- 1State Key Laboratory of Protein and Plant Gene Research, Beijing, China
- 2Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
- 3College of Life Sciences, Peking University, Beijing, China
- 4Department of Vegetable Sciences, Beijing Key Laboratory of Growth and Developmental Regulation for Protected Vegetable Crops, China Agricultural University, Beijing, China
- 5National Center of Plant Gene Research, Beijing, China
In our previous efforts to understand the regulatory mechanisms of cucumber unisexual flower development, we observed a stamen-specific down-regulation of the ethylene receptor CsETR1 in stage 6 female flowers of cucumber (Cucumis sativus L.). This down-regulation is correlated with the primordial anther-specific DNA damage that characterizes inappropriate stamen development in cucumber female flowers. To understand how CsETR1 is down regulated in the stamen, we characterized a cucumber MADS box gene homologous to Arabidopsis AP3, CsAP3. We demonstrated that CsAP3 is functionally equivalent to the Arabidopsis B-class MADS gene AP3. However, three novel characteristics of CsAP3 were found. These include firstly, binding and activating CsETR1 promoter in vitro and in vivo; secondly, containing a GV repeat in its C-terminus, which is conserved in cucurbits and required for the transcription activation; and thirdly, decreased expression as the node number increases, which is similar to that found for CsETR1. These findings revealed not only the conserved function of CsAP3 as a B-class floral identity gene, but also its unique functions in regulation of female flower development in cucumber.
Introduction
Unisexual flowers are an efficient way to promote outcrossing and increase the genetic diversity of flowering plants (Bai and Xu, 2013). Understanding the mechanism of unisexual flower development is also important in crop improvement, especially for the crops and vegetables bearing unisexual flowers, such as maize (Zea mays L.) and cucumber (Cucumis sativus L.). For these plants, effective ways of regulating unisexual flower development could increase the efficiency of seed and fruit production.
Cucumber is a well-established model system for unisexual flower development (Perl-Treves, 1999; Bai and Xu, 2013). Since the late 1950’s, two approaches have been taken to decipher the regulatory mechanism of unisexual flower development. The physiological approach led to Witter and Bokovac (1958) reporting that ethylene plays key roles in promoting female flowers. Based on genetic approaches, it was proposed that interactions of three Mendelian genes, F, M, and A, determine whether a bisexual floral bud develops into a male or female flower (Galun, 1961; Kubicki, 1969a,b; Perl-Treves, 1999). Recently, all three genes have been cloned and found to encode 1-aminocyclopropane-1-carboxylic acid synthase, the rate-limiting enzyme in ethylene biosynthesis. F encodes CsACS1G, M encodes CsACS2, and A encodes CsACS11 (Kamachi et al., 1997; Trebitsh et al., 1997; Mibus and Tatlioglu, 2004; Boualem et al., 2008, 2009, 2015; Li et al., 2009; Zhang et al., 2015). This progress confirms at the molecular level that ethylene plays a key role in unisexual flower development in cucumber, consistent with the correlation of ethylene production and genotypes of cucumber unisexual flowers reported by Yamasaki et al. (2001). However, little is known about how these ACS genes affect unisexual flower development.
To understand how a bisexual floral bud develops into either male or female flower, we systematically analyzed the morphogenetic process of cucumber unisexual flowers and revealed that the morphological divergence started from stage 6 flowers (Bai et al., 2004). We further found that the earliest observable biochemical difference is DNA damage at stamen primordia of stage 7 female flowers, which correlated with the arrest of stamen development (Hao et al., 2003). Since physiological investigations have demonstrated the effects of ethylene in promoting female flowers, we investigated whether the DNA damage at the arrested stamens is induced by ethylene. After demonstrating ethylene-induced DNA damage in cucumber protoplasts, we found that the expression of an ethylene receptor gene CsETR1 is stamen-specifically down regulated in stage 6 female floral buds (Wang et al., 2010). Considering the ethylene receptors act as negative regulators of ethylene signal (Hua and Meyerowitz, 1998), stamen-specific down-regulation of the ethylene receptor gene CsETR1 may increase the sensitivity of the stamen to ethylene. Together with the finding of preferential function of an ethylene-inducible DNase gene CsCaN in the stamen primordia of female floral buds (Gu et al., 2011), we proposed that down-regulation of CsETR1 induces stamen-specific DNA damage, leading to an arrest in stamen development in female flowers (Bai and Xu, 2012, 2013). However, it is unknown how CsETR1 is specifically down regulated in the stamens of cucumber female flowers.
One of the most significant breakthroughs in plant developmental biology in the past three decades is the identification of genes determining the developmental identity of floral organs, summarized as the “ABC model” (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994). These findings suggested that floral organ identities are determined by interactions of a group of MADS box genes, i.e., “A” class genes (AP2 in Arabidopsis and their homologs in other plants) alone specify sepal identity; the combination of “A” and “B” class genes (AP3/PI in Arabidopsis, GLO/DEF in Antirrhinum and their homologs in other plants) specifies petal identity; the combination of “B” and “C” class genes (AG in Arabidopsis, PLE in Antirrhinum and their homologs in other plants) specifies stamen identity; and “C” class genes alone specify carpel identity (Bowman et al., 1989, 1991; Carpenter and Coen, 1990; Schwarz-Sommer et al., 1990; Sommer et al., 1990; Yanofsky et al., 1990; Jack et al., 1992; Goto and Meyerowitz, 1994). From the perspective of the “ABC model”, it is reasonable to expect that the stamen-specific down-regulation of CsETR1 in cucumber female flowers could be mediated by a cucumber homolog of the B or C class genes that are responsible for stamen organ identity determination in other species.
To explore whether B or C class genes are involved in the stamen-specific down-regulation of CsETR1 expression, detailed functional analyses of these genes are required. When we started this investigation, one B class gene CUM26 and two C class genes, namely CUM1 and CUM10, were reported in cucumber (Kater et al., 1998, 2001). None of them seems involved in stamen-specific DNA damage and arrest. As no cucumber genome was available until Huang et al. (2009), to identify candidate genes involved in down-regulating CsETR1 expression in the stamens of female flowers, we performed suppression subtractive hybridization (SSH). We identified a MADS box gene of unknown function named CsMADS1 that was differentially expressed in stamens of stage 5 and stage 6 female flowers (Gu et al., 2011).
In this work, we analyzed the characteristics of CsMADS1 in detail. Based on sequence similarity, expression patterns and genetic complementary tests, we demonstrated that this MADS box gene encodes a homolog of Arabidopsis AP3 and therefore renamed it as CsAP3 for clarity. In addition, we found that CsAP3 can bind and activate the promoter of CsETR1, that this activation ability required a GV repeat in its C-terminus, and that CsAP3 expression is decreased in the flowers at higher nodes, consistent with the expression pattern of CsETR1 (Wang et al., 2010). These findings revealed that while the CsAP3 has an equivalent function in organ identity determination to Arabidopsis AP3, it has novel characteristics required for the stamen-specific regulation of CsETR1 expression. These novel characteristics provided a new example for functional divergence of ABC genes.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of the monoecious cucumber (Cucumis sativus L.) line Zhongnong No.5 were purchased from the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (CAASs). The seeds of hermaphrodite line H34 were kindly provided by Prof. Run Cai of Shanghai Jiaotong University. The seeds were sown in soil and plants were grown in a growth room under a day/night light regime of 16/8 h and a temperature of 25/18°C for sample collection.
Plants used for examination of gene expression in the floral buds at various nodes were grown in a greenhouse of the Institute of Vegetables and Flowers, CAAS. Two methods were used to collect floral samples at various nodes. Method 1 was as follows: when plants had 8-9 true leaves and floral buds of the 8th node were around stage 6–8 (when male and female buds could be easily distinguished by eye), floral buds of the 8th nodes and below were collected, separately for male and female, from five plants; floral buds 9–12 were collected from another five plants when the plants had 10–12 true leaves and the floral buds of 12th node were around stage 6–8; floral buds of 15th node and above contained in shoot tips were collected by simply collecting shoot tips of an additional five plants (Supplementary Figure S1A). Method 2 was as follows: five plants were grown to more than 15 nodes. Male and female floral buds around stage 6–8 were collected and grouped according to 8th node and below, 9–12th node, and 15th node and above (Supplementary Figure S1B).
Arabidopsis seeds were treated with 10% NaClO for 10 min and then washed by sterilized water for five times. The seeds were spread on the MS media. Arabidopsis plants were grown under long-day conditions (16-h-light/8-h-dark cycle) at 22°C and transgenic plants were generated using the floral-dip method (Clough and Bent, 1998).
Construction of Transgenic Plants
The ap3–13 mutant was kindly provided by Tom Jack of Dartmouth College. The CsMADS1 complementation line was constructed by transforming the ap3–13 heterozygous plants with P35S:CsMADS1 plasmid (for detailed information see Supplementary Figure S2).
The dual transgenic Arabidopsis lines used for examining the activity of CsAP3 on the CsETR1 promoter were constructed by transforming pER10 or pX6 vector containing CsAP3 into the transgenic Arabidopsis containing the PCsETR1:GUS construct (Supplementary Figure S3). After 14 days of selection on MS containing hygromycin B and kanamycin surviving seedlings were transferred to the MS medium containing 17-β-estradiol. After an induction period of 48 h, RNA was extracted and real-time qPCR was performed to monitor the expression of CsAP3 and GUS. The lines in which the expression of CsAP3 can be induced were used to detect the GUS expression, and those in which the expression of CsAP3 cannot be induced were used as negative controls (for detailed information see Supplementary Figure S4). The induction system was previously reported (Zuo et al., 2000, 2001). pER10 and pX6 vectors were provided by Nam-Hai Chua.
Phylogenetic Analyses
The protein sequence of Arabidopsis thaliana AP3 was used to search for MADS homologs in available genome sequences of 11 representative species1,2 ,3 (Supplementary Table S1). Multiple sequence alignment was performed using ClustalX (Plate-Forme de Bio-Informatique, Illkirch Cedex, France), and phylogenetic analysis of the whole sequences was performed using neighbor-joining (NJ) with the complete deletion option, the Jones–Taylor–Thornton (JTT) model, Gamma Distribution (G) of 1, and 1000 bootstrap replicates in MEGA version 5 (Tamura et al., 2011)4.
Real-Time qPCR Analyses
Total RNA was isolated with Trizol Reagent (Invitrogen #15596-026), digested with DNase I (Takara #2270A) and reverse transcribed to cDNA with Superscript-III reverse transcriptase (Invitrogen # 18080-044) according to the user manual. qPCR analyses were performed using SYBR Premix Ex Taq Mix (Takara # RR420A) and ABI7500 according to the manufacturer’s instructions. The relative expression was calculated according the comparative CT methods (Schmittgen and Livak, 2008). CsACTIN2 (Csa6M484600) and CsTUB1 (Csa4M000580) for cucumber and UBIQUITIN (AT4G05320) for Arabidopsis were used as reference. Primers were designed using Primer Primier 5 and are listed in Supplementary Table S2. For each sample, at least three biological replicates were analyzed and for each PCR, at least three technical replicates were used to calculate the CT value.
In situ and Whole-Mount In situ Hybridization Analyses
Shoot tips containing flower buds at various stages were fixed in 4% paraformaldehyde (PFA) overnight at 4°C. After fixation, tissues were washed, dehydrated, and embedded in wax for sectioning and in situ hybridization as described (Zhang et al., 2013). Tissues for whole-mount in situ hybridization were not embedded, and after dehydration, the procedure was performed according to the methods previously reported (Hejatko et al., 2006). CsAP3-specific regions were amplified with corresponding primer pairs (see Supplementary Table S2) and transcribed in vitro as probes using the Digoxigenin RNA labeling kit (Roche # 11175025910).
EMSA Assays
The full-length CDS regions of CsAP3, CUM26, AP3, and PI were cloned into pCold™ TF vector (TaKaRa #3365) and the verified constructs were transformed into BL21 (DE3) E. coli to express the corresponding proteins. IPTG was added into the LB medium when the OD600 of the BL21 cells was 0.4 to 0.6, and then the cultures were incubated at 16°C for 8 to 10 h. The proteins were purified with Ni-NTA Agarose (QIAGEN #30210) according to the user manual after the induction by IPTG. The purified proteins were analyzed by SDS-PAGE and quantified with the Bradford methods (Kruger, 1994). DNA fragments labeled with the Biotin 3′ End DNA Labeling Kit (Thermo #89818) were used as probes and unlabelled DNA fragments were used as competitor. CArG boxes were designed to be in the middle of the probes. EMSA assays were performed according to the instructions of the LightShift® Chemiluminescent EMSA Kit (Thermo #20148). The probe sequences for EMSA are listed in Supplementary Table S2.
ChIP-qPCR
ChIP was performed according to the protocols previously reported (Liu et al., 2013). The AP3 antibody, which recognizes a region conserved between CsAP3 and AP3, was purchased from Santa Cruz Biotechnology (sc-12639). Chromatin was extracted from flowers of cucumber and Arabidopsis and immunoprecipitated DNA fragments were used as templates for real-time qPCR with specific primers (See Supplementary Table S2). Leaf and IgG were used as negative controls. The same initial amounts of tissue and antibody were used for the leaf control and the same amounts of antibody and IgG was used for the IgG control. The internal reference for Arabidopsis was TUBULIN2 (Liu et al., 2007). To find a proper internal reference for cucumber ChIP-qPCR, we examined the cucumber homolog of Arabidopsis TUBULIN and ACTIN, CsTUBULIN, and CsACTIN. After comparing the ChIP-qPCR results for relative fold enrichment of CsTUBULIN and CsACTIN, we found that they were almost the same. Based on these results, we used CsTUBULIN as the internal reference for cucumber.
Nuclear and Cytoplasmic Protein Extraction
The indicated plant tissues were ground into fine powder with liquid nitrogen, pre-chilled mortars, and pestles. Then, the samples were resuspended in nuclear isolation buffer (Saleh et al., 2008) at 2 g tissue/25 mL buffer. After brief mixing, the homogenized slurry was filtered through double-layer MiraCloth (CALBIOCHEM #475855), then centrifuged at 11000 g for 20 min at 4°C. The nuclear material could be seen at the bottom of the tube as a white tight pellet overlaid by chlorophyll. The supernatants were saved as the cytoplasmic extract and cold nucleus lysis buffer (Saleh et al., 2008) was used to resuspend the pellet (nuclei). Nuclear protein was obtained from the extract by brief sonication and centrifugation at 13800 g for 20 min at 4°C.
Transcription Activity Assay in Yeast
The cDNAs encoding the C-terminal regions of CsAP3, AP3, CsAP3_C_m (without the GV repeat) and AP3_C_m (with the GV repeat inserted into AP3) were cloned into pGBKT7 vectors and then transformed into S. cerevisiae strain AH109. Colonies were first screened on SD medium without Trp. Six independent positive clones were picked arbitrarily, and transferred to SD medium without Trp, His, Ade for high stringency selection. Vector construction, transformation procedures, and screening methods were according to Protocol No. PT3955-1 (Clontech Laboratories).
Dual Luciferase Transient Expression Assay
The cDNAs encoding CsAP3, AP3, CsAP3_m (without the GV repeat) and AP3_m (with the GV repeat inserted into AP3) were cloned into the vector pGreenII-62SK. The 2,000 bp upstream of the CsETR1 coding region were cloned into the vector pGreenII-0800-LUC (Hellens et al., 2005). The verified constructs were transformed into Agrobacterium tumefaciens strain GV3101 and the positive clones were incubated in LB medium until the OD600 was about 0.5. The cell culture was collected by centrifugation and re-suspended in IFB buffer (10 mM MES, 10 mM MgCl2, 150 μM acetosyringone), placed in the darkness for 3 h. The cell culture mixture containing the effector and reporter construct strains at a ratio of 9:1 was inoculated into tobacco (Nicotiana benthamiana) leaves. After about 48 h, leaf disks were collected, and the activity of firefly luciferase and Renillia luciferase were analyzed. Leaf disks inoculated with reporter and empty effector constructs were used as background controls. Firefly and Renillia luciferase assays were performed according to the instructions of the Dual-Luciferase® Reporter Assay System (Promega #E1910) using a GloMaxR 20/20 Luminometer. For each sample, at least eight biological replicates were used.
Results
CsAP3 is a Homolog of Arabidopsis AP3
We previously identified a MADS box-containing EST with differential expression in stamens of various stages. Using 5′RACE (Rapid Amplification of cDNA Ends) and 3′RACE, we got the full length CDS and then deposited it to NCBI under the name of CsMADS1 (accession number AY944060) (Gu et al., 2011). Later it was numerated as Csa3M865440 in the cucumber genome project (Huang et al., 2009). CsMADS1 contains 732 bp encoding 244 amino acids. This sequence is highly similar to Arabidopsis AP3, with 58% identity at the amino acid level and contains all of the conserved domains found in typical MADS proteins (Supplementary Figure S5). Phylogenetic analysis with available information from sequenced organisms (Supplementary Table S1) grouped CsMADS1 into the “B” class, sub-clustered with Arabidopsis AP3, and separate from cucumber CUM26 (CsAAD02250) and Arabidopsis PI (Figure 1). Based on this information, we further analyzed the functions of this newly identified cucumber MADS box gene.
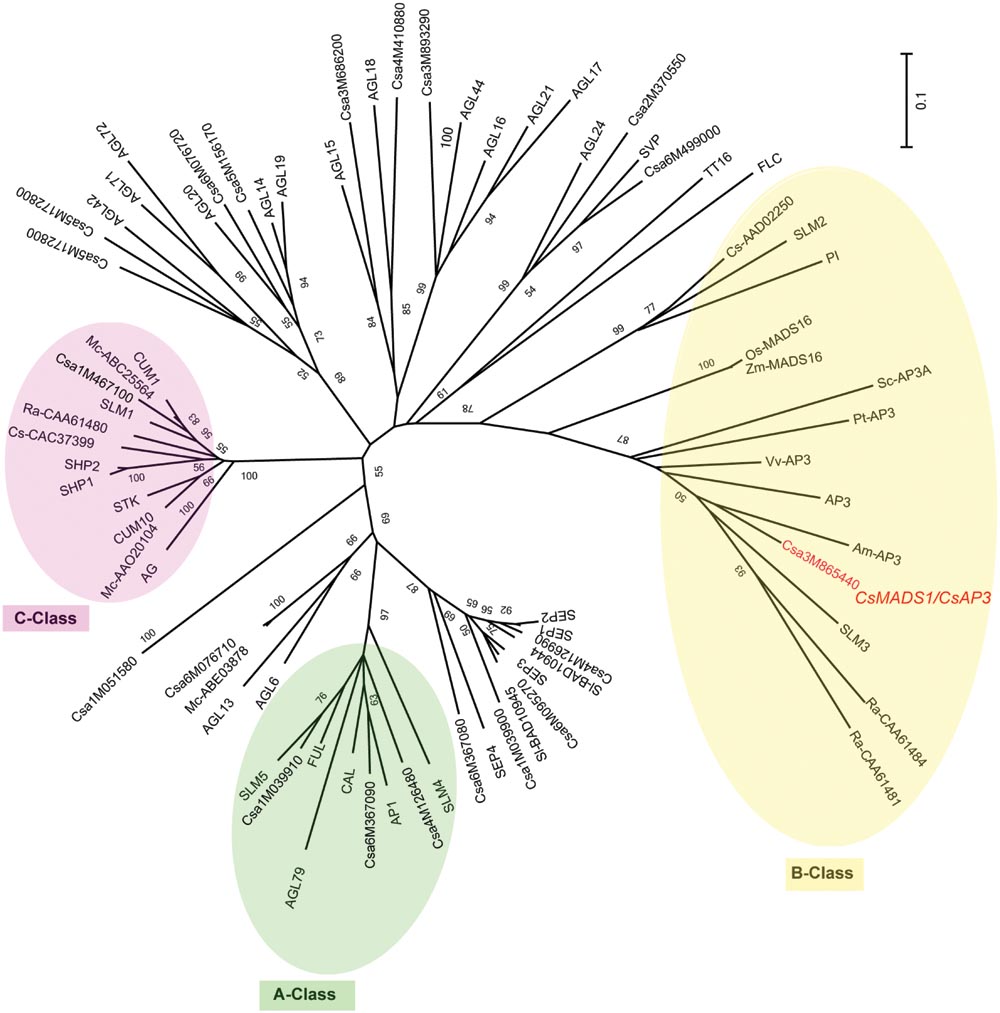
FIGURE 1. Unrooted phylogenetic analysis of the MADS gene family. An unrooted neighbor-joining (NJ) tree based on protein sequence data was constructed (see Materials and Methods). >50% bootstrap values are marked on the tree. A Class proteins are highlighted with a green circle; B Class proteins with a yellow circle; C Class proteins with a pink circle. Csa3M865440 is CsMADS1 (renamed herein CsAP3), and grouped with B Class protein AP3, but not PI. Am (Antirrhinum majus), At (Arabidopsis thaliana), Cs/Csa (Cucumis sativus), Mc (Momordica charantia), Os (Oryza sativa), Pt (Populus trichocarpa), Ra (Rumex acetosa), Sl (Silene latifolia), Sc (Silene conica), Vv (Vitis vinifera), Zm (Zea mays).
CsMADS1 was preferentially expressed in the 2nd and 3rd whorls of floral organs (Figures 2A,B,E–P; Supplementary Figures S6A–K,M), similar to AP3 in Arabidopsis (Jack et al., 1992). However, expression of CsMADS1 was dramatically decreased from stage 7–8 in stamens of female flowers (Figures 2P–R; Supplementary Figure S6L), when the stamen development is clearly inhibited (Hao et al., 2003; Bai et al., 2004). The decreased expression in female flowers is different from AP3 of Arabidopsis and from CsMADS1 expression in the stamen of male cucumber flowers.
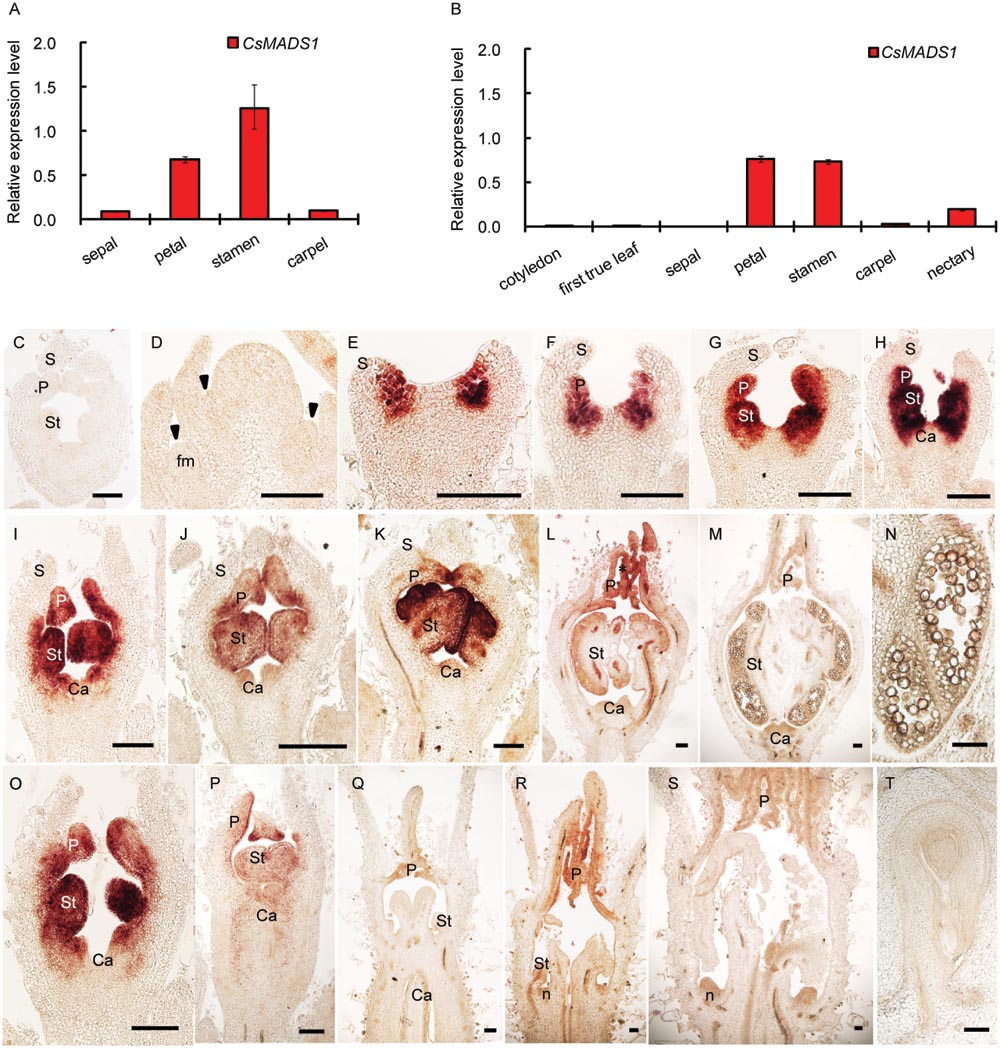
FIGURE 2. Expression pattern of CsMADS1/CsAP3. (A–B) Real-time qPCR analysis showing that CsMADS1/CsAP3 expression was high in stamens and petals of hermaphrodite H34 floral buds at stage 6–8 (A) and monoecious female floral buds at stage 7–8 (B), but low in other organs. (C–T) Detection of CsAP3 with in situ hybridization. (C) Flower bud hybridized with CsAP3 sense probe. (D–H) Longitudinal sections of flower buds at stages 1–5. Arrowheads indicate newly initiated floral buds (early stage 1). (I–M) Longitudinal sections of stages 6–10 male flower buds. (N) Longitudinal section of a stamen of the stage 10 male flower bud. (O–S) CsAP3 expression in female flower buds at stages 6, 7, 8–2, 8–3, and 9, respectively. (T) Longitudinal section of an ovule of a female bud at stage 11. fm, floral meristem; S, sepal; P, petal; St, stamen; Ca, carpel; n, nectary. Scale bars = 100 μm.
To clarify whether CsMADS1 is functionally equivalent to AP3 in Arabidopsis, we carried out a genetic complementation test by transforming an Arabidopsis ap3 mutant (ap3–13) with CsMADS1 (Supplementary Figure S7). CsMADS1 could successfully complement the ap3 mutant phenotype (Figure 3), demonstrating that CsMADS1 is functionally equivalent to AP3 in terms of organ identity determination. Based on these results, we confirmed that CsMADS1 is the homolog of AP3 and re-designated CsMADS1 as CsAP3.
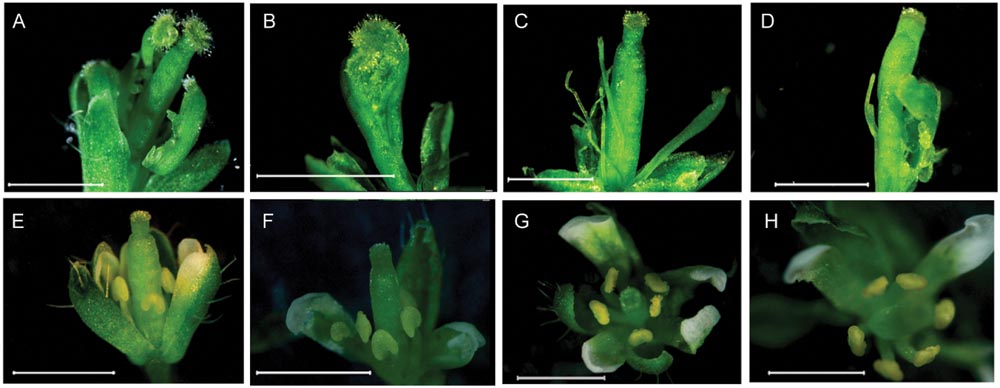
FIGURE 3. Rescue of the Arabidopsis ap3 phenotype with CsMADS1. The Arabidopsis ap3 mutant was transformed with a construct driving constitutive expression of CsMADS1. (A–D) Floral phenotypes of ap3 homozygotes. (E–H) Floral phenotypes of ap3 homozygotes were rescued with transgenic CsMADS1 but with some remaining variations. (E,F) Independent lines 19 and 24. (G,H) Independent line 33.
CsAP3 Binds the CArG Box in the CsETR1 Promoter but AP3 Does Not Bind the ETR1 CArG Box
To examine whether CsAP3 is involved in the regulation of CsETR1 transcription, we first analyzed the promoter of CsETR1, and found typical CArG boxes predicted to be MADS family protein binding sites (Schwarz-Sommer et al., 1992; Treisman and Ammerer, 1992; Riechmann et al., 1996) (Supplementary Table S3). Then, we carried out ChIP-qPCR assays using an antibody capable of recognizing CsAP3 protein (Supplementary Figures S8A,B) and found that the CsETR1 promoter regions containing CArG boxes were significantly enriched in the flower samples but not in leaf samples, which showed no CsAP3 gene expression (Figures 4A,B). When using IgG as another negative control, we got the similar results (Supplementary Figure S8C). These results indicated that CsAP3 binds the CsETR1 promoter in vivo.
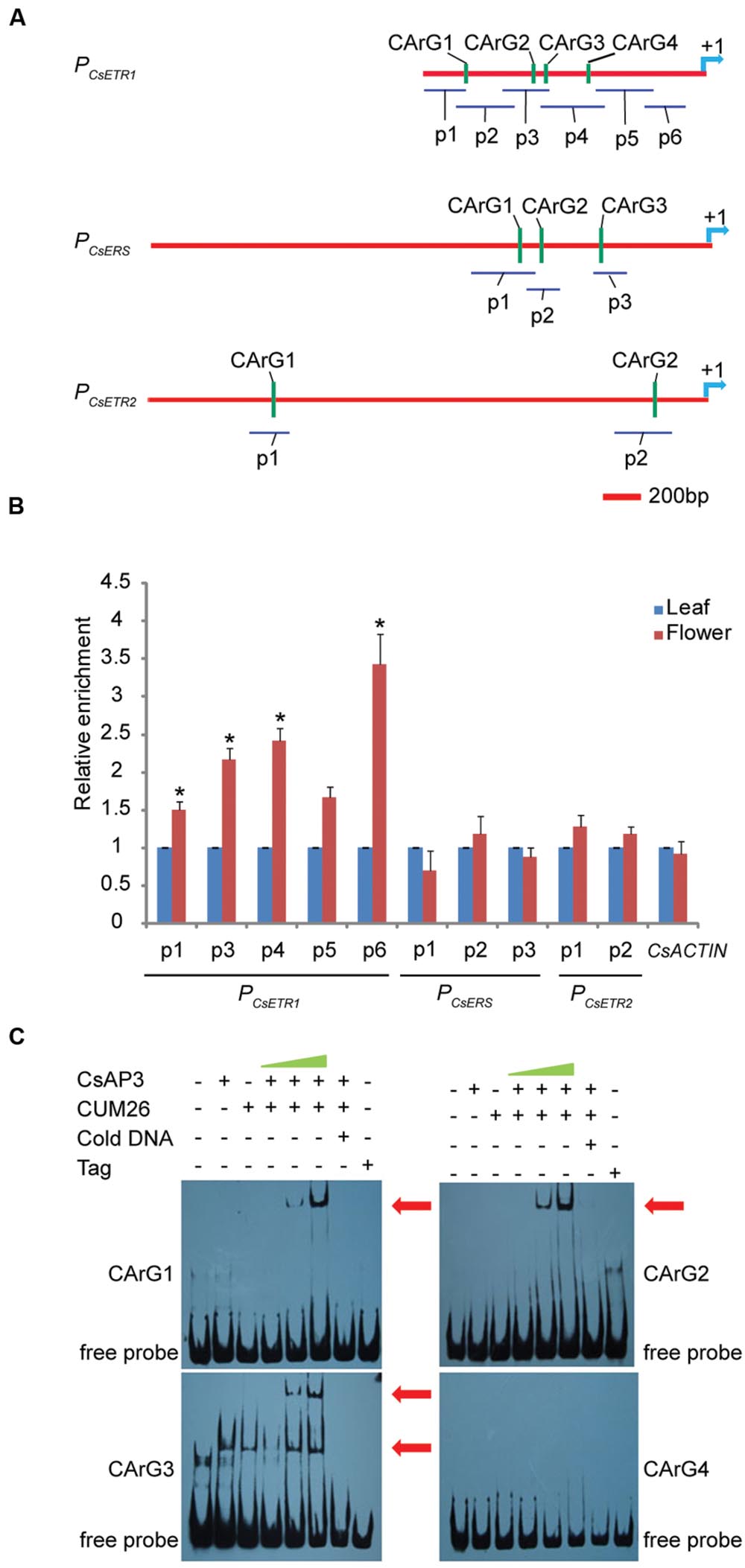
FIGURE 4. CsAP3 can bind the CsETR1 promoter in vivo and in vitro (A) Diagram showing the positions of fragments used for examination of binding activity of CsAP3 to the CsETR1 promoter in vivo by ChIP-qPCR (p1–p6) and in vitro by EMSA (CArG1–CArG4). Primers used in ChIP-qPCR for amplifying the fragments containing CArG box of CsERS and CsETR2 promoters are also shown. The position +1 denotes the adenine of the translational start codon (ATG). (B) ChIP-qPCR showing that CsAP3 significantly enriched fragments of CsETR1 promoter (p1–p4) that contain a CArG box but not p5 (p2 failed to amplify under various conditions). CsAP3 also significantly enriched fragment p6 although it contains no predicted CArG box. CsAP3 did not significantly enrich fragments containing CArG boxes of the CsERS or CsETR2 promoter. The ChIP results are presented as enrichment relative to CsTUBULIN. Error bars indicate SD (n = 3). ∗p < 0.05 in Student’s t-test compared to negative control. (C) EMSA showing that CsAP3 directly bound fragments of the CsETR1 promoter containing CArG boxes. Increasing concentrations of proteins were used and 500-fold molar excess unlabeled DNA fragments were added as competitor. The shifted bands are indicated with red arrowheads.
To examine whether CsAP3 can bind the promoters of two other ethylene receptor genes, we examined the distribution of CArG boxes on the promoters of CsETR2 and CsERS and carried out ChIP-qPCR assays using the above antibody and primers covering these CArG boxes (Figure 4A). None of the examined CsETR2 or CsERS promoter segments was enriched (Figure 4B; Supplementary Figure S8C). These results showed that CsAP3 can selectively bind the promoter of CsETR1, but not CsETR2 or CsERS, suggesting that such binding might be significant in function.
To confirm the binding activity of CsAP3 to the CsETR1 promoter, we further carried out Electrophoretic Mobility Shift Assays (EMSAs) with recombinant CsAP3 (Supplementary Figure S9A). Considering that AP3 functions coordinately with PI (Immink et al., 2010; Smaczniak et al., 2012b), CUM26 protein (Supplementary Figure S9A), the cucumber homolog of Arabidopsis PI (Kater et al., 2001), were used in the EMSA system together with CsAP3. Synthesized DNA fragments containing the CArG box of CsETR1 promoter were used as probe as listed in Supplementary Figure S9B. CsAP3 interacted with CArG boxes 1-3, but not CArG box 4, in the presence of CUM26 (Figure 4C). These results demonstrated that CsAP3 proteins bind the CsETR1 promoter directly.
To clarify whether the binding of CsAP3 upon the CsETR1 promoter is correlated with unisexual flower development in cucumber, we examined if Arabidopsis AP3 can bind ETR1 promoter as Arabidopsis bears only perfect/bisexual flowers. We performed ChIP-qPCR assays with AP3 antibody and the ETR1 promoter, which also contains CArG boxes (Figure 5A). The AP3 antibody could effectively enrich segments containing CArG boxes in AP3 and AP1 genes as previously reported (Sundstrom et al., 2006), demonstrating that the ChIP-qPCR assay worked in Arabidopsis. However, no CArG box-containing segments of the ETR1 promoter were enriched by the AP3 antibody (Figures 5A,B). This result is consistent with recently published ChIP-seq data, in which no ETR1-related DNA sequence was enriched by AP3 antibody (Wuest et al., 2012). To further clarify the interaction between AP3 and CArG boxes in the ETR1 promoter, we carried out EMSA. Figure 5C shows that CArG boxes were not bound by AP3 in the presence of PI in vitro. These results indicate that unlike the CsAP3, AP3 cannot bind the ETR1 promoter, and imply that the CsAP3 binding to the CsETR1 promoter may play roles in cucumber unisexual flower development.
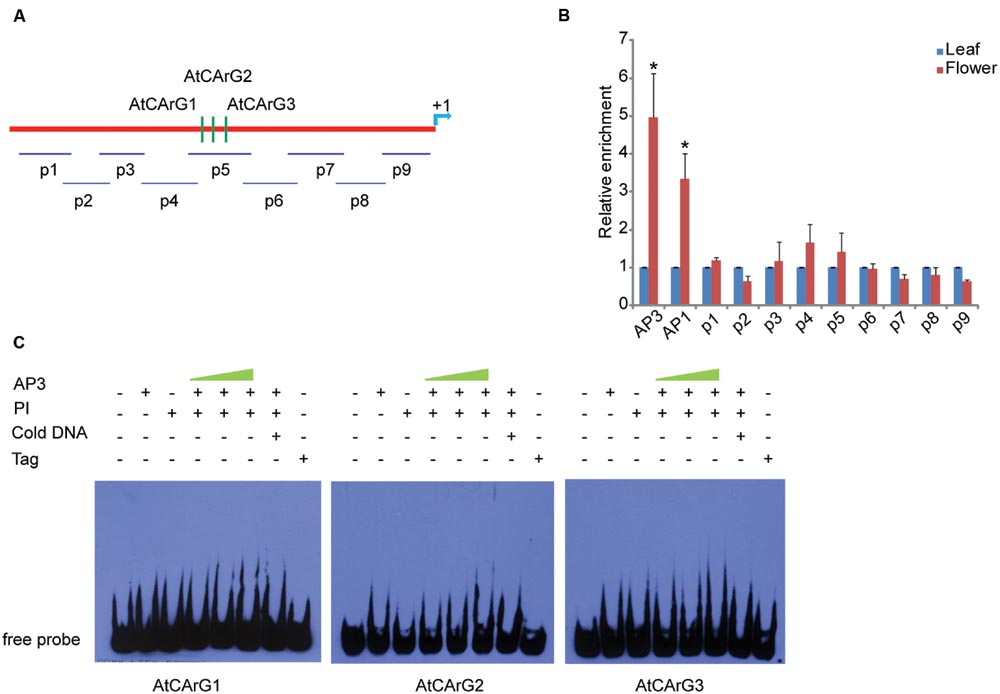
FIGURE 5. AP3 cannot bind the ETR1 promoter effectively in vivo or in vitro (A) Diagram of the positions of fragments used for examination of binding activity of AP3 to the ETR1 promoter [1448 bp upstream of ATG, according to Wang et al. (2003)] in vivo by ChIP-qPCR (p1–p9) and in vitro by EMSA (CArG1–CArG3). The position +1 denotes the adenine of the translational start codon (ATG). (B) ChIP-qPCR showing that AP3 did not enrich the fragments of the ETR1 promoter. Confirmed binding sites of AP3 and AP1 (Sundstrom et al., 2006) were used as positive controls. The ChIP results are presented as relative enrichment to TUBULIN2. Error bars indicate SD (n = 3). ∗p < 0.05 and ∗∗p < 0.01, respectively, in Student’s t-test compared to negative control. (C) EMSA assays showed that AP3 cannot directly bind fragments of the ETR1 promoter containing putative CArG boxes.
CsAP3 Contains an Additional C-terminal GV Repeat that is Required for Transcriptional Activation
We asked whether CsAP3 binding to the CsETR1 promoter could lead to transcriptional regulation of CsETR1. As there is currently no reliable protocol to routinely construct transgenic cucumber, we used transgenic Arabidopsis as an alternative. We constructed dual transgenic Arabidopsis lines containing a CsETR1 promoter-driven GUS gene and an estrogen-inducible G1090 promoter-driven CsAP3 gene (derived from pER10 or pX6, Supplementary Figure S3). If CsAP3 were able to activate the CsETR1 promoter, we expected to observe increased GUS expression upon estrogen-induced CsAP3 expression in the double transgenic lines. Since not all the estrogen-treated plants resulted in induced CsAP3 expression, we used those lines with no CsAP3 induction as internal negative controls. In the lines in which the CsAP3 mRNA was induced by estrogen and CsAP3 protein accumulated, GUS expression is indeed increased (Figures 6A,C). By contrast, in the lines in which the CsAP3 was not induced, GUS expression did not change (Figure 6B). These data suggested that CsAP3 can activate the CsETR1 promoter.
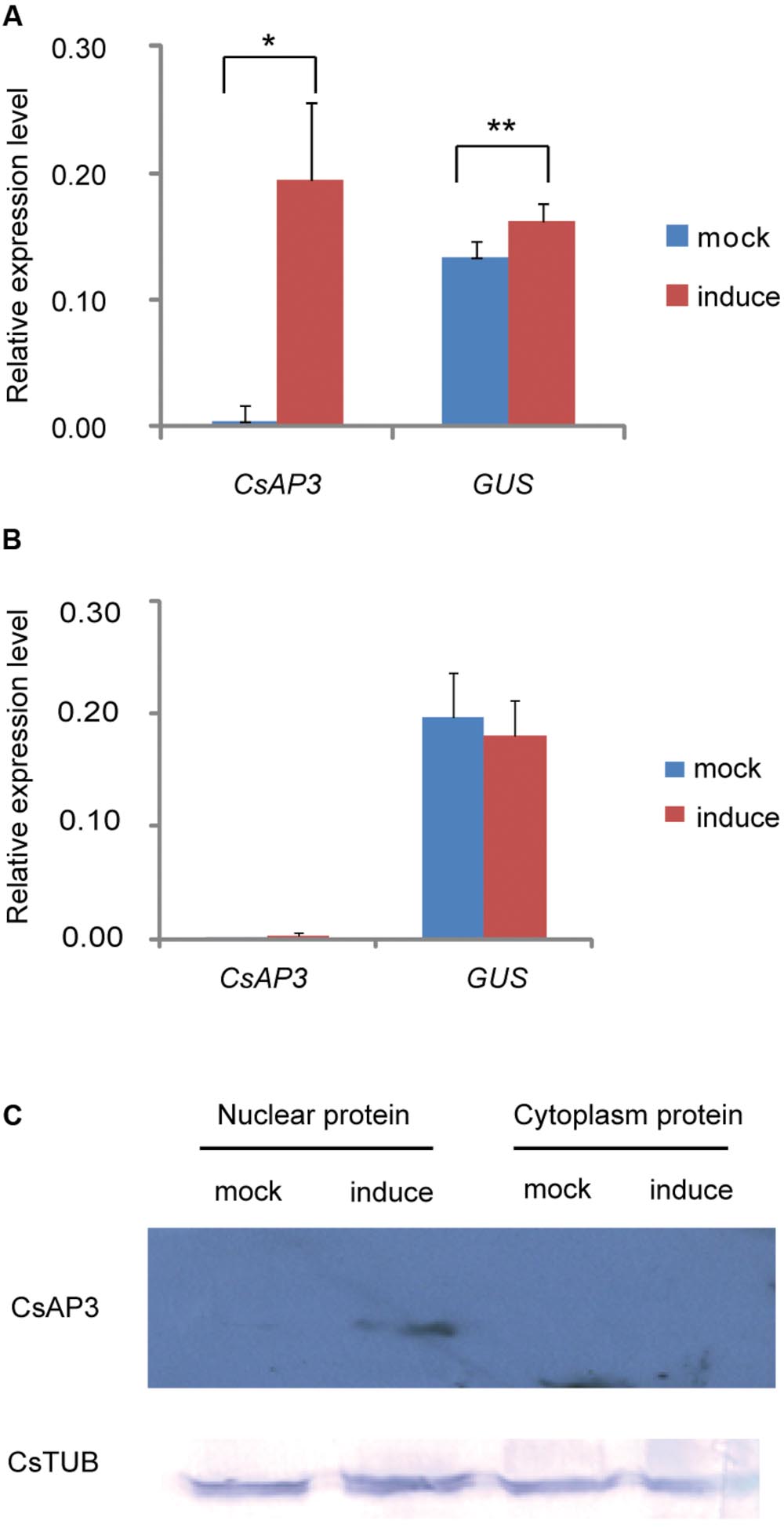
FIGURE 6. CsAP3 can activate the CsETR1 promoter in transgenic Arabidopsis. (A) When expression of CsAP3 was induced by the presence of estrogen, GUS expression was activated (red bar) to significantly higher levels than the background detected in the mock conditions, i.e., in the absence of estrogen (blue bars) (B) In the lines in which the expression of CsAP3 could not be induced in the presence of estrogen, the GUS expression (red bar) was not activated above background levels (blue bar). (C) CsAP3 protein accumulated in the nuclear fraction upon application of estrogen, detected by immunoblotting with the AP3 antibody. CsTUB was used as the loading control, detected by immunoblotting with TUBULIN antibody. Data show mean ± SEM. n = 10 in (A) and n = 6 in (B). ∗p < 0.05 and ∗∗p < 0.01, respectively, in Student’s t-test.
Based on the complementation of the ap3 mutant phenotype, CsAP3 is functionally equivalent to Arabidopsis AP3 in stamen identity determination. However, it has additional functionality in binding and activating the CsETR1 promoter. To clarify the reason, we carried out a detailed sequence comparison between the two proteins. While CsAP3 contains typical MADS and other domains, which are highly conserved and explain its functional equivalence as a B-class protein (Supplementary Figures S5), we found an additional 8-amino acid “GV repeat” (GVGVGIGG) from 185–192 aa of CsAP3 (Figure 7A, red framed). To clarify whether such a “GV repeat” might have functional significance despite occurring in the highly variable C-terminal region, we carried out a sequence comparison with available AP3 homologs from representative plant species. These plants included 6 cucurbits in addition to cucumber, namely Momordica charantia, Luffa cylindrical, Cucurbita maxima, Cucurbita pepo, Cucumis melo, Citrullus lanatus; and 7 non-cucurbits, in addition to Arabidopsis, Antirrhinum majus, Nicotiana tabacum, Vitis vinifera, Euptelea pleiosperma, Akebia trifoliate, Oryza sativa, and Zea mays. The GV repeat was identified in all cucurbits with little variation, but not in any of non-cucurbits even though two of these species, Akebia trifloiata and Zea mays, bear unisexual flowers (Supplementary Figure S10). It is known that all the examined cucurbits plants bear unisexual flowers and their female flowers are promoted by ethylene (Rudich, 1969; Byers et al., 1972; Hossain et al., 1995; Sugiyama et al., 1998; Manzano et al., 2009, 2014; Martinez et al., 2014). The sequence comparison strongly suggests that the GV repeat might have functional relevance to a common feature shared by the cucurbits.
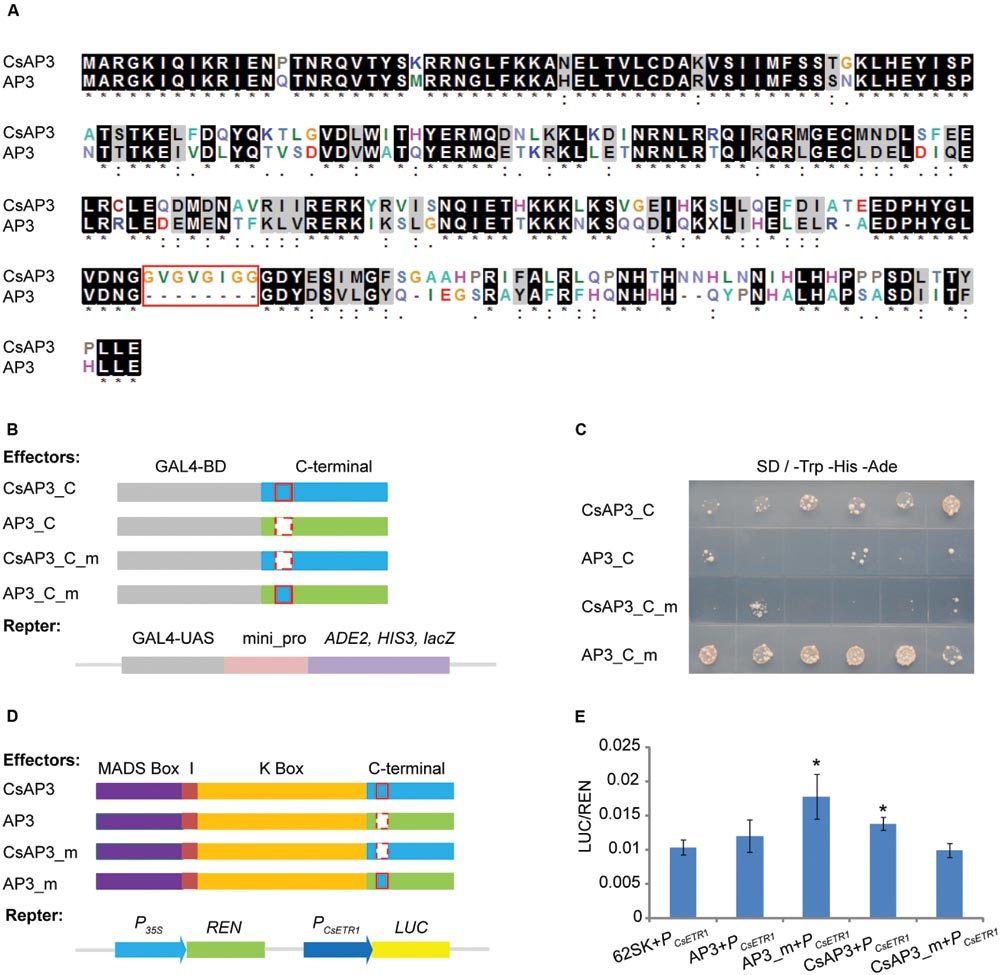
FIGURE 7. The GV-repeat sequence confers transcriptional activation activity to the C-terminal regions of AP3 and CsAP3. (A) ClustalX alignment of CsAP3 and AP3 protein sequences. Amino acid residues displaying 100% identity or similarity in the two homologs are shaded black and gray. The red rectangle shows the GV-repeat sequence in CsAP3. (B) Diagram of effector and reporter vector constructs for the transcriptional activity assay in yeast. Light blue and green show the C-terminal regions of CsAP3 and AP3, respectively. Red rectangles with solid and dashed lines indicate with and without the GV repeat, respectively. (C) Transcriptional activity assay in yeast. Yeast with CsAP3_C or AP3_C_m, both of which contain the GV repeat, could activate the reporter gene ADE2 and HIS3. However, CsAP3_C_m or AP3_C, both lacking the GV repeat, could not. The spots from left to right are independent clones of the same type of transformant. (D) Diagram of effector and reporter vector constructs used in tobacco transient dual luciferase assays. Red rectangles with solid and dashed lines indicate with and without the GV repeat, respectively. (E) Dual luciferase transient expression assay in tobacco leaves. Compared to the negative control (62SK + PCsETR1), AP3_m and CsAP3 up-regulated the LUC/REN ratios significantly but AP3 and CsAP3_m did not. Error bars indicate SEM (n = 8). ∗∗p < 0.01, in Student’s t-test.
To test whether the GV repeats in the C-terminal region, conserved in cucurbits, play any role in CsAP3 activation of the CsETR1 promoter, we conducted transcription activity assays of the CsAP3 and AP3 C-terminus. Figures 7B,C shows that the C-terminus of CsAP3 (with the GV repeats) had transcriptional activity but AP3 (without the GV repeats) did not. Importantly, the C-terminus of CsAP3 lost the transcriptional activation activity when the GV repeats were deleted and the C-terminus of AP3 acquired transcriptional activation activity with the insertion of the GV repeats. These findings demonstrated that the GV repeat is required for transcriptional activity. This characteristic was confirmed in tobacco transient activation assays using full-length CsAP3, AP3 and their mutated forms, by adding the GV repeat to AP3 and removing it from CsAP3 respectively, with CsETR1 promoter as a target (Figures 7D,E). The latter experiment not only verified the transcriptional activation activity of the GV repeat in the CsAP3 C-terminus, uncovering a functional difference between CsAP3 and AP3, but also strengthened the previous conclusion that CsAP3 activates the CsETR1 promoter.
The Expression of CsAP3 Is Decreased at the Upper Nodes
The above results indicated that CsAP3 activates the CsETR1 promoter, implying that higher CsAP3 expression would lead to the higher CsETR1 expression. This correlation of expression levels between CsAP3 and CsETR1 was indeed found in our examination of CsAP3 expression (Figure 2) and the expression examination of CsETR1 shown in Figure 4 of Wang et al. (2010). We found that CsAP3 expression was higher at stage 8 in male flowers with a well-developed stamen, and lower in female flowers with the inhibited stamens (Figure 2). Consistent with these results, Wang et al. (2010) showed that the expression of CsETR1 was lower in the stamens than that in the carpels of stage 6 female flowers and that in the stamens of stage 6 male flowers. This agreement in expression patterns supports the regulatory role of CsAP3 upon CsETR1.
We previously reported that CsETR1 expression in stamen primordia of female floral buds decreased as the node number increased, consistent with the phenomenon that in monoecious cucumber, male flowers are produced in lower nodes and female flower are produced in higher nodes (Wang et al., 2010). If CsAP3 indeed plays role in regulating CsETR1 expression in cucumber, CsAP3 expression should also decrease in later nodes as that of CsETR1 does. Accordingly, we examined CsAP3 expression in stage 6–8 male and female floral buds collected at nodes 8 and below, nodes 9 to 12 and the shoot tip above node 15, with CUM26 and CUM10 expression used for comparison. To avoid interference from sampling methods, we collected samples in two different ways. In one way, we collected floral buds at stage 6-8 from plants grown to the designated nodes (Method 1, Supplementary Figure S1A). Using this method, CsAP3 and CUM26, the two cucumber B class genes, were found to be expressed higher in male flowers than in female flowers (Figure 8). CUM10, the cucumber “C” class gene, was expressed more highly in female flowers than in male flowers (Figure 8C). These expression patterns of the three genes are in line with the respective organ development in unisexual flowers.
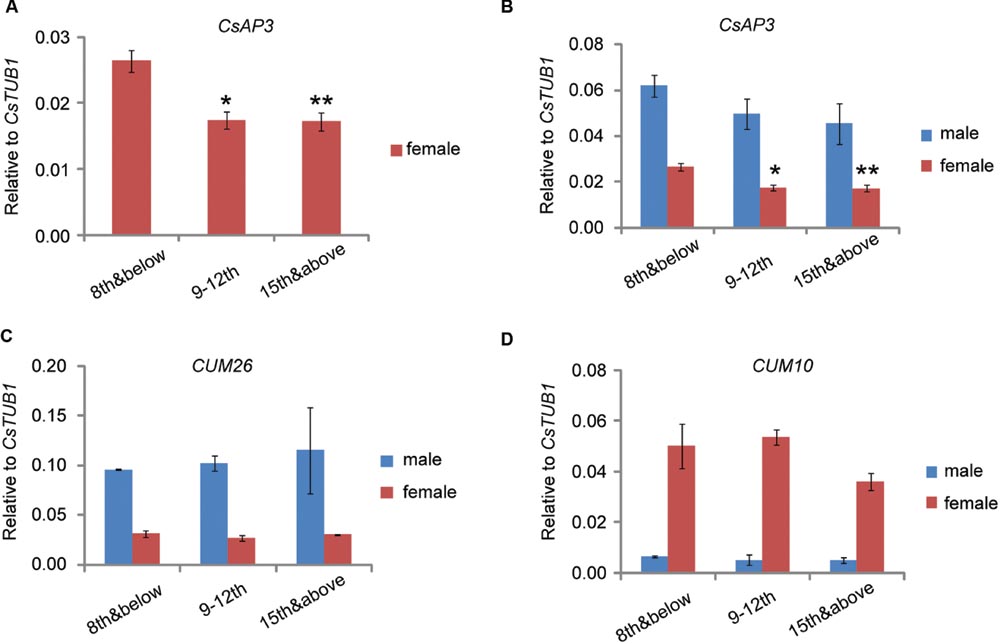
FIGURE 8. CsAP3 expression decreases as the node number increases. Expression levels of CsAP3 (A,B), CUM26 (C), and CUM10 (D) in male and female floral buds collected at nodes 8 and below, nodes 9–12, and the shoot tip. The expression of CsAP3 decreases in higher nodes, not only in male, but more significantly in female flowers (A). The data in this figure are from buds collected from plants grown to the indicated number of nodes (Materials and Methods 1, Supplementary Figure S1A), but the trend of CsAP3 expression in floral buds was independent of the method of sample collection (See Supplementary Figures S1B–E). Data are presented as mean ± SD (n = 3). ∗p < 0.05 and ∗∗p < 0.01, respectively, in Student’s t-test in comparison to the 8th and below value for the same sex floral buds. (A) is the magnified part of CsAP3 expression in female flower from (B).
One phenomenon of particular interest was that although CsAP3 had relatively lower expression in all female flowers, it exhibited a significant decrease in floral buds as the node number increased (Figures 8A,B). No such differences of CUM26 and CUM10 expression were observed in male or female flowers (Figures 8C,D). Similar results were obtained when we collected floral buds at stage 6–8 from the designated nodes in plants grown to more than 15 nodes (Method 2, Supplementary Figure S1B–E). Together with the lower expression of both CsAP3 and CsETR1 in the stamens of female compared to male flowers, the concordant decreases in expression of CsAP3 and CsETR1 in upper nodes suggests that CsAP3 might regulate CsETR1 transcription.
Discussion
To find potential regulators responsible for the stamen-specific down-regulation of CsETR1 in female flowers of cucumber, we characterized the previously cloned CsMADS1 gene that is preferentially expressed in stamen (Gu et al., 2011). We demonstrated that the CsMADS1 gene is the Arabidopsis AP3 homolog in cucumber and renamed it as CsAP3, based on analysis of sequence similarity, expression pattern and functional complementation. We further demonstrated that CsAP3 can bind CArG box-containing regions of the CsETR1 promoter in vitro (based on EMSA) and in vivo (based on ChIP-qPCR), and that it activates the CsETR1 promoter in the inducible assay system in transgenic Arabidopsis plants. Furthermore, we identified a C-terminal GV repeat in cucurbit AP3 homologs and showed that such a GV repeat is responsible for the ability of CsAP3 to transcriptionally activate CsETR1. Based on these findings together with the fact that expressions of both CsAP3 and CsETR1 decrease as the node numbers increase, we propose that CsAP3 likely plays a role in the regulation of the stamen-specific down-regulation of CsETR1 in female cucumber flowers.
Although the lack of effective cucumber transformation methods prevented functional demonstration of the role of CsAP3 in regulating CsETR1 transcription in planta, our work revealed two novel characteristics of CsAP3, in addition to the one that CsAP3 binds and activates the CsETR1 promoter.
One such previously unknown characteristic is the GV repeat found in the C-terminus of CsAP3. B-class MADS box genes are required for stamen organ identity determination and are functionally conserved in various angiosperm species (Causier et al., 2010; Smaczniak et al., 2012a). There are four major domains in MADS box proteins (MADS, I, K, and C). The MADS box is a DNA-binding domain, whereas I and K are dimerization domains (Kaufmann et al., 2005; Smaczniak et al., 2012a). By contrast, the C domain is highly variable and little is known about its function. It has been reported that the C domain has transcriptional activation activity in AP1 and SEP3 but not in AP3, PI, and AG (Honma and Goto, 2001) and the C domain can enhance/stabilize interactions mediated by the K domain (Fan et al., 1997; Kaufmann et al., 2005; Immink et al., 2010). Others have reported that the C domain of AP3 and PI is dispensable for organ identity function (Piwarzyk et al., 2007; Benlloch et al., 2009; Smaczniak et al., 2012b). In database searches, we found the GV repeat in genes from various species including bacteria, fungi, animals and plants (Supplementary Table S4). Although simple sequence repeats in proteins have been described and investigated in various organisms (Urbanus et al., 2010; Dornelas et al., 2011), no functional information regarding the GV repeat was available before its role in transcriptional activation was discovered in this work. Since the GV repeat is highly conserved in cucurbits, it will be interesting to elucidate whether the GV repeat was co-opted exclusively into the AP3 homologs of cucumber relatives and how it occurred. This finding opens up new opportunities to understand the divergence of MADS box genes in sequences and related developmental functions during evolution, particularly in ethylene-related unisexual flower development.
The other novel characteristic discovered herein is the decreased expression of CsAP3 with the node number increase. Although Schultz and Haughn (1991) reported that floral morphology in the lfy mutant changes with node number in inflorescences, no experiment has been conducted to explore if such a phenotype results from a gradient of gene expression. In monoecious cucumber, more male flowers are produced at lower nodes and more female flowers at upper nodes (Currence, 1932; Dax-Fuchs et al., 1978). The decreased expression of CsETR1 and CsAP3 at upper nodes and the regulatory relationship between CsAP3 and CsETR1 seems to properly explain this trend, considering the correlation of stamen-specific down-regulation of CsETR1 and female flower development (Wang et al., 2010). While it is not yet known how the expression of CsAP3 is regulated, the phenomenon that gene expression changes with node number may not be specific to CsAP3 and its target CsETR1, and its significance may not be restricted to unisexual flower development in cucumber.
Our data also show that the activation activity of CsAP3 on transcription of CsETR1 is minor although statistically significant. Considering the monoecious property of cucumber and that all three major genes responsible for the unisexual flower development in cucumber encode ACS, the moderate effect of CsAP3 upon CsETR1 may be crucial for the floral bud at a bipotential stage. The plant would need to sense the internal and external environment sensitively and flexibly and maintain a beneficial balance of male and female ratio of flowers within a single plant. In fact, the connection of a minor decrease in expression of the ethylene receptor with critical developmental events exists not only in cucumber as ethylene-induced stamen-specific DNA damage, but also in the Aponogeton madagascariensis (lace plant), where it results in perforations in leaves through programmed cell death (Rantong et al., 2015). These phenomena highlight that in some situations, moderate regulation of gene expression may play important roles in biological processes.
Author Contributions
J-JS, FL, D-HW, X-FL, XL, NL, H-TG, and CZ, conducted the experiments; J-JS, FL, J-CL, C-XH, S-WH, Z-XZ, Z-HX, and S-NB, designed the experiments; J-JS, and S-NB, wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tom Jack of Dartmouth College, Chi-Kuang Wen of the Institute of Shanghai Plant Physiology and Ecology, Chinese Academy of Sciences (CAS), Jin-Song Zhang of the Institute of Genetics and Developmental Biology (CAS), and Hong-Wei Guo of College of Life Sciences, PKU, for sharing their mutant seeds with us; Nam-Hai Chua of Rockefeller University for sharing his pER10 and pX6 constructs with us; Mei-Qin Chen of College of Life Sciences, PKU, for sharing her experience with whole-mount in situ hybridization; Jia-Ming Zhang of the Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences for his help with sample collection in Hainan Province; and Guang-Yuan Rao of College of Life Sciences, PKU, Anna Kultunow of CSRIO, and Bill Lucas of UC Davis for kind comments and suggestions on the project. This work was supported by grants from ICGEB (CRP/CHN07-02), NSFC (10721403, 90717113), and MST (2011ZX08009-003-003).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01181
Footnotes
- ^http://www.phytozome.net
- ^http://blast.ncbi.nlm.nih.gov
- ^http://cucumber.genomics.org.cn
- ^http://www.megasoftware.net
References
Bai, S. L., Peng, Y. B., Cui, J. X., Gu, H. T., Xu, L. Y., Li, Y. Q., et al. (2004). Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220, 230–240. doi: 10.1007/s00425-004-1342-2
Bai, S. N., and Xu, Z. H. (2012). Bird-nest puzzle: can the study of unisexual flowers such as cucumber solve the problem of plant sex determination? Protoplasma 249(Suppl. 2), S119–S123. doi: 10.1007/s00709-012-0396-4
Bai, S. N., and Xu, Z. H. (2013). Unisexual cucumber flowers, sex and sex differentiation. Int. Rev. Cell Mol. Biol. 304, 1–55. doi: 10.1016/B978-0-12-407696-9.00001-4
Benlloch, R., Roque, E., Ferrandiz, C., Cosson, V., Caballero, T., Penmetsa, R. V., et al. (2009). Analysis of B function in legumes: PISTILLATA proteins do not require the PI motif for floral organ development in Medicago truncatula. Plant J. 60, 102–111. doi: 10.1111/j.1365-313X.2009.03939.x
Boualem, A., Fergany, M., Fernandez, R., Troadec, C., Martin, A., Morin, H., et al. (2008). A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321, 836–838. doi: 10.1126/science.1159023
Boualem, A., Troadec, C., Camps, C., Lemhemdi, A., Morin, H., Sari, M. A., et al. (2015). A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 350, 688–691. doi: 10.1126/science.aac8370
Boualem, A., Troadec, C., Kovalski, I., Sari, M. A., Perl-Treves, R., and Bendahmane, A. (2009). A conserved ethylene biosynthesis enzyme leads to andromonoecy in two cucumis species. PLoS ONE 4:e6144. doi: 10.1371/journal.pone.0006144
Bowman, J. L., Drews, G. N., and Meyerowitz, E. M. (1991). Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3, 749–758. doi: 10.1105/tpc.3.8.749
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. doi: 10.1105/tpc.1.1.37
Byers, R. E., Baker, L. R., Sell, H. M., Herner, R. C., and Dilley, D. R. (1972). Ethylene: a natural regulator of sex expression of Cucumis melo L. Proc. Natl. Acad. Sci. U.S.A. 69, 717–720. doi: 10.1073/pnas.69.3.717
Carpenter, R., and Coen, E. S. (1990). Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev. 4, 1483–1493. doi: 10.1101/gad.4.9.1483
Causier, B., Schwarz-Sommer, Z., and Davies, B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21, 73–79. doi: 10.1016/j.semcdb.2009.10.005
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Currence, T. (1932). Nodal sequence of flower type in cucumber. Proc. Am. Soc. Hortic. Sci. 29, 477–479.
Dax-Fuchs, E., Atsmon, D., and Halevy, A. H. (1978). Vegetative and floral bud abortion in cucumber plants: hormonal and environmental effects. Sci. Hortic. 9, 317–327. doi: 10.1016/0304-4238(78)90041-9
Dornelas, M. C., Patreze, C. M., Angenent, G. C., and Immink, R. G. (2011). MADS: the missing link between identity and growth? Trends Plant Sci. 16, 89–97. doi: 10.1016/j.tplants.2010.11.003
Fan, H. Y., Hu, Y., Tudor, M., and Ma, H. (1997). Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 12, 999–1010. doi: 10.1046/j.1365-313X.1997.12050999.x
Galun, E. (1961). Study of the inheritance of sex expression in the cucumber, the interactions of major genes with modifying genetic and non-genetic factors. Genetica 32, 134–163. doi: 10.1007/BF01816091
Goto, K., and Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Gu, H. T., Wang, D. H., Li, X., He, C. X., Xu, Z. H., and Bai, S. N. (2011). Characterization of an ethylene-inducible, calcium-dependent nuclease that is differentially expressed in cucumber flower development. New Phytol. 192, 590–600. doi: 10.1111/j.1469-8137.2011.03825.x
Hao, Y. J., Wang, D. H., Peng, Y. B., Bai, S. L., Xu, L. Y., Li, Y. Q., et al. (2003). DNA damage in the early primordial anther is closely correlated with stamen arrest in the female flower of cucumber (Cucumis sativus L.). Planta 217, 888–895. doi: 10.1007/s00425-003-1064-x
Hejatko, J., Blilou, I., Brewer, P. B., Friml, J., Scheres, B., and Benkova, E. (2006). In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 1, 1939–1946. doi: 10.1038/nprot.2006.333
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13. doi: 10.1186/1746-4811-1-13
Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. doi: 10.1038/35054083
Hossain, M. A., Islam, M., and Ali, M. (1995). Sexual crossing between two genetically female plants and sex genetics of kakrol (Momordica dioica Roxb.). Euphytica 90, 121–125.
Hua, J., and Meyerowitz, E. M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271. doi: 10.1016/S0092-8674(00)81425-7
Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275–1281. doi: 10.1038/ng.475
Immink, R. G., Kaufmann, K., and Angenent, G. C. (2010). The ‘ABC’ of MADS domain protein behaviour and interactions. Semin. Cell Dev. Biol. 21, 87–93. doi: 10.1016/j.semcdb.2009.10.004
Jack, T., Brockman, L. L., and Meyerowitz, E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. doi: 10.1016/0092-8674(92)90144-2
Kamachi, S., Sekimoto, H., Kondo, N., and Sakai, S. (1997). Cloning of a cDNA for a 1-aminocyclopropane-1-carboxylate synthase that is expressed during development of female flowers at the apices of Cucumis sativus L. Plant Cell Physiol. 38, 1197–1206. doi: 10.1093/oxfordjournals.pcp.a029106
Kater, M. M., Colombo, L., Franken, J., Busscher, M., Masiero, S., Van Lookeren Campagne, M. M., et al. (1998). Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10, 171–182. doi: 10.1105/tpc.10.2.171
Kater, M. M., Franken, J., Carney, K. J., Colombo, L., and Angenent, G. C. (2001). Sex determination in the monoecious species cucumber is confined to specific floral whorls. Plant Cell 13, 481–493. doi: 10.2307/3871401
Kaufmann, K., Melzer, R., and Theissen, G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Kruger, N. J. (1994). The Bradford method for protein quantitation. Methods Mol. Biol. 32, 9–15. doi: 10.1385/0-89603-268-X:9
Kubicki, B. (1969a). Investigations on sex determination in cucumber (Cucumis sativs L.) V. Genes controlling intensity of femaleness. Genet. Pol. 10, 69–85.
Kubicki, B. (1969b). Investigations on sex determination in cucumber (Cucumis sativus L.). VII. Andromonoecism and hermaphroditism. Genet. Pol. 10, 101–120.
Li, Z., Huang, S., Liu, S., Pan, J., Zhang, Z., Tao, Q., et al. (2009). Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics 182, 1381–1385. doi: 10.1534/genetics.109.104737
Liu, C., Li, L. C., Chen, W. Q., Chen, X., Xu, Z. H., and Bai, S. N. (2013). HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes. Plant Cell 25, 257–269. doi: 10.1105/tpc.112.107045
Liu, C., Zhou, J., Bracha-Drori, K., Yalovsky, S., Ito, T., and Yu, H. (2007). Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134, 1901–1910. doi: 10.1242/dev.003103
Manzano, S., Martínez, C., Domínguez, V., Avalos, E., Garrido, D., Gómez, P., et al. (2009). A major gene conferring reduced ethylene sensitivity and maleness in Cucurbita pepo. J. Plant Growth Regul. 29, 73–80. doi: 10.1007/s00344-009-9116-5
Manzano, S., Martinez, C., Garcia, J. M., Megias, Z., and Jamilena, M. (2014). Involvement of ethylene in sex expression and female flower development in watermelon (Citrullus lanatus). Plant Physiol. Biochem. 85, 96–104. doi: 10.1016/j.plaphy.2014.11.004
Martinez, C., Manzano, S., Megias, Z., Barrera, A., Boualem, A., Garrido, D., et al. (2014). Molecular and functional characterization of CpACS27A gene reveals its involvement in monoecy instability and other associated traits in squash (Cucurbita pepo L.). Planta 239, 1201–1215. doi: 10.1007/s00425-014-2043-0
Mibus, H., and Tatlioglu, T. (2004). Molecular characterization and isolation of the F/f gene for femaleness in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 109, 1669–1676. doi: 10.1007/s00122-004-1793-7
Perl-Treves, R. (1999). “Male to female conversion along the cucumber shoot: approaches to studying sex genes and flora development in Cucumis sativus,” in Sex Determination in Plants, ed. C. Ainsworth (Oxford: BIOS Scientific Publishers), 189–216.
Piwarzyk, E., Yang, Y., and Jack, T. (2007). Conserved C-terminal motifs of the Arabidopsis proteins APETALA3 and PISTILLATA are dispensable for floral organ identity function. Plant Physiol. 145, 1495–1505. doi: 10.1104/pp.107.105346
Rantong, G., Evans, R., and Gunawardena, A. H. (2015). Lace plant ethylene receptors, AmERS1a and AmERS1c, regulate ethylene-induced programmed cell death during leaf morphogenesis. Plant Mol. Biol. 89, 215–227. doi: 10.1007/s11103-015-0356-4
Riechmann, J. L., Krizek, B. A., and Meyerowitz, E. M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. U.S.A. 93, 4793–4798. doi: 10.1073/pnas.93.10.4793
Rudich, J. (1969). Increase in femaleness of three cucurbits by treament with Ethrel, an ethylene-releasing compound. Planta 86, 69–76. doi: 10.1007/BF00385305
Saleh, A., Alvarez-Venegas, R., and Avramova, Z. (2008). An efficient chromatin immunoprecipitation (ChIP) pr(Bai and Xu, 2012)otocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3, 1018–1025. doi: 10.1038/nprot.2008.66
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schultz, E. A., and Haughn, G. W. (1991). LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3, 771–781. doi: 10.1105/tpc.3.8.771
Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H., and Sommer, H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250, 931–936. doi: 10.1126/science.250.4983.931
Schwarz-Sommer, Z., Hue, I., Huijser, P., Flor, P. J., Hansen, R., Tetens, F., et al. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11, 251–263.
Smaczniak, C., Immink, R. G., Angenent, G. C., and Kaufmann, K. (2012a). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098. doi: 10.1242/dev.074674
Smaczniak, C., Immink, R. G., Muino, J. M., Blanvillain, R., Busscher, M., Busscher-Lange, J., et al. (2012b). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. U.S.A. 109, 1560–1565. doi: 10.1073/pnas.1112871109
Sommer, H., Beltran, J. P., Huijser, P., Pape, H., Lonnig, W. E., Saedler, H., et al. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 9, 605–613.
Sugiyama, K., Kanno, T., and Morishita, M. (1998). Evaluation method of female flower bearing ability in watermelon using silver thiosulfate (STS). J. Jpn. Soc. Hortic. Sci. 67, 185–189. doi: 10.2503/jjshs.67.185
Sundstrom, J. F., Nakayama, N., Glimelius, K., and Irish, V. F. (2006). Direct regulation of the floral homeotic APETALA1 gene by APETALA3 and PISTILLATA in Arabidopsis. Plant J. 46, 593–600. doi: 10.1111/j.1365-313X.2006.02720.x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Trebitsh, T., Staub, J. E., and O’Neill, S. D. (1997). Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (F) locus that enhances female sex expression in cucumber. Plant Physiol. 113, 987–995. doi: 10.1104/pp.113.3.987
Treisman, R., and Ammerer, G. (1992). The SRF and MCM1 transcription factors. Curr. Opin. Genet. Dev. 2, 221–226. doi: 10.1016/S0959-437X(05)80277-1
Urbanus, S. L., Dinh, Q. D., Angenent, G. C., and Immink, R. G. (2010). Investigation of MADS domain transcription factor dynamics in the floral meristem. Plant Signal. Behav. 5, 1260–1262. doi: 10.4161/psb.5.10.12949
Wang, W., Hall, A. E., O’malley, R., and Bleecker, A. B. (2003). Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc. Natl. Acad. Sci. U.S.A. 100, 352–357. doi: 10.1073/pnas.02370851000237085100
Wang, D. H., Li, F., Duan, Q. H., Han, T., Xu, Z. H., and Bai, S. N. (2010). Ethylene perception is involved in female cucumber flower development. Plant J. 61, 862–872. doi: 10.1111/j.1365-313X.2009.04114.x
Weigel, D., and Meyerowitz, E. M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. doi: 10.1016/0092-8674(94)90291-7
Witter, S. H., and Bokovac, M. J. (1958). The effect of gibberellins on economic crops. Econ. Bot. 12, 213–255. doi: 10.1007/BF02859769
Wuest, S. E., O’Maoileidigh, D. S., Rae, L., Kwasniewska, K., Raganelli, A., Hanczaryk, K., et al. (2012). Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. U.S.A. 109, 13452–13457. doi: 10.1073/pnas.1207075109
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Yamasaki, S., Fujii, N., Matsuura, S., Mizusawa, H., and Takahashi, H. (2001). The M locus and ethylene-controlled sex determination in andromonoecious cucumber plants. Plant Cell Physiol. 42, 608–619. doi: 10.1093/pcp/pce076
Zhang, X., Zhou, Y., Ding, L., Wu, Z., Liu, R., and Meyerowitz, E. M. (2013). Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25, 83–101. doi: 10.1105/tpc.112.107854
Zhang, Z., Mao, L., Chen, H., Bu, F., Li, G., Sun, J., et al. (2015). Genome-wide mapping of structural variations reveals a copy number variant that determines reproductive morphology in cucumber. Plant Cell 27, 1595–1604. doi: 10.1105/tpc.114.135848
Zuo, J., Niu, Q. W., and Chua, N. H. (2000). Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273. doi: 10.1046/j.1365-313x.2000.00868.x
Keywords: cucumber, unisexual flower development, MADS box genes, ethylene, stamen arrestment
Citation: Sun J-J, Li F, Wang D-H, Liu X-F, Li X, Liu N, Gu H-T, Zou C, Luo J-C, He C-X, Huang S-W, Zhang X-L, Xu Z-H and Bai S-N (2016) CsAP3: A Cucumber Homolog to Arabidopsis APETALA3 with Novel Characteristics. Front. Plant Sci. 7:1181. doi: 10.3389/fpls.2016.01181
Received: 13 April 2016; Accepted: 22 July 2016;
Published: 04 August 2016.
Edited by:
Stefan De Folter, Center for Research and Advanced Studies of the National Polytechnic Institute (CINVESTAV), MexicoReviewed by:
Vagner Benedito, West Virginia University, USAJuan José Ripoll, University of California, San Diego, USA
Miguel A. Perez-Amador, Institute of Molecular and Cellular Biology of Plants – Consejo Superior de Investigaciones Científicas, Spain
Copyright © 2016 Sun, Li, Wang, Liu, Li, Liu, Gu, Zou, Luo, He, Huang, Zhang, Xu and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu-Nong Bai, shunongb@pku.edu.cn
†These authors have contributed equally to this work.
 Jin-Jing Sun1,2,3†
Jin-Jing Sun1,2,3† Feng Li
Feng Li Dong-Hui Wang
Dong-Hui Wang Xiao-Feng Liu
Xiao-Feng Liu Chao-Xing He
Chao-Xing He San-Wen Huang
San-Wen Huang Xiao-Lan Zhang
Xiao-Lan Zhang Shu-Nong Bai
Shu-Nong Bai