- National Center for Soybean Improvement, Key Laboratory of Biology and Genetics and Breeding for Soybean, Ministry of Agriculture, State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China
Phytophthora root rot (PRR), caused by Phytophthora sojae, has negative effects on soybean yield in China and can be controlled by identifying germplasm resources with resistance genes. In this study, the resistance locus RpsHN in the soybean line Meng8206 was mapped using two mapping populations. Initial mapping was realized using two recombinant inbred line (RIL) populations and included 103 F6:8 RILs derived from a cross of Meng8206 × Linhedafenqing, including 2600 bin markers, and 130 F6:8 RILs derived from a cross of Meng8206 × Zhengyang148, including 2267 bin markers. Subsequently, a 159 F2:3 secondary population derived from a cross of Meng8206 × Linmeng6-46, were used to fine map this locus using SSR markers. Finally, the resistance locus from Meng8206 was fine mapped to a 278.7 kb genomic region flanked by SSR markers SSRSOYN-25 and SSRSOYN-44 at a genetic distance of 1.6 and 1.0 cM on chromosome 3 (Chr. 03). Real-time RT-PCR analysis of the possible candidate genes showed that three genes (Glyma.03g04260, Glyma.03g04300, and Glyma.03g04340) are likely involved in PRR resistance. These results will serve as a basis for cloning, transferring of resistant genes and breeding of P. sojae-resistant soybean cultivars through marker-assisted selection.
Introduction
Phytophthora root rot (PRR), caused by the soil-borne oomycete pathogen Phytophthora sojae, is the second highest yield-suppressing disease (Schmitthenner, 1985; Wrather and Koenning, 2009). This disease has resulted in significant economic losses worldwide (Tyler, 2007). In Heilongjiang Province of China, P. sojae is widespread but unevenly distributed. It has been estimated that more than 150,000 ha of the soybean grown in fields become infected annually (Tian et al., 2016).
Cultivating Phytophthora-resistant soybean cultivars can reduce the incidence of PRR. There are two types of resistant cultivars: race-specific resistance and partial resistance (Sugimoto et al., 2012). Partial resistance is controlled by multiple genes and exhibits broad-spectrum and durable resistance to a range of pathogen species (Kou and Wang, 2010). However, in plants, partial resistance may be lost under high disease pressure to P. sojae (Dorrance, 2003). One of the most effective methods to control plant diseases is the development of varieties with vertical resistance genes (Flor, 1971).
To our knowledge, 22 race-specific resistance soybean cultivars containing single genes have been identified and reported: L88-8470 (Rps1a), L77-1863 (Rps1b), L75-3735 (Rps1c), L93-3312 (Rps1d), L77-1794 (Rps1k), L76-1988 (Rps2), L83-570 (Rps3a), L91-8347 (Rps3b), L92-7857 (Rps3c), L85-2352 (Rps4), L85-3059 (Rps5), L89-1581 (Rps6), L93-3258 (Rps7), PI 399073 (Rps8), Ludou4 (Rps9), Wandou15 (Rps10), PI 594527 (Rps11), PI 567139B (RpsUN1 and RpsUN2), Yudou25 (RpsYu25), Yudou29 (RpsYD29), Waseshiroge (unnamed Rps gene), and Nannong 10-1 (RpsJS) (Sugimoto et al., 2011, 2012; Sun et al., 2011, 2014; Wu et al., 2011; Zhang et al., 2013a,b; Ping et al., 2015; Li et al., 2016). Twenty-three genes/alleles identified in the soybean cultivars listed above were located on six chromosomes. The genes/alleles Rps1 (including five alleles Rps1-a, Rps1-b, Rps1-c, Rps1-d, and Rps1-k), Rps7, Rps9, RpsUN1, RpsYu25 and an unnamed Rps gene (Rps1?) on chromosome 3, Rps3 (including three alleles Rps3a, Rps3b, Rps3c) and Rps8 on chromosome 13, Rps2 and RpsUN2 on chromosome 16, and Rps4, Rps5 and Rps6 on chromosome 18 were detected by linkage analysis and genetic mapping (Kilen et al., 1974; Mueller et al., 1978; Athow et al., 1980; Ploper et al., 1985; Buzzell and Anderson, 1992; Cregan et al., 1999; Gordon et al., 2006; Sugimoto et al., 2011; Sun et al., 2011; Wu et al., 2011). RpsYD29 was mapped to a 204.8-kb region on chromosome 3, and two nucleotide-binding site and leucine-rich repeat (NBS-LRR) type genes Glyma03g04030.1 and Glyma03g04080.1 were identified (Zhang et al., 2013b). Rps1k has an NBS-LRR structure that is typical of resistance proteins. However, the physical location of Rps1k is unknown in the reference genome of ‘Williams 82’ (Gao et al., 2005; Gao and Bhattacharyya, 2008). RpsJS, a fine mapping gene located in a 138.9-kb region with 14 candidate genes on chromosome 18, and three genes Glyma18g51930, Glyma18g51950 and Glyma18g51960 were characterized as NBS-LRR type genes (Sun et al., 2014). Rps11 mapped to a 225.3-kb region on chromosome 7, and Rps10 mapped to a 311-kb region on chromosome 17 (Zhang et al., 2013a; Ping et al., 2015). Furthermore, the Rps10 mapping region contained two candidate genes, Glyma17g28950.1 and Glyma17g28970.1, annotated as serine/threonine (Ser/Thr) protein kinases.
Another measure for PRR resistance is pyramid-breeding. Pyramiding resistance genes may increase the resistance of soybean cultivars to many pathogen races, and pyramiding genes could be rapidly achieved using molecular markers (Lohnes and Schmitthenner, 1997). Based on the methods of controlling PRR, the identification of a novel Rps gene in soybean cultivars is needed to study resistance, and the development of new molecular markers is needed for marker-assisted selection (MAS).
The germplasm Meng8206 (ZDD11436) is a soybean line developed from Yangtze-Huai region of China, studied in drought-tolerance and cyst nematode-tolerance (Duan et al., 2008; Wang et al., 2015). The objectives of the present study were to analyze the inheritance of Meng8206 resistance, identify resistance loci and manipulate predicted candidate genes.
Materials and Methods
Plant Materials and P. Sojae Isolates
Two F6:8 recombinant inbred line (RIL) populations were used for initial mapping: 103 RILs and 130 RILs were constructed from a cross between Meng8206 × Linhedafenqing and Meng8206 × Zhengyang148, respectively. An F2:3 secondary population was used for fine mapping: 159 lines were constructed from a cross between Meng8206 × Linmeng6-46 (Supplementary Figure 1). The soybean lines Meng8206, Linhedafenqing, Zhengyang148 and Linmeng6-46 were obtained from National Center for Soybean Improvement, Nanjing Agricultural University, Nanjing, China. Meng8206 was also obtained from the Chinese National Soybean GeneBank (CNSGB).
To clarify the response type of Meng8206 to P. sojae, 15 differentials were used, and each cultivar had an independent Rps gene. The 15 differential cultivars included Harlon (Rps1a), Harosoy13XY (Rps1b), Williams79 (Rps1c), PI103091 (Rps1d), Williams82 (Rps1k), L76-1988 (Rps2), Chapman (Rps3a), PRXI46-36 (Rps3b), PRXI 45-48 (Rps3c), L85-2352 (Rps4), L85-3059 (Rps5), Harosoy62XY (Rps6), Harosoy (Rps7), Yudou25 (RpsYu25), and LuDou4 (Rps9). In addition, Williams (no known Rps gene) was a susceptible variety used as an inoculation reference.
P. sojae Isolates and Disease Evaluation
A total of eight P. sojae isolates (Supplementary Table 1) with different virulence capabilities were provided by Professor Yuanchao Wang of Nanjing Agricultural University and maintained on V8 juice agar medium (10% V8 vegetable juice, 0.02% CaCO3 and 1.0% Bacto-agar). These isolates were used to evaluate the resistance identified among the parents and 15 differential cultivars. P. sojae HeN08-35 (virulence formula is 3a, 3c, 4, 5, 6 and 7) was used to evaluate two mapping populations.
A modified hypocotyl inoculation technique was utilized for disease evaluation in this experiment (Sun et al., 2011, 2014). All materials were planted in plastic pots containing vermiculite; the mycelia from 7-day-old seedlings were maintained on V8 juice agar and subsequently inoculated onto wounded hypocotyls. After inoculation, the seedlings were placed in a high humidity mist chamber for 48 h and subsequently transferred to a greenhouse at 25°C with a 14-h light/10-h dark photoperiod for 5 days. Two F6:8 RILs and F2:3 family reactions were evaluated at 5 days post-inoculation (DPI) and recorded as the percentage of dead seedlings. Each family had 30 plants scored. The standard criterion of each family is as follows: if the percentage of dead seedlings >80%, then this family was recorded as homozygous susceptible (S); if the percentage of dead seedlings <20%, then this family was recorded as homozygous resistant (R); and if the percentage of dead seedlings is between 21 and 79%, then this family was recorded as heterozygous resistant (Rs) (Gordon et al., 2006; Ping et al., 2015).
SNP Genotyping and Bin Map Construction
The genomic DNA was extracted from the young leaves of two RIL populations according to Zhang and Wang (2004) and used to construct the genomic DNA library after Taq I digestion according to Baird et al. (2008). The 400- to 700-bp DNA fragments were sequenced using the Illumina HiSeq 2000 standard protocol for MSG (multiplexed shotgun genotyping), and 90-mer paired-end reads were generated (Andolfatto et al., 2011). SOAP2 (Li et al., 2009) software was used for aligning the sequenced reads to the Williams 82 reference genome. SNP calling and genotyping were conducted using RealSFS software (Yi et al., 2010) based on the Bayesian estimation. Subsequently, using a three-standard filter, 50 < depth < 2500, a probability of site mutation ≥95%, and every SNP loci separated by at least 5 bp, we obtained high confidence SNPs.
Bin maps were constructed using a sliding window approach. The sliding window contained 15 SNPs. As the window slides, the genotypes are called and recombination breakpoints are determined. The same genotype across the entire RIL population was recognized as a single recombination bin (Huang et al., 2009).
DNA Preparation of F2 Individuals and Pooling for Bulk Segregation Analysis
Plant genomic DNA was extracted from young leaves using the CTAB method with minor modifications (Allen et al., 2006). Resistant and susceptible bulks for the bulk segregation analysis (BSA) were, respectively, formed using the plant genomic DNA of 10 resistant and 10 susceptible F2 individuals (Michelmore et al., 1991), and the DNA concentration for the two bulks was greater than 50 ng/μl.
SSR Marker Development and PCR
According to the initial mapping physical position, the sequence was downloaded from Phytozome Glyma1.01, and simple repeat sequences were assessed using SSR Hunter 1.32. New SSR markers were designed using Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, USA). In addition, some markers from Soybase3 and the published paper (Zhang et al., 2013b) were used. PCR was conducted according to Sun et al. (2014).
Data Analysis and Genetic Linkage Analysis
A goodness-of-fit to the Mendelian segregation ratio was calculated using Chi-square (χ2) analysis to examine the segregation patterns of the phenotypes and selected SSR markers. The resistance locus for initial mapping was detected using composite interval mapping (CIM) in QTL Cartographer 2.5(threshold value 2.5) (Wang et al., 2012). The linkage map of RpsHN for fine mapping was constructed using Joinmap 4.0 linkage analysis software (van Ooijen, 2006). The linkage groups were analyzed with a log-likelihood (LOD) threshold of 3.0.
Expression Analysis of Candidate Genes
Meng8206 (R) and LinMeng6-46 (S) seedlings were cultivated for 7 days and subsequently inoculated with isolate HeN08-35. Approximately 1-cm samples of the treated hypocotyl tissues were collected at five time points. Total RNA was extracted from the plants using the RNA Simple Total RNA kit (TIANGEN, China). cDNA was synthesized using the Prime ScriptTM RT Reagent Kit (TaKaRa, Japan) using a standard protocol. The experiment was repeated three times.
The CDS sequences for the candidate genes were obtained from Phytozome4. The primers for qRT-PCR were designed using Primer Premier 5.0. In addition, the housekeeping gene Actin was used as a control. These primers are shown in Supplementary Table 2, and qRT-PCR was conducted using a Light Cycler 480 instrument.
Results
Phenotype Reaction of the Parents to P. sojae Isolates
The 8 different isolates were applied to evaluate 4 parents and 15 differentials. The results showed that Meng8206 was resistant to the HeN08-35 isolate and was susceptible to the other 7 isolates (Supplementary Table 3). The phenotype reaction of Meng8206 was different for each of the 15 differentials, conferred through an independent Rps gene. The three parents Zhengyang148, Linhedafenqing and Linmeng6-46 were susceptible to all selected P. sojae isolates, and their phenotype reactions were the same as Williams. In addition, we also analyzed the genetic diversity and phenotypic relationships of the four parents and 15 differentials to 8 P. sojae using cluster tree analysis in the NTSYS program (Figure 1). When the coefficient was more than 0.8, Meng8206 existed independently as a subgroup, suggesting that Meng8206 may possess a novel Rps gene.
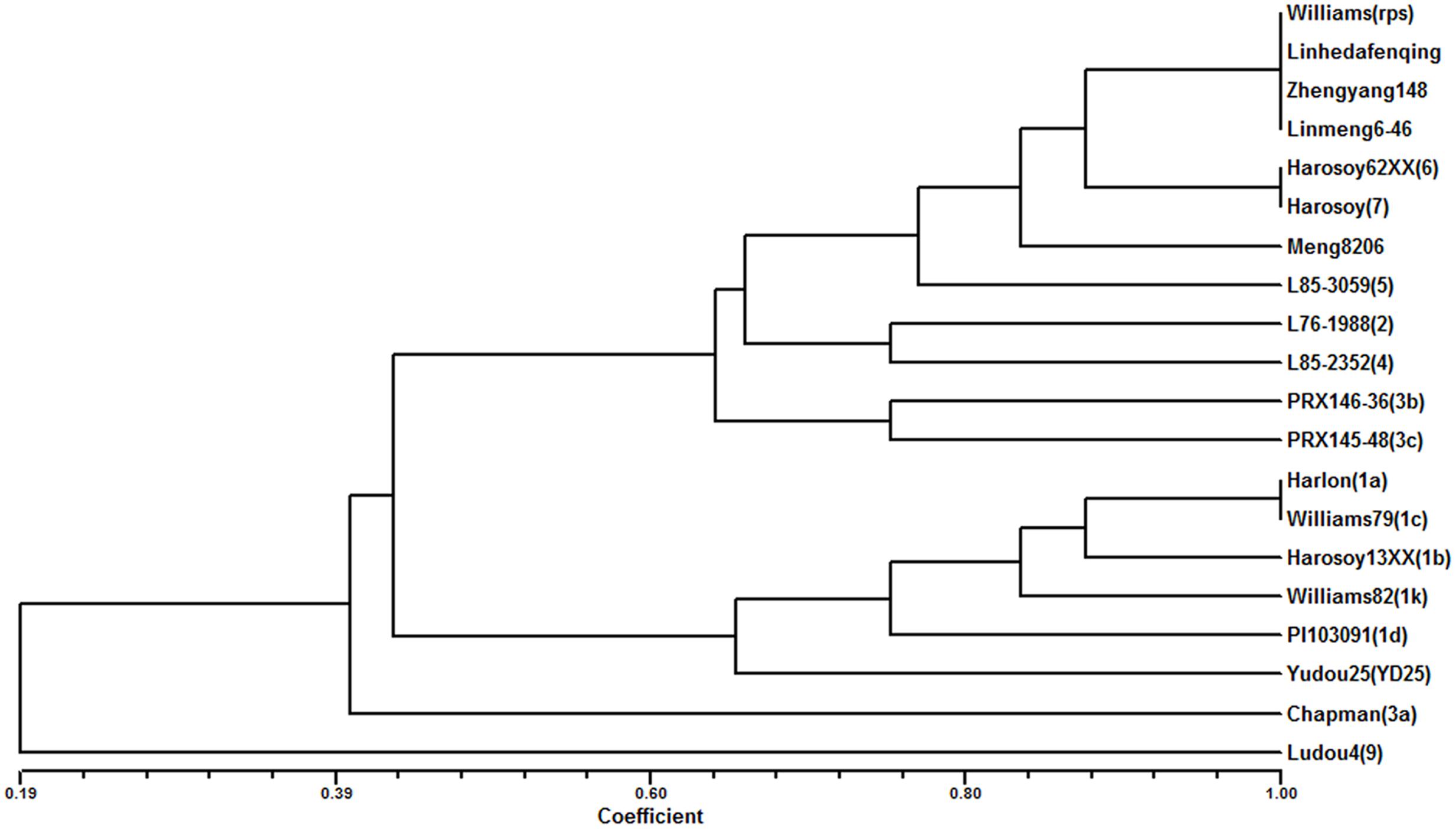
FIGURE 1. Dendrogram reflected by UPGMA cluster analysis of the reaction to 8 P. sojae among Meng8206, Zhengyang148, Linhedafenqing, and LinMeng6-46, 15 differentials each had an independent Rps gene: Harlon (Rps1a), Harosoy13XX (Rps1b), Williams79 (Rps1c), PI103091 (Rps1d), Williams82 (Rps1k), L76-988 (Rps2), Chapman (Rps3a), PRX146-36 (Rps3b), PRX145-48 (Rps3c), L85-2352 (Rps4), L85-3059 (Rps5), Harosoy62XX (Rps6), Harosoy (Rps7), YuDou25 (RpsYu25), LuDou4 (Rps9) and the susceptible cultivar Williams.
Inheritance of Resistance to P. sojae HeN08-35
In initial mapping populations, among the 103 F6:8 RILs derived from a cross of Meng8206 × Linhedafenqing, 48 RILs were homozygous resistant (R), 55 RILs were homozygous susceptible (S) to P. sojae isolate HeN08-35, and the actual segregation ratio was consistent with expected ratio 1:1 (χ2 = 0.49 and p = 0.49) (Table 1). This result suggested that Meng8206 resistance was controlled by a single gene. Thus, this locus was temporarily designated RpsHN. Among the 130 F6:8 RILs derived from a cross of Meng8206 × Zhengyang148, 44 RILs were homozygous resistant (R), and 86 RILs were homozygous susceptible (S) to P. sojae HeN08-35. The phenotype data showed no clearly inheritance mechanisms of quality traits. Thus, it was decided to perform QTL analysis for mapping of PRR resistance in this population.
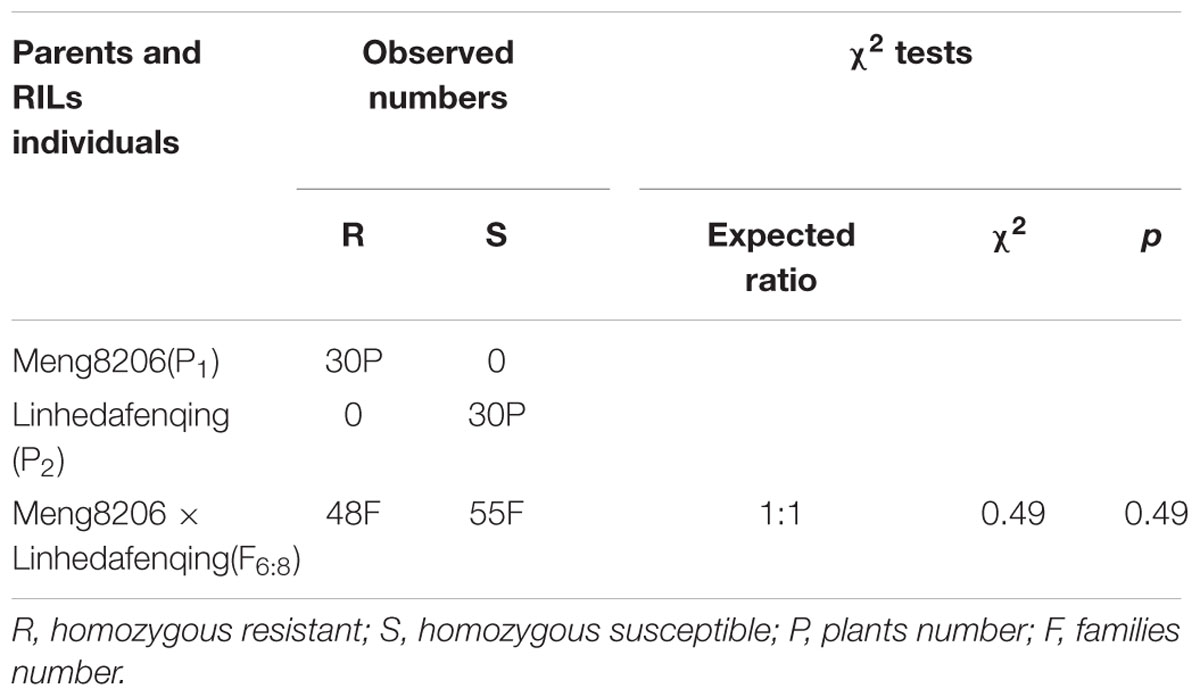
TABLE 1. Segregation analysis of resistance to P. sojae HeN08-35 in 103 RILs of Meng8206 × Linhedafenqing.
In the fine mapping population, among the 159 F2:3 individuals derived from a cross of Meng8206 × Linmeng6-46, 38 were homozygous resistant (R), 69 were segregating individuals Rs, 52 were homozygous susceptible (S) to P. sojae HeN 08-35, and the actual segregation ratio was consistent with the expected ratio 1:2:1 (χ2 = 4.77, p = 0.09) (Table 2). This result suggested that Meng8206 resistance was controlled by a single dominant gene RpsHN.

TABLE 2. Segregation analysis of resistance to P. sojae isolates HeN08-35 in 159 F2:3 families of Meng8206 × Linmeng6-46.
Initial Mapping of the RpsHN Gene
To analyze the resistance locus of Meng8206 in the two RIL populations, a bins map was constructed using a sliding window approach. A total of 2600 bins were identified in the RIL population derived from a cross of Meng8206 × Linhedafenqing; the genetic distance was 2626.00 cM, and the average genetic distance between markers was 1.01 cM. A total of 2267 bins were identified in the RIL population derived from a cross of Meng8206 × Zhengyang148; the genetic distance was 2584.38 cM, and the mean genetic distance between markers was 1.14 cM.
For the Meng8206 × Linhedafenqing RIL population, a resistance locus was detected on Chr03 with an LOD score of 56.89 using CIM. This locus was located between marker bin249 and bin250, at nucleotide positions 3,515,595 and 4,237,477, respectively, determined through a BLAST search in Glyma1.0. The additive effect of this locus was 0.51 and resistant allelic effect came from Meng8206 (Supplementary Figures 2A,B and Table 3). Interestingly, only a significant resistance locus with an LOD score of 22.66 was also identified on Chr. 03 in the Meng8206 × Zhengyang148 RILs population, and this locus was located between marker bin282 and bin283, at nucleotide positions 3,564,629 and 4,734,455, respectively. The additive effect of this locus was 0.34 and resistant allelic effect came from Meng8206 (Supplementary Figures 2C,D and Table 3). Based on the physical location of the marker, an intersection was detected in the two RILs populations. Because Meng8206 was the same resistance parent in the two RIL populations, the two loci was the same locus.
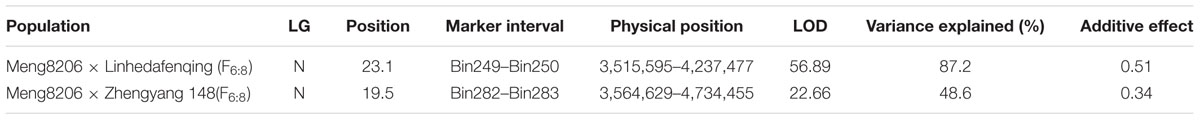
TABLE 3. QTL mapping of RpsHN in the F6:8 RILs of Meng8206 × Linhedafenqing and Meng8206 × Zhengyang148.
Fine Mapping of the RpsHN Locus
We conducted fine mapping on the region between markers bin249 and bin283. The physical distance of bin249 and bin283 is approximately 1218.8 kb (Figures 2B,C). In this region, 33 and 6 SSR markers were selected according to Song et al. (2010) and Zhang et al. (2013b), respectively. The two markers satt009 and satt1k2a showed polymorphisms between Meng8206 and Linmeng6-46 using the BSA method.
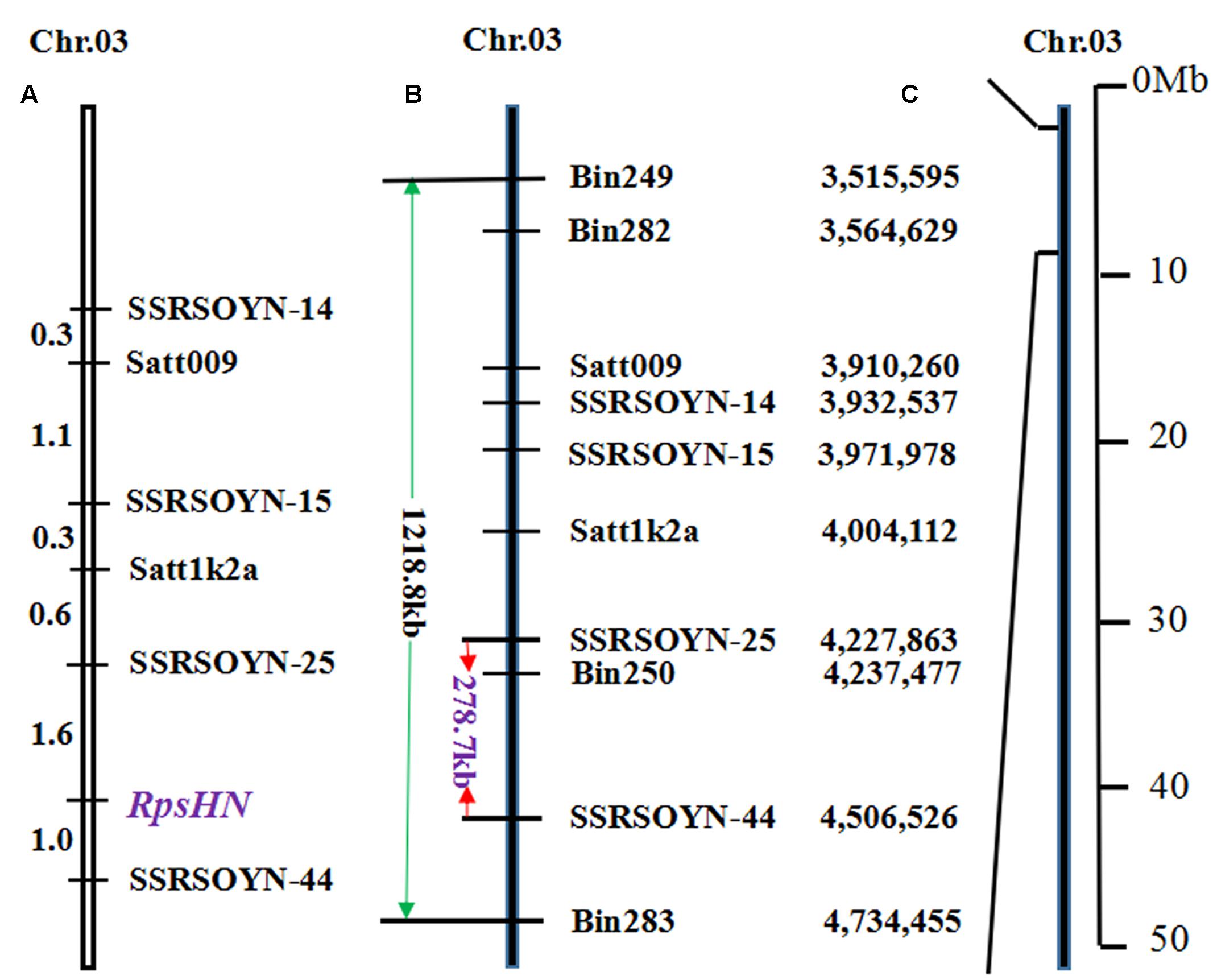
FIGURE 2. Genetic linkage map and physical map of RpsHN on chromosome 3. Genetic distance (cM) is shown on the left, and markers are shown on the right. (A) Linkage map of RpsHN from the present study. (B) Physical map of SSR and bins markers from the present study. The green line refers to the physical distance between bin249 and bin283, and the red line refers to the fine mapping region. (C) Physical position of the mapped region of RpsHN on chromosome 3 (Lin et al., 2013).
In addition, a total of 183 repeat motifs (SSR loci) in this region were identified using SSR hunter and used for the fine mapping of RpsHN. Four SSR markers, SSRSOYN-14, SSRSOYN-15, SSRSOYN-25 and SSRSOYN-44, showed polymorphisms between Meng8206 and Linmeng6-46 using the BSA method (Supplementary Table 4). Together with satt009 and satt1k2a, six polymorphic marker segregation patterns were revealed by analyzing 159 F2:3 families, consistent with the 1:2:1 ratio (Supplementary Table 5).
A genetic map, including six SSR markers and RpsHN was constructed, and RpsHN was closely linked to the SSR markers SSRSOYN-25 and SSRSOYN-44 at genetic distances of 1.6 and 1.0 cM, respectively (Figure 2A).
Candidate Gene Prediction
The genomic region of Williams 82 was delimited by the markers SSRSOYN-25 and SSRSOYN-44. A BLAST search showed that the physical distance of SSRSOYN-25 and SSRSOYN-44 are at nucleotide position 4,227,863 and 4,506,526 in Glyma1.0, is appropriately 278.7 kb (Figure 2B). A total of eight genes were annotated according to the Glyma 1.0 (Supplementary Table 6). Among these genes, Glyma.03g04260 and Glyma.03g04300 encoded NB-ARC domain-containing disease resistance protein. Glyma.03g04340 encodes serine/threonine protein kinase (STK), which is involved in plant disease resistance. These genes were predicted as possible candidate genes.
To confirm whether Glyma.03g04260, Glyma.03g04300 and Glyma.03g04340 were induced under the treatment of P. sojae, the expression patterns of three genes were examined using qRT-PCR analysis in Meng8206 and Linmeng6-46. As shown in Figure 3, compared with the control (0 h), the expression of Glyma.03g04260 was down-regulated at 12, 36, and 48 h after treatment in the resistant line Meng8206 and the susceptible line Linmeng6-46, and the expression of Glyma.03g04300 did not significantly change in Linmeng6-46 and was up-regulated in Meng8206 at 12, 36, and 48 h after treatment. Glyma.03g04340, compared with Glyma.03g04300, had opposite expression levels. These results showed that three genes were induced by P. sojae HeN08-35. Thus, Glyma.03g04260, Glyma.03g04300 and Glyma.03g04340 were considered as potential candidate genes.
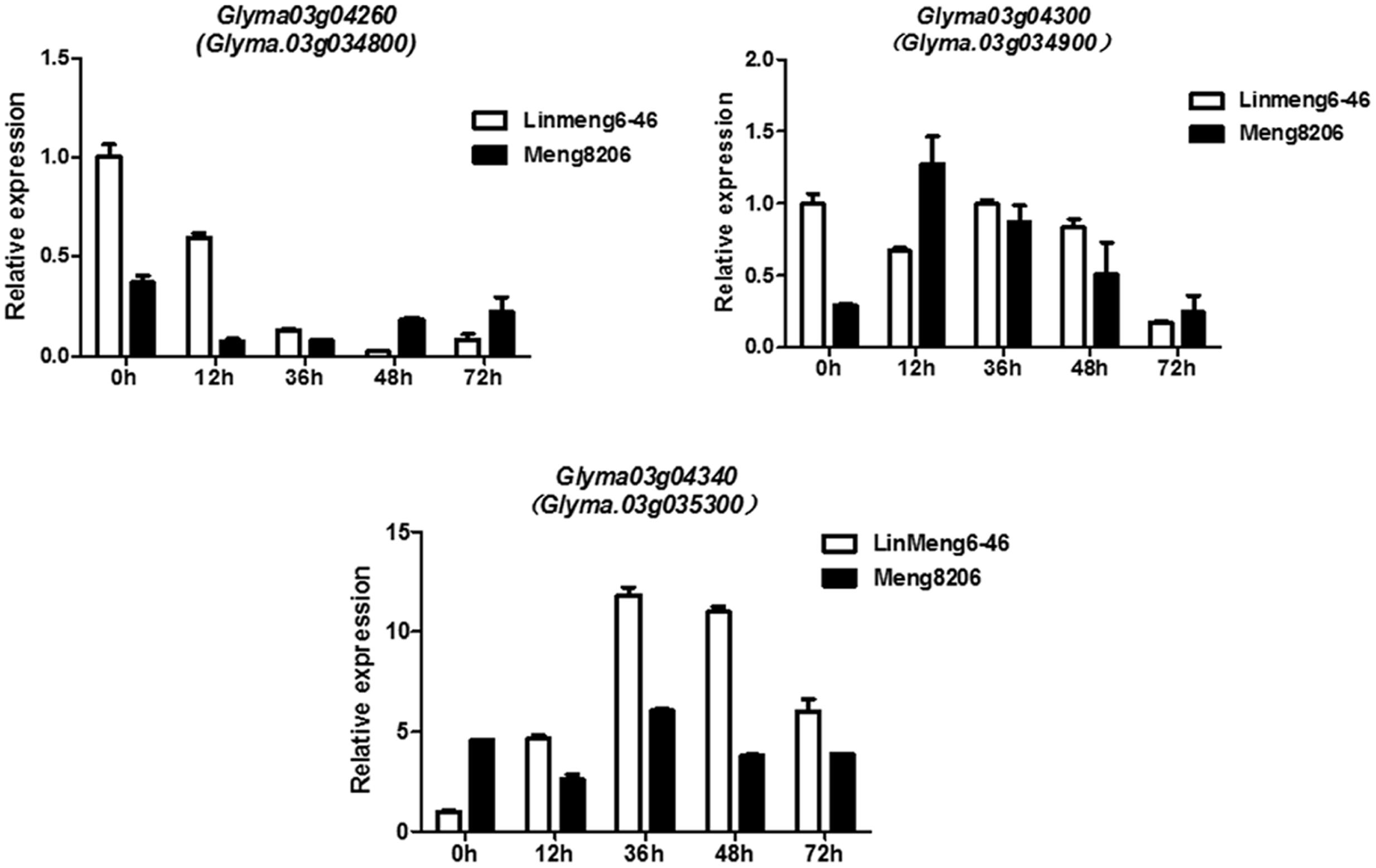
FIGURE 3. Relative expression levels of Glyma.03g04260 (Glyma.03g034800), Glyma.03g04300 (Glyma.03g034900) and Glyma.03g04340 (Glyma.03g035300) in Linmeng6-46 and Meng8206. Seven-day-old soybean seedlings were inoculated with isolate HeN08-35. The sampling times were 0, 12, 36, 48, and 72 hours post-inoculation (hpi).
Discussion
Soybean [Glycine max (L.) Merr.] is one of the most important oil crops in China. Many cultivars/lines have been studied for resistance to P. sojae and the identification of resistance loci (Zhang et al., 2014; Huang et al., 2016). In the present study, we identified the loci of the Meng8206 for resistance to P. sojae HeN08-35 using two mapping populations. Based on the phenotype reaction types of Meng8206 and the physical position of RpsHN on chromosome 3, we inferred that RpsHN is a novel gene tightly linked to Rps1 or a new allele at the Rps1 locus. Three genes, Glyma.03g04260, Glyma.03g04300 and Glyma.03g04340, were considered as potential candidate genes.
Soybean line Meng8206 was evaluated using eight isolates with different virulence formulas in this study. The results showed that Meng8206 was resistant to the HeN08-35 isolate and was susceptible to the other seven isolates. The parent Meng8206 showed resistance to HeN08-35 isolate as shown in Supplementary Figure 3. We proposed Meng8206 contains at least one novel locus resistant to HeN08-35. Fortunately, we used the F6:8 RIL populations derived from a cross of Meng8206 × Linhedafenqing and Meng8206 × Zhengyang148 to map the locus, and found that only a single resistance locus was detected on Chr03. Subsequently, this locus was verified by secondary populations derived from a cross of Meng8206 × Linmeng6-46. To F6:8 RIL population (Meng8206 × Zhengyang148), only a significant resistance locus was detected between marker bin282 and bin283 which fitted the expected 1:1 segregation ratio by χ2 test. Segregation distortion of phenotype may be due to the variation in genetic background of the progenies.
Previous studies have shown that 11 Rps genes were mapped to the N group, Rps1 allele genes (including five alleles Rps1a, Rps1b, Rps1c, Rps1d, and Rps1k), Rps7, Rps9, Rps1?, RpsUN1 and RpsYD29. RpsHN clustered in a subgroup different from the Rps1 alleles, RpsYu25 and Rps9 subgroups. Rps7 was located above marker satt009 (Bernard and Cremeens, 1981; Wu et al., 2011), while RpsHN was located below marker satt009; thus, the RpsHN locus is different from Rps7. The Rps gene in Waseshiroge (Sugimoto et al., 2011) was located “below” Satt009 and was flanked by Satt009 (0.9 cM), while RpsHN was located “below” Satt009 (3.6 cM) and was located “above” SSRSOYN-44 and was flanked by SSRSOYN-44(1.0 cM). So the Rps gene from Waseshiroge may be located close to Satt009 (nucleotide position 3,919,203), and RpsHN located close to SSRSOYN-44 (nucleotide position 4,506,526) (Figure 2A). Because Willimas82 was acted as reference sequence, RpsHN may be different from the Rps gene in Waseshiroge. RpsUN1 (Li et al., 2016) locus mapped between BARCSOYSSR_03_0233 and BARCSOYSSR_03_0246 which were unfortunately not polymorphic between the two parents Meng8206 and Linmeng6-46. Landrace PI 567139B (RpsUN1) was resistant to P. sojae pmg (17)-1 (pathotypes corresponding to races 17) and Meng8206 (RpsHN) was susceptible to P. sojae P7063 (pathotypes corresponding to races 17) (Supplementary Table 3), two mapping parents had different resistance reaction. So we think RpsUN1 may be different from RpsHN. RpsYD29 (Zhang et al., 2013b) and RpsHN were separated by the SSR marker satt1k2a. These results indicated that RpsHN may be a new gene difference from Rps1 or a new allele gene of Rps1.
Two types of soybean resistance genes to P. sojae have successfully been cloned thus far, NBS-LRR types and protein kinases. Four coiled-coil (CC)-NBS-LRR type genes were BAC-cloned in the Rps1k fine mapping region (Gao et al., 2005; Gao and Bhattacharyya, 2008). Two serine/threonine protein kinase type genes were cloned in the Rps10 fine mapping region (Zhang et al., 2013a). In the present study, two NB-ARC type genes and a protein kinase-type gene were considered as potential candidate genes. Three candidate genes Glyma.03g04260, Glyma.03g04300 and Glyma.03g04340 can be further studied for resistance pathways and functions.
Conclusion
We identified putatively a novel resistance gene, RpsHN, which can be used for breeding cultivars for Phytophthora resistance. The tightly linked SSR markers SSRSOYN-25 and SSRSOYN-44, as the functional markers, could contribute to the MAS breeding program.
Author Contributions
HX and TZ conceived the research. HX and JN designed the research. JN, NG, JS, LL, YC, SL, JH, and JZ performed the experiments and analyzed the data. JN drafted the manuscript. HX and TZ revised the paper.
Funding
This work was supported by the Special Fund for Agro-scientific Research in the Public Interest (Grant No. 201303018), Genetically Modified Organisms Breeding Major Projects (Grant No. 2014ZX08004-002), Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT13073), Modern Agro-industry Technology Research System of China (CARS-004-PS10), The National Natural Science Foundation of China (Grant No. 31271750), The National Natural Science Foundation of China (Grant No. 31571691).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank Prof. Yuanchao Wang (Nanjing Agricultural University) for providing the isolates of P. sojae.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00538/full#supplementary-material
Footnotes
- ^ http://www.phytozome.net/soybean
- ^ http://www.bio-soft.net/dna/SSRHunter.htm
- ^ http://soybase.org
- ^ http://www.phytozome.net/soybean
References
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Andolfatto, P., Davison, D., Erezyilmaz, D., Hu, T. T., Mast, J., Sunayama-Morita, T., et al. (2011). Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 21, 610–617. doi: 10.1101/gr.115402.110
Athow, K. L., Laviolette, F. A., Mueller, E. H., and Wilcox, J. R. (1980). A new major gene for resistance to Phytophthora megasperma var. sojae in soybean. Phytopathology 70, 977–980. doi: 10.1094/Phyto-70-977
Baird, N. A., Etter, P. D., Atwood, T. S., Currey, M. C., Shiver, A. L., Lewis, Z. A., et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376. doi: 10.1371/journal.pone.0003376
Bernard, R. L., and Cremeens, C. R. (1981). An allele at the rps1 locus from the variety ‘Kingwa’. Soybean Genet. Newsl. 8, 40–42.
Buzzell, R. I., and Anderson, T. R. (1992). Inheritance and race reaction of a new soybean Rps1 allele. Plant Dis. 76, 600–601. doi: 10.1094/PD-76-0600
Cregan, P. B., Jarvik, T., Bush, A. L., Shoemaker, R. C., Lark, K. G., Kahler, A. L., et al. (1999). An integrated genetic linkage map of the soybean genome. Crop Sci. 39, 1464–1490.
Dorrance, A. E. (2003). Effect of partial resistance on Phytophthora stem rot incidence and yield of soybean in Ohio. Plant Dis. 87, 308–312. doi: 10.1094/PDIS.2003.87.3.308
Duan, Y. X., Zhou, B., Chen, L. J., and Wu, H. (2008). Representative analysis of the establishment of a core collection focused on race 3 of soybean cyst nematode. Soybean Sci. 27, 366–372. doi: 10.1017/S1479262110000493
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Gao, H., and Bhattacharyya, M. K. (2008). The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol. 8:29. doi: 10.1186/1471-2229-8-29
Gao, H., Narayanan, N. N., Ellison, L., and Bhattacharyya, M. K. (2005). Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol. Plant Microbe Interact. 18, 1035–1045. doi: 10.1094/MPMI-18-1035
Gordon, S. G., St Martin, S. K., and Dorrance, A. E. (2006). 8 maps to a resistance gene rich region on soybean molecular linkage group F. Crop Sci. 46, 168–173.
Huang, J., Guo, N., Li, Y., Sun, J., Hu, G., Zhang, H., et al. (2016). Phenotypic evaluation and genetic dissection of resistance to Phytophthora sojae in the Chinese soybean mini core collection. BMC Genet. 17, 85. doi: 10.1186/s12863-016-0383-4
Huang, X., Feng, Q., Qian, Q., Zhao, Q., Wang, L., Wang, A., et al. (2009). High-throughput genotyping by whole-genome resequencing. Genome Res. 19, 1068–1076. doi: 10.1101/gr.089516.108
Kilen, T. C., Hartwig, E. E., and Keeling, B. L. (1974). Inheritance of a second major gene for resistance to Phytophthora rot in soybeans. Crop Sci. 14, 260–262. doi: 10.2135/cropsci1974.0011183X001400020027x
Kou, Y., and Wang, S. (2010). Broad-spectrum and durability: understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 13, 181–185. doi: 10.1016/j.pbi.2009.12.010
Li, L., Feng, L., Wang, W., Ping, J., Fitzgerald, J. C., Zhao, M., et al. (2016). Fine mapping and candidate gene analysis of two loci conferring resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 129, 2379–2386. doi: 10.1007/s00122-016-2777-0
Li, R., Yu, C., Li, Y., Lam, T. W., Yiu, S. M., Kristiansen, K., et al. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Lin, F., Zhao, M., Ping, J., Johnson, A., Zhang, B., Abney, T. S., et al. (2013). Molecular mapping of two genes conferring resistance to Phytophthora sojae in a soybean landrace PI 567139B. Theor. Appl. Genet. 126, 2177–2185. doi: 10.1007/s00122-013-2127-4
Lohnes, D. G., and Schmitthenner, A. F. (1997). Position of the Phytophthora resistance gene Rps7 on the soybean molecular map. Crop Sci. 37, 555–556. doi: 10.2135/cropsci1997.0011183X003700020040x
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Mueller, E. H., Athow, K. L., and Laviolette, F. A. (1978). Inheritance of resistance to four physiologic races of Phytophthora megasperma var. sojae. Phytopathology 68, 1318–1322. doi: 10.1094/phyto-68-1318
Ping, J., Fitzgerald, J. C., Zhang, C., Lin, F., Bai, Y., Wang, D., et al. (2015). Identification and molecular mapping of Rps11, a novel gene conferring resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 129, 445–451. doi: 10.1007/s00122-015-2638-2
Ploper, L. D., Athow, K. L., and Laviolette, F. A. (1985). A new allele at the Rps, locus for resistance to Phytophthora megasperma f.sp.glycinea in soybean. Phytopathology 75, 690–694. doi: 10.1007/s00122-013-2073-1
Schmitthenner, A. F. (1985). Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 69, 362–368. doi: 10.1094/PD-69-362
Song, Q. J., Jia, G. F., Zhu, Y. L., Grant, D., Nelson, R. T., Hwang, E. Y., et al. (2010). Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci. 50, 1950–1960. doi: 10.2135/cropsci2009.10.0607
Sugimoto, T., Kato, M., Yoshida, S., Matsumoto, I., Kobayashi, T., Kaga, A., et al. (2012). Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breed. Sci. 61, 511–522. doi: 10.1270/jsbbs.61.511
Sugimoto, T., Yoshida, S., Kaga, A., Hajika, M., Watanabe, K., Aino, M., et al. (2011). Genetic analysis and identification of DNA markers linked to a novel Phytophthora sojae resistance gene in the Japanese soybean cultivar Waseshiroge. Euphytica 182, 133–145. doi: 10.1007/s10681-011-0525-8
Sun, J., Li, L., Zhao, J., Huang, J., Yan, Q., Xing, H., et al. (2014). Genetic analysis and fine mapping of RpsJS, a novel resistance gene to Phytophthora sojae in soybean [Glycine max (L.) Merr]. Theor. Appl. Genet. 127, 913–919. doi: 10.1007/s00122-014-2266-2
Sun, S., Wu, X. L., Zhao, J. M., Wang, Y. C., Tang, Q. H., Yu, D. Y., et al. (2011). Characterization and mapping of RpsYu25, a novel resistance gene to Phytophthora sojae. Plant Breed. 130, 139–143. doi: 10.1111/j.1439-0523.2010.01794.x
Tian, M., Zhao, L., Li, S., Huang, J., Sui, Z., Wen, J., et al. (2016). Pathotypes and metalaxyl sensitivity of Phytophthora sojae and their distribution in Heilongjiang, China 2011–2015. J. Gen. Plant Pathol. 82, 132–141. doi: 10.1007/s10327-016-0654-y
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
van Ooijen, J. W. (2006). JoinMap 4 Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen: Kyazma B.V.
Wang, S., Basten, C. J., Gaffney, P., and Zeng, Z.-B. (2012). Windows QTL Cartographer 2.5. Raleigh, NC: North Carolina State University.
Wang, W., Jiang, W., Zhang, J., Miao, L., Zhao, T., Gai, J., et al. (2015). Selection of drought-tolerant soybean and evaluation of the drought tolerance indices. Crop Sci. 34, 808–818.
Wrather, A., and Koenning, S. (2009). Effects of diseases on soybean yields in the United States 1996 to 2007. Plant Health Prog. doi: 10.1094/PHP-2009-0401-01-RS
Wu, X. L., Zhang, B. Q., Sun, S., Zhao, J. M., Yang, F., Guo, N., et al. (2011). Identification, genetic analysis and mapping of resistance to Phytophthora sojae of Pm28 in soybean. Agric. Sci. China 10, 1506–1511. doi: 10.1016/s1671-2927(11)60145-4
Yi, X., Liang, Y., Huerta-Sanchez, E., Jin, X., Cuo, Z. X., Pool, J. E., et al. (2010). Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78. doi: 10.1126/science.1190371
Zhang, J., Sun, S., Wang, G., Duan, C., Wang, X., Wu, X., et al. (2014). Characterization of Phytophthora resistance in soybean cultivars/lines bred in Henan province. Euphytica 196, 375–384. doi: 10.1007/s10681-013-1040-x
Zhang, J., Xia, C., Duan, C., Sun, S., Wang, X., Wu, X., et al. (2013a). Identification and candidate gene analysis of a novel phytophthora resistance gene Rps10 in a Chinese soybean cultivar. PLoS ONE 8:e69799. doi: 10.1371/journal.pone.0069799
Zhang, J., Xia, C., Wang, X., Duan, C., Sun, S., Wu, X., et al. (2013b). Genetic characterization and fine mapping of the novel Phytophthora resistance gene in a Chinese soybean cultivar. Theor. Appl. Genet. 126, 1555–1561. doi: 10.1007/s00122-013-2073-1
Keywords: soybean, Phytophthora root rot, fine mapping, resistance gene, bins and SSR markers, linkage map
Citation: Niu J, Guo N, Sun J, Li L, Cao Y, Li S, Huang J, Zhao J, Zhao T and Xing H (2017) Fine Mapping of a Resistance Gene RpsHN that Controls Phytophthora sojae Using Recombinant Inbred Lines and Secondary Populations. Front. Plant Sci. 8:538. doi: 10.3389/fpls.2017.00538
Received: 30 January 2017; Accepted: 27 March 2017;
Published: 11 April 2017.
Edited by:
Jacqueline Batley, University of Western Australia, AustraliaReviewed by:
Leah McHale, Ohio State University at Columbus, USAMark Gijzen, Agriculture and Agri-Food Canada, Canada
Copyright © 2017 Niu, Guo, Sun, Li, Cao, Li, Huang, Zhao, Zhao and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han Xing, hanx@njau.edu.cn Tuanjie Zhao, tjzhao@njau.edu.cn
 Jingping Niu
Jingping Niu Na Guo
Na Guo Jutao Sun
Jutao Sun Tuanjie Zhao
Tuanjie Zhao Han Xing
Han Xing