- Institute for Plant Physiology, Justus Liebig University, Giessen, Germany
Phytochromes are the principle photoreceptors in light-regulated plant development, primarily acting via translocation of the light-activated photoreceptor into the nucleus and subsequent gene regulation. However, several independent lines of evidence indicate unambiguously that an additional cytoplasmic signaling mechanism must exist. Directional responses in filament tip cells of the moss Physcomitrella patens are steered by phy4 which has been shown to interact physically with the blue light receptor phototropin at the plasma membrane. This complex might perceive and transduce vectorial information leading to cytoskeleton reorganization and finally a directional growth response. We developed yeast two-hybrid procedures using photochemically functional, full-length phy4 as bait in Physcomitrella cDNA library screens and growth assays under different light conditions, revealing Pfr-dependent interactions possibly associated with phytochrome cytoplasmic signaling. Candidate proteins were then expressed in planta with fluorescent protein tags to determine their intracellular localization in darkness and red light. Of 14 candidates, 12 were confirmed to interact with phy4 in planta using bimolecular fluorescence complementation. We also used database information to study their expression patterns relative to those of phy4. We discuss the likely functional characteristics of these holophytochrome-interacting proteins (HIP’s) and their possible roles in signaling.
Introduction
Plant phytochromes absorb predominantly red (R; ∼660 nm) and far-red (FR; ∼710–730 nm) light, thereby steering physiological responses including seed germination, de-etiolation, shade avoidance, and flowering. The R-absorbing Pr dark state is restricted to the cytoplasm and is physiologically inactive. In light it is converted to the physiologically active FR-absorbing Pfr state, which is then translocated from the cytoplasm to the nucleus where it regulates the transcription of numerous genes via direct interaction with master transcription factors such as the PIF’s, leading to the long-term responses mentioned above. However, several physiological responses cannot be explained by gene regulation. Firstly, responses like R/FR-dependent ion fluxes (Tanada, 1968) or cytoplasmic streaming (Takagi et al., 2003) occur within seconds, far too fast for such a mechanism. Secondly, the directional responses in higher plants mediated by the blue light (B) absorbing photoreceptor phototropin are modulated by phytochromes: although this might derive from phytochrome effects on gene expression, several of these responses are still seen in mutants in which nuclear translocation is defective, implying that Pfr is also physiologically active in the cytoplasm (Rösler et al., 2007; Kami et al., 2012). Indeed, Arabidopsis phyA and phyB Pfr bind and phosphorylate PKS1, a cytoplasmic protein, whereas phyB regulates translation of PORA by Pfr-dependent binding to PENTA1, also a cytoplasmic protein (Fankhauser et al., 1999; Paik et al., 2012). The fact that phytochromes can connect to signal transduction systems independently of transcription/translation is even more obvious in bryophytes. Phototropic growth of moss filament tip cells is steered by R in a FR-reversible manner, indicating that the photoreceptor is phytochrome. This cannot result from transcription/translation regulation because directional information would inevitably be lost in the process. Furthermore, gene targeting in Physcomitrella and Ceratodon showed that the response is steered by a specific phytochrome, phy4 (Mittmann et al., 2004, 2009). Thus phy4 must be associated with a signaling mechanism which faithfully transmits the vectorial information given by the incident light. Indeed, we found that phy4 interacts with phototropin at the plasma membrane (Jaedicke et al., 2012), suggesting a physical link to the machinery controlling cytoskeletal organization and tip growth (Meske and Hartmann, 1995; Meske et al., 1996). Chloroplast translocation too is regulated vectorially by phy4 (Kadota et al., 2000; Mittmann et al., 2004). Interestingly, in both the fern Adiantum and the alga Mougeotia chloroplast movements are also steered vectorially in a R/FR-reversible manner, indicative of phytochrome action – but in these cases via neochrome, a chimeric photoreceptor comprising a phytochrome sensory module attached to a phototropin. Remarkably, transgenic neochrome in Arabidopsis rescues phototropism in phototropin-deficient mutants – but the response is then seen in both B and R (Nozue et al., 1998; Kanegae et al., 2006; Kanegae and Kimura, 2015). The field of phytochrome cytoplasmic signaling has been reviewed recently (Hughes, 2013).
It remains unclear, however, how phytochrome cytoplasmic signals are transmitted. Even assuming that the phytochrome simply hijacks the phototropin-associated machinery is futile as little is known about phototropin signaling beyond NPH3 (Roberts et al., 2011). We therefore decided to search for Physcomitrella phy4 partners directly, describing our initial results here. We developed Y2H procedures using full-length, photochemically functional, R-activated phy4 holophytochrome as bait in cDNA library screens: to our knowledge this is the first report of such a procedure. The apparent interaction was checked with the full-length CDS model and R/FR reversibility tested. Candidate interactors were then investigated in planta regarding their intracellular localization in darkness (D) and R by fusing fluorescent tags to the N- and C-termini of the respective gene products. Finally in planta interaction with phy4 was investigated using bimolecular fluorescence complementation (split-YFP) methods. We thereby identified 14 putative holophytochrome interacting proteins (HIP’s), 12 of which were confirmed in split-YFP. Their possible roles in phytochrome cytoplasmic signaling are discussed.
Materials and Methods
Cloning and Y2H Procedures
The MatchMaker GAL4 Two-Hybrid System 3 (Clontech) was used to screen for potential cytoplasmic signaling partners of Physcomitrella phy4 (Pp3c27_7830V1.1). Full-length phy4 bait was cloned from first strand Physcomitrella cDNA into pGBKT7 and pBAC, a derivate of pBRIDGE (both Clontech), to yield hybrid proteins with the BD attached to the N- and C-termini (BD:phy4 and phy4:BD), respectively. A pre-transformed oligo dT-primed cDNA library in the prey vector pGADT7 was kindly provided by Hans Sommer (MPI Cologne) in yeast strain AH109. Single, double and screening transformations were performed as described (Agatep et al., 1998; Gietz and Woods, 2002). The library was either transformed with 40 μg BD:phy4 in a 40x scale or mated to Y187 yeast pre-transformed with phy4:BD. For mating, the bait strain was grown overnight, harvested and mixed in a in a 2:1 ratio with cells of the thawn library aliquot in 2x YPDA medium and incubated for 24 h at 40 rpm and 30°C. Transformation and mating mixtures were plated either with water or 0.5x YPDA on -Trp/-Leu/-His TSD medium with 2.5 mM 3-AT or with 0.25 mM 3-AT and 30 μM PCB, respectively. PCB was extracted from Spirulina and purified as described (Jaedicke et al., 2012). Plates were incubated for 14 days at 30°C in D or in 0.7 μmol m-2 s-1 Rc (660 nm LEDs). Fresh PCB was added 2–3 times during the incubation period. Positives were picked and transferred to a -Trp/-Leu DSD (double SD) masterplate and also further selected on TSD with 3-AT and on -Trp/-Leu/-His/-Ade QSD (quadruple SD) in the case of double transformation and TSD with PCB in case of library mating. DNA extracts were transformed into E. coli, the cloned pGADT7 DNA extracted, the inserts sequenced and finally identified by BLAST searches at NCBI1 and Cosmoss2. Based on EST and our own sequencing data, the likely best gene model was selected and the corresponding amino acid sequences were BLASTed against Arabidopsis thaliana and Viridiplantae non-redundant protein databases at NCBI in order to identify putative homologs in higher plants.
AD:HIP4 and AD:HIP6 constructs were cloned by Phusion amplification of cDNA, subcloning into pCR bluntII TOPO (Invitrogen) and ClaI/SacI cloning into pGADT7. All other constructs of candidate phy4-interacting partners were obtained by Gateway cloning. Full length entry clones were generated by Phusion amplification of cDNA in a 2-step PCR whereby attB1 and attB2 sites were attached to the product. For N-terminal and C-terminal tags, the CDS was amplified with and without stop codons, respectively. Gel-purified PCR products were cloned into pDONR207 with BP-clonase (Invitrogen). PRL1 and PLP entry clones were created by TA-cloning of PCR products into pCR8GW TOPO (Invitrogen). The resulting full length entry clones were used to create expression constructs for Y2H assays and cytological analyses via the LR reaction. Inserts were recombined into AD: and :AD prey vectors pGADT7g (Uetz and Grigoriev, 2005) and pGADCg (Stellberger et al., 2010), respectively, and transformed into yeast together with BD:phy4 and phy4:BD holophytochrome baits for semi-quantitative growth assays in order to verify the interaction and its potential light-dependency. For this, 2 × 105 doubly transformed yeast cells were spotted on TSD medium with 3-AT and 30 μM PCB and incubated for 5 days in continuous 0.7 μmol m-2 s-1 (Rc) or 12 min/h red light (660 nm LEDs, 4 μmol m-2 s-1) pulses (Rp) or red followed by 12 min/h far-red (740 nm LEDs, 5 μmol m-2 s-1) pulses (Rp+FRp) or darkness (D). Various controls were included to ensure that appropriate interactions were being monitored (see also Jaedicke et al., 2012). Constitutive dimerization was demonstrated by BD:phy4 and phy4:BD baits combined with the AD:phy4 prey. As negative controls and for the adjustment of appropriate 3-AT concentrations, BD:phy4 and phy4:BD baits were combined with the empty pGADT7 vector (from this 2.5 mM or 1 mM 3-AT were used for doubly transformed yeast growth assays). As a control for successful holophytochrome formation, BD:phyA (in pGBKT7) and phyA:BD (in pBAC) baits were combined with AD:FHY1 (1 mM 3-AT) as this Y2H interaction is Pfr-dependent (Hiltbrunner et al., 2005).
For intracellular localization studies, each partner CDS was Gateway-cloned into modified destination vectors p2CGW7-2 × 35S, p2GWC7-2 × 35S, p2FGW7-2 × 35S, and p2GWF7-2 × 35S (Karimi et al., 2002), creating N- or C-terminal CFP- or GFP-fusions, respectively. For in planta interaction studies, the CDS was recombined into pSAT4-DEST-n(1-174)eYFP-CI(YFPN:) and pSAT4(A)-DEST-n(1-174)eYFP-N1(:YFPN) or in pSAT5-DEST-C(175-end)eYFP-CI (YFPC:) and pSAT5(A)-DEST-c(175-end)EYFP-N1 (:YFPC) destination vectors (Citovsky et al., 2006). phy4 pSAT constructs were created as described (Jaedicke et al., 2012). Negative controls for interaction were provided by empty YFPC and YFPN vectors in combination with the HIP pSAT expression plasmids.
In Planta Localization and Interaction Studies
Biolistic transfection of Physcomitrella filaments was performed as described (Jaedicke et al., 2012). For subcellular protein localization studies, transfected filaments were incubated for 2 days in D and observed via fluorescence microscopy without pre-treatment and following 1 h R pre-treatment (660 nm LEDs, 3 μmol m-2 s-1). For split-YFP studies, plant material was incubated for 3 days in D and then subjected to confocal microscopy with and without analogous pre-treatments.
Transformed cells were identified by R fluorescence of the co-transfected nuclear marker mCherry:VirD2NLS using a Leica Z16 apo fluorescence macroscope. For localization and split-YFP studies, an automated DM6000b fluorescence microscope and a TCS SP2 AOBS confocal laser scanning microscope (both Leica) were used, respectively. For filter cube and look-up table details, see Jaedicke et al. (2012). Single scans of YFP fluorescence were recorded with 4x line and 6x frame average. In order to distinguish YFP signals from possible mCherry bleed-through, YFP emission detection was narrowed to 525–535 nm. The red mCherry- and chlorophyll autofluorescence and the transmission images were taken with 4x line and 2x frame average. Adobe Photoshop CS5 was used for image processing (enhancement of brightness and contrast).
In Silico Analyses
The protein sequences derived from the relevant HIP gene models were further analyzed to identify domains and thus potential functions. Co- and possible synexpression with phy4 was investigated using data from eFP, Phytozome and Genevestigator with the help of ClustVis3,4,5,6 .
Results and Discussion
Y2H Library Screens Revealed 14 Putative HIP’s
We developed Y2H methods to screen for phy4-interacting proteins that might be involved in the phy4 cytoplasmic signaling pathway. Importantly, instead of photochemically impotent fragments, we established the use of functional, full-length holophytochrome as bait by feeding the transgenic yeast cells with PCB to allow auto assembly in vivo, as evidenced by R/FR reversible interaction of Arabidopsis phyA with FHY1 (Supplementary Figure S1) and, subsequently, of phy4 with various putative partners. We generated bait constructs with the BD N- and C-terminally fused to full-length phy4, carefully characterizing their homodimerization and autoactivation properties. The N-terminal fusion thereby showed a higher autoactivation of the HIS3-reporter than the C-terminally fused bait. Accordingly, 3-AT (a competitive inhibitor of the HIS3 gene product) was included at 2.5 and 1 mM, respectively. Both phy4 baits showed constitutive homodimerization together with the phy4 prey fused N-terminally to the AD (Supplementary Figure S1). We screened the pre-transformed cDNA library with both apo-BD:phy4 and holo-phy4:BD in Rc via sequential double transformation and mating, respectively. Functionality of the mating protocol for holophytochrome formation was confirmed by mating Y187 phyA:BD to AH109 AD:FHY1 and selecting on PCB supplemented medium in Rc (Supplementary Figure S1). The yeast mating protocol evoked substantially less autoactivation than the transformation protocol, necessitating minimal 3-AT (0.25 mM only for phy4:BD). Therefore we expected also weak or temporary interactions with phy4 Pfr to be identifiable.
The 4.2 × 105 clones were screened with phy4:BD bait, corresponding to about fourfold coverage of the Physcomitrella transcriptome. 1 × 104 clones were additionally screened with BD:phy4 bait. The 69 and 75 clones, respectively, survived two rounds of selection and were sequenced, following which 54 and 23 yeast clones excluding duplicates were subjected to BLAST analyses at Cosmoss to identify the corresponding loci in the Physcomitrella genome (see Supplementary Tables S1 and S2). Annotation was improved by domain analysis and by BLAST searches of the respective protein sequences against the Viridiplantae and Arabidopsis non-redundant protein sequence databases. On this basis, genes which were considered unlikely to be involved in signaling were not considered further, leaving candidates putatively involved in signaling processes, posttranslational modification, transcription/translation and likely membrane, transport- or cytoskeleton-associated proteins. We subsequently analyzed 11 and 4 candidates, respectively, by cloning the full-length cDNA into AD: and :AD prey plasmids and combining them in yeast with full-length photoactive BD:phy4 and phy4:BD bait in semiquantitative growth assays under different light conditions (Supplementary Figure S2). Only one now failed to interact with BD:phy4 and phy4:BD (data not shown), perhaps because a serendipitous binding site shown by the partial cDNA expression product was lost in the full length configuration. The remaining 14 candidates showed interaction with BD:phy4 and/or phy4:BD full-length holophytochrome bait either constitutively or, more commonly, in a Rp-enhanced (FRp-reversible) manner. These were thus provisionally termed holophytochrome-interacting proteins (HIP1–14).
Verification and Characterization of HIP’s
The cellular localization of each putative HIP was subsequently studied in planta using Physcomitrella filaments transfected with appropriate fluorescent protein hybrid constructs in D and after R pre-treatment. As a rule the same cells were observed before and after illumination. Finally, interaction in planta between each putative HIP and phy4 was studied using bimolecular fluorescence complementation (in D and after R pre-treatment) with split-YFP hybrid proteins, whereby the cellular localization of the interaction itself was documented. In all cases, hybrids in which the additional domains were fused either to the N- or the C-termini of the putative HIP and phy4 were generated, thereby accounting both for possible functional artifacts arising from the fusion and for topological differences that might affect the interaction readout. Respective split-YFP negative controls are shown in Supplementary Figure S3. Twelve out of the 14 putative HIP’s were thereby proven to interact in planta. The two putative HIP’s which failed to show an in planta split-YFP signal might interact too weakly for YFP reconstitution or represent false positives from Y2H. The first of these, Pp3c19_20830V1.1 (initially named HIP2) represents a likely pirin-related protein (see HIP1 below). Also depending on the configuration, this protein localized to the cytoplasm or to cytoplasm and nucleus, respectively (see Supplemetary Figure S5). The second, Pp3c1_11190C1.1 (Cosmoss Version 3 accession number; initially named HIP10), encodes a protein with an N-terminal ankyrin- and a C-terminal BTB/POZ (Boad-complex, Tamtrac, Bric a brac/POx virus and ZF)-domain (see Supplemetary Figure S4). CFP-fusions were localized either in cytosolic speckles or uniformly in the cytoplasm and nucleus, depending on the FP-configuration. Our results for the 12 HIP’s confirmed in planta are described below.
HIP1 (Pp3c2_10320V1.1)
Independent library screens revealed two different members of the pirin iron-containing subgroup of the cupin superfamily, (HIP1, 38.7 kDa) and Pp3c19_20830V1.1 (initially HIP2, 33.9 kDa). Semiquantitative studies of full length CDS models showed strong and weaker binding to phy4 holophytochrome in Y2H, respectively. Indeed except in the case of HIP1–BD:phy4, the interaction was R/FR reversible (Supplementary Figure S2). However, as Pp3c19_20830 showed insignificant split-YFP-signals (Supplementary Figure S5B) it was excluded as a genuine HIP. In Physcomitrella HIP1 was distributed in the cytoplasm and nucleus (Figure 1A). In planta interaction was verified for HIP1 in the phy4:YFPC–YFPN:HIP1 configuration (Figure 1B) although the result was ambiguous because the signal was detectable either in cytoplasm and nucleus or in cytosolic speckles and nucleus in different cells both in D and following R pre-treatment.
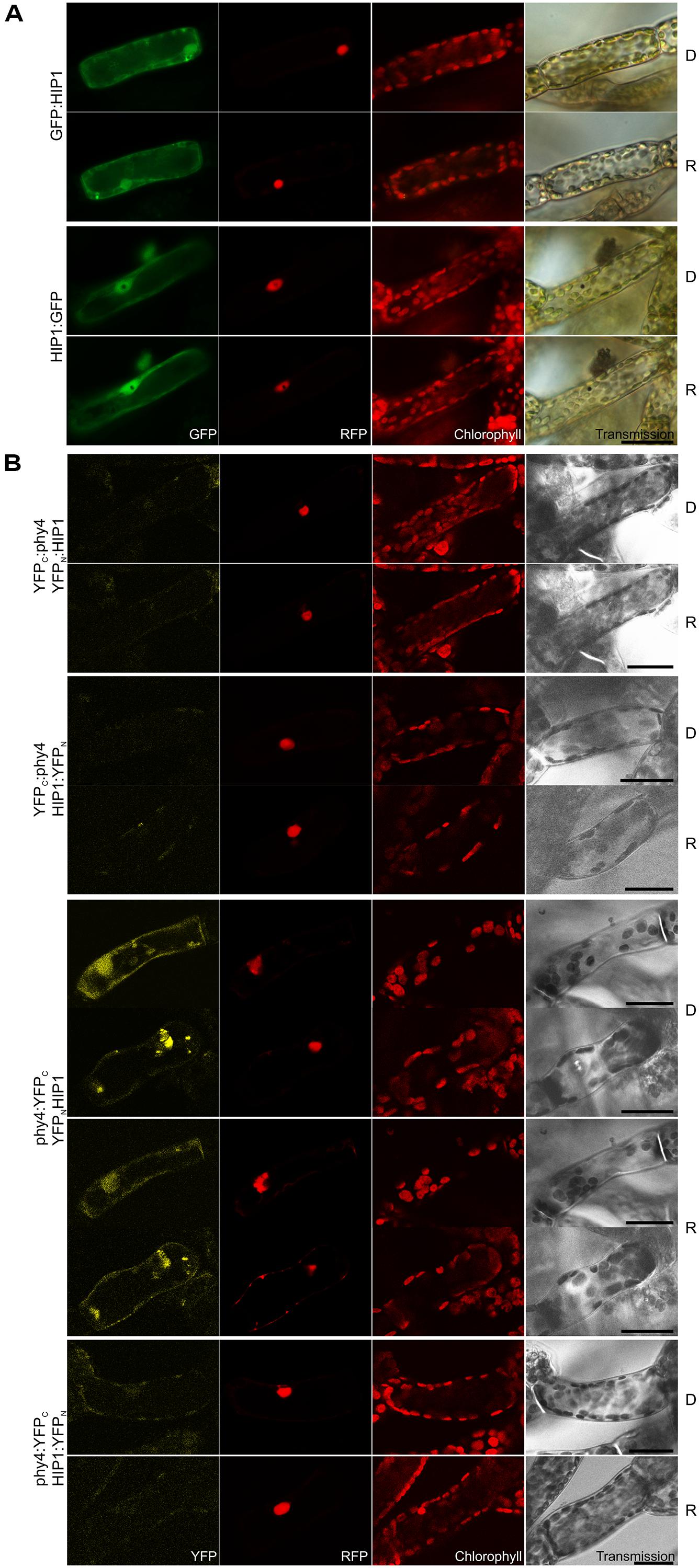
FIGURE 1. HIP1 (Pp3c2_10320V1.1) intracellular localization (A) and split-YFP-studies of HIP1–phy4 interaction (B) each without (D) and with red pre-treatment (R) using fluorescence- and confocal microscopy, respectively. (A) Green fluorescence of N- and C-terminal HIP1:GFP fusions monitored by GFP (column 1) and was visible in nucleus and cytoplasm in both cases; Red fluorescence of the co-transfected mCherry:NLS nuclear marker (RFP) is shown in column 2; Chlorophyll autofluorescence (chlorophyll) and a differential interference contrast image (DIC) is shown in columns 3 and 4, respectively. For each FP configuration cells were photographed before (D) and after red light pre-treatment (R). The apparent “hole” in the nucleus of the HIP1:GFP fluorescence images and the corresponding black dot in the DIC images represents a large gold particle. (B) All possible split-YFP configurations were analyzed without (D) and with red pre-treatments (R) via confocal microscopy as shown in column 1: YFPC:phy4–YFPN:HIP1 (rows 1+2), YFPC:phy4–HIP1:YFPN (rows 3 and 4), phy4:YFPC–YFPN:HIP1 (rows 5–8) and phy4:YFPC–HIP1:YFPN (rows 9 and 10). Analogous to panel (A), columns depict (from left to right) YFP fluorescence, RFP fluorescence from mCherry:NLS, chlorophyll autofluorescence and a transmission image. Two different signal patterns were observed for phy4:YFPC–YFPN:HIP1, thus representative images are shown for each. Other configurations yielded only background signals alongside channel bleed-through. Scale bars 30 μm.
HIP1 is apparently expressed under all conditions and in all cell types although weekly in leaflets (see Supplementary Materials). The data provide little evidence for synexpression with phy4.
Holophytochrome-interacting protein 1 (like Pp3c19_20830) harbors an N-terminal cupin domain including the potential metal-coordinating residues H116, H118, H160, and E162 corresponding to H57, H59, H101, and E103 in the human sequence (Pang et al., 2004; Adams and Jia, 2005). The pirin-typical C-terminal domain (which resembles the cupin domain while lacking the metal-coordinating residues) is also present in HIP1 (but not Pp3c19_20830). Conserved in mammals, fungi, plants and prokaryotes, four and three genes encoding pirin-like proteins exist in Arabidopsis and Physcomitrella, respectively (see Summary Table 1 in the Supplementary Material).
Pirin functions are quite diverse. Human, Arabidopsis and E. coli pirins have been shown to possess quercetinase activity (Adams and Jia, 2005). Quercetin – amongst other things – plays a role in UV acclimation (Hectors et al., 2014) and has been shown to inhibit polar auxin transport (Fischer et al., 1997; Kuhn et al., 2011; Buer et al., 2013) and thus might play a role in tropic responses (Lewis et al., 2011). Indeed, light-grown prn1- Arabidopsis seedlings showed disoriented hypocotyl growth, whereas etiolated mutant seedlings showed enhanced UV-induced quercetin levels (Orozco-Nunnelly et al., 2014). In Arabidopsis quercetin is localized in and around the nucleus, in the endomembrane system and at the plasma membrane (Peer et al., 2001). Although this would correlate with the phy4–phototropin complex at the plasma membrane (Jaedicke et al., 2012) our data do not imply that HIP1 is localized at the plasma membrane (Figure 1). Diverse physiological effects have been associated with pirins in plants, including pathogen susceptibility, apoptosis, ABA-suppression of seed germination and B-induced LHC expression, possibly in association with G-proteins (Orzaez et al., 2001; Lapik and Kaufman, 2003; Warpeha et al., 2007; Zhang et al., 2014). Phytochrome signaling via G-proteins is controversial, however (Neuhaus et al., 1993; Okamoto et al., 2001; Jones et al., 2003; Urano et al., 2013). The Physcomitrella genome codes for at least one G-protein but this has not been studied to-date.
HIP3 (Pp3c10_4820V1.1)
Holophytochrome-interacting protein 3 (37.4 kDa) carries CHY, CTCHY, and C3HC4 (referring to a specialized cysteine-histidine pattern) RING-type ZF domains. Y2H analysis with the full-length CDS showed weak but R-enhanced interaction with phy4 in all conformations. phy4:BD–ADg:HIP3 showed interaction in Rc but not in Rp (Supplementary Figure S2). In planta, although no particular changes in localization were seen upon irradiation, HIP3 showed cytosolic or nuclear and cytosolic localization in approximately half of the observed cells, respectively (CFP:HIP3) or cytosolic and nuclear (HIP3:CFP) localization, depending on the position of the CFP tag (see Figure 2A). Cytosolic localization is interesting because both fusion proteins might be small enough (37 kDa + 27 kDa tag) to enter the nucleus passively, at least as monomers. Conventional NLS and NES motifs are not apparent in the HIP3 sequence. In planta interaction was seen for phy4:YFPN–HIP3:YFPC in the nucleus and the cytoplasm, consistent with the localization data, the other combinations yielding only background signals alongside channel bleed-through (see Figure 2B). Interestingly, the split-YFP signal only appeared after R pre-irradiation, implying Pfr-dependent interaction in planta as in Y2H.
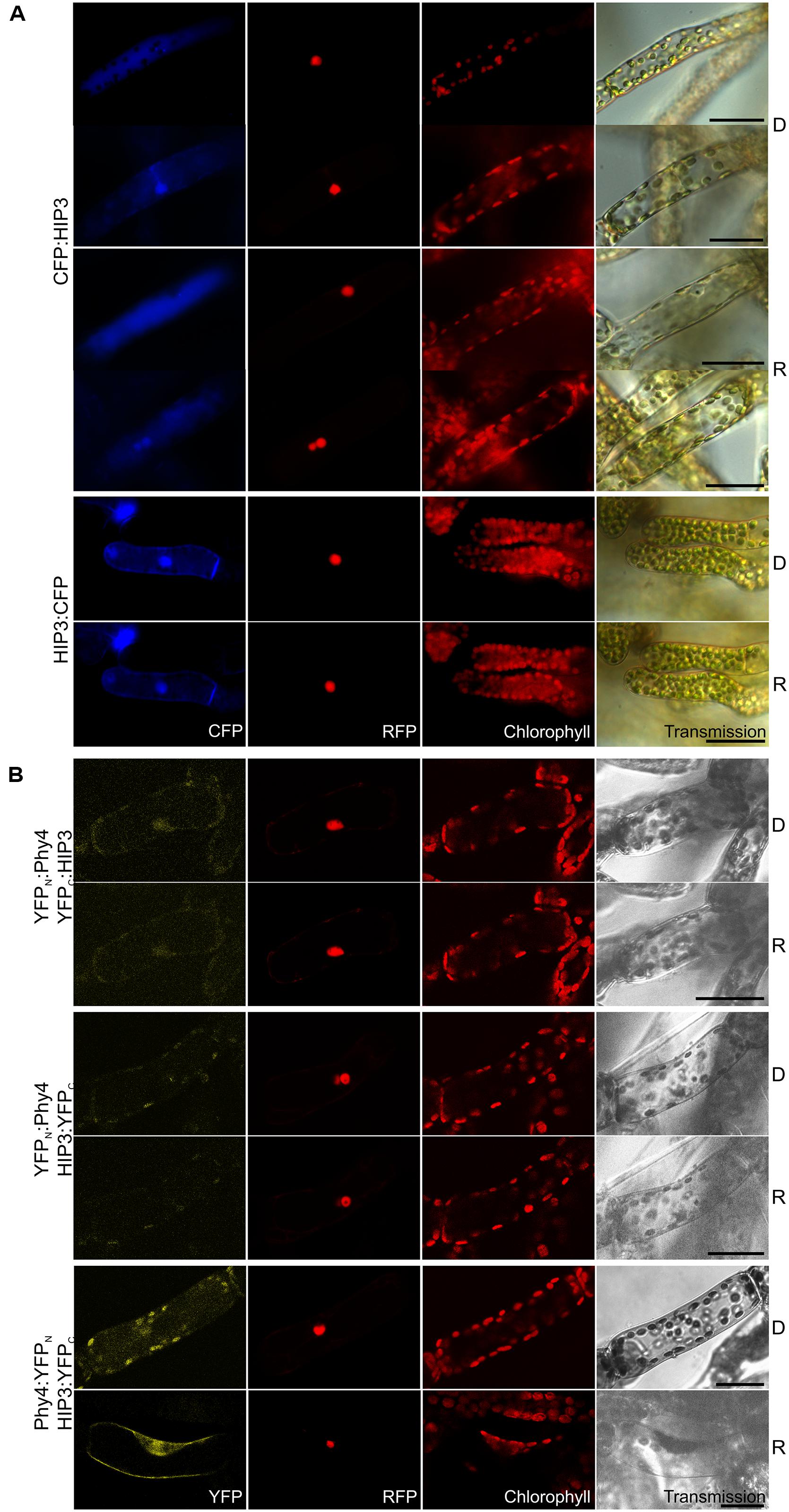
FIGURE 2. HIP3 (Pp3c10_4820V1.1) intracellular localization (A) and split-YFP-studies of HIP3–phy4 interaction (B) each without (D) and with red pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) CFP:HIP3 (rows 1–4) and HIP3:CFP (rows 5 and 6). CFP:HIP3 localization was ambiguous, being observable either in the nucleus and cytoplasm (rows 2 and 4) or in the cytoplasm only (rows 1 and 3) in both conditions, whereas HIP3:CFP showed nuclear and cytoplasmic localization (column 1, CFP). (B) Split-YFP analyses of YFPN:phy4–YFPC:HIP3 (rows 1 and 2), YFPN:phy4–HIP3:YFPC (rows 3 and 4) and phy4:YFPN–HIP3:YFPC (rows 5 and 6) configurations. Significant nucleo-cytoplasmic signals were only seen for phy4:YFPN–HIP3:YFPC after R pre-treatment (column 1, YFP; D and R here from separate experiments). All other configurations/conditions yielded only background signals alongside channel bleed-through. Scale bars 30 μm.
Holophytochrome-interacting protein 3 is apparently expressed under all conditions and in all cell types except in dehydrated material (see Supplementary Materials). The data provide little evidence for synexpression with phy4.
Zinc finger-proteins are known to bind diverse ligands including DNA, RNA, lipids, and proteins (Laity et al., 2001; Matthews and Sunde, 2002), thereby executing functions in a variety of cellular processes such as transcription, translation, cytoskeleton organization, protein folding, and others. The CHY-ZF is named due to its conserved motif CxHY, the function of which is still unknown (Leng et al., 2003). The 40–60 residues comprising RING-type ZF’s, however, are considered to bind two zinc atoms and to mediate protein–protein interactions (Freemont, 1993; Borden and Freemont, 1996; Saurin et al., 1996). RING-type ZFs can also serve as E3-ubiquitin ligases in protein degradation and are thus commonly involved in gene regulation via repressor destruction in plants. BLAST searches indeed revealed high similarity between HIP3 and 4–6 other Physcomitrella and at least seven Arabidopsis proteins. For example, Arabidopsis RING FINGER AND CHY ZINC FINGER DOMAIN-CONTAINING PROTEIN 1 (AT5G22920) shows 63% identity in the 250 residue region of similarity. This nuclear-localized E3 ubiquitin ligase is involved in the regulation of stomatal opening, its activity being regulated via phosphorylation by SnRK2.6 (Ding et al., 2015). CHY and CTCHY and RING-type ZF protein (AT5G25560), a PGPD 14-like protein (Kosarev et al., 2002) of unknown function, shows 57% identity except at the N-terminus. Similarly, MYB30-INTERACTING E3 LIGASE 1 (AT5G18650) with 59% identity also except in the 64 residue N-terminal region) degrades the MYB30 transcription factor, a negative regulator of defense responses in Arabidopsis (Marino et al., 2013). Several known Arabidopsis phytochrome-interacting and/or signaling molecules are known to possess RING-ZF domains, for example RFI2, which plays a role in phytochrome-controlled seedling de-etiolation (Chen and Ni, 2006) or VOZ1, a phyB partner controlling flowering (Yasui et al., 2012). The central regulator of photomorphogenesis, the E3 ubiquitin ligase COP1 also contains a C3HC4-type RING-ZF motif. Moreover, the PENTA protein involved in translational regulation of PORA (the key light-regulated enzyme in chlorophyll biosynthesis) by interaction with phyB is also a C3H-type ZF protein (Paik et al., 2012).
HIP4/PRL1 (Pp3c16_8560E1.1)
Holophytochrome-interacting protein 4 (53.9 kDa) is encoded by PRL1 sharing 62% sequence identity with its homolog in Arabidopsis. In Y2H studies full length HIP4/PRL1 interacted with holo phy4 in all four bait–prey configurations. Three interactions were R-enhanced and partially FR-reversible whereas BD:phy4–HIP4:ADg interacted constitutively (Supplementary Figure S2). Independently of the light treatment, HIP4:GFP localized to both nucleus and cytoplasm while CFP:HIP4 was concentrated in the nucleus (Figure 3A). Constitutive interaction was confirmed in planta in all combinations except phy4:YFPC–HIP4:YFPN (Figure 3B), the reconstituted split-YFP signal being apparent in both nucleus and cytoplasm for phy4:YFPC–YFPN:HIP4 and, less clearly, YFPC:phy4–HIP4:YFPN, corresponding to the localization of HIP4:GFP. YFPC:phy4–YFPN:HIP4 interacted in the cytoplasm.
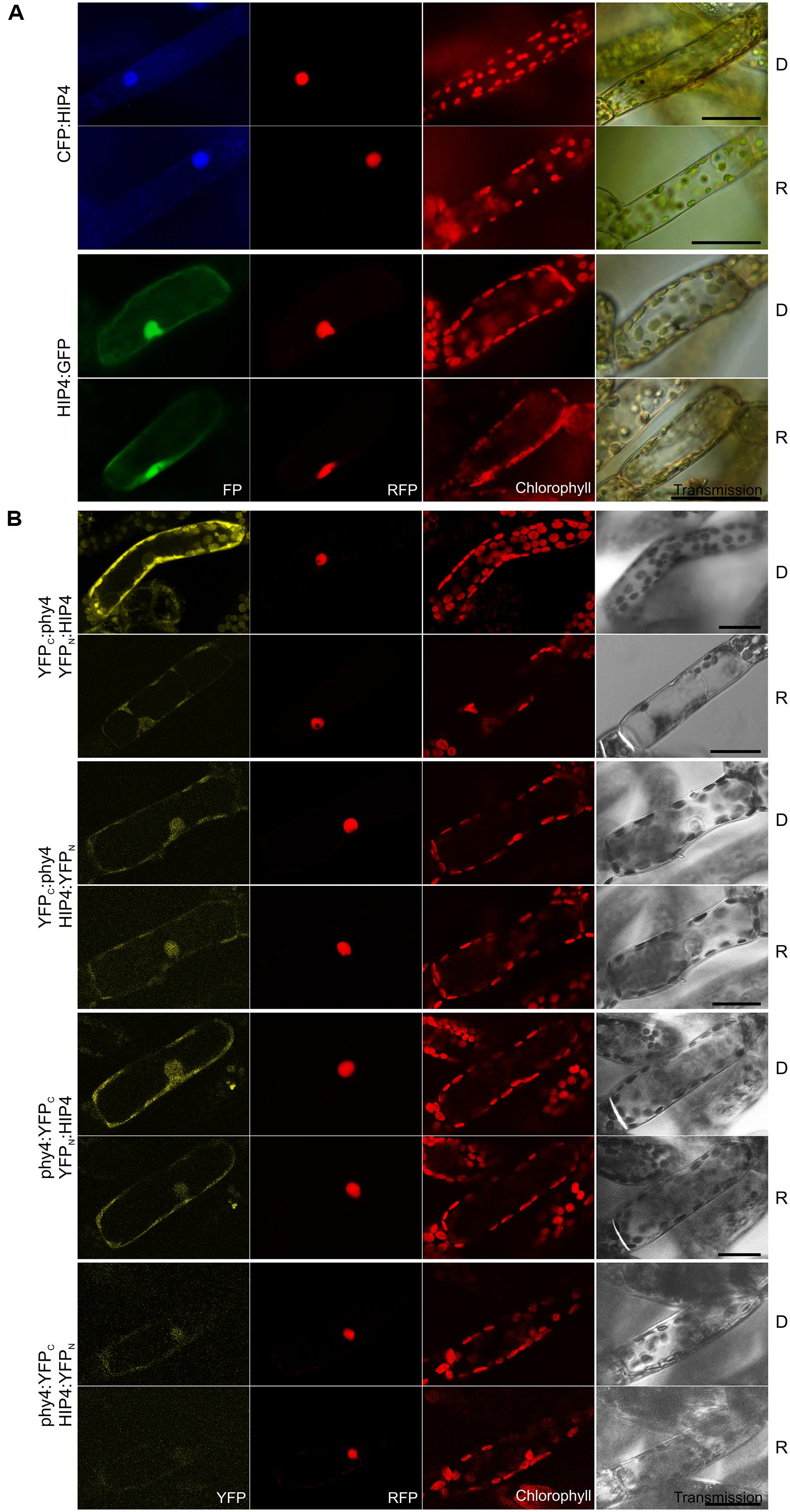
FIGURE 3. HIP4 (Pp3c16_8560E1.1) intracellular localization (A) and split-YFP-studies of HIP4–phy4 interaction (B) each without (D) and with red pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) CFP:HIP4 (rows 1 and 2) was mostly nuclear and HIP4:GFP (rows 3 and 4) was distributed in nucleus and cytoplasm (column 1, FP). (B) All possible split-YFP configurations were analyzed in D and R, respectively: YFPC:phy4–YFPN:HIP4 (rows 1 and 2), YFPC:phy4–HIP4:YFPN (rows 3 and 4), phy4:YFPC–YFPN:HIP4 (rows 5 and 6) and phy4:YFPC–HIP4:YFPN (rows 7 and 8). Significant signals were observed for YFPC:phy4–YFPN:HIP4 (cytoplasmic signal), phy4:YFPC–YFPN:HIP4 (nucleo-cytoplasmic signal), and weakly for YFPC:phy4–HIP4:YFPN (maybe nucleo-cytoplasmic signal) in both D and R (column 1, YFP). phy4:YFPC–HIP4:YFPN yielded only background signals alongside channel bleed-through. Scale bars 30 μm.
Holophytochrome-interacting protein 4 is apparently expressed under all conditions and in all cell types (see Supplementary Materials). The Genevestigator data for both treatments and anatomy provide some evidence for synexpression with phy4. This is not apparent for the other data, however.
PLEIOTROPIC REGULATORY LOCUS 1 is well-conserved amongst land plants, fungi, and animals including mammals. Two homologs are present in both Physcomitrella and Arabidopsis (HIP4/PRL1 & Pp3c5_15520V1.1 and PRL1 & PRL2, respectively). Arabidopsis PRL1 is an α-importin-interacting protein localizing both to the nucleus and associating with the nuclear envelope- and ER membranes (Nemeth et al., 1998). It harbors seven WD40 domains and a variant of the SV40-type NLS in the C-terminal region (residues 238–241), which is not constitutively recognized but rather dependent on phosphorylation of the NLS or the importin (Nemeth et al., 1998). Our localization studies of HIP4/PRL1 indicate that at least R does not affect recognition of the NLS and subsequent nuclear import since we do not observe any change in localization upon light treatment (Figure 3A). Although N-terminally fused CFP:HIP4 was exclusively nuclear, HIP4:GFP was seen in both the nucleus and cytoplasm, whereas YFPC:phy4–YFPN:HIP4 was exclusively cytoplasmic (Figure 3B). Perhaps phy4 binds the C-terminus of HIP4/PRL1, thereby hampering NLS recognition and effecting cytoplasmic localization.
The PRL1 WD40 domains probably constitute β-propellers, together forming a platform for protein–protein interactions. Such platforms are often involved in signal transduction and transcriptional regulation, as in the case of COP1 in repression of photomorphogenic development. Arabidopsis PRL1 seems to be involved in numerous responses, as the name implies. Initially reported to play a role in cell polarity determination by acting on the assembly of the actin cytoskeleton in fission yeast (Xia et al., 1996), PRL1 was later suggested to play a central role in integrating light signals with cytokinin- and sugar response pathways in plants (Nemeth et al., 1998). Indeed prl1- hypocotyls are shorter than the wild type in darkness but not in light, representing a weak COP phenotype, consistent with PRL1 acting as a positive regulator of brassinosteroid synthesis (Szekeres et al., 1996; Nemeth et al., 1998). It was shown to promote BR-biosynthesis by interacting with different SnRK’s (see also HIP3 above) in light and dark, possibly by targeting them for degradation (Bhalerao et al., 1999; Lee et al., 2008; Flores-Perez et al., 2010). PRL1 is also involved in root meristem regulation possibly by restricting transcription of the homeobox transcription factor WOX5 to the quiescent center (Ji et al., 2015). Thus PRL1 in Arabidopsis might either represent a convergence center for integrating light, sugar, hormone, and other responses or be involved in distinct signaling pathways in different cells. SnRK photoregulation would be a possible function of HIP4/PRL1 in Physcomitrella, as SNF1 homologs are present (Thelander et al., 2004, 2007). Light, sugar and the phytohormones cytokinin and auxin are all closely involved in the developmental switches in Physcomitrella whereby filament cells develop into chloronemata, caulonemata, or gametophore buds (Thelander et al., 2004, 2007; Decker et al., 2006; von Schwartzenberg, 2006).
HIP5 (Pp3c7_3040V1.1)
Holophytochrome-interacting protein 5 (54.4 kDa) bears four kelch repeats toward the C-terminus. It has one homolog in Physcomitrella (Pp3c11_19970V1.1, see Summary Table 1 in the Supplementary Material). The semi-quantitative Y2H interaction assay confirmed R-enhanced and FR-reversible interaction in all phy4–HIP5 configurations tested with the exception of BD:phy4–HIP5:ADg which showed no interaction (Supplementary Figure S2). HIP5 seems to be constitutively present in nucleus and cytoplasm irrespective of the position of the fluorescent tag (Figure 4A). Interestingly, an older and N-terminally shorter version of the HIP5 gene model showed exclusively cytoplasmic localization as N-terminal GFP-fusion, implying that the putative NLS resides at the very N-terminus (in the first 129 amino acids; not shown). phy4–HIP5 interaction was confirmed in planta, but in contrast to the Y2H data, interaction was not affected by light treatments (Figure 4B). Analogously to the HIP5 localization data, nuclear and cytoplasmic distribution was seen for phy4:YFPC–HIP5:YFPN and perhaps also for YFPC:phy4–YFPN:HIP5 (although this fluorescence might represent RFP bleed-through in the YFP channel as the signal was very weak). Interestingly, the YFP-signal from the reconstituted phy4:YFPC–YFPN:HIP5 located exclusively to the cytoplasm, although GFP:HIP5 was apparent in both cytoplasm and nucleus.
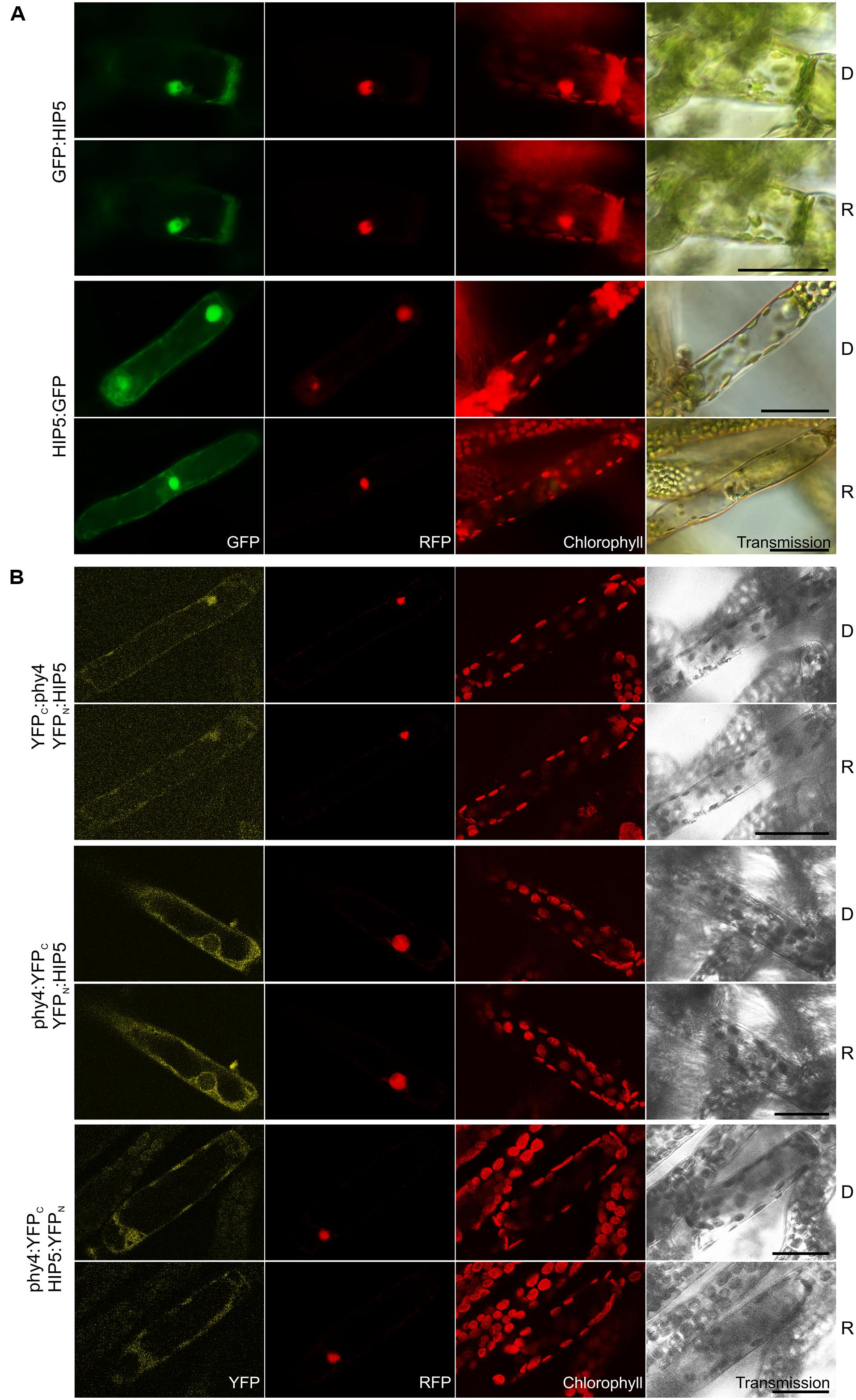
FIGURE 4. HIP5 (Pp3c7_3040V1.1) intracellular localization (A) and split-YFP-studies of HIP5–phy4 interaction (B) each without (D) and with red pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) GFP:HIP5 (rows 1 and 2) and HIP5:GFP (rows 3 and 4) were distributed in the cytoplasm and in the nucleus (column 1, GFP). (B) Three split-YFP configurations were examined and showed different signal patterns/strengths (column 1, YFP): YFPC:phy4–YFPN:HIP5 (rows 1 and 2; maybe nucleo-cytoplasmic distribution), phy4:YFPC–YFPN:HIP5 (rows 3 and 4; cytoplasmic distribution) and phy4:YFPC–HIP5:YFPN (rows 5 and 6; nucleo-cytoplasmic distribution). The apparent YFPC:phy4–YFPN:HIP5 signal might also represent RFP bleed-through in the YFP channel since the signal was very weak. Scale bars 30 μm.
Holophytochrome-interacting protein 5 is apparently expressed under all conditions and in all cell types (see Supplementary Materials). The Genevestigator data for treatments, anatomy and development provide some evidence for synexpression with phy4. This is not apparent for the eFP and Phytozome data, however.
The ∼50-residue kelch motif forms a blade-like fold comprising 4–6 anti-parallel β-sheets, often repeated to form a β-propeller structure frequently involved in protein–protein interactions. In fungi, various kelch repeat proteins associate with microtubules and actin nucleating proteins at cell termini, thereby establishing cell polarity (Mata and Nurse, 1997; Feierbach et al., 2004) whereas others are involved in polarized growth (Takeshita et al., 2008). We speculate therefore that HIP5 might play a similar role in phy4-directed vectorial growth. Indeed, amongst both fungal and Arabidopsis BLAST hits was a scruin – an actin crosslinker, implying a role in cytoskeleton organization consistent with the role of microfilaments in filament tip cell phototropism (Meske and Hartmann, 1995; Meske et al., 1996). On the other hand, HIP5 shares ∼35% sequence identity with many Arabidopsis F-box/kelch-repeat (FBK) proteins such as galactose oxidase/kelch repeat superfamily protein (AT1G14330), MIR2111-5p target protein (AT3G27150) and SKIP11 (AT2G02870), although these sequences rather align with the C-terminal region and thus the kelch repeats of HIP5 (64–71% of the query), whereas the first 135–175 amino acids of HIP5 are dissimilar to the Arabidopsis proteins. All three Arabidopsis FBK proteins are chloroplast- and/or nuclear-localized, interacting with SKP1-like proteins (Schumann et al., 2011). FBK kelch repeats for example are responsible for binding of target proteins to the SCF ubiquitin ligase complex in the degradation pathway (Cardozo and Pagano, 2004; Hua and Vierstra, 2011). The Arabidopsis and Physcomitrella genomes contain ca. 100 and 71 FBK genes, respectively (Schumann et al., 2011), several of which are known to be involved in photoresponses. The PAS/LOV-domain-containing ZEITLUPE, FKF1, LKP2 FBK’s are themselves photoreceptors (Sawa et al., 2007; Demarsy and Fankhauser, 2009; Takase et al., 2011), whereas AFR and CFK1 are positive regulators downstream of phyA and phyB, respectively (Wei et al., 1994a,b; Harmon and Kay, 2003; Franciosini et al., 2013). CFK1 seems to be absent from Physcomitrella although the COP9 signallosome with which CFK1 interacts is thought to be universal in eukaryotes (Wei et al., 1998), thus HIP5 might be involved in a similar regulatory system. Most FBK’s are nuclear although some are located in both cytoplasm and nucleus – as is the 26S proteasome (Schumann et al., 2011). Pp3c2_13250, Pp3c1_22300, Pp3c10_4990, and Pp3c14_4360, on the other hand, might represent Arabidopsis AFR homologs with 29–26% identity covering 72–100% of the query sequence.
HIP6 (Pp3c6_4480V1.1)
Holophytochrome-interacting protein 6 (43.7 kDa) harbors a predicted transmembrane domain and a p-loop containing sulfotransferase domain at the N- and C-terminus, respectively. Interaction of HIP6 with phy4 in Y2H was constitutive in the BD:phy4–AD:HIP6 and phy4:BD–AD:HIP6 configurations, whereas phy4:BD–HIP6:ADg showed R/FR-reversible interaction (Supplementary Figure S2). In planta both N- and C-terminal fusion configurations localized exclusively to the cytoplasm, with HIP6:GFP concentrating in discrete regions (cytoplasmic foci; Figure 5A). This distribution was also observed for phy4:YFPC–YFPN:HIP6 interaction, whereas other split-YFP configurations (phy4:YFPC–HIP6:YFPN and perhaps YFPC:phy4–YFPN:HIP6) rather showed cytoplasmic and weaker nuclear localization (Figure 5B; although narrower emission detection implied in the latter case that the apparent YFP nuclear signal might have arisen from RFP bleed-through; data not shown). Light pre-treatments had little effect on in planta interaction (also in the case of phy4:YFPC–HIP6:YFPN corresponding to phy4:BD–HIP6:ADg in Y2H). In neither Y2H nor split-YFP in planta was significant interaction seen between N-terminally tagged phy4 and C-terminally tagged HIP6.
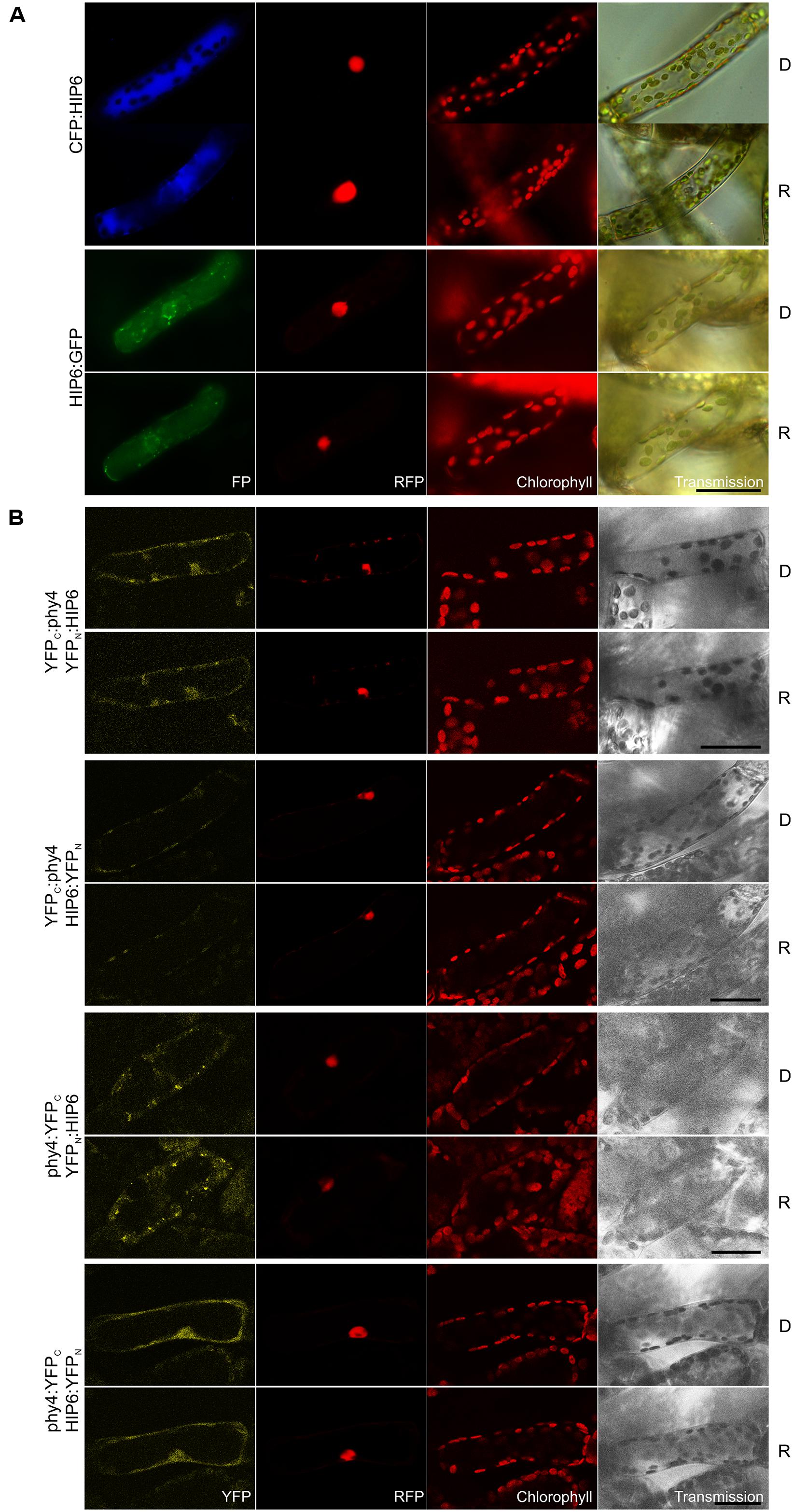
FIGURE 5. HIP6 (Pp3c6_4480V1.1) intracellular localization (A) and split-YFP-studies of HIP6–phy4 interaction (B) each without (D) and with red pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) Intracellular localizations of CFP:HIP6 (rows 1 and 2) and HIP6:GFP (rows 3 and 4) were both localized in the cytoplasm, with HIP6:GFP showing subcellular foci (column 1, FP). (B) All four split-YFP configurations were investigated: YFPC:phy4–YFPN:HIP6 (rows 1 and 2), YFPC:phy4–HIP6:YFPN (rows 3 and 4), phy4:YFPC–YFPN:HIP6 (rows 5 and 6), and phy4:YFPC–HIP6:YFPN (rows 7 and 8). Significant YFP signals were apparent only for phy4:YFPC–YFPN:HIP6 (cytoplasm and cytoplasmic foci) and phy4:YFPC–HIP6:YFPN (cytoplasm and nucleus; column 1, YFP), whereas apparent YFPC:phy4–YFPN:HIP6 nuclear fluorescence might represent RFP bleed-through and YFPC:phy4–HIP6:YFPN only shows chloroplast bleed-through. Scale bars 30 μm.
Holophytochrome-interacting protein 6 is apparently expressed under all conditions and in all cell types (see Supplementary Materials). The Genevestigator and Phytozome data provide some evidence for synexpression with phy4.
The p-loop domain is widespread in nature, using conserved motifs GXXXXGK(T/S) and ZZZ(D/E) (where X and Z represent arbitrary and hydrophobic residues, respectively) with a β-sheet followed by a glycine-rich loop and an α-helix to co-ordinate the ATP or GTP β-γ-phosphate moiety (Walker et al., 1982; Saraste et al., 1990; Milner-White et al., 1991; Ramakrishnan et al., 2002). Thus, HIP6 belongs to the superfamily of p-loop-containing nucleoside triphosphate hydrolases. This large group is involved in processes like replication, transcription, DNA repair, translation, signal transduction, protein localization, and -transport, signal-sequence recognition, membrane transport, chromosome partitioning, and activation of metabolites (Saraste et al., 1990; Vetter and Wittinghofer, 1999; Koonin et al., 2000). Apart from the common motifs and the p-loop structure itself, the sequences (even the core of the p-loop) and structures are extremely diverse, hampering (phylogenetic and) functional classification (Pathak et al., 2014). So far, only a few plant p-loop NTPases have been functionally characterized. Some are thought to be involved in pathogen defense responses and others in salinity stress tolerance (Fenyk et al., 2012; Cheung et al., 2013).
Plant sulfotransferases (SOT’s) are either soluble or transmembrane proteins that transfer the sulfate group from a 3′-phosphoadenosine-5′-phosphosulfate donor to proteins or smaller substrates including brassinosteroids, jasmonates, flavonoids, or salicylic acid (Klein and Papenbrock, 2004; Hirschmann et al., 2014). Intriguingly, plant SOT’s sulfate the flavonol quercetin (Varin et al., 1992), discussed above in connection with pirins. Quercetin and quercetin sulfates are thought to regulate root development by altering basipetal auxin transport, elongation growth and gravitropism (Faulkner and Rubery, 1992; Lewis et al., 2011). Whereas three HIP6 homologs are apparent in Physcomitrella (see Summary Table 1 in the Supplementary Material), 22 SOT’s with different sulfotransfer-domain-types and diverse expression patterns have been identified in Arabidopsis. The hydroxyl-jasmonate sulfating SOT15 is transcriptionally controlled by the circadian clock and PIF4 and -5 (Yamashino et al., 2013) whereas SOT18 seems to be regulated by the phytochrome-controlled transcription factor HY5 (Huseby et al., 2013). Various Arabidopsis SOT’s are thought to be functional in such diverse processes as defense and stress responses, regulation of hormone synthesis or activity, signaling and developmental regulation (Rouleau et al., 1999; Gidda et al., 2003; Marsolais et al., 2007; Hirschmann et al., 2014). Mutation of Arabidopsis tyrosyl-protein sulfotransferase has pleiotropic defects such as in cotyledon and leaf size and pigmentation, flower number and root development (Komori et al., 2009) the latter being associated with disturbed auxin distribution due to altered PIN and auxin-biosynthesis gene expression (Zhou et al., 2010). The function of HIP6 is therefore unclear, especially as it shares highest similarity to the newly identified Arabidopsis SOT’s 19–21 (see Supplementary Materials) whose substrate specificity is unknown (Klein and Papenbrock, 2009; Hirschmann et al., 2014).
HIP7 (Pp3c17_9390V1.1)
The singleton HIP7 (31.5 kDa) is likely to represent a Ser/Thr protein kinase. In Y2H studies full length HIP7 interaction with phy4 was comparatively weak but nevertheless R-enhanced (Supplementary Figure S2), strongest growth being observed for the phy4:BD–ADg:HIP7 configuration. HIP7 fusions with CFP were intracellularly distributed in the nucleus and the cytoplasm (Figure 6A) with CFP:HIP7 in particular showing cytosolic foci in ∼40% of the cells observed (data not shown). In split-YFP, nuclear and cytoplasmic signals were seen for YFPC:HIP7 in combination with both YFPN:phy4 and phy4:YFPN, in accordance with the localization data (Figure 6B). Since the split-YFP signals were significantly weaker than those from the RFP:NLS nuclear marker, we checked for RFP bleed-through by narrowing detection to 10 nm around the emission maximum of YFP. As the fluorescence pattern was unchanged, we consider the signal to represent a bona fide nuclear interaction. Interestingly, the phy4:YFPN–YFPC:HIP7-interaction appeared to be R-dependent, the signal only being observable after illumination, implying Pfr-dependent binding in planta.
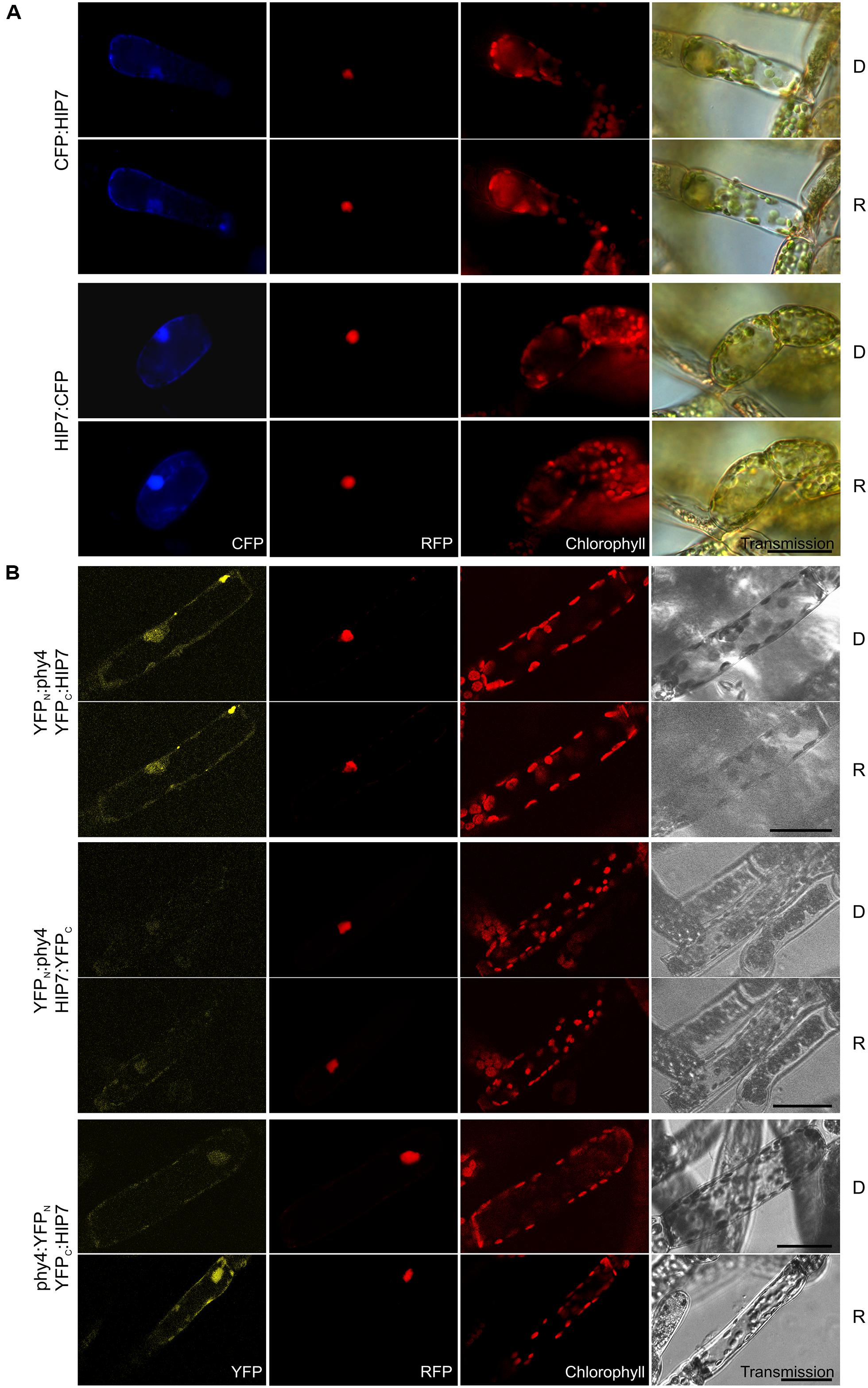
FIGURE 6. HIP7 (Pp3c17_9390V1.1) intracellular localization (A) and split-YFP-studies of HIP7–phy4 interaction (B) each without (D) and with R pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) Intracellular localization of CFP:HIP7- (rows 1 and 2) and HIP7:CFP-fusions (rows 3 and 4) was observed, showing nuclear and cytoplasmic signal patterns (column 1, CFP). CFP:HIP7 in addition showed cytosolic foci in ∼40% of the cells observed. (B) Three of four possible split-YFP configurations were analyzed: YFPN:phy4–YFPC:HIP7 (rows 1 and 2), YFPN:phy4–HIP7:YFPC (rows 3 and 4) and phy4:YFPN–YFPC:HIP7 (rows 5 and 6). YFPN:phy4–YFPC:HIP7, and phy4:YFPN–YFPC:HIP7 yielded significant split-YFP-signals mostly in nucleus and cytoplasm (column 1, YFP). In the latter case significant signals were only seen after R-pre-treatment (D and R conditions here being shown from independent experiments). Scale bars 30 μm.
Holophytochrome-interacting protein 7 is apparently expressed under all conditions and in all cell types (see Supplementary Materials). Some Genevestigator data groups imply synexpression with phy4 as does the Phytozome data.
HIP7 shows low but significant similarity to various Arabidopsis Ser/Thr-protein kinases such as HT1 (AT1G62400), a kinase with an armadillo repeat domain (AT5G18700), a MAP kinase (AT1G73660), and a PAS-domain-containing protein tyrosine kinase (AT3G06620) with ∼40% query coverage and ∼30% sequence identity). HIP7-based BLAST searches of Viridiplantae proteins yielded many with somewhat more concordant similarity, most of which have been judged to be HT1 relatives. Arabidopsis HT1 is a protein kinase involved in the regulation of stomatal aperture (Hashimoto et al., 2006). Although stomatal opening is generally seen as a phototropin-mediated B effect, R is effective in maintaining stomatal opening, a response specifically impaired in ht1- (Matrosova et al., 2015). Although the effect can largely be explained by photosynthetic CO2-depletion, a separate photoreceptor is probably also involved. Indeed, end-of-day FR treatment accelerated stomatal closure in Phaseolus (Holmes and Klein, 1985). Mosses develop stomata in the epidermis of the sporophyte: whereas they show responses to ABA similar to those of higher plants (Chater et al., 2011), light effects have not been studied. HIP7 might in any case be involved in other signaling pathways involving post-translational phosphorylation. Indeed, Arabidopsis HT1 is also expressed in stem, flower and even root tissues, implying diverse but still unknown cellular functions.
HIP8 (Pp3c11_25550V1.1)
Holophytochrome-interacting protein 8 (55.4 kDa) harbors a predicted protein kinase catalytic domain and several calcium-binding EF-hands and thus probably constitutes a CDPK. Y2H implied only weak interaction with phy4, but the signal was enhanced in Rc and stronger with BD:phy4 than with phy4:BD (Supplementary Figure S2). Expression of N- and C-terminal FP-fusions resulted in cytoplasmic and nuclear fluorescence signals approximately with equal signal intensities in both compartments with no apparent effect of light pre-treatment (Figure 7A). In planta interaction was verified for all combinations except YFPC:phy4–HIP8:YFPN (Figure 7B). Cytoplasmic interaction was clearly enhanced by R pre-treatments in phy4:YFPN–YFPC:HIP8, although in phy4:YFPN–HIP8:YFPC this was less pronounced. Although this implies that either the phy4–HIP8 complex is enhanced in or translocates to the cytoplasm in R, this behavior was only seen when the tag was fused to phy4 C-terminus: YFPC:phy4–YFPN:HIP8 interaction was exclusively cytoplasmic.
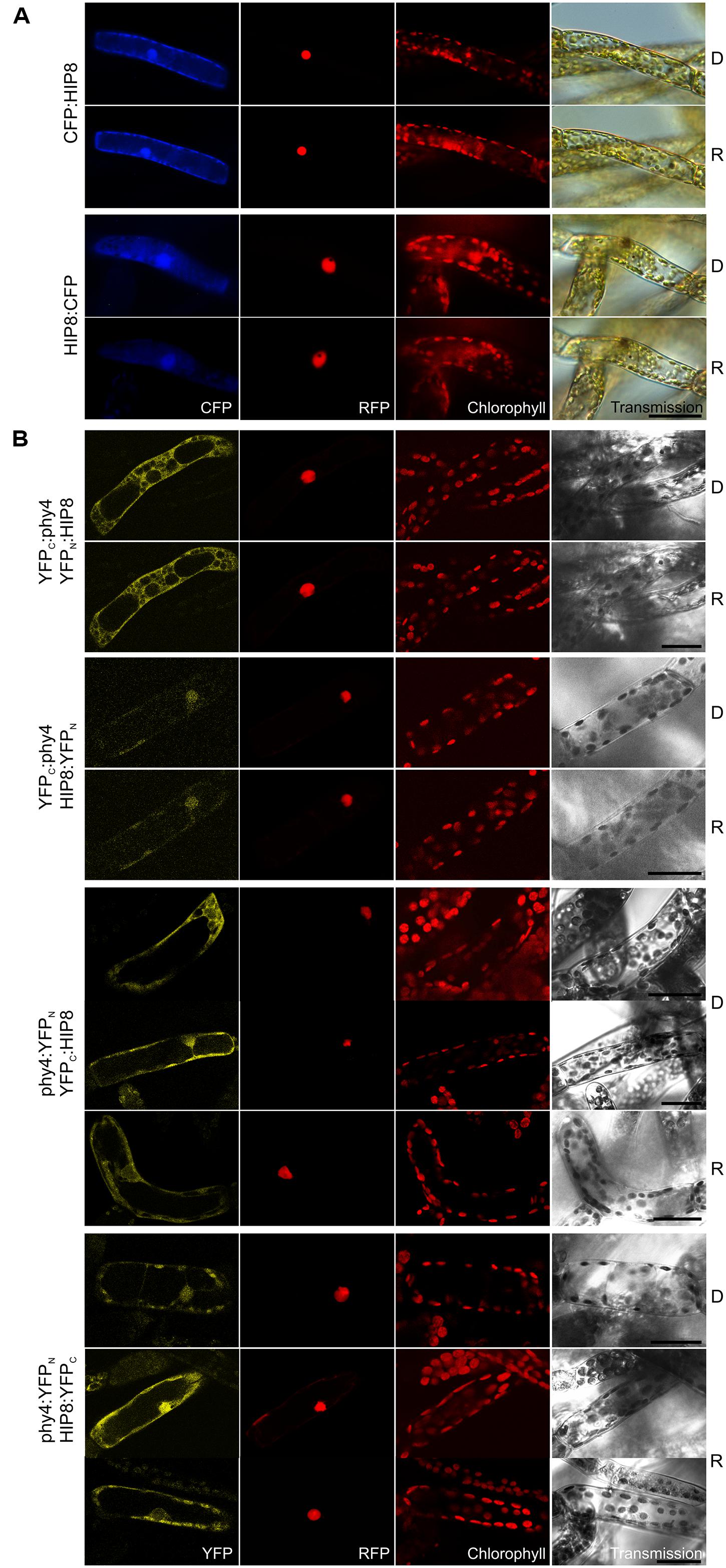
FIGURE 7. HIP8 (Pp3c11_25550V1.1) intracellular localization (A) and split-YFP-studies of HIP8–phy4 interaction (B) each without (D) and with R pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) CFP:HIP8- (rows 1 and 2) and HIP8:CFP-fluorescence (rows 3 and 4) was observed in nucleus and cytoplasm (column 1, CFP). (B) All four split-YFP configurations were studied, however, in the case of YFPN:phy4–YFPC:HIP8 and YFPN:phy4–HIP8:YFPC the negative controls showed significant signals (thus these configurations were replaced by YFPC:phy4–YFPN:HIP8 and YFPC:phy4–HIP8:YFPN). Analyses of YFPC:phy4–YFPN:HIP8 (rows 1 and 2), YFPC:phy4–HIP8:YFPN (rows 3 and 4), phy4:YFPN–YFPC:HIP8 (rows 5–7; two different signal patterns in D) and phy4:YFPN–HIP8:YFPC (rows 8–10; two different signal patterns in R) resulted in either nucleo-cytoplasmic or exclusively cytoplasmic signals in all configurations except YFPC:phy4–HIP8:YFPN (column 1, YFP). Scale bars 30 μm.
Holophytochrome-interacting protein 8 is apparently expressed under all conditions and in all cell types but shows strongly increased transcript levels during leaflet dedifferentiation (see Supplementary Materials). The Genevestigator data for treatments provide some evidence for synexpression with phy4 although this is not apparent for the other datasets.
The eukaryotic second messenger Ca2+ is involved in plant responses to various abiotic and biotic stimuli (Kudla et al., 2010), primary targets being Ca2+-channels and transporters (Wheeler and Brownlee, 2008). Ca2+ from the apoplast or the vacuole thereby briefly increases the local concentration near the plasma membrane or organelle membrane – a phenomenon known as the calcium signature. Prior to the discovery of Pfr-dependent nuclear translocation and its interaction with the PIF family of transcription factors in the nucleus, Ca2+ had been suggested to be a key player in phytochrome signaling, showing crucial roles in chloroplast development and LHC expression, phyA-mediated light regulation of asparagine synthase, R-induced stem protoplast swelling and phyA- and PIF3-mediated regulation of succinate dehydrogenase (Neuhaus et al., 1993, 1997; Bowler et al., 1994; Long and Iino, 2001; Eprintsev et al., 2013). Indeed, there is evidence that phytochromes initiate Ca2+ and protein phosphorylation signals via CDPK’s (Datta et al., 1985; Li et al., 1991). Furthermore, Ca2+ addition to Cucumis cotyledon extracts can mimic the R/FR effects on phosphorylation and at least two CDPK’s were present whose activities were modulated by phytochromes (Vidal et al., 2007). CDPK’s possess a N-terminal variable domain, a protein kinase-, an autoinhibitory and a C-terminal calmodulin-like domain, the latter typically consisting of two pairs of EF-hand motifs (Rutschmann et al., 2002; Christodoulou et al., 2004). CDPKs directly bind Ca2+, leading to a conformational change and kinase de-repression, thereby converting the Ca2+ signal into phosphorylation of metabolic enzymes, ion channels and transcription factors and thus into diverse downstream cellular responses (reviewed in Hamel et al., 2014). Kinase activity can also be regulated by (auto)phosphorylation/dephosphorylation, phospholipids and 14-3-3-proteins (reviewed in Cheng et al., 2002), however. Interestingly, the holo phy4:BD-screen also identified a 14-3-3 protein (see HIP14). Overall domain organization seems to be conserved in all CDPK’s, and land plant CDPK’s usually cluster in four distinct clades (Hamel et al., 2014). Comparison of the HIP8 amino acid sequence with the Viridiplantae protein database indeed revealed numerous orthologs with high levels of similarity, the Arabidopsis and Physcomitrella genomes encoding 34 and 25 CDPK’s, respectively (Hamel et al., 2014). The only moss representative characterized to-date seems to be associated with nutrient starvation (Mitra and Johri, 2000). Several Arabidopsis CDPK’s such as CPK1 - the most similar to HIP8 (covering 91% of the query with 65% identity) – are thought to be involved in ABA signaling, regulation of stomatal aperture, and stress responses (Mori et al., 2006; Ma and Wu, 2007; Zhu et al., 2007; Geiger et al., 2010; Zou et al., 2010; Brandt et al., 2012; Boudsocq and Sheen, 2013; Liese and Romeis, 2013). Although CPK1 is membrane-/peroxysome-localized and thus probably involved in oxidative stress and lipid metabolism (Dammann et al., 2003), this is not the case for HIP8 which thus probably has different functions. Arabidopsis CPK17 and perhaps other CDPKs on the other hand are involved in Ca2+-mediated pollen tube tip growth (Myers et al., 2009; Hamel et al., 2014). As Physcomitrella filament tip growth shows many similarities to that of pollen tubes including a tip-focused calcium gradient (Meske and Hartmann, 1995; Meske et al., 1996; see HIP13), we speculate that HIP8 might play a related role in phy4-steered phototropism. On the other hand, whereas Arabidopsis CPK17 is plasma membrane-localized (Hamel et al., 2014), this seems not to be the case for HIP8 (Figure 7). However, in both algae and higher plants several CDPKs are associated with the cortical actin cytoskeleton and cytoplasmic streaming (Putnam-Evans et al., 1989; McCurdy and Harmon, 1992). Light-induced cytoplasmic streaming is also mediated by phytochrome in the angiosperm Vallisneria. This Ca2+-and actin-dependent response is of particular interest regarding phytochrome cytoplasmic signaling as it is restricted to the area of irradiation and begins within 3 s of irradiation (Takagi et al., 2003), thereby ruling out a transcription-translation-based transduction mechanism. If the functions of actin-associated CDPKs and HIP8 are indeed related, it might represent the molecular link between the directional signal sensed by phytochrome and cytoskeleton-steered tip cell growth/bending response.
HIP9 (Pp1s254_10V6.2)
Holophytochrome-interacting protein 9 (63.9 kDa) harbors a predicted rhomboid peptidase S54 domain and predicted transmembrane regions. Y2H studies with phy4 showed weak but R-enhanced interaction with full length HIP9 in all configurations tested (Supplementary Figure S2). In planta localization studies showed cytoplasmic and perhaps plasma membrane (CFP:HIP9) or cytoplasmic and nuclear localization with no sign of membrane association (HIP9:CFP; Figure 8A). Although the predicted protein size (64 kDa + 27 kDa tag) exceeds the nuclear pore exclusion limit, the C-terminal FP-fusion was able to enter the nucleus. Indeed, two NLS in the N-terminal region are predicted by NucPred7 (Brameier et al., 2007), so nuclear import is not unreasonable. Interaction was confirmed in planta only in the phy4:YFPN–HIP9:YFPC configuration following R pre-treatment (in harmony with the Y2H data; Figure 8B). In this case, fluorescence was generally distributed between cytoplasm and nucleus although occasionally a signal at the plasma membrane was apparent.
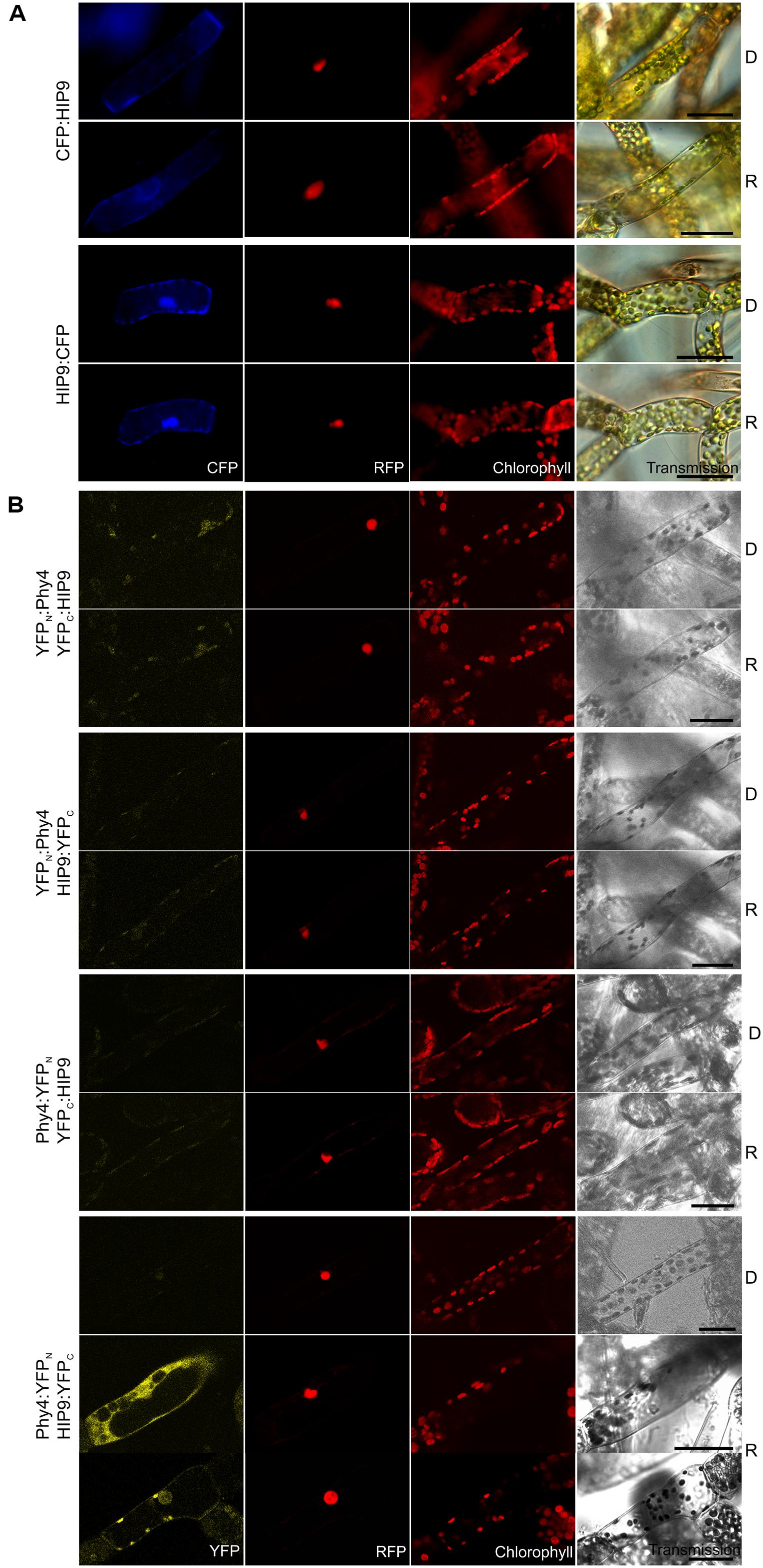
FIGURE 8. HIP9 (Pp1s254_10V6.2) intracellular localization (A) and split-YFP-studies of HIP9–phy4 interaction (B) each without (D) and with R pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) CFP:HIP9- (rows 1 and 2) and HIP9:CFP-fluorescence (rows 3 and 4) was observed in the cytoplasm or the nucleus and cytoplasm, respectively (column 1, CFP). (B) All possible split-YFP configurations were investigated: YFPN:phy4–YFPC:HIP9 (rows 1 and 2), YFPN:phy4–HIP9:YFPC (rows 3 and 4), phy4:YFPN–YFPC:HIP9 (rows 5 and 6) and phy4:YFPN–HIP9:YFPC (rows 7–9; two distinct patterns in R). However, two different signal patterns (nucleus and cytoplasm or perhaps nucleus and plasma membrane) were only observed for phy4:YFPN–HIP9:YFPC after R pre-treatment (column 1, YFP; D and R images here from separate experiments). Scale bars 30 μm.
Holophytochrome-interacting protein 5 is apparently expressed under all conditions and in all cell types (see Supplementary Materials). The data provided no evidence for synexpression with phy4, however.
Rhomboid domain proteases are ubiquitous intramembrane serine proteases (Urban et al., 2001; Koonin et al., 2003; Urban and Wolfe, 2005). According to crystal structures, RBL’s possess seven transmembrane helices forming a conical cavity accessible to the aqueous surroundings, enabling cleavage of a transmembrane helix with the help of serine and histidine residues forming a catalytic dyad (Kanaoka et al., 2005; Baker et al., 2007; Ben-Shem et al., 2007). The Arabidopsis and Physcomitrella genomes encode 17 and 16 RBL’s, respectively (Li et al., 2015), including the homolog (Pp3c22_8560V1.1, see Summary Table 1 in the Supplementary Material). Of those in Arabidopsis, four show poorly conserved active sites, two localizing to the Golgi apparatus suggesting a role in secretion, while others localize to mitochondria, plastids, and perhaps also the endoplasmic reticulum and the plasma membrane, implying different functions (Kanaoka et al., 2005; Lemberg and Freeman, 2007; Kmiec-Wisniewska et al., 2008; Knopf et al., 2012; Thompson et al., 2012). Transcription factors represent potential targets, ∼10% of Arabidopsis transcription factors requiring release from membranes to become functional (Kim and Park, 2007; Seo et al., 2008). RBL’s have also been shown to be involved in Ca2+-mediated stress responses, fertility and flower development, structural and functional maintenance of chloroplast and mitochondrion integrity and mitochondrial retrograde signaling (Aguilar-Hernandez et al., 2011; Thompson et al., 2012; Ng et al., 2013). HIP9 is most similar to RBL 9 (At5g38510), predicted to be inactive due to the lack both catalytic dyad amino acids but retaining the glycine within the GxSx rhomboid protease motif, perhaps representing a modified active site (Lemberg and Freeman, 2007). The substrates and functions of this RBL group are not known, however. Thus whether HIP9 is indeed an RBL membrane protein with switchable protease activity is doubtful. In particular, rather than membrane integration, our data implies at most a weak association with the plasma membrane. Whereas this would correlate with the phy4–phot interaction at the plasma membrane (Jaedicke et al., 2012), an involvement of HIP9 in that signaling complex remains no more than a possibility.
HIP11 (Pp3c20_16210V1.1)
Holophytochrome-interacting protein 11, (20.2 kDa) shows low similarity to the Arabidopsis pfkB-like carbohydrate kinase family protein AT5G19150. A likely homolog in Physcomitrella is Pp3c7_8840V1.1. Holo-BD:phy4 interacted weakly with ADg:HIP11 and HIP11:ADg in Y2H, whereas interactions of holo-phy4:BD with HIP11 were light-enhanced but fully FR-reversible only in case of ADg:HIP11 (Supplementary Figure S2). GFP:HIP11 and HIP11:GFP mostly showed cytoplasmic/perinuclear and rather unstructured cytoplasmic localization, respectively (Figure 9A), although in a subset of cells cytoplasmic and nuclear localization could be observed in both cases (not shown). In planta interaction was verified in the phy4:YFPN–YFPC:HIP11 configuration, the split-YFP signal being apparent in the cytoplasm and perinuclear region (Figure 9B) in accordance with the localization data.
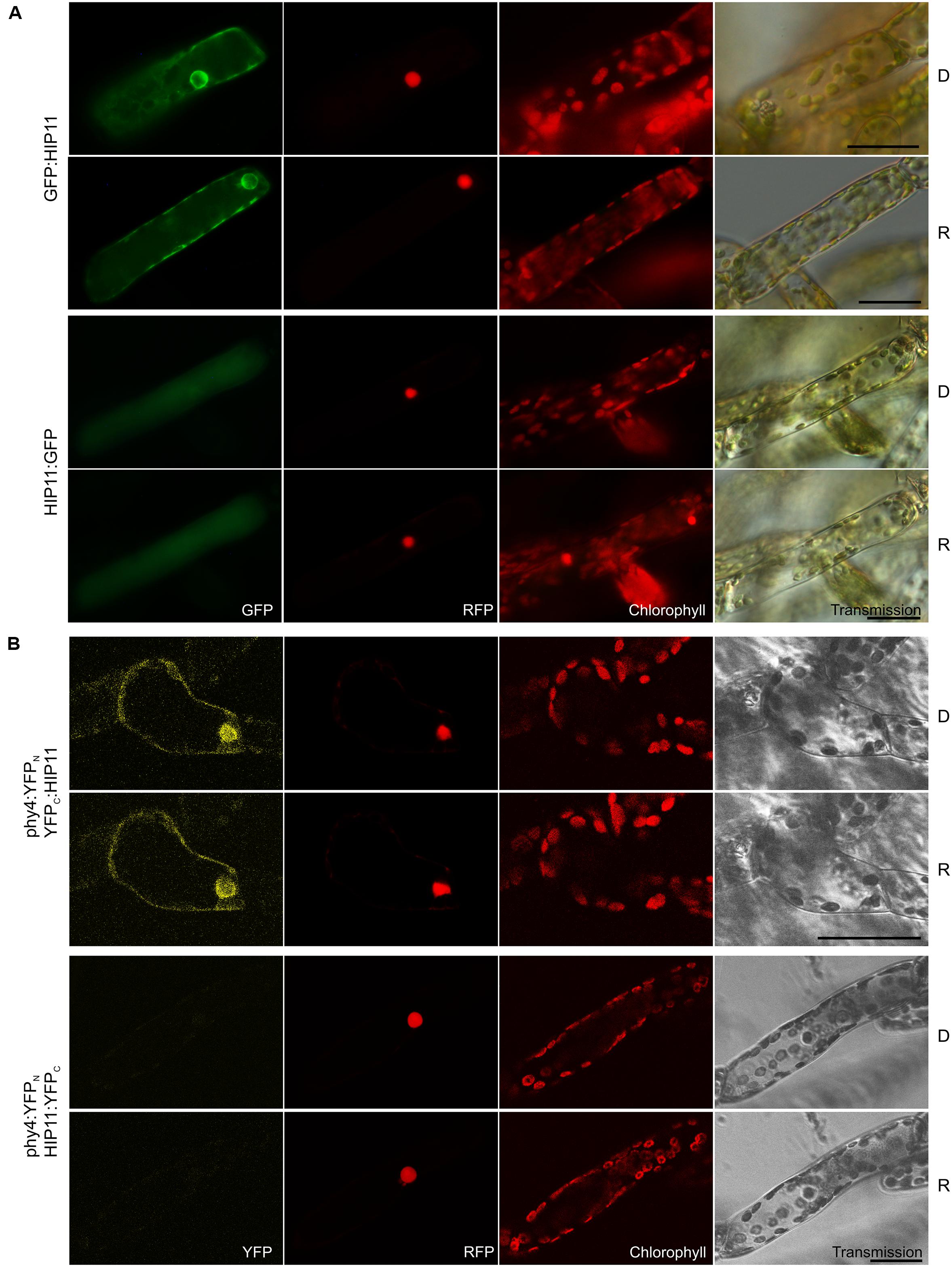
FIGURE 9. HIP11 (Pp3c20_16210V1.1) intracellular localization (A) and split-YFP-studies of HIP11–phy4 interaction (B) each without (D) and with R pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) GFP:HIP11 (rows 1 and 2) and HIP11:GFP (rows 3 and 4) showed cytoplasmic and perinuclear or cytoplasmic localization, respectively (column 1, GFP). In a subset of cells cytoplasmic and nuclear localization could be observed in both cases (not shown). (B) Corresponding to the Y2H interaction behavior, split-YFP configurations in combination with C-terminally fused phy4 were analyzed: phy4:YFPN–YFPC:HIP11 (rows 1 and 2) and phy4:YFPN–HIP11:YFPC (rows 3 and 4). (Peri)nuclear and cytoplasmic signals were observed in the former case alone (column 1, YFP). Scale bars 30 μm.
Holophytochrome-interacting protein 11 is apparently expressed very differently under different conditions and particularly weakly at early developmental stages (such as in spores and protonemata). There is little evidence for synexpression with phy4.
Although HIP11 was predicted by InterPro to carry an N-terminal transmembrane region (residues 7–25), we found no direct evidence of membrane-association. On the other hand, its small size and an NLS (RRRR, residues 112–115, Brameier et al., 2007) predicted by NucPred are consistent with the occasional nuclear localization we observed. As BLAST searches revealed only 31% identity within the 36% query coverage with respect to Arabidopsis AT5G19150 (whose function is unknown), HIP11 and Pp3c7_8840V1.1 might be functionally unique to mosses.
HIP12 (Pp3c13_15620V1.1)
Holophytochrome-interacting protein 12 (17.5 kDa) is the evolutionarily and structurally conserved eukaryotic translation initiation factor 5A (eIF-5A). In Y2H studies, interaction of HIP12/eIF-5A with phy4 was relatively weak but nevertheless light-induced, strong and FR-reversible and constitutive in cases of BD:phy4–ADg:HIP12/BD:phy4–HIP12:ADg, phy4:BD–ADg:HIP12, and phy4:BD–HIP12:ADg, respectively (Supplementary Figure S2). Irrespective of light pre-treatments or FP position, HIP12/eIF-5A was localized in the cytoplasm and nucleus (Figure 10A), presumably because the 18 kDa + 27 kDa FP chimera would be small enough to enter the nucleus passively, although in some HIP12:GFP-cells nuclear fluorescence was much weaker than in the cytoplasm (data not shown). Interaction was confirmed in planta although the site of localization was strongly dependent on the construct used (see Figure 10B). Whereas phy4:YFPN–HIP12:YFPC-interaction was seen in both nucleus and cytoplasm or cytoplasm only, YFPN:phy4–YFPC:HIP12 was apparent only in the cytoplasm. YFPC:phy4–HIP12:YFPN on the other hand failed to show significant interaction. phy4:YFPN–YFPC:HIP12 was omitted from the figure as the negative control :YFPN–YFPC:HIP12 showed substantial fluorescence signals, implying an artifactual interaction.
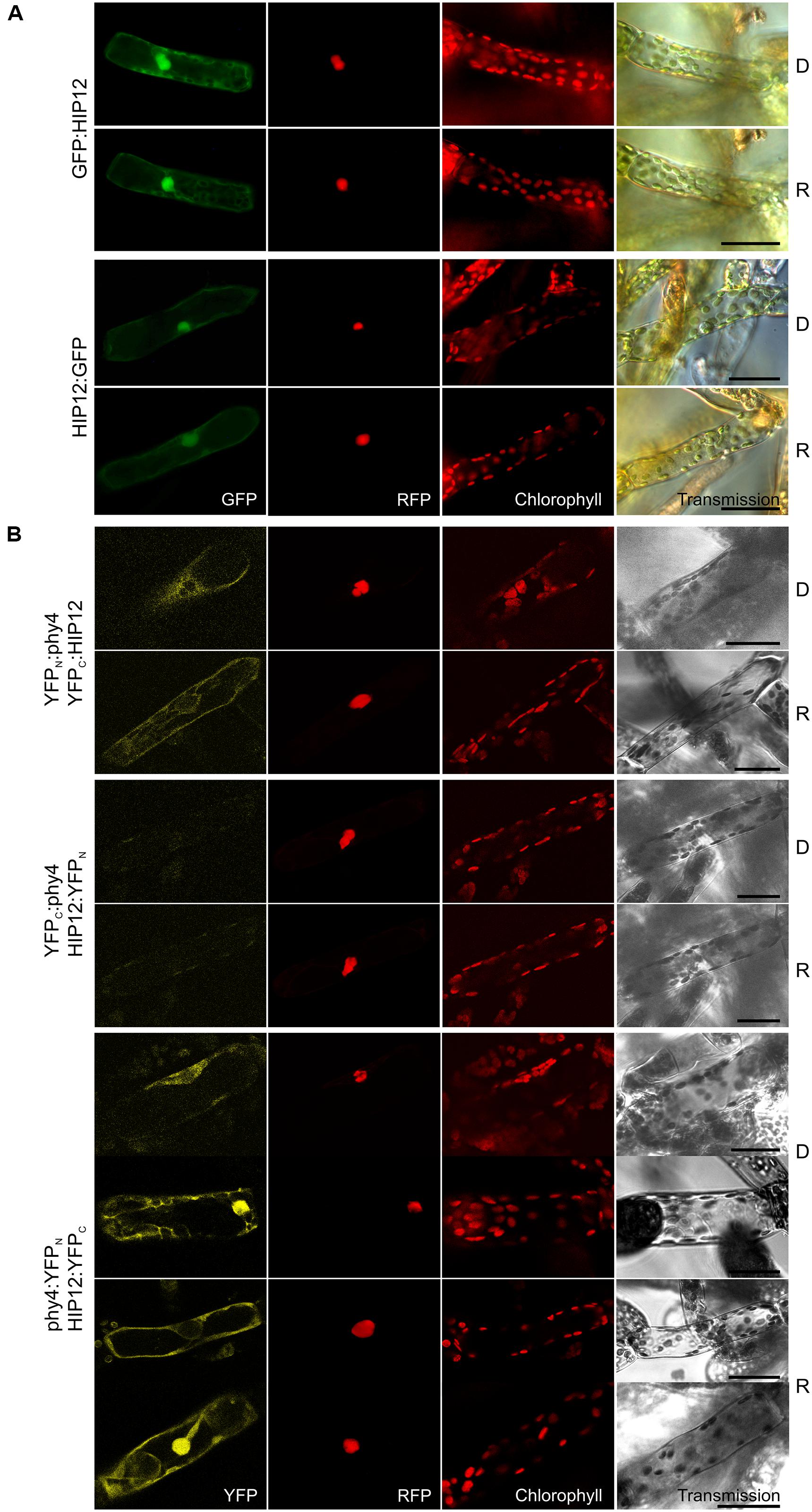
FIGURE 10. HIP12 (Pp3c13_15620V1.1) intracellular localization (A) and split-YFP-studies of HIP12–phy4 interaction (B) each without (D) and with R pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) N-and C-terminally GFP-tagged HIP12 localized to nucleus and cytoplasm (rows 1 and 2; and 3 and 4, respectively; column 1, GFP). In some cases the HIP12:GFP signal was confined largely to the cytoplasm (not shown). (B) All four split-YFP configurations were tested, however, YFPN:phy4–HIP12:YFPC and phy4:YFPN–YFPC:HIP12 were excluded because the respective negative controls showed significant signals. Thus the following configurations are shown: YFPN:phy4–YFPC:HIP12 (rows 1 and 2), YFPC:phy4–HIP12:YFPN (rows 3 and 4) and phy4:YFPN–HIP12:YFPC (rows 5–8; two distinct patterns in both conditions). All configurations except YFPC:phy4–HIP12:YFPN yielded significant cytoplasmic or nucleo-cytoplasmic signals (column 1, YFP). Scale bars 30 μm.
Holophytochrome-interacting protein 12 is apparently strongly expressed in under most conditions and in most cell types except leaflets (see Supplementary Materials). According to Genevestigator HIP12 expression is only weakly affected by different treatments but increases strongly on dedifferentiation of the leaflet cells (see Supplementary Materials). The Genevestigator treatment data alone provided evidence for synexpression with phy4.
Basic local alignment search tool analysis revealed a likely HIP12 homolog in Physcomitrella (Pp3c12_3540V1.1) and three in Arabidopsis. Despite its name, eIF-5A is now generally considered to be involved in translation elongation rather than initiation (Kemper et al., 1976; Gregio et al., 2009; Patel et al., 2009; Saini et al., 2009; Tong et al., 2009). The association of phy4 with eIF-5A and thus translation provides an interesting possible connection to the reported association of phyB Pfr with the cytoplasmic translational regulator PENTA1 and associated inhibition of PORA translation (Paik et al., 2012). Interestingly, in fission yeast eIF-5A has been associated with organization of the cytoskeleton, cell polarity and bud formation (Weir and Yaffe, 2004; Zanelli and Valentini, 2005; Chatterjee et al., 2006), providing a potential link to steering of Physcomitrella tip cell growth by phy4 (see also HIP13 below).
The complex localization behavior of HIP12 we observed in planta correlates with observations in other systems. Mammalian eIF-5A is localized in the cytoplasm and perinuclear region where it interacts with ribosomes attached to the endoplasmic reticulum, forming a network-like structure (Shi et al., 1996) resembling the pattern we observed (see Figure 10). Post-translational conversion of a lysine residue to hypusine (Chen and Liu, 1997; Park et al., 1997) regulates eIF5-A’s activity and localization, however, the precursor being found in both cytoplasm and nucleus whereas the hypusinated eIF-5A localizes predominantly to the cytoplasm (Park et al., 1982; Chen, 1983; Torrelio et al., 1984; Park, 1989; Lee et al., 2009). In maize, phosphorylation of the terminal serine changes eIF-5A-2 localization, unphosphorylatable mutants localizing to nucleus and cytoplasm, while phospho-mimic mutants sequester to the nucleus (Lebska et al., 2010; Lewandowska-Gnatowska et al., 2011). Indeed it was suggested that the phyA-associated phosphatase PP2A (see HIP14 below) is responsible for the dephosphorylation. In Arabidopsis several isoforms have been identified and associated with xylem formation, abiotic stress tolerance and responses to cytokinin including senescence (Chamot and Kuhlemeier, 1992; Wang et al., 2001, 2012; Thompson et al., 2004; Feng et al., 2007; Liu et al., 2008; Ren et al., 2013). HIP12 is most similar to eIF-5A-3, however, which is preferentially expressed in the phloem but whose function is not known (Ma et al., 2010).
HIP13 (Pp3c1_23670V1.1)
Holophytochrome-interacting protein 13 (49.3 kDa) is the eukaryotic/archaeal translation elongation factor subunit EF1α. Y2H studies showed constitutive interaction with HIP13/EF1α in case of BD:phy4 and rather weak but R-enhanced interaction with phy4:BD, respectively (Supplementary Figure S2). Irrespective of FP-fusion configuration or light treatment, HIP13/EF1α localized to the cytoplasm (Figure 11A) and perinuclear region. Accordingly, most split-YFP signals also constitutively appeared in the cytoplasm and perinuclear region (YFPC:phy4–YFPN:HIP13 and phy4:YFPN-YFPC:HIP13, Figure 11B). In particular, YFPC:phy4–YFPN:HIP13 cytoplasmic signals resembled cytoskeleton-like substructures. phy4:YFPC–HIP13:YFPN signals were additionally seen in the nucleus, however. As the negative controls YFPC: or YFPN: with HIP13:YFPN or HIP13:YFPC, respectively, produced substantial signals in the cytoplasm and/or nucleus, the respective combinations with N-terminally fused phy4 were categorized as false positive signals and excluded from discussion.
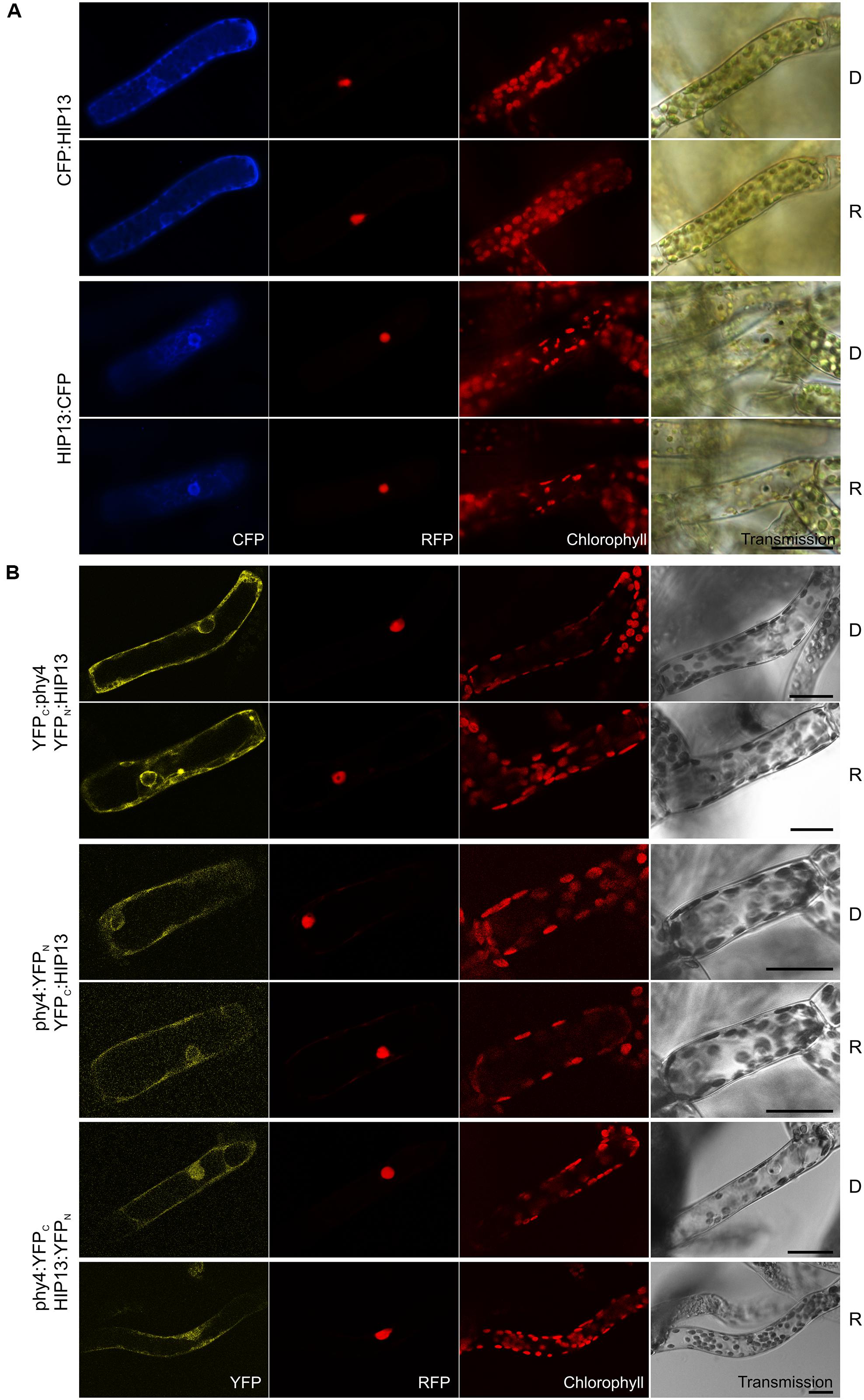
FIGURE 11. HIP13 (Pp3c1_23670V1.1) intracellular localization (A) and split-YFP studies of HIP13–phy4 interaction (B) each without (D) and with R pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) Both CFP:HIP13 (rows 1 and 2) and HIP13:CFP (rows 3 and 4) were localized in the cytoplasm and perinuclear region (column 1, CFP). (B) All four split-YFP configurations were tested, however, YFPC:phy4–HIP13:YFPN and phy4:YFPC–YFPN:HIP13 were excluded because the respective negative controls showed significant signals. Thus the following configurations are shown: YFPC:phy4–YFPN:HIP13 (rows 1 and 2), phy4:YFPN–YFPC:HIP13 (rows 3 and 4) and phy4:YFPC–HIP13:YFPN (rows 5 and 6), all yielding significant cytoplasmic signals with the exception of phy4:YFPC–HIP13:YFPN the signal which could be observed in nucleus and cytoplasm (column 1, YFP). Scalebars 30 μm.
Holophytochrome-interacting protein 13 is apparently expressed strongly under most conditions and in most cell types but weakly in spores and leaflets (see Supplementary Materials). There is little evidence for synexpression with phy4.
Translation elongation factor 1α is an extremely conserved multifunctional protein harboring an N-terminal RAS-like GTPase (also called p-loop NTPase) and C-terminal GTP EFTU-D2 and -D3 domains. The RAS-like GTPase domain comprises several nucleotide-binding sites whereas the D3-domain is responsible for actin binding (Gross and Kinzy, 2005). As the name implies, EF1α plays an essential role in translational elongation, thus interaction with phy4 might relate to phytochrome-regulated PENTA1-mediated phytochrome regulation of translation (Paik et al., 2012) as discussed for HIP12. On the other hand, EF1α has been shown to bind and bundle actin filaments in various species (Demma et al., 1990; Dharmawardhane et al., 1991; Bassell et al., 1994; Collings et al., 1994). EF1α constitutes 1–2% of total protein in normal growing cells (Slobin, 1980; Dharmawardhane et al., 1991), more than half of which is estimated to be associated with actin (Edmonds et al., 1995). Interaction with actin is dependent on both EF1α-GTP binding and phosphorylation status (Dharmawardhane et al., 1991; Yang and Boss, 1994; Izawa et al., 2000). EF1α activates phosphatidylinositol 4-kinase (Yang et al., 1993) thereby regulating the capping and severing of actin filaments proteins. It also serves as a substrate for Rho-associated and Ca2+-dependent protein kinases (Yang and Boss, 1994; Izawa et al., 2000). Moreover, EF1α binds and severs microtubules in a Ca2+/calmodulin-dependent manner in various organisms (Durso and Cyr, 1994; Kaur and Ruben, 1994; Shiina et al., 1994). Cytoskeleton-binding of EF1α probably links the translation machinery to the spatial organization of the cell whereby EF1α regulates transport and anchorage of mRNA and the compartmentalization of translation in eukaryotic cells (Gross and Kinzy, 2005).
Four and ∼11 EF1α-homologs exist in Arabidopsis and Physcomitrella, respectively (Ransom-Hodgkins, 2009). EF1α’s promiscuous actions in the cell, in particular its association with the cytoskeleton and possibly Ca2+ signaling, make it an interesting candidate for a role in phy4-mediated phototropism in filament tip cells. In unilateral R the Pfr distribution at the plasma membrane somehow generates a Ca2+-gradient at the illuminated side of the cell, shifting the site of F-actin synthesis (Hartmann and Weber, 1988; Meske and Hartmann, 1995; Meske et al., 1996). As a result, a directionally translocated actin cap forms, defining the new center of outgrowth corresponding to polarized vesicle transport along the actin bundles (Schnepf, 1986; Meske et al., 1996). More recently, profilin 2 has been shown to be involved in actin turnover and in actin cytoskeleton-mediated vesicular trafficking together with EF1α in Arabidopsis root cells (Takac et al., 2011). Indeed, profilin is essential for tip cell growth in Physcomitrella (Vidali et al., 2007). Profilin together with actin and CHUP1 are also central players in chloroplast movement responses in Arabidopsis (Schmidt von Braun, 2008; Schmidt von Braun and Schleiff, 2008). Chloroplast photorelocation is a cytoplasmic directional response mediated by phytochrome in mosses. Given that CHUP1 is also present in Physcomitrella, a machinery involving phytochrome, profilin, EF1α and the actin-filament network can be envisaged. It should be recalled, however, that the equivalent response in Arabidopsis is mediated by the blue light receptor phototropin, phytochrome playing a modulating role (DeBlasio et al., 2003; Luesse et al., 2010).
HIP14 (Pp3c3_8540C1.1)
Holophytochrome-interacting protein 14 (29.1 kDa) is a 14-3-3 protein family member. Y2H analysis of phy4 interaction with the full-length protein model confirmed interaction only with the phy4:BD configuration (Supplementary Figure S2). In planta GFP:HIP14 localized to cytoplasm and nucleus, whereas the HIP14:GFP configuration was exclusively cytoplasmic showing a cytoskeleton-like pattern (Figure 12A) This pattern was not apparent in the GFP:HIP14 configuration. The HIP14-phy4 interaction was verified in planta: phy4:YFPN–YFPC:HIP14 on the one hand showed a strong nuclear and cytoplasmic signal, reflecting that of GFP:HIP14, and in addition showed an exclusively cytoplasmic, cytoskeleton-like pattern in a subset of cells (Figure 12B). The reconstituted phy4:YFPN–HIP14:YFPC signal, like that of HIP14:GFP, was exclusively cytoplasmic although we did not observe cytoskeleton-like patterns.
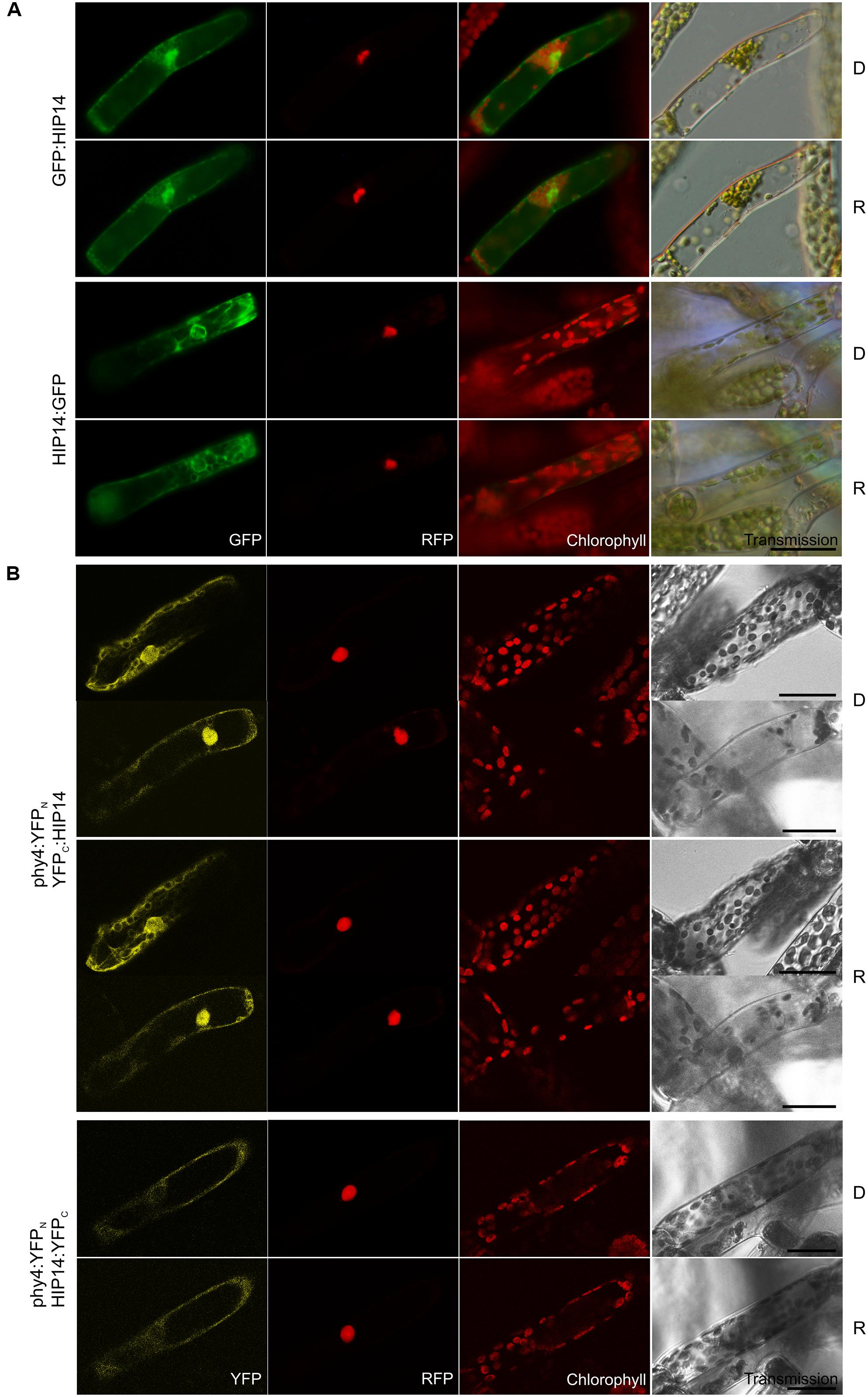
FIGURE 12. HIP14 (Pp3c3_8540C1.1) intracellular localization (A) and split-YFP-studies of HIP14–phy4 interaction (B) each without (D) and with R pre-treatment (R) using fluorescence- and confocal microscopy, respectively. Overall figure structure is analogous to Figure 1. (A) GFP:HIP14 (rows 1 and 2) and HIP14:GFP (rows 3 and 4) localized to cytoplasm and nucleus and to cytoplasm and perinuclear region showing a cytoskeleton-like pattern, respectively (column 1, GFP). (B) Corresponding to the Y2H interaction behavior, split-YFP configurations in combination with C-terminally fused phy4 were analyzed: phy4:YFPN–YFPC:HIP14 (rows 1–4; two distinct signal patterns in both conditions) and phy4:YFPN–HIP14:YFPC (rows 5 and 6). All configurations tested yielded significant cytoplasmic or nucleo-cytoplasmic signals (column 1, YFP). Scale bars 30 μm.
Holophytochrome-interacting protein 14 is apparently expressed very differently under different conditions and in different cell types with little evidence for synexpression with phy4 (see Supplementary Materials).
Ubiquitous in eukaryotic cells (Isobe et al., 1991; Yaffe, 2002), 14-3-3 proteins possess a conserved central region involved in dimerization and interaction with client proteins via an amphipathic groove. 14-3-3-targets are mostly phosphothreonine and phosphoserine proteins harboring (R/K)(XX/S)X(S/T)pXP motifs, the conformation of which is affected by 14-3-3 binding, leading to changes in the target’s catalytic activity, binding to down-stream signaling components and/or intracellular localization (see de Boer et al., 2013). ∼10 other 14-3-3 proteins are encoded in the Physcomitrella genome. Comparison with the 13 14-3-3 family members in Arabidopsis showed HIP14 to be most similar to isoforms GF14 ψ and ω which are localized predominantly in the trichome nucleus and cytoplasm with cytoskeleton association (Paul et al., 2005), similarly to our in planta localization data for HIP14:GFP and split-YFP-data for phy4:YFPN–YFPC:HIP14 (see Figures 12A,B). Despite their sequence similarity, plant 14-3-3 proteins execute specific functions on account of cell- or stimulus-specific expression, differential phosphorylation or intracellular localization (Paul et al., 2005). Altogether >750 14-3-3 protein targets have been identified in Arabidopsis, thus their functions are diverse, including regulation of hormone signaling and responses to external stimuli such as light (see Denison et al., 2011). Certain 14-3-3 isoforms mediate stomatal opening via interaction with light-activated phototropins and subsequently binding to the phosphorylated/activated plasma membrane H+-ATPase (Kinoshita et al., 2003; Ueno et al., 2005; Sullivan et al., 2009; Hayashi et al., 2010, 2011; Tseng et al., 2012), while others may regulate chloroplast movement perhaps also by interaction with phototropin (Sullivan et al., 2009; Tseng et al., 2012). Additionally, 14-3-3 proteins are involved in photoperiodic control of flowering, probably by direct interaction with key players in floral induction such as CONSTANS and FT (florigen; Pnueli et al., 2001; Mayfield et al., 2007; Purwestri et al., 2009). Numerous 14-3-3 clients are involved in light signaling including RPT2, phot2, PP2A, COP1 and -8, CONSTANS-LIKE 1, and PIF3, as well as several other gibberellic acid/phytochrome signaling proteins (Shin et al., 2011). 14-3-3 κ- and χ- single and double null mutants resembled PIF3-overexpressors (Mayfield et al., 2007; Adams et al., 2014), implying a role of certain 14-3-3’s in plant phytochrome action perhaps by acting as antagonists/regulators of PIF3-dependent light-signaling. Further 14-3-3 clients included PRL1 (see HIP4 above) and phyB (Shin et al., 2011), providing intriguing corollaries to the present study. Moreover, whereas in Rc wild-type Arabidopsis seedlings show poor gravitropic orientation, 14-3-3 ν- null mutants showed enhanced gravitropism, rather like those of phyB- (Mayfield et al., 2007). Indeed, 14-3-3 protein association also with phototropin and cryptochrome signaling might imply involvement in different photoreceptor signaling complexes (see Paul et al., 2008), as was suggested for PKS1, NPH3, and PP2A (see Jaedicke et al., 2012). We speculate that the HIP14–phy4 interaction in Physcomitrella might be part of such a complex.
Comparison of phy4 and HIP Expression Data
Generally, partner proteins must be co-expressed (expressed in the same cells at the same times and under the same conditions) unless the absence of one or other is physiologically relevant. Furthermore, the expression levels of functionally associated genes are often correlated (synexpression). We therefore collected available expression data from eFP, Phytozome and Genevestigator for phy4 and HIP1-14 to investigate these aspects. phy4 is expressed to some extent in all analyzed developmental stages and conditions (see Supplementary Materials). Light effects are fairly small (∼twofold differences). Pairwise comparison of HIP and phy4 expression showed correlation implying synexpression in several cases (HIP4–8 and -12). We also performed a hierarchical clustering analysis of all three datasets according to condition and gene similarities to identify HIP’s with expression patterns similar to phy4 (see Supplementary Figure S6). The Genevestigator treatment effects on phy4 were most similar to HIP4 and -6 followed by -7 and -5. The most similar expression level patterns were for HIP12, -4, and -9 followed by -11 and -13 according to the Phytozome treatment data. The expression patterns of HIP4, -6, and -7 followed by -3 were most similar to phy4 according to the eFP developmental data and to HIP3 and -6 followed by -4, -7, and -9 according to Genevestigator anatomy data. Taken together, these comparisons imply synexpression of phy4 in particular with HIP4, -6, and -7, providing indirect support for their possible involvement in phy4 action.
Conclusion
In summary, phy4 steers directional growth in moss filaments, thereby necessarily signaling in the cytoplasm rather than via transcriptional regulation in the nucleus. How this cytoplasmic signal arises is not known, but it might derive from the physical interaction of phy4 with phototropin at plasma membrane (Jaedicke et al., 2012). In the present study we identified and characterized a number of novel HIP’s, all of which are at least partly located in the cytoplasm as are their interactions with phy4. Most are conserved in higher plants. Our findings and their possible significance are discussed in more detail below.
In addition to the Arabidopsis phyA-FHY1 positive control, most of the HIP’s characterized in this work showed significant R-enhanced, FR-reversible binding in Y2H, indicating that at least the phytochrome-typical R/FR photochromicity and associated structural changes were significant in the growth readout. Notably, no FR-enhanced interaction was seen, in harmony with the established dogma that in plants Pr has no physiological function. phy4–phy4 homodimerization, presumably resulting from interactions between DHp-like domains, was also demonstrated. We therefore consider it likely that the full-length holophytochrome hybrid baits employed in our screens were functional and folded similarly to the native photoreceptor. This is rather surprising in the case of the four candidates identified in the BD:phy4 screen because it is unclear how the presumptive knot around the N-terminal PAS domain (Wagner et al., 2005; Essen et al., 2008) can be formed in the presence of the 17 kDa BD extension. Perhaps the BD and/or the loop through which it passes remain/s largely unfolded even after translation of the GAF domain has begun, allowing the knot to form normally. Alternatively, the extended peptide chain might fold back on itself to form a hairpin which alone protrudes through the loop. It is also possible that the holoprotein remains functional even if knot formation is blocked.
Despite our earlier discovery of the interaction between phy4 and phototropin at the plasma membrane in planta, we had quite expected PIF-like proteins to be found in the Y2H screens. This was not the case, however. One explanation might be that, in contrast to fragments, full-length holophytochrome bait is unattractive for such prey in the yeast nucleus. The selection for full-length cDNA’s in the construction of the library we used might also be a significant factor (61% of the prey sequences from the initial screen represented the full-length CDS according to the corresponding gene model). In the case of Arabidopsis phyB for example, the interaction seen with the PHL N-terminus was absent for full-length PHL (Endo et al., 2013). Alternatively, PIF-like transcription factors might not be substrates for phy4. However, as different Y2H screens with Arabidopsis phyA fragments found different partners, it is not surprising that we also found others for Physcomitrella phy4. In any case, even though in the initial screen 10 of the 108 sequences identified were duplicated at least once, the library was not exhaustively screened, so rare phy4 interactors are quite likely to have been missed.
Of the 14 putative HIP’s (full-length CDS gene models showing robust Y2H interaction with phy4 in at least one of the four possible hybrid configurations) only two failed to show split-YFP interaction in planta. This high rate of confirmation is probably partly because our Y2H screening protocol exploited mating rather than double transformation, significantly reducing background activation and thus the likelihood of false positives while enhancing sensitivity. We also suspect that our use of functional Pfr as bait in the initial screens increased the number of true interactors found. On the other hand, despite phy4–HIP interaction being apparent in both Y2H and in planta, false positives are still possible.
We emphasize that even a proven physical interaction does not necessarily represent a signaling route. Consequently, it is unlikely that all the HIP’s identified in this work are involved in phy4 signaling. None has been studied experimentally and most had not even been annotated. Summarizing our analyses of the HIP sequences:
(1) Of the HIP’s with possible associations with general signaling mechanisms, HIP1 is a likely pirin (a protein group suggested to associate phyA with G protein signaling), HIP4 and -5 probably form β-propeller structures and thus, like HIP3, might represent an E3 ubiquitin ligase, HIP7 is a likely signaling protein kinase and HIP8 is probably a Ca2+-dependent protein kinase. HIP13 too might be associated with Ca2+ signaling.
(2) HIP12 and -13 are likely to be involved in translation regulation. This correlates interestingly with phyB regulation of PORA translation by Pfr-dependent interaction with PENTA (Paik et al., 2012). Like HIP3, PENTA is a cytosolic zinc-finger protein.
(3) Various HIP’s might be associated with posttranslational protein modification. HIP7, -8, and perhaps -11 are likely Ser/Thr protein kinases, HIP6 is a likely sulfotransferase and HIP9 might represent an endopeptidase.
(4) HIP4, -5, -12, -13, and -14 are associated in various ways with the cytoskeleton. This is of particular significance because phy4-mediated vectorial effects (photo- and polarotropism in filament tip cells as well as chloroplast translocation – see Mittmann et al., 2004) are associated with microfilaments and their organization (Meske and Hartmann, 1995; Meske et al., 1996). In particular the 14-3-3 protein HIP14 is interesting in this regard.
These and other characteristics of the various HIP’s identified in this study are hypothetical, providing in many cases no more than hints as to their physiological roles. Obviously, the most appropriate way to clarify their true functions is to knockout the relevant loci by homologous recombination (Schaefer and Zrÿd, 1997) as was successful for phy4 itself (Mittmann et al., 2004). This work has been initiated and we have established different physiological assays to study effects on vectorial responses the results of which will clarify the putative involvement of HIP’s in phy4 signaling.
As discussed in the Section “Introduction,” whereas there is no doubt that the cytoplasmic signal retaining vectorial information derives from phytochromes in lower plants, it is unclear whether a related function exists in higher plants. Similarly, whereas phy4–phot interaction at the plasma membrane has been established and is likely to be associated with vectorial signaling in mosses, the situation in higher plants is less clear. We hope that useful clues regarding the latter and/or the evolution of phytochrome signaling will appear with elucidation of the moss system.
Author Contributions
AE, KM, and JH designed research and; AE and KM performed experiments. AE and KM analyzed the data, AE and JH wrote and all authors critically read and corrected the manuscript.
Funding
We gratefully acknowledge the financial support of the Deutsche Forschungsgemeinschaft (Hu702/5 to JH).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tanja Gans and Melanie Bingel for excellent technical assistance, Rabea Meyberg and Natalie Kempel for a subset of the localization data, Anne Holz and Adriaan Dorresteijn for use of the confocal microscope, Mathias Zeidler for numerous valuable suggestions and advice (all University of Gießen), Stefan Rensing and Pierre-François Perroud (University of Marburg, Germany) for helpful advice concerning expression data analysis, Hans Sommer (MPI for Plant Breeding Research, Cologne, Germany) for the pre-transformed Y2H cDNA library, Peter Uetz and Jürgen Haas (University of Karlsruhe, Germany) for the Gateway Y2H destination vectors, and Nebion (Zurich, Switzerland) for providing us with the Genevestigator data.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00613
Abbreviations
AD, GAL4 transcription factor activation domain; AFR, ATTENUATED FAR RED RESPONSE; B, blue; BD, GAL4 transcription factor DNA binding domain; BLAST, basic local alignment search tool; CDPK, calcium-dependent protein kinase; CDS, coding DNA sequence; CFK1, COP9 INTERACTING F-BOX KELCH 1; CFP, cyan fluorescent protein; CHUP1, CHLOROPLAST UNUSUAL POSITIONING; CO, CONSTANS; COP, CONSTITUTIVELY PHOTOMORPHOGENIC; D, dark; DSD, double synthetic dropout; EF1α, translation elongation factor 1α ; eIF-5A, eukaryotic translation initiation factor 5A; FBK, F-box/kelch; FHY1, FAR-RED-ELONGATED HYPOCOTYL 1; FKF1, FLAVIN-BINDING KELCH-REPEAT F-BOX 1; FP, fluorescent protein; FR, far-red; FT, FLOWERING LOCUS T (florigen); GFP, green fluorescent protein; GPA1, G-protein α-subunit; HIP, holophytochrome-interacting protein; HT1, HIGH LEAF TEMPERATURE 1; LKP2, LOV-KELCH PROTEIN 2; NES, nuclear export signal/sequence; NLS, nuclear localization signal/sequence; NPH3, NON-PHOTOTROPIC HYPOCOTYL 3; PCB, phycocyanobilin; Pfr, far-red absorbing form of phytochrome; PHL, PHYTOCHROME-DEPENDENT LATE-FLOWERING; phot, phototropin holoprotein; phy, phytochrome holoprotein; PIF, phytochrome-interacting factor; PKS1, phytochrome kinase substrate 1; PORA, protochlorophyllide oxidoreductase A; PP2A, PROTEIN PHOSPHATASE 2A; Pr, red absorbing form of phytochrome; PRL1, PLEIOTROPIC REGULATORY LOCUS 1; PRN1, pirin 1; QSD, quadruple synthetic dropout; R, Rc/Rp, red, continuous red, pulsed red; RBL, rhomboid-like protein; RFI2, RED AND FAR RED INSENSITIVE 2; RFP, red fluorescent protein; RPT2, ROOT PHOTOTROPISM 2; SCF, SKP1p-Cdc53p (cullin)-RBX1-F-box; SnRK, SNF1-related protein kinase; SOT, sulfotransferase; TSD, triple synthetic dropout; VOZ1, VASCULAR PLANT ONE ZF 1; W, white; Y2H, yeast two-hybrid; YFP, YFPN/YFPC, yellow fluorescent protein, N-terminal region, C-terminal region; YPDA, yeast peptone dextrose adenine; ZF, zinc finger.
Footnotes
- ^www.ncbi.nlm.nih.gov
- ^www.cosmoss.org
- ^bar.utoronto.ca/efp_physcomitrella/cgi-bin/efpWeb.cgi
- ^phytozome.jgi.doe.gov
- ^www.genevestigator.com
- ^biit.cs.ut.ee/clustvis/
- ^www.sbc.su.se/~maccallr/nucpred/
References
Adams, E., Diaz, C., Hong, J. P., and Shin, R. (2014). 14-3-3 proteins participate in light signaling through association with PHYTOCHROME INTERACTING FACTORs. Int. J. Mol. Sci. 15, 22801–22814. doi: 10.3390/ijms151222801
Adams, M., and Jia, Z. (2005). Structural and biochemical analysis reveal pirins to possess quercetinase activity. J. Biol. Chem. 280, 28675–28682. doi: 10.1074/jbc.M501034200
Agatep, R., Kirkpatrick, R. D., Parchaliuk, D. L., Woods, R. A., and Gietz, R. D. (1998). Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol protocol. Tech. Tips Online 3, 133–137. doi: 10.1016/S1366-2120(08)70121-1
Aguilar-Hernandez, H. S., Santos, L., Leon-Galvan, F., Barrera-Pacheco, A., Espitia-Rangel, E., De Leon-Rodriguez, A., et al. (2011). Identification of calcium stress induced genes in amaranth leaves through suppression subtractive hybridization. J. Plant Physiol. 168, 2102–2109. doi: 10.1016/j.jplph.2011.06.006
Baker, R. P., Young, K., Feng, L., Shi, Y., and Urban, S. (2007). Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc. Natl. Acad. Sci. U.S.A. 104, 8257–8262. doi: 10.1073/pnas.0700814104
Bassell, G. J., Powers, C. M., Taneja, K. L., and Singer, R. H. (1994). Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J. Cell Biol. 126, 863–876. doi: 10.1083/jcb.126.4.863
Ben-Shem, A., Fass, D., and Bibi, E. (2007). Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl. Acad. Sci. U.S.A. 104, 462–466. doi: 10.1073/pnas.0609773104
Bhalerao, R. P., Salchert, K., Bakó, L., Ökrész, L., Szabados, L., Muranaka, T., et al. (1999). Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. U.S.A. 96, 5322–5327. doi: 10.1073/pnas.96.9.5322
Borden, K. L., and Freemont, P. S. (1996). The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6, 395–401. doi: 10.1016/S0959-440X(96)80060-1
Boudsocq, M., and Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Bowler, C., Neuhaus, G., Yamagata, H., and Chua, N.-H. (1994). Cyclic GMP and calcium mediate phytochrome phototransduction [published erratum appears in Cell 1994 Nov 18;79(4):743]. Cell 77, 73–81. doi: 10.1016/0092-8674(94)90236-4
Brameier, M., Krings, A., and Maccallum, R. M. (2007). NucPred—Predicting nuclear localization of proteins. Bioinformatics 23, 1159–1160. doi: 10.1093/bioinformatics/btm066
Brandt, B., Brodsky, D. E., Xue, S., Negi, J., Iba, K., Kangasjarvi, J., et al. (2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U.S.A. 109, 10593–10598. doi: 10.1073/pnas.1116590109
Buer, C. S., Kordbacheh, F., Truong, T. T., Hocart, C. H., and Djordjevic, M. A. (2013). Alteration of flavonoid accumulation patterns in transparent testa mutants disturbs auxin transport, gravity responses, and imparts long-term effects on root and shoot architecture. Planta 238, 171–189. doi: 10.1007/s00425-013-1883-3
Cardozo, T., and Pagano, M. (2004). The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751. doi: 10.1038/nrm1471
Chamot, D., and Kuhlemeier, C. (1992). Differential expression of genes encoding the hypusine-containing translation initiation factor, eIF-5A, in tobacco. Nucleic Acids Res. 20, 665–669. doi: 10.1093/nar/20.4.665
Chater, C., Kamisugi, Y., Movahedi, M., Fleming, A., Cuming, A. C., Gray, J. E., et al. (2011). Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr. Biol. 21, 1025–1029. doi: 10.1016/j.cub.2011.04.032
Chatterjee, I., Gross, S. R., Kinzy, T. G., and Chen, K. Y. (2006). Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol. Genet. Genomics 275, 264–276. doi: 10.1007/s00438-005-0086-4
Chen, K. Y. (1983). An 18000-dalton protein metabolically labeled by polyamines in various mammalian cell lines. Biochim. Biophys. Acta 756, 395–402. doi: 10.1016/0304-4165(83)90350-1
Chen, K. Y., and Liu, A. Y. (1997). Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol. Signals 6, 105–109. doi: 10.1159/000109115
Chen, M., and Ni, M. (2006). RED AND FAR-RED INSENSITIVE 2, a RING-domain zinc finger protein, mediates phytochrome-controlled seedling deetiolation responses. Plant Physiol. 140, 457–465. doi: 10.1104/pp.105.073163
Cheng, S.-H., Willmann, M. R., Chen, H.-C., and Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129, 469–485. doi: 10.1104/pp.005645
Cheung, M. Y., Li, M. W., Yung, Y. L., Wen, C. Q., and Lam, H. M. (2013). The unconventional P-loop NTPase OsYchF1 and its regulator OsGAP1 play opposite roles in salinity stress tolerance. Plant Cell Environ. 36, 2008–2020. doi: 10.1111/pce.12108
Christodoulou, J., Malmendal, A., Harper, J. F., and Chazin, W. J. (2004). Evidence for differing roles for each lobe of the calmodulin-like domain in a calcium-dependent protein kinase. J. Biol. Chem. 279, 29092–29100. doi: 10.1074/jbc.M401297200
Citovsky, V., Lee, L. Y., Vyas, S., Glick, E., Chen, M. H., Vainstein, A., et al. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362, 1120–1131. doi: 10.1016/j.jmb.2006.08.017
Collings, D. A., Wasteneys, G. O., Miyazaki, M., and Williamson, R. E. (1994). Elongation factor 1 alpha is a component of the subcortical actin bundles of characean algae. Cell Biol. Int. 18, 1019–1024. doi: 10.1006/cbir.1994.1025
Dammann, C., Ichida, A., Hong, B., Romanowsky, S. M., Hrabak, E. M., Harmon, A. C., et al. (2003). Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 132, 1840–1848. doi: 10.1104/pp.103.020008
Datta, N., Chen, Y. R., and Roux, S. J. (1985). Phytochrome and calcium stimulation of protein phosphorylation in isolated pea nuclei. Biochem. Biophys. Res. Commun. 128, 1403–1408. doi: 10.1016/0006-291X(85)91096-4
de Boer, A. H., Van Kleeff, P. J., and Gao, J. (2013). Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasma 250, 425–440. doi: 10.1007/s00709-012-0437-z
DeBlasio, S. L., Mullen, J. L., Luesse, D. R., and Hangarter, R. P. (2003). Phytochrome modulation of blue light-induced chloroplast movements in Arabidopsis. Plant Physiol. 133, 1471–1479. doi: 10.1104/pp.103.029116
Decker, E. L., Frank, W., Sarnighausen, E., and Reski, R. (2006). Moss systems biology en route: phytohormones in Physcomitrella development. Plant Biol. (Stuttg.) 8, 397–405. doi: 10.1055/s-2006-923952
Demarsy, E., and Fankhauser, C. (2009). Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 12, 69–74. doi: 10.1016/j.pbi.2008.09.002
Demma, M., Warren, V., Hock, R., Dharmawardhane, S., and Condeelis, J. (1990). Isolation of an abundant 50,000-dalton actin filament bundling protein from Dictyostelium amoebae. J. Biol. Chem. 265, 2286–2291.
Denison, F. C., Paul, A. L., Zupanska, A. K., and Ferl, R. J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22, 720–727. doi: 10.1016/j.semcdb.2011.08.006
Dharmawardhane, S., Demma, M., Yang, F., and Condeelis, J. (1991). Compartmentalization and actin binding properties of ABP-50: the elongation factor-1 alpha of Dictyostelium. Cell Motil. Cytoskeleton 20, 279–288. doi: 10.1002/cm.970200404
Ding, S., Zhang, B., and Qin, F. (2015). Arabidopsis RZFP34/CHYR1, a Ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27, 3228–3244. doi: 10.1105/tpc.15.00321
Durso, N. A., and Cyr, R. J. (1994). A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor-1 alpha. Plant Cell 6, 893–905. doi: 10.1105/tpc.6.6.893
Edmonds, B. T., Murray, J., and Condeelis, J. (1995). pH regulation of the F-actin binding properties of Dictyostelium elongation factor 1 alpha. J. Biol. Chem. 270, 15222–15230. doi: 10.1074/jbc.270.25.15222
Endo, M., Tanigawa, Y., Murakami, T., Araki, T., and Nagatani, A. (2013). PHYTOCHROME-DEPENDENT LATE-FLOWERING accelerates flowering through physical interactions with phytochrome B and CONSTANS. Proc. Natl. Acad. Sci. U.S.A. 110, 18017–18022. doi: 10.1073/pnas.1310631110
Eprintsev, A. T., Fedorin, D. N., and Igamberdiev, A. U. (2013). Ca(2)(+) is involved in phytochrome A-dependent regulation of the succinate dehydrogenase gene sdh1-2 in Arabidopsis. J. Plant Physiol. 170, 1349–1352. doi: 10.1016/j.jplph.2013.04.006
Essen, L.-O., Mailliet, J., and Hughes, J. (2008). The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714. doi: 10.1073/pnas.0806477105
Fankhauser, C., Yeh, K. C., Lagarias, J. C., Zhang, H., Elich, T. D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541. doi: 10.1126/science.284.5419.1539
Faulkner, I. J., and Rubery, P. H. (1992). Flavonoids and flavonoid sulphates as probes of auxin-transport regulation in Cucurbita pepo hypocotyl segments and vesicles. Planta 186, 618–625. doi: 10.1007/BF00198044
Feierbach, B., Verde, F., and Chang, F. (2004). Regulation of a formin complex by the microtubule plus end protein tea1p. J. Cell Biol. 165, 697–707. doi: 10.1083/jcb.200403090
Feng, H., Chen, Q., Feng, J., Zhang, J., Yang, X., and Zuo, J. (2007). Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death. Plant Physiol. 144, 1531–1545. doi: 10.1104/pp.107.098079
Fenyk, S., Campillo Ade, S., Pohl, E., Hussey, P. J., and Cann, M. J. (2012). A nucleotide phosphatase activity in the nucleotide binding domain of an orphan resistance protein from rice. J. Biol. Chem. 287, 4023–4032. doi: 10.1074/jbc.M111.314450
Fischer, C., Speth, V., Fleig-Eberenz, S., and Neuhaus, G. (1997). Induction of zygotic polyembryos in wheat: influence of auxin polar transport. Plant Cell 9, 1767–1780. doi: 10.1105/tpc.9.10.1767
Flores-Perez, U., Perez-Gil, J., Closa, M., Wright, L. P., Botella-Pavia, P., Phillips, M. A., et al. (2010). Pleiotropic regulatory locus 1 (PRL1) integrates the regulation of sugar responses with isoprenoid metabolism in Arabidopsis. Mol. Plant 3, 101–112. doi: 10.1093/mp/ssp100
Franciosini, A., Lombardi, B., Iafrate, S., Pecce, V., Mele, G., Lupacchini, L., et al. (2013). The Arabidopsis COP9 SIGNALOSOME INTERACTING F-BOX KELCH 1 protein forms an SCF ubiquitin ligase and regulates hypocotyl elongation. Mol. Plant 6, 1616–1629. doi: 10.1093/mp/sst045
Freemont, P. S. (1993). The RING finger. A novel protein sequence motif related to the zinc finger. Ann. N. Y. Acad. Sci. 684, 174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x
Geiger, D., Scherzer, S., Mumm, P., Marten, I., Ache, P., Matschi, S., et al. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. U.S.A. 107, 8023–8028. doi: 10.1073/pnas.0912030107
Gidda, S. K., Miersch, O., Levitin, A., Schmidt, J., Wasternack, C., and Varin, L. (2003). Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 278, 17895–17900. doi: 10.1074/jbc.M211943200
Gietz, R. D., and Woods, R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. doi: 10.1016/S0076-6879(02)50957-5
Gregio, A. P., Cano, V. P., Avaca, J. S., Valentini, S. R., and Zanelli, C. F. (2009). eIF5A has a function in the elongation step of translation in yeast. Biochem. Biophys. Res. Commun. 380, 785–790. doi: 10.1016/j.bbrc.2009.01.148
Gross, S. R., and Kinzy, T. G. (2005). Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12, 772–778. doi: 10.1038/nsmb979
Hamel, L. P., Sheen, J., and Seguin, A. (2014). Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci. 19, 79–89. doi: 10.1016/j.tplants.2013.10.009
Harmon, F. G., and Kay, S. A. (2003). The F box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr. Biol. 13, 2091–2096. doi: 10.1016/j.cub.2003.11.019
Hartmann, E., and Weber, M. (1988). Storage of the phytochrome-mediated phototropic stimulus of moss protonematal cells. Planta 175, 39–49. doi: 10.1007/BF00402880
Hashimoto, M., Negi, J., Young, J., Israelsson, M., Schroeder, J. I., and Iba, K. (2006). Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nat. Cell Biol. 8, 391–397. doi: 10.1038/ncb1387
Hayashi, M., Inoue, S., Takahashi, K., and Kinoshita, T. (2011). Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+-ATPase in stomatal guard cells. Plant Cell Physiol. 52, 1238–1248. doi: 10.1093/pcp/pcr072
Hayashi, Y., Nakamura, S., Takemiya, A., Takahashi, Y., Shimazaki, K., and Kinoshita, T. (2010). Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol. 51, 1186–1196. doi: 10.1093/pcp/pcq078
Hectors, K., Van Oevelen, S., Geuns, J., Guisez, Y., Jansen, M. A., and Prinsen, E. (2014). Dynamic changes in plant secondary metabolites during UV acclimation in Arabidopsis thaliana. Physiol. Plant. 152, 219–230. doi: 10.1111/ppl.12168
Hiltbrunner, A., Viczian, A., Bury, E., Tscheuschler, A., Kircher, S., Toth, R., et al. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15, 2125–2130. doi: 10.1016/j.cub.2005.10.042
Hirschmann, F., Krause, F., and Papenbrock, J. (2014). The multi-protein family of sulfotransferases in plants: composition, occurrence, substrate specificity, and functions. Front. Plant Sci. 5:556. doi: 10.3389/fpls.2014.00556
Holmes, M. G., and Klein, W. H. (1985). Evidence for phytochrome involvement in light-mediated stomatal movement in Phaseolus vulgaris L. Planta 166, 348–353. doi: 10.1007/BF00401172
Hua, Z., and Vierstra, R. D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334. doi: 10.1146/annurev-arplant-042809-112256
Hughes, J. (2013). Phytochrome cytoplasmic signaling. Annu. Rev. Plant Biol. 64, 377–402. doi: 10.1146/annurev-arplant-050312-120045
Huseby, S., Koprivova, A., Lee, B. R., Saha, S., Mithen, R., Wold, A. B., et al. (2013). Diurnal and light regulation of sulphur assimilation and glucosinolate biosynthesis in Arabidopsis. J. Exp. Bot. 64, 1039–1048. doi: 10.1093/jxb/ers378
Isobe, T., Ichimura, T., Sunaya, T., Okuyama, T., Takahashi, N., Kuwano, R., et al. (1991). Distinct forms of the protein kinase-dependent activator of tyrosine and tryptophan hydroxylases. J. Mol. Biol. 217, 125–132. doi: 10.1016/0022-2836(91)90616-E
Izawa, T., Fukata, Y., Kimura, T., Iwamatsu, A., Dohi, K., and Kaibuchi, K. (2000). Elongation factor-1 alpha is a novel substrate of rho-associated kinase. Biochem. Biophys. Res. Commun. 278, 72–78. doi: 10.1006/bbrc.2000.3772
Jaedicke, K., Lichtenthäler, A. L., Meyberg, R., Zeidler, M., and Hughes, J. (2012). A phytochrome-phototropin light signaling complex at the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 109, 12231–12236. doi: 10.1073/pnas.1120203109
Ji, H., Wang, S., Li, K., Szakonyi, D., Koncz, C., and Li, X. (2015). PRL1 modulates root stem cell niche activity and meristem size through WOX5 and PLTs in Arabidopsis. Plant J. 81, 399–412. doi: 10.1111/tpj.12733
Jones, A. M., Ecker, J. R., and Chen, J. G. (2003). A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol. 131, 1623–1627. doi: 10.1104/pp.102.017624
Kadota, A., Sato, Y., and Wada, M. (2000). Intracellular chloroplast photorelocation in the moss Physcomitrella patens is mediated by phytochrome as well as by a blue-light receptor. Planta 210, 932–937. doi: 10.1007/s004250050700
Kami, C., Hersch, M., Trevisan, M., Genoud, T., Hiltbrunner, A., Bergmann, S., et al. (2012). Nuclear phytochrome A signaling promotes phototropism in Arabidopsis. Plant Cell 24, 566–576. doi: 10.1105/tpc.111.095083
Kanaoka, M. M., Urban, S., Freeman, M., and Okada, K. (2005). An Arabidopsis Rhomboid homolog is an intramembrane protease in plants. FEBS Lett. 579, 5723–5728. doi: 10.1016/j.febslet.2005.09.049
Kanegae, T., Hayashida, E., Kuramoto, C., and Wada, M. (2006). A single chromoprotein with triple chromophores acts as both a phytochrome and a phototropin. Proc. Natl. Acad. Sci. U.S.A. 103, 17997–18001. doi: 10.1073/pnas.0603569103
Kanegae, T., and Kimura, I. (2015). A phytochrome/phototropin chimeric photoreceptor of fern functions as a blue/far-red light-dependent photoreceptor for phototropism in Arabidopsis. Plant J. 83, 480–488. doi: 10.1111/tpj.12903
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY (TM) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Kaur, K. J., and Ruben, L. (1994). Protein translation elongation factor-1 alpha from Trypanosoma brucei binds calmodulin. J. Biol. Chem. 269, 23045–23050.
Kemper, W. M., Merrick, W. C., Redfield, B., Liu, C. K., and Weissbach, H. (1976). Purification and properties of rabbit reticulocyte elongation factor 1. Arch. Biochem. Biophys. 174, 603–612. doi: 10.1016/0003-9861(76)90389-1
Kim, S. G., and Park, C. M. (2007). Membrane-mediated salt stress signaling in flowering time control. Plant Signal. Behav. 2, 517–518. doi: 10.4161/psb.2.6.4645
Kinoshita, T., Emi, T., Tominaga, M., Sakamoto, K., Shigenaga, A., Doi, M., et al. (2003). Blue-light- and phosphorylation-dependent binding of a 14-3-3 protein to phototropins in stomatal guard cells of broad bean. Plant Physiol. 133, 1453–1463. doi: 10.1104/pp.103.029629
Klein, M., and Papenbrock, J. (2004). The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J. Exp. Bot. 55, 1809–1820. doi: 10.1093/jxb/erh183
Klein, M., and Papenbrock, J. (2009). Kinetics and substrate specificities of desulfo-glucosinolate sulfotransferases in Arabidopsis thaliana. Physiol. Plant. 135, 140–149. doi: 10.1111/j.1399-3054.2008.01182.x
Kmiec-Wisniewska, B., Krumpe, K., Urantowka, A., Sakamoto, W., Pratje, E., and Janska, H. (2008). Plant mitochondrial rhomboid, AtRBL12, has different substrate specificity from its yeast counterpart. Plant Mol. Biol. 68, 159–171. doi: 10.1007/s11103-008-9359-8
Knopf, R. R., Feder, A., Mayer, K., Lin, A., Rozenberg, M., Schaller, A., et al. (2012). Rhomboid proteins in the chloroplast envelope affect the level of allene oxide synthase in Arabidopsis thaliana. Plant J. 72, 559–571. doi: 10.1111/j.1365-313X.2012.05090.x
Komori, R., Amano, Y., Ogawa-Ohnishi, M., and Matsubayashi, Y. (2009). Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 15067–15072. doi: 10.1073/pnas.0902801106
Koonin, E. V., Makarova, K. S., Rogozin, I. B., Davidovic, L., Letellier, M. C., and Pellegrini, L. (2003). The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 4:R19.
Koonin, E. V., Wolf, Y. I., and Aravind, L. (2000). Protein fold recognition using sequence profiles and its application in structural genomics. Adv. Protein Chem. 54, 245–275. doi: 10.1016/S0065-3233(00)54008-X
Kosarev, P., Mayer, K. F., and Hardtke, C. S. (2002). Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 3, RESEARCH0016. doi: 10.1186/gb-2002-3-4-research0016
Kudla, J., Batistic, O., and Hashimoto, K. (2010). Calcium signals: the lead currency of plant information processing. Plant Cell 22, 541–563. doi: 10.1105/tpc.109.072686
Kuhn, B. M., Geisler, M., Bigler, L., and Ringli, C. (2011). Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol. 156, 585–595. doi: 10.1104/pp.111.175976
Laity, J. H., Lee, B. M., and Wright, P. E. (2001). Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46. doi: 10.1016/S0959-440X(00)00167-6
Lapik, Y. R., and Kaufman, L. S. (2003). The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein alpha-subunit GPA1 and regulates seed germination and early seedling development. Plant Cell 15, 1578–1590. doi: 10.1105/tpc.011890
Lebska, M., Ciesielski, A., Szymona, L., Godecka, L., Lewandowska-Gnatowska, E., Szczegielniak, J., et al. (2010). Phosphorylation of maize eukaryotic translation initiation factor 5A (eIF5A) by casein kinase 2: identification of phosphorylated residue and influence on intracellular localization of eIF5A. J. Biol. Chem. 285, 6217–6226. doi: 10.1074/jbc.M109.018770
Lee, J. H., Terzaghi, W., Gusmaroli, G., Charron, J. B., Yoon, H. J., Chen, H., et al. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20, 152–167. doi: 10.1105/tpc.107.055418
Lee, S. B., Park, J. H., Kaevel, J., Sramkova, M., Weigert, R., and Park, M. H. (2009). The effect of hypusine modification on the intracellular localization of eIF5A. Biochem. Biophys. Res. Commun. 383, 497–502. doi: 10.1016/j.bbrc.2009.04.049
Lemberg, M. K., and Freeman, M. (2007). Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 17, 1634–1646. doi: 10.1101/gr.6425307
Leng, R. P., Lin, Y., Ma, W., Wu, H., Lemmers, B., Chung, S., et al. (2003). Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112, 779–791. doi: 10.1016/S0092-8674(03)00193-4
Lewandowska-Gnatowska, E., Szymona, L., Lebska, M., Szczegielniak, J., and Muszynska, G. (2011). Phosphorylation of maize eukaryotic translation initiation factor on Ser2 by catalytic subunit CK2. Mol. Cell. Biochem. 356, 241–244. doi: 10.1007/s11010-011-0952-9
Lewis, D. R., Ramirez, M. V., Miller, N. D., Vallabhaneni, P., Ray, W. K., Helm, R. F., et al. (2011). Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 156, 144–164. doi: 10.1104/pp.111.172502
Li, H., Dauwalder, M., and Roux, S. J. (1991). Partial purification and characterization of a Ca(2+)-dependent protein kinase from pea nuclei. Plant Physiol. 96, 720–727. doi: 10.1104/pp.96.3.720
Li, Q., Zhang, N., Zhang, L., and Ma, H. (2015). Differential evolution of members of the rhomboid gene family with conservative and divergent patterns. New Phytol. 206, 368–380. doi: 10.1111/nph.13174
Liese, A., and Romeis, T. (2013). Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta 1833, 1582–1589. doi: 10.1016/j.bbamcr.2012.10.024
Liu, Z., Duguay, J., Ma, F., Wang, T. W., Tshin, R., Hopkins, M. T., et al. (2008). Modulation of eIF5A1 expression alters xylem abundance in Arabidopsis thaliana. J. Exp. Bot. 59, 939–950. doi: 10.1093/jxb/ern017
Long, C., and Iino, M. (2001). Light-dependent osmoregulation in pea stem protoplasts. photoreceptors, tissue specificity, ion relationships, and physiological implications. Plant Physiol. 125, 1854–1869. doi: 10.1104/pp.125.4.1854
Luesse, D. R., Deblasio, S. L., and Hangarter, R. P. (2010). Integration of Phot1, Phot2, and PhyB signalling in light-induced chloroplast movements. J. Exp. Bot. 61, 4387–4397. doi: 10.1093/jxb/erq242
Ma, F., Liu, Z., Wang, T. W., Hopkins, M. T., Peterson, C. A., and Thompson, J. E. (2010). Arabidopsis eIF5A3 influences growth and the response to osmotic and nutrient stress. Plant Cell Environ. 33, 1682–1696. doi: 10.1111/j.1365-3040.2010.02173.x
Ma, S. Y., and Wu, W. H. (2007). AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 65, 511–518. doi: 10.1007/s11103-007-9187-2
Marino, D., Froidure, S., Canonne, J., Ben Khaled, S., Khafif, M., Pouzet, C., et al. (2013). Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun. 4, 1476. doi: 10.1038/ncomms2479
Marsolais, F., Boyd, J., Paredes, Y., Schinas, A. M., Garcia, M., Elzein, S., et al. (2007). Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760). Planta 225, 1233–1244. doi: 10.1007/s00425-006-0413-y
Mata, J., and Nurse, P. (1997). Tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89, 939–949. doi: 10.1016/S0092-8674(00)80279-2
Matrosova, A., Bogireddi, H., Mateo-Penas, A., Hashimoto-Sugimoto, M., Iba, K., Schroeder, J. I., et al. (2015). The HT1 protein kinase is essential for red light-induced stomatal opening and genetically interacts with OST1 in red light and CO2 -induced stomatal movement responses. New Phytol. 208, 1126–1137. doi: 10.1111/nph.13566
Matthews, J. M., and Sunde, M. (2002). Zinc fingers–folds for many occasions. IUBMB Life 54, 351–355. doi: 10.1080/15216540216035
Mayfield, J. D., Folta, K. M., Paul, A. L., and Ferl, R. J. (2007). The 14-3-3 Proteins mu and upsilon influence transition to flowering and early phytochrome response. Plant Physiol. 145, 1692–1702. doi: 10.1104/pp.107.108654
McCurdy, D. W., and Harmon, A. C. (1992). Calcium-dependent protein kinase in the green alga Chara. Planta 188, 54–61. doi: 10.1007/BF00198939
Meske, V., and Hartmann, E. (1995). Reorganisation of microfilaments in protonemal tip cells of the moss Ceratodon purpureus during the phototropic response. Protoplasma 188, 59–69. doi: 10.1007/BF01276796
Meske, V., Rupert, V., and Hartmann, E. (1996). Structural basis for the red light induced repolarisation of tip growth in caulonemal cells of Ceratodon purpureus. Protoplasma 192, 189–198. doi: 10.1007/BF01273891
Milner-White, E. J., Coggins, J. R., and Anton, I. A. (1991). Evidence for an ancestral core structure in nucleotide-binding proteins with the type A motif. J. Mol. Biol. 221, 751–754. doi: 10.1016/0022-2836(91)80170-Y
Mitra, D., and Johri, M. M. (2000). Enhanced expression of a calcium-dependent protein kinase from the moss Funaria hygrometrica under nutritional starvation. J. Biosci. 25, 331–338. doi: 10.1007/BF02703786
Mittmann, F., Brücker, G., Zeidler, M., Repp, A., Abts, T., Hartmann, E., et al. (2004). Targeted knockout in Physcomitrella reveals direct actions of phytochrome in the cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 101, 13939–13944. doi: 10.1073/pnas.0403140101
Mittmann, F., Dienstbach, S., Weisert, A., and Forreiter, C. (2009). Analysis of the phytochrome gene family in Ceratodon purpureus by gene targeting reveals the primary phytochrome responsible for photo- and polarotropism. Planta 230, 27–37. doi: 10.1007/s00425-009-0922-6
Mori, I. C., Murata, Y., Yang, Y., Munemasa, S., Wang, Y. F., Andreoli, S., et al. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 4:e327. doi: 10.1371/journal.pbio.0040327
Myers, C., Romanowsky, S. M., Barron, Y. D., Garg, S., Azuse, C. L., Curran, A., et al. (2009). Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 59, 528–539. doi: 10.1111/j.1365-313X.2009.03894.x
Nemeth, K., Salchert, K., Putnoky, P., Bhalerao, R., Koncz-Kalman, Z., Stankovic-Stangeland, B., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12, 3059–3073. doi: 10.1101/gad.12.19.3059
Neuhaus, G., Bowler, C., Hiratsuka, K., Yamagata, H., and Chua, N.-H. (1997). Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 16, 2554–2564. doi: 10.1093/emboj/16.10.2554
Neuhaus, G., Bowler, C., Kern, R., and Chua, N.-H. (1993). Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell 73, 937–952. doi: 10.1016/0092-8674(93)90272-R
Ng, S., Ivanova, A., Duncan, O., Law, S. R., Van Aken, O., De Clercq, I., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25, 3450–3471. doi: 10.1105/tpc.113.113985
Nozue, K., Kanegae, T., Imaizumi, T., Fukuda, S., Okamoto, H., Yeh, K. C., et al. (1998). A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc. Natl. Acad. Sci. U.S.A. 95, 15826–15830. doi: 10.1073/pnas.95.26.15826
Okamoto, H., Matsui, M., and Deng, X. W. (2001). Overexpression of the heterotrimeric G-protein alpha-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell 13, 1639–1652. doi: 10.2307/3871391
Orozco-Nunnelly, D. A., Muhammad, D., Mezzich, R., Lee, B. S., Jayathilaka, L., Kaufman, L. S., et al. (2014). Pirin1 (PRN1) is a multifunctional protein that regulates quercetin, and impacts specific light and UV responses in the seed-to-seedling transition of Arabidopsis thaliana. PLoS ONE 9:e93371. doi: 10.1371/journal.pone.0093371
Orzaez, D., De Jong, A. J., and Woltering, E. J. (2001). A tomato homologue of the human protein PIRIN is induced during programmed cell death. Plant Mol. Biol. 46, 459–468. doi: 10.1023/A:1010618515051
Paik, I., Yang, S., and Choi, G. (2012). Phytochrome regulates translation of mRNA in the cytosol. Proc. Natl. Acad. Sci. U.S.A. 109, 1335–1340. doi: 10.1073/pnas.1109683109
Pang, H., Bartlam, M., Zeng, Q., Miyatake, H., Hisano, T., Miki, K., et al. (2004). Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J. Biol. Chem. 279, 1491–1498. doi: 10.1074/jbc.M310022200
Park, M. H. (1989). The essential role of hypusine in eukaryotic translation initiation factor 4D (eIF-4D). Purification of eIF-4D and its precursors and comparison of their activities. J. Biol. Chem. 264, 18531–18535.
Park, M. H., Cooper, H. L., and Folk, J. E. (1982). The biosynthesis of protein-bound hypusine (N epsilon -(4-amino-2-hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (N epsilon -(4-aminobutyl)lysine). J. Biol. Chem. 257, 7217–7222.
Park, M. H., Lee, Y. B., and Joe, Y. A. (1997). Hypusine is essential for eukaryotic cell proliferation. Biol. Signals 6, 115–123. doi: 10.1159/000109117
Patel, P. H., Costa-Mattioli, M., Schulze, K. L., and Bellen, H. J. (2009). The Drosophila deoxyhypusine hydroxylase homologue nero and its target eIF5A are required for cell growth and the regulation of autophagy. J. Cell Biol. 185, 1181–1194. doi: 10.1083/jcb.200904161
Pathak, E., Atri, N., and Mishra, R. (2014). Analysis of P-loop and its flanking region subsequence of diverse NTPases reveals evolutionary selected residues. Bioinformation 10, 216–220. doi: 10.6026/97320630010216
Paul, A. L., Folta, K. M., and Ferl, R. J. (2008). 14-3-3 proteins, red light and photoperiodic flowering: a point of connection? Plant Signal. Behav. 3, 511–515. doi: 10.4161/psb.3.8.5717
Paul, A. L., Sehnke, P. C., and Ferl, R. J. (2005). Isoform-specific subcellular localization among 14-3-3 proteins in Arabidopsis seems to be driven by client interactions. Mol. Biol. Cell 16, 1735–1743. doi: 10.1091/mbc.E04-09-0839
Peer, W. A., Brown, D. E., Tague, B. W., Muday, G. K., Taiz, L., and Murphy, A. S. (2001). Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol. 126, 536–548. doi: 10.1104/pp.126.2.536
Pnueli, L., Gutfinger, T., Hareven, D., Ben-Naim, O., Ron, N., Adir, N., et al. (2001). Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13, 2687–2702. doi: 10.2307/3871528
Purwestri, Y. A., Ogaki, Y., Tamaki, S., Tsuji, H., and Shimamoto, K. (2009). The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 50, 429–438. doi: 10.1093/pcp/pcp012
Putnam-Evans, C., Harmon, A. C., Palevitz, B. A., Fechheimer, M., and Cormier, M. J. (1989). Calcium-dependent protein kinase is localized with F-actin in plant cells. Cell Motil. Cytoskeleton 12, 12–22. doi: 10.1002/cm.970120103
Ramakrishnan, C., Dani, V. S., and Ramasarma, T. (2002). A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide-binding and other proteins. Protein Eng. 15, 783–798. doi: 10.1093/protein/15.10.783
Ransom-Hodgkins, W. D. (2009). The application of expression analysis in elucidating the eukaryotic elongation factor one alpha gene family in Arabidopsis thaliana. Mol. Genet. Genomics 281, 391–405. doi: 10.1007/s00438-008-0418-2
Ren, B., Chen, Q., Hong, S., Zhao, W., Feng, J., Feng, H., et al. (2013). The Arabidopsis eukaryotic translation initiation factor eIF5A-2 regulates root protoxylem development by modulating cytokinin signaling. Plant Cell 25, 3841–3857. doi: 10.1105/tpc.113.116236
Roberts, D., Pedmale, U. V., Morrow, J., Sachdev, S., Lechner, E., Tang, X., et al. (2011). Modulation of phototropic responsiveness in Arabidopsis through ubiquitination of phototropin 1 by the CUL3-Ring E3 ubiquitin ligase CRL3(NPH3). Plant Cell 23, 3627–3640. doi: 10.1105/tpc.111.087999
Rösler, J., Klein, I., and Zeidler, M. (2007). Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc. Natl. Acad. Sci. U.S.A. 104, 10737–10742. doi: 10.1073/pnas.0703855104
Rouleau, M., Marsolais, F., Richard, M., Nicolle, L., Voigt, B., Adam, G., et al. (1999). Inactivation of brassinosteroid biological activity by a salicylate-inducible steroid sulfotransferase from Brassica napus. J. Biol. Chem. 274, 20925–20930. doi: 10.1074/jbc.274.30.20925
Rutschmann, F., Stalder, U., Piotrowski, M., Oecking, C., and Schaller, A. (2002). LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 129, 156–168. doi: 10.1104/pp.000869
Saini, P., Eyler, D. E., Green, R., and Dever, T. E. (2009). Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121. doi: 10.1038/nature08034
Saraste, M., Sibbald, P. R., and Wittinghofer, A. (1990). The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434. doi: 10.1016/0968-0004(90)90281-F
Saurin, A. J., Borden, K. L., Boddy, M. N., and Freemont, P. S. (1996). Does this have a familiar RING? Trends Biochem. Sci. 21, 208–214. doi: 10.1016/S0968-0004(96)80017-X
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Schaefer, D. G., and Zrÿd, J.-P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11, 1195–1206. doi: 10.1046/j.1365-313X.1997.11061195.x
Schmidt von Braun, S. (2008). Chup1 – A Chloroplast Movement Protein and its Interactions. Ph.D. dissertation, Ludwig Maximilian University of Munich, Munich.
Schmidt von Braun, S., and Schleiff, E. (2008). The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta 227, 1151–1159. doi: 10.1007/s00425-007-0688-7
Schnepf, E. (1986). Cellular polarity. Ann. Rev. Plant Physiol. 37, 23–47. doi: 10.1146/annurev.pp.37.060186.000323
Schumann, N., Navarro-Quezada, A., Ullrich, K., Kuhl, C., and Quint, M. (2011). Molecular evolution and selection patterns of plant F-box proteins with C-terminal kelch repeats. Plant Physiol. 155, 835–850. doi: 10.1104/pp.110.166579
Seo, P. J., Kim, S. G., and Park, C. M. (2008). Membrane-bound transcription factors in plants. Trends Plant Sci. 13, 550–556. doi: 10.1016/j.tplants.2008.06.008
Shi, X. P., Yin, K. C., Zimolo, Z. A., Stern, A. M., and Waxman, L. (1996). The subcellular distribution of eukaryotic translation initiation factor, eIF-5A, in cultured cells. Exp. Cell Res. 225, 348–356. doi: 10.1006/excr.1996.0185
Shiina, N., Gotoh, Y., Kubomura, N., Iwamatsu, A., and Nishida, E. (1994). Microtubule severing by elongation factor 1 alpha. Science 266, 282–285. doi: 10.1126/science.7939665
Shin, R., Jez, J. M., Basra, A., Zhang, B., and Schachtman, D. P. (2011). 14-3-3 proteins fine-tune plant nutrient metabolism. FEBS Lett. 585, 143–147. doi: 10.1016/j.febslet.2010.11.025
Slobin, L. I. (1980). The role of eucaryotic factor Tu in protein synthesis. The measurement of the elongation factor Tu content of rabbit reticulocytes and other mammalian cells by a sensitive radioimmunoassay. Eur. J. Biochem. 110, 555–563. doi: 10.1111/j.1432-1033.1980.tb04898.x
Stellberger, T., Hauser, R., Baiker, A., Pothineni, V. R., Haas, J., and Uetz, P. (2010). Improving the yeast two-hybrid system with permutated fusions proteins: the Varicella Zoster Virus interactome. Proteome Sci. 8, 8. doi: 10.1186/1477-5956-8-8
Sullivan, S., Thomson, C. E., Kaiserli, E., and Christie, J. M. (2009). Interaction specificity of Arabidopsis 14-3-3 proteins with phototropin receptor kinases. FEBS Lett. 583, 2187–2193. doi: 10.1016/j.febslet.2009.06.011
Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., et al. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. doi: 10.1016/S0092-8674(00)81094-6
Takac, T., Pechan, T., Richter, H., Muller, J., Eck, C., Bohm, N., et al. (2011). Proteomics on brefeldin A-treated Arabidopsis roots reveals profilin 2 as a new protein involved in the cross-talk between vesicular trafficking and the actin cytoskeleton. J. Proteome Res. 10, 488–501. doi: 10.1021/pr100690f
Takagi, S., Kong, S. G., Mineyuki, Y., and Furuya, M. (2003). Regulation of actin-dependent cytoplasmic motility by type II phytochrome occurs within seconds in Vallisneria gigantea epidermal cells. Plant Cell 15, 331–345. doi: 10.1105/tpc.007237
Takase, T., Nishiyama, Y., Tanihigashi, H., Ogura, Y., Miyazaki, Y., Yamada, Y., et al. (2011). LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 67, 608–621. doi: 10.1111/j.1365-313X.2011.04618.x
Takeshita, N., Higashitsuji, Y., Konzack, S., and Fischer, R. (2008). Apical sterol-rich membranes are essential for localizing cell end markers that determine growth directionality in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 19, 339–351. doi: 10.1091/mbc.E07-06-0523
Tanada, T. (1968). A rapid photoreversible response of barley root tips in the presence of 3-indoleacetic acid. Proc. Natl. Acad. Sci. U.S.A. 59, 376–380. doi: 10.1073/pnas.59.2.376
Thelander, M., Nilsson, A., Olsson, T., Johansson, M., Girod, P. A., Schaefer, D. G., et al. (2007). The moss genes PpSKI1 and PpSKI2 encode nuclear SnRK1 interacting proteins with homologues in vascular plants. Plant Mol. Biol. 64, 559–573. doi: 10.1007/s11103-007-9176-5
Thelander, M., Olsson, T., and Ronne, H. (2004). Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J. 23, 1900–1910. doi: 10.1038/sj.emboj.7600182
Thompson, E. P., Smith, S. G., and Glover, B. J. (2012). An Arabidopsis rhomboid protease has roles in the chloroplast and in flower development. J. Exp. Bot. 63, 3559–3570. doi: 10.1093/jxb/ers012
Thompson, J. E., Hopkins, M. T., Taylor, C., and Wang, T. W. (2004). Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends Plant Sci. 9, 174–179. doi: 10.1016/j.tplants.2004.02.008
Tong, Y., Park, I., Hong, B. S., Nedyalkova, L., Tempel, W., and Park, H. W. (2009). Crystal structure of human eIF5A1: insight into functional similarity of human eIF5A1 and eIF5A2. Proteins 75, 1040–1045. doi: 10.1002/prot.22378
Torrelio, B. M., Paz, M. A., and Gallop, P. M. (1984). Cellular proliferation and hypusine synthesis. Exp. Cell Res. 154, 454–463. doi: 10.1016/0014-4827(84)90169-1
Tseng, T. S., Whippo, C., Hangarter, R. P., and Briggs, W. R. (2012). The role of a 14-3-3 protein in stomatal opening mediated by PHOT2 in Arabidopsis. Plant Cell 24, 1114–1126. doi: 10.1105/tpc.111.092130
Ueno, K., Kinoshita, T., Inoue, S., Emi, T., and Shimazaki, K. (2005). Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol. 46, 955–963. doi: 10.1093/pcp/pci104
Uetz, P., and Grigoriev, A. (2005). “The yeast interactome,” in Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics, Vol. 5, eds L. B. Jorde, P. F. R. Little, M. J. Dunn, and S. Subramaniam (Chichester: John Wiley & Sons Ltd), 2033–2051.
Urano, D., Chen, J.-G., Botella, J. R., and Jones, A. M. (2013). Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 3, 120186. doi: 10.1098/rsob.120186
Urban, S., Lee, J. R., and Freeman, M. (2001). Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–182. doi: 10.1016/S0092-8674(01)00525-6
Urban, S., and Wolfe, M. S. (2005). Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc. Natl. Acad. Sci. U.S.A. 102, 1883–1888. doi: 10.1073/pnas.0408306102
Varin, L., Deluca, V., Ibrahim, R. K., and Brisson, N. (1992). Molecular characterization of two plant flavonol sulfotransferases. Proc. Natl. Acad. Sci. U.S.A. 89, 1286–1290. doi: 10.1073/pnas.89.4.1286
Vetter, I. R., and Wittinghofer, A. (1999). Nucleoside triphosphate-binding proteins: different scaffolds to achieve phosphoryl transfer. Q. Rev. Biophys. 32, 1–56. doi: 10.1017/S0033583599003480
Vidal, D., Gil, M. T., Alvarez-Florez, F., Moysset, L., and Simon, E. (2007). Protein kinase activity in Cucumis sativus cotyledons: effect of calcium and light. Phytochemistry 68, 438–445. doi: 10.1016/j.phytochem.2006.11.001
Vidali, L., Augustine, R. C., Kleinman, K. P., and Bezanilla, M. (2007). Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19, 3705–3722. doi: 10.1105/tpc.107.053413
von Schwartzenberg, K. (2006). Moss biology and phytohormones – cytokinins in Physcomitrella. Plant Biol. (Stuttg.) 8, 382–388. doi: 10.1055/s-2006-923962
Wagner, J. R., Brunzelle, J. S., Forest, K. T., and Vierstra, R. D. (2005). A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331. doi: 10.1038/nature04118
Walker, J. E., Saraste, M., Runswick, M. J., and Gay, N. J. (1982). Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951.
Wang, L., Xu, C., Wang, C., and Wang, Y. (2012). Characterization of a eukaryotic translation initiation factor 5A homolog from Tamarix androssowii involved in plant abiotic stress tolerance. BMC Plant Biol. 12:118. doi: 10.1186/1471-2229-12-118
Wang, T. W., Lu, L., Wang, D., and Thompson, J. E. (2001). Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eukaryotic translation initiation factor 5A from tomato. J. Biol. Chem. 276, 17541–17549. doi: 10.1074/jbc.M008544200
Warpeha, K. M., Upadhyay, S., Yeh, J., Adamiak, J., Hawkins, S. I., Lapik, Y. R., et al. (2007). The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 143, 1590–1600. doi: 10.1104/pp.106.089904
Wei, N., Chamovitz, D. A., and Deng, X. W. (1994a). Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development [published erratum appears in Cell 1994 Oct 7;79(1):following 179]. Cell 78, 117–124. doi: 10.1016/0092-8674(94)90578-9
Wei, N., Kwok, S. F., Von Arnim, A. G., Lee, A., Mcnellis, T. W., Piekos, B., et al. (1994b). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6, 629–643. doi: 10.2307/3869868
Wei, N., Tsuge, T., Serino, G., Dohmae, N., Takio, K., Matsui, M., et al. (1998). The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 8, 919–922. doi: 10.1016/S0960-9822(07)00372-7
Weir, B. A., and Yaffe, M. P. (2004). Mmd1p, a novel, conserved protein essential for normal mitochondrial morphology and distribution in the fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell 15, 1656–1665. doi: 10.1091/mbc.E03-06-0371
Wheeler, G. L., and Brownlee, C. (2008). Ca2+ signalling in plants and green algae–changing channels. Trends Plant Sci. 13, 506–514. doi: 10.1016/j.tplants.2008.06.004
Xia, G., Ramachandran, S., Hong, Y., Chan, Y. S., Simanis, V., and Chua, N. H. (1996). Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. Plant J. 10, 761–769. doi: 10.1046/j.1365-313X.1996.10040761.x
Yaffe, M. B. (2002). How do 14-3-3 proteins work?– Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513, 53–57. doi: 10.1016/S0014-5793(01)03288-4
Yamashino, T., Kitayama, M., and Mizuno, T. (2013). Transcription of ST2A encoding a sulfotransferase family protein that is involved in jasmonic acid metabolism is controlled according to the circadian clock- and PIF4/PIF5-mediated external coincidence mechanism in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 77, 2454–2460. doi: 10.1271/bbb.130559
Yang, W., and Boss, W. F. (1994). Regulation of phosphatidylinositol 4-kinase by the protein activator PIK-A49. Activation requires phosphorylation of PIK-A49. J. Biol. Chem. 269, 3852–3857.
Yang, W., Burkhart, W., Cavallius, J., Merrick, W. C., and Boss, W. F. (1993). Purification and characterization of a phosphatidylinositol 4-kinase activator in carrot cells. J. Biol. Chem. 268, 392–398.
Yasui, Y., Mukougawa, K., Uemoto, M., Yokofuji, A., Suzuri, R., Nishitani, A., et al. (2012). The phytochrome-interacting vascular plant one-zinc finger1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 24, 3248–3263. doi: 10.1105/tpc.112.101915
Zanelli, C. F., and Valentini, S. R. (2005). Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics 171, 1571–1581. doi: 10.1534/genetics.105.048082
Zhang, B., Tremousaygue, D., Denance, N., Van Esse, H. P., Horger, A. C., Dabos, P., et al. (2014). PIRIN2 stabilizes cysteine protease XCP2 and increases susceptibility to the vascular pathogen Ralstonia solanacearum in Arabidopsis. Plant J. 79, 1009–1019. doi: 10.1111/tpj.12602
Zhou, W., Wei, L., Xu, J., Zhai, Q., Jiang, H., Chen, R., et al. (2010). Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22, 3692–3709. doi: 10.1105/tpc.110.075721
Zhu, S. Y., Yu, X. C., Wang, X. J., Zhao, R., Li, Y., Fan, R. C., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19, 3019–3036. doi: 10.1105/tpc.107.050666
Keywords: phytochrome, signal transduction, phototropin, Physcomitrella patens, yeast two-hybrid, split-YFP, bimolecular fluorescence complementation, protein–protein interaction
Citation: Ermert AL, Mailliet K and Hughes J (2016) Holophytochrome-Interacting Proteins in Physcomitrella: Putative Actors in Phytochrome Cytoplasmic Signaling. Front. Plant Sci. 7:613. doi: 10.3389/fpls.2016.00613
Received: 01 January 2016; Accepted: 21 April 2016;
Published: 12 May 2016.
Edited by:
Enamul Huq, The University of Texas at Austin, USAReviewed by:
Andreas Hiltbrunner, University of Freiburg, GermanyMargaret Ahmad, University of Paris, France
Copyright © 2016 Ermert, Mailliet and Hughes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jon Hughes, jon.hughes@uni-giessen.de
 Anna Lena Ermert
Anna Lena Ermert Katharina Mailliet
Katharina Mailliet Jon Hughes
Jon Hughes