- 1Department of Life Science, Sogang University, Seoul, South Korea
- 2Department of Plant Molecular Systems Biotechnology, Kyung Hee University, Yongin, South Korea
Salt stress can severely reduce crop yields. To understand how rice (Oryza sativa) plants respond to this environmental challenge, we investigated the genes involved in conferring salt tolerance by screening T-DNA tagging lines and identified OsSta2-D (Oryza sativa Salt tolerance activation 2-Dominant). In that line, expression of OsSta2 was enhanced by approximately eightfold when compared with the non-transformed wild type (WT). This gene was highly expressed in the callus, roots, and panicles. To confirm its role in stress tolerance, we generated transgenic rice that over-expresses OsSta2 under a maize ubiquitin promoter. The OsSta2-Ox plants were salt-tolerant at the vegetative stage, based on our calculations of chlorophyll fluorescence (Fv/Fm), fresh and dry weights, chlorophyll concentrations, and survival rates. Under normal paddy field conditions, the Ox plants were somewhat shorter than the WT control but had improved agronomic traits such as higher total grain yield. They were also more tolerant to osmotic stress and hypersensitive to abscisic acid. Based on all of these results, we suggest that OsSta2 has important roles in determining yields as well as in conferring tolerance to salt stresses.
Introduction
For more than half of the world’s people, rice (Oryza sativa) is a major food crop. Global demand for this grain will rise as populations continue to grow. Diverse environmental stresses cause plants to respond at the molecular level by altering the expression of different sets of regulatory or signaling genes as well as genes that encode proteins related to stress tolerance (Apse and Blumwald, 2002; Seki et al., 2003; Shinozaki et al., 2003; Wang et al., 2003; Kumar et al., 2013; Fahad et al., 2015; Kazan, 2015; Parihar et al., 2015; Petrov et al., 2015). Drought and salt stresses are common environmental factors that restrict rice productivity (Yeo and Flowers, 1984; Xoconostle-Cázares et al., 2010; Das et al., 2015; Fita et al., 2015). On high-salinity soils, annual grain yields can be reduced by 30–50% (Eynard et al., 2005). Significant progress has been made in understanding the mechanism(s) for salt tolerance in many plant species, including rice (Kumar et al., 2013; Deinlein et al., 2014; Parihar et al., 2015). Under salt stress, cells can be protected and normal plant growth maintained through cellular responses such as cytosolic calcium release, ionic imbalances in the vacuole, stress signal transduction, and expression of several regulatory genes (Kasuga et al., 1999; Kader and Lindberg, 2010; Ismail et al., 2014). Because all of these responses indicate that various species utilize a common set of signaling pathways and genes, researchers can exploit this to engineer plants with greater salt tolerance.
Transcription factors (TFs) such as AP2/ERF, bZIP, MYB, NAC, zinc-finger, MYC, and WRKY are important because they can regulate the downstream expression of many stress-responsive genes (Bhatnagar-Mathur et al., 2008; Joshi et al., 2016; Wang et al., 2016). Transgenic application of TFs is a useful approach for developing plants that are more tolerant to abiotic stresses. Among them, AP2/ERFs have multiple roles in plants, controlling processes such as, leaf epidermal cell identity; the development of leaf petioles, flowers, and embryos; and fruit ripening (Elliott et al., 1996; Moose and Sisco, 1996; van der Graaff et al., 2000; Boutilier et al., 2002; Wang et al., 2007; Krizek, 2009; Licausi et al., 2013).
The AP2/ERF proteins are also involved in plant responses to biotic stress. For example, ERF proteins modulate the expression of many pathogenesis-related genes by binding to GCC box (AGCCGCC) (Ohme-Takagi and Shinshi, 1995; Solano et al., 1998; Fujimoto et al., 2000; Gu et al., 2002; Onate-Sanchez and Singh, 2002; Onate-Sanchez et al., 2007; Liu et al., 2012; Zhao et al., 2012; Jisha et al., 2015; Mishra et al., 2015). Proteins such as ERN1, -2, -3, and EFD from Medicago truncatula regulate the development of legume root nodules to establish symbiosis with nitrogen-fixing bacteria (Andriankaja et al., 2007; Middleton et al., 2007). Likewise, the miR172-AP2-1 node acts as a key regulator of nitrogen fixation in the symbiotic relationship of Phaseolus vulgaris–Rhizobium etli (Nova-Franco et al., 2015).
Apart from their role in biotic stress responses, AP2/ERF proteins also participate in response to abiotic stresses such as drought, salt, and cold (Nakano et al., 2006; Xu et al., 2011; Mizoi et al., 2012; Licausi et al., 2013; Fu et al., 2014; Jisha et al., 2015). These proteins contain a conserved AP2/ERF domain (Riechmann et al., 2000; Sharoni et al., 2011; Licausi et al., 2013). One of the best-studied is a group of CBF/DREBs that activate the expression of many stress-related genes and improve drought, salt, and cold tolerance (Stockinger et al., 1997; Liu et al., 1998; Kasuga et al., 1999; Sakuma et al., 2006; Lata and Prasad, 2011; Schmidt et al., 2013; Zhuang et al., 2013; Rong et al., 2014; Yang et al., 2014, 2016; Duan et al., 2015).
Rice (O. sativa ssp. japonica) has at least 139 AP2/ERF family genes (Nakano et al., 2006), and various environmental stresses induce their expression (Dubouzet et al., 2003; Cao et al., 2006; Fukao et al., 2006; Xu et al., 2006; Liu et al., 2007; Hattori et al., 2009). For example, genes for the AP2/ERF proteins SNORKEL1 and SNORKEL2 promote the accumulation of gibberellic acid in deep-water rice and rapid stem elongation under flooding conditions as an escape strategy (Hattori et al., 2009). In contrast, the AP2/ERF protein SUB1A-1 in submergence-tolerant rice varieties is part of a quiescence strategy that prevents shoot elongation and increases their rate of survival (Xu et al., 2006). Constitutive expression in rice of AP2/ERF genes such as DREB1A, HARDY (from Arabidopsis), HvCBF4 (from Hordeum vulgare), and TERF1 (from Solanum lycopersicon) enhances tolerance to abiotic stress (Oh et al., 2005, 2007; Karaba et al., 2007; Gao et al., 2008), while overexpression of the rice AP2/ERF gene AP37 increases drought tolerance at the vegetative stage and leads to higher grain yields (Oh et al., 2009). Overexpression in rice of TSRF1, another AP2/ERF protein, also improves tolerance to osmotic stress and drought (Zhang et al., 2004, 2007; Quan et al., 2010). Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice (Schmidt et al., 2013) while the rice ERF TF factor OsERF922 negatively regulates resistance to the development of salt tolerance (Liu et al., 2012). Furthermore, overexpression of rice OsEREBP1 increases tolerance to both biotic and abiotic stresses (Jisha et al., 2015). Based on these earlier reports, rice functional genomics, including reverse and forward genetics methods, is now an important research field for identifying novel genes involved in plant stress responses and tolerance. These genes can become new targets for genetic engineering of rice and other crops to improve tolerance.
In this study, we characterized a gene that is induced by several types of stress. Overexpression of OsSta2 made rice plants more tolerant to oxidative and salt stresses at the seedling and vegetative stages, respectively. This overexpression also helped improve overall agronomical traits under normal paddy field conditions.
Materials And Methods
Plant Materials
Rice (O. sativa ssp. japonica cv. Dongjin) seeds were surface-sterilized and germinated in a wet paper towel for 2 days. The resultant seedlings were cultured in a walk-in growth chamber (Koencon, South Korea) under conditions of 30°C [day/22°C (night) and a 12-h photoperiod (Lee et al., 2015)].
Abiotic Stress Treatments and Assays of Stress Tolerance
Gene expression was analyzed using rice seedlings that had been hydroponically cultured in Yoshida solution (Yoshida et al., 1976). At 8 DAG, they were exposed to various types of stress for 0, 1, 3, 6, 12, or 24 h. The treatments included drought (water removal), salt (300 mM NaCl), cold (4°C), or abscisic acid (100 μM ABA). After the treatment period, 100 mg leaf tissue was collected for RNA extraction.
To test the extent of tolerance in our transgenic rice lines, we sowed seeds in a soil box. At 8 DAG, the seedlings underwent drought stress when water was withheld for 30–40 h until the leaves wilted. To induce salt stress, 8 DAG seedlings were transferred to either 100 mM NaCl for 7 days or 250 mM NaCl solution for 72 h. To examine their response to a low temperature, we incubated 8 DAG seedlings for 48–72 h at 4°C (Koencon, South Korea). At the end of each treatment period, the plants were returned to normal growing conditions for 6 days of recovery before their phenotypes were recorded and their survival rates were calculated. For all treatments, dry weights were determined after the plants had been dried at 80°C for 2 days.
To examine osmotic stress tolerance and ABA sensitivity, we germinated surface-sterilized, de-hulled rice seeds on a half-strength MS medium for 5 days before transferring the seedlings to a half-strength MS medium supplemented with 0 or 200 mM mannitol, or with 0, 5, or 10 μM ABA. Seedlings were oriented vertically and their growth was observed 7 days after this transfer (Kim H. et al., 2012). The stress tolerance assay also included an examination of chlorophyll fluorescence. Briefly, the fifth leaves from 12 DAG seedlings were removed and incubated in 500 mM NaCl for 48 h, then either air-dried for 3 h (28°C; 110 μmol m-2 s-1) or incubated at 4°C in deionized water for up to 48 h (4°C; 110 μmol m-2 s-1). The Fv/Fm values, which represent the photochemical efficiency of PSII in a dark-adapted state (Fv, variable fluorescence; Fm, maximum fluorescence) were calculated with data obtained by using a Mini-PAM-II Photosynthesis Yield Analyzer (Walz, Germany). A leaf disk assay was conducted to examine salt tolerance. Healthy and fully expanded leaves (∼60 DAG) were washed in deionized autoclaved water before 1-cm-diameter disks were cut and floated for 24 h in 30-mL solutions of various concentrations of NaCl (100, 200, or 250 mM) (Tuteja et al., 2013). The effects of salt stress were represented as phenotypic changes and quantifications of chlorophyll (Arnon, 1949). Briefly, 1-cm disks were ground and extracted with 80% acetone. Absorption was measured at 645 and 663 nm with a spectrophotometer (Shimadzu, Japan).
Screening of Activation Tagging Lines for Salt Tolerance
Rice T-DNA tagging mutants were screened for salt tolerance (100 or 250 mM NaCl) by using a mixed pool of approximately 5,000 individuals from the T2 generation of a T-DNA ATL (Jeong et al., 2002, 2006). After 2–7 days of induced salt stress, followed by 6 days of recovery, a mutant line showing enhanced tolerance (based on a high survival rate) was identified and further characterized.
Inverse PCR (IPCR) was performed by Cla1 cutting in our pGA2715-tagged lines (Jeong et al., 2002; Jung et al., 2003), the primers for the 1st and 2nd IPCR included in Supplementary Table S2. Samples were amplified for 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 5 min. Aliquots from the primary PCR products were used for the secondary PCR reaction and then the PCR products were directly sequenced. Genomic sequences containing the tagging sequence were retrieved from Rice GE Database1.
Gene Expression Analysis by RT-PCR and qRT-PCR
Total RNA was isolated from rice leaf samples with an RNeasy Mini Plant Kit (Qiagen, Germany) and cDNAs were synthesized with RT Complete Kits (Biofact, South Korea), according to the manufacturers’ instructions. Primers were designed with Gene Runner software2 and NCBI primer blast3. Primer pairs (Supplementary Table S1) were used at concentration of 5–10 picomoles. In addition, 3 μL of cDNA (6 ng of total RNA) was used as template. All RT-PCRs were performed at an initial 95°C for 5 min, followed by 25–35 cycles of 95°C for 30 s, 58°C for 30–60 s, and 72°C for 30–60 s. The PCR products were visualized on a 0.8% agarose gel. The qRT-PCR analysis utilized a SYBER® FAST Universal qPCR Kit (Kapa, South Africa) and a LightCycler® 96 (Roche Life Science, Germany). The qPCR procedures were performed at 95°C for 3 min, followed by 40 amplification cycles of 95°C for 3 s, 60°C for 20 s, and 72°C for 20 s. A melting curve was obtained through a protocol involving 95°C for 5 s, 65°C for 1 min, and 97°C for 1 min; followed by cooling at 40°C for 10 min. Relative expression levels were calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001), using RAc1 as an internal control.
In silico Analysis of the OsSta2 Promoter
Promoter sequences (approximately 2 kb long) upstream of the ATG start codon were analyzed from Oryzabase (Kurata and Yamazaki, 2006), and cis-elements in those promoters were searched in the PLACE database4 (Higo et al., 1999).
Generation of OsSta2 Overexpression Lines
For construction of the OsSta2-overexpression (-Ox) vector, OsSta2 cDNA (J065129D08) was obtained from KOME5. The cDNA was placed between the SacI and BamHI sites by subcloning and then cloned in to the pGA3426 binary vector with a maize ubiquitin promoter and the nos terminator (Kim et al., 2009). Scutellum-derived calli of ‘Dongjin’ rice were transformed by Agrobacterium-mediated co-cultivation methods. 5 days scutella were used for transformation experiments. Subculture was done for 4 days in 2N6 medium (Hiei et al., 1994; Koh et al., 2007). The transgenic plants were then transferred to a confined paddy field for further growth. For segregation analysis of the transgenic lines, seeds were germinated in a half-strength MS medium supplemented with hygromycin (50 mg L-1). The number of surviving seedlings was recorded after incubation at 30°C for 5 days. Lines in which the survival rate was 100% were considered transgene homozygotes.
Investigation of Agricultural Traits
Rice plants were grown from May until the grain was harvested at the end of October. These experiments were conducted annually for 4 years, 2012 through 2015, at the LMO paddy field of Kyungpook National University and Kyung Hee University, South Korea (Permit number, RDA- A-2011-039). To analyze the agricultural traits of rice, we sampled eight plants from each of three independent lines and recorded the numbers of tillers and panicles per plant, the numbers of spikelets and filled grains per panicle, lengths of the panicles and culms, and 1,000-grain weights.
A-2011-039). To analyze the agricultural traits of rice, we sampled eight plants from each of three independent lines and recorded the numbers of tillers and panicles per plant, the numbers of spikelets and filled grains per panicle, lengths of the panicles and culms, and 1,000-grain weights.
Statistical Analysis
Mean values (±SE) were determined from the data set for three replications. Differences between stress treatments were examined with LSD and χ2 tests, and were considered statistically significant at P < 0.05.
Results
Isolation of a Salt Stress-Tolerant Activation Tagging Line
Screening a mixed pool of the T2 generation of PFG T-DNA tagging mutants (Lee et al., 2004; Jeong et al., 2006), we identified Line PFG_3A-05272.R, which had enhanced tolerance to treatments with 100 mM and 250 mM NaCl (Figure 1A and Table 1). Molecular analysis by inverse PCR revealed that the T-DNA was tagged between LOC_Os02g43820 and LOC_Os02g43830 (Figure 1B). However, expression of only LOC_Os02g43820 was induced, by eightfold, when compared with the wild type (WT) (Figure 1C). This gene was named Oryza sativa Salt tolerance activation 2-Dominant, or OsSta2-D (AK241246). Its deduced amino acids contain a 775-bp open reading frame that yields a 56-amino acid protein. Potential stress-related cis-acting elements like, W-box, GT1, MYB, MYC, GATA box, ABRE and ERd1, etc., were found in the 2 kb upstream region of OsSta2 (Buchel et al., 1999; Finkelstein and Lynch, 2000; Chen et al., 2002; Xue, 2002; Yang and Poovaiah, 2002; Itzhaki et al., 1994; Xie et al., 2005) (Supplementary Figure S1 and Table S1).
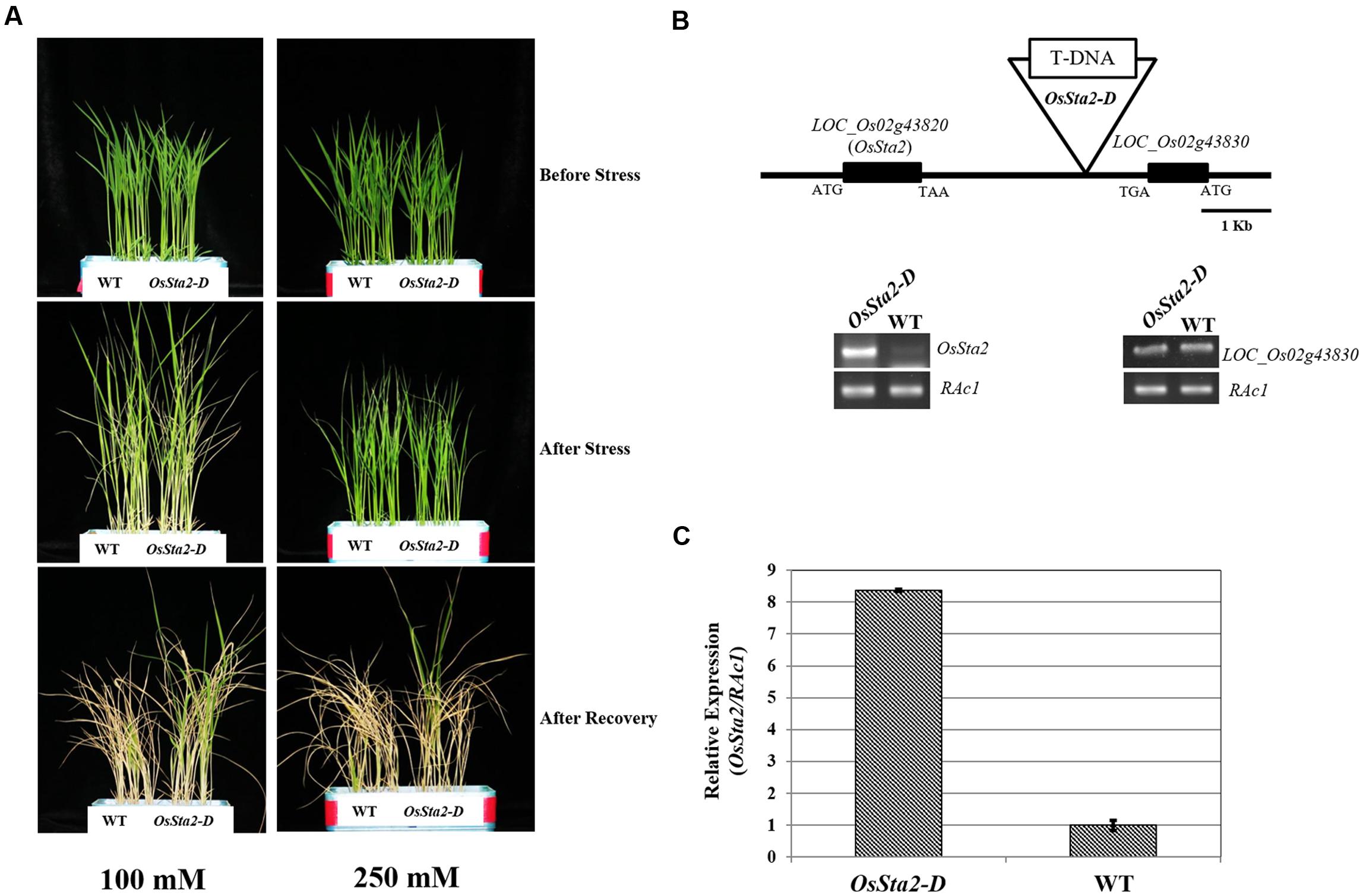
FIGURE 1. Identification of salt-tolerant activation tagging line (ATL) OsSta2-D. (A) Visual phenotypes of salt response by ATL and WT after 8 DAG seedlings were exposed to either 100 mM NaCl for 7 days or 250 mM NaCl for 48 h. All seedlings were returned to normal conditions for 6 days of recovery. (B) Schematic diagram of ATL of OsSta2 and RT-PCR of LOC_Os02g43830 (OsSta2) and LOC_Os02g43830. (C) qRT-PCR of ATL showing OsSta2 mRNA levels isolated from young seedlings. Error bars represent standard error of three replicates.
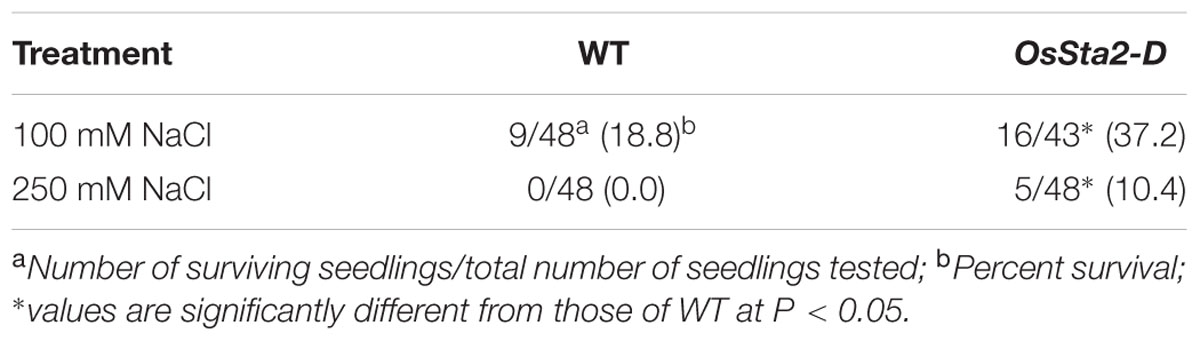
TABLE 1. Level of salt tolerance in WT rice and OsSta2-D lines, based on survival rates of 8 DAG seedlings exposed to salinity treatment (100 mM NaCl for 7 days or 250 mM NaCl for 48 h) and then returned to normal growing conditions for 6 days of recovery.
Expression Analysis of OsSta2
Expression of OsSta2 was examined by RT-PCR and validated by qRT-PCR. Although the gene was detected in all tissue types, transcripts were more abundant in the panicles, callus, and 7 DAG roots, while levels were relatively low in 7 DAG shoots (Figure 2A). Expression increased by approximately twofold after 12 h of salt stress (Figure 2B), and after 24 h of drought or ABA treatment (Figures 2C,D), but was not induced under cold stress (Figure 2E).
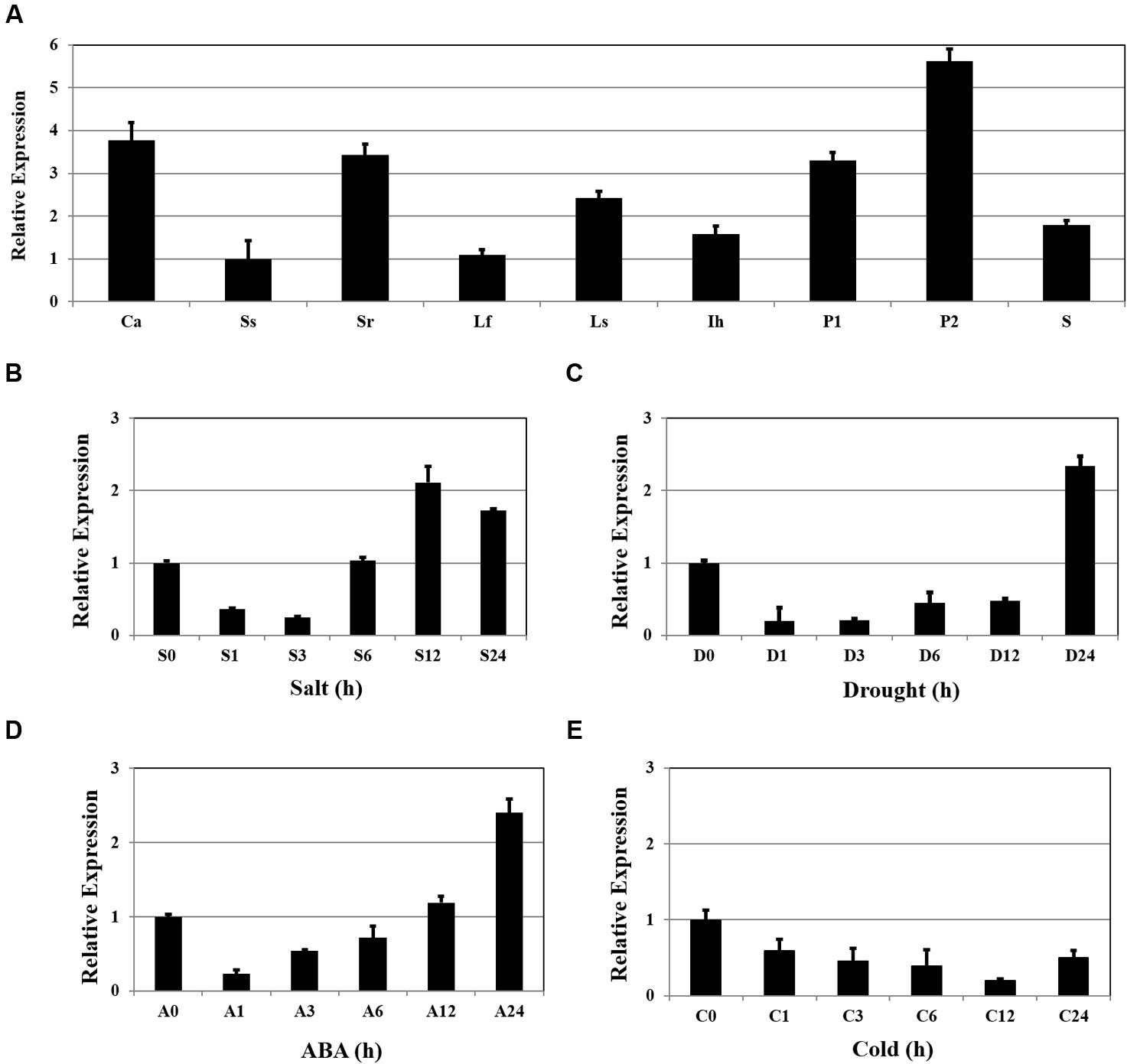
FIGURE 2. Patterns of OsSta2 expression under stress conditions and in developing tissues. (A) qRT-PCR analysis of OsSta2 throughout various stages of development. Ca, callus; Ss, 7 DAG shoot; Sr, 7-day-old root; Lf, mature leaf; Ls, flag leaf sheaths; Ih, highest internodes at pre-heading stage; P1, 3- to 8-cm-long panicles; P2, mature panicles; S, mature seed at 3 days after pollination (DAP). RAc1 was used as reference gene. qRT-PCR analysis of OsSta2 detected in stress-treated seedlings after 0, 1, 3, 6, or 24 h of exposure to salt (B), drought (C), ABA (D), or cold (E). Error bars represent standard error of three replicates.
Generation of OsSta2 Transgenic Rice
A full-length cDNA sequence (J065129D08) obtained from KOME was incorporated under the maize ubiquitin promoter in the pGA3426 vector (Figure 3A). pGA3426 vector has T7 terminator in T-DNA which have been used for expression of foreign gene (Jeon et al., 2000). We could not clone the full-length cDNA as reported by Fu et al. (2007), and could not even detect any transcript spanning the putative AP2 domain (Supplementary Table S2 and Figure S4). The cassette was transformed into ‘Dongjin’ rice and 21 independent transgenic lines were generated. The insertion of OsSta2 was confirmed by PCR analyses of the genomic DNA (Supplementary Figure S2). From those primary transgenic plants, we chose five lines with normal seed formation and used them for T1 production in the confined paddy field. Seeds were harvested and subjected to selection on a hygromycin-containing medium for segregation analysis. Three T2 overexpression lines (Ox13, Ox19, and Ox20) that over-expressed OsSta2 were identified by RT-PCR and validated by qRT-PCR analysis (Figure 3B). Different generations of overexpression lines were used for different set of experiments (Supplementary Figure S5). Southern blot analysis was done to check the copy number integration in three independent overexpression lines by digesting 4 μg DNA with HindIII, EcoRI and BamHI restriction enzyme (Supplementary Figure S6). None of those independent lines differed morphologically from each other when grown under normal conditions either in a soil box or on half-strength MS media (Figures 3C,D).
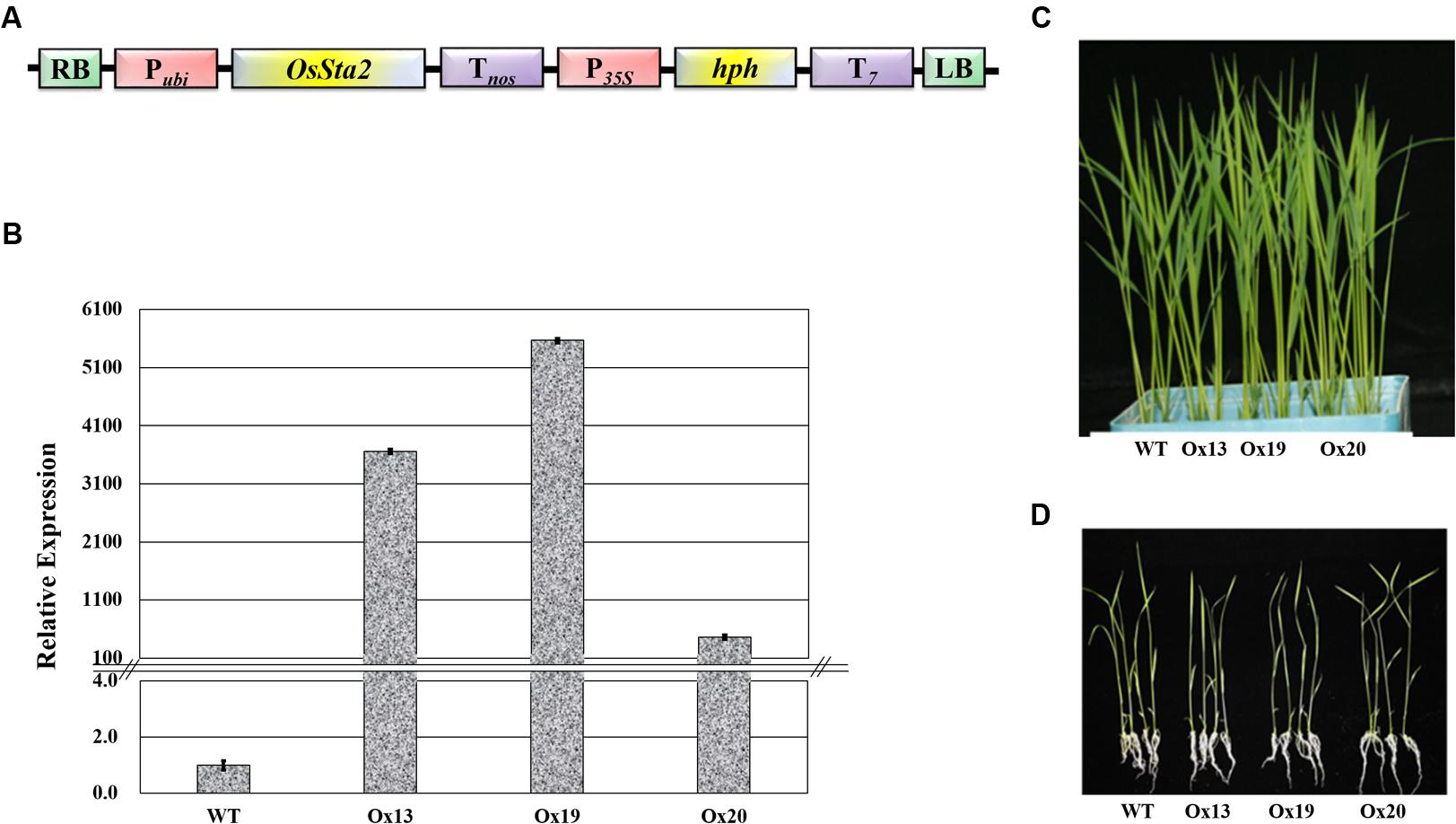
FIGURE 3. Production of transgenic rice over-expressing OsSta2. (A) Schematic diagram of Pubi::OsSta2 construct used for transformation; P35S, CaMV 35S promoter; Pubi, maize ubiquitin promoter; T7, terminator sequence of Transcript 7; Tnos, terminator sequence of nopaline synthase; hph, hygromycin phosphotransferase; RB and LB, right and left border sequences of Ti plasmid from A. tumefaciens. (B) qRT-PCR analysis of OsSta2 in transgenic overexpression (Ox) rice lines. Actin was loading control. (C) Visual phenotype for 8 DAG seedlings from OsSta2-Ox line grown in soil box. (D) Visual phenotype of 14 DAG seedlings grown in half-strength MS medium.
Overexpression of OsSta2 in Transgenic Rice Plants Enhances their Salt Tolerance at the Vegetative Stage
To investigate whether overexpression of OsSta2 can confer salt tolerance at the vegetative stage, we exposed rice seedlings to 100 mM NaCl for 7 days and found that 10.1–22.7% of the OsSta2-Ox transgenics survived versus 7.2% of the WT plants. Similar results were obtained after treatment with 250 mM NaCl for 72 h, with 25.8–34.5% survival for the transgenics versus 11.6% survival for the WT (Figures 4A–C). After the recovery period, fresh and dry weights were 2–11% higher for the OsSta2-Ox plants than for the WT (Figure 4 and Supplementary Figure S3). Drought tolerance was not improved in the transgenics at the vegetative stage.
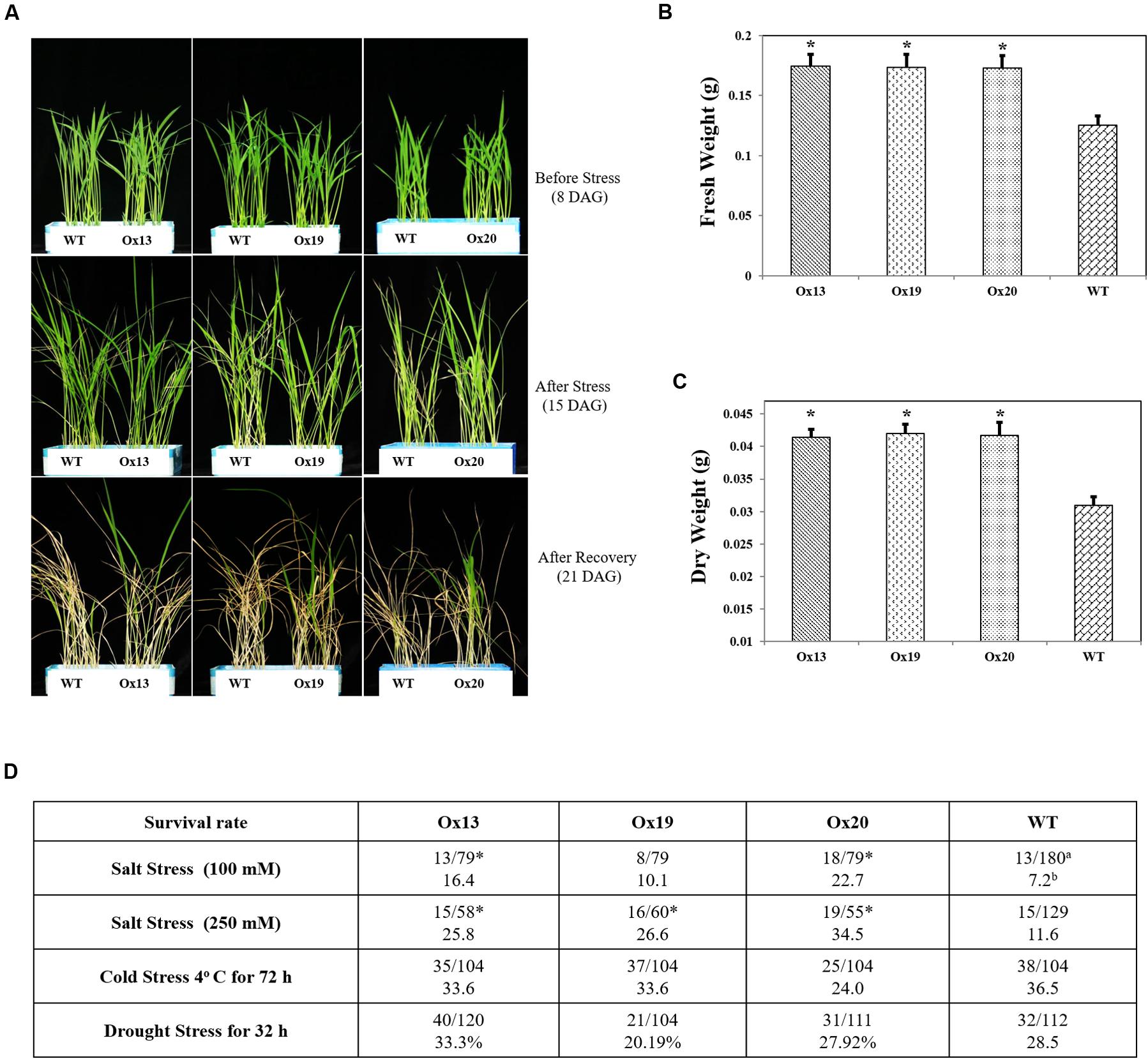
FIGURE 4. Enhanced salt tolerance in transgenic rice over-expressing OsSta2 at vegetative stage. (A) Visual phenotype of salt response by Ox transgenic plants and WT after 8 DAG seedlings were exposed to 100 mM NaCl for 7 days before returning to normal conditions for 6 days of recovery. (B) Fresh weights after recovery. (C) Weights after plants were dried for 2 days. (D) Stress tolerance of OsSta2-Ox lines based on 8 DAG seedlings treated with drought (water withheld for 32 h), salt (100 mM NaCl treatment for 7 days or 250 mM NaCl treatment for 72 h), or cold (4°C for 72 h). Percent survival was calculated after recovery period. aNumber of surviving seedlings/total number of seedlings tested; bPercent survival; ∗, results are significantly different between Ox line and WT at P < 0.05.
Leaf disk assays performed under various concentrations of NaCl revealed that less chlorophyll was lost from the Ox lines than from the WT plants (Figures 5A,B). For example, in response to 100 mM NaCl, the WT samples contained 5.8 mg of chlorophyll per g of leaf tissue versus 19.6–21.5 mg per g in the transgenics, i.e., 14–16% more than in the WT. Similar results were obtained in response to 200 or 250 mM NaCl. There, chlorophyll concentrations in the WT ranged from 3.0 to 6.2 mg per g, which was 9–12% lower than the 12.0–20.0 mg g measured in the Ox lines. These higher levels of chlorophyll in the transgenics demonstrated that OsSta2 expression was positively correlated with improved salt tolerance (Figure 5C).
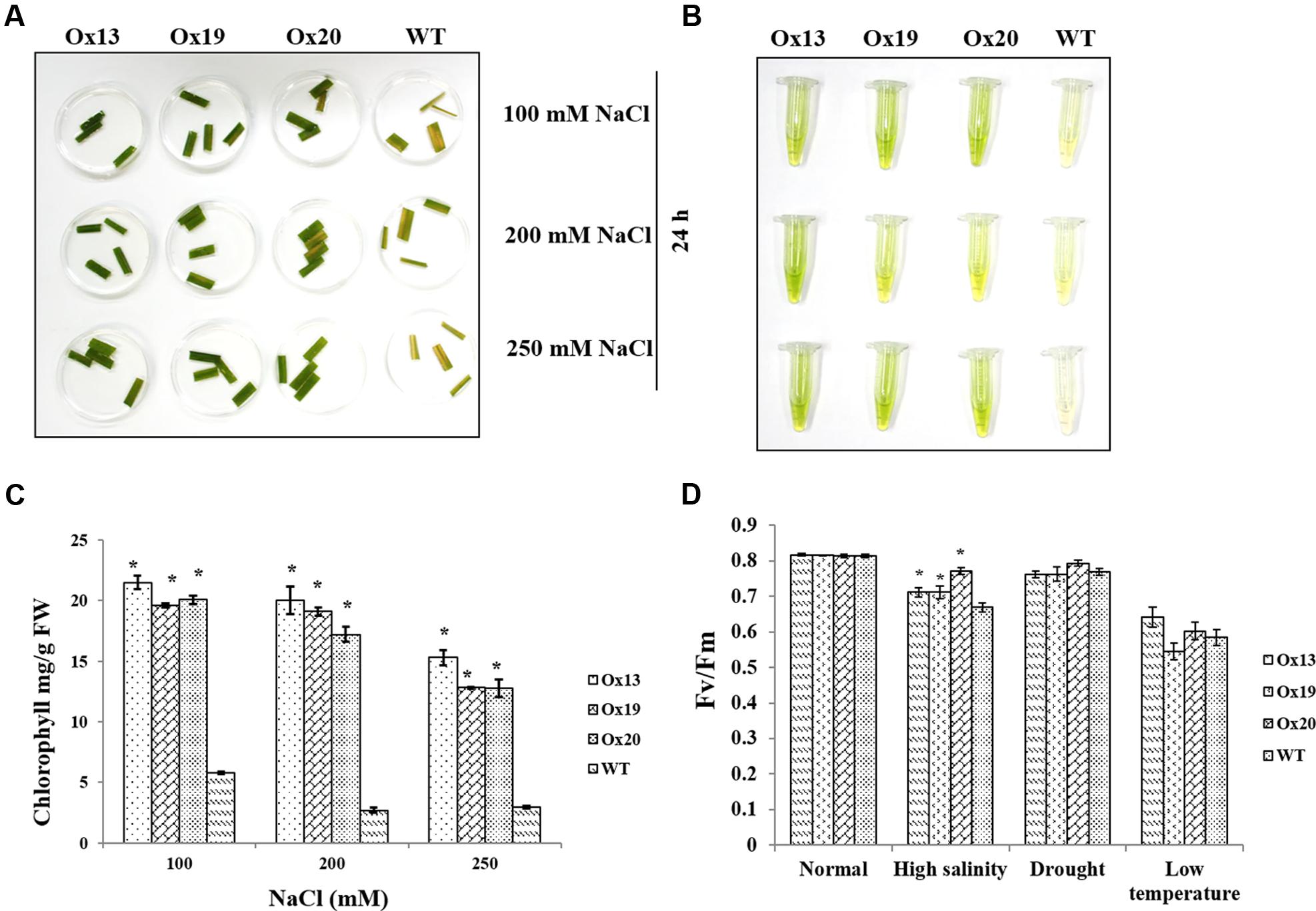
FIGURE 5. Comparison of photosynthetic rates under stress conditions. (A) Leaf disk assays of homozygous T4 OsSta2-Ox plants, WT, and ATL under salinity stress (100, 200, or 250 mM NaCl) after 24 h. (B,C) Estimates of chlorophyll concentration (mg per g fresh weight) in homozygous T4 OsSta2. (D) Changes in chlorophyll fluorescence (Fv/Fm) in leaves sampled from rice plants grown in soil for 12 days before being exposed to high salinity (500 mM NaCl for 48 h), drought (lack of water in Petri plate for 3 h), or low-temperature stress (4°C for 48 h) under continuous light (150 μmol m-2 s-1). Three independent homozygous T4 lines of OsSta2-Ox plants and WT controls were tested. ∗, differences between Ox line and WT are significant at P < 0.05.
Under high-salinity stress, Fv/Fm values for WT plants were reduced from 0.81 to 0.66 which were 18.52% reduction. Fv/Fm values for transgenics plants were reduced from 0.81 to 0.70 which were 13.58% reduction and were 5% better than that of WT plants (Figure 5D). In contrast, under drought or low-temperature stress, Fv/Fm values were similar between the OsSta2-Ox and WT plants.
Overexpression of OsSta2 Increases Grain Yields
Three independent homozygous lines of OsSta2-Ox, together with the WT control, were grown in a paddy field. Mature transgenic plants showed semi-dwarfism but this phenotypic difference from the WT was not apparent at the four-leaf stage. Culms were 7–9% shorter from the Ox plants, i.e., 79–82 cm versus 86 cm for the WT stems (Figure 6). However, the Ox plants produced more tillers than the WT control, and grain yields were higher from those transgenics under normal field conditions. In particular, the grain filling rates were 17 and 23% higher for Ox13 and Ox19, respectively, than for the WT, and total grain weights were increased by 5–8% over the WT total. Filling rates did not differ significantly between Ox20 and the WT, suggesting that OsSta2 expression was lower in that transgenic line. Nevertheless, the total grain weight was 10% higher in Ox20 than in the WT, perhaps because plants of the former type produced 8% more spikelets per panicle. Taken together, these results again showed that overexpression of OsSta2 can improve grain yields significantly.
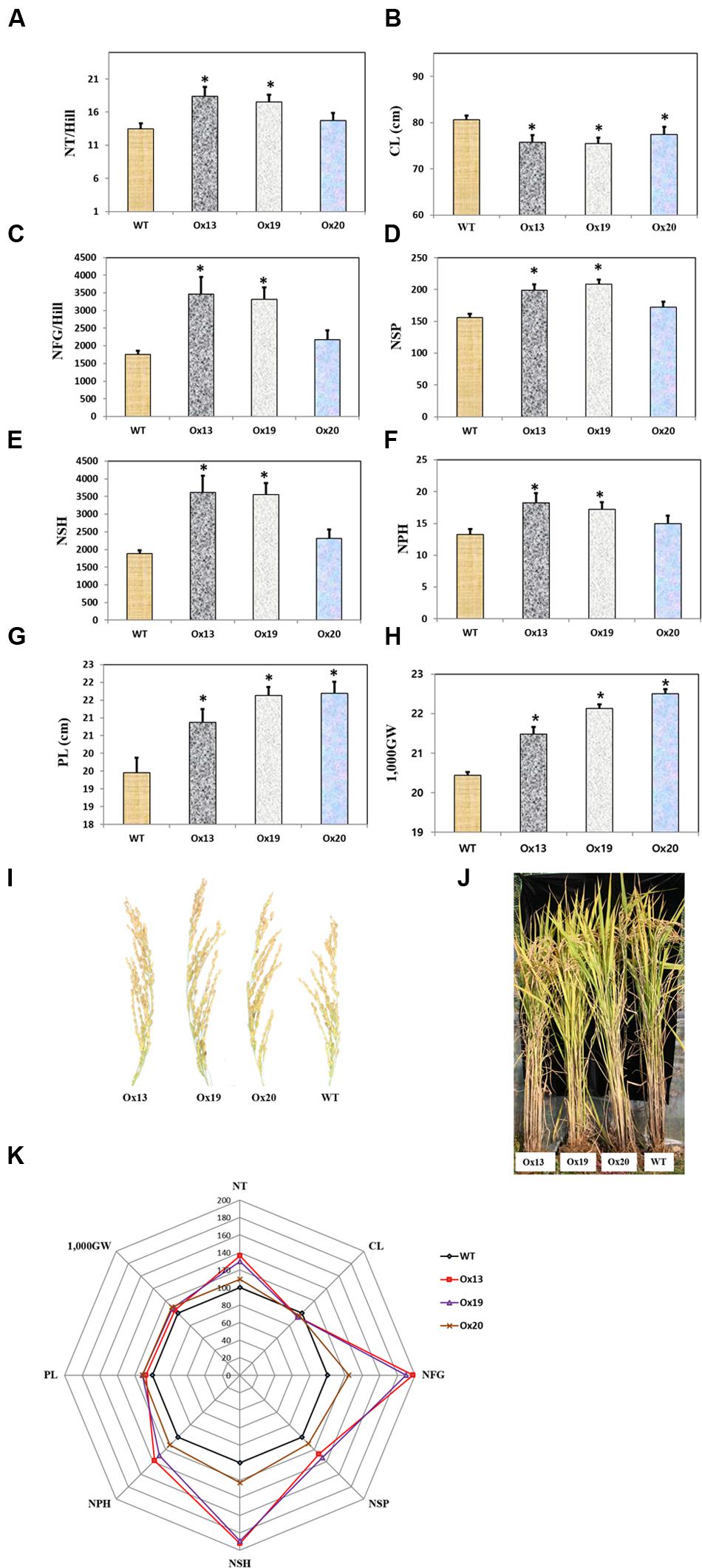
FIGURE 6. Agronomic traits of field-grown rice plants under normal conditions. (A) Number of tillers per hill. (B) Culm length. (C) Number of filled grains per hill. (D) Number of spikelets per panicle. (E) Number of spikelets per hill. (F) Number of panicles per hill. (G) Panicle Length. (H) 1,000-grain weight. (I) Morphology of mature rice panicle. (J) Morphology of mature paddy field-grown rice plant. (K) Spider plot of agronomic traits of three independent homozygous T5 lines of OsSta2-Ox and corresponding WT controls under normal conditions was drawn using Microsoft Excel software. Each data point represents percentage of mean values (n = 8). Mean value for WT controls was set at ‘100%’ as reference. CL, Culm length; NFG, number of filled grains; NSH, number of spikelets per hill; NSP, number of spikelets per panicle; NP, number of panicles per hill; NT, Numbers of tillers per hill; 1,000GW, 1,000-grain weight. Bars indicate standard error of 8 replicates. ∗, differences between Ox line and WT are significant at P < 0.05.
OsSta2-Ox Transgenic Plants are Hypersensitive to ABA
Although the growth of WT shoots was repressed by ABA, this inhibitory effect was more significant in shoots from Ox plants (Figure 7). For example, after treatment with 5 μM ABA, relative shoot lengths from Ox plants were 61.76–63.89% shorter than those measured from plants not exposed to ABA. By comparison, the shoots from ABA-treated WT plants were 56.2% shorter than those of the WT control (untreated) plants. A similar pattern was found in response to 10 μM ABA (i.e., Ox shoots from ABA-treated plants were 71.4–74.1% shorter than those from untreated transgenics while shoots from ABA-treated WT plants were 69.5% shorter than their untreated counterparts). These results suggested that OsSta2 is hypersensitive to ABA and is involved in its signaling pathway.
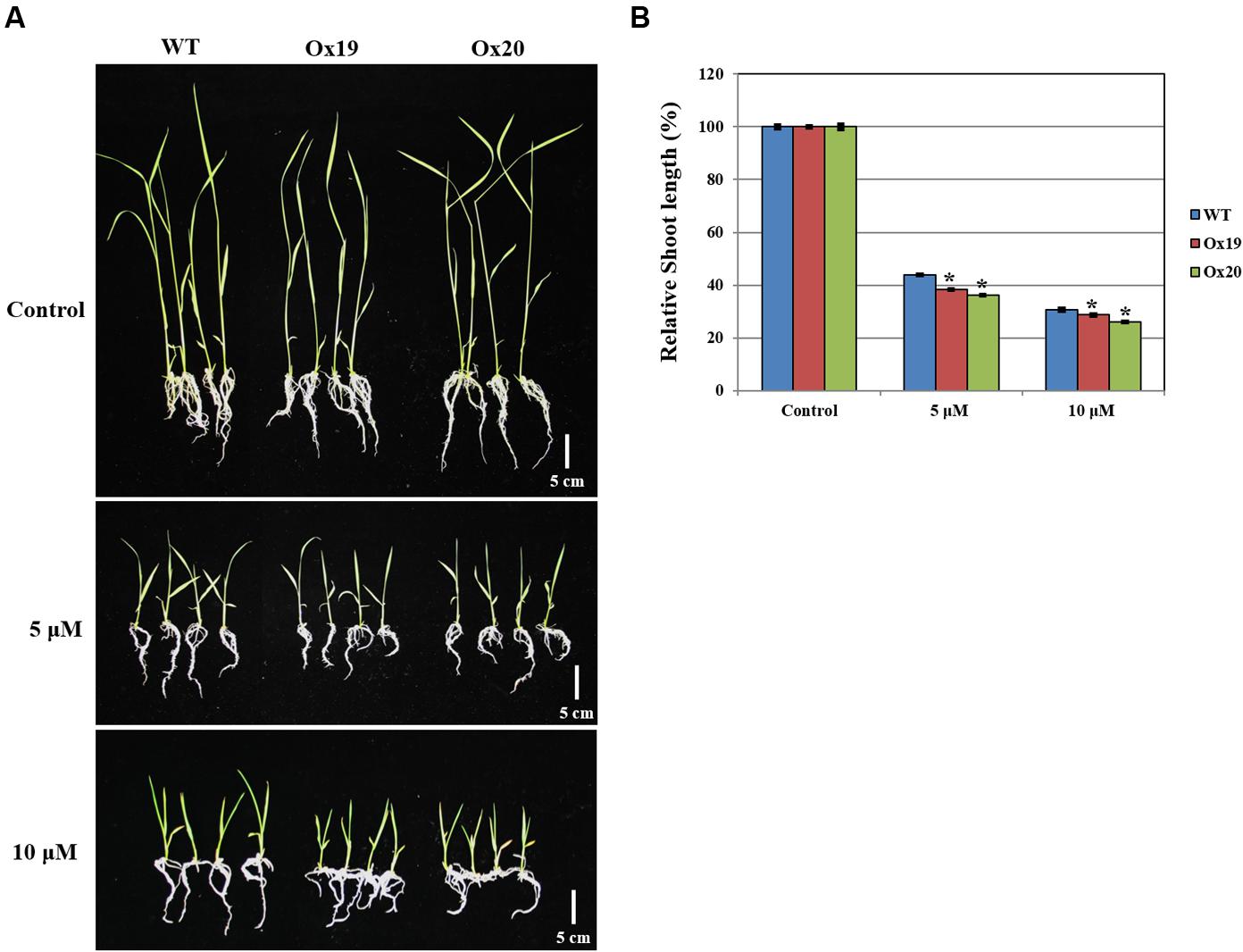
FIGURE 7. Analysis of ABA sensitivity. OsSta2-Ox rice showing ABA-sensitive phenotypes at early seedling stage. At 5 DAG, two independent Ox lines and WT control were transferred to half-strength MS media supplemented with different ABA concentrations. (A) Growth inhibition by ABA treatment. Photographs show representative seedlings 7 days after transfer. (B) Relative shoot lengths of control and Ox lines. Results are mean ± SE (n = 9 seedlings per experiment). Bar = 5 cm. ∗, differences between Ox line and WT are significant at P < 0.05.
OsSta2-Ox Transgenic Plants are more Tolerant than the WT to Osmotic Stress
When grown in a half-strength MS medium supplemented with 200 mM mannitol, the shoots from Ox plants were 46–50% shorter than those from the untreated transgenics while shoots from mannitol-treated WT plants were 54% shorter than those from the untreated WT (Figure 8). This demonstrated that OsSta2 helps confer tolerance to osmotic stress.
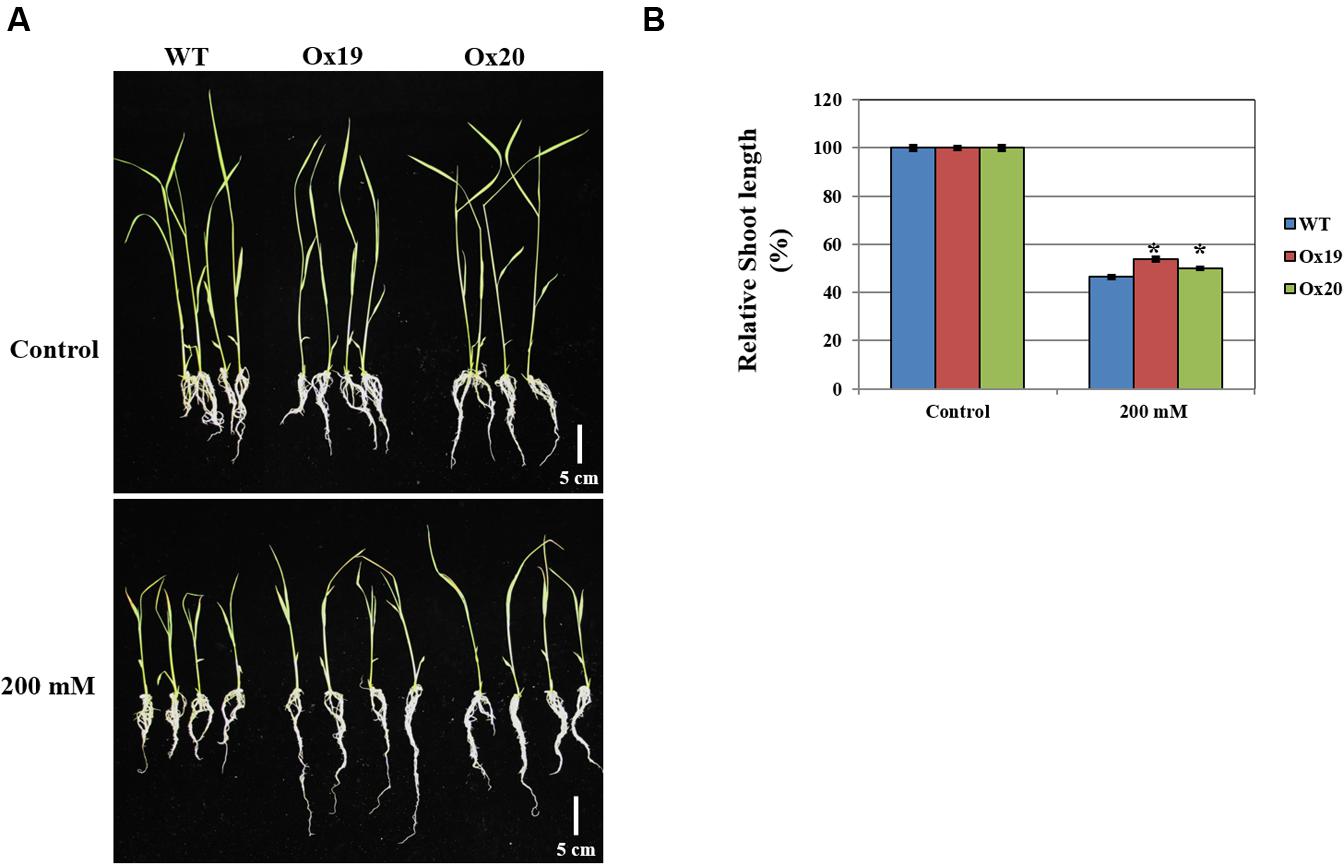
FIGURE 8. Analysis of tolerance to osmotic stress. OsSta2-Ox rice showing normal phenotypes at early seedling stage. At 5 DAG, two independent Ox lines and WT control were transferred to half-strength MS media supplemented with 200 mM mannitol. (A) Growth in presence of mannitol. Photographs show representative seedlings 7 days after transfer. (B) Relative shoot lengths of WT control and Ox lines. Results are mean ± SE (n = 9 seedlings per experiment). Bar = 5 cm. ∗, differences between Ox line and WT are significant at P < 0.05.
Discussion
Rice is a salt-sensitive crop at the germination stage, but becomes more tolerant as plants progress from young seedlings to the vegetative stage (Heenan et al., 1988; Lutts et al., 1995). We used various assays to monitor activation or overexpression of OsSta2 and to determine how this gene can confer enhanced salt tolerance at the seedling stage. Fujimoto et al. (2000) have reported that AtERF5 (At5g47230), acts as a functional activator of GCC box-mediated transcription. AtERF5 also plays a role as a positive regulator of JA/ethylene-mediated defenses against Botrytis cinerea (Moffat et al., 2012). However, no previous research confirmed its function under abiotic stresses, although a role for ERF TFs has been suggested (Nakano et al., 2006). We found here that OsSta2 could respond to salt, drought, and ABA treatments because its promoter region contains multiple stress-related cis-elements, i.e., ABREs, DRE/CRT, and a MYB/MYC recognition site. Those same elements also occur in the promoter region of stress-responsive genes that are regulated by DREB, ERF, and MYB/MYC TFs, respectively (Urao et al., 1993; Baker et al., 1994; Abe et al., 1997, 2003; Guan et al., 2000; Simpson et al., 2003; Tran et al., 2004; Kaplan et al., 2006). All of these findings provide evidence of the role that OsSta2 has in conferring salt tolerance.
Grain yield is an important parameter when investigating the effects of abiotic challenges. Overexpression of stress-related genes can alter the productivity and overall architecture of rice plants (Oh et al., 2009; Jeong et al., 2010; Xia et al., 2012; Alam et al., 2014; Yoon et al., 2016). Therefore, it was important that we examine grain yields using stable transgenic lines that did not segregate under paddy field conditions. This approach facilitated our identification of a segregating line of transgenic rice plants up to the T3 generation, even if they were homozygous for a particular transgene. To determine how yields were increased in OsSta2-Ox rice under normal conditions, we relocated T4 homozygous lines in 2014 and T5 homozygous lines in 2015 to the paddy field. Those lines had been pre-screened for segregation in the field from 2011 to 2013. Grain production was significantly improved in the Ox plants when compared with the WT, mainly because the former type of plant had more tillers and panicles, and its panicles were longer than those of the WT.
During its response to osmotic stress, plants utilize the ABA signaling transduction pathway to initiate the expression of defense genes (Chinnusamy et al., 2004; Singh et al., 2015). Overexpression of some stress-related genes, e.g., OsZIP72 and OsABI5, results in abiotic-stress tolerance and causes the transgenic plants to be hypersensitive to exogenous ABA (Zou et al., 2008; Lu et al., 2009; Kim et al., 2014). We also found that OsSta2-Ox plants showed increased responsiveness to exogenous ABA, which suggested that this gene has a role in the ABA pathway during the stress response. Therefore, we proposed that OsSta2 has a role in the ABA signaling pathway and that this response to salinity is ABA-dependent.
When plants recognize and respond to stress in an ABA-hypersensitive manner, the processes associated with physiological processes may retard growth because necessary resources are instead being directed toward mechanisms for protection. This can occur even under normal environmental conditions because higher levels of transcripts for genes related to abiotic-stress tolerance can inhibit plant development, especially when such genes are constitutively over-expressed. This is particularly true for genes associated with ABA signaling because that phytohormone has important regulatory roles (Sreenivasulu et al., 2012). For example, rice plants that constitutively over-express DREB1A grow more slowly under standard conditions (Kasuga et al., 1999; Nakashima et al., 2007). This might explain why our OsSta2-Ox plants showed slight retardation when grown to maturity in the paddy field. However, no such inhibition was noted when young OsSta2-Ox seedlings were grown either in soil boxes in a chamber or on plates containing a half-strength MS medium. To partially overcome this problem when conducting experiments, researchers utilize promoters that are stress-inducible, such as OsDhn1, rd29A, and OsPOX1 (Kasuga et al., 1999; Wang et al., 2005; Kim S.H. et al., 2012; Lee et al., 2013; Kumar et al., 2014).
As with salt stress, OsSta2-Ox plants were also more tolerant to osmotic stress, maintaining a much healthier growth pattern (as reflected in shoot length parameters) than the WT seedlings in response to mannitol treatment. Similar results have been described previously (Zou et al., 2012; Kumar et al., 2013; Kim et al., 2014; You et al., 2014; Singh et al., 2015). Hence, we can conclude that OsSta2-Ox plants exhibit ABA-dependent salt tolerance via osmotic stress signaling.
The extent to which Ox lines are salt-tolerant also depends on the level of OsSta2 expression and its involvement in the tolerance pathway (Ashraf, 2009). Because salinity-promoted oxidative stress is peripheral to ionic and osmotic stresses, it might not be possible to achieve adequate salt tolerance through the manipulation of OsSta2 alone but it might entail strong interactions with other stress-related genes (Ashraf, 2009; Kumar et al., 2013). Therefore, further exploration of such genetic inter-relationships is necessary if we are to produce crop plants that are more tolerant to environmental challenges.
Conclusion
We have demonstrated that overexpression of OsSta2 enhances the tolerance of transgenic rice plants to salt and osmotic stresses. This is manifested by an increase in tiller numbers and grain yields. However, additional analyses of gene expression and how they finely regulate plant processes are required in the future.
Author Contributions
MK and S-RK design experiments and wrote manuscript. MK performed all the experiments. All the authors approved final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Priscilla Licht for critical editing of the manuscript. This research was supported by grants from Sogang University and the Next-Generation BioGreen 21 Program (PJ011927), Rural Development Administration, South Korea.
Abbreviations
ABA, abscisic acid; ATL, activation tagging line; DAG, days after germination; LSD, least significant difference; MS, Murashige and Skoog.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00316/full#supplementary-material
Footnotes
- ^ http://signal.salk.edu/cgi-bin/RiceGE5
- ^ http://www.generunner.net/
- ^ http://www.ncbi.nlm.nih.gov/tools/primer-blast/
- ^ http://tenor.dna.affrc.go.jp/
- ^ https://dbarchive.biosciencedbc.jp/en/kome/desc.html
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. doi: 10.1105/tpc.9.10.1859
Alam, M. M., Tanaka, T., Nakamura, H., Ichikawa, H., Kobayashi, K., Yaeno, T., et al. (2014). Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol. J. 13, 85–96. doi: 10.1111/pbi.12239
Andriankaja, A., Boisson-Dernier, A., Frances, L., Sauviac, L., Jauneau, A., Barker, D. G., et al. (2007). AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19, 2866–2885. doi: 10.1105/tpc.107.052944
Apse, M. P., and Blumwald, E. (2002). Engineering salt tolerance in plants. Curr. Opin. Biotechnol. 13, 146–150. doi: 10.1016/S09581669(02)00298-7
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Ashraf, M. (2009). Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 27, 84–93. doi: 10.1016/j.biotechadv.2008.09.003
Baker, S., Wilhelm, K., and Thomashow, M. (1994). The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Bhatnagar-Mathur, P., Vadez, V., and Sharma, K. (2008). Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep. 27, 411–424. doi: 10.1007/s00299-007-0474-9
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749. doi: 10.1105/tpc.001941
Buchel, A. S., Brederode, F. T., Bol, J. F., and Linthorst, H. J. M. (1999). Mutation of GT-1 binding sites in the Pr-1A promoter influences the level of inducible gene expression in vivo. Plant Mol. Biol. 40, 387–396. doi: 10.1023/A:1006144505121
Cao, Y., Song, F., Goodman, R. M., and Zheng, Z. (2006). Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J. Plant Physiol. 163, 1167–1178. doi: 10.1016/j.jplph.2005.11.004
Chen, W., Provart, N. J., Glazebrook, J., Katagiri, F., Chang, H. S., Eulgem, T., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. doi: 10.1105/tpc.010410
Chinnusamy, V., Schumaker, K., and Zhu, J. K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J. Exp. Bot. 55, 225–236. doi: 10.1093/jxb/erh005
Das, P., Nutan, K. K., Singla-Pareek, S. L., and Pareek, A. (2015). Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice. Front. Plant Sci. 6:712. doi: 10.3389/fpls.2015.00712
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., Schroeder, J. I., et al. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Duan, Y. B., Li, J., Qin, R. Y., Xu, R. F., Li, H., Yang, Y. C., et al. (2015). Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 90, 49–62. doi: 10.1007/s11103-015-0393-z
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Elliott, R. C., Betzner, A. S., Huttner, E., Oakes, M. P., Tucker, W. Q., Gerentes, D., et al. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168. doi: 10.1105/tpc.8.2.155
Eynard, A., Lal, R., and Wiebe, K. (2005). Crop response in salt-affected soils. J. Sustain. Agric. 27, 5–50. doi: 10.1300/J064v27n01-03
Fahad, S., Hussain, S., Matloob, A., Khan, F. A., Khaliq, A., Saud, S., et al. (2015). Phytohormones and plant responses to salinity stress: a review. Plant Growth Regul. 75, 391–404. doi: 10.1007/s10725-014-0013-y
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Fita, A., Rodriguez-Burruezo, A., Boscaiu, M., Prohens, J., and Vicente, O. (2015). Breeding and domesticating crops adapted to drought and salinity: a new paradigm for increasing food production. Front. Plant Sci. 6:978. doi: 10.3389/fpls.2015.00978
Fu, M., Kang, H. K., Son, S. H., Kim, S. K., and Nam, K. H. (2014). A Subset of RAV transcription factors modulates drought and salt stress responses aba-independently in Arabidopsis. Plant Cell Physiol. 55, 1892–1904. doi: 10.1093/pcp/pcu118
Fu, X. Y., Zhang, Z., Peng, R. H., Xiong, A. S., Liu, J. G., Wu, L. J., et al. (2007). Isolation and characterization of a novel cDNA encoding ERF/AP2-type transcription factor OsAP25 from Oryza sativa L. Biotechnol. Lett. 29, 1293–1299. doi: 10.1007/s10529-007-9370-1
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Fukao, T., Xu, K., Ronald, P. C., and Bailey-Serres, J. (2006). A variable cluster of ethylene-responsive-like factors regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18, 2021–2034. doi: 10.1105/tpc.106.043000
Gao, S., Zhang, H., Tian, Y., Li, F., Zhang, Z., Lu, X., et al. (2008). Expression of TERF1 in rice regulates expression of stress-responsive genes and enhances tolerance to drought and high-salinity. Plant Cell Rep. 27, 1787–1795. doi: 10.1007/s00299-008-0602-1
Gu, Y. Q., Wildermuth, M. C., Chakravarthy, S., Loh, Y. T., Yang, C., He, X., et al. (2002). Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14, 817–831. doi: 10.1105/tpc.000794
Guan, L. M., Zhao, J., and Scandalios, J. G. (2000). Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J. 22, 87–95. doi: 10.1046/j.1365-313x.2000.00723.x
Hattori, Y., Nagai, K., Furukawa, S., Song, X. J., Kawano, R., Sakakibara, H., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030. doi: 10.1038/nature08258
Heenan, D. P., Lewin, L. G., and McCaffery, D. W. (1988). Salinity tolerance in rice varieties at different growth stages. Aust. J. Exp. Agric. 28, 343–349. doi: 10.1071/EA9880343
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Ismail, A., Takeda, S., and Nick, P. (2014). Life and death under salt stress: same players, different timing? J. Exp. Bot. 65, 2963–2979. doi: 10.1093/jxb/eru159
Itzhaki, H., Maxson, J. M., and Woodson, W. R. (1994). An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc. Natl. Acad. Sci. 91, 8925–8929.
Jeon, J. S., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S. H., et al. (2000). Leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884. doi: 10.1105/tpc.12.6.871
Jeong, D. H., An, S., Kang, H. G., Moon, S., Han, J. J., Park, S., et al. (2002). T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130, 1636–1644. doi: 10.1104/pp.014357
Jeong, D. H., An, S., Park, S., Kang, H. G., Park, G. G., Kim, S. R., et al. (2006). Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 45, 123–132. doi: 10.1111/j.1365-313X.2005.02610.x
Jeong, J. S., Kim, Y. S., Baek, K. H., Jung, H., Ha, S. H., Choi, Y. D., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. doi: 10.1104/pp.110.154773
Jisha, V., Dampanaboina, L., Vadassery, J., Mithofer, A., Kappara, S., and Ramanan, R. (2015). Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 10:e0127831. doi: 10.1371/journal.pone.0127831
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7:1029. doi: 10.3389/fpls.2016.01029
Jung, K. H., Hur, J., Ryu, C. H., Choi, Y., Chung, Y. Y., Miyao, A., et al. (2003). Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol. 44, 463–472. doi: 10.1093/pcp/pcg064
Kader, M. A., and Lindberg, S. (2010). Cytosolic calcium and pH signaling in plants under salinity stress. Plant Signal. Behav. 5, 233–238. doi: 10.4161/psb.5.3.10740
Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478.
Kaplan, B., Davydov, O., Knight, H., Galon, Y., Knight, M. R., Fluhr, R., et al. (2006). Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18, 2733–2748. doi: 10.1105/tpc.106.042713
Karaba, A., Dixit, S., Greco, R., Aharoni, A., Trijatmiko, K. R., Marsch Martinez, N., et al. (2007). Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc. Natl. Acad. Sci. 104, 15270–15275. doi: 10.1073/pnas.0707294104
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Kazan, K. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229. doi: 10.1016/j.tplants.2015.02.001
Kim, H., Hwang, H., Hong, J. W., Lee, Y. N., Ahn, I. P., Yoon, I. S., et al. (2012). A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63, 1013–1024. doi: 10.1093/jxb/err338
Kim, H., Lee, K., Hwang, H., Bhatnagar, N., Kim, D. Y., Yoon, I. S., et al. (2014). Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 65, 453–464. doi: 10.1093/jxb/ert397
Kim, S. H., Choi, H. S., Cho, Y. C., and Kim, S. R. (2012). Cold-responsive regulation of a flower-preferential class III peroxidase gene, OsPOX1, in rice (Oryza sativa L.). J. Plant Biol. 55, 123–131. doi: 10.1007/s12374-011-9194-3
Kim, S. R., Lee, D. Y., Yang, J. I., Moon, S., and An, G. (2009). Cloning vectors for rice. J. Plant Biol. 52, 73–78. doi: 10.1007/s12374-008-9008-4
Kim, S. Y., Chung, H. J., and Thomas, T. L. (1997). Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 11, 1237–1251. doi: 10.1046/j.1365-313X.1997.11061237.x
Koh, S., Lee, S. C., Kim, M. K., Koh, J. H., Lee, S., An, G., et al. (2007). T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 65, 453–466. doi: 10.1007/s11103-007-9213-4
Krizek, B. (2009). AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol. 150, 1916–1929. doi: 10.1104/pp.109.141119
Kumar, K., Kumar, M., Kim, S. R., Ryu, H., and Cho, Y. G. (2013). Insights into genomics of salt stress response in rice. Rice 6:27. doi: 10.1186/1939-8433-6-27
Kumar, M., Lee, S. C., Kim, J. Y., Kim, S. J., Aye, S. S., and Kim, S. R. (2014). Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 57, 383–393. doi: 10.1007/s12374-014-0487-1
Kurata, N., and Yamazaki, Y. (2006). Oryzabase. An integrated biological and genome information database for rice. Plant Physiol. 140, 12–17. doi: 10.1104/pp.105.063008
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lee, S. C., Han, S. K., and Kim, S. R. (2015). Salt- and ABA-inducible OsGASR1 is involved in salt tolerance. J. Plant Biol. 58, 96–101. doi: 10.1007/s12374-014-0497-z
Lee, S. C., Kim, J. Y., Kim, S. H., Kim, S. J., Lee, K., Han, S. K., et al. (2004). Trapping and characterization of cold-responsive genes from T-DNA tagging lines in rice. Plant Sci. 166, 69–79. doi: 10.1016/j.plantsci.2003.08.008
Lee, S. C., Kim, S. H., and Kim, S. R. (2013). Drought inducible OsDhn1 promoter is activated by OsDREB1A and OsDREB1D. J. Plant. Biol. 56, 115–121. doi: 10.1007/s12374-012-0377-3
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, D., Chen, X., Liu, J., Ye, J., and Guo, Z. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3911. doi: 10.1093/jxb/ers079
Liu, J. G., Zhang, Z., Qin, Q. L., Peng, R. H., Xiong, A. S., Chen, J. M., et al. (2007). Isolated and characterization of a cDNA encoding ethylene-responsive element binding protein (EREBP)/AP2-type protein, RCBF2, in Oryza sativa L. Biotechnol. Lett. 29, 165–173. doi: 10.1007/s10529-006-9214-4
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP / AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopez-Molina, L., and Chua, N. H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. doi: 10.1093/pcp/41.5.541
Lu, G., Gao, C., Zheng, X., and Han, B. (2009). Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229, 605–615. doi: 10.1007/s00425-008-0857-3
Luo, H., Song, F., Goodman, R. M., and Zheng, Z. (2005). Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant. Biol. 7, 459–468. doi: 10.1055/s-2005-865851
Lutts, S., Kinet, J. M., and Bouharmont, J. (1995). Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J. Exp. Bot. 46, 1843–1852. doi: 10.1093/jxb/46.12.1843
Middleton, P. H., Jakab, J., Penmetsa, R. V., Starker, C. G., Doll, J., Kalo, P., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19, 1221–1234. doi: 10.1105/tpc.106.048264
Mishra, S., Phukan, U., Tripathi, V., Singh, D., Luqman, S., and Shukla, R. (2015). PsAP2, an AP2/ERF family transcription factor from Papaver somniferum, enhances abiotic and biotic stress tolerance in transgenic tobacco. Plant Mol. Biol. 89, 173–186. doi: 10.1007/s11103-015-0361-7
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Moffat, C. S., Ingle, R. A., Wathugala, D. L., Saunders, N. J., Knight, H., and Knight, M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7:e35995. doi: 10.1371/journal.pone.0035995
Montgomery, J., Goldman, S., Deikman, J., Margossian, L., and Fischer, R. L. (1993). Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proc. Natl. Acad. Sci. U.S.A. 90, 5939–5943.
Moose, S. P., and Sisco, P. H. (1996). Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 10, 3018–3027. doi: 10.1101/gad.10.23.3018
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nakashima, K., Tran, L. S., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Nova-Franco, B., Íñiguez, L. P., Valdés-López, O., Alvarado-Affantranger, X., Leija, A., Fuentes, S. I., et al. (2015). The micro-RNA72c-APETALA2-1 node as a key regulator of the common bean-Rhizobium etli nitrogen fixation symbiosis. Plant Physiol. 168, 273–291. doi: 10.1104/pp.114.255547
Oh, S. J., Kim, Y. S., Kwon, C. W., Park, H. K., Jeong, J. S., and Kim, J. K. (2009). Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 150, 1368–1379. doi: 10.1104/pp.109.137554
Oh, S. J., Kwon, C. W., Choi, D. W., Song, S. I., and Kim, J. K. (2007). Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol. J. 5, 646–656. doi: 10.1111/j.1467-7652.2007.00272.x
Oh, S. J., Song, S. I., Kim, Y. S., Jang, H. J., Kim, S. Y., Kim, M., et al. (2005). Arabidopsis CBF3 / DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 138, 341–351. doi: 10.1104/pp.104.059147
Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. doi: 10.1105/tpc.7.2.173
Onate-Sanchez, L., Anderson, J. P., Young, J., and Singh, K. B. (2007). AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 143, 400–409. doi: 10.1104/pp.106.086637
Onate-Sanchez, L., and Singh, K. B. (2002). Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol. 128, 1313–1322. doi: 10.1104/pp.010862
Parihar, P., Singh, S., Singh, R., Singh, V. P., and Prasad, S. M. (2015). Effect of salinity stress on plants and its tolerance strategies: a review. Environ. Sci. Pollut. Res. 22, 4056–4075. doi: 10.1007/s11356-014-3739-1
Petrov, V., Hille, J., Mueller-Roeber, B., and Gechev, T. S. (2015). ROS-mediated abiotic stress-induced programmed cell death in plants. Plant Physiol. 6:69. doi: 10.3389/fpls.2015.00069
Quan, R., Hu, S., Zhang, Z., Zhang, H., Zhang, Z., and Huang, R. (2010). Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 8, 476–488. doi: 10.1111/j.1467-7652.2009.00492.x
Quinn, J. M., Eriksson, M., Moseley, J. L., and Merchant, S. (2002). Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol. 128, 463–471. doi: 10.1104/pp.010694
Reyes, J. C., Muro Pastor, M. I., and Florencio, F. J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134, 1718–1732. doi: 10.1104/pp.103.037788
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rong, W., Qi, L., Wang, A., Ye, X., Du, L., Liang, H., et al. (2014). The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12, 468–479. doi: 10.1111/pbi.12153
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Schmidt, R., Mieulet, D., Hubberten, H. M., Obata, T., Hoefgen, R., Fernie, A. R., et al. (2013). Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131. doi: 10.1105/tpc.113.113068
Seki, M., Kamei, A., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2003). Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr. Opin. Biotechnol. 14, 194–199. doi: 10.1016/S0958-1669(03)00030-2
Sharoni, A. M., Nuruzzaman, M., Satoh, K., Shimizu, T., Kondoh, H., Sasaya, T., et al. (2011). Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 52, 344–360. doi: 10.1093/pcp/pcq196
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant. Biol. 6, 410–417.
Simpson, S. D., Nakashima, K., Narusaka, Y., Seki, M., Shinozaki, K., and Yamaguchi Shinozaki, K. (2003). Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene, function in induction by dehydration stress and dark-induced senescence. Plant J. 33, 259–270. doi: 10.1046/j.1365-313X.2003.01624.x
Singh, A., Jha, S. K., Bagri, J., and Pandey, G. K. (2015). ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE 10:e0125168. doi: 10.1371/journal.pone.0125168
Solano, R., Stepanova, A., Chao, Q., and Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENERESPONSE- FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Sreenivasulu, N., Harshavardhan, V. T., Govind, G., Seiler, C., and Kohli, A. (2012). Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506, 265–273. doi: 10.1016/j.gene.2012.06.076
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat / DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94, 1035–1040.
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Tuteja, N., Sahoo, R. K., Garg, B., and Tuteja, R. (2013). OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64). Plant J. 76, 115–127. doi: 10.1111/tpj.12277
Urao, T., Yamaguchi-Shinozaki, K., Urao, S., and Shinozaki, K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5, 1529–1539. doi: 10.1105/tpc.5.11.1529
van der Graaff, E., Dulk Ras, A. D., Hooykaas, P. J., and Keller, B. (2000). Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development 127, 4971–4980.
Wang, A., Tan, D., Takahashi, A., Li, T. Z., and Harada, T. (2007). MdERFs, two ethylene-response factors involved in apple fruit ripening. J. Exp. Bot. 58, 3743–3748. doi: 10.1093/jxb/erm224
Wang, H., Wang, H., Shao, H., and Tang, X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7:67. doi: 10.3389/fpls.2016.00067
Wang, W., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Wang, Y., Ying, J., Kuzma, M., Chalifoux, M., Sample, A., McArthur, C., et al. (2005). Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 43, 413–424. doi: 10.1111/j.1365-313X.2005.02463.x
Xia, K., Wang, R., Ou, X., Fang, Z., Tian, C., Duan, J., et al. (2012). OsTIR1 and OsAFB2 downregulation via osmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 7:e30039. doi: 10.1371/journal.pone.0030039
Xie, Z., Zhang, Z. L., Zou, X., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Xoconostle-Cázares, B., Ramírez-Ortega, F. A., Flores-Elenes, L., and Ruiz-Medrano, R. (2010). Drought tolerance in crop plants. Am. J. Plant Physiol. 5, 241–256. doi: 10.3923/ajpp.2010.241.256
Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang Rodriguez, R., Heuer, S., et al. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708. doi: 10.1038/nature04920
Xu, Z. S., Chen, M., Li, L. C., and Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Xue, G. P. (2002). Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res. 30:e77. doi: 10.1093/nar/gnf076
Yang, H., Yu, C., Yan, J., Wang, X., Chen, F., Zhao, Y., et al. (2014). Overexpression of the Jatropha curcas JcERF1 gene coding an AP2/ ERF-Type transcription factor increases tolerance to salt in transgenic tobacco. Biochemistry 79, 1226–1236. doi: 10.1134/S0006297914110108
Yang, T., and Poovaiah, B. W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277, 45049–45058. doi: 10.1074/jbc.M207941200
Yang, Y., Dong, C., Li, X., Du, J., Qian, M., Sun, X., et al. (2016). A novel AP2/ERF transcription factor from Stipa purpurea leads to enhanced drought tolerance in Arabidopsis thaliana. Plant Cell Rep. 35, 2227–2239. doi: 10.1007/s00299-016-2030-y
Yeo, A. R., and Flowers, T. J. (1984). “Mechanisms of salinity resistance in rice and their role as physiological criteria in plant breeding,” in Salinity Tolerance in Plants: Strategies for Crop Improvement, eds R. C. Staples and G. A. Toenniesen (New York, NY: Wiley), 151–170.
Yoon, D. H., Lee, S. S., Park, H. J., Lyu, J. I., Chong, W. S., Liu, J. R., et al. (2016). Overexpression of OsCYP19-4 increases tolerance to cold stress and enhances grain yield in rice (Oryza sativa). J. Exp. Bot. 67, 69–82. doi: 10.1093/jxb/erv421
Yoshida, S., Forno, D. A., Cock, J. H., and Gomez, K. A. (1976). Laboratory Manual for Physiological Studies of Rice. Manila: International Rice Research Institute, 61–66.
You, J., Zong, W., Hu, H., Li, X., Xiao, J., and Xiong, L. (2014). A STRESS RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 166, 2100–2114. doi: 10.1104/pp.114.251116
Zhang, H., Li, W., Chen, J., Yang, Y., Zhang, Z., Wang, X. C., et al. (2007). Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol. Biol. 63, 63–71. doi: 10.1007/s11103-006-9072-4
Zhang, H., Zhang, D., Chen, J., Yang, Y., Huang, Z., Huang, D., et al. (2004). Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol. Biol. 55, 825–834. doi: 10.1007/s11103-004-2140-8
Zhao, Y., Wei, T., Yin, K. Q., Chen, Z., Gu, H., Qu, L. J., et al. (2012). Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol. 195, 450–460. doi: 10.1111/j.1469-8137.2012.04160.x
Zhuang, J., Jiang, H. H., Wang, F., Peng, R. H., Yao, Q. H., and Xiong, A. S. (2013). A rice OsAP23, functioning as an AP2/ERF transcription factor, reduces salt tolerance in transgenic Arabidopsis. Plant. Mol. Biol. Rep. 31, 1336–1345. doi: 10.1007/s11105-013-0610-3
Zou, J., Liu, C., Liu, A., Zou, D., and Chen, X. (2012). Over expression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice. J. Plant Physiol. 169, 628–635. doi: 10.1016/j.jplph.2011.12.014
Keywords: abiotic stress, salt stress, drought stress, osmotic stress, rice, ABA, agronomic trait, tiller number
Citation: Kumar M, Choi J, An G and Kim S-R (2017) Ectopic Expression of OsSta2 Enhances Salt Stress Tolerance in Rice. Front. Plant Sci. 8:316. doi: 10.3389/fpls.2017.00316
Received: 16 November 2016; Accepted: 21 February 2017;
Published: 10 March 2017.
Edited by:
Zhulong Chan, Huazhong Agricultural University, ChinaReviewed by:
Mirza Hasanuzzaman, Sher-e-Bangla Agricultural University, BangladeshRatna Karan, University of Florida, USA
Rohit Joshi, Jawaharlal Nehru University, India
Copyright © 2017 Kumar, Choi, An and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seong-Ryong Kim, sungkim@sogang.ac.kr
 Manu Kumar
Manu Kumar Juyoung Choi
Juyoung Choi Gynheung An2
Gynheung An2 Seong-Ryong Kim
Seong-Ryong Kim