- 1Department of Agricultural and Environmental Science, University of Bari Aldo Moro, Bari, Italy
- 2Department of Soil, Plant and Food Sciences, University of Bari Aldo Moro, Bari, Italy
Aldehyde Oxidase (AO) enzyme (EC 1.2.3.1) catalyzes the final steps of carotenoid catabolism and it is a key enzyme in the abscisic acid (ABA) biosynthesis. AO isoforms are located in the cytosolic compartment of tissues in many plants, where induce the oxidation of aldehydes into carboxylic acid, and in addition, catalyze the hydroxylation of some heterocycles. The goal of the present study was to characterize the AO genes involved in the accumulation of carotenoid pigments in wheat grain, an important quantitative trait controlled by multiple genes. The cDNAs corresponding to the four AO isoforms from Arabidopsis thaliana and five AO isoforms from Brachypodium distachyon were used as query in 454 sequence assemblies data for Triticum aestivum cv. Chinese Spring (https://urgi.versailles.inra.fr/blast/blast.php) to obtain the partial or whole orthologous wheat AO sequences. Three wheat isoforms, designated AO1, AO2, and AO3 were located on the chromosome groups 2, 5, and 7, respectively, and mapped on two consensus wheat maps by SNP markers located within the AO gene sequences. To validate the possible relationships between AO3 genes and carotenoid accumulation in wheat, the expression levels of AO-A3 and AO-B3 gene were determined during the kernel maturation stage of two durum wheat cultivars, Ciccio and Svevo, characterized by a low and high carotenoid content, respectively. Different AO-A3 gene expression values were observed between the two cultivars indicating that the AO-A3 allele present in Ciccio was more active in carotenoid degradation. A gene marker was developed and can be used for marker-assisted selection in wheat breeding programs.
Introduction
Yellow pigment concentration (YPC) in wheat is a quantitative trait controlled by a complex genetic system and influenced by environmental factors (Qin et al., 2016). The yellow color of grain and flour is mainly due to carotenoid accumulation in the pericarp and endosperm. Carotenoids are precursors of the vitamin A, with high nutritional relevance for human diet (Della Penna and Pogson, 2006; Britton, 2009), and substrates for the synthesis of apocarotenoids, compounds derived from oxidative cleavage and further modifications (Wurtzel et al., 2012). Apocarotenoids include retinol (vitamin A) and the hormones abscisic acid (ABA) and strigolactones (Rosati et al., 2010).
The carotenoid biosynthesis has been almost completely clarified in Arabidopsis thaliana, rice, maize and in some ornamental plants (Hirschberg, 2001; Chaudhary et al., 2010; Ruiz-Sola and Rodriguez-Concepcion, 2012; da Silva Messias et al., 2014; Moise et al., 2014). Briefly, the first committed step of biosynthesis is mediated by the phytoene synthase (PSY) enzyme for the condensation of two molecules of geranylgeranyl diphosphate to produce phytoene. Then, the phytoene goes through a series of desaturation reactions by phytoene desaturase (PDS), zeta-carotene isomerase (Z-ISO), zeta-carotene desaturase (ZDS), and carotenoid isomerase (CRTISO) enzymes, producing lycopene molecules. Double lycopene cyclization can produce precursor of lutein (branch β-ε) by lycopene ε-cyclase (LYCE) or β-carotene (branch β-β) by lycopene ß-cyclase (LYCB) (Qin et al., 2016).
Several QTL located on almost all wheat chromosomes have been detected by linkage mapping using biparental populations, genome wide association study (GWAS) and “candidate genes” approaches using germplasm collections (see review by Colasuonno et al., 2017). Numerous studies have attested the central role of some key genes in the carotenoid pathway and their association to QTL for YPC, such as the (CRTISO) gene on chromosomes 1A (Cazzonelli and Pogson, 2010), violaxanthin de-epoxidase (VDE) on 2B (Tsilo et al., 2011; Roncallo et al., 2012), lycopene epsilon-cyclase (LYCE) on chromosome group 3 (Howitt et al., 2009; Crawford and Francki, 2013), phytoene synthase (PSY1 and PSY2) on chromosomes 7A and 5A (Pozniak et al., 2007; Li et al., 2009).
Oxidative cleavage enzymes involved in carotenoid degradation, sequestration, and storage can also influence the accumulation of carotenoid pigments in tissues (Gonzalez et al., 2013). β-carotene and some of its derivatives can be modified by carotenoid cleavage dioxygenases (CCDs) resulting in the production of strigolactones (Alder et al., 2012). Moreover, a family of carotenoid 9-cis-epoxycarotenoid dioxygenases (NCEDs), which catalyzes the cleavage of carotenoids at specific double bonds, was shown to be active in the production of apocarotenoids (Auldridge et al., 2006). In particular, the ABA, derived from the conversion of violaxanthin by aldehyde oxidase (AO), is involved in regulating plant responses to various environmental stresses (e.g., drought and salinity stress) and long-distance signaling within the plants (Davies et al., 2005; Figure 1).
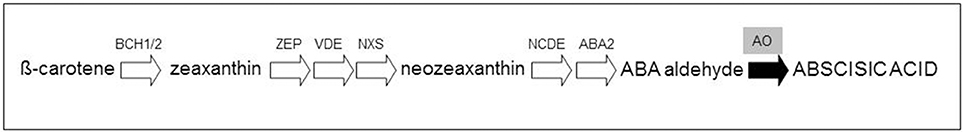
Figure 1. Simplified representation of the abscisic acid pathway. The conversion of β-carotene to abscisic acid is catalyzed by the β-carotene hydroxylases (BCH1/2), zeaxanthin epoxidase (ZEP), violaxanthin de-epoxidase (VDE), neoxanthin synthase (NXS), 9-cis-epoxycarotenoid dioxygenase (NCDE) and short-chain alcohol dehydrogenase (ABA2). The last step involves the aldehyde oxidese (AO), responsible of the synthesis of abscisic acid (black arrow).
A major QTL for YPC, accounting for up to 60% of the phenotypic variation, was detected on the long arm of chromosome 7A in different experiments carried out on diversified genetic materials using biparental populations (He et al., 2008, 2009; Zhang and Dubcovsky, 2008). Subsequently, two different QTL were identified on 7AL (Zhang and Dubcovsky, 2008; Blanco et al., 2011; Crawford et al., 2011): the first one on the distal bin 7AL18-0.90-1.00 associated with allelic variation at Psy1 (He et al., 2008, 2009; Zhang et al., 2009), and the second one on the bin 7AL21-0.74-0.86 linked with allelic variations at AO (Colasuonno et al., 2014, 2017).
The members of the AO enzyme (EC 1.2.3.1) play a role into last steps of carotenoid catabolism by oxidation of abscisic aldehyde in (ABA) (Seo et al., 2000a; Figure 1). Indeed, AO enzyme exists in multiple forms expressed in leaves, roots and seeds, depending on the species (Seo et al., 1998, 2000a,b; Xiong et al., 2001). AO proteins have been physiologically studied in Arabidopsis thaliana (Seo et al., 2000b), tomato (Min et al., 2000), barley (Omarov et al., 2003), maize (Qin et al., 2013), and partially in common wheat (Gallè et al., 2013).
The objectives of the present study were to: (a) identify the AO gene family in wheat and map each member on two wheat high-density consensus maps; (b) characterize AO3 transcription level by qRT-PCR in kernel tissue of two durum wheat cultivars differing for carotenoid content; (c) develop AO3 markers suitable for MAS to be used in breeding programs.
Materials and Methods
Plant Materials
A recombinant inbred line (RIL) mapping population, previously developed from the cross Svevo × Ciccio and genotyped using the SNP iSelect array (Colasuonno et al., 2014), was evaluated for the abscisic aldehyde oxidase 3 (AO-A3) gene and yellow pigment content. The elite durum wheat cultivars Svevo and Ciccio were different for qualitative and quantitative traits, such as grain yield components, grain protein content, grain yellow pigments and adaptive traits. Regression analysis, carried out in the Svevo x Ciccio RIL population grown in four environments (Valenzano 2006, Foggia 2006, Valenzano 2007, and Foggia 2007) and implemented in QGene, was used to underlined the association between the abscisic aldehyde oxydase 3 (AO-A3) gene and yellow pigment content. The marker-trait association was considered significant when –log10(P) ≥ 3.0. Phenotypic variation explained by the gene marker (R2) and additive effects were investigated. Positive and negative signs on the effect values indicated the contribution of Svevo and Ciccio, respectively, toward higher trait value.
Aldehyde Oxidase (AO) Isolation and Characterization
The sequences of AO genes from Arabidopsis thaliana and Brachypodiun distachyon were downloaded from the TAIR database (http://arabidopsis.org/) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Bdistachyon), respectively, and used as query in 454 sequence assemblies data for Triticum aestivum cv. Chinese Spring (https://urgi.versailles.inra.fr/blast/blast.php) to obtain partial or whole orthologous wheat AO sequences, including the promoter region.
The three wheat AO sequences included in the contigs 2AL_6408908, 5BL_10864426 and 7AL_4455104 were blasted against the available dataset of SNP marker sequences reported by Wang et al. (2014), and SNPs with ≥ 80% identity were considered within the AO genes. The high-density linkage maps described by Maccaferri et al. (2015) for durum wheat and by Wang et al. (2014) for common wheat were used as reference maps for determining a detailed map position of AO genes.
Wheat AO gene and promoter structure prediction was conducted with the FGENESH program (http://www.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind). Each predicted protein was considered into the next step of analysis.
Further BLASTn analysis for three representative isoforms was extended to Wheat 61k GeneChip in PLEXdb database (http://www.plantgdb.org) for obtaining information on each transcription pattern variation during different wheat developmental stages (Experiment TA3).
The bioinformatics analysis regarding the promoter region was conducted with PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to reveal cis-acting regulatory elements and Plant MITE database (http://pmite.hzau.edu.cn/MITEblast/blast.html) to identify transposon regions.
Quantitative Real-Time PCR
In order to pick primer combinations to be used for AO3 gene expression analysis, cDNA sequences of AO-A3 (included in the contig 7AL-4455104) and AO-B3 (accession number Ta_AK331622 from NCBI database) were aligned to highlight dissimilarities between the two homoeologous genes. Specific primer pairs for each gene were designed in a region of the second exon since a number of polymorphisms were detected between the A and B genomes (Table S1).
The genetic material used for the AO expression analysis was represented by the durum wheat cultivars Ciccio and Svevo characterized by low and high values of YPC, respectively. Kernel tissues from each cultivar were harvested, frozen in liquid nitrogen and stored at −80°C until RNA extraction. Total RNA was extracted with RNeasy Plant Mini Kit (QIAGEN®) and checked on 1.5% denaturing agarose gel. All RNA samples were lead to the same concentration (1 μg) for subsequent treatment with DNase I recombinant (Roche Applied Science, Mannheim, Germany), in order to remove genomic DNA, and then were reverse-transcribed into double stranded cDNA with Trascriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Mannheim, Germany). Data were normalized using three reference genes: Cell Division Control AAA-Superfamily of ATPases (CDC), ADP-Ribosilation Factor (ADP-RF), and RNase L Inhibitor-like protein (RLI) (Paolacci et al., 2009; Giménez et al., 2011). These genes have a stability value around 0.035 when evaluated with NormFinder software (Andersen et al., 2004).
Quantitative Real-Time PCR analyses were carried out using EVA® GREEN in the CFX96TM Real time PCR Systems (BIO-RAD). The PCR cycle was: 95°C for 3 min followed by 40 cycles of 95°C for 10 s and at 60°C for 30 s. Amplification efficiency (98% to 100%) for the primer set was determined by amplification of cDNA with a series of six scalar dilution (1:5) per reaction. Each 10 μl PCR reaction contained 1 μl of a 1:5 dilution cDNA, 5 μl of EvaGreen Mix 10X (Bio-Rad), and 500 nM of each primer. All experiments were performed in Hard-Shell 96-well skirted PCR plates (HSP9601) with Microseal R “B” Adhesive Seals (MSB-1001) from Bio-Rad. Fluorescence signals were collected at each polymerization step. The specificity of the amplicons was confirmed by the presence of a single band of expected size for each primer pair in agarose gel (2% w/v), by a single peak melting curves of the PCR products and by sequencing of the amplified fragments (3,500 Genetic Analyzer, Applied Biosystems). qRT-PCR data for both genes were derived from the mean values of three independent amplification reactions carried out on five different plants harvested in the same phenotypic stage (biological replicates). All calculations and analyses were performed using CFX Manager 2.1 software (Bio-Rad Laboratories) using the ΔCt method, which used the relative quantity (RQ) calculated with a ratio of the RQ of the target gene to the relative expression of the reference gene (including the three reference targets in each sample). Standard deviations were used to normalize values for the highest or lowest individual expression levels (CFX Manager 2.1 software user manual, Bio-Rad Laboratories). The Student's t-test was used to underline significant differences between control and treated samples for the two considered AO3 genes.
Development of AO3 Gene Markers for MAS
Primers were designed using Primer3 (http://frodo.wi.mit.edu/primer3/) software on the basis of the 7AL_4455104 contig sequence including the SNP marker IWB59875 previously resulted associated to carotenoid content (Colasuonno et al., 2014). The primer sequences PG42-for (TCTACACACCACGAGACCTT) and PG48-rev (GCAACTCAGCGATCCAACAATAT) amplified a 635 bp fragment.
PCR products derived from the durum cvs. Ciccio and Svevo were loaded on agarose gel to confirm amplicon length, and injected onto the WAVE® system (Transgenomic, Omaha, NE, USA) to undergo check peak intensity of the amplicons. The temperature required for successful resolution of heteroduplex molecules was determined using the DHPLC Navigator™ Software (Transgenomic, Omaha, NE, USA), which considered the specific amplicon sequence and size to calculate the denaturation curve. PCR product was cleaned using ExoSAP-IT (USB, Cleveland, OH, USA) according to manufacturer's instructions, then sequenced using BigDye terminator sequencing kit on a ABI-3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The detection of molecular marker was performed on DHPLC device in “mutation detection” under “Rapid DNA” run mode.
Results
Isolation of Genomic Sequences of AO Genes in Wheat
The cDNAs corresponding to the four AO isoforms from A. thaliana (AT5G20960, AT3G43600, AT2G27150, and AT1G04580, designated, respectively O1, AO2, AO3, and AO4) and the five AO isoforms from B. distachyon (XM010230033, XM_003557870, XM_003559293, and XM_003559295, all designated AO2, and XM_003561213 (designated AO) were used as query in 454 sequence assemblies data for T. aestivum cv. Chinese Spring (https://urgi.versailles.inra.fr/blast/blast.php) to obtain the partial or whole orthologous wheat AO sequences. The search into the Chinese Spring sequence database released several AO sequence fragments included in 13 different contigs located on the wheat chromosome groups 2, 5, and 7 (Table 1). The definitive assignment of AO sequences to the wheat A, B and D homoeologous chromosomes and the accurate map position was performed based on the best blastn hit (percentage identity) with the available dataset of SNP marker sequences reported by Wang et al. (2014) (Table 1). The Recommended Rules for Gene Symbolization reported in the Wheat Catalog (McIntosh et al., 2005) were used for AO nomenclature. In particular, six SNPs corresponding to AO1 genes mapped on chromosome group 5 (AO-B1 and AO-D1), 12 SNPs within AO2 mapped on chromosome group 2 (AO-A2, AO-B2, and AO-D2), and 16 SNP markers mapped on chromosome group 7 (AO-A3, AO-B3, and AO-D3). Out of 34 SNPs corresponding to AO gene sequences, five and nine markers were located on the consensus durum (Maccaferri et al., 2015) and bread wheat maps (Wang et al., 2014), respectively (Figure 2).
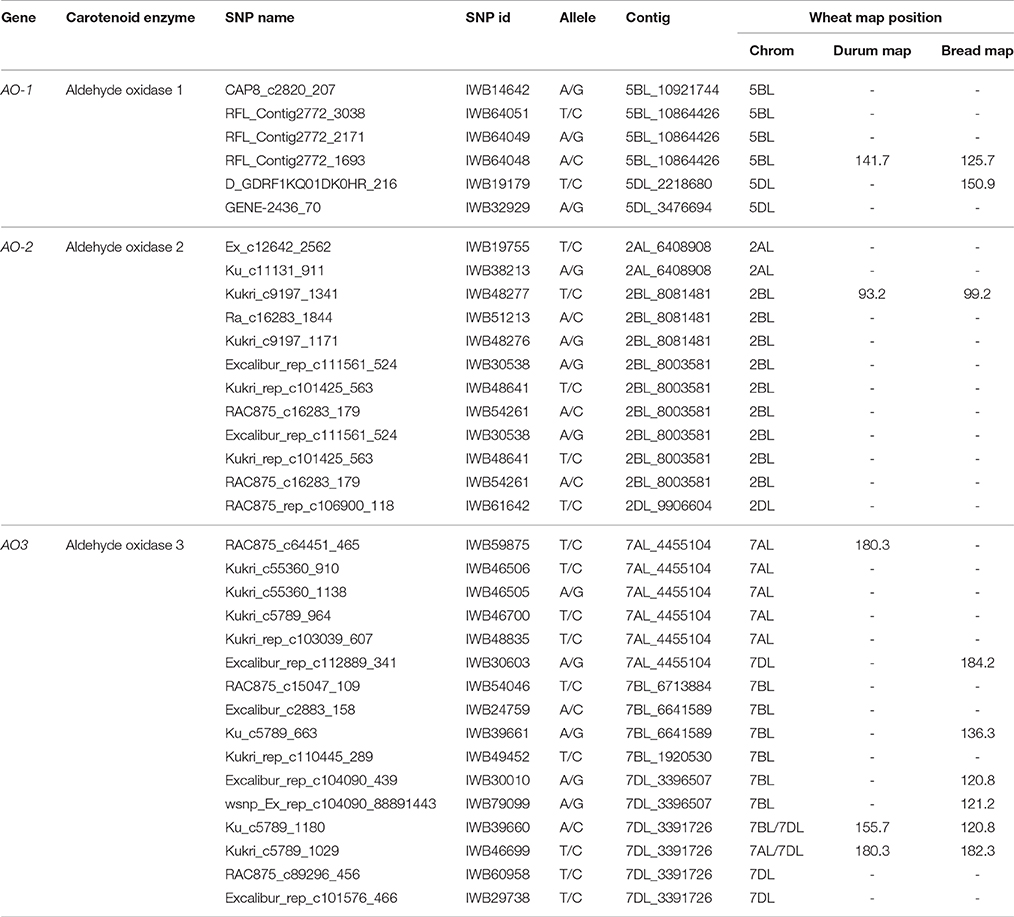
Table 1. List of aldehyde oxidase genes in wheat with corresponding contig number, SNP markers, chromosome localization and map position on the durum (Maccaferri et al., 2015) and bread wheat (Wang et al., 2014) consensus maps.
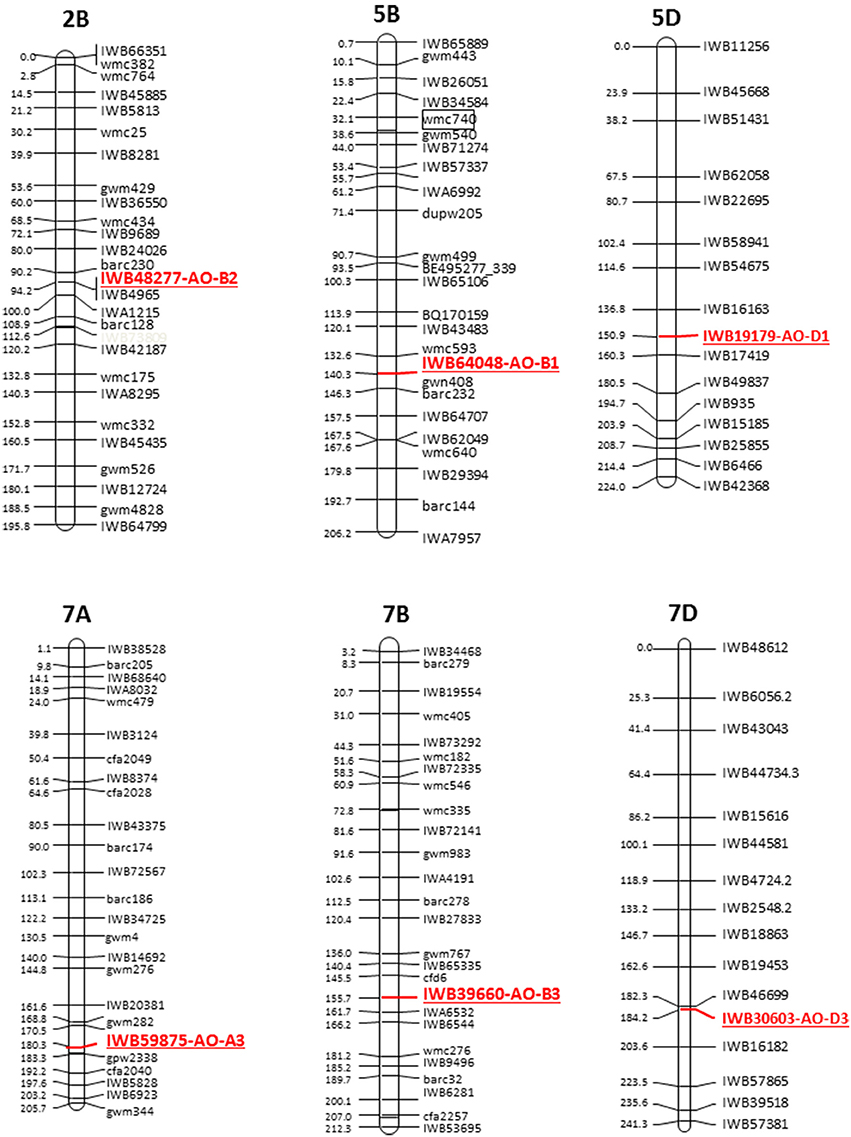
Figure 2. Schematic representation of the durum wheat linkage map and AO markers. Each linkage map derives from the durum consensus map (Maccaferri et al., 2015) and has been represented by a SNP marker every about 20 cM. SSR markers have been also inserted every about 20 cM to compare the consensus SNP map with published SSR-based maps.
Characterization of Wheat AO3 Gene Sequences
The genomic sequence and structure of AO-A3, AO-B3, and AO-D3 genes was investigated in the cv Chinese Spring in order to study the possible relationships between AO3 genes and carotenoid accumulation in the wheat grain (Colasuonno et al., 2014). Relying on the Softberry calculation regarding the 7AL-4455104 contig, the AO-A3 genomic sequence was 6,749 bp long with 42% GC content. The predicted gene sequence included an mRNA of 3,786 bp and a protein length of 1,262 amino acids. For AO-B3, the partial sequence included in the contig 7BL_6713884 was implemented by the complete EST sequence AK331622 (from NCBI database) and allowed to obtain a cDNA of 4,279 bp. The prediction of AO-D3 gene, considering the 7DL_3391726 contig, presented a genomic sequence of 6,215 bp with an mRNA length of 4,038 bp and a protein 1,346 amino acids. A similar intron/exons structure was predicted between the wheat AO3 genes composed by 10 exons and 9 introns sharing an identity of > 80% among homoeologues and 97% between AO-B3 and AO-D3. The Fgenesh++ gene prediction pipeline highlighted a lack of similarity for the first and last exons between AO-A3 and AO-B3/AO-D3. Only the last exon displayed high identity (100%) between AO-B3 and AO-D3 isoforms.
Furthermore, the wheat AO3 cDNA alignment showed a smaller region (81 bp) in exon 6 for AO-A3 than in exon 6 for AO-B3 and AO-D3, and a corresponding longer region in the adjacent intron. This difference suggested an alternative splicing site and the sequencing analysis of cv. Chinese Spring cDNAs confirmed this length polymorphism among genomes.
BLAST analysis using Phytozome v.7 software (http://www.phytozome.net) with B. distachyon and O. sativa genomes allowed the comparison of wheat AO3 genes with the orthologous genes located on chromosome 1 of Brachypodium (locus name: Bradi1g52740) and chromosome 7 of rice (locus name: Os07g18154). The Brachypodium AO consist in a sequence of 12,336 bp with a CDS of 4,053 bp, whereas in rice genome AO had a sequence of 12,959 bp with a CDS of 2,535 nucleotides. Comparison between wheat and Brachypodiun genomic AO sequences showed identities of about 84%, and an identity of 89% between the two CDS. A similar intron/exons structure was observed between AO3-A1 from wheat and AO from Brachypodium composed by 10 exons and nine introns, except for the first and last exons (Figure 3). An identity of 79% was found aligning the AO-A3 genomic sequence with the rice AO sequence.
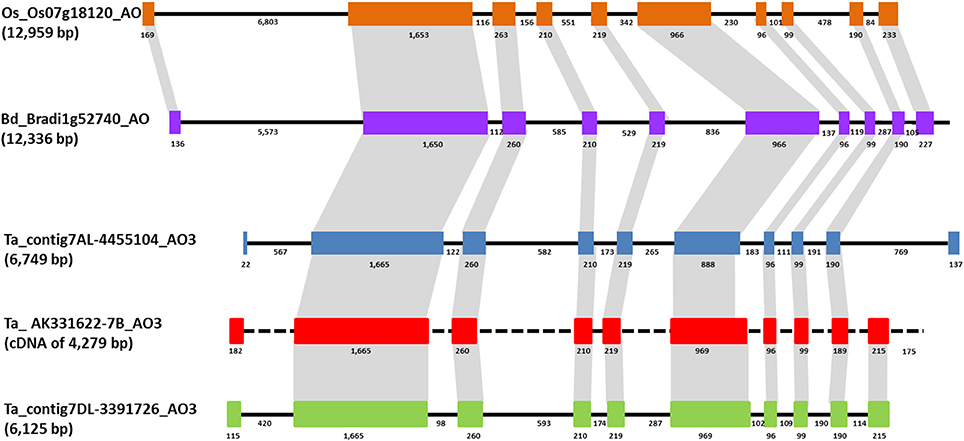
Figure 3. Comparison of AO3 gene structures in rice, Brachypodium, and wheat is shown based on colored boxes highlighting conserved exons. Intron and exon sizes are shown as well as the whole gene (in brackets the total length). Rice and Brachypodium AO share the same structure with ten exons of conserved sizes and nine introns. Brachypodium and wheat AO3 show an high similarity in sequence and structure for eight exons. Black dashed line indicates the absence of intron sequence since only a cDNA sequence has been found for AO-B3.
In order to investigate the role of promoter region, a bioinformatics analysis was conducted in public database and a region of 1,600 bp (from contig7AL_4455104) upstream of the AO-A3 was identified. The analysis through Softberry showed clearly the presence of a promoter regions upstream the gene (data showed in Supplemental Material S2). Moreover, PlantCARE revealed the presence of 32 elements that are characteristic of promoters. These elements include the A-box, ARE, Box4, BoxI, Box-W1, CAAT-box, CAT-box, CATT-motif, CCAAT-box, CCGTCC-box, CGTCA-motif, CTAG-motif, G-box, GAG-motif, GARE-motif, GC-motif, GCN4_motif, GT1-motif, I-box, P-box, Skn-1_motif, Sp1, TATA-box, TCA-element, TCCACCT-motif, TCT-motif, TGACG-motif, TGG-motif, Wbox, WUN-motif, boxE, boxes, and circadian.
Further investigation through the Plant MITE database revealed the presence of two transposon regions. The first one was located at 336 bp and belonged to MITE family DTC, the other one at 901 bp position corresponded to MITE family DTT. The promoter region sequences in the cvs Ciccio and Svevo relieved absence of significant polymorphisms. According to this, the differences on the expression levels among cultivars could be due to a different regulatory mechanisms in action inducing different genes.
Expression Profile of AO Genes in Wheat
The possible relationships between AO genes and carotenoid accumulation was investigated in the wheat grain. The AO-A3 and AO-B3 gene expression levels were determined in two durum wheat cultivars, Ciccio and Svevo, characterized by a low and high YPC, respectively. Total RNA was extracted from kernels, and quantitative real-time PCR was conducted with specific primers in order to analyze individually the two homeologous AO3 genes. High expression levels were observed for AO-A3 and AO-B3 during seed maturation stage in Ciccio, while low amounts were detected in Svevo (Figure 4). Significant different expression values (P < 0.001, t-test) were observed between the two cultivars only for AO-A3. The data suggested that the AO-A3 allele present in Ciccio (cultivar characterized by a low content of YPC) was more active into carotenoid degradation, while the Svevo (high YPC) allele was not fully involved in the carotenoid catabolism.
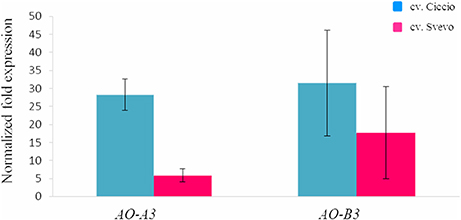
Figure 4. Comparison of the expression levels of AO-A3 and AO-B3 genes in kernel tissue of cv. Svevo and cv. Ciccio by qRT-PCR. The y ax shows the normalized fold expression. The error bars indicate the ± SE of the mean.
In order confirm our data and to understand the expression trend of wheat AO genes, a check on data available in the PLEXdb database was carried out considering the cDNAs included in the largest contigs (2AL_6408908, 5BL_10864426, and 7AL-4455104) chosen as representative of each wheat homoeologous group (Figure 5). The Wheat 61k GeneChip showed different expression pattern of AO genes during developmental stages underlining how AO2 and AO1 resulted constantly expressed in all stages with maximum levels of 8.93 RMA normalization for AO2 in seedling root (phase 4). Instead, AO3 showed significantly high expression levels (values higher than the mean values ± 2 SD) especially in last phases of embryo and endosperm kernel formation (phase 9, 12, and 13) indicating a major role in the last developmental stage of wheat seeds.
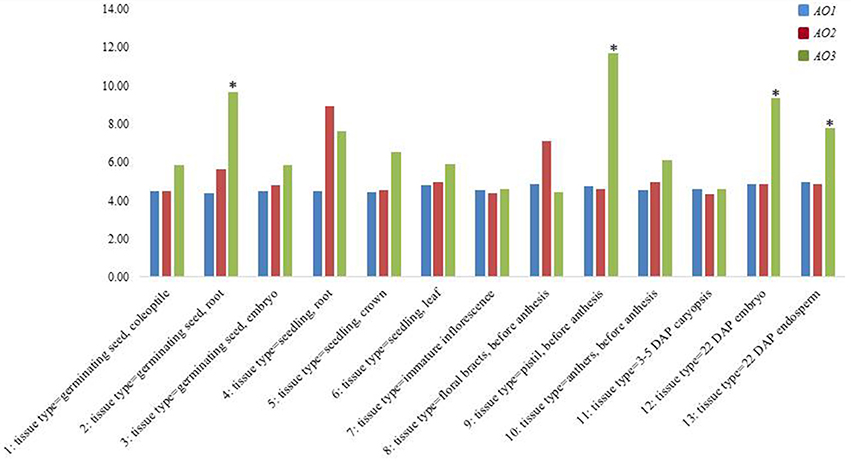
Figure 5. Expression analysis from PLEXdb database of all wheat AO genes in a wide range of tissues and developmental stages in wheat (1–13). The asterisk signals indicate, respectively the values higher or lower than the mean values ± 2 SD.
Development of a Functional AO3 Marker
A considerable emphasis has been placed on the AO-A3 gene for the development of molecular markers to be used for MAS in wheat breeding. Due to the availability of SNP data corresponding to this isoform, the IWB59875 marker mapped on chromosome 7A and resulted associated to carotenoid content (Colasuonno et al., 2014) was chosen for setting up a SNP-based method suitable for wheat breeding. Before proceeding into the analysis of DNA fragments, PCR primers specific for AO-A3 gene were designed respecting the DHPLC conditions, such as the distance of 50 bp from the SNP site and a fragment size ≥ 200 and ≤ 800 bp. After that, the DHPLC technique was optimized for the run conditions on each amplicon, considering the optimal temperature (55°C). According to the basic principle of the DHPLC technique, if the sample contained the DNA from a homozygous genotype, the chromatogram showed a single peak derived from only homoduplex molecules. The homoduplex originated from Ciccio was practically undistinguishable from that of Svevo, having the same retention time. On the contrary, when the sample was a mix of two cultivars, and a SNP was present, the chromatogram exhibited two peaks of similar area, one corresponding to the coeluting homoduplex molecules (imputable to both Ciccio or Svevo with themselves), the other to the early elution of the heteroduplex molecules (due to Ciccio DNA combined with that of Svevo). As shown in Figure 6, the DHPLC allowed detecting two chromatographic peaks corresponding to heteroduplex and homoduplex molecules derived by the presence of the T/C substitution (detected by the IWB59875 marker) in the two durum varieties. The regression analysis conducted between the AO-A3 gene marker and YPC confirmed the association with the trait with an high LOD scores in all the four environment considered and in the mean across them. The phenotypic variation explained by the gene marker (R2) ranged from 18.0 to 41.0% and the effect due to the Ciccio alleles was reported for all the environment analyzed in Table 2.
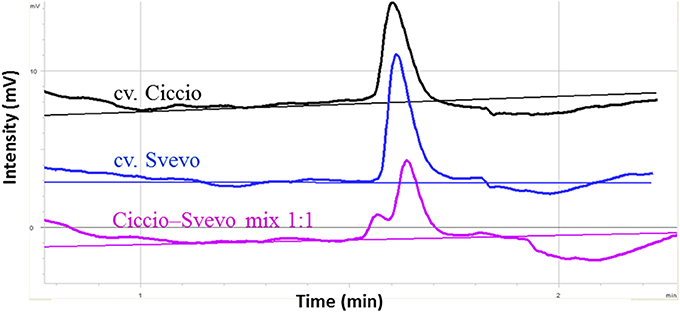
Figure 6. Optimization of DHPLC analysis for SNP detection between cvs. Ciccio and Svevo at locus IWB59875. The chromatograms correspond to the elution profiles of homoduplex molecules (cv Ciccio in black, cv Svevo in dark blue), and of homoduplexes plus heteroduplexes derived from 1:1 mixed DNA (pink line).
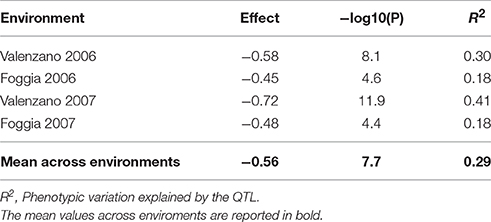
Table 2. Regression analysis between the SNP marker IWB59875 for the gene abscisic aldehyde oxydase 3 (AO-A3) and yellow pigment content evaluated in the Svevo × Ciccio RIL population grown in four environments.
Discussion
High levels of carotenoid pigments in wheat kernels have important positive implications for human health since they are antioxidant compounds and precursor of vitamin A. Knowing the role of the main carotenoid genes in wheat and the specific alleles present in each cultivar can allow to develop superior cultivars through marker-assisted breeding programmes.
Several studies reported many QTL for carotenoid content spread all over the wheat genome (reviewed by Colasuonno et al., 2017). Among them, two different QTL were mapped on the long arm of chromosome group 7, co-localized with phytoene synthase 1 (Psy1) and aldehyde oxidase 3 (AO3) genes, respectively (Zhang and Dubcovsky, 2008; Blanco et al., 2011; Colasuonno et al., 2014, 2017). While the Psy1 involvement in YPC has been deeply studied, the role of AO3 gene in the carotenoid accumulation needs to be elucidated. AO isoforms are key enzymes for ABA biosynthesis (Fang et al., 2008; Zdunek-Zastocka, 2010). Plant AO family is composed by proteins with high similarity in sequences, but different subunit composition and substrate preferences. AO isoforms have been largely characterized in Arabidopsis and resulted composed by 4 isoforms (Seo et al., 2000a). While significant information exists on AO families in Arabidopsis, lettuce, tomato, peas, Brassica, rice and maize, a complete picture of this family is missing in wheat (Zdunek-Zastocka, 2010).
Overall, 34 SNPs within AO wheat sequences were identified, making significant advancement on the localization of AO genes on wheat chromosome groups. Indeed, the recent availability of the high-resolution consensus map of durum (Maccaferri et al., 2015) and bread wheat maps (Wang et al., 2014) allowed to determine the precise map position. The AO3 mapping was consistent with results reported by Colasuonno et al. (2014, 2017), who identified the chromosomal locations based on survey sequence from the International Wheat Genome Sequencing Consortium (http://www.wheatgenome.org/). Instead, the map positions of AO2 and AO1 genes were reported for the first time: AO2 localized on chromosome group 5 and the AO1 on chromosome group 2.
The present study considered the AO gene expression in wheat. AO isoforms were found expressed in root tissues under osmotic stress (Gallè et al., 2013). The AO2 and AO1 genes resulted expressed in all stages with maximum levels for AO2 in seedling root. While, the AO3 showed elevated expression levels during kernel maturation with a possible involvement in carotenoids pathway since they are accumulate at last developmental stage.
The first connection between carotenoid content and the AO3 genes emerged form a genetic study on QTL in a RIL population derived crossing the durum wheat cvs Ciccio and Svevo (Colasuonno et al., 2014). Previously, other studies (Zhang and Dubcovsky, 2008; Blanco et al., 2011) indicated the presence of this second QTL for carotenoids, using different genetic materials. Then again, Colasuonno et al. (2017) showed that AO-A3 gene was significant associated to carotenoid variation using GWAS and the “candidate gene” approaches.
Based on the evidence presented, we have envisaged that mutations in AO3 induce activity-loss in carotenoid catabolism from violaxanthin/neoxanthin to (ABA) and allows an accumulation of carotenoid compounds. The transcriptional level of the AO-A3 and AO-B3 genes in the cvs Ciccio and Svevo was in accordance with this hypothesis, demonstrating how AO-A3 gene resulted in significant low expression levels in Svevo, cultivar with high content of carotenoids. This could be explained by the presence of an AO-A3 allele in the cv Svevo not fully activated for carotenoid catabolism and ABA biosynthesis. The AO3 analysis need to be further examined to evaluate how the ABA concentration differs in relation to allele functionality. Indeed, in Arabidopsis and maize (Wurtzel et al., 2012; Gonzalez et al., 2013) the carotenoid degradation is important in determining total carotenoid accumulation.
In addition, the regression analysis conducted in the RIL population confirmed the association between the AO-A3 gene marker and YPC. Besides, the development of a molecular gene marker for AO-A3 through the most sensible and cost-effective technique (such as the DHPLC) demonstrated for the first time its applicability on marker assisted selection programmes (MAS). Giancaspro et al. (2016) used the same technology as tool for SNP detection based on genotyping arrays in food science.
The present work characterizes suitable AO3 genes providing a new insight into the regulation of carotenoid accumulation. Although the gene expression analysis revealed differences between the two cultivars, no polymorphisms were observed in the promoter regions, suggesting the presence of complex gene regulation mechanisms. The factors influencing pigment content are complex (Howitt and Pogson, 2006; Lachman et al., 2017). Further investigations needed in order to understand the carotenoid pigment regulation system in wheat.
Author Contributions
PC, AB, and AG designed the research; IM and ML performed the research. PC and RS wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by grants from Puglia Region, Italy, project PSR “SAVEGRAIN.”
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00863/full#supplementary-material
References
Alder, A., Jamil, M., Marzorati, M., Bruno, M., Vermathen, M., Bigler, P., et al. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. doi: 10.1126/science.1218094
Andersen, C. L., Jensen, J. L., and Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Auldridge, M. E., McCarty, D. R., and Klee, H. J. (2006). Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9, 315–321. doi: 10.1016/j.pbi.2006.03.005
Blanco, A., Colasuonno, P., Gadaleta, A., Mangini, G., Schiavulli, A., Simeone, R., et al. (2011). Quantitative trait loci for yellow pigment concentration and individual carotenoid compounds in durum wheat. J. Cereal Sci. 54, 255–264. doi: 10.1016/j.jcs.2011.07.002
Britton, G. (2009). “Vitamin A and vitamin A deficiency,” in Carotenoids, Vol. 5, eds G. Britton, S. Liaaen-Jensen, and H. Pfander (Basel: Birkhauser Verlag; Nutrition and Health), 173–190.
Cazzonelli, C. I., and Pogson, B. J. (2010). Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15, 266–274. doi: 10.1016/j.tplants.2010.02.003
Chaudhary, N., Nijhawan, A., Khurana, J. P., and Khurana, P. (2010). Carotenoid biosynthesis genes in rice: structural analysis, genome-wide expression profiling and phylogenetic analysis. Mol. Genet. Genomics 283, 13–33. doi: 10.1007/s00438-009-0495-x
Colasuonno, P., Gadaleta, A., Giancaspro, A., Nigro, D., Giove, S., Incerti, O., et al. (2014). Development of a high-density SNP-based linkage map and detection of yellow pigment content QTLs in durum wheat. Mol. Breed. 34, 1563–1578. doi: 10.1007/s11032-014-0183-3
Colasuonno, P., Lozito, M. L., Marcotuli, I., Nigro, D., Giancaspro, A., Mangini, G., et al. (2017). The carotenoid biosynthetic and catabolic genes in wheat and their association with yellow pigments. BMC Genom. 18:122. doi: 10.1186/s12864-016-3395-6
Crawford, A. C., and Francki, M. G. (2013). Lycopene-ε-cyclase (e-LCY3A) is functionally associated with quantitative trait loci for flour b* colour on chromosome 3A in wheat (Triticum aestivum L.). Mol. Breed. 31, 737–741. doi: 10.1007/s11032-012-9812-x
Crawford, A. C., Stefanova, K., Lambe, W., McLean, R., Wilson, R., Barclay, I., et al. (2011). Functional relationships of phytoene synthase 1 alleles on chromosome 7A controlling flour colour variation in selected Australian wheat genotypes. Theor. Appl. Genet. 123, 95–108. doi: 10.1007/s00122-011-1569-9
da Silva Messias, R., Galli, V., Dos Anjos, E., Silva, S. D., and Rombaldi, C. V. (2014). Carotenoid biosynthetic and catabolic pathways: gene expression and carotenoid content in grains of maize landraces. Nutrients 6, 546–563. doi: 10.3390/nu6020546
Davies, W., Kudoyarova, G., and Hartung, W. (2005). Long-distance ABA signaling and its relation to other signaling pathways in the detection of soil drying and the mediation of the plants response to drought. J. Plant Growth Regul. 24, 285–295. doi: 10.1007/s00344-005-0103-1
Della Penna, D., and Pogson, B. J. (2006). Vitamin synthesis in plants: tocopherols and carotenoids. Annu. Rev. Plant Biol. 57, 711–738. doi: 10.1146/annurev.arplant.56.032604.144301
Fang, J., Chai, C., Qian, Q., Li, C., Tang, J., Sun, L., et al. (2008). Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 54, 177–189. doi: 10.1111/j.1365-313X.2008.03411.x
Gallè, A., Csiszar, J., Benyo, D., Laskay, G., Leviczky, T., Erdei, L., et al. (2013). Isohydric and anisohydric strategies of wheat genotypes under osmotic stress: biosynthesis and function of ABA in stress responses. J. Plant Physiol. 170, 1389–1399. doi: 10.1016/j.jplph.2013.04.010
Giancaspro, A., Colasuonno, P., Zito, D., Blanco, A., Pasqualone, A., and Gadaleta, A. (2016). Varietal traceability of bread “Pane Nero di Castelvetrano” by denaturing high pressure liquid chromatography analysis of single nucleotide polymorphisms. Food Control 59, 809–817. doi: 10.1016/j.foodcont.2015.07.006
Giménez, M. J., Pistón, F., and Atienza, S. G. (2011). Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 233, 163–173. doi: 10.1007/s00425-010-1290-y
Gonzalez, J. S., Ha, S. H., Magallanes-Lundback, M., Gilliland, L. U., Zhou, A., Lipka, A. E., et al. (2013). Carotenoid cleavage dioxygenase 4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25, 4812–4826. doi: 10.1105/tpc.113.119677
He, X. Y., He, Z. H., Ma, W., Appels, R., and Xia, X. C. (2009). Allelic variants of phytoene synthase 1 (Psy1) genes in Chinese and CIMMYT wheat cultivars and development of functional markers for flour colour. Mol. Breed. 23, 553–563. doi: 10.1007/s11032-009-9255-1
He, X. Y., Zhang, Y. L., He, Z. H., Wu, Y. P., Xiao, Y. G., Ma, C. X., et al. (2008). Characterization of phytoene synthase 1 gene (Psy1) located on common wheat chromosome 7A and development of a functional marker. Theor. Appl. Genet. 116, 213–221. doi: 10.1007/s00122-007-0660-8
Hirschberg, J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4, 210–218. doi: 10.1016/S1369-5266(00)00163-1
Howitt, C. A., Cavanagh, C. R., Bowerman, A. F., Cazzonelli, C., Rampling, L., Mimica, J. L., et al. (2009). Alternative splicing, activation of cryptic exons and amino acid substitutions in carotenoid biosynthetic genes are associated with lutein accumulation in wheat endosperm. Funct. Integr. Genom. 9, 363–376. doi: 10.1007/s10142-009-0121-3
Howitt, C. A., and Pogson, B. J. (2006). Carotenoid accumulation and function in seeds and nongreen tissues. Plant Cell Environ. 29, 435–445. doi: 10.1111/j.1365-3040.2005.01492.x
Lachman, J., Martinek, P., Kotíková, Z., Orsák, M., and Šulc, M. (2017). Genetics and chemistry of pigments in wheat grain-A review. J. Cereal Sci. 74, 145–154. doi: 10.1016/j.jcs.2017.02.007
Li, F., Tzfadia, O., and Wurtzel, E. T. (2009). The phytoene synthase gene family in the Grasses. Plant Signal. Behav. 3, 208–211. doi: 10.4161/psb.4.3.7798
Maccaferri, M., Ricci, A., Salvi, S., Milner, S., Noli, E., Martelli, P., et al. (2015). A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 13, 648–663. doi: 10.1111/pbi.12288
McIntosh, R. A., Devos, K. M., Dubcovsky, J., Rogers, W. J., Morris, C. F., Appels, R., et al. (2005). Catalogue of Gene Symbols for Wheat 2005. Available online at: http://wheat.pw.usda.gov/ggpages/wgc/2005upd.html
Min, X., Okada, K., Brockmann, B., Koshiba, T., and Kamiya, Y. (2000). Molecular cloning and expression patterns of three putative functional aldehyde oxidase genes and isolation of two aldehyde oxidase pseudogenes in tomato. Biochim. Biophys. Acta 1493, 337–341. doi: 10.1016/S0167-4781(00)00190-1
Moise, A. R., Al-Babili, S., and Wurtzel, E. T. (2014). Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 114, 164–193. doi: 10.1021/cr400106y
Omarov, R., Dräger, D., Tischner, R., and Lips, H. (2003). Aldehyde oxidase isoforms and subunit composition in roots of barley as affected by ammonium and nitrate. Physiol. Plantarum 117, 337–342. doi: 10.1034/j.1399-3054.2003.00043.x
Paolacci, A., Tanzarella, O., Porceddu, E., and Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10:11. doi: 10.1186/1471-2199-10-11
Pozniak, C. J., Knox, R. E., Clarke, F. R., and Clarke, J. M. (2007). Identification of QTL and association of a phytoene synthase gene with endosperm colour in durum wheat. Theor. Appl. Genet. 114, 525–537. doi: 10.1007/s00122-006-0453-5
Qin, S., Zhang, Z., Ning, T., Ren, S., Su, L., and Li, Z. (2013). Abscisic acid and aldehyde oxidase activity in maize ear leaf and grain relative to post-flowering photosynthetic capacity and grain-filling rate under different water/nitrogen treatments. Plant Physiol. Biochem. 70, 69–80. doi: 10.1016/j.plaphy.2013.04.024
Qin, X., Fischer, K., Yu, S., Dubcovsky, J., and Tian, L. (2016). Distinct expression and function of carotenoid metabolic genes and homoeologs in developing wheat grains. BMC Plant Biol. 16:155. doi: 10.1186/s12870-016-0848-7
Roncallo, P. F., Cervigni, G. L., Jensen, C., Miranda, R., Carrera, A. D., Helguera, M., et al. (2012). QTL analysis of main and epistatic effects for flour color traits in durum wheat. Euphytica 185, 77–92. doi: 10.1007/s10681-012-0628-x
Rosati, C., Diretto, G., and Giuliano, G. (2010). Biosynthesis and engineering of carotenoids and apocarotenoids in plants: state of the art and future prospects. Biotechnol. Genet. Eng. Rev. 26, 139–162. doi: 10.5661/bger-26-139
Ruiz-Sola, M. A., and Rodriguez-Concepcion, M. (2012). Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 10:e0158. doi: 10.1199/tab.0158
Seo, M., Akaba, S., Oritani, T., Delarue, M., Bellini, C., Caboche, M., et al. (1998). Higher activity of an aldehyde oxidase in the auxin-overproducing superrootl mutant of Arabidopsis thaliana. Plant Physiol. 116, 687–693. doi: 10.1104/pp.116.2.687
Seo, M., Koiwai, H., Akaba, S., Komano, T., Oritani, T., Kamiya, Y., et al. (2000a). Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J. 23, 481–488. doi: 10.1046/j.1365-313x.2000.00812.x
Seo, M., Peeters, A. J. M., Koiwai, H., Oritani, T., Marion-Poll, A., Zeevaart, J. A. D., et al. (2000b). The Arabidopsis aldehyde oxidase 3 (AA03). gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. U.S.A. 97, 12908–12913. doi: 10.1073/pnas.220426197
Tsilo, T. J., Hareland, G. A., Chao, S., and Anderson, J. A. (2011). Genetic mapping and QTL analysis of flour colour and milling yield related traits using recombinant inbred lines in hard red spring wheat. Crop Sci. 51:237. doi: 10.2135/cropsci2009.12.0711
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90,000 SNP array. Plant Biotechnol. J. 2, 787–796. doi: 10.1111/pbi.12183
Wurtzel, E. T., Cuttriss, A., and Vallabhaneni, R. (2012). Maize provitamin a carotenoids, current resources, and future metabolic engineering challenges. Front. Plant Sci. 3:29. doi: 10.3389/fpls.2012.00029
Xiong, L., Ishitani, M., Lee, H., and Zhu, J. K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13, 2063–2083. doi: 10.1105/tpc.13.9.2063
Zdunek-Zastocka, E. (2010). The activity pattern and gene expression profile of aldehyde oxidase during the development of Pisum sativum seeds. Plant Sci. 179, 543–548. doi: 10.1016/j.plantsci.2010.08.005
Zhang, K. P., Chen, G. F., Zhao, L., Liu, B., Xu, X.-B., and Tian, J. C. (2009). Molecular genetic analysis of flour colour using a doubled haploid population in bread wheat (Triticum aestivum L.). Euphytica 165, 471–484. doi: 10.1007/s10681-008-9756-8
Keywords: wheat, carotenoid genes, aldehyde oxidase, SNP, yellow pigments
Citation: Colasuonno P, Marcotuli I, Lozito ML, Simeone R, Blanco A and Gadaleta A (2017) Characterization of Aldehyde Oxidase (AO) Genes Involved in the Accumulation of Carotenoid Pigments in Wheat Grain. Front. Plant Sci. 8:863. doi: 10.3389/fpls.2017.00863
Received: 22 February 2017; Accepted: 09 May 2017;
Published: 24 May 2017.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Antonio Ferrante, Università degli Studi di Milano, ItalyZhongfu Ni, China Agricultural University, China
Copyright © 2017 Colasuonno, Marcotuli, Lozito, Simeone, Blanco and Gadaleta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pasqualina Colasuonno, pattybiotec@yahoo.it
Agata Gadaleta, agata.gadaleta@uniba.it
 Pasqualina Colasuonno
Pasqualina Colasuonno Ilaria Marcotuli
Ilaria Marcotuli Maria L. Lozito
Maria L. Lozito Rosanna Simeone2
Rosanna Simeone2 Antonio Blanco
Antonio Blanco Agata Gadaleta
Agata Gadaleta