- 1Council of Scientific and Industrial Research – Institute of Himalayan Bioresource Technology, Palampur, India
- 2Academy of Scientific and Innovative Research, New Delhi, India
Universal stress proteins (USPs) are known to be expressed in response to various abiotic stresses in a wide variety of organisms, such as bacteria, archaebacteria, protists, algae, fungi, plants, and animals. However, in plants, biological function of most of the USPs still remains obscure. In the present study, Arabidopsis USP gene (AtUSP) showed induction in response to abscisic acid (ABA) and various abiotic stresses viz. heat, dehydration, salt, osmotic, and cold stresses. Additionally, in silico analysis of AtUSP promoter identified several cis-elements responsive to phytohormones and abiotic stresses such as ABRE, ERE, DRE, and HSE, etc. To functionally validate the AtUSP promoter, the 1115 bp region of promoter was characterized under phytohormone and abiotic stress treatments. Deletion analysis of promoter was carried out by cloning the full length promoter (D0) and its three 5′ deletion derivatives, D1 (964 bp), D2 (660 bp), and D3 (503 bp) upstream of the β-glucuronidase (GUS) reporter gene, which were then stably transformed in Arabidopsis plants. The AtUSP promoter (D0) showed minimal activity under non-stress conditions which was enhanced in response to phytohormone treatments (ABA and ACC) and abiotic stresses such as dehydration, heat, cold, salt, and osmotic stresses. The seedlings harboring D1 and D2 deletion fragments showed constitutive GUS expression even under control condition with increased activity almost under all the treatments. However, D3 seedlings exhibited complete loss of activity under control condition with induction under ACC treatment, dehydration, heat, oxidative, salt, and osmotic stresses. Thus, present study clearly showed that AtUSP promoter is highly inducible by phytohormones and multiple abiotic stresses and it can be exploited as stress inducible promoter to generate multi-stress tolerant crops with minimal effects on their other important traits.
Introduction
During their life cycle, plants have to cope with various abiotic stresses, such as drought, salinity, high or low temperature, oxidative stress, etc. To counter these stresses, plants have evolved various stress tolerance and avoidance mechanisms. These mechanisms involve signal transduction cascades that respond to external stimuli by activating stress-responsive genes, which in turn bring about some morphological, physiological, and metabolic changes that help plants to survive these adverse conditions (Ahuja et al., 2010). In the field conditions, plants are often exposed to multiple stresses, simultaneously (Mittler, 2006), thus, there is a need to engineer plants with capability to survive and grow well under multiple abiotic stresses. In past, several studies have identified large subset of multiple abiotic stress-responsive genes, which might play important role in crosstalk between various stress signaling pathways (Kilian et al., 2007; Matsui et al., 2008).
The universal stress proteins (USPs) were first discovered in bacteria to be overexpressed in response to various stresses (Nystrom and Neidhardt, 1992). Accumulating evidences suggest that USPs are induced during multiple stresses and play important role in survival under abiotic stresses. For example, Nachin et al. (2005) showed the role of USP in survival of bacteria when exposed to various stress conditions, such as nutrient starvation and exposure to heat and oxidative stress. Furthermore, some studies showed induction of USPs in pathogenic microbes when exposed to immunological stresses of hosts, wherein they play important role in survival (Liu et al., 2007; Gomes et al., 2011). USPs are conserved among the archaebacteria, eubacteria, protozoa, fungi, plants, and metazoans (Kerk et al., 2003; Kvint et al., 2003; Isokpehi et al., 2011a). In Escherichia coli, six USP paralogs have been identified namely, UspA, UspC, UspD, UspE, UspF, and UspG (Kvint et al., 2003). Loss of function of UspA in bacteria was shown to have detrimental effect on bacterial growth during stationary phase (Nystrom and Neidhardt, 1993). Broadly, USPs have been classified into two groups; first group includes USPs with structure similar to UspA of Haemophilus influenzae with no ATP binding activity, while second group includes proteins with structural similarity to UspFG-type proteins of Methanococcus jannaschii that possess ATP binding activity. The USP proteins may be small, with single USP domain or may be large containing two tandem repeats of USP domains or may be present along with other functional domains such as Na+/H+ exchanger, amino acid permease, and protein kinase (Nachin et al., 2005). Kerk et al. (2003) identified 44 genes encoding USPA domain containing proteins in Arabidopsis and performed their phylogenetic analysis which revealed that they have evolved from 1MJH like ancestor. Since then, several USP proteins have been reported to play important roles in stress tolerance in various plants, such as Arabidopsis, rice, cotton, tomato, pigeonpea, and Salicornia brachiata (Sauter et al., 2002; Lenman et al., 2008; Merkouropoulos et al., 2008; Maqbool et al., 2009; Loukehaich et al., 2012; Udawat et al., 2014, 2016; Gonzali et al., 2015; Jung et al., 2015; Sinha et al., 2016). However, the precise function of most of the plant USPs has not been deciphered, so far.
From lower to higher organisms, the regulation of gene expression is the major factor that determines the adaptive capacity of an organism to various environmental stresses (Gasch et al., 2000; Balazi and Oltvai, 2005). In plants, spatial and temporal expression of specific genes is required to coordinate growth, development, and responses to various abiotic stresses. This tight regulation of gene expression occurs at the two levels, transcriptional and post-transcriptional. Multiple cis-acting regulatory elements present in the promoters of the genes are a type of transcriptional gene regulatory elements that are required for the specific expression pattern of a gene. Till date, various stress-responsive cis-elements have been identified in the promoters of stress inducible genes that allow their stress specific expression. The promoters of multiple stress-responsive genes have a number of regulatory elements that respond to multiple stresses, such as ABRE (abscisic acid-responsive element), LTRE (low temperature-responsive element), MYC and DRE (dehydration-responsive element) are the potential targets for regulating stress inducible expression of transgenes in transgenic plants (Maruyama et al., 2012). Plant responses to abiotic stresses, such as drought, salinity, cold, and heat are largely mediated by different hormonal signaling pathways that might act synergistically or antagonistically. Abscisic acid (ABA), ethylene and salicylic acid are the major phytohormones that act as link between plant responses to different abiotic stresses (Li et al., 2004; Fujita et al., 2006). Overexpression of stress-responsive genes under the control of CaMV35S promoter results in strong expression of genes that provide plants with stress tolerance. However, it might result in undesirable phenotypes, such as retarded and stunted growth and low seed yield (Liu et al., 1998). Several studies have shown that expression of stress-responsive genes driven under stress inducible promoter is far stronger than under CaMV35S promoter and that enhances plant stress tolerance with least effect on growth and yield of plant under normal conditions (Kasuga et al., 1999). Thus, in order to engineer multiple stress tolerant crop plants, it is necessary to identify and characterize multiple stress-responsive promoters, which may be used to regulate expression of transgenes. Since, many plants share similar transcriptional machineries and regulatory elements, therefore, Arabidopsis can be used as model system to characterize multiple stress-responsive promoters. Several of the bacterial UspA characterized till date provide resistance to multiple stresses (Nystrom and Neidhardt, 1992; Nachin et al., 2005; Liu et al., 2007). Till date, several plant USPs have been reported to be responsive to more than one stress. The SpUSP from wild tomato has been reported to be induced under ABA, ethylene, drought, salt, heat, wounding, oxidative, and cold stresses (Loukehaich et al., 2012). The expression of SbUSP gene from Salicornia brachiata has been shown to be induced by salt, drought, cold, and heat stress (Udawat et al., 2014). The promoter of cotton USP has been found to be responsive to dehydration, ABA, salt, heavy metals, gibberellic acid, and dark condition (Zahur et al., 2009). The ability of USPs to respond and provide tolerance against multiple stresses, suggests that their promoters might be good candidates to drive multiple stress-responsive expression of transgenes in transgenic plants.
In the present study, AtUSP expression was found to be induced under phytohormone and various abiotic stresses. Recently, overexpression of AtUSP has been shown to confer heat and oxidative stress tolerance in Arabidopsis (Jung et al., 2015). Therefore, the 1115 bp region upstream of the translation start site of AtUSP was cloned and functionally characterized in Arabidopsis through deletion analysis. Full length, AtUSP promoter showed least activity under non-stress conditions while its expression was highly induced by ABA, ACC, dehydration, heat, cold, salt, and osmotic stress. Full length AtUSP promoter showed tissue specific β-glucuronidase (GUS) expression which was lost in the subsequent deleted promoter fragments. This study showed that AtUSP promoter is multiple stress-responsive and can be used for guiding multiple stress-responsive expression of transgene in transgenic plants.
Materials and Methods
Plant Materials and Growth Conditions
Two week-old seedlings of wild type (WT; ecotype, Col-0) and transgenic Arabidopsis thaliana grown on ½ strength Murashige and Skoog (MS; Murashige and Skoog, 1962) medium supplemented with 1% sucrose and 0.8% agar at 22–23°C with 16/8h photoperiod and light intensity of 100 μmolm-2s-1 were used for AtUSP expression studies and promoter characterization under hormone and abiotic stress treatments, respectively.
Stress Treatments
For expression and promoter analysis, 2 week-old WT and transgenic seedlings were subjected to different abiotic stress and hormone treatments. ABA (100 μM), ACC (ethylene precursor; 1-aminocyclopropane-1-carboxylic acid, 100 μM), salt (150 mM, NaCl), osmotic (300 mM, mannitol) and oxidative (50 μM methyl viologen) treatments were given by placing the seedlings on filter paper saturated with respective chemical in the ½ MS solution for 8 h (Tarte et al., 2015). Dehydration treatment was given by air drying seedlings on Whatman 3 MM paper for 30 min. Heat and cold treatments were given by incubating the seedlings on ½ MS medium at temperature of 37°C and 4°C for 8 h, respectively. Seedlings placed on filter paper saturated with ½ MS solution without any treatment were considered as control and each treatment was given in triplicate. After stress treatment, seedlings were harvested, frozen in liquid nitrogen and stored at –80°C till further use.
RNA Isolation and Quantitative Real Time PCR Analysis
Total RNA was isolated from the frozen plant samples using iRIS method as described previously (Ghawana et al., 2011). First strand cDNA synthesis was carried out from 1 μg of total RNA using Verso cDNA synthesis kit (Thermo Scientific, USA). Real time primers for AtUSP (At3g53990) and GUS genes (Supplementary File S1) were designed using Primer Express 3.0.1 software (Applied Biosystems). The β-actin was used as internal control and the amplification specificity was determined by melt curve analysis. The expression analysis was performed with three biological and three technical replicates and relative gene expression was analyzed using 2-ΔΔCT method as described previously (Livak and Schmittgen, 2001).
Cloning of AtUSP Promoter and Its Deleted Fragments
A 1115 bp region of AtUSP promoter was amplified from genomic DNA of WT Arabidopsis and cloned into pGEM-T easy vector. Specific primers were designed with SalI and NcoI sites in the forward and reverse primers, respectively, to amplify full length promoter (D0) excluding the coding sequence. Based on the position of different stress-responsive elements identified through in silico analysis, three 5′ promoter deletions were made by sequentially deleting 151, 455, and 612 bp from the 5′region and were named as D1, D2, and D3, respectively. Same reverse primer with NcoI site was used to amplify D0, D1, D2, and D3 fragments with SalI site in the D0 and EcoRI site in the D1, D2, and D3 forward primers (Supplementary File S2). All these fragments were cloned into promoter-less vector pCAMBIA1391z fused with GUS gene and their schematic representation is depicted in Supplementary Figure S1.
Sequence Analysis
The AtUSP promoter sequence was analyzed using PLACE1 and PLANTCARE2 databases to identify various cis-regulatory elements present in the AtUSP promoter.
Agrobacterium-Mediated Transformation of Arabidopsis Plants
Four-week old Arabidopsis thaliana (WT) plants grown at 22–23°C with 16/8h photoperiod and light intensity of 100 μmolm-2s-1 were used for Agrobacterium-mediated transformation using vacuum infiltration method. For this, the unopened buds were dipped in the Agrobacterium tumefaciens (GV3101 strain) suspension in YEP medium with 30% sucrose and 0.2% Silvet-L77 solution by inverting the pots and applying the vacuum of 400 mm Hg for 5 min. The transformed plants were kept in the horizontal position in the dark for 24 h and then grown until the T0 generation seeds were harvested. T0 generation seeds were screened on 1X MS medium (1% sucrose and 0.8% agar) with hygromycin selection (20 μg/ml). The true transformants that were hygromycin resistant and able to reach the four leaf stage were transferred to pots containing a 1:1:1 mixture of vermiculite:perlite:coco peat. These plants were grown to maturity until the T1 generation seeds were harvested. Similarly, T1 and T2 seeds were germinated and plants were grown to maturity to harvest T2 and T3 generation seeds, respectively. Genomic DNA of eight T3 transgenic lines of Arabidopsis transformed with empty vector pCAMBIA1391z and recombinant vectors with D0, D1, D2, and D3 promoter fragments were isolated and analyzed for the presence of respective promoter fragment using PCR amplification (Supplementary Figure S2). The positive T3 transgenic Arabidopsis lines were used for functional characterization of AtUSP promoter and transgenic lines harboring empty vector pCAMBIA1391z were used as negative control.
Histochemical Assay of GUS Activity
Histochemical GUS assay was carried out using the method as described previously (Jefferson, 1987). Two week old Arabidopsis seedlings with or without treatment were kept in GUS staining buffer containing 1 mM 5-bromo-4-chloro-3-indolyl β-D-glucuronidase (HiMedia), 100 mM sodium phosphate (pH 7.5), 0.5 mM potassium ferricyanide and 0.5 mM potassium ferrocyanide, 10 mM ETDA and 0.1 % (v/v) Triton-X 100 and a vacuum of 400 mm of Hg was applied for 5 min. Seedlings were incubated at 37°C overnight and cleared with 70% ethanol in order to remove chlorophyll for clear visualization of blue color of GUS stain. The stained seedlings were then observed and photographed under Carl-Zeiss Stereo DiscoveryV12 with AxioVision software.
Quantitative Assay of GUS Activity
Quantitative assay was also performed as described by Jefferson (1987) using 4-methylumbelliferyl-β-D-glucuronide as substrate (MUG, Sigma). Protein was extracted from stress treated seedlings frozen in liquid nitrogen by grinding in GUS extraction buffer [50 mM sodium phosphate (pH 7.0), 10 mM β-mercaptoethanol, 10 mM EDTA, 0.1% (w/V) N-lauroyl sarcosine (SLS, Sigma), and 0.1% (v/v) Triton X-100). The tissue extract was centrifuged for 15 min at 4°C at 13000 rpm. The 10 μL of the protein extract was mixed with 390 μL of aliquoted GUS assay solution containing 22 mg MUG in 50 ml extraction buffer and incubated at 37°C for 1 h. The 100 μL of sample was taken from each tube at 0, 30, and 60 min incubation time, and 4.9 ml of 0.2 M Na2CO3 was added to stop the reaction. Fluorescence was measured with a fluorometer. Protein concentration was determined according to Bradford reagent using BSA standard (Bradford, 1976). Standard curve was prepared using 4-methylumbelliferone (4-MU, Sigma). GUS activity was expressed as nmol MU/min/mg protein.
Statistical Analysis
The data is given in the form of mean of the values with standard deviation of replicates. Statistical analysis was carried out using Student’s t-test and p-values of <0.05 and <0.01 between treated and untreated samples were considered as significant and highly significant and marked with ∗ and ∗∗, respectively.
Results
Expression Analysis of AtUSP in Response to Hormone Treatment and Various Abiotic Stresses
The expression level of AtUSP gene was analyzed by qRT-PCR in 2-week old seedlings of A. thaliana (WT) in response to phytohormone treatments (ABA and ACC) and abiotic stresses such as dehydration, salt, osmotic, oxidative, heat, and cold stress (Figure 1). The expression of AtUSP was highly induced under ABA treatment and all the abiotic stress conditions, except for the oxidative stress. The highest induction was observed after heat exposure (22.9-fold) followed by cold (17.7-fold), and dehydration (16.4-fold). Salt and osmotic stresses also led to induction of AtUSP transcript levels by 2.3- and 3.7-fold, respectively (Figure 1; Supplementary File S3). ABA treatment resulted in 8.7-fold induction, while ACC treatment led to no significant change in the AtUSP expression. AtUSP gene expression was downregulated by 1.7-fold in response to oxidative stress.
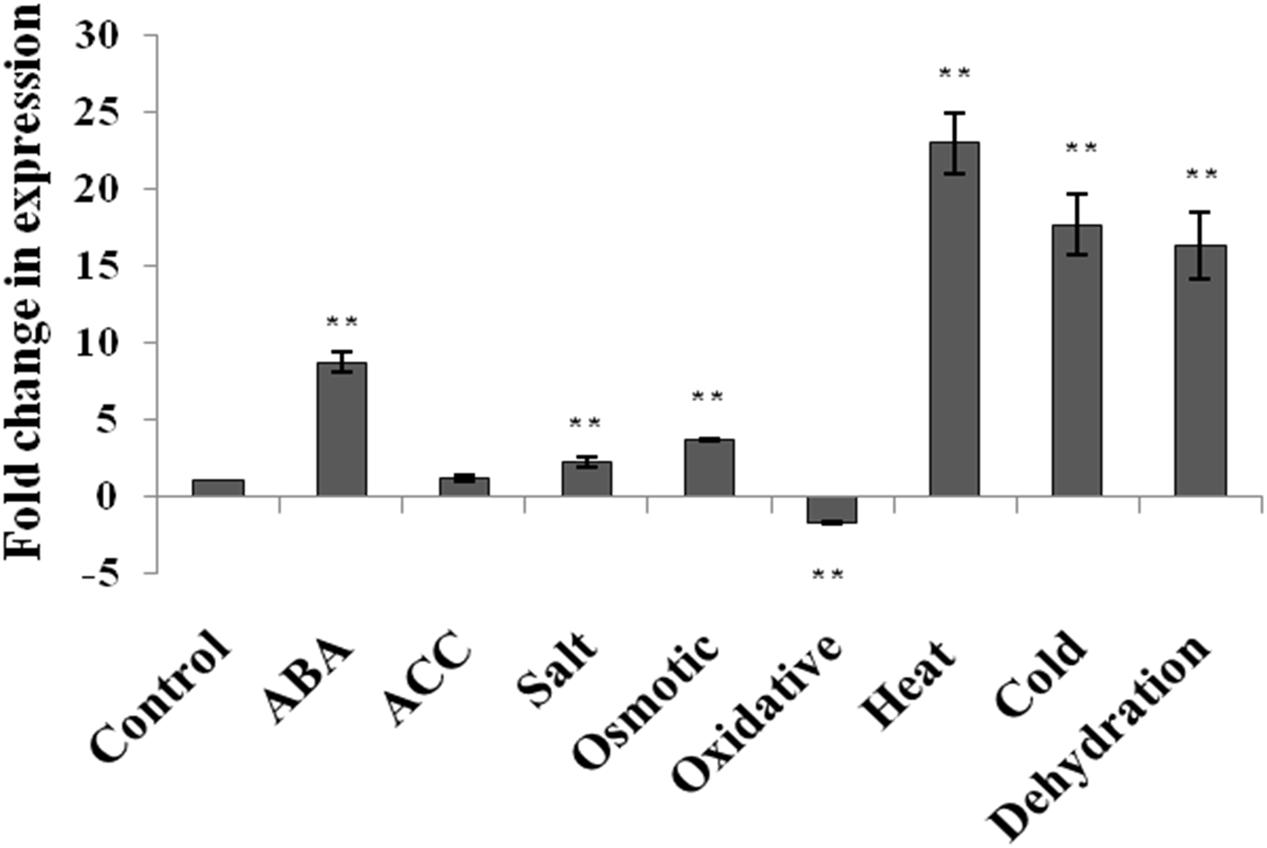
FIGURE 1. Expression analysis of AtUSP in response to various phytohormones and abiotic stresses in 2 week old seedlings of Arabidopsis thaliana. Data are represented as mean ± SD of replicates and statistical significance was determined by Student’s t-test (∗P < 0.05 and ∗∗P < 0.01).
Cloning of AtUSP Promoter and Its Sequence Analysis
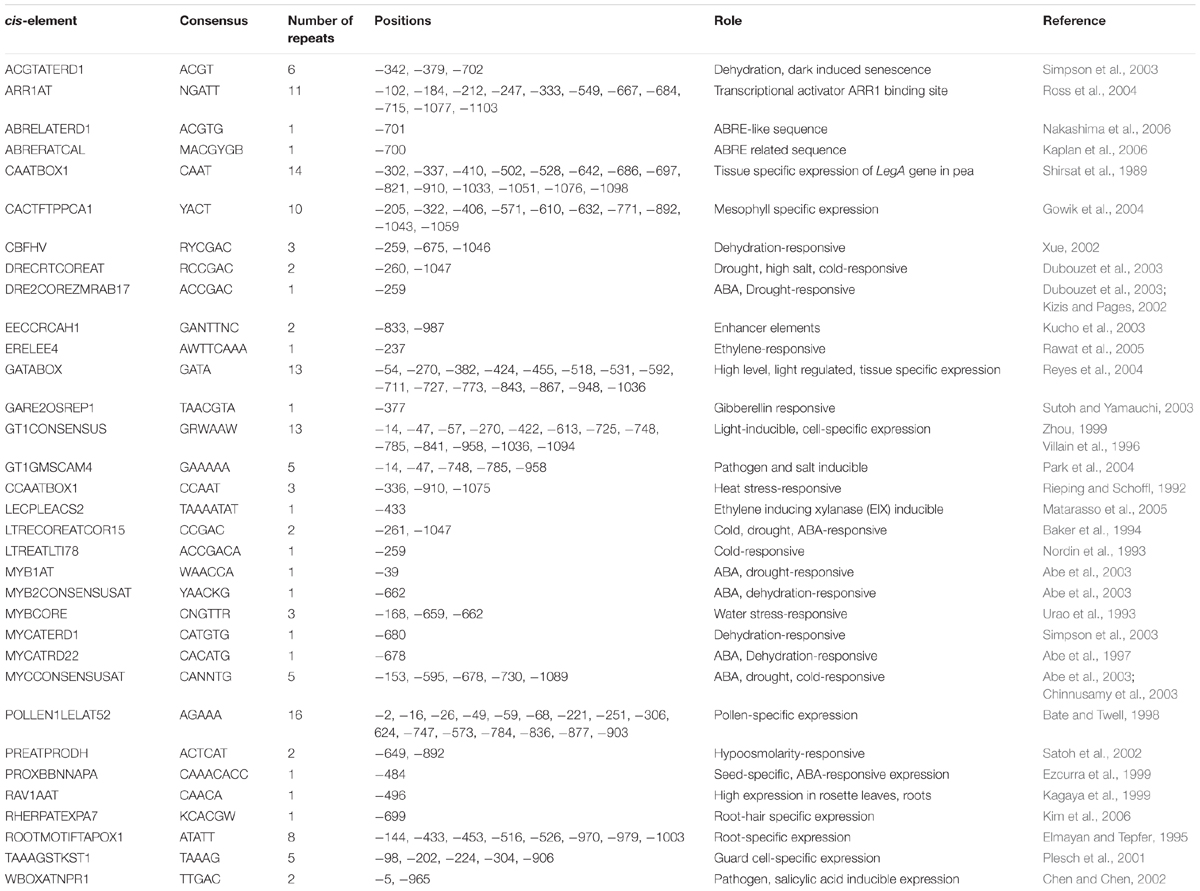
Since AtUSP was found to be multiple abiotic stress-responsive, its 1115 bp region upstream to translation start site was selected to characterize its promoter. This region was amplified from the genomic DNA of WT Arabidopsis and cloned in pGEM-T easy vector. The promoter sequence was analyzed using PLACE and PLANTCARE databases that led to identification of different putative cis-acting regulatory elements present in the AtUSP promoter (Table 1). The sequence motifs for ABA (ABRELATERD1 and ABRERATCAL), ethylene (ERELEE4), and gibberellin (GARE2OSREP1) were present at -700, -701, -238, and -377 positions, respectively (Figure 2A). Two motifs for salicylic acid (WBOXATNPR1) were present at positions -5 and -965. Five MYCCONSENSUSAT motifs at -153, -595, -678, -730, -1089, and one motif of each MYCATRD22 and MYBCONSENSUSAT were present at positions -678 and -662, respectively. Additionally, three motifs of MYBCORE elements known to be water stress-responsive were also present at -168, -659, and -662 positions. Five motifs of GT1GMSCAM4 responsible for pathogen and salt inducible expression were also found to be present in AtUSP promoter. It also contained two motifs of PREATPRODH known to be hypo-osmolarity responsive. AtUSP promoter had a number of cis-acting responsive elements for tissue specific expression such as CAATBOX1 and GATABOX present in several repeats (Figure 2A). It also contained certain motifs required for organ-specific expression, for example, PROXBBNNAPA for seed-specific expression, CACTFTPPCA1 for mesophyll cell-specific expression, RAV1AAT for high expression in rosette leaves and roots, RHERPATEXPA7 for root-hair specific expression, ROOTMOTIFTAPOX1 for root-specific expression, POLLEN1LELAT52 for pollen-specific expression, and TAAAGSTKST1 for guard cell-specific expression.
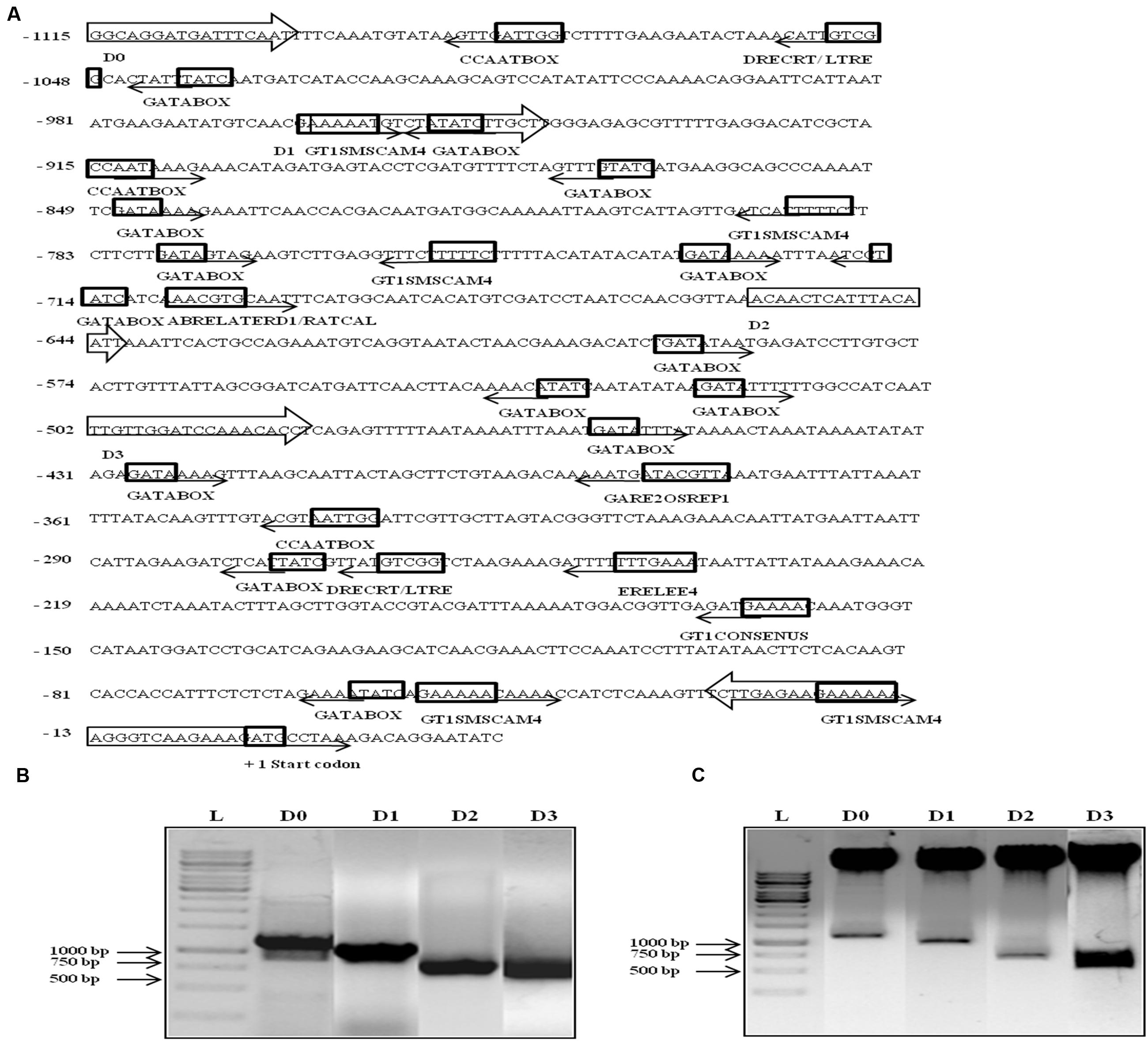
FIGURE 2. In silico analysis, amplification, and cloning of full length promoter of AtUSP (D0) and its 5′ deletion derivatives (D1, D2, and D3) into promoterless vector pCAMBIA1391z. (A) Various putative cis-acting elements identified in full length (1115 bp) AtUSP promoter and its three 5′ deletion derivatives with arrows indicating the direction of forward and reverse primers. Arrows at the cis-acting elements indicate the position of elements on either + strand (forward direction) or - strand (reverse direction). +1 indicates the translation start site. (B) Amplification of full length (1115 bp) AtUSP promoter and its three deletion derivatives. (C) Restriction digestion of cloned D0, D1, D2, and D3 fragments in promoterless vector pCAMBIA1391z.
Development of Transgenic Arabidopsis Harboring AtUSP Promoter Constructs
The 1115 bp region just upstream of AtUSP translation start site was cloned in the promoterless vector pCAMBIA1391z at SalI/NcoI sites and named as D0 construct containing the full length AtUSP promoter. Then, three deleted fragments were amplified by sequentially deletion of 151, 455, and 612 bp from the 5′ side of the full length promoter (Figure 2B). These fragments were then individually cloned into the promoterless vector pCAMBIA1391z at EcoRI/NcoI sites and were designated as D1 (964 bp), D2 (660 bp), and D3 (503 bp) constructs (Figure 2C). The D0, D1, D2, and D3 constructs were then transformed into Arabidopsis (WT) by Agrobacterium-mediated transformation using vacuum infiltration method. True transformants were first screened on 1X MS agar plates containing hygromycin and then further checked for the presence of desired fragment by PCR amplification using forward primer of each promoter fragment and reverse primer from GUS gene.
Stress Induction of the GUS Gene Expression Driven by AtUSP Promoter at the Transcript Level
The GUS transcript level was measured using qRT-PCR in the 2 week old T3 seedlings of D0, D1, D2, and D3 transgenic Arabidopsis lines under non-stress and stress conditions (Figures 3A–I; Supplementary File S4). Under control conditions, the GUS gene showed 12.4- and 6.4-fold induction in the D1 and D2 lines, respectively, while 1.4-fold reduction in D3 lines with respect to D0 lines (Figure 3A). The D0 lines exhibited 2.5- and 4.5-fold GUS gene induction in response to ABA and ACC treatments, respectively. The D0 lines showed 7.2-, 6.6-, 5.0-, 4.0-, and 2.5-fold induction in the GUS gene expression under, dehydration, heat, cold, salt and osmotic stress, respectively (Figures 3E–I). While in case of oxidative stress, GUS expression was unaltered in D0 lines (Figure 3D). The D1 lines showed GUS gene induction only under cold (4.1-fold) stress with respect to D1 lines under non-stress conditions (Figure 3G). The GUS expression was downregulated in response to ABA, ACC, oxidative and heat stress in the D1 lines (Figures 3B–D,F). In case of D2 lines, the GUS expression was induced (1.9-fold) only under cold stress (Figure 3G), while, GUS expression was downregulated in response to ABA, ACC, oxidative dehydration, salt and osmotic stress (Figures 3B–E,H,I). The D3 seedlings showed induction in GUS expression in response to ACC treatment (2.7-fold), oxidative (1.6-fold), dehydration (3.1-fold), heat (2.8-fold), and salt stress (1.8-fold) with respect to D3 seedlings under non-stress conditions (Figures 3C–F,H). The GUS expression was downregulated by 3.4-fold in the D3 seedlings only under ABA treatment (Figure 3B).
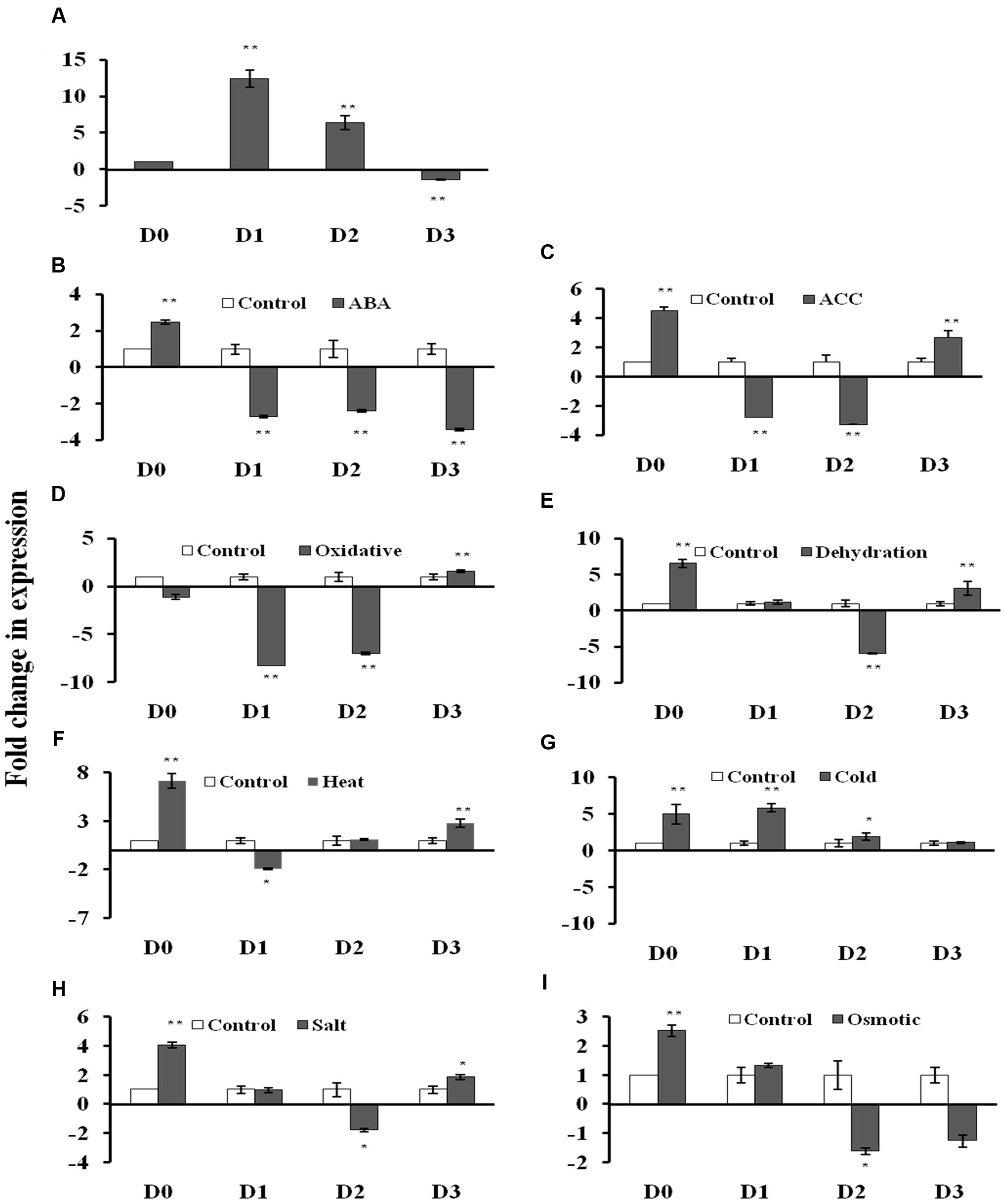
FIGURE 3. β-glucuronidase (GUS) expression analysis in transgenic A. thaliana containing four different AtUSP:GUS constructs under control condition and various phytohormones and abiotic stress treatments. (A) control condition, (B) abscisic acid (ABA) (100 μM), (C) ACC (100 μM), (D) oxidative stress (50 μM, methyl viologen), (E) Dehydration (air drying), (F) Heat (37°C), (G) Cold (4°C), (H) Salt (150 mM, NaCl), (I) Osmotic (300 mM, mannitol). Data are represented as mean ± SD of replicates and statistical significance was determined by Student’s t-test (∗P < 0.05 and ∗∗P < 0.01).
Histochemical GUS Assay for Phytohormone Treatment and Abiotic Stress-Responsiveness
Since AtUSP gene was found to be induced under multiple abiotic stresses, T3 generation of transgenic plants harboring AtUSP promoter and its three 5′ deleted fragments were analyzed for GUS induction under hormone treatments (such as ABA and ACC) and abiotic stresses (such as dehydration, salt, osmotic, heat, cold, and oxidative stress using histochemical staining and fluorimetric GUS assay (Figures 4 and 5). Under control conditions, strong GUS stain was detected in roots, while, a slight GUS staining was also detected in the region surrounding the mid-rib of the leaves, and hypocotyl of the D0 seedlings (Figure 4). In contrast, a strong GUS stain was observed in the entire leaf and root of the D1 and D2 seedlings under control condition. However, GUS stain was not detected in the D3 seedlings under control condition. Under ABA treatment, dark GUS stain was detected in the leaf tips, stomata, and roots of the D0 seedlings. However, in the D1 seedlings, GUS expression was found to be in the entire leaf, hypocotyls, and roots under ABA treatment. The GUS staining in D2 seedlings in response to ABA treatment was comparable to GUS stain found in untreated D2 seedlings which was evident by comparable GUS activities observed in the D2 seedlings under both the conditions (Figures 4 and 5). However, GUS staining was completely absent in D3 seedlings. ACC treatment resulted in slight GUS staining in the region surrounding mid-rib of leaves, hypocotyls, and roots in the D0 seedlings. Similar GUS staining pattern was observed in the leaves and roots of the D1 and D2 seedlings. ACC treated D3 seedlings also exhibited GUS staining in the leaves and roots. Dehydration stress also resulted in similar GUS staining pattern as that observed in case of ABA treatment. Strong GUS expression was observed in the leaf tip and stomata as in case of ABA treated D0 lines. The D1 and D2 seedlings also showed GUS expression following dehydration. However, in contrast to ABA treatment, D3 seedlings also exhibited GUS staining in response to dehydration, which was further confirmed by GUS activity assay with 1.9-fold increase in GUS activity observed under dehydration (Figure 5; Supplementary File S5). In D0 and D1 seedlings, GUS stain was detected in the region surrounding the leaf tips, hypocotyl, stomata, and roots under heat stress. Similar GUS staining was detected in D1 seedlings except for roots as GUS stain was detected only in root apex. Comparatively, weaker expression was observed in D2 and D3 seedling under heat stress, where GUS stain was detected in the entire leaf, hypocotyls, and roots. With cold treatment, GUS stain was detected only at the leaf tip, hypocotyl, stomata, and roots in the D0 seedlings. The tissue specific cold induction in GUS expression was observed in D1 seedlings with stronger expression in the region surrounding the leaf tip, stomata, hypocotyl with weak GUS expression in the roots. There was complete loss of GUS staining in the D3 seedlings under cold stress. Oxidative stress resulted in the weak GUS expression at the leaf tips, stomata, and roots of D0 and D1 seedlings as compared to control conditions. Interestingly, significant GUS activity (2.8-fold) was detected in D3 seedlings when subjected to oxidative stress (Figure 5). Under salt stress, GUS stain can be detected in the region surrounding the leaf tip, mid-rib region, hypocotyl, and in the entire root of the D0 seedlings. Similar GUS stain was also detected in the D1 seedlings with lower staining in the roots. Strong GUS staining was also observed in the salt treated D2 and D3 seedlings. Osmotic stress also resulted in similar GUS expression pattern in D0 seedlings as observed under salt stress, except in roots where GUS staining was found only in the root tip. Slight induction in GUS expression was observed in the D1 and D2 seedlings, while, weaker induction was still observed in D3 seedlings (Figure 4).
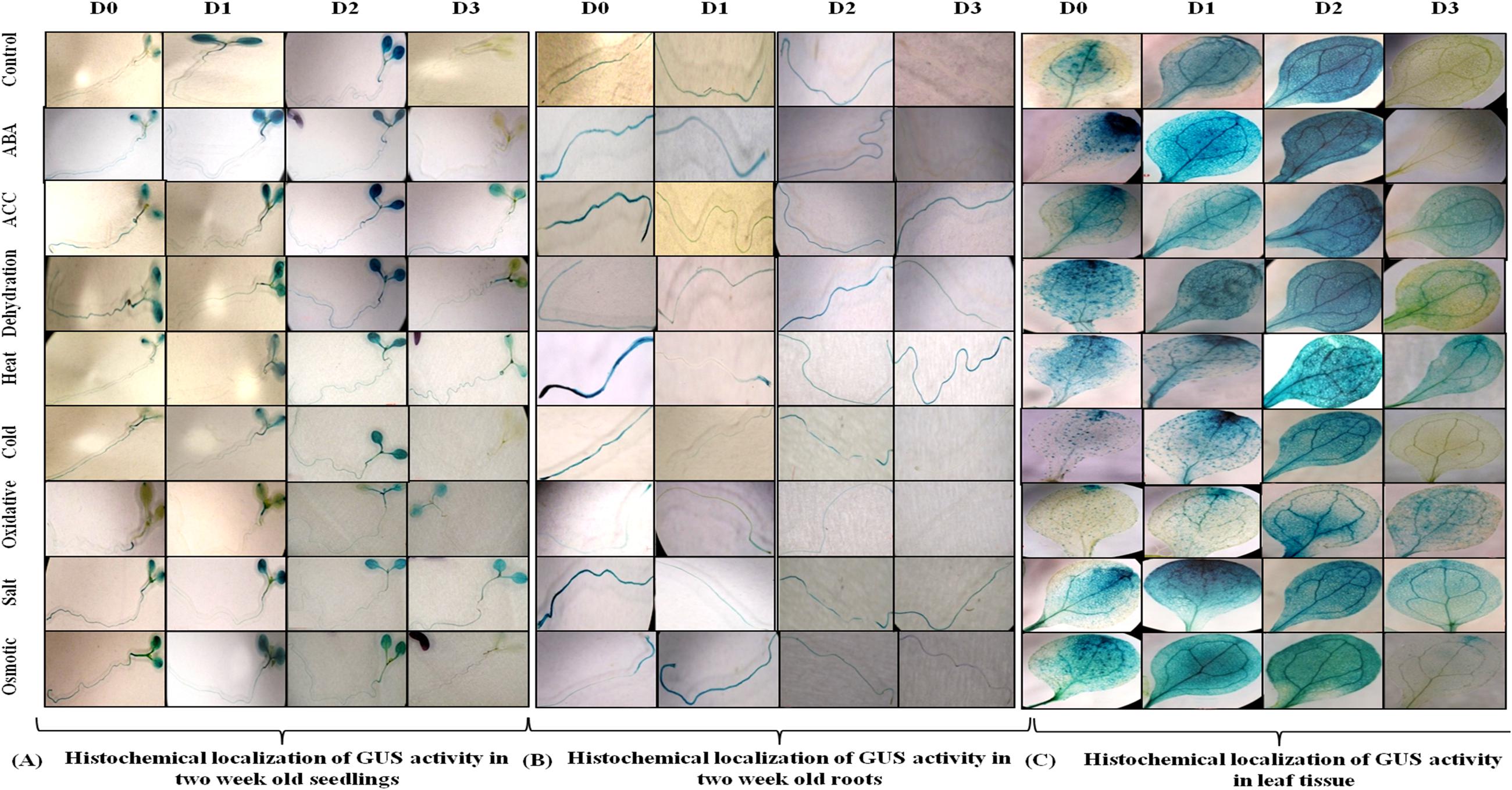
FIGURE 4. Histochemical localization of GUS activity. (A) In 2 week old seedlings (B) in root tissue and (C) in leaf tissue of transgenic Arabidopsis carrying full length AtUSP promoter (D0) and its three 5′ deletion derivatives (D1, D2, D3) in response to various phytohormones and abiotic stress treatments.
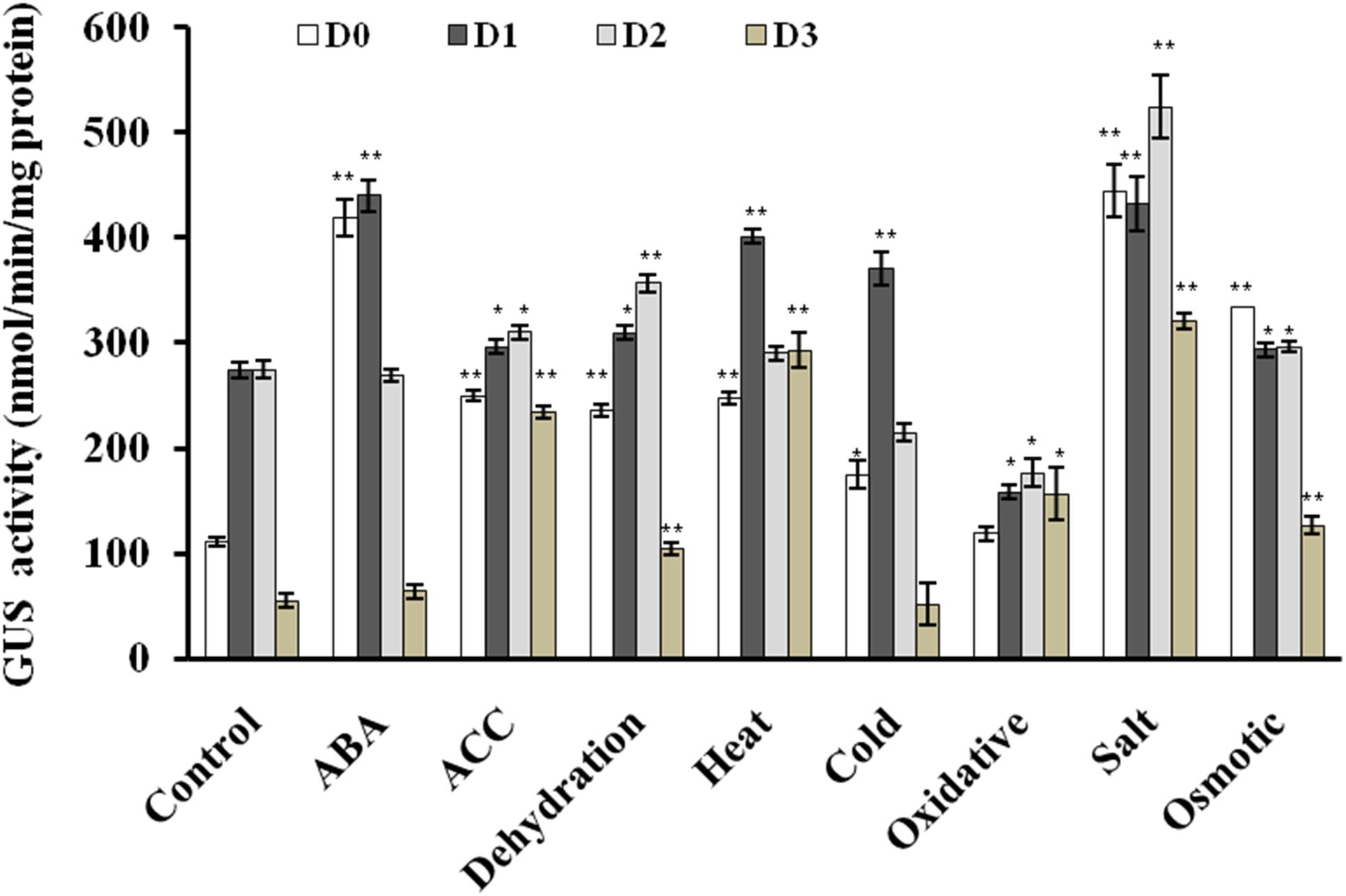
FIGURE 5. Quantitative fluorometric assay for GUS activity in 2 week old seedlings in response to various phytohormones and abiotic stress treatments. The experiment was carried out in three replicates. Data are represented as mean ± SD of replicates and statistical significance was determined by Student’s t-test (∗P < 0.05 and ∗∗P < 0.01).
Discussion
Universal stress proteins, well known for their universal expression under different stresses in numerous plants are considered as potential targets for developing stress tolerant plants. Expression of several plant USP genes has been shown to be induced under various abiotic stresses, but their precise molecular function is still unknown (Sauter et al., 2002; Maqbool et al., 2009; Loukehaich et al., 2012; Gonzali et al., 2015; Jung et al., 2015; Sinha et al., 2016; Udawat et al., 2016). Two plant USPs, i.e., SpUSP from wild tomato and SbUSP from S. brachiata have been reported to be induced in response to multiple abiotic stresses (Loukehaich et al., 2012; Udawat et al., 2014). In addition, cotton USP promoter also showed enhanced activity under ABA, dehydration, and salt treatments (Zahur et al., 2009). These studies suggest that promoters of USP genes have the potential to be used as stress inducible promoter for the development of multiple stress tolerant crops. In present study, AtUSP gene was found to be induced under phytohormone and abiotic stress treatments. AtUSP gene was highly induced by abiotic stresses such as dehydration, cold, heat, salt, osmotic stress and phytohormone, ABA. Our results are in accordance with a previous microarray study which reported that AtUSP was upregulated under drought stress (Isokpehi et al., 2011b). In another report, AtUSP protein level was enhanced in response to cold acclimation that was responsible for enhanced freezing tolerance (Kawamura and Uemura, 2003). Recently, AtUSP was shown to have chaperone like function which is regulated by ROS level and provide resistance to heat shock and oxidative stress (Jung et al., 2015).
Based on the multiple stress-responsive characteristic of AtUSP, full length promoter (1115 bp) and its three 5′ deletion derivatives were generated and used to drive expression of GUS reporter gene by cloning them in a promoterless vector pCAMBIA1391z. These constructs were then transformed into WT A. thaliana by Agrobacterium tumefaciens-mediated transformation. The 1115 bp region of AtUSP promoter contained several cis-regulatory elements required for tissue specific, hormone, and stress-responsive expression. AtUSP promoter contained MYCCONSENSUSAT, MYCATRD22, MYBCORE, and MYBCONSENSUSAT elements which were reported to be also present in the promoter of dehydration-responsive RD22 that is known to play role in ABA, drought and cold signaling (Abe et al., 1997, 2003). Several repeats of CAATBOX1 and GATABOX motifs required for tissue specific expression were also present (Shirsat et al., 1989). ABA-responsive elements, such as ABRELATERD1, ABRERATCAL, ethylene-responsive element (ERELEE4), and gibberellic acid-responsive element (GARE2OSREP1) were also present in the AtUSP promoter, which are responsible for ABA, ethylene and gibberellic acid induced expression, respectively (Sutoh and Yamauchi, 2003; Rawat et al., 2005; Kaplan et al., 2006; Nakashima et al., 2006). GT1GMSCAM4 motifs known for pathogen and salt induced expression (Park et al., 2004) were also present. The AtUSP promoter analysis was in agreement to earlier report, which suggested that the ∼0.5 kb region of promoter of multiple stress-responsive genes tend to contain large number of cis-regulatory elements responsive to multiple stresses with several unique elements (Walther et al., 2007). For D0 and D3 seedlings, the GUS transcript level strongly correlated with the respective GUS activity. For D1 and D2 seedlings, there was no strong correlation between the GUS transcript and the corresponding GUS activity under different phytohormone and stress treatments. This might be due to the fact that we compared GUS expression in the D1 and D2 seedlings under various stresses with respect to their respective controls and moreover, the basal expression levels of GUS gene in the D1 and D2 seedlings were already high under control conditions. GUS staining significantly correlated with the measured GUS activity in the non-stressed and stressed transgenic seedlings. GUS histochemical staining as well as GUS activity assay clearly revealed that D0 seedlings possess minimal activity under non-stressed condition and oxidative stress. However, the D1 and D2 seedlings showed much stronger GUS expression with respect to D0 seedlings under non-stressed condition. The intense spread of GUS stain in the entire leaf tissue of the D1 and D2 seedlings can be due to the deletion of four motifs of CAAT box and one motif of GATABOX that have been shown to be required for tissue specific expression (Shirsat et al., 1989; Reyes et al., 2004). The complete loss of GUS expression in the D3 seedlings under control conditions might be due to deletion of 10 motifs of GATABOX required for high level transcription, RAV1AAT responsible for high expression in leaves and five motifs of ROOTMOTIFTAPOX1 required for root-specific expression (Elmayan and Tepfer, 1995; Kagaya et al., 1999; Reyes et al., 2004). Under control conditions, strong GUS expression in the roots of D0 seedlings might be due to the presence of eight ROOTMOTIFTAPOX1 motifs which is responsible for strong expression in roots (Elmayan and Tepfer, 1995). The full length AtUSP (D0) promoter was strongly activated by ABA, ACC, dehydration, salt, osmotic, heat, and cold stresses. These results strengthen our qRT-PCR based gene expression analysis which also highlighted the multiple stress-responsive characteristic of AtUSP. Recently, Jung et al. (2015), showed that AtUSP possess molecular chaperone like function which is regulated by ROS homeostasis. They also showed that AtUSP plays an essential role in heat and oxidative stress tolerance in Arabidopsis. ABA accumulation has been shown to act as trigger to mediate stress tolerance during dehydration, salt, and cold stresses (Finkelstein and Lynch, 2000). Therefore, this might provide the possible explanation for similar GUS expression pattern observed in response to ABA and dehydration. Plant responses include both ABA-dependent and ABA-independent mechanisms that help to overcome abiotic stress conditions. ABA-dependent pathway includes many ABA-inducible genes that contain ABRE elements in their promoters (Mundy et al., 1990). AtUSP promoter also contains two motifs of ABRE-related sequences that might be responsible for ABA-mediated stress induction of GUS activity in D0 and D1 seedlings. Although, higher GUS activity was observed in the D1 seedlings in response to ABA, however, at the transcript level, GUS expression was downregulated in the D1 seedlings under ABA treatment, which might be attributed to higher GUS expression observed in the D1 seedlings under non-stress conditions (Figure 3A). ABA induction of GUS activity was lost in the D2 and D3 seedlings with deletion of two ABRE motifs. ABA-independent signaling involves dehydration-responsive elements in promoter region that are bound by DRE binding transcription factors (DREBs/CBFs) to induce the expression of downstream genes. Several studies have shown that overexpression of DREB/CBF lead to drought, salinity, and freezing tolerance in crops (Liu et al., 1998; Datta et al., 2012; Pierre et al., 2012). AtUSP promoter also contains several DRE elements that might be responsible for its multiple stress-responsive characteristic. D3 seedlings also showed weak induction with respect to non-stress conditions in response to dehydration which might be due to the fact that it still contains dehydration-responsive elements such as MYCCONSENSUSAT, LTRECOREATCOR15, DRECRTCOREAT, CBFHV, and DRE2COREZMRAB17 which have been shown to be part of ABA-independent signaling. Strong induction of GUS expression in D0 and D1 seedlings under osmotic stress in comparison to their respective controls can be attributed to presence of all the motifs of ABRE, MYB2CONSENSUSAT, MYCCONSENSUSAT, DRECRTCOREAT, and DRE2COREZMRAB17 which are directly or indirectly involved in osmotic stress response. Ethylene has been shown to play a positive role in mediating the plant salt tolerance (Cao et al., 2006). Similar GUS expression pattern was observed in the ACC and salt treated transgenic seedlings. Weak GUS induction observed in the D3 seedlings upon salt treatment even with the deletion of GT1GMSCAM4 might be due to the presence of ERE (ERELEE4) element. Similar weak induction can be seen in case of ACC treatment of D3 seedlings which reinforced the fact that ethylene plays a positive role in plant salt tolerance (Cao et al., 2006). Stronger induction in GUS activity was observed in the D0 and D1 seedlings while weak induction was observed in case of D2 seedlings under heat stress as compared to their respective controls. Heat stress induction of GUS activity was also observed in the D3 seedlings as this construct still retained one of the three heat stress-responsive elements responsible for heat induced expression (Rieping and Schoffl, 1992). Strong cold induction in GUS expression was observed only in D0 and D1 seedlings. The deletion of three motifs of MYCCONSENSUSAT in the D2 and D3 constructs might have resulted in the loss of cold induction in GUS expression in the D2 and D3 seedlings. Comparatively, lower GUS stain was detected in the D0, D1, and D2 seedlings under oxidative stress as compared to their respective controls. However, GUS expression was found to be increased in case of D3 seedlings as compared to non-stress condition. This might be due to the deletion of a negative regulatory region present in the promoter sequence in between -1115 and -504 bp region in the D3 seedlings for oxidative stress. This negative regulatory region might be responsible for the observed downregulation of AtUSP gene under oxidative stress.
Conclusion
The present study reveals the multiple stress-responsive characteristic of AUSP gene and its promoter was functionally characterized under phytohormone and abiotic stress treatments. The GUS expression and histochemical studies showed that AtUSP promoter can respond to phytohormones such as ABA, ethylene, and several abiotic stresses such as dehydration, salt, osmotic, heat, and cold stress. Therefore, the D0 promoter fragment can be considered as potential multiple stress inducible promoter to derive the expression of transgenes that may confer stress tolerance to the plants.
Author Contributions
AS conceived and designed the experiments. MB and PG performed the experiments. MB and AS performed the data analysis. AS and SK secured the funds to support this research. MB wrote the manuscript. MB and AS revised and finalized the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MB and PG thank Council of Scientific and Industrial Research for providing junior and senior research fellowships. This manuscript represents CSIR–IHBT communication number 4101.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01957/full#supplementary-material
Footnotes
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. doi: 10.1105/tpc.9.10.1859
Ahuja, I., De Vos, R. C., Bones, A. M., and Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15, 664–674. doi: 10.1016/j.tplants.2010.08.002
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Balazi, G., and Oltvai, Z. N. (2005). Sensing your surroundings: how transcription-regulatory networks of the cell discern environmental signals. Sci. STKE 2005:e20. doi: 10.1126/stke.2822005pe20
Bate, N., and Twell, D. (1998). Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 37, 859–869. doi: 10.1023/A:1006095023050
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cao, W. H., Liu, J., Zhou, Q. Y., Cao, Y. R., Zheng, S. F., Du, B. X., et al. (2006). Expression of tobacco ethylene receptor NTHK1 alters plant responses to salt stress. Plant Cell Environ. 29, 1210–1219. doi: 10.1111/j.1365-3040.2006.01501.x
Chen, C., and Chen, Z. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129, 706–716. doi: 10.1104/pp.001057
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Datta, K., Baisakh, N., Ganguly, M., Krishnan, S., Yamaguchi, Shinozaki K., and Datta, S. K. (2012). Overexpression of Arabidopsis and rice stress genes inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol. J. 10, 579–586. doi: 10.1111/j.1467-7652.2012.00688.x
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Elmayan, T., and Tepfer, M. (1995). Evaluation in tobacco of the organ specificity and strength of the rolD promoter, domain A of the 35S promoter and the 35S2 promoter. Transgenic Res. 4, 388–396. doi: 10.1007/BF01973757
Ezcurra, E. M., Wycliffe, P., Stalberg, K., and Rask, L. (1999). Interaction between composite elements in the napA promoter: both the B-box ABA-responsive complex and the RY/G complex are necessary for seed-specific expression. Plant Mol. Biol. 40, 699–709. doi: 10.1023/A:1006206124512
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., et al. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257. doi: 10.1091/mbc.11.12.4241
Ghawana, S., Paul, A., Kumar, H., Kumar, A., Singh, H., Bhardwaj, P. K., et al. (2011). An RNA isolation system for plant tissues rich in secondary metabolites. BMC Res. Notes 4:85. doi: 10.1186/1756-0500-4-85
Gomes, C. S., Izar, B., Pazan, F., Mohamed, W., Mraheil, M. A., Mukherjee, K., et al. (2011). Universal stress proteins are important for oxidative and acid stress resistance and growth of Listeria monocytogenes EGD-e In Vitro and In Vivo. PLoS ONE 6:e24965. doi: 10.1371/journal.pone.0024965
Gonzali, S., Loreti, E., Cardarelli, F., Novi, G., Parlanti, S., Pucciariello, C., et al. (2015). Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 1:15151. doi: 10.1038/nplants.2015.151
Gowik, U., Burscheidt, J., Akyildiz, M., Schlue, U., Koczor, M., Streubel, M., et al. (2004). cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16, 1077–1090. doi: 10.1105/tpc.019729
Isokpehi, R. D., Mahmud, O., Mbah, A. N., Simmons, S. S., Avelar, L., Rajnarayanan, R. V., et al. (2011a). Developmental regulation of genes encoding universal stress proteins in Schistosoma mansoni. Gene Regul. Syst. Bio. 5, 61–74. doi: 10.4137/GRSB.S7491
Isokpehi, R. D., Simmons, S. S., Cohly, H. H., Ekunwe, S. I., Begonia, G. B., and Ayensu, W. K. (2011b). Identification of drought-responsive universal stress proteins in viridiplantae. Bioinform. Biol. Insights 5, 41–58. doi: 10.4137/BBI.S6061
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. doi: 10.1007/BF02667740
Jung, Y. J., Melencion, S. M., Lee, E. S., Park, J. H., Alinapon, C. V., Oh, H. T., et al. (2015). Universal stress protein exhibits a redox-dependent chaperone function in Arabidopsis and enhances plant tolerance to heat shock and oxidative stress. Front. Plant Sci. 6:1141. doi: 10.3389/fpls.2015.01141
Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. doi: 10.1093/nar/27.2.470
Kaplan, B., Davydov, O., Knight, H., Galon, Y., Knight, M. R., Fluhr, R., et al. (2006). Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18, 2733–2748. doi: 10.1105/tpc.106.042713
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1002/9780470515778
Kawamura, Y., and Uemura, M. (2003). Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J. 36, 141–154. doi: 10.1046/j.1365-313X.2003.01864.x
Kerk, D., Bulgrien, J., Smith, D. W., and Gribskov, M. (2003). Arabidopsis proteins containing similarity to the universal stress protein domain of bacteria. Plant Physiol. 131, 1209–1219. doi: 10.1104/pp.102.016006
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313x.2007.03052.x
Kim, D. W., Lee, S. H., Choi, S. B., Won, S. K., Heo, Y. K., Cho, M., et al. (2006). Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18, 2958–2970. doi: 10.1105/tpc.106.045229
Kizis, D., and Pages, M. (2002). Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J. 30, 679–689. doi: 10.1046/j.1365-313X.2002.01325.x
Kucho, K., Yoshioka, S., Taniguchi, F., Ohyama, K., and Fukuzawa, H. (2003). cis-acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 133, 783–793. doi: 10.1104/pp.103.026492
Kvint, K., Nachin, L., Diez, A., and Nystrom, T. (2003). The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6, 140–145. doi: 10.1016/S1369-5274(03)00025-0
Lenman, M., Sorensson, C., and Andreasson, E. (2008). Enrichment of phosphoproteins and phosphopeptide derivatization identify universal stress proteins in elicitor-treated Arabidopsis. Mol. Plant Microbe Interact. 21, 1275–1284. doi: 10.1094/MPMI-21-10-1275
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.2307/3870648
Liu, W. T., Karavolos, M. H., Bulmer, D. M., Allaoui, A., Hormaeche, R. D., Lee, J. J., et al. (2007). Role of the universal stress protein UspA of Salmonella in growth arrest, stress and virulence. Microb. Pathog. 42, 2–10. doi: 10.1016/j.micpath.2006.09.002
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loukehaich, R., Wang, T., Ouyang, B., Ziaf, K., Li, H., Zhang, J., et al. (2012). SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 63, 5593–5606. doi: 10.1093/jxb/ers220
Maqbool, Z. M., Husnain, T., and Riazuddin, S. (2009). GUSP1 and GUSP2, two drought-responsive genes in Gossypium arboretum have homology to universal stress proteins. Plant Mol. Biol. Rep. 27, 109–114. doi: 10.1007/s11105-008-0049-0
Maruyama, K., Todaka, D., Mizoi, J., Yoshida, T., Kidokoro, S., Matsukura, S., et al. (2012). Identification of cis-acting promoter elements in cold- and dehydration induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. doi: 10.1093/dnares/dsr040
Matarasso, N., Schuster, S., and Avni, A. (2005). A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell 17, 1205–1216. doi: 10.1105/tpc.105.030775
Matsui, A., Ishida, J., Morosawa, T., Mochizuki, Y., Kaminuma, E., Endo, T. A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. doi: 10.1093/pcp/pcn101
Merkouropoulos, G., Andreasson, E., Hess, D., Boller, T., and Peck, S. C. (2008). An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP kinases 3 and 6. J. Biol. Chem. 283, 10493–10499. doi: 10.1074/jbc.M800735200
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. doi: 10.1016/j.tplants.2005.11.002
Mundy, J., Yamaguchi-Shinozaki, K., and Chua, N. H. (1990). Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc. Natl. Acad. Sci. U.S.A. 87, 1406–1410. doi: 10.1073/pnas.87.4.1406
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nachin, L., Nannmark, U., and Nystrom, T. (2005). Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187, 6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. doi: 10.1007/s11103-005-2418-5
Nordin, K., Vahala, T., and Palva, E. T. (1993). Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 21, 641–653. doi: 10.1007/BF00014547
Nystrom, T., and Neidhardt, F. C. (1992). Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 6, 3187–3198. doi: 10.1111/j.1365-2958.1992.tb01774.x
Nystrom, T., and Neidhardt, F. C. (1993). Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J. Bacteriol. 175, 3949–3956. doi: 10.1128/JB.187.18.6253-6254
Park, H. C., Kim, M. L., Kang, Y. H., Jeon, J. M., Yoo, J. H., Kim, M. C., et al. (2004). Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 135, 2150–2161. doi: 10.1104/pp.104.041442
Pierre, C. S., Crossa, J. L., Bonnett, D., Yamaguchi-Shinozaki, K., and Reynolds, M. P. (2012). Phenotyping transgenic wheat for drought resistance. J. Exp. Bot. 63, 1799–1808. doi: 10.1093/jxb/err385
Plesch, G., Ehrhardt, T., and Mueller-Roeber, B. (2001). Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J. 28, 455–464. doi: 10.1046/j.1365-313X.2001.01166.x
Rawat, R., Xu, Z. F., Yao, K. M., and Chye, M. L. (2005). Identification of cis-elements for ethylene and circadian regulation of the Solanum melongena gene encoding cysteine proteinase. Plant Mol. Biol. 57, 629–643. doi: 10.1007/s11103-005-0954-7
Reyes, J. C., Muro-Pastor, M. I., and Florencio, F. J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134, 1718–1732. doi: 10.1104/pp.103.037788
Rieping, M., and Schoffl, F. (1992). Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol. Gen. Genet. 231, 226–232. doi: 10.1007/BF00279795
Ross, E. J., Stone, J. M., Elowsky, C. G., Arredondo-Peter, R., Klucas, R. V., and Sarath, G. (2004). Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J. Exp. Bot. 55, 1721–1731. doi: 10.1093/jxb/erh211
Satoh, R., Nakashima, K., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiol. 130, 709–719. doi: 10.1104/pp.009993
Sauter, M., Rzewuski, G., Marwedel, T., and Lorbiecke, R. (2002). The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 53, 2325–2331. doi: 10.1093/jxb/erf096
Shirsat, A., Wilford, N., Croy, R., and Boulter, D. (1989). Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol. Gen. Genet. 215, 326–331. doi: 10.1007/BF00339737
Simpson, S. D., Nakashima, K., Narusaka, Y., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 33, 259–270. doi: 10.1046/j.1365-313X.2003.01624.x
Sinha, P., Pazhamala, T. L., Singh, V. K., Saxena, R. K., Krishnamurthy, L., Azam, S., et al. (2016). Identification and validation of selected universal stress protein domain containing drought-responsive genes in Pigeonpea (Cajanus cajan L.). Front. Plant. Sci. 6:1065. doi: 10.3389/fpls.2015.01065
Sutoh, K., and Yamauchi, D. (2003). Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 34, 635–645. doi: 10.1046/j.1365-313X.2003.01753.x
Tarte, V. N., Seok, H. Y., Woo, D. H., Le, D. H., Tran, H. T., Baik, J. W., et al. (2015). Arabidopsis Qc-SNARE gene AtSFT12 is involved in salt and osmotic stress responses and Na(+) accumulation in vacuoles. Plant Cell Rep. 34, 1127–1138. doi: 10.1007/s00299-015-1771-3
Udawat, P., Jha, R. K., Sinha, D., Mishra, A., and Jha, B. (2016). Overexpression of a cytosolic abiotic stress responsive universal stress protein (SbUSP) mitigates salt and osmotic stress in transgenic tobacco plants. Front. Plant. Sci. 7:518. doi: 10.3389/fpls.2016.00518
Udawat, P., Mishra, A., and Jha, B. (2014). Heterologous expression of an uncharacterized universal stress protein gene (SbUSP) from the extreme halophyte, Salicornia brachiata, which confers salt and osmotic tolerance to E. coli. Gene 536, 163–170. doi: 10.1016/j.gene.2013.11.020
Urao, T., Yamaguchi-Shinozaki, K., Urao, S., and Shinozaki, K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5, 1529–1539. doi: 10.2307/3869736
Villain, P., Mache, R., and Zhou, D. X. (1996). The mechanism of GT element-mediated cell type-specific transcriptional control. J. Biol. Chem. 271, 32593–32598. doi: 10.1074/jbc.271.51.32593
Walther, D., Brunnemann, R., and Selbig, J. (2007). The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PLoS Genet. 3:e11. doi: 10.1371/journal.pgen.0030011
Xue, G. P. (2002). Characterization of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res. 30:e77. doi: 10.1093/nar/gnf076
Zahur, M., Maqbool, A., Ifran, M., Barozai, M. Y., Rashid, B., Riazuddin, S., et al. (2009). Isolation and functional analysis of cotton universal stress protein promoter in response to phytohormones and abiotic stresses. Mol. Biol. (Mosk) 43, 628–635. doi: 10.1134/S0026893309040086
Keywords: abiotic stress, Arabidopsis, universal stress protein, AtUSP, phytohormones, promoter, deletion analysis
Citation: Bhuria M, Goel P, Kumar S and Singh AK (2016) The Promoter of AtUSP Is Co-regulated by Phytohormones and Abiotic Stresses in Arabidopsis thaliana. Front. Plant Sci. 7:1957. doi: 10.3389/fpls.2016.01957
Received: 20 October 2016; Accepted: 09 December 2016;
Published: 26 December 2016.
Edited by:
Avinash Mishra, Central Salt & Marine Chemicals Research Institute (CSIR), IndiaReviewed by:
Charu Lata, National Botanical Research Institute (CSRI), IndiaVivekanand Tiwari, Weizmann Institute of Science, Israel
Ivelin Pantchev, Sofia University, Bulgaria
Copyright © 2016 Bhuria, Goel, Kumar and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anil K. Singh, anils13@gmail.com
†Present address: Anil K. Singh, ICAR-Indian Institute of Agricultural Biotechnology, PDU Campus, IINRG, Namkum, Ranchi, India
 Monika Bhuria
Monika Bhuria Parul Goel
Parul Goel Sanjay Kumar1,2
Sanjay Kumar1,2 Anil K. Singh
Anil K. Singh