- The Genetic Engineering International Cooperation Base of Chinese Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Chinese Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China
Plant 14-3-3 proteins are involved in diverse biological processes, but for the model monocotyledonous species, Brachypodium distachyon, their roles in abiotic stress tolerance are not well understood. In this study, a total of eight Bd14-3-3 genes were identified from B. distachyon and these were designated respectively as BdGF14a–BdGF14g. The qRT-PCR analyses of 3-month-old plants of B. distachyon showed that these genes were all expressed in the stems, leaves, and spikelets. By contrast, most of the plants had relatively lower transcriptional levels in their roots, except for the BdGF14g gene. The different expression profiles of the Bd14-3-3s under various stress treatments, and the diverse interaction patterns between Bd14-3-3s and BdAREB/ABFs, suggested that these gene products probably had a range of functions in the stress responses. The NaCl-induced Bd14-3-3 gene, BdGF14d, was selected for overexpression in tobacco. BdGF14d was found to be localized throughout the cell and it conferred enhanced tolerance to salt in the transgenic plants. Lowered contents of malondialdehyde, H2O2, and Na+, and lower relative electronic conductance (Rec%), yet greater activities of catalase and peroxidase, were observed in the overexpressing plants. Higher photosynthetic rate, transpiration rate, stomatal conductance, and water use efficiency were measured in the transgenic lines. Following abscisic acid (ABA) or NaCl treatment, stomatal aperture in leaves of the BdGF14d-overexpression plants was significantly lower than in leaves of the wild type (WT) controls. The stress-related marker genes involved in the ABA signaling pathway, the reactive oxygen species (ROS)-scavenging system, and the ion transporters were all up-regulated in the BdGF14d-overexpressing plants as compared with WT. Taken together, these results demonstrate that the Bd14-3-3 genes play important roles in abiotic stress tolerance. The ABA signaling pathway, the ROS-scavenging system, and ion transporters were all involved in enhancing the tolerance to salt stress in the BdGF14d-overexpression plants.
Introduction
Many processes in the life cycles of plant require specific signal transduction to achieve vital biological functions. A common way to transfer such information is via the interaction between proteins, as modified by reversible phosphorylation (Ferl, 2004; Chevalier et al., 2009). However, since reversible phosphorylation of a protein alone is insufficient to complete the interaction directly, 14-3-3s, as crucial adaptor proteins are of great importance in plants because they bind phosphorylated peptides in the regulation of a diversity of processes (Denison et al., 2011), notably primary metabolism (Comparot et al., 2003; Diaz et al., 2011) and light (Sullivan et al., 2009; Taoka et al., 2011) and hormone signaling pathways (Ishida et al., 2004; Schoonheim et al., 2007; Gokirmak et al., 2010; Wang H. et al., 2011). In recent years, an increasing number of studies have shown that 14-3-3s also play important roles in how plants respond to a variety of biotic (Li et al., 2014; Teper et al., 2014) and abiotic (Chen et al., 2006; Sun et al., 2014; Zhou et al., 2014; He et al., 2015) stresses via the binding of the phosphorylated target proteins (Obsil and Obsilova, 2011).
The 14-3-3 proteins occur as homo- or hetero-dimers: each subunit forms a highly conserved amphipathic groove consisting of little more than nine antiparallel α-helices for ligand binding, as indicated by the crystal structure solved from tobacco (Wurtele et al., 2003; Ferl, 2004). The structural characteristics of these 14-3-3 proteins equip them for the regulation of various environmental signaling pathways, such as those related to drought, high salinity, and extreme temperatures.
Many recent studies have reported that 14-3-3s are implicated in abiotic responses as determined by an expression assay (Aksamit et al., 2005; Chen et al., 2006; Yao et al., 2007; Denison et al., 2011), while other studies have testified to their importance through investigations of transgenic plants. Cotton (Gossypium hirsutum) and Arabidopsis thaliana plants overexpressing the 14-3-3 gene AtGF14λ (Yan et al., 2004) and A. thaliana plants overexpressing the 14-3-3 gene AtGRF9 (He et al., 2015) each showed improved drought tolerance. On the other hand, a 14-3-3λ and 14-3-3κ double mutant showed enhanced salt resistance (Zhou et al., 2014). Potato (Solanum tuberosum) plants overexpressing 14-3-3 genes showed higher antioxidant activity than wild type (WT) plants; the converse result was found for transgenic potato plants in which the expression of 14-3-3 proteins was repressed (Lukaszewicz et al., 2002). In the case of Brachypodium distachyon, which is emerging as a monocotyledonous plant model, the roles of 14-3-3 proteins in stress responses are much less well understood (Draper et al., 2001).
In our study, a total of eight 14-3-3 genes in B. distachyon were identified and their cDNAs were cloned. Here, we present results on the expression profiles, and on the interactions between Bd14-3-3 proteins and BdAREB/ABF transcriptional factors (TFs) in B. distachyon, and we also show that transgenic tobacco (Nicotiana tabacum) plants overexpressing BdGF14d are tolerant to salt stress.
Materials and Methods
Identification and Cloning of the Bd14-3-3 Gene Family from B. distachyon
For reference, a total of 13 complete amino acid sequences of Arabidopsis 14-3-3s were downloaded from TAIR1. The databases of Phytozome v11.02 and the plantGDB3 were both used to extensively search for Bd14-3-3s; all the putative 14-3-3 sequences were submitted to Pfam4 to validate their 14-3-3 conserved domains, and then they were uploaded to NCBI5 to perform BLAST analyses. All the accession numbers of the obtained Bd14-3-3 genes are listed in Supplementary Table S1 and all the genes were amplified using gene-specific primers (Supplementary Table S2). The exon–intron gene structures were analyzed on BdGDB6. Sequence alignments among the Bd14-3-3 family and Arabidopsis and rice plants were undertaken using Clustal W. The results were edited by GeneDoc and the phylogenetic tree was constructed with the MEGA6 software tool using the neighbor-joining method (Chen et al., 2012). The promoter sequences were downloaded from PLAZA7 and the cis-elements in the promoters were analyzed on PlantCARE8.
Analysis of the Gene Expression Profiles by qRT-PCR
Seeds of the B. distachyon inbred line, Bd21 were germinated in distilled water under a photoperiod of 16-h light/8-h darkness. For RNA extraction, leaf samples of 2-week-old seedlings treated with 200 mM NaCl, 20% PEG6000, 10 mM H2O2, and 100 μM abscisic acid (ABA) were collected at 0, 1, 3, 6, 12, and 24 h after treatments. Different organs of 3-month-old plants of B. distachyon were also sampled, namely the roots, stems, leaves, and spikelets. Total RNA was extracted using a plant tissue total RNA extraction kit (Zomanbio, Beijing, China), the first cDNA chain was synthesized using the PrimeScriptTM RT reagent Kit with a gDNA Eraser (Takara, Japan). The qRT-PCR analysis was performed using the SuperReal PreMix Plus (SYBR Green, Tiangen, Beijing, China) on a qRT-PCR machine (Bio-Rad, Hercules, CA, USA). The primer sequences used for the expression analysis are given in Supplementary Table S3.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed according to reported methods (Sun et al., 2015). Eight BdAREB/ABFs were isolated during the identification of the Bd14-3-3s. The Bd14-3-3s and BdAREB/ABFs were incorporated into pGBKT7 and pGADT7 eukaryotic expression vectors. The primers used for the cloning of the BdAREB/ABFs and the vector constructions are shown in Supplementary Tables S4 and S5.
Generation of Transgenic Lines, and Salt-Stress Tolerance Assays
The complete cDNA sequence of BdGF14d was introduced into the eukaryotic expression vector pBI121 as driven by the CaMV 35S promoter. The transformations of tobacco by the Agrobacterium tumefaciens strain EHA105 containing recombinant plasmids or empty vectors, respectively, were performed as described elsewhere (Huang et al., 2015). Three overexpressed lines were screened on 100 mg/L kanamycin and confirmed by qRT-PCR tests using gene-specific primers were selected for salt-tolerance assay (Figure 5A). Seven-day-old seedlings of the WT and vector control (VC) plants and of the overexpressing lines were transferred from plates to a 1/2 MS (Murashige–Skoog) solid medium containing 150 or 200 mM NaCl. Their root lengths were measured after 2-weeks of treatments. Two-week-old seedlings of the five lines cultured on plates were planted in soil and grown under normal conditions for an additional 3 weeks. A 500 mM NaCl solution was used to treat the seedlings every 2 days for 20 days, after which plant physiological indices were measured. Measurement of relative electronic conductance (Rec%) was performed as described previously (Yang et al., 2017). The contents of H2O2 and malondialdehyde (MDA) and the activity of peroxidase (POD) and catalase (CAT) were determined using spectrophotometric methods and commercially available detection kits9. Measurements of Na+ and K+ contents were carried out as described for Arabidopsis (Sun et al., 2015). Photosynthetic rate, transpiration rate, and stomatal conductance were measured using the LI-6400 portable photosynthesis system (LI-COR Inc., Lincoln, NE, USA), and the water use efficiency (WUE) was calculated as described (Msanne et al., 2011).
Stomatal Aperture Measurements
The stomatal aperture in leaves was measured as described preciously (Grondin et al., 2015). Six-week-old tobacco leaves of the transgenic lines and the WT plants were sampled and treated with 200 mM NaCl and 50 μM ABA.
qRT-PCR Analyses of Related Marker Genes
Two-week-old transgenic plants and WT lines cultured on plates were transferred to MS solid medium containing 200 mM NaCl and grown for 7 days, after which entire seedlings were sampled for RNA extraction. The primers used for the qRT-PCR of the specific marker genes involved in salt tolerance are listed in Supplementary Table S6 (Deng et al., 2013a,b).
Subcellular Localization Analysis of BdGF14d
The subcellular localization of BdGF14d protein was investigated as described for tobacco plants (Hu et al., 2015). The recombinant plasmid 35S::BdGF14d-GFP was used to perform the assay.
Statistical Analysis
All data were analyzed by reported methods (Wang et al., 2015).
Results
Cloning of the 14-3-3 Genes from B. distachyon
A total of eight Bd14-3-3 genes were identified from B. distachyon through the BLAST searches and the Pfam analyses; they were designated as BdGF14a, BdGF14b, BdGF14c1, BdGF14c2, BdGF14d, BdGF14e, BdGF14f, and BdGF14g, respectively, based on the nomenclature in rice (Chen et al., 2006) and their chromosomal location, which was inconsistent with previous report (Cao et al., 2016) that we identified an alternative splicing in BdGF14c. BdGF14c1 and BdGF14c2 had the same sequences of 1–244 amino acid residues, but the last 18 amino acid residues for BdGF14c1 and the last six amino acid residues for BdGF14c2 were different, due to the alternative splicing patterns of the BdGF14c gene (Figure 1). All the Bd14-3-3s cDNAs were cloned using gene-specific primers (Supplementary Table S2) from B. distachyon and a multiple full-length amino acid sequence alignment, a phylogenetic analysis against Arabidopsis and rice, and an exon–intron structural annotation were performed. The results indicated that the Bd14-3-3 family members in B. distachyon were highly conserved during evolution (Figure 1A), in accordance with their highly similar amino acid alignment (Figure 1C). The exon–intron structure analysis showed that most of the Bd14-3-3s had three to five exons and two to four introns, apart from BdGF14e, which was composed of seven exons and six introns (Figure 1B). According to their different gene structures, BdGF14a, BdGF14b, BdGF14c1, BdGF14c2, BdGF14d, BdGF14f, and BdGF14g were classified into the non-𝜀 group, whereas BdGF14e was classified into the 𝜀 group, based on the taxonomy in Arabidopsis (DeLille et al., 2001). To explore the putative cis-elements involved in the abiotic stress response in promoters, 1 kb sequences upstream of the Bd14-3-3s CDS was downloaded and the ensuing abiotic stress-responsive cis-elements were analyzed. It was found that all the Bd14-3-3 genes except BdGF14d had one or two ABA-responsive elements (ABREs); the BdGF14b, BdGF14c1, BdGF14c2, and BdGF14f contained a low-temperature responsive element (LTR); and the BdGF14a, BdGF14b, BdGF14c1, BdGF14c2, and BdGF14g harbored a heat-stress responsive element (HSE) (Supplementary Table S1). Taken together, these results indicated, that the Bd14-3-3 genes could participate in these abiotic stresses.
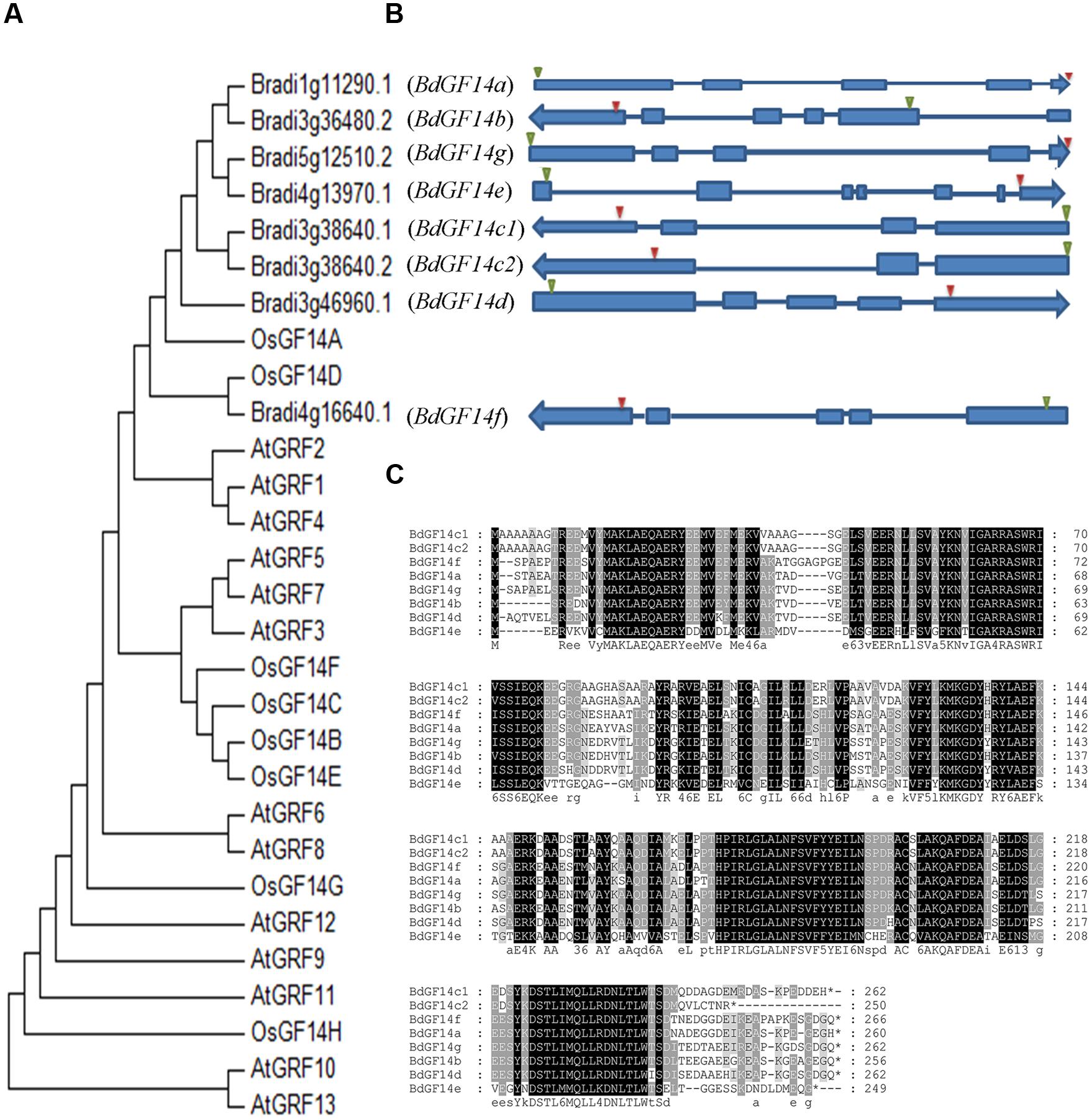
FIGURE 1. Phylogenetic and gene structure analyses of the Bd14-3-3s. (A) Phylogenetic tree of 14-3-3s from B. distachyon, rice, and Arabidopsis plants constructed using MAGA6. (B) Gene structures of Bd14-3-3s. Solid blue bars and strings represent the exons and introns, respectively; green and red triangles represent the translation starts and stops, respectively. Arrows indicate transcriptional directions on the chromosomes. (C) Amino acid sequence alignments of the Bd14-3-3 family.
Interactions between Bd14-3-3s and BdAREB/ABF TFs Proteins
Recent work has shown that the interactions between basic leucine zipper (bZIP) and the 14-3-3 proteins can influence numerous plant biological processes, such as the ABA-mediated signaling involved in abiotic stress responses. The AREB/ABF genes involved in ABA signaling that belong to subgroup A of bZIPs were cloned to verify the interactions between BdAREB/ABF TFs and the Bd14-3-3 proteins. The results demonstrated that all the Bd14-3-3s showed interactions with two to six BdAREB/ABF TFs (Figure 2), except for BdGF14c2 which was self-activated (data not shown). Among these, all the Bd14-3-3 family members were able to interact with BdbZIP62, but they interacted with neither BdbZIP56-2 nor BdFDL36. BdGF14c1 and BdGF14e exhibited the same interaction mode, in that both of them interacted with the same six BdAREB/ABF TFs. BdGF14d interacted with only two BdAREB/ABF TF members. The other Bd14-3-3 proteins, BdGF14a, BdGF14b, BdGF14f, and BdGF14g, showed interactions with four, three, five, and three BdAREB/ABF TFs, respectively. As signal transduction in plants always occurs via protein–protein interactions, these results suggested that the Bd14-3-3 proteins likely participated in the ABA-mediated signaling pathway by interacting with the BdAREB/ABF TF members.
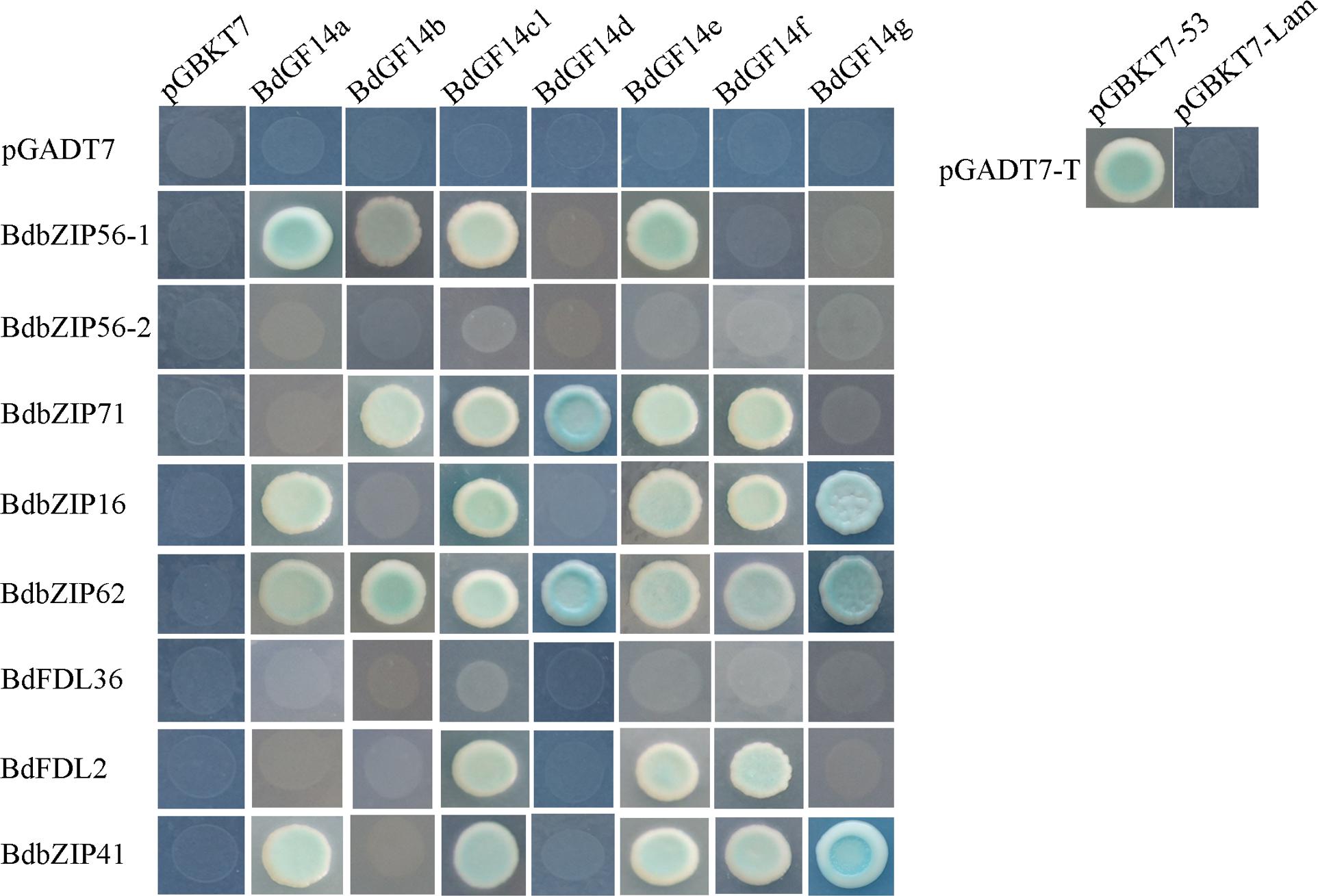
FIGURE 2. Yeast two-hybrid analyses between the Bd14-3-3s and the BdAREB/ABF TFs. The yeast strain AH109, transferred along with the described plasmid combinations, was spotted onto Trp-/Leu-/His-/Ade-medium containing x-α-gal. The Bd14-3-3s were cloned into pGBKT7, and the BdAREB/ABFs were cloned into pGADT7 vectors. Transformants containing pGADT7-T and pGBKT7-53 vs. pGBKT7-Lam represent the positive vs. negative controls, respectively.
Expression Profiles of Bd14-3-3s in Different Plant Organs
To investigate the biological function of Bd14-3-3s, the expression profiles of these Bd14-3-3s in the roots, stems, leaves, and spikelets of 3-month-old plants of B. distachyon were investigated. The results indicated that most members of this gene family except BdGF14g had relatively lower expression levels in the roots than in the leaves (Figure 3). Besides this differences, all the Bd14-3-3 genes exhibited global expression in the stems, leaves and spikelets. Notably, BdGF14a and BdGF14e showed significantly higher expression levels in the spikelets, pointing to the putative functional involvement of these two genes in seed development.
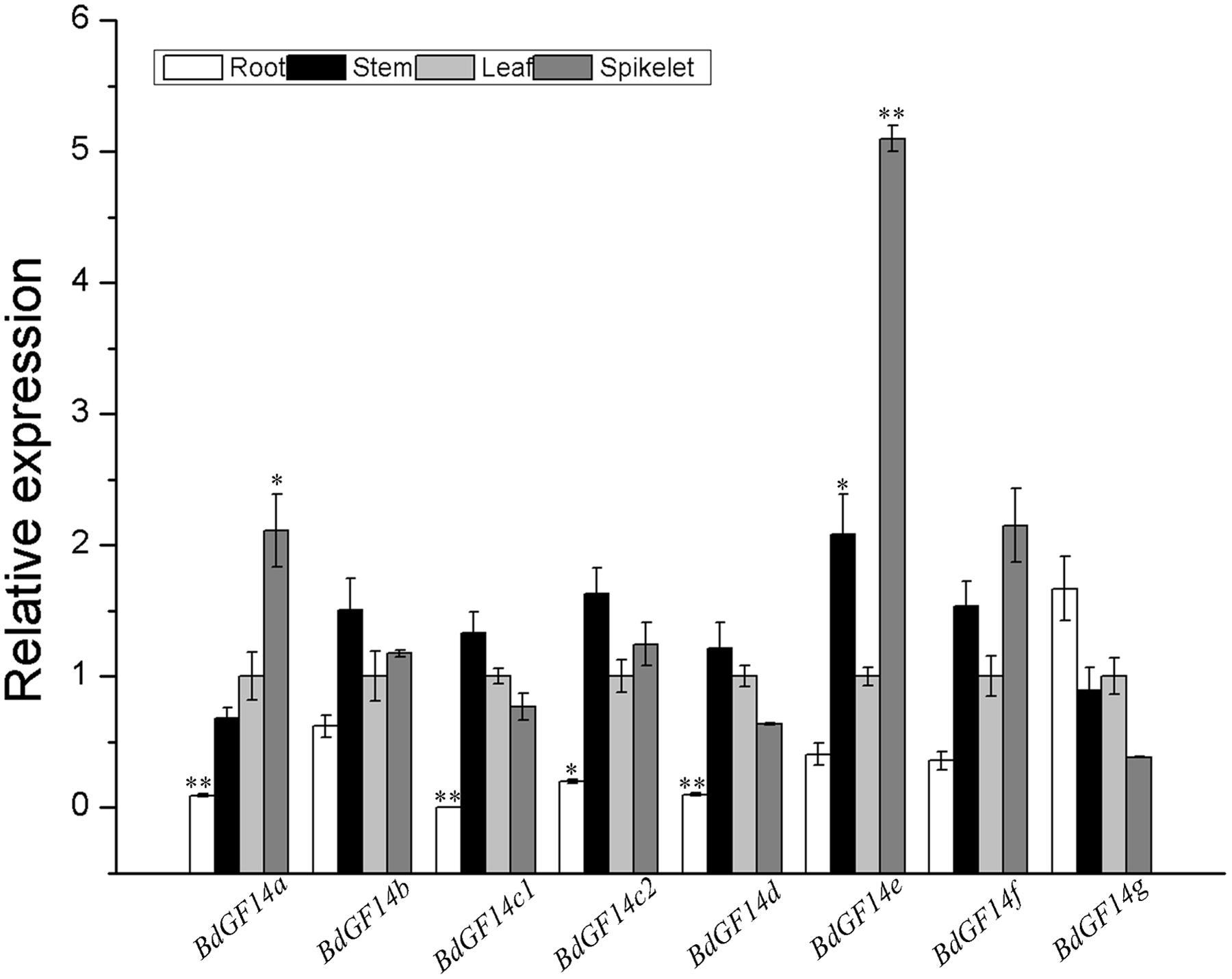
FIGURE 3. Expression of the Bd14-3-3s in different plant organs. Roots, stems, leaves, and spikelets of 3-month-old plants of B. distachyon were sampled to extract RNA for expression analysis. The y-axis represents the relative expression of the genes. All bars are means (±SE) calculated from three replicates; asterisks denote significant differences between leaves and the other organs (∗P < 0.05; ∗∗P < 0.01).
Expression Profiles of Bd14-3-3s in Response to Abiotic Stresses
Plant 14-3-3 proteins play important roles in response to various abiotic stresses (Aksamit et al., 2005; Sun et al., 2014; He et al., 2015). Therefore, qRT-PCR analyses were performed to determine the expression patterns of the 14-3-3 gene family under conditions of abiotic stress PEG6000, NaCl, and the signaling molecules ABA and H2O2. Considering that the fluctuation of the Bd14-3-3s expression was influenced by photoperiod in the control, the results revealed that only BdGF14d showed distinct up-regulation in response to the NaCl treatment, whereas the other genes exhibited different degrees of down-regulation (Figure 4A). Under the PEG treatment, BdGF14e was up-regulated; however, BdGF14f and BdGF14g were up-regulated at 3 h after 20% PEG6000 was added (Figure 4B). In response to H2O2, BdGF14b showed evidence of up-regulation, whereas BdGF14c1 and BdGF14d exhibited down-regulation; the other genes demonstrated moderate variation (Figure 4C). Under the ABA treatment, BdGF14e and BdGF14f showed evidence of up-regulation, and BdGF14b and BdGF14g were up-regulated rapidly at 1 h after treatment but were then down-regulated at 12 h; the other genes exhibited no significant variation in response to ABA (Figure 4D). The expression patterns of the Bd14-3-3 gene family suggested that different 14-3-3 isoforms played specific roles in the response to diverse abiotic stresses, and that, correspondingly, the respective 14-3-3 proteins might participate in the signaling transduction network of this response.
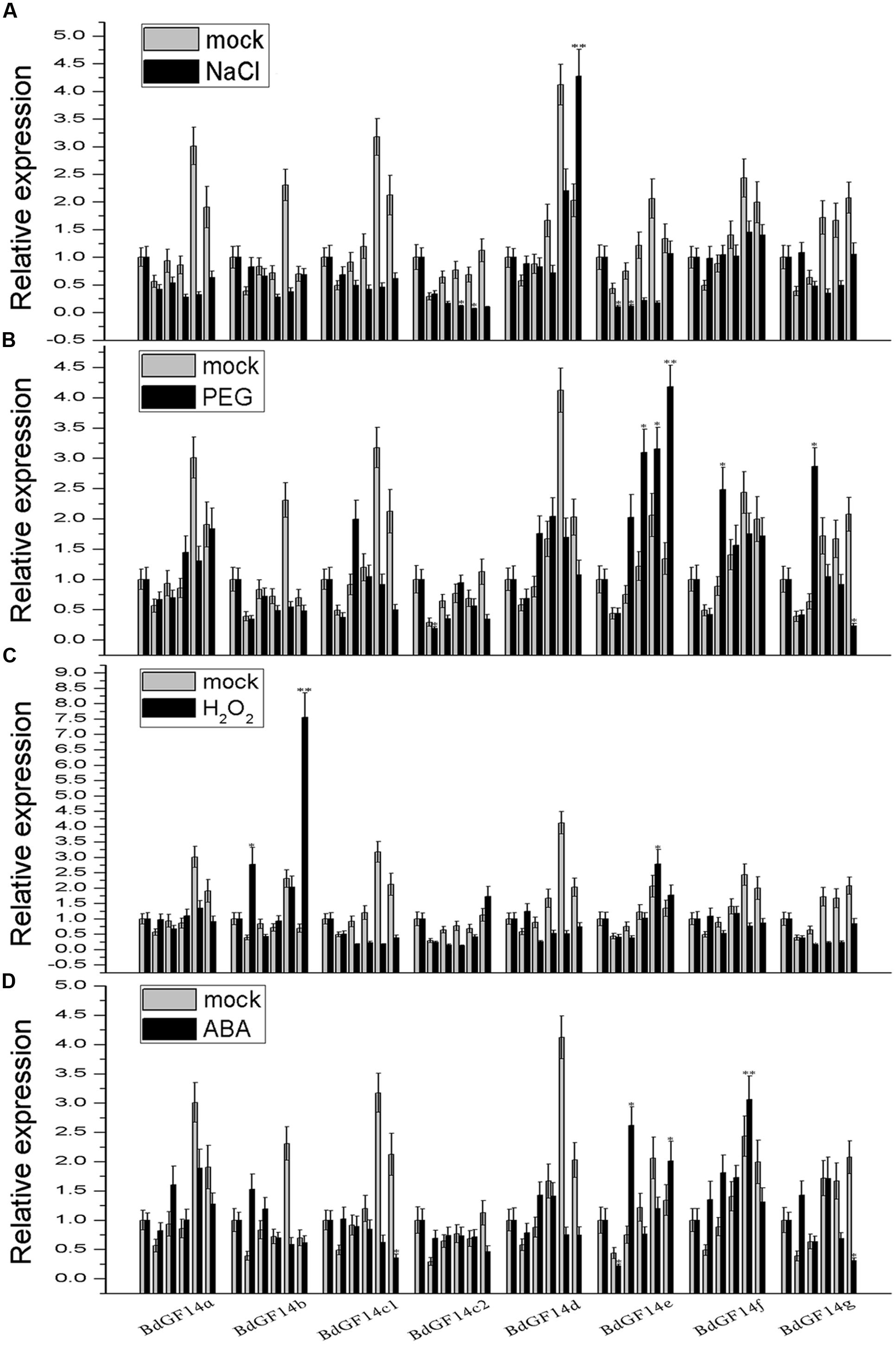
FIGURE 4. Expression profiles of Bd14-3-3s under applied stress treatments. Leaves of B. distachyon were collected at 0, 1, 3, 6, 12, and 24 h after being treated with 200 mM NaCl (A), 20% PEG (B), 10 mM H2O2 (C), and 100 μM ABA (D), respectively, and RNA was extract for expression analyses. Mock indicates the control (no treatment applied). Bars are means (±SE), all the experiments were repeated three times. Asterisks denote significant differences between 0 h and different time points (∗P < 0.05; ∗∗P < 0.01).
Overexpression of BdGF14d Enhances Salt Tolerance in Transgenic Tobacco Plants
Although functional redundancy is common in a multigene family such as 14-3-3s, BdGF14d was the only gene in the Bd14-3-3s family to respond to NaCl, based on the expression profiles. Hence, BdGF14d was selected for a salt tolerance study by overexpressing it in transgenic tobacco plants. Three transgenic lines screened on 100 mg/L kanamycin and confirmed by qRT-PCR analyses using gene-specific primers were selected for this salt tolerance assay (Figure 5A). To evaluate the performance of the BdGF14d-overexpression seedlings under extensive salt stress, the 7-day-old seedlings of the WT, VC, and transgenic lines that germinated on a MS medium were transferred to 1/2 MS medium containing 100 and 150 mM NaCl to grow for a further 2 weeks. The BdGF14d-overexpression lines developed distinctly longer roots than WT under both levels of the NaCl salt treatment 100 and 150 mM (Figure 5B); yet they showed no apparent difference from WT and VC lines under normal conditions (Figure 5C). The 3-week-old seedlings of the three BdGF14d-overexpression and WT, VC lines were treated with 500 mM NaCl for 35 days in soil and significantly higher survival rates (52.1, 71.7, and 51.3%, respectively) than WT (15.2%) and VC (20.7%) were observed. Moreover, the leaves of transgenic seedlings maintained their growing status, whereas those of the WT and VC plants had already wilted or died (Figure 5D), yet there was no apparent difference between WT, VC and transgenic lines under normal conditions (Figure 5E).
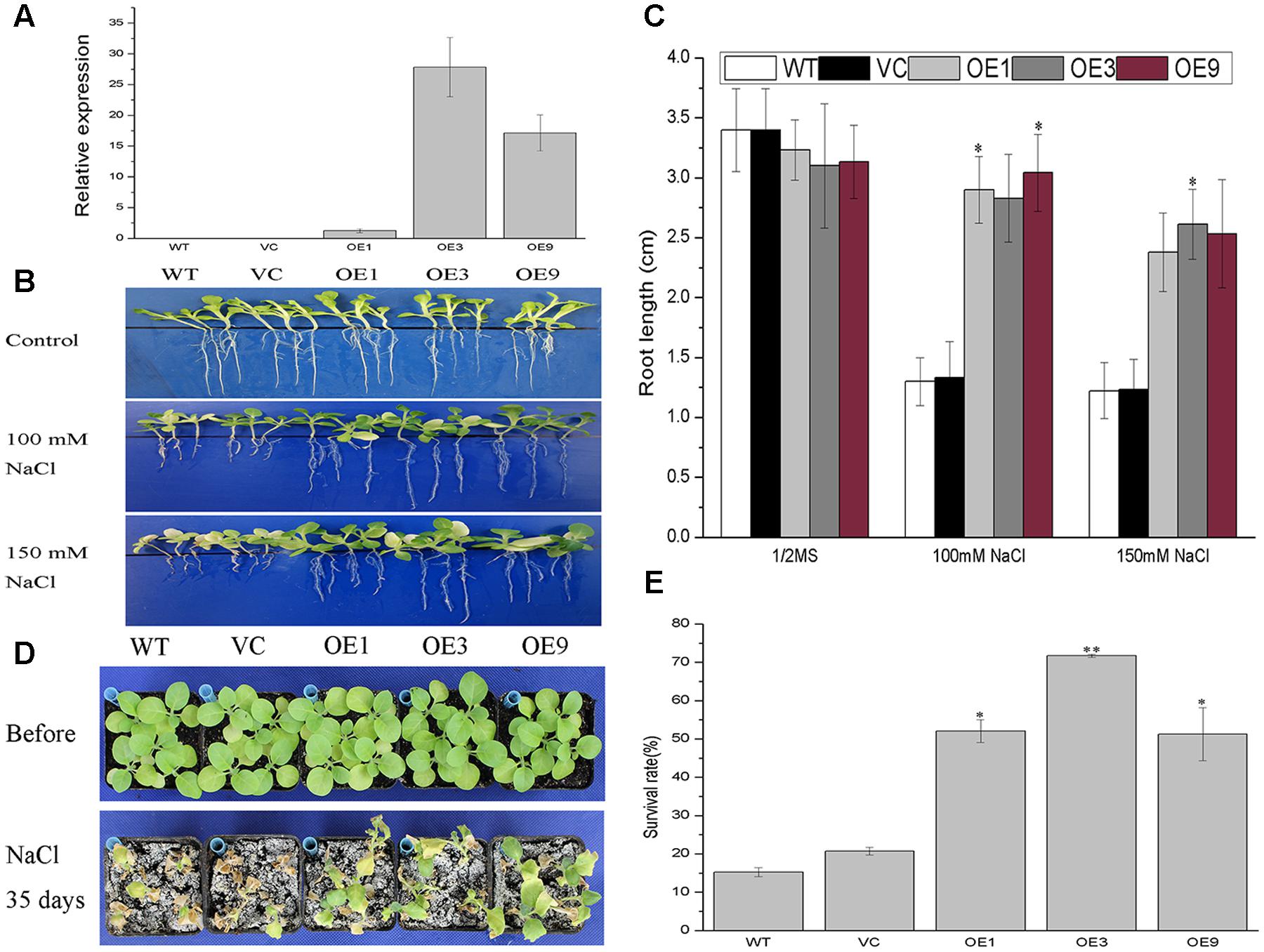
FIGURE 5. Improved tolerance to salt in the BdGF14d-overexpression plants. (A) The relative expression levels of BdGF14d in transgenic tobacco plants. The Ntubiquitin was used as internal reference. (B) Seven-day-old seedlings of WT, VC, and transgenic lines germinated on MS were transferred to 1/2 MS containing 100 mM and 150 mM NaCl to grow for a further 2 weeks. (C) The root lengths were measured and statistically analyzed. (D) Two-week-old seedlings growing on MS medium were planted in soil to grow under normal condition for 3 weeks and 500 mM NaCl solution was used to treat the seedlings for a further 35 days. (E) The respective survival rates were calculated. All the experiments were repeated three times. Bars are means (±SE); asterisks denote significant differences between WT and the transgenic lines (∗P < 0.05; ∗∗P < 0.01).
Salt stress will promote the accumulation of reactive oxygen species (ROS), which leads to oxidative damage (Hossain and Dietz, 2016). To investigate the salt tolerance mechanism in the BdGF14d-overexpression seedlings in physiological terms, the related indices of MDA, Rec%, and H2O2 these reflect membrane and intracellular oxidative damage and the antioxidant enzyme system of CAT and POD were determined in transgenic lines and control plants after NaCl treatment or under normal growing conditions. The results showed that the BdGF14d-overexpression plants had obviously lower contents of MDA and H2O2 and lower Rec% after the NaCl treatment (Figures 6A–C). In the transgenic lines, the activity of CAT was significantly higher than in WT under the salt stress and normal condition; likewise, POD activity of transgenic lines was clearly higher under normal condition. Interestingly, the POD activity of overexpressing plants showed a slight increase than that of WT after the salt treatment (Figures 6G,H). The above results suggested that in the BdGF14d-overexpression plants tolerance to salt was enhanced by activating the antioxidant enzymes to mitigate oxidative damage.
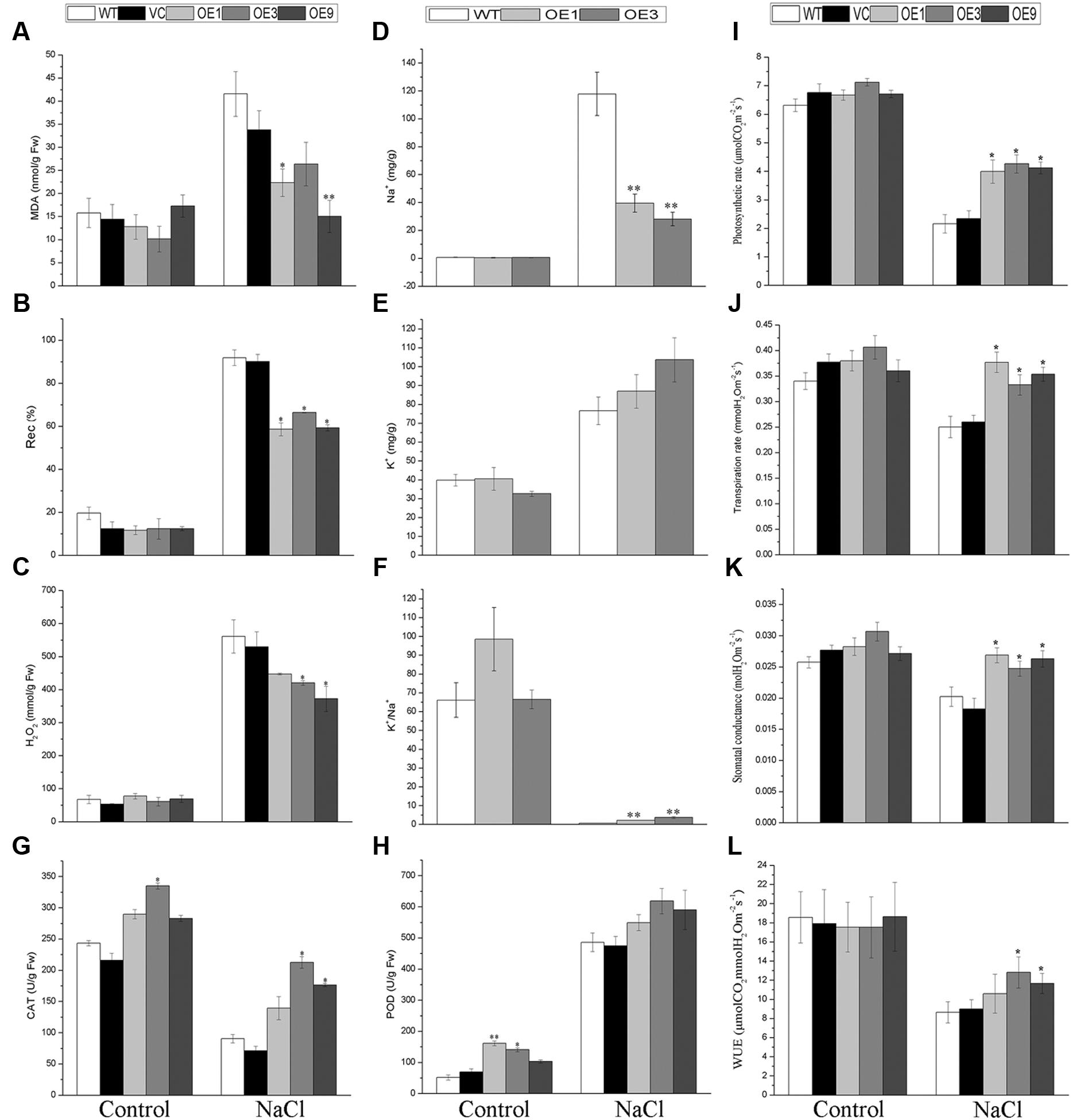
FIGURE 6. Plant physiological indices of transgenic lines and WT. Two-week-old WT and transgenic lines geminated on MS were planted in soil to grow for 3 weeks; all seedlings were treated with a 500 mM NaCl solution every 2 weeks for a further 20 days. Leaves of the five lines with or without salt applied were sampled for measurements of MDA (A), Rec% (B), H2O2 (C), CAT (G), POD (H), photosynthetic rate (I), transpiration rate (J), stomatal conductance (K), WUE (L), and for their contents of Na+ (D) and K+ (E). The ratio of K+/Na+ was also calculated (F). Bars are means (±SE); and asterisks denote significant differences between WT and the transgenic lines (∗P < 0.05; ∗∗P < 0.01). All the experiments were repeated three times.
To determine the accumulation of Na+ and K+, these two cations were measured in the leaves of transgenic and WT plants with or without salt stress applied. The results showed that, under salt stress, the transgenic lines accumulated significantly less Na+ and possessed distinctly higher K+/Na+ ratio in leaves than did the WT plants (Figures 6D,F). By contrast, the contents of K+ showed no significant variation in all plants tested (Figure 6E).
To explore the WUE of WT and the overexpressing plants under normal conditions and under NaCl treatment, their photosynthetic rate, transpiration rate, and stomatal conductance were measured. The results indicated that the BdGF14d-overexpression plants had a higher photosynthetic rate, transpiration rate, stomatal conductance, and WUE than did the WT plants under salt stress; however, no significant differences were observed between the two plant types under normal condition (Figures 6I–L).
Overexpression of BdGF14d Promotes Stomatal Closure under NaCl and ABA Conditions
Salt stress facilitates the accumulation of endogenous ABA, which is a vital regulator of stomatal closure to reduce the amount of Na+ transported from roots to shoots and to minimize water loss via transpiration (Kim et al., 2010; Campos et al., 2016). The differences in morphology and physiology observed above between the transgenic lines and WT plants under salt stress prompted us to investigate whether the overexpressing BdGF14d in tobacco plants affected the stomatal closure in seedlings treated with NaCl or ABA. No apparent differences were observed when the transgenic and WT plants were dipped in a bathing solution without NaCl or ABA in the light. However, there were stark differences in stomatal aperture when detached leaves were treated with either NaCl or ABA (Figure 7). Specifically, under either treatment, the degree of stomatal closure in leaves was significantly greater in the BdGF14d-overexpression plants than in the WT plants. These results suggested that overexpressing BdGF14d accelerated stomatal closure under NaCl stress and that this mechanism to enhance salt tolerance was dependent upon ABA signaling.
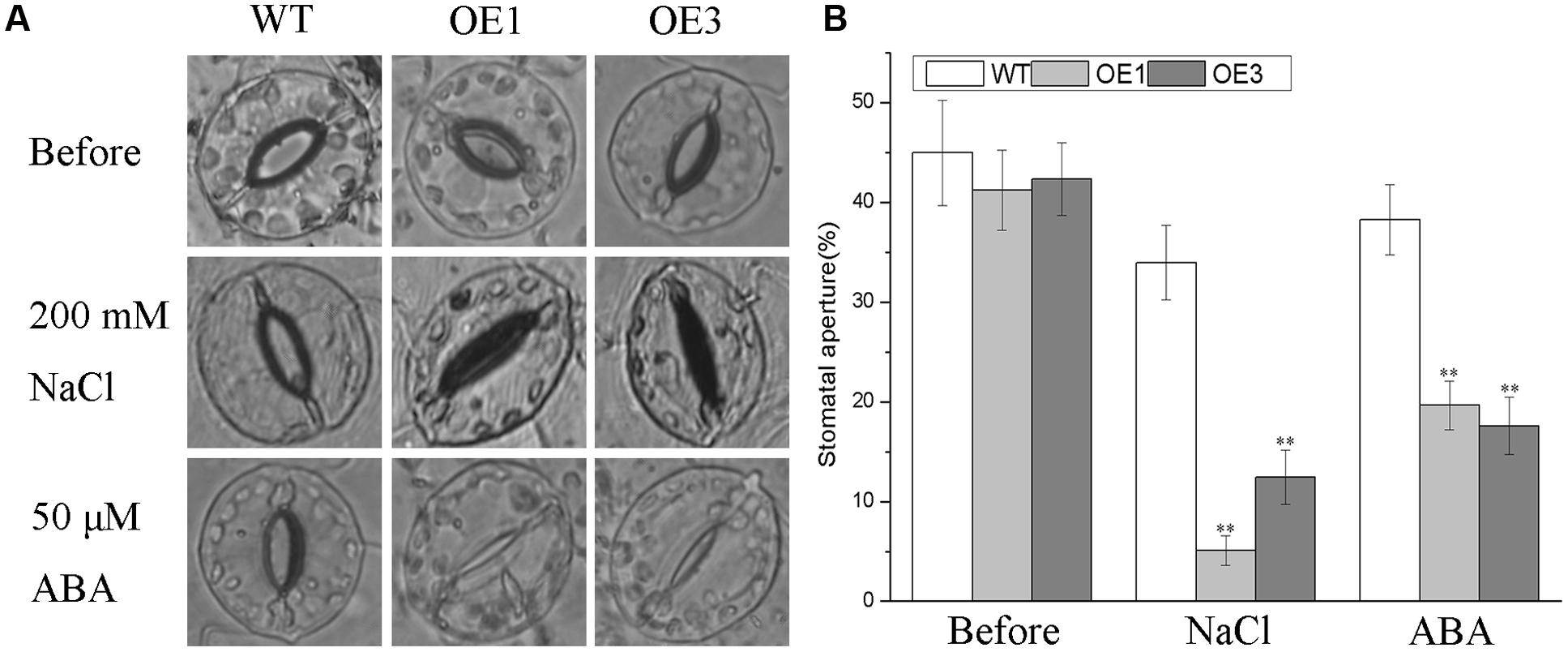
FIGURE 7. Analysis of stomatal aperture in transgenic lines and WT plants under the NaCl and ABA treatments. Detached leaves of WT and transgenic lines were dipped in stomatal opening buffer under light for 6 h. A total of 200 mM NaCl and 50 μM ABA were added to treat the leaves for 1 and 2 h, respectively (A) and the stomatal apertures were calculated (B). Bars are means (±SE); asterisks denote significant differences between WT and the transgenic lines (∗∗P < 0.01).
The qRT-PCR Analyses of Marker Genes Involved under Salt Stress
To further determine the effects of BdGF14d in the overexpressing plants, related marker genes involved in the response to salt stress were amplified by qRT-PCR, with or without the NaCl treatment. A crucial ABA biosynthetic gene NtNCED1, a key target of the ABA signaling gene, NtABF2, and an ABA-induced gene TobLTP1 were all significantly up-regulated under salt stress in transgenic lines, relative to WT, albeit no distinct differences were observed without salt applied. The ROS-scavenging genes NtCAT and, NtPOX2, and the stress defense gene NtERD10C, showed obvious up-regulation in the BdGF14d-overexpression plants as compared with WT under the salt treatment. The expression levels of the plasma membrane Na+/H+ antiporter gene NtSOS1 and of both tonoplast Na+/H+ antiporters genes, NtNHX2 and NtNHX4, showed evident up-regulation in the overexpressed lines as compared with the WT plants under salt (Figure 8). These expression results suggested that BdGF14d appeared to confer salt tolerance to transgenic plants via changes in ABA signaling, ROS-scavenging, ion transporters, and cellular protective proteins (such as NtERD10Cs).
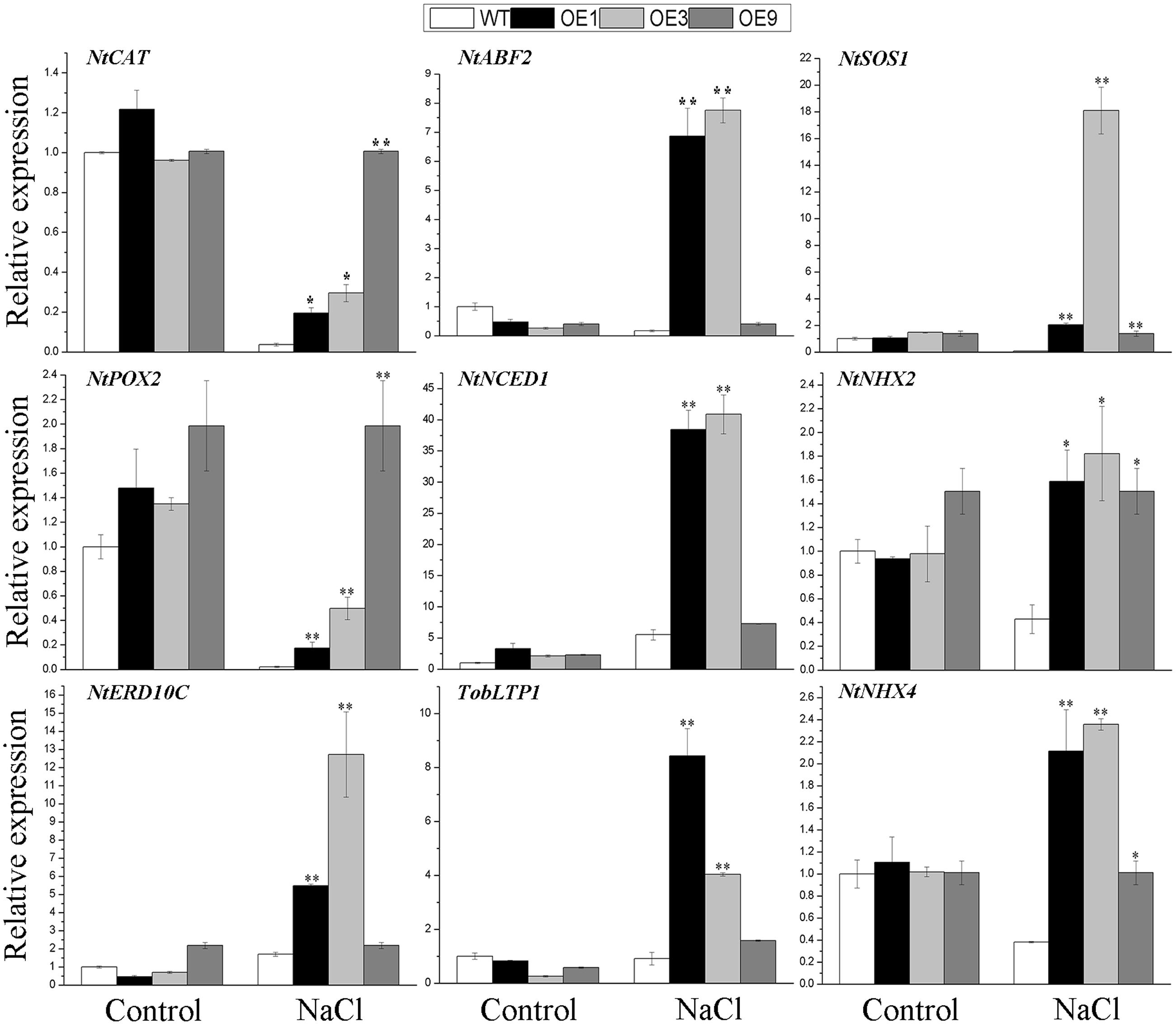
FIGURE 8. The qRT-PCR analyses of related marker genes of the transgenic lines and WT under salt stress. Two-week-old seedlings that germinated on MS medium were transferred to MS containing 200 mM NaCl to grow for 7 days. Entire seedlings of the five lines with or without NaCl applied were sampled to extract RNA for expression analyses. Bars are means (±SE); asterisks denote significant differences between WT and the transgenic lines (∗P < 0.05; ∗∗P < 0.01).
BdGF14d is Localized throughout the Cell
To determine the subcellular localization of the BdGF14d protein, the leaf epidermal cells of tobacco plants were injected with A. tumefaciens that contained the recombinant plasmid pBI121-BdGF14d-GFP. The results showed that the BdGF14d was localized throughout the cell (Figure 9).
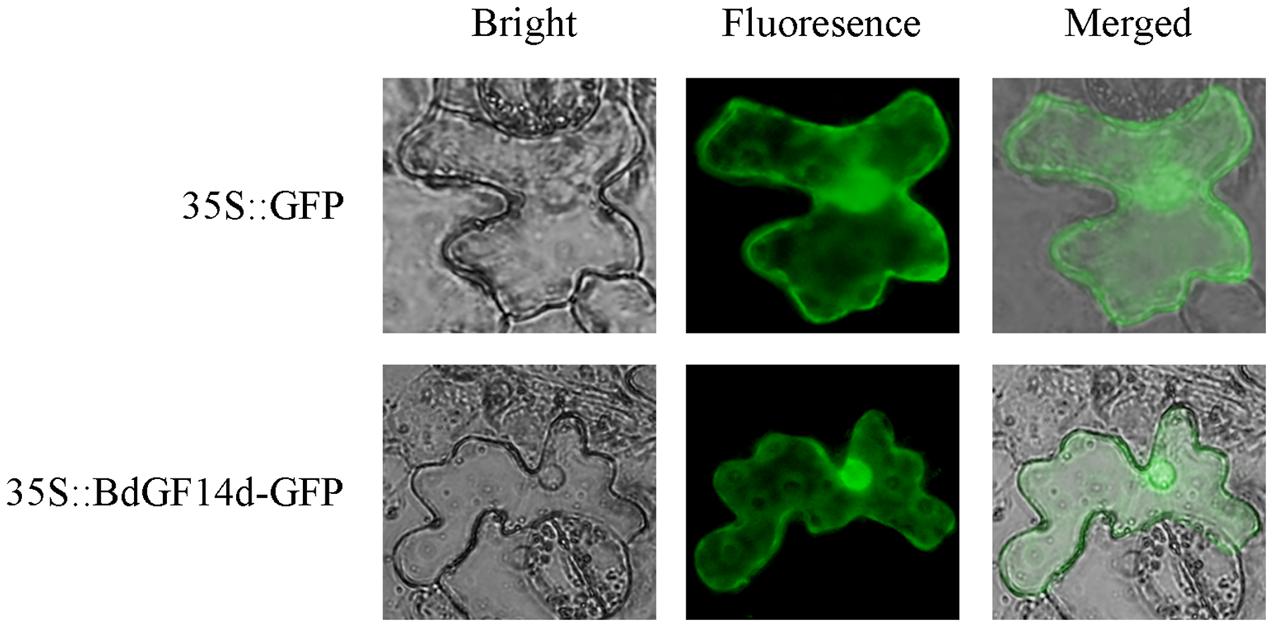
FIGURE 9. Subcellular localization of the BdGF14d protein. BdGF14d and GFP were inserted into the pBI121 vector, and the recombinant plasmid pBI121-BdGF14d-GFP and vector control pBI121 were introduced into A. tumefaciens for injection into the epidermal cells of tobacco leaves.
Discussion
The scaffolding protein 14-3-3 family plays a vital role in signaling networks in eukaryotic cells by binding the phosphorylated target proteins to form dynamic protein complexes, which directly decides the interaction strength of proteins through (de)phosphorylation (Tzivion et al., 2001; Sehnke et al., 2002). An increasing number of studies have reported the importance of 14-3-3 proteins in the tolerance of abiotic stress, but their exact roles in the B. distachyon plant species, a newly emerging monocotyledon model, have not yet been explored.
In this study, eight 14-3-3 genes in B. distachyon were identified and preliminary functional analyses were successfully carried out. An alternative splicing of BdGF14c1 containing different exon–intron gene structures was found, this was designated as BdGF14c2, which differed from the other seven 14-3-3 genes identified in a previous report (Cao et al., 2016). The promoter cis-elements analyses of the eight Bd14-3-3 genes suggested their potential functions involved in response to abiotic stresses and in ABA signaling. Expression profiles of the Bd14-3-3s in the different organs showed relatively lower expression levels in roots but higher levels in the spikelets (Figure 3), suggesting that the Bd14-3-3s likely participated in seed development. Further, the expression profiles of the Bd14-3-3 gene family under abiotic stresses and ABA treatment showed isoform-specific up- or down-regulation (Figure 4), thus implying their various roles in the abiotic stress responses of B. distachyon. The crucial roles of the 14-3-3 proteins in the light-signaling pathway have been clarified extensively in Arabidopsis (Sullivan et al., 2009; Gao et al., 2014) and rice (Taoka et al., 2011), and certain light-responsive elements were identified in promoters of the Bd14-3-3s (data not shown). Hence, when these are taken into consideration, up-regulation of the expression levels of the Bd14-3-3s following exposure to light was expected under controlled condition. After taking into account the fluctuating expression levels of the Bd14-3-3 in control plants, the significant up-regulation of BdGF14d when plants were treated with NaCl, and likewise that of BdGF14e, BdGF14f, and BdGF14g with PEG, of BdGF14b with H2O2, and of BdGF14e and, BdGF14f with ABA, together indicates that Bd14-3-3s seem to be involved in diverse abiotic stress and ABA responses.
Previous studies have demonstrated that the 14-3-3 proteins can exert critical effects in plant response to abiotic stresses (Yan et al., 2004; Sun et al., 2014; He et al., 2015). In our study, the BdGF14d-overexpression tobacco plants had significantly higher tolerance to salt stress than did WT plants. This result was confirmed by the several physiological index measurements and verified further by the qRT-PCR analyses of the related marker genes in the transgenic lines and WT plants. ABA clearly plays a pivotal role in the diverse adaptive stress responses to unfavorable environmental stimuli including high salinity, drought and extreme temperatures as an important plant hormone (Fujita et al., 2011; de Boer et al., 2013). In Arabidopsis, the phosphorylated ABF3 combines with 14-3-3 to stabilize its conformation and thereby facilitate ABA signaling (Sirichandra et al., 2010), and in barley plants the 14-3-3s responded to ABA and controlled ABA action (Schoonheim et al., 2007).
To better explore the putative role of ABA in the improved salt tolerance of transgenic plants, we carried out (1) a yeast two-hybrid assay between the Bd14-3-3s and BdAREB/ABF TFs in B. distachyon, (2) a stomatal aperture test under the ABA treatment, and (3) qRT-PCR analyses of related marker genes known to be involved in ABA signaling. The interactions between BdGF14d and an ABA-insensitive 5 (ABI5)-like protein, BdbZIP62, and a transcription factor responsible for ABA regulation 1 (TRAB1)-like protein BdbZIP71 involved in ABA signaling in B. distachyon, were both confirmed by the yeast two-hybrid assay, thus suggesting that BdGF14d was indeed involved in the ABA signaling pathway (Figure 2). The adaptive responses to abiotic stresses as mediated by ABA include stomatal closure and the transcription of a number of related genes involved in the stress tolerance process (Choi et al., 2000; An et al., 2008; Wang R.S. et al., 2011; Geilfus et al., 2015; Campos et al., 2016). The 14-3-3 proteins seem to participate in ABA-mediated stomatal closure (Cotelle and Leonhardt, 2016): there was less stomatal aperture in leaves of the BdGF14d-overexpression plants than in those of WT under the ABA treatment (Figure 7); this implicates BdGF14d in ABA-mediated stomatal closure. The higher expression levels of three ABA signaling related genes NtNCED1 that controls the biosynthesis of ABA, NtABF2 that facilitates various ABA-mediated signal transductions, and TobLTP1 that responds to ABA (Zhang et al., 2005; Deng et al., 2013b; Hu et al., 2015) found in the BdGF14d-overexpression plants (vs. WT; Figure 8), suggests that BdGF14d promoted ABA signaling transduction. From the above set of results, we infer that the BdGF14d probably enhanced tolerance to salt in the transgenic plants via ABA signaling.
Excess ROS accumulation induced by diverse abiotic stresses generates oxidative damage in the plasma membrane and intracellularly, and so plants have evolved effective antioxidant enzyme systems for ROS detoxification (Liu and He, 2016; Rao and Chaitanya, 2016). The relatively higher activity of the antioxidant enzymes confer upon maize an improved tolerance to environmental stimuli (Stepien and Klobus, 2005). Maintaining a strong antioxidant defense system is of great importance to plants for enhanced tolerance to high salt condition (Hu et al., 2012; Hossain and Dietz, 2016). In the present study, there was relatively higher activity of CAT and POD, which both belong to the antioxidant enzyme, in the BdGF14d-overexpression lines than in the WT plants (Figures 6G,H). Similarly, NtCAT and NtPOX2 also exhibited higher expression levels in the transgenic lines when under salt stress (Figure 8). These results revealed that the BdGF14d-overexpression plants had improved salt tolerance that was linked to an enhanced ROS-scavenging system.
High salinity inhibits the growth and reproduction of plants via excessive osmotic stress and ion damage (Deinlein et al., 2014; Lv et al., 2014). However, plants have evolved physiological mechanisms, such as the inhibition of stomatal openings to limit the transportation of Na+ from roots to leaves by transpiration, as well as the reduction of Na+ accumulation in the leaf cytoplasm by excreting Na+ out and compartmentalizing excessive Na+ into vacuoles, so that they can continue to function while under salt stress (Zhu, 2003; Zhang and Shi, 2013). Increased activity of the plasma membrane Na+/H+ antiporter, SOS1, and of the vacuolar Na+/H+ antiporters, NHXs, confers plants salt tolerance to plants by transporting excess Na+ from the cytoplasm out of the cell and into vacuoles, respectively, to reduce the toxic levels of Na+ (Li et al., 2009; Barragan et al., 2012; Zhang and Shi, 2013; Roy et al., 2014). Further, an increased abundance of 14-3-3s enhances the activity of PM H+-ATPase, thus generating an H+ electrochemical gradient that provides the driving force for SOS1 to mitigate abiotic stresses (Bunney et al., 2002; Kerkeb et al., 2002; Fuglsang et al., 2007; Pallucca et al., 2014). To resist salt stress in particular, the transcriptional levels of the vacuolar Na+/H+ exchangers NHXs are up-regulated by ABA to facilitate stomatal closure and intracellular ionic homeostasis (Apse et al., 1999; Zhu, 2003; Barragan et al., 2012). In the present study, the higher expression levels of NtSOS1, NtNHX2, and NtNHX4 (Figure 8) and the lower accumulation of Na+ (Figure 6D) in the BdGF14d-overexpressing plant leaves (vs. WT), together, indicate that the regulation of ion transporters by BdGF14d was involved in the improvement of salt stress tolerance.
The photosynthetic rate and WUE in the BdGF14d-overexpression plants were higher than in WT plants under salt stress; but interestingly, the transpiration rate and stomatal conductance were also higher in the transgenic lines (Figures 6I–L). This outcome was probably due to the complicated regulatory mechanism of the whole plant and the dynamic physiological changes across the different growth stages of seedlings.
Conclusion
Eight Bd14-3-3 genes were identified and cloned from B. distachyon plants. The different expression profiles of the Bd14-3-3 gene family under the different stress treatments tested, coupled to the diverse interaction patterns between Bd14-3-3s and BdAREB/ABF TFs, suggest various roles for these genes in abiotic stress responses involving ABA. Overexpression of BdGF14d, a NaCl-induced member of the Bd14-3-3 gene family, enhanced tolerance to salt stress in the transgenic tobacco plants. The ROS-scavenging system and ion transporters contributed to salt resistance, while ABA signaling was involved in the enhanced salt tolerance of the BdGF14d-overexpression plants. Together, the findings provide valuable information for elucidating the roles and complex mechanisms of plant 14-3-3s in response to diverse abiotic stresses.
Author Contributions
GH, GY, JC, and YH designed the experiments and wrote the paper. YH performed all experiments and analyzed the data. YZ and LC helped to identify the genes. YZ helped to conduct the experiments. CW, QL, FZ, QW, and KL participated in partial experiments. All authors read and approved the manuscript.
Funding
The work was supported by National Genetically Modified New Varieties of Major Projects of China (2016ZX08010004-004), the National Natural Science Foundation of China (No. 31570261, 31601995), and Education Department of Hubei Province (B2016295).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank the Analytical and Testing Center of Wuhan University for providing atomic absorption spectroscopy assay.
Abbreviations
ABA, abscisic acid; ABI5, abscisic acid insensitive 5; AREB/ABF, ABA responsive element binding factor; bZIP, basic leucine zipper; CaMV, Cauliflower mosaic virus; CAT, catalase; CDS, coding sequence; ERD10C, early responsive to dehydration 10C; GA, gibberellin; GFP, green fluorescent protein; HSE, heat stress responsive element; LTR, low-temperature responsive element; MDA, malondialdehyde; MS, Murashige–Skoog; NCED1, 9-cis-epoxycarotenoid dioxygenase 1; NHX, Na+/H+ antiporter; OE, overexpression; POD, peroxidase; qRT-PCR, quantitative real-time polymerase chain reaction; Rec%, relative electronic conductance; ROS, reactive oxygen species; TFs, transcription factors; TobLTP1, tobacco lipid transfer protein 1; TRAB1, transcription factor responsible for ABA regulation 1; UTR, untranslated region; VC, vector control; WT, wild type; WUE, water use efficiency.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00340/full#supplementary-material
Footnotes
- ^ http://www.arabidopsis.org
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://www.plantgdb.org
- ^ http://pfam.xfam.org
- ^ https://www.ncbi.nlm.nih.gov
- ^ http://www.plantgdb.org/BdGDB
- ^ http://bioinformatics.psb.ugent.be/plaza
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html
- ^ www.njjcbio.com
References
Aksamit, A., Korobczak, A., Skala, J., Lukaszewicz, M., and Szopa, J. (2005). The 14-3-3 gene expression specificity in response to stress is promoter-dependent. Plant Cell Physiol. 46, 1635–1645. doi: 10.1093/pcp/pci179
An, Z., Jing, W., Liu, Y., and Zhang, W. (2008). Hydrogen peroxide generated by copper amine oxidase is involved in abscisic acid-induced stomatal closure in Vicia faba. J. Exp. Bot. 59, 815–825. doi: 10.1093/jxb/erm370
Apse, M. P., Aharon, G. S., Snedden, W. A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. doi: 10.1126/science.285.5431.1256
Barragan, V., Leidi, E. O., Andres, Z., Rubio, L., De Luca, A., Fernandez, J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24, 1127–1142. doi: 10.1105/tpc.111.095273
Bunney, T. D., van den Wijngaard, P. W. J., and de Boer, A. H. (2002). 14-3-3 Protein regulation of proton pumps and ion channels. Plant Mol. Biol. 50, 1041–1051. doi: 10.1023/a:1021231805697
Campos, J. F., Cara, B., Perez-Martin, F., Pineda, B., Egea, I., Flores, F. B., et al. (2016). The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation. Plant Biotechnol. J. 14, 1345–1356. doi: 10.1111/pbi.12498
Cao, H., Xu, Y., Yuan, L., Bian, Y., Wang, L., Zhen, S., et al. (2016). Molecular characterization of the 14-3-3 gene family in Brachypodium distachyon L. reveals high evolutionary conservation and diverse responses to abiotic stresses. Front. Plant Sci. 7:1099. doi: 10.3389/fpls.2016.01099
Chen, F., Li, Q., Sun, L. X., and He, Z. H. (2006). The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 13, 53–63. doi: 10.1093/dnares/ds1001
Chen, L., Hu, W., Tan, S., Wang, M., Ma, Z., Zhou, S., et al. (2012). Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS ONE 7:e46744. doi: 10.1371/journal.pone.0046744
Chevalier, D., Morris, E. R., and Walker, J. C. (2009). 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu. Rev. Plant Biol. 60, 67–91. doi: 10.1146/annurev.arplant.59.032607.092844
Choi, H.-I., Hong, J.-H., Ha, J.-O., Kang, J.-Y., and Kim, S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730.
Comparot, S., Lingiah, G., and Martin, T. (2003). Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. J. Exp. Bot. 54, 595–604. doi: 10.1093/jxb/erg057
Cotelle, V., and Leonhardt, N. (2016). 14-3-3 proteins in guard cell signaling. Front. Plant Sci. 6:1210. doi: 10.3389/fpis.2015.01210
de Boer, A. H., van Kleeff, P. J., and Gao, J. (2013). Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasma 250, 425–440. doi: 10.1007/s00709-012-0437-z
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
DeLille, J. M., Sehnke, P. C., and Ferl, R. J. (2001). The arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 126, 35–38. doi: 10.1104/pp.126.1.35
Deng, X., Hu, W., Wei, S. Y., Zhou, S. Y., Zhang, F., Han, J. P., et al. (2013a). TaCIPK29, a CBL-interacting protein kinase gene from wheat, confers salt stress tolerance in transgenic tobacco. PLoS ONE 8:e69881. doi: 10.1371/journal.pone.0069881
Deng, X., Zhou, S., Hu, W., Feng, J., Zhang, F., Chen, L., et al. (2013b). Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol. Plant. 149, 367–377. doi: 10.1111/ppl.12046
Denison, F. C., Paul, A. L., Zupanska, A. K., and Ferl, R. J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22, 720–727. doi: 10.1016/j.semcdb.2011.08.006
Diaz, C., Kusano, M., Sulpice, R., Araki, M., Redestig, H., Saito, K., et al. (2011). Determining novel functions of Arabidopsis 14-3-3 proteins in central metabolic processes. BMC Syst. Biol. 5:192. doi: 10.1186/1752-0509-5-192
Draper, J., Mur, L. A. J., Jenkins, G., Ghosh-Biswas, G. C., Bablak, P., Hasterok, R., et al. (2001). Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555. doi: 10.1104/pp.010196
Ferl, R. J. (2004). 14-3-3 proteins: regulation of signal-induced events. Physiol. Plant. 120, 173–178. doi: 10.1111/j.0031-9317.2004.0239.x
Fuglsang, A. T., Guo, Y., Cuin, T. A., Qiu, Q., Song, C., Kristiansen, K. A., et al. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19, 1617–1634. doi: 10.1105/tpc.105.035626
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Gao, J., van Kleeff, P. J., Oecking, C., Li, K. W., Erban, A., Kopka, J., et al. (2014). Light modulated activity of root alkaline/neutral invertase involves the interaction with 14-3-3 proteins. Plant J. 80, 785–796. doi: 10.1111/tpj.12677
Geilfus, C. M., Mithofer, A., Ludwig-Muller, J., Zorb, C., and Muehling, K. H. (2015). Chloride-inducible transient apoplastic alkalinizations induce stomata closure by controlling abscisic acid distribution between leaf apoplast and guard cells in salt-stressed Vicia faba. New Phytol. 208, 803–816. doi: 10.1111/nph.13507
Gokirmak, T., Paul, A. L., and Ferl, R. J. (2010). Plant phosphopeptide-binding proteins as signaling mediators. Curr. Opin. Plant Biol. 13, 527–532. doi: 10.1016/j.pbi.2010.06.001
Grondin, A., Rodrigues, O., Verdoucq, L., Merlot, S., Leonhardt, N., and Maurel, C. (2015). Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 27, 1945–1954. doi: 10.1105/tpc.15.00421
He, Y., Wu, J., Lv, B., Li, J., Gao, Z., Xu, W., et al. (2015). Involvement of 14-3-3 protein GRF9 in root growth and response under polyethylene glycol-induced water stress. J. Exp. Bot. 66, 2271–2281. doi: 10.1093/jxb/erv149
Hossain, M. S., and Dietz, K. J. (2016). Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front. Plant Sci. 7, 15. doi: 10.3389/fpls.2016.00548
Hu, W., Yan, Y., Hou, X., He, Y., Wei, Y., Yang, G., et al. (2015). TaPP2C1, a group F2 protein phosphatase 2C gene, confers resistance to salt stress in transgenic tobacco. PLoS ONE 10:e0129589. doi: 10.1371/journal.pone.0129589
Hu, W., Yuan, Q., Wang, Y., Cai, R., Deng, X., Wang, J., et al. (2012). Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol. 53, 2127–2141. doi: 10.1093/pcp/pcs154
Huang, Q., Wang, Y., Li, B., Chang, J., Chen, M., Li, K., et al. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15, 268. doi: 10.1186/s12870-015-0644-9
Ishida, S., Fukazawa, J., Yuasa, T., and Takahashi, Y. (2004). Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell 16, 2641–2651. doi: 10.1105/tpc.104.024604
Kerkeb, L., Venema, K., Donaire, J. P., and Rodriguez-Rosales, M. P. (2002). Enhanced H+/ATP coupling ratio of H+-ATPase and increased 14-3-3 protein content in plasma membrane of tomato cells upon osmotic shock. Physiol. Plant. 116, 37–41. doi: 10.1034/j.1399-3054.2002.1160105.x
Kim, T. H., Bohmer, M., Hu, H., Nishimura, N., and Schroeder, J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61, 561–591. doi: 10.1146/annurev-arplant-042809-112226
Li, H., Liu, D., He, H., Zhang, N., Ge, F., and Chen, C. (2014). Molecular cloning of a 14-3-3 protein gene from Lilium regale Wilson and overexpression of this gene in tobacco increased resistance to pathogenic fungi. Sci. Hortic. 168, 9–16. doi: 10.1016/j.scienta.2013.12.034
Li, H. T., Liu, H., Gao, X. S., and Zhang, H. (2009). Knock-out of Arabidopsis AtNHX4 gene enhances tolerance to salt stress. Biochem. Biophys. Res. Commun. 382, 637–641. doi: 10.1016/j.bbrc.2009.03.091
Liu, Y., and He, C. (2016). Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep. 35, 995–1007. doi: 10.1007/s00299-016-1950-x
Lukaszewicz, M., Matysiak-Kata, I., Aksamit, A., Oszmianski, J., and Szopa, J. (2002). 14-3-3 protein regulation of the antioxidant capacity of transgenic potato tubers. Plant Sci. 163, 125–130. doi: 10.1016/s0168-9452(02)00081-x
Lv, D., Subburaj, S., Cao, M., Yan, X., Li, X., Appel, R., et al. (2014). Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol. Cell. Proteomics 13, 632–652. doi: 10.1074/mcp.M113.030171
Msanne, J., Lin, J., Stone, J. M., and Awada, T. (2011). Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234, 97–107. doi: 10.1007/s00425-011-1387-y
Obsil, T., and Obsilova, V. (2011). Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 22, 663–672. doi: 10.1016/j.semcdb.2011.09.001
Pallucca, R., Visconti, S., Camoni, L., Cesareni, G., Melino, S., Panni, S., et al. (2014). Specificity of epsilon and non-epsilon isoforms of Arabidopsis 14-3-3 proteins towards the H+-ATPase and other targets. PLoS ONE 9:e90764. doi: 10.1371/journal.pone.0090764
Rao, D. E., and Chaitanya, K. V. (2016). Photosynthesis and antioxidative defense mechanisms in deciphering drought stress tolerance of crop plants. Biol. Plant. 60, 201–218. doi: 10.1007/s10535-016-0584-8
Roy, S. J., Negrao, S., and Tester, M. (2014). Salt resistant crop plants. Curr. Opin. Biotechnol. 26, 115–124. doi: 10.1016/j.copbio.2013.12.004
Schoonheim, P. J., Sinnige, M. P., Casaretto, J. A., Veiga, H., Bunney, T. D., Quatrano, R. S., et al. (2007). 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J. 49, 289–301. doi: 10.1111/j.1365-313X.2006.02955.x
Sehnke, P. C., DeLille, J. M., and Ferl, R. J. (2002). Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell 14, S339–S354. doi: 10.1105/tpc.010430
Sirichandra, C., Davanture, M., Turk, B. E., Zivy, M., Valot, B., Leung, J., et al. (2010). The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS ONE 5:e13935. doi: 10.1371/journal.pone.0013935
Stepien, P., and Klobus, G. (2005). Antioxidant defense in the leaves of C3 and C4 plants under salinity stress. Physiol. Plant. 125, 31–40. doi: 10.1111/j.1399-3054.2005.00534.x
Sullivan, S., Thomson, C. E., Kaiserli, E., and Christie, J. M. (2009). Interaction specificity of Arabidopsis 14-3-3 proteins with phototropin receptor kinases. FEBS Lett. 583, 2187–2193. doi: 10.1016/j.febslet.2009.06.011
Sun, T., Wang, Y., Wang, M., Li, T., Zhou, Y., Wang, X., et al. (2015). Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 15, 269. doi: 10.1186/s12870-015-0657-4
Sun, X., Luo, X., Sun, M., Chen, C., Ding, X., Wang, X., et al. (2014). A Glycine soja 14-3-3 protein GsGF14o participates in stomatal and root hair development and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 55, 99–118. doi: 10.1093/pcp/pct161
Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Teper, D., Salomon, D., Sunitha, S., Kim, J. G., Mudgett, M. B., and Sessa, G. (2014). Xanthomonas euvesicatoria type III effector XopQ interacts with tomato and pepper 14-3-3 isoforms to suppress effector-triggered immunity. Plant J. 77, 297–309. doi: 10.1111/tpj.12391
Tzivion, G., Shen, Y. H., and Zhu, J. (2001). 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 20, 6331–6338. doi: 10.1038/sj.onc.1204777
Wang, H., Yang, C., Zhang, C., Wang, N., Lu, D., Wang, J., et al. (2011). Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21, 825–834. doi: 10.1016/j.devcel.2011.08.018
Wang, R. S., Pandey, S., Li, S., Gookin, T. E., Zhao, Z., Albert, R., et al. (2011). Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12:216. doi: 10.1186/1471-2164-12-216
Wang, X., Zeng, J., Li, Y., Rong, X., Sun, J., Sun, T., et al. (2015). Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 6:615. doi: 10.3389/fpls.2015.00615
Wurtele, M., Jelich-Ottmann, C., Wittinghofer, A., and Oecking, C. (2003). Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 22, 987–994. doi: 10.1093/emboj/cdg104
Yan, J., He, C., Wang, J., Mao, Z., Holaday, S. A., Allen, R. D., et al. (2004). Overexpression of the Arabidopsis 14-3-3 protein GF14λ in cotton leads to a “stay-green” phenotype and improves stress tolerance under moderate drought conditions. Plant Cell Physiol. 45, 1007–1014.
Yang, D.-Y., Ma, N.-N., Zhuang, K.-Y., Zhu, S.-B., Liu, Z.-M., and Yang, X.-H. (2017). Overexpression of tomato SlGGP-LIKE gene improves tobacco tolerance to methyl viologen-mediated oxidative stress. J. Plant Physiol. 209, 31–41. doi: 10.1016/j.jplph.2016.10.013
Yao, Y., Du, Y., Jiang, L., and Liu, J. Y. (2007). Molecular analysis and expression patterns of the 14-3-3 gene family from Oryza sativa. J. Biochem. Mol. Biol. 40, 349–357.
Zhang, J. L., and Shi, H. (2013). Physiological and molecular mechanisms of plant salt tolerance. Photosynth. Res. 115, 1–22. doi: 10.1007/s11120-013-9813-6
Zhang, X., Zhang, Z., Chen, J., Chen, Q., Wang, X. C., and Huang, R. (2005). Expressing TERF1 in tobacco enhances drought tolerance and abscisic acid sensitivity during seedling development. Planta 222, 494–501. doi: 10.1007/s00425-005-1564-y
Zhou, H., Lin, H., Chen, S., Becker, K., Yang, Y., Zhao, J., et al. (2014). Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26, 1166–1182. doi: 10.1105/tpc.113.117069
Keywords: Bd14-3-3s, BdGF14d, salt tolerance, ABA signaling, ROS-scavenging system, B. distachyon
Citation: He Y, Zhang Y, Chen L, Wu C, Luo Q, Zhang F, Wei Q, Li K, Chang J, Yang G and He G (2017) A Member of the 14-3-3 Gene Family in Brachypodium distachyon, BdGF14d, Confers Salt Tolerance in Transgenic Tobacco Plants. Front. Plant Sci. 8:340. doi: 10.3389/fpls.2017.00340
Received: 22 November 2016; Accepted: 27 February 2017;
Published: 13 March 2017.
Edited by:
Zhulong Chan, Huazhong Agricultural University, ChinaReviewed by:
Asish Kumar Parida, CSIR-Central Salt and Marine Chemicals Research Institute, IndiaXinbo Chen, Hunan Agricultural University, China
Copyright © 2017 He, Zhang, Chen, Wu, Luo, Zhang, Wei, Li, Chang, Yang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junli Chang, cjl@hust.edu.cn Guangxiao Yang, ygx@hust.edu.cn Guangyuan He, hegy@hust.edu.cn
†These authors have contributed equally to this work.
 Yuan He†
Yuan He† Guangxiao Yang
Guangxiao Yang Guangyuan He
Guangyuan He