- 1Division of Plant Science, National Center for Soybean Biotechnology, University of Missouri, Columbia, MO, USA
- 2Departement de Phytologie, Laval University, Quebec, QC, Canada
- 3Division of Plant Sciences and Interdisciplinary Plant Group, Christopher S. Bond Life Sciences Center, University of Missouri, Columbia, MO, USA
Soybean, one of the most important crops worldwide, is severely affected by abiotic stress. Drought and flooding are the major abiotic stresses impacting soybean yield. In this regard, understanding water uptake by plants, its utilization and transport has great importance. In plants, water transport is mainly governed by channel forming aquaporin proteins (AQPs). Tonoplast intrinsic proteins (TIPs) belong to the plant-specific AQP subfamily and are known to have a role in abiotic stress tolerance. In this study, 23 soybean TIP genes were identified based on the latest soybean genome annotation. TIPs were characterized based on conserved structural features and phylogenetic distribution. Expression analysis of soybean TIP genes in various tissues and under abiotic stress conditions demonstrated tissue/stress-response specific differential expression. The natural variations for TIP genes were analyzed using whole genome re-sequencing data available for a set of 106 diverse soybean genotypes including wild types, landraces and elite lines. Results revealed 81 single-nucleotide polymorphisms (SNPs) and several large insertions/deletions in the coding region of TIPs. Among these, non-synonymous SNPs are most likely to have a greater impact on protein function and are candidates for molecular studies as well as for the development of functional markers to assist breeding. The solute transport function of two TIPs was further validated by expression in Xenopus laevis oocytes. GmTIP1;5 was shown to facilitate the rapid movement of water across the oocyte membrane, while GmTIP2;5 facilitated the movement of water and boric acid. The present study provides an initial insight into the possible roles of soybean TIP genes under abiotic stress conditions. Our results will facilitate elucidation of their precise functions during abiotic stress responses and plant development, and will provide potential breeding targets for modifying water movement in soybean.
Introduction
The need for sustainable production of food is a critical issue for human and environmental health due to the continuously growing global population. Soybean, being a source of edible oil and protein rich meal, is considered a promising crop to fulfill the increasing food demand (Krishnamurthy and Shivashankar, 1975). Soybean seed contains over 40% protein and 20% oil. However, stresses imposed by environmental factors greatly affect soybean yield and quality. Water is one of the important factors causing severe yield losses either with excess availability from flooding or with limitation resulting from drought. Plants combat such stresses by regulating water distribution at different levels, such as the vascular system and the permeability of plasma membranes. Aquaporins (AQPs), a class of channel forming proteins, facilitate transport of water and many other solutes across cellular membranes (Javot and Maurel, 2002; Maurel et al., 2002; Tyerman et al., 2002). AQPs are integral membrane proteins that belong to the major intrinsic protein (MIP) family. In higher plants, MIPs are classified into five subfamilies, including plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), NOD 26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs), and uncategorized intrinsic proteins (XIPs) (Chaumont et al., 2001; Kaldenhoff and Fischer, 2006; Danielson and Johanson, 2008).
It has been shown that the expression of TIP genes varies in different tissue, hormone and abiotic treatments. ZmTIP2-3 transcripts was detected only in maize root tissues and was induced by salt and water stresses (Lopez et al., 2004). Cotton GhTIP1:1 transcripts mainly accumulated in roots and hypocotyls under normal conditions, but were dramatically up-regulated in cotyledons and down-regulated in roots within a few hours after cotton seedlings were cold-treated (Li et al., 2009). Several root-specific RB7-type TIP genes have been identified from Arabidopsis thaliana (Yamamoto et al., 1990), Solanum tuberosum (Heinrich et al., 1996), Petroselinum crispum (Roussel et al., 1997), Helianthus annuus (Sarda et al., 1999), Mesembryanthemum crystallinum (Kirch et al., 2000). Recently, one strawberry RB7-type TIP gene, FaRB7 also exhibited a root-specific expression pattern (Vaughan et al., 2006).
Tonoplast intrinsic proteins genes also have been reported to be involved in the elevation of abiotic stress tolerance in several plant species. Notably, the TIP gene TsTIP1;2 from Thellungiella salsuginea provided increased tolerance against drought, salt and oxidative stresses when ectopically expressed in Arabidopsis (Wang et al., 2014). Similarly, increased salinity tolerance was achieved with the heterologous expression of tomato SITIP2;2 in Arabidopsis (Xin et al., 2014). In soybean, Zhang et al. (2016) observed that the expression pattern of GmTIP2;3 was affected by PEG and ABA, and overexpressing GmTIP2;3 in yeast cells improved osmotic stress tolerance. However, another TIP2 gene, GsTIP2;1 cloned from Glycine soja, resulted in reduced tolerance to salt and dehydration stress when overexpressed in Arabidopsis (Wang et al., 2011). Such contrasting results indicate diverse regulation of TIPs within the subfamily that may be due to tissue-specific expression patterns, since GmTIP2;3 was found to be highly expressed in roots whereas GsTIP2;1 showed comparatively higher expression in leaves. Higher water movement by GmTIP2;3 in roots can be correlated with efficient water uptake, leading to enhanced osmotic stress tolerance. In contrast, water movement regulated by GsTIP2;1 seems to increase water loss through transpiration. Similar observations have been reported in rice, where unbalanced expression of most aquaporins in leaves compared to root is thought to result in rapid depletion of leaf water and subsequent inhibition of photosynthesis (Nada and Abogadallah, 2014).
Slow wilting is an important physiological parameter to study water stress tolerance in soybean. Genetic variation observed for the slow wilting trait has proven very useful for improving yield under drought conditions (Sloane et al., 1990). Several genotypes display heritable variation for the slow wilting phenotype. For instance, Sloane et al. (1990) observed delayed wilting in PI 416937 and PI 471938 soybean lines under drought conditions in the field. Recently, two new genotypes, PI 567731 and PI 567690, have been identified for the slow wilting trait under field conditions (Pathan et al., 2014). Interestingly, a study conducted with slow wilting soybean line PI416937 has revealed association between AQPs and hydraulic conductance (Sadok and Sinclair, 2010a,b, 2012). Devi et al. (2015) reported down-regulation of AQPs under high vapor pressure deficit (VPD) conditions in PI 416937, which may be due to the reduced uptake of water, resulting in conservation for later availability. Recently, our study has identified several differentially expressed AQP genes among the slow wilting and fast wilting soybean lines (Prince et al., 2015). These results prompted us to undertake detailed studies of candidate AQPs thought to be involved in soybean abiotic stress tolerance. (Verkman et al., 2014; Madeira et al., 2016).
In the present study, a comprehensive analysis of the soybean TIP gene family was carried out including phylogenetic relationships, chromosomal location, gene duplication status, gene structure, conserved motif, expression profiling under abiotic stress, and natural variation in soybean wild types, landraces and elite lines. The water transport function of two GmTIP proteins was validated through oocyte experiments. Xenopus laevis oocytes have very low background activity that helps to achieve a high signal-to-noise ratio as required to study transporters. Therefore, Xenopus oocytes have been routinely used for the evaluation of solute permeability by several different transporters (Osawa et al., 2006; Verkman et al., 2014). More particularly, AQPs are frequently analyzed using X. laevis oocytes (Maurel et al., 1993; Deshmukh et al., 2015). These data will contribute to future studies to functionally characterize TIP proteins in soybean.
Materials and Methods
Identification and Structural Organization of Tonoplast Intrinsic Protein (TIP) Genes
The Arabidopsis TIP2;1 amino acid sequence was used as query to perform a database search using BLASTP against predicted proteins in the G. max Wm82.a2.v1 genome derived from Phytozome databases. Sequence with at least 50% identity with the query sequence was classified as candidate GmTIPs. BLAST hits with less than a 100 bitscore were removed. Manual curation was then performed to match TIPs identified in the present study with those reported earlier by Deshmukh et al. (2013). The genomic sequences, CDS, and protein sequences for all GmTIPs were retrieved from Phytozome (V111). Novel TIP genes identified with the recent version of the soybean genome annotation were characterized for the aromatic/arginine (Ar/R) selectivity filters (SFs), Froger’s residues, Asn-Pro-Ala (NPA) motifs and the spacing between NPAs. The exon/intron organizations of GmTIPs was visualized with the Gene Structure Display Server program (Hu et al., 2015; GSDS2).
Identification of Conserved Protein Motif and Channel Structure Prediction
The protein sequences were analyzed to identify conserved protein motifs (Motif scan) using the ‘Multiple EM for Motif Elicitation’ (MEME) program (Bailey et al., 2006). Transmembrane domains in newly identified TIPs were predicted using TOPCONS software tools3. Protein structures were modeled based on the structure of Arabidopsis TIP2;14. The Pore Walker tool was used to predict the pore feature5 (Pellegrini-Calace et al., 2009).
Phylogenetic Tree Analysis
Multiple sequence alignments were conducted with the amino acid sequence of GmTIPs. Subsequently, a phylogenetic tree was constructed using the Maximum-Likelihood method provided in the MEGA 6.0 software tool (Tamura et al., 2013). The reliability of an inferred tree was confirmed with bootstrap analysis performed with 1,000 replications.
Chromosomal Distribution and Gene Duplications in GmTIPs
The chromosomal location of all GmTIP genes was obtained through BLASTN searches against the G. Max Wm82.a2.v1 genome database in Phytozome. GmTIPs were located on soybean chromosomes based on physical positions. To further analyze gene duplication events, synonymous substitution (Ks) and non-synonymous substitution (Ka) rates were downloaded from the Plant Genome Duplication Database (PGDD) database6. The date of duplication events was subsequently estimated according to the equation T = Ks/2λ, in which the mean synonymous substitution rate (λ) for soybean is 6.1 × 10-9 (Lynch and Conery, 2000).
Expression Profiling Using RNA-seq Datasets
The RNA-seq data generated by Libault et al. (2010) for nine different tissues including flower, leaves, nodules, pod, root, root hair, seed, shoot apical meristem, and stem were used to analyze expression patterns of GmTIPs. Expression profiling of GmTIPs in leaf tissues of PI 567690 (drought tolerant) and Pana (drought susceptible) grown under drought conditions, and PI 408105A (flooding tolerant) and S99-2281 (PI 654356, flooding susceptible) grown under flooding conditions, was extracted from earlier reported data (Mutava et al., 2015; Prince et al., 2015; Syed et al., 2015). Briefly, at the V5 stage (five unfolded trifoliate leaves), drought stress was imposed by withdrawing water for 21 days and flooding stress was imposed by overwatering for 15 days. Similarly, RNA-seq data for Williams 82 plants subjected to very mild stress (VMS), mild stress (MS), and severe stress (SS) conditions, as well as recovery from severe stress after re-watering (SR), was used to study expression of GmTIPs (Song et al., 2016). Hierarchical clustering of expression data was performed using dCHIP software (Li and Wong, 2001).
Analysis of Synonymous and Non-synonymous SNP Variants in 106 Soybean Lines
All single-nucleotide polymorphism (SNP) datasets located in exonic regions were extracted from whole genome re-sequencing (sequencing depth approximately 15X) data (Valliyodan et al., 2016) as described by Patil et al. (2016). Annotation and effect prediction for the SNPs and other variants were performed using SnpEff7 SNPs were further classified into synonymous and non-synonymous categories.
Xenopus laevis Oocyte Assay
Full length cDNAs of soybean aquaporins were cloned into the Xenopus expression vector pOO2, which contains 5′ and 3′ UTR regions of the X. laevis β-globin gene, including an extended polyA tract for improved mRNA stability and expression (Ludewig et al., 2002). cRNA was synthesized using the SP6 Ambion mMessage mMachine kit with linearized pOO2 constructs as templates. Oocytes were isolated and maintained as described (Pike et al., 2009) and injected with 46 ng of cRNA per oocyte. Water transport was evaluated by perfusing oocytes with hypoosmotic ND96 solution and monitoring swelling over time (Maurel et al., 1993; Durbak et al., 2014). In addition, boron transport was evaluated by replacing the NaCl-containing ND96 with an isoosmotic boron-ND96 solution and monitoring swelling over time (Durbak et al., 2014).
Results
Genome-Wide Identification of Tonoplast Intrinsic Proteins in Soybean
A total of twenty-three TIP genes was identified based on a comprehensive phylogenetic tree analysis within the GmMIP gene family (Deshmukh et al., 2013; Zhang et al., 2013). Here TIP genes were reanalyzed based on a recently released version of the soybean genome annotation (Wm82.a2.v1). The number of TIPs identified in soybean is in good agreement with earlier studies performed with a previous genome annotation (Deshmukh et al., 2013; Zhang et al., 2013). However, the transcript of GmTIP4;2 (Glyma04G08830) reported in earlier studies seems to have been based on a mispredicted gene model and was therefore removed from the Wm82.a2.v1 annotation. Another TIP gene, Glyma12g01490, earlier reported as a pseudo-gene because of transcript truncation, was determined to generate a full-length transcript (new ID Glyma.12G012300) in the new version. Two other TIP pseudogenes reported in earlier studies were not present in the new genome version. Therefore, one gene was removed and one added, resulting in a corrected set of 23 TIPs. The gene names, gene IDs and locations in the genome are listed in Supplementary Table S1. The newly identified TIP gene (Glyma12G01490) was named GmTIP5;2 according to the clustering in the phylogenetic tree.
Phylogenetic Relationship and Gene Structure
To get a better understanding of the evolutionary history, an unrooted phylogenetic tree was constructed using the Maximum-Likelihood (ML) method on the basis of multiple sequence alignment of the 23 soybean TIP proteins (Figure 1A). According to the ML phylogenetic tree, the TIP family is divided into five subgroups designated as Group 1–Group 5. Group 1, the largest clade, contains nine members, representing 39.1% of the total TIP genes. Group 4 constitutes the smallest clade with only one member. To gain further insights into the evolutionary relationships among GmTIP genes, the exon/intron structures of individual GmTIP genes were predicted based on the alignment of CDS sequences with corresponding genomic DNA sequences. As illustrated in Figure 1B, 20 out of 23 GmTIP genes have three exons, while the remaining three only possess two exons. Genes within the same clade demonstrated similar exon/intron distribution patterns in terms of exon/intron length, with the exception of GmTIP5;2 containing a short second exon.
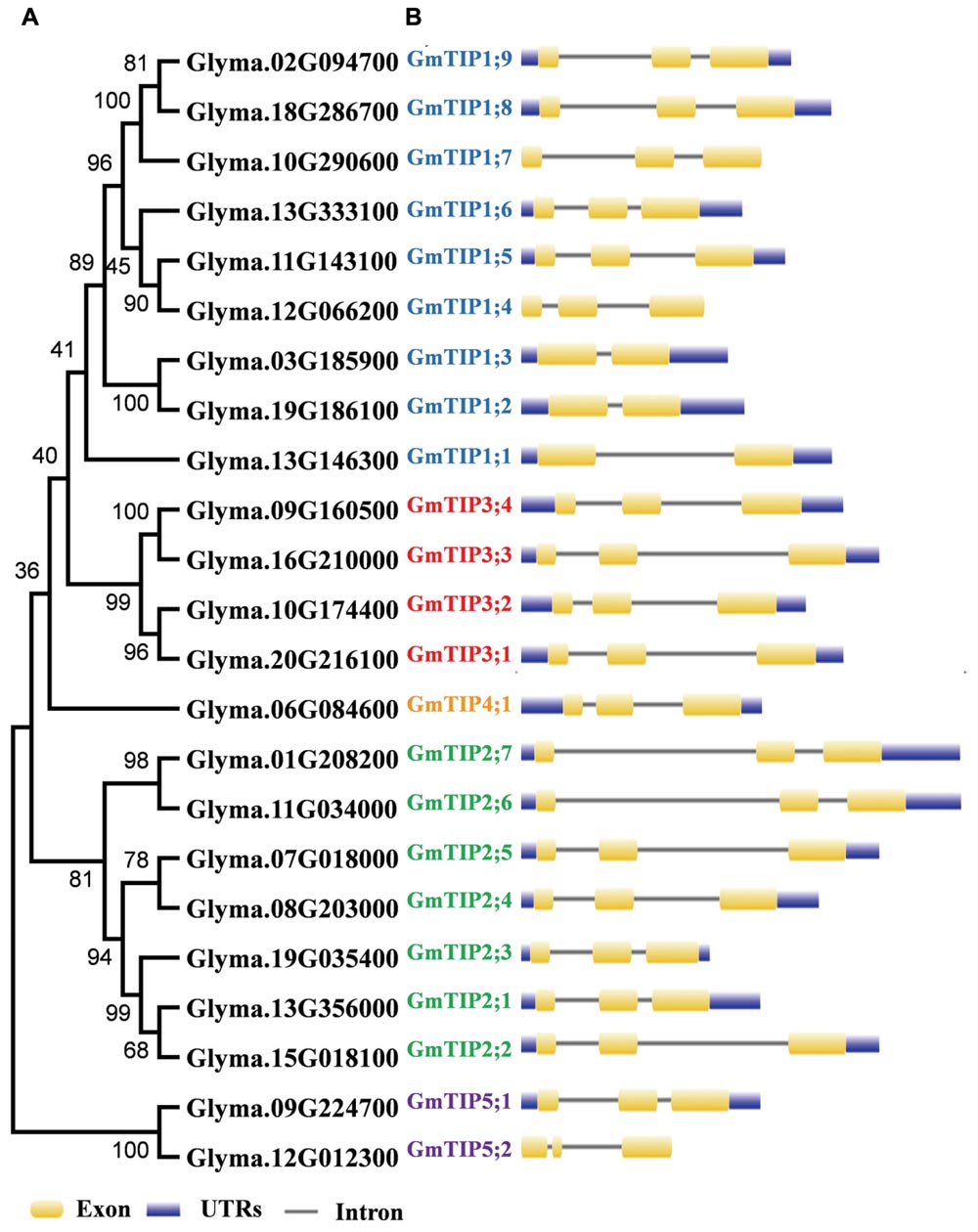
FIGURE 1. Phylogenetic relationship and exon-intron structure of soybean Tonoplast intrinsic proteins (TIPs). (A) The unrooted tree was constructed via alignment of full-length amino acid sequences from soybean using MEGA6 software by the neighbor-joining method. (B) Lengths of the exons and introns of each TIP gene are displayed proportionally. Exons and introns are indicated by yellow rectangles and thin lines, respectively. The untranslated regions (UTRs) are indicated by blue rectangles.
Chromosomal Location and Duplication of Soybean TIP Genes
The 23 TIP genes were unevenly distributed on 16 of the 20 soybean chromosomes (Figure 2), with three GmTIPs on chromosome 13, and two GmTIPs each on chromosomes 9, 10, 11, 12, and 19. The remaining chromosomes only contained one TIP each.
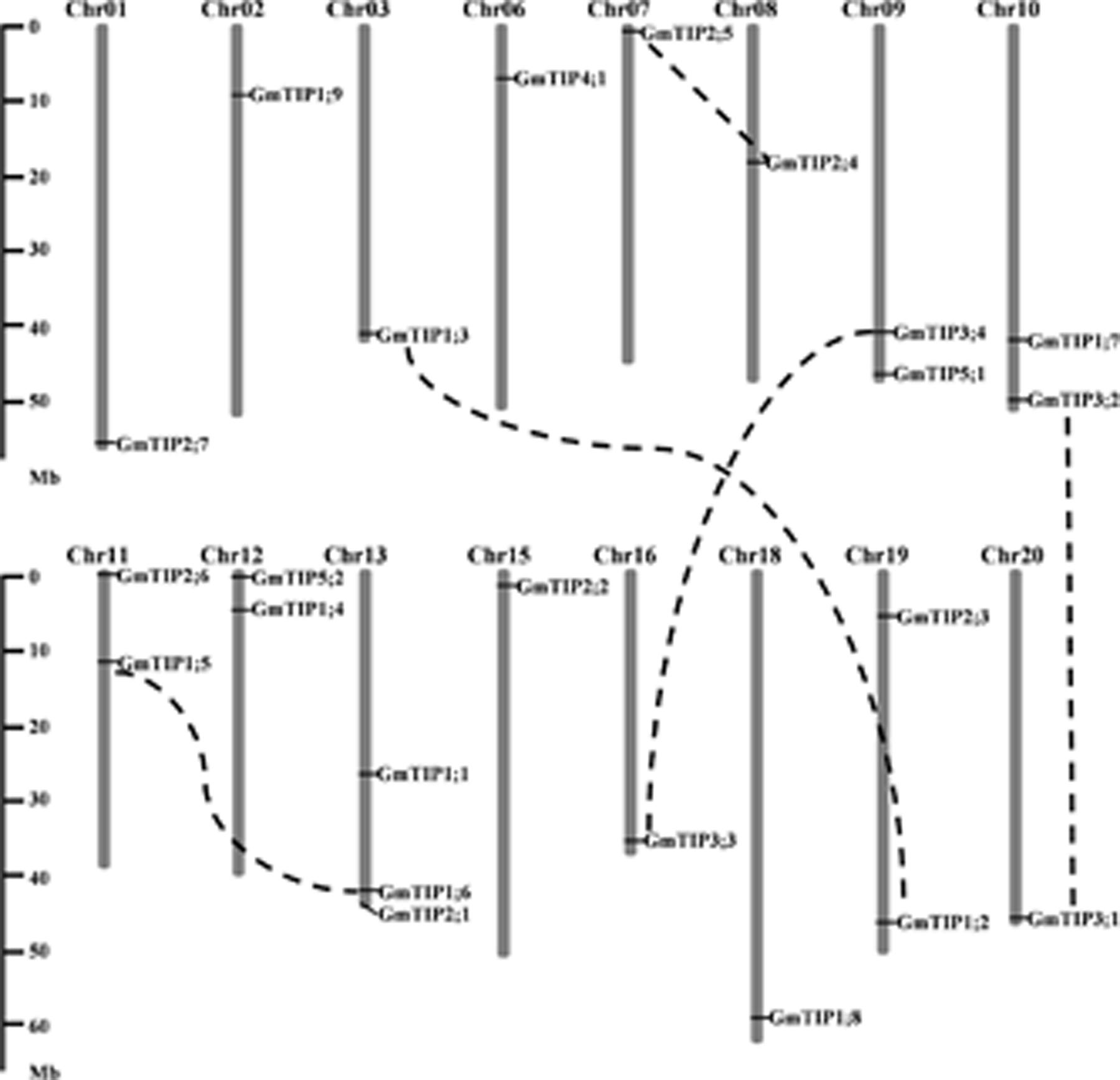
FIGURE 2. Chromosomal locations and gene duplication events of soybean TIP gene family members. The segmentally duplicated gene pairs are linked by black dotted lines. The left scale represents physical distance along the chromosomes in megabases (Mb). Chromosome numbers are shown at the top of each vertical gray bar.
During evolution, the soybean genome has undergone two rounds of whole genome duplication (Schmutz et al., 2010). In order to examine the duplication patterns of soybean TIP genes, the PGDD was searched to identify segmentally duplicated pairs, and tandem duplication was identified based on the gene loci. No tandem duplication was found in this gene subfamily. To identify duplicated pairs, synonymous (Ks) and non-synonymous substitution (Ka) distance values were calculated and the Ka/Ks ratios were used to evaluate the duplication time. The Ka/Ks ratio for each segmentally duplicated gene pair varied from 0.06 to 0.28 (Supplementary Table S2). This analysis suggests that all mutations in paralogous GmTIP genes are neutral or disadvantageous, as their Ka/Ks ratios were less than 1. We found that the five closest branches of soybean TIP genes experienced duplication during the soybean whole genome duplication period, while the others were duplicated 67 Mya or earlier. The duplicated GmTIPs exist in the form of sister pairs in the phylogenetic tree (Figure 1) and are shown linked by dotted lines in Figure 2.
Sequence and Conserved Domain Analysis in GmTIP Gene Family
The protein size of TIP members varied from 238 to 256 amino acids with 78–99% identity in each subgroup. All soybean GmTIP proteins contain two NPA motifs, and the amino acids representing the Ar/R selectivity filter (SF) and Froger’s residues are highly conserved. The Ar/R SF in the newly identified GmTIP5;2 consists of the amino acid sequence S-V-G-C, and the spacing between the NPA domains is 110 amino acids, which is consistent with other members of the GmTIP5 subfamily (Supplementary Table S3). Similarly, the sequence A-A-Y-W is a common feature for both GmTIP5;1 and GmTIP5;2.
Tertiary protein structure predicted for TIPs showed six conserved transmembrane domains. However, the three dimensional geometry of the pore structure obtained with PoreWalker software (Pellegrini-Calace et al., 2009) showed substantial variation in pore size and constrictions in the pore. The tertiary structure and pore morphology for GmTIP1;5 and GmTIP2;5 are shown in Figure 3.
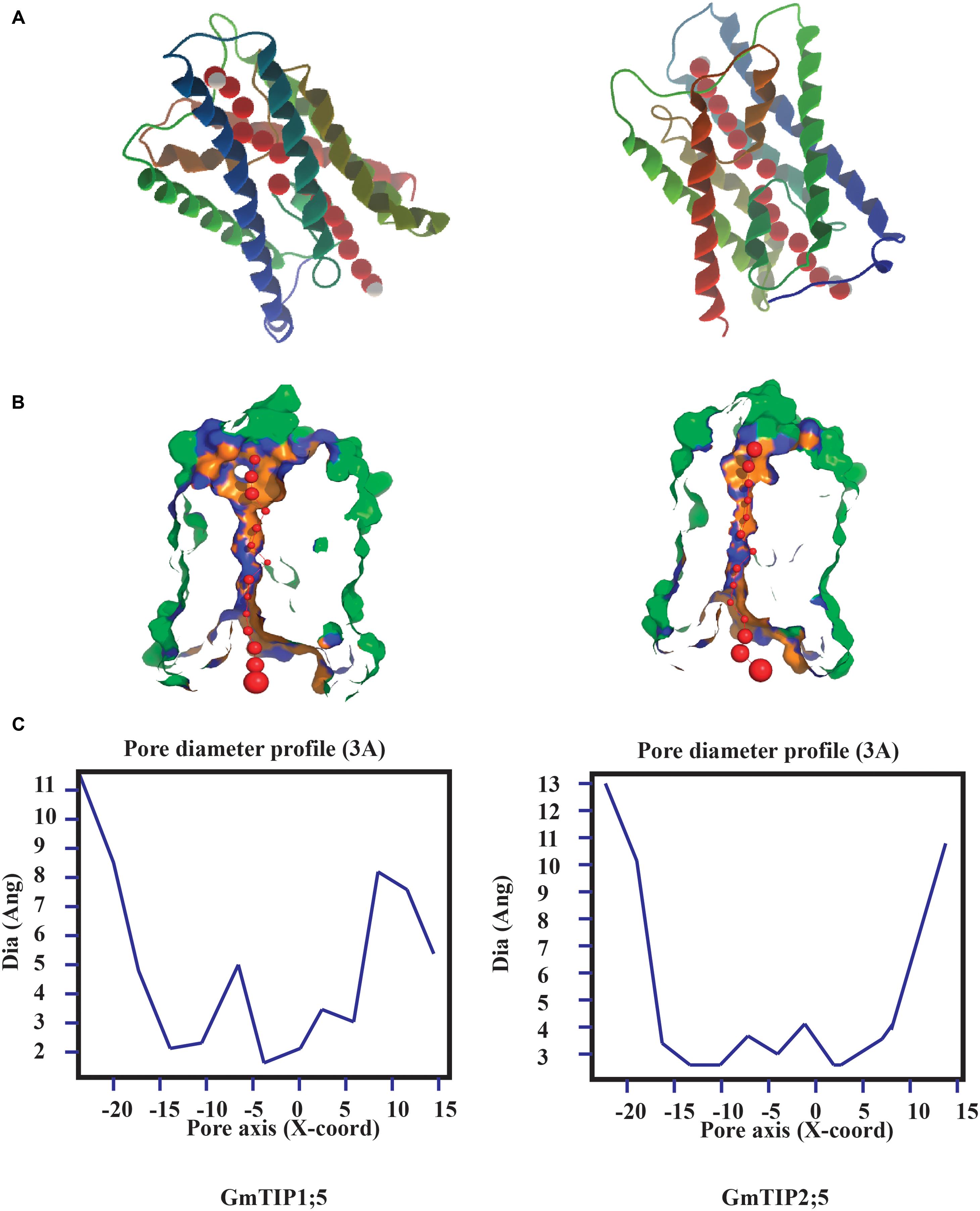
FIGURE 3. Protein tertiary structure showing pore morphology of GmTIP1;5 and GmTIP2;5. (A) Tertiary structures comprised of six transmembrane domains and water molecules (red) passing through the pores of GmTIP1;5 (left) and GmTIP2;5 (right) visualized with CLC genomic workbench. (B) Cross sections of the proteins showing pore. (C) Pore diameter profile of GmTIP1;5 (left) and GmTIP2;5 (right) at 3 Å steps corresponding to the pore shape in (B). Pore axis (X-Coord): the position along the pore axis is shown as x-coordinate in Å. Dia (Ang): pore diameter value in Å.
The characterization of these aquaporins as TIP genes was based on predicted amino acid similarities with known TIPs from other plant species, with location in the tonoplast of the corresponding proteins needing experimental verification. However, the predicted localization of members of the GmTIP subfamily was very diverse, including cytosol, plasma membrane, endoplasmic reticulum, vacuole, mitochondria, and chloroplast (Zhang et al., 2013). We further searched for conserved motifs in GmTIP proteins with the MEME program to gain additional insights into their diversity. As shown in Figure 4, 10 conserved motifs designated as motif 1 to motif 10 were found. All GmTIPs possess motifs 1, 2, and 5. Most of the GmTIPs contain motifs 1 to 8, whereas motif 9 and motif 10 were exclusively present in the GmTIP5 subgroup. Motif 3 was not present in GmTIP1;1, GmTIP4;1, and GmTIP5;2. Motif 4 was not present in GmTIP1;4, GmTIP5;1, and GmTIP5;2.
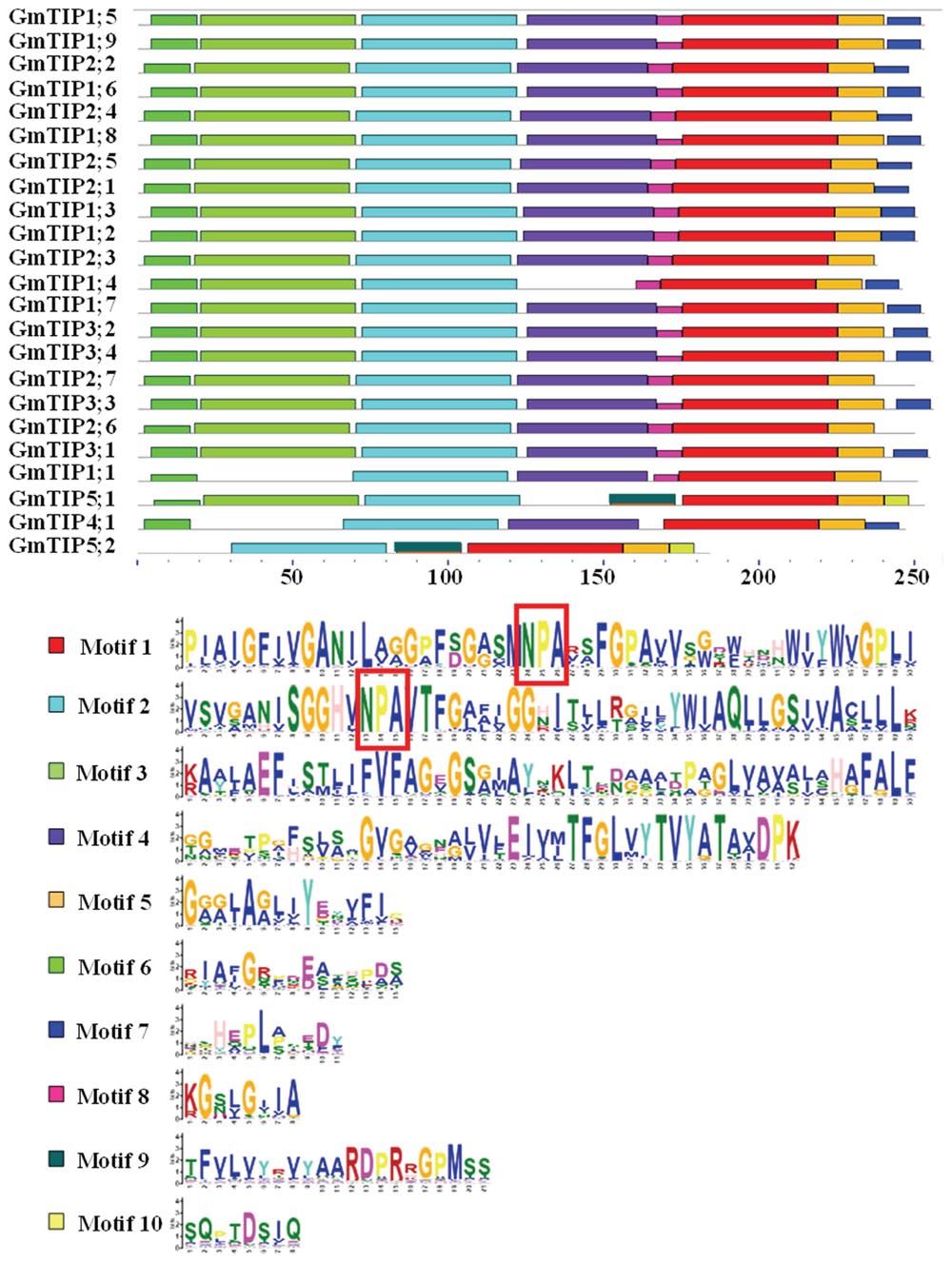
FIGURE 4. Identification and distribution of conserved motifs in soybean TIP protein sequences. Distribution of conserved motifs in soybean TIP members. All motifs were identified by Multiple EM for Motif Elicitation (MEME) using the complete amino acid sequences of GmTIP proteins. Different motifs are indicated by different colored boxes numbered 1–10. The annotation of each motif is listed at the bottom. Motif 1 and motif 2 contain the NPA domain.
Differential Expression of Soybean TIPs in Soybean Tissues and Under Abiotic Stress Conditions
Very diverse expression patterns for GmTIPs were observed in the transcriptome data representing nine different tissues, namely flower, leaf, nodule, pod, root, root hair, seed, shoot apical meristem, and stem tissue (Libault et al., 2010). Most of the GmTIPs showed tissue specific expression (Figure 5). For example, GmTIP1;1, 1;2, 1;3, 1;4, 2;6, 2;7, and 4;1 were highly expressed in root, but have relatively lower expression levels in other tissues. Similarly, GmTIP1;5, 1;6, 2;4, and 2;5 were highly expressed in stem, but have a relatively lower expression level in other tissues. No gene showed higher expression levels in leaves and root hairs, while most phylogenetically paired genes showed similar expression patterns. The GmTIP3 subgroup was only expressed in seed tissue, although this pattern broadened under stress conditions (see below). Interestingly, two phylogenetic gene pairs displayed different expression patterns (GmTIP2;1/GmTIP2;2, GmTIP1;8/GmTIP1;9), indicating neofunctionalization.
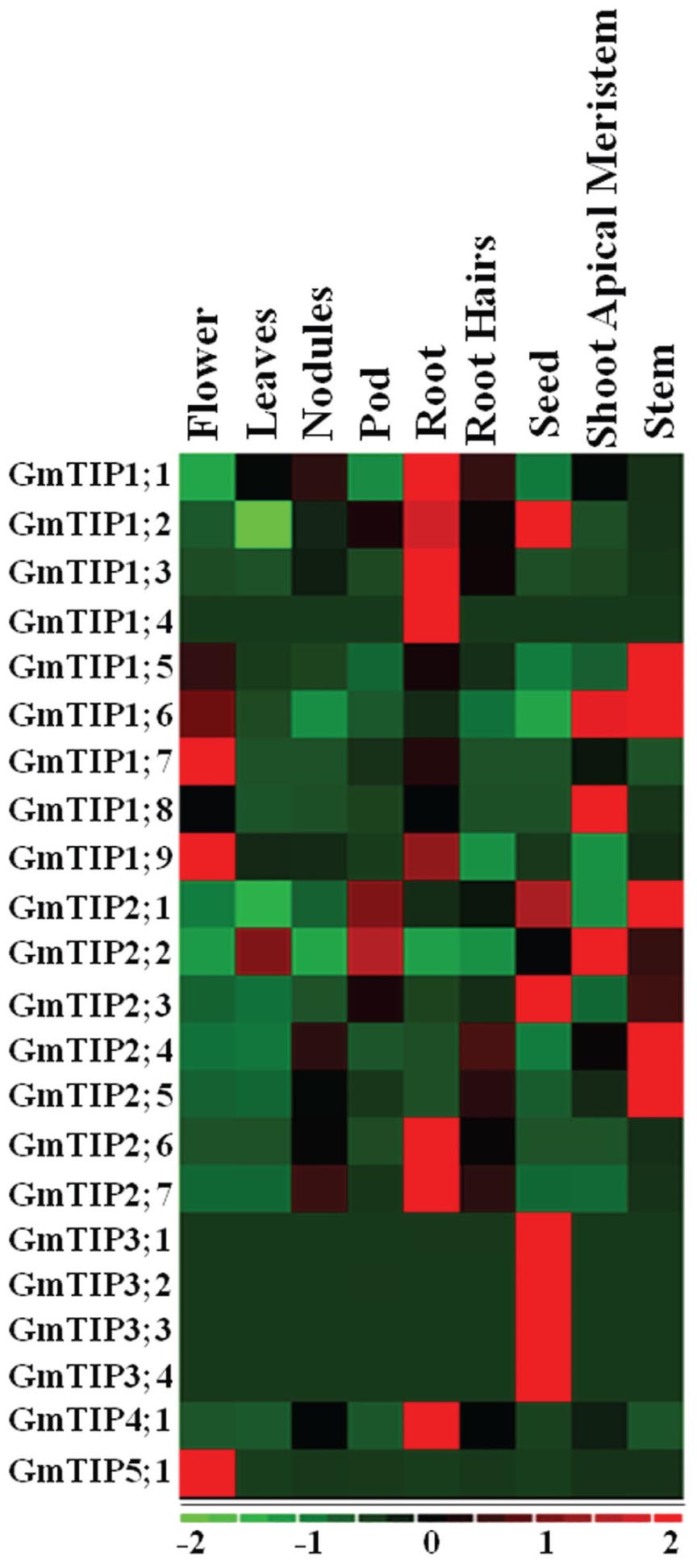
FIGURE 5. Heatmap of expression profiles of the soybean GmTIP gene family in nine tissues. Relative tissue expression levels of GmTIPs based on RNA-seq data were used to construct the expression patterns of soybean genes. The expression data (Reads Per Kilobase Million) values were median-centered and normalized for each gene in different tissue before transforming to color scale. The color bar at the bottom shows the range of expression values from highest expression level (red) to lowest expression level (green), 0 is the median expression level (Black).
The tissue specific expression pattern of GmTIPs was investigated in the Williams 82 genotype under varying water-deficit conditions (Figure 6A). GmTIP3;2, 3;3, and 3;4 were up-regulated in shoots under serious drought condition, even after water-recovery, and down-regulated in all other tissues and conditions. Interestingly, the whole GmTIP2 subfamily was down-regulated under very mild drought stress in leaf tissue and up-regulated after severe drought stress and water-recovery, except one (GmTIP2;2). These results indicated that a gene subfamily may share similar regulatory elements, and also have similar physiological functions. Most of the root-specific expressed genes were induced under varying water-deficit conditions.
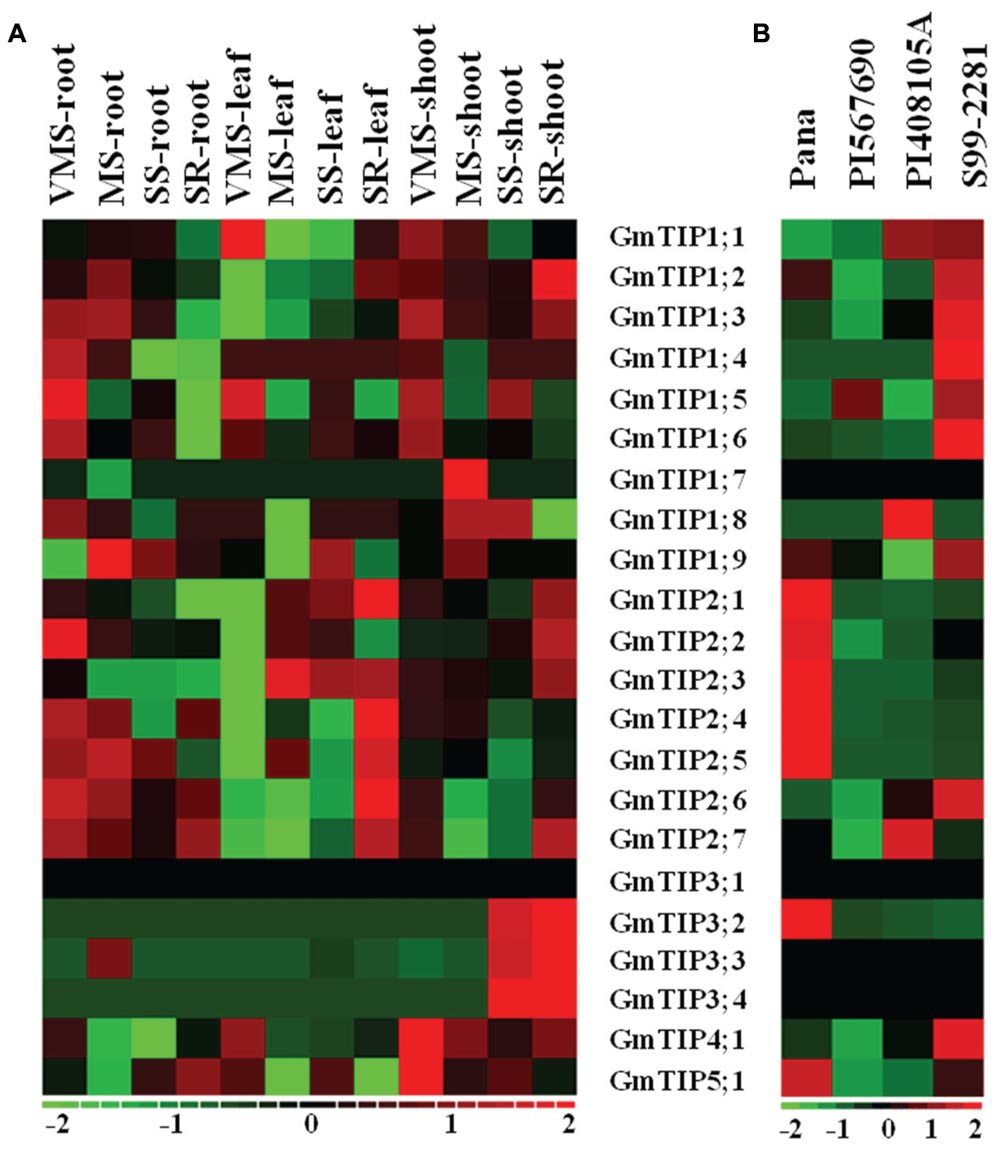
FIGURE 6. Expression profiles of soybean TIP genes under different abiotic stress conditions or in different germplasms. (A) Heatmap representation of expression patterns of soybean TIP genes across the root, leaf, and shoot tissue in Williams 82 under varying water-deficit stress conditions (VMS: very mild stress; MS: mild stress; SS: severe stress; SR: water recovery after severe stress). (B) Expression profiles of the soybean TIP genes in leaves of Pana (fast wilting, drought vs. control), PI 567690 (slow wilting, drought vs. control), PI 408105A (flooding tolerant, flooding vs. control), S99-2281(flooding sensitive, flooding vs. control). The expression data values were median-centered and normalized for each gene before transforming to the color scale (log2-transformed ratios). The color bar at the bottom shows the range of expression values from increased expression (red) to decreased expression (green), 0 means no gene expression pattern changed (Black).
To further investigate the expression profiles of TIP genes under drought and flooding conditions, the expression patterns of TIP gene families were extracted from two RNA-seq datasets (Prince et al., 2015 and unpublished data) (Figure 6B). PI 567690 is a slow wilting soybean line, and Pana is a fast wilting line under drought conditions (Pathan et al., 2014; Mutava et al., 2015). GmTIP2;1, 2;2, 2;3, 2;4, 2;5, 3;2, and 5;1 mRNA levels were induced in leaves of Pana, but decreased in leaves of PI 567690 after drought stress. PI 408105A is a flooding tolerant line, and S99-2281 is a flooding sensitive line (Mutava et al., 2015). GmTIP1;2, 1;3, 1;4, 1;5, 1;6, 1;9, 2;6, and 4;1 mRNA levels were induced in leaves of S99-2281, but decreased or were not induced in leaves of PI 408105A after flooding stress. Conversely, GmTIP1;8 and 2;7 were induced in PI 408105A and decreased in S99-2281. These results indicated that GmTIP genes could play a role under drought or flooding stress.
The expression pattern of the newly identified GmTIP5;2 could not be determined because all reads from the above RNA-seq datasets were aligned to the previous version of the G. max reference genome, Gmax1.1, and Phytozome v9.0. GmTIP3;1 was not induced or decreased in any tissues (Williams 82) or PI lines under abiotic stress conditions (Figures 6A,B). Also GmTIP3;1, GmTIP3;4, GmTIP3;3, and GmTIP1;7, did not show any response to abiotic stress in the PI or cultivar lines (Figure 6B).
Analysis of SNP Variation of the GmTIP Gene Family in 106 Soybean Lines
Single-nucleotide polymorphisms located in the coding regions of GmTIPs were identified to investigate the genetic variation within this gene family in diverse soybean lines (Supplementary Table S4). A total of 81 SNPs were observed in the 23 GmTIP genes. These SNPs exhibited an uneven distribution: the GmTIP5 subfamily contains the highest number of SNPs compared to other subfamilies, whereas no SNPs were found in GmTIP2;1 and GmTIP2;3. Out of 81 SNPs, one SNP located in GmTIP5;1 generates a premature stop codon. Thirty-six SNPs were non-synonymous and were distributed in the coding regions of 14 GmTIPs. Some deletions were found in non-synonymous SNP locations, which cause protein-coding changes. Not a single non-synonymous SNP was identified within the remaining seven genes (GmTIP1;2, GmTIP1;5, GmTIP1;9, GmTIP2;4, GmTIP2;5, GmTIP2;6, and GmTIP4;1). SNPs unique to G. soja lines (PI# highlighted red) are summarized in Supplementary Table S4. Only 18 unique SNPs were identified in seven G. soja lines, and eight out of these 18 SNPs were non-synonymous. The pattern of SNP distributions correlates well with the phylogenetic distribution of TIPs (Figure 1; Supplementary Table S2). This sequence information enabled us to identify novel GmTIP alleles from soybean lines that will serve as valuable breeding resources.
Genetic variations were further investigated in four soybean slow wilting lines (PI 471938, PI 416937, PI 567690, and PI 567731) to establish associations between SNPs and the slow wilting trait. However, no unique SNPs were identified within coding sequences in these four slow wilting lines as compared with other non-slow wilting lines. The effects of non-synonymous SNP on the pore shape were further analyzed in GmTIP5;1 and GmTIP5;2, which have more non-synonymous SNPs in the protein coding region than other GmTIPs. Interestingly, we find that two non-synonymous SNPs (Gm09_41743311 and Gm09_41743354) in GmTIP5;1 do not influence the channel 3D shape. However, three non-synonymous SNPs (Gm12_895269, Gm12_895302, and Gm12_895476) in GmTIP5;2 have an effect on the pore shape. Different pore diameter profiles were obtained between Williams 82 genotype and some of the other sequenced genotypes (Figure 7). For example, the structure of GmTIP5;2 in PI 567690 was different than that in Williams 82. These results indicate that the slow wilting trait in these lines may be governed by different molecular mechanisms, such as GmTIP expression level differences or is due to their different 3D structures.
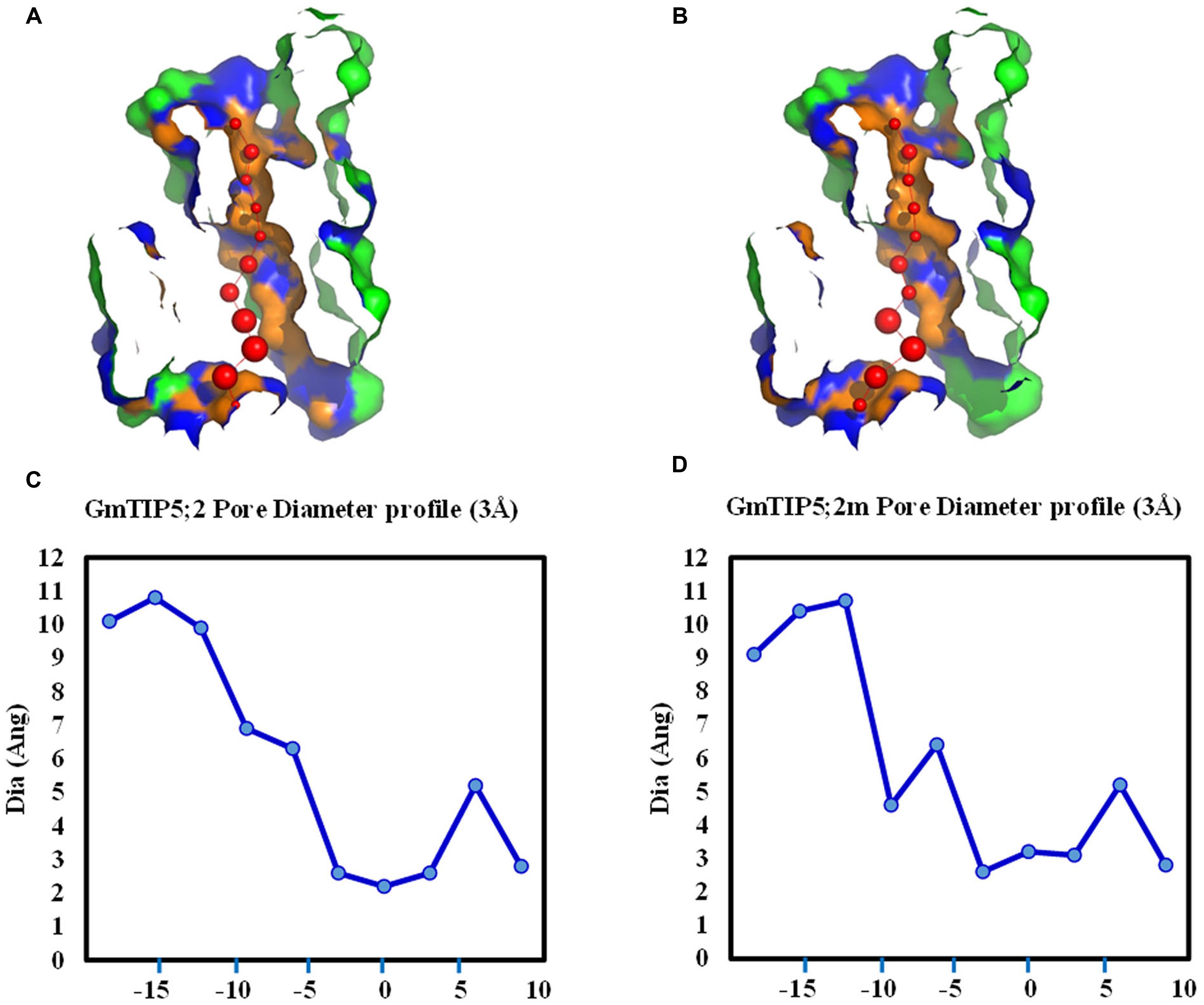
FIGURE 7. Variation in pore morphology of GmTIP5;2 and GmTIP5;2m. GmTIP5;2m contains two amino acid changed (148: A<>G; 159: T<>R) due to two SNP (Gm12_895269 and Gm_895302). (A) and (B) Cross sections of the proteins showing pore of GmTIP5;2 (A) and GmTIP5;2m (B). (C) and (D) Pore diameter profile of GmTIP5;2 (C) and GmTIP5;2m (D) at 3 Å steps. Pore axis (X-Coord): the position along the pore axis is shown as x-coordinate in Å. Dia (Ang): pore diameter value in Å.
Cloning and Functional Characterization of GmTIP1;5 and GmTIP2;5 in Xenopus Oocytes
No non-synonymous SNP was found in GmTIP1;5 and GmTIP2;5 in 106 soybean germplasm; however, they were shown to be differentially expressed in response to drought (GmTIP1;5 and GmTIP2;5) and flooding (GmTIP1;5) in different varieties of soybeans with contrasting phenotypes and were mainly expressed in stem where they could be highly important in water transport during abiotic stress. Further in-depth analysis of GmTIP1;5 and GmTIP2;5 using X. laevis oocytes was performed to establish a link between differential expression patterns and trait development. GmTIP2;5 and GmTIP1;5-expressing oocytes showed a rapid rate of swelling for the first 5 min after being subjected to a hypoosmotic solution (Figures 8A–C). After 5 min, the rate of swelling in GmTIP2;5 expressing oocytes declined as they began to burst. In contrast, GmTIP1;5 expressing oocytes continued to increase in diameter at a rate greater than the controls for 20 min. GmTIP2;5-expressing oocytes swelled more rapidly than GmTIP1;5-expressing oocytes during the first 5 min. Mock-injected oocytes displayed a slower rate of swelling, probably due to diffusion of water across the plasma membrane. It was concluded from these results that GmTIP2;5 and GmTIP1;5 facilitate the transport of water and are aquaporins.
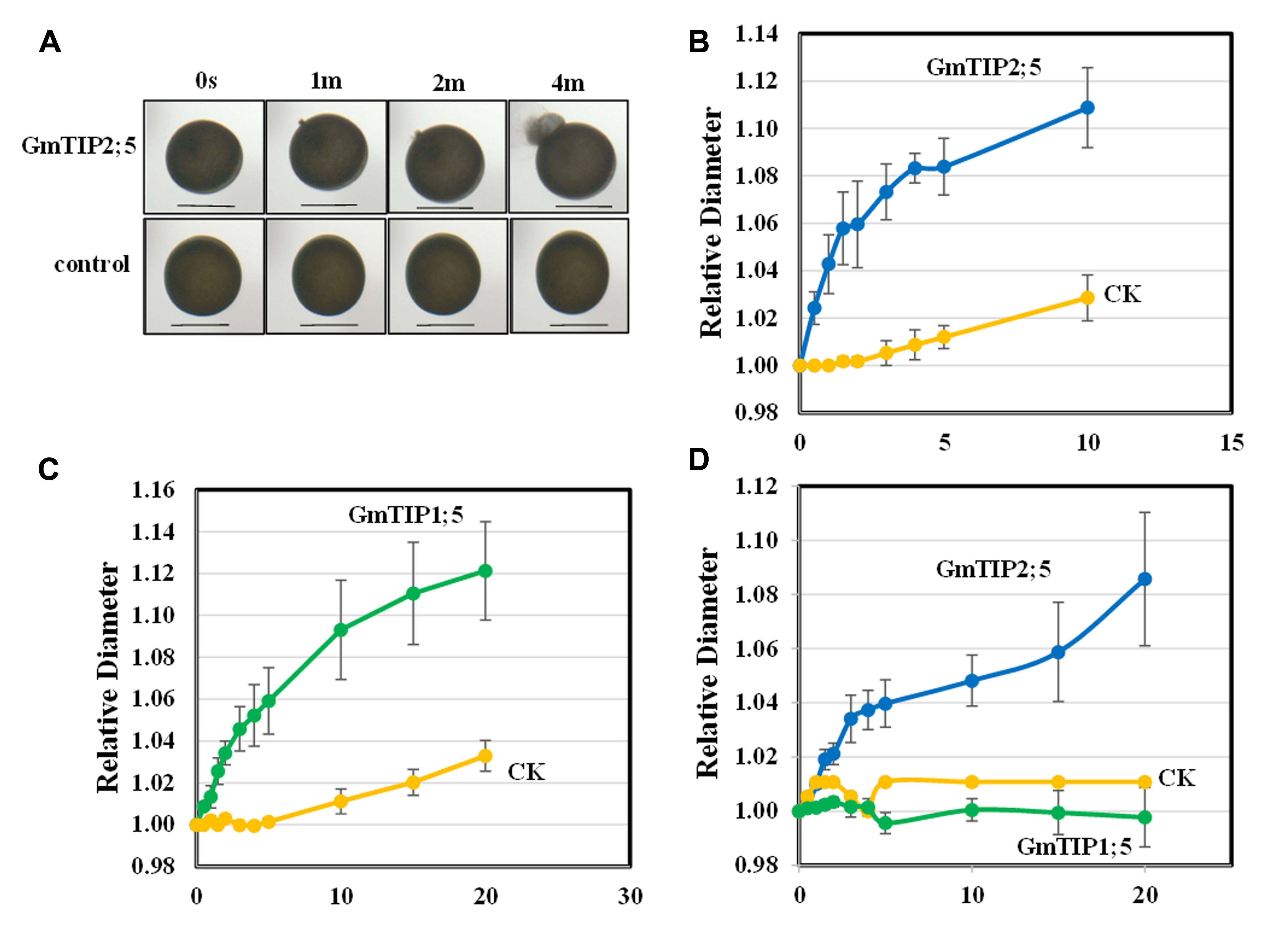
FIGURE 8. Oocyte Swelling Assays with GmTIP1;5 and GmTIP2;5. The relative diameter of GmTIP-expressing oocytes following exposure to hypoosmotic or boron-containing isosmotic solutions was measured to characterize the osmotic permeability of GmTIP1;5 and GmTIP2;5-containing membranes. (A) Water transport assay with Xenopus leaevis oocytes in hypoosmotic solution showed that GmTIP2;5-mediated water uptake leads to rapid swelling and bursting (top) compared to the control (bottom). Scale bars = 1 mm. (B) Rate of oocyte swelling in hypoosmotic solution in mock-injected controls (yellow line) vs. GmTIP2;5-expressing oocytes (blue line). Oocytes were observed to burst after 10 min. (C) Rate of oocyte swelling in hypoosmotic solution in mock injected controls (yellow line) vs. GmTIP1;5-expressing oocytes (green line). Results are reported as means ± SEM (N = 5 oocytes). (D) Rate of oocyte swelling in isosmotic boric acid solution in mock injected controls (yellow line) vs. GmTIP2;5-expressing oocytes (blue line) and GmTIP1;5 (green line). The volume of each oocyte was measured for 20 min or until it burst.
Some aquaporins transport boron and other solutes in addition to water (Durbak et al., 2014). To test if these GmTIPs can transport boron, we applied an isoosmotic solution containing 200 mM boric acid in place of 96 mM NaCl. The GmTIP2;5-expressing oocytes showed significantly increased swelling with boric acid compared to the control (Figure 8D). In contrast, control and GmTIP1;5-expressing oocytes did not swell. These results indicated that GmTIP2;5 can transport water as well as boric acid.
Discussion
GmTIPs Play Important Roles in Abiotic Stress
Here, 23 full-length aquaporin-coding sequences belonging to the TIP subfamily were identified in the soybean genome. The number of TIPs in the soybean genome is much higher than those identified in species like Arabidopsis, maize, rice, and sorghum (Regon et al., 2014). Several studies have shown differential expression of plant aquaporins in response to environmental stresses in several different species (Alexandersson et al., 2005; Zhu et al., 2005; Ligaba et al., 2011; Cohen et al., 2013; Li et al., 2014). Therefore, it is important to investigate the interaction between the expression of GmTIPs and abiotic stress. This study explored the expression patterns of GmTIP genes in soybean PI 567690 and Pana lines. The soybean genotype PI 567690 exhibits significantly lower wilting and less yield loss under drought condition than the elite cultivar Pana (Pathan et al., 2014). The difference in gene expression between PI 567690 and Pana had been investigated by RNA-seq (Prince et al., 2015), and our preliminary data indicated that the PI 567690 genotype has a limited transpiration response when exposed to a high vapor pressure deficit (unpublished data). In the present analysis, we found that the expression level of seven TIP genes was reduced in slow-wilting soybean lines but increased in the fast-wilting soybean lines after drought stress (Figure 6B). At the same time, the expression level of eight different TIP genes was down-regulated or unchanged in the waterlogging tolerant varieties, but upregulated in the waterlogging susceptible varieties. Only one gene was found to be up-regulated in slow-wilting lines under drought conditions, and only three genes were found to be up-regulated in the water-logging tolerant lines. These results are consistent with the hypothesis that due to lessened water transport in soybean leaves, the genotype PI 567690 exhibited more resistance to drought stress. We therefore conclude that GmTIPs may play an important role in soybean both in drought and flooding tolerance. However, the gene regulation patterns in root tissue of water-logging resistant lines should be further explored.
The expression patterns of GmTIPs under a given condition varied among different tissues and was complex (Figures 6A,B). GmTIP1;5 and GmTIP2;5 showed a higher expression in stem, but lower expression in other tissues. Interestingly, GmTIP2;5 transcript was up-regulated under varying water-deficit stress in root, but down-regulated after water-recovery. This is the TIP that showed a very rapid water uptake in Xenopus oocytes. GmTIP1;5, which showed a slower, but steady uptake in Xenopus oocytes, was mainly induced under the VMS condition in root, leaf and shoot, not in other water-deficit conditions. Furthermore, GmTIP1;5 is the only one that showed upregulation in slow wilting lines under drought condition among all TIP genes. In addition, only one synonymous SNP was located in the coding sequences of GmTIP1;5 and two synonymous SNPs were located in the coding sequence of GmTIP2;5. Therefore, observed variations in gene expression seem to have prominent roles in functional variation. Further investigation of the water transport function of these two genes in soybean would help to establish a link between differential expression patterns and trait development.
To gain further insight into the possible physiological functions of the members of this large family and aid the development of genetic engineering in soybean, the TIP that directs tissue-specific expression pattern and functions in water transport would be highly desirable. Overall, a more comprehensive mechanism will emerge as multiple aquaporin transport functions and integration of the different stress signals are determined at the whole plant level. Further characterization of the GmTIP genes involved in abiotic stress resistance genotypes may aid in developing tolerant germplasm and cultivars.
Water Permeability of GmTIPs
All G. max TIP aquaporins showed the canonical double NPA motif and a group ar/R SF that are conserved across different species. These results indicate that GmTIPs may play similar roles in regulating water absorption. As such, it is important to validate representative candidate genes for water transport by using other methods such as heterologous expression in Xenopus oocytes. Basic GmTIP gene family information and phylogenetic sequence analysis in the present study provide a list of candidate genes that may play important roles in soybean drought and flooding tolerance. The function of two of these GmTIP genes has been demonstrated to be involved in water transport. However, AQPs are not only water transporters but also solute transporters (Maurel et al., 2008). Most AQPs can transport glycerol, urea, boric acid or arsenic as we showed with boron and GmTIP2;5. Yet even though a protein has been predicted to be an aquaporin, it may not transport water. Thus GmTIP candidate genes must be functionally characterized.
Aquaporins can be further functionally characterized with transport inhibitors. One such inhibitor, AgNO3 or silver sulfadiazine, has been reported to inhibit aquaporin water transport by binding to cysteine or histidine residues, resulting in blockage of the pore (Daniels et al., 1996; Ishibashi et al., 1997; Niemietz and Tyerman, 2002). Interestingly, the transpiration rate of the slow-wilting cultivar PI416937 is insensitive to silver treatment, and only a small change in AQP abundance following silver treatment was found in this line as compared to sensitive genotypes (Sadok and Sinclair, 2010b). All these results suggest that low AQP abundance may underlie the low leaf hydraulic conductivity of PI 416937 and its limited transpiration rates under high vapor pressure deficit (Sadok and Sinclair, 2010a, 2012; Devi et al., 2015, 2016). The effects of silver ions on water transport in Xenopus oocytes expressing GmTIP1;5, GmTIP2;5, or other AQPs could help us to identify key candidate genes linked with the soybean slow-wilting trait. For example, we hypothesize that GmTIP5;1 would be a good target gene for engineering reduced gene expression for improved water stress tolerance, since under severe water-deficit conditions GmTIP5;1 was induced in shoot and leaf tissues but showed decreased expression in the slow wilting line and flooding tolerant lines under stress conditions.
Utilization of Natural Variation of GmTIPs
Analysis of whole genome resequencing data provides an immense opportunity to mine natural variants in diverse germplasm (Patil et al., 2016; Valliyodan et al., 2016). The soybean germplasm, both wild and cultivated species, provides a wide range of abiotic stress tolerance. This study initiated characterization of GmTIPs by summarizing and analyzing all available SNP information, including whether SNPs lead to synonymous or non-synonymous substitutions, in 106 soybean lines (Valliyodan et al., 2016). These 106 soybean lines represent diverse lines (wild types, landraces and elite lines), including four slow wilting soybean lines (PI 416937, PI 471938, PI 567690, and PI 567731). Aside from the expression level, genetic variation (natural or induced) also may impact the functionality of aquaporin genes and cause a variation in phenotypes. To our knowledge, this is the first report of natural variation in soybean for TIP genes. With this information, the correlation between the SNP haplotype and the phenotype (slow wilting score) for these 106 lines can be calculated to further characterize the association between the GmTIP genetic background and the slow-wilting trait. Furthermore, the association analysis can be extended to the whole MIP gene family and the gene promoter region to better understand the mechanism of slow wilting at the genomic level.
It has been reported that plant plasma membrane aquaporins are deactivated by dephosphorylation under conditions of drought stress, or by protonation of a conserved histidine residue following a drop in cytoplasmic pH due to anoxia during flooding (Tournaire-Roux et al., 2003; Törnroth-Horsefield et al., 2006). Analyzing the detailed effects of all non-synonymous SNPs on the existing crystal structure of plant AQPs (for examples: AtTIP2;1, SoPIP2;1) (Törnroth-Horsefield et al., 2006) in some interesting soybean lines may enable us to better understand the structure changes and mechanisms of water transport regulation.
Conclusion
In this study, the soybean TIP gene family was identified based on the new soybean genome annotation. Twenty-three TIP members were assigned to five subfamilies based on sequence similarity and phylogenetic relationship. The modulation of expression profiles of these 23 GmTIP genes was examined with deep transcriptome sequencing under drought or flooding conditions and during plant development. The potential roles of these genes in transport of substrates were also discussed. The identified natural variations in this gene family from 106 soybean germplasms will benefit future gene functional analysis and utilization. We also demonstrated that GmTIP1;5 and GmTIP2;5 can function as water transporters in oocytes. All results presented here represent an important resource for designing experiments for functional validation of candidate genes in plant development and abiotic stress responses, and for developing soybean germplasm with improved abiotic stress tolerance.
Author Contributions
LS and RD designed the experiments, analyzed data, and prepared the manuscript. NN, SMP, LS and WG worked on the oocytes experiments. GP, RD, and BV analyzed the natural variation SNP data. RD and LS worked on defining pore-lining residues and solute prediction. SJP and RM were involved in the gene expression pattern analysis. HN conceived and supervised the project. All authors have read, revised, and approved the manuscript.
Funding
This project was funded by the Missouri Soybean Merchandising Council and United Soybean Board.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Amanda Durbak and Dr. Paula McSteen, University of Missouri, for help with oocyte swelling assays. We thank Theresa Musket and Kevin Ngo for proof-reading and editing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01564
Footnotes
- ^ https://phytozome.jgi.doe.gov
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://topcons.cbr.su.se/
- ^ http://swissmodel.expasy.org/interactive
- ^ http://www.ebi.ac.uk/thornton-srv/software/PoreWalker/
- ^ http://chibba.agtec.uga.edu/
- ^ http://snpeff.sourceforge.net
References
Alexandersson, E., Fraysse, L., Sjövall-Larsen, S., Gustavsson, S., Fellert, M., Karlsson, M., et al. (2005). Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59, 469–484. doi: 10.1007/s11103-005-0352-1
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Chaumont, F., Barrieu, F., Wojcik, E., Chrispeels, M. J., and Jung, R. (2001). Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125, 1206–1215. doi: 10.1104/pp.125.3.1206
Cohen, D., Bogeat-Triboulot, M. B., Vialet-Chabrand, S., Merret, R., Courty, P. E., Moretti, S., et al. (2013). Developmental and environmental regulation of aquaporin gene expression across Populus species: divergence or redundancy? PLoS ONE 8:e55506. doi: 10.1371/journal.pone.0055506
Daniels, M. J., Chaumont, F., Mirkov, T. E., and Chrispeels, M. J. (1996). Characterization of a new vacuolar membrane aquaporin sensitive to mercury at a unique site. Plant Cell 8, 587–599. doi: 10.1105/tpc.8.4.587
Danielson, J. A., and Johanson, U. (2008). Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 8:45. doi: 10.1186/1471-2229-8-45
Deshmukh, R. K., Vivancos, J., Guérin, V., Sonah, H., Labbé, C., Belzile, F., et al. (2013). Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol. Biol. 83, 303–315. doi: 10.1007/s11103-013-0087-3
Deshmukh, R. K., Vivancos, J., Ramakrishnan, G., Guérin, V., Carpentier, G., Sonah, H., et al. (2015). A precise spacing between the NPA domains of aquaporins is essential for silicon permeability in plants. Plant J. 83, 489–500. doi: 10.1111/tpj.12904
Devi, M. J., Sinclair, T. R., and Taliercio, E. (2015). Comparisons of the effects of elevated vapor pressure deficit on gene expression in leaves among two fast-wilting and a slow-wilting soybean. PLoS ONE 10:e0139134. doi: 10.1371/journal.pone.0139134
Devi, M. J., Sinclair, T. R., and Taliercio, E. (2016). Silver and zinc inhibitors influence transpiration rate and aquaporin transcript abundance in intact soybean plants. Environ. Exp. Bot. 122, 168–175. doi: 10.1016/j.envexpbot.2015.10.006
Durbak, A. R., Phillips, K. A., Pike, S., O’Neill, M. A., Mares, J., Gallavotti, A., et al. (2014). Transport of boron by the tassel-less1 aquaporin is critical for vegetative and reproductive development in maize. Plant Cell 26, 2978–2995. doi: 10.1105/tpc.114.125898
Heinrich, T., Potter, R., and Jones, M. G. K. (1996). PotRB7: a gene equivalent to TobRB7 from potato. Plant Physiol. 112, 862–862.
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 3, 1296–1297. doi: 10.1093/bioinformatics/btu817
Ishibashi, K., Kuwahara, M., Kageyama, Y., Tohsaka, A., Marumo, F., and Sasaki, S. (1997). Cloning and functional expression of a second new aquaporin abundantly expressed in testis. Biochem. Biophys. Res. Commun. 237, 714–718. doi: 10.1006/bbrc.1997.7219
Javot, H., and Maurel, C. (2002). The role of aquaporins in root water uptake. Ann. Bot. 90, 301–313. doi: 10.1093/aob/mcf199
Kaldenhoff, R., and Fischer, M. (2006). Functional aquaporin diversity in plants. Biochim. Biophys. Acta 1758, 1134–1141. doi: 10.1016/j.bbamem.2006.03.012
Kirch, H. H., Vera-Estrella, R., Golldack, D., Quigley, F., Michalowski, C. B., Barkla, B. J., et al. (2000). Expression of water channel proteins in Mesembryanthemum crystallinum. Plant Physiol. 123, 111–124. doi: 10.1104/pp.123.1.111
Krishnamurthy, K., and Shivashankar, K. (1975). Soybean production in Karnataka. UAS Technical Series. Bangalore: University of Agricultural Science.
Li, C., and Wong, W. H. (2001). Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. U.S.A. 98, 31–36. doi: 10.1073/pnas.011404098
Li, D. D., Tai, F. J., Zhan, Z. T., Li, Y., Zheng, Y., Wu, Y. F., et al. (2009). A cotton gene encodes a tonoplast aquaporin that is involved in cell tolerance to cold stress. Gene 438, 26–32. doi: 10.1016/j.gene.2009.02.023
Li, G., Santoni, V., and Maurel, C. (2014). Plant aquaporins: role in plant physiology. Biochim. Biophys. Acta 1840, 1574–1582. doi: 10.1016/j.bbagen.2013.11.004
Libault, M., Farmer, A., Joshi, T., Takahashi, K., Langley, R. J., Franklin, L. D., et al. (2010). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99. doi: 10.1111/j.1365-313X.2010.04222.x
Ligaba, A., Katsuhara, M., Shibasaka, M., and Djira, G. (2011). Abiotic stresses modulate expression of major intrinsic proteins in barley (Hordeum vulgare). C. R. Biol. 334, 127–139. doi: 10.1016/j.crvi.2010.11.005
Lopez, F., Bousser, A., Sissoëff, I., Hoarau, J., and Mahé, A. (2004). Characterization in maize of ZmTIP2-3, a root-specific tonoplast intrinsic protein exhibiting aquaporin activity. J. Exp. Bot. 55, 539–541. doi: 10.1093/jxb/052
Ludewig, U., von Wireìn, N., and Frommer, W. B. (2002). Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J. Biol. Chem. 277, 13548–13555. doi: 10.1074/jbc.M200739200
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Madeira, A., Moura, T. F., and Soveral, G. (2016). Detecting aquaporin function and regulation. Front. Chem. 4:3. doi: 10.3389/fchem.2016.00003
Maurel, C., Javot, H., Lauvergeat, V., Gerbeau, P., Tournaire, C., Santoni, V., et al. (2002). Molecular physiology of aquaporins in plants. Int. Rev. Cytol. 215, 105–148. doi: 10.1016/S0074-7696(02)15007-8
Maurel, C., Reizer, J., Schroeder, J. I., and Chrispeels, M. J. (1993). The vacuolar membrane protein γ-TIP creates water specific channels in Xenopus oocytes. EMBO J. 12, 2241–2247.
Maurel, C., Verdoueq, L., Luu, D., and Santoni, V. (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59, 595–624. doi: 10.1146/annurev.arplant.59.032607.092734
Mutava, R. N., Prince, S. J., Syed, N. H., Song, L., Valliyodan, B., Chen, W., et al. (2015). Understanding abiotic stress tolerance mechanisms in soybean: a comparative evaluation of soybean response to drought and flooding stress. Plant Physiol. Biochem. 86, 109–120. doi: 10.1016/j.plaphy.2014.11.010
Nada, R. M., and Abogadallah, G. M. (2014). Aquaporins are major determinants of water use efficiency of rice plants in the field. Plant Sci. 227, 165–180. doi: 10.1016/j.plantsci.2014.08.006
Niemietz, C. M., and Tyerman, S. D. (2002). New potent inhibitors of aquaporins: silver and gold compounds inhibit aquaporins of plant and human origin. FEBS Lett. 531, 443–447. doi: 10.1016/S0014-5793(02)03581-0
Osawa, H., Stacey, G., and Gassmann, W. (2006). ScOPT1 and AtOPT4 function as proton-coupled oligopeptide transporters with broad but distinct substrate specificities. Biochem. J. 393, 267–275. doi: 10.1042/BJ20050920
Pathan, S. M., Lee, J. D., Sleper, D. A., Fritschi, F. B., Sharp, R. E., Carter, T. E. Jr., et al. (2014). Two soybean plant introductions display slow leaf wilting and reduced yield loss under drought. J. Agron. Crop Sci. 200, 231–236. doi: 10.1111/jac.12053
Patil, G., Do, T., Vuong, T. D., Valliyodan, B., Lee, J. D., Chaudhary, J., et al. (2016). Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 6:19199. doi: 10.1038/srep19199
Pellegrini-Calace, M., Maiwald, T., and Thornton, J. M. (2009). PoreWalker: a novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structure. PLoS Comput. Biol. 5:e1000440. doi: 10.1371/journal.pcbi.1000440
Pike, S., Patel, A., Stacey, G., and Gassmann, W. (2009). Arabidopsis OPT6 is an oligopeptide transporter with exceptionally broad substrate specificity. Plant Cell Physiol. 50, 1923–1932. doi: 10.1093/pcp/pcp136
Prince, S. J., Joshi, T., Mutava, R. N., Syed, N., Joao Vitor Mdos, S., Patil, G., et al. (2015). Comparative analysis of the drought-responsive transcriptome in soybean lines contrasting for canopy wilting. Plant Sci. 240, 65–78. doi: 10.1016/j.plantsci.2015.08.017
Regon, P., Panda, P., Kshetrimayum, E., and Panda, S. K. (2014). Genome-wide comparative analysis of tonoplast intrinsic protein (TIP) genes in plants. Funct. Integr. Genomics 14, 617–629. doi: 10.1007/s10142-014-0389-9
Roussel, H., Bruns, S., Gianinazzi-Pearson, V., Hahlbrock, K., and Franken, P. (1997). Induction of a membrane intrinsic protein-encoding mRNA in arbuscular mycorrhiza and elicitor-stimulated cell suspension cultures of parsley. Plant Sci. 126, 203–210. doi: 10.1016/S0168-9452(97)00106-4
Sadok, W., and Sinclair, T. R. (2010a). Genetic variability of transpiration response of soybean [(L.) Merr.] Shoots to Leaf Hydraulic Conductance Inhibitor AgNO3. Crop Sci. 50, 1423–1430. doi: 10.2135/cropsci2009.10.0575
Sadok, W., and Sinclair, T. R. (2010b). Transpiration response of ‘slow-wilting’and commercial soybean (Glycine max (L.) Merr.) genotypes to three aquaporin inhibitors. J. Exp. Bot. 61, 821–829. doi: 10.1093/jxb/erp350
Sadok, W., and Sinclair, T. R. (2012). Zinc treatment results in transpiration rate decreases that vary among soybean genotypes. J. Plant Nutr. 35, 1866–1877. doi: 10.1080/01904167.2012.706683
Sarda, X., Tousch, D., Ferrare, K., Cellier, F., Alcon, C., Dupuis, J. M., et al. (1999). Characterization of closely related delta-TIP genes encoding aquaporins which are differentially expressed in sunflower roots upon water deprivation through exposure to air. Plant Mol. Biol. 40, 179–191. doi: 10.1023/A:1026488605778
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Sloane, R. J., Patterson, R. P., and Carter, T. E. (1990). Field drought tolerance of a soybeanplant introduction. Crop Sci. 30, 118–123. doi: 10.2135/cropsci1990.0011183X003000010027x
Song, L., Prince, S., Valliyodan, B., Joshi, T., Maldonado dos Santos, J. V., Wang, J., et al. (2016). Genome-wide transcriptome analysis of soybean primary root under varying water-deficit conditions. BMC Genomics 17:57. doi: 10.1186/s12864-016-2378-y
Syed, N. H., Prince, S. J., Mutava, R. N., Patil, G., Li, S., Chen, W., et al. (2015). Core clock, SUB1, and ABAR genes mediate flooding and drought responses via alternative splicing in soybean. J. Exp. Bot. 66, 7129–7149. doi: 10.1093/jxb/erv407
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Törnroth-Horsefield, S., Wang, Y., Hedfalk, K., Johanson, U., Karlsson, M., Tajkhorshid, E., et al. (2006). Structural mechanism of plant aquaporin gating. Nature 439, 688–694. doi: 10.1038/nature04316
Tournaire-Roux, C., Sutka, M., Javot, H., Gout, E., Gerbeau, P., Luu, D. T., et al. (2003). Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425, 393–397. doi: 10.1038/nature01853
Tyerman, S. D., Niemietz, C. M., and Bramley, H. (2002). Plant aquaporins: multifunctional water and solute channels with expanding roles. Plant Cell Environ. 25, 173–194. doi: 10.1046/j.0016-8025.2001.00791.x
Valliyodan, B., Dan, Q., Patil, G., Zeng, P., Huang, J., Dai, L., et al. (2016). Landscape of genomic diversity and trait discovery in soybean. Sci. Rep. 6:23598. doi: 10.1038/srep23598
Vaughan, S. P., James, D. J., Lindesy, K., and Massiah, A. J. (2006). Characterization of FaRB7, a near root-specific gene from strawberry (Fragaria × ananassa Duch.) and promoter activity analysis in homologous and heterologous hosts. J. Exp. Bot. 57, 3901–3910. doi: 10.1093/jxb/erl185
Verkman, A. S., Anderson, M. O., and Papadopoulos, M. C. (2014). Aquaporins: important but elusive drug targets. Nat. Rev. Drug Discov. 13, 259–277. doi: 10.1038/nrd4226
Wang, L. L., Chen, A. P., Zhong, N. Q., Liu, N., Wu, X. M., Wang, F., et al. (2014). The Thellungiella salsuginea tonoplast aquaporin TsTIP1;2 functions in protection against multiple abiotic stresses. Plant Cell Physiol. 55, 148–161. doi: 10.1093/pcp/pct166
Wang, X., Li, Y., Ji, W., Bai, X., Cai, H., Zhu, D., et al. (2011). A novel Glycine soja tonoplast intrinsic protein gene responds to abiotic stress and depresses salt and dehydration tolerance in transgenic Arabidopsis thaliana. J. Plant Physiol. 168, 1241–1248. doi: 10.1016/j.jplph.2011.01.016
Xin, S., Yu, G., Sun, L., Qiang, X., Xu, N., and Cheng, X. (2014). Expression of tomato SlTIP2;2 enhances the tolerance to salt stress in the transgenic Arabidopsis and interacts with target proteins. J. Plant Res. 127, 695–708. doi: 10.1007/s10265-014-0658-7
Yamamoto, Y. T., Cheng, C. L., and Conkling, M. A. (1990). Root-specific genes from tobacco and Arabidopsis homologous to an evolutionarily conserved gene family of membrane channel proteins. Nucleic Acids Res. 18, 7449–7449. doi: 10.1093/nar/18.24.7449
Zhang, D., Tong, J., He, X., Xu, Z., Xu, L., Wei, P., et al. (2016). A novel soybean intrinsic protein gene, GmTIP2;3, involved in responding to osmotic stress. Front. Plant Sci. 6:1237. doi: 10.3389/fpls.2015.01237
Zhang, D. Y., Ali, Z., Wang, C. B., Xu, L., Yi, J. X., Xu, Z. L., et al. (2013). Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS ONE 8:56312. doi: 10.1371/journal.pone.0056312
Keywords: aquaporin, tonoplast intrinsic proteins (TIPs), water transporter, soybean, abiotic stress, expression, SNP
Citation: Song L, Nguyen N, Deshmukh RK, Patil GB, Prince SJ, Valliyodan B, Mutava R, Pike SM, Gassmann W and Nguyen HT (2016) Soybean TIP Gene Family Analysis and Characterization of GmTIP1;5 and GmTIP2;5 Water Transport Activity. Front. Plant Sci. 7:1564. doi: 10.3389/fpls.2016.01564
Received: 29 July 2016; Accepted: 04 October 2016;
Published: 21 October 2016.
Edited by:
Janin Riedelsberger, University of Talca, ChileReviewed by:
Martin Hajduch, Slovak Academy of Sciences, SlovakiaAndrea Ariani, University of California, Davis, USA
Copyright © 2016 Song, Nguyen, Deshmukh, Patil, Prince, Valliyodan, Mutava, Pike, Gassmann and Nguyen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Henry T. Nguyen, nguyenhenry@missouri.edu
 Li Song
Li Song Na Nguyen
Na Nguyen Rupesh K. Deshmukh
Rupesh K. Deshmukh Gunvant B. Patil
Gunvant B. Patil Silvas J. Prince
Silvas J. Prince Babu Valliyodan
Babu Valliyodan Raymond Mutava1
Raymond Mutava1 Sharon M. Pike
Sharon M. Pike Walter Gassmann
Walter Gassmann Henry T. Nguyen
Henry T. Nguyen