- State Key Laboratory of Crop Stress Biology in Arid Areas, College of Horticulture, Northwest A&F University, Yangling, China
RNA silencing functions as a major natural antiviral defense mechanism in plants. RNA-dependent RNA polymerases (RDRs) that catalyze the synthesis of double-stranded RNAs, are considered as a fundamental element in RNA silencing pathways. In Arabidopsis thaliana, RDR1, 2 and 6 play important roles in anti-viral RNA silencing. Expression of RDR1 can be elevated following plant treatment with defense hormones and virus infection. RDR1 has been studied in several crop species, but not in pepper (Capsicum annuum L.). Here, a RDR1 gene was isolated from Capsicum annuum L., designated as CaRDR1. The full-length cDNA of CaRDR1 was 3,351 bp, encoding a 1,116-amino acid protein, which contains conserved regions, such as the most remarkable motif DLDGD. The transcripts of CaRDR1 could be induced by salicylic acid (SA), abscisic acid (ABA), H2O2, and tobacco mosaic virus (TMV). Silencing of CaRDR1 in pepper resulted in increased susceptibility to TMV as evident by severe symptom, increased of TMV-CP transcript, higher malondialdehyde (MDA) content and lower antioxidant enzymes activities compared with that of control plants. CaRDR1-overexpressing in Nicotiana benthamiana showed mild disease symptom and reduced TMV-CP transcripts than that of empty vector (EV) following TMV inoculation. The RNA silencing related genes, including NbAGO2, NbDCL2, NbDCL3, and NbDCL4 elevated expression in overexpressed plants. Alternative oxidase (AOX), the terminal oxidase of the cyanide (CN)-resistant alternative respiratory pathway, catalyze oxygen-dependent oxidation of ubiquinol in plants. It has an important function in plant defense against TMV. In addition, CaRDR1 overexpression promoted the expression of NbAOX1a and NbAOX1b. In conclusion, these results suggest that CaRDR1 plays a positive role in TMV resistance by regulating antioxidant enzymes activities and RNA silencing-related genes expression to suppress the replication and movement of TMV.
Introduction
Pepper is an important vegetable crop with a wide variety of uses. In 2014, pepper production was 36.1 million tons including green fruit and dried pods harvested in 3.6 million hectares all over the world (http://www.fao.org). Virus is the most seriously threatened in pepper production, results in crop losses under field conditions. In order to defend against viral infection, plants have evolved complicated mechanisms. RNA silencing acts as an important antiviral defense mechanism (Baulcombe, 2004; Ding and Lu, 2011). A whole RNA silencing comprises three procedures, initiation, maintenance, and signal amplification. The plant dicer-like (DCL) nucleases, argonaute (AGO) proteins, RNA-dependent RNA polymerases (RDRs) are the central functional components of the RNA silencing-based antiviral defense (Baulcombe, 2004). DCLs deal with cutting double-strand RNAs into 21–24 nt small RNAs (Carmell and Hannon, 2004). AGO containing RNA-induced silencing complexes (RISCs) are incorporated into these small RNAs in RNA degradation, translational inhibition, or heterochromatin formation (Bologna and Voinnet, 2014). In Arabidopsis, DCL2, DCL3, and DCL4 target viral genomes to yield virus derived small interfering RNAs (viRNAs) of 22-, 24,- and 21-nts, respectively. Antiviral immunity is conferred by DCL4-dependent, 21-nt viRNAs with DCL2 acting as a DCL4 surrogate (Blevins et al., 2006; Bouché et al., 2006; Fusaro et al., 2006; Diaz-Pendon et al., 2007; Donaire et al., 2008). 24-nt viRNAs produced by DCL3 might be related to the perception of non-cell autonomous silencing signals (Brosnan et al., 2007; Diaz-Pendon et al., 2007). Whereas, AGO1 and AGO2 proteins are specifically involved in antiviral defense by catalyzing viral RNA cleavage (Jaubert et al., 2011).
RNA-dependent RNA polymerases (RDRs) are essential for synthesis of double-stranded RNAs (dsRNAs), which eventually cleave into small RNAs, to originate a new turn of RNA silencing (Sijen et al., 2001; Wassenegger and Krczal, 2006; Qi et al., 2009; Voinnet, 2009; Wang et al., 2010). RDRs have been identified in a wide range of plants. The first plant-encoded RDR was isolated from tomato (Solanum lycopersicum), and named as SlRDR1 (Schiebel et al., 1998). Silencing of SlRDR1 using virus-induced gene silencing (VIGS) significantly reduces plant defense against tobacco mosaic virus (TMV) in tomato (Liao et al., 2015). In Arabidopsis, salicylic acid (SA) treatment and viral infection can induce a RDR homolog. AtRDR1 knockout mutants accumulate higher levels of viral RNAs than those of wild-type (WT) plants after infection with tobamovirus (Yu et al., 2003). The promoter of AtRDR1 has an extensive scope response to diverse stresses and is sensitive to ABA and SA. Analysis of promoter activity has revealed that AtRDR1 is primarily expressed in the vascular tissue system, specifically in phloem cell layers of roots (Xu et al., 2013). However, it showed TMV inoculation-induced transient upregulation of AtRDR1 expression was attributed to wounding-induced injury, but not a direct consequence of infection (Hunter et al., 2013). Similar to tomato, NtRDRP1 was isolated from tobacco, and NtRDRP1 transcript could be induced by viral infection or SA treatment. The transgenic NtRDRP1 antisense transgenic plants accumulate more virus RNA and develop symptoms that both in inoculated leaves and upper uninoculated leaves (Xie et al., 2001). A RDR1 homolog, NbRdRP1m, isolated from N. benthamiana, contains a 72-nt insert in the 5′ position of the ORF (Yang et al., 2004). NbRdRP1m could also be induced by SA treatment and TMV infection. N. benthamiana plants overexpression with a SA-induced RDR1 gene from Medicago truncatula exhibit resistance to TMV, turnip vein-clearing virus and sunn hemp mosaic virus (Yang et al., 2004). Moreover, tobacco plants overexpression with NtRDR1 from tobacco show hyper susceptibility to plum pox poty virus and other viruses, alike to RDR6-silenced plants (Ying et al., 2010). In potato (Solanum tuberosum), SA induces expression of StRDR1, however, knockdown of StRDR1 gene does not increase susceptibility to the viruses, such as TMV, PVX, and PVY (Hunter et al., 2016). OsRDR1 in rice (Oryza sativa) is required for Bromovirus–mediated RNA silencing (Chen et al., 2010). In addition, maize ZmRdRP1 can be induced by exogenous SA, methyl jasmonate (MeJA) treatment and sugarcane mosaic virus infection (He et al., 2010). Similarly, SA and fungal (Rhizoctonia solani Kuhn) infection induce GhRDR1 in cotton (Gossypium hirsutum; Gao et al., 2009). These results energetically put forward an important role of RDR1 in plant antiviral defense, nonetheless, the functions of RDR1 still remain disputed in different species.
Despite the discrepancy in published results, it is well-recognized that plant RDR1 is involved in viRNAs biogenesis and viRNAs-mediated antiviral defense (Qi et al., 2009; Qu, 2010). However, the role of RDR1 in pepper virus defense still remains unknown. In the present study, a RDR1 orthologous gene in pepper designated as CaRDR1, was identified, and its spatial expression patterns were characterized. CaRDR1 was predominantly expressed in pepper stem, where played a major role in nutrient transport and virus spread. Its expression was upregulated by exogenous SA, ABA, H2O2, and TMV infection. Silencing of CaRDR1 increased the transcript level of TMV-CP and reduced the resistance of pepper to TMV. Overexpression of CaRDR1 in N. benthamiana resulted in less accumulation of TMV-CP transcripts following TMV infection. In addition, the expression levels of RNA silencing related genes, such as NbAGO2, NbDCL2, NbDCL3, and NbDCL4 were upregulated by TMV inoculation in CaRDR1 expressing N. benthamiana. These results strongly suggest that CaRDR1 plays a positive role in TMV resistance in pepper.
Materials and Methods
Plant Material Growth Conditions and Treatment
Pepper (Capsicum annuum L.) lines P79 and P54 used in the present study were pepper inbred lines from our lab. They showed different response to TMV-U1 (Figures S1A,B, Presentation S1). The pepper seedlings were grown in a plant growth chamber under a 16/8 h light/dark period at 25/20°C. Eight-week-old seedlings of pepper used for relative expression of CaRDR1 in root, stem and leaf. Meanwhile, N. benthamiana were grown under a 16/8 h light/dark period at 25/20°C. Eight -week-old seedlings were used for TMV inoculation.
For hormone treatments, leaves of 8-week-old seedlings of pepper were sprayed with 2 mM SA, 100 μM methyl jasmonate (MeJA), 100 μM ABA and 10 mM H2O2 until surface run-off (Hunter et al., 2013; Cai et al., 2015). SA and H2O2 were dissolved in distilled water, MeJA and ABA were first dissolved in absolute ethanol to prepare a 100 mM stock solution, then diluted with sterile water to a final concentration of 100 μM. The control plants were sprayed with corresponding solvent.
TMV-U1 strain was provided by Guizhou Tobacco Science Research Institute (Shi and Guo, 2012; Ge et al., 2015). TMV-U1 was mechanically inoculated on two or three lower leaves of 8-week-old seedlings by rubbing the virus (0.01 M sodium phosphate buffer, pH 7.0) with carborundum (Kim et al., 2005). Mock inoculation was performed with the buffer only. Accumulation of TMV was confirmed by RT-PCR. The primers used for the experiments were listed in Table S1.
Cloning of CaRDR1
Total RNA was extracted from pepper leaves using Omega plant RNA kit, and cDNA was synthesized using M-MuLV reverse transcriptase (Thermo Scientific, USA). The cDNA samples were amplified by PCR: 95°C for 5 min, 35 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 3.5 min, and then 72°C for 10 min. The primers were listed in Table S1.
Sequence Alignment and Phylogenetic Analysis
Through BLAST analysis in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using the sequence of CaRDR1 protein, the related RDR1 proteins amino acid sequence in various species were gained. The multiple sequence alignments of CaRDR1 and related RDR1 proteins were performed using ClustalW in the MEGA5 software package, and the boxes were drawn using the BoxShade web site (http://www.ch.embnet.org/software/BOX_form.html). The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with Poisson model and 1000 bootstrap replicates test through MEGA5 software (Saitou and Nei, 1987; Tamura, 2011). The sequence information of the proteins used for phylogenetic tree construction was listed in Table S2.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using Omega plant RNA kit, and cDNA was synthesized using PrimeScript RT reagent Kit (Takara, Dalian, China). Quantitative real-time RT-PCR (qRT-PCR) was performed using SYBR® Premix Ex TaqTM from TaKaRa (China) on iQ5 Real-Time PCR Detection System (BIO-RAD Corp., Hercules, California, USA). Pepper ubiquitin-conjugating protein (Ubi-3) was used as the reference gene (Wan et al., 2011). Three biological replicates were performed for qRT-PCR assay. Relative expression levels of genes were determined using the comparative threshold method (2−ΔΔCt; Livak and Schmittgen, 2001). The primers for qRT-PCR were listed in Table S1.
Virus Induced Gene Silencing (VIGS)
Vectors for VIGS had previously been described (Liu et al., 2002a). For pTRV2:CaRDR1, a 383-bp cDNA fragment of CaRDR1 gene was PCR-amplified using primers shown in Table S1. The fully expanded cotyledons of pepper plants were co-infiltrated with Agrobacterium tumefaciens strain GV3101 carrying each TRV derivative (Li et al., 2014; Jing et al., 2016). The plants were maintained at 18–22°C in a plant growth chamber with a 16/8 h light/dark period. The efficacy of gene silence was confirmed by RT-PCR.
Vector Construction and Generation of Transgenic N. benthamiana
The pepper CaRDR1 gene was inserted into the binary vector 35S:PBI121. Then recombinant plasmid CaRDR1-PBI121 and plasmid PBI121 were introduced into Agrobacterium tumefaciens strain GV3101. N. benthamiana was transformed via Agrobacterium-mediated leaf transformation according the protocols of Horsch et al. (1985) and Hou et al. (2015). The plants obtained were PCR-confirmed to select positive transgenic lines (T0), and the seeds from T0 were collected for future research.
Determination of the Malondialdehyde (MDA) Content and Antioxidant Enzyme Activity
The content of MDA was measured using Thiobarbituric acid-reactive substances (TBARS) concentration (War et al., 2012). The extraction of antioxidant enzymes were executed as described by Liao et al. (2012). Leaves (0.5 g) were blended in 10 mL of 25 mM phosphate buffer (pH 7.8) with 0.2 mM EDTA and 2% (w/v) PVP. The homogenate was centrifuged at 12,000 g for 20 min at 4°C. The supernatant was collected for the enzyme activity. Superoxide dismutase (SOD) activity was measured by inhibiting the photochemical reduction of nitroblue tetrazolium (Stewart and Bewley, 1980). Catalase (CAT) activity was monitored the decrease of the absorbance at 240 nm and the activity of peroxidase was assayed using guaiacol by monitoring the absorbance at 470 nm (Madhusudhan et al., 2009).
Statistical Analysis
SAS software is used for statistical analysis. The values are represented as the mean ± standard errors of three independent experiments. Significant differences of the data were by univariate ANOVA analysis with the least significant difference (LSD) at P < 0.05.
Results
Cloning and Characterization of CaRDR1 in Pepper
Through BLAST analysis in Pepper Genome Platform (http://peppergenome.snu.ac.kr/), we identified a putative RDR homolog named as CaRDR1 (Capan11g001709). CaRDR1 was cloned using cDNA extracted from pepper leaves of line P79. The full-length CaRDR1 cDNA consists of 3,351 bp and encodes 1116 amino acids, in which molecular mass of 127.4 kDa and isoelectric point of 8.51 were detected. The full-length of CaRDR1 genomic sequence contains 5,217 bp, including four exons and three introns (Figure 1A). The sequence alignment of the amino acid residues of CaRDR1 compared with other members of the RDR1 family was performed using ClustalW in the MEGA5 software package (Tamura, 2011). CaRDR1 in pepper was 60.69, 86.56, 86.12, 87.99, and 88.90% identical to the RDR1s from Arabidopsis, tabacco, Nicotiana glutinosa, tomato and potato, respectively. The RNA-dependent RNA polymerase catalytic domain of CaRDR1 was located from His (370 aa position) to Val (892 aa position) and the RNA recognition motif (RRM) started from Ile (5 aa position) to Ile (62 aa position; Figure 1B). Furthermore, the RDR1s from various species share a signature motif DLDGD. In addition, CaRDR1 also contained the conserved components which are typical structures in RDR1 family (Wassenegger and Krczal, 2006; Bologna and Voinnet, 2014; Figure 1B).
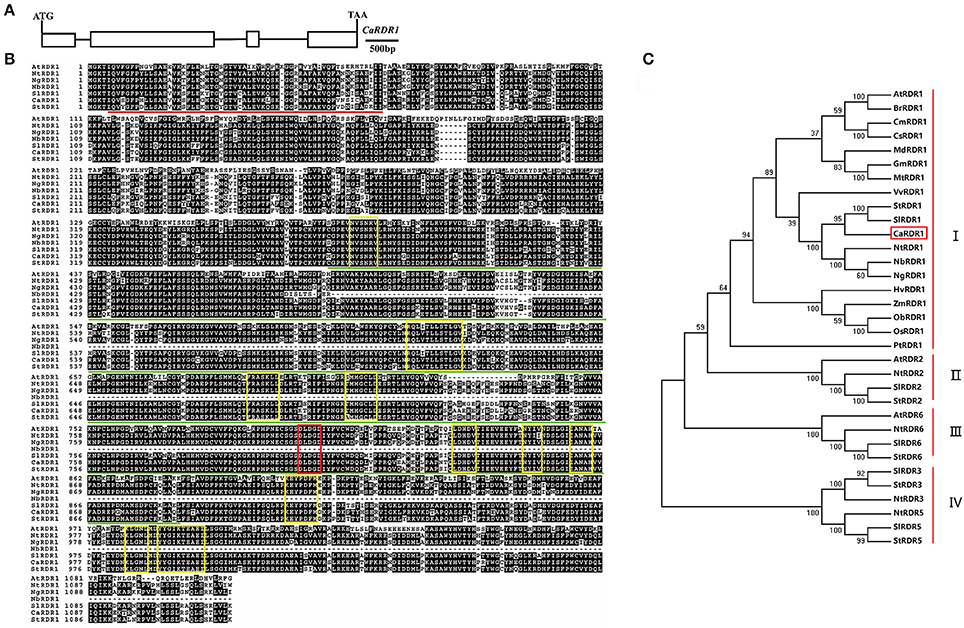
Figure 1. Sequence alignment and phylogenetic analyses of CaRDR1 and its homologs in various species. (A) Structural analysis of CaRDR1 in pepper. Exons and introns were shown in box and line, respectively. Ca, Capsicum annuum L. (B) Sequence alignment of the amino acid of CaRDR1 with other RDR1 proteins. The identical and similar residues were shown in black and gray, respectively. The highly conserved regions for RDR1s were highlighted in yellow box, and signature DLDGD was marked with red box. The highly conserved RNA recognition motif (RRM) and RNA-dependent RNA polymeras (RdRP) domains were indicated respectively in red and green lines. At, Arabidopsis thaliana; Nt, Nicotiana tobacum; Ng, Nicotiana glutinosa; Nb, Nicotiana benthamiana; Sl, Solanum lycopersicum; St, Solanum tuberosum. (C) Phylogenetic analyses of CaRDR1 and its homologs using MEGA5 software based on the neighbor joining method. CaRDR1 from pepper was indicated in red boxes.
In order to understand the evolutionary relationship between CaRDR1 and other RDRs homologs from various species, we performed phylogenetic analysis using MEGA5 software (Figure 1C). The results revealed that RDRs could be classified into four main groups. RDR1s from pepper, tomato, potato and tobacco, which belonged to solanaceae family, were fall into the same clade, suggesting that they might have similar functions. These observations revealed that CaRDR1 was a RDR1 homolog in pepper.
Expression Pattern of CaRDR1 Gene
To further elucidate the function of CaRDR1, its expression pattern analysis was performed in roots, stems, leaves, flowers, green fruits, and red fruits of pepper P79 using qRT-PCR. As shown in Figure 2, CaRDR1 transcript was expressed in all tissues examined, which was consistent with the vital roles of RDR1 in plants. While the highest expression of CaRDR1 was detected in the stems. Similar to NbRDR1m of N. benthamiana (Yang et al., 2004), transcript levels of CaRDR1 in stems, roots and flowers were higher than that in leaves (Figure 2). It is to be noted that AtRDR1 of Arabidopsis is mainly expressed in tissues with mature vascular system (Xu et al., 2013). These data indicated that CaRDR1 might have an important role in limiting pathogen spread.
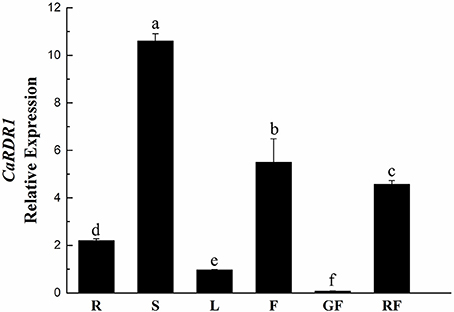
Figure 2. qRT-PCR analyses relative expression of CaRDR1 in different tissues of pepper. Three biological replicates were performed for this experiment and the pepper Ubi3 gene was used as the reference gene. Error bars indicate the standard errors. Letters indicate the significant differences (P < 0.05) between samples. R, roots; S, stems; L, leaves; F, flowers; GF, green fruits; GF, red fruits.
CaRDR1 Was Induced by Phytohormones Treatment and TMV Infection
Since RDRs have been shown as a major constituents for the siRNAs production involved in plant response to stresses (Xie et al., 2001; Hannon, 2002; Yu et al., 2003; Baulcombe, 2004; Ding and Lu, 2011), we analyzed the response of CaRDR1 gene to exogenous SA, MeJA, ABA, and H2O2 in both P79 and P54, which had different response to TMV infect (Figures S1A,B). The results showed that CaRDR1 was induced by SA treatment in both P79 and P54 plants. The response of CaRDR1 to SA increased then reaching the maximum induction at 12 h with a 42-fold increase and then declined in P79 (Figure 3A). Surprisingly, the expression patterns of CaRDR1 were almost similar in P54 plants, where the transcript levels of CaRDR1 peaked at 12 h with a 25-fold increase (Figure S1C). After ABA treatment, CaRDR1 relative expression reached a maximum induction of 2.4-fold at 12 h (Figure 3D). The response of CaRDR1 to H2O2 increased gradually, reached a peak at 48 h of 2.5-fold (Figure 3E), however, it was not induced by MeJA (Figure 3C). To our surprise, the transcripts of CaRDR1 did not significantly response to MeJA, ABA, and H2O2 in P54 (data not show).
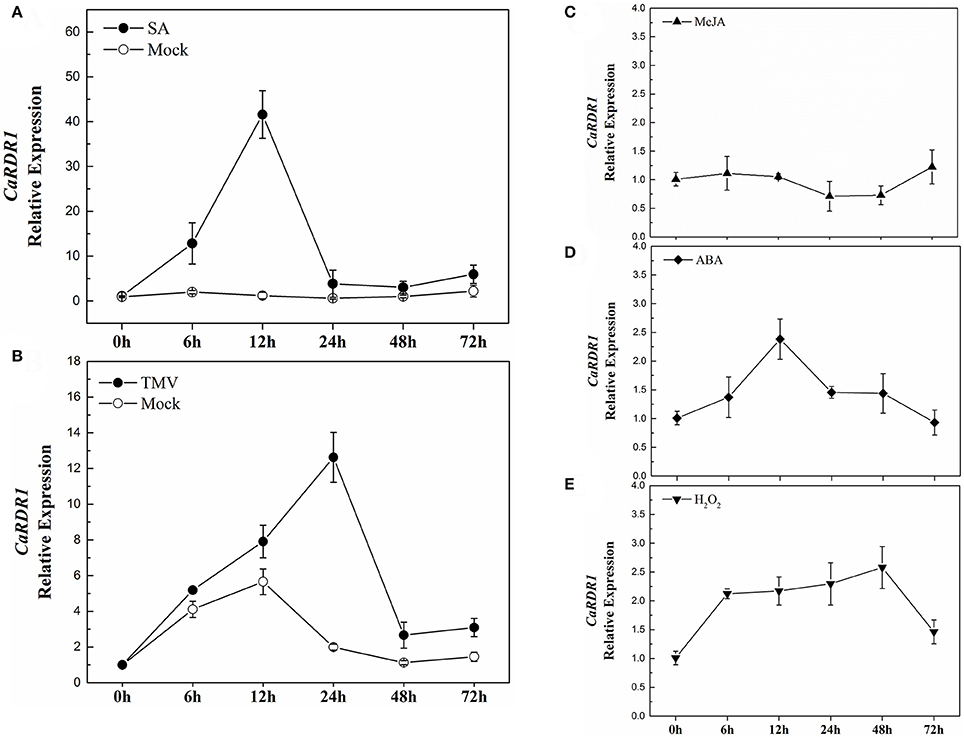
Figure 3. qRT-PCR analyses of CaRDR1 expression as influenced by exogenous phytohormones and TMV inoculation in P79. (A) Effect of 2 mM SA on the expression of CaRDR1 in pepper leaves. (B) Effect of TMV on the expression of CaRDR1 in pepper leaves. (C) Effect of 100 μM MeJA on the expression of CaRDR1 in pepper leaves. (D) Effect of 100 μM ABA on the expression of CaRDR1 in pepper leaves. (E) Effect of 10 mM H2O2 on the expression of CaRDR1 in pepper leaves. The pepper Ubi3 was used as the reference gene, and three biological replicates were performed for these experiments. Error bars indicate the standard errors.
Furthermore, to examine the response of CaRDR1 to TMV, at the 8-week-old, pepper leaves were inoculated with TMV-U1 train using mechanical inoculation technique, subsequently, the transcript level of CaRDR1 was analyzed. Leaves inoculated with phosphate buffer were used as controls. As shown in Figure 3B, CaRDR1 was up-regulated upon TMV infection, and the expression level of CaRDR1 showed a 12-fold-increase after TMV inoculation for 24 h in P79, the highest transcript level was detected at 24 h when the transcript level of CaRDR1 was induced to 9-fold in P54 (Figure S1D). These results indicated that CaRDR1 might be involved in SA-modulated plant virus defense and participated in pepper interaction with TMV.
Silencing of CaRDR1 Reduced the TMV Resistance of Pepper
To determine the CaRDR1 loss-of-function phenotype in pepper TMV defense, VIGS was performed to generate CaRDR1-silenced plants by using recombinant tobacco rattle virus (TRV) construct (Liu et al., 2002b) containing the specific 383 bp cDNA sequence. And the empty vector (EV) was also injected to pepper, that acted as the Control. qRT-PCR analysis revealed that the transcript level of the CaRDR1 was reduced by 80% in the silenced plants, suggesting that CaRDR1 was effectively knocked down by VIGS (Figure 4B, Figure S2A). The WT and TRV:00 plants of P79 showed no symptom, but the upper un-inoculated leaves of CaRDR1-silenced plants exhibited chlorisis and mosaic 15 days post TMV inoculated (Figure 4A). The upper un-inoculated leaves of WT and TRV:00 plants showed mosaic of P54, TRV:CaRDR1 ones exhibited shrinking and mosaic (Figure 4B). Furthermore, the TMV-CP expression in leaves from the CaRDR1-silenced lines was increased by 3-fold as compared to that in the control plants after 7d of TMV incubation in P79, and 1.5-fold as compared to control in P54 (Figure 4C, Figure S2B). Moreover, some biochemical indices were examined in the CaRDR1 silenced plants, and we found that loss-of-function of CaRDR1 could increase MDA production after TMV infection. For example, the MDA content was induced by TMV inoculation in both silenced lines and control plants, but the higher MDA accumulation was detected in CaRDR1 silenced plants at all time points tested after TMV infection (Figure 4D, Figure S2C). These results suggested that plasma membrane damage was more serious in CaRDR1-silenced plants in both resistant and susceptible plants. Furthermore, after TMV infection, the SOD and POD activities were initially elevated and then decreased at 8 dpi in both silenced and control plants, but the activity of CAT was increased gradually. However, the activities of these three antioxidant enzymes in CaRDR1-silenced plants were significantly lower than that of control plants, indicating that knockdown of CaRDR1 inhibited antioxidant enzyme activity under TMV inoculation (Figures 4E–G, Figures S2D–F). The patterns of biochemical indices were similar in P79 and P54, but the MDA accumulation was higher in P54 and CaRDR1-silenced plants than that of P79 plants at all time points tested after TMV infection (Figures S2C–F). The activities of these three antioxidant enzymes were highest in P79 control plants after TMV infection. Since SOD, POD, and CAT are directly involved in scavenging reactive oxygen species (ROS) (Chen et al., 1993; Montalbini et al., 1995; Agnieszka et al., 2009), CaRDR1 modulated changes in their activities might indicate a positive role of CaRDR1 in TMV defense of pepper.
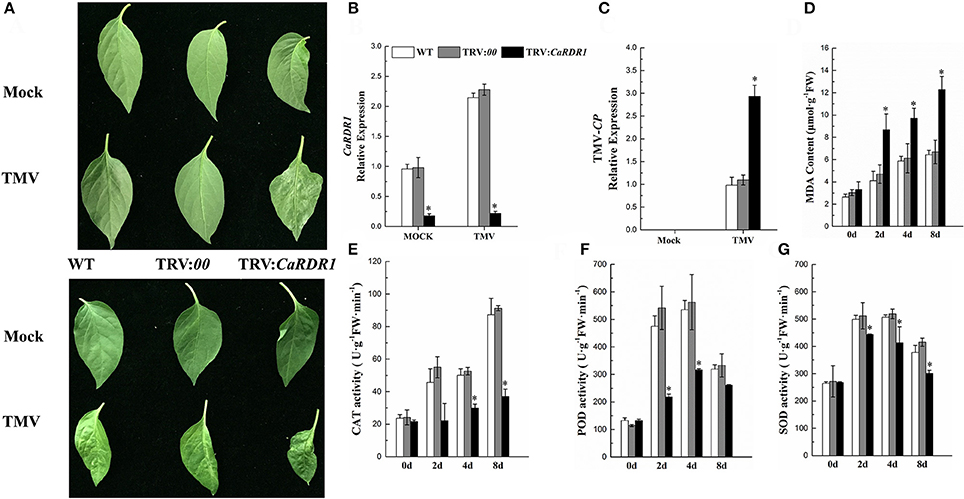
Figure 4. Silencing of CaRDR1 attenuated the TMV resistance of pepper. (A) Phenotype of upper un-inoculated leaves from WT, empty vector (TRV:00), and CaRDR1-silenced (TRV:CaRDR1) plants, P79 (upper) and P54 (down) at 15 dpi with TMV. (B,C) qRT-PCR was used to determine the relative level of CaRDR1 (B) and TMV-CP (C) transcript of WT, empty vector (TRV: 00), and CaRDR1-silenced (TRV: CaRDR1) plants at 7 days post-inoculation (dpi). (D) The malondialdehyde (MDA) content. (E) catalase (CAT) activities (F) peroxidase (POD) activities (G) superoxide dismutase (SOD) activities. Error bars indicate the standard errors. Asterisks indicate the significant differences (P < 0.05) between WT, TRV:00 and TRV: CaRDR1 lines.
Overexpression of CaRDR1 Protected N. benthamiana from TMV-Induced Damage
As silencing of CaRDR1 reduced TMV defense of pepper, we hypothesized that overexpression of CaRDR1 might enhance TMV resistance. Therefore, we generated individual CaRDR1 overexpressing (CaRDR1-OE) transgenic N. benthamiana lines and EV line. We used real-time RT-PCR analysis to identify lines that expressed the transgene at high levels (Figure S3). It was observed that developing leaves from both CaRDR1-OE (OE-3, OE-4, OE-6) and EV plants became chlorotic and shrinking 10 days post-inoculation (dpi). Meanwhile, EV control plants displayed stem and leaf necrosis, however, such symptom was not observed on plants transformed with CaRDR1. By 20 dpi, EV plants were dead or near death, whereas CaRDR1-OE plants showed continued growth with yellowing and mosaic symptoms in young leaves (Figure 5). qRT-PCR was used to analyze the TMV-CP expression in leaves from the CaRDR1-OE lines. At 3 dpi, the transgenic lines expressed an increased transcript level of CaRDR1 but a decreased transcript level of CP RNA compared with that of EV line (Figure 6A). By 7 dpi, accumulation of TMV-CP transcript was remarkably increased both in the transgenic and EV lines, but EV line showed a 3.5-fold increased transcript of TMV-CP compared to that of CaRDR1-OE lines (Figure 6A). In addition, NbTOM1, which is required for efficient multiplication of Tobamoviruses, was down-regulated in CaRDR1-OE plants as compared with EV plants (Figure 6B). Likewise, from 1 to 3 dpi, a decreased MDA content was detected in leaves of CaRDR1-OE lines compared to EV plants (Figure 6C). As plants possess antioxidants that can scavenge ROS to protect cells from oxidative damage, the activities of SOD, POD and CAT were investigated in both CaRDR1-OE and EV plants after TMV inoculation. The activities of CAT and POD in the CaRDR1-OE lines kept higher than in the EV plants, but the TMV treatment resulted in non-significant increase of the activities of CAT in both CaRDR1-OE and EV ones (Figures 6D,E). The CaRDR1-OE lines exhibited high SOD activity in un-treatment plants, but non-significant difference with EV lines under TMV infection. qPCR analysis showed that the TMV infection increased the relative mRNA abundance of NbAOX1a in CaRDR1-OE of 2.5-fold compared to EV plants at 3 dpi. NbAOX1b did not responded to TMV (Figures 6G,H).

Figure 5. Phenotype analysis of wild type, empty vector and CaRDR1-overexpressed (OE-3, OE-4, and OE-6) plants at 0, 10, and 20 dpi with TMV.
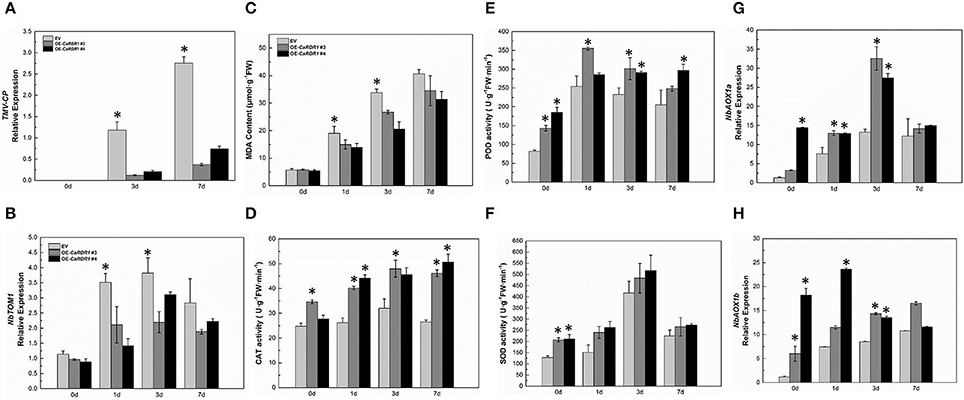
Figure 6. Overexpression CaRDR1 enhanced the TMV resistance of N. benthamiana. (A) qRT-PCR was used to determine the relative level of TMV–CP of empty vector and CaRDR1-OE plants at 0, 3, and 7 dpi. (B) qRT-PCR was used to determine the relative level of NbTOM in upper un-inoculated leaves of empty vector and CaRDR1-OE plants at 0, 1, 3, and 7 dpi. (C) The MDA content (D–F) CAT (D), SOD (E) and POD (F) activities measurement in leaves of empty vector and CaRDR1-OE plants at 0, 1, 3, and 7 dpi. (G,H) qRT-PCR was used to determine the relative level of NbAOX1a (G) and NbAOX1b (H) transcript in empty vector and CaRDR1-OE plants at 0, 1, 3, and 7 dpi. Three biological replicates were performed for these experiments. The N. benthamiana NbEF1α gene was used as the reference gene. Error bars indicate the standard errors. Asterisks indicate the significant differences (P < 0.05) between EV and CaRDR1-OE lines.
NbAGO1, NbAGO2, NbDCL2, NbDCL3, NbDCL4, NbRDR6, and NbRDR1 participate in RNA silencing-mediated virus defense conjointly (Nakasugi et al., 2013). Since our results reveal that CaRDR1 is positively correlated with TMV resistance, we then intended to clarify whether CaRDR1 affects the expression profiles of RNA silencing genes. The expression patterns of NbAGO1, NbAGO2, NbDCL2, NbDCL3, NbDCL4, and NbRDR6 were monitored in CaRDR1-OE and EV plants following TMV inoculation. The overexpression of CaRDR1 showed up-regulated of NbAGO2. TMV inoculation induced up-regulation of NbAGO1, NbAGO2, NbDCL2, and NbDCL4. The expression of NbAGO1 was two times higher in CaRDR1-OE plants than that in EV plants at 1 dpi (Figure 7A). TMV inoculation gradually increased transcript of NbAGO2 over time, which remained consistently higher in CaRDR1-OE plants than that in EV plants (Figure 7B). The magnitude of NbDCL2/4 were also higher in CaRDR1-OE plants during TMV infection. The highest transcript levels were detected at 3 dpi, when the transcript of NbDCL2/4 were induced to 2- and 3-fold, respectively in CaRDR1-OE plants compared to EV (Figures 7C,E). In contrast, expression of NbRDR6 was downregulated in CaRDR1-OE plants, particularly at 3 dpi (Figure 7F). The data presented here indicate that CaRDR1 might play an important role in regulating these RNA silencing related genes upon TMV inoculation. Taken together, these results suggested that CaRDR1 functions positively in TMV resistance by up-regulating AGOs and DCLs in N. benthamiana.
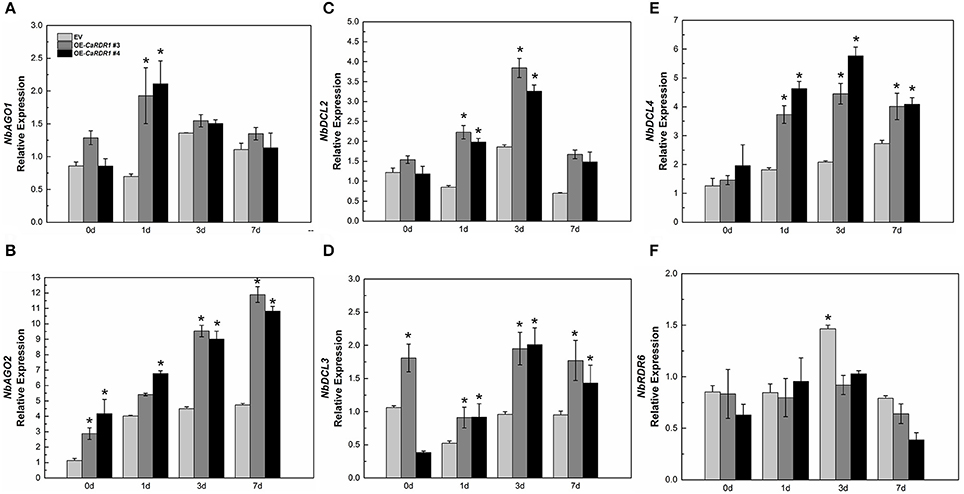
Figure 7. RNA silencing related genes expression in EV and CaRDR1-OE plants after TMV inoculation (A–F). (A) NbAGO1 (B) NbAGO2 (C) NbDCL2 (D) NbDCL3 (E) NbDCL4 (F) NbRDR6. The leaf samples were obtained from empty vector and CaRDR1-OE plants at 0, 1, 3, and 7 dpi. Three biological replicates were performed for these experiments. The N. benthamiana NbEF1α gene was used as the reference gene. Error bars indicate the standard errors. Asterisks indicate the significant differences (P < 0.05) between EV and CaRDR1-OE lines.
Discussion
In the present study, a CaRDR1 gene was cloned from pepper. The expression of CaRDR1 was induced by SA, ABA, H2O2, and TMV. Further, down-regulated the transcripts of CaRDR1 through VIGS caused severe symptom and more virus RNA accumulation in pepper. Ectopic expression CaRDR1 in N. benthamiana suppressed the lethal damage of TMV, in addition enhanced the expression of RNA silencing-related genes. The data suggested that CaRDR1 might act as a positive regulator in the interaction of pepper and TMV.
RNA silencing functions as a major natural antiviral defense mechanism in plants, but there are little studies of this process in pepper. RDRs synthesize dsRNA intermediates and play an important role in the initiation and amplification of RNA silencing (Baulcombe, 2004; Ding and Lu, 2011). There are six RDRs in Arabidopsis thaliana, AtRDR1, AtRDR2, AtRDR3a, AtRDR3b, AtRDR3c, and AtRDR6 (Wassenegger and Krczal, 2006). Several recent researches indicate that plant RDR1 is involved in antiviral defense (Diaz-Pendon et al., 2007; Qi et al., 2009; Qu, 2010). RDR1 orthologs have been identified in many species, such as Arabidopsis (AtRDR1), tomato (SlRDR1), Nicotiana spp. (NtRDR1, NbRDR1, and NgRDR1), potato (StRDR1), rice (OsRDR1), and maize (ZmRDR1; Schiebel et al., 1998; Xie et al., 2001; Yu et al., 2003; Yang et al., 2004; Wassenegger and Krczal, 2006; Liu et al., 2009; Hunter et al., 2013). Nonetheless, the functions of RDR1 still remain disputed in different hosts and viruses. In this study, a CaRDR1 gene was identified from pepper. Similar to other RDR1s, CaRDR1 also has a signature catalytic motif DLDGD (Figure 1B), which is likely to form the nucleotidyl transferase active site partially via a coordinated divalent cation (Iyer et al., 2003; Wassenegger and Krczal, 2006). Phylogenetic analysis indicated that RDR1 proteins in the solanaceae family fall into the same clade (Figure 1C). Investigated its expression pattern in different plant parts, it was found that CaRDR1 was predominantly expressed in pepper stem (Figure 2). As the previous research, NbRdRP1m in N. benthamiana expressed higher in stem than leaf, moreover, AtRDR1 was expressed in tissues with mature vascular system in phloem (Yang et al., 2004; Xu et al., 2013). The vascular system of plants has been shown to play an important role in virus spread (Lough and Lucas, 2006; Petricka et al., 2012). Considering the positive role of CaRDR1 in defense against TMV, we proposed that the high CaRDR1 expression in the stem might limit pathogen spread, leading to an enhancement in pepper resistance against virus.
Plant hormones play a vital role in plant immunity. In pathogens defense, plants produce a complicated mixture of SA, JA, and ABA to modulate plant defense response to invading pathogens (Kunkel and Brooks, 2002; Chen et al., 2013; He et al., 2017). CaRDR1 was response to multiple plant hormones in this study (Figure 3), especially induced by SA like other RDR1 orthologs (Yang et al., 2004; Hunter et al., 2013; Liao et al., 2013). Interestingly, the expression of CaRDR1 could not be induced by MeJA treatment. Nonetheless, NgRDR1 in Nicotiana glutinosa was up-regulated by MeJA (Liu et al., 2009). The interactions between SA and JA signaling are complex, the primary mode of interaction between them appears to be mutual antagonism (Kunkel and Brooks, 2002). It is suggested that CaRDR1 involved in pepper TMV resistance cooperated with SA rather than with JA. CaRDR1 also could be induced by ABA and H2O2, which suggests that CaRDR1 may interact with SA, H2O2, and ABA signal pathways. The different expression patterns of RDRs upon treatments with different signaling molecules indicate the complexity of the RDR family in higher plants, and also suggest that different RDRs may participate in different signal pathways and have different functions.
CaRDR1 was induced by TMV infection in both TMV resistant and susceptible pepper genotypes. While knockdown of CaRDR1 in leaves by VIGS increased the transcript level of TMV-CP and reduced the resistance to TMV of pepper (Figure 4). This is consistent with previous reports that NtRDRP1 antisense transgenic tobacco accumulated significantly higher levels of viral and displayed more severe disease symptoms (Xie et al., 2001). The AtRDR1 knockout mutant accumulated higher persistent viral RNAs levels after infected by a tobamovirus and a tobravirus in Arabidopsis (Yu et al., 2003). However, suppression the transcripts of StRDR1 in potato did not increase the susceptibility of potato to PVX, PVY, and TMV (Hunter et al., 2016). MDA is one of the important lipid peroxidation products involved in the plant defense singling under many stresses (Gechev et al., 2002; War et al., 2012). While our data displayed that down-regulation of CaRDR1 led to an increasing of MDA content, and overexpression of CaRDR1 led to MDA content decreased (Figures 4D, 6C). Increased MDA content are often associated with oxidative stress (Gechev et al., 2002; Madhusudhan et al., 2009). It suggested that down-regulation of CaRDR1 subsequent damage to the plasma membrane. Attenuated antioxidant enzymes activities were obtained in CaRDR1-silencied plants compared to the control ones under TMV infection (Figures 4E–G). It has been reported that TMV infection could induce POD activity but suppress CAT activity in tobacco, bell pepper and tomato plants (Madhusudhan et al., 2009). POD activity increased in RDR1 silenced tobacco lines 3 days after inoculation with PVY (Rakhshandehroo et al., 2012). ROS perform multiple roles during plant defense responses to microbial attack by acting directly in the initial defense (Klessig et al., 2000). On the other hand, ROS accumulating overly shown to be toxic. Antioxidant enzymes regulate the content of ROS depends on its role in virus defense. Therefore, the activities of antioxidant enzymes may be influenced by the period of measurement. The VIGS-silencing experiment implied that the role of RDR1 in the defense response was different among crops. There are some disparities among the RDR gene family in different plants species. The redundant and overlap of RDRs functions might be one explanation for these results.
TMV-U1 strain causes the death of N. benthamiana plants. CaRDR1 overexpression showed moderate disease symptom and delayed the lethality of TMV-U1 on N. benthamiana plants (Figure 5). Furthermore, CaRDR1-OE transgenic N. benthamiana showed less TMV-CP transcripts than EV ones (Figure 6A). These results agreed with previous studies that MtRDR1 overexpression provided protection against TMV infection, which eventually kept tissue adjacent to the apical meristem free from TMV particles (Yang et al., 2004; Lee et al., 2016). TOM1 interacts with the helicase domain of TMV replicase to form the replication complex, and supports tobamovirus multiplication on an early stage of infection with TOM3 (Hagiwara et al., 2003; Asano et al., 2005). In addition, the expression of NbTOM1 which required for efficient multiplication of Tobamoviruses decreased in CaRDR1-OE lines (Figure 6B), suggesting that CaRDR1 not only limited pathogen spread, but also suppressed TMV replication. CaRDR1 overexpression alleviated the virus damage on the plasma membrane by less MDA accumulation (Figure 6C). The activity of CAT was higher in CaRDR1-OE lines than in EV plants (Figure 6D). Curiously, we observed that CaRDR1 transgenic plants compared with WT plants had greater POD and SOD activities without the TMV infection (Figures 6E,F). The CaMV35S promoter was used in CaRDR1 transgenic, it suggested that the constitutive overexpression of CaRDR1 influenced the action mechanism of the antioxidant enzymes. Plant mitochondria possess a second terminal oxidase, termed the alternative oxidase (AOX). AOX is a terminal oxidase of the plant mitochondrial electron transport chain, which has an important function in plant defense against TMV (Chivasa et al., 1997; Fu et al., 2010; Lee et al., 2011; Liao et al., 2012). The expression of NbAOX1a and NbAOX1b were distinctly higher in CaRDR1-overexpressing lines, and TMV up-regulated the transcripts of NbAOX1a indicating a potential involvement of CaRDR1 in the AOX-mediated defense response to TMV (Figures 6G,H). There is crosstalk between the functional RDR1-mediated signal pathway and the AOX pathway. ROS might be involved in the crosstalk between RDR1 and AOX. Overexpression of CaRDR1 may altered AOX gene expression by influencing the action mechanism of antioxidant enzymes and the content of ROS. It is suggested NbAOX1a work as an assist for antioxidant systems after TMV challenge, restricting the accumulation of ROS, which might injure the plant membrane system.
The RNA silencing pathways components are encoded by gene families of the DCLs, AGOs, and RDRs. They accomplish targeted RNA degradation, translational repression and heterochromatin modification (Eamens et al., 2008). In this study, expression levels of NbAGO1, NbAGO2, NbDCL2, NbDCL3, and NbDCL4 were relatively higher in transgenic N. benthamiana (Figure 7). NbAGO1 and NbAGO2 might participate in the interaction between N. benthamiana and TMV with RDR1. This agreed with a previous report that southern rice black-streaked dwarf virus (SRBSDV) infection significantly increased the expression of OsAGO1d, OsAGO2, OsRDR1, and OsRDR6 (Xu and Zhou, 2017). RDR mediated the amplification process of RNA silencing, which is likely to be essential in virus defense to ensure the viral RNAs silencing keep pace with the replication of viral RNA (Baulcombe, 2004). Previous research showed that dsRNA was synthesized by host RDR1 or RDR6, recognized by DCL4 or DCL2, and processed into the secondary viral siRNAs to direct more potent antiviral silencing (Wang et al., 2010). It suggested the up-regulated NbDCL2 and NbDCL4 might be consociation with the overexpression CaRDR1 to suppress the virus accumulation in N. benthamiana. Surprisingly, the expression of NbRDR6 was reduced in CaRDR1-OE plants. NtRDR1 might have a dual role, on one hand, contributing to SA mediated antiviral defense, on the other hand, suppressing the RDR6-mediated antiviral RNA silencing (Ying et al., 2010). AtRDR6 was involved in cucumber mosaic virus (CMV) defense (Dalmay et al., 2000; Mourrain et al., 2000). NbRDR6 had been discovered having effects on limiting the virus systemic spread during the Potato potexvirus X (PVX) infection (Schwach et al., 2005). In this study, CaRDR1 might have a combination with NbAGO2, NbDCL2/3/4 and complementary effect with NbRDR6. Owing to the apparent function of CaRDR1 within different regulatory networks, it is difficult to define the detailed and specific individual pathway in TMV response. Therefore, further study is essential to gain a more thorough knowledge of roles of CaRDR1 in these pathways and to elucidate the regulatory mechanisms more precisely.
Conclusions
CaRDR1 exhibited a high degree of identity with other RDR1s of Solanaceae. The transcripts of CaRDR1 was induced by TMV and SA. CaRDR1 also responded to H2O2 and ABA. When CaRDR1 was silenced in pepper plants, the silenced plants showed increased TMV-CP transcript and MDA content but decreased antioxidant enzymes activities. In contrast, CaRDR1-OE plants showed mild symptom, decreased TMV-CP transcripts and elevated expression of RNA silencing related genes, including NbAGO2, NbDCL2, and NbDCL4. CaRDR1 was likely to limit pathogen spread and suppress TMV replication to protect plant from TMV attack.
Author Contributions
YL and LQ designed the study. LQ, NM, and YangZ performed the experiments. LQ, TM, and GZ analyzed the data, LQ wrote original manuscript, YL and YanZ revised the manuscript, and YL gave the final approval of the version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by The National Key Technology R&D Program of China (2013BAD01B04-14). The TMV-U1 train was kindly provided by Yushuang Guo of Guizhou Tobacco Science Research Institute.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01068/full#supplementary-material
Table S1. The sequences of primers used in this study.
Table S2. The sequence information of the proteins used for phylogenetic tree construction.
Figure S1. qRT-PCR analyses of CaRDR1 transcript as influenced by exogenous SA treatment and TMV inoculation in P79 and P54 pepper genotypes. (A) Disease incidence (%) of TMV in P79 and P54 (B) Disease index of TMV in P79 and P54 (C) Effect of SA on the expression of CaRDR1 in pepper leaves. (D) Effect of TMV on the expression of CaRDR1 in pepper leaves. The pepper Ubi3 was used as the reference gene, and three biological replicates were performed for these experiments. Err bars indicate the standard errors. Asterisks indicate the significant differences (P < 0.05) between P79 and P54.
Figure S2. Silencing of CaRDR1 attenuated the TMV resistance of pepper in P79 and P54 pepper genotypes. (A,B) qRT-PCR was used to determine the relative level of CaRDR1 in P54 (A) and TMV −CP (B) transcript in un-inoculated leaves of empty vector (TRV: 00) and CaRDR1-silenced (TRV: CaRDR1) plants at 7 days post-inoculation (dpi). (C) The MDA content measurement in inoculated leaves of empty vector (TRV: 00) and CaRDR1 -silenced (TRV: CaRDR1) plants. (D,F) POD (D), CAT (E), and SOD (F) activities measurement in in inoculated leaves of empty vector (TRV: 00) and CaRDR1 silencing (TRV: CaRDR1) plants. Three biological replicates were performed for these experiments. Error bars indicate the standard errors. Asterisks indicate the significant differences (P < 0.05) between TRV: 00 and TRV: CaRDR1 lines.
Figure S3. Molecular characterization of transgenic plants. (A) PCR confirmation of transgenic N. benthamiana plants. Lanes: EV PBI121 vector; M molecular size marker; V CaRDR1-PBI121 vector; WT, untransformed wild type; OE-2,3,4,6,7,9 independent transgenic lines. (B) qRT-PCR analysis of CaRDR1 mRNA levels from leaves of 4-week-old transgenic N. benthamiana. Three biological replicates were performed for this experiment and the N. benthamiana NbEF1α gene was used as the reference gene Err bars indicate the standard errors. Asterisks indicate the significant differences (P < 0.05) between WT, EV and CaRDR1-OE lines.
Presentation S1. Disease evaluation in pepper plants.
Abbreviations
ABA, abscisic acid; AGO, argonaute; AOX, alternative oxidase; CAT, catalase; CMV, cucumber mosaic virus; DCL, dicer-like; EV, empty vector; LSD, least significant difference; MDA, malondialdehyde; POD, peroxidase; qRT-PCR, quantitative real-time PCR; PVX, potato potexvirus X; RDR, RNA-dependent RNA polymerases; ROS, reactive oxygen species; RRM, RNA recognition motif; SA, salicylic acid; SOD, superoxide dismutase; SRBSDV, southern rice black-streaked dwarf virus; TBARS, thiobarbituric acid-reactive substances; TMV, tobacco mosaic virus; TMV-CP, tobacco mosaic virus-coat protein; TRV, tobacco rattle virus; VIGS, virus-induced gene silencing; viRNAs, virus derived small interfering RNAs
References
Agnieszka, B., Dorota, K. P., Magdalena, K., Jacek, H., Karolina, S. T., Liliana, S., et al. (2009). Role of polyisoprenoids in tobacco resistance against biotic stresses. Physiol. Plant. 135, 351–364. doi: 10.1111/j.1399-3054.2009.01204.x
Asano, M., Satoh, R., Mochizuki, A., Tsuda, S., Yamanaka, T., Nishiguchi, M., et al. (2005). Tobamovirus-resistant tobacco generated by RNA interference directed against host genes. FEBS Lett. 579, 4479–4484. doi: 10.1016/j.febslet.2005.07.021
Blevins, T., Rajeswaran, R., Shivaprasad, P. V., Beknazariants, D., Siammour, A., Park, H. S., et al. (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34, 6233–6246. doi: 10.1093/nar/gkl886
Bologna, N. G., and Voinnet, O. (2014). The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65, 473–503. doi: 10.1146/annurev-arplant-050213-035728
Bouché, N., Lauressergues, D., Gasciolli, V., and Vaucheret, H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25, 3347–3356. doi: 10.1038/sj.emboj.7601217
Brosnan, C. A., Mitter, N., Christie, M., Smith, N. A., Waterhouse, P. M., and Carroll, B. J. (2007). Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 14741–14746. doi: 10.1073/pnas.0706701104
Cai, H., Yang, S., Yan, Y., Xiao, Z., Cheng, J., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Carmell, M. A., and Hannon, G. J. (2004). RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 11, 214–218. doi: 10.1038/nsmb729
Chen, H., Tamai, A., Mori, M., Ugaki, M., Tanaka, Y., Samadder, P. P., et al. (2010). Analysis of rice RNA-dependent RNA polymerase 1 (OsRDR1) in virus-mediated RNA silencing after particle bombardment. J. Gen. Plant Pathol. 76, 152–160. doi: 10.1007/s10327-010-0226-5
Chen, L., Zhang, L., Li, D., Wang, F., and Yu, D. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 1963–1971. doi: 10.1073/pnas.1221347110
Chen, Z., Ricigliano, J. W., and Klessig, D. F. (1993). Purification and characterization of a soluble salicylic acid-binding protein from tobacco. Proc. Natl. Acad. Sci. U.S.A. 90, 9533–9537. doi: 10.1073/pnas.90.20.9533
Chivasa, S., Murphy, A. M., Naylor, M., and Carr, J. P. (1997). Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell 9, 547–557. doi: 10.1105/tpc.9.4.547
Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D. C. (2000). An RNA-Dependent RNA polymerase gene in arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. doi: 10.1016/S0092-8674(00)80864-8
Diaz-Pendon, J. A., Li, F., Li, W. X., and Ding, S. W. (2007). Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19, 2053–2063. doi: 10.1105/tpc.106.047449
Ding, S. W., and Lu, R. (2011). Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr. Opin. Virol. 1, 533–544. doi: 10.1016/j.coviro.2011.10.028
Donaire, L., Barajas, D., Martinez-Garcia, B., Martinez-Priego, L., Pagan, I., and Llave, C. (2008). Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J. Virol. 82, 5167–5177. doi: 10.1128/JVI.00272-08
Eamens, A., Wang, M. B., Smith, N. A., and Waterhouse, P. M. (2008). RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiol. 147, 456–468. doi: 10.1104/pp.108.117275
Fu, L. J., Shi, K., Gu, M., Zhou, Y. H., Dong, D. K., Liang, W. S., et al. (2010). Systemic induction and role of mitochondrial alternative oxidase and nitric oxide in a compatible tomato-Tobacco mosaic virus interaction. Mol. Plant Microbe Interact. 23, 39–48. doi: 10.1094/MPMI-23-1-0039
Fusaro, A. F., Matthew, L., Smith, N. A., Curtin, S. J., Dedic-Hagan, J., Ellacott, G. A., et al. (2006). RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7, 1168–1175. doi: 10.1038/sj.embor.7400837
Gao, Q., Liu, Y., Wang, M., Zhang, J., Gai, Y., Zhu, C., et al. (2009). Molecular cloning and characterization of an inducible RNA-dependent RNA polymerase gene, GhRdRP, from cotton (Gossypium hirsutum L.). 36, 47–56. doi: 10.1007/s11033-007-9150-y
Ge, Y. H., Huanping, L., Zheng, J., Chen, X. J., and Geng, Z. L. (2015). Anti tobacco mosaic virus activity of triterpenoidsaponin from Phytolacca americana L. Chinese J. Anim. Nutr. 17, 300–306. doi: 10.3969/j.issn.1008-7303.2015.03.08
Gechev, T., Gadjev, I., Breusegem, F. V., Inzé, D., Dukiandjiev, S., Toneva, V., et al. (2002). Hydrogen peroxide protects tobacco from oxidative stress by inducing a set of antioxidant enzymes. Cell. Mol. Life Sci. 59, 708–714. doi: 10.1007/s00018-002-8459-x
Hagiwara, Y., Komoda, K., Yamanaka, T., Tamai, A., Meshi, T., Funada, R., et al. (2003). Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 22, 344–353. doi: 10.1093/emboj/cdg033
He, J., Dong, Z., Jia, Z., Wang, J., and Wang, G. (2010). Isolation, expression and functional analysis of a putative RNA-dependent RNA polymerase gene from maize (Zea mays L.). Mol. Biol. Rep. 37, 865–874. doi: 10.1007/s11033-009-9692-2
He, Y., Zhang, H., Sun, Z., Li, J., Hong, G., Zhu, Q., et al. (2017). Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 214, 388–399. doi: 10.1111/nph.14376
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., and Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Hou, Y., Meng, K., Ye, H., Ban, Q., Wang, B., Suo, J., et al. (2015). The persimmon 9-lipoxygenase gene DkLOX3 plays positive roles in both promoting senescence and enhancing tolerance to abiotic stress. Front. Plant Sci. 6:01073. doi: 10.3389/fpls.2015.01073
Hunter, L. J. R., Brockington, S. F., Murphy, A. M., Pate, A. E., Gruden, K., MacFarlane, S. A., et al. (2016). RNA-dependent RNA polymerase 1 in potato (Solanum tuberosum) and its relationship to other plant RNA-dependent RNA polymerases. Sci. Rep. 6:11. doi: 10.1038/srep23082
Hunter, L. J., Westwood, J. H., Heath, G., Macaulay, K., Smith, A. G., Macfarlane, S. A., et al. (2013). Regulation of RNA-dependent RNA polymerase 1 and isochorismate synthase gene expression in Arabidopsis. PLoS ONE 8:e66530. doi: 10.1371/journal.pone.0066530
Iyer, L. M., Koonin, E. V., and Aravind, L. (2003). Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 3:1. doi: 10.1186/1472-6807-3-1
Jaubert, M., Bhattacharjee, S., Mello, A. F. S., Perry, K. L., and Moffett, P. (2011). ARGONAUTE2 Mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 156, 1556–1564. doi: 10.1104/pp.111.178012
Jing, H., Li, C., Ma, F., Ma, J. H., Khan, A., Wang, X., et al. (2016). Genome-wide identification, expression diversication of dehydrin gene family and characterization of CaDHN3 in pepper (Capsicum annuum L.). PLoS ONE 11:e0161073. doi: 10.1371/journal.pone.0161073
Kim, D. H., Cho, J. D., Kim, J. H., Kim, J. S., and Cho, E. K. (2005). Ultrastructural characteristics of necrosis and stunt disease in red pepper by the mixed infections of Tobacco mosaic virus-U1 or Pepper mild mottle virus and Pepper mottle virus. Plant Pathol. J. 21, 252–257. doi: 10.5423/PPJ.2005.21.3.252
Klessig, D. F., Durner, J., Noad, R., Navarre, D. A., Wendehenne, D., Kumar, D., et al. (2000). Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. U.S.A. 97, 8849–8855. doi: 10.1073/pnas.97.16.8849
Kunkel, B. N., and Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/S1369-5266(02)00275-3
Lee, W. S., Fu, S. F., Li, Z., Murphy, A. M., Dobson, E. A., Garland, L., et al. (2016). Salicylic acid treatment and expression of an RNA-dependent RNA polymerase 1 transgene inhibit lethal symptoms and meristem invasion during tobacco mosaic virus infection in Nicotiana benthamiana. BMC Plant Biol. 16:15. doi: 10.1186/s12870-016-0705-8
Lee, W. S., Fu, S. F., Verchotlubicz, J., and Carr, J. P. (2011). Genetic modification of alternative respiration in Nicotiana benthamiana affects basal and salicylic acid-induced resistance to Potato virus X. BMC Plant Biol. 11:41. doi: 10.1186/1471-2229-11-41
Li, C., Zhang, Z. C., Ghebremariam, K. M., Wang, L. H., Wu, L., and Liang, Y. (2014). A novel method for the evaluation of virus-induced gene silencing efficiency. Genet. Mol. Res. 13, 9443–9452. doi: 10.4238/2014.November.11.9
Liao, Y. W. K., Liu, Y. R., Liang, J. Y., Wang, W. P., Zhou, J., Xia, X. J., et al. (2015). The relationship between the plant-encoded RNA-dependent RNA polymerase 1 and alternative oxidase in tomato basal defense against Tobacco mosaic virus. Planta 241, 641–650. doi: 10.1007/s00425-014-2207-y
Liao, Y. W. K., Shi, K., Fu, L. J., Zhang, S., Li, X., and Dong, D. K. (2012). The reduction of reactive oxygen species formation by mitochondrial alternative respiration in tomato basal defense against TMV infection. 235, 225–238. doi: 10.1007/s00425-011-1483-z
Liao, Y. W. K., Sun, Z. H., Zhou, Y. H., Shi, K., Li, X., Zhang, G. Q., et al. (2013). The role of hydrogen peroxide and nitric oxide in the induction of plant-encoded RNA-dependent RNA polymerase 1 in the basal defense against tobacco mosaic virus. PLoS ONE 8:76090. doi: 10.1371/journal.pone.0076090
Liu, Y., Gao, Q., Wu, B., Ai, T., and Guo, X. (2009). NgRDR1, an RNA-dependent RNA polymerase isolated from Nicotiana glutinosa, was involved in biotic and abiotic stresses. Plant Physiol. Biochem. 47, 359–368. doi: 10.1016/j.plaphy.2008.12.017
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002a). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002b). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. Cell Mol. Biol. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lough, T. J., and Lucas, W. J. (2006). Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57, 203–232. doi: 10.1146/annurev.arplant.56.032604.144145
Madhusudhan, K. N., Srikanta, B. M., Shylaja, M. D., Prakash, H. S., and Shetty, H. S. (2009). Changes in antioxidant enzymes, hydrogen peroxide, salicylic acid and oxidative stress in compatible and incompatible host-tobamovirus interaction. J. Plant Interact. 4, 157–166. doi: 10.1080/17429140802419516
Montalbini, P., Buonaurio, R., and Kumar, N. N. U. (1995). Peroxidase activity and isoperoxidase pattern in tobacco leaves infected with Tobacco Necrosis Virus and other viruses inducing necrotic and non-necrotic alterations. J. Phytopathol. 143, 295–301. doi: 10.1111/j.1439-0434.1995.tb00263.x
Mourrain, P., Béclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. doi: 10.1016/S0092-8674(00)80863-6
Nakasugi, K., Crowhurst, R. N., Bally, J., Wood, C. C., Hellens, R. P., and Waterhouse, P. M. (2013). De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS ONE 8:e59534. doi: 10.1371/journal.pone.0059534
Petricka, J. J., Winter, C. M., and Benfey, P. N. (2012). Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590. doi: 10.1146/annurev-arplant-042811-105501
Qi, X. P., Bao, F. S., and Xie, Z. X. (2009). Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE 4:e4971. doi: 10.1371/annotation/8d1a816e-b366-4833-b558-724ec28d1b87
Qu, F. (2010). Antiviral role of plant-encoded RNA-dependent RNA polymerases revisited with deep sequencing of small interfering RNAs of virus origin. Mol. Plant Microbe Interact. 23, 1248–1252. doi: 10.1094/MPMI-06-10-0124
Rakhshandehroo, F., Behboodi, B. S., and Mohammadi, M. (2012). Changes in peroxidase activity and transcript level of the MYB1 gene in transgenic tobacco plants silenced for the RDR-1 gene after systemic infection with potato virus Yo. J. Phytopathol. 160, 187–194. doi: 10.1111/j.1439-0434.2012.01882.x
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schiebel, W., Pélissier, T., Riedel, L., Thalmeir, S., Schiebel, R., Kempe, D., et al. (1998). Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10, 2087–2101. doi: 10.1105/tpc.10.12.2087
Schwach, F., Vaistij, F. E., Jones, L., and Baulcombe, D. C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852. doi: 10.1104/pp.105.063537
Shi, Y. W., and Guo, Y. S. (2012). Prokaryotic expression and purification of TMV CP gene isolated in Guizhou. Guizhou Agric. Sci. 40, 18–20. doi: 10.3969/j.issn.1001-3601.2012.12.006
Sijen, T., Fleenor, J., Simmer, F., Thijssen, K. L., Parrish, S., Timmons, L., et al. (2001). On the Role of RNA Amplification in dsRNA-triggered gene silencing. Cell 107, 465–476 doi: 10.1016/S0092-8674(01)00576-1
Stewart, R. R., and Bewley, J. D. (1980). Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol. 65, 245–248. doi: 10.1104/pp.65.2.245
Tamura, K. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wan, H., Yuan, W., Ruan, M., Ye, Q., Wang, R., Li, Z., et al. (2011). Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 416, 24–30. doi: 10.1016/j.bbrc.2011.10.105
Wang, X. B., Wu, Q. F., Ito, T., Cillo, F., Li, W. X., Chen, X. M., et al. (2010). RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 484–489. doi: 10.1073/pnas.0904086107
War, A. R., Paulraj, M. G., War, M. Y., and Ignacimuthu, S. (2012). Differential defensive response of groundnut germplasms to Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae). J. Plant Interact. 7, 45–55. doi: 10.1080/17429145.2011.587898
Wassenegger, M., and Krczal, G. (2006). Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 11, 142–151. doi: 10.1016/j.tplants.2006.01.003
Xie, Z., Fan, B., Chen, C., and Chen, Z. (2001). An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 98, 6516–6521. doi: 10.1073/pnas.111440998
Xu, D., and Zhou, G. (2017). Characteristics of siRNAs derived from Southern rice black-streaked dwarf virus in infected rice and their potential role in host gene regulation. Virol. J. 14:27. doi: 10.1186/s12985-017-0699-314.
Xu, T., Zhang, L., Zhen, J., Fan, Y., Zhang, C., and Wang, L. (2013). Expressional and regulatory characterization of Arabidopsis RNA-dependent RNA polymerase 1. Planta 237, 1561–1569. doi: 10.1007/s00425-013-1863-7
Yang, S. J., Carter, S. A., Cole, A. B., Cheng, N. H., and Nelson, R. S. (2004). A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. U.S.A. 101, 6297–6302. doi: 10.1073/pnas.0304346101
Ying, X. B., Dong, L., Zhu, H., Duan, C. G., Du, Q. S., Lv, D. Q., et al. (2010). RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana. Plant Cell 22, 1358–1372. doi: 10.1105/tpc.109.072058
Keywords: pepper, CaRDR1, tobacco mosaic virus (TMV), resistance, Nicotiana benthamiana
Citation: Qin L, Mo N, Zhang Y, Muhammad T, Zhao G, Zhang Y and Liang Y (2017) CaRDR1, an RNA-Dependent RNA Polymerase Plays a Positive Role in Pepper Resistance against TMV. Front. Plant Sci. 8:1068. doi: 10.3389/fpls.2017.01068
Received: 23 March 2017; Accepted: 02 June 2017;
Published: 28 June 2017.
Edited by:
Corina Vlot, Helmholtz Zentrum München, GermanyReviewed by:
Shu Yuan, Sichuan Agricultural University, ChinaYuichiro Watanabe, University of Tokyo, Japan
Copyright © 2017 Qin, Mo, Zhang, Muhammad, Zhao, Zhang and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Liang, liangyan@nwsuaf.edu.cn
 Lei Qin
Lei Qin Ning Mo
Ning Mo Yang Zhang
Yang Zhang Yan Zhang
Yan Zhang Yan Liang
Yan Liang