- 1State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 2Faculty of Biology, Vietnam National University of Science, Hanoi, Vietnam
The “elevational shift” scenario has been proposed as a model to explain the response of cold-adapted organisms to Quaternary climatic fluctuations in Europe and North America. However, the elevational shift model has not been well-explored in eastern Asia, which is more topographically complex than the other Northern Hemisphere biogeographic regions. Here, we evaluated the role of elevational shift in the closely related firs, or Abies Mill., of subtropical and temperate China. These firs are typical alpine trees with sensitivity to climate change. We tested the elevational shift hypothesis in firs of China using phylogeographic methods and ecological niche models. Our phylogeographic analyses comprised mitochondrial and chloroplast polymorphisms surveyed across 479 individuals from 43 populations representing 11 species. M1 of the 11 mitotypes and C1 of the 25 chlorotypes were inferred as the ancestral haplotype, and they had the widest distribution. The results of our phylogeographic survey revealed multiple centers of genetic diversity in distinct geographic regions and no latitudinal trend. Moreover, our results showed range expansions for seven taxa during the last glacial (64.9–18.2 or 32.5–9.1 kya), and this was consistent with the Quaternary fossil record of Abies in China. Taken together, our findings support a historical biogeographic pattern in firs of glacial expansions, probably through corridors at lower elevation, and interglacial fragmentations, through isolation at higher elevation peaks. Therefore, Abies in China probably undergoes elevational shift in response to climate change. Facing the forecasting global warming, the risk of several critically endangered firs was further enhanced as these species would have little escape space in situ to higher altitudes. According to our ENMs, we proposed an ex situ conservation strategy in the southern Hengduan Mountains region of south western China.
Introduction
Quaternary climate oscillations played an important role in population genetic structure and geographical distribution of species (Davis and Shaw, 2001). Depending on species' niche requirements, there are two common scenarios concerning the impact of Quaternary glacial-interglacial cycles on the spatial distribution of biodiversity (Hewitt, 2004). One is the “latitudinal shift” scenario for thermophilous, low elevation temperate species. Under this scenario, the historical interglacial periods facilitated range expansions from glacial refugia at low latitudes to higher latitudes (Schmitt and Rákosy, 2007; Andreia et al., 2016; Nicolas et al., 2017). Such latitudinal shifts led either to higher genetic diversity at lower latitudes than at higher ones (Hewitt, 1996) or to lower genetic diversity at lower latitudes than at higher ones (Petit et al., 2003; Lascoux et al., 2004; Cheddadi et al., 2009). The other inter-glacial cycle scenario is “elevational shift.” Under this scenario, cold-adapted organisms, such as alpine (or arctic) plants, responded to Quaternary climatic fluctuations by shifting along an altitudinal gradient (Galbreath et al., 2009; Awad et al., 2014; Bystriakova et al., 2014; Inoue and Berg, 2016). Such species not only migrated to lower latitudes but also to lower elevations, they had relatively wider distributions during glacial maxima but became restricted to higher mountains or sky islands during interglacial periods. Thus, elevational shifts lead to the actual pattern of multiple centers of genetic diversity, and to negligible trends in latitudinal diversity (Hewitt, 1996). Examples of species that experienced elevational shift include Abies cilicica (Awad et al., 2014) and Asplenium fontanum (Bystriakova et al., 2014).
The elevational and latitudinal scenarios have been previously documented in Europe (e.g., Bystriakova et al., 2014; Ruosch et al., 2016) and North America (e.g., Dechaine and Martin, 2004, 2005; Inoue and Berg, 2016). Nevertheless, few studies have sought to clarify elevational and latitudinal shifts in eastern Asia (Qiu et al., 2011; Liu et al., 2014). During the Quaternary glaciations, most regions in eastern Asia remained ice-free, with ice cover occurring as valley and piedmont glaciers and ice caps only in regions with high peaks (Shi et al., 1984, 1987; Rutter, 1995; Hewitt, 2000, 2004; Shi, 2002), which is in contrast to the extreme glaciation across Europe and North America. Latitudinal shifts have been detected in several warm-adapted species (e.g., Qiu et al., 2011; Liu et al., 2014), such as in Juglans mandshurica and Euptelea pleiospermum (Wang et al., 2016; Wei et al., 2016). However, few studies have been conducted on arctic-alpine species despite their potential importance for a comprehensive understanding of the impact of global warming on cold-adapted species.
Firs are the dominant trees in the boreal and temperate montane forests of the Northern Hemisphere (Cheng and Fu, 1978; Farjon, 2001). These plants have stringent biological requirements for humidity and temperature stability, which have made them good paleoclimatic indicators (Xu et al., 1980; Shi, 1996). The climatic requirements of firs are highly conserved phylogenetically (Xiang et al., 2000, 2004, 2009, 2015; Fan, 2006), and makes them a good model system to study historical elevational shifts as well as responses to ongoing climate changes.
The genus Abies is discontinuously distributed in the temperate and subtropical montane forests of the northern hemisphere, with subtropical and temperate China as the well-known center of diversity (Cheng and Fu, 1978; Farjon, 2001). Subtropical and temperate China is known for their complex topography which provides varied microhabitats for living organisms (Wu, 1980; Ying et al., 1993; Axelrod et al., 1996; Ying, 2001). There are 13–20 Abies species in these adjacent regions with, an elevation from ca. 1,500 to 4,300 m (Cheng and Fu, 1978; Farjon, 2001; Rushforth, pers. comm.). Using morphological characters, Abies of China has been divided into as many as three questionably distinct subsections (Liu, 1971; Farjon and Rushforth, 1989), though all species form one clade according to recent molecular phylogenetic studies (Xiang et al., 2000, 2004, 2009, 2015; Shao and Xiang, 2015). Such a monophyletic group is suitable to study diversification processes by recovering intraspecific divergences and phylogeographic patterns for multiple-species (Liepelt et al., 2010; Liu et al., 2012). Thus, these Abies species provide an excellent system to investigate elevational shift through comparative phylogeography and ecological niche modeling (Ying, 2001; Qian et al., 2003; Liu et al., 2014).
In the present study, we conducted phylogeographic analyses and historical ecological niche prediction on Chinese subalpine and temperate firs, with the aims to: (i) evaluate the significance of the elevational shift scenario. If this were the case, we would expect to detect historical expansions during glacials and a resulting phylogeographic pattern. (ii) explore the genetic diversity structure and demographic history of the endangered firs with emphasis on the conservation under the conditions of ongoing climate change.
Materials and Methods
Population Sampling
We collected DNA materials for 475 individuals from 42 populations of Chinese subalpine and temperate firs, representing 10 species (Figure S1, Table S2). Four individuals of one population of A. fansipanensis QP Xiang, LK Fu, and Nan Li from Mt Fan Si Pan in northern Vietnam are included in the present study as a typical example of a highly endangered fir with only limited individuals (Xiang, 1997). Within each population, we sampled needles from eleven–twelve individuals growing at least 100 m apart except for six populations (pop. 2, 9, 14, 22, 36, and 43; Figure S1), where we could only sample four–eight individuals. We sampled two to 13 populations each of the following seven species: A. chensiensis Tiegh., A. delavayi Franch., A. fabri (Mast.) Craib, A. forrestii Coltm.-Rog., A. georgei Hand.-Mazz., A. recurvata Masters, and A. squamata Masters. Among them, 181 individuals from 17 populations (Nos. 1–13 and 33–36) of A. chensiensis and A. recurvata have been characterized for mtDNA and cpDNA sequence variation in a previous study (Shao and Xiang, 2015). To address the conservation implications of four other highly endangered fir species, we sampled only one population of each of them due to their limited availability and restrict geographic distributions: A. fanjingshanensis WL Huang, YL Tu, and SZ Fang (IUCN: endangered), A. yuanbaoshanensis YJ Lu and LK Fu (IUCN: endangered), A. ziyuanensis LK Fu and SL Mo (IUCN: endangered) and A. fansipanensis (IUCN: critically endangered; IUCN, 2016). Our sampling also included the following outgroup taxa: A. bracteata endemic to North America as outgroup in the mitotype analyses, and A. alba, A. nordmanniana (from Euro-Mediterranean), and A. mariesii from Japan as outgroup taxa in the chlorotype analyses. Our outgroup selection was based on a previous phylogenetic study (Xiang et al., 2015); only one individual was sampled for each outgroup. We present all sampling efforts in Table S2 and indicate samples that resulted from this study vs. a prior one comprising 181 individuals from 17 populations of A. chensiensis and A. recurvata (Nos. 1–13 and 33–36). Population sampling in the wild was strictly observed according to international rules (Nagoya, 2010).
DNA Extraction, PCR, and Sequencing
We extracted total genomic DNA using a modified cetyltrimethyl ammonium bromide (CTAB) protocol (Rogers and Bendich, 1988) or DNA secure Plant Kit (Tiangen). We used the DNA extractions to isolate two mtDNA fragments, nad1 intron 2 and nad5 intron 4, and one cpDNA intergenic spacer, trnS/G, following procedures in Shao and Xiang (2015). We performed sequencing reactions using a DYEnamic ET Terminator Kit (Amersham Pharmacia Biotech), and detected labeled sequencing products on a MegaBACE 1000 (Amersham Biosciences, Buckinghamshire, UK). We deposited all newly generated sequences in GenBank under accession numbers KP635976–KP636041.
Population Structure and Demographic Analyses
We estimated molecular diversity indices, including the average pairwise differences per base pair between sequences (π) (Nei and Li, 1979), and the haplotype diversity (h) (Nei, 1987) for each taxon and population using DNASP version 4.10 (Rozas et al., 2003). Using h and π, we detected patterns of spatial genetic diversity using the GDivPAL function in Spads 1.0 (Dellicour and Mardulyn, 2014). We also calculated haplotype networks among mitotypes and chlorotypes, individually, in NETWORK version 4.2.0.1, with gaps treated as single events (Bandelt et al., 1999).
We used the program PERMUT (Pons and Petit, 1996) to calculate the average gene diversity within populations (HS), total gene diversity (HT) and two measures of population differentiation (GST and NST). When NST estimates were significantly higher than the GST-values, we inferred a phylogeographic structure. We performed an analysis of molecular variance (AMOVA) using the program ARLEQUIN 3.5 (Excoffier and Lischer, 2010) to determine the amount of genetic variation within and among the taxa and populations. In order to test the correlation between genetic and geographic distances, the mantel test was performed (Mantel, 1967; Nei and Li, 1979). We identified population groups that were geographically homogenous and maximally differentiated from each other using spatial analysis of molecular variance (SAMOVA; Dupanloup et al., 2002). We detected distinct populations (K) via a simulated annealing approach to maximize among population variance (FCT), and we chose the K between 2 and 10 where FCT began to plateau.
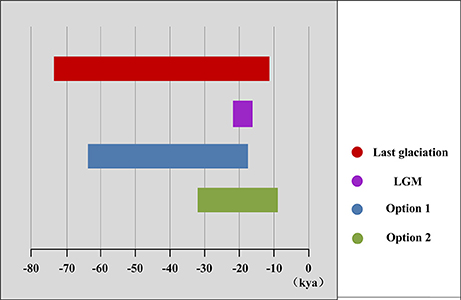
We performed mismatch distribution analyses in the program ARLEQUIN 3.5 to detect historical population expansion events. When we detected an expansion event, we estimated its age using the formula s = 2 ut (Rogers and Harpending, 1992) where t is the age of the expansion, u = μkg (μ is the substitution rate per nucleotide site per year, k is the average sequence length, and g is the generation time in years). In this study, we estimated expansions using a range of neutral substitution rates for cpDNA (μ = 2.61 × 10−10–4.02 × 10−10 ss−1 year−1) (Gernandt et al., 2008), an average haplotype sequence length (1,111 bp) in Pinaceae (Brown et al., 2004). In Pinaceae, the minimum estimate of generation time was 25 years (Brown et al., 2004). However, it should be mentioned that sexual maturity and effective generation time (when seedlings are actually recruited) of fir species in wild populations was also likely to be around 50 years (Cheng and Fu, 1978; Fan, 2006). Here, we calculated the historical expansion time under the two different minimum estimate of generation time (option 1: g = 25 years; option 2: g = 50 years).
Ecological Niche Modeling
We performed ecological niche modeling (ENM) of Chinese subalpine and temperate firs in MAXENT (version 3.2.1; Phillips et al., 2006). The ENMS were not performed on species level for the extremely limited reliable occurrence records except for A. chensinesis (Fan, 2006; Shao and Xiang, 2015). For input into MAXENT, we obtained species occurrence data from the Chinese Virtual Herbarium (CVH, www.cvh.org.cn) and personal observations, including 112 records. These records involved 36 A. chensiensis records, 15 A. delavayi records, 11 A. fabri records, 8 A. forrestii records, 9 A. georgei records, 20 A. recurvata records, seven A. squamata records, 1 A. fanjingshanensis record, 1 A. yuanbaoshanensis record, 3 A. ziyuanensis records, and 1 A. fansipanensis record.
We used 19 BIOCLIM variables as environmental data for ENM representing five different time periods. Past and current data for these variables were available from the WorldClim database (www.worldclim.org). The data from WorldClim included baseline climate (BioClim layers for the period 1,950–2,000) at a spatial resolution of 30 arc s), data for the last glacial maximum (LGM; ~21,000 years BP) with spatial resolution of 2.5 arc min resolution simulated by CCSM model and MIROC model, and the last interglacial (LIG; ~120,000–140,000 years BP) period with spatial resolution in 30 arc s resolution. We obtained predictions for future climatic conditions in 2,050 and 2,080, from the International Center for Tropical Agriculture (www.ccafs-climate.org) based on the HadCM3. The HadCM3 model shows two scenarios (A2, and B2), which were represented in 2.5 arc min resolution Many climatic variables were highly correlated (|Pearson's R| ≥ 0.8). Therefore, our final ENMs were constructed using seven variables (mean diurnal range of temperature, temperature seasonality, max temperature of warmest month, mean temperature of driest quarter, mean temperature of coldest quarter, precipitation of driest, and warmest quarter) with limited covariance.
To construct ENMs, we conducted ten replicate runs with these parameters: default convergence threshold, maximum iterations (1,500), and 30% of the sites for model training (Waltari et al., 2007; Jezkova et al., 2009). We evaluated model performance using the area under the receiver operating characteristic curve (AUC) calculated by MAXENT.
Pollen Data Collection
We initially surveyed more than 200 Quaternary fossil pollen records from both publications and database sources involving distinct species and time span, following Cao et al. (2013). During the last glaciation, Abies spp. occurred in 10 pollen records with reliable chronologies and sufficiently high sampling resolutions, covering the subtropical and temperate China (Liu and Ye, 1977; Zhou et al., 1978; Zhang et al., 1997; Sun and Li, 1999; Wu et al., 2002; Xu et al., 2002; Feng et al., 2007; Tang et al., 2007; Cook et al., 2011; see details in Table S1).
Results
Mitochondrial Genealogy
The length of the mtDNA fragments, nad1 intron 2 and nad5 intron 4, was conservative; 675 bp and 220 bp, respectively. The fragments contained seven point mutations and four indels resulting in 11 distinct mitotypes (M1–11; Table S3). Our results show that M1 has the widest distribution and is likely the ancestral haplotype according to the NETWORK analysis. M1 was the dominant haplotype in the central Hengduan Mountains region (Figure 1) and also occurred in Vietnam and southeast China which harbored four rare and critically endangered species, A. fanjingshanensis, A. fansipanensis, A. yuanbaoshanensis, and A. ziyuanensis (Pop. 40–43; Figure 1).
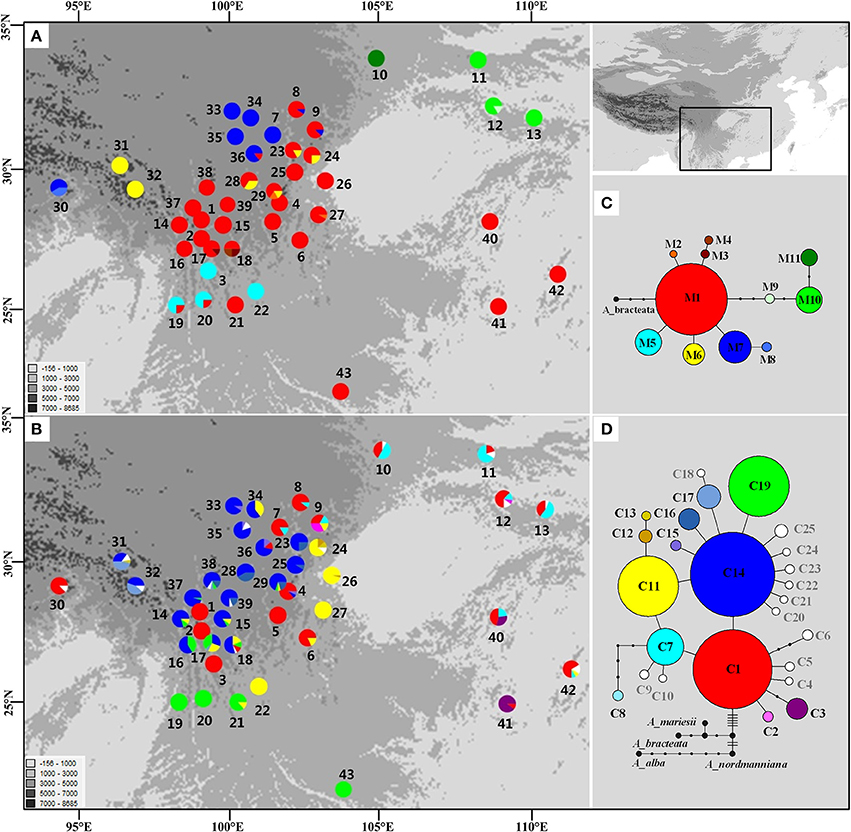
Figure 1. Distributions and networks of mitotypes (A,C) and chlorotypes (B,D) recorded in the Chinese subtropical and temperate firs. Private chlorotypes are shown in white. See Table S2 for population code numbers.
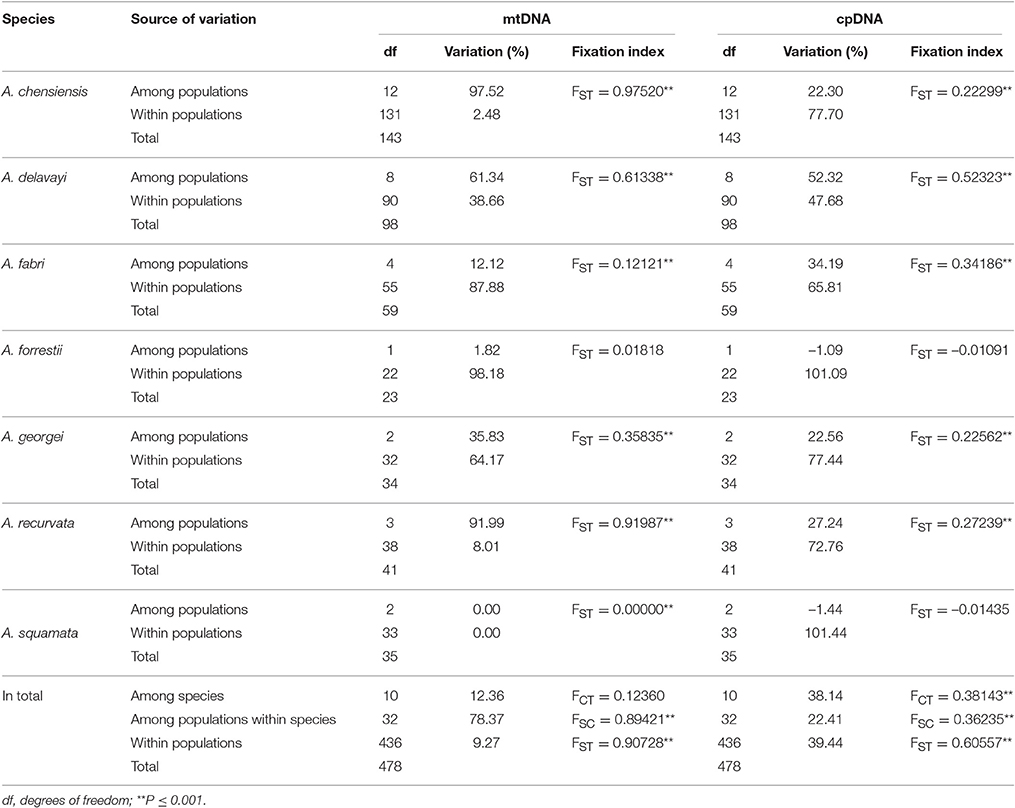
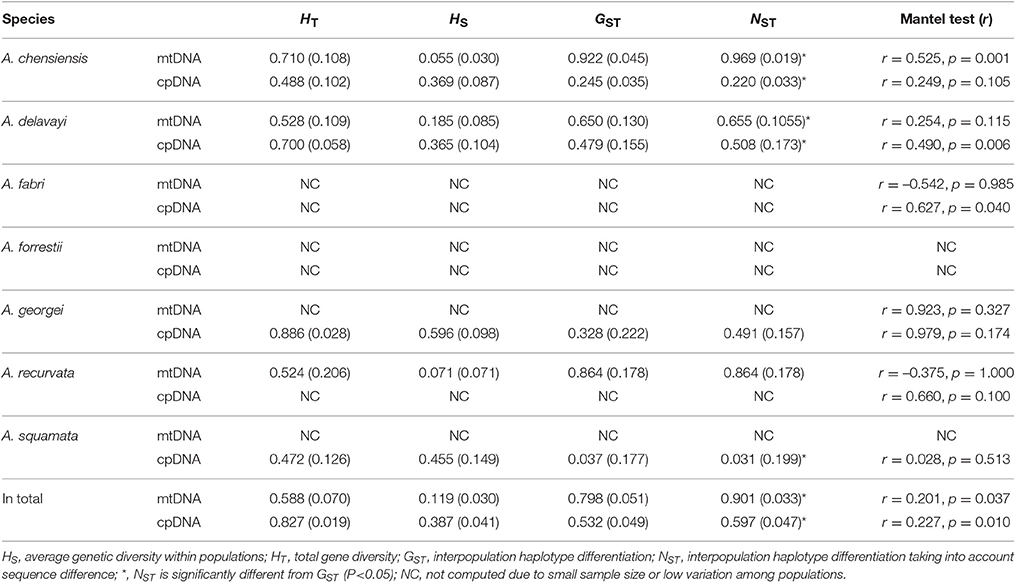
The estimates of molecular diversity π and h, are summarized in Table S2. Diversity of mitochondria was high in several distinct geographic regions including the Hengduan Mountains, the Qinling-Daba Mountains, and Southeast China according to the results from Spads (green dots in Figures 2A,B). Within each species, the distribution of mitochondria diversity also didn't show any latitudinal trend (Table S2; Figures S4–S6). Our AMOVA analyses revealed that 12.36% of the total mtDNA sequence variation occurred among taxa, whereas 78.37% of the variation occurred among populations within taxa (Table 1). The mtDNA sequence variation showed HS much lower than HT. Furthermore, mitotypes exhibited strongly isolated geographic distributions and a significant phylogeographic structure (GST < NST; P < 0.05; Table 2). Genetic diversity was significantly correlated with geographic distances (r = 0.201, p = 0.037) according to our Mantel test (Table 2). However, no significant geographic division was revealed by the SAMOVA analyses.
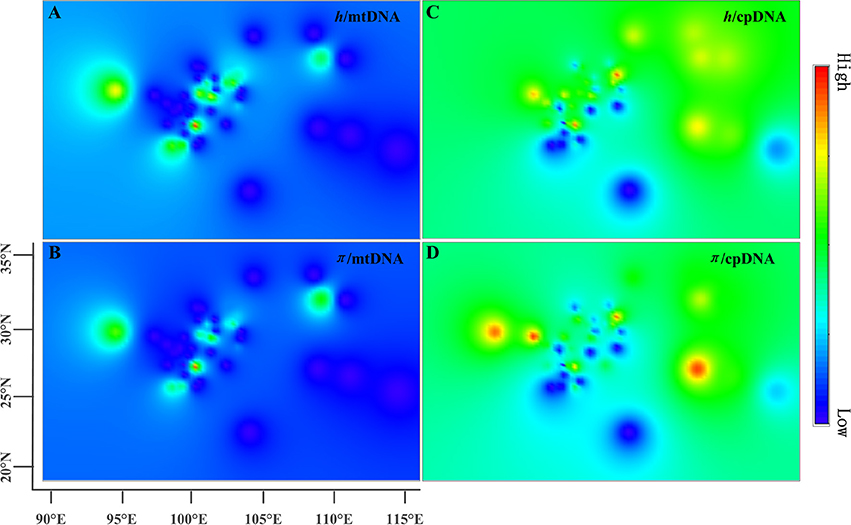
Figure 2. Genetic diversity across the range of the Chinese subtropical and temperate firs. The X-axis represents longitude and the Y-axis latitude. (A) pattern of mtDNA haplotype diversity (h); (B) pattern of mtDNA nucleotide diversity (π); (C) pattern of cpDNA haplotype diversity (h); (D) pattern of cpDNA nucleotide diversity (π).
Chloroplast Genealogy
In the chloroplast DNA data, we detected six indels and 17 substitutions resulting in 25 chlorotypes (C1–25; Table S4). Five chlorotypes had frequencies above 5% (C1, C7, C11, C14, and C19). Fifteen chlorotypes (C2, C4–6, C8–10, C12, C18, and C20–25) were fixed in single species, while the remaining ten were shared among species (Figure 1B). The chlorotype C1 possessed the widest geographic distribution and may be ancestral haplotype according to the NETWORK analysis (Figure 1).
The cpDNA exhibited higher genetic diversity than the mtDNA according to the Spads analysis (Figure 2; cpDNA in green). The cpDNA possessed multiple centers of diversity according to the indices h and π (Figures 2C,D; yellow and red dots). Within each species, the distribution of cpDNA diversity also didn't show any latitudinal trend (Table S2; Figures S4–S6). Of the variation among chlorotypes, 38.14% was among taxa and and 39.44% was within populations (Table 1). Chlorotypes exhibited significant phylogeographic structure (GST < NST; P < 0.05), and HS was significantly lower than HT (Table 2). However, we did not detect a significant division among populations according to SAMOVA analysis, but the Mantel test revealed a significant correlation between genetic and geographic distances (r = 0.227, P = 0.010; Table 2).
Population Demographic History
We performed mismatch distribution analyses on Chinese subalpine and temperate firs taken together and each species individually using cpDNA sequence data. We analyzed seven species for mismatched distributions (A. chensiensis, A. delavayi, A. fabri, A. forrestii, A. georgei, A. recurvate, and A. squamata) and excluded the four rare and endangered species (A. fanjingshanensis, A. fansipanensis, A. yuanbaoshanensis, and A. ziyuanensis) because of the small sample size (No. pop = 1).
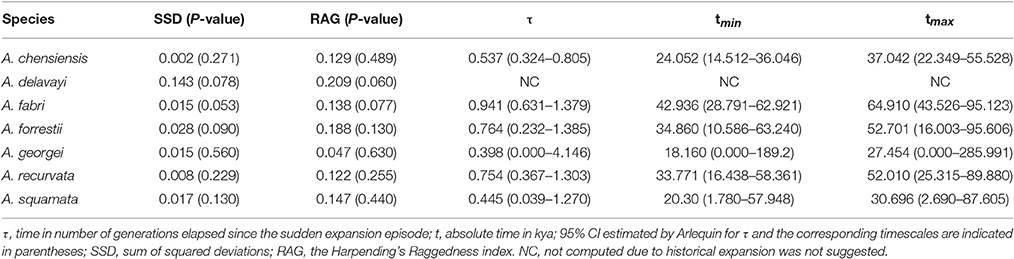
The mismatch distribution results rejected the demographic expansion hypothesis for the Chinese subalpine and temperate firs as a whole (PSSD = 0.000; Figure S2). However, the expansion hypothesis was supported for all of the seven species analyzed separately except for A. delavayi (PSSD = 0.078; Figure S2). We determined that the demographic expansion of the six species probably occurred in 64.9–18.2 kya (option 1: g = 25 years) or 32.5–9.1 kya (option 2: g = 50 years; Figure 3, Table 3).
Ecological Niche Modeling
The value of the AUC test (mean ± SD) for the ecological niche modeling, averaged across all 10 runs, was very high (0.96 ± 0.005). Models for the present (Figure 4A) and last interglacial (Figure 4B) showed contracted ranges compared to the LGM (Figure 4C), indicating losing of suitable habitat in south, central, and east China. The future predictions of suitable habitat were narrower than for the present day, and ranges were further reduced between 2,050 (Figure 4D) and 2,080 (Figure 4E). Except the Models for the LGM, the suitable habitats of the four highly endangered firs in southeast China were always absent (Figure 4). As for other seven species, the suitable habitats were slightly changed in all time periods bun not in the LIG. During the LIG when globe warming, suitable habitat of fir species distributed in northern part of the Hengduan Mountains region was contracted heavily, such as A. recurvata, A. fabri, A. forrestii, and A. squamata (Figure 4, Figure S1).
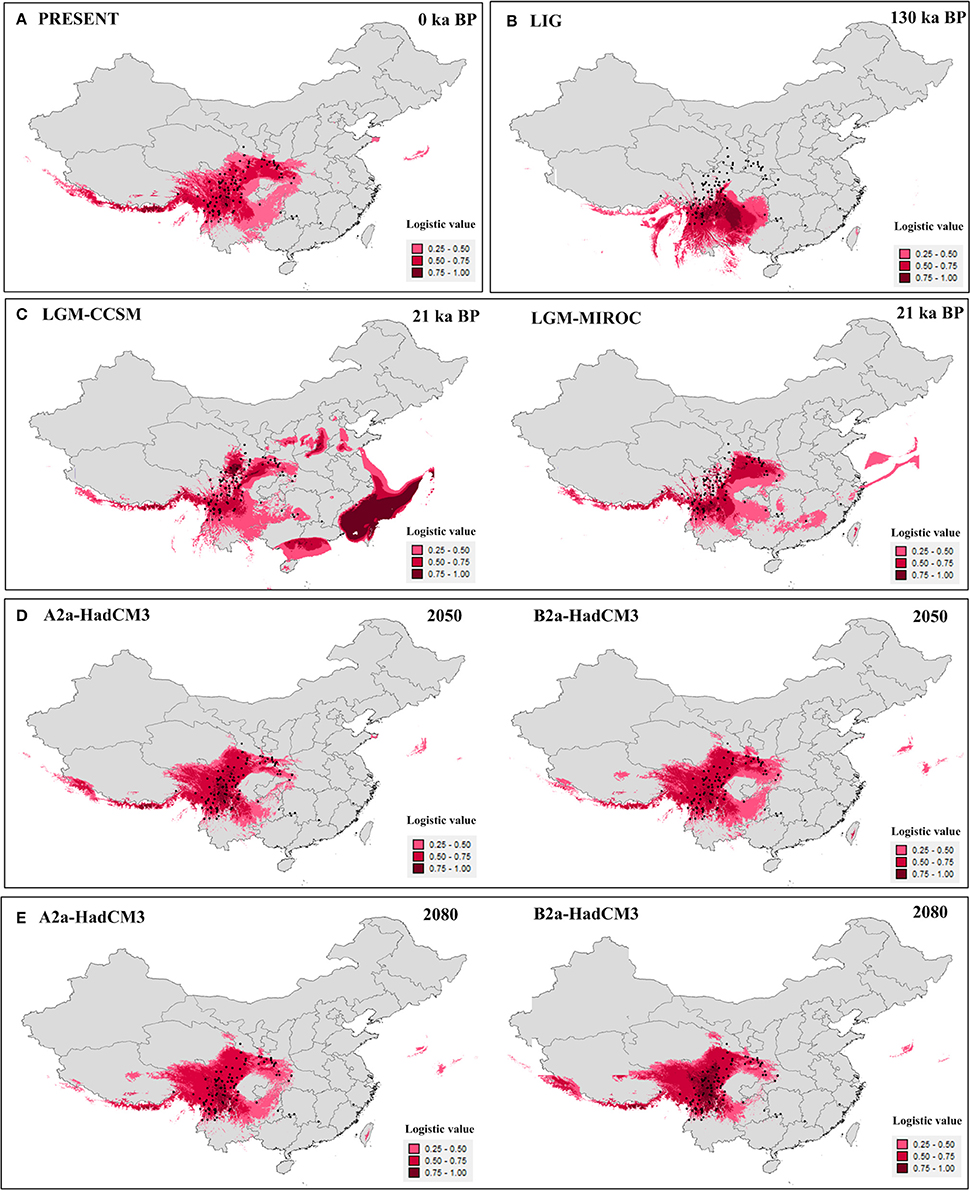
Figure 4. Results of ecological niche modeling of Chinese subtropical and temperate firs. (A) Predicted distribution probability (in logistic value) for current climatic conditions. (B) Average projection of the model to the last interglacial (c. 120–140 kyr BP). (C) Average projections of the model to the last glacial maximum [c. 21 kyr before present (BP)] using climatic variables under the Community Climate System Model (CCSM) and Model for Interdisciplinary Research on Climate (MIROC). (D,E) Average projections of the model to the future [2,050 (D) and 2,080 (E)] using climatic variables under the HadCM3 model according to two distinct scenarios (A2 and B2). Black dots present extant occurrence points.
Pollen-Based Distribution during the Last Glaciation
Palynological data indicated that Abies were much more common in lowland forests of central (no. 2,3,8) and eastern (no. 1,4,5,7,9) China during the last glaciation (Figure S3). This pattern depicted wider distribution during the last glaciation than today, Except for three pollen records from northeastern China (no. 1, 4, and 8), the pollen-based distribution pattern was highly consistent with the ENMs for the LGM by CCSM model (Figure 4C).
Discussion
The Elevation Shift of the Chinese Subtropical and Temperate Firs during the Quaternary
Previous studies concerning arctic-alpine species from Europe and North America such as Sedum lanceolatum (Dechaine and Martin, 2005), Asplenium fontanum (Bystriakova et al., 2014), and Abies cilicica (Awad et al., 2014), indicate glacial expansion of elevational shift scenario. This “elevation shift” scenario had not been well-documented in arctic-alpine species of East Asia, and places constraints on its application to biodiversity conservation. By combining molecular phylogeography and ENM, we explored how the Chinese subtropical and temperate firs (typical alpine species) in eastern Asia responded to Quaternary climate change.
Two distinct expansion models have been proposed by Hewitt (1996) to distinguish elevational shift from latitudinal shift. The pioneer expansion model of latitudinal shift is characterized by decrease of genetic diversity along latitudes during interglacial expansion (Hewitt, 2004; Yan et al., 2012; Andreia et al., 2016; Nicolas et al., 2017). The phalanx expansion model of elevational shift supposes glacial expansions without significant reduction of genetic diversity along latitudes, resulting in multiple centers of diversity but without decreased genetic diversity along latitude (Galbreath et al., 2009; Awad et al., 2014; Bystriakova et al., 2014; Inoue and Berg, 2016). Based on current phylogeographical patterns, the phalanx expansion model via elevational shift was potentially active in Chinese subtropical and temperate firs. Firstly, multiple centers of fir diversity were discovered in the Hengduan Mountains region, Qinling-Daba Mountains region and in Southeast China, indicating no latitudinal decreasing trends (Figure 2). A similar pattern was also found within each species (Table S2; Figures S4–S6). Secondly, relatively low genetic diversity of mtDNA and cpDNA were found both within populations and species across Chinese subalpine and temperate firs (Table S2). Such low genetic diversity or strong homogeneity could be the result of rapid recolonization following severe bottleneck effects, accompanied by climate-driven elevation shifts along a local elevation gradient (Hewitt, 2000; Jaramillo-Correa et al., 2004; Jiang et al., 2011). This result is consistent with similar processes reported in the Mediterranean region and North America (Ziegenhagen et al., 2005; Jaramillo-Correa et al., 2008; Wang et al., 2011). In addition, the population diversity structure of Abies in subtropical and temperate China is similar to that of congeners in North America and Europe especially by having differentiation among populations (mtDNA: 90.73%; cpDNA: 60.56%; Table 1) significantly greater than the averages of conifers (mtDNA: 76.4%; cpDNA: 16.5%; reviewed in Petit et al., 2005) (Liepelt et al., 2002; Fady-welterlen, 2005; Jaramillo-Correa et al., 2008; Fady and Conord, 2010; Wang et al., 2011). Greater differentiation of conifers among populations may result from recent habitat fragmentations or severe bottleneck effects due to global warming since the Holocene epoch (Hewitt, 2004; Jaramillo-Correa et al., 2004; Jiang et al., 2011). Thus, the phylogeographical pattern of Chinese subtropical and temperate firs could be considered potential evidence of elevational shift, in general, and of the phalanx model in particular.
More directly evidences were further revealed by the expansion time estimation and ecological niche modeling. We estimated that the ranges of most firs (i.e., taken individually, except A. delavayi) expanded during approximately 64.9–18.2 kya (option 1: g = 25 years) or 32.5–9.1 kya (option 2: g = 50 years; Table 3; Figure S2). These two timing of the expansion were highly coincident with the last glaciation in China, which was 73–10.4 kya with a maximum during 21–18 kya (Yi et al., 2004, 2005, 2007; Zhao et al., 2011). Specifically, range expansion of Abies in subtropical and temperate China might have commenced shortly after the beginning of the last glacial cycle and continued throughout the last glacial maximum (option 1) or since the middle of the last glaciation till the end (option 2; Figure 3). Both the two options indicated the glacial expansions via elevational shift.
Curiously, we did not detect evidence for recent, historical range expansion in all subtropical and temperate China and Vietnam firs considered together or in A. delavayi. For A. delavayi, distribution fragmentation could be easily inferred by its typical high-altitude distribution, extending from the southern part of Hengduan Mountains region to north Vietnam. For the whole Chinese subtropical and temperate firs, multiple asynchronous expansions, multiple independent refugia in situ and complex topography in this region might be responsible for the undetectable in mismatch distribution analyses (Cun and Wang, 2010; Yan et al., 2012; Qin et al., 2013). Such analyses seemed to underestimate the possibility of historical expansions in some situations. Nevertheless, our historical distributional projections showed that firs of subtropical and temperate China and Vietnam as a whole underwent an expansion from the LIG to the LGM; that is, during a period of global cooling. Distributional projections for the present and future show ongoing range contractions (Figure 4) coincident with continued global warming. Although such analysis was not conducted at the species level due to insufficient occurrence records except for A. chensiensis, we believed that this tendency should be well-represented in respective taxa because of their conservative ecological niche requirement (Cheng and Fu, 1978; Farjon, 2001; Fan, 2006). The accuracy of ENMs was further validated by the high similarity between the LGM-CCSM prediction (Figure 4C) and pollen-based distribution during the last glaciation (Figure S3). Two prior studies modeled future geographic distributions of six fir species of China, Japan, and Pakistan, and determined that those species would all lose their suitable habitat in the lowlands region in response to ongoing global warming (Tanaka et al., 2012; Ali et al., 2014).
Many plant species have strong preferences for a narrow range of climatic conditions. Therefore, past climatic conditions can be inferred from the palynological records of plants (Tinner and Lotter, 2006). Palynological data indicate that Abies were much more common in lowland forests of central and eastern China during the last glaciation (Figure S3) when the mean annual temperature was 5–10°C lower than today (Xu et al., 1980; Shi, 1996; Cao et al., 2013). Moreover, the abundance of Abies pollen in central China decreased with the onset of the interglacial period, which was warmer and less humid than the glacial one. The current mountain top distribution of several highly endangered fir species (e.g., A. beshanzuensis, A. fanjingshanensis, A. yuanbaoshanensis, and A. ziyuanensis) is living evidence of interglacial fragmentation (Cheng and Fu, 1978; Xiang, 2001; Jiang et al., 2015). Even though deforestation in China had been significant in past decades, these fir species were not a main target due both to their distribution along mountain tops or along the tree line away from human settlements, and to the low-quality wood properties (Fan, 2006). In summary, our results including genetic diversity, mismatch distribution analyses, ecological niche models, combining evidence from other lines involving fossil records and the current distribution patterns, led to a conclusion that the Chinese subtropical and temperate firs experienced elevational shift during the Quaternary climate change.
Implications for the Conservation of the Endangered Firs
Our results show that Chinese subtropical and temperate firs have undergone major habitat fragmentation and genetic isolation and differentiation since the Holocene. The main goal of biological conservation is to maintain as many of the important genetic building blocks of the species as possible so that the evolutionary progress is not constrained (Fraser and Bernatchez, 2001; D'Amen et al., 2013). Thus, defining conservation units with evolutionary differences is necessary for these economically and ecologically valuable threatened firs (e.g., de Querioz, 1998; Fraser and Bernatchez, 2001).
In subtropical and temperate China and Vietnam, four species of Abies are listed as critically endangered due to their restricted distribution on isolated mountain tops and small population sizes: A. beshanzuensis, A. fanjingshanensis (Pop. 40), A. yuanbaoshanensis (Pop. 41) and A. ziyuanensis (Pop. 42) in warm, temperate China and A. fansipanensis (Pop. 43) in Vietnam (Fu, 1992; Xiang, 2001). Abies beshanzuensis was not included in this study because its population comprises only three individuals and is too ecologically fragile for sampling. The other four species all possessed the M1 ancestral mitotype but had high cpDNA diversity (Figure 1). This observation implied a possible shared ancestral gene pool which was fragmented during interglacials (Jaramillo-Correa et al., 2006; Loehr et al., 2006; Dubreuil et al., 2010). Specifically, the total number of cpDNA haplotypes was three, two, four, and one in A. fanjingshanensis (C1, C3, C7), A. yuanbaoshanensis (C1, C3), A. ziyuanensis (C1, C5, C7, C11), and A. fansipanensis (C19), respectively (Figure 1). These distinct chloroplast haplotypes in each species highlight the distinct evolutionary histories of the species (Liepelt et al., 2009; Jay et al., 2012), thus each may represent separate conservation units.
For the species sensitive to environmental change, conservation planning must take into consideration the climatic envelope of the species and its present geographic distribution (Root et al., 2003; Thomas et al., 2004; Forest et al., 2007). The climatic envelope of Abies has been shrinking in geographic space since the last ice age and will probably continue to do so under present global warming conditions (Figure 4). The reasons are various, such as pest and disease damage, human disturbance, and habitat destruction. Unfortunately, Abies may have limited opportunity to escape global warming through elevational shift, because most species already occur at or nearly at the maximum available altitude: For example, A. beshanzuenesis occurs on Baishanzu Mountain (1,857 m) at 1,750 m, A. fanjingshanensis is restricted to 2,050–2,390 m on Fanjing Mountain (2,494 m), A. yuanbaoshanensis is distributed on Yuanbao Mountain (2,081 m) at 1,900–2,000 m, and A. ziyuanensis is located on Shunhuang Mountain (1,882 m) at 1,700 m (Fan, 2006; Li et al., 2012). While some species of Abies can move upwards, they may not be able to do so at a fast enough rate (Huck et al., 2012; Moritz and Agudo, 2013). Based on the meta-analyses, Chen et al. (2011) proposed that the distributions of many terrestrial organisms currently shifted to higher elevations at a median rate of 11.0 m per decade or to higher latitudes at a median rate of 16.9 km per decade to accomodate global warming. Such a high rate is difficult to achieve for conifers, which have a two (or longer) year reproductive cycle (Liu, 1971; Farjon and Rushforth, 1989; Xiang, 2001).
In situ conservation is the current govermentnal strategy for these Abies species, but it is clearly not a permanent solution. In fact, there is not any suitable habitat for these highly endangered firs in actual or 2,050/2,080 models (Figure 4). Moreover, a massive suitable habitat contraction occurred in the northern part of the Hengduan Mountains region during the LIG (Figure 4B). Facing the forecasted global warming, the suitable habitats of A. recurvata, A. fabri, A. forrestii, and A. squamata were believed to be heavily contracted (Figure 4B, Figure S1). Thus, we propose that these species urgently require ex situ management in botanical gardens in areas of suitable climate with dedicated breeding programs. According to our ENMs (Figure 4), the suitable habitats for breeding firs were stable in the southern Hengduan Mountains region during each time periods. At present, more than 30 National Nature Reserves are located in the Hengduan Mountains, and also some relevant state-grade research institutes, such as the Kunming Institute of Botany, Chinese Academy of Sciences (Chen et al., 2009). These specific gardens and their existing programs would strengthen the development of ex situ conservation.
Data Accessibility
The matrix of plastid DNA alignments is included as Tables S3, S4 of the Supporting Information, respectively. Original sequences of the three plastid regions are available in the GenBank database (Accession nos. KP635976–KP636041), details see Table S5.
Author Contributions
Conceived and designed the experiments: QX. Performed the experiments and analyzed the data: YS. Contributed reagents/materials/analysis tools: XZ and LP. Wrote the paper: YS and QX.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Chong Wu, Ran Wei, Jiu-Qiong Sun, and Xue-Ping Wei for their assistance with collection of materials. We are also indebted to Liu Liu, Hai-Lang Zhou, and Yan-Yan Liu for their help with data analyses. We acknowledge Yan-Yan Guo for helping with marker selection and sequencing. This study is supported by the National Natural Science Foundation of China Grant (NSFC no. 30870167, 31370260 and 31600163), and the National Foundation for Science and Technology Development, Ministry of Science and Technology, Vietnam (NSFC no. 106.11-2012.30).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00578/full#supplementary-material
Figure S1. Sampling locations of 43 populations of Chinese subtropical and temperate firs. Each taxon is represented by distinct color and shape.
Figure S2. The mismatch distributions for seven fir species in subtropical and temperate China based on cpDNA. A. chensiensis, A. delavayi, A. fabri, A. forrestii, A. georgei, A. recurvata, A. squamata, and the Chinese subtropical and temperate firs as a whole are presented in (A–H), respectively.
Figure S3. The extant distribution of Abies in China and 10 pollen records during the last glaciation. The red shows the present geographical ranges and the black triangle indicates the distribution of 10 pollen records in the subtropical and temperate China.
Figure S4. Distributions and networks of mitotypes and chlorotypes recorded in Abies chensiensis. Private chlorotypes are shown in white. See Table S2 for population code numbers.
Figure S5. Distributions and networks of mitotypes and chlorotypes recorded in Abies delavayi. Private chlorotypes are shown in white. See Table S2 for population code numbers.
Figure S6. Distributions and networks of mitotypes and chlorotypes recorded in Abies fabri. Private chlorotypes are shown in white. See Table S2 for population code numbers.
Table S1. Ten fossil records of Abies spp. in eastern and central China during the last glaciation.
Table S2. Details of sample locations and sample sizes of 43 populations and outgroups. For each population, estimates of haplotype diversity (h), nucleotide diversity (n) and frequencies of haplotypes for mitochondrial DNA and chloroplast DNA sequences are also indicated.YN, Yunnan; SC, Sichuan; GS, Gansu; SX, Shanxi; CQ, Chongqing; HB, Hubei; GZ, Guizhou; HN, Hunan; GX, Guangxi; N, samples size. *, highly endangered species. #, outgroups.
Table S3. Variable sites of the 11 mitotypes detected in Chinese subalpine and temperate firs.
Table S4. Variable sites of the 25 chlorotypes detected in Chinese subalpine and temperate firs. #, AAGA; *, ACGGA; &, TATCTA; α, ATTATTG; β, CTATTGTAAAGATATTAGTTGCTCAAGG; γ, ACTAC.
Table S5. Species names and GenBank accession numbers of DNA sequences used in this study. * means newly generated sequences.
References
Ali, K., Ahmad, H., Khan, N., and Jury, S. (2014). Future of Abies pindrow in Swat district, Northern Pakistan. J. Forestry Res. 25, 211–214. doi: 10.1007/s11676-014-0446-1
Andreia, C. T., Fabiano, S., Caroline, T., Fernanda, C., Nicole, M. V., Michel, J. F. B., et al. (2016). Phylogeography and ecological niche modelling in Eugenia uniflora (Myrtaceae) suggest distinct vegetational responses to climate change between the southern and the northern Atlantic Forest. Bot. J. Linn. Soc. 182, 670–688. doi: 10.1111/boj.12473
Awad, L., Fady, B., Khater, C., Roig, A., and Cheddadi, R. (2014). Genetic structure and diversity of the endangered fir tree of Lebanon (Abies cilicica Carr.): implications for conservation. PLoS ONE 9:e90086. doi: 10.1371/journal.pone.0090086
Axelrod, D. I., Al-Shehbaz, I., and Raven, P. H. (1996). “History of the modern flora of China,” in Proceedings of the First International Symposium on Floristic Characteristics and Diversity of East Asian plants, eds A. L. Zhang and S. G.Wu (Beijing: China Higher Education Press), 43–55.
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median–joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Brown, G. R., Gill, G. P., Kuntz, R. J., Langley, C. H., and Neale, D. B. (2004). Nucleotide diversity and linkage disequilibrium in loblolly pine. Proc. Natl. Acad. Sci. U.S.A. 101, 15255–15260. doi: 10.1073/pnas.0404231101
Bystriakova, N., Stephen, W. A., Stephen, J. R., Michael, G., Johannes, C. V., and Schneider, H. (2014). Present, past and future of the European rock fern Asplenium fontanum: combining distribution modelling and population genetics to study the effect of climate change on geographic range and genetic diversity. Ann. Bot. 113, 453–465. doi: 10.1093/aob/mct274
Cao, X. Y., Ni, J., Herzschuh, U., Wang, Y. B., and Zhao, Y. (2013). A late Quaternary pollen dataset from eastern continental Asia for vegetation and climate reconstructions: set up and evaluation. Rev. Palaeobot. Palynol. 194, 21–37. doi: 10.1016/j.revpalbo.2013.02.003
Cheddadi, R., Fady, B., François, L., Hajar, L., Suc, J. P., Huang, K., et al. (2009). Putative glacial refugia of Cedrus atlantica deduced from Quaternary pollen records and modern genetic diversity. J. Biogeogr. 36, 1361–1371. doi: 10.1111/j.1365-2699.2008.02063.x
Chen, I. C., Hill, J. K., Ohlemüller, R., Roy, D. B., and Thomas, C. D. (2011). Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026. doi: 10.1126/science.1206432
Chen, Y. H., Tang, Z. R., and Fang, J. Y. (2009). Distribution of nature reserves and status of biodiversity protection in China. Biodivers. Sci. 17, 664–674. doi: 10.3724/SP.J.1003.2009.08324
Cheng, W. C., and Fu, L. K. (1978). “Abies Mill,” in Flora Reipublicae Popularis Sinica, Vol. 7, eds W. C. Cheng and L. K. Fu (Beijing: Science Press), 55–95.
Cook, C. G., Jones, R. T., Langdon, P. G., Leng, M. J., and Zhang, E. (2011). New insights on Late Quaternary Asian palaeomonsoon variability and the timing of the Last Glacial Maximum in southwestern China. Quat. Sci. Rev. 30, 808–820. doi: 10.1016/j.quascirev.2011.01.003
Cun, Y. Z., and Wang, X. Q. (2010). Plant recolonization in the Himalaya from the southeastern Qinghai–Tibetan Plateau: geographical isolation contributed to high population differentiation. Mol. Phylogenet. Evol. 56, 972–982. doi: 10.1016/j.ympev.2010.05.007
D'Amen, M., Zimmermann, N. E., and Pearman, P. B. (2013). Conservation of phylogeographical lineages under climate change. Glob. Ecol. Biogeogr. 22, 93–104. doi: 10.1111/j.1466-8238.2012.00774.x
Davis, M. B., and Shaw, R. G. (2001). Range shifts and adaptive responses to Quaternary climate change. Science 292, 673–679. doi: 10.1126/science.292.5517.673
de Querioz, K. (1998). “The general lineage concept of species, species criteria, and the process of speciation,” in Endless Forms: Species and Speciation, eds D. J. Howard and S. H. Berlocher (Oxford: Oxford University Press), 57–75.
Dechaine, E. G., and Martin, A. P. (2004). Historic cycles of fragmentation and expansion in Parnassius smintheus (Papilionidae) inferred using mitochondrial DNA. Evolution 58, 113–127. doi: 10.1111/j.0014-3820.2004.tb01578.x
Dechaine, E. G., and Martin, A. P. (2005). Marked genetic divergence among sky island populations of Sedum lanceolatum (Crassulaceae) in the rocky mountains. Am. J. Bot. 92, 477–486. doi: 10.3732/ajb.92.3.477
Dellicour, S., and Mardulyn, P. (2014). Spads 1.0: a toolbox to perform spatial analyses on DNA sequence data sets. Mol. Ecol. Resour. 14, 647–651. doi: 10.1111/1755-0998.12200
Dubreuil, M., Riba, M., Gonza'lez-Martı'nez, S. C., Vendramin, G. G., Sebastiani, S., and Mayol, M. (2010). Genetic effects of chronic habitat fragmentation revisited: strong genetic structure in a temperate tree, Taxus baccata (Taxaceae), with great dispersal capability. Am. J. Bot. 97, 303–310. doi: 10.3732/ajb.0900148
Dupanloup, I., Schneider, S., and Excoffier, L. (2002). A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 11, 2571–2581. doi: 10.1046/j.1365-294X.2002.01650.x
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fady, B., and Conord, C. (2010). Macroecological patterns of species and genetic diversity in vascular plants of the Mediterranean basin. Div. Distrib. 16, 53–64. doi: 10.1111/j.1472-4642.2009.00621.x
Fady-welterlen, B. (2005). Is there really more biodiversity in Mediterranean forest ecosystems. Taxon 54, 905–910. doi: 10.2307/25065477
Farjon, A. (2001). World Checklist and Bibliography of Conifers, 2nd Edn. London: The Royal Botanic Gardens, Kew.
Farjon, A., and Rushforth, K. D. (1989). A classification of Abies Miller (Pinaceae). Notes R. Bot. Gard. Edinb. 46, 59–79.
Feng, Z. D., Tang, L. Y., Ma, Y. Z., Zhai, Z. X., Wu, H. N., Li, F., et al. (2007). Vegetation variations and associated environmental changes during marine isotope stage 3 in the western part of the Chinese Loess Plateau. Palaeogeogr. Palaeoclimatol. Palaeoecol. 246, 278–291. doi: 10.1016/j.palaeo.2006.10.004
Forest, F., Grenyer, R., Rouget, M., Davies, T. J., Cowling, R. M., Faith, D. P., et al. (2007). Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757–760. doi: 10.1038/nature05587
Fraser, D. J., and Bernatchez, L. (2001). Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol. Ecol. 10, 2741–2752. doi: 10.1046/j.1365-294X.2001.t01-1-01411.x
Fu, L. K. (1992). China Plant Red Data Book: Rare and Endangered Plants, Vol. 1. Beijing: Science Press.
Galbreath, K. E., Hafner, D. J., and Zamudio, K. R. (2009). When cold is better: climate-driven elevation shifts yield complex patterns of diversification and demography in an alpine specialist (American pika, Ochotona Princeps). Evolution 63, 2848–2863. doi: 10.1111/j.1558-5646.2009.00803.x
Gernandt, D. S., Magallón, S., López, G. G., Flores, O. Z., Willyard, A., and Liston, A. (2008). Use of simultaneous analyses to guide fossil based calibrations of Pinaceae phylogeny. Int. J. Plant Sci. 169, 1086–1099. doi: 10.1086/590472
Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276. doi: 10.1111/j.1095-8312.1996.tb01434.x
Hewitt, G. M. (2000). The genetic legacy of the quaternary ice ages. Nature 405, 907–913. doi: 10.1038/35016000
Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. Ser. B 359, 183–195. doi: 10.1098/rstb.2003.1388
Huck, S., Büdel, B., and Schmitt, T. (2012). Ice-age isolation, postglacial hybridization and recent population bottlenecks shape the genetic structure of Meum athamanticum in Central Europe. Flora 207, 399–407. doi: 10.1016/j.flora.2012.03.005
Inoue, K., and Berg, D. J. (2016). Predicting the effects of climate change on population connectivity and genetic diversity of an imperiled freshwater mussel, Cumberlandia monodonta (Bivalvia: Margaritiferidae), in riverine systems. Glob. Change Biol. 23, 94–107. doi: 10.1111/gcb.13369
IUCN (2016). The IUCN Red List of Threatened Species. Version 2016-3. Available online at: http://www.iucnredlist.org
Jaramillo-Correa, J. P., Aguirre-Planter, E., Khasa, D. P., Eguiarte, L. E., Piñero, D., Furnier, G. R., et al. (2008). Ancestry and divergence of subtropical montane forest isolates: molecular biogeography of the genus Abies (Pinaceae) in southern Mexico and Guatemala. Mol. Ecol. 17, 2476–2490. doi: 10.1111/j.1365-294X.2008.03762.x
Jaramillo-Correa, J. P., Beaulieu, J., and Bousquet, J. (2004). Variation in mitochondrial DNA reveals multiple distant glacial refugia in black spruce (Piceamariana), a transcontinental North American conifer. Mol. Ecol. 13, 2735–2747. doi: 10.1111/j.1365-294X.2004.02258.x
Jaramillo-Correa, J. P., Beaulieu, J., Ledig, F. T., and Bousquet, J. (2006). Decoupled mitochondrial and chloroplast DNA population structure reveals Holocene collapse and population isolation in a threatened Mexican-endemic conifer. Mol. Ecol. 15, 2787–2800. doi: 10.1111/j.1365-294X.2006.02974.x
Jay, F., Manel, S., Alvarez, N., Durand, E. Y., Thuiller, W., Holderegger, R., et al. (2012). Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol. Ecol. 21, 2354–2368. doi: 10.1111/j.1365-294X.2012.05541.x
Jezkova, T., Jaeger, J. R., Marshall, Z. R., and Riddle, B. R. (2009). Pleistocene impacts on the phylogeography of the desert pocket mouse (Chaetodipus penicillatus). J. Mammal. 90, 306–320. doi: 10.1644/08-MAMM-A-243.1
Jiang, X. L., Shao, Y. Z., Wang, W. J., and Liu, B. D. (2015). The discovery, current status and future prospects of endangered firs in China. Bull. Biol. 50, 9–11.
Jiang, Z. Y., Peng, Y. L., Hu, X. X., Zhou, Y. F., and Liu, J. Q. (2011). Cytoplasmic DNA variation in and genetic delimitation of Abies nephrolepis and Abies holophylla in northeastern China. Can. J. For. Res. 41, 1555–1561. doi: 10.1139/x11-069
Lascoux, M., Palme, A. E., Cheddadi, R., and Latta, R. G. (2004). Impact of ice ages on the genetic structure of trees and shrubs. Philos. Trans. R. Soc. Lond. B 359, 197–207. doi: 10.1098/rstb.2003.1390
Li, X. X., Tao, C., Wang, Q. X., and Cui, G. F. (2012). Characteristics of geographic distribution of four critically endangered species of Abies in subtropical China and its relationship with climate. Chin. J. Plant Ecol. 36, 1154–1164. doi: 10.3724/SP.J.1258.2012.01154
Liepelt, S., Bialozyt, R., and Ziegenhagen, B. (2002). Wind–dispersed pollen mediates postglacial gene flow among refugia. Proc. Natl. Acad. Sci. U.S.A. 99, 14590–14594. doi: 10.1073/pnas.212285399
Liepelt, S., Cheddadi, R., de Beaulieu, J. L., Fady, B., Gömöry, D., Hussendörfer, E., et al. (2009). Postglacial range expansion and its genetic imprints in Abies alba (Mill.) — a synthesis from palaeobotanic and genetic data. Rev. Palaeobot. Palynol. 153, 139–149. doi: 10.1016/j.revpalbo.2008.07.007
Liepelt, S., Mayland-Quellhorst, E., Lahme, M., and Ziegenhagen, B. (2010). Contrasting geographical patterns of ancient and modern genetic lineages in Mediterranean Abies species. Plant Syst. Evol. 284:141. doi: 10.1007/s00606-009-0247-8
Liu, T. S. (1971). A Monograph of the Genus Abies. Taipei: Department of Forestry, College of Agriculture, National Taiwan University.
Liu, J. L., and Ye, P. Y. (1977). Studies on the Quaternary sporo-pollen assemblage from shanghai and zhejiang with reference to its stratigraphic and palaeoclimatic sighficance. Acta Palaeontol. Sin. 16, 1–10.
Liu, J. Q., Sun, Y. S., Ge, X. J., Gao, L. M., and Qiu, Y. X. (2012). Phylogeographical studies of plants in China: advances in the past and directions in the future. J. Syst. Evol. 50, 267–275. doi: 10.1111/j.1759-6831.2012.00214.x
Liu, L., Hao, Z. Z., Liu, Y. Y., Wei, X. X., Cun, Y. Z., and Wang, X. Q. (2014). Phylogeography of Pinus armandii and its relatives: heterogeneous contributions of geography and climate changes to the genetic differentiation and diversification of Chinese white pines. PLoS ONE 9:e85920. doi: 10.1371/journal.pone.0085920
Loehr, J., Worley, K., Grapputo, A., Carey, J., Veitch, A., and Coltman, D. W. (2006). Evidence for cryptic glacial refugia from North American mountain sheep mitochondrial DNA. J. Evol. Biol. 19, 419–430. doi: 10.1111/j.1420-9101.2005.01027.x
Mantel, N. (1967). The detection of disease clustering and a generalization regression approach. Cancer Res. 27, 209–220.
Moritz, C., and Agudo, R. (2013). The future of species under climate change: resilience or decline? Science 341, 504–508. doi: 10.1126/science.1237190
Nagoya (2010). Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity. The Secretariat of the Convention on Biological Diversity, Toronto: ON.
Nei, M., and Li, W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273. doi: 10.1073/pnas.76.10.5269
Nicolas, V., Martinez-Vargas, J., and Hugot, J. P. (2017). Molecular data and ecological niche modelling reveal the evolutionary history of the common and Iberian moles (Talpidae) in Europe. Zool. Scr. 46, 12–26. doi: 10.1111/zsc.12189
Petit, R., Aguinagalde, I., de Beaulieu, J. L., Bittkau, C., Brewer, S., Cheddadi, R., et al. (2003). Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300, 1563–1565. doi: 10.1126/science.1083264
Petit, R. J., Duminil, J., Fineschi, S., Hampe, A., Salvini, D., and Vendramin, G. G. (2005). Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 14, 689–701. doi: 10.1111/j.1365-294X.2004.02410.x
Phillips, S. J., Anderson, R. P., and Schapire, R. E. (2006). Maximum entropy modeling of species geographical distributions. Ecol. Modell. 190, 231–259. doi: 10.1016/j.ecolmodel.2005.03.026
Pons, O., and Petit, R. J. (1996). Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237–1245.
Qian, H., Song, J. S., Krestov, P., Guo, Q. F., Wu, Z. M., Shen, X. S., et al. (2003). Large scale phytogeographical patterns in East Asia in relation to latitudinal and climatic gradients. J. Biogeogr. 30, 129–141. doi: 10.1046/j.1365-2699.2003.00807.x
Qin, A. L., Wang, M. M., Cun, Y. Z., Yang, F. S., Wang, S. S., Ran, J. H., et al. (2013). Phylogeographic evidence for a link of species divergence of Ephedra in the Qinghai-Tibetan Plateau and adjacent regions to the Miocene Asian aridification. PLoS ONE 8:e56243. doi: 10.1371/journal.pone.0056243
Qiu, Y. X., Fu, C. X., and Comes, H. P. (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and enviromrntal change in the world's most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244. doi: 10.1016/j.ympev.2011.01.012
Rogers, A. R., and Harpending, H. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569.
Rogers, S. O., and Bendich, A. J. (1988). Extraction of DNA from plant tissues. Plant Mol. Biol. 6, 1–10. doi: 10.1007/978-94-017-5294-7_6
Root, T. L., Price, J. T., Hall, K. R., Schneider, S. H., Rosenzweig, C., and Pounds, J. A. (2003). Fingerprints of global warming on wild animals and plants. Nature 421, 57–60. doi: 10.1038/nature01333
Rozas, J., Sánchez–DelBarrio, J. C., Messeguer, X., and Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. doi: 10.1093/bioinformatics/btg359
Ruosch, M., Spahni, R., Joos, F., Henne, P. D., Knaap, W. O., Tinner, W., et al. (2016). Past and future evolution of Abies alba forests in Europe – comparison of a dynamic vegetation model with palaeo data and observations. Glob. Chang. Biol. 22, 727–740. doi: 10.1111/gcb.13075
Schmitt, T., and Rákosy, L. (2007). Changes of traditional agrarian landscapes and their conservation implications: a case study of butterflies in Romania. Div. Distrib. 13, 855–862. doi: 10.1111/j.1472-4642.2007.00347.x
Shao, Y. Z., and Xiang, Q. P. (2015). Species delimitation and phylogeography of Abies chensiensis complex: inferred from morphological and molecular data. Bot. J. Linn. Soc. 177, 175–188. doi: 10.1111/boj.12235
Shi, N. (1996). Development spruce and fir in north China during the Pliocene and the early Pleistocene: palaeoclimatic implications. Q. Sci. 4, 319–328.
Shi, Y. F. (2002). Characteristics of late Quaternary monsoonal glaciation on the Tibetan Plateau and in East Asia. Q. Int. 97–98, 79–91. doi: 10.1016/S1040-6182(02)00053-8
Shi, Y. F., Cui, Z. J., and Li, J. J. (1987). Reassessment of Quaternary glaciation problems in East China. Earth Sci. 2, 45–54.
Shi, Y. F., Zheng, B. X., Su, Z., and Mu, Y. Z. (1984). Study of Quaternary glaciation in Mts. Tomur-Hantengri area, Tian Shan. J. Glaciol. Geocryol. 6, 1–14.
Sun, X. J., and Li, X. (1999). A pollen record of the last 37 ka in deep sea core 17940 from the northern slope of the South China Sea. Mar. Geol. 156, 227–244. doi: 10.1016/S0025-3227(98)00181-9
Tanaka, N., Nakao, K., Tsuyama, I., Higa, M., Nakazono, E., and Matsui, T. (2012). Predicting the impact of climate change on potential habitats of fir (Abies) species in Japan and on the East Asian continent. Proc. Environ. Sci. 13, 455–466. doi: 10.1016/j.proenv.2012.01.039
Tang, L. Y., Li, C. H., An, C. B., and Wang, W. G. (2007). Vegetation history of the western Loess Plateau of China during the last 40ka based on pollen record. Acta Palaeontol. Sin. 46, 45–61. doi: 10.3969/j.issn.0001-6616.2007.01.003
Thomas, C. D., Cameron, A., Green, R. E., Bakkenes, M., Beaumont, L. J., Collingham, Y. C., et al. (2004). Extinction risk from climate change. Nature 427, 145–148. doi: 10.1038/nature02121
Tinner, W., and Lotter, A. F. (2006). Holocene expansions of Fagus silvatica and Abies alba in central europe: where are we after eight decades of debate? Quat. Sci. Rev. 25, 526–549. doi: 10.1016/j.quascirev.2005.03.017
Waltari, E., Hijmans, R. J., Peterson, A. T., Nyári, Á. S., Perkins, S. L., and Guralnick, R. P. (2007). Locating Pleistocene refugia: comparing phylogeographical and ecological niche model predictions. PLoS ONE 2:e563. doi: 10.1371/journal.pone.0000563
Wang, J., Abbott, R. J., Peng, Y. L., Du, F. K., and Liu, J. Q. (2011). Species delimitation and biogeography of two fir species (Abies) in central China: cytoplasmic DNA variation. Heredity 94, 375–394. doi: 10.1038/hdy.2011.22
Wang, W. T., Xu, B., Zhang, D. Y., and Bai, W. N. (2016). Phylogeography of postglacial range expansion in Juglans mandshurica (Juglandaceae) reveals no evidence of bottleneck, loss of genetic diversity, or isolation by distance in the leading-edge populations. Mol. Phylogenet. Evol. 102, 255–264. doi: 10.1016/j.ympev.2016.06.005
Wei, X. Z., Sork, V. L., Meng, H. J., and Jiang, M. X. (2016). Genetic evidence for central-marginal hypothesis in a Cenozoic relict tree species across its distribution in China. J. Biogeogr. 43, 2173–2185. doi: 10.1111/jbi.12788
Wu, G. X., Qin, J. G., Deng, B., and Li, C. X. (2002). Palynomorphs in the first paleosol layer in the Yangtze Delta and their palaeoenvironmental implication. Chin. Sci. Bull. 47, 1837–1841. doi: 10.1360/02tb9401
Xiang, Q. P. (1997). Abies fansipanensis—A new species of the genus Abies from Vietnam. Acta Phytotaxonmica Sin. 35, 356–359.
Xiang, Q. P. (2001). A preliminary survey on the distribution of rare and endangered plants of Abies in China. Guihaia 21, 113–117. doi: 10.3969/j.issn.1000-3142.2001.02.007
Xiang, Q. P., Wei, R., Shao, Y. Z., Yang, Z. Y., Wang, X. Q., and Zhang, X. C. (2015). Phylogenetic relationships, possible ancient hybridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Mol. Phylogenet. Evol. 82, 1–14. doi: 10.1016/j.ympev.2014.10.008
Xiang, Q. P., Xiang, Q. Y., Guo, Y. Y., and Zhang, X. C. (2009). Phylogeny of Abies (Pinaceae) inferred from nrITS sequence data. Taxon 58, 141–152.
Xiang, Q. P., Xiang, Q. Y., Liston, A., and Zhang, X. C. (2004). Phylogenetic relationships in Abies (Pinaceae): evidence from PCR-RFLP of the nuclear ribosomal DNA internal transcribed spacer region. Bot. J. Linn. Soc. 145, 425–435. doi: 10.1111/j.1095-8339.2004.00286.x
Xiang, Q. P., Xiang, Q. Y., Liston, A., Fu, L. K., and Fu, D. Z. (2000). Length variation of the nuclear ribosomal DNA internal transcribed spacer in the genus Abies with references to its systematic utility in Pinaceae. Acta Bot. Sin. 42, 946–951. doi: 10.3321/j.issn:1672-9072.2000.09.012
Xu, Q. H., Yang, X. L., Ke, Z. M., Liang, W. D., and Yang, Z. J. (2002). Environment changes in Yanshan Mountain area during the latest Pleistocene. Geogr. Territorial Res. 18, 69–72. doi: 10.3969/j.issn.1672-0504.2002.02.019
Xu, R., Kong, Z. C., and Du, N. Q. (1980). Spruce–fir forest in pleistocene China and its significance in Quaternary Research. Q. Res. 5, 48–56.
Yan, H. F., Zhang, C. Y., Wang, F. Y., Hu, C. M., Ge, X. J., and Hao, G. (2012). Population expanding with the phalanx model and lineages split by environmental heterogeneity: a case study of Primula obconica in subtropical China. PLoS ONE 7:e41315. doi: 10.1371/journal.pone.0041315
Yi, C. L., Cui, Z. J., and Xiong, H. G. (2005). Numerical periods of Quaternary glaciations in China. Q. Sci. 25, 609–619. doi: 10.3321/j.issn:1001-7410.2005.05.010
Yi, C. L., Liu, K. X., Cui, Z. J., Jiao, K. Q., Yao, T. D., and He, Y. Q. (2004). AMS radiocarbon dating of late Quaternary glacial landforms, the source area of the Urumqi River, Tien Shan: a pilot study of 14C dating on inorganic carbon. Q. Int. 121, 99–107. doi: 10.1016/j.quaint.2004.01.026
Yi, C. L., Zhu, Z. Y., Wei, L., Cui, Z. J., Zheng, B. X., and Shi, Y. F. (2007). Advances in numerical dating of Quaternary glaciations in China. Zeitschrift Fur Geomorphol. 51, 153–175. doi: 10.1127/0372-8854/2007/0051S2-0153
Ying, T. S. (2001). Species diversity and distribution pattern of seed plants in China. Biodivers. Sci. 9, 393–398. doi: 10.3321/j.issn:1005-0094.2001.04.011
Ying, T. S., Zhang, Y. L., and Boufford, D. E. (eds.). (1993). The Endemic Genera of Seed Plants of China. Beijing: Science Press.
Zhang, J. H., Kong, Z. C., and Du, N. Q. (1997). An analysis of different sedimentary environment influence on pollen deposit in Beijing. Acta Sedimentol. Sin. 15, 57–63.
Zhao, J., Shi, Y., and Wang, J. (2011). Comparison between Quaternary glaciations in China and the Marine Oxygen Isotope Stage (MIS): an improved schema. Acta Geogr. Sin. 66, 867–884. doi: 10.11821/xb201107001
Zhou, K. S., Yan, F. H., Liang, X. l., and Ye, Y. Y. (1978). Pollen analysis of late quaternary in Beijing plain and its significance. Sci. Grol. Sinica 1, 57–66.
Keywords: conservation, ecological niche modeling, endangered species, phylogeography, Quaternary
Citation: Shao Y-Z, Zhang X-C, Phan LK and Xiang Q-P (2017) Elevation Shift in Abies Mill. (Pinaceae) of Subtropical and Temperate China and Vietnam—Corroborative Evidence from Cytoplasmic DNA and Ecological Niche Modeling. Front. Plant Sci. 8:578. doi: 10.3389/fpls.2017.00578
Received: 18 December 2016; Accepted: 30 March 2017;
Published: 18 April 2017.
Edited by:
Alessio Mengoni, University of Florence, ItalyReviewed by:
Dragos Postolache, National Research and Development Institute in Forestry “Marin Dracea”, RomaniaDavid Vieites, Consejo Superior de Investigaciones Científicas, Spain
Bruno Fady, Institut National de la Recherche Agronomique, France
Copyright © 2017 Shao, Zhang, Phan and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiao-Ping Xiang, qpxiang@ibcas.ac.cn
 Yi-Zhen Shao
Yi-Zhen Shao Xian-Chun Zhang1
Xian-Chun Zhang1 Loc Ke Phan
Loc Ke Phan