- 1Department of Biotechnology, University of Verona, Verona, Italy
- 2Department of Agriculture, Food, Natural Resources, Animals and Environment, University of Padova, Padova, Italy
A small set of TTG2-like homolog proteins from different species belonging to the WRKY family of transcription factors were shown to share a similar mechanism of action and to control partially conserved biochemical/developmental processes in their native species. In particular, by activating P-ATPases residing on the tonoplast, PH3 from Petunia hybrida promotes vacuolar acidification in petal epidermal cells whereas TTG2 from Arabidopsis thaliana enables the accumulation of proanthocyanidins in the seed coat. In this work we functionally characterized VvWRKY26 identified as the closest grapevine homolog of PhPH3 and AtTTG2. When constitutively expressed in petunia ph3 mutant, VvWRKY26 can fulfill the PH3 function in the regulation of vacuolar pH and restores the wild type pigmentation phenotype. By a global correlation analysis of gene expression and by transient over-expression in Vitis vinifera, we showed transcriptomic relationships of VvWRKY26 with many genes related to vacuolar acidification and transport in grapevine. Moreover, our results indicate an involvement in flavonoid pathway possibly restricted to the control of proanthocyanidin biosynthesis that is consistent with its expression pattern in grape berry tissues. Overall, the results show that, in addition to regulative mechanisms and biological roles shared with TTG2-like orthologs, VvWRKY26 can play roles in fleshy fruit development that have not been previously reported in studies from dry fruit species. This study paves the way toward the comprehension of the regulatory network controlling vacuolar acidification and flavonoid accumulation mechanisms that contribute to the final berry quality traits in grapevine.
Introduction
Grape berry needs a relatively long period to develop during which its tissues undergo several physiological and biochemical changes leading to the final composition and quality of ripe fruit. The primary and secondary metabolites mainly responsible for grape quality at ripening are compartmentalized in the vacuolar lumen where the physiological conditions favor their stabilization. During early berry development, both mesocarp and skin cells accumulate and store high concentrations of organic acids in their vacuoles, as malic and tartaric acids, which have remarkably low pH values (Ruffner, 1982). The acidification mechanism has been described in fruit of many species, including grapevine (Milner et al., 1995; Müller et al., 1996; Suzuki et al., 1999; Terrier et al., 2001). A combined proton pumping system constituted by V-PPases and V-ATPases influences the electrochemical potential gradient and allows the active transport of sugars, ions and organic acids across the tonoplast.
Another mechanism recently identified in Petunia hybrida for vacuole acidification requires PhPH1 and PhPH5 encoding two interacting P-ATPase transmembrane transporters located on the tonoplast. The functioning of both these pumps is essential for creating the proper acidic conditions in petal epidermal cells where anthocyanins are stored, whereas the expression of PhPH5 alone is required for proanthocyanidin (PA) deposition in the seed coat (Verweij et al., 2008; Faraco et al., 2014). In Arabidopsis the AtTT13, homolog of PhPH5, functions as a proton pump in the tonoplast of seed coat endothelium cells and generates the driving force for the vacuolar uptake of PA precursors (Appelhagen et al., 2015). There is little information on the existence of a similar acidification mechanism in fleshy fruits. The lack of expression of the PhPH5/AtTT13 citrus homolog was proposed as being responsible for the low acidity in the juice of Faris “sweet” lemon (Citrus limon) variety (Aprile et al., 2011). The homologs of both PhPH1 and PhPH5/AtTT13 were identified in grapevine and were shown to replace the function of the respective endogenous genes when expressed in petunia (Faraco et al., 2014; Li et al., 2016), suggesting the existence of a similar acidification mechanism in grapevine.
In petunia and Arabidopsis the expression of such P-ATPases is controlled by a MYB-bHLH-WDR (MBW) complex constituted by a MYB transcription factor (PhPH4 and AtMYB5/AtTT2, respectively), a bHLH transcription factor (PhAN1 and AtTT8, respectively) and a WD-regulatory protein (PhAN11 and AtTTG1, respectively) (Quattrocchio et al., 2006; Gonzalez et al., 2009; Li et al., 2009; Faraco et al., 2014).
The WRKY transcription factor PhPH3 has recently been characterized in petunia as an additional key regulator of vacuolar acidification, acting downstream and in concert with the MBW complex and controlling the expression of PhPH1 and PhPH5 (Verweij et al., 2016). The ph3 mutants show an increase of petal homogenate pH that causes a shift in petal color, an abnormal seed development and female sterility (Verweij et al., 2016).
The functional homolog of PhPH3 in Arabidopsis is AtTTG2 that controls the seed PA and mucilage deposition, and the development of endothelium and trichomes (Johnson et al., 2002; Ishida et al., 2007). Similarly to PhPH3, AtTTG2 interacts with the MBW complex for the control of the vacuolar transport of PAs during seed coat development by regulating the expression of AtTT13 and also by directly activating the expression of TRANSPARENT TESTA 12 (AtTT12), a MATE transporter of glycosilated epicathechin PA precursors (Debeaujon et al., 2001; Gonzalez et al., 2016). Another functional homolog of AtTTG2 was identified in Brassica napus. BnTTG2 was shown to affect trichome development when over-expressed in B. napus plants and was capable of rescuing the trichome phenotype of Arabidopsis TTG2 mutant (Li et al., 2015).
PhPH3, AtTTG2 and BnTTG2 are all members of the plant-specific WRKY family whose regulatory functions have mainly been associated with the response to abiotic and biotic stresses but also with various developmental and physiological processes (Rushton et al., 2010; Schluttenhofer and Yuan, 2015). These transcription factors have a 60 amino acidic sequence, called WRKY domain, containing the conserved sequence WRKYGQK and a zinc-finger motif, and is responsible for recognition of W-boxes in the promoters of target genes (Ciolkowski et al., 2008). The WRKY proteins are classified in groups I–III based on both the number of WRKY domains and the features of their zinc-finger motif (Eulgem et al., 2000). PhPH3, AtTTG2 and BnTTG2 proteins are all members of group Ia with two WRKY domains and a C2H2 zinc-finger motif (Johnson et al., 2002; Xie et al., 2005; Li et al., 2015; Verweij et al., 2016).
In Vitis vinifera the WRKY family is composed of 59 members (Wang M. et al., 2014) and the few proteins functionally characterized so far seem to be mostly involved in plant response to biotic or abiotic stress (Marchive et al., 2007, 2013; Li et al., 2010; Liu et al., 2011; Zhu et al., 2012; Jiang et al., 2015; Merz et al., 2015). In this study, we characterized VvWRKY26, the closest grapevine homolog to PhPH3 and AtTTG2. We showed that VvWRKY26 is able to complement the petunia ph3 mutation. Moreover, from detailed analyses of expression in grapevine organs and the study of VvWRKY26 transiently over-expressing grapevine plantlets we provided evidence for a role of VvWRKY26 in the regulation of vacuolar transport and flavonoid accumulation in grape berry.
Materials and Methods
Bioinformatics
To identify the grapevine WRKY sequence homolog to PhPH3, the predicted proteome of the near-homozygous PN40024 genotype of Vitis vinifera “Pinot noir” 12X V1 version (http://genomes.cribi.unipd.it) was queried using the conserved amino acid sequence of PhPH3 (Verweij et al., 2016).
VvWRKY26 protein was aligned against the full amino acid sequences of AtTTG2 (AT2G37260), BnTTG2 (AJD07412) and PhPH3 (AMR43368) using Muscle.
The phylogenetic tree was constructed from a Muscle alignment of a portion of the protein sequences that includes the C-terminal WRKY domains up to the stop codon. The final phylogenetic tree was obtained using the neighbor-joining method and bootstrap analysis (1000 replicates) and Mega6 software (Tamura et al., 2013). The following GenBank accession numbers were used: AtWRKY2 (AT5G56270), AtWRKY4 (AT1G13960), AtWRKY6 (AT1G62300), AtWRKY7 (AT4G24240), AtWRKY18 (AT4G31800), AtWRKY23 (AT2G47260), AtWRKY35 (AT2G34830), AtWRKY36 (AT1G69810), AtWRKY42 (AT4G04450), AtWRKY62 (AT5G01900), AtWRKY70 (AT3G56400), AtTTG2 (AT2G37260), VpWRKY1 (ACY69975), VpWRKY2 (ADD70008), VpWRKY3 (AEN71143), VvWRKY01 (AAT46067), VtWRKY11 (B2G283), VvWRKY33 (AHG99400), MrWRKY30 (Jiang et al., 2015), GhWRKY3 (ADI52618.1), PcWRKY1 (AAD55974.1), PcWRKY3 (AAC49528), PcWRKY4 (AAG35658), OsWRKY6 (DAA05071), OsWRKY76 (DAA05141), WIZZ (BAA87058.1), CaWRKY1 (AAO86686), CjWRKY1 (BAF41990).
The online MEME Suite (http://meme-suite.org/doc/overview.html; Bailey et al., 2009) was used to discover protein motifs with an expected value lower than 2 × 10−12 using the following search parameters: four to twelve residues (minimum to maximum length), any number of repetitions, and 10 motifs maximum.
Plant Material and Growth Conditions
All petunia plants used in this study derived from the Amsterdam University collection. Complementation analysis of VvWRKY26 was conducted out in a line mutated in PH3 locus that is heterozygous for the allele ph3B2267FP harboring 7 bp footprint left by the excision of the transposon dTpH1 and for the allele ph3R49 featuring the deletion of part of the coding sequence (Verweij et al., 2016). The ph3 mutant, the wild type R27 and the transgenic lines were cultivated under normal glasshouse conditions. For the transient over-expression of VvWRKY26 in grapevine, plantlets of Vitis vinifera cv. Sultana were micropropagated in vitro and cultivated in a growth chamber at 25°C with a 16-h photoperiod.
Gene Cloning and Genetic Transformation
The VvWRKY26 cDNA sequence was amplified by PCR from grapevine post fruit set seed cDNA of cv. Corvina using Pfu DNA polymerase (Promega) and the primers listed in Supplementary Table 1. The generated PCR fragment was purified and directionally cloned into the Gateway entry vector pENTR/D-TOPO (Invitrogen) reflecting the presence of a 5′-CACC-3′ leader sequence in each forward primer. The product was verified by sequencing and transferred into the binary over-expression vector pK7GW2,0 (Laboratory of Plant Systems Biology, PSB; Ghent University, Belgium; https://gateway.psb.ugent.be/vector/show/pK7WG2/search/index/overexpression/any) by site-specific LR recombination.
For stable transformation of P. hybrida, the 35S:VvWRKY26 construct was inserted into Agrobacterium tumefaciens strain EHA105 by electroporation. P. hybrida plants were transformed using the leaf disc method (van der Meer, 1999) and regenerated transgenic shoots were transferred to soil and hardened off in a temperature-controlled glasshouse.
For transient transformation of Vitis vinifera cv. Sultana, the 35S:VvWRKY26 construct was transferred to Agrobacterium tumefaciens strain C58C1 by electroporation. As control, Agrobacterium was also transformed with a pK7WG2.0 vector containing a non-coding sequence. Six in vitro 5 weeks old plants of grapevine cv. Sultana were immersed in each bacterial suspension and vacuum infiltrated (2 × 2 min at 90 kPa). After agroinfiltration, plantlets were rinsed with sterile water and allowed to recover in vitro for 6 days before collecting material for RNA extraction and transcriptomic analysis.
Measurement of Petunia Petal Extract pH
Petal limb tissues collected from 10 distinct flowers, representing 10 biological replicates, were collected from the ph3 mutant, the wild type R27 and each transgenic line. The pH of petal extracts was measured by grinding the petal limbs in 6 mL of distilled water, as described by Quattrocchio et al. (2006). The actual pH values measured for each line showed some variation over time, possibly reflecting the variable environmental conditions in the glasshouse, but the differences between transgenic and control lines were constant. The absolute pH values could therefore reliably be compared between samples measured in the same experiment.
Anthocyanin Determination in Petunia Petals
Petal limb tissues collected from three distinct flowers, representing three biological replicates, were collected from the ph3 mutant, the wild type R27 and each transgenic line. Powdered petal samples were extracted in 8 volumes (w/v) of methanol acidified with 0.1% (v/v) hydrochloric acid in an ultrasonic bath at 40 kHz for 15 min at room temperature. Total amount of anthocyanins was determined by spectrophotometer at λ = 540 nm using malvidin 3-glucoside as standard.
RNA Extraction and Expression Analyses by qPCR
For molecular characterization of petunia plants, total RNA was isolated from flowers using TRIzol® Reagent (Invitrogen) following the manufacturer's instructions. For molecular characterization of grapevine cv. Sultana plantlets, total RNA was isolated from 20 to 40 mg of young and well expanded ground leaves using Spectrum™ Plant Total RNA kit (Sigma-Aldrich) according to the manufacturer's instructions.
All RNA samples were quantified with the NanoDrop 2000 instrument (Thermo Scientific Technologies) and 1 μg aliquots were treated with DNase I (Promega) and then reverse transcribed using Super-Script™ III Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions.
The expression profiles analyzed in petunia petals and grapevine organs were determined by qPCR as described by Zenoni et al. (2011), using the SYBR Green PCR master mix (Applied Biosystems) and a Mx3000P real time PCR system (Stratagene). Each expression value, relative to an actin internal control in petunia experiments and VvUBIQUITIN1 in grapevine, was determined using three biological triplicates. In all qPCR analyses, gene expression data (cycle threshold values) were used to quantify relative gene expression by using the efficiency corrected method described in Pfaffl (2001). The primer sequences used for qPCR analysis are listed in Supplementary Table 1.
Microarray Analyses
For microarray analysis RNA quality and integrity were determined using Bioanalyzer Chip RNA 7500 series II (Agilent).
For microarray analysis on petunia petals the cDNA synthesis, labeling, hybridization and washing reaction were done according to the NimbleGen Arrays User's Guide (V 3.2). Hybridization was performed on a NimbleGen microarray 111012_PAX_PT_expr_HX12 chip (Roche, NimbleGen Inc.), representing 27,627 predicted genes on the basis of the Petunia axillaris genome. The microarray was scanned using a ScanArray 4000XL (Perkin-Elmer) at 532 nm (Cy-3 absorption peak) and GenePix Pro7 software (Molecular Devices) according to the manufacturer's instructions. Images were analyzed using NimbleScan v2.5 software (Roche), which produces Pair Files containing the raw signal intensity data for each probe and Calls Files with normalized expression data derived from the average of the intensities of the probes for each gene. The normalized gene expression data were finally converted in log2 values to process the data. A Pearson's Correlation analysis was conducted to evaluate the robustness of the three biological replicates in each sample. A gene was considered to be expressed if the normalized expression value was higher than the value obtained by averaging the fluorescence of a negative control present on the chip, for at least two or three biological replicates. A Significance Analysis of Microarrays (SAM) was implemented using TMeV software (http://mev.tm4.org).
For microarray analysis on Sultana leaves transiently over-expressing VvWRKY26, the cDNA synthesis, labeling, hybridization and washing reactions were performed according to the Agilent Microarray-Based Gene Expression Analysis Guide (V 6.5). A new Agilent custom microarray was designed on the 4-pack 44K format (Agilent Sure Print HD 4X44K 60-mer G2514F-048771). This custom microarray was created by using an Agilent's web-based application able to create custom microarray designs and oligolibraries (https://earray.chem.agilent.com/earray/), as described by Dal Santo et al. (2016). Scanning and Feature Extraction was performed by using an Agilent Scanner following the settings and parameters indicated in the instruction manual. After the extraction was completed successfully, the QC report for each extraction was analyzed to assess the quality of the overall hybridization procedure. A data-matrix was prepared selecting from each single sub-array outcome file the gProcessedSignalvalues, which are the raw fluorescence intensities of each probe. The data were normalized on the 75th percentile and a correlation analysis was then conducted to assess the consistency of the biological triplicates. Correlation matrixes were prepared using R software and Pearson's Correlation Coefficient (PCC) as the statistical metric. Data reported in the normalized data-matrix were used to determine the genes differently expressed in different samples by performing a T-test (TMeV).
Correlation Analysis of Expression
Correlation analysis of expression was performed exploiting the grapevine co-expression database VTCdb (http://vtcdb.adelaide.edu.au/Home.aspx) using the VIT_08s0040g03070 relative to VvWRKY26 and selecting HRR co-expression measure (Wong et al., 2013).
Berry Fixation, Embedding, and in situ Hybridization (ISH)
Berries of Vitis vinifera cv. Corvina clone 48 were collected from grapevines grown in a commercial vineyard at Montorio (45° 27′ 17″ North, 11° 03′ 14″ East, Verona, Italy) in the 2010/2011 growing season. The samples were collected at 8 weeks before véraison (corresponding to fruit set stage) and at 1 week after véraison. After harvest, the samples were fixed with 2% formaldehyde and 0.25% glutaraldehyde in PBS buffer (pH 7.5) under vacuum overnight.
Fixed berries were rinsed five times in PBS and then dehydrated in four consecutive solutions containing increasing ethanol concentrations (25, 50, 75, and 100%). Post fixation steps were performed with four overnight treatments in ethanol solutions containing increasing xylene concentrations (25, 50, 75, and 100%). The embedding was performed progressively substituting the xylene with Paraplast Plus (Thermo Fisher Scientific). Tissue sections (7 μm thick) were cut with a 2035 Leica microtome (Leica Microsystems GmbH, Wetzlar, Germany), floated on warm water, and immobilized on poly-L-lysine coated slides to aid handling. The slides were then air-dried at 37°C and used for ISH. All the following ISH steps were done using an adaptation of a universal liquid handling robot, Freedom EVO 100, from Tecan and performed as described by Begheldo et al. (2013).
The antisense and sense VvWRKY26 probes 749 bp in length were selected by PCR on leaf cDNA of Vitis vinifera cv. Corvina with the primer sets listed in Supplementary Table 1.
Results
VvWRKY26 Phylogenetic Analysis
A previous search for WRKY genes in the genome of the near-homozygous PN40024 genotype of Vitis vinifera cv. Pinot noir (Jaillon et al., 2007) led to the identification of 59 sequences belonging to this family (Wang M. et al., 2014). Among these, sequence VIT_08S0040G03070 named VvWRKY26, located on chromosome 8, was identified as the closest gene to AtTTG2 by Wang M. et al. (2014). In this study, we performed a Blast search in the grapevine PN40024 12X V1 genome showing that VvWRKY26 is also the closest grapevine gene to PhPH3, as recently shown by Verweij et al. (2016).
The full-length coding region of VvWRKY26 was amplified from cv. Corvina post fruit set seed cDNA, cloned and submitted to GenBank under the accession number KX823961.
VvWRKY26 contains a 1434 bp open reading frame encoding a protein of 477 amino acidic residues with a predicted mass of 52.27 kDa and a calculated pI = 8.84. The deduced amino acid sequence is 99% similar to the predicted one of cv. Pinot noir with only one different amino acid at position 324 (Supplementary Figure 1).
A phylogenetic analysis of 32 WRKY proteins, including characterized WRKY TFs of grapevine and of different plant species, showed that VvWRKY26 belongs to the cluster of Group I WRKYs as PhPH3, AtTTG2 and BnTTG2, as also shown by Verweij et al. (2016) (Figure 1). The cluster contains other grapevine WRKY factors that are mainly involved in response to biotic and abiotic stresses, such as VvWRKY33 and VpWRKY2 (Li et al., 2010; Merz et al., 2015) and VvWRKY01 (VvWRKY2) related to lignin synthesis (Guillaumie et al., 2010). In the same cluster are AtWRKY2 and AtWRKY4 that play roles in seed germination, in pollen development (Jiang and Yu, 2009; Guan et al., 2014) and in sugar metabolism (Hammargren et al., 2008), respectively, and PcWRKY1 and GhWRKY3 involved in the response against plant pathogens (Eulgem et al., 1999; Turck et al., 2004; Guo et al., 2011). The other grapevine WRKY sequences included in this phylogenetic analysis all belong to different clusters and groups, further underlying functional differences with VvWRKY26.
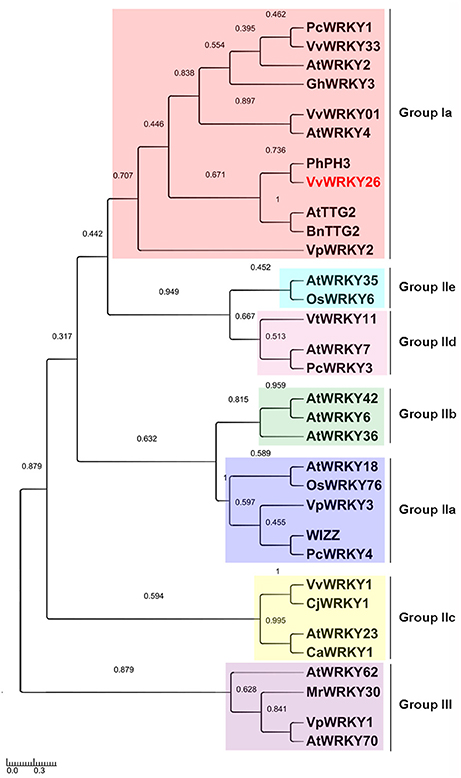
Figure 1. Phylogenetic tree including WRKYs belonging to different species. The analysis, based on alignments of the C-terminal WRKY domain, was performed using the neighbor-joining method by the Mega version 6 program (Tamura et al., 2013). The scale bar represents the number of substitutions per site and the numbers next to the nodes are bootstrap values from 1000 replicates. The accession numbers are reported in the Materials and methods section.
VvWRKY26 is characterized by the presence of four introns and shares the position of the first three with the homologs PhPH3, AtTTG2 and BnTTG2 (Figure 2A; Johnson et al., 2002; Li et al., 2015; Verweij et al., 2016). Only for VvWRKY26 and PhPH3 is there an additional intron in the region corresponding to the C-terminal WRKY domain (Verweij et al., 2016).
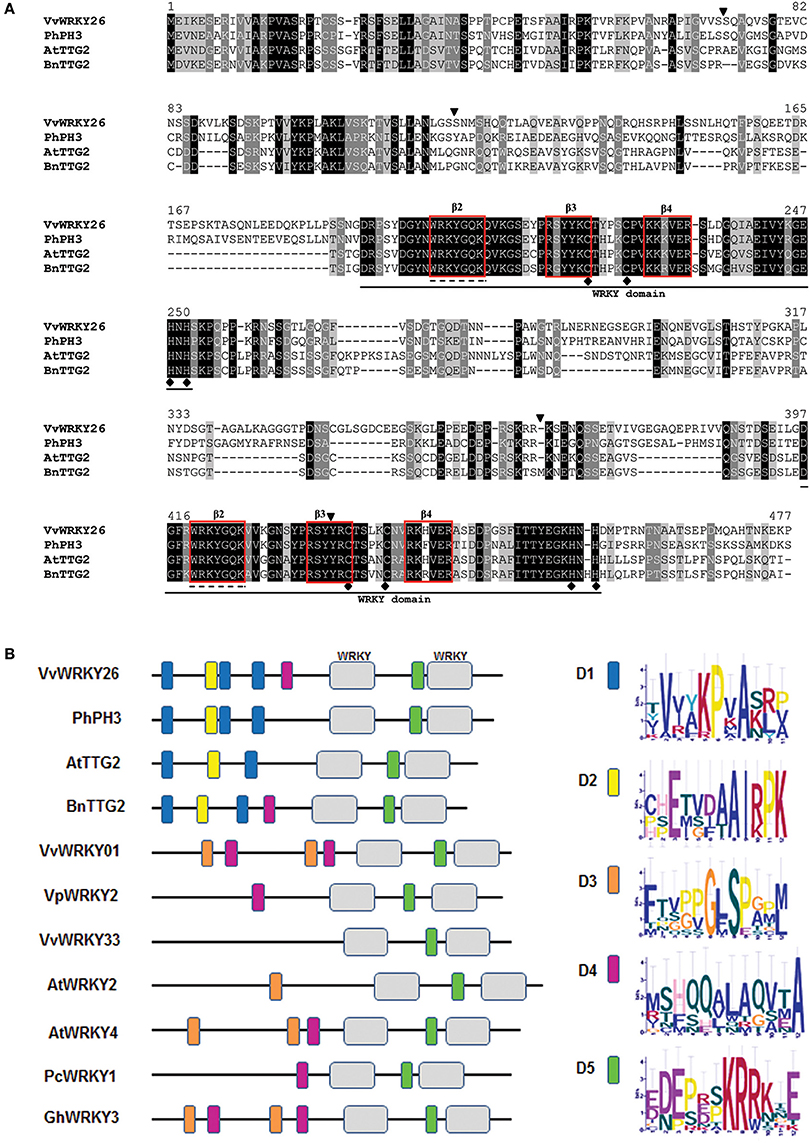
Figure 2. Analyses of primary protein structures. (A) Alignment of VvWRKY26, PhPh3, AtTTG2, and BnTTG2 predicted sequences. Identical, conserved and similar residues are shown in black, light gray and dark gray, respectively. The black lines below the alignment locate the two WRKY domains, with the conserved sequences WRKYGQK and the C2H2 zinc-finger motifs underlined by dashed lines and black dots, respectively. Triangles above the alignment indicate the position of the introns. The first three introns are conserved in all three species, while the last one close to the 3′ UTR is present only in petunia and grapevine. Red squares individuate β2, β3, and β4 in the WRKY domains. (B) Protein domain organization of Group I WRKY factors represented by colored boxes identified by MEME Suite. The consensus sequence of the motifs is reported.
The alignment of the amino acidic sequences revealed that, among all, VvWRKY26 is closer to PhPH3 with 54% sequence similarity compared with 48 and 46% to AtTTG2 and BnTTG2, respectively (Figure 2A). Analysis of the protein primary structure revealed the presence of two WRKY domains in correspondence to amino acids 200 and 401, classifying VvWRKY26 as a group Ia member of the WRKY family, as PhPH3, AtTTG2, and BnTTG2 (Figures 1, 2A; Xie et al., 2005; Verweij et al., 2016). In all these four WRKY proteins both WRKY domains are highly conserved and contain the amino acid sequence WRKYGQK at the N-terminal ends together with the C2H2-type zinc-finger motifs C–X4–C–X22–H–X1–H (Johnson et al., 2002; Li et al., 2015; Verweij et al., 2016). As proposed for AtWRKY1 using a crystal structural model (Brand et al., 2013), three strands β2, β3, and β4 that are necessary for the specific protein-DNA interaction can be identified within the WRKY domains of the four proteins (Figure 2A).
By further analysing the Group I WRKY factors from Figure 1 by MEME bioinformatic tool, we highlighted other conserved protein motifs (Figure 2B). Among these, the D1 and D2 domains located at the N-terminus are specific of the TTG2-like WRKY proteins (Figure 2B; Supplementary Figure 2). Moreover, the domain D5 containing a putative nuclear localization signal KRRKxE is common to all sequences and was previously identified also by Wang M. et al. (2014)
Overall, our analyses allowed the identification of VvWRKY26 as the grapevine homolog of both petunia PH3 and Arabidopsis TTG2, and highlighted unique domains shared by TTG2-like WRKYs.
Heterologous Expression of VvWRKY26 in Petunia ph3 Mutant
To gain information on the role of VvWRKY26 and ascertain its ability to fulfill the function of PhPH3 we performed complementation analysis in a P. hybrida ph3 mutant. The coding sequence of VvWRKY26 was expressed under the control of the constitutive 35S promoter in the mutated petunia line. The ph3 mutation affects the vacuolar acidification in petal epidermal cells, resulting in a higher pH and a shift of the anthocyanin absorption spectrum (Verweij et al., 2016). Therefore, the corollas of the ph3 mutant line present a pale pink pigmentation in comparison to the strong red color of the wild type line R27 (Figure 3A).
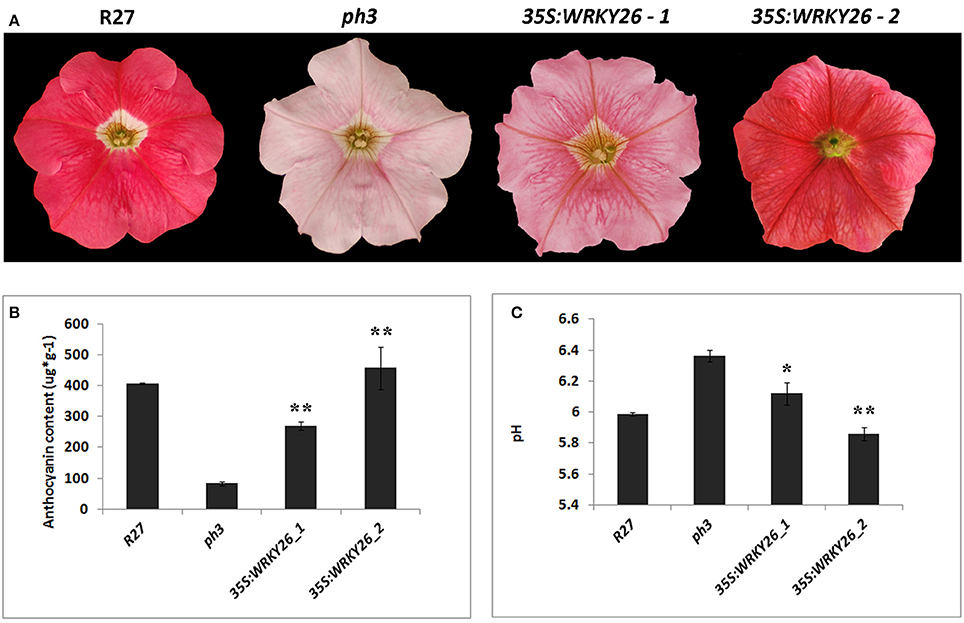
Figure 3. Complementation analysis in ph3 petunia mutant with 35S:VvWRKY26. (A) Phenotype of untransformed flowers of the wild type R27 and mutant ph3 lines compared to transgenic flowers of two lines expressing VvWRKY26. (B) Total anthocyanin content (μg*g−1 fresh weight) of petal limb extracts from untransformed R27 and ph3 lines and transgenic plants determined by spectrophotometry at 540 nm. Purified malvidin 3-glucoside was used as a standard. Data represent the mean of three biological replicates ± SE. Asterisks indicate significant difference against the ph3 mutant line (**P > 0.01). (C) The pH values of crude petal limb extracts from untransformed R27 and ph3 lines and transgenic plants. Each pH value is the mean of 10 biological replicates ± SE. Asterisks indicate significant difference against the ph3 mutant line (*P > 0.05; **P > 0.01).
The transformation yielded 6 PCR positive plants with a range of petal color phenotypes, e.g. from evenly red-pigmented corollas similar to the R27 wild type line, to pale pink like the mutant ph3 flowers (Figure 3A). For further phenotypic characterization, we selected two independent lines (lines 4 and 2) showing intermediate (strong pink) and fully complemented (red) petal phenotypes, respectively, corresponding to low and high expression of 35S:VvWRKY26 as determined by qPCR (Figure 3A; Supplementary Figure 3). We also observed a strong pigmentation on the petal veins in both transgenic lines that was independent of the complementation level (Figure 3A). No other phenotypic effects were observed on transgenic petunia plants in comparison with the ph3 mutant line.
The pH analysis of petal crude extracts revealed that both transgenic lines presented significantly lower pH compared to the ph3 mutant and that only the fully red complemented line 2 presented pH values similar to the wild type R27 (Figure 3B), suggesting that VvWRKY26 fully restored the ph3 mutation fulfilling the function of the endogenous PH3 in the regulation of vacuolar acidification.
We analyzed the anthocyanin content of petal extracts for each line. As shown in Figure 3C, surprisingly the ph3 mutant was affected in the anthocyanin synthesis, accumulating significantly less anthocyanin content than the wild type R27 line. On the contrary, both transgenic lines expressing VvWRKY26 presented anthocyanin levels comparable to the wild type R27 line, in a consistent manner with the levels of transgene expression (Figure 3C) suggesting that VvWRKY26 can also contribute to the control of anthocyanin synthesis.
In petunia the mutation in PH3 gene causes seed abortion and female sterility indicating a role as yet uncharacterized in seed development (Verweij et al., 2016). In our complementation analysis, we observed that the expression of VvWRKY26 partially overcame the sterility of the ph3 line, in fact the self-fertilization of VvWRKY26 over-expressing petunias yielded few capsules with seeds showing wild type phenotype (data not shown). However, because of the sterility of ph3 mutant plants, the screening and comparison of the seed phenotypes of the self-pollinated transgenic and mutant plants was impossible.
Transcriptomic Analysis of Petunia Petals Expressing VvWRKY26
To identify potential target genes of VvWRKY26, we compared the petal transcriptomes of 35S:VvWRKY26 plants and untransformed ph3 mutant. Three independent transgenic lines showing fully complemented petal phenotype and comparable expression level of the transgene were chosen as biological replicates (lines 1, 2, and 5; Supplementary Figure 3). Total RNA was extracted from petals of each sample and hybridized against a NimbleGen microarray 111012_PAX_PT_expr_HX12 chip containing 27,627 probes representing the Petunia axillaris transcriptome (Zenoni et al., 2011). We identified 546 genes differentially expressed by implementing a Significance Analysis of Microarrays (SAM) and using a False Discovery Rate (FDR) of 0.1% (Microarray Gene Expression Omnibus database accession GSE86890). All transcripts were annotated using the functional information content of the Petunia axillaris transcriptome (Supplementary File 1; Zenoni et al., 2011). The most relevant differentially expressed genes were selected by considering the subset of up-regulated genes in each transgenic line with a fold change (FC) value ≥ 2 compared with mutant petals. Putative functional annotations were manually improved by BLAST analysis and those with no similarity to known sequences (no hit) were removed from the subset (Supplementary File 1).
The ectopic expression of VvWRKY26 affects genes belonging to a wide range of cellular processes (Supplementary File 1). In particular, we observed the modulation of many genes putatively encoding transmembrane transporters (Table 1), such as an uncharacterized ammonium transporter (PETAX069902_Contig1) and the potassium channels KAT2 (PETAX007908_Contig1) and HAK5 (PETAX015455_Contig1) involved in stomatal opening and in the response to low K+ concentration, respectively (Sharma et al., 2013; Nieves-Cordones et al., 2014). Electron carriers and three lipoxygenases putatively involved in vacuolar trafficking and homeostasis were also up-regulated by the ectopic expression of VvWRKY26 (Table 1). Interestingly, a transcript corresponding to a nodulin N21-like (PETAX090339_Contig 1) gene was one of the most induced genes with an FC of 49.78. The function of this class of genes is not yet understood, but the presence of several transmembrane domains in the proteins and structural homologies with bacterial multidrug exporters suggest a role in transport (Guillaumie et al., 2008).
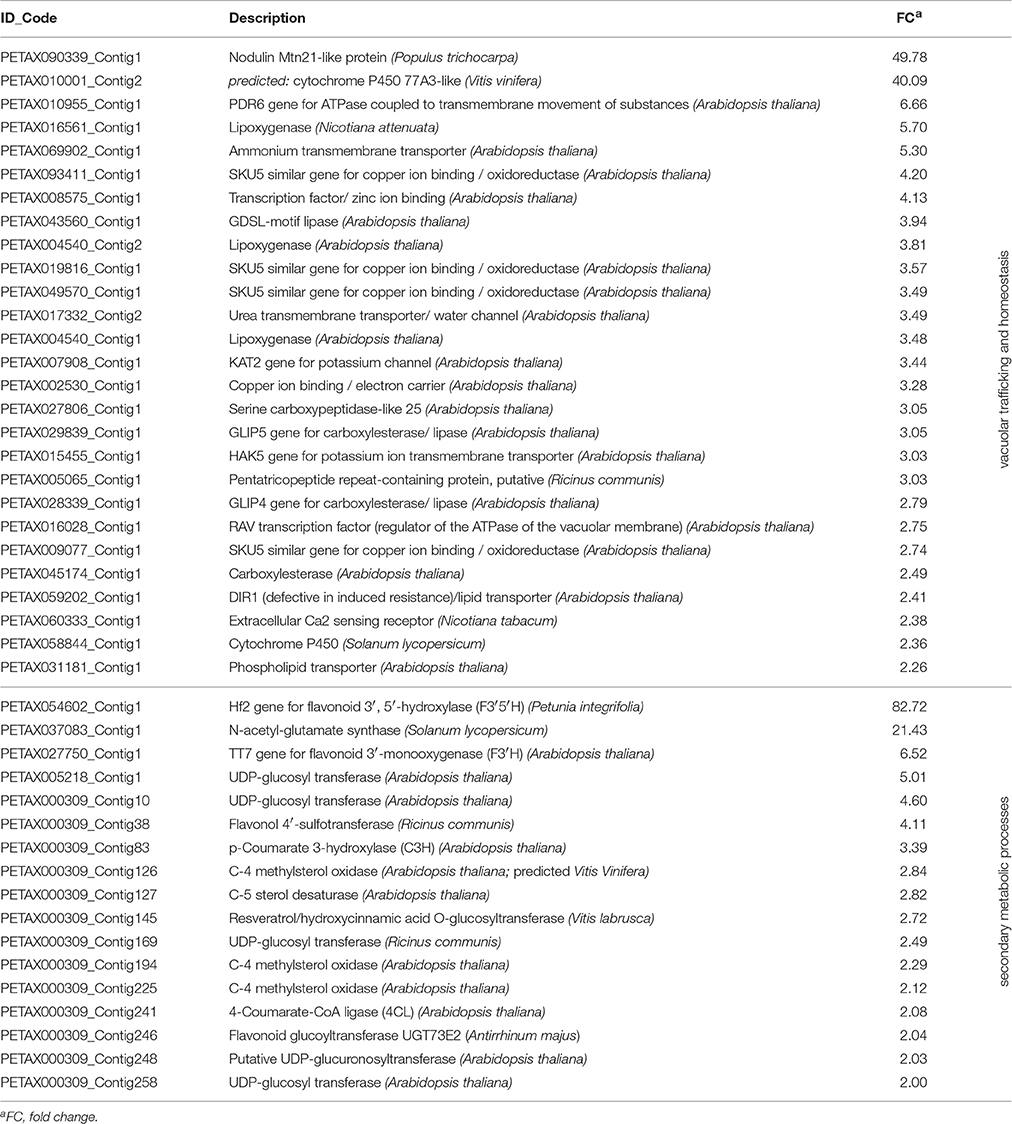
Table 1. Subset of up-regulated genes in ph3/35S:VvWRKY26 petunia petals involved in secondary metabolism and in vacuolar trafficking and homeostasis.
Related to the flavonoid pathway, the microarray analysis revealed a strong induction of the Petunia HF2 gene encoding for the FLAVONOID 3′, 5′-HYDROXYLASE (F3′5′H, PETAX054602_Contig1) and, to a lesser extent, of the AtTT7 homolog for FLAVONOID-3′-MONOOXYGENASE (F3′H, PETAX027750_Contig1). The encoded enzymes are responsible for the hydroxylation of (colorless) dihydroflavonols in the 3′ and 5′ positions for the production of the tri- and di-substituted anthocyanin derivatives, respectively (Table 1; Bogs et al., 2006). Also the expression of JAF13 (PETAX002230_Contig1), encoding for a bHLH transcription factor specifically involved in the control of anthocyanin synthesis (Quattrocchio et al., 1998), increased in VvWRKY26 expressing plants, although with a low fold change (FC = 2.11; Supplementary File 1). Other genes involved in secondary metabolic processes resulted modulated (Table 1). Among these, noteworthy are the 4-COUMARATE-CoA LIGASE (4CL, PETAX000309_Contig241) and p-COUMARATE 3-HYDROXYLASE (C3H, PETAX000309_Contig83) of the phenylpropanoid pathway.
Ultimately, even cell wall metabolism resulted as being affected by VvWRKY26, as revealed by the modulation of several pectinesterases, an expansin (PETAX032024_Contig1) and an endoβ-1,4-glucanase (PETAX019178_Contig1) (Supplementary File 1).
Our microarray analysis was validated comparing by qPCR the expression level of two modulated genes, N21 and F3′H in VvWRKY26 complemented lines and in the ph3 mutated lines (Figure 4A).
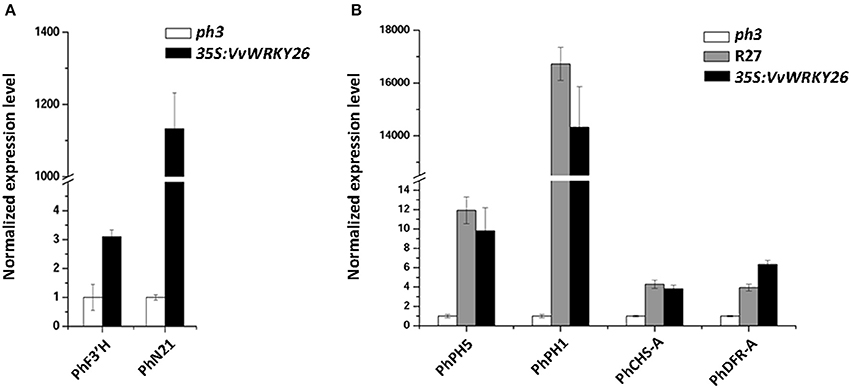
Figure 4. Expression analyses in petunia petals by qPCR. (A) Expression analysis of F3′H and N21 in the ph3 mutant and in VvWRKY26 expressing lines as confirmation of the microarray results. (B) Expression analysis of structural genes related to vacuolar acidification (PhPH5 and PhPH1) and to anthocyanin synthesis (PhCHS-A and PhDFR-A) in the untransformed R27 and ph3 lines and VvWRKY26 expressing plants. In all analyses the data correspond to the mean ± SE of three biological replicates (corresponding to lines 1, 2, and 5; Supplementary Figure 2) relative to an ACTIN housekeeping control and normalized against the ph3 mutant value. Abbreviations correspond to: PhF3′H, FLAVONOID-3′-MONOOXYGENASE; PhN21, NODULIN MTN21-LIKE PROTEIN; PhPH5, H+ P3A-ATPASE; PhPH1, P3B-ATPASE; PhCHS-A, CHALCONE SYNTHASE A; PhDFR-A, DIHYDROFLAVONOL 4-REDUCTASE A.
These transcriptomic findings indicate that the heterologous expression of VvWKY26 impacted multiple metabolic processes in petunia and, above all, those related to transport and flavonoid synthesis. However, it did not provide an exhaustible explanation for the phenotypic changes observed in petals specifically regarding vacuolar acidification and anthocyanin biosynthesis. We therefore analyzed by qPCR the expression level of the pH structural genes PH5 and PH1 and of the anthocyanin-related genes CHS-A and DFR-A in the transgenic line with highest levels of VvWRKY26 expression (line 2), in the wild type R27 and in the ph3 mutant lines (Figure 4B). The analysis revealed that the ectopic expression of VvWRKY26 in ph3 background induced the expression of PH5, PH1, DFR-A and CHS-A in petunia petals at comparable levels with the wild type R27 restoring the ph3 mutation. qPCR data highlighted a difference, not detected by microarray analysis, probably due to its lower sensitivity, in the DFR-A and CHS-A expression level between mutant and complemented petunia lines. Otherwise, the expression of the pH-related genes PH5 and PH1 was not detected by microarray because they are not represented among chip probes.
These results support multiple roles for VvWRKY26 in petunia indicating that, above all, it is likely involved in the regulatory network underlying both pigmentation and vacuolar acidification.
VvWRKY26 Expression Analysis in Grapevine
The expression of VvWRKY26 was determined by consulting the global gene expression map of Vitis vinifera cv. Corvina (Figure 5A; Fasoli et al., 2012). VvWRKY26 is expressed in many grapevine organs with the highest levels in inflorescence, flower, petal and carpel, in young leaf, in rachis and tendril and in all stages of bud development, except for winter–bud. In berry tissues (whole pericarp, flesh and skin) and seed, VvWRKY26 transcripts were mainly detected at the earliest stages of development (fruit set and post fruit set) and declined thereafter. The VvWRKY26 expression pattern was confirmed by qPCR in a reduced set of Corvina organs, as berry pericarp throughout development (fruit set, post fruit set, véraison, mid-ripening, ripening), seed (post fruit set), young leaf, tendril and in young and well-developed inflorescence (Figure 5B).
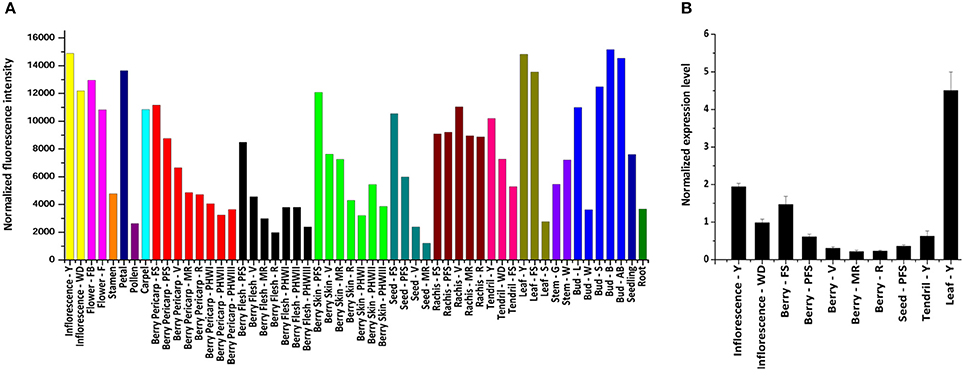
Figure 5. VvWRKY26 expression profile in grapevine. (A) VvWRKY26 expression profile in the V. vinifera cv. Corvina atlas in 45 organs/tissues during development. (B) Expression analysis of VvWRKY26 by qPCR in selected organs/stages of development of cv. Corvina Data, relative to VvUBIQUITIN1 control, are the mean of three biological replicates ± SE. For both analyses the abbreviations after organ correspond to: FS, fruit set; PFS, post fruit set; V, véraison; MR, mid-ripening; R, ripening; Bud - L, latent bud; Bud - W, winter bud; Bud - S, bud swell; Bud - B, bud burst; Bud - AB, bud after burst; Inflorescence - Y, young; Inflorescence - WD, well-developed; Flower - FB, flowering begins; Flower - F, flowering; Tendril - Y, young; Tendril - WD, well-developed; Tendril - FS, mature; Leaf - Y, young; Leaf - FS, mature; Leaf - S, senescing leaf; Stem - G, green; Stem - W, woody.
In an attempt to gain insights into its role during berry development, we localized in detail the expression sites of VvWRKY26 in berry tissues of Corvina at fruit set and post véraison by performing in situ hybridization (Figure 6). Hybridization with the VvWRKY26 sense probe as control showed no background staining or signal (Figures 6A–C). The analysis revealed that the expression of VvWRKY26 in seed is mainly localized in the cellular layer constituting the inner integument in both early (Figures 6D,E) and late (Figure 6G) developmental stages. In berry VvWRKY26 expression was mainly localized in cells surrounding the vascular bundles of the mesocarp at fruit set (Figures 6F,G). A similar and even clearer signal was also detected later in development (post véraison), revealing mRNA localization specifically in the phloematic cells of vasculature (Figure 6H). Moreover, only at post véraison a hybridization signal was also detected in the epidermal cell layer of the exocarp (Figure 6I).
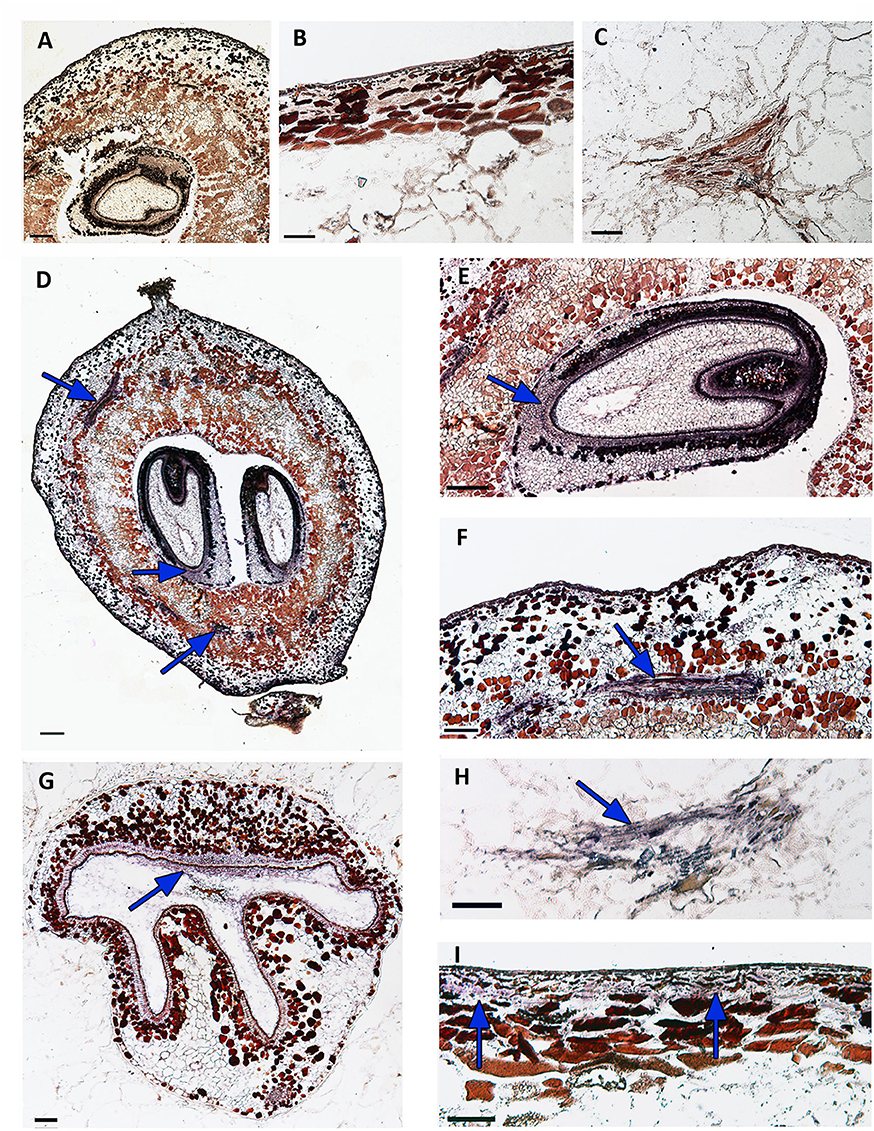
Figure 6. Localization of VvWRKY26 transcripts by in situ hybridization in berry at fruit set and post véraison stages. Sections of berry hybridized with a VvWRKY26 RNA sense probe as negative control did not show any significant signals (A–C). The antisense probe detected VvWRKY26 transcripts, resulting in a violet-gray coloration (D–I). (A–C) represent the hybridization of longitudinal section of berry at post véraison, a magnification of skin and of a vascular bundle, respectively, with a VvWRKY26 RNA sense probe (negative control). (D–F), representing the hybridization of the berry at fruit set, report the signals in the longitudinal section of the whole berry (D), in the inner integument of the seed coat (E) and in cells surrounding the vascular bundles (F). At this developmental stage no signal was detected in the skin. (G–I), representing the hybridization of the berry at post véraison, report the signals in the inner integument of a seed coat (transversal section, G), in the phloematic cells (H) and in the epidermal cell layer of the skin (I).
Our results indicated that VvWRKY26 is widely expressed in grapevine organs mainly during early developmental stages. In berry its expression is localized in the vascular bundles of the mesocarp, while in seed in the inner integument of the seed coat.
Correlation Analysis of Gene Expression by the Grapevine Database VTCdb
In order to discover genes co-expressed with VvWRKY26 in berry and possibly representing its targets or collaborators, we investigated the grapevine co-expression database VTCdb, encompassing over 800 public microarray datasets related to Vitis vinifera and representing the transcriptome changes in a wide range of biological and environmental conditions (Wong et al., 2013). The use of such a platform allowed to visualize the expression profile of genes highly correlated with VvWRKY26 transcriptional pattern over a wide range of experimental and/or environmental conditions.
Considering a PCC > 0.7, we generated a list of 114 co-expressed genes (Supplementary File 2), that were automatically annotated using V1 version of the 12X grapevine genome and manually classified in functional categories based on their known or putative function (Supplementary Figure 4). The most highly represented category was constituted by transport-related genes (20.2%) (Table 2). Interestingly, these included the homologs of PhPH5/AtTT13 (VIT_09S0002G00130) and PhPH1 (VIT_07S0191G00260) involved in vacuolar acidification (Verweij et al., 2008; Faraco et al., 2014; Appelhagen et al., 2015). We also identified three genes related to K+ transport as the homolog of the Arabidopsis NHX1, an anion exchanger involved in vacuolar K+ fluxes mediating stomatal aperture (VIT_14S0128G00020; Andrés et al., 2014), the K+ efflux antiporter 3 (KEA3, VIT_00S0282G00020) belonging to the cation/proton antiporters-2 superfamily in Arabidopsis (Kunz et al., 2014) and the K+ uptake permease 6 (KUP6, VIT_14S0066G01580) (Kim et al., 1998). In the same category there are two members of the nucleobase-ascorbate transporter (NAT) family, NAT6 (VIT_17S0000G07850) and NAT11 (VIT_00S0813G00010), putatively involved in long-distance transport due to their preferential expression in vascular tissues of many Arabidopsis organs (Maurino et al., 2006) and the homologs of PHT2-1 (VIT_00S0291G00060) and PHT1-9 (VIT_18S0122G00780), representing two Arabidopsis inorganic phosphate transporters (Versaw and Harrison, 2002; Lapis-Gaza et al., 2014). Among the other co-expressed genes, we identified two MATE transporters (VIT_08S0056G01120; VIT_08S0056G01000), an ABC transporter (VIT_13S0074G00690) and an anthocyanin membrane protein 1 (ANM1, VIT_08S0007G03570), whose detailed functions are still unknown.
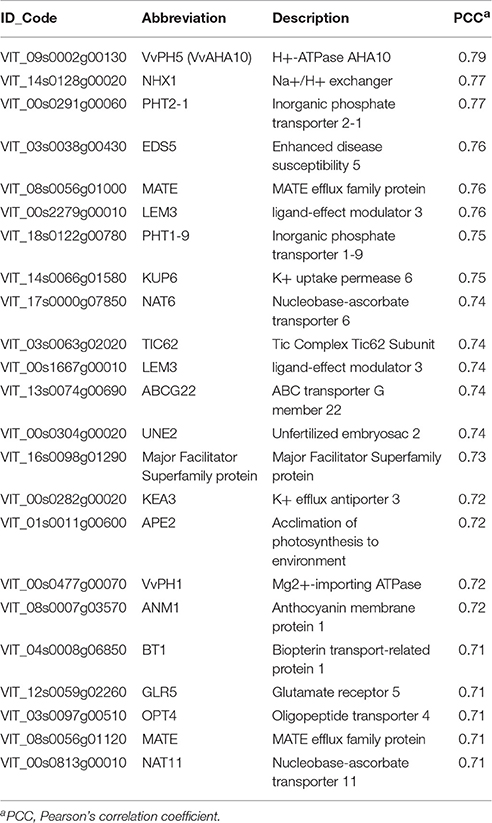
Table 2. List of transport-related genes resulting as highly correlated with VvWRKY26 (PCC > 0.7) in the correlation analysis of global gene expression performed using the grapevine transcriptomic platform VTCdb.
Among the genes not grouped in this category but highly co-expressed with VvWRKY26 we found the R2R3-MYB repressor MYBC2-L1 (VIT_01S0011G04760), recently demonstrated to influence pigmentation and cell vacuolar pH when expressed in petunia petals, and anthocyanin and PA biosynthesis in grapevine (Cavallini et al., 2014, 2015; Huang et al., 2014). We also found MYC1 (VIT_07S0104G00090), a bHLH factor involved in the regulation of flavonoid synthesis (Hichri et al., 2010; Matus et al., 2010).
Another gene worth mentioning for the highest correlation of expression with VvWRKY26 is UV RESISTANCE LOCUS 8 (UVR8), a photoreceptor that induces changes in gene expression after absorption of UV-B rays mediating the photomorphogenic response (Binkert et al., 2014; Carbonell-Bejerano et al., 2014; Liu et al., 2015; Yin et al., 2015; Loyola et al., 2016).
Other two highly represented groups are related to cellular homeostasis and transcription factor activity, constituting 10.5% and 8.8% of the 114 genes, respectively. Many other functional categories were identified as almost equally represented. However, apart from the transport-related genes, it was difficult to find a putative functional association between all the other co-expressed genes.
The high correlation of expression of genes related to transport and vacuolar metabolism provides a first insight into the possible role exerted by VvWRKY26 in grapevine and indicates putative functional similarity with the petunia ortholog PH3. Moreover, the identification of many functional categories suggests that VvWRKY26 might also function in other physiological or stress-related pathways and therefore perform multiple functions.
Transcriptomic Analysis of Vitis vinifera cv. Sultana Leaves Transiently Over-expressing VvWRKY26
To identify potential target genes of VvWRKY26 in the native species we transfected six grapevine cv. Sultana plantlets with a 35S:VvWRKY26 construct by vacuum Agrobacterium-mediated infection. As control, six plantlets were transformed with a construct containing a non-coding sequence. Six days after infection the over-expression of VvWRKY26 that occurred was verified by RT-PCR and qPCR analysis (Supplementary Figures 5A–B). Using an Agilent platform we compared the leaf transcriptomes of three selected over-expressing and control lines showing comparable high and low VvWRKY26 expression level, respectively (Supplementary Figure 5B; Microarray Gene Expression Omnibus database accession GSE86891). Performing a t-test analysis with a Pearson's correlation value of 0.05, we identified 983 differentially expressed genes that were annotated using the V1 version of the 12X draft release of the grapevine genome and by manual BLAST analysis (Supplementary File 3). As putative targets of VvWRKY26 we focused on up-regulated genes with a fold change ≥ 2 that we distributed into 18 Gene Ontology functional categories (Supplementary File 3; Supplementary Figure 6). Those genes with no similarity to known sequences or function (no hit or unknown protein) were removed from the subset.
The transient over-expression of VvWRKY26 affects a wide range of cellular processes (Supplementary File 3). Secondary metabolism and Transport categories were nonetheless well represented (Table 3). As observed in petunia, among genes related to transport, we found two genes encoding for ammonium transporters (VIT_07S0031G02950 and VIT_07S0031G02990), and two nodulins (VIT_17S0000G00830 and VIT_04S0044G00380). In addition, different ion transporters, a MATE efflux protein (VIT_08S0007G08200) and an ABC transporter (VIT_01S0011G04670) were modulated in their expression level, according to the proposed role of VvWRKY26 in transport. Moreover, carboxyesterases, lipoxygenases and patatins, putatively involved in lipid metabolism resulted as slightly up-regulated by the transgene supporting a role of VvWRKY26 in membrane remodeling and in maintaining vacuolar homeostasis.
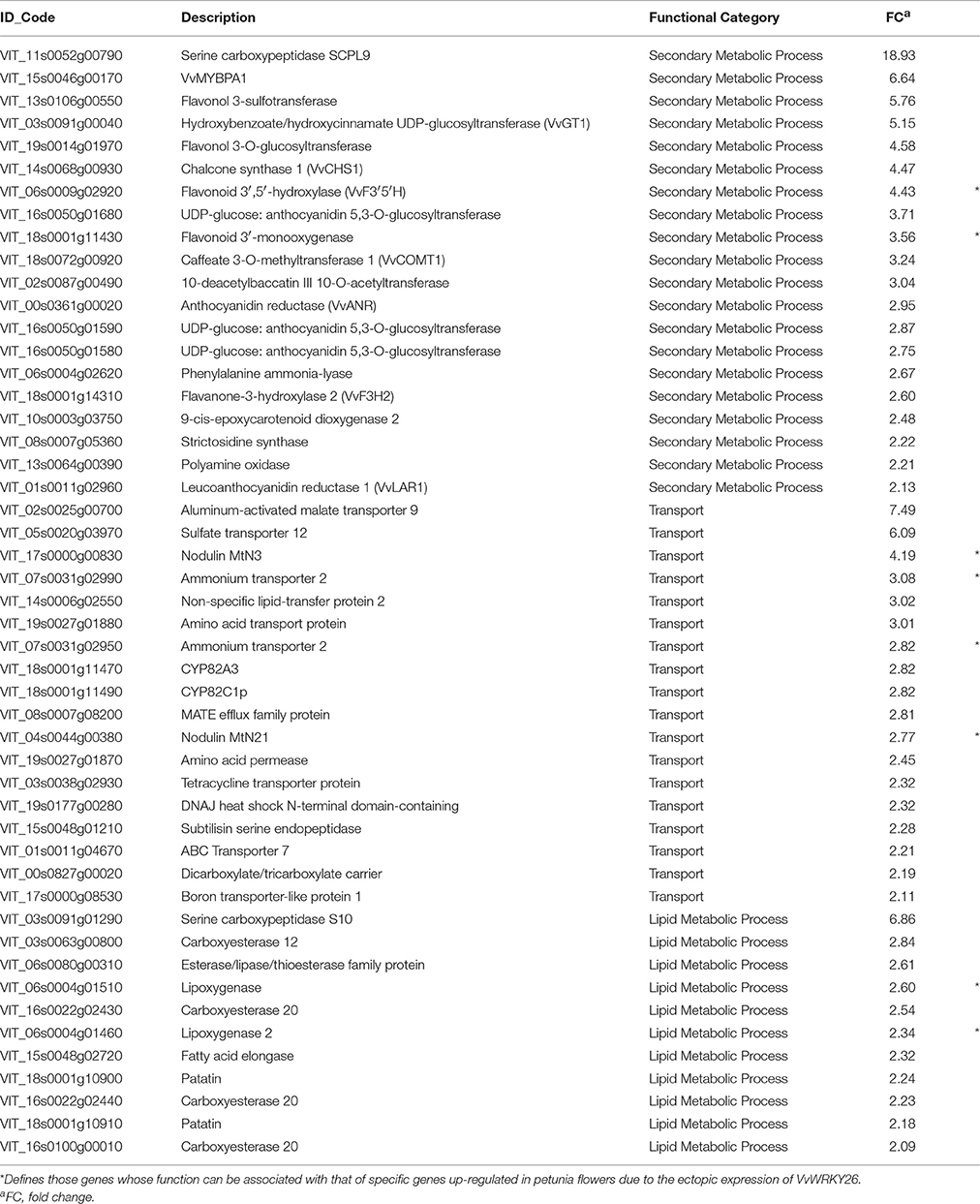
Table 3. Sub-set of up-regulated genes in VvWRKY26 over-expressing Sultana plantlets involved in Secondary metabolism, Transport, and Lipid metabolism.
The Secondary metabolism category was widely represented by genes encoding enzymes acting in flavonoid biosynthesis (Table 3). The transcription factor MYBPA1 (VIT_15S0046G00170), the regulator of PA biosynthesis, resulted as being one of the most induced genes by 35S:VvWRKY26 (Bogs et al., 2007). Consistently, we observed the modulation of several structural flavonoid genes like CHS1 (VIT_14S0068G00930), F3H2 (VIT_18S0001G14310), F3′5′H (VIT_06S0009G02920) and F3′H (VIT_18S0001G11430). Moreover, the transcriptional analysis revealed the up-regulation of many genes specifically involved in proanthocyanidin synthesis, i.e., ANTHOCYANIDIN REDUCTASE (ANR, VIT_00S0361G00020), LEUCOANTHOCYANIDIN REDUCTASE 1 (LAR1, VIT_01S0011G02960), and the recently characterized HYDROXYBENZOATE/HYDROXYCINNAMATE UDP-GLUCOSYLTRANSFERASE (GT1, VIT_03S0091G00040) (Bogs et al., 2005; Khater et al., 2012). The laccase encoding transcript VIT_18s0122g00250, putatively involved in the oxidative polymerization of flavan-3-ols and strongly induced in 35S:VvWRKY26 plantlets, may also be included in this proanthocyanidin-related gene list. Lastly, SERINE CARBOXYPEPTIDASE SCPL9 (VIT_11S0052G00790) resulted as significantly induced with an FC of 18.92. This gene was also recently identified in the transcriptomic screening of grapevine hairy roots over-expressing MYBPA1 and MYBPA2 (Khater et al., 2012). Based on these results we suggest that in grapevine VvWRKY26 functions as a regulator of the flavonoid pathway specifically targeting the proanthocyanidin branch.
In our analysis we also found several genes belonging to the stress response category and different senescence-associated proteins, supporting a role of VvWRKY26 in plant response to abiotic stresses as previously proposed (Guo et al., 2014; Wang L. et al., 2014).
Our microarray analysis was validated by qPCR comparing the expression level of MYBPA1, CHS1, PH5, and PH1, in control and VvWRKY26 transiently over-expressing lines (Figure 7).
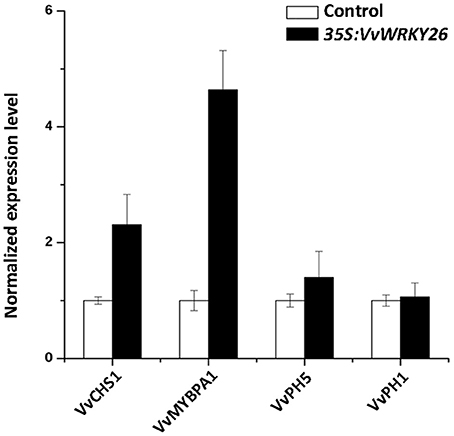
Figure 7. Expression analysis of CHS1, MYBPA1, PH5, and PH1 by qPCR in the control lines and VvWRKY26 expressing Sultana plantlets. The data correspond to the mean ± SE of three biological replicates relative to the VvUBIQUITIN1 control and normalized against each control value. Abbreviations correspond to: VvCHS1, CHALCONE SYNTHASE 1; VvMYBPA1, MYB regulator of PA biosynthesis; VvPH5, H+ P3A-ATPASE; VvPH1, P3B-ATPASE.
Taken together, the results obtained by VvWRKY26 over-expression in grapevine partially confirmed the functional analysis conducted in petunia, suggesting its involvement in transport, vacuole-associated metabolisms and flavonoid biosynthesis, specifically in control of the PA pathway.
Discussion
VvWRKY26 is the Grapevine Ortholog of phPH3, AtTTG2, and BnTTG2
The WRKY proteins represent one of the largest families of transcription factors in the plant kingdom. Since the first identification in sweet potato (Ishiguro and Nakamura, 1994), the regulative functions of members of this family have mainly been associated with plant response to biotic and abiotic stresses. However, a remarkable number of WRKY proteins have also important roles in development and in the control of secondary metabolic pathways (Schluttenhofer and Yuan, 2015). This is the case of the well-characterized TTG2 of Arabidopsis that is a regulator of seed and trichome development (Johnson et al., 2002; Garcia et al., 2005; Ishida et al., 2007; Gonzalez et al., 2016). Two WRKY factors extremely close to AtTTG2 have recently been characterized, PH3 of P. hybrida, a novel regulator of vacuolar acidification in petals (Verweij et al., 2016), and TTG2 of Brassica napus involved in response to salt stress and in trichome development (Li et al., 2015).
In this study, we isolated and characterized VvWRKY26 as the closest grapevine homolog to Arabidopsis TTG2 and petunia PH3. Our phylogenetic analysis showed that VvWRKY26 together with AtTTG2, PhPH3 and BnTTG2 form a distinct clade of WRKYs belonging to group Ia, defined by the presence of two WRKY domains and the C2H2-type zinc-finger motifs (Wang M. et al., 2014). Moreover, our phylogenetic tree based on the sequence similarity of the C-terminal WRKY domains also reflects the classification in subgroups established for the number of the WRKY domain in the protein and the type of zinc-finger motif (Eulgem et al., 2000; Zhang and Wang, 2005; Rushton et al., 2010), indicating a high degree of amino acidic conservation of the C-terminal WRKY domain within each subgroup. The importance of the C-terminal WRKY domain relies on its exclusive strong ability to recognize and bind the cognate W-boxes in the promoter of the target genes, in contrast to the N-terminal domain, that was shown to bind the DNA with low affinity (Brand et al., 2013). However, the N-terminal region is characterized by unique domains shared by members of the TTG2-like clade, suggesting that it may contribute to give specificity in the regulatory function of these WRKY factors.
VvWRKY26 showed high homology with PhPH3, sharing very similar β2, β3, and β4 strands necessary for the interaction with DNA as proposed by Brand et al. (2013). Moreover, both coding sequences harbor an intron between the WRKYGQK motif and the zinc finger that is typical of the C-terminal WRKY domain of group I WRKY proteins (Eulgem et al., 2000; Brand et al., 2013). This feature distinguishes AtTTG2 and BnTTG2 from the grapevine and petunia homologs. Interestingly, BnTTG2 can fulfill AtTTG2 regulative functions in trichome development when expressed in Arabidopsis (Li et al., 2015) and, on its part, AtTTG2 mimics PhPH3 in the control of vacuolar acidification when expressed in petunia (Verweij et al., 2016).
Moreover, both AtTTG2 and PhPH3 are part of a similar regulative and transcriptional network, in which the respective MBW complexes activate their expression and also require their interaction to activate the downstream pathways (Gonzalez et al., 2016; Verweij et al., 2016). In this study we provide evidence that VvWRKY26 is able to fulfill the functions of PhPH3 when expressed in petunia ph3 mutant, and functional analogies were also found between VvWRKY26 and the homolog AtTTG2 regarding proanthocyanidin biosynthesis as indicated by the induction of numerous PA-related genes in Sultana plants over-expressing VvWRKY26. These observations suggest that all members of the TTG2-like clade are functionally orthologs and are likely part of a similar regulation mechanism, although they control pathways that are partially distinct in their endogenous species. The functional orthology might also depend on the promoter sequence of the target genes and therefore is achieved by both remarkable conservation within WRKY proteins between species and the cognate binding site (C/T)TGAC(T/C) of the W-boxes (Pesch et al., 2014). For Group I WRKY factors, it has been proposed that two different and consecutive W-box motifs in the promoter sequence might be necessary for the binding of both WRKY domains (Brand et al., 2013). Future identification of the target genes in grapevine and analysis of the DNA regulative regions will be helpful to elucidate the recognition mechanism of VvWRKY26 and to highlight analogies of the regulative mechanisms within the TTG2-like protein cluster.
Evidence of an Involvement of VvWRKY26 in Vacuolar Acidification and Transport across the Tonoplast
The sequencing of the grapevine genome and the consequent availability of about 30,000 predicted gene sequences (Jaillon et al., 2007) prompted the exploitation of regeneration and transformation tools to gain information about gene function in grapevine. However, due to the recalcitrant nature of many cultivars to the agrobacterium-mediated stable transformation and the long regeneration time of the transgenic plantlets, the stable transformation of grapevine is still time-consuming and difficult to perform. The ectopic expression of a grapevine gene in a heterologous species such as P. hybrida, representing a fast growing model, has previously proven to be a useful alternative for functional characterization (Bogs et al., 2006; Cavallini et al., 2014, 2015; Provenzano et al., 2014; Li et al., 2016). Moreover, many well-characterized mutants for genes involved in different biochemical and developmental pathways are available for this model species. In our case, the petunia line mutated in PH3, and thus showing a higher pH of corolla crude extracts and reduced anthocyanin content, was used to confirm the functional orthology between PhPH3 and VvWRKY26. In fact, ectopic expression of VvWRKY26 caused an increase of PH5 and PH1 expression, the structural genes for vacuolar acidification, with the subsequent activation of pumping mechanisms leading to the decrease of pH in petal homogenates to wild type levels. Consistently, in our correlation analysis of gene expression using VTCdb, VvWRKY26 gene expression was highly correlated with the grapevine homologs of the petunia PH1 and PH5, whose functions as P-ATPase pumps were inferred by heterologous expression in petunia (Provenzano, 2011; Li et al., 2016).
However, the transient over-expression of VvWRKY26 in grapevine cv. Sultana did not induce PH1 and the expression of PH5 resulted as only weakly increased, as shown by microarray and qPCR analyses. The high expression of the endogenous VvWRKY26 in young leaves, as reported in the grapevine transcriptomic atlas (Fasoli et al., 2012) and by our qPCR, may have masked the effect of transgene over-expression. Alternatively, VvWRKY26 may require other co-factors for the activation of the downstream pathways, as recently demonstrated for the orthologs AtTTG2 and PhPH3 both cooperating with the respective WD protein of the MBW regulatory complex (Li et al., 2016; Verweij et al., 2016). It was previously shown that the grapevine MYB5a and MYB5b, participating in a MBW complex (Deluc et al., 2008; Hichri et al., 2010), are able to regulate cell pH when expressed in petunia petals (Cavallini et al., 2014). VvWRKY26 might require to be co-expressed with MYB5a and MYB5b to exert its biological function.
In the list of genes co-expressed with VvWRKY26 from the VTCdb and in the lists of genes induced by the heterologous and ectopic expression of VvWRKY26 in petunia and grapevine, respectively, the transport-related category was always highly represented indicating that VvWRKY26 might also impact the transport of anions/cations and other solutes across the tonoplast. In particular we identified many genes related to K+ transport and uptake. By exploiting VTCdb, the homolog of the Arabidopsis NHX1, a tonoplast-located K+/ H+exchanger required for K+ uptake into the vacuole of guard cells for stomatal opening was found (Andrés et al., 2014). In petunia the over-expression of Ipomoea nil NHX1 counteracts the acidifying effect of PH1 and PH5 indicating that these NHX transporters influence the vacuolar pH in the opposite manner to the H+ ATPase pumps (Faraco et al., 2014). We also identified KUP6, a member of the K+ uptake permease family. In grapevine two KUP genes VvKUP1 and VvKUP2 have been characterized for their role in K+ accumulation likely associated with berry growth during development (Davies et al., 2006). Interestingly, in petunia 35S:VvWRKY26 induces two genes encoding for the Arabidopsis homolog AtKAT2, an inward rectifying K+ channel involved in stomatal opening (Sharma et al., 2013), and AtHAK5, a K+ transporter active in roots at very low external K+ concentration (Nieves-Cordones et al., 2014). Considering also the many additional transport-related genes, the information gained exploiting transcriptomic-based approaches suggested that VvWRKY26 participates in different transport regulatory mechanisms and controls the expression of many different genes generally contributing to maintaining vacuolar homeostasis. VvWRKY26 would thus act as regulator of the vacuolar pH and ion transport, controlling both H+ pumps that acidify the vacuolar lumen as well as additional targets that counterbalance the positive charge accumulation and dissipate the electrochemical gradient across the tonoplast.
Evidence of an Involvement of VvWRKY26 in Flavonoid Biosynthesis
The constitutive expression of VvWRKY26 in petunia ph3 line resulted in the induction of the flavonoid structural genes CHS-A, F3′H, F3′5′H and DFR-A, and in the restoration of the wild type anthocyanin content that was impaired in ph3 mutant. This suggests that, in addition to the proposed role in vacuolar acidification and transport, VvWRKY26 might also be implicated in the control of the flavonoid biosynthetic pathway. The HF2 gene encoding F3′5′H resulted as being the most up-regulated in the microarray analysis performed on ph3/35S:VvWRKY26 complemented petunias. This enzyme is responsible for the hydroxylation of dihydroflavonols leading to the synthesis of the tri-substituted delfinidin-, malvidin- and petunidin-derivatives conferring a pink/purple pigmentation to the corolla. The genetic background of the ph3 mutant plant is however characterized by mutations in HF1 and HF2 loci causing a non-functional F3′5′H proteins (Matsubara et al., 2006). Therefore, despite the HF2 induction by VvWRKY26, petals of the fully complemented 35S:VvWRKY26 petunia plants have a reddish color typical of the cyanidin-based anthocyanin profile of the R27 background. The microarray analysis of petunia petals expressing VvWRKY26 also revealed the weak induction of a bHLH regulator of the flavonoid pathway, JAF13, whose precise role in the pigmentation of petunia corollas still has to be fully elucidated (Quattrocchio et al., 1998; Montefiori et al., 2015).
A role of VvWRKY26 as a flavonoid regulator, which seems to narrow to the PA synthesis, can also be inferred from the analysis of the grapevine plantlets transiently over-expressing the transcription factor. The induction of MYBPA1 (Bogs et al., 2007) and of its targets, i.e. the structural genes ANR, LAR1, and GT1 (Bogs et al., 2005; Khater et al., 2012), indicates the ability of VvWRKY26 to influence the PA branch by controlling the expression of this specific PA regulator, although a direct control of the biosynthetic steps by VvWRKY26 cannot be ruled out. Our hypothesis is also reinforced by the up-regulation of CHS1, a chalcone synthase previously associated to PAs (Harris et al., 2013) and of several serine-carboxypeptidases, some of which, shown to be induced by MYBPA1 (Terrier et al., 2009). Finally, the up-regulation of both F3′5′H and F3′H genes in the leaves of the over-expressing grapevines, that were also found upregulated in ph3/35S:VvWRKY26 petunias, confirms that VvWRKY26 regulates the hydroxylation steps of flavonoids in the endogenous species. The Arabidopsis homolog TTG2 was also shown to be a key regulator of PA accumulation, and ttg2 mutants have a reduced deposition of PAs in the seed coat. AtTTG2 drives the PA vacuolar storage by directly regulating the ATPase proton pump TT13 and the MATE vacuolar transporter TT12 (Appelhagen et al., 2015; Gonzalez et al., 2016).
Roles of VvWRKY26 in Grapevine
The function of VvWRKY26 studied through the aforementioned approaches together with the analysis of its expression pattern in grapevine organs at various developmental stages allow us to speculate about the biochemical and developmental processes regulated in planta. We particularly focused on berry where VvWRKY26 expression was high at early stages of fruit development and progressively declined at later stages. This general expression pattern is consistent with the hypothesized role in the regulation of berry acidification and PA biosynthesis, because the pH of berry homogenate is low at the first berry developmental stages and increases thereafter, and most of the PAs accumulate in berry tissues before the onset of ripening. In situ hybridization showed VvWRKY26 expression in vascular bundles of berry pericarp at fruit set and during ripening, supporting its role in transport through development. The correlation with many transporters repeatedly encountered in our study suggests a specific involvement in the exchange of ions such as K+ between vessels and berry flesh. Grapes are very rich in potassium that is an essential macronutrient for growth and development. It is the main cation in must and wine and its concentration in the berry at harvest may affect pH and thus impact potential wine quality.
The high expression of VvWRKY26 at the earliest stages of berry development also relies on its specific mRNA localization in the inner integument of the seed coat. This localization is consistent with the PA accumulation profile in the seed coat (Bogs et al., 2007), and the induction of many PA-related genes by VvWRKY26 over-expression in Sultana plantlets corroborates its involvement in the deposition of PAs in seed tissues. The heterologous expression of VvWRKY26 in petunia partially overcame the sterility in the ph3 line, suggesting that both PhPH3 and VvWRKY26 might somehow be implicated in fertility determinacy. Such a role could be related to its regulative function on PA synthesis. In fact, it has been proposed that the Arabidopsis homolog TTG2 determines primarily cell elongation of the integument during seed development likely through the control of tannin synthesis that accumulates in these cell layers (Garcia et al., 2005). Dilkes et al. (2008) demonstrated that the role of AtTTG2 in seed endosperm development and PA synthesis has determinant implications in controlling the lethality of progenies derived by interploidy crosses. It is therefore plausible to hypothesize that VvWRKY26 functions as PA regulator and may also affect seed development and plant fertility in grapevine.
Clear signals of VvWRKY26 mRNAs were specifically detected in the outer layers of berry skin after véraison when anthocyanins start to accumulate, indicating that VvWRKY26 may contribute to the synthesis of anthocyanins, a role that would mimic what has been observed in petunia corollas. The contribution of VvWRKY26 to the regulative mechanism of anthocyanin accumulation in grape and its role in relation to the known MBW complex of regulators (Walker et al., 2007; Hichri et al., 2010; Matus et al., 2010) controlling anthocyanin biosynthesis in grape berry skin deserve more attention.
Finally, considering that “beyond fruit” the expression of VvWRKY26 was detected in many plant organs, mainly during the early development, it must be postulated that its biochemical/developmental regulative roles are widespread in most organs of the grapevine. Future experiments are needed to address this issue.
Final Remarks
In this work we introduced VvWRKY26 as a new member of the TTG2-like protein clade. We shed light on VvWRKY26 multiple regulative functions in grapevine, suggesting above all that the high homology with AtTTG2 and PhPH3 can reflect similar regulative functions. Results obtained in the native species allowed us to propose VvWRKY26 as a putative regulator of vacuolar transport and acidification in grapevine but also to glimpse its involvement in other processes including flavonoid biosynthesis with a particular role in PA deposition. We demonstrated that VvWRKY26 could participate in the vacuolar acidification pathway through the regulation of an ATPase pump system likely conserved, at least partly, in grapevine, petunia and Arabidopsis. In grapevine seeds and berry skin, vacuolar acidification possibly represents a driving force for PA transport and accumulation in vacuoles, but in berry pulp this mechanism could play a key role in determining final berry juice acidity. Further investigations of these aspects would represent an important advance in the possibility to control the final berry quality traits for winemaking.
Author Contributions
AA and EC coordinated the study, discussed the results and wrote the manuscript. AA generated grapevine plantlets over-expressing VvWRKY26, performed both petunia and grapevine transcriptomic analyses, and performed berry fixation and embedding for in situ hybridization. EC performed the phylogenetic analysis, expression and correlation analyses, and characterized petunia ph3/35S:VvWRKY26 complemented lines. SZ interpreted the microarray data, supervised the study and drafted the manuscript. LF generated petunia ph3/35S:VvWRKY26 complemented lines. MB performed in situ hybridization. BR supervised in situ hybridization and contributed in drafting the manuscript. GT designed and supervised the study, interpreted the data and drafted the manuscript.
Funding
This work benefited from the networking activities coordinated under the EU-funded COST ACTION FA1106 “An integrated systems approach to determine the developmental mechanisms controlling fleshy fruit quality in tomato and grapevine”, from FUR granted by the University of Verona to SZ and GBT and from “Ricerca scientifica Ex-60%” to BR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Mario Pezzotti (University of Verona, Italy) for supporting this study and for critical discussions, Francesca Quattrocchio and Ronald Koes (University of Amsterdam, The Netherlands) for providing the petunia ph3 mutant line and for helpful discussions, Flavia Guzzo (University of Verona, Italy) for helping in the preparation of tissue slides for ISH, and Fabio Finotti (University of Verona, Italy) for taking care of the plants.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01979/full#supplementary-material
References
Andrés, Z., Pérez-Hormaeche, J., Leidi, E. O., Schlücking, K., Steinhorst, L., McLachlan, D. H., et al. (2014). Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc. Natl. Acad. Sci. U.S.A. 111, e1806–1814. doi: 10.1073/pnas.1320421111
Appelhagen, I., Nordholt, N., Seidel, T., Spelt, K., Koes, R., Quattrochio, F., et al. (2015). TRANSPARENT TESTA 13 is a tonoplast P3A -ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J. 82, 840–849. doi: 10.1111/tpj.12854
Aprile, A., Federici, C., Close, T. J., De Bellis, L., Cattivelli, L., and Roose, M. L. (2011). Expression of the H+-ATPase AHA10 proton pump is associated with citric acid accumulation in lemon juice sac cells. Funct. Integr. Genomics 11, 551–563. doi: 10.1007/s10142-011-0226-3
Bailey, L. T., Bodén, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Begheldo, M., Ditengou, F. A., Cimoli, G., Trevisan, S., Quaggiotti, S., Nonis, A., et al. (2013). Whole-mount in situ detection of microRNAs on Arabidopsis tissues using Zip Nucleic Acid probes. Anal. Biochem. 434, 60–66. doi: 10.1016/j.ab.2012.10.039
Binkert, M., Kozma-Bognár, L., Terecskei, K., De Veylder, L., Nagy, F., and Ulm, R. (2014). UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26, 4200–4213. doi: 10.1105/tpc.114.130716
Bogs, J., Downey, M. O., Harvey, J. S., Ashton, A. R., Tanner, G. J., and Robinson, S. P. (2005). Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 139, 652–663. doi: 10.1104/pp.105.064238
Bogs, J., Ebadi, A., McDavid, D., and Robinson, S. P. (2006). Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol. 140, 279–291. doi: 10.1104/pp.105.073262
Bogs, J., Jaffé, F. W., Takos, A. M., Walker, A. R., and Robinson, S. P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143, 1347–1361. doi: 10.1104/pp.106.093203
Brand, L. H., Fischer, N. M., Harter, K., Kohlbacher, O., and Wanke, D. (2013). Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 4, 9764–9778. doi: 10.1093/nar/gkt732
Carbonell-Bejerano, P., Diago, M. P., Martínez-Abaigar, J., Martínez-Zapater, J. M., Tardáguila, J., and Nú-ez-Olivera, E. (2014). Solar ultraviolet radiation is necessary to enhance grapevine fruit ripening transcriptional and phenolic responses. BMC Plant 14:183. doi: 10.1186/1471-2229-14-183
Cavallini, E., Matus, J. T., Finezzo, L., Zenoni, S., Loyola, R., Guzzo, F., et al. (2015). The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470. doi: 10.1104/pp.114.256172
Cavallini, E., Zenoni, S., Finezzo, L., Guzzo, F., Zamboni, A., Avesani, L., et al. (2014). Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant Cell Physiol. 55, 517–534. doi: 10.1093/pcp/pct190
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., and Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Dal Santo, S., Commisso, M., D'Incà, E., Anesi, A., Stocchero, M., Zenoni, S., et al. (2016). The terroir concept interpreted through grape berry metabolomics and transcriptomics. J. Vis. Exp. 116:e54410. doi: 10.3791/54410
Davies, C., Shin, R., Liu, W., Thomas, M. R., and Schachtman, D. P. (2006). Transporters expressed during grape berry (VitisviniferaL.). development are associated with an increase in berry size and berry potassium accumulation. J. Exp. Bot. 57, 3209–3216. doi: 10.1093/jxb/erl091
Debeaujon, I., Peeters, A. J. M., Leon-Kloosterziel, K. M., and Korneef, M. (2001). The TRANSPARENT TESTA 12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–871. doi: 10.1105/tpc.13.4.853
Deluc, L., Bogs, J., Walker, A. R., Ferrier, T., Decendit, A., Merillon, J. M., et al. (2008). The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 147, 2041–2053. doi: 10.1104/pp.108.118919
Dilkes, B. P., Spielman, M., Weizbauer, R., Watson, B., Burkart-Waco, D., et al. (2008). The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol. 6:e308. doi: 10.1371/journal.pbio.0060308
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., Rushton, P. J., Schmelzer, E., Hahlbrock, K., and Somssich, I. E. (1999). Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699. doi: 10.1093/emboj/18.17.4689
Faraco, M., Spelt, C., Bliek, M., Verweij, W., Hoshino, A., Espen, L., Prinsi, B., et al. (2014). Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color. Cell Rep. 6, 32–43. doi: 10.1016/j.celrep.2013.12.009
Fasoli, M., Dal Santo, S., Zenoni, S., Tornielli, G. B., Farina, L., Zamboni, A., et al. (2012). The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24, 3489–3505. doi: 10.1105/tpc.112.100230
Garcia, D., Fitz Gerald, J. N., and Berger, F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17, 52–60. doi: 10.1105/tpc.104.027136
Gonzalez, A., Brown, M., Hatlestad, G., Akhavan, N., Smith, T., Hembd, A., et al. (2016). TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 419, 54–63. doi: 10.1016/j.ydbio.2016.03.031
Gonzalez, A., Mendenhall, J., Huo, Y., and Lloyd, A. (2009). TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 325, 412–421. doi: 10.1016/j.ydbio.2008.10.005
Guan, Y., Meng, X., Khanna, R., LaMontagne, E., Liu, Y., and Zhang, S. (2014). Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 10:e1004384. doi: 10.1371/journal.pgen.1004384
Guillaumie, S., Goffner, D., Barbier, O., Martinant, J. P., Pichon, M., and Barrière, Y. (2008). Expression of cell wall related genes in basal and ear internodes of silking brown-midrib-3, caffeic acid O-methyltransferase (COMT) down-regulated, and normal maize plants. BMC Plant Biol. 8:71. doi: 10.1186/1471-2229-8-71
Guillaumie, S., Mzid, R., Mechin, V., Léon, C., Hichri, I., Destrac-Irvine, A., et al. (2010). The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol. Biol. 72, 215–234. doi: 10.1007/s11103-009-9563-1
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., et al. (2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 65, 1513–1528. doi: 10.1093/jxb/eru007
Guo, R., Yu, F., Gao, Z., An, H., Cao, X., and Guo, X. (2011). GhWRKY3, a novel cotton (Gossypium hirsutum L.) WRKY gene, is involved in diverse stress responses. Mol. Biol. Rep. 38, 49–58. doi: 10.1007/s11033-010-0076-4
Hammargren, J., Rosenquist, S., Jansson, C., and Knorpp, C. (2008). A novel connection between nucleotide and carbohydrate metabolism in mitochondria: sugar regulation of the Arabidopsis nucleoside diphosphate kinase 3a gene. Plant Cell Rep. 27, 529–534. doi: 10.1007/s00299-007-0486-5
Harris, N. N., Luczo, J. M., Robinson, S. P., and Walker, A. R. (2013). Transcriptional regulation of the three grapevine chalcone synthase genes and their role in flavonoid synthesis in Shiraz. Aust. J. Grape Wine Res. 19, 221–229. doi: 10.1111/ajgw.12026
Hichri, I., Heppel, S. C., Pillet, J., Léon, C., Czemmel, S., Delrot, S., et al. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523. doi: 10.1093/mp/ssp118
Huang, Y. F., Vialet, S., Guiraud, J. L., Torregrosa, L., Bertrand, Y., Cheynier, V., et al. (2014). A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 201, 795–809. doi: 10.1111/nph.12557
Ishida, T., Hattori, S., Sano, R., Inoue, K., Shirano, Y., Hayashi, H., et al. (2007). Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19, 2531–2543. doi: 10.1105/tpc.107.052274
Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 50 upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/BF00282746
Jiang, W., Wu, J., Zhang, Y., Yin, L., and Lu, J. (2015). Isolation of a WRKY30 gene from Muscadiniarotundifolia (Michx) and validation of its function under biotic and abiotic stresses. Protoplasma 252, 1361–1374. doi: 10.1007/s00709-015-0769-6
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Jiang, W., and Yu, D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9:96. doi: 10.1186/1471-2229-9-96
Johnson, C. S., Kolevski, B., and Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. doi: 10.1105/tpc.001404
Khater, F., Fournand, D., Vialet, S., Meudec, E., Cheynier, V., and Terrier, N. (2012). Identification and functional characterization of cDNAs coding for hydroxybenzoate/hydroxycinnamate glucosyltransferases co-expressed with genes related to proanthocyanidin biosynthesis. J. Exp. Bot. 63, 1201–1214. doi: 10.1093/jxb/err340
Kim, E. J., Kwak, J. M., Uozumi, N., and Schroeder, J. I. (1998). AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10, 51–62. doi: 10.1105/tpc.10.1.51
Kunz, H. H., Gierth, M., Herdean, A., Satoh-Cruz, M., Kramer, D. M., Spetea, C., et al. (2014). Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 7480–7485. doi: 10.1073/pnas.1323899111
Lapis-Gaza, H. R., Jost, R., and Finnegan, P. M. (2014). Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1-8 and PHT1-9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 14:334. doi: 10.1186/s12870-014-0334-z
Li, H., Xu, Y., Xiao, Y., Zhu, Z., Xie, X., Zhao, H., et al. (2010). Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta 232, 1325–1337. doi: 10.1007/s00425-010-1258-y
Li, Q., Yin, M., Li, Y., Fan, C., Yang, Q., Wu, J., et al. (2015). Expression of Brassica napus TTG2, a regulator of trichome development, increases plant sensitivity to salt stress by suppressing the expression of auxin biosynthesis genes. J. Exp. Bot. 66, 5821–5836. doi: 10.1093/jxb/erv287
Li, S. F., Milliken, O. N., Pham, H., Seyit, R., Napoli, R., Preston, J., et al. (2009). The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21, 72–89. doi: 10.1105/tpc.108.063503
Li, Y., Provenzano, S., Bliek, M., Spelt, C., Appelhagen, I., Machado de Faria, L., et al. (2016). Evolution of tonoplast P-ATPase transporters involved in vacuolar acidification. New Phytol. 211, 1092–1107. doi: 10.1111/nph.14008
Liu, H., Yang, W., Liu, D., Han, Y., Zhang, A., and Li, S. (2011). Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol. Biol. Rep. 38, 417–427. doi: 10.1007/s11033-010-0124-0
Liu, L., Gregan, S., Winefield, C., and Jordan, B. (2015). From UVR8 to flavonol synthase: UV-B-induced gene expression in Sauvignon blanc grape berry. Plant Cell Environ. 38, 905–919. doi: 10.1111/pce.12349
Loyola, R., Herrera, D., Mas, A., Wong, D. C., Höll, J., Cavallini, E., Amato, A., et al. (2016). The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J. Exp. Bot. 67, 5429–5445. doi: 10.1093/jxb/erw307
Marchive, C., Léon, C., Kappel, C., Coutos-Thévenot, P., Corio-Costet, M. F., Delrot, S., et al. (2013). Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS ONE 8:e54185. doi: 10.1371/journal.pone.0054185
Marchive, C., Mzid, R., Deluc, L., Barrieu, F., Pirrello, J., Gauthier, A., et al. (2007). Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J. Exp. Bot. 58, 1999–2010. doi: 10.1093/jxb/erm062
Matsubara, K., Chen, S., Lee, J., Kodama, H., Kokubun, H., Watanabe, H., et al. (2006). PCR-Based markers for the genotype identification of Flavonoid-3′5′-hydroxylase genes governing floral anthocyanin biosynthesis in commercial petunias. Breed. Sci. 56, 389–397. doi: 10.1270/jsbbs.56.389
Matus, J. T., Poupin, M. J., Cañón, P., Bordeu, E., Alcalde, J. A., and Arce-Johnson, P. (2010). Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitisvinifera L.). Plant Mol. Biol. 72, 607–620. doi: 10.1007/s11103-010-9597-4
Maurino, V. G., Grube, E., Zielinski, J., Schild, A., Fischer, K., and Flügge, U. I. (2006). Identification and expression analysis of twelve members of the nucleobase-ascorbate transporter (NAT) gene family in Arabidopsis thaliana. Plant Cell Physiol. 47, 1381–1393. doi: 10.1093/pcp/pcl011
Merz, P. R., Moser, T., Höll, J., Kortekamp, A., Buchholz, G., Zyprian, E., et al. (2015). The transcription factor VvWRKY33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmoparaviticola. Physiol. Plant. 153, 365–380. doi: 10.1111/ppl.12251
Milner, I. D., Ho, L. C., and Hall, J. L. (1995). Properties of proton and sugar transport at the tonoplast of tomato (Lycopersicon esculentum) fruit. Physiol. Plant. 94, 399–410. doi: 10.1111/j.1399-3054.1995.tb00945.x
Montefiori, M., Brendolise, C., Dare, A. P., Lin-Wang, K., Davies, K. M., Hellens, R. P., et al. (2015). In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 66, 1427–1436. doi: 10.1093/jxb/eru494
Müller, M. L., Irkens-Kiesecker, U., Rubinstein, B., and Taiz, L. (1996). On the mechanism of hyperacidifcation in lemon. J. Biol. Chem. 271, 1916–1924. doi: 10.1074/jbc.271.4.1916
Nieves-Cordones, M., Alemán, F., Martínez, V., and Rubio, F. (2014). K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 171, 688–695. doi: 10.1016/j.jplph.2013.09.021
Pesch, M., Dartan, B., Birkenbihl, R., Somssich, I. E., and Hülskamp, M. (2014). Arabidopsis TTG2 regulates TRY expression through enhancement of activator complex-triggered activation. Plant Cell 26, 4067–4083. doi: 10.1105/tpc.114.129379
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Provenzano, S. (2011). The Genetics of Anthocyanin Production, Accumulation and Display: A Comparative Study in Different Species. Ph.D. Thesis, Vrije Universiteit Amsterdam VU University Amsterdam.
Provenzano, S., Spelt, C., Hosokawa, S., Nakamura, N., Brugliera, F., Demelis, L., et al. (2014). Genetic control and evolution of anthocyanin methylation. Plant Physiol. 165, 962–977. doi: 10.1104/pp.113.234526
Quattrocchio, F., Verweij, C. W., Kroon, A., Spelt, C., Mol, J., and Koes, R. (2006). PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18, 1274–1291. doi: 10.1105/tpc.105.034041
Quattrocchio, F., Wing, J. F., van der Woude, K., Mol, J. N. M., and Koes, R. (1998). Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13, 475–488. doi: 10.1046/j.1365-313X.1998.00046.x
Ruffner, H. P. (1982). Metabolism of tartaric and malic acids in Vitis: a review - Part B. Vitis 21, 346–358.
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. X. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schluttenhofer, C., and Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Sharma, T., Dreyer, I., and Riedelsberger, J. (2013). The role of K+ channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front. Plant Sci. 4:224. doi: 10.3389/fpls.2013.00224
Suzuki, Y., Maeshima, M., and Yamaki, S. (1999). Molecular cloning of vacuolar H+ -pyrophosphatase and its expression during the development of pear fruit. Plant Cell Physiol. 8, 900–904. doi: 10.1093/oxfordjournals.pcp.a029620
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Terrier, N., Sauvage, F. X., Ageorges, A., and Romieu, C. (2001). Changes in acidity and in proton transport at the tonoplast of grape berries during development. Planta 213, 20–28. doi: 10.1007/s004250000472
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verriès, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Turck, F., Zhou, A., and Somssich, I. E. (2004). Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to Its native promoter and the defense-related gene PcPR1-1 in Parsley. Plant Cell 16, 2573–2585. doi: 10.1105/tpc.104.024810
van der Meer, I. M. (1999). Agrobacterium-mediated transformation of Petunia leaf disks. Methods Mol. Biol. 111, 327–334.
Versaw, W. K., and Harrison, M. J. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14, 1751–1766. doi: 10.1105/tpc.002220
Verweij, C. W., Spelt, C., Di Sansebastiano, G. P., Vermeer, J., Reale, L., Ferranti, F., et al. (2008). An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat. Cell Biol. 10, 1456–1462. doi: 10.1038/ncb1805
Verweij, C. W., Spelt, C. E., Bliek, M., de Vries, M., Wit, N., Faraco, M., et al. (2016). Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in Arabidopsis. Plant Cell 28, 786–803. doi: 10.1105/tpc.15.00608
Walker, A. R., Lee, E., Bogs, J., McDavid, D. A., Thomas, M. R., and Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. doi: 10.1111/j.1365-313X.2006.02997.x
Wang, L., Zhu, W., Fang, L., Sun, X., Su, L., Liang, Z., et al. (2014). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 14:103. doi: 10.1186/1471-2229-14-103
Wang, M., Vannozzi, A., Wang, G., Liang, Y. H., Tonielli, G. B., Zenoni, S., Cavallini, E., et al. (2014). Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 1:14016. doi: 10.1038/hortres.2014.16
Wong, D. C. J., Sweetman, C., Drew, D. P., and Ford, C. M. (2013). VTCdb: a gene co-expression database for the crop species Vitis vinifera (grapevine) BMC Genomics 14:882. doi: 10.1186/1471-2164-14-882
Xie, Z., Zhang, Z. L., Zou, X., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Yin, R., Arongaus, A. B., Binkert, M., and Ulm, R. (2015). Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell 27, 202–213. doi: 10.1105/tpc.114.133868
Zenoni, S., D'Agostino, N., Tornielli, G. B., Quattrocchio, F., Chiusano, M. L., Koes, R., et al. (2011). Revealing impaired pathways in the an11 mutant by high-throughput characterization of Petunia axillaris and Petunia inflata transcriptomes. Plant J. 68, 11–27. doi: 10.1111/j.1365-313X.2011.04661.x
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. doi: 10.1186/1471-2148-5-1
Keywords: flavonoids, grapevine, petunia, vacuolar acidification, WRKY
Citation: Amato A, Cavallini E, Zenoni S, Finezzo L, Begheldo M, Ruperti B and Tornielli GB (2017) A Grapevine TTG2-Like WRKY Transcription Factor Is Involved in Regulating Vacuolar Transport and Flavonoid Biosynthesis. Front. Plant Sci. 7:1979. doi: 10.3389/fpls.2016.01979
Received: 08 November 2016; Accepted: 13 December 2016;
Published: 05 January 2017.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Lingfei Xu, Northwest A&F University, ChinaJean-Marc Routaboul, Institut National de la Recherche Agronomique, France
Copyright © 2017 Amato, Cavallini, Zenoni, Finezzo, Begheldo, Ruperti and Tornielli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giovanni Battista Tornielli, giovannibattista.tornielli@univr.it
†These authors have contributed equally to this work.
 Alessandra Amato
Alessandra Amato Erika Cavallini
Erika Cavallini Sara Zenoni
Sara Zenoni Laura Finezzo1
Laura Finezzo1 Maura Begheldo
Maura Begheldo Benedetto Ruperti
Benedetto Ruperti Giovanni Battista Tornielli
Giovanni Battista Tornielli