known for being involved in both SA and JA signaling pathways (Dong et al., 2003; Schluttenhofer et al., 2014). The deduced SA- and JA-related AdWRKY and AiWRKY genes were used as query sequences to identify cultivated peanut WRKY genes (AhWRKY) using local BLASTN against the peanut transcriptome database and the peanut genome (unpublished data). Ultimately, we found 13 AhWRKY genes, named AhWRKY1 to AhWRKY13 (Tables S6, S7), were deduced to be potentially involved in both SA and JA signaling pathways in cultivated peanut.
We examined the transcriptional levels of these 13 genes in five different tissues by qRT-PCR and found each gene could be detected in at least one of the five tested tissues (Figure 3). Previous studies demonstrated that most WRKY genes expressed constitutively (Huang et al., 2012; Wei et al., 2012b; Dou et al., 2014; Jiang et al., 2014). The expression of AhWRKY1, 2, 3, 4, 5, 6, 7, 9, 11, and 13 genes could be detected in all five tissues (Figure 3). The expression of AhWRKY5, 8 and 10 genes were not detected in root; the expression of AhWRKY5 and 12 genes was not detected in seed. All 13 AhWRKY genes were expressed in stem, leaf and flower (Figure 3). AhWRKY2, 3, 4, 6, 8, 10, and 13 genes were mainly expressed in seed, indicating their function in seed development. Ding et al. (2015) showed that most cotton WRKY genes were highly expressed in all developmental stages of seed. These genes could be good candidates for Aspergillus flavus resistance, because Fountain et al. (2015) found that WRKY genes may play important roles in maize kernels against A. flavus infection.
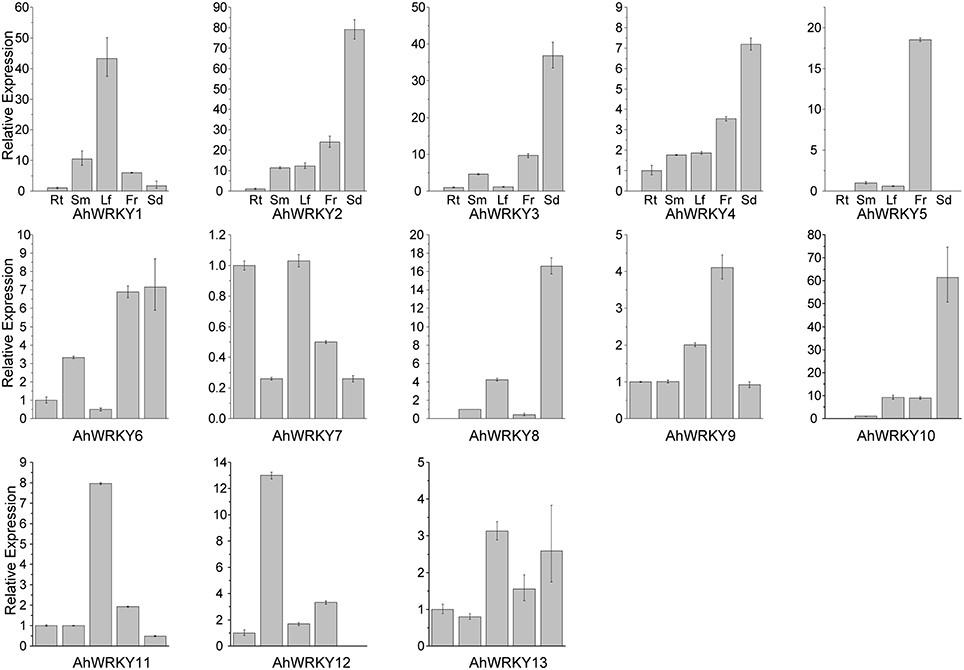
Figure 3. The expression pattern of AhWRKY genes in different tissues. The abbreviation Rt, Sm, Lf, Fr, and Sd in the tissue label indicated root, stem, leaf, flower and seed, respectively.
Then we asked if gene activities of 13 AhWRKY genes were affected by SA and MeJA treatments. As shown in Figure 4, AhWRKY1, 7, 8, 9, 10, 11, 12, and 13 genes were down-regulated at all five time points under SA treatment. However, AhWRKY5, 6, and 12 genes was up-regulated and the AhWRKY1, 7, 8, and 13 genes were down-regulated at all five time points under MeJA treatment (Figure 5). AhWRKY12 genes showed an opposite response to SA and MeJA treatment, while AhWRKY1, 7, 8, and 13 genes responded similar to these two hormones. Previous studies showed shat SA and MeJA signaling pathways often play an opposite role in defense, but synergistic effect between these phytohormones had also been reported (Mur et al., 2006). Lai et al. (2008) found that AtWRKY4 played positive role in plant resistance to necrotrophic pathogens, while overexpression of AtWRKY4 increased the plant susceptibility to biotrophic pathogen. However, AtWRKY70 (Li et al., 2004) and CaWRKY27 (Dang et al., 2014) regulated simultaneously SA and MeJA signaling pathways and acted as a positive regulator in response to pathogen.
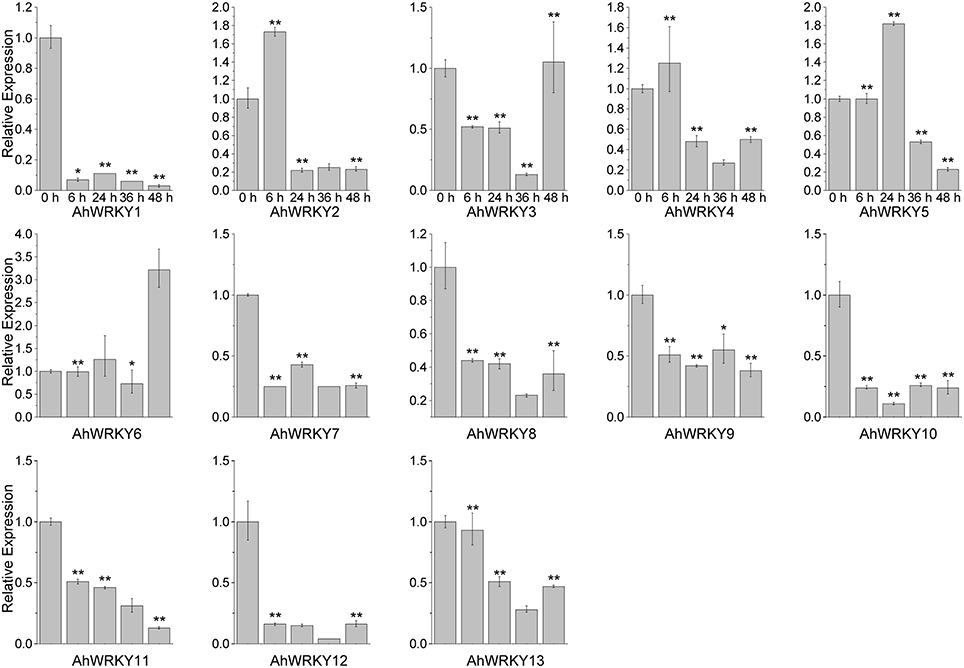
Figure 4. The expression pattern of AhWRKY genes after SA treatment. The Y-axis indicates the relative expression level; X-axis (0, 6, 24, 36, and 48 h) indicated hours of SA treatment. The standard errors are plotted using vertical lines. * and ** mean significant difference at P < 0.05 and P < 0.01, respectively.
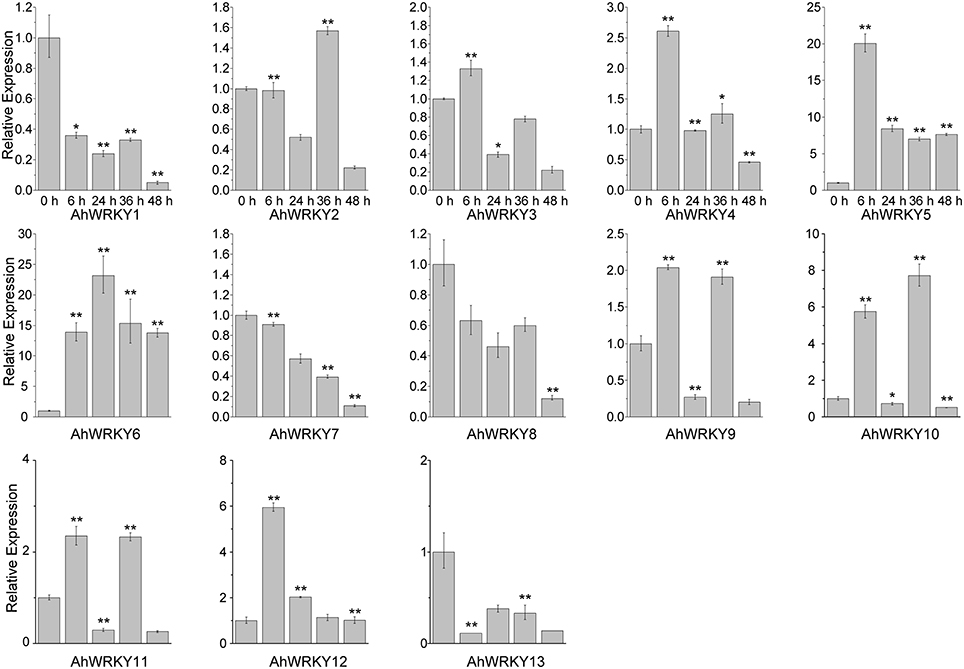
Figure 5. The expression pattern of AhWRKY genes after MeJA treatment. The Y-axis indicates the relative expression level; X-axis (0, 6, 24, 36, and 48 h) indicated hours of MeJA treatment. The standard errors are plotted using vertical lines. * and ** mean significant difference at P < 0.05 and P < 0.01, respectively.
AhWRKY1, 7, 8, 12, and 13 genes are orthologous genes of AtWRKY53, 40, 53, 21, and 3, respectively (Table S6). AtWRKY53 showed a strong increase in expression within the first 2 h but the transcript level greatly reduced at later time under SA treatment (Yu et al., 2001; Kalde et al., 2003). Miao and Zentgraf (2007) found that 4 h MeJA treatment reduced expression of AtWRKY53. AtWRKY40 gene was not considered as a positive regulator for systemic acquired resistance (Wang et al., 2006). Knocking out the AtWRKY40 gene led to the increased resistant to Golovinomyces orontii (Shen et al., 2007) and Pseudomonas syringe (Xu et al., 2006). Kalde et al. (2003) found that the expression of AtWRKY21 was induced by SA. CrWRKY36, orthologous gene of AtWRKY21, was up-regulated after 2 h MeJA treatment (Schluttenhofer et al., 2014). AtWRKY3 gene was involved in SA and JA signaling pathways. AtWRKY3 gene played a negative role in SA signaling pathway and mediated resistance to biotrophic pathogens, while it played a positive role in MeJA mediated resistance to necrotrophic pathogen (Lai et al., 2008). Interestingly, the expression of AhWRKY3 and 12 (paralogous genes), orthologous genes of AtWRKY21, responded oppositely to SA and MeJA treatments. Orthologous genes shared a conserved ancestral gene function in different species, while function of paralogous genes diversified through gene duplication (Koonin, 2005; Gabaldón and Koonin, 2013).
Conclusions
Similar number of WRKY proteins was identified in A. duranensis and A. ipaënsis. Orthologous gene pairs were found on the identical or similar locus on chromosomes in these two species. Our results showed that segmental duplication events played a major role in AdWRKY and AiWRKY evolution. Peanut WRKY proteins were under strong purifying pressure. Similar or opposite response of peanut WRKY genes to SA and JA treatments was observed. Our results could help to select appropriate candidate genes for further characterization of their pathogen resistant functions in Arachis species.
Author Contributions
HS wrote the manuscript and performed the laboratory assays. PW performed the phylogenetic analysis and reviewed the manuscript. JL provided the value comments and revised the grammar of the manuscript. CZ provided help in analysis of qRT-PCR. YB helped to revise the manuscript. XW served as the principal investigator, facilitated the project, and assisted in manuscript preparation and revision.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Kenta Shirasawa, Kazusa DNA research institute Chiba Japan, for his critical review and comments. This study was supported by National High Tech Project, NSFC Project (2011BAD35B04, 2013AA102602, 31500217), Young Talents Training Program of Shandong Academy of Agricultural Sciences and Shandong Province Germplasm Innovation and Utilization Project.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00534
Figure S1. Phylogenetic tree of AtWRKY and AdWRKY domains. The phylogenetic tree was constructed using MEGA 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
Figure S2. Phylogenetic tree of AtWRKY and AiWRKY domains. The phylogenetic tree was constructed using MEGA 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
Figure S3. Phylogenetic tree of AdWRKY and AiWRKY proteins. The phylogenetic tree was constructed using MEGA 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
Figure S4. Phylogenetic tree of AdWRKY and GmWRKY proteins. The phylogenetic tree was constructed using MEGA 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
Figure S5. Phylogenetic tree of AiWRKY and GmWRKY proteins. The phylogenetic tree was constructed using MEGA 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
Figure S6. Phylogenetic tree of AtWRKY and AiWRKY genes involved in both response to SA and MeJA. The phylogenetic tree was constructed using MEGA 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
Figure S7. Phylogenetic tree of AtWRKY and AiWRKY genes involved in both response to SA and MeJA. The phylogenetic tree was constructed using MEGA 6.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates.
Table S1. Arachis duranensis WRKY genes identified in the V14167.a1.M1.
Table S2. Arachis ipaënsis WRKY genes identified in the K30076.a1.M1.
Table S3. Analysis of positive selection among different codons of AdWRKY and AiWRKY using site model. ω, Ka/Ks value; ω0, Ka/Ks value in 0 category; ω1, Ka/Ks value in the first category; ω2, Ka/Ks value in the second category; p, the number of free parameters for the ω ratios; p0, the number of free parameters for the ω0 ratios; p1, the number of free parameters for the ω1 ratios; p2, the number of free parameters for the ω2 ratios; q, the parameters of the beta distribution.
Table S4. Tests for selection among codons of AdWRKY using branch-site models. ω, Ka/Ks value; ωb0, Ka/Ks value in background under model 0; ωb1, Ka/Ks value in background under model 1; ωb2a, Ka/Ks value in background under model 2a; ωb2b, Ka/Ks value in background under model 2b; ωf0, Ka/Ks value in foreground under model 0; ωf1, Ka/Ks value in foreground under model 1; ωf2a, Ka/Ks value in foreground under model 2a; ωf2b, Ka/Ks value in foreground under model 2b; p, the number of free parameters for the ω ratios; p0, the number of free parameters for the ω ratios under model 0; p1, the number of free parameters for the ω ratios under model 1; p2a, the number of free parameters for the ω ratios under model 2a; p2b, the number of free parameters for the ω ratios under model 2b.
Table S5. Tests for selection among codons of AiWRKY using branch-site models. ω, Ka/Ks value; ωb0, Ka/Ks value in background under model 0; ωb1, Ka/Ks value in background under model 1; ωb2a, Ka/Ks value in background under model 2a; ωb2b, Ka/Ks value in background under model 2b; ωf0, Ka/Ks value in foreground under model 0; ωf1, Ka/Ks value in foreground under model 1; ωf2a, Ka/Ks value in foreground under model 2a; ωf2b, Ka/Ks value in foreground under model 2b; p, the number of free parameters for the ω ratios; p0, the number of free parameters for the ω ratios under model 0; p1, the number of free parameters for the ω ratios under model 1; p2a, the number of free parameters for the ω ratios under model 2a; p2b, the number of free parameters for the ω ratios under model 2b.
Table S6. The information of AhWRKY genes.
Table S7. qRT-PCR primers in this study.
References
Altenhoff, A. M., and Dessimoz, C. (2012). Inferring orthology and paralogy. Methods Mol. Biol. 855, 259–279. doi: 10.1007/978-1-61779-582-4_9
Bencke-Malato, M., Cabreira, C., Wiebke-Strohm, B., Bücker-Neto, L., Mancini, E., Osorio, M. B., et al. (2014). Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 14:236. doi: 10.1186/s12870-014-0236-0
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K. S., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. doi: 10.1038/ng.3517
Bertioli, D. J., Seijo, G., Freitas, F. O., Valls, J. F. M., Leal-Bertioli, S. C. M., and Moretzsohn, M. C. (2011). An overview of peanut and its wild relatives. Plant Genet. Resour. C 9, 134–149. doi: 10.1017/S1479262110000444
Brand, L. H., Fischer, N. M., harter, K., Kohlbacher, O., and Wanke, D. (2013). Elucidating the evolutionary conserved DNA-binding specificities of wrky transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 41, 9764–9778. doi: 10.1093/nar/gkt732
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chang, I. F., Curran, A., Woolsey, R., Quilici, D., Cushman, J. C., Mittler, R., et al. (2009). Proteomic profiling of tandem affinity pyrified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9, 2967–2985. doi: 10.1002/pmic.200800445
Chang, S., and Puryear, J. J. C. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. doi: 10.1007/BF02670468
Cheong, Y. H., Chang, H. S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. doi: 10.1104/pp.002857
Dang, F., Wang, Y., She, J., Lei, Y., Liu, Z., Eulgem, T., et al. (2014). Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 150, 397–411. doi: 10.1111/ppl.12093
Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100, 8024–8029. doi: 10.1073/pnas.1230660100
Ding, M., Chen, J., Jiang, Y., Lin, L., Cao, Y., Wang, M., et al. (2015). Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol. Genet. Genomics 290, 151–171. doi: 10.1007/s00438-014-0904-7
Ding, Z. J., Yan, J. Y., Xu, X. Y., Yu, D. Q., Li, G. X., Zhang, S. Q., et al. (2014). Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 79, 13–27. doi: 10.1111/tpj.12538
Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. doi: 10.1023/A:1020780022549
Dou, L., Zhang, X., Pang, C., Song, M., Wei, H., Fan, S., et al. (2014). Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics 289, 1103–1121. doi: 10.1007/s00438-014-0872-y
Dutilh, B. E., van Noort, V., van der Heijden, R. T. J. M., Boekhout, T., Snel, B., and Huynen, M. A. (2007). Assessment of phylogenomic and orthology approaches for phylogenetic inference. Bioinformatics 23, 815–824. doi: 10.1093/bioinformatics/btm015
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., and Somssich, I. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Fountain, J. C., Raruang, Y., Luo, M., Brown, R. L., Guo, B., and Chen, Z. Y. (2015). Potential roles of WRKY transcription factors in regulating host defense responses during Aspergillus flavus infection of immature maize kernels. Physiol. Mol. Plant P 89, 31–40. doi: 10.1016/j.pmpp.2014.11.005
Gabaldón, T., and Koonin, E. V. (2013). Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 14, 360–366. doi: 10.1038/nrg3456
Gao, Q. M., Venugopal, S., Navarre, D., and Kachroo, A. (2011). Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155, 464–476. doi: 10.1104/pp.110.166876
Grunewald, W., Karimi, M., Wieczorek, K., Van de Cappelle, E., Wischnitzki, E., Grundler, F., et al. (2008). A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 148, 358–368. doi: 10.1104/pp.108.119131
Herbert, T. T., and Stalker, H. T. (1981). Resistance to peanut stunt virus in cultivated and wild Arachis species. Peanut Sci. 8, 45–47. doi: 10.3146/i0095-3679-8-1-12
Huang, S., Gao, Y., Liu, J., Peng, X., Niu, X., Fei, Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics 287, 495–513. doi: 10.1007/s00438-012-0696-6
Jiang, Y., Duan, Y., Yin, J., Ye, S., Zhu, J., Zhang, F., et al. (2014). Genome-wide identification and characterization of the Populusx WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 65, 6629–6644. doi: 10.1093/jxb/eru381
Jiang, Y. J., Liang, G., and Yu, D. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5, 1375–1388. doi: 10.1093/mp/sss080
Johnson, C.S., Kolevski, B., and Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. doi: 10.1105/tpc.001404
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Journot-Catalino, N., Somssich, I. E., Roby, D., and Kroj, T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302. doi: 10.1105/tpc.106.044149
Kalde, M., Barth, M., Somssich, I. E., and Lippok, B. (2003). Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant Microbe. Interact. 16, 295–305. doi: 10.1094/MPMI.2003.16.4.295
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Bio. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kayum, M. A., Jung, H. J., Park, J. I., Ahmed, N. U., Saha, G., Yang, T. J., et al. (2015). Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa. Mol. Genet. Genomics 290, 79–95. doi: 10.1007/s00438-014-0898-1
Kochert, G., Stalker, H., Gimenes, M., Galgaro, M., Lopes, C., and Moore, K. (1996). RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut, Arachis hypogaea (Leguminosae). Am. J. Bot. 83, 1282–1291.
Koonin, E. V. (2005). Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39, 309–338. doi: 10.1146/annurev.genet.39.073003.114725
Lai, Z. B., Vinod, K. M., Zheng, Z. Y., Fan, B. F., and Chen, Z. X. (2008). Roles of Arabidosis WRKY3 and WRKY4 transcription factors in plant responses to pathogens BMC Plant Biol. 8:68. doi: 10.1186/1471-2229-8-68
Li, C., Li, D., Shao, F., and Lu, S. (2015). Molecular cloning and expression analysis of WRKY transcription factor genes in Salvia miltiorrhiza. BMC Genomics 16:200. doi: 10.1186/s12864-015-1411-x
Li, J., Brader, G., and Palva, E. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Ling, J., Jiang, W., Zhang, Y., Yu, H., Mao, Z., Gu, X., et al. (2011). Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 12:471. doi: 10.1186/1471-2164-12-471
Liu, D., Leib, K., Zhao, P., Kogel, K. H., and Langen, G. (2014). Phylogenetic analysis of barley WRKY proteins and characterization of HvWRKY1 and-2 as repressors of the pathogen-inducible gene HvGER4c. Mol. Genet. Genomics 289, 1331–1345. doi: 10.1007/s00438-014-0893-6
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene wxpression sata using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, M., Dennis, E. S., Berger, F., Peacock, W. S., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 17531–17536. doi: 10.1073/pnas.0508418102
Luo, X., Sun, X., Liu, B., Zhu, D., Bai, X., Cai, H., et al. (2013). Ectopic expression of a WRKY homolog from Glycine soja alters flowering time in Arabidopsis. PLoS ONE 8:e73295. doi: 10.1371/journal.pone.0073295
McHale, L., Tan, X., Koehl, P., and Michelmore, R. W. (2006). Plant NBS-LRR proteins: adaptable guards. Genome Biol. 7:212. doi: 10.1186/gb-2006-7-4-212
Miao, Y., and Zentgraf, U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by a jasmonic and salicylic acid equilibrium. Plant Cell 19, 819–830. doi: 10.1105/tpc.106.042705
Mur, L. A., Kenton, P., Atzorn, R., Miersch, O., and Wasternack, C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. doi: 10.1104/pp.105.072348
Nagy, E. D., Guo, Y., Tang, S., Bowers, J. E., Okashah, R. A., Taylor, C. A., et al. (2012). A high-density genetic map of Arachis duranensis, a diploid ancestor of cultivated peanut. BMC Genomics 13:469. doi: 10.1186/1471-2164-13-469
Nei, M., and Rooney, A. P. (2005). Concerted and birth- and -death evolution of multigene families. Annu. Rev. Genet. 39, 121–152. doi: 10.1146/annurev.genet.39.073003.112240
Pande, S., and Narayana Rao, J. (2001). Resistance of wild Arachis species to late leaf spot and rust in greenhouse trials. Plant Dis. 85, 851–855. doi: 10.1094/PDIS.2001.85.8.851
Ramos, M. L., Fleming, G., Chu, Y., Akiyama, Y., Gallo, M., and Ozias-Akins, P. (2006). Chromosomal and phylogenetic context for conglutin genes in Arachis based on genomic sequence. Mol. Genet. Genomics 275, 578–592. doi: 10.1007/s00438-006-0114-z
Ren, X., Chen, Z. Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q. A., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors:genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rinerson, C. I., Rabara, R. C., Tripathi, P., Shen, Q. J., and Rushton, P. J. (2015). The evolution of WRKY transcription factors. BMC Plant Biol. 15, 66. doi: 10.1186/s12870-015-0456-y
Robatzek, S., and Somssich, I. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. doi: 10.1101/gad.222702
Ross, C., Liu, Y., and Shen, Q. (2007). The WRKY gene family in Rice (Oryza sativa). J. Integr. Plant Biol. 49, 827–842. doi: 10.1111/j.1672-9072.2007.00504.x
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Rushton, P., Somssich, I., Ringler, P., and Shen, Q. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schluttenhofer, C., Pattanaik, S., Patra, B., and Yuan, L. (2014). Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement. BMC Genomics 15:502. doi: 10.1186/1471-2164-15-502
Seijo, G., Lavia, G. I., Fernandez, A., Krapovickas, A., Ducasse, D. A., Bertioli, D. J., et al. (2007). Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. Am. J. Bot. 94, 1963–1971. doi: 10.3732/ajb.94.12.1963
Seijo, J., Lavia, G., Fernandez, A., krapovickas, A., Ducasse, D., Moscone, E., et al. (2004). Physical mapping of the 5S and 18S-25S rRNA genes by FISH as evidence that Arachis duranensis and A. ipaënsis are the wild diploid progenitors of A. hypogaea (Leguminosae). Am. J. Bot. 91, 1294–1303. doi: 10.3732/ajb.91.9.1294
Shen, Q. H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103. doi: 10.1126/science.1136372
Simpson, C. E. (2001). Use of wild Arachis species/introgression of genes into A. hypogaea L. Peanut Sci. 28, 114–116. doi: 10.3146/i0095-3679-28-2-12
Skibbe, M., Qu, N., Galis, I., and Baldwin, I. (2008). Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20, 1984–2000. doi: 10.1105/tpc.108.058594
Song, H., and Nan, Z. (2014). Genome-wide indentification and analysis of WRKY transcription factors in Medicago truncatula (In Chinese). Hereditas (Beijing) 36, 152–168 doi: 10.3724/SP.J.1005.2014.0152
Song, H., Wang, P., Hou, L., Zhao, S., Zhao, C., Xia, H., et al. (2016). Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 7:9. doi: 10.3389/fpls.2016.00009
Song, H., Wang, P., Nan, Z., and Wang, X. (2014). The WRKY transcription factor genes in Lotus japonicus. Int. J. Genomics 2014:420128. doi: 10.1155/2014/420128
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, 609–612. doi: 10.1093/nar/gkl315
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Bio. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, J., Wang, F., Hou, X. L., Wang, Z., and Huang, Z. N. (2014). Genome-wide fractionation and identification of WRKY transcription factors in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol. Biol. Rep. 32, 781–795. doi: 10.1007/s11105-013-0672-2
van Verk, M. C., Bol, J. F., and Linthorst, H. J. M. (2011). WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 11:89. doi: 10.1186/1471-2229-11-89
van Verk, M. C., Pappaioannou, D., and Neeleman, L. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995. doi: 10.1104/pp.107.112789
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogens 2:e123. doi: 10.1371/journal.ppat.0020123
Wang, X., Zeng, J., Li, Y., Rong, X., Sun, J., Sun, T., et al. (2015). Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 6:615. doi: 10.3389/fpls.2015.00615
Wei, K., Chen, J., Chen, Y., Wu, L., and Xie, D. (2012a). Multiple-strategy analyses of ZmWRKY subgroups and functional exploration of ZmWRKY genes in pathogen responses. Mol. Biosyst. 8, 1940–1949. doi: 10.1039/C2MB05483C
Wei, K. F., Chen, J., Y. F. C., and Wu, L. J., Xie, D. X. (2012b). Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 19, 153–164. doi: 10.1093/dnares/dsr048
Wu, K. L., Guo, Z. J., Wang, H. H., and Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xia, H., Zhao, C., Hou, L., Li, A., Zhao, S., Bi, Y., et al. (2013). Transcriptome profiling of peanut gynophores revealed global reprogramming of gene expression during early pod development in darkness. BMC Genomics 14:517. doi: 10.1186/1471-2164-14-517
Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interaction betwenn pathogen-induced Arabidosis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. doi: 10.1105/tpc.105.037523
Yao, Q. Y., Xia, E. H., Liu, F. H., and Gao, L. Z. (2015). Genome-wide identification and comparative expression analysis reveal a rapid expansion and functional divergence of duplicated genes in the WRKY gene family of cabbage, Brassica oleracea var. capitata. Gene 557, 35–42. doi: 10.1016/j.gene.2014.12.005
Yang, X., Deng, C., Zhang, Y., Cheng, Y., Huo, Q., and Xue, L. (2015). The WRKY transcription factor genes in eggplant (Solanum melongena L.) and turkey berry (Solanum torvum Sw.). Int. J. Mol. Sci. 16, 7608–7626. doi: 10.3390/ijms16047608
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Bio. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yin, G., Xu, H., Xiao, S., Qin, Y., Li, Y., Yan, Y., et al. (2013). The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 13:148. doi: 10.1186/1471-2229-13-148
Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1539. doi: 10.1105/TPC.010115
Zentella, R., Zhang, L. Z., Park, M., Thomas, S. G., Endo, A., Murase, K., et al. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057. doi: 10.1105/tpc.107.054999
Zhang, J. Z. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/s0169-5347(03)00033-8
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. doi: 10.1186/1471-2148-5-1
Zhang, Z. L., Xie, Z., Zou, X., Casaretto, J., Ho, T., and Shen, Q. (2004). A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134, 1500–1513. doi: 10.1104/pp.103.034967
Zhu, X., Liu, S., Meng, C., Qin, L., Kong, L., and Xia, G. (2013). WRKY transcription factors in wheat and their induction by biotic and abiotic stress. Plant Mol. Biol. Rep. 31, 1053–1067. doi: 10.1007/s11105-013-0565-4
Zou, C. S., Jiang, W. B., and Yu, D. Q. (2010). Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 61, 3901–3914. doi: 10.1093/jxb/erq204
Keywords: Arachis, disease resistant, gene duplication, phylogenetic relationship, WRKY
Citation: Song H, Wang P, Lin J-Y, Zhao C, Bi Y and Wang X (2016) Genome-Wide Identification and Characterization of WRKY Gene Family in Peanut. Front. Plant Sci. 7:534. doi: 10.3389/fpls.2016.00534
Received: 30 December 2015; Accepted: 04 April 2016;
Published: 26 April 2016.
Edited by:
Janila Pasupuleti, International Crops Research Institute for the Semi Arid Tropics, IndiaReviewed by:
Steven B. Cannon, Agricultural Research Service, USAMarc Libault, University of Oklahoma, USA
Copyright © 2016 Song, Wang, Lin, Zhao, Bi and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingjun Wang, xingjunw@hotmail.com
 Hui Song
Hui Song Pengfei Wang
Pengfei Wang Jer-Young Lin2
Jer-Young Lin2 Chuanzhi Zhao
Chuanzhi Zhao Xingjun Wang
Xingjun Wang