- 1State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China
- 2Institute of Industrial Crops, Jiangsu Academy of Agricultural Science, Nanjing, China
- 3Jiangsu Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing, China
The Stress Associated RNA-binding protein 1 (SRP1) repressed by ABA, salt and cold encodes a C2C2-type zinc finger protein in Arabidopsis. The knock-out mutation in srp1 reduced the sensitivity of seed to ABA and salt stress during germination and post-germinative growth stages. In contrast, SRP1-overexpressing seedlings were more sensitive to ABA and salt compared to wild type plants. In the presence of ABA, the transcript levels of ABA signaling and germination-related genes including ABI3. ABI5. EM1 and EM6 were less induced in srp1 compared to WT. Interestingly, expression of ABI2 encoding a protein phosphatase 2C protein were significantly up-regulated in srp1 mutants. By in vitro analysis, SRP1 was identified as a novel RNA-binding protein directly binding to 3′UTR of ABI2 mRNA. Moreover, transient expression assay proved the function of SRP1 in reducing the activity of luciferase whose coding sequence was fused with the ABI2 3’UTR. Together, it is suggested that SRP1 is involved in the ABA signaling by post-transcriptionally repressing ABI2 expression in Arabidopsis.
Introduction
Post-transcriptional regulatory events, such as RNA processing, splicing and transport, are crucial to plant growth, development and response to environmental stresses (Ambrosone et al., 2012; Jung et al., 2013). RNA-binding proteins (RBPs) play important roles in post-transcriptional regulation by directly binding to single/double strand RNA (ssRNA/dsRNA) molecules (Ambrosone et al., 2012). RBPs are widely distributed among plant species. More than 200 putative RBP genes have been identified in Arabidopsis thaliana, and ∼250 in rice (Oryza sativa L.) (Lorković, 2009; Cook et al., 2011; Ambrosone et al., 2012). In the past fifteen years, the researches on the roles of RBPs in the regulation of plant development and stress responses have been gradually increased (Ambrosone et al., 2012; Jung et al., 2013).
RBPs contain several common motifs such as RNA recognition motif (RRM), K homology (KH) domain (Lorković and Barta, 2002), ZnF domain (mainly CCCH type) (Kim et al., 2007; Qu et al., 2014), DEAD/DEAH box (Owttrim, 2006), Pumilio/FBF (PUF) domain, double-stranded RNA-binding domain (DS-RBD) (Tam et al., 2010), Piwi/Argonaute/Zwiklle (PAZ) domain (Song et al., 2003), and the auxiliary domains (glycine-rich, arginine-rich, arginine-glycine, or serine-arginine repeats) (Alba and Pagès, 1998; Ambrosone et al., 2012). The abscisic acid (ABA) is an important plant hormone playing critical roles in many aspects of plant growth and stress responses. Several ABA-regulated RBPs have been reported to be involved in plant stress responses so far (Ambrosone et al., 2012). The RBPs, such as cap binding complex (CBC) (Papp et al., 2004), AAPK interacting protein 1 (AKIP1) (Lamond and Spector, 2003), glycine-rich RNA-binding protein (GRP) (Wang et al., 2012), and ABA-regulated RNA-binding protein (ARP1) (Jung et al., 2013) are all involved in ABA. Cap Binding Protein 20 (CBP20) is the subunit of CBC and the cbp20 is hypersensitive to ABA (Papp et al., 2004). The recent study has revealed the splicing function of CBP20, and the loss function of it induces the improper splicing of Indeterminate Domain 14 (IDD14) transcription factor and results in the hypersensitivity to salt stress (Kong et al., 2014). RZ-1 is a kind of GRP proteins. RZ-1a, one member of RZ-1 orthologs, is reported to has negative impact on seed germination (Kim et al., 2007). The homologous proteins of RZ-1a, RZ-1b and RZ-1c have been revealed to play critical roles in regulating pre-mRNA splicing (Wu et al., 2015). ARP1 is a putative RBP and modulates several germination responsive genes under ABA (Jung et al., 2013).
Our previous work has identified a novel protein family (SRZ family) with C2C2-type zinc finger motifs from rice (Oryza sativa L.) (Huang et al., 2008). The SRZ1 contains three C2C2-type zinc finger motifs (DWXCX1-4CX3NX6CX2C, X means any residues), and it is down-regulated by ABA and abiotic stresses (Huang et al., 2008). However precise functions of SRZ proteins remain to be revealed. In present study, we analyzed the function of Stress Associated RNA-binding protein 1 (SRP1), a SRZ1 homolog in Arabidopsis, and showed that SRP1 was a novel RNA binding protein involved in the ABA signaling by binding to 3′UTR region of ABI2 and triggering the instability of ABI2 mRNA.
Materials and Methods
Plant Materials
The Arabidopsis Columbia (Col-0) ecotype wild-type (WT), srp1-1 (SAIL_187_C10), srp1-2 (SAIL_312_E06), aba3-1 (CS157), and 35S::SRP1 transgenic lines were used in this work. The mutants were obtained from Arabidopsis Biological Resource Center1 (ABRC). 35S::SRP1 transgenic lines were generated by Agrobacterium-mediated transformation in this study. The seeds were sown on 1/2 Murashige and Skoog (MS) medium with 3% (w/v) sucrose and 0.8% agar in a light homoeothermic incubator (24 °C) with 16 h light/8 h dark. After 3 days, the germinated seeds were transferred into pot (R = 3 cm, H = 8 cm) filled with nutritional soil and vermiculite (1:1) and grown under 50 μmol m-2 s-1 light at 22°C and 16 h light/8 h dark in a greenhouse. The seeds harvested at the same time were used for the experiment.
Germination and Post-Germination Growth Assays
One hundred seeds each line were sown in the plate (10 cm × 10 cm × 1 cm) with 1/2 MS medium containing different concentrations of ABA or NaCl in triplicates. The plates with seeds were stored at 4°C in dark for four days, and subsequently incubated in the growth chamber (22°C) with 16 h light/8 h dark. Germination and green cotyledon rates were scored at the indicated times after the incubation. Statistical analysis was performed with three biological replicates. Seeds were counted as germinated when the radicle tip had fully penetrated the seed coat (radicle protrusion) as previously described (Liu et al., 2013).
Generation of SRP1 Transgenic Line and Mutant Identification
The coding sequence of SRP1 was cloned into binary vector pCAMBIA1300s which was further transferred into Agrobacterium tumefaciens strain EHA105. The Agrobacterium-mediated transformation in Arabidopsis was conducted as previously described (Clough and Bent, 1998). Transgenic plants were screened in hygromycin (25 mg/L)-containing medium and the positive seedlings were randomly selected to be verified by the primers for the hygromycin resistance gene (Supplementary Table 1). Mutants srp1-1 (SAIL_187_C10) and srp1-2 (SAIL_312_E06) for the SRP1 gene were identified using the primers LP, RP and pCSA110LB for amplification of T-DNA insertion. The mutants or the transgenic line were further verified by semi-quantitative RT-PCR or qRT-PCR analysis. The primers were listed in Supplementary Table 1.
Real-time PCR
Total RNA were prepared from WT, srp1 and transgenic lines as previously described (Gutierrez et al., 2009). Approximately 5 μg of RNA were treated with DNase I using a DNA free Kit (Thermo Fisher Scientific, Waltham, MA, USA), and cDNA was synthesized by reverse transcribing total RNA using SuperScript III reverse transcriptase (Invitrogen, Foster City, CA, USA) with oligo (dT)12 primer according to the manufacturer’s instructions. Transcript levels of ABA signaling-related genes RD29A (AT5G52310), RD29B (AT5G52300), ABI1 (AT4G26080), ABI2 (AT5G57050), ABI3 (AT3G24650), ABI5 (AT2G36270), EM1 (AT3G51810), and EM6 (AT2G40170) were assessed by real-time quantitative reverse transcription PCR (qRT-PCR). Arabidopsis Actin7 was used as the internal control for the qRT-PCR, respectively (Liu et al., 2007; Yelagandula et al., 2014). The primers were listed in Supplementary Table 1.
Subcellular Localization
The transient expression vector was constructed by cloning SRP1 into pA7-GFP to fuse with GFP. The pA7-GFP vector was used as a control. The vectors were expressed transiently in Arabidopsis leaf protoplasts using the PEG (polyethylene glycol) method (Abel and Theologis, 1994). DAPI (4′,6-diamidino-2-phenylindole) was used to dye the nucleus. The visualization of SRP1-GFP or GFP was observed using the Leica SP2 confocal laser scanning microscope.
RNA Electrophoretic Mobility Shift Assay
The SRP1 coding sequence was cloned into the expression vector pGEX-2T to generate GST-labeled SRP1 protein. The fusion protein was prepared according to the GST Gene Fusion System Handbook (Amersham Biosciences, Piscataway, NJ, USA). The 3′UTR sequences of ABI2/3 dsDNA were cloned with the primers containing the T7 promoter sequence (Rohde et al., 2000; Ohta et al., 2003). The 3′UTRs of ABI2/3 ssRNA was labeled using T7 RNA polymerase and biotin-16 UTP according to the instruction manual (Thermo Fisher Scientific, Waltham, MA, USA). The binding reactions were performed by incubating biotin-labeled RNA with recombinant protein at 23°C for 30 min in 10 μl of binding buffer containing 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.5), 50 mM NaCl, 5 mM 2-mercaptoethanol, 1 mM DL-Dithiothreitol (DTT), 5 μM ZnCl2 and 4 mM MgCl2 as described (Qu et al., 2014). Electrophoresis was performed at 4°C in 8% native TRIS borate-acrylamide (29:1) gels, which were transferred to N+ nylon membrane (Roche, Basel, Swiss). The membrane was exposed using the Chemiluminescent Biotin-labeled Nucleic Acid Detection Kit (Thermo Fisher Scientific, Waltham, MA, USA), and scanned by ChemiDoc XRS+ imager (Bio-Rad, Irvine, CA, USA). For competition assays, biotin-labeled RNA and recombinant protein were incubated prior to the addition of unlabeled competitor RNA. The sequences of ABI2 and ABI3 3′UTRs were listed in Supplementary Table 2.
Transactivation Activity Assay in Protoplast System
In protoplast system analyzing the effect of SRP1 on the stability of mRNA, the control reporter vector 3× DRE-LUC was constructed by inserting three copies of dehydration response element (DRE, TACCGACAT) and a minimal TATA region of 35S promoter of Cauliflower Mosaic Virus (CaMV) into the upstream of the firefly luciferase (LUC) in pGL3-basic vector. The reporter vectors 3× DRE-LUC-ABI2 3′UTR and 3× DRE-LUC-ABI3 3′UTR were constructed by insertion of ABI2 and ABI3 3′UTR into the downstream of luciferase coding sequence. Effector vectors 35S-DREB1A and 35S-SRP1 were constructed by insertion of the DREB1A (AT4G25480) or SRP1 coding region into PA7 vector (Narusaka et al., 2003). The pPTRL (Renilla reniformis Luciferase driven by 35S promoter) was used as the internal control. The effectors, reporters and internal control were co-transfected into Arabidopsis protoplasts. After culturing for 16 h, the luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and quantitated by the GloMax 20/20 luminometer (Promega, Madison, WI, USA). Statistical analysis was performed with three biological replicates.
To analyze the transactivation activity of SRP1, the assay in Arabidopsis protoplast system was performed as previously described (Fang et al., 2015). The reporter vector was constructed by inserting four copies of GAL4 binding element and a minimal TATA region of 35S promoter of CaMV into the upstream of the firefly LUC in pPCV814 vector. The control effector vector 35S-Gal4DB was constructed by inserting the GAL4DB into pRTL2-GUS vector. The SRP1 was cloned into control effector vector to generate 35S-Gal4DB-SRP1 as the effector. The pPTRL was used as the internal control. The effectors, reporters and internal control were co-transfected into Arabidopsis protoplasts. After culturing for 16 h, the luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and quantitated by the GloMax 20/20 luminometer (Promega, Madison, WI, USA). Statistical analysis was performed with three biological replicates.
Transactivation Activity Assay in Yeast
The transactivation activity assays in the yeast were performed using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech, Mountain View, CA, USA). BD-SRP1 was constructed by cloning the full-length CDS of SRP1 into pGBKT7 to fuse with the GAL4 binding domain. The construct and the empty vectors were transformed into the yeast strain AH109. The yeast cells were plated onto synthetic dextrose (SD)/-Trp medium to confirm the transformation. At the same time, the yeast cells were plated onto SD/-His/-Trp/-Leu containing 3 mM 3-amino-1,2,4-triazole medium to screen for transactivation activity.
Results
Identification and Characterization of SRP1
Previously we reported a stress repressive zinc finger protein SRZ1 in rice (Huang et al., 2008). In present study, we studied the function of SRP1, the homolog of SRZ1 in Arabidopsis. The SRP1 (AT2G17975) gene possesses a translated region of 807-bp that encodes a protein containing 268 amino acids, with a calculated molecular weight of 30.24 kDa. On the basis of conserved domain analysis, four C2C2-type ZnF motifs were found in SRP1 and homologous to mammalian RanBP2-type ZnF domains (Nguyen et al., 2011; O’Connell et al., 2012; Vandevenne et al., 2014) (Figure 1A). The homologous proteins of SRP1 were found widely distributing among plant species, and they contained three or four C2C2-type ZnF motifs (Figures 1B,C). In Arabidopsis, four orthologs of SRP1 were identified, but SRP1 was unique among those five homologous proteins for the extra ZnF motif in N-terminal.
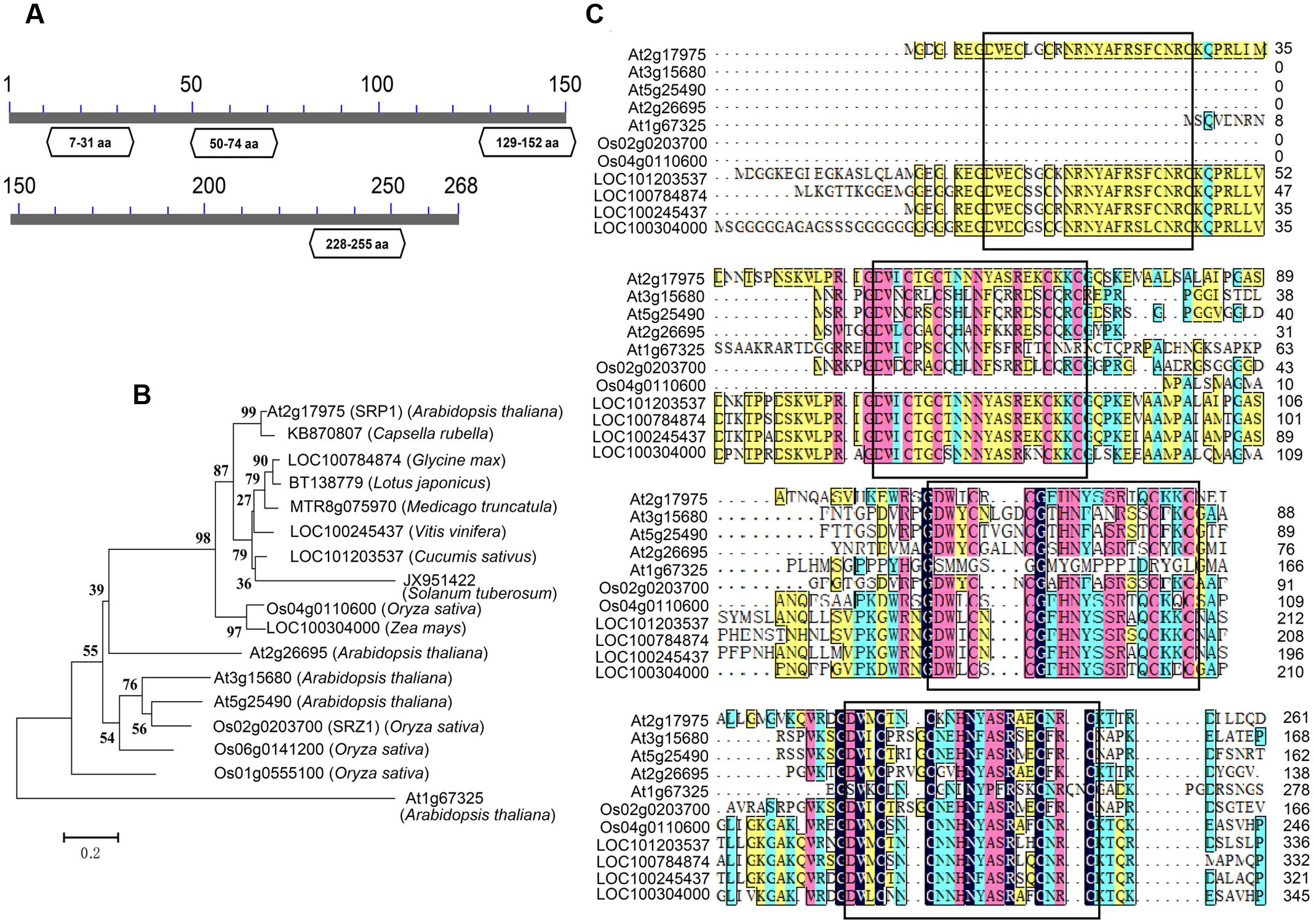
FIGURE 1. Characterization of SRP1. (A) The conserved domain analysis of SRP1. The conserved domains in SRP1 were identified in NCBI (http://www.ncbi.nlm.nih.gov). The zinc finger domains are shown in boxes with domain positions. The numbers represent the amino acids. (B) Phylogenetic analysis of C2C2 type zinc-finger proteins. Phylogenetic analysis of SRP1 and other plant C2C2 type zinc-finger proteins was performed using the neighbor-joining method wrapped in MEGA. Numbers at the nodes indicate bootstrap values (1000 replicates). (C) Sequence alignment of SRP1 and other plant C2C2-type zinc-finger proteins. The zinc finger motifs are boxed, respectively.
SRP1 Is Down-regulated by ABA and Abiotic Stress
The expression of SRP1 was analyzed in the WT seedlings treated with ABA, salt stress and cold stress. SRP1 expression was down-regulated under the treatment of ABA within 24 h and gradually decreased along with the treatment time (Figures 2A,B). The expression of SRP1 was also repressed by salt or cold stresses (Figures 2C,D). Compared with WT, the expression of SRP1 in aba3-1 was up-regulated (Figure 2E). The aba3-1 is ABA deficient mutant, and the level of endogenous ABA in aba3-1 is reduced as the conversion of ABA-aldehyde to ABA is blocked (Léon-Kloosterziel et al., 1996). These results indicate the expression of SRP1 is repressed by abiotic stress and negatively regulated by ABA. It is of interest to investigate the involvement of SRP1 in ABA signaling as it is significantly down-regulated by ABA (Figure 2B).
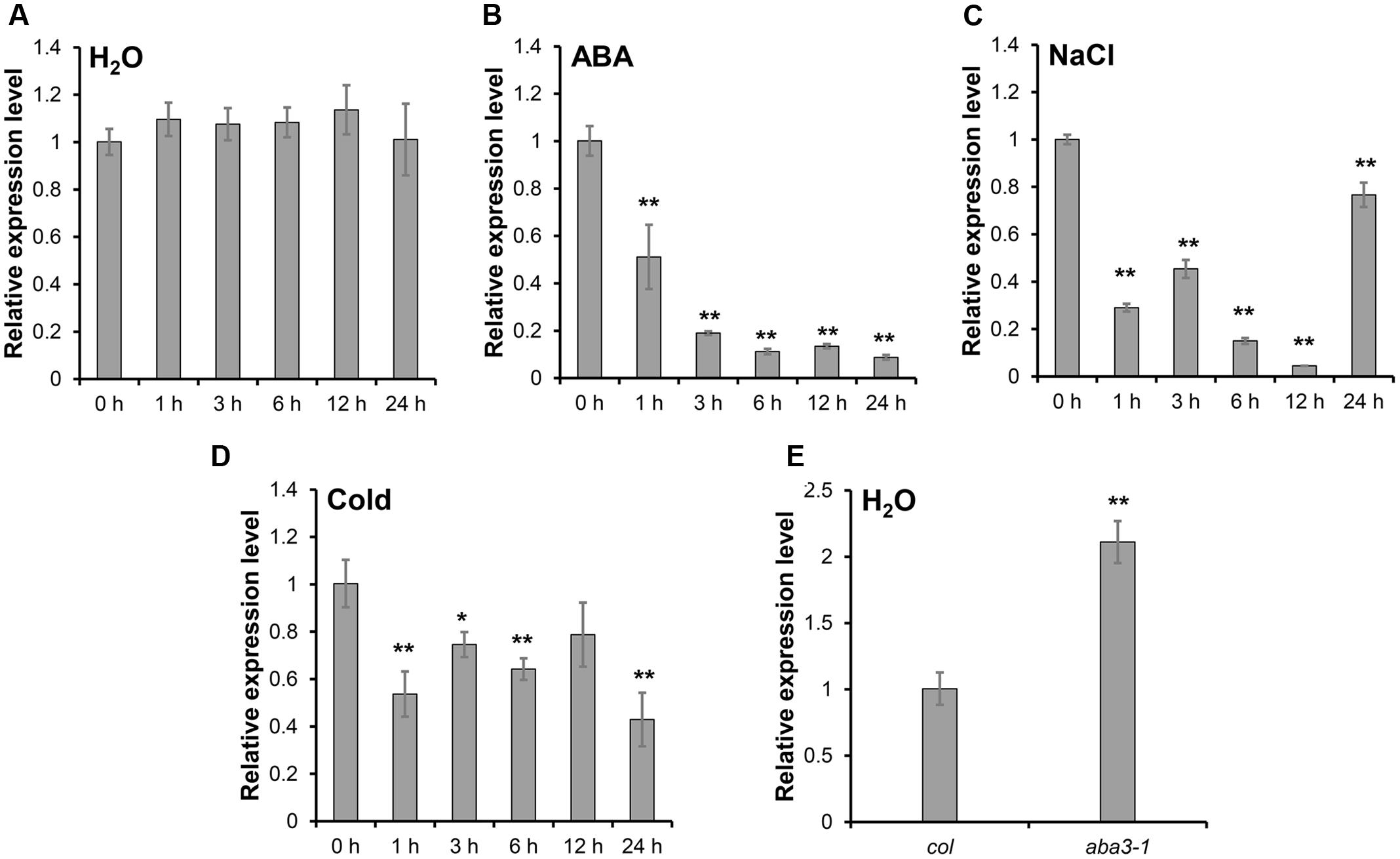
FIGURE 2. The expression pattern of SRP1 under abiotic stresses. The seeds were germinated in the 1/2 MS medium. The 7-days-old seedlings were transferred into the control or stress conditions to analyze the expression pattern of SRP1 by qRT-PCR. “∗”, “∗∗” represent significant differences relative to each control and P-value < 0.05 or P-value < 0.01, based on student’s t-test. Each value was the mean ± SD of three biological determinations. (A) The expression of SRP1 in WT. (B) The expression of SRP1 in WT treated with 10 μM ABA. (C) The expression of SRP1 in WT treated with 100 mM NaCl. (D) The expression of SRP1 in WT treated with cold stress (4 °C). (E) The expression of SRP1 in aba3-1.
SRP1 Modulates Seed Germination under ABA Treatment
The SRP1 T-DNA insertion mutants srp1-1 and srp1-2 (Figure 3A) were verified via genomic PCR (Figure 3B) and semi-quantitative RT-PCR (Figure 3C). The 35S::SRP1 plants (T4 generation) were used in subsequent analysis. By qRT-PCR verification, it was found that SRP1 was highly expressed in the 35S::SRP1 plant (Figure 3D).
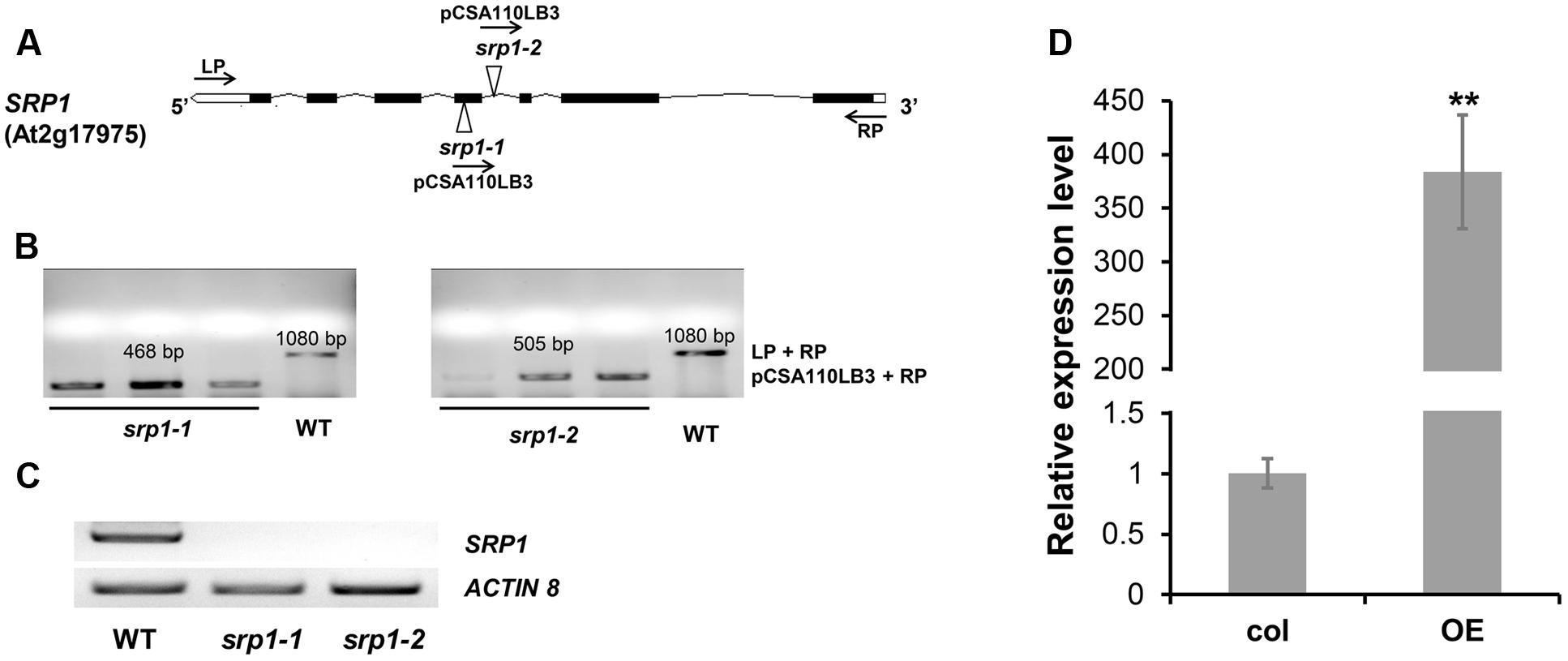
FIGURE 3. Identification of srp1 and 35S::SRP1. (A) Position of the T-DNA insertions in the SRP1 genomic region. Black boxes and lines depict the exons and introns, respectively. White boxes depict the UTR regions and the Triangles depict the T-DNA insert location. The arrows depict the primers used in the genomic PCR identification. (B) The genomic PCR identification of T-DNA insertion. The homozygous mutants were verified by amplifying the overlapping region of T-DNA insertion and the gene. (C) The semi-quantitative RT-PCR analysis of the mutants. Actin 8 was used as the internal control. (D) The qRT-PCR analysis of 35S::SRP1 (OE). “∗∗” represents significant difference relative to each control and P-value < 0.01, based on student’s t-test. Each value was the mean ± SD of three biological determinations.
The seeds of the WT, srp1-1. srp1-2 and 35S::SRP1 plants were germinated on 1/2 MS medium supplemented with or without ABA to analyze the sensitivity to ABA. The germination rates among WT, srp1-1. srp1-2, and 35S::SRP1 plants were similar on the medium under control condition (Supplementary Figure 1A). As shown in Figure 4A, the germination rates among WT, srp1-1 and srp1-2 were similar on the 1/2 MS medium supplemented with 1 μM ABA. However, the germination rates of srp1-1 and srp1-2 seeds were significantly higher than WT on the third day when 1.5 or 2 μM exogenous ABA was applied. The germination rates of mutant seeds with 2.5 μM ABA treatment were significantly higher than WT from the third to the sixth day. The germination rate of WT was reduced by 4.97, 29.36, 44.64, or 52.06%, respectively, on the third day when 1, 1.5, 2, or 2.5 μM exogenous ABA was applied. However, the germination rate of srp1-1 was reduced by 0.05, 10.74, 24.38, or 29.33%, while srp1-2 was reduced by 2.05, 10.29, 24.28, or 32.10%. The difference between the mutants and WT was increased with the ABA concentration (Figure 4B). In contrast, the 35S::SRP1 line exhibited significantly lower germination rate than WT under ABA treatment (Figure 4A).
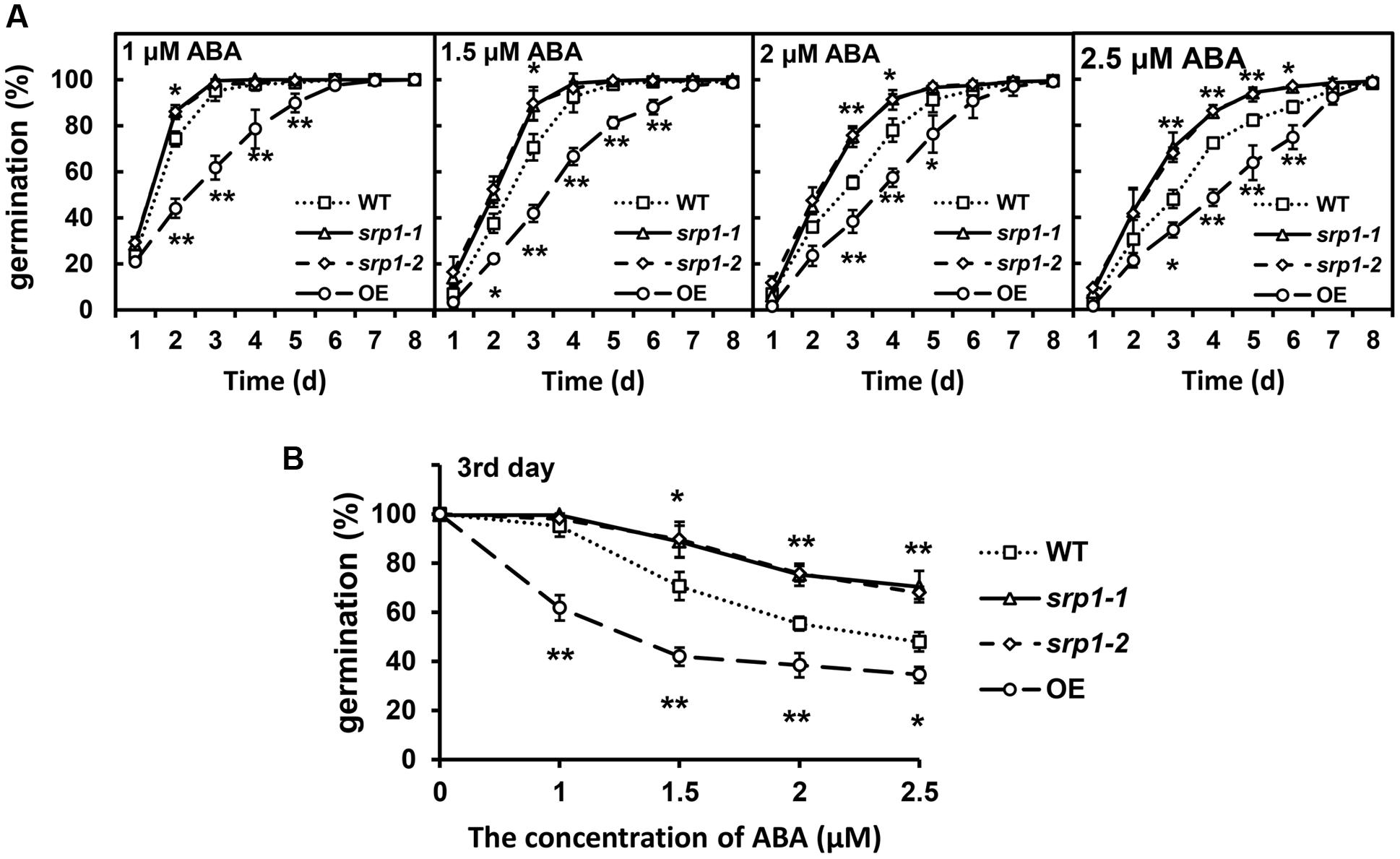
FIGURE 4. Seed germination under ABA treatment. (A) The germination of srp1 mutants and OE lines in 1/2 MS medium supplemented with 1, 1.5, 2, and 2.5 μM ABA. The time indicates the days after vernalization. (B) The germination of srp1 mutants and OE lines in 1/2 MS medium supplemented with different concentration of ABA on the 3rd day after vernalization. “∗”, “∗∗” represent significant differences relative to each control and P-value < 0.05 or P-value < 0.01, based on student’s t-test. Each value was the mean ± SD of three biological determinations.
SRP1 Modulates Post-germinative Growth under ABA
The green cotyledon rates of the WT, srp1-1. srp1-2, and 35S::SRP1 plants were also analyzed to find out the sensitivity of Arabidopsis to ABA during the post-germinative growth stage. The green cotyledon rates among WT, srp1-1. srp1-2, and 35S::SRP1 plants were similar under control condition (Supplementary Figure 1B). Seven days after the incubation on the 1/2 MS medium supplemented with 1 μM ABA, the cotyledon greening efficiencies of srp1-1 and srp1-2 were significantly higher than WT, while 35S::SRP1 line was significantly lower (Figure 5A). The green cotyledon rates of srp1-1 and srp1-2 were significantly higher than WT from the seventh day to the eleventh day when 1 μM exogenous ABA was applied, and the difference between mutants and WT was extremely significant from the ninth day to the eleventh day (Figure 5B). The green cotyledon rates of mutants and WT treated with 1.5, 2, or 2.5 μM exogenous ABA were also analyzed but no significant difference was observed (Figure 5C). Moreover, the green cotyledon rates of mutants and WT were decreased along with the ABA concentration. In contrast, the 35S::SRP1 plants treated with 1 μM exogenous ABA exhibited significantly lower green cotyledon rate than WT from the seventh day after sowing (Figure 5B). These results indicated that the srp1 mutants were more insensitive to exogenous ABA than the WT during the post-germinative growth, while the 35S::SRP1 plant was more sensitive.
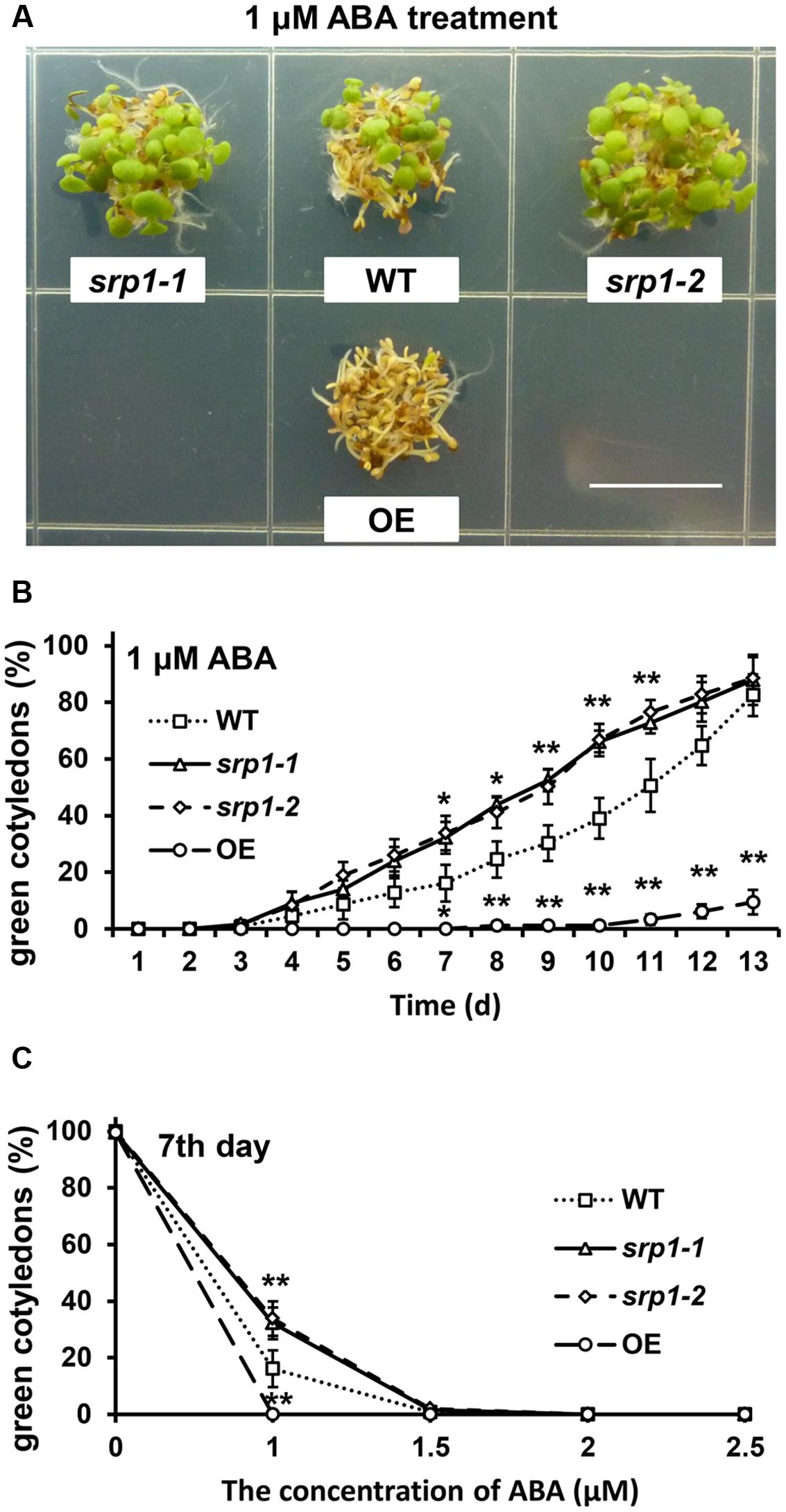
FIGURE 5. Post-germinative growth under ABA treatment. (A) The green cotyledons of srp1 mutants and OE lines in 1/2 MS medium supplemented with 1 μM ABA. Scale = 1 cm. (B) The green cotyledon rate of srp1 mutants and OE lines in 1/2 MS medium supplemented with 1 μM ABA. The time indicates the days after vernalization. (C) The green cotyledon rates of srp1 mutants and OE lines in 1/2 MS medium supplemented with 1, 1.5, 2, and 2.5 μM ABA on the 7th day after vernalization. “∗”, “∗∗” represent significant differences relative to each control and P-value < 0.05 or P-value < 0.01, based on student’s t-test. Each value was the mean ±SD of three biological determinations.
SRP1 Modulates Seed Germination and Post-germinative Growth under NaCl
In the presence of 100 mM NaCl, the germination rates of srp1-1 and srp1-2 seeds were significantly higher than WT in the first two days (Figure 6A), and the cotyledon greening efficiencies of srp1-1 and srp1-2 were significantly higher than WT on the third day (Figures 6B,C). In contrast, the 35S::SRP1 line exhibited significantly lower germination and green cotyledon rate than WT under NaCl treatment (Figure 6).
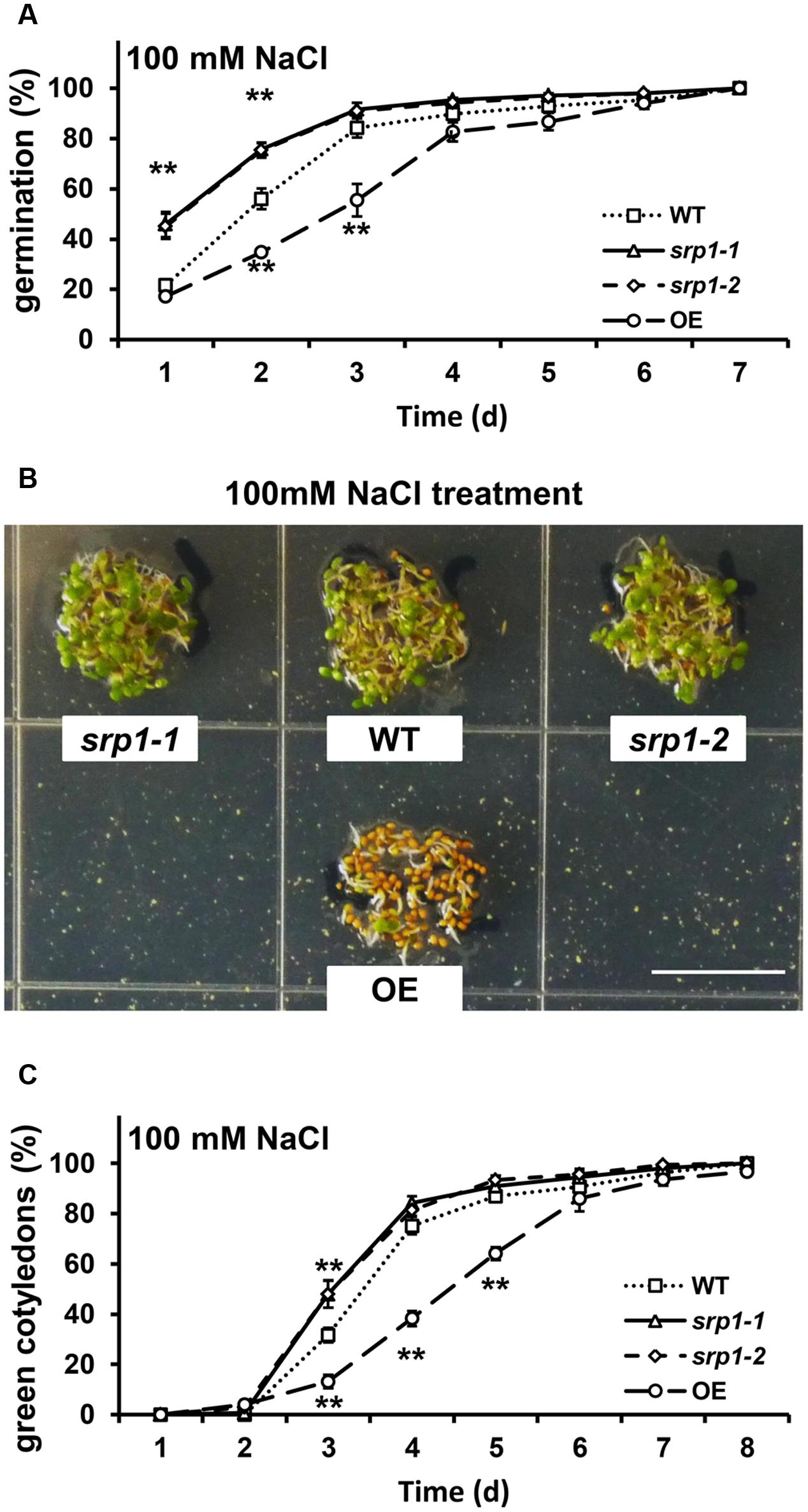
FIGURE 6. Seed germination and post-germinative growth under NaCl treatment. (A) The germination of srp1 mutants and OE lines in 1/2 MS medium supplemented with 100 mM NaCl. (B) The green cotyledons of srp1 mutants and OE lines in 1/2 MS medium supplemented with 100 mM NaCl. Scale = 1 cm. (C) The green cotyledon rate of srp1 mutants and OE lines in 1/2 MS medium supplemented with 100 mM NaCl. The time indicates the days after vernalization. “∗∗” represents significant difference relative to each control and P-value < 0.01, based on student’s t-test. Each value was the mean ± SD of three biological determinations.
SRP1 Modulates the Expression of ABA Signaling-related Genes
The contents of endogenous ABA in srp1 and WT were analyzed and we did not observe the marked differences among the mutant, 35S::SRP1 plant and WT (Supplementary Figure 2). The transcript abundances of ABA signaling and germination-related gene RD29A, RD29B, ABA INSENSITIVE 1 (ABI1), ABA INSENSITIVE 2 (ABI2), ABA INSENSITIVE 3 (ABI3), ABA INSENSITIVE 5 (ABI5), LATE EMBRYOGENESIS ABUNDANT 1 (EM1). and LATE EMBRYOGENESIS ABUNDANT 1 (EM6) were analyzed in the srp1 mutants. As shown in Figures 7A,B, the transcript levels of RD29A and RD29B were repressed by SRP1 knock-out under the control condition, but the transcriptions of ABI1 and ABI2 were enhanced. The transcript levels of RD29A. RD29B. ABI3. ABI5. EM1, and EM6 were significantly less induced in srp1 mutants than in WT under ABA treatment (Figures 7C,D). Interestingly, the transcript level of ABI2 was significantly increased in the srp1 mutants under control and ABA conditions, while that of ABI1 was slightly increased under control condition. In addition, the expressions of those genes in 35S::SRP1 plant were opposite. These results indicate the involvement of SRP1 in ABA signaling.
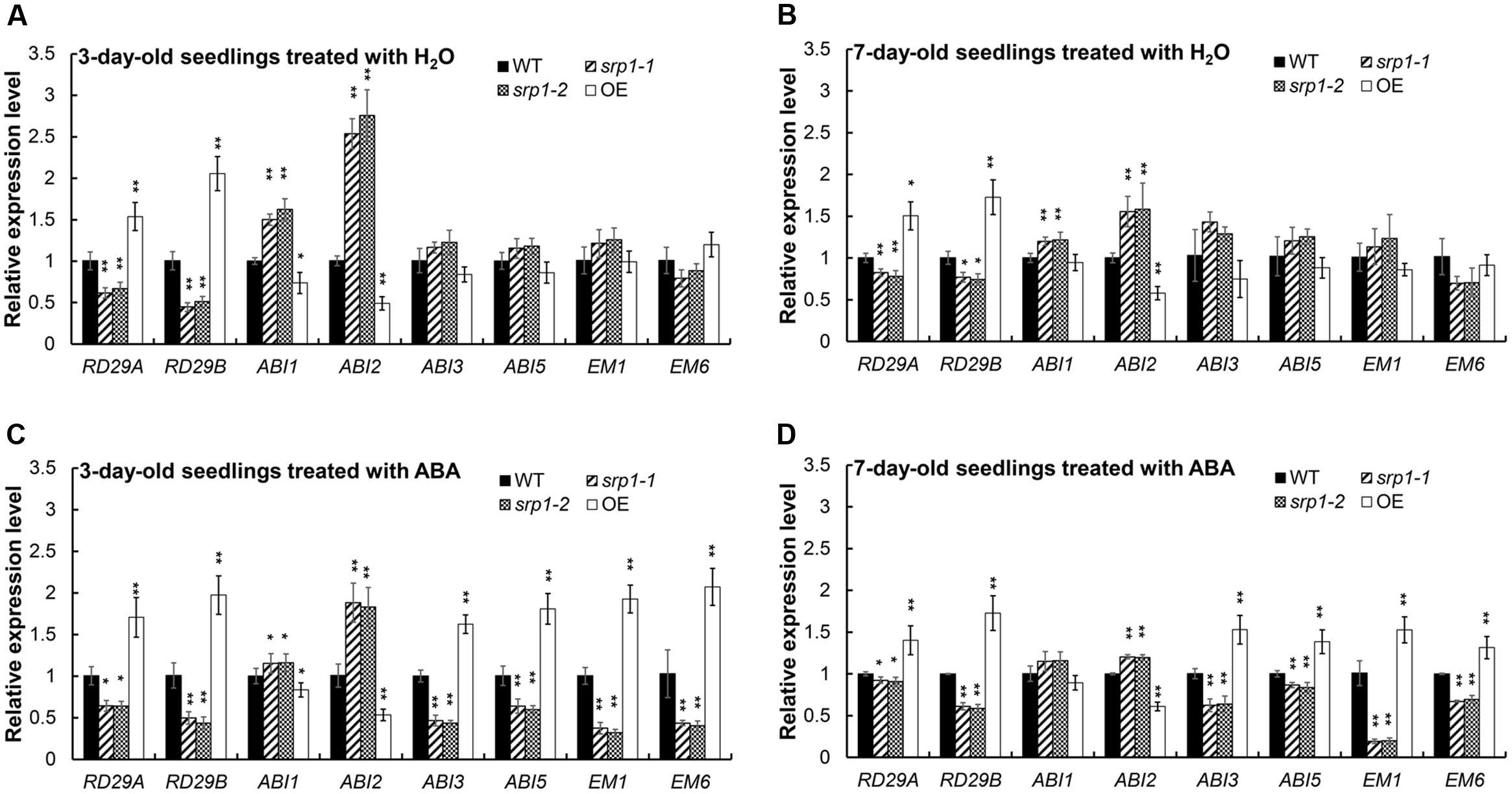
FIGURE 7. The transcript abundances of the genes involved in ABA signaling. (A) Transcript abundances of RD29A, RD29B, ABI1, ABI2, ABI3, ABI5, EM1, and EM6 in 3-day-old seedlings grown in MS medium for 24 h were analyzed by qRT-PCR. (A,B) Transcript abundances of RD29A, RD29B, ABI1, ABI2, ABI3, ABI5, EM1 and EM6 in 7-day-old seedlings grown in MS medium for 24 h were analyzed by qRT-PCR. (C) Transcript abundances of RD29A, RD29B, ABI1, ABI2, ABI3, ABI5, EM1, and EM6 in 3-day-old seedlings grown in MS medium containing 2.5 μM ABA for 24 h were analyzed by qRT-PCR. (D) Transcript abundances of RD29A, RD29B, ABI1, ABI2, ABI3, ABI5, EM1, and EM6 in 7-day-old seedlings grown in MS medium containing 2.5 μM ABA for 24 h were analyzed by qRT-PCR. “∗”, “∗∗” represent significant differences relative to each control and P-value < 0.05 or P-value < 0.01, based on student’s t-test. Each value was the mean ± SD of three biological determinations.
SRP1 Possess the Dual Function in ABI2 3′UTR RNA Binding and Transcriptional Activation
It has been demonstrated that AU rich element (ARE) could be bound by RBP (Lai and Blackshear, 2001), and the core sequence of ARE is AUUUA. In this study, the distributions of AUUUA sequence in ABA signaling-related genes were analyzed. Interestingly, four AUUUA sequences were found in the 3′UTR of ABI2 while zero or fewer AUUUA sequences were found in other genes (Figure 8A). As shown in Figure 8B, the biotin labeled ABI2 3′UTR mRNA lagged in the wells containing SRP1 protein. Moreover, the signals of lagging probes were reduced along with the addition of ABI2 3′UTR mRNA without labeling. By contrast, the biotin labeled ABI3 3′UTR mRNA did not lag in the wells containing SRP1 protein (Figure 8B). These results indicate the activity of SRP1 in binding to ABI2 3′UTR rather than ABI3 3′UTR. In addition, SRP1 was found with the transcriptional activation activity in yeast GAL4 system (Figure 8C). To further confirm the result, we examined the transcriptional activation activity of SRP1 in Arabidopsis protoplast system. As shown in Figure 8D, SRP1 could increase the luciferase activity by four-fold, indicating the transcriptional activation role of SRP1. The SRP1-green fluorescent protein (GFP) fusion protein was transiently expressed in Arabidopsis protoplasts. The SRP1 was revealed to be localized in nucleus of Arabidopsis protoplast cells (Figure 8E). The result suggested the function of SRP1 was performed in the nucleus.
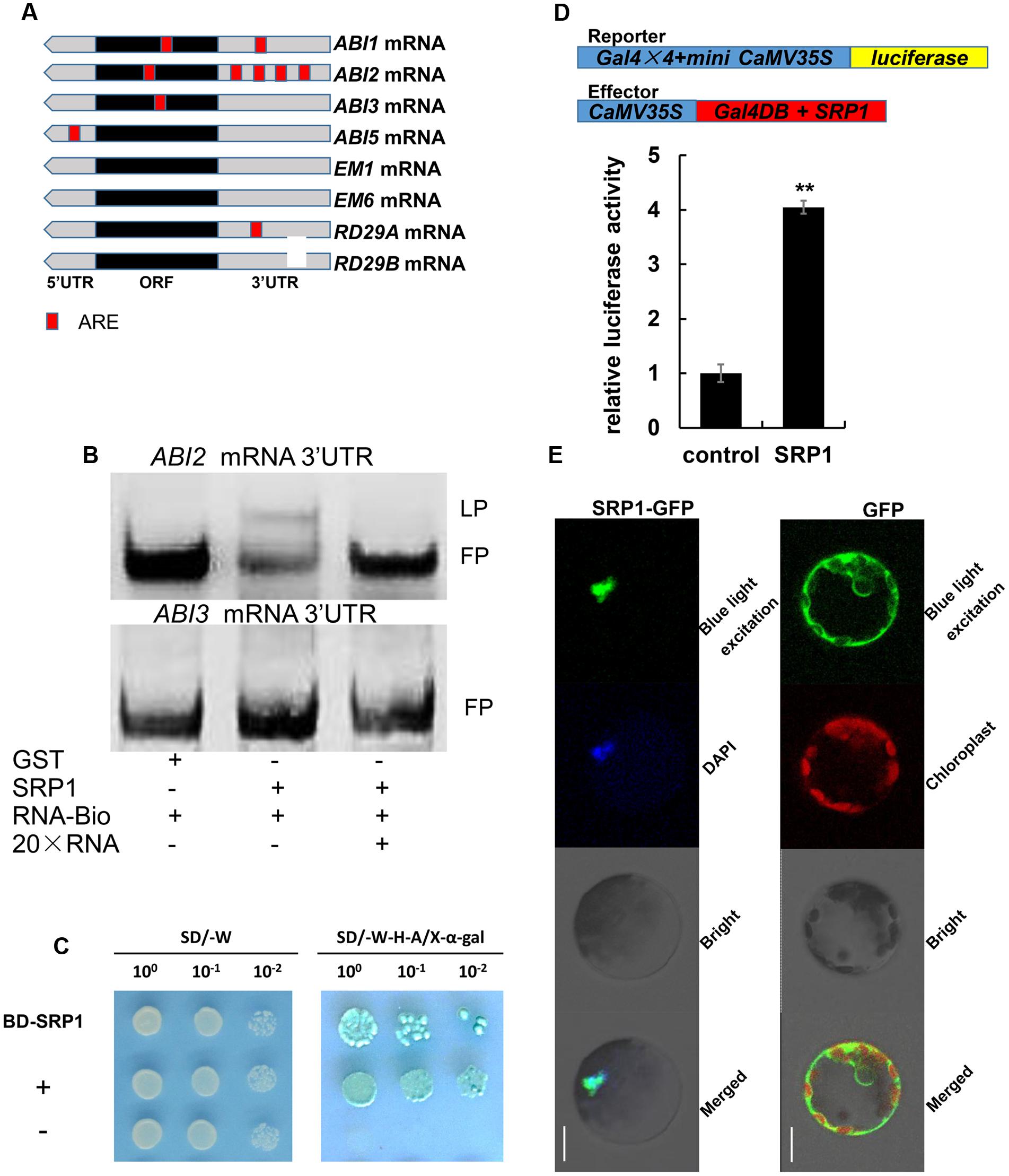
FIGURE 8. The ABI2 mRNA binding and transcriptional activity of SRP1. (A) The core sequence of ARE element (AUUUA) in ABA signaling-related genes. The red boxes depict the location of AUUUA sequence. The gray and black boxes depict the UTR and ORF, respectively. (B) The RNA-EMSA analysis of SRP1 binding ability to ABI2/3 3′UTR. GST protein was used as the negative control. The RNA (ABI2/3 3′UTR) labeled with biotin was used as the probe. The unlabeled RNA (ABI2/3 3′UTR) was used as the competitor. LP, lagged probe. FP, free probe. (C) Transactivation analysis of SRP1 in the yeast GAL4 system. SRP1-pGBKT7 was transformed into yeast to analyze the transcriptional activity (BD-SRP1). The autonomously activated SRP1-pGBKT7 indicated the transcriptional activity of SRP1. The transformation of empty pGBKT7 was used as the negative control (–) while the co-transformation of pGBKT7-53 and pGADT7-T was used as the positive control (+). (D) Transactivation analysis of SRP1 in Arabidopsis protoplast system. 35S-Gal4DB was used as the negative control (control). The 35S-SRP1 was used as the effector to test transactivation activity of SRP1 (SRP1). Relative activity was normalized by the activity of Renilla reniformis luciferase. “∗∗” represents significant difference relative to each control and P-value < 0.01, based on student’s t-test. Each value was the mean ±SD of three biological determinations. (E) Subcellular localization of SRP1. The subcellular localization of SRP1 was analyzed in Arabidopsis leaf protoplasts. The green fluorescent protein SRP1-GFP fusion product was transiently expressed in Arabidopsis leaf protoplasts. The nucleus was dyed with DAPI.
SRP1 Affects the Stability of mRNA Fused with ABI2 3′UTR
The effect of SRP1 on ABI2 3′UTR-containing mRNA stability was examined in Arabidopsis protoplast cells to confirm whether the binding of SRP1 to ABI2 3′UTR mediated mRNA silencing (Figure 9). In this experiment, two reporter vectors, LUC-ABI2 3′UTR and LUC-ABI3 3′UTR were constructed by inserting the ABI2 or ABI3 3′UTR into the downstream of luciferase coding sequence in the control reporter LUC (Figure 9A). All of the reporter vectors were trans-activated by dehydration-responsive element-binding protein 1A (DREB1A), but the luciferase activity was reduced if an additional ABI2 3′UTR was fused with luciferase gene (Figure 9B). Importantly, the luciferase activity of LUC-ABI2 3′UTR was reduced by the addition of SRP1 while the luciferase activities of LUC and LUC-ABI3 3′UTR were enhanced (Figure 9B). Considering the transcriptional activity of SRP1 observed in yeast and protoplast systems, all three of these vectors were supposed to be enhanced. Hence, the decreased luciferase activity of LUC-ABI2 3′UTR confirmed the role of SRP1 in reducing the stability of mRNA with 3′UTR of ABI2.
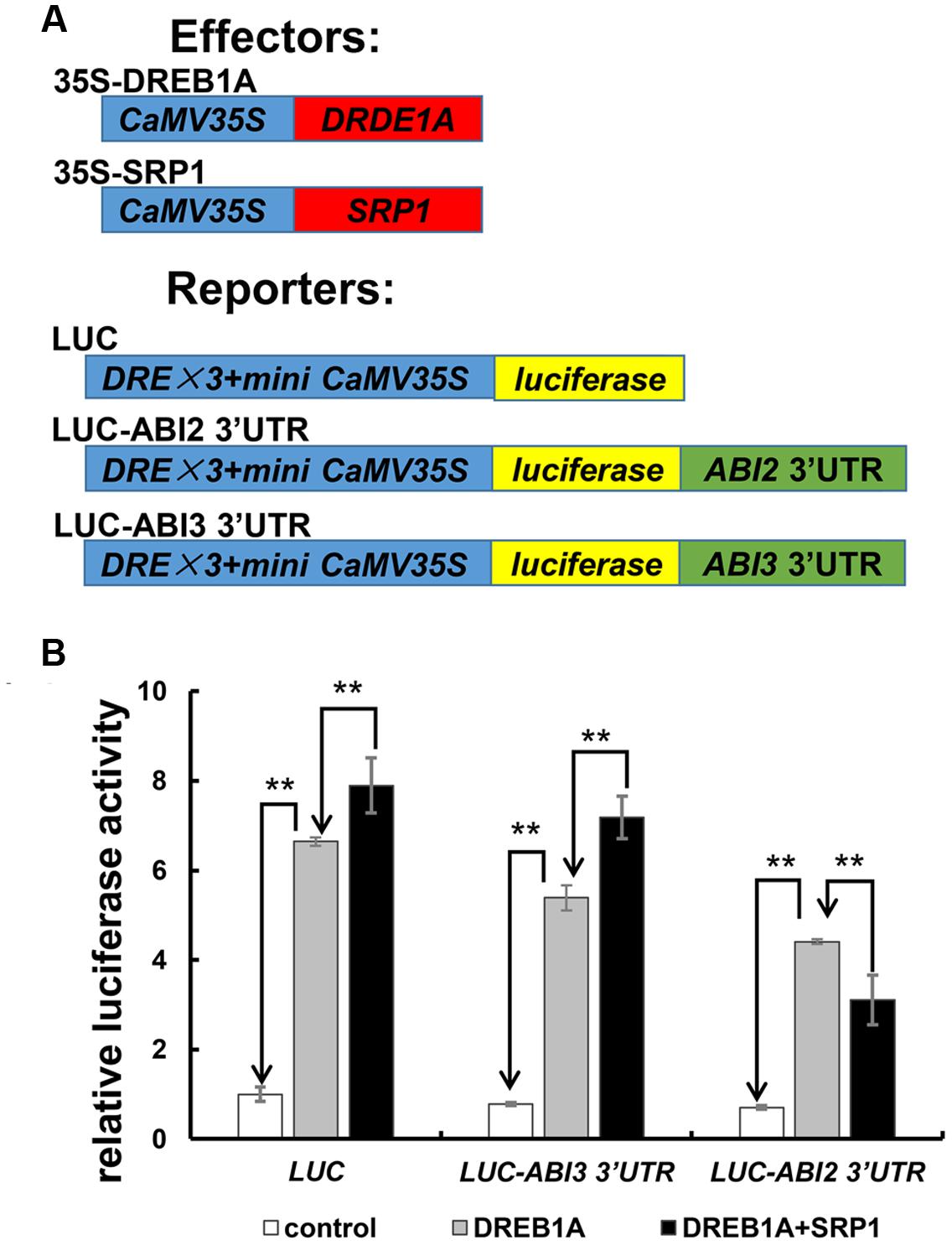
FIGURE 9. The effect of SRP1 on the stability of mRNA with ABI2 3′UTR. (A) Effectors and reporters used in transient expression analysis. 35S-DREB1A and 35S-SRP1 was used as the effectors. LUC effector was constructed by inserting three copies of DRE and a mini 35S into the upstream of the Luciferase in pGL3-basic vector. LUC-ABI2/ABI3 3′UTR effector was constructed by inserting the 3′UTR of ABI2/ABI3 into the downstream of Luciferase in LUC. (B) The effect of SRP1 on the activities of reporter vectors (LUC, LUC-ABI2 3′UTR and LUC-ABI3 3′UTR). The empty PA7 vector was used as the negative control (control). As SRP1 possessed the transactivation activity, 35S-DREB1A effector was used as the positive control to enhance the expression of reporter (DREB1A). The 35S-DREB1A, 35S-SRP1 and reporters were co-transfected into Arabidopsis protoplasts to test the effect of SRP1 on the stability of mRNA (DREB1A + SRP1). Relative activity was normalized by the activity of Renilla reniformis luciferase. “∗∗” represents significant difference relative to each control and P-value < 0.01, based on student’s t-test. Each value was the mean ±SD of three biological determinations.
Discussion
The post-transcriptional regulation in ABA signaling is critical but not fully known. In present study, a novel RBP was found regulating ABA signaling at post-transcriptional level. Several RBPs containing CCCH-type zinc finger motif have been characterized, such as atRZ-1a, OsTZF1 and AtTZF1 (Kim et al., 2007; Ambrosone et al., 2012; Qu et al., 2014). The CCCH-type ZnF motif is required to achieve high-affinity RNA-binding (Qu et al., 2014). SRP1 contains four C2C2-type ZnF motifs, and it has been poorly studied in plant. The mammal protein RanBP2 containing the similar motif has been reported to recognize and bind to the ssRNA (Nguyen et al., 2011). Unlike RanBP2 in mammals, SRP1 only contains four zinc finger motifs while RanBP2 is a big protein containing multiple functional domains besides zinc finger motifs. Other similar zinc finger motif containing proteins in mammal, such as ZnF Ran binding domain-containing protein 2 (ZRANB2) and RNA-binding motif 5 (RBM5), are reported to have the ssRNA-binding activity (O’Connell et al., 2012; Vandevenne et al., 2014). The database searching indicates that SRP1 is plant-specific and widely distributed in plant species, implying they may play a specific role in plants through the C2C2-type ZnF motifs.
Many RBPs have been shown with the involvements in abiotic stress (Ambrosone et al., 2012). The mRNA expression of a glycine-rich RBP atRZ1-a is down-regulated by ABA (Kim et al., 2005), and the expression of RRM containing RBP ARP1 is also down-regulated by ABA (Jung et al., 2013). With the treatment of ABA, transgenic Arabidopsis plants overexpressing atRZ-1a display retarded germination and seedling growth compared with the WT plants while atrz-1a mutant germinates earlier and grows faster than the WT plants (Kim et al., 2007). Similarly, the expression of SRP1 was down-regulated by ABA and abiotic stress. The srp1 knock-out mutants showed ABA insensitivity, while 35S::SRP1 plant showed ABA hypersensitivity during the seed germination and post-germinative growth stages (Figures 4 and 5). These results support the notion that SRP1 is a positive component of ABA-triggered responses in seed germination and seedling growth.
ABI proteins play important roles in ABA signaling during the germination. ABI1. ABI2. ABI3, and ABI5 are induced by ABA, and the knock-out mutants of them are insensitive to ABA during the germination stage (Suzuki et al., 2003). With the treatment of ABA, these ABI genes were differentially regulated in srp1 knock-out mutants in comparison with WT. Among them, ABI3 and ABI5 were significantly less induced by ABA in the mutants than WT. In contrast, ABI1 was slightly more induced by ABA in the mutants than WT, while the expression of ABI2 was significantly increased. Among these proteins, ABI3 is a B3 domain transcription factor playing an important role in inhibiting the germination under ABA treatment (Rohde et al., 2000). ABI1 and ABI2 are protein phosphatase 2C (PP2C) proteins negatively regulating ABA response (Schweighofer et al., 2004; Hubbard et al., 2010), and the induction of them by ABA is feedback inhibited by ABI3 (Suzuki et al., 2003). It seems that, the enhanced expression of ABI1 and ABI2 in srp1 negatively regulates the seed sensitivity to ABA. The basic leucine zipper (bZIP)-type transcription factor ABI5, activated by ABI3, is an positive regulator in ABA signaling (Nakabayashi et al., 2005). Moreover, the expressions of EM1. EM6. RD29A and RD29B were less induced in the srp1 mutants. Previous studies have revealed that RD29A and RD29B are up-regulated by ABA and ABI3 (Nakashima et al., 2006). ABI5 induces expressions of EM1 and EM6 which can inhibit the embryo development (Finkelstein and Lynch, 2000). These results suggest that the deficiency of SRP1 decreases the seed sensitivity to ABA by inhibiting the expressions of ABI3 and ABI5. Subsequently, EM1 and EM6 expression levels are decreased and the germination is delayed.
ABI2 plays important roles in both ABA and salt stress responses. The abi2-1 exhibits increased tolerance to salt shock and ABA insensitivity by disrupting the interaction between SOS2 and ABI2 (Ohta et al., 2003). HDA6 mutant, axe1-5, is hypersensitive to ABA and salt stress, and the expression of ABI2 in that is decreased while treated with ABA or salt stress (Chen et al., 2010). Contrary to the phenotype of axe1-5 under ABA, the srp1 mutants showed reduced salt sensitivity during the seed germination and post-germinative growth stages (Figure 6), which was most probably due to the increased transcript level of ABI2.
Post-transcriptional regulation involves mRNA transport, localization, stability or translation through the interactions between RBPs and mRNAs (Tam et al., 2010; Qu et al., 2014). A well-documented example is AU rich element (ARE)-interacting protein hTTP, which can bind to the 3′UTR of mRNAs encoding regulators involved in innate immune responses and promote the degradation of them (Lai and Blackshear, 2001; Barreau et al., 2005). The 3′UTR of ABI2 mRNA contains four core sequences of ARE (AUUUA), while no AUUUA sequence is present in the 3′UTR of ABI3. In accordance with this, SRP1 could interact with 3′UTR of ABI2 rather than 3′UTR of ABI3. Correspondingly, SRP1 could cause the instability of the mRNA fused with ABI2 3′UTR rather than ABI3 3′UTR. These evidences support the hypothesis that SRP1 functions as an RBP triggering the instability of ABI2 mRNA in ABA signaling by binding to its 3′UTR. Unlike the pre-mRNA splicing related CBP20 and RZ-1, SRP1 possesses function by regulating the stability of mRNA.
SRP1 was localized in the nucleus of Arabidopsis, and the function of it most probably performed there. Several RNase proteins are localized in same organelle, such as RNase III DICER-Like1 (DCL1), RNase D, RNase P, RNase Z, etc (Dubrovsky et al., 2003; Glazov et al., 2003; Song et al., 2007; Gobert et al., 2013). Those RNase proteins can cleave the special RNA sequences and play important roles in processing of RNA. DCL1 is essential for the accumulation of microRNA (Song et al., 2007). RNase D is a 3′ to 5′ exonuclease required for the processing of RNA (Glazov et al., 2003). RNase P plays the critical role in removing 5′-leader sequences from tRNA precursors (Gobert et al., 2013). RNase Z is important for the generation of mature tRNA 3′ end (Dubrovsky et al., 2003). The further study is necessary to reveal whether SRP1 possesses the cleaving function as nucleus-localized RNase or cooperates with them.
Our work demonstrates that SRP1 is an RBP functioning in ABA signaling and exerts its roles by modulating the levels of ABI genes (Figure 10). Among these genes, ABI2 is directly post-transcriptionally modulated by SRP1. More studies are required to reveal the relationships between SRP1 and other ABI genes, e.g., ABI3 and ABI5. The transcriptional activation activity of SRP1 implies the dual roles of SRP1 in transcriptional and post-transcriptional regulations on target genes. Conclusively, the present study identified a novel plant-specific RNA-binding protein involving ABA signaling by post-transcriptionally repressing ABI2.
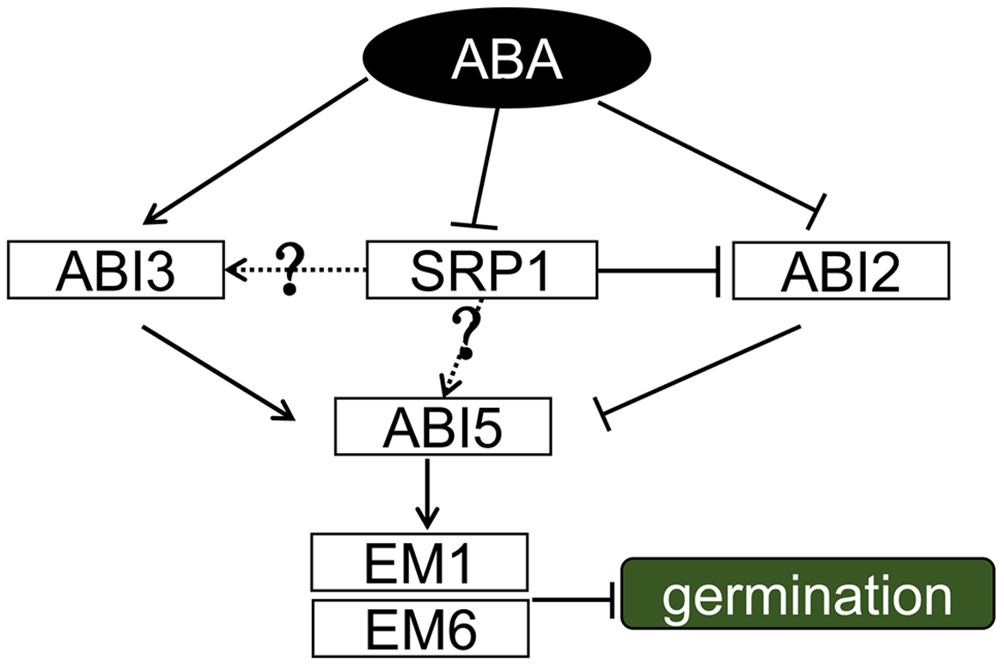
FIGURE 10. A proposed working model for SRP1. During ABA signaling, ABA induced expression of ABI3/5 and inhibited the activities of ABI2. ABI2 was post-transcriptional modulated by SRP1, and the degradation of ABI2 mRNA was repressed when SRP1 is down-regulated under ABA. ABI3 and ABI5 were down-regulated in srp1. It was assumed that the down-regulation of SRP1 under ABA results in the suppression of ABI3 and ABI5, but more evidence was necessary to demonstrate it. The solid lines depict the relationships proved in this or previous studies, and the dotted lines and question marks depict the unproved relationships.
Author Contributions
JH planned the project. JX, YC, RM, XY, HF, XH, EX, and LQ performed the experiments. JX, HZ, and JH analyzed the data. JX and JH wrote the article.
Funding
This work was supported by the Natural Science Foundation of China (31571627, 91335106), the National Key Technology R&D Program (2015BAD01B01), Natural Science Foundation of Jiangsu Province (BK20141362) and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank Dr. Magnus Holm and Dr. Donging Xu from Department of Biological and Environmental Sciences of Gothenburg University for their valuable suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00024/full#supplementary-material
Footnotes
References
Abel, S., and Theologis, A. (1994). Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J. 5, 421–427. doi: 10.1111/j.1365-313X.1994.00421.x
Alba, M., and Pagès, M. (1998). Plant proteins containing the RNA-recognition motif. Trends Plant Sci. 3, 15–21. doi: 10.1016/S1360-1385(97)01151-5
Ambrosone, A., Costa, A., Leone, A., and Grillo, S. (2012). Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci. 182, 12–18. doi: 10.1016/j.plantsci.2011.02.004
Barreau, C., Paillard, L., and Osborne, H. B. (2005). AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 33, 7138–7150. doi: 10.1093/nar/gki1012
Chen, L. T., Luo, M., Wang, Y. Y., and Wu, K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cook, K. B., Kazan, H., Zuberi, K., Morris, Q., and Hughes, T. R. (2011). RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 39, D301–D308. doi: 10.1093/nar/gkq1069
Dubrovsky, E. B., Dubrovskaya, V. A., Levinger, L., Schiffer, S., and Marchfelder, A. (2003). Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res. 32, 255–262. doi: 10.1093/nar/gkh182
Fang, H., Meng, Q., Xu, J., Tang, H., Tang, S., Zhang, H., et al. (2015). Knock-down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol. Biol. 87, 441–458. doi: 10.1007/s11103-015-0294-1
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Glazov, E., Phillips, K., Budziszewski, G. J., Meins, F. Jr., and Levin, J. Z. (2003). A gene encoding an RNase D exonuclease-like protein is required for post-transcriptional silencing in Arabidopsis. Plant J. 35, 342–349. doi: 10.1046/j.1365-313X.2003.01810.x
Gobert, A., Pinker, F., Fuchsbauer, O., Gutmann, B., Boutin, R., Roblin, P., et al. (2013). Structural insights into protein-only RNase P complexed with tRNA. Nat. Commun. 4:1353. doi: 10.1038/ncomms2358
Gutierrez, L., Bussell, J. D., Păcurar, D. I., Schwambach, J., Păcurar, M., and Bellini, C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119–3132. doi: 10.1105/tpc.108.064758
Huang, J., Wang, M. M., Jiang, Y., Wang, Q. H., Huang, X., and Zhang, H. S. (2008). Stress repressive expression of rice SRZ1 and characterization of plant SRZ gene family. Plant Sci. 174, 227–235. doi: 10.1016/j.plantsci.2007.11.010
Hubbard, K. E., Nishimura, N., Hitomi, K., Getzoff, E. D., and Schroeder, J. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24, 1695–1708. doi: 10.1101/gad.1953910
Jung, H. J., Kim, M. K., and Kang, H. (2013). An ABA-regulated putative RNA-binding protein affects seed germination of Arabidopsis under ABA or abiotic stress conditions. J. Plant Physiol. 170, 179–184. doi: 10.1016/j.jplph.2012.09.002
Kim, Y. O., Kim, J. S., and Kang, H. (2005). Cold-inducible zinc finger containing glycine rich RNA binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 42, 890–900. doi: 10.1111/j.1365-313X.2005.02420.x
Kim, Y. O., Pan, S., Jung, C. H., and Kang, H. (2007). A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol. 48, 1170–1181. doi: 10.1093/pcp/pcm087
Kong, X., Ma, L., Yang, L., Chen, Q., Xiang, N., Yang, Y., et al. (2014). Quantitative proteomics analysis reveals that the nuclear cap-binding complex proteins arabidopsis CBP20 and CBP80 modulate the salt stress response. J. Proteome Res. 13, 2495–2510. doi: 10.1021/pr4012624
Lai, W. S., and Blackshear, P. J. (2001). Interactions of CCCH zinc finger proteins with mRNA: tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. J. Biol. Chem. 276, 23144–23154. doi: 10.1074/jbc.M100680200
Lamond, A. I., and Spector, D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612. doi: 10.1038/nrm1172
Léon-Kloosterziel, K. M., Gil, M. A., Ruijs, G. J., Jacobsen, S. E., Olszewski, N. E., Schwartz, S. H., et al. (1996). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10, 655–661. doi: 10.1046/j.1365-313X.1996.10040655.x
Liu, J. X., Srivastava, R., Che, P., and Howell, S. H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19, 4111–4119. doi: 10.1105/tpc.106.050021
Liu, X., Zhang, H., Zhao, Y., Feng, Z., Li, Q., Yang, H. Q., et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 15485–15490. doi: 10.1073/pnas.1304651110
Lorković, Z. J. (2009). Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 14, 229–236. doi: 10.1016/j.tplants.2009.01.007
Lorković, Z. J., and Barta, A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635. doi: 10.1093/nar/30.3.623
Nakabayashi, K., Okamoto, M., Koshiba, T., Kamiya, Y., and Nambara, E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 41, 697–709. doi: 10.1111/j.1365-313X.2005.02337.x
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. doi: 10.1007/s11103-005-2418-5
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148.
Nguyen, C. D., Mansfield, R. E., Leung, W., Vaz, P. M., Loughlin, F. E., Grant, R. P., et al. (2011). Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J. Mol. Biol. 407, 273–283. doi: 10.1016/j.jmb.2010.12.041
O’Connell, M. R., Vandevenne, M., Nguyen, C. D., Matthews, J. M., Gamsjaeger, R., Segal, D. J., et al. (2012). Modular assembly of RanBP2-type zinc finger domains to target single-stranded RNA. Angew. Chem. Int. Ed. Engl. 51, 5371–5375. doi: 10.1002/anie.201200866
Ohta, M., Guo, Y., Halfter, U., and Zhu, J. K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. U.S.A. 100, 11771–11776. doi: 10.1073/pnas.2034853100
Owttrim, G. W. (2006). RNA helicases and abiotic stress. Nucleic Acids Res. 34, 3220–3230. doi: 10.1093/nar/gkl408
Papp, I., Mur, L. A., Dalmadi, A., Dulai, S., and Koncz, C. (2004). A mutation in the cap binding protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol. Biol. 55, 679–686. doi: 10.1007/s11103-004-1680-2
Qu, J., Kang, S. G., Wang, W., Musier-Forsyth, K., and Jang, J. C. (2014). The Arabidopsis thaliana tandem zinc finger 1 (AtTZF1) protein in RNA binding and decay. Plant J. 78, 452–467. doi: 10.1111/tpj.12485
Rohde, A., Kurup, S., and Holdsworth, M. (2000). ABI3 emerges from the seed. Trends Plant Sci. 5, 418–419. doi: 10.1016/S1360-1385(00)01736-2
Schweighofer, A., Hirt, H., and Meskiene, I. (2004). Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9, 236–243. doi: 10.1016/j.tplants.2004.03.007
Song, J. J., Liu, J., Tolia, N. H., Schneiderman, J., Smith, S. K., Martienssen, R. A., et al. (2003). The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10, 1026–1032. doi: 10.1038/nsb1016
Song, L., Han, M. H., Lesicka, J., and Fedoroff, N. (2007). Arabidopsis pri-miRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. U.S.A. 104, 5437–5442. doi: 10.1073/pnas.0701061104
Suzuki, M., Ketterling, M. G., Li, Q.-B., and McCarty, D. R. (2003). Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 132, 1664–1677. doi: 10.1104/pp.103.022475
Tam, P. P., Barrette-Ng, I. H., Simon, D. M., Tam, M. W., Ang, A. L., and Muench, D. G. (2010). The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 10:44. doi: 10.1186/1471-2229-10-44
Vandevenne, M., O’Connell, M. R., Helder, S., Shepherd, N. E., Matthews, J. M., Kwan, A. H., et al. (2014). Engineering specificity changes on a RanBP2 zinc finger that binds single-stranded RNA. Angew. Chem. Int. Ed. Engl. 53, 7848–7852. doi: 10.1002/anie.201402980
Wang, S., Wang, R., Liang, D., Ma, F., and Shu, H. (2012). Molecular characterization and expression analysis of a glycine-rich RNA-binding protein gene from Malus hupehensis Rehd. Mol. Biol. Rep. 39, 4145–4153. doi: 10.1007/s11033-011-1197-0
Wu, Z., Zhu, D., Lin, X., Miao, J., Gu, L., Deng, X., et al. (2015). RNA-binding proteins At RZ-1B and At RZ-1C play a critical role in regulation of pre-mRNA splicing and gene expression during Arabidopsis development. Plant Cell 23, 2819–2824. doi: 10.1105/tpc.15.00949
Keywords: abiotic stress, ABA signaling, ABI2, Arabidopsis thaliana, RNA-binding protein, zinc finger protein
Citation: Xu J, Chen Y, Qian L, Mu R, Yuan X, Fang H, Huang X, Xu E, Zhang H and Huang J (2017) A Novel RNA-Binding Protein Involves ABA Signaling by Post-transcriptionally Repressing ABI2. Front. Plant Sci. 8:24. doi: 10.3389/fpls.2017.00024
Received: 25 October 2016; Accepted: 05 January 2017;
Published: 24 January 2017.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Fan Chen, Institute of Genetics and Developmental Biology (CAS), ChinaHunseung Kang, Chonnam National University, South Korea
Diqiu Yu, Xishuangbanna Tropical Botanical Garden (CAS), China
Copyright © 2017 Xu, Chen, Qian, Mu, Yuan, Fang, Huang, Xu, Zhang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji Huang, huangji@njau.edu.cn
 Jianwen Xu
Jianwen Xu Yihan Chen
Yihan Chen Luofeng Qian1,3
Luofeng Qian1,3 Xi Huang
Xi Huang Ji Huang
Ji Huang