- 1College of Horticulture and Landscape Architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, China
- 2College of Horticulture, South China Agricultural University, Guangzhou, China
- 3Vegetable Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 4Pepper Institute, Zunyi Academy of Agricultural Sciences, Zunyi, China
- 5Guizhou Provincial College-based Key Lab for Tumor Prevention and Treatment with Distinctive Medicines, Zunyi Medical University, Zunyi, China
Dof (DNA-binding One Zinc Finger) transcription factor family is unique to plants and has diverse roles associated with plant-specific phenomena, such as light, phytohormone and defense responses as well as seed development and germination. Although, genome-wide analysis of this family has been performed in many species, information regarding Dof genes in the pepper, Capsicum annuum L., is extremely limited. In this study, exhaustive searches of pepper genome revealed 33 potential CaDofs that were phylogenetically clustered into four subgroups. Twenty-nine of the 33 Dof genes could be mapped on 11 chromosomes, except for chromosome 7. The intron/exon organizations and conserved motif compositions of these genes were also analyzed. Additionally, phylogenetic analysis and classification of the Dof transcription factor family in eight plant species revealed that S. lycopersicum and C. annuum as well as O. sativa and S. bicolor Dof proteins may have evolved conservatively. Moreover, comprehensive expression analysis of CaDofs using a RNA-seq atlas and quantitative real-time polymerase chain reaction (qRT-PCR) revealed that these genes exhibit a variety of expression patterns. Most of the CaDofs were expressed in at least one of the tissues tested, whereas several genes were identified as being highly responsive to heat and salt stresses. Overall, this study describes the first genome-wide analysis of the pepper Dof family, whose genes exhibited different expression patterns in all primary fruit developmental stages and tissue types, as in response to abiotic stress. In particular, some Dof genes might be used as biomarkers for heat and salt stress. The results could expand our understanding of the roles of Dof genes in pepper.
Introduction
The DNA-binding one zinc finger (Dof) protein is a representative of the plant-specific members of transcription factors (TFs). All Dof transcription factors share a common DNA-binding domain (C2C2-Dof) that is highly conserved in the N-terminal region composed of approximately 52 amino acid residues. It was predicted that the C2C2-Dof motif may form a single zinc finger that is essential for binding a conversed target DNA sequence with a 5′-(T/A)AAAG-3′ core (Yanagisawa and Schmidt, 1999). The C-terminal region of Dof proteins is highly variable. This unstable C-terminal domain could act as either a transcriptional activator or repressor in the control of the expression of many structural genes, and execute different regulatory functions (Guo et al., 2009; Cominelli et al., 2011; Corrales et al., 2014).
The Dof TFs have been reported to be involved in a wide spectrum of biological processes, such as light-responsiveness (Yanagisawa and Sheen, 1998; Park et al., 2003; Ward et al., 2005), seed development, maturation, and germination (Diaz et al., 2002, 2005; Gualberti et al., 2002; Papi et al., 2002; Isabel-LaMoneda et al., 2003; Dong et al., 2007; Gabriele et al., 2010; Gaur et al., 2011; Santopolo et al., 2015). Meanwhile there were evidenced that some Dof TFs participated in plant hormone and stress responses as well (Skirycz et al., 2006; Corrales et al., 2014; Xu et al., 2014; Jiang et al., 2015; Song et al., 2016). In Arabidopsis, more than 10 Dof proteins, including the OBF BINDING PROTEIN (OBP1, OBP2, and OBP3), Dof Affecting Germination (DAG1 and DAG2), and Cycling Dof Factors (CDF1-5) (Imaizumi et al., 2005; Skirycz et al., 2008; Fornara et al., 2009) have been functionally characterized. Among them, OBP1 might be involved in glutathione S-transferases 6 (GST6) expression and respond to plant hormones and stress (auxin, salicylic acid, and H2O2) signals (Pan et al., 2014). OBP1 could also control cell division by regulating the expression of several cell cycle-associated genes (Skirycz et al., 2008). OBP2 (AtDof1.1) is involved in the regulation of glucosinolate biosynthesis in Arabidopsis (Skirycz et al., 2006). OBP3 plays important roles in plant growth and development (Kang and Singh, 2000) and is characterized as a novel component of light signaling (Ward et al., 2005). The DAG1 and DAG2 proteins are also involved in light-dependent seed germination in Arabidopsis (Gualberti et al., 2002; Papi et al., 2002; Gabriele et al., 2010). CDFs play an important role in photoperiodic flowering in Arabidopsis through by binding directly to the C2C2-Dof sites in the CONSTANS (CO) promoter to repress CO transcription (Imaizumi et al., 2005). Combining loss-of-function mutations in four of these genes (CDF1, 2, 3, and 5) causes photoperiod-insensitive early flowering by increasing the CO mRNA level (Fornara et al., 2009; Corrales et al., 2014).
The Dof gene family has also been comprehensively identified in several plants based on the completion of an increasing number of genome sequencing projects. Thirty-six and 30 Dof genes have been found in the model plant Arabidopsis and rice, respectively (Yanagisawa, 2002; Lijavetzky et al., 2003), 27 in Brachypodium distachyon (Hernando-Amado et al., 2012), 31 in wheat (Shaw et al., 2009), 26 in barley (Moreno-Risueno et al., 2007), 28 in sorghum (Kushwaha et al., 2011), 78 in soybeans (Guo and Qiu, 2013), 34 in tomatoes (Cai et al., 2013), 35 in potatoes (Venkatesh and Park, 2015), 76 in Chinese cabbage (Ma et al., 2015), 38 in pigeonpeas (Malviya et al., 2015), and 36 in cucumber (Wen et al., 2016). According to the sequence similarity, the Dofs could be organized into four groups or subfamilies (A, B, C, and D), and groups B, C, and D could be further subdivided into subgroups (Lijavetzky et al., 2003; Ma et al., 2015; Feng et al., 2016; Wen et al., 2016).
Pepper (Capsicum spp.) is one of the most important and widely cultivated vegetable crops belonging to the family Solanaceae, which also includes potatoes, tomatoes, and tobacco, eggplants, etc. Lately, the pepper genome was sequenced (Kim et al., 2014; Qin et al., 2014). In addition, a large number of RNA sequencing reads derived from several tissues such as root, shoot, leaf, flower, and fruit are also available. These datasets provide a framework for the identification and functional characterization of gene family from a global view for pepper improvement and basic research. This study aimed to identify all potential Dof genes encoded in the pepper genome. Further, some routine bioinformatics analyses were performed including gene structures, chromosomal distribution and phylogenetic analysis. Finally, functional prediction was performed based on the gene expression analysis in different organs and developmental stages, and in response to heat and salinity stresses. The results gained herein will provide an important foundation for future studies on gene cloning and functional characterization of Dofs in pepper.
Materials and Methods
Plant Materials and Stress Treatments
Seeds of the pepper cultivar “Zunla-1” (Capsicum annuum L.) were provided by the Pepper Institute, the Zunyi Academy of Agricultural Sciences in China. The seeds were first sterilized and germinated in an incubator (28°C), as previously described (Qin et al., 2014). The germinated seeds were then sown in pots and grown under a 16 h day/8 h night cycle (28°C/21°C day/night temperature cycle) until the seedlings developed six leaves. Uniformly developed plants were then exposed to heat (38°C) and NaCl (300 mM) treatments for 3, 6, and 12 h. For all treatments, leaves were harvested from the same position of the seedlings with three biological replicates. For each replicate, leaves from eight plants were put together and rapidly frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Identification of Dof Genes in Pepper
The conserved Dof domain (PF02701) based on a Hidden Markov Model (HMM) was firstly downloaded from the Pfam database (Pfam 27.0, http://Pfam.sanger.ac.uk/). Then the HMM profile of the Dof domain was used to do BLASTP search in two pepper genome databases (http://peppersequence.genomics.cn/page/species/index.jsp, release 2.0 cv. Zunla-1; http://peppergenome.snu.ac.kr/, cv. CM334) (Kim et al., 2014; Qin et al., 2014) with an expected value (e-value) cut-off of 0.01. All protein sequences obtained were confirmed for the presence of an intact C2C2-Dof domain by ScanProsite (http://www.expasy.ch/tools/scanprosite/) and SUPERFAMILY 1.75 (http://supfam.org/SUPERFAMILY/hmm.html). In order to obtain the integrated catalog of Dof genes in pepper, the output results from two databases were combined and filtered the redundant sequences. The isoelectric points and protein molecular weights of all non-redundant sequences were obtained with the help of the proteomics and sequence analysis tools on the ExPASy proteomics server (http://expasy.org/).
Chromosomal Location and Gene Structure Analysis
The chromosomal location information of each pepper Dof was obtained via BLASTP search against the pepper genome database which has been built by our group (Qin et al., 2014) (cv. Zunla-1) with default parameters. The exon and intron structures of individual Dof gene were illustrated using the Gene Structure Display Server (GSDS 2.0, http://gsds.cbi.pku.edu.cn/index.php) by aligning the cDNA sequences with the corresponding genomic DNA sequences.
Conserved Motif Analysis
Functional motifs or domains of Dof protein sequences were analyzed using PROSITE (http://prosite.expasy.org/) and the Conserved Domain database (http://www.ncbi.nlm.nih.gov/cdd/). MEME (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) (Version 4.9.1; Bailey et al., 2009) was used to identify motifs in candidate sequences. MEME was run online using the following parameters: Distribution of motif occurrences: Any number of repetitions, Number of different motifs: 25, Minimum/Maximum number of sites: 5/100, Minimum/Maximum motif width: 6/100.
Phylogenetic Analyses
Multiple alignments of the full-length protein sequences were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The phylogenetic tree was constructed and drawn by using MEGA 6.06 program (http://www.megasoftware.net/) (Tamura et al., 2013) by the neighbor-joining (NJ) method with 1000 bootstrap replicates. Only clades with a test value higher than 50 were selected for the consensus tree.
RNA Isolation and Quantitative Real-Time PCR
Total RNA from leaves was isolated using the Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. RNA was treated with RNase-free DNase I (Takara, Dalian, China) to remove any contamination of genomic DNA. First-strand cDNA synthesis was carried out with approximately 1 μg RNA using the PrimeScript reverse transcription kit (Takara, Dalian, China) and random primers, according to the manufacturer's procedure. Primers with melting temperatures of 58–60°C, lengths of 20–27 bp, and product lengths of 100–250 bp were designed using Primer Premier 5.0 software. All primer sequences are listed in Additional File 6.
qRT-PCR was performed on an CFX96 instrument (Bio-Rad, Alfred Nobel Drive Hercules, CA, USA) using SYBR Green qPCR kits (Kapa Biosystems, Boston, Massachusetts, USA), according to the manufacturer's instructions. The constitutive actin gene served as the endogenous control is AY572427 described previously (Lee et al., 2009). PCR was done in 20 μL volumes containing a 250 nM concentration of each primer, 40 ng of cDNA and 10 μL of KAPA SYBR® FAST Universal 2X qPCR Master Mix (KAPA SYBR® DNA polymerase is an engineered version of Taq DNA polymerase, designed specifically for real-time PCR using SYBR Green I chemistry). The PCR amplification condition included an initial heat-denaturing step at 95°C for 5 min; then 40 cycles at 95°C for 5 s and at 58°C for 50 s. Fluorescence was measured at the end of each cycle. A melting-curve analysis was performed by heating the PCR product from 65 to 90°C. Expression level of each CaDof gene was calculated using the 2−ΔΔCt method, as previously described (Livak and Schmittgen, 2001). SPSS 19.0 (SPSS Inc., USA) was used for statistical analysis, and the Dunnett's t-test was used to detect significant difference between all stress treatments and their controls.
Pepper RNA-seq Data Analysis
For expression profiling analysis of pepper Dof genes, we utilized the Illumina RNA-seq data that were previously generated by pepper genome sequencing (Qin et al., 2014). The expression level of each gene was measured by fragments per kilobase of exon model per million reads mapped (FPKM) values (Additional File 4). Heat maps for above-mentioned genes were generated, which have positive RPKM values in at least one or more of the samples. For the developmental stage dataset, RPKM values were log2 transformed before generating heat maps. The heat map was generated by BAR Heatmapper Plus (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi) online.
Results
Pepper Dof Transcription Factor Gene Family Isolation
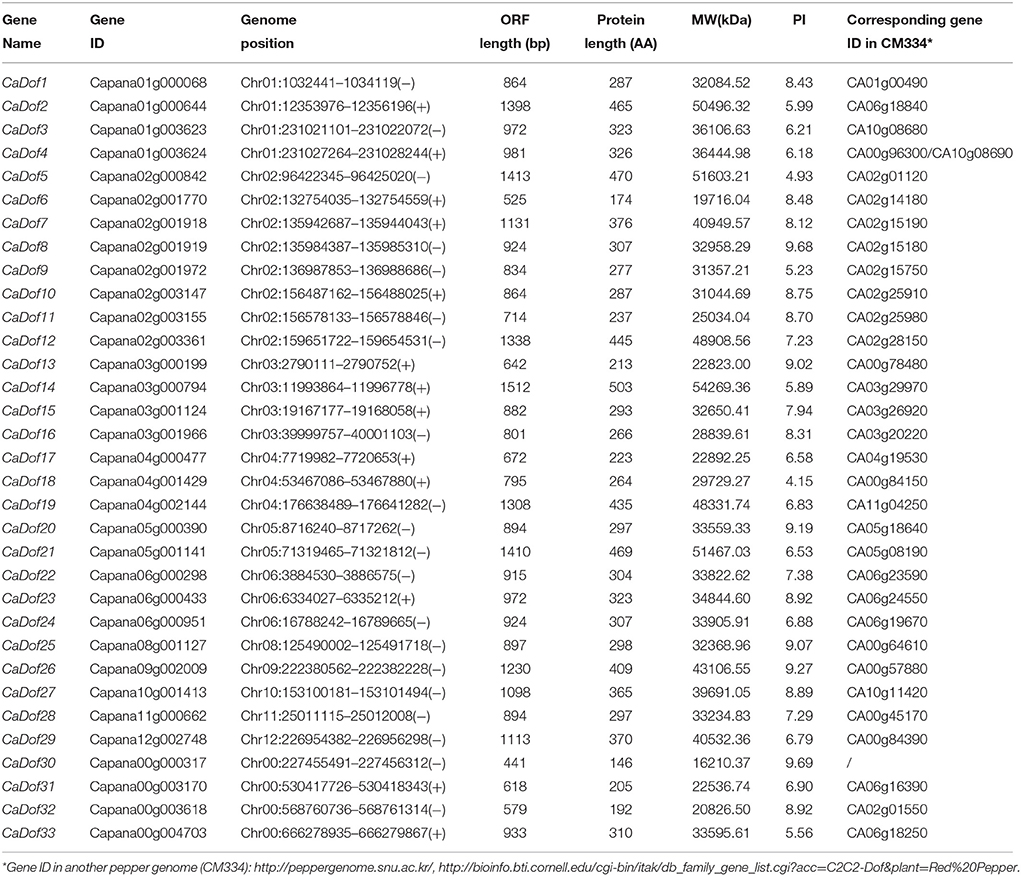
A total of 33 non-redundant Dof genes were identified (Additional File 1). All had a typical DNA binding domain of 52 residues spanning a single C2/C2 zinc finger structure (Figure 1). Pepper Dof genes were designated as CaDof1-CaDof33 based on the positions of their corresponding genes on chromosomes 1–12 from top to bottom. The full length coding sequences of the CaDof genes ranged from 441 bp (CaDof30) to 1512 bp (CaDof14). The size of deduced Dof proteins varied between 146 and 503 amino acids (aa) with an average of approximately 317 aa. The molecular weight (Mw) varied from 16.2 to 54.3 kDa, and the theoretical pI of these genes ranged from 4.15 (CaDof18) to 9.69 (CaDof30) (Table 1).

Figure 1. Multiple sequence alignment of Dof domain in the pepper. (A) Sequence representation LOGO derived from multiple sequence alignment of the Dof motifs. (B) Multiple sequence alignment of pepper Dof motifs.
Chromosomal Localization and Gene Structure of CaDof Genes
The physical locations of the CaDof genes on pepper chromosomes were identified (Figure 2). The results showed that 29 of the 33 CaDof genes could be located on the 11 chromosomes, except chromosome 7, with an obviously non-uniform distribution. However, four members (CaDof30-CaDof33) could not be anchored on any of the pepper chromosomes. We arranged them on a pseudo-chromosome, designated as Chr00, which was concatenated by the unplaced 3134 scaffolds (705 Mb in total) (Qin et al., 2014). Chromosome 2 contained the largest number of CaDof genes (8 members). In contrast, only one CaDof gene was found on each of chromosomes 8, 9, 10, 11, and 12.
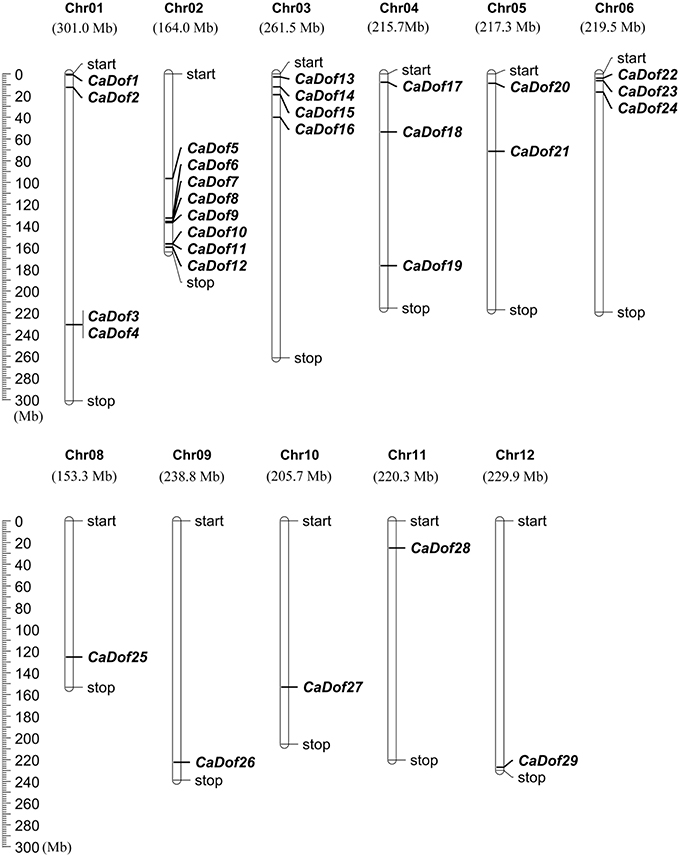
Figure 2. Distribution of CaDof genes in pepper chromosomes. Twenty nine CaDof genes were mapped to the 11 linkage groups (Chr01 through Chr12, except Chr07), whereas four CaDofs were mapped on a pseudo-chromosome, designated as Chr00.
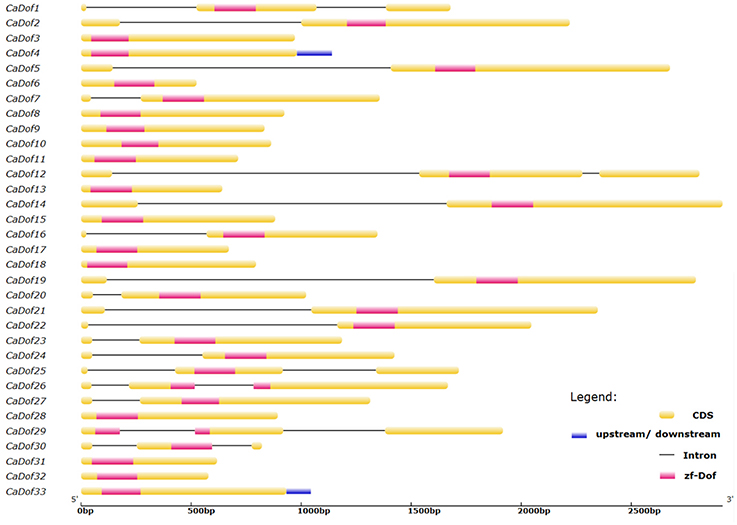
In order to gain further insight into their evolutionary imprints, the exon-intron structure of each member of the CaDof family was analyzed. The number of introns of the CaDof genes ranged from 0 to 2 (Figure 3). Fifteen (45.45%) of the CaDof genes were intronless, whereas 12 (36.36%) of them contained one intron, and were generally located upstream of the DNA binding domain. Among them, six genes (CaDof1, CaDof12, CaDof25, CaDof26, CaDof29, and CaDof30) (18.18%) contained two introns.
Phylogenetic Analysis and Classification of the Dof Transcription Factor Family
In order to evaluate the evolutionary relationships among CaDof proteins, an unrooted phylogenetic tree was constructed based on the alignment of the full length of protein sequences by the neighbor-joining method with 1000 bootstrap replicates. Pepper Dof transcription factor family could be further divided into four major groups (from I to IV) with 50% bootstrap values (Figure 4A). Group I had the most members (10 genes, or 30.3%), followed by groups II and III, each comprised eight CaDof genes, whereas group IV contained the fewest gene members (7 genes, or 21.2%). Furthermore, some CaDofs within the same group shared similar exon-intron structure patterns in terms of intron number. For instance, almost all CaDof genes in group III had no intron, except for CaDof7. The two introns CaDof genes were mainly in group I (CaDof and CaDof25) and group II (CaDof26, CaDof29, and CaDof30). These similar structure features may be related to their functions in pepper genome.
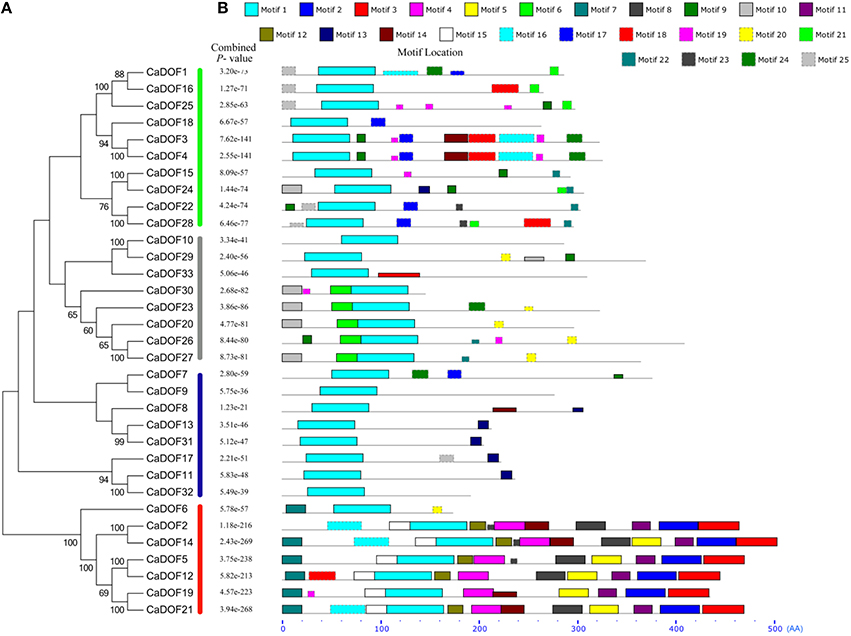
Figure 4. Phylogenetic tree constructed by MEGA 6 (A) and motif locations identified by MEME program (B). Height of motif “block” is proportional to −log (p-value); combined p-values are shown in middle. Sequence logos and detailed information for each motif are shown in (B).
To further reveal the diversification of Dof genes in pepper, putative motifs were predicted using the program MEME, and 25 distinct motifs were identified. The schematic distribution of the 25 motifs among the different gene groups is shown in Figure 4B. The identified multilevel consensus sequence for the motifs is shown in Additional File 2. Motif 1 observed in all CaDof proteins was the conserved Dof domain. As expected, members who had similar motif compositions could be clustered into one class, suggesting functional similarities among the Dof proteins within the same subfamily. Class II showed two conserved motifs (motif 6 and motif 10). Class III contained special motif 13. In class IV 10 motifs (9, 11, 16, 17, 3, 6, 5, 13, 2, and 4) were conserved, among which motif 22 might be characteristic of class IV. Motif 19 was the conserved motif in class I, with other variable motifs being 10, 15, 20, 12, 23, and 18. The motif distribution also confirmed that the Dof genes were conserved during evolution. The varieties of motif distributions in different subgroups implied sources of functional differentiation in Dof genes in the evolutionary processes.
To further explore the evolutionary relationships within the pepper Dof family and those from other species, a total of 207 Dof protein sequences (Additional File 3) from eight species (four dicots including Arabidopsis thaliana, Vitis vinifera, Solanum lycopersicum, and Capsicum annuum, two monocots including Oryza sativa and Sorghum bicolor, Chlamydomonas reinhardtii and Physcomitrella patens subsp. patens) were used to construct a joint tree (Figure 5). The phylogenetic tree showed that Dof genes can be categorized into seven subgroups (groups 1–7) based on sequence similarities with a high bootstrap value (>50%). Among these, subgroup 1 constituted the largest clade containing 42 members, followed by subgroup 4 (40 members), and subgroup 2 (35 members); subgroup 3 contained only 17 members, which was the smallest clade. Subgroup 3 was primarily made up of the lower plant P. patens, whereas subgroup 7 contained the monocot-specific group of rice and sorghum.
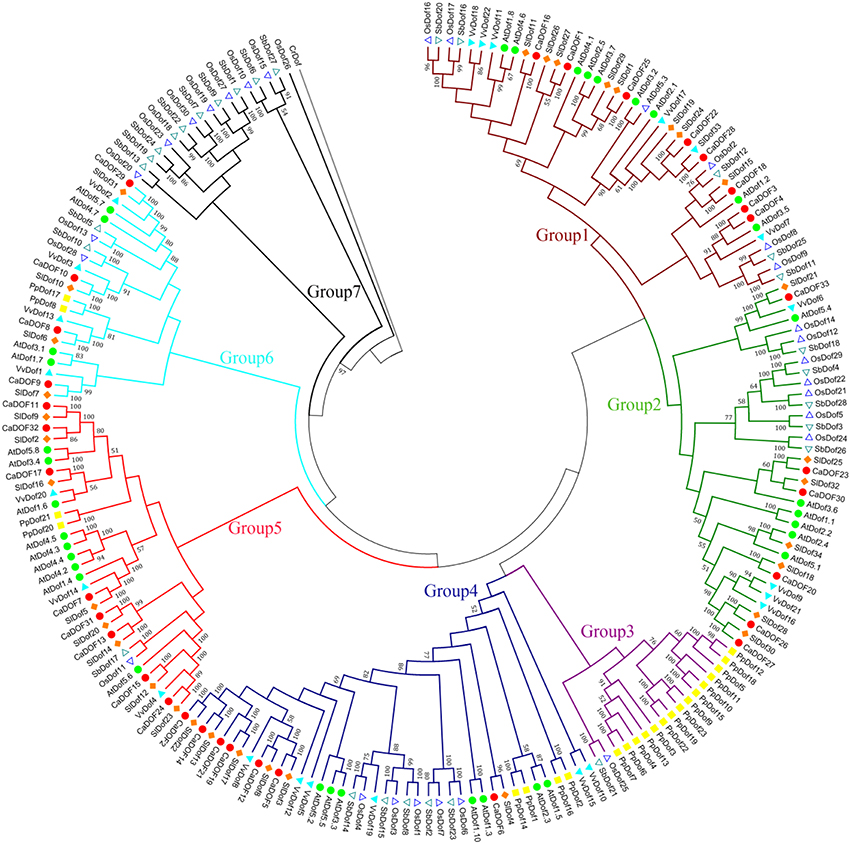
Figure 5. Phylogenetic tree of the Dof transcription factors in eight representative species. Arabidopsis thaliana Dofs (AtDof1.1-5.8) and Oryza sativa Dofs (OsDof1-30) were obtained from Yanagisawa (2002), Solanum lycopersicum Dofs (SlDof1-34) were obtained from Cai et al. (2013), Sorghum bicolor Dofs (SbDof1-28) were obtained from Kushwaha et al. (2011). The putative orthologs from V. vinifera, Chlamydomonas reinharditii and Physcomitrella patents were assigned to corresponding CaDof proteins with the E < 0.01, which were extracted by a Blast P search from Phytozome v9.1 (http://www.phytozome.net/). The tree rooted using CrDof as an outgroup. These 207 Dof protein sequences (Additional File 3) were aligned. Bootstraping values are indicated as percentages (when > 50%) along the branches.
The Dof phylogenetic tree also showed essentially the same clustering patterns in S. lycopersicum (tomato) and C. annuum (pepper), two members of the Solanaceae family. In total, 29 pairs of Dof proteins from S. lycopersicum and C. annuum were clustered as pairs, indicating that they might be orthologous. For example, the pair of SlDof31 and CaDof29 and another pair of SlDof9 and CaDof11 are highly similar, indicating that some consensus in domain may have existed before the pepper–tomato divergence. The same results appeared between O. sativa and S. bicolor, which is consistent with the notion that both belonged to the grass family. The phylogenetic similarity found in S. lycopersicum and C. annuum, and O. sativa and S. bicolor Dof proteins suggests that they may have evolved conservatively.
Differential Expression of CaDof Genes in Various Tissues and Developing Fruits
In a previous study, we obtained a 90.5Gb Illumina RNA-Seq atlas of pepper genes from 46 libraries representing all primary developmental stages and tissue types, including various fruits in different developmental stages (Qin et al., 2014). Thus, we utilized these data to investigate the transcription levels of the CaDof genes in the root, stem, leaf, bud, and flower, as well as in the developing fruits of the pepper cultivar Zunla-1 (Additional File 4).
Among all CaDof genes, transcripts of 31 CaDofs were found in at least one of the 14 different tissues and developmental fruit stages. However, the other two CaDofs (CaDof3 and CaDof4) were not detected in these RNA-seq libraries because they were at too low of a level. As shown in the heat map representation (Figure 6), most of the CaDof s exhibited divergent expression profiles in the 14 tissues examined. Most CaDof genes showed the highest expression level in roots and stems. Some Dof genes had very high transcript abundance in one or two tissues, but their expressions were almost negligible in other tissues. In particular, CaDof6, CaDof14, CaDof16, and CaDof28 exhibited a higher expression level in the root, whereas CaDof18, CaDof29, and CaDof32 could not be detected. Almost all CaDofs were expressed in the stem, except CaDof32, especially CaDof28, CaDof10, CaDof14, and CaDof16 exhibited relatively higher transcript abundance. CaDof10, CaDof14, and CaDof28 mRNA were highly expressed in leaves, whereas CaDof18 and CaDof32 had low expression. In both buds and opening flowers, CaDof28 and CaDof33 were dominantly expressed. Interestingly, the highest transcript abundance of CaDof18 was in the bud, whereas in other tissues its expression level was nearly neglected.
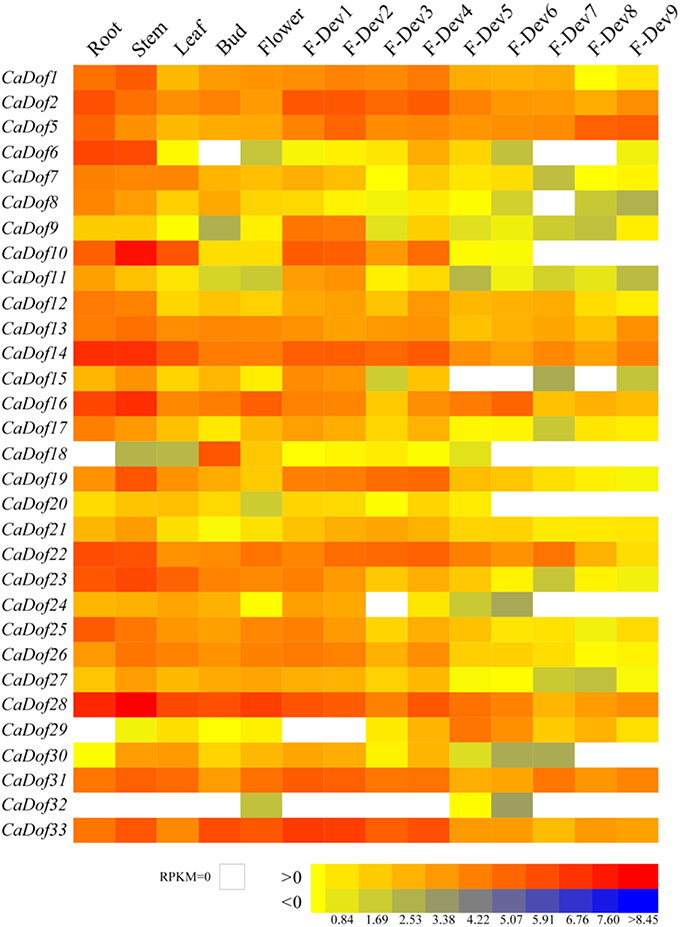
Figure 6. Heat map representation of CaDof genes across different tissues and developmental stages. The Illumina RNA-seq data were used to assess CaDofs transcript accumulation in total RNA samples extracted from root, stem, leaf, buds, flower, and fruit. The developmental fruit included nine stages, such as six pre-breaker stages (0–1 cm, 1–3 cm, 3–4 cm, 4–5 cm, and mature green fruit, Dev1–5), the breaker stage (fruit turning red, Dev6), and three post-breaker stages (3, 5, and 7 days after breaker, Dev7-9). The FPKM values were log2 transformed and heat map generated using BAR Heat Mapper Plus software. Bar at the bottom represents log2 transformed values. Genes highly or weakly expressed in the tissues are colored red and blue, respectively.
In previous study, the developmental stages of pepper fruit were divided into nine stages including five color pre-breaker stages (Dev1–5, fruit size from 0–1 cm, 1–3 cm, 3–4 cm, 4–5 cm, and mature green fruit), the breaker stage (fruit turning red, Dev6), and three post-breaker stages (Dev7–9, including 3, 5, and 7 days after breaker, respectively; Qin et al., 2014). The expression pattern of CaDof genes during fruit development was analyzed. Most of the CaDof genes, including CaDof9, CaDof10, CaDof11, CaDof19, CaDof23, CaDof26, CaDof27, and CaDof33 exhibited a gradually down-regulated expression profile during fruit development. During the fruit ripening stages (Dev 6–9, the color breaker stages), numerous CaDofs had very low expression or expression was negligible, including as CaDof6, CaDof8, CaDof10, CaDof11, CaDof15, CaDof18, CaDof20, CaDof24, CaDof30, and CaDof32. Interestingly, the expression level of CaDof5 was up-regulated from Dev5 to Dev9. However, the relative mRNA levels of CaDof2, CaDof9, CaDof10, CaDof15, and CaDof19 were significantly increased from flower to early fruit (Dev1) development, especially CaDof9, CaDof10, and CaDof11, and their expression levels we increased by more than 20 fold. These genes may be involved in pepper fruit formation. Therefore, further study of these CaDofs is vitally important and might be offer new insights into the understanding of the molecular mechanism of pepper fruit development and ripening.
Expression Profiling of CaDof Genes under Heat and Salt Stress
Functions of pepper Dof genes in response to abiotic stress are unknown. In order to elucidate the roles of CaDof genes in respone to abiotic stresses, we analyzed their expression profiles under abiotic stressors, including heat (38°C) and salinity (300 mM NaCl) treatment by using qRT- PCR in seedling leaf tissues. qRT- PCR primers are listed in Additional File 5.
Among all CaDofs, we successfully designed and verified 20 primer pairs representing 61% of the putative CaDofs, except for CaDof3, CaDof4, CaDof6, CaDof8, CaDof12, CaDof17, CaDof18, CaDof19, CaDof21, CaDof22, CaDof29, CaDof30, and CaDof32. As expected, most of the verified CaDof genes were activated and showed higher expression levels by both stressors (Additional File 6). Under heat stress, 18 of the 20 CaDof genes displayed maximum expression at 6 h, and progressively declined thereafter until 12 h (Figure 7). Some genes were slightly up-regulated, such as CaDof1, CaDof7, CaDof9, CaDof11, CaDof14, CaDof20, and CaDof24, etc. Some were strongly induced, increasing more than 10 fold (such as CaDof10, CaDof13, CaDof16, CaDof25, and CaDof28). The greatest increase in expression (nearly 13 fold) occurred in CaDof10 at 6 h of heat treatment. The results indicated that the expression of CaDofs were responsive to early heat stress.
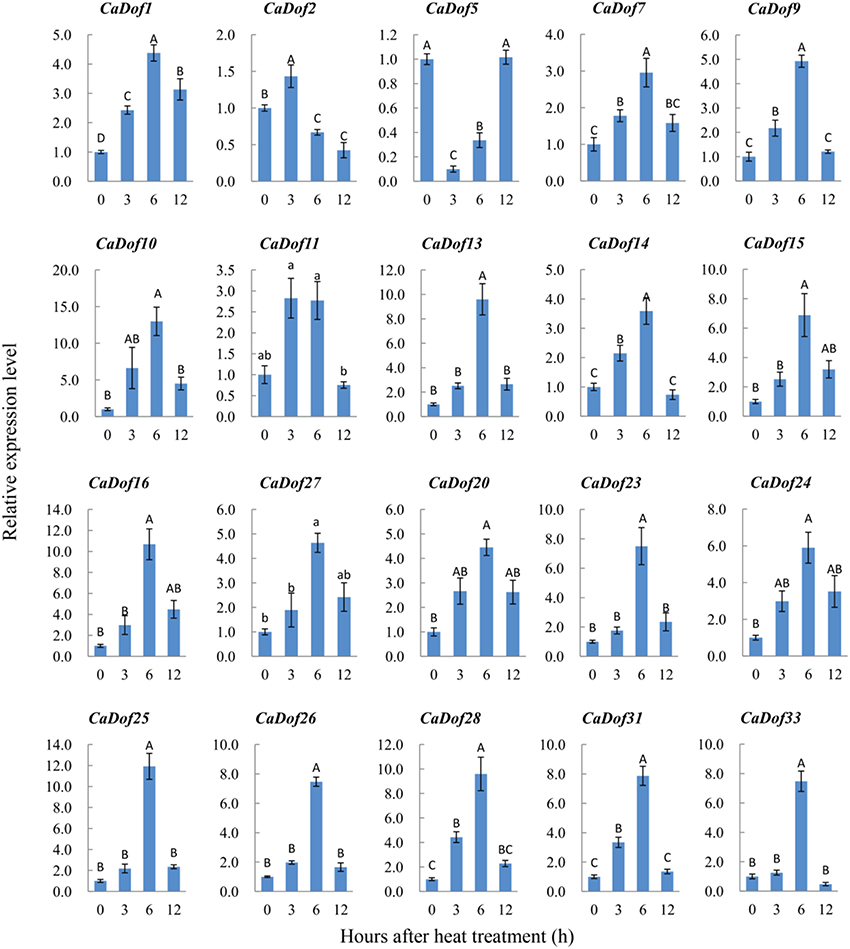
Figure 7. Expression profiles of CaDof genes in response to heat stress treatment. qRT-PCR analyses were used to assess CaDof transcript levels in the leaves sampled at 0, 3, 6, and 12 h after exposure to heat (38°C) in six seedlings leaves.
Similar to their response to heat stress, salt stress caused up-regulation of most CaDof genes. For example, CaDof10, CaDof13, CaDof15, CaDof16, CaDof24, and CaDof25 were significantly up-regulated, whereas the transcripts of CaDof1, CaDof9, and CaDof28 were slightly up-regulated (Figure 8). However, several genes, including CaDof7 CaDof11, CaDof20, and CaDof26 were not sensitive to salt stress. Not all of the CaDofs exhibited high transcript accumulation under salt stress at 6 h. Two genes (CaDof2 and CaDof31) and five others (CaDof1, CaDof15, CaDof16, CaDof24, and CaDof27) had the highest expression levels at 3 h and 12 h after treatment, respectively. It is worth mentioning that one gene (CaDof5) exhibited different responses to the two stressors. CaDof5 was markedly down-regulated under heat and salt stress treatment at 3 h, whereas it was up-regulation again at 12 h.
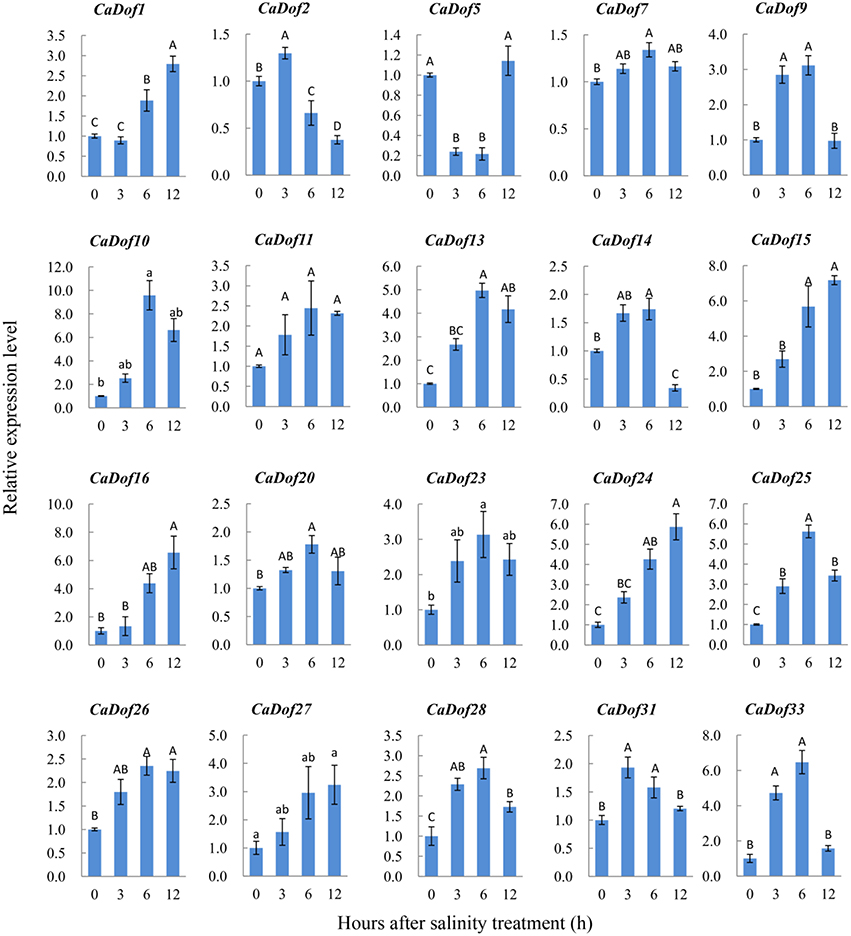
Figure 8. Expression profiles of CaDof genes in response to salt stress treatment. qRT-PCR analyses were used to assess CaDof transcript levels in the leaves sampled at 0, 3, 6, and 12 h after exposure to NaCl (300 mM) treatment in six seedlings leaves.
Discussion
Increasing evidence indicates that the Dof genes play important roles in a series of plant-specific physiological phenomena. To date, most of the research on the functions of the Dof genes has been focused in Arabidopsis (Yanagisawa, 2002; Lijavetzky et al., 2003), while it has been extremely limited in other non-model plants like pepper. In this study, we conducted a broad study of the Dof genes in pepper, including investigation of their structure, chromosomal organization, evolutionary relationships and expression profiles in different tissues and under heat or salt stress conditions. To our knowledge, this is the first comprehensive analysis of Dof genes in pepper plants.
Characteristics of Dof Family Genes in Pepper
This study revealed 33 potential Dof genes in the pepper genome. The number is quite conserved among Arabidopsis (36) (Yanagisawa, 2002), rice (30) (Lijavetzky et al., 2003), tomato (34) (Cai et al., 2013; Corrales et al., 2014), and potato (35) (Venkatesh and Park, 2015). However, it is significantly lower than that present in soybeans (78) (Guo and Qiu, 2013) and Chinese cabbage (76) (Ma et al., 2015). As we know, the genome size of pepper is almost quadrupled larger than that of tomato (3.26 GB vs. 850 Mb), and nearly seven times as large as Chinese cabbage (3.26 Gb vs. 485 Mb). Obviously, the number of Dof TFs in these species is not proportional with the genome size. This is because pepper did not undergo any whole genome duplication (WGD) after its divergence from the common Solanaceae ancestor (The Tomato Genome Consortium, 2012), confirming again the speculation that proliferation of TEs primarily contributed to pepper genome expansion (Qin et al., 2014). Besides, phylogenetic analysis showed that subgroup 7 clusters independently from those in monocot including rice and sorghum only, suggesting a potential functional diversity between dicot and monocot plants.
The intron-exon divergence was closely related to the evolutionary relationship of plants. The intron-exon organizations and intron numbers of Dof genes in pepper genome were quite similar to Arabidopsis (Yanagisawa, 2002), rice (Yanagisawa, 2002) and tomato (Cai et al., 2013). Moreover, the motif analysis indicated that the conserved C2C2-Dof domain was uniformly observed in all CaDof proteins, suggesting that CaDof TFs were evolutionarily high conserved in plants.
Potential Roles of CaDof Genes in the Tissue Differentiation and Organ Development
Previous efforts have been made to explore the association of Dof genes with plant tissue differentiation and organ development (Gupta et al., 2015; Venkatesh and Park, 2015). In the present study, we firstly explored the expression evidence for all putative CaDof genes in different tissues (root, stem, leaf, flower, and fruit) by using RNA-Seq data. The CaDof genes showed differentially expressed in various analyzed tissues as reported in other plants (Cai et al., 2013; Ma et al., 2015; Song et al., 2016). CaDof6, CaDof14, CaDof16, and CaDof28 exhibited relatively high expression levels in the root, indicating that they could play a role in the development of the plant root. Almost all CaDofs were expressed in the stem, especially CaDof28, CaDof10, CaDof14, and CaDof16 showed predominantly higher transcript abundance. It is well known that roots and stems contained abundant vascular tissue. In Arabidopsis, more than half of the members of Dof family are expressed in the vascular system (Le Hir and Bellini, 2013). Among them, AtDof2.4 and AtDof5.8 were proposed to function in the early stage but different processes for vascular development (Konishi and Yanagisawa, 2007; Gardiner et al., 2010). The CaDof28 homolog, Dof5.6/HCA2 induces the formation of inter-fascicular cambium and regulates vascular tissue development (Guo et al., 2009). We infer that CaDof28 could have similar functions during the pepper vascular tissue development.
Moreover, we identified one gene (CaDof18) that was preferentially expressed in early stage of flower. AtDOF4.7 participates in the transcriptional regulation of floral organ abscission via an effect on cell wall hydrolase gene expression (Wei et al., 2010). Cycling DOF Factor1 (CDF1) had been demonstrated that may repress transcription of CONSTANS (CO) and thus show floral delay in A. thaliana (Lucas-Reina et al., 2015). In rice, 16 Dof genes were found expressing during the grain filling process also expressed at the flowering stage indicating that these genes are involved in the regulation of genes which are required throughout the seed development (Gaur et al., 2011). It may be conjectured that CaDof18 might also play critical roles in floral development.
Fruit development and ripening is a complex and highly controlled biological process that is controlled by transcriptional regulatory networks involving many TFs, such as MADS-box, NAC, as well as EIN3/EIL, etc. (Feng et al., 2016). However, few Dofs had been well characterized their potential roles in this process. In banana, MaDof10, 23, 24, and 25 were ethylene-inducible and their transcript levels increased during fruit ripening. MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes (Feng et al., 2016). In potato, the StCDF1 was reported to control the tuber formation by repressing potato CO1/2 expression and thus allowing tuber induction (Kloosterman et al., 2013). In our study, the expression of CaDof29 gradually raised from fruit developmental stage one to stage five, and reached peak in mature green fruit (Dev5), suggesting its importance in initial stage of fruit development. Its homolog, SCAP1 (AtDof5.7) has been proven to control the final stage of guard cell differentiation by regulating the expression of multiple genes responsible for stomatal maturation and function (Negi et al., 2013). Another gene CaDof5, being homologous to Arabidopsis CDF3, showed up-regulated during fruit ripening, especially in mature red fruit (Dev8 and Dev9). We speculate that it could involve in pepper fruit ripening. However, many others, such as CaDof10, CaDof11, CaDof19, CaDof26, CaDof27, and CaDof33 exhibited a gradually down-regulated expression profile during fruit development. They might play a different role during fruit development.
Potential Roles of CaDofs in Response to Abiotic Stresses
Previous studies have shown that Dof genes play important roles in plants in responding to various biotic and abiotic stresses (Noguero et al., 2013; Gupta et al., 2015). In Arabidopsis, the OBP2 (AtDof1.1) expression level increased two to three fold within 4–6 h after MeJA treatment and mechanical wounding (Skirycz et al., 2006). In tomato, SlCDF1–5, homologs of Arabidopsis CDFs, were reported to be differentially induced in response to osmotic, salt, heat, and low-temperature stresses (Corrales et al., 2014). In Chinese cabbage, most of the BraDof genes were up-regulated quickly by salt, drought, heat, and cold stress treatments (Ma et al., 2015). In potato, most of the StDof genes are up-regulated in various abiotic stresses including drought, high salinity, and ABA. Furthermore, StDof genes show either ABA-dependent or -independent expression pattern (Venkatesh and Park, 2015). In chrysanthemum, CmDofs showed either up-regulated or down-regulated when exposed to plant hormones and abiotic stress.
In the present study, the expression profiles of 20 CaDofs exposed to heat and salt stresses were also investigated by using qRT-PCR in pepper seedlings. Consistent with previous results, majority of the CaDof genes were responsive to these two stressors, exhibiting first increased and then decreased expression profiles. Interestingly, three homologs of Arabidopsis CDFs, CaDof2, CaDof5, and CaDof14 showed different expression patterns. Maximum induction was observed under heat and salt treatment at 3 or 6 h for CaDof2 and CaDof14 respectively, with decay at 12 h. However, CaDof5 was markedly down-regulated under heat and salt stress treatment at 3 h, whereas it got to peak again at 12 h. The similar result was confirmed in tomato (Corrales et al., 2014). These results manifest the potential positive roles of CaDof genes in plant adaptation to abiotic stresses; however, much more work is needed to verify their roles.
Conclusion
In this study, 33 putative Dof transcription factor genes were identified in the pepper genome. Further, the 33 Dof transcription factors were characterized according to the conserved amino acid residues within the Dof domain, the conserved motifs and gene organization, phylogenetic analysis, and global expression profile among different tissues and under different stresses (heat and salt) by using high-throughput sequencing and qRT-PCR. The results obtained from this study provide useful clueto understand the molecular basis of the Dof gene family in the pepper and other plants in the family of Solanaceae. In particular, the expression profiling analysis of these genes will provide an important foundation for future studies assessing their functions.
Author Contributions
Conceived and designed the experiments: ZW, KH, and CQ. Performed the experiments: ZW, JC, XX, GL, XL, XC, and XT. Analyzed the data: ZW, JC, and CQ. Wrote the paper: ZW and CQ. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work was supported grants from the National Natural Science Foundation of China (31201639, 31372076), the Guangdong Natural Science Foundation of China (2014A030313593, S2011030001410), the Zunyi City Natural Science Foundation of China (No. 201201), the Guizhou Province and Zunyi City Science and Technology Cooperation Project of China (No. 201307, 201542), the Zunyi County Technology Cooperation Project (SSX201407) and Key Lab Construction Project of the Educational Department of Guizhou Province (Guizhou Education Cooperation KY [2014] 212).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00574
Additional file 1. The identified nucleotide and amino acid sequences of pepper Dof genes.
Additional file 2. Multilevel consensus sequence identified by MEME among CaDof genes in this study.
Additional file 3. The amino acid sequences of Dof genes in Arabidopsis (At), rice (Os), and tomatoes (Sl) used in phylogenetic tree construction.
Additional file 4. RNA-Seq RPKM expression matrix for all primary developmental stages and tissue types of Zunla-1.
Additional file 5. Primers used for qRT-PCR.
Additional file 6. The heat map representation of CaDof genes in response to heat and salt stress.
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Cai, X., Zhang, Y., Zhang, C., Zhang, T., Hu, T., Ye, J., et al. (2013). Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Integr. Plant Biol. 55, 552–566. doi: 10.1111/jipb.12043
Cominelli, E., Galbiati, M., Albertini, A., Fornara, F., Conti, L., Coupland, G., et al. (2011). DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biol. 11:162. doi: 10.1186/1471-2229-11-162
Corrales, A.-R., Nebauer, S. G., Carrillo, L., Fernández-Nohales, P., Marqués, J., Renau-Morata, B., et al. (2014). Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 65, 995–1012. doi: 10.1093/jxb/ert451
Diaz, I., Martinez, M., Isabel-LaMoneda, I., Rubio-Somoza, I., and Carbonero, P. (2005). The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J. 42, 652–662. doi: 10.1111/j.1365-313X.2005.02402.x
Diaz, I., Vicente-Carbajosa, J., Abraham, Z., Martínez, M., Isabel-La Moneda, I., and Carbonero, P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29, 453–464. doi: 10.1046/j.0960-7412.2001.01230.x
Dong, G., Ni, Z., Yao, Y., Nie, X., and Sun, Q. (2007). Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol. Biol. 63, 73–84. doi: 10.1007/s11103-006-9073-3
Feng, B. H., Han, Y. C., Xiao, Y. Y., Kuang, J. F., Fan, Z. Q., Chen, J. Y., et al. (2016). The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J. Exp. Bot. 67, 2263–2275. doi: 10.1093/jxb/erw032
Fornara, F., Panigrahi, K. C., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Gabriele, S., Rizza, A., Martone, J., Circelli, P., Costantino, P., and Vittorioso, P. (2010). The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 61, 312–323. doi: 10.1111/j.1365-313X.2009.04055.x
Gardiner, J., Sherr, I., and Scarpella, E. (2010). Expression of DOF genes identifies early stages of vascular development in Arabidopsis leaves. Int. J. Dev. Biol. 54, 1389–1396. doi: 10.1387/ijdb.093006jg
Gaur, V. S., Singh, U. S., and Kumar, A. (2011). Transcriptional profiling and in silico analysis of Dof transcription factor gene family for understanding their regulation during seed development of rice Oryza sativa L. Mol. Biol. Rep. 38, 2827–2848. doi: 10.1007/s11033-010-0429-z
Gualberti, G., Papi, M., Bellucci, L., Ricci, I., Bouchez, D., Camilleri, C., et al. (2002). Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14, 1253–1263. doi: 10.1105/tpc.010491
Guo, Y., Qin, G., Gu, H., and Qu, L. J. (2009). Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 21, 3518–3534. doi: 10.1105/tpc.108.064139
Guo, Y., and Qiu, L. J. (2013). 7-Genome-wide analysis of the dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 8:e76809. doi: 10.1371/journal.pone.0076809
Gupta, S., Malviya, N., Kushwaha, H., Nasim, J., Bisht, N. C., Singh, V. K., et al. (2015). Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 241, 549–562. doi: 10.1007/s00425-014-2239-3
Hernando-Amado, S., González-Calle, V., Carbonero, P., and Barrero-Sicilia, C. (2012). The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol. 12:202. doi: 10.1186/1471-2229-12-202
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Isabel-LaMoneda, I., Diaz, I., Martinez, M., Mena, M., and Carbonero, P. (2003). SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. Plant J. 33, 329–340. doi: 10.1046/j.1365-313X.2003.01628.x
Jiang, Q., Wang, F., Tan, H. W., Li, M. Y., Xu, Z. S., Tan, G. F., et al. (2015). De novo transcriptome assembly, gene annotation, marker development, and miRNA potential target genes validation under abiotic stresses in Oenanthe javanica. Mol. Genet. Genomics 290, 671–683. doi: 10.1007/s00438-014-0953-y
Kang, H. G., and Singh, K. B. (2000). Characterization of salicylic acid-responsive, arabidopsis Dof domain proteins: overexpression of OBP3 leads to growth defects. Plant J. 21, 329–339. doi: 10.1046/j.1365-313x.2000.00678.x
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kloosterman, B., Abelenda, J. A., Gomez Mdel, M., Oortwijn, M., de Boer, J. M., Kowitwanich, K., et al. (2013). Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495, 246–250. doi: 10.1038/nature11912
Konishi, M., and Yanagisawa, S. (2007). Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiol. Biochem. 45, 623–629. doi: 10.1016/j.plaphy.2007.05.001
Kushwaha, H., Gupta, S., Singh, V. K., Rastogi, S., and Yadav, D. (2011). Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol. Biol. Rep. 38, 5037–5053. doi: 10.1007/s11033-010-0650-9
Lee, S., Hong, J. C., Jeon, W. B., Chung, Y. S., Sung, S., Choi, D., et al. (2009). The salicylic acid-induced protection of non-climacteric unripe pepper fruit against Colletotrichum gloeosporioides is similar to the resistance of ripe fruit. Plant Cell Rep. 28, 1573–1580. doi: 10.1007/s00299-009-0756-5
Le Hir, R., and Bellini, C. (2013). The plant-specific dof transcription factors family: new players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 4:164. doi: 10.3389/fpls.2013.00164
Lijavetzky, D., Carbonero, P., and Vicente-Carbajosa, J. (2003). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 3:17. doi: 10.1186/1471-2148-3-17
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lucas-Reina, E., Romero-Campero, F. J., Romero, J. M., and Valverde, F. (2015). An evolutionarily conserved DOF-CONSTANS module controls plant photoperiodic signaling. Plant Physiol. 168, 561–574. doi: 10.1104/pp.15.00321
Ma, J., Li, M. Y., Wang, F., Tang, J., and Xiong, A. S. (2015). Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 16:33. doi: 10.1186/s12864-015-1242-9
Malviya, N., Gupta, S., Singh, V. K., Yadav, M. K., Bisht, N. C., Sarangi, B. K., et al. (2015). Genome wide in silico characterization of Dof gene families of pigeonpea (Cajanus cajan (L) Millsp.). Mol. Biol. Rep. 42, 535–552. doi: 10.1007/s11033-014-3797-y
Moreno-Risueno, M. A., Martínez, M., Vicente-Carbajosa, J., and Carbonero, P. (2007). The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol. Genet. Genomics 277, 379–390. doi: 10.1007/s00438-006-0186-9
Negi, J., Moriwaki, K., Konishi, M., Yokoyama, R., Nakano, T., Kusumi, K., et al. (2013). A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 23, 479–484. doi: 10.1016/j.cub.2013.02.001
Noguero, M., Atif, R. M., Ochatt, S., and Thompson, R. D. (2013). The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
Pan, Y., Li, Q., Wang, Z., Wang, Y., Ma, R., Zhu, L., et al. (2014). Genes associated with thermosensitive genic male sterility in rice identified by comparative expression profiling. BMC Genomics 15:1114. doi: 10.1186/1471-2164-15-1114
Papi, M., Sabatini, S., Altamura, M. M., Hennig, L., Schäfer, E., Costantino, P., et al. (2002). Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol. 128, 411–417. doi: 10.1104/pp.010488
Park, D. H., Lim, P. O., Kim, J. S., Cho, D. S., Hong, S. H., and Nam, H. G. (2003). The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 34, 161–171. doi: 10.1046/j.1365-313X.2003.01710.x
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Santopolo, S., Boccaccini, A., Lorrai, R., Ruta, V., Capauto, D., Minutello, E., et al. (2015). DOF AFFECTING GERMINATION 2 is a positive regulator of light-mediated seed germination and is repressed by DOF AFFECTING GERMINATION 1. BMC Plant Biol. 15:72. doi: 10.1186/s12870-015-0453-1
Shaw, L. M., McIntyre, C. L., Gresshoff, P. M., and Xue, G. P. (2009). Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct. Integr. Genomics 9, 485–498. doi: 10.1007/s10142-009-0130-2
Skirycz, A., Radziejwoski, A., Busch, W., Hannah, M. A., Czeszejko, J., Kwasniewski, M., et al. (2008). The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J. 56, 779–792. doi: 10.1111/j.1365-313X.2008.03641
Skirycz, A., Reichelt, M., Burow, M., Birkemeyer, C., Rolcik, J., Kopka, J., et al. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47, 10–24. doi: 10.1111/j.1365-313X.2006.02767.x
Song, A., Gao, T., Li, P., Chen, S., Guan, Z., Wu, D., et al. (2016). Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium. Front. Plant Sci. 7:199. doi: 10.3389/fpls.2016.00199
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
The Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Venkatesh, J., and Park, S. W. (2015). Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 94, 73–85. doi: 10.1016/j.plaphy.2015.05.010
Ward, J. M., Cufr, C. A., Denzel, M. A., and Neff, M. M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17, 475–485. doi: 10.1105/tpc.104.027722
Wei, P. C., Tan, F., Gao, X. Q., Zhang, X. Q., Wang, G. Q., Xu, H., et al. (2010). Overexpression of AtDOF4.7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis. Plant Physiol. 153, 1031–1045. doi: 10.1104/pp.110.153247
Wen, C.-L., Cheng, Q., Zhao, L., Mao, A., Yang, J., Yu, S., et al. (2016). Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 6:23072. doi: 10.1038/srep23072
Xu, X., Shen, Y., Fu, J., Lu, L., and Li, J. (2014). De novo assembly of the grass carp Ctenopharyngodon idella transcriptome to identify miRNA targets associated with motile aeromonad septicemia. PLoS ONE 9:e112722. doi: 10.1371/journal.pone.0112722
Yanagisawa, S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560. doi: 10.1016/S1360-1385(02)02362-2
Yanagisawa, S., and Schmidt, R. J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17, 209–214.
Keywords: pepper, DNA-binding one zinc finger, phylogenetic analysis, expression analysis, heat stress, salinity stress
Citation: Wu Z, Cheng J, Cui J, Xu X, Liang G, Luo X, Chen X, Tang X, Hu K and Qin C (2016) Genome-Wide Identification and Expression Profile of Dof Transcription Factor Gene Family in Pepper (Capsicum annuum L.). Front. Plant Sci. 7:574. doi: 10.3389/fpls.2016.00574
Received: 12 January 2016; Accepted: 13 April 2016;
Published: 29 April 2016.
Edited by:
Sergio Lanteri, University of Turin, ItalyReviewed by:
Agata Gadaleta, University of Bari, ItalyAiping Song, Nanjing Agricultural University, China
Copyright © 2016 Wu, Cheng, Cui, Xu, Liang, Luo, Chen, Tang, Hu and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kailin Hu, hukailin@scau.edu.cn;
Cheng Qin, qincheng1001@163.com
 Zhiming Wu
Zhiming Wu Jiaowen Cheng2
Jiaowen Cheng2 Cheng Qin
Cheng Qin