- 1College of Plant Sciences & Technology, Huazhong Agricultural University, Wuhan, China
- 2Shanghai Agrobiological Gene Center, Shanghai, China
Drought is one of the vitally critical environmental stresses affecting both growth and yield potential in rice. Drought resistance is a complicated quantitative trait that is regulated by numerous small effect loci and hundreds of genes controlling various morphological and physiological responses to drought. For this study, 270 rice landraces and cultivars were analyzed for their drought resistance. This was done via determination of changes in plant height and grain yield under contrasting water regimes, followed by detailed identification of the underlying genetic architecture via genome-wide association study (GWAS). We controlled population structure by setting top two eigenvectors and combining kinship matrix for GWAS in this study. Eighteen, five, and six associated loci were identified for plant height, grain yield per plant, and drought resistant coefficient, respectively. Nine known functional genes were identified, including five for plant height (OsGA2ox3, OsGH3-2, sd-1, OsGNA1, and OsSAP11/OsDOG), two for grain yield per plant (OsCYP51G3 and OsRRMh) and two for drought resistant coefficient (OsPYL2 and OsGA2ox9), implying very reliable results. A previous study reported OsGNA1 to regulate root development, but this study reports additional controlling of both plant height and root length. Moreover, OsRLK5 is a new drought resistant candidate gene discovered in this study. OsRLK5 mutants showed faster water loss rates in detached leaves. This gene plays an important role in the positive regulation of yield-related traits under drought conditions. We furthermore discovered several new loci contributing to the three investigated traits (plant height, grain yield, and drought resistance). These associated loci and candidate genes significantly improve our knowledge of the genetic control of these traits in rice. In addition, many drought resistant cultivars screened in this study can be used as parental genotypes to improve drought resistance of rice by molecular breeding.
Introduction
Rice is one of the most important staple foods and plays an important role to ensure food safety. However, rice production consumes copious amounts of fresh water. In China, 49% of all fresh water resources are used for rice production (Zhang, 2007). Due to environmental damage and anomalous climatic variation, drought has become one of the main limiting factors for global grain production (Pennisi, 2008). Throughout Asia, 20% of all rice production areas are affected by drought per year (Gowda et al., 2011). This strengthens the disparity between supply and demand of rice production. Therefore, enhancing the drought resistance of rice is vital to reduce the effect of drought on rice production.
Drought, which is a soil water deficit, can result in insufficient moisture for a plant to adequately grow and complete its life cycle. Water shortage will seriously affect rice growth and lead to a series of physiological and biochemical changes. Many studies focused on plant drought resistance. These studies reported that drought usually leads to a delayed flowering time, restrained plant growth, and ultimately loss of yield in rice (Liu et al., 2005; Yue et al., 2005; Zou et al., 2007). The reduction in yield is due to a reduced spikelet number or reduced fertility of spikelet caused by drought stress (Ekanayake et al., 1989; Liu et al., 2010). In addition, drought can cause the change of antioxidant system and osmotic regulatory system in rice, accumulating antioxidants, and osmotic regulation substances (Ouyang et al., 2010).
With the development of molecular genetic technologies, more and more studies focused on the discovery of drought resistant genes and their function. These results are useful for breeders to develop drought resistant cultivars. QTL (quantitative trait loci) mapping used to be the main method to discover drought resistance loci. For example, many drought resistance-related QTLs were identified by using a recombinant inbred line (RIL) population derived from a cross of lowland indica rice Zhenshan 97 and upland japonica rice IRAT109 in previous studies (Yue et al., 2006; Liu et al., 2010). Based on the above drought resistance-related QTLs, 17 near-isogenic lines (NILs) were constructed and phenotypic variations of these NILs were investigated under drought and normal conditions, among them, qFSR4 was fine mapped for spikelet number, flag leaf width and root volume, further analysis showed that NARROW LEAF 1 (NAL1) regulating leaf width was located in the qFSR4 interval (Qi et al., 2008; Ding et al., 2011). Some studies have shown that NARROW LEAF 1 can affect root development and drought resistance in rice (Fujita et al., 2013; Cho et al., 2014). In addition, root traits are very important traits for drought resistance in plant. Dro1, a major QTL involved in deep rooting of rice was cloned by fine-mapping, it can increase rice yield under drought conditions (Uga et al., 2011, 2013). And qRT9, a QTL controlling root thickness and root length was mapped an 11.5 kb interval in upland rice (Li et al., 2015). Although linkage analysis for drought resistance has made some achievements in rice, many QTLs/genes of drought resistance remain hidden. Due to a rapid development of sequencing techniques, genome-wide association studies (GWAS) became a new method for gene mining of target traits. Compared to QTL analysis, GWAS is based on natural populations, and can detect multiple alleles at the same site (Flint-Garcia et al., 2003). GWAS have widely been used in human genetic studies as well as in plants, such as Arabidopsis, maize, sorghum, and rice (Atwell et al., 2010; Huang et al., 2011; Tian et al., 2011; Li et al., 2012; Riedelsheimer et al., 2012; Morris et al., 2013; Chen et al., 2014; Wen et al., 2014; Yang et al., 2014, 2015; Wang et al., 2015; Yano et al., 2016). Identification of the allelic variation underpinning the phenotypic diversity in rice will result in enormous practical implications for rice breeding.
Genome-wide association study (GWAS) has been widely used for the genetic analysis of rice. Huang et al. (2010) constructed the first high-density haplotype map in rice via re-sequencing of 517 landraces and cultivars, and they identified 37 strong association loci for 14 agronomic traits via GWAS. Their study was considered as the coming of age for GWAS in rice (Clark, 2010). The genetic architecture of rice metabolism was dissected via GWAS, and five candidate genes were identified or annotated (Chen et al., 2014). Recently, a high-throughput phenotyping platform was developed for GWAS, achieving good results and further promoting GWAS for the dissection of genetic mechanisms for other traits (Yang et al., 2014).
In previous study, GWAS has been used to dissect drought resistance in maize (Lu et al., 2010; Wang et al., 2016; Zhang et al., 2016). However, a systematic examination of drought resistance in rice via GWAS was still called for. In this study, we conduct GWAS on a natural rice population under well watered and drought stress conditions, exploring the drought resistance of rice. The collection contains a wide range of genetic variation (Wu et al., 2015), highlighting its suitability for mapping studies aimed at detecting genetic variation and segregation in a diverse set of rice varieties. Using this strategy, 18, 5, and 6 loci could be associated with plant height, grain yield per plant, and drought resistant coefficient. Nine known genes were identified for three traits. The function of these genes is either directly or indirectly related to the three target traits. We also discovered new loci that contribute to these three traits and that were missed by previous studies. Among these, OsRLK5 was preliminarily confirmed to positively regulate yield-related traits under drought condition.
Materials and Methods
Plant Material and Field Experiment
The plant material consisted of 270 accessions of rice landraces and cultivars, collected from Asia, Africa, and America. This population has previously been used in GWAS for mesocotyl elongation and ratio of deep rooting (Lou et al., 2015; Wu et al., 2015). All accessions were tested under two water regimes: well watered and drought stress, in a drought resistance screening facility (Luo, 2010) at the Baihe Experimental Station of the Shanghai Agrobiological Gene Center (31°15′N, 121°10′E, 4 m altitude) in 2011 and 2012. The plant material was arranged by single factor randomized block design. Seeds derived from a single plant, from which the genomic DNA was extracted for sequencing, was used for field trials. Staged sowing was used according to growth durations. There were 22 hills per row with a space of 18 cm between rows and 16 cm between hills. Fertilizer application and pest control were identical to normal field management. The method for drought treatment was the same as reported in our previous study (Liu et al., 2005). Drought stress was implemented at the early booting stage and lasted for a total of 35 days. During the treatment, drip irrigation was provided to keep the plants growing well in the well-water treatment regime every day and stop watering in drought stress regime.
Measurement of Soil Water Content, Plant Traits, and Statistical Analysis
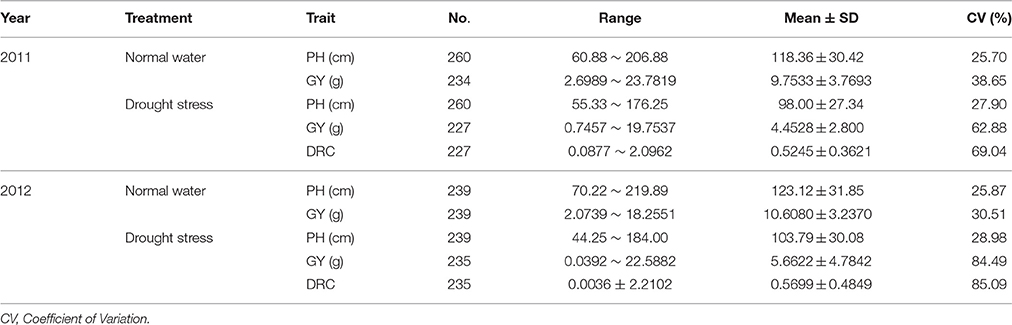
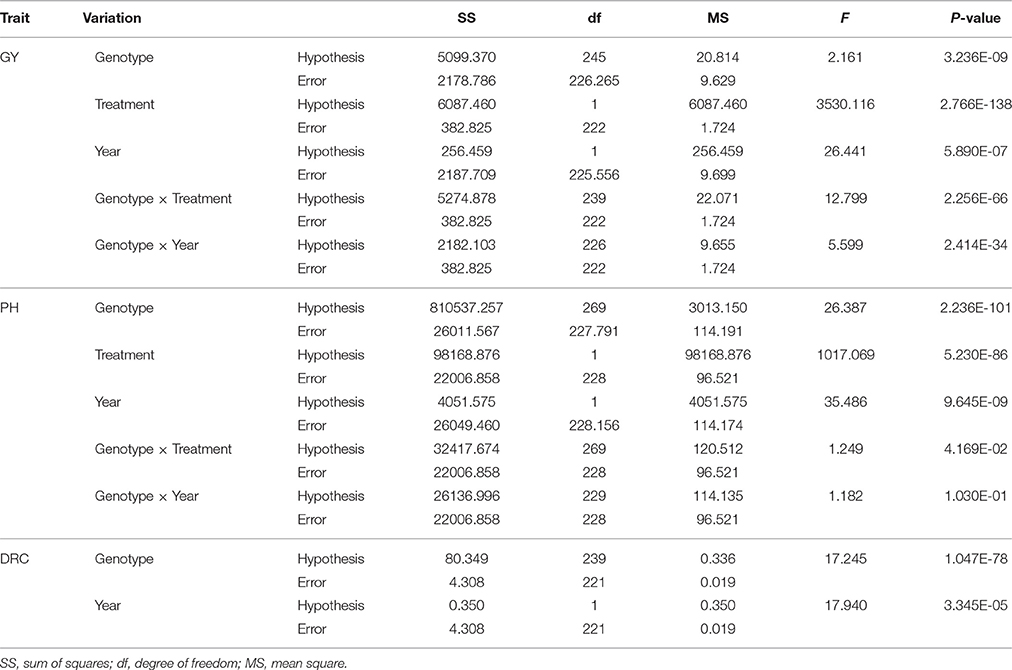
Soil moisture content was measured every 3 days for both drought stress regime and irrigational regime during drought stress. Plant height (PH) was investigated before harvest and grain yield per plant (GY) was investigated after harvest. The drought resistant coefficient (DRC) was calculated as the ratio of the grain yield per plant under drought stress regime to the grain yield per plant under water regime. Statistic analysis was conducted using SPSS software (version 19.0). Mixed model was used for ANOVA of phenotypic data. Genotype and treatment were treated as fixed factor while year was treated as a random factor. Type Π sums of squares were used for ANOVA for unbalanced data in Table 2 (Langsrud, 2003).
Genotype
Genomic DNA (gDNA) was extracted from a single plant and used for sequencing. A total of 270 accessions were genotyped via re-sequencing and using an Illumina HiSeq2000. Among these, genotypic data of 101 accessions were generated by Shanghai Agrobiological Gene Center, and the genotypic data of the remaining 169 accessions were provided by the Huazhong Agricultural University (Chen et al., 2014; Yang et al., 2014). Paired-end sequence reads were mapped to a rice reference genome sequence of japonica cv. Nipponbare (MSU v6.1) using the software BWA, then used for SNP identification, following the procedures described by Wu et al. (2015).
GWAS Analysis
The genome-wide association mapping was conducted via the efficient mixed-model association (EMMA) method using the GAPIT software package in R (Lipka et al., 2012). For this study, a total of 1019,883 SNP markers were used for GWAS. 144,995 SNPs, with less than 10% missing data in this natural population, were used for kinship calculation among individuals and principal component analysis (PCA) to adjust the population structure. The genome-wide threshold was set at p = 9.81E-07, calculated via the formula: 1/total number of SNPs, which was widely used in plant GWAS studies (Wen et al., 2014; Wang et al., 2015; Bai et al., 2016). We furthermore evaluated the extent of local LD (linkage disequilibrium) for each significant SNP. The extended region, where LD between nearly SNPs and lead SNP (with the lowest p-value) decayed to r2 = 0.6, was defined as the local LD-based QTL interval (Yano et al., 2016). To test whether the significant associated loci respond to drought stress, we performed a two-way ANOVA for each significant locus to test the significance of the interactive effect between locus and water status (normal water and drought stress). The set of loci with significant interactions with the water status were defined as QTL responsive to drought stress (Zhang et al., 2016).
The Deletion Mutants
The CRISPR/Cas9 method was used to generate deletion mutants for the verification of candidate gene function. The mutant of OsRLK5 was obtained via the CRISPR/Cas9 method as described by Zhang et al. (2014), using the target sequence GAAAGATCCCGAAGTGGATATGG. We obtained homozygous individuals with 1 bp deletion within target sequences (GAAAGATCCCGAAGT-GATATGG) at the CDS region.
The mutant of OsGNA1 was gained via the CRISPR/Cas9 method as described by Ma et al. (2015), with the target sequence of GGGGCACGTCGAGGACGTCGTGG. Several individual homozygous or heterozygous plants were obtained. The homozygous mutants had the target sequence GGGGCACGTCGAGGAC-TCGTGG, and those homozygous and heterozygous mutants with 1 bp deletion at the CDS region were used in further experiments. The primers used for identification of mutants and haplotype analysis of OsGNA1 were listed in Table S1.
Sequence Analysis of OsGNA1
We sequenced the promoter and coding region of OsGNA1 in the natural population. Sequence alignment was conducted with ClustalX 1.83. Cluster analysis was conducted via MEGA 6.0 (Tamura et al., 2013). LD analysis was conducted using Haploview 4.2 (http://www.broad.mit.edu/mpg/haploview/; Barrett et al., 2005).
Results
The Change of Soil Water Content during Drought Stress in 2011 and 2012
Drought treatment lasted for a total of 35 days. In this period, the absolute water content of the soil declined from 23.03 to 7.78%. Under the drip irrigation regime in 2011, values were always higher than 18.88%. The absolute water content of the soil declined from 22.98 to 8.07% under drought stress regime, while always remaining higher than 19.01% under the drip irrigation regime in 2012 (Figure S1). The change of soil water content showed a similar trend in both years.
Phenotype Statistics and Analysis of Variance of Traits
The descriptive statistics for three traits are shown in Table 1. The coefficient of variation among different genotypes ranged from 25.70 to 28.98, 30.51 to 84.49, and 69.04 to 85.09% for PH, GY, and DRC, respectively. The ANOVA results revealed significant differences of all three traits for genotypes, treatments, and years (Table 2). These results indicate large phenotypic variation in this natural population, possibly increasing the chance to discover candidate genes for related traits via genetic analysis.
Genome-Wide Association Study
Principal component analysis (PCA) analysis revealed two divergent groups belonging to two subspecies of cultivated rice (Figure S2), the indica subspecies and the japonica subspecies, respectively. This is consistent with known information of the rice germplasm (Huang et al., 2010).
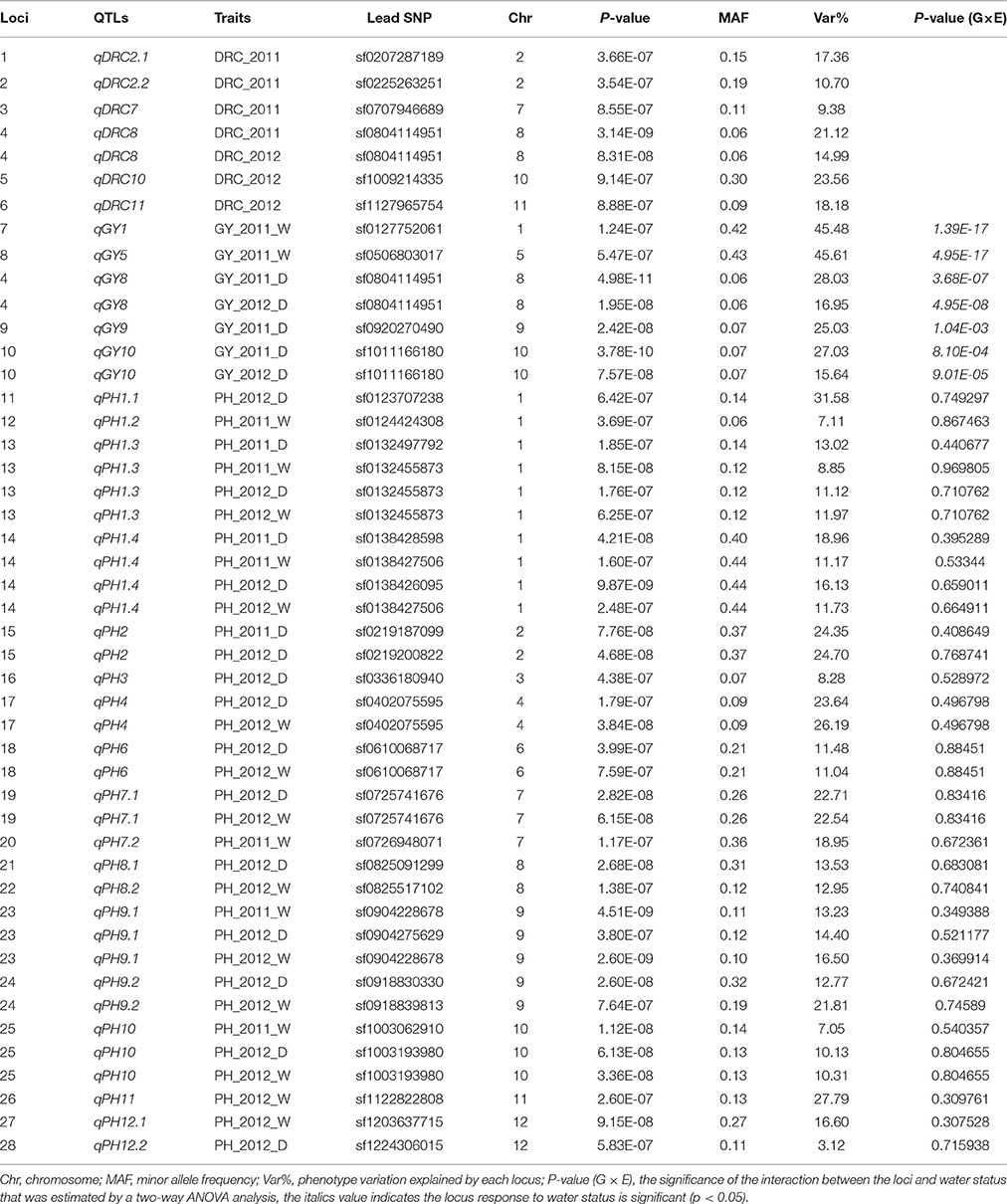
We controlled population structure by setting top two eigenvectors and combining kinship matrix for GWAS in this study. A total of 18, 5, and 6 loci were detected for plant height, grain yield per plant, and drought resistant coefficient, respectively (Table 3).
Genome-wide association study (GWAS) for plant height: 13 associated loci were detected under normal water conditions and 13 associated loci under drought stress while eight loci were detected under both conditions (Figure 1, Table 3). These QTLs explained a range from 3.12 to 31.58% of phenotypic variation. The associated SNP sf0123707238 on chromosome 1 provided the largest contribution to PH variation. qPH1.3 and qPH1.4 were detected under both normal water and drought stress conditions in both years, explaining the range from 8.85 to 18.96% of the phenotypic variation. Additionally, two-way ANOVA revealed that all loci that were detected for plant height showed no significant interaction between locus and water status, indicating that these QTLs of plant height were not responsive to drought stress.
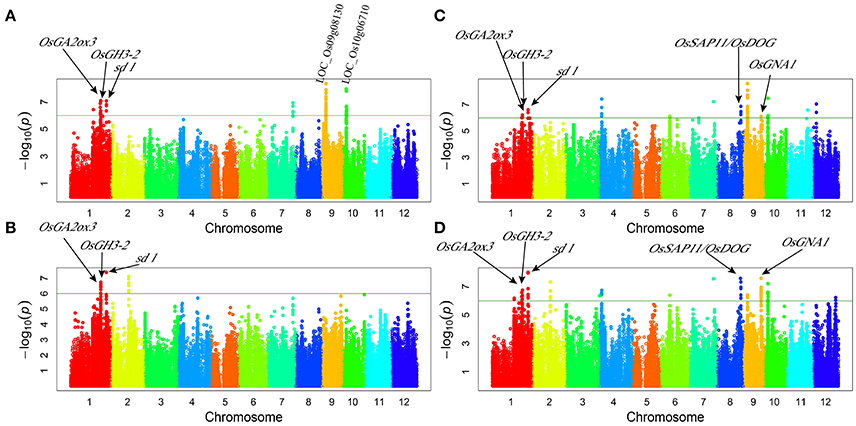
Figure 1. Genome-wide manhattan plots of association mapping for plant height. (A) plant height under normal water condition in 2011; (B) plant height under drought stress condition in 2011; (C) plant height under normal water condition in 2012; (D) plant height under drought stress condition in 2012.
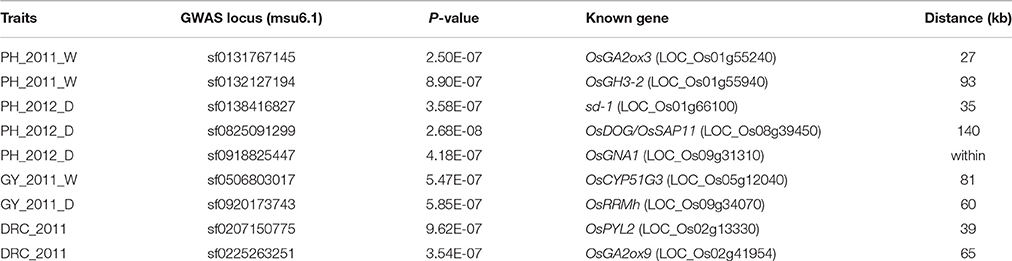
The peak signal of the GWAS loci often appeared in the region near but not within the known genes (Huang et al., 2010). We found the similar cases of five known genes involved in plant height: OsGA2ox3 (Lo et al., 2008), OsGH3-2 (Du et al., 2012), sd-1 (Spielmeyer et al., 2002), OsDOG/OsSAP11 (Giri et al., 2011; Liu et al., 2011), and OsGNA1 (Jiang et al., 2005). These genes were located within or nearby four QTLs intervals and were identified for PH via GWAS (Figure 1, Table 4). Additionally, according to genome annotation information (MSU 6.1) and KEGG pathway database, LOC_Os09g08130 and LOC_Os10g06710 related to the indole alkaloid biosynthesis pathway, located near qPH9.1 and qPH10, respectively (Table 3).
Genome-wide association study (GWAS) for grain yield: a total of five QTLs were detected for grain yield per plant. Three QTLs (qGY8, qGY9, and qGY10) were detected under drought stress and two QTLs (qGY1 and qGY5) were detected under normal water conditions (Figure 2, Table 3). These QTLs explained the range from 15.64 to 45.61% of phenotypic variation. The associated SNP sf0506803017 on chromosome 5 contributed the highest to GY variation (Table 3). qGY8 and qGY10 were detected for GY under drought stress in both 2011 and 2012. In addition, the two-way ANOVA revealed all loci detected for grain yield showing a significant interaction between locus and water status, indicating that these QTLs of grain yield were likely responsive to drought stress.
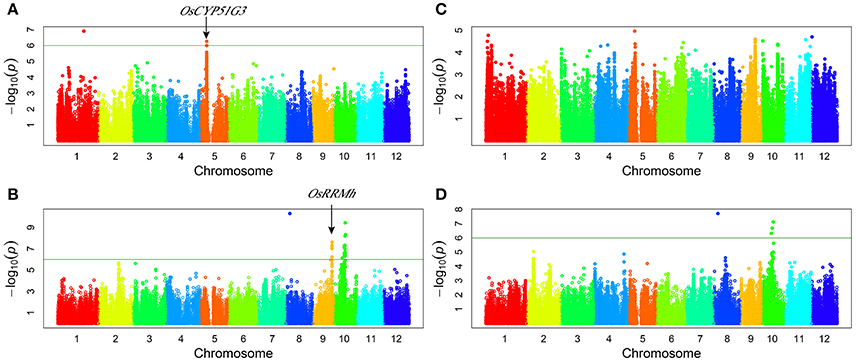
Figure 2. Genome-wide manhattan plots of association mapping for grain yield per plant. (A) grain yield under normal water condition in 2011; (B) grain yield under drought stress condition in 2011; (C) grain yield under normal water condition in 2012; (D) grain yield under drought stress condition in 2012.
OsCYP51G3 (LOC_Os05g12040) was found to be located near the lead SNP sf0506803017 (Figure 2A, Table 4) and involved in steroid biosynthesis, regulating plant height and seed setting rate (Xia et al., 2015). OsRRMh (LOC_Os09g34070) was 157 kb away from the significant association signal (sf0920270490) (Figure 2B, Table 4) and has been reported to regulate fertility rate and number of spikelet per panicle (Liu and Cai, 2013).
Genome-wide association study (GWAS) for drought resistant coefficient: six QTLs were detected for DRC, explaining a range of the phenotypic variation from 9.38 to 23.56% (Figure 3, Table 3). Since the DRC reflected relative yield between drought stress and normal water condition, these six QTLs for DRC should be involved in the response to drought stress. Two known genes are close to the association intervals (Figure 3), i.e., OsPYL2 (Tian et al., 2015) near the lead SNP sf0207287189 and OsGA2ox9 (Lo et al., 2008) near the lead SNP sf0225263251.
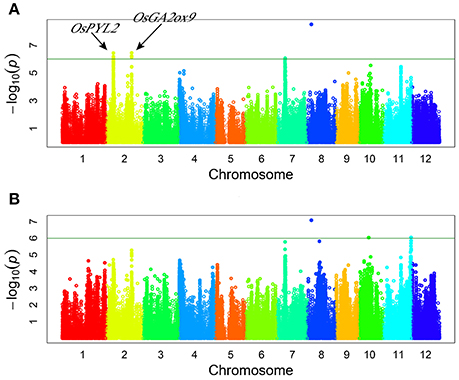
Figure 3. Genome-wide manhattan plots of association mapping for drought resistant coefficient. (A) DRC in 2011; (B) DRC in 2012.
Candidate Genes Involved in Drought-Response Loci
The purpose of rice drought resistance breeding is to improve grain yield under drought conditions. DRC reflects the yield changes (mostly decreases) of plants suffering from drought stress. A total of 162 candidate genes were discovered based on 10 GWAS loci (QTL interval, r2 of LD > 0.6), involved in response to drought stress as well as RNA-seq data under contrasting water status (unpublished data) (Table S2). Among them, 63 genes were up-regulated more than 2-fold and 99 genes were down regulated to less than half their expression. The expression levels of most candidate genes under drought stress conditions tended to return to normal levels after re-watering.
OsGNA1 plays an important role for plant height and root length. It is noticeable that the sf0918825447 at the promoter of OsGNA1 (LOC_Os09g31310) was detected for plant height under drought stress conditions with a p-value of 4.18E-07 in 2012. It was 5 kb away from the lead SNP (sf0918830330) for plant height under drought stress. It remains unclear whether OsGNA1 regulates plant height, although it has been shown to regulate root development (Jiang et al., 2005). To confirm whether OsGNA1 regulates plant height, we generated a gRNA construct and introduced it into the Nipponbare to knock out the OsGNA1 gene via a CRISPR/Cas 9 strategy (Ma et al., 2015). Several homozygous or heterozygous mutant plants were obtained. The seedling height and root length of both heterozygous and homozygous mutants were significant shorter compared to non-mutant transgenic plants (Figure 4). These results demonstrate that OsGNA1 positively regulates plant height and root length. Elevated expression levels were beneficial to plant height and root length.
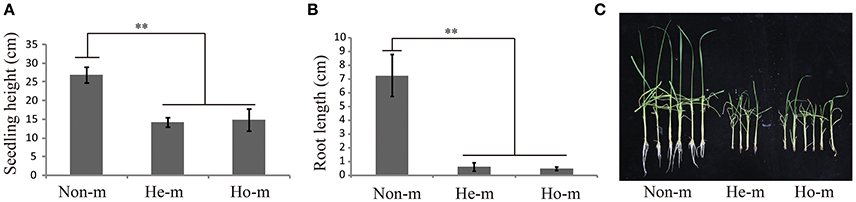
Figure 4. Analysis of seeding height and root length for the mutant of OsGNA1. (A) seedling height in T0 mutant plants. (B) root length in T0 mutant plants. Values represent mean ± SD. Non-m, non-mutant (n = 13), He-m, heterozygous mutants (n = 4), Ho-m, homozygous mutants (n = 6). “**” represent significances at p < 0.01.
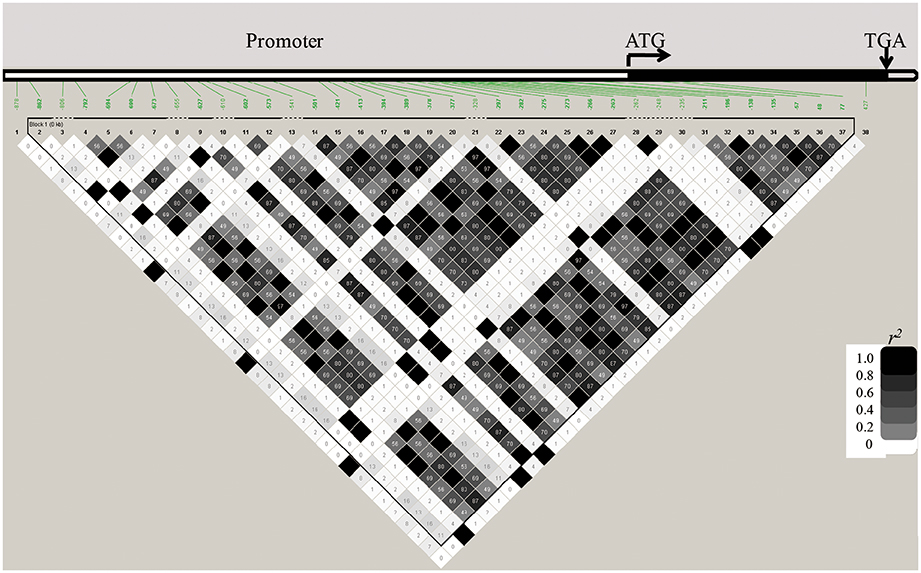
We sequenced OsGNA1 from −891 to 748 bp in 184 accessions of the natural population and identified 38 polymorphic loci (36 SNPs and 2 indels) (Table S3). Among them, one indel (+48) and two SNPs (+77, +427) at the CDS region caused insertion/deletion of two amino acids, substitution of Pro (CCG) to Leu (CTG), and Pro (CCG) to Ser (TCG), respectively. Cluster analysis divided 184 accessions into five groups (Figure 5). LD analysis of 38 polymorphism loci is depicted in Figure 6. We performed haplotype analyses based on 38 polymorphism loci of OsGNA1 and classified the genotype into five haplotypes (Hap) (Table S3). Hap1 and Hap5 mainly contained the indica subpopulation, while Hap2 mainly contained the japonica subpopulation. At adult stage under drought stress Hap2 had the lowest PH, and Hap5 had the highest PH and highest DRC. At seedling stage, Hap5 had the longest root length (Table S3). The one amino acid mutation caused by +427 SNP only existed in Hap5 (Table S3).
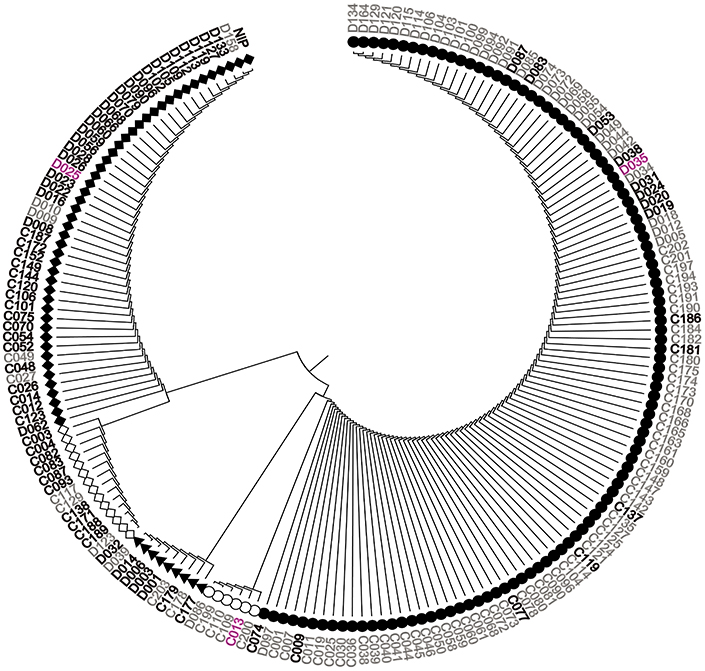
Figure 5. Cluster analysis of OsGNA1. Black font represents japonica, gray font represents indica, purple font represents aus. NIP = Nipponbare.
Mutation of OsRLK5 reduced the tolerance of plants to drought stress. OsRLK5 (LOC_Os02g13430) was a differentially expressed gene in the qDRC2.1 interval (Table S2), we generated a gRNA construct that was introduced into the Nipponbare to knock out OsRLK5 via CRISPR/Cas 9 technology (Zhang et al., 2014). Rate of water-loss of detached leaves and grain yield related traits were identified in both homozygous mutants and wild-type plants. As depicted in Figure 7, the mutant plants lost water faster than the wild-type plants. Compared to wild-type plants, plant height, panicle length, panicle neck length, seed-setting rate, spikelet numbers per panicle, and grain yield per plant were significantly reduced in the mutant plants under drought stress conditions (Figure 8). These results demonstrate that OsRLK5 is responsible for the GY under drought stress.
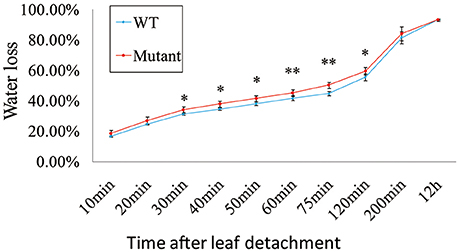
Figure 7. Water loss in the mutant of OsRLK5. Values represent mean ± SD, n = 5. “*” and “**” represent significances at p < 0.05, p < 0.01, respectively.
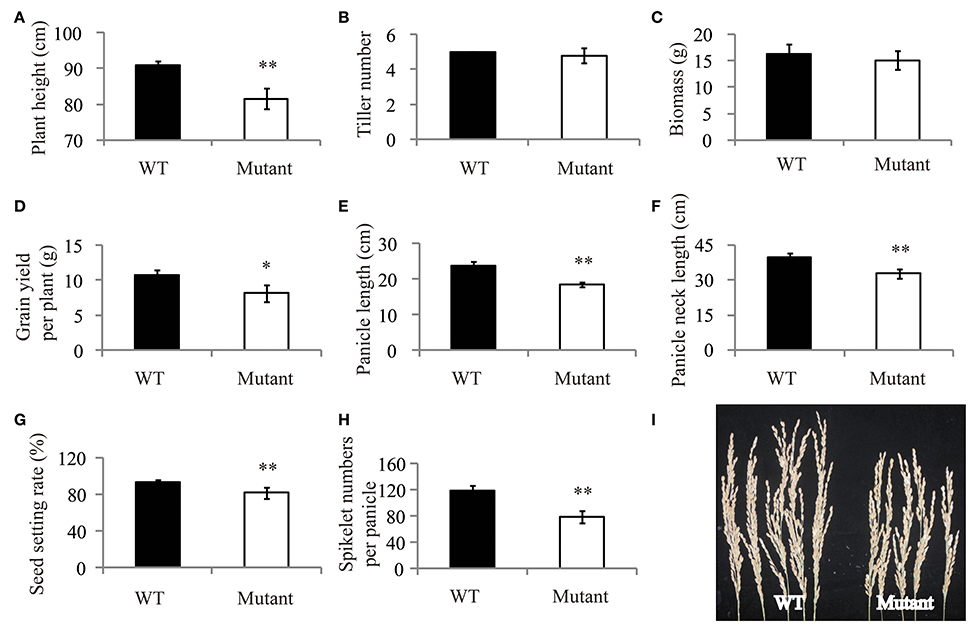
Figure 8. Drought resistance assay of OsRLK5 under drought stress condition. Plant height (A), tiller number (B), biomass (C), grain yield per plant (D), panicle length (E), panicle neck length (F), seed setting rate (G), and spikelet numbers per panicle (H) were measured under drought stress condition. (I), the panicle of wild-type Nipponbare (WT) and transgenic plant (Mutant). Values represent mean ± SD, n = 5. “*” and “**” represent significances at p < 0.05, p < 0.01, respectively.
Additionally, using genome-wide association study for eigenvectors (EigenGWAS; Chen et al., 2016), five SNPs in the loci of OsRLK5 were significantly detected for the third eigenvector, implying this gene was under selection within this population. None or few SNPs were detected for all three largest eigenvectors for OsGNA1 and eigenvector 1 and 2 for OsRLK5 (Table S4). Roughly, eigenvector 1 is responding to the population stratification of indica and japonica subspecies. Further analysis is needed to understand the selective effect on OsRLK5 gene among subgroups determined by eigenvector 3.
Discussion
Drought is one of the main abiotic stresses affecting the production of most field crops. Drought stress can occur at any crop growth stage and can affect productivity to variable degrees depending on the onset time, duration, and intensity of drought. Drought stress typically suppresses plant growth at the vegetable stage and has been reported to directly reduce production at the reproductive stage (Yue et al., 2005, 2006). Agronomists and breeders have made significant progress in improving the drought resistance of crops, however, the genetic and molecular basis for drought resistance in crops remains largely unclear. In this study, drought resistance of the rice germplasm was evaluated using contrasting soil moisture regimes. Combining high density SNPs and phenotypes of this germplasm, we conducted GWAS for plant height, grain yield per plant, and drought resistant coefficient. Five, two, and two known genes for the same three traits were found within or close to the associated loci, respectively. These associated loci and candidate genes could enhance the knowledge for understanding the mechanisms underlying rice drought resistance and could be used in improvement of drought resistance of rice via molecular breeding. In addition, many drought resistant germplasm accessions were obtained in this study that should be valuable in rice breeding program facing water scarcity.
Compared to linkage analysis, GWAS greatly accelerated both speed and accuracy of locating QTL or candidate genes. For example, OsSPL13 has been quickly identified based on genome-wide association study for the grain size and expression profiling in panicles between small-grain and large-grain varieties. Further analysis indicated that OsSPL13 positively regulates cell size in the grain hull, resulting in enhanced grain length and yield in rice (Si et al., 2016). In this study, nine reported functional genes were found to be near the significant GWAS loci (Table 4). The function of these genes is consistent with their associated traits. The discovery of the novel drought resistant gene OsRLK5 and the new functions of OsGAN1 in this study further support GWAS as a viable approach to quickly identify new genes that are contributing to the complex traits.
Numerous factors affect plant height of rice. Gibberellin (GA) is one of the most important determinants (Monna et al., 2002; Sasaki et al., 2002; Spielmeyer et al., 2002). In this study, several candidate genes (SD-1, OsGA2ox3, and OsDOG), controlling rice plant height were involved in synthesis or signal transduction of GA. For example, SD-1 encodes a gibberellin 20-oxidase, a key enzyme in the biosynthesis of gibberellin catalyzing (GA53 → GA44 → GA19 → GA20) (Monna et al., 2002; Sasaki et al., 2002; Spielmeyer et al., 2002). Interestingly, the GWAS approach also found that SD-1 is important for plant height under drought stress condition (Figure 1), suggesting that SD-1 may be involved in drought resistance. OsGA2ox3 was also involved in the GA metabolic pathway, and it has been reported that OsGA2ox3 regulates plant height and tiller number (Lo et al., 2008). Another candidate gene for rice height, OsDOG, has been reported to act as a new dynamic equilibrium regulator for the gibberellin metabolism. It has furthermore been reported that OsDOG negatively regulates cell elongation and plant height in rice (Liu et al., 2011). OsSAP11 was an A20/AN1 zinc-finger gene and contained stress-associated proteins. Over-expression of OsSAP11 can enhance the tolerance to water deficit and salt stress in transgenic Arabidopsis plants (Giri et al., 2011). OsDOG/OsSAP11 was identified within the important locus for plant height, especially as a significant signal with lower p-value under drought stress conditions.
Auxin plays an important role in plant height. In this study, OsGH3-2 was identified at the qPH1.2 interval, encoding an enzyme catalyzing the IAA conjunction to amino acids. OsGH3-2 has been reported to be involved in the regulation of plant height, root and stomatal development, as well as in the modulation of ABA level and drought resistance (Du et al., 2012). Two more candidate genes (LOC_Os09g08130 and LOC_Os10g06710) that were detected for plant height are also related to the auxin metabolism. LOC_Os09g08130 was annotated as indole-3-glycerol phosphate synthase and was located upstream of the indole alkaloid biosynthesis pathway. LOC_Os10g06710 was annotated as amidase, involved in the metabolic pathway of indole acetic acid. Indole acetic acid is an important phytohormone and plays a crucial role in cell division, differentiation, and elongation, in root development and plant height regulation (Petersson et al., 2009; Du et al., 2012; Lu et al., 2015).
In particular, a significant SNP (sf0918825447) with p-value of 4.18E-07 for plant height is located at the promoter region of OsGNA1. The associated peak signal was detected with a lower p-value under drought stress condition compared to normal water condition, implying its contribution to drought resistance. A previous study demonstrated OsGNA1 to play an important role in root development. Compared to wild type rice plants, mutant gna1 plants exhibited reduced root elongation, shorter taproot, lateral root and root hairs (Jiang et al., 2005). Similarly, the mutant of OsGNA1 exhibited shorter root length and seedling height compared to wild type plants in this study. These results suggest that OsGNA1 can regulate plant height and drought resistance. A haplotype analysis revealed that Hap5 was likely to improve drought resistance in rice because Hap5 had the highest PH at the adult stage under drought stress and had the longest maximum root length at seedling stage. Moreover, Hap5 had the highest DRC (Table S3).
In this study, the candidate genes for drought responsive associated loci were discovered together with a set of RNA-Seq data for rice plants under contrasting moisture regimes (unpublished). A total of 162 genes showed significantly different expression levels in leaves between drought treated and normal water treated plants. Two functional genes, OsPYL2 and OsGA2ox9 were discovered in this study. OsGA2ox9 was involved in GA metabolic pathways, and a previous study reported it to control plant height, tiller number, and root length in rice (Lo et al., 2008). Roots play an important role for absorbing water and nutrients. Plants can benefit from longer roots for water uptake from deeper soil layers as drought stress occurs. These above results imply that OsGA2ox9 was likely affecting drought resistance via regulation of root growth in rice. OsPYL2 was reported to be an ABA receptor (Tian et al., 2015). ABA is involved in the adaptation of plants to abiotic stress. The combination of ABA and the ABA receptor can activate the ABA signal transduction and trigger the corresponding physiological reaction, including stomatal closure (Cutler et al., 2010; Kim et al., 2010). Thus, OsPYL2 is likely to regulate stomatal behavior to reduce water loss, then to enhance the drought resistance. Furthermore, our preliminary research verified the function of OsRLK5 to be positively related to the leaf water content and grain yield under drought stress (Figures 7, 8).
Two known genes (OsCYP51G3 and OsRRMh) were mapped for grain yield in this study, and the functions of these genes correlate well with the grain yield trait. OsCYP51G3 has been reported to mediate the biosynthesis of phytosterols and brassinosteroids, and to regulate seed setting rate (Xia et al., 2015). OsRRMh plays an important role during the transition from vegetable to reproductive phase in rice. Compared to the wild-type, the OsRRMh RNAi lines exhibit enlarged panicles while over-expression of OsRRMh lines lead to lower fertility rates and less number of spikelets per panicle (Liu and Cai, 2013).
Genome-wide association study (GWAS) based on large-scale re-sequencing provides a powerful tool to discover genetic variants that can be used for crop improvement, including improvement of drought resistance. GWAS has been successfully used at a high resolution to uncover associations that involve complex traits from inbred lines of field-grown maize and rice landraces and cultivars with genetic variants (Huang et al., 2010; Lu et al., 2010; Chen et al., 2014; Wen et al., 2014; Wu et al., 2015; Yano et al., 2016). Using high density SNPs at a whole-genome level, GWAS provides high-resolution genetic mapping that can narrow down the associated regions to candidate genes. Our study demonstrates that known genes as well as novel genes and loci can be dissected via GWAS analysis. Moreover, candidate genes discovered in this study provide useful information for further studies in drought resistance of crops. Functional identification of these candidate genes will improve our understanding of the mechanisms of crop responses to drought stress.
Author Contributions
LL, HL, HM, and XM designed this study. XM, KX, SC, TL, and XL performed the experiments. XM, FF, and HW analyzed the data. XM, HL, and LL drafted the manuscript.
Funding
This work was supported by grants from the National Program for Basic Research of China (2012CB114305); the National High-Tech Research and Development Program of China (2014AA10A603); the Shanghai Municipal Commission of Science and Technology (15DZ2290700; 16ZR1431200); the Project of Subject Construction, Shanghai Academy of Agricultural Sciences, Grant No. SAAS-2016(02); the Youth Talent Development Project supported by the Shanghai Municipal Agriculture Commission (No. 2015-1-6).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the National Key Laboratory of Crop Genetic Improvement at the Huazhong Agricultural University for providing the seeds and genome sequence data of the rice germplasm collection.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01801/full#supplementary-material
References
Atwell, S., Huang, Y. S., Vilhjalmsson, B. J., Willems, G., Horton, M., Li, Y., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631. doi: 10.1038/nature08800
Bai, X., Zhao, H., Huang, Y., Xie, W., Han, Z., Zhang, B., et al. (2016). Genome-wide association analysis reveals different genetic control in panicle architecture between indica and japonica rice. Plant Genome 9, 1–10. doi: 10.3835/plantgenome2015.11.0115
Barrett, J. C., Fry, B., Maller, J., and Daly, M. J. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. doi: 10.1093/bioinformatics/bth457
Chen, G. B., Lee, S., Zhu, Z., Benyamin, B., and Robinson, M. (2016). EigenGWAS: finding loci under selection through genome-wide association studies of eigenvectors in structured populations. Heredity (Edinb). 117, 51–61. doi: 10.1038/hdy.2016.25
Chen, W., Gao, Y., Xie, W., Gong, L., Lu, K., Wang, W., et al. (2014). Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 46, 714–721. doi: 10.1038/ng.3007
Cho, S., Yoo, S., Zhang, H., Lim, J., and Paek, N. (2014). Rice NARROW LEAF 1 Regulates leaf and adventitious root development. Plant Mol. Biol. Rep. 32, 270–281. doi: 10.1007/s11105-013-0675-z
Clark, R. M. (2010). Genome-wide association studies coming of age in rice. Nat. Genet. 42, 926–927. doi: 10.1038/ng1110-926
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Ding, X., Li, X., and Xiong, L. (2011). Evaluation of near-isogenic lines for drought resistance QTL and fine mapping of a locus affecting flag leaf width, spikelet number, and root volume in rice. Theor. Appl. Genet. 123, 815–826. doi: 10.1007/s00122-011-1629-1
Du, H., Wu, N., Fu, J., Wang, S., Li, X., Xiao, J., et al. (2012). A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 63, 6467–6480. doi: 10.1093/jxb/ers300
Ekanayake, I. J., De Datta, S. K., and Steponkus, P. L. (1989). Spikelet sterility and flowering response to rice water stress at anthesis. Ann. Bot. 63, 257–264.
Flint-Garcia, S. A., Thornsberry, J. M., and Buckler, E. S. T. (2003). Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54, 357–374. doi: 10.1146/annurev.arplant.54.031902.134907
Fujita, D., Trijatmiko, K. R., Tagle, A. G., Sapasap, M. V., Koide, Y., Sasaki, K., et al. (2013). NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc. Natl. Acad. Sci. U.S.A. 110, 20431–20436. doi: 10.1073/pnas.1310790110
Giri, J., Vij, S., Dansana, P. K., and Tyagi, A. K. (2011). Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 191, 721–732. doi: 10.1111/j.1469-8137.2011.03740.x
Gowda, V. R. P., Henry, A., Yamauchi, A., Shashidhar, H. E., and Serraj, R. (2011). Root biology and genetic improvement for drought avoidance in rice. Field Crops Res. 122, 1–13. doi: 10.1016/j.fcr.2011.03.001
Huang, X., Wei, X., Sang, T., Zhao, Q., Feng, Q., Zhao, Y., et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967. doi: 10.1038/ng.695
Huang, X., Zhao, Y., Wei, X., Li, C., Wang, A., Zhao, Q., et al. (2011). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39. doi: 10.1038/ng.1018
Jiang, H., Wang, S., Dang, L., Chen, H., Wu, Y., Jiang, X., et al. (2005). A novel short-root gene encodes a glucosamine-6-phosphate acetyltransferase required for maintaining normal root cell shape in rice. Plant Physiol. 138, 232–242. doi: 10.1104/pp.104.058248
Kim, T. H., Bohmer, M., Hu, H., Nishimura, N., and Schroeder, J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61, 561–591. doi: 10.1146/annurev-arplant-042809-112226
Langsrud, O. (2003). ANOVA for unbalanced data, use type II instead of type III sums of squares. Stat. Comput. 13, 163–167. doi: 10.1023/A:1023260610025
Li, H., Peng, Z., Yang, X., Wang, W., Fu, J., Wang, J., et al. (2012). Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 45, 43–50. doi: 10.1038/ng.2484
Li, J., Han, Y., Liu, L., Chen, Y., Du, Y., Zhang, J., et al. (2015). qRT9, a quantitative trait locus controlling root thickness and root length in upland rice. J. Exp. Bot. 66, 2723–2732. doi: 10.1093/jxb/erv076
Lipka, A. E., Tian, F., Wang, Q., Peiffer, J., Li, M., Bradbury, P. J., et al. (2012). GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399. doi: 10.1093/bioinformatics/bts444
Liu, D., and Cai, X. (2013). OsRRMh, a Spen-like gene, plays an important role during the vegetative to reproductive transition in rice. J. Integr. Plant Biol. 55, 876–887. doi: 10.1111/jipb.12056
Liu, G., Mei, H., Liu, H., Yu, X., Zou, G., and Luo, L. (2010). Sensitivities of rice grain yield and other panicle characters to late-stage drought stress revealed by phenotypic correlation and QTL analysis. Mol. Breed. 25, 603–613. doi: 10.1007/s11032-009-9356-x
Liu, H., Zhou, G., Liu, G., Hu, S., Li, M., Yu, X., et al. (2005). Correlation analysis and QTL identification for canopy temperature, leaf water potential and spikelet fertility in rice under contrasting moisture regimes. Sci. Bull. 50, 317–326. doi: 10.1007/BF02897572
Liu, Y., Xu, Y., Xiao, J., Ma, Q., Li, D., Xue, Z., et al. (2011). OsDOG, a gibberellin-induced A20/AN1 zinc-finger protein, negatively regulates gibberellin-mediated cell elongation in rice. J. Plant Physiol. 168, 1098–1105. doi: 10.1016/j.jplph.2010.12.013
Lo, S. F., Yang, S. Y., Chen, K. T., Hsing, Y. I., Zeevaart, J. A., Chen, L. J., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 20, 2603–2618. doi: 10.1105/tpc.108.060913
Lou, Q., Chen, L., Mei, H., Wei, H., Feng, F., Wang, P., et al. (2015). Quantitative trait locus mapping of deep rooting by linkage and association analysis in rice. J. Exp. Bot. 66, 4749–4757. doi: 10.1093/jxb/erv246
Lu, G., Coneva, V., Casaretto, J. A., Ying, S., Mahmood, K., Liu, F., et al. (2015). OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 83, 913–925. doi: 10.1111/tpj.12939
Lu, Y., Zhang, S., Shah, T., Xie, C., Hao, Z., Li, X., et al. (2010). Joint linkage-linkage disequilibrium mapping is a powerful approach to detecting quantitative trait loci underlying drought tolerance in maize. Proc. Natl. Acad. Sci. U.S.A. 107, 19585–19590. doi: 10.1073/pnas.1006105107
Luo, L. J. (2010). Breeding for water-saving and drought-resistance rice (WDR) in China. J. Exp. Bot. 61, 3509–3517. doi: 10.1093/jxb/erq185
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A Robust CRISPR/Cas9 System for Convenient, high-efficiency multiplex genome editing in monocot and dicot Plants. Mol. Plant. 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Monna, L., Kitazawa, N., Yoshino, R., Suzuki, J., Masuda, H., Maehara, Y., et al. (2002). Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 9, 11–17. doi: 10.1093/dnares/9.1.11
Morris, G. P., Ramu, P., Deshpande, S. P., Hash, C. T., Shah, T., Upadhyaya, H. D., et al. (2013). Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. U.S.A. 110, 453–458. doi: 10.1073/pnas.1215985110
Ouyang, S. Q., Liu, Y. F., Liu, P., Lei, G., He, S. J., Ma, B., et al. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62, 316–329. doi: 10.1111/j.1365-313X.2010.04146.x
Pennisi, E. (2008). The blue revolution, drop by drop, gene by gene. Science 320, 171–173. doi: 10.1126/science.320.5873.171
Petersson, S. V., Johansson, A. I., Kowalczyk, M., Makoveychuk, A., Wang, J. Y., Moritz, T., et al. (2009). An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell. 21, 1659–1668. doi: 10.1105/tpc.109.066480
Qi, J., Qian, Q., Bu, Q., Li, S., Chen, Q., Sun, J., et al. (2008). Mutation of the rice Narrow leaf 1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147, 1947–1959. doi: 10.1104/pp.108.118778
Riedelsheimer, C., Lisec, J., Czedik-Eysenberg, A., Sulpice, R., Flis, A., Grieder, C., et al. (2012). Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl. Acad. Sci. U.S.A. 109, 8872–8877. doi: 10.1073/pnas.1120813109
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. doi: 10.1038/416701a
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Spielmeyer, W., Ellis, M. H., and Chandler, P. M. (2002). Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. U.S.A. 99, 9043–9048. doi: 10.1073/pnas.132266399
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tian, F., Bradbury, P. J., Brown, P. J., Hung, H., Sun, Q., Flint-Garcia, S., et al. (2011). Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43, 159–162. doi: 10.1038/ng.746
Tian, X., Wang, Z., Li, X., Lv, T., Liu, H., Wang, L., et al. (2015). Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice 8, 28. doi: 10.1186/s12284-015-0061-6
Uga, Y., Okuno, K., and Yano, M. (2011). Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 62, 2485–2494. doi: 10.1093/jxb/erq429
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Wang, Q., Xie, W., Xing, H., Yan, J., Meng, X., Li, X., et al. (2015). Genetic architecture of natural variation in rice chlorophyll content revealed by a genome-wide association study. Mol. Plant. 8, 946–957. doi: 10.1016/j.molp.2015.02.014
Wang, X., Wang, H., Liu, S., Ferjani, A., Li, J., Yan, J., et al. (2016). Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48, 1233–1241. doi: 10.1038/ng.3636
Wen, W., Li, D., Li, X., Gao, Y., Li, W., Li, H., et al. (2014). Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 5, 3438. doi: 10.1038/ncomms4438
Wu, J., Feng, F., Lian, X., Teng, X., Wei, H., Yu, H., et al. (2015). Genome-wide Association Study (GWAS) of mesocotyl elongation based on re-sequencing approach in rice. BMC Plant Biol. 15, 218. doi: 10.1186/s12870-015-0608-0
Xia, K., Ou, X., Tang, H., Wang, R., Wu, P., Jia, Y., et al. (2015). Rice microRNA osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol. 208, 790–802. doi: 10.1111/nph.13513
Yang, W., Guo, Z., Huang, C., Duan, L., Chen, G., Jiang, N., et al. (2014). Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 5, 5087. doi: 10.1038/ncomms6087
Yang, W., Guo, Z., Huang, C., Wang, K., Jiang, N., Feng, H., et al. (2015). Genome-wide association study of rice (Oryza sativa L.) leaf traits with a high-throughput leaf scorer. J. Exp. Bot. 66, 5605–5615. doi: 10.1093/jxb/erv100
Yano, K., Yamamoto, E., Aya, K., Takeuchi, H., Lo, P. C., Hu, L., et al. (2016). Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 48, 927–934. doi: 10.1038/ng.3596
Yue, B., Xiong, L., Xue, W., Xing, Y., Luo, L., and Xu, C. (2005). Genetic analysis for drought resistance of rice at reproductive stage in field with different types of soil. Theor. Appl. Genet. 111, 1127–1136. doi: 10.1007/s00122-005-0040-1
Yue, B., Xue, W., Xiong, L., Yu, X., Luo, L., Cui, K., et al. (2006). Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics 172, 1213–1228. doi: 10.1534/genetics.105.045062
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. doi: 10.1111/pbi.12200
Zhang, Q. (2007). Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. U.S.A. 104, 16402–16409. doi: 10.1073/pnas.0708013104
Zhang, X., Warburton, M. L., Setter, T., Liu, H., Xue, Y., Yang, N., et al. (2016). Genome-wide association studies of drought-related metabolic changes in maize using an enlarged SNP panel. Theor. Appl. Genet. 129, 1449–1463. doi: 10.1007/s00122-016-2716-0
Keywords: GWAS, plant height, grain yield, drought resistance, candidate genes
Citation: Ma X, Feng F, Wei H, Mei H, Xu K, Chen S, Li T, Liang X, Liu H and Luo L (2016) Genome-Wide Association Study for Plant Height and Grain Yield in Rice under Contrasting Moisture Regimes. Front. Plant Sci. 7:1801. doi: 10.3389/fpls.2016.01801
Received: 19 August 2016; Accepted: 15 November 2016;
Published: 29 November 2016.
Edited by:
Guo-Bo Chen, Evergreen Landscape and Architecture Studio, ChinaReviewed by:
Liyuan Zhou, Zhejiang University, ChinaHaiming Xu, Zhejiang University, China
Kejian Wang, China National Rice Research Institute, China
Zhenyu Gao, China National Rice Research Institute, China
Copyright © 2016 Ma, Feng, Wei, Mei, Xu, Chen, Li, Liang, Liu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyan Liu, lhy@sagc.org.cn
Lijun Luo, lijun@sagc.org.cn
 Xiaosong Ma
Xiaosong Ma Fangjun Feng
Fangjun Feng Haibin Wei2
Haibin Wei2 Lijun Luo
Lijun Luo