- 1National Engineering Research Center of Plant Space Breeding, South China Agricultural University, Guangzhou, China
- 2Department of Rice Breeding, Jiamusi Rice Research Institute of Heilongjiang Academy of Agricultural Sciences, Jiamusi, China
- 3Department of Plant Breeding, College of Agricultural, South China Agricultural University, Guangzhou, China
- 4Plant Protection Research Institute Guangdong Academy of Agricultural Sciences/Guangdong Provincial key Laboratory of High Technology for Plant Protection, Guangzhou, China
Although adenosine monophosphate (AMP) binding domain is widely distributed in multiple plant species, detailed molecular functions of AMP binding proteins (AMPBPs) in plant development and plant-pathogen interaction remain unclear. In the present study, we identified an AMPBP OsAAE3 from a previous analysis of early responsive genes in rice during Magnaporthe oryzae infection. OsAAE3 is a homolog of Arabidopsis AAE3 in rice, which encodes a 4-coumarate-Co-A ligase (4CL) like protein. A phylogenetic analysis showed that OsAAE3 was most likely 4CL-like 10 in an independent group. OsAAE3 was localized to cytoplasm, and it could be expressed in various tissues. Histochemical staining of transgenic plants carrying OsAAE3 promoter-driven GUS (β-glucuronidase) reporter gene suggested that OsAAE3 was expressed in all tissues of rice. Furthermore, OsAAE3-OX plants showed increased susceptibility to M. Oryzae, and this finding was attributable to decreased expression of pathogen-related 1a (PR1) and low level of peroxidase (POD) activity. Moreover, OsAAE3 over-expression resulted in increased content of H2O2, leading to programmed cell-death induced by reactive oxygen species (ROS). In addition, OsAAE3 over-expression repressed the floret development, exhibiting dramatically twisted glume and decreased fertility rate of anther. Meanwhile, the expressions of lignin biosynthesis genes were significantly decreased in OsAAE3-OX plants, thereby leading to reduced lignin content. Taken together, OsAAE3 functioned as a negative regulator in rice blast resistance, floret development, and lignin biosynthesis. Our findings further expanded the knowledge in functions of AMBPs in plant floret development and the regulation of rice-fungus interaction.
Introduction
Adenosine monophosphate (AMP) binding domain-containing proteins widely exist in various plant species, and this family consists of members with diverse functions, including luciferases, peptide antibiotic synthetases, acetyl-CoA synthetases (ACSs), acyl-CoA synthetases, 4-coumarate-CoA ligases (4CLs), and various other closely-related synthetases (Shockey et al., 2000; Stremmel et al., 2001; Shockey and Browse, 2011). Members of this superfamily catalyze the initial adenylation of a carboxylate to form an acyl-AMP intermediate, followed by a secondary partial reaction, most commonly the formation of a thioester (Can et al., 2014). ACSs play a crucial role in both de novo synthesis and modification of existing lipids, and the resulting products also participate in the regulation of plant growth and development (Sasaki and Nagano, 2004; Souza et al., 2008). AMP-binding domain-containing 4CLs are critical enzymes in the phenylpropanoid metabolism pathway, which drive the carbon flow from primary metabolism to different branches of secondary metabolism in plant. 4CLs in Arabidopsis have overlapping and distinct roles in phenylpropanoid metabolism. 4CL1 accounts for the majority of the total 4CL activity, and loss of 4CL1 leads to reduced lignin content without growth defect (Soltani et al., 2006). 4CL3 is expressed in a broad range of cell types, and it probably has an extra function in flavonoid metabolism. In addition, free fatty acids released from the plastids become metabolically available when they are converted to their corresponding Co-A thioesters (Li et al., 2015). This activation is induced by long-chain acyl-coenzyme asynthetases (LACSs). LACS4 and LACS9 double-mutants have shown to strongly reduce biosynthesis of endoplasmic reticulum-derived lipid precursors, which are necessary substrates for glycolipid synthesis in the plastids (Jessen et al., 2011). The expression of rice OsBIABP1 is activated by M. oryzae infection, which may be a defense-related AMP-binding protein (AMPBP) that is involved in the regulation of defense response through SA and/or JA/ET signaling pathways. However, functions of those genes remain unexplored in rice (Zhang et al., 2009).
As a polyphenolic polymer, lignin is accumulated and deposited in cell wall, and this accumulation enhances the ability of the cell wall and provides mechanized protection for the plasma membrane-wrapped protoplasm (Zhao, 2016). However, the lignin deposition is highly dependent on the cell type, tissue, developmental stage and plant species. As part of the normal differentiation and function of specific cell types, lignification also serves as an integral feature of restriction to plant non-woody tissues (Barros et al., 2015). Lignin biosynthesis can be triggered as a response to various biotic and abiotic stresses in cells. Evidence has clearly illustrated that lignin biosynthesis genes play crucial roles in basal defense and normal growth of plants (Wang and Balint-Kurti, 2016). PALs can catalyze the lignin precursor phenylalanine and transform it into cinnamic acid in lignin biosynthesis pathway (Pascual et al., 2016). OsPAL4 is able to improve broad-spectrum disease resistance in rice by increasing the expression of OsPAL2 and repressing the expression of unlinked OsPAL6 (Tonnessen et al., 2015). Cinnamyl alcohol dehydrogenase (CAD) catalyzes the last step of monolignol biosynthesis. OsCAD2 is largely responsible for monolignol biosynthesis in rice stem, while mutant plant exhibits drastically reduced CAD activity and undetectable sinapyl alcohol dehydrogenase activity (Zhang et al., 2006; Hirano et al., 2012). 4CL mediates the activation of a number of hydroxycinnamic acids for the biosynthesis of monolignols and other phenolic secondary metabolites in higher plants. Suppression of Os4CL3 expression results in significant lignin reduction, impaired plant growth, decreased panicle fertility and other morphological changes (Gui et al., 2011).
Rice blast is caused by the ascomycetous fungus Magnaporthe oryzae, which is one of the most serious and devastating epiphytic diseases in rice production worldwide (Ashkani et al., 2015). Currently, more than 24 major R genes conferring resistance to M. oryzae have been identified in rice, including Pi-ta (Jia et al., 2016), Pi-k (Wu et al., 2014), and Pb1 (Inoue et al., 2013), and modulation of these R genes significantly maintains and improves the grain yield and quality in modern rice cultivars. These major R genes mainly encode the nucleotide-binding site-leucine-rich repeat (NBS-LRR) proteins that recognize diverse effectors (Avirulence proteins, Avr) and activate the downstream immunity response (DeYoung and Innes, 2006; Marone et al., 2013). Meanwhile, the rice-blast system has been developed as a model to study the mechanism of pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) in plant-fungus interaction (Andolfo and Ercolano, 2015; Stael et al., 2015). To date, the underlying molecular mechanism of rice resistance to diseases has been illustrated in multiple levels, including transcriptome, proteome, post-transcriptional modification and epigenetic regulation (Miah et al., 2013; Xu et al., 2015; Li et al., 2016; Sharma et al., 2016). Various regulatory factors mediating the blast resistance in rice have been identified by a combination of biochemical, genetic and high-throughput sequencing approaches. However, functional roles of single gene in complex defense network of rice blast still need to be further elucidated.
In the present study, we identified and characterized an Arabidopsis AAE3 (Foster et al., 2012) homolog in rice. Arabidopsis AAE3 encodes a specific cytoplasmic oxalyl-CoA synthetase containing the conserved AMP-binding domain, which is required for oxalate degradation, normal seed development and defense against an oxalate-producing fungal pathogen. Here, the cytoplasmic 4CL like protein OsAAE3 (LOC_Os04g58710) was identified from an analysis of transcriptome and proteome profile. In leaf tissue, increased OsAAE3 activity was significantly correlated with decreased resistance to rice blast and reduced lignin content. Furthermore, our results also showed that OsAAE3 possessed multiple potential roles in metabolism and plant anther development.
Materials and Methods
Plant Materials and Growth Conditions
Oryzae sativa japonica cultivar Pik-H4 NILs was used as the wild-type rice strain in this study. Pik-H4 NIL contains the Pik-H4 resistance gene (an allele of Pik locus) in the susceptible cultivar LTH background. The M. oryzae race GDYJ7, one of the primary M. oryzae races found in Guangdong Province, China, is incompatible with Pik-H4.
Sixth-leaf-stage rice seedlings were used in the present study, which were grown under natural light in a greenhouse at 26°C for inoculation of the rice blast fungus. Freshly prepared M. oryzae spores (1 × 105 conidia/mL 0.02% v/v gelatin) were sprayed onto the rice leaves using an air sprayer. Inoculated plants were kept in a humidity chamber at 28°C, and rice leaves were harvested for RNA extraction at 0, 12, 24, 36, and 48 h after inoculation.
Subcellular Localization Analysis
The full-length OsAAE3 cDNA insert without a stop codon was amplified by PCR. Amplified fragments were digested with XbaI/BamHI and cloned between the cauliflower mosaic virus (CaMV) 35S promoter and the GFP gene. Rice protoplast was isolated from 12-day-old rice seedling stem and sheath. Briefly, 30 rice seedlings were cut into approximately 0.5 mm strips, and then incubated in an enzyme solution (1.5% Cellulase RS, 0.75% Macerozyme R-10, 0.6 M mannitol, 10 mM MES at pH 5.7, 10 mM CaCl2, and 0.1% BSA) for 4–5 h in the dark with gentle shaking (60–80 rpm). After washing twice with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES at pH 5.7), last protoplast was resuspended in MMG solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES at pH 5.7). The resulting OsAAE3-GFP fusion construct and empty GFP vector were transiently co-expressed in rice protoplasts by 40% PEG induction. Fluorescence was examined using laser-scanning confocal microscope (Zhang et al., 2011) (Model LSM 780; Carl Zeiss, Jena, Germany).
GUS Assay
We first cloned about 2 kb promoter region of OsAAE3 from rice genomic DNA. The amplified sequence was inserted into the NcoI/BamHI sites of pCAMBIA1305 vector. The resulting construct was introduced into agrobacterium strain EHA105 and transformed to wild-type (Pik-H4 NIL) calli as described previously. The tissues of the transgenic plants were washed three times with 100 mM NaPO4 buffer (pH 7.0), and incubated with a staining solution [100 mM NaPO4 (pH 7.0), 10 mM EDTA, 2 mM 5-bromo-4-chloro-3-indolyl-b-GlcA, 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, and 0.2% Triton X-100] for 20 min to 24 h at 37°C (Jefferson et al., 1987).
Total RNA Extraction and Real-Time PCR Analysis
Total RNA was extracted from 100 mg of fourth-leaf-stage rice seedling with Trizol Reagent (Invitrogen, Beijing, China), and purified RNA was reversely transcribed into cDNA using PrimeScript RT reagent Kit (Takara, Dalian, China) according to the manufacturer's instructions. The cDNA was quantified by real-time PCR using a 20 μL reaction system by SYBR Premix ExTaq™ (TaKaRa, Dalian, China) on an ABI StepOne Plus system. Table S3 lists the primer sequences used for PCR analysis. Differences in gene expression were expressed as fold change relative to control and calculated using the 2−ΔΔCT method. Each measurement was carried out in triplicate, and the error bars represent SE of the mean of fold changes for three biological replicates.
Generation of the OsAAE3-OX Transgenic Plants
The full-length of OsAAE3 cDNA was isolated by RT-PCR from the leaves of fourth-leaf-stage rice plants using the cDNA F/R primers (Table S3) encompassing the translation start and stop codons. This cDNA insert was digested with BamHI and cloned between the maize ubiquitin promoter and the Nos terminator in the plant expression vector pOX containing the hygromycin resistance gene as a selection maker. Prof. Yaoguang Liu (South China Agricultural University, Guangzhou 510642, China) provided the plant binary vector pOX. pOX-OsAAE3 was then introduced into agrobacterium strain EHA105 and then transformed to wild-type (Pik-H4 NIL) calli as described previously (Hiei et al., 1994). Transgenic rice plants were regenerated from the transformed calli on selection medium containing 50 mg/L hygromycin and 250 mg/L cefotaxime. OsAAE3 levels in the transgenic rice plants were further confirmed with real-time PCR.
Measure POD Activities and H2O2 Content in Fresh Leaves
The enzyme extracts of POD were prepared following the method of Cai et al. (2008) with some modifications. Briefly, 300 mg fresh leaves were frozen and ground in liquid N2, and the powder was mixed with 4 mL 0.05 M PBS (pH 7.8) and transferred into 5-ml tube. After thawing, the tubes were centrifuged at 8000 rpm/min for 15 min, and the supernatant containing the total peroxidase (POD) was collected. The POD activity was measured as the rate of decomposition of H2O2 by POD, with guaiacol as the hydrogen donor, by spectrophotometrically measuring the rate of color development at 436 nm due to guaiacol oxidation (Cai et al., 2008).
Hydrogen peroxide was performed using the Ferric Xylenol Orange method as described previously. Actually, fresh leaf tissue was ground in cold acetone and filtered to remove cellular debris. The supernatants were extracted with CCl4-CHCl3 solution. Then the extract was transferred into a new tube containing 250 μM ferrous ammonium sulfate, 100 μM sorbitol and 100 μM xylenol orange in 25 mM H2SO4. The mixture reacted 30 min in the dark at room temperature, and the absorbance was detected at 560 nm (Gay et al., 1999).
Lignin Content Assay
Briefly, 1 g of fresh leaves was homogenized in 5 mL cold 95% ethanol and centrifuged at 5000 rpm/min for 30 min, and the precipitate was washed by ethanol-hexane solution (1:2, V/V) for three times. After thoroughly dried, the washed precipitate was placed in a glass reaction vial (15 mL) with 5 mL of 25% (v/v) acetyl bromide in acetic acid, sealed with Teflon lined caps, and heated at 70°C for 30 min. After digestion, the vial's contents were quantitatively transferred to a 10-mL volumetric flask containing 0.9 mL of 2 M NaOH, 5 mL of acetic acid and 0.1 mL of 7.5 M hydroxylamine, and the flask was filled to 10 mL with acetic acid. After reaction solution was centrifuged at 1000 g for 7 min, the absorption values of supernatant were determined at 280 nm. According to the standard curve, lignin contents were calculated (Xie et al., 2011).
Results
OsAAE3 Expression Induced by M. oryzae
We have previously compared the global gene expression in resistance line Pik-H4 NILs with the susceptible cultivar LTH after M. oryzae inoculation via a transcriptome-proteome analysis. We identified 61 and 69 genes that were up-regulated and down-regulated in Pik-H4 NILs line, respectively (Table S1). Based on our transcriptome-proteome analysis in Pik-H4 NILs line, Pik-H4 modulates multiple genes involved in diverse biological processes, including defense-related hormone biosynthesis, disease resistance, response stress, photosynthesis, and signal transduction (Figure 1A).
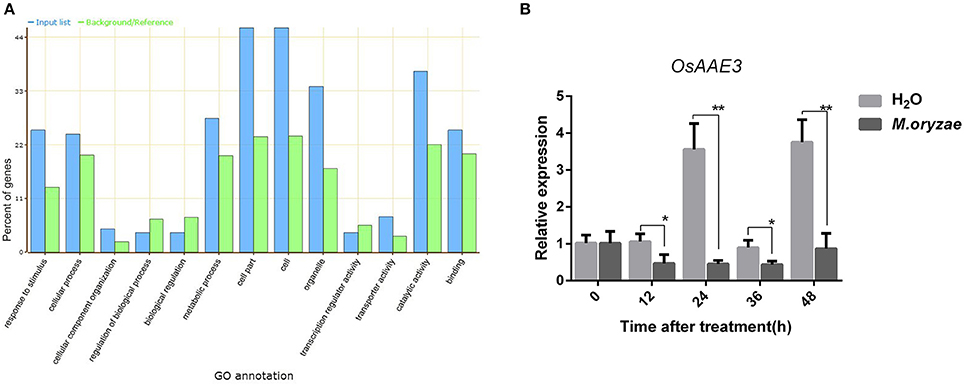
Figure 1. OsAAE3 expression was down-regulated by M. oryzae. (A) GO annotation of differential genes expression in Pik-H4 NILs by transcription-proteomics analysis. (B) OsAAE3 expression induced by M. oryzae after 48 h, Sterile H2O was used as controls. Values shown are means ± SD from three independent experiments, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with control group.
An AMPBP3 (4CL-like protein), named OsAAE3 (LOC_Os04g58710), was identified from the transcriptome and proteome analysis of early responsive genes in rice during M. oryzae infection (Table S1). We then examined the expression pattern of OsAAE3 over a time course of 48 h after inoculation with M. oryzae by quantitative RT-PCR (qRT-PCR). The OsAAE3 expression at the mRNA level was significantly decreased at 12 h and reached its lowest level at 24 h, and then it was maintained at a relatively low level from 24 to 48 h after inoculation with M. oryzae in wild-type plants (Figure 1B). In contrast, the OsAAE3 expression presented circadian rhythmicity pattern during the whole stage after spraying of water.
Identification and Characterization of OsAAE3
The genome sequence and the cDNA fragment encoding of OsAAE3 were isolated from rice using gene-specific primers based on the sequence (LOC_Os04g58710) of the rice genome database (Rice Genome Annotation Project). The genome sequence of OsAAE3 was 2221 bp, full-length coding sequence (CDS) was 1557 bp, which harbored two exons and one intron, and it encoded a protein of 519 amino acid residues with a deduced molecular weight of 54.47 kDa. This result was similar to our DNA sequence analysis (Figures 2A–C).
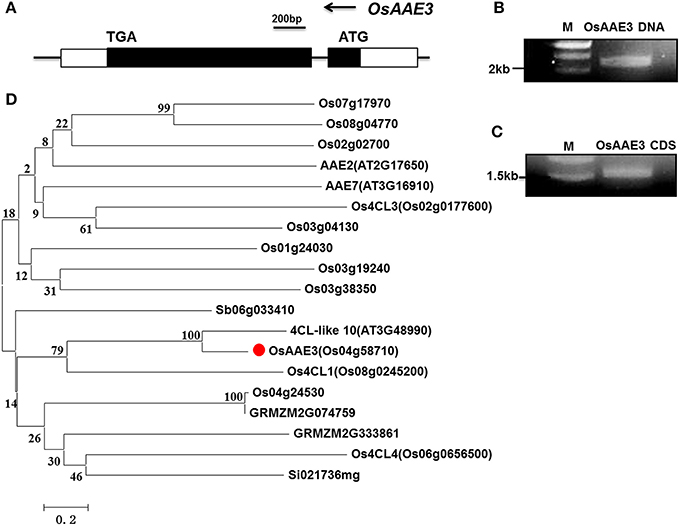
Figure 2. Identification and characterization of OsAAE3. (A) Schematic gene structure of OsAAE3 in rice genome. (B,C) The OsAAE3 DNA and CDS fragments were examined by agarose gel electrophoresis. (D) Comparative phylogenetic analysis of the OsAAE3 proteins in plants. Sequences were aligned using ClustalX. The evolutionary history was inferred using a Neighbor-Joining phylogenetic tree generated with the software MEGA6. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (2000 replicates) is shown next to each branch. Putative OsAAE3 (Os04g58710) members in A. thaliana (AT2G17650, AT3G16910, AT3G48990), O. sativa (Os07g17970, Os08g04770, Os02g02700, Os02g0177600, Os03g04130, Os01g24030, Os03g19240, Os03g38350, Os08g0245200, Os04g24530, Os06g0656500), Sorghum bicolor (Sb06g033410), Zea mays (GRMZM2G074759, GRMZM2G333861), Setaria italica (Si021736 mg).
According to the prediction from Pfam database and comparison with other AMPBPs, amino acid sequence analysis showed that an AMP-binding domain was located from 34 to 439 aa in the OsAAE3 sequence. Furthermore, we identified orthologous protein sequence of OsAAE3 from several plant models, including rice, Arabidopsis, maize, sorghum and soybean. A phylogenetic analysis based on those sequences showed that OsAAE3 was most likely 4CL-like 10 in an independent group (Figure 2D). As indicated from the sequence alignment, there were 90% similarities between OsAAE3 and 4CL-like 10 (Figure S1). Therefore, it is of great interest to verify whether OsAAE3 was indeed involved in monolignol catabolism in the same way as its homolog in Arabidopsis.
OsAAE3 Is Localized to the Cytosol
The AMP-binding domain of OsAAE3 suggested that it was probably localized to the cytosol like its homolog AAE3 in Arabidopsis. Meanwhile, the PSORT database revealed the multi-organelle localization of OsAAE3, including the mitochondrial inner membrane, plasma membrane, Golgi body, and mitochondrial intermembrane space, but the available prediction scores were unreliable. To further investigate the subcellular localization of OsAAE3, we constructed an OsAAE3-GFP fusion protein driven by the CaMV 35S promoter, and the empty GFP was used as the negative control. The resulting vectors 35S:OsAAE3-GFP and GFP were transiently co-transformed into rice protoplast cells with the PEG-mediated procedure. Interestingly, the OsAAE3-GFP fusion protein exhibited similar pattern to the empty GFP control, and the OsAAE3-GFP signal was strongly detected in the cytoplasm of rice protoplast cells (Figure 3). Therefore, the transient expression assay indicated that OsAAE3 encoded a cytoplasmic synthetase like 4CL that could possibly catalyze the lignin degradation.
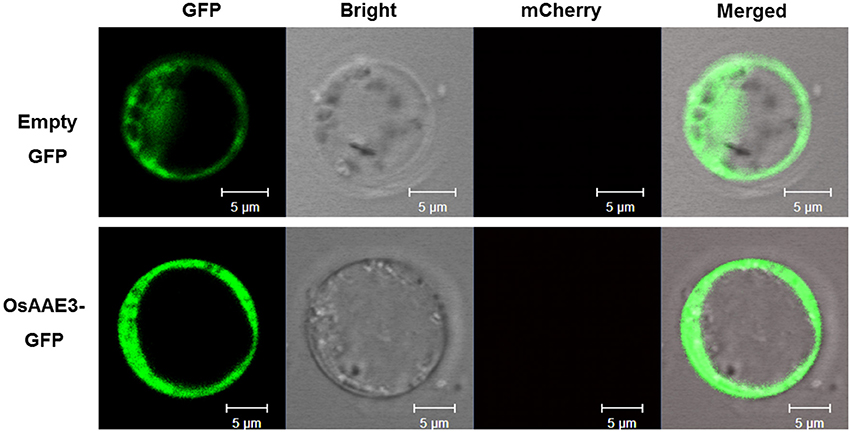
Figure 3. Subcellular localization assay of OsAAE3. Subcellular localization indicated that OsAAE3-GFP signal was strongly detected in the cytoplasm of rice protoplast cell. OsAAE3-GFP and GFP signals are green, Scale bar is 5 μm.
Expression Pattern Analysis of OsAAE3
To evaluate the expression pattern of OsAAE3 in different tissues, total RNA was extracted from root, stem, leaf sheath, leaf blade, young panicle and glume of rice at the heading stage. Semi-quantitative PCR and real-time PCR (RT-PCR) were performed to determine the relative expression of OsAAE3. Semi-quantitative PCR results suggested that the full-length coding sequence of OsAAE3 could be easily amplified from all of tissues without tissue specificity (Figure 4B). OsAAE3 was constitutively expressed in various types of tissues, but its highest expression was detected in leaf blade, followed by root, young panicle, glume, stem, and leaf sheath. Such finding was confirmed by RT-PCR experiment (Figure 4C).
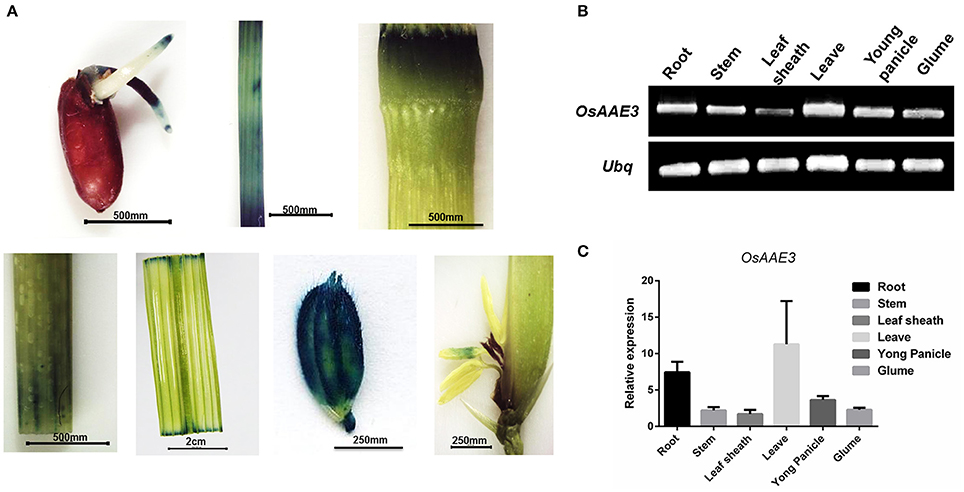
Figure 4. Expression pattern analysis of OsAAE3. (A) Activity determination of the OsAAE3 promoter. GUS staining of germinated seed, stem, node, shoot, blade leaf, glume, and anther. (B,C) Semi-quantitative PCR and real-time PCR determined the relative expression of OsAAE3 in root, stem, leaf sheath, leave, young panicle, and glume.
We further cloned the promoter sequence of OsAAE3 into GUS (β-glucuronidase) reporter system, and then the constructed promoter (OsAAE3)-GUS fusion vector was transformed into rice callus to assess the promoter activity. GUS staining analysis indicated that the OsAAE3 promoter activity was detected in the root, epidermis cells of leaves, stem vascular cells, glume vascular cells and anther (Figure 4A). Taken together, OsAAE3 was expressed in all tissues of the plant.
Generation of the OsAAE3-OX Transgenic Plants
To determine the detailed molecular roles of OsAAE3 in rice physiological and biochemical reactions, we constructed the transgenic rice plants over-expressing OsAAE3 (OsAAE3-OX) under the control of ubiquitin promoter (Figure 5B). The OsAAE3 expression at the mRNA level was significantly increased in transgenic plants (OsAAE3-OX) compared with wild-type plants, and these data were validated using qRT-PCR and semi-quantitative PCR (Figures 5C,D). Over-expression of OsAAE3 repressed the plant growth, showing dwarfing, rolling, and narrow leaves as well as abnormal glumes (Figure 5A and Figure S2). Therefore, based on the observed abnormal phenotypes of OsAAE3-OX plants, we believed that OsAAE3 served as a negative regulator in the regulation of plant development and growth.
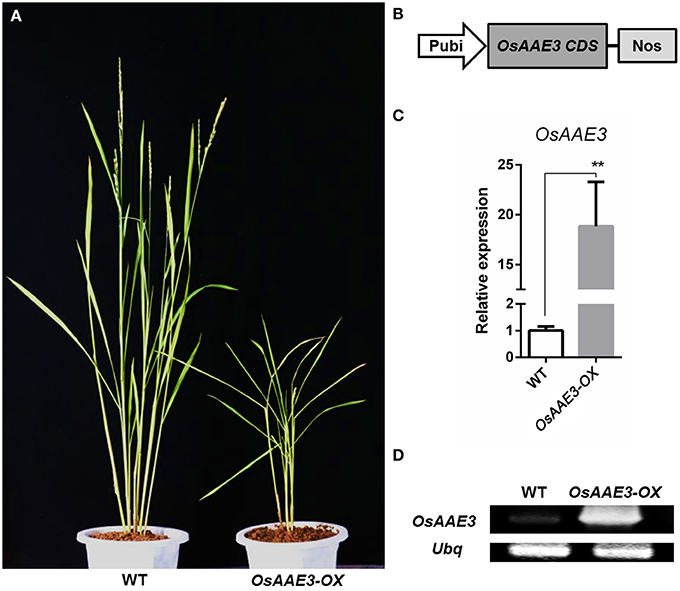
Figure 5. OsAAE3 transgenic plants Identification. (A) The morphology of wild-type and OsAAE3-OX transgenic plants (T1 generation). The photograph was taken about 60 days after heading of OsAAE3 transgenic plants (single line) and wild-type plants. (B) Schematic structure of OsAAE3-OX transgenic vectors construction. (C,D) Expression analysis of OsAAE3 overexpression line measured by semi-quantitative PCR and real-time PCR compared with wild-type. Values shown are means ± SD from three independent experiments, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT.
Over-Expression of OsAAE3 Reduces the Rice Blast Resistance
We next examined resistance of the transgenic plants to M. oryzae in order to elucidate the molecular basis of OsAAE3 in disease resistance in rice. Disease symptoms in plants were quantified at 7 days after inoculation, and the OsAAE3-OX plants exhibited reduced resistance to M. oryzae compared with wild-type plants (Figure 6A). This result indicated that over-expression of OsAAE3 affected the basal level of resistance to M. oryzae in rice. In addition, the disease spots on the leaf surface were intensively associated with expressions of pathogen-related (PR) genes. We further investigated expressions of three PR genes in OsAAE3-OX and wild-type plants by RT-PCR. The data demonstrated that the expressions of PR1a (Mitsuhara et al., 2008) and PR10 (Choi et al., 2015) were down-regulated in OsAAE3-OX plants compared with wild-type pants under the normal growth condition, but the PR10 expression was not significantly decreased in OsAAE3-OX line (Figure 6B). However, the expression of PR1b (Agrawal et al., 2000) was significantly increased in OsAAE3-OX plants (Figure 6C). Therefore, we concluded that blast induced by OsAAE3 over-expression was attributable to the reduced PR1a protein that down-regulated the innate defense response upon fungal invasion.
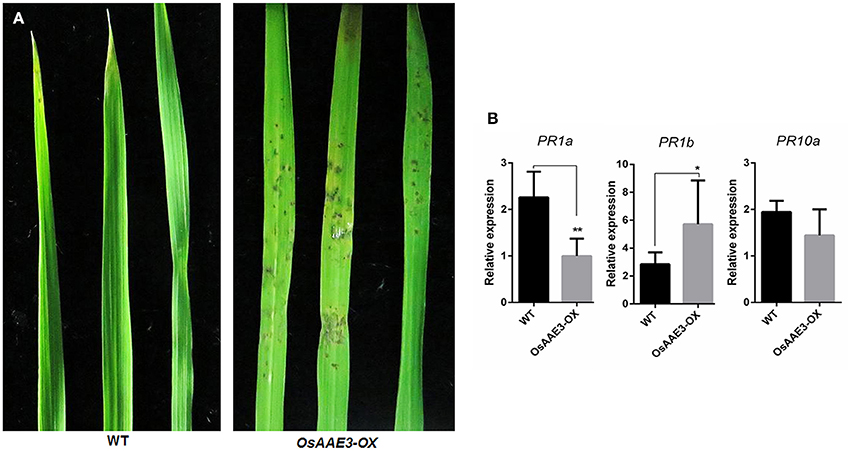
Figure 6. OsAAE3 reduced the rice blast resistance. (A) Photographs of blast fungus-inoculated fourth leaves of OsAAE3 transgenic plants and wild-type at the six-leaf stage. A conidial suspension of blast fungus race GDYJ7 was sprayed on the leaf surfaces, and local lesions were observed 5 days later. (B) Relative expression of PR1a, PR1b, and PR10 in OsAAE3-OX and wild-type plants. Values shown are means ± SD from three independent experiments, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT.
The OsAAE3 ortholog encode the peroxisomal co-enzyme synthetase in maize, sorghum and brachypodium. PODs mainly participate in a broad range of physiological processes, including lignin synthesis, reactive oxygen species (ROS) metabolism and programmed cell-death (PCD) (Kalsoom et al., 2015). ROS are essential to protect plants from various environmental stresses, while excessive accumulation of ROS causes damage to plant cells. Therefore, it remains unclear whether OsAAE3 had effects on the POD activity during the ROS accumulation. We further extracted the total PODs from fresh leaves of OsAAE3-OX and wild-type plants when they grew under common conditions, and then examined the POD activity. The results suggested that the POD activity was ~50% lower in OsAAE3-OX plants compared with wild-type plants (Figure 7B). It has also been proved that intracellular POD activity is inhibited by the accumulated H2O2 level. Furthermore, RT-PCR data indicated that the expressions of POD synthesis-related genes at the mRNA level were significantly decreased in OsAAE3-OX plants (Figure 7A). In addition, we showed that the H2O2 level was significantly increased in OsAAE3-OX plants compared with wild-type plants (Figure 7C). These findings suggested that the over-expression of OsAAE3 induced high concentration of H2O2 via repressing the POD activity, leading to hypersensitive response (HR) and PCD.
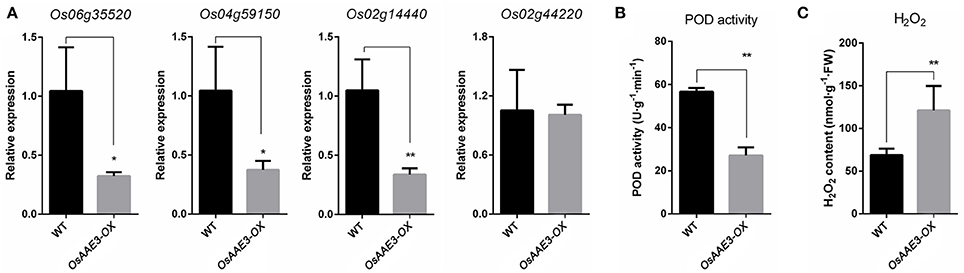
Figure 7. OsAAE3 negatively regulates POD activity during the ROS accumulation. (A) Relative expression of POD synthesis-related genes in OsAAE3-OX and wild-type plants. Values shown are means ± SD from three independent experiments, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT. (B,C) POD activity assay and H2O2 content assay in OsAAE3-OX and wild-type plants. Values shown are means ± SD (n = 10), and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT.
Over-Expression of OsAAE3 Suppresses the Floret Development
Floret is one of most important organ for rice development, which not only determines the growth time from the vegetative stage to the reproductive stage, but also offers sufficient seeds to extend the life of this species. In the present study, the over-expression of OsAAE3 triggered several phenotypic alterations in the leaf, root, tiller number, flowering time, and flower development. The OsAAE3-OX plants obviously displayed an abnormal floret structure. The panicle heads and stalks hardly grew out from the leaf sheath at the heading stage, and each individual floret of the spikelet showed multiple abnormal and twisted glumes as well as increased number of empty glumes (Figures 8A,B). The interior palea was part of glume, and its development was intensively depressed and even disappeared. The spikelet also had an extra lodicule without expansion, leading to greatly impaired floret development.
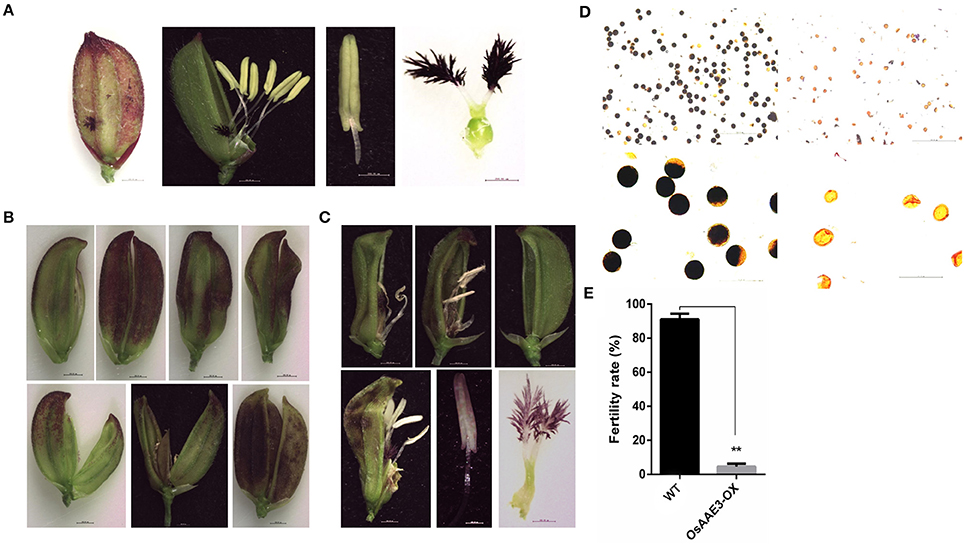
Figure 8. OsAAE3 suppresses the floret development. (A) Flower phenotype of wild-type plant at heading stage. (B,C) Microscopic analysis of OsAAE3-OX plants flower morphology at the heading stage. (D) Potassium iodide staining of pollen grains from the wild type and OsAAE3 transgenic plants. (E) Fertility rate analysis of wild-type and OsAAE3-OX.
Moreover, we examined the morphology of the floret structure in the OsAAE3-OX plants by microscopic analysis. The results revealed that the pistils and anthers also had abnormal morphology at the reproductive stage, and the stalk base of pistils was not expanded after flowering. The anther stalk suffered the external pressure from the twisted glume and showed the twisty phenotype (Figure 8C). Compared with wild-type plants, the anther of OsAAE3-OX plants was deficient and had a pale yellow color, showing wrinkled surface with larger pores (Figure 8C). Furthermore, we found that the basic viability of the pollen was affected by over-expression of OsAAE3 (Figure 8D). The fertility rate of the OsAAE3-OX plants was significantly decreased to nearly 5% compared with 90% in wild-type plants (Figure 8E). These results suggested that OsAAE3 over-expression exerted a great influence on the fertility of pollen via suppressing the anther development.
OsAAE3 Represses the Accumulation of Total Lignin
OsAAE3-encoding protein contains a 4CL like domain, and the 4CL proteins function as key enzymes of phenylpropanoid metabolism in higher plants (Voelker et al., 2010). The question remains unclear whether OsAAE3 regulated the lignin biosynthesis in rice. We first measured expressions of a subset of critical lignin synthesis genes in rice, including 4CL5 (Gui et al., 2011), PAL, PAL4, and Gh2. The results indicated that expressions of PAL, PAL4, and Gh2 at the mRNA level were significantly decreased in OsAAE3-OX plants, while the expression of 4CL5 was obviously increased compared with wild-type plants (Figure 9A). Moreover, examination of PAL activity in wild-type and OsAAE3-OX showed that PAL activity was obviously decreased in OsAAE3 overexpression line (Figure 9D), directly suggesting that OsAAE3 repressed the PAL activity. Therefore, we concluded that OsAAE3 over-expression promoted the expressions of phenylpropanoid catbolic genes and repressed the expressions of lignin biosynthesis genes.
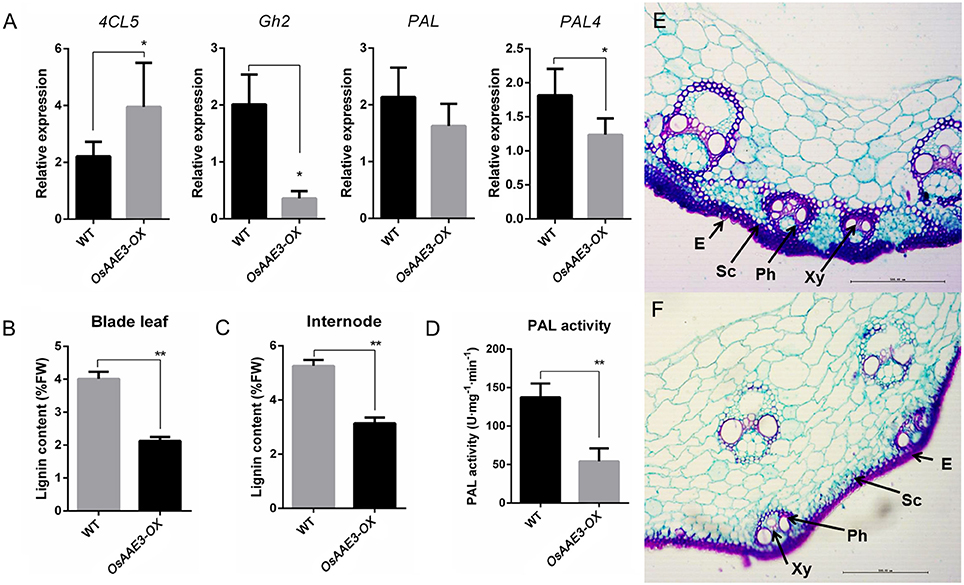
Figure 9. OsAAE3 represses the accumulation of total lignin. (A) Lignin biosynthesis-related genes expression in wild type and OsAAE3-OX. Values shown are means ± SD from three independent experiments, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT. (B,C) Total lignin content assay of blade leaf and internode in wild-type and OsAAE3-OX. Values shown are means ± SD (n = 10), and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT. (D) PAL activity assay in wild-type and OsAAE3-OX. Values shown are means ± SD (n = 5), and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT. (E,F) Lignin staining in wild-type and OsAAE3-OX. E, Epidermis; Ph, phloem; Sc, sclerenchyma cells; Xy, xylem.
Furthermore, we examined the content of total lignin in rice fresh blade leaf and internode at 2 months of age. The data demonstrated that OsAAE3 over-expression affected the lignin biosynthesis (Figures 9B,C). In addition, the rice culms were sectioned and stained for lignin with safranin O/fast green regents. According to the structure of stained cell wall, we found that the lignin content was lower in OsAAE3-OX plants (Figures 9E,F). Taken together, up-regulation of OsAAE3 obviously provoked the accumulation of 4CLs in cells, which predominantly disrupted the lignin biosynthesis.
Discussion
OsAAE3 Is Closely Related to 4CL
OsAAE3 belonged to the large superfamily of AMPBPs in rice, while the amino acid sequence of AAE3 was not closely related to any other members. The most closely related proteins are 4CL like proteins in Arabidopsis, which exhibited 90% sequence similarity to AAE3, suggesting that OsAAE3 had probably evolved into a subsidiary enzyme of 4CL (Figure S1). On the contrary, this clearly stated that OsAAE3 was different from Os4CLs in biological function according to observed phenotype of OsAAE3-OX plants. 4CL ligates coenzyme A (CoA) with hydroxycinnamic acids, such as 4-coumaric and caffeic acids, into hydroxycinnamoyl-CoA thioesters, and these reactions accelerate the process of monolignol biosynthesis for lignification in plant cell walls (Chen et al., 2013). Although the major functions of 4CLs have been broadly explored in Arabidopsis and rice, the detailed roles of 4CL-like proteins remain unclear (Taylor-Teeples et al., 2015). We believed that OsAAE3 was most likely a 4CL, which mediated the phenylpropanoid metabolite. The phenylpropanoid pathway plays a critical role in the plant innate immunity system, which is responsible for offense-related compound biosynthesis, such as the flavonoids, total phenolics, and defense lignin (Zhang and Liu, 2015; Le Roy et al., 2016). Therefore, repression of the phenylpropanoid pathway through OsAAE3 activation resulted in improved plant disease susceptibility. Both the expressions of PAL1 and PAL4 at the mRNA level were decreased in OsAAE3-OX plants, clearly supporting this hypothesis that OsAAE3 was involved in the regulation of phenylpropanoid metabolism.
OsAAE3 Negatively Regulates the Rice Blast Resistance
Fungus-induced rice blast is a potential threat to the rice production, which is characterized by sporadic and unpredictable outbreaks, leading to significant yield loss (Takatsuji, 2014). However, to protect themselves from pathogen invasion, plants have to effectively integrate extracellular and intracellular signals to activate the physiological and biochemical responses by enhancing the hormone defense pathway, switching off the plant growth and regulating the expressions of immunity-related genes and proteins (Lozano-Durán and Zipfel, 2015).
Interestingly, M. oryzae down-regulated the OsAAE3 expression according to the database from transcriptome-proteome analysis, and we further confirmed such result by RT-PCR (Figure 1). However, OsAAE3 over-expression improved the disease susceptibility (Figure 5A), therefore, we supposed that OsAAE3 was a negative regulator in the complex immunity network of rice. Under the normal growth condition, plants are able to synthesize sufficient lignin for whole plant growth. In this case, OsAAE3 utilized its 4CL-like structure to expand the chemical element phenylpropanoid and promote the lignin biosynthesis, and the balance between the phenylpropanoid metabolism and lignin biosynthesis was mediated by OsAAE3 (Figures 9A,D). Therefore, rice had to switch off the OsAAE3 expression under the M. oryzae invasion, and reduced OsAAE3 activities were conducive to transformation of phenylpropanoid into defense-related compounds, such as plant hormone salicylic acid, phytoalexins and total phenolics (Liu et al., 2015). However, OsAAE3 over-expression drastically disturbed the balance, and a large number of disease spots were gradually spread along with plant growth. In addition, OsAAE3 over-expression also down-regulated the expressions of two PR genes, including PR1a and PR10 (Figure 5B), which are tightly correlated with the onset of defense responses against a variety of fungal, viral, and bacterial pathogens (Liu et al., 2015).
Generally speaking, the disease spots on the leaf surface are correlated with the HR, and ROS accumulation may trigger the HR (Pottosin et al., 2014). Actually, plants scavenge destructive free radicals relying on robust antioxidant organelle class III peroxidases (Prxs) (Francoz et al., 2015). Prxs also serve as an important class of enzymes responsible for the stress-induced formation and degradation of ROS (Marjamaa et al., 2009). However, our survey indicated that OsAAE3 over-expression depressed the POD activities and reduced the expressions of POD synthesis-related genes, thereby lowering POD activities and increasing the content of H2O2, which might be the major cause of cell death after M. oryzae inoculation (Figure 7).
Table S2 shows that we identified three potential proteins that also presented in the results of transcription-proteomics analysis, including the AMP-binding protein (LOC_Os04g58710), RRM recognition motif protein (LOC_Os03g25960), and protein of unknown function DUF1296 domain containing protein (LOC_Os01g47430). In addition, we also found that the Pik-H4 was most likely Pb1 to interact with WRKY45 (Inoue et al., 2013), and Pik-H4 was also associated with OsBIHD1 to balance the relation between the growth and blast resistance. These results suggested that the components Pik-H4-OsAAE3 were activated after recognizing the Avr-Pik. As a consequence, the Pik-H4 dissociated from the receptor complex, entered the nucleus and interacted with multiple transcription factors, and those transcription factors precisely controlled the switch of downstream target genes expression. Therefore, we believed that the OsAAE3 abundance was regulated by the transcription factor under the pathogen invasion.
OsAAE3 Involves in Lignin Synthesis Pathway
A large part of host protection against invasion by fungal pathogens relies on a defensive system that is highly coordinated to prevent the spread of pathogens (Ye and Ma, 2016). Furthermore, enhanced deposition of lignin can provide a structural barrier against pathogen spread, and the toxic phenolic precursors produced during lignin biosynthesis or polymerization can directly inhibit pathogen multiplication and movement (Miedes et al., 2014). As a homolog of OsAAE3 in Arabidopsis, AAE3 encodes the oxalyl-CoA synthetase that is required for multiple physiology reactions, including oxalate degradation, seed development and defense against an oxalate-producing fungal pathogen. Here, our findings suggested that OsAAE3 exerted a negative influence on lignin biosynthesis (Figure 9), and the content of lignin was significantly reduced in OsAAE3-OX plants. Meanwhile, GUS activity assay displayed that OsAAE3 was expressed in the anther cell, and we speculated that OsAAE3 over-expression caused the high concentration of H2O2. The accumulation of ROS might activate the process of PCD, and the fertility rate was obviously decreased in OsAAE3-OX plants compared with wild-type plants (Yi et al., 2016). Based on our histological observation of anther's section, we speculated that less accumulation of lignin repressed the tapetum development when mature pollen grain appeared, and this could be another reason of decreased fertility rate in OsAAE3-OX plants.
The questions remain unclear whether OsAAE3 was involved in the oxalate degradation like its homolog in Arabidopsis, and whether OsAAE3 induced dual-directional regulation between immunity network and plant growth pathways. If the answers were positive, there should be other transcriptional factors, which might regulate the OsAAE3 expression after the immune activation by various pathogens. Our future work will focus on OsAAE3-modulated oxalate degradation, and we will identify the transcriptional regulators of OsAAE3. Taken together, our current study further expanded our knowledge in functions of AMBPs in plant development and the regulation of rice blast resistance.
Author Contributions
ZC, JW, TG, and HW conceived and designed the experiments. HL, ZG, WL, SK, SD, and FG performed the experiments. HL and DS analyzed the data and wrote the paper. MH, WX, GY, and YL revised the paper. All authors read and approved the final version of the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the “863” High Technology Research and Development Program of China (863 Program) (2011AA10A101) and the Natural Science Foundation of Guangdong Province of China (2014A030313463).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.02041/full#supplementary-material
References
Agrawal, G. K., Rakwal, R., and Jwa, N. S. (2000). Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem. Biophys. Res. Commun. 278, 290–298. doi: 10.1006/bbrc.2000.3781
Andolfo, G., and Ercolano, M. R. (2015). Plant innate immunity multicomponent model. Front. Plant Sci. 6:987. doi: 10.3389/fpls.2015.00987
Ashkani, S., Rafii, M. Y., Shabanimofrad, M., Miah, G., Sahebi, M., Azizi, P., et al. (2015). Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front. Plant Sci. 6:886. doi: 10.3389/fpls.2015.00886
Barros, J., Serk, H., Granlund, I., and Pesquet, E. (2015). The cell biology of lignification in higher plants. Ann. Bot. 115, 1053–1074. doi: 10.1093/aob/mcv046
Cai, K., Gao, D., Luo, S., Zeng, R., Yang, J., and Zhu, X. (2008). Physiological and cytological mechanisms of silicon-induced resistance in rice against blast disease. Physiol. Plant. 134, 324–333. doi: 10.1111/j.1399-3054.2008.01140.x
Can, M., Armstrong, F. A., and Ragsdale, S. W. (2014). Structure, function, and mechanism of the nickel metalloenzymes, CO dehydrogenase, and acetyl-CoA synthase. Chem. Rev. 114, 4149–4174. doi: 10.1021/cr400461p
Chen, H. C., Song, J., Williams, C. M., Shuford, C. M., Liu, J., Wang, J. P., et al. (2013). Monolignol pathway 4-coumaric acid:coenzyme A ligases in Populus trichocarpa: novel specificity, metabolic regulation, and simulation of coenzyme A ligation fluxes. Plant Physiol. 161, 1501–1516. doi: 10.1104/pp.112.210971
Choi, C., Hwang, S. H., Fang, I. R., Kwon, S. I., Park, S. R., Ahn, I., et al. (2015). Molecular characterization of Oryza sativa WRKY6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 208, 846–859. doi: 10.1111/nph.13516
DeYoung, B. J., and Innes, R. W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249. doi: 10.1038/ni1410
Foster, J., Kim, H. U., Nakata, P. A., and Browse, J. (2012). A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell 24, 1217–1229. doi: 10.1105/tpc.112.096032
Francoz, E., Ranocha, P., Nguyen-Kim, H., Jamet, E., Burlat, V., and Dunand, C. (2015). Roles of cell wall peroxidases in plant development. Phytochemistry 112, 15–21. doi: 10.1016/j.phytochem.2014.07.020
Gay, C., Collins, J., and Gebicki, J. M. (1999). Hydroperoxide assay with the ferric-xylenol orange complex. Anal. Biochem. 273, 149–155. doi: 10.1006/abio.1999.4208
Gui, J., Shen, J., and Li, L. (2011). Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol. 157, 574–586. doi: 10.1104/pp.111.178301
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282.
Hirano, K., Aya, K., Kondo, M., Okuno, A., Morinaka, Y., and Matsuoka, M. (2012). OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm. Plant Cell Rep. 31, 91–101. doi: 10.1007/s00299-011-1142-7
Inoue, H., Hayashi, N., Matsushita, A., Xinqiong, L., Nakayama, A., Sugano, S., et al. (2013). Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 110, 9577–9582. doi: 10.1073/pnas.1222155110
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Jessen, D., Olbrich, A., Knüfer, J., Krüger, A., Hoppert, M., Polle, A., et al. (2011). Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J. 68, 715–726. doi: 10.1111/j.1365-313X.2011.04722.x
Jia, Y., Zhou, E., Lee, S., and Bianco, T. (2016). Coevolutionary dynamics of rice blast resistance gene Pi-ta and Magnaporthe oryzae avirulence gene AVR-Pita 1. Phytopathology 106, 676–683. doi: 10.1094/PHYTO-02-16-0057-RVW
Kalsoom, U., Bhatti, H. N., and Asgher, M. (2015). Characterization of plant peroxidases and their potential for degradation of dyes: a review. Appl. Biochem. Biotechnol. 176, 1529–1550. doi: 10.1007/s12010-015-1674-3
Le Roy, J., Huss, B., Creach, A., Hawkins, S., and Neutelings, G. (2016). Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 7:735. doi: 10.3389/fpls.2016.00735
Li, Y., Kim, J. I., Pysh, L., and Chapple, C. (2015). Four isoforms of arabidopsis 4-coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 169, 2409–2421. doi: 10.1104/pp.15.00838
Li, Z. Y., Xia, J., Chen, Z., Yu, Y., Li, Q. F., Zhang, Y. C., et al. (2016). Large-scale rewiring of innate immunity circuitry and microRNA regulation during initial rice blast infection. Sci. Rep. 6:25493. doi: 10.1038/srep25493
Liu, J., Osbourn, A., and Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Lozano-Durán, R., and Zipfel, C. (2015). Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20, 12–19. doi: 10.1016/j.tplants.2014.09.003
Marjamaa, K., Kukkola, E. M., and Fagerstedt, K. V. (2009). The role of xylem class III peroxidases in lignification. J. Exp. Bot. 60, 367–376. doi: 10.1093/jxb/ern278
Marone, D., Russo, M. A., Laidò, G., De Leonardis, A. M., and Mastrangelo, A. M. (2013). Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int. J. Mol. Sci. 14, 7302–7326. doi: 10.3390/ijms14047302
Miah, G., Rafii, M. Y., Ismail, M. R., Puteh, A. B., Rahim, H. A., Islam, K., et al. (2013). A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int. J. Mol. Sci. 14, 22499–22528. doi: 10.3390/ijms141122499
Miedes, E., Vanholme, R., Boerjan, W., and Molina, A. (2014). The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5:358. doi: 10.3389/fpls.2014.00358
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics 279, 415–427. doi: 10.1007/s00438-008-0322-9
Pascual, M. B., El-Azaz, J., de la Torre, F. N., Cañas, R. A., Avila, C., and Canovas, F. M. (2016). Biosynthesis and metabolic fate of phenylalanine in conifers. Front. Plant Sci. 7:1030. doi: 10.3389/fpls.2016.01030
Pottosin, I., Velarde-Buendia, A. M., Bose, J., Zepeda-Jazo, I., Shabala, S., and Dobrovinskaya, O. (2014). Cross-talk between reactive oxygen species and polyamines in regulation of ion transport across the plasma membrane: implications for plant adaptive responses. J. Exp. Bot. 65, 1271–1283. doi: 10.1093/jxb/ert423
Sasaki, Y., and Nagano, Y. (2004). Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 68, 1175–1184. doi: 10.1271/bbb.68.1175
Sharma, T. R., Das, A., Thakur, S., Devanna, B. N., Singh, P. K., Jain, P., et al. (2016). Oscillating transcriptome during rice-magnaporthe interaction. Curr. Issues Mol. Biol. 19, 99–120. doi: 10.21775/9781910190357.10
Shockey, J., and Browse, J. (2011). Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J. 66, 143–160. doi: 10.1111/j.1365-313X.2011.04512.x
Shockey, J., Schnurr, J., and Browse, J. (2000). Characterization of the AMP-binding protein gene family in Arabidopsis thaliana: will the real acyl-CoA synthetases please stand up? Biochem. Soc. Trans. 28, 955–957. doi: 10.1042/bst0280955
Soltani, B. M., Ehlting, J., and Douglas, C. J. (2006). Genetic analysis and epigenetic silencing of At4CL1 and At4CL2 expression in transgenic Arabidopsis. Biotechnol. J. 1, 1124–1136. doi: 10.1002/biot.200600140
Souza, C. A., Barbazuk, B., Ralph, S. G., Bohlmann, J., Hamberger, B., and Douglas, C. J. (2008). Genome-wide analysis of a land plant-specific acyl:coenzyme A synthetase (ACS) gene family in Arabidopsis, poplar, rice and Physcomitrella. New Phytol. 179, 987–1003. doi: 10.1111/j.1469-8137.2008.02534.x
Stael, S., Kmiecik, P., Willems, P., Van Der Kelen, K., Coll, N. S., Teige, M., et al. (2015). Plant innate immunity–sunny side up? Trends Plant Sci. 20, 3–11. doi: 10.1016/j.tplants.2014.10.002
Stremmel, W., Pohl, L., Ring, A., and Herrmann, T. (2001). A new concept of cellular uptake and intracellular trafficking of long-chain fatty acids. Lipids 36, 981–9.
Takatsuji, H. (2014). Development of disease-resistant rice using regulatory components of induced disease resistance. Front. Plant Sci. 5:630. doi: 10.3389/fpls.2014.00630
Taylor-Teeples, M., Lin, L., de Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tonnessen, B. W., Manosalva, P., Lang, J. M., Baraoidan, M., Bordeos, A., Mauleon, R., et al. (2015). Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 87, 273–286. doi: 10.1007/s11103-014-0275-9
Voelker, S. L., Lachenbruch, B., Meinzer, F. C., Jourdes, M., Ki, C., Patten, A. M., et al. (2010). Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol. 154, 874–886. doi: 10.1104/pp.110.159269
Wang, G. F., and Balint-Kurti, P. J. (2016). Maize homologs of CCoAOMT and HCT, two key enzymes in lignin biosynthesis, form complexes with the NLR Rp1 protein to modulate the defense response. Plant Physiol. 171, 2166–2177. doi: 10.1104/pp.16.00224
Wu, W., Wang, L., Zhang, S., Li, Z., Zhang, Y., Lin, F., et al. (2014). Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant Microbe Interact. 27, 759–769. doi: 10.1094/MPMI-02-14-0046-R
Xie, X., Zhang, X., Dong, Z., and Guo, H. (2011). Dynamic changes of lignin contents of MT-1 elephant grass and its closely related cultivars. Biomass Bioenergy 35, 1732–1738. doi: 10.1016/j.biombioe.2011.01.018
Xu, X. H., Wang, C., Li, S. X., Su, Z. Z., Zhou, H. N., Mao, L. J., et al. (2015). Friend or foe: differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNAseq and metabolite profiling. Sci. Rep. 5:13624. doi: 10.1038/srep13624
Ye, W., and Ma, W. (2016). Filamentous pathogen effectors interfering with small RNA silencing in plant hosts. Curr. Opin. Microbiol. 32, 1–6. doi: 10.1016/j.mib.2016.04.003
Yi, J., Moon, S., Lee, Y. S., Zhu, L., Liang, W., Zhang, D., et al. (2016). Defective tapetum cell death 1 (DTC1) regulates ROS levels by binding to metallothionein during tapetum degeneration. Plant Physiol. 170, 1611–1623. doi: 10.1104/pp.15.01561
Zhang, K., Qian, Q., Huang, Z., Wang, Y., Li, M., Hong, L., et al. (2006). GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol. 140, 972–983. doi: 10.1104/pp.105.073007
Zhang, X. C., Yu, X., Zhang, H. J., and Song, F. M. (2009). Molecular characterization of a defense-related AMP-binding protein gene, OsBIABP1, from rice. J. Zhejiang Univ. Sci. B 10, 731–739. doi: 10.1631/jzus.B0920042
Zhang, X., and Liu, C. J. (2015). Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 8, 17–27. doi: 10.1016/j.molp.2014.11.001
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30. doi: 10.1186/1746-4811-7-30
Keywords: 4CL-like, Rice blast, AMPBP, Lignin, ROS
Citation: Liu H, Guo Z, Gu F, Ke S, Sun D, Dong S, Liu W, Huang M, Xiao W, Yang G, Liu Y, Guo T, Wang H, Wang J and Chen Z (2017) 4-Coumarate-CoA Ligase-Like Gene OsAAE3 Negatively Mediates the Rice Blast Resistance, Floret Development and Lignin Biosynthesis. Front. Plant Sci. 7:2041. doi: 10.3389/fpls.2016.02041
Received: 02 September 2016; Accepted: 20 December 2016;
Published: 10 January 2017.
Edited by:
Hua Lu, University of Maryland, Baltimore County, USAReviewed by:
Xiping Wang, Northwest A&F University, ChinaZonghua Wang, Fujian Agriculture and Forestry University, China
Copyright © 2017 Liu, Guo, Gu, Ke, Sun, Dong, Liu, Huang, Xiao, Yang, Liu, Guo, Wang, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiafeng Wang, bcjfwang@gmail.com
Zhiqiang Chen, chenlin@scau.edu.cn
 Hao Liu
Hao Liu Zhenhua Guo2
Zhenhua Guo2 Dayuan Sun
Dayuan Sun