- 1Key Laboratory of Ion Beam Bioengineering and Bioenergy Forest Research Center of State Forestry Administration, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei, China
- 2Institute of Technical Biology & Agriculture Engineering, Science Island Branch of Graduate School, University of Science and Technology of China, Hefei, China
- 3School of Pharmacy, Anhui Medical University, Hefei, China
- 4The Sericultural Research Institute, Anhui Academy of Agricultural Science, Hefei, China
- 5School of Forestry and Landscape Architecture, Anhui Agricultural University, Hefei, China
Chinese tallow (Sapium sebiferum L.) is a promising landscape and bioenergy plant. Measuring gene expression by quantitative real-time polymerase chain reaction (qRT-PCR) can provide valuable information on gene function. Stably expressed reference genes for normalization are a prerequisite for ensuring the accuracy of the target gene expression level among different samples. However, the reference genes in Chinese tallow have not been systematically validated. In this study, 12 candidate reference genes (18S, GAPDH, UBQ, RPS15, SAND, TIP41, 60S, ACT7, PDF2, APT, TBP, and TUB) were investigated with qRT-PCR in 18 samples, including those from different tissues, from plants treated with sucrose and cold stresses. The data were calculated with four common algorithms, geNorm, BestKeeper, NormFinder, and the delta cycle threshold (ΔCt). TIP41 and GAPDH were the most stable for the tissue-specific experiment, GAPDH and 60S for cold treatment, and GAPDH and UBQ for sucrose stresses, while the least stable genes were 60S, TIP41, and 18S respectively. The comprehensive results showed APT, GAPDH, and UBQ to be the top-ranked stable genes across all the samples. The stability of 60S was the lowest during all experiments. These selected reference genes were further validated by comparing the expression profiles of the chalcone synthase gene in Chinese tallow in different samples. The results will help to improve the accuracy of gene expression studies in Chinese tallow.
Introduction
Chinese tallow (Sapium sebiferum L.), which belongs to the Euphorbiaceae family is native to eastern Asia (Esser, 2002). It has been cultured for biofuel production and as an ornamental plant (Jeffrey and Padley, 1991). The fruits produce a highly saturated fatty acid in the tallow layer and highly unsaturated oil in the seed (Bolley and McCormack, 1950; Boldor et al., 2010). Tallow is applied for manufacturing candles, soap, cloth, and fuel, while the seed's oil can be used for making varnishes and native paints (Brooks et al., 1987; Jeffrey and Padley, 1991). It is estimated that the tree produces 4,700 l of oil ha−1 year−1. The average commercial yields far exceed those of traditional oil seed crops (Boldor et al., 2010). Nowadays, the tree is extensively propagated for ornamental purposes. With a moderate autumn chill, it can turn flaming red, plum purple, yellow orange, or mixed colors (Gao et al., 2016). Color is one of the most important characters of ornamental plants and directly affects their ornamental and economic value; creating novelty colors has been the pursuit of plant breeders (Zhao and Tao, 2015). Anthocyanins are water-soluble flavonoids that accumulate in vacuoles, which are responsible for plant pigments. Increasing and reducing anthocyanin content are likely to make the color change (Chalker-Scott, 1999; Winkel-Shirley, 2001). Moreover, anthocyanin biosynthesis is a response to biotic and abiotic stressors, including drought, temperature extremes, nitrogen and phosphorus deficiencies, wounding, bacterial, and fungal infections (Chalker-Scott, 1999). Therefore, anthocyanins are usually considered to be a stress symptom and enhance plant tolerance to the stress factors.
It has long been suggested that sugars are involved in anthocyanin synthesis in the reproductive organ or vegetative tissues (Mita et al., 1997; Baier et al., 2004). In Vitis vinifera cells, the production of anthocyanin was increased in the sucrose-containing medium, which is quite similar to Arabidopsis thaliana (Gollop et al., 2001; Lloyd and Zakhleniuk, 2004; Solfanelli et al., 2006). The sugar signal associated with anthocyanin accumulation is intimately linked to several signaling factors such as light and hormones, but the mechanisms are not well-understood (Das et al., 2012). Moderate low-temperature stress has been demonstrated as an important factor in increasing anthocyanin in Arabidopsis, maize (Zea mays), sorghum (Sorghum bicolor), and apple (Malus domestica) (Shichijo et al., 1993; Graham, 1998; Chalker-Scott, 1999; Rodriguez et al., 2014; Wang N. et al., 2016). In Arabidopsis, cold stress (4°C) induced the accumulation of anthocyanin in the leaves and stems (Leyva et al., 1995). Low temperature also induced anthocyanin accumulation in petunia (Petunia hybrida) flowers (Shvarts et al., 1997). In Chinese tallow, we also found that cold stress can induce anthocyanin biosynthesis in the leaves (Data not shown). However, the mechanism by which low-temperature stress induces anthocyanin accumulation in Chinese tallow remains unclear.
To promote gene function studies in Chinese tallow, accurately quantifying key genes expression will facilitate a better understanding of gene function in anthocyanin biosynthesis. Techniques such as Northern blot (Kevil et al., 1997), in situ hybridization (Ogilvie et al., 1990), RNase protection assay (Carey et al., 2013), serial analysis of gene expression (SAGE) (Velculescu et al., 1997), or quantitative real-time PCR (qRT-PCR) are often used to quantify gene expression (Kevil et al., 1997). Nowadays, qRT-PCR has become the preferred method due to its high sensitivity, specificity, and accuracy in detecting the target gene expression (Bustin et al., 2005). Moreover, qRT-PCR is fast, easy to use, and highly reproducible. It requires only a minimal amount of RNA and prevents the use of radioactivity (Radonic et al., 2004). The prerequisite for reliable gene expression analysis by qRT-PCR is the selection of appropriate reference genes, which can minimize the variations caused by it (Huggett et al., 2005). The reference genes are normally housekeeping genes, which are stably expressed in various samples across different experimental conditions or treatments (Thellin et al., 1999, 2009). In past decades, a series of reference genes, such as β-actin (ACT), ubiquitin (UBQ), 18S ribosomal RNA (18S rRNA), and glyceraldehyde-3-phoshate dehydrogenase (GAPDH), have been widely used for gene expression analysis in plants (Bustin, 2002; Kim et al., 2003; Brunner et al., 2004; Czechowski et al., 2005). However, no reference genes are universal across different species and experimental conditions. For example, GAPDH has been regarded as a stable reference gene in coffee (Coffea) (Goulao et al., 2012), bermudagrass (Cynodon dactylon) (Chen et al., 2015), and physic nut (Jatropha curcas) (Zhang et al., 2013) under a series of experimental conditions. Nevertheless, GAPDH was the least stable reference gene in petunia (Mallona et al., 2010), tea plant (Camellia sinensis) (Wu et al., 2016), peach (Prunus persica) (Tong et al., 2009), and papaya (Carica papaya) (Zhu et al., 2012). Therefore, the selection of appropriate reference genes under different experimental conditions is essential for improving the accuracy and reliability of qRT-PCR. Typically, a set of candidate reference genes is evaluated in a pilot experiment with several statistical algorithms, such as geNorm (Vandesompele et al., 2002), BestKeeper (Pfaffl et al., 2004), NormFinder (Andersen et al., 2004), and the delta cycle threshold (ΔCt) (Silver et al., 2006) to identify the most suitable reference genes for qRT-PCR analysis.
Until now, no systematic validation of reference genes has been performed in Chinese tallow (Wang Y. et al., 2016). Therefore, we evaluated the expression stability of 12 genes (18S, GAPDH, UBQ, RPS15, SAND, TIP41, 60S, ACT7, PDF2, APT, TBP, and TUB) among the tissues of the seedlings and under sucrose and low- temperature stress treatments by qRT-PCR. To validate the reliability of the reference genes, the expression levels of the chalcone synthase gene in Chinese tallow (SsCHS) were analyzed. Chalcone synthase (CHS, EC 2.3.1.74) is a key enzyme in the first step for the biosynthesis of flavonoid and anthocyanin pigments in plants. It yields naringenin chalcone by condensing one p-coumaroyl-COA and three malonyl-CoA. In plants, CHS is activated by a wide range of environmental and developmental stimuli. Many studies have shown that the CHS gene's expression can be induced by light/UV light and in response to circadian clock, phytopathogens, elicitors or wounding, resulting in the enhanced production of flavonoids. In Arabidopsis, cold stress and sucrose can both directly induce AtCHS expression in leaves after treatment (Leyva et al., 1995). SsCHS is a homolog of the CHS gene from Arabidopsis. Thus, it would be expected to have similar expression patterns to AtCHS under abiotic and biotic stress conditions. Our results give an accurate and suitable reference gene selection for gene expression normalization, providing basic results for future gene expression studies on Chinese tallow tree.
Materials and Methods
Plant Materials and Stress Treatments
The seeds of Chinese tallow were collected from the experimental field at the Hefei Institutes of Physical Science (31°54′N, 117°10′E) of the Chinese Academy of Sciences (CASHIPS) located in Hefei, Anhui Province. Seeds were pre-treated with NaOH for 2 h to scrub the outer cellular tallow layer surrounding the seed coat, then stored in sand at room temperature for 3 months for germination (Li et al., 2011). Germinated seeds were transferred into the growth chamber at 25 ± 2°C under a 14/10 h (light/dark cycle) photoperiod with an irradiance of 40 μmol m−2 s−1 provided by cool white fluorescent lamps (Philips, India). Each group of three pots of 3-week-old seedlings with consensus growth status was selected and treated with sucrose and low temperature, respectively. For the low temperature treatment, seedlings were transferred into 4°C with the rest of the growing conditions being the same. For the sucrose treatment, 200 mM of sucrose (Sangon, China) solution was directly applied to the leaves. The leaves were collected at 0, 3, 6, 12, 24, 48, and 72 h after treatment according to the experiments conducted in Arabidopsis (Leyva et al., 1995; Solfanelli et al., 2006). The tissue samples from the root, stem, cotyledon, young leaf, and mature leaf were collected from the 3-week-old Chinese tallow seedlings. All samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction. Three biological replicates were collected for each sample.
Total RNA Extraction and cDNA Synthesis
Total RNA was extracted using the E.Z.N.A.® Plant RNA Kit (Omega Bio-tec, USA) according to the manufacturer's protocol. The quantity and quality of the RNA were analyzed by 1.2% (w/v) agarose gel electrophoresis and a ScanDrop® Spectrophotometer (Analytik Jena AG, Jena, Germany). cDNA was synthesized by the TransScript® RT Reagent Kit (TransGen Biotech, Peking, China) according to the manufacturer's protocol. The cDNA was diluted 10-fold with RNAase-free water for qRT-PCR.
Primer Design and qRT-PCR Analysis
The qRT-PCR primers (Table 1) were designed using the software Primer Premier 5. To verify the sequences of the reference genes, all the amplicons of the 12 candidate genes were analyzed by electrophoresis and sequencing. Each PCR reaction contained 10 μL of TransStart Top Green qPCR SuperMix (TransGen Biotech, Peking, China), 200 nM of each primer, and 2 μL of diluted cDNA in a total volume of 20 μL. No template controls (NTC) had water instead of cDNA as a template. The qRT-PCR reactions were performed with the qTOWER 2.0 Real-time PCR Detection System (Analytik Jena AG, Jena, Germany). The reactions were conducted under the following conditions: 30 s at 94°C for the initial denaturation, followed by 45 cycles of 5 s at 94°C, 15 s at the optimal temperature for each primer pair (Table 1), and 10 s at 72°C for PCR amplification. After 45 cycles, the dissociation curve was determined to confirm the specificity of each primer again by heating the product from 60 to 95°C. The normalized reporter (Rn) threshold was automatically selected to obtain the cycle threshold (Ct) values. The amplification efficiency of each set of primers was tested prior to the expression studies and calculated as E = −1 + 10 (−1/slope) (Pfaffl, 2001), where the slope was derived from a standard curve generated by five-fold serial dilutions of cDNA obtained from young leaves.
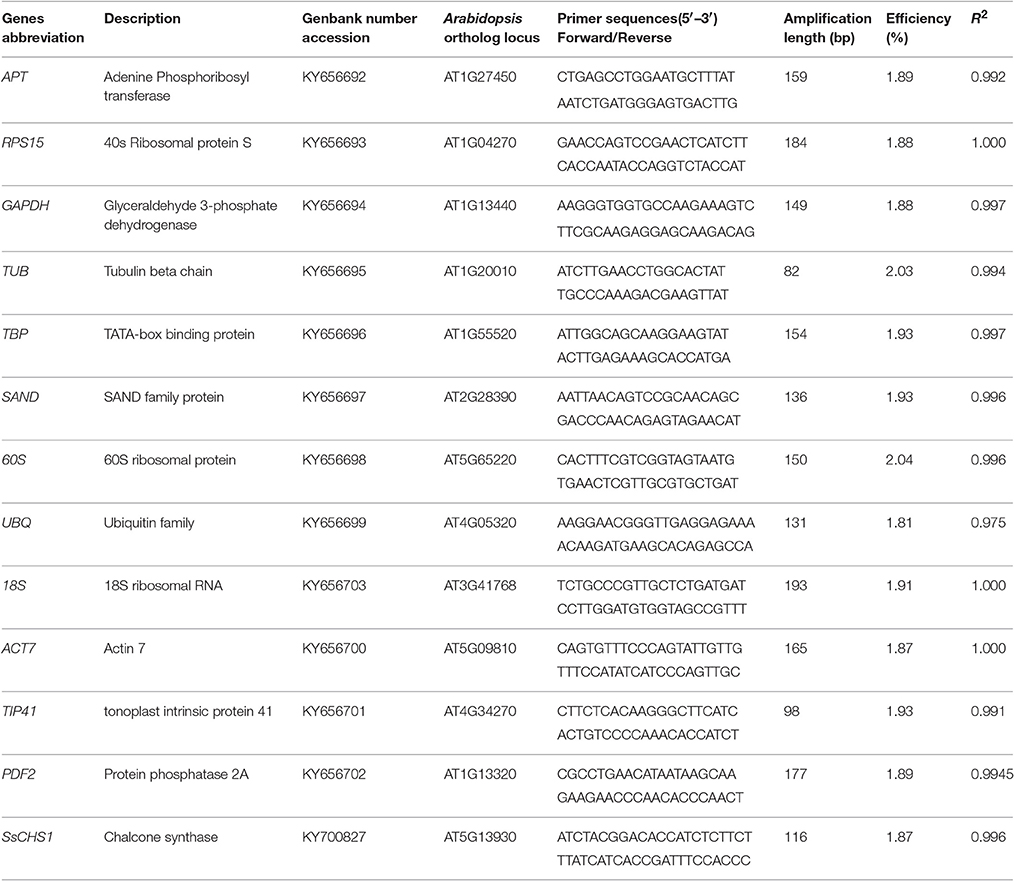
Table 1. Genes description, amplification length, and PCR efficiency for Sapium sebiferum RGs selection.
Data Analysis of Gene Expression Stability
Four algorithms, geNorm v. 3.5, BestKeeper, NormFinder, and ΔCt were used to rank the stability of the candidate reference genes. The pairwise variation (Vn/Vn+1) between two sequential normalization factors was obtained by geNorm software to determine the optimal number of reference genes needed for normalization. The recommended cut-o? threshold was 0.15, below which an additional control gene was not required for normalization. RefFinder (http://150.216.56.64/referencegene.php) was used to rank the stability of these candidates by comparing and integrating all four algorithms.
Validation of Reference Genes
To validate the reliability of the reference genes, the expression levels of the CHS gene from Chinese tallow were analyzed via the top-ranking reference genes, as recommended by RefFinder, alone or with a combination for data normalization. In comparison, the least stable reference genes were also used. The statistical tests of gene expression data were calculated by one-way ANOVA. Statistical significance was considered at *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
RNA Quality, PCR Specificity, and Amplification Efficiency of the Candidate Reference Genes
To detect the RNA's quality and quantity, OD260, OD280, and OD230 were measured by the ScanDrop® Spectrophotometer. The OD260/OD280 ratios were between 1.8 and 2.0, and the OD260/OD230 ratios were higher than 2.0 (Table S1). This suggests high purity and freedom from organic contamination of the RNA sample. The purity and integrity of the RNA were also checked by agarose gel electrophoresis, and three RNA bands and no genomic DNA was observed (Figure S1). In this study, we selected 12 candidate reference genes through the homology search from the Chinese tallow transcriptome according to the stable reference genes reported in Arabidopsis. The primers for qRT-PCR were designed according to unigene sequences obtained from Chinese tallow tree transcriptome (Table 1). The sizes of all PCR products were from 82 to 193 bp (Table 1). All primer pairs showed a single major peak in the melting curve analysis (Figure S2) and a single band at the expected size by 2% agarose gel electrophoresis (Figure S3). No signal or band was detected in the negative control. The amplification efficiency of the primers varied from 0.81 to 1.07, and the correlation coefficients (R2) of the standard curve ranged from 0.9753 to 0.9999 (Table 1).
Ct Value Distribution and Expression Profile of the 12 Reference Genes
The expression levels of the 12 selected reference genes were confirmed with the threshold cycle values. All qRT-PCR assays were performed on three biological replicates. Most of the genes' Ct values varied from 17 to 25 (Figure 1). The 18S gene showed the highest expression level in different samples, with the lowest Ct values being from 8.39 ± 0.46 to 12.7 ± 0.43. The APT gene showed the lowest expression level in the root, with the highest Ct values of 25.76 ± 0.2 (Table S2).
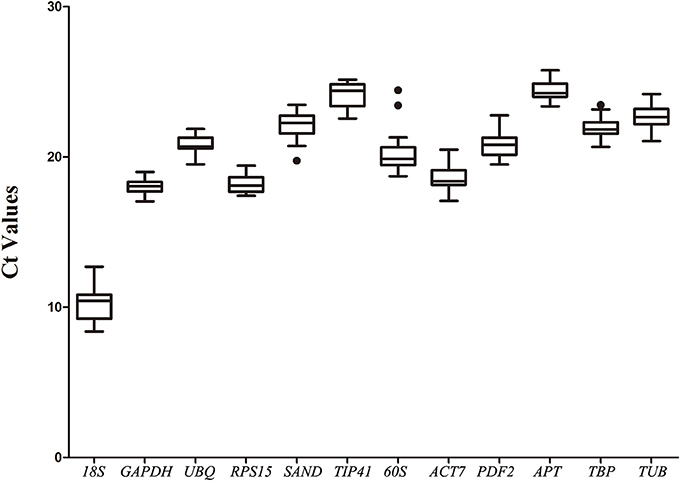
Figure 1. Expression levels of 12 candidate reference genes across all experimental samples. The box graph indicates the interquartile range, the median, and maximum/minimum values. Dots indicate outliers.
GeNorm Analysis
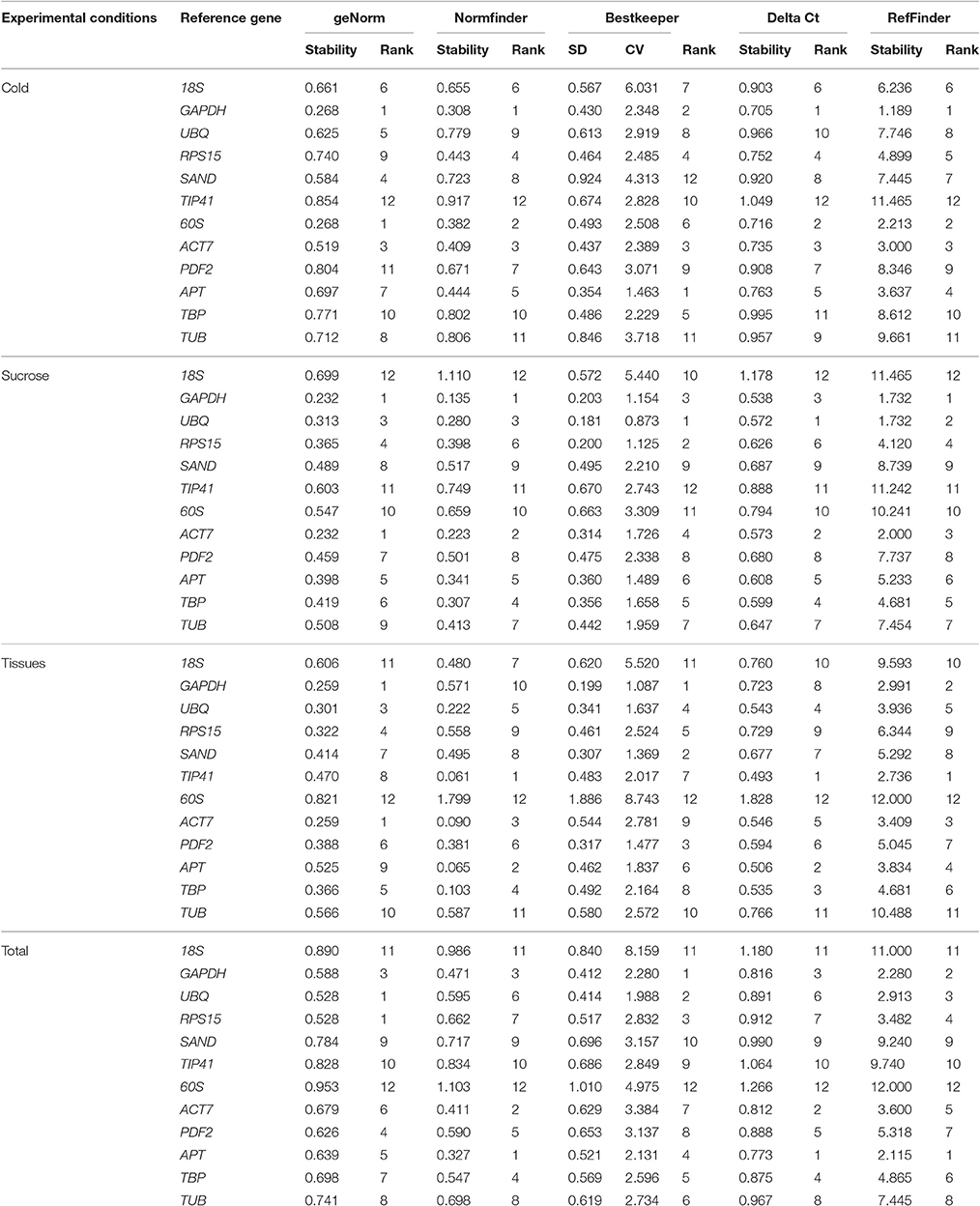
The geNorm software used in this study was used to evaluate the stability of the 12 candidate reference genes by calculating the gene expression stability (M). Genes with the lowest M value were considered to be most stable. As shown in Table 2, the M values of almost all genes were no more than the recommended threshold of 1.5. Cold stress treatment revealed that GAPDH and 60S were the most stable genes. Sucrose stress results and tissue-specific experiments analysis showed that GAPDH and ACT7 were the most stable genes. When the total samples were taken into account, the most stable genes were RPS15 and UBQ, while 60S was the least stable. The optimal number of reference gene analysis was based on average pairwise variation Vn/n+1 with a cut-off score of 0.15,below which the inclusion of an additional reference gene was not required. Of the three groups, V2/3 all had a score of < 0.15 (Figure 2). Notably, two reference genes were sufficient for accurate normalization. In consideration of the data from the whole groups, three genes were suitable for all samples in this study (Figure 2).
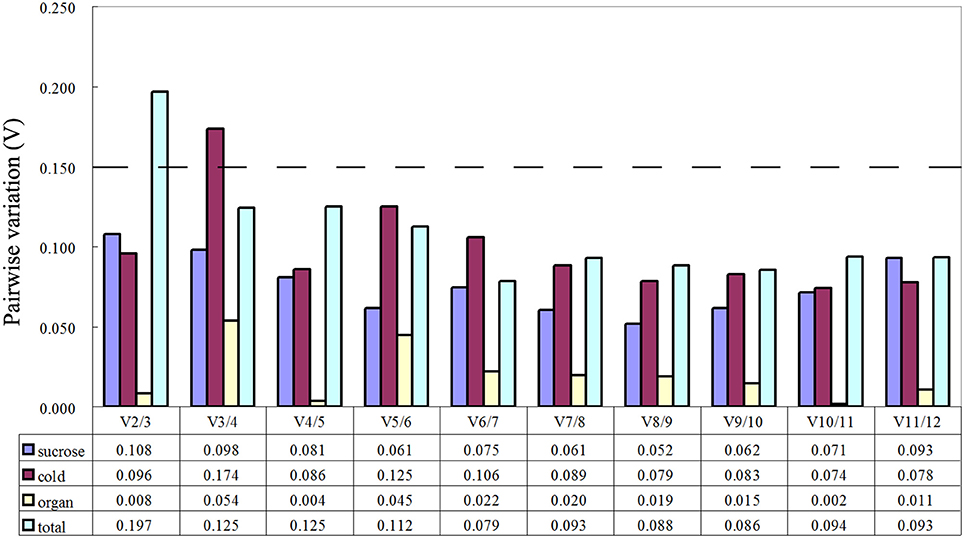
Figure 2. Determination of the optimal number of reference genes for normalization by pairwise variation (V) using geNorm. The pairwise variation (Vn/Vn+1) was calculated between normalization factors NFn and NFn+1, by geNorm to determine the optimal number of reference genes for qRT-PCR data normalization.
Normfinder Analysis
Normfinder was employed to verify a more stable gene based on intra- group and inter- group variation. Similar to the GeNorm results, the M-value was also negatively correlated with its stability. In tissue-specific experiments, TIP41 was the most stable gene. The cold-stress results and sucrose-stress treatment experiment both showed that GAPDH was the most stable gene. However, considering the M -value in all the above mentioned samples, the most stable reference gene was APT (Table 2).
Bestkeeper Algorithm
Bestkeeper ranks the genes' stability by computing the standard deviation (SD) value and coefficient of variance (CV). The lowest CV and SD (CV ± SD) means the most stable reference gene. SD values higher than 1 can be excluded (Table 2). GAPDH was the most stable gene for the tissue-specific experiment, while UBQ showed remarkably stable expression in the sucrose-stress treatment experiments. In the cold-stress treatment experiments, APT was identified as the most stable gene. GAPDH was the most stable gene for all the experiments.
ΔCt Method
The ΔCt method identifies the most stable genes by relative pair-wise comparisons. UBQ was the most stable gene in the sucrose treatment while GAPDH was the most stable in the cold treatment. APT was the most stable gene for the tissue-specific experiment and entire dataset (Table 2).
Comprehensive Stability Analysis of Reference Ggenes by Reffinder
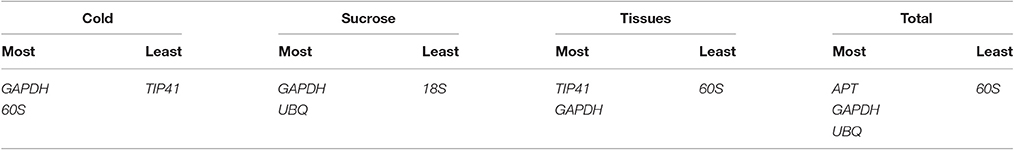
Data generated by the four algorithms were further compared with the web-based comprehensive tool RefFinder. The results of the merged data revealed that the two best RGs for normalization were GAPDH and 60S for the cold treatment, GAPDH and UBQ for the sucrose treatment, and GAPDH and TIP41 for the tissue-specific experiments. Considering all the experiments, the combination of UBQ, GAPDH, and APT as reference genes was the best choice. Based on the number of RGs suggested by geNorm and the ranking list suggested by RefFinder, the best combination of RGs for each treatment is shown in Table 3.
Expression Analysis of SsCHS Genes for Reference Gene Validation
To confirm the validation of the reference genes, the SsCHS gene was examined under different experimental conditions with qRT-PCR. The results showed that similar to the expression pattern of AtCHS studied in Arabidopsis, the gene expression level of SsCHS was significantly induced by cold- and sucrose-stress treatments when normalized to the most stable reference genes. However, the gene expression values were quite different when different reference genes (most stable, a combination of most stable, least stable) were employed for normalization. Under the cold treatment, when the least stable gene, TIP41, was used for normalization, the maximum relative expression level of SsCHS was up-regulated 26.33 fold at 24 h, but when the most stable gene, GAPDH, was applied, the maximum relative expression level was up-regulated 114.22 at 72 h, which was more consistent with the maximum value of 91.02 at 72 h calculated with the combination of 60S and GAPDH (Figure 3A). Under the sucrose treatment, when normalized with the most stable (GAPDH) and combination of reference genes (GAPDH and UBQ), the SsCHS expression patterns were basically the same as the level of the product continuous induced. However, the result was different without upregulation when normalized with the least stable gene 18S (Figure 3B). For tissue-type analysis, when stable reference genes were used for normalization, SsCHS was expressed highly in the root, followed by cotyledon, young leaf, stem, and old leaf. However, the expression level of SsCHS in the root was overestimated when using the least stable reference gene (60S) for normalization (Figure 3C). These findings suggest that the choice of reliable reference genes is essential for the accurate normalization of target gene expression levels.
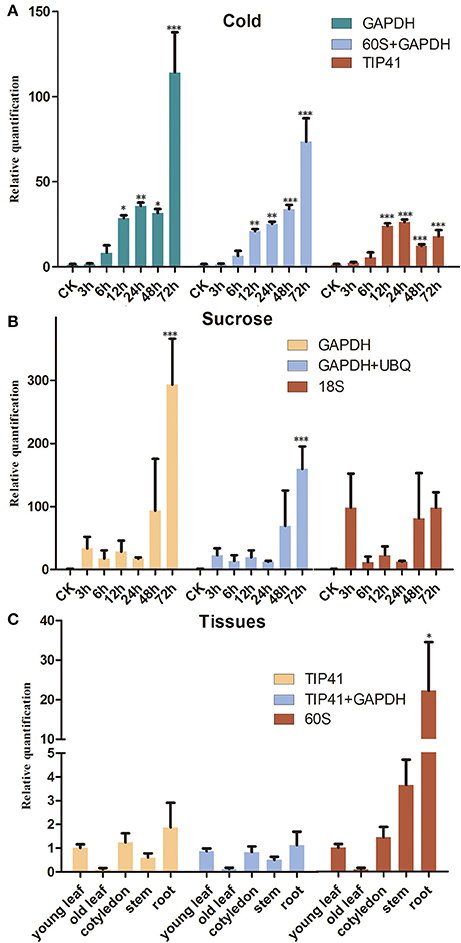
Figure 3. Relative quantification of SsCHS expression using the validated reference gene(s). (A) Leaves were collected from 3-week-old seedlings subjected to cold-stress after 0, 3, 6, 12, 24, 48, and 72 h of treatment. (B) Leaves were collected from 3-week-old seedlings subjected to sucrose-stress after 0, 3, 6, 12, 24, 48, and 72 h of treatment. (C) Different tissues were collected from 3-week-old seedlings. * indicates statistically significant (p < 0.05); **Indicates greatly statistically significant (p < 0.01); ***Indicates greatly statistically significant (p < 0.001). The results are depicted as the mean ± SD (n = 3).
Discussion
qRT-PCR is broadly used for the measurement of target gene expression. According to the “Minimum Information for publication of Quantitative real-time PCR Experiments” (MIQE) guidelines, using reference genes as the internal control is the most suitable normalization method for qRT-PCR analysis (Bustin et al., 2009). However, no single reference gene can be taken for quantifying the target genes expression levels under all conditions (Pfaffl et al., 2004; Huis et al., 2010). To obtain accurate results, it is essential to select suitable reference genes which were stably expressed in different tissues or under specific experiment conditions.
In this study, we report the systematic analysis of 12 reference genes, commonly used in other plant gene expression studies, in Chinese tallow under cold stress, sucrose stress, and in different tissues of the seedlings. The 12 genes were chosen in this paper based on previous studies, including both commonly used reference genes as well as new reference genes. In past studies, 18S was taken as a reference gene for testing the expression of Diacylglycerol acyltransferases (DGATs) in Chinese tallow (Wang Y. et al., 2016). 18S is generated from a large, common precursor RNA (pre-rRNA). It has been exploited as a reference gene for a long time. However, in many species, such as Oxytropis ochrocephala Bunge and rubber tree, it was the least stable reference gene (Long et al., 2015; Zhuang et al., 2015). In addition, candidate genes showing high-level variation should be avoided as internal controls (Wang et al., 2015). Our results showed that the expression level of 18S was too high and unstable compared with other reference genes, which is consistent with the results from papaya under many experimental conditions (Zhu et al., 2012).
In addition, four available methods (ΔCt, BestKeeper, NormFinder, and geNorm) were utilized for evaluating the stability of the expression levels of 12 candidate reference genes in five tissues and under two experimental treatments. The results showed that the least stable genes were nearly the same, while the most stable genes varied. For example, under the cold treatment, ΔCt, NormFinder, and geNorm all ranked GAPDH as the most stable gene, while BestKeeper ranked APT as the most stable. Then we used the RefFinder tool to compare and rank the 12 candidate reference genes based on the integrated results from each above mentioned program.
Many studies have proved that the accuracy of qRT-PCR can be improved by using more than one reference gene. In this study, two reference genes are needed for more accurate normalization under two stress treatment and in different plant tissues evaluated by geNorm software. The final ranking showed that GAPDH and 60S were the most stable genes through the cold treatment. GAPDH and UBQ performed best as reference genes under the sucrose treatment. GAPDH has been demonstrated as a stable RG in physic nut, coffee and carrot (Daucus carota) under a series of experimental condition (Goulao et al., 2012; Zhang et al., 2013; Tian et al., 2015). However, it was the least stable reference gene in petunia, tea plant, peach, and papaya (Tong et al., 2009; Mallona et al., 2010; Zhu et al., 2012; Wu et al., 2016). 60S was suggested as the most stable internal control in safflower (Carthamus tinctorius) and populus (Populus euphratica) under cold stress (Wang et al., 2014). UBQ was a stable reference gene proposed in different vegetative and reproductive tissues of sexual and apomictic Boechera (Pellino et al., 2011). It was also employed as internal controls for gene expression analysis in a wide variety of stages of longan somatic embryogenesis cultured under different temperatures (Lin and Lai, 2010). TIP41 and GAPDH were selected as the stable reference genes in the tissues (young leaf, old leaf, stem, cotyledon, and root) of 3-week-old seedlings of Chinese tallow. TIP41 was used as a reference gene for tissues (leaves, stems, cotyledons, hypocotyls, and roots) in peanut (Arachis hypogaea) (Chi et al., 2012) and bermudagrass leaves under heat stress (Chen et al., 2015).
To evaluate the suitability of the selected reference genes in this study, the expression levels of the CHS gene were examined in Chinese tallow. CHS encodes the first dedicated enzyme-CHS in the anthocyanin biosynthesis pathway, which catalyzes the stepwise condensation of three molecules of malonyl-CoA to one molecule of 4-coumaroyl-CoA to yield naringenin chalcone (Dooner et al., 1991). Anthocyanin is widely synthesized in seed plants to provide coloration, protection against various stresses, and components for cellular activities (Mol et al., 1998). In Chinese tallow, one of the most important colored tree species in China, anthocyanin is the primary substance that contributes to the leaf color (Feild et al., 2001). In addition, anthocyanin promotes the tree's ability to adapt to the environment. The study revealed that low-temperature-induced anthocyanin accumulation in red orange vesicles reached values eight times higher than those kept at 25°C after treatment for 75 days. Furthermore, qRT-PCR confirmed that the expression of PAL, CHS, DFR, and UFGT was strongly induced by low temperature, since the expression levels of all transcripts increased at least 40-fold compared with the control samples (Lo Piero et al., 2005). In Arabidopsis, sucrose treatment can strongly up-regulate the expression of key genes involved in anthocyanin biosynthetic pathways and anthocyanin content (Solfanelli et al., 2006).
Here, in Chinese tallow seedlings, SsCHS was remarkably continuously induced by the cold and sucrose treatments. For tissue-specific analysis, SsCHS was highly expressed in the root with a relatively lower expression level in old leaves when the most stable reference genes were used for normalization. Anthocyanins accumulated in non-photosynthetic tissues, were postulated to act as modulators of reactive oxygen signaling cascades involved in plant growth and development. These results conclusively provide the information required for selecting reliable reference genes in gene expression studies and ensure more accurate and reliable gene expression quantification in Chinese tallow.
Author Contributions
XC and YM conceived and designed the experiments. XC, YM, and JN performed the experiments. XC and YM analyzed the data. SH, WL, ML, YW, JH, and QW contributed reagents/materials. XC, JN, and WZ wrote the manuscript. LW edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study is supported by National Natural Science Foundation of China (11375232), Special Fund for Forest Scientific Research in the Public Welfare (201404110), the Science and Technology Service program of Chinese Academy of Sciences (KFJ-EW-STS-083), the Grant of the President Foundation of Hefei Institutes of Physical Science of Chinese Academy of Sciences (YZJJ201502) as well as the Key deployment project of Chinese Academy of Sciences (KFZD-SW-105).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00637/full#supplementary-material
Figure S1. Agarose gel (1.2 %) electrophoresis of the RNA.
Figure S2. Melting curves of 12 reference genes showing single peaks.
Figure S3. Agarose gel (2%) electrophoresis showing amplification of a single PCR product of the expected size; M represents DL2000 DNA marker.
Table S1. RNA purity.
Table S2. Cycle threshold (Ct) values for twelve candidate reference genes.
References
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Baier, M., Hemmann, G., Holman, R., Corke, F., Card, R., Smith, C., et al. (2004). Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol. 134, 81–91. doi: 10.1104/pp.103.031674
Boldor, D., Kanitkar, A., Terigar, B. G., Leonardi, C., Lima, M., and Breitenbeck, G. A. (2010). Microwave assisted extraction of biodiesel feedstock from the seeds of invasive chinese tallow tree. Environ. Sci. Technol. 44, 4019–4025. doi: 10.1021/es100143z
Bolley, D. S., and McCormack, R. H. (1950). Utilization of the seed of the Chinese tallow tree. J. Am. Oil Chem. Soc. 27, 84–87. doi: 10.1007/BF02634394
Brooks, G., Morrice, N. A., Ellis, C., Aitken, A., Evans, A. T., and Evans, F. J. (1987). Toxic phorbol esters from Chinese tallow stimulate protein kinase C. Toxicon 25, 1229–1233. doi: 10.1016/0041-0101(87)90141-3
Brunner, A. M., Yakovlev, I. A., and Strauss, S. H. (2004). Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 4:14. doi: 10.1186/1471-2229-4-14
Bustin, S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29, 23–39. doi: 10.1677/jme.0.0290023
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Bustin, S. A., Benes, V., Nolan, T., and Pfaffl, M. W. (2005). Quantitative real-time RT-PCR - a perspective. J. Mol. Endocrinol. 34, 597–601. doi: 10.1677/jme.1.01755
Carey, M. F., Peterson, C. L., and Smale, S. T. (2013). The RNase protection assay. Cold Spring Harb. Protoc. 2013:pdb.prot071910. doi: 10.1101/pdb.prot071910
Chalker-Scott, L. (1999). Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 70, 1–9. doi: 10.1111/j.1751-1097.1999.tb01944.x
Chen, Y., Tan, Z., Hu, B., Yang, Z., Xu, B., Zhuang, L., et al. (2015). Selection and validation of reference genes for target gene analysis with quantitative RT-PCR in leaves and roots of bermudagrass under four different abiotic stresses. Physiol. Plant. 155, 138–148. doi: 10.1111/ppl.12302
Chi, X., Hu, R., Yang, Q., Zhang, X., Pan, L., Chen, N., et al. (2012). Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol. Genet. Genomics 287, 167–176. doi: 10.1007/s00438-011-0665-5
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
Das, P. K., Shin, D. H., Choi, S. B., and Park, Y. I. (2012). Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol. Cells 34, 501–507. doi: 10.1007/s10059-012-0151-x
Dooner, H. K., Robbins, T. P., and Jorgensen, R. A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25, 173–199. doi: 10.1146/annurev.ge.25.120191.001133
Feild, T. S., Lee, D. W., and Holbrook, N. M. (2001). Why leaves turn red in autumn. The role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiol. 127, 566–574. doi: 10.1104/pp.010063
Gao, R., Su, Z., Yin, Y., Sun, L., and Li, S. (2016). Germplasm, chemical constituents, biological activities, utilization, and control of Chinese tallow (Triadica sebifera (L.) Small). Biol. Invasions 18, 809–829. doi: 10.1007/s10530-016-1052-2
Gollop, R., Farhi, S., and Perl, A. (2001). Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Sci. 161, 579–588. doi: 10.1016/S0168-9452(01)00445-9
Goulao, L. F., Fortunato, A. S., and Ramalho, J. C. (2012). Selection of reference genes for normalizing quantitative Real-Time PCR gene expression data with multiple variables in Coffea spp. Plant Mol. Biol. Rep. 30, 741–759. doi: 10.1007/s11105-011-0382-6
Graham, T. L. (1998). Flavonoid and flavonol glycoside metabolism in Arabidopsis. Plant Physiol. Biochem. 36, 135–144. doi: 10.1016/S0981-9428(98)80098-3
Huggett, J., Dheda, K., Bustin, S., and Zumla, A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6, 279–284. doi: 10.1038/sj.gene.6364190
Huis, R., Hawkins, S., and Neutelings, G. (2010). Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 10:71. doi: 10.1186/1471-2229-10-71
Jeffrey, B. S. J., and Padley, F. B. (1991). Chinese vegetable tallow-characterization and contamination by stillingia oil. J. Am. Oil Chem. Soc. 68, 123–127. doi: 10.1007/BF02662332
Kevil, C. G., Walsh, L., Laroux, F. S., Kalogeris, T., Grisham, M. B., and Alexander, J. S. (1997). An improved, rapid Northern protocol. Biochem. Biophys. Res. Commun. 238, 277–279. doi: 10.1006/bbrc.1997.7284
Kim, B. R., Nam, H. Y., Kim, S. U., Kim, S. I., and Chang, Y. J. (2003). Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 25, 1869–1872. doi: 10.1023/A:1026298032009
Leyva, A., Jarillo, J. A., Salinas, J., and Martinezzapater, J. M. (1995). Low-temperature induces the accumulation of phenylalanine ammonialyase and chalcone synthase messenger-rnas of Arabidopsis thaliana in a light-dependent manner. Plant Physiol. 108, 39–46. doi: 10.1104/pp.108.1.39
Li, S., Liu, J., Tian, S., and Yin, T. (2011). Study on the causes of Sapium sebiferum seed dormancy and the methods for dormancy breaking. J. Nanjing For. Univ. Nat. Sci. Ed. 35, 1–4. doi: 10.3969/j.issn.1000-2006.2011.05.001
Lin, Y. L., and Lai, Z. X. (2010). Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 178, 359–365. doi: 10.1016/j.plantsci.2010.02.005
Lloyd, J. C., and Zakhleniuk, O. V. (2004). Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J. Exp. Bot. 55, 1221–1230. doi: 10.1093/jxb/erh143
Long, X., He, B., Gao, X., Qin, Y., Yang, J., Fang, Y., et al. (2015). Validation of reference genes for quantitative real-time PCR during latex regeneration in rubber tree. Gene 563, 190–195. doi: 10.1016/j.gene.2015.03.026
Lo Piero, A. R., Puglisi, I., Rapisarda, P., and Petrone, G. (2005). Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J. Agric. Food Chem. 53, 9083–9088. doi: 10.1021/jf051609s
Mallona, I., Lischewski, S., Weiss, J., Hause, B., and Egea-Cortines, M. (2010). Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 10:4. doi: 10.1186/1471-2229-10-4
Mita, S., Murano, N., Akaike, M., and Nakamura, K. (1997). Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for beta-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J. 11, 841–851. doi: 10.1046/j.1365-313X.1997.11040841.x
Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217. doi: 10.1016/S1360-1385(98)01242-4
Ogilvie, A. D., Wood, N. C., Dickens, E., Wojtacha, D., and Duff, G. W. (1990). In situ hybridisation. Ann. Rheum. Dis. 49(Suppl. 1), 434–439.
Pellino, M., Sharbel, T. F., Mau, M., Amiteye, S., and Corral, J. M. (2011). Selection of reference genes for quantitative real-time PCR expression studies of microdissected reproductive tissues in apomictic and sexual Boechera. BMC Res. Notes 4:303. doi: 10.1186/1756-0500-4-303
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Radonic, A., Thulke, S., Mackay, I. M., Landt, O., Siegert, W., and Nitsche, A. (2004). Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862. doi: 10.1016/j.bbrc.2003.11.177
Rodriguez, V., Butron, A., Rady, M. O. A., Soengas, P., and Revilla, P. (2014). Identification of quantitative trait loci involved in the response to cold stress in maize (Zea mays L.). Mol. Breed. 33, 363–371. doi: 10.1007/s11032-013-9955-4
Shichijo, C., Hamada, T., Hiraoka, M., Johnson, C. B., and Hashimoto, T. (1993). Enhancement of red-light-induced anthocyanin synthesis in sorghum first internodes by moderate low temperature given in the pre-irradiation culture period. Planta 191, 238–245. doi: 10.1007/BF00199755
Shvarts, M., Weiss, D., and Borochov, A. (1997). Temperature effects on growth, pigmentation and post-harvest longevity of petunia flowers. Sci. Hortic. 69, 217–227. doi: 10.1016/S0304-4238(97)00015-0
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. doi: 10.1186/1471-2199-7-33
Solfanelli, C., Poggi, A., Loreti, E., Alpi, A., and Perata, P. (2006). Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 140, 637–646. doi: 10.1104/pp.105.072579
Thellin, O., ElMoualij, B., Heinen, E., and Zorzi, W. (2009). A decade of improvements in quantification of gene expression and internal standard selection. Biotechnol. Adv. 27, 323–333. doi: 10.1016/j.biotechadv.2009.01.010
Thellin, O., Zorzi, W., Lakaye, B., De Borman, B., Coumans, B., Hennen, G., et al. (1999). Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75, 291–295. doi: 10.1016/S0168-1656(99)00163-7
Tian, C., Jiang, Q., Wang, F., Wang, G. L., Xu, Z. S., and Xiong, A. S. (2015). Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot Leaves. PLoS ONE 10:e0117569. doi: 10.1371/journal.pone.0117569
Tong, Z., Gao, Z., Wang, F., Zhou, J., and Zhang, Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10:71. doi: 10.1186/1471-2199-10-71
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Velculescu, V. E., Zhang, L., Zhou, W., Polyak, K., Basrai, M., Bassett, D., et al. (1997). Serial analysis of gene expression (SAGE). Am. J. Hum. Genet. 61:A36.
Wang, H. L., Chen, J., Tian, Q., Wang, S., Xia, X., and Yin, W. (2014). Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol. Plant. 152, 529–545. doi: 10.1111/ppl.12206
Wang, H. L., Li, L., Tang, S., Yuan, C., Tian, Q., Su, Y., et al. (2015). Evaluation of appropriate reference genes for reverse transcription-quantitative PCR studies in different tissues of a desert poplar via comparision of different algorithms. Int. J. Mol. Sci. 16, 20468–20491. doi: 10.3390/ijms160920468
Wang, N., Zhang, Z. Y., Jiang, S. H., Xu, H. F., Wang, Y. C., Feng, S. Q., et al. (2016). Synergistic effects of light and temperature on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Organ Cult. 127, 217–227. doi: 10.1007/s11240-016-1044-z
Wang, Y., Peng, D., Zhang, L., Tan, X., Yuan, D., Liu, X., et al. (2016). Overexpression of SsDGAT2 from Sapium sebiferum (L.) roxb increases seed oleic acid level in Arabidopsis. Plant Mol. Biol. Rep. 34, 638–648. doi: 10.1007/s11105-015-0954-y
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Wu, Z.-J., Tian, C., Jiang, Q., Li, X.-H., and Zhuang, J. (2016). Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci. Rep. 6:19748. doi: 10.1038/srep19748
Zhang, L., He, L. L., Fu, Q. T., and Xu, Z. F. (2013). Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using real-time quantitative, PCR. Int. J. Mol. Sci. 14, 24338–24354. doi: 10.3390/ijms141224338
Zhao, D., and Tao, J. (2015). Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 6:261. doi: 10.3389/fpls.2015.00261
Zhu, X., Li, X., Chen, W., Chen, J., Lu, W., Chen, L., et al. (2012). Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS ONE 7:e44405. doi: 10.1371/journal.pone.0044405
Keywords: Chinese tallow (Sapium sebiferum), qRT-PCR, reference genes, gene expression, sucrose and cold stress
Citation: Chen X, Mao Y, Huang S, Ni J, Lu W, Hou J, Wang Y, Zhao W, Li M, Wang Q and Wu L (2017) Selection of Suitable Reference Genes for Quantitative Real-time PCR in Sapium sebiferum. Front. Plant Sci. 8:637. doi: 10.3389/fpls.2017.00637
Received: 12 December 2016; Accepted: 10 April 2017;
Published: 04 May 2017.
Edited by:
Roger Deal, Emory University, USAReviewed by:
Palakolanu Sudhakar Reddy, International Crops Research Institute for the Semi-Arid Tropics, IndiaZheng Xiao, Jiangsu Academy of Agricultural Sciences, China
Copyright © 2017 Chen, Mao, Huang, Ni, Lu, Hou, Wang, Zhao, Li, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lifang Wu, lfwu@ipp.ac.cn
†These authors have contributed equally to this work.
 Xue Chen
Xue Chen Yingji Mao1,2†
Yingji Mao1,2† Jun Ni
Jun Ni Lifang Wu
Lifang Wu