- 1College of Horticulture, South China Agricultural University, Guangzhou, China
- 2Vegetable Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
Glucosinolates are Brassicaceae-specific secondary metabolites that act as crop protectants, flavor precursors, and cancer-prevention agents, which shows strong evidences of anticarcinogentic, antioxidant, and antimicrobial activities. MYB28, the R2R3-MYB28 transcription factor, directly activates genes involved in aliphatic glucosinolate biosynthesis. In this study, the MYB28 homology (BoaMYB28) was identified in Chinese kale (Brassica oleracea var. alboglabra Bailey). Analysis of the nucleotide sequence indicated that the cDNA of BoaMYB28 was 1257 bp with an ORF of 1020 bp. The deduced BoaMYB28 protein was a polypeptide of 339 amino acid with a putative molecular mass of 38 kDa and a pI of 6.87. Sequence homology and phylogenetic analysis showed that BoaMYB28 was most closely related to MYB28 homologs from the Brassicaceae family. The expression levels of BoaMYB28 varies across the tissues and developmental stages. BoaMYB28 transcript levels were higher in leaves and stems compared with those in cotyledons, flowers, and siliques. BoaMYB28 was expressed across all developmental leaf stages, with higher transcript accumulation in mature and inflorescence leaves. Over-expression and RNAi studies showed that BoaMYB28 retains the basic MYB28 gene function as a major transcriptional regulator of aliphatic glucosinolate pathway. The results indicated that over-expression and RNAi lines showed no visible difference on plant morphology. The contents of aliphatic glucosinolates and transcript levels of aliphatic glucosinolate biosynthesis genes increased in over-expression lines and decreased in RNAi lines. In over-expression lines, aliphatic glucosinolate contents were 1.5- to 3-fold higher than those in the wild-type, while expression levels of aliphatic glucosinolate biosynthesis genes were 1.5- to 4-fold higher than those in the wild-type. In contrast, the contents of aliphatic glucosinolates and transcript levels of aliphatic glucosinolate biosynthesis genes in RNAi lines were considerably lower than those in the wild-type. The results suggest that BoaMYB28 has the potential to alter the aliphatic glucosinolates contents in Chinese kale at the genetic level.
Introduction
Glucosinolates (GS) are nitrogen- and sulfur-rich plant amino acid-derived secondary metabolites found in Arabidopsis thaliana and the Brassicaceae family, such as broccoli, cabbage, cauliflower, kale, and turnip (Fahey et al., 1997; Kliebenstein et al., 2001; Wittstock and Halkier, 2002; Herr and Büchler, 2010). Glucosinolates play important roles in the plant defense system against insects and microbial infections (Manici et al., 1997). Glucosinolates and their hydrolytic products also contribute to the special flavors and odors of the Brassicaceae (Ishida et al., 2014). Based on the precursor amino acids used and R group modifications, glucosinolates are classified into three major chemical classes, namely aliphatic, aromatic, and indoic glucosinolates (Sønderby et al., 2010). Approximately 130 different aliphatic glucosinolates are known to occur in plants (Agerbirk and Olsen, 2012; Augustine and Bisht, 2015).
The aliphatic glucosinolate biosynthetic pathway has been elucidated in A. thaliana, including the genes responsible for side-chain elongation, core structure formation and secondary modification (Fahey et al., 2001; Grubb and Abel, 2006; Sønderby et al., 2010). MYB28, MYB29, and MYB76, which belong to subgroup-12 of the R2R3-MYB superfamily, act as direct transcriptional regulators of aliphatic glucosinolate biosynthesis (Gigolashvili et al., 2008; Baskar and Park, 2015). MYB28 has been shown to be the key regulator followed by MYB29, while MYB76 plays a minor and accessory role in the aliphatic glucosinolate pathway (Gigolashvili et al., 2009).
MYB28 positively regulates genes involved in aliphatic glucosinolate biosynthesis, including side-chain elongation (MAM1 and MAM3) and core structure formation (CYP79F1, CYP79F2, CYP83A1, ST5b, and ST5c) (Gigolashvili et al., 2007; Augustine et al., 2013; Baskar and Park, 2015). In A. thaliana, AtMYB28 over-expression lines showed a significant increase in steady-state mRNA levels of aliphatic glucosinolate biosynthesis genes in leaves and inflorescences. In contrast, expression levels of aliphatic glucosinolate biosynthesis genes in AtMYB28 RNAi lines were lower than those in the wild-type (Gigolashvili et al., 2007). MYB28 controls the accumulation of long-chain and short-chain aliphatic glucosinolates (Chen et al., 2006; Hirai et al., 2007; Kim et al., 2013; Miao et al., 2013). In AtMYB28 over-expression lines, aliphatic glucosinolate contents were 2- to 7-fold higher than those in the wild-type, while glucoiberin and glucoalyssin levels were 2- to 3-fold higher than those in the wild-type. In contrast, in AtMYB28 RNAi lines, glucoraphanin, glucoiberin, and glucoalyssin levels were considerably lower than those in the wild-type. However, glucoerucin levels were neither increased nor decreased in over-expression or RNAi lines (Gigolashvili et al., 2007).
The expression pattern of MYB28 varies across tissues and developmental stages (Gigolashvili et al., 2007; Augustine et al., 2013; Baskar and Park, 2015). Expression levels of MYB28 in seedlings, stems, and siliques were higher than those in roots and primary leaf tissues of Brassica juncea. Brassica nigra showed a similar tissue-specific expression pattern (Augustine et al., 2013). In Brassica rapa ssp. pekinensis, MYB28 transcript levels are highest in inflorescences and stems (Kim et al., 2013). MYB28 is expressed throughout leaf development in B. juncea, with MYB28 expression being higher in mature and inflorescence leaves than in primary and young leaves (Augustine et al., 2013).
Chinese kale (Brassica oleracea var. alboglabra Bailey) is a rich source of antioxidants and anticarcinogenic compounds, including glucosinolates, carotenoids, vitamin C, and phenolic compounds (Choi et al., 2014). The market demand for Chinese kale is increasing due to its high nutritional value. Chinese kale is widely distributed in southern China, the Taiwan region, Japan, and Southeast Asia and has spread quickly to Europe and America (Sun et al., 2012a). Bolting stems are the most common edible parts of Chinese kale, as they are tender and crispy with good flavor. Consumption of tender rosette leaves and sprouts has also increased in southern China in recent years (Sun et al., 2011; Qian et al., 2015). In Chinese kale, the total glucosinolates contents varied extensively among the sprouts, rosette leaves and bolting stems, with highest content in the sprouts (Sun et al., 2011, 2012a,b). Compared with other glucosinolate, gluconapin was the most predominant glucosinolate in Chinese kale (Sun et al., 2011; Qian et al., 2015).
Although the role of MYB28 is very well known in other close relatives of the Chinese kale. The role and molecular characteristics of MYB28 in the aliphatic glucosinolate biosynthetic pathway of Chinese kale are unknown. Our study provides information on the molecular mechanisms of BoaMYB28 in aliphatic glucosinolate biosynthesis in Chinese kale. The objectives of the study are to help breeders to develop cultivars with high beneficial aliphatic glucosinolate levels and low anti-nutritional aliphatic glucosinolates levels. The present study explores the molecular characterization of MYB28 in Chinese kale for the first time. Gene structure prediction, sequence alignment and phylogenetic analysis are investigated in detail. Differential expression profiles of BoaMYB28 have been carried out. To the best of our knowledge, this is the first report of the effect of BoaMYB28 transformation on glucosinolate composition and content in A. thaliana and Chinese kale. We have also analyzed contents of aliphatic glucosinolate and expression levels of the aliphatic glucosinolate biosynthesis genes in A. thaliana and Chinese kale transgenic lines.
Materials and Methods
Plant Materials and Growth Conditions
Chinese kale seeds were germinated in plastic pots, and the seedlings were cultured in the field at 22–25°C at South China Agricultural University (Guangzhou, China). Bolting stems were harvested when plants were fully grown, with inflorescences as high as the apical leaves used for cDNA sequence cloning. For spatial expression pattern analysis, cotyledon (7 days), leaf (15 days), stem (30 days), flower (anthesis), and silique (15 days post-anthesis) were excised from 10 plants. For temporal expression pattern analysis, primary leaf (15 days), young leaf (30 days), mature leaf (60 days), and inflorescence leaf (anthesis) were excised from 10 plants.
Arabidopsis thaliana seeds (ecotype Col-0) were sterilized and plated on agar-solidified half-strength Murashige and Skoog (MS) medium containing 2.15 g/l MS salts and 0.5% sucrose, pH 5.8. Wild-type seeds were cold-treated at 4°C for 3 days in the dark and grown in a culture chamber at 21°C under 16/8 h light/dark cycle. Seedlings were transferred to soil and grown at 25°C under 16/8 h light/dark regime and at 40% humidity.
Plant materials were harvested in the early morning, weighed when fresh and washed with distilled water. All samples were immediately transported to the laboratory within 10 min, frozen in liquid nitrogen and lyophilised in an ultralow -80°C freezer for RNA isolation and/or for aliphatic glucosinolate content analysis.
RNA Extraction and cDNA Synthesis
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) followed by treatment with DNase (Promega, MA, United States) to remove possible DNA contamination. RNA quality and content were determined by gel electrophoresis and biophotometer (Eppendorf, Germany). First-strand cDNA was synthesized from 1 μg total RNA with an oligo-dT primer according to the instructions of the Reverse Transcriptase M-MLV Kit (Takara, Japan).
Molecular Cloning of BoaMYB28
The full-length coding sequence of BoaMYB28 was isolated from Chinese kale. The primers were designed based on a sequence identified in the BRAD Brassica database (Cheng et al., 2011). PCR amplification was performed using the primers listed in Supplementary Table S1. The PCR program was as follows: 95°C for 5 min; 35 cycles of 95°C for 1 min, 58°C for 1 min, and 72°C for 1 min; with a final extension at 72°C for 10 min. Five independent PCRs were used for isolating the gene. PCR products were ligated into the pMD18-T vector (Takara, Japan) and transformed into Escherichia coli DH5α. The BoaMYB28 sequence was submitted to GenBank and was given the accession number KP723785.
Expression Analysis of BoaMYB28 and Aliphatic Glucosinolate Biosynthesis Genes
The expression patterns of the target genes were assayed by quantitative real-time PCR (qRT-PCR) using the primers listed in Supplementary Table S1. The reaction was performed using the MyiQ Real-Time PCR Detection System (Bio-Rad) platform using the SYBR Green Master Mix (Roche, Swiss) in a total 20 μl volume. The relative gene expression was calculated by the 2-ΔΔCt method.
The amplification protocol was as follows: 95°C for 2 min; 35 cycles at 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s; with a 5 min final extension at 72°C. β-actin was selected as the endogenous control gene for relative quantification. PCR reactions were performed from three independent biological replicates with three technical replicates for each. Data was analyzed using Bio-Rad CFX Manager software.
Transformation of BoaMYB28 in A. thaliana and Chinese Kale and Southern Blot
For generation of the BoaMYB28 over-expression and RNAi construct, the coding sequence of MYB28 was cloned into the plasmid vector, pFGC5941 (GenBank number AY310901) (Kerschen et al., 2004). The pFGC5941 vector contained the herbicide resistance gene (bar), which was used as selectable marker gene for generating transgenic lines (Supplementary Figures S10, S11). The different constructs were transformed into Agrobacterium strain GV3101 by the freeze-thaw method and further into A. thaliana by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on half-strength MS medium (0.5% sucrose, 0.8% agar, pH 5.8) (Gigolashvili et al., 2008).
Chinese kale seeds were surface sterilized with 70% ethanol for 5 min followed by 10% sodium hypochlorite for 10 min and then rinsed three times in sterile distilled water. The sterilized seeds were germinated for 1 week in half-strength MS medium with 30 g/l sucrose and 8 g/l agar at pH 5.8 (Murashige and Skoog, 1962). The hypocotyl explants were cultured for 3 days in MS medium with 1 mg/l 6-benzylaminopurine (BAP) and 1 mg/l naphthaleneacetic acid (NAA). The precultured hypocotyls were soaked in the overnight-cultured Agrobacterium tumefaciens strain EHA105 suspension for 5 min. The explants were cultured for 3 days in MS medium with the addition of 1 mg/l BAP and 1 mg/l NAA.
After washing any remaining bacteria away with sterile water, the explants were cultured for 1 week in MS medium with 500 mg/l cefotaxime sodium (Cef), 1 mg/l BAP, and 1 mg/l NAA. The explants were transferred to selective shoot regeneration medium containing MS basal medium with 5 mg/l phosphinothericin (ppt), 500 mg/l Cef, 1 mg/l BAP, and 1 mg/l NAA. After culturing for 2 weeks, the shoot buds were sub-cultured on MS medium with 10 mg/l ppt, 500 mg/l Cef, 1 mg/l BAP, and 1 mg/l NAA. The elongated green shoots were transferred to MS medium with 250 mg/l Cef and 2 mg/l NAA for rooting. After culturing for 4–5 weeks, plantlets with well-developed roots were hardened off and transplanted into the greenhouse for further analysis and recovery of seeds (Qian et al., 2015).
Southern blot was used to confirm that MYB28 was integrated into A. thaliana and Chinese kale genomes (Zang et al., 2009). Aliquots of DNA (25 μg) were digested overnight with EcoRI, fractionated by 0.8% (w/v) agarose gel electrophoresis and transferred to a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech, United Kingdom). The probe used for Southern analysis was bar, a selectable marker gene. Probe labeling, pre-hybridisation, hybridisation and detection were performed according to the protocol of DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics GmbH, Germany).
Extraction and Determination of Glucosinolates in A. thaliana and Chinese Kale
Extraction and determination of aliphatic glucosinolates was performed in 5-week-old rosette leaves of transgenic and wild-type A. thaliana. Glucosinolates were extracted and analyzed as previously described (Gigolashvili et al., 2008). Freeze-dried samples were extracted twice with 70% methanol with the addition of glucotropaeolin (Applichem) as an internal standard. The methanol extracts were applied to a DEAE Sephadex A-25 column (acetic acid activated). The columns were sealed and desulphated overnight using purified Helix pomatia sulfatase (type H1, Sigma, Deisenhofen, Germany). HPLC analysis of desulphoglucosinolates was carried out as previously described (Gigolashvili et al., 2008). Detection of desulphoglucosinolates was performed at 229 nm by applying the response factors as described previously (Gigolashvili et al., 2007).
Glucosinolates of Chinese kale were extracted and analyzed as previously described (Sun et al., 2012a). The freeze-dried samples (500 mg) were mixed with 2 ml of 75% methanol at 80°C for 15 min and centrifuged at 10,000 × g for 5 min. The supernatant was further washed with 2 ml of 75% methanol and centrifuged at 10,000 × g for 5 min. The supernatant sample was applied onto DEAE-Sephadex A-25 column (acetic acid activated) and rinsed three times with 2 ml 0.02 M sodium acetate. The column was washed three times with 20 mM acetic acid and twice with water. The supernatant was mixed with 200 μl arylsulfatase solution. After incubation for 16 h at 35°C, the desulphoglucosinolates were eluted with 4 ml of Milli-Q water and filtered through a 0.45-μm membrane filter. HPLC analysis of desulphoglucosinolates was carried out as previously described (Qian et al., 2015). Glucosinolates were extracted using ortho-nitrophenyl-β-D-galactopyranoside (Sigma) as an internal standard.
Bioinformatics Analysis
The amino acid sequence was predicted using DNAman. Multiple sequence alignments of amino acid sequences were performed using ClustalW. Phylogenetic trees were generated using MEGA based on the NJ (neighbor-joining) method. The confidence value for the phylogeny analysis was calculated using the bootstrap method. Molecular weight and isoelectric point of the protein were calculated with ProtParam (Brown et al., 2003). The conserved domains were predicted by CDD (Brown et al., 2003). The secondary structures and tertiary structures were predicted by SOPMA and SWISS MODEL (Brown et al., 2003). The transmembrane domains were predicted by TMHMM 2.0. The topology was determined using TMpred software. Signal peptide prediction was performed using SignalP 4.1. The hydrophilic and hydrophobic regions of the protein were predicted using ProtScale. Putative N-glycosylation sites were predicted using NetNGlyc 1.0 (Yu et al., 2004). Putative phosphorylation sites were predicted using NetPhos 2.0. YinOYang 1.2 Server was used to predict the putative O-glycosylation sites.
Statistical Analysis
The data were analyzed with the SPSS software package version 11.5 (SPSS Inc. Chicago, IL, United States). Data were analyzed for statistical significance by one-way ANOVA model. The means were determined by the Least Significant Differences (LSD) test at p = 0.05.
Results
Molecular Cloning and Structural Analysis of BoaMYB28
We got the reference sequence from BRAD Brassica database. BRAD Brassica database is a new database which focuses on the genetics and genomics of the Brassica plants1. We designed the cloning primers by Primer 6.0 and determined the full-length of the target gene by PCR amplification. Two MYB28 sequences (MYB28-1 and MYB28-2) were cloned from different varieties of Chinese kale. The cDNAs of the two MYB28 sequences were 98.57% identical with a total 18 different nucleotides over 1257 bp (Supplementary Figure S1). The deduced amino acid sequences were 100% identical (Supplementary Figure S2). We submitted only one sequence (MYB28-1) to GenBank (BoaMYB28). Analysis of the nucleotide sequence indicated that the BoaMYB28 cDNA was 1257 bp with an ORF of 1020 bp (Supplementary Figure S3). The full-length cDNA contained a 138-bp 5′-untranslated region and 99-bp 3′-untranslated region. The deduced MYB28 protein sequence was 339 amino acids in length with a putative molecular mass of 38 kDa and a pI of 6.87. Homology searches against the Conserved Domain Database (CDD) revealed that BoaMYB28 contained Myb-like DNA-binding domains (positions 14–61 and 67–112) and belonged to the SANT (SWI3, ADA2, N-CoR, and TFIIIB) superfamily. Secondary structure prediction by SDPMA showed that BoaMYB28 contained 37.46% alpha helices, 45.72% random coils, 7.37% extended strand, and 9.44% beta turn (Supplementary Figure S4). A similar result was obtained after constructing the BoaMYB28 three-dimensional structure using SWISS-MODEL (Supplementary Figure S5).
Sequence Alignment and Phylogenetic Analysis of BoaMYB28
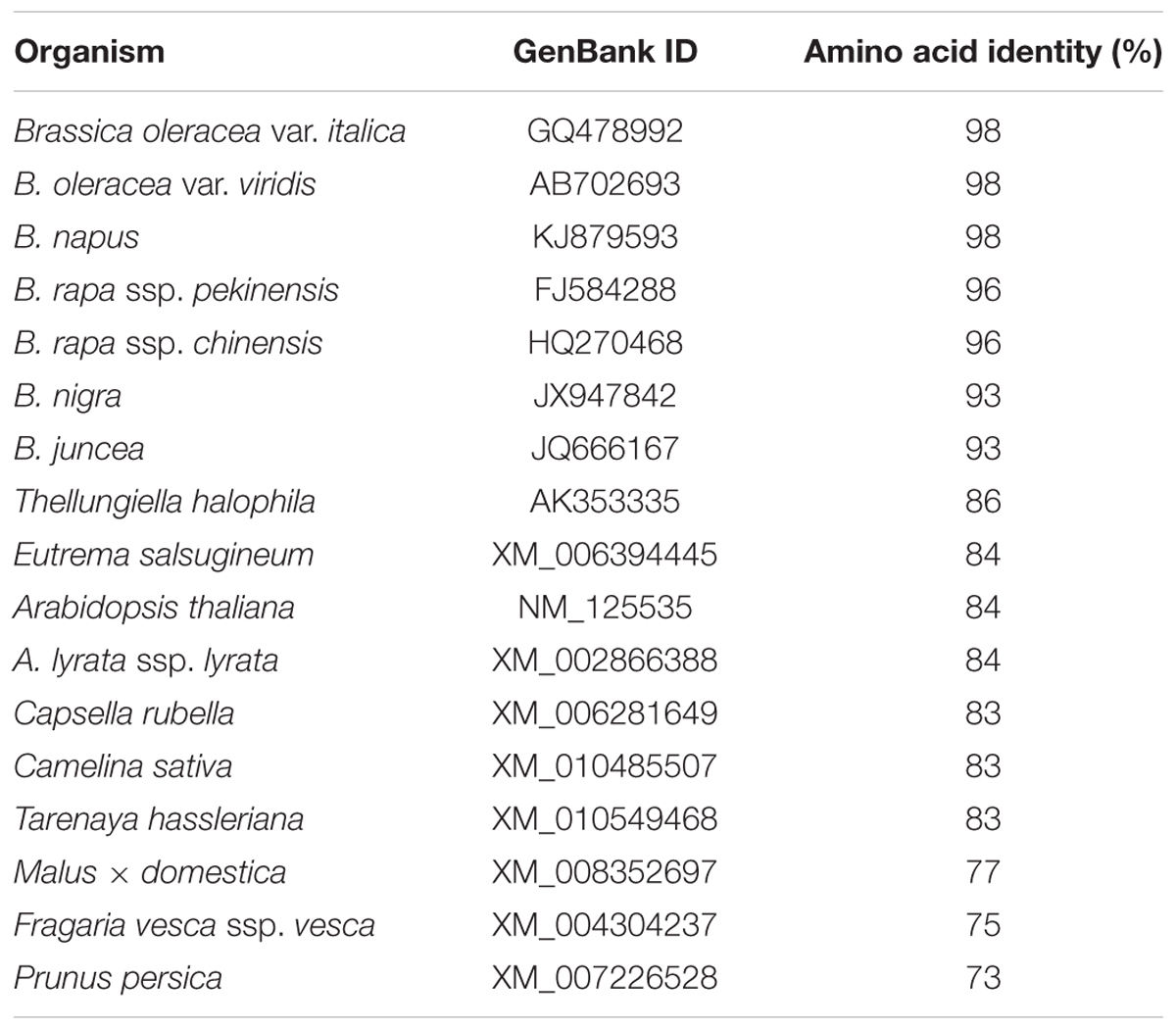
Alignment of the deduced amino acid sequence of BoaMYB28 revealed 73–98% identity with MYB28 proteins from 17 selected plant species. BoaMYB28 showed the higher level of sequence identity with MYB28 proteins from other members of the genus Brassica and lower levels of sequence identity with MYB28 proteins form other genera (Table 1).
In order to evaluate the molecular evolutionary relationships, the NJ method was used to construct a phylogenetic tree of MYB28 protein sequences (Figure 1). The distances between different genera were relatively large while those within the same genus were relatively small. Multiple sequence alignment showed that BoaMYB28 was phylogenetically closer to MYB28 proteins from other Brassica vegetables than other genera. The phylogenetic relationships were in accordance with the classification and evolutionary status via morphological and biochemical characteristics.
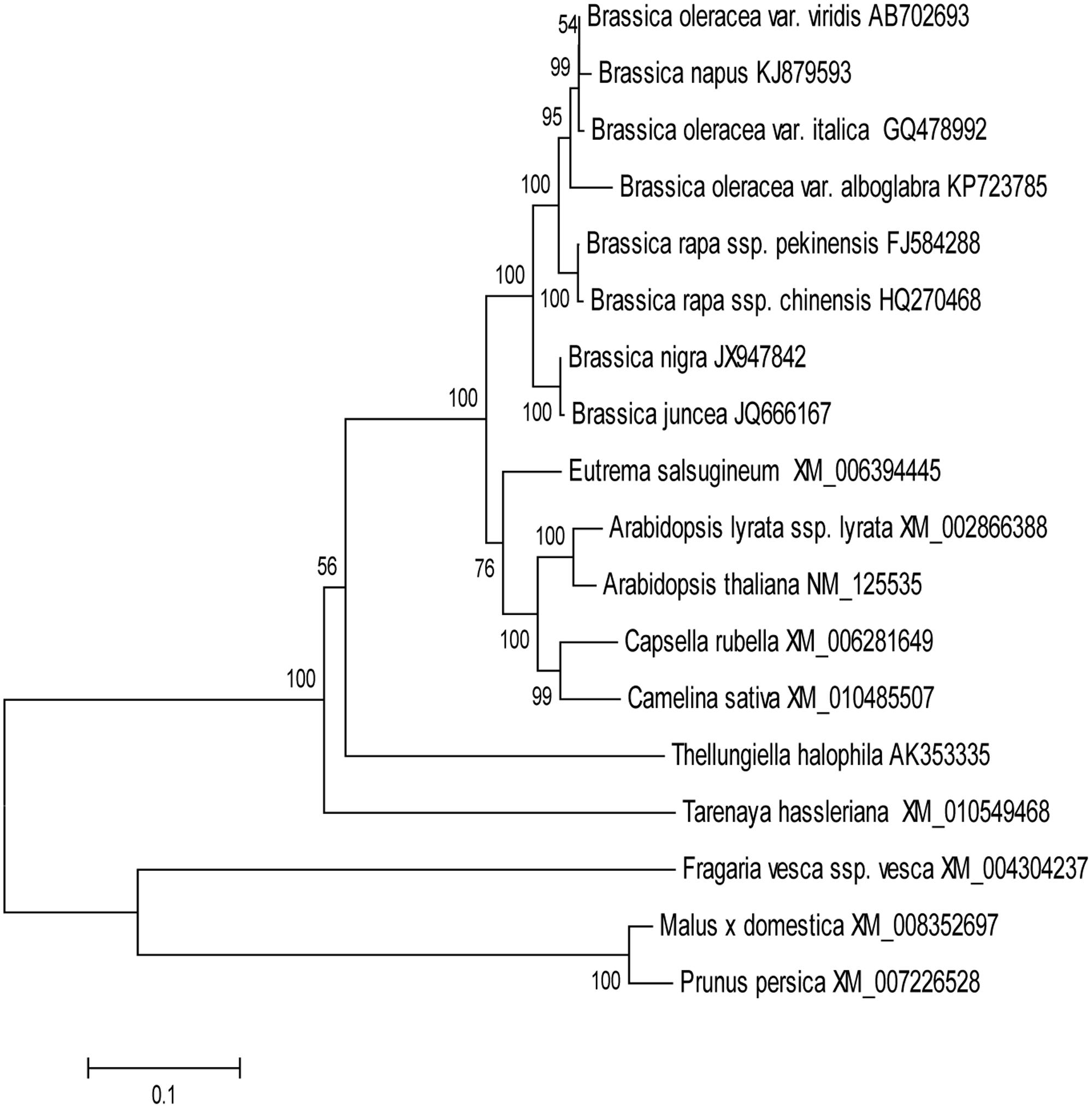
FIGURE 1. The phylogenetic tree of MYB28 proteins. The phylogenetic trees were generated based on the NJ (neighbor-joining) sequences distance method and constructed using MEGA 6.06 software. Numbers were bootstrap values for 1000 trails.
Structural Analysis of BoaMYB28
Bioinformatic tools were used to conduct structural analysis, such as identification of putative structural domains in BoaMYB28 proteins. The TMHMM results suggested that BoaMYB28 had no obvious transmembrane domain, which implied that BoaMYB28 was neither a membranous acceptor nor was located in the membrane. TMpred analysis predicted a lack of a transmembrane helix in BoaMYB28, and SignalP analysis also revealed a lack of a signal peptide. We examined the hydrophobic/hydrophilic nature of BoaMYB28 using ProtScale software, with a predicted maximum value of 1.5 and a minimum value of -3.578. Hydrophilic amino acid residues predominated in the peptide chain, indicating that BoaMYB28 might be a hydrophilic protein. NetNGlyc analysis identified three single N-glycosylation sites at positions 134, 177, and 276. Prediction of phosphorylation sites revealed 28 potential serine, 9 potential threonine, and 2 potential tyrosine targets were present in BoaMYB28. YinOYang analysis revealed that 12 potential O-glycosylation sites exist in BoaMYB28.
Spatial and Temporal Expression Patterns of BoaMYB28
qRT-qPCR was used to analyze the BoaMYB28 mRNA expression levels in five tissues (cotyledon, leaf, stem, flower, and silique) and four developmental stages of leaf (primary leaf, young leaf, mature leaf, and inflorescence leaf). BoaMYB28 was expressed in all of the collected tissues under normal growth conditions (Figure 2). However, BoaMYB28 expression levels differed among the tissues. BoaMYB28 was abundantly expressed in tissues that synthesize glucosinolates, such as leaves and stems, whereas cotyledons, flowers, and siliques showed lower transcript accumulation. BoaMYB28 had the highest transcript abundance in stems and the lowest transcript accumulation in siliques. The expression level of BoaMYB28 in stems was 9.52 times higher than that in siliques.
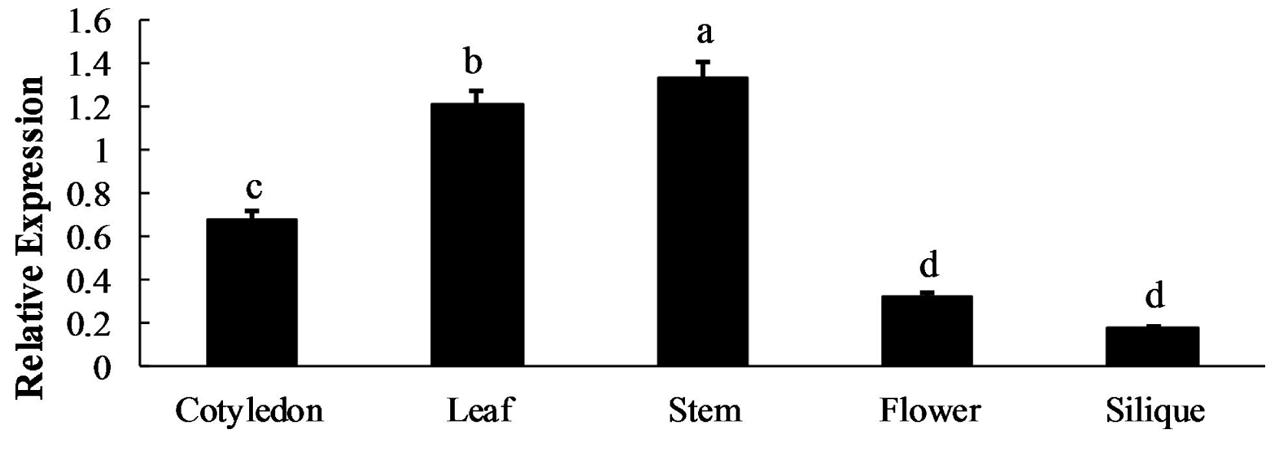
FIGURE 2. Spatial expression patterns of BoaMYB28. The tissues are defied as: cotyledon (7 days), leaf (15 days), stem (30 days), flower (anthesis), and silique (15 days post-anthesis). The data were analyzed using a one-way analysis of variance. β-actin mRNA expression was detected as the internal control. The same letters indicate no significant differences, while different letters indicate a statistically significant difference in expression. Each bar represents the mean (± standard error) of three independent biological replicates.
The temporal expression of BoaMYB28 was further determined at different developmental stages of Chinese kale leaves, tissues where glucosinolate synthesis predominantly occurs. During the developing stages of the leaves, BoaMYB28 transcript levels were higher in the mature and inflorescence leaves than those in the primary and young leaves (Figure 3). BoaMYB28 had the highest transcript abundance in the mature leaves and the lowest transcript abundance in the primary leaves. The expression level of BoaMYB28 in the mature leaves was 4.2 times higher than that in the primary leaves.
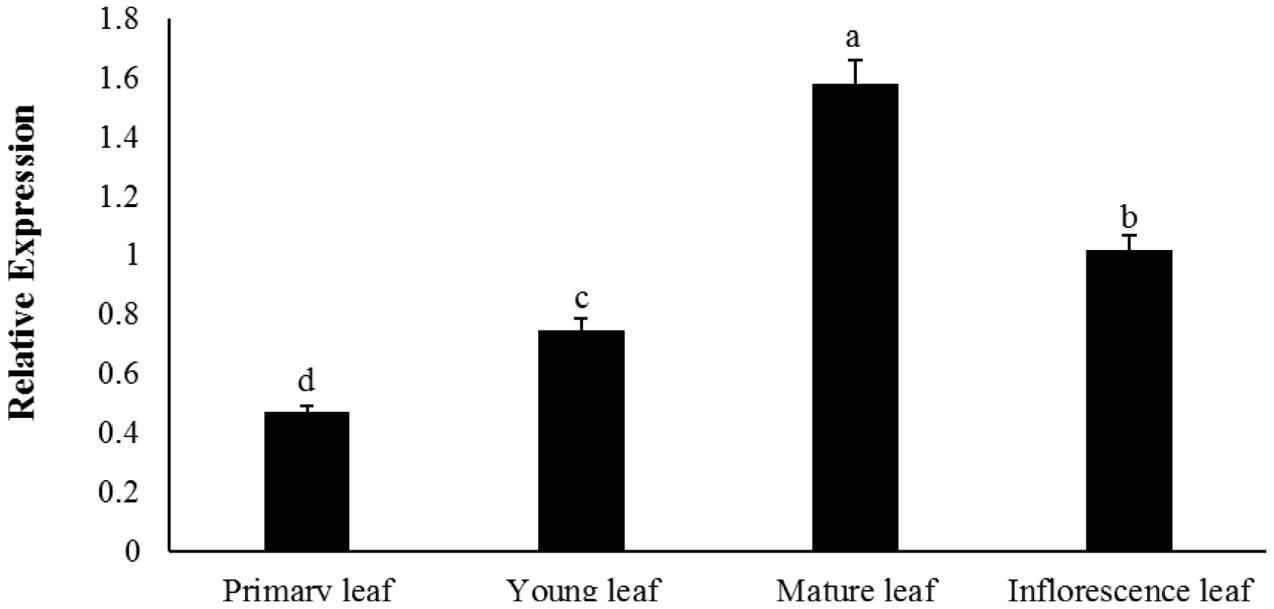
FIGURE 3. Temporal expression patterns of BoaMYB28 at different developmental stages of leaves. The stages are defied as: primary leaf (15 days), young leaf (30 days), mature leaf (60 days), and inflorescence leaf (anthesis). The data were analyzed using a one-way analysis of variance. β-actin mRNA expression was detected as the internal control. The same letters indicate no significant differences, while different letters indicate a statistically significant difference in expression. Each bar represents the mean (± standard error) of three independent biological replicates.
Glucosinolates Contents in Transgenic Plants
In order to illustrate that aliphatic glucosinolates contents can be controlled by BoaMYB28, several transgenic over-expression and RNAi plants were created using Agrobacterium tumefaciens-mediated transformation. About 30 primary transformants were used for BoaMYB28 over-expression constructs of A. thaliana. About 60 primary transformants were used for both BoaMYB28 over-expression and RNAi constructs of Chinese kale. Three representative over-expressing lines (OE.Ara1, OE.Ara2, and OE.Ara3) of A. thaliana were chosen for further analysis. Three representative over-expressing lines (OE.Boa1, OE.Boa2, and OE.Boa3) and RNAi lines (RNAi.Boa1, RNAi.Boa2, and RNAi.Boa3) of Chinese kale were chosen for further analysis. T2 transformants were used for phenotypic and molecular analysis. Southern blot and qRT-PCR analysis confirmed that BoaMYB28 had been integrated into A. thaliana and Chinese kale genomes (Supplementary Figures S6, S7 and Figures 4, 5). BoaMYB28 transgenic lines of A. thaliana (OE.Ara1) and Chinese kale (OE.Boa1 and RNAi.Boa1) showed no visible differences in plant morphology (Supplementary Figures S8, S9).
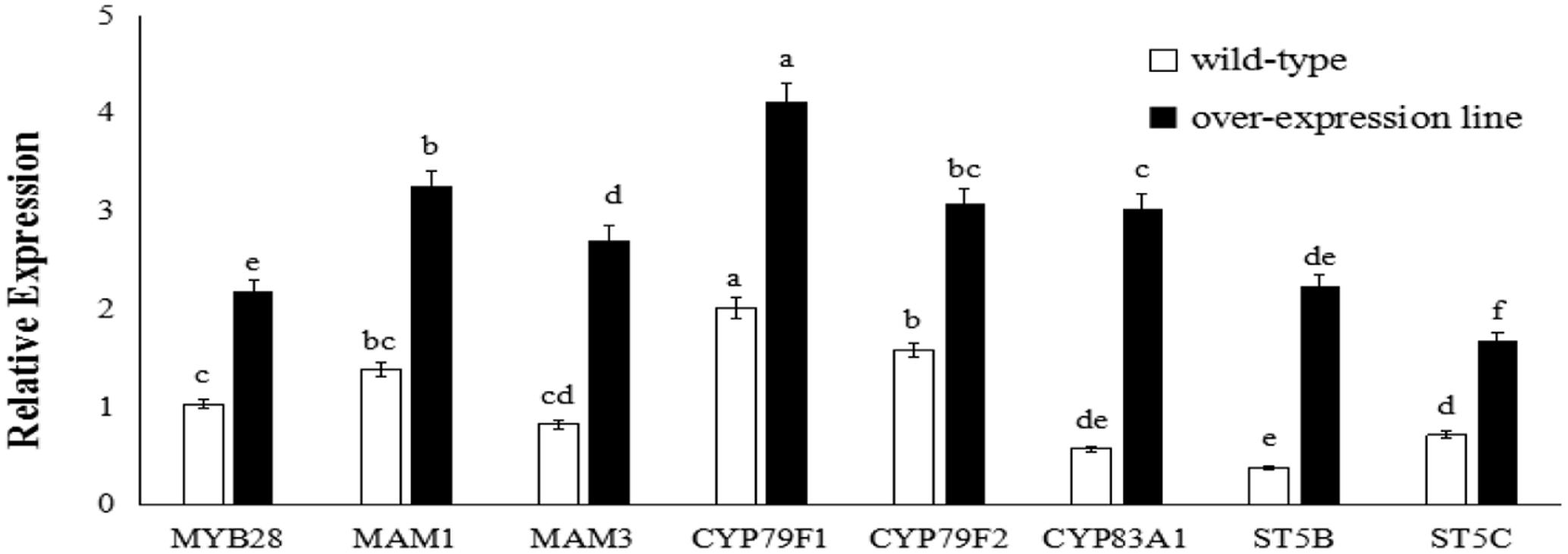
FIGURE 4. Transcript levels of aliphatic glucosinolate biosynthesis genes in BoaMYB28 transgenic lines of Arabidopsis thaliana. Total RNA was prepared from rosette leaves of 5-week-old plants. Each PCR assay was repeated three times with two independent sets of plants. The data were analyzed using a one-way analysis of variance. β-actin mRNA expression was detected as the internal control. The same letters indicate no significant differences, while different letters indicate a statistically significant difference in expression. Each bar represents the mean (± standard error) of three independent biological replicates.
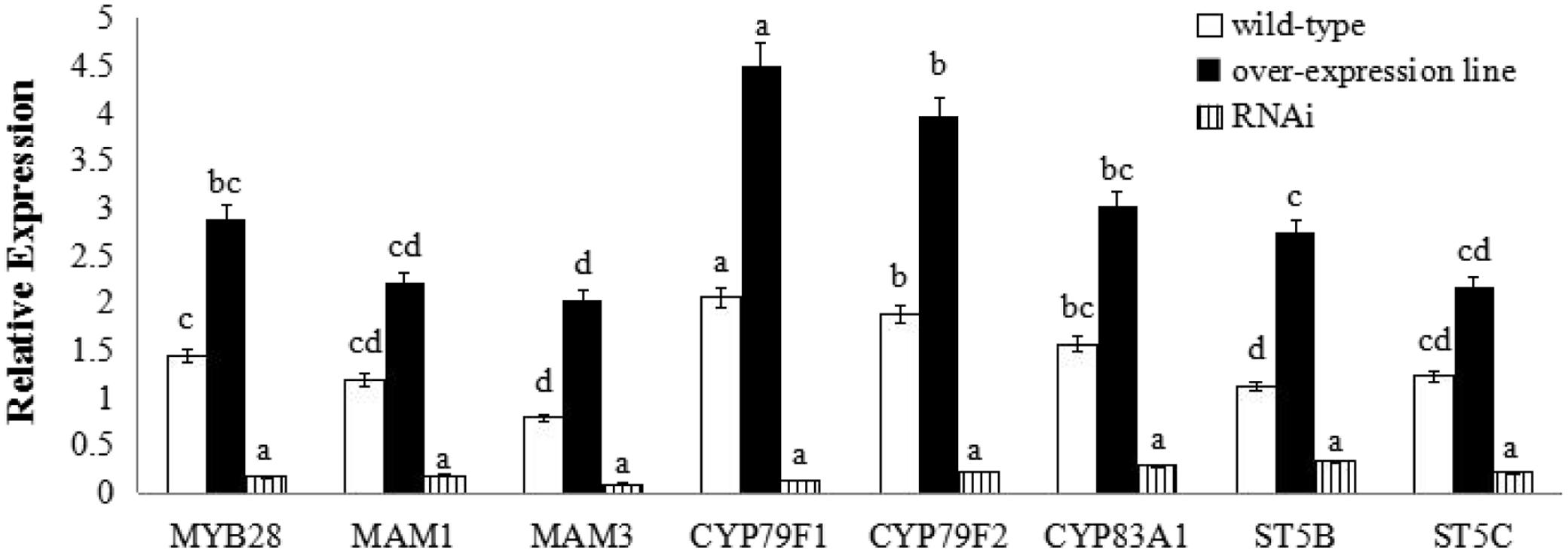
FIGURE 5. Transcript levels of aliphatic glucosinolate biosynthesis genes in BoaMYB28 transgenic lines of Chinese kale. Total RNA was prepared from bolting stems. Each PCR assay was repeated three times with two independent sets of plants. The data were analyzed using a one-way analysis of variance. β-actin mRNA expression was detected as the internal control. The same letters indicate no significant differences, while different letters indicate a statistically significant difference in expression. Each bar represents the mean (± standard error) of three independent biological replicates.
Over-expression lines of A. thaliana exhibited increased levels of both short- and long-chain aliphatic glucosinolates compared to those of the wild-type (Table 2). Levels of 4-methylsulfinylbutyl-GS (4MSOB), the major aliphatic glucosinolate in A. thaliana, were found to be increased by 1.5- to 1.8-fold in over-expression lines. Levels of other short-chain aliphatic glucosinolates, such as 3-methylsulfinylpropyl-GS (3MSOP) and 5-methylsulfinylpentyl-GS (5MSOP), were increased by 1.7- to 2.3-fold. Accumulation of the long-chain aliphatic glucosinolate, 8-methylsulfinyloctyl-GS (8MSOO), also increased by 1.4- to 1.9-fold compared with that of the wild-type plants. The levels of indolic glucosinolates such as indolyl-3-methyl-GS (I3M), 1-methoxyindolyl-3-methyl-GS (1MOI3M), 4-methoxyindolyl-3-methyl-GS (4MOI3M), and 4-hydroxyindolyl-3-methyl-GS (4HOI3M), were found to be unaltered in the transgenic lines.
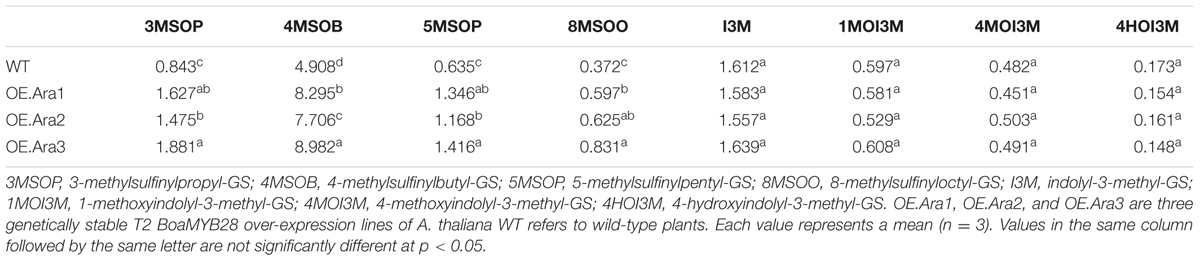
TABLE 2. The composition and content of glucosinolates (μmol⋅g-1 DW) in rosette leaves of A. thaliana among different transgenic lines and the wild-type.
Levels of the main aliphatic glucosinolates, such as 2-hydroxy-3-butenyl-GS (2HO3BE), 2-propenyl-GS (2PE), 3-butenyl-GS (3BE), and 4MSOB were analyzed in the bolting stems of transgenic Chinese kale. The over-expression lines contained approximately 2- to 3-fold higher levels of 4MSOB compared to that of the wild-type (Table 3). Levels of 3BE, the alkenyl product of 4MSOB, were increased by 1.6- to 2.3-fold in over-expression lines. The level of 2HO3BE, the hydroxyl product of 3BE, was also increased by 1.9- to 2.6-fold. Accumulation of 2PE increased by 1.4- to 2.0-fold compared with that of the wild-type. The levels of 2HO3BE, 2PE, 3BE, and 4MSOB significantly decreased in the RNAi lines (Table 3). Levels of indolic glucosinolates such as I3M, 1MOI3M, 4MOI3M, and 4HOI3M were not affected in the transgenic lines.
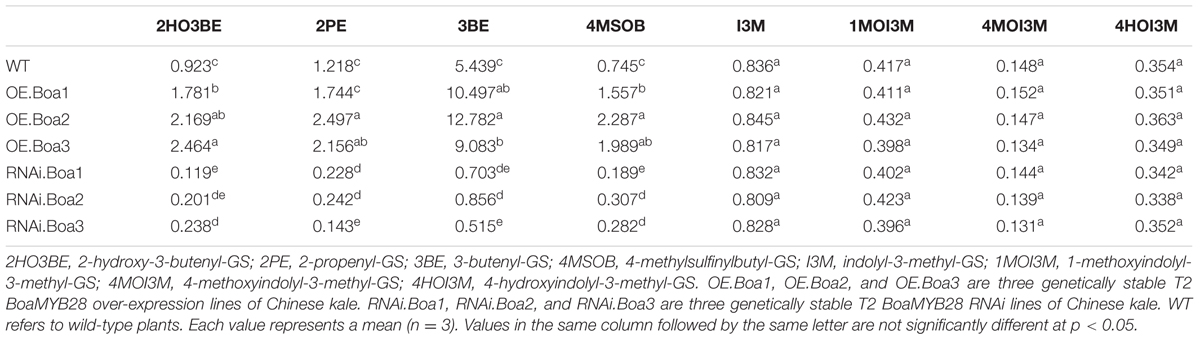
TABLE 3. The composition and content of glucosinolates (μmol⋅g-1 DW) in bolting stems of Chinese kale among different transgenic lines and wild-type.
Expression Profiles of Glucosinolate Biosynthesis Genes in Transgenic Plants
BoaMYB28 transgenic lines of A. thaliana (OE.Ara1) and Chinese kale (OE.Boa1 and RNAi.Boa1) were used for the qRT-PCR (Figures 4, 5). Increased transcript levels of genes involved in aliphatic glucosinolate biosynthesis corresponded well with the increased transcript level of BoaMYB28 in the over-expression lines. In over-expression lines of A. thaliana, the expression levels of MYB28, side-chain elongation genes and core structure formation genes were 1.63-fold, 1.75- to 2.23-fold, and 1.39- to 4.02-fold higher than those in the wild-type. The expression levels of MYB28, side-chain elongation genes and core structure formation genes in over-expression lines of Chinese kale were 1.45-fold, 1.62- to 2.34-fold, and 1.21- to 3.18-fold higher than those in the wild-type. In contrast, a significant decrease in the expression levels of the target genes was observed in BoaMYB28 RNAi lines. The results indicated that BoaMYB28 was a positive regulator of aliphatic glucosinolate biosynthetic pathway genes in Chinese kale.
BoaMYB28 up-regulated the aliphatic glucosinolate biosynthesis genes. MAM1 and MAM3 are involved in methionine chain elongation, the first phase of glucosinolate biosynthesis (Textor et al., 2007; Redovniković et al., 2012). MAM1 catalyses the production of short-chained aliphatic glucosinolates, whereas MAM3 is involved in the biosynthesis of long-chained aliphatic glucosinolates (Textor et al., 2007). The transcript levels of MAM1 and MAM3 in over-expression lines of A. thaliana were 1.82-fold and 2.05-fold higher than those in the wild-type, while the expression levels of MAM1 and MAM3 in over-expression lines of Chinese kale were 1.57-fold and 1.93-fold higher than those in the wild-type. However, the expression level of MAM1 was higher than that of MAM3 in over-expression lines of A. thaliana and Chinese kale. Although the expression level of MAM1 was higher than that of MAM3, the change interval of MAM3 was larger than MAM1. Consequently, MAM3 was more tightly regulated by BoaMYB28 compared with MAM1.
CYP79F1 and CYP79F2 are involved in biosynthesis of core structure of aliphatic glucosinolates (Chen et al., 2003). CYP79F1 is responsible for the generation of short-chained aliphatic glucosinolates, while CYP79F2 is capable of mediating the biosynthesis of long-chained aliphatic glucosinolates (Tantikanjana et al., 2004; Miao et al., 2013). In this study, CYP79F1 and CYP79F2 were differentially regulated by BoaMYB28. The transcript levels of CYP79F1 and CYP79F2 in over-expression lines of A. thaliana were 1.74-fold and 1.53-fold higher than those in the wild-type, while the expression levels of CYP79F1 and CYP79F2 in over-expression lines of Chinese kale were 1.98-fold and 1.62-fold higher than those in the wild-type. The expression level of CYP79F1 was higher than that of CYP79F2 in over-expression lines of A. thaliana and Chinese kale. The results indicated that expression of CYP79F1 was more tightly controlled by BoaMYB28 compared with CYP79F2.
Other core structure formation genes (CYB83A1, ST5B, and ST5C) also showed the same and dramatic change in BoaMYB28 transgenic lines of A. thaliana and Chinese kale, which was similar with the variation of BoaMYB28. The results indicated that CYB83A1, ST5B, and ST5C were positively regulated by BoaMYB28.
Discussion
Glucosinolates and their hydrolytic products play important biological roles as flavor precursors, crop protectants and cancer-preventing compounds in cruciferous and other vegetables. The majority of genes and intermediates involved in aliphatic glucosinolate metabolism have been identified in A. thaliana (Hansen et al., 2001; Diebold et al., 2002; Gigolashvili et al., 2007, 2008; Li et al., 2013).
MYB28 homologs have been isolated from Brassica crops such as B. juncea, B. rapa, and B. nigra (Augustine et al., 2013). In the present study, we identified MYB28 for the first time in Chinese kale. Four MYB28 homologs (BjuMYB28-1, -2, -3, -4) have been isolated from B. juncea (Augustine et al., 2013), while three MYB28 homologs (BrMYB28.1,.2,.3) have been isolated from B. rapa ssp. pekinensis (Baskar and Park, 2015). In the current study, two MYB28 sequences (MYB28-1 and MYB28-2) were cloned from different varieties of Chinese kale. We submitted only one sequence of MYB28 (MYB28-1) to GenBank (BoaMYB28). It is necessary to further study whether if there are another member of MYB28 in Chinese kale.
In order to investigate the evolutionary origin further, an amino acid sequence alignment of MYB28 proteins from other plants was constructed. The amino acid sequence encoded by BoaMYB28 shared close evolutionary ancestry with MYB28 homologs from other Brassica plants. BoaMYB28 exhibited a close genetic relationship with MYB28 proteins from other Brassica species. B. oleracea var. italica, B. oleracea var. viridis and Chinese kale (B. oleracea var. alboglabra Bailey) are all varieties of B. oleracea. BoaMYB28 had the highest level of sequence identity with B. oleracea var. italica (98%) and B. oleracea var. viridis (98%). The results indicated that MYB28 genes are evolutionarily conserved, retaining a very high level of sequence conservation while they have evolved via duplication (paralogs) and hybridisation (homologs) in two relatively simpler varieties of B. oleracea. The evolutionary relationships and classification of species can be approximated based on phylogenetic analysis (Ma et al., 2015). Phylogenetic analysis showed that BoaMYB28 was most closely related to MYB28 homologs from the Brassicaceae family, such as B. oleracea var. italica and B. oleracea var. viridis.
In A. thaliana, MYB28 expression was detected mainly in generative organs and mature leaves (Gigolashvili et al., 2007). A similar observation was made in B. nigra, B. juncea, and B. rapa (Augustine et al., 2013). Expression levels of BoaMYB28 were high in the commercial organs of Chinese kale, which are known for the accumulation of aliphatic glucosinolates. These results could facilitate tissue-specific engineering of improved aliphatic glucosinolate traits in Chinese kale. MYB28 expression was high at the onset of the reproductive phase in B. juncea, which is a critical time of flowering and seed formation (Augustine et al., 2013). In B. rapa ssp. pekinensis, the expression level of MYB28 was exclusively high in 2-month-old outer leaves (Baskar and Park, 2015). BoaMYB28 was expressed throughout leaf development, with high transcript accumulation in the mature and inflorescence leaves. The temporal expression pattern of BoaMYB28 suggested a role in aliphatic glucosinolate formation and distribution in Chinese kale leaves.
BoaMYB28 has the same basic gene function as other MYB28 genes that have been previously reported from other Brassica crops (Augustine et al., 2013). The unchanged growth phenotype of transgenic plants clearly demonstrated that BoaMYB28 did not affect plant growth. Over-expression and RNAi of BoaMYB28 in transgenic plants demonstrated that BoaMYB28 positively regulated genes involved in the aliphatic glucosinolate biosynthetic pathway and controlled the accumulation of both short- and long-chain aliphatic glucosinolates. In over-expression lines of Chinese kale, the growth rates of expression levels of CYP79F1 and CYP79F2 were higher than those of MAM1 and MAM3, which indicated that core structure formation genes was more tightly regulated by BoaMYB28 compared with side-chain elongation genes in Chinese kale. Increases and decreases in aliphatic glucosinolate levels corresponded well with altered transcript levels of BoaMYB28 in the transgenic lines. The altered aliphatic glucosinolate contents in transgenic lines of A. thaliana and Chinese kale correlated with altered mRNA levels of the genes involved in side-chain elongation (MAM1 and MAM3) and core structure formation (CYP79F1, CYP79F2, CYP83A1, ST5b, and ST5c) of aliphatic glucosinolates. The levels of indolic glucosinolates remained unchanged in the transgenic lines, indicating that BoaMYB28 specifically regulates aliphatic glucosinolate accumulation without affecting indolic glucosinolate levels.
Conclusion
In conclusion, we have isolated the full-length cDNA of BoaMYB28 from Chinese kale for the first time. Spatial and temporal gene expression patterns were detected, which may facilitate further studies on the regulatory metabolisms of aliphatic glucosinolate biosynthesis in Chinese kale. Aliphatic glucosinolate accumulation and expression profiles of aliphatic glucosinolate biosynthesis genes in transgenic lines of A. thaliana and Chinese kale were also investigated. This research results may be utilized to increase beneficial aliphatic glucosinolates and decrease anti-nutritional aliphatic glucosinolates for health benefits.
Author Contributions
Designed the study: LY and JL. Provided the materials: HC, BC, and GC. Performed the experiment and analyzed the data: LY. Wrote the paper: LY and JL. All authors read and approved the final manuscript.
Funding
This work was supported by the key project of Guangdong Science and Technology Section (2013B051000069 and 2014B020202005) and the key project of Guangzhou Science Technology and Innovation Commission (201508030021).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01083/full#supplementary-material
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Agerbirk, N., and Olsen, C. E. (2012). Glucosinolate structures in evolution. Phytochemistry 77, 16–45. doi: 10.1016/j.phytochem.2012.02.005
Augustine, R., and Bisht, N. C. (2015). Biotic elicitors and mechanical damage modulate glucosinolate accumulation by co-ordinated interplay of glucosinolate biosynthesis regulators in polyploid Brassica juncea. Phytochemistry 117, 43–50. doi: 10.1016/j.phytochem.2015.05.015
Augustine, R., Majee, M., Gershenzon, J., and Bisht, N. C. (2013). Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea. J. Exp. Bot. 64, 4907–4921. doi: 10.1093/jxb/ert280
Baskar, V., and Park, S. W. (2015). Molecular characterization of BrMYB28 and BrMYB29 paralogous transcription factors involved in the regulation of aliphatic glucosinolate profiles in Brassica rapa ssp. pekinensis. C. R. Biol. 338, 434–442. doi: 10.1016/j.crvi.2015.04.001
Brown, P. D., Tokuhisa, J. G., Reichelt, M., and Gershenzon, J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62, 471–481. doi: 10.1016/S0031-9422(02)00549-6
Chen, S., Glawischnig, E., Jørgensen, K., Naur, P., Jørgensen, B., Olsen, C. E., et al. (2003). CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. 33, 923–937. doi: 10.1046/j.1365-313X.2003.01679.x
Chen, Y., Yang, X., He, K., Liu, M., Li, J., Gao, Z., et al. (2006). The MYB transcription factor family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Cheng, F., Liu, S., Wu, J., Fang, L., Sun, S., Liu, B., et al. (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11:136. doi: 10.1186/1471-2229-11-136
Choi, S.-H., Park, S., Lim, Y., Kim, S.-J., Park, J.-T., and An, G. (2014). Metabolite profiles of glucosinolates in cabbage varieties (Brassica oleracea var. capitata) by season, color, and tissue position. Hortic. Environ. Biotechnol. 55, 237–247. doi: 10.1007/s13580-014-0009-6
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Diebold, R., Schuster, J., Daschner, K., and Binder, S. (2002). The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol. 129, 540–550.
Fahey, J. W., Zalcmann, A. T., and Talalay, P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56, 5–51. doi: 10.1016/S0031-9422(00)00316-2
Fahey, J. W., Zhang, Y., and Talalay, P. (1997). Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc. Natl. Acad. Sci. U.S.A. 94, 10367–10372. doi: 10.1073/pnas.94.19.10367
Gigolashvili, T., Berger, B., and Flügge, U.-I. (2009). Specific and coordinated control of indolic and aliphatic glucosinolate biosynthesis by R2R3-MYB transcription factors in Arabidopsis thaliana. Phytochem. Rev. 8, 3–13. doi: 10.1007/s11101-008-9112-6
Gigolashvili, T., Engqvist, M., Yatusevich, R., Müller, C., and Flügge, U. I. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177, 627–642. doi: 10.1111/j.1469-8137.2007.02295.x
Gigolashvili, T., Yatusevich, R., Berger, B., Müller, C., and Flügge, U. I. (2007). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51, 247–261. doi: 10.1111/j.1365-313X.2007.03133.x
Grubb, C. D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11, 89–100. doi: 10.1016/j.tplants.2005.12.006
Hansen, C. H., Wittstock, U., Olsen, C. E., Hick, A. J., Pickett, J. A., and Halkier, B. A. (2001). Cytochrome p450 CYP79F1 from arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem. 276, 11078–11085. doi: 10.1074/jbc.M010123200
Herr, I., and Büchler, M. W. (2010). Dietary constituents of broccoli and other cruciferous vegetables: implications for prevention and therapy of cancer. Cancer Treat. Rev. 36, 377–383. doi: 10.1016/j.ctrv.2010.01.002
Hirai, M. Y., Sugiyama, K., Sawada, Y., Tohge, T., Obayashi, T., Suzuki, A., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 6478–6483. doi: 10.1073/pnas.0611629104
Ishida, M., Hara, M., Fukino, N., Kakizaki, T., and Morimitsu, Y. (2014). Glucosinolate metabolism, functionality and breeding for the improvement of Brassicaceae vegetables. Breed. Sci. 64, 48–59. doi: 10.1270/jsbbs.64.48
Kerschen, A., Napoli, C. A., Jorgensen, R. A., and Muller, A. E. (2004). Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566, 223–228. doi: 10.1016/j.febslet.2004.04.043
Kim, Y., Li, X., Kim, S. J., Kim, H. H., Lee, J., Kim, H., et al. (2013). MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis). Molecules 18, 8682–8695. doi: 10.3390/molecules18078682
Kliebenstein, D. J., Kroymann, J., Brown, P., Figuth, A., and Pedersen, D. (2001). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126, 811–825. doi: 10.1104/pp.126.2.811
Li, Y., Tang, X., Hirai, M. Y., and Yan, X. (2013). Response of aliphatic glucosinolate biosynthesis to signaling molecules in MAM gene knockout mutants of Arabidopsis. Plant Biotechnol. 30, 403–406. doi: 10.5511/plantbiotechnology.13.0523a
Ma, J., Xu, G., Wan, L., and Wang, N. (2015). Molecular cloning, sequence analysis and tissue-specific expression of Akirin2 gene in Tianfu goat. Gene 554, 9–15. doi: 10.1016/j.gene.2014.09.030
Manici, L. M., Lazzeri, L., and Palmieri, S. (1997). In vitro fungitoxic activity of some glucosinolates and their enzyme-derived products toward plant pathogenic fungi. J. Agric. Food Chem. 45, 2768–2773. doi: 10.1021/jf9608635
Miao, H., Wei, J., Zhao, Y., Yan, H., Sun, B., Huang, J., et al. (2013). Glucose signalling positively regulates aliphatic glucosinolate biosynthesis. J. Exp. Bot. 64, 1097–1109. doi: 10.1093/jxb/ers399
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Qian, H., Sun, B., Miao, H., Cai, C., Xu, C., and Wang, Q. (2015). Variation of glucosinolates and quinone reductase activity among different varieties of Chinese kale and improvement of glucoraphanin by metabolic engineering. Food Chem. 168, 321–326. doi: 10.1016/j.foodchem.2014.07.073
Redovniković, I. R., Textor, S., Lisnić, B., and Gershenzon, J. (2012). Expression pattern of the glucosinolate side chain biosynthetic genes MAM1 and MAM3 of Arabidopsis thaliana in different organs and developmental stages. Plant Physiol. Biochem. 53, 77–83. doi: 10.1016/j.plaphy.2012.01.015
Sønderby, I. E., Geu-Flores, F., and Halkier, B. A. (2010). Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci. 15, 283–290. doi: 10.1016/j.tplants.2010.02.005
Sun, B., Liu, N., Zhao, Y., Yan, H., and Wang, Q. (2011). Variation of glucosinolates in three edible parts of Chinese kale (Brassica alboglabra Bailey) varieties. Food Chem. 124, 941–947. doi: 10.1016/j.foodchem.2010.07.031
Sun, B., Yan, H., Liu, N., Wei, J., and Wang, Q. (2012a). Effect of 1-MCP treatment on postharvest quality characters, antioxidants and glucosinolates of Chinese kale. Food Chem. 131, 519–526. doi: 10.1016/j.foodchem.2011.09.016
Sun, B., Yan, H., Zhang, F., and Wang, Q. (2012b). Effects of plant hormones on main health-promoting compounds and antioxidant capacity of Chinese kale. Food Res. Int. 48, 359–366. doi: 10.1016/j.foodres.2012.04.021
Tantikanjana, T., Mikkelsen, M. D., Hussain, M., Halkier, B. A., and Sundaresan, V. (2004). Functional analysis of the tandem-duplicated P450 genes SPS/BUS/CYP79F1 and CYP79F2 in glucosinolate biosynthesis and plant development by Ds transposition-generated double mutants. Plant Physiol. 135, 840–848. doi: 10.1104/pp.104.040113
Textor, S., de Kraker, J. W., Hause, B., Gershenzon, J., and Tokuhisa, J. G. (2007). MAM3 catalyses the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 144, 60–71. doi: 10.1104/pp.106.091579
Wittstock, U., and Halkier, B. A. (2002). Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 7, 263–270. doi: 10.1016/S1360-1385(02)02273-2
Yu, C. S., Lin, C. J., and Hwang, J. K. (2004). Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. doi: 10.1110/ps.03479604
Keywords: Chinese kale, aliphatic glucosinolate, transcription factors, over-expression, RNAi
Citation: Yin L, Chen H, Cao B, Lei J and Chen G (2017) Molecular Characterization of MYB28 Involved in Aliphatic Glucosinolate Biosynthesis in Chinese Kale (Brassica oleracea var. alboglabra Bailey). Front. Plant Sci. 8:1083. doi: 10.3389/fpls.2017.01083
Received: 06 December 2016; Accepted: 06 June 2017;
Published: 21 June 2017.
Edited by:
Jacqueline Batley, University of Western Australia, AustraliaReviewed by:
Rupesh Kailasrao Deshmukh, Laval University, CanadaFei Gao, University of Arkansas, United States
Katarzyna Gacek, Plant Breeding and Acclimatization Institute, Poland
Copyright © 2017 Yin, Chen, Cao, Lei and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianjun Lei, jjlei@scau.edu.cn
 Ling Yin
Ling Yin Hancai Chen2
Hancai Chen2 Bihao Cao
Bihao Cao Jianjun Lei
Jianjun Lei