- 1College of Horticulture, Qingdao Agricultural University, Qingdao, China
- 2College of Horticulture, Nanjing Agricultural University, Nanjing, China
Among the Rosaceae species, the gametophytic self-incompatibility (GSI) is controlled by a single multi-allelic S locus, which is composed of the pistil-S and pollen-S genes. The pistil-S gene encodes a polymorphic ribonuclease (S-RNase), which is essential for identifying self-pollen. However, the S-RNase system has not been fully characterized. In this study, the self-S-RNase inhibited the Ca2+-permeable channel activity at pollen tube apices and the selectively decreased phospholipase C (PLC) activity in the plasma membrane of Pyrus pyrifolia pollen tubes. Self-S-RNase decreased the Ca2+ influx through a PLC-mediated signaling pathway. Phosphatidylinositol-specific PLC has a 26-amino acid insertion in pollen tubes of the ‘Jinzhuili’ cultivar, which is a spontaneous self-compatible mutant of the ‘Yali’ cultivar. ‘Yali’ plants exhibit a typical S-RNase-based GSI. Upon self-pollination, PLC gene expression is significantly higher in ‘Jinzhuili’ pollen tubes than that in ‘Yali’ pollen tubes. Moreover, the PLC in pollen tubes can only interact with one of the two types of S-RNase from the style. In the Pyrus x bretschneideri Rehd, the PLC directly interacted with the S7-RNase in the pollen tube, but not with the S34-RNase. Collectively, our results reveal that the effects of S-RNase on PLC activity are required for S-specific pollen rejection, and that PLC-IP3 participates in the self-incompatibility reaction of Pyrus species.
Introduction
Self-incompatibility (SI) is an important mechanism that prevents self-fertilization and maintains genetic diversity in flowering plants (Silva and Goring, 2001). Gametophytic self-incompatibility (GSI) is the most common SI system (Franklin-Tong and Franklin, 2003), which has been described in >60 families of flowering plants (Kao and McCubbin, 1996). Only two GSI systems have been elucidated in detail at the molecular level (Franklin-Tong and Franklin, 2003; De Franceschi et al., 2012). One GSI mechanism, so far found only in the Papaveraceae, has a small secreted peptide–the S-protein (PrsS: Papaver rhoeas style S) as its pistil S-component. PrpS (P. rhoeas pollen S) is the Papaver pollen S determinant (Wheeler et al., 2009), but the function of PrpS is unclear yet (Fujii et al., 2016). Pollen growth can be inhibited within minutes by placing it on the stigma. An important finding is that the S-protein system includes a Ca2+-dependent signaling network. Self-incompatibility triggers an influx of extracellular Ca2+ into incompatible pollen tubes (Franklin-Tong et al., 2002), and the apical Ca2+ gradient rapidly disappears with the cessation of pollen tube growth during GSI (Franklin-Tong et al., 1997). The other GSI mechanism, which has been detected in the Solanaceae, Rosaceae and Scrophulariaceae families, has S-RNase as the pistil S-component and an F-box protein (SLF) as the pollen S-component (Zhang and Hiratsuka, 1999; Yang et al., 2007). In all species exhibiting GSI, incompatible pollen tubes will not grow beyond one-third of the length of the style. The self-S-RNase enters pollen tubes with the assistance of ABC transporters in apple (Meng et al., 2014) and self-S-RNase degrades ribosomal RNA (Quinet et al., 2014). However, RNA degradation is not the only SI event. S-RNase specifically induces tip-localized reactive oxygen species disruption, actin cytoskeleton depolymerisation, and nuclear DNA degradation in incompatible pear pollen tubes (Liu et al., 2007; Wang et al., 2010). Nevertheless, it is unclear whether S-RNase triggers alterations of pollen tube elements to inhibit incompatible pollen tube growth in a direct or indirect way. It has been suggested that pollen rejection systems and incompatibility systems are interconnected, and should be regarded as different parts of a single system that provides fine control over plant fertilization (Cruz-Garcia et al., 2003). Free cytosolic Ca2+ ([Ca2+]i) in pollen tubes is a well-established second messenger responsible for transducing directional growth signals (Malhó et al., 1994; Steinhorst and Kudla, 2013). A tip-focused apical [Ca2+]i gradient is a feature of growing pollen tubes (Pierson et al., 1996). Moreover, IP3 (inositol 1, 4, 5-triphosphate)-mediated [Ca2+]i signals are precisely controlled in both time and space, resulting in a signaling system that is simultaneously ubiquitous, versatile, and specific (Mak et al., 2000). Inositol 1, 4, 5-trisphosphate and other inositol lipid-related compounds have been proposed as important regulators of pollen tube growth (Zonia and Munnik, 2004; Monteiro et al., 2005). In addition, phosphatidylinositol-specific phospholipase C (PI-PLC) regulates tip growth by controlling IP3-gated Ca2+ fluxes and altering the spectrum of PI lipids in the tube apex (Dowd et al., 2006; Heilmann and Ischebeck, 2016). Thus, there are close links between PLC, IP3 production, and Ca2+ gradient during pollen tube growth.
Self- and non-self-recognitions are vital to the survival of all living organisms ranging from bacteria to humans. There are many biological events in the process of recognition, including immune defense and mate choice. Self- and non-self-discriminations are involved in mate choice (Li et al., 2017). Pollen–stigma interactions are similar to host–pathogen interactions (Hodgkin et al., 1988; Kovaleva et al., 2002; Kovaleva and Zakharova, 2003; Wu et al., 2008; Wilkins et al., 2014). When plants are under the stress of biotic and abiotic factors, the activity of PLC is rapidly activated, and the expression of PLC gene was up-regulated (Vossen et al., 2010).
In the Maloideae of Rosaceae, S-locus F-box genes appear to be involved in the determination of pollen S specificity. However, despite the fact that several models have been developed to explain how the two components of the S-locus interact, the mechanism by which SLF/SFB (S-haplotype-specific F-box) and S-RNase interact to trigger the SI reaction remains uncertain (Akagi et al., 2016; Ashkani and Rees, 2016). Moreover, the speculative GSI models based on SLF are often paradoxical. More and more data suggest that other loci have crucial influence on the sexual (in) compatible phenotype (Hegedűs et al., 2012). As has been proven by the finding of unexpected diversity in molecular mechanisms adopted for SI, further studies are likely to uncover novel forms of molecular interactions (Fujii et al., 2016). The purpose of this study is to explore the effect of S-RNase on Ca2+ channels, which has been identified in our published paper (Qu et al., 2007) on phospholipase C activity and its gene expression in the apical pollen tube. In doing so, we assessed PLC function in GSI, and developed a new model to explain our results.
Results
S-RNase Effects on Ca2+ Gradients and Currents
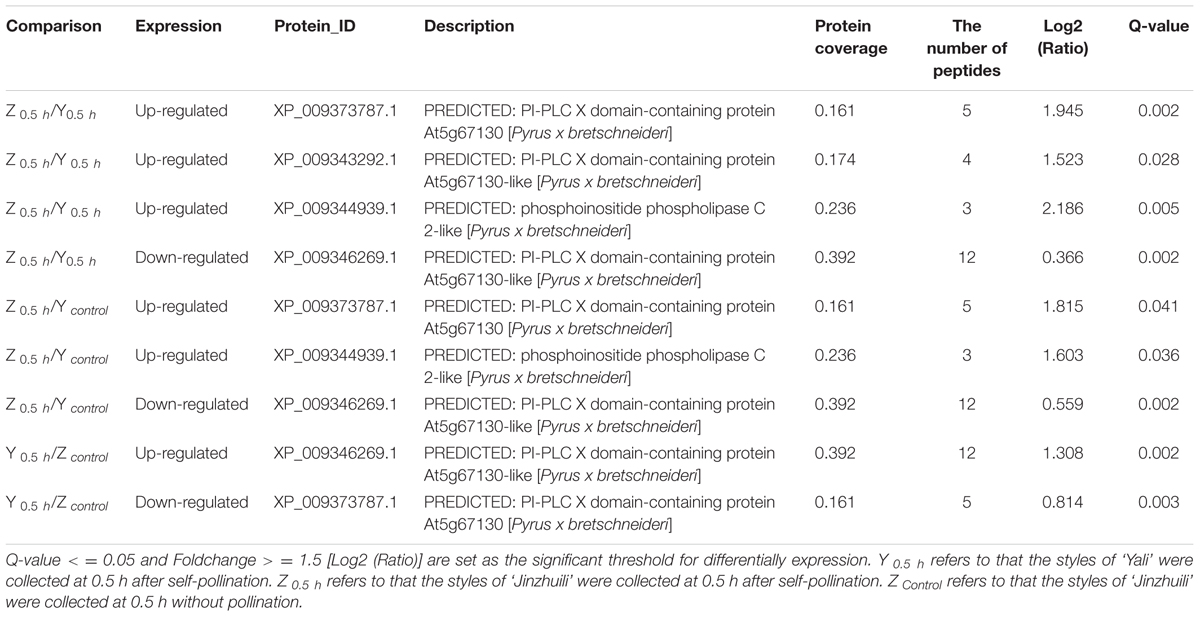
When fluo-4/AM was loaded into ‘Housui’ pollen tubes at room temperature, the normally growing pollen tubes preserved the Ca2+ gradient at the tube apex (Figures 1A,B). It had no effect on the apical Ca2+ gradient by adding 1.0 μg μl-1 non-self-S-RNase (‘Imamuraaki’) (Figures 1C,D). However, after adding 1.0 μg μl-1 self-S-RNase (‘Housui’), it decreased [Ca2+]i at the pollen tube tips and sub-tips (Figures 1E,F). These results indicate that self-S-RNase can decrease Ca2+ concentrations in pollen tube apices and disrupt Ca2+ gradients.
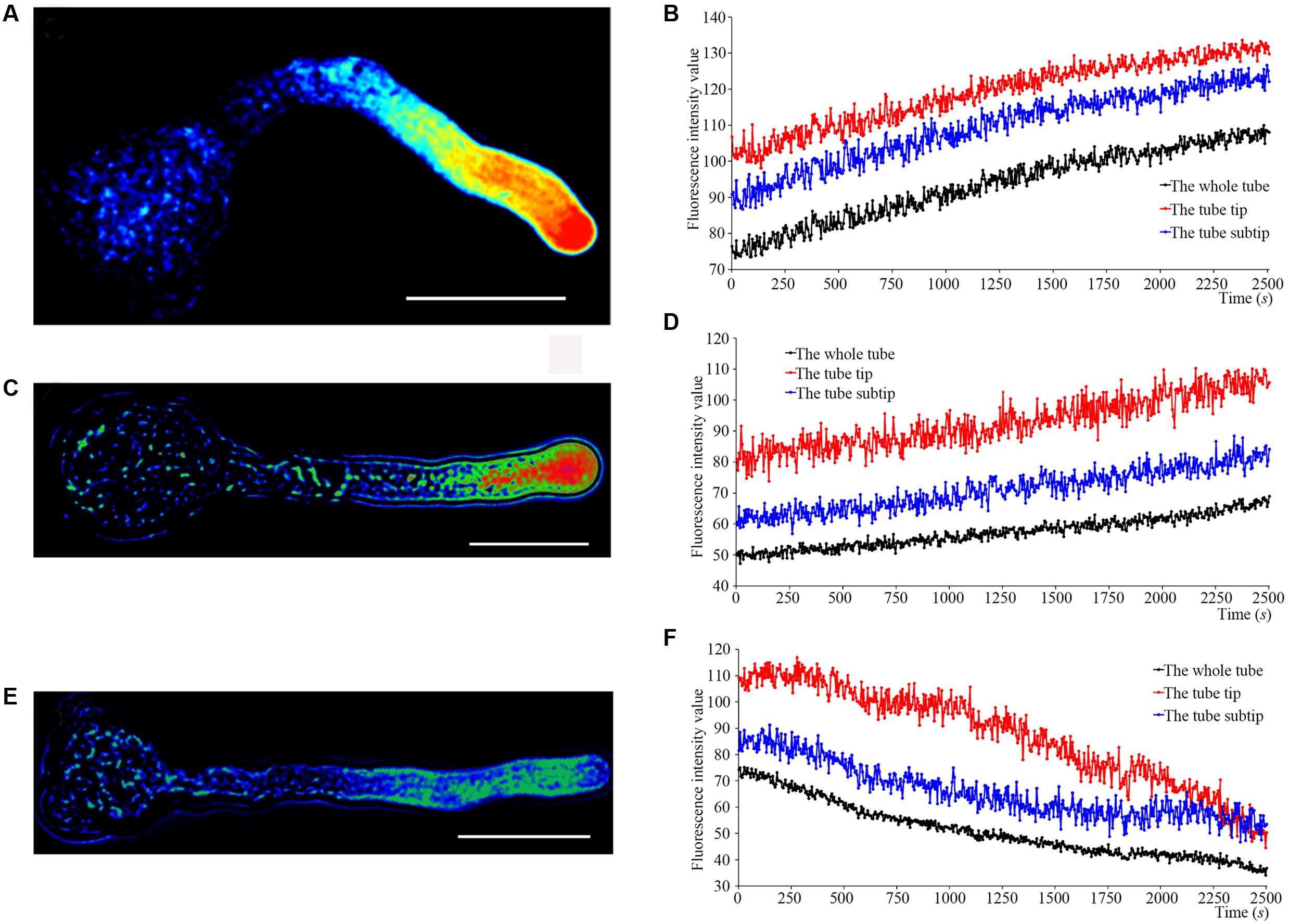
FIGURE 1. S-RNase affects the Ca2+ gradient at the pollen tube tip of ‘Housui.’ After adding S-RNase to the pollen tube culture medium, we observed the changes of Ca2+ gradient at apical pollen tube. (A,B) Variation in [Ca2+]c in an untreated pollen tube (control). The image is of a pollen tube stained with fluo-4/AM. (C,D) Variation in [Ca2+]c in a pollen tube treated with 1.0 μg μl-1 non-self-S-RNase (‘Imamuraaki’). (E,F) Variation in [Ca2+]c in a pollen tube treated with 1.0 μg μl-1 self-S-RNase (‘Housui’). Scar bar is 60 μm in (A,C,E).
We determined Ca2+ currents in ‘Housui’ pollen tube apex protoplasts with whole-cell recordings 30 min after adding 1.0 μg μl-1 (final concentration) ‘Imamuraaki’ non-self-S-RNase into the bath solution, and replacement with 0.4 or 1.0 μg μl-1 ‘Housui’ self-S-RNase. Compared to controls with 10 mM Ca2+, non-self-S-RNase had no effect on Ca2+ currents. However, in the presence of ‘Housui’ self-S-RNase, the calcium ion current presented very significant decrease (Figure 2A). The current amplitudes of 1.0 μg μl-1 non-self-S-RNase were >4-fold bigger than the amplitudes of 1.0 μg μl-1 self-S-RNase at a reference voltage of -200 mV (Figure 2B). These results suggest that self-S-RNase was selective in its regulation of Ca2+ channel activity in pollen tube protoplast membranes.
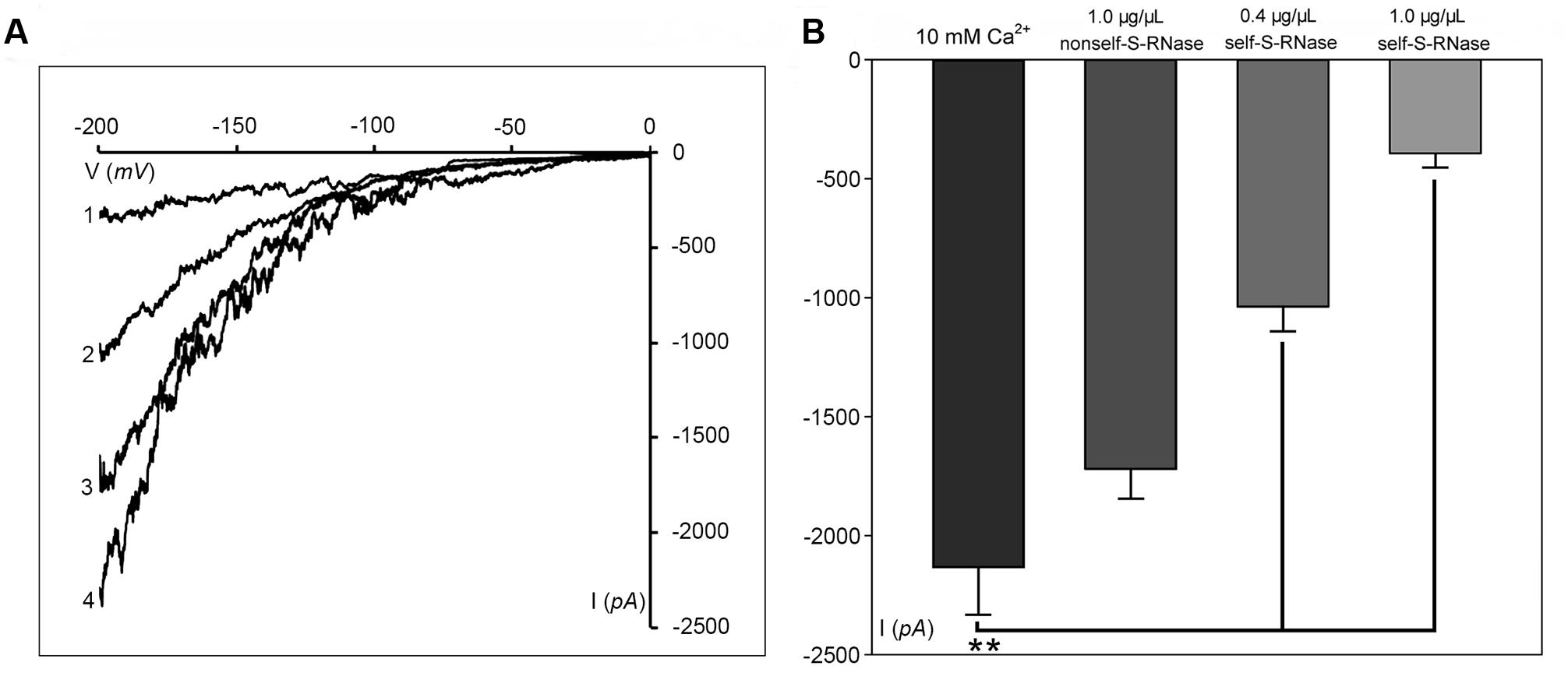
FIGURE 2. S-RNase affects Ca2+ currents in the pollen tube tip plasma membrane. (A) The representative recording currents trace from ramp voltage clamping. Different treatment with (1) 1.0 μg μl-1 self-S-RNase (‘Housui’), (2) 0.4 μg μl-1 self-S-RNase, or (3) 1.0 μg μl-1 non-self-S-RNase (‘Imamuraaki’), (4) 10 mM Ca2+ in bath solutions. (B) Summary of experiments in (A). Comparing the current at the standard of –200 mV. Comparison between self-S-RNase (0.4 and 1.0 μg μl-1, both n = 14) with non-self-S-RNase (1.0 μg μl-1, n = 14). The data are presented as the mean ± standard error. ∗∗p < 0.01.
IP3 Regulation of Ca2+ Channel Activity in Pollen Tubes
Phosphatidylinositol-specific phospholipase C controls the levels of PIP2 (i.e., substrate; phosphatidylinositol 4,5-bisphosphate) and IP3 (i.e., cleavage product), both of which are strong tip growth regulator candidates (Helling et al., 2006). We recorded the Ca2+ currents in pollen tube apices with our established method (Qu et al., 2007). When U-73122, an inhibitor of PLC activity (Mogami et al., 1997; Evdonin et al., 2004), was added to protoplasts through a pipette at a final concentration of 10 μM, the Ca2+ currents significantly decreased (p < 0.01). The current amplitudes, which were assessed at a standard reference voltage of -200 mV, were approximately sixfold smaller than those of the controls (Figure 3A). After adding U-73343-a biologically inactive analog of U-73122, it did not show significant difference from the control (p > 0.01) (Figure 3A). We observed that compared to controls, IP3 increased Ca2+ currents when the final concentration was 10 or 100 pM in the pipette solution, with a greater effect at 100 pM (Figure 3B). Moreover, U-73122, with a final concentration of 10 μM in culture medium, also disrupted tip-focused Ca2+ gradients of pollen tubes, as revealed by fluo-4/AM staining (Figures 3C,D). These results suggest that hyperpolarization-activated Ca2+ channels at the tips of pollen tubes were regulated by IP3, and that PLC regulated the tip-focused Ca2+ gradient of pollen tubes.
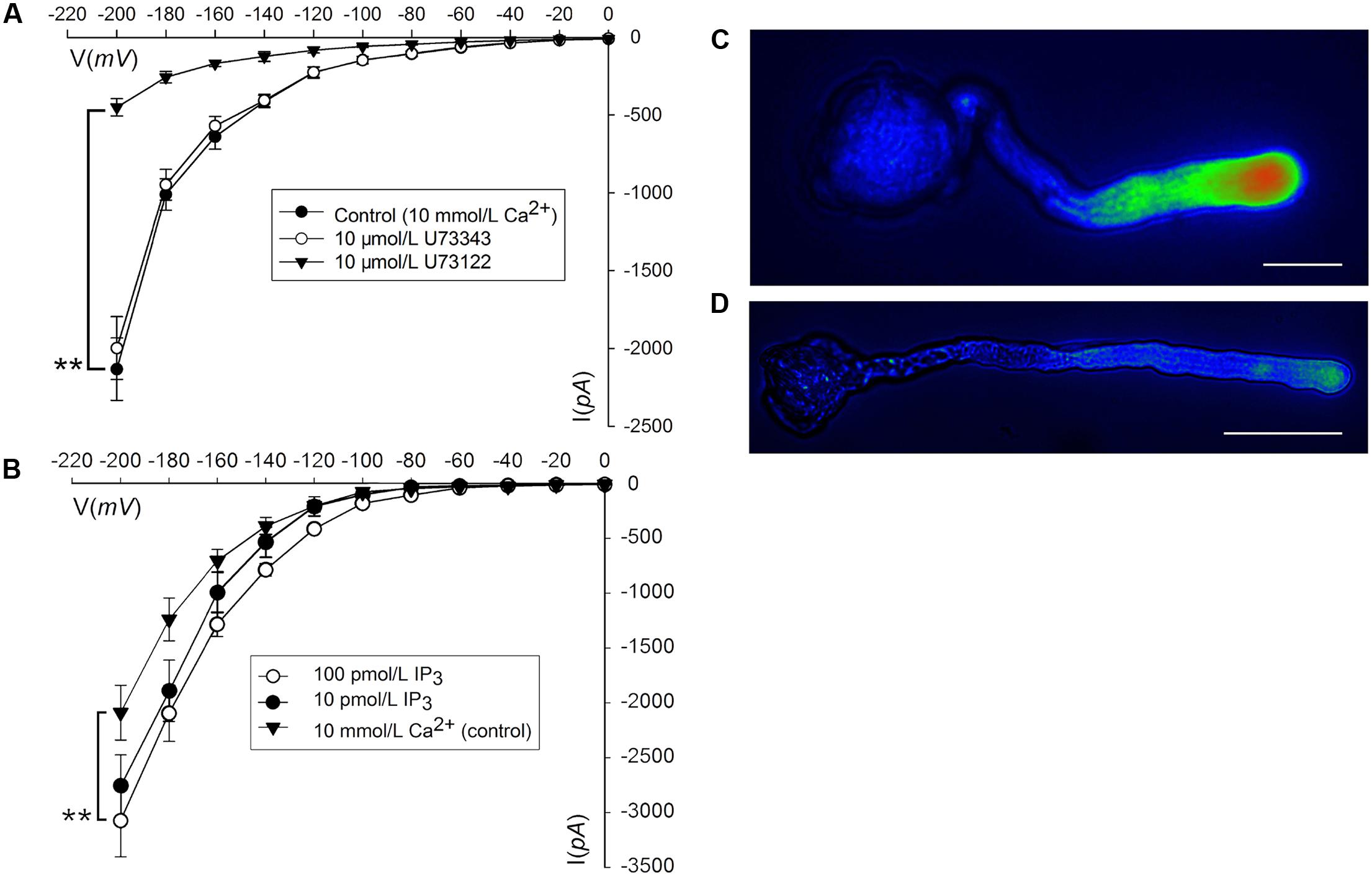
FIGURE 3. IP3-regulated hyperpolarization-activated Ca2+ channels and tip-focused Ca2+ gradient at the pollen tube apex. Ca2+ currents in ‘Housui’ pollen tube protoplasts were recorded by using a step pattern with basal pipette solutions of U-73122, U-73343, or IP3 at various concentrations. The image of pollen tube was stained with fluo-4/AM. (A) Ca2+ currents were compared among basal pipette solutions with and without U73343 or U73122 based on I/V. Data of pollen tube apex protoplasts were recorded for each concentration and the means ± standard errors were calculated. We also compared the current at –200 mV. U-73122 (10 μM, n = 11), U-73343 (10 μM, n = 9), and Control (10 mM Ca2+, n = 15). ∗∗p < 0.01. (B) Effects of IP3 in pipettes were recorded by using ramp voltage clamping, and they were compared with the effects on Control (10 mM Ca2+, n = 15). We also compared the current at –200 mV with pipette solutions containing IP3 at two different concentrations (100 and 10 pM, n = 18 and 14) respectively. ∗∗p < 0.01. (C) Image of the tip-focused Ca2+ gradient in a pollen tube exhibiting normal growth. (D) The Ca2+ gradient was eliminated when 10 μM (final concentration) U-73122 was added to the culture medium. Scar bar is 35 μm in (C) and 50 μm in (D).
S-RNase-Specific Decreases in PLC Activity
We collected plasma membrane fractions (containing cell-membrane Phospholipase C) from ‘Housui’ pollen tubes (Figure 4A), and analyzed the influence of ‘Housui’ self-S-RNase and ‘Imamuraaki’ non-self-S-RNase on its activity. PLC activity was monitored in vitro by incubating plasma membrane fractions with 3H-PIP2 and measuring the labeled product in the soluble fraction (presumably IP3) (Figure 4B). It remarkably decreased PLC activity by adding 10 μM (final concentration) U-73122. However, 1 μg μl-1 (final concentration) self-S-RNase also severely inhibited PLC activity, while non-self-S-RNase of the same final concentration only had minimal effect on the PLC activity (Figure 4B). These results suggest that self-S-RNase inhibited PLC activity in self-pollen tubes.
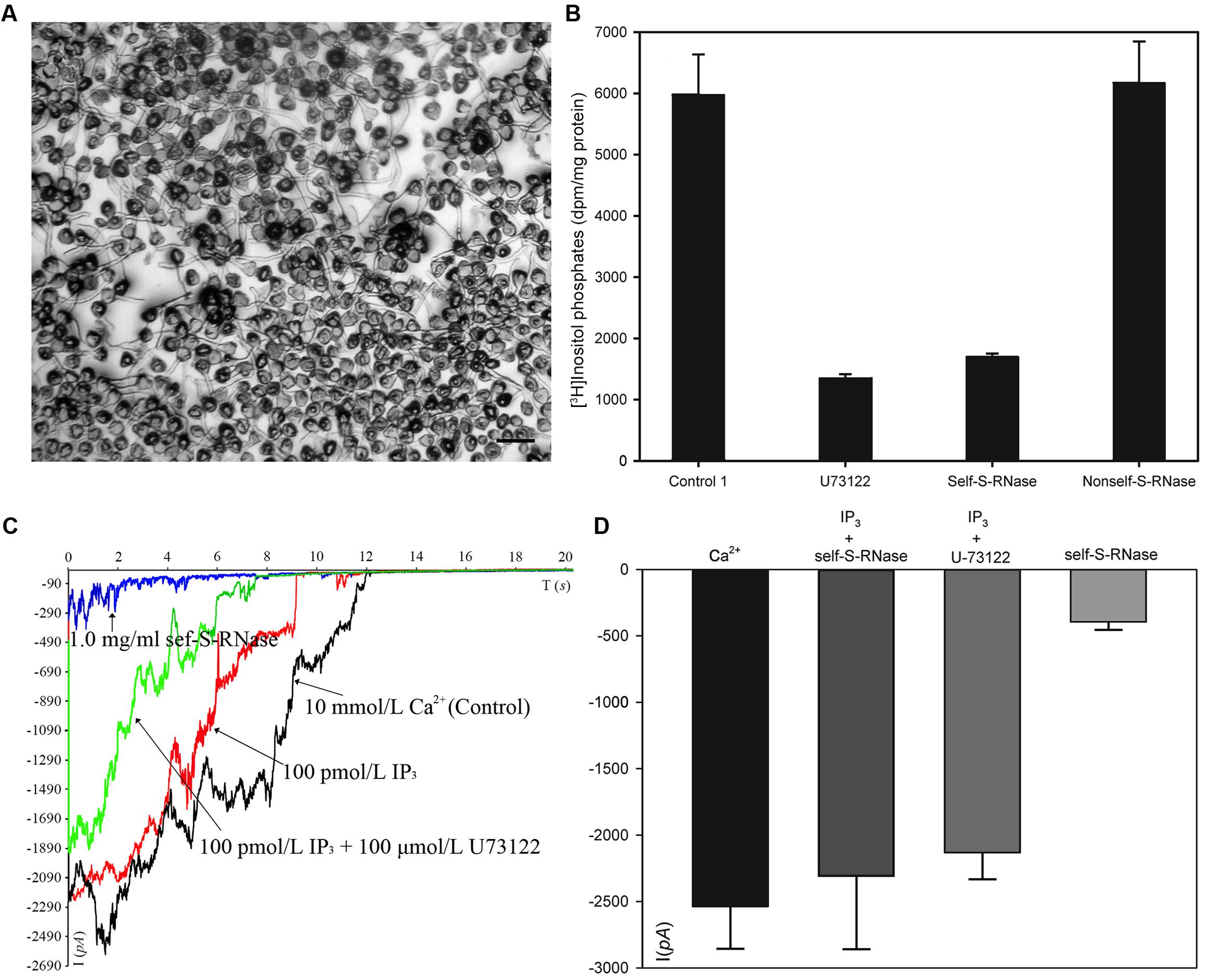
FIGURE 4. S-RNase affected PLC activity in pollen tubes and IP3-regulated Ca2+ channels of pollen tube apices. (A) Membrane was extracted from pollen tubes (‘Housui’) grown for 3 h with a germination rate of >80%. The scale bar corresponds to 200 μm. (B) [3H]-IP3 was measured after membrane protein cleavage to [3H] PI 4,5-P2PIP2 (control). The presented data are the averages of three or four independent experiments, and are presented as the mean ± standard error. (C) Ca2+ currents were recorded in response to a hyperpolarizing voltage ramp from 0 to –200 mV (ramp speed 9.84 mV s-1). The black curve represents the current when the extracellular solution contained 10 mmol/L Ca2+ (control). The red curve refers to the current when the pipette solutions contained 100 pmol/L IP3 and bath solution contained 1.0 mg ml-1 self-S-RNase (‘Housui’) at the same time. The green curve represents the current when the pipette solutions contained a mixture of 100 pmol/L IP3 and 10 μmol/L U-73122. The blue curve represents the current when the extracellular solution contained 1.0 mg ml-1 self-S-RNase. (D) The currents were compared at –200 mV for the three treatments.
When the basal pipette solution contained 100 pM IP3, Ca2+ currents in ‘Housui’ protoplasts (whole-cell configuration) were fully recovered from the suppression caused by the presence of 1 μg μl-1 ‘Housui’ self-S-RNase in the bath solution. Furthermore, compared to the control, no significant differences were found in current when the pipette solution contained a mixture of 10 μM U-73122 and 100 pM IP3. These results indicate that IP3 enabled the recovery of Ca2+ currents previously suppressed by U-73122 (Figures 4C,D). The data suggest that self-S-RNase suppressed the activity of Ca2+ channels at pollen tube tips by inhibiting PLC activity and decreasing IP3 levels in protoplasts.
PCL Interacts with S-RNase
‘Dangshansuli’ (Pyrus x bretschneideri Rehd.) is the most important commercial Asian pear cultivar, with a production volume of 4 million tons per year. The draft genome of the ‘Dangshansuli’ was the first comprehensive fine pear genome (Wu et al., 2012b). Its S-genotype is S7S34. To further identify possible PLC and S-RNase interactions, we performed yeast two-hybrid experiments. The PLC coding region was subcloned into the pGBKT7 vector, and it was tested against S7-RNase and S34-RNase pGADT7 fusion constructs. Full-length PLC interacted with full-length S7-RNase, but not with S34-RNase (Figure 5 and Supplementary Figure S1). To further gain insights into the relationship between PLC and S-RNase, we transiently expressed GFP-PLC and RFP-S-RNase in tobacco leaves. Based on the fluorescence microscopy, we found that when fused to GFP, the same PLC fragment used in the yeast-two-hybrid experiment (without the membrane-associated C2 region) mainly localized near the nucleus, and the signal overlapped with the DsRed fluorescence emitted by the RFP-S7-RNase (Figure 6A). However, it did not result in fluorescence superposition by expressing RFP-S34-RNase and GFP-PLC in tobacco cells (Figure 6B). These results suggest that PLC interacted with the products of multiple alleles of one S-RNase, rather than the products of two S-RNases.
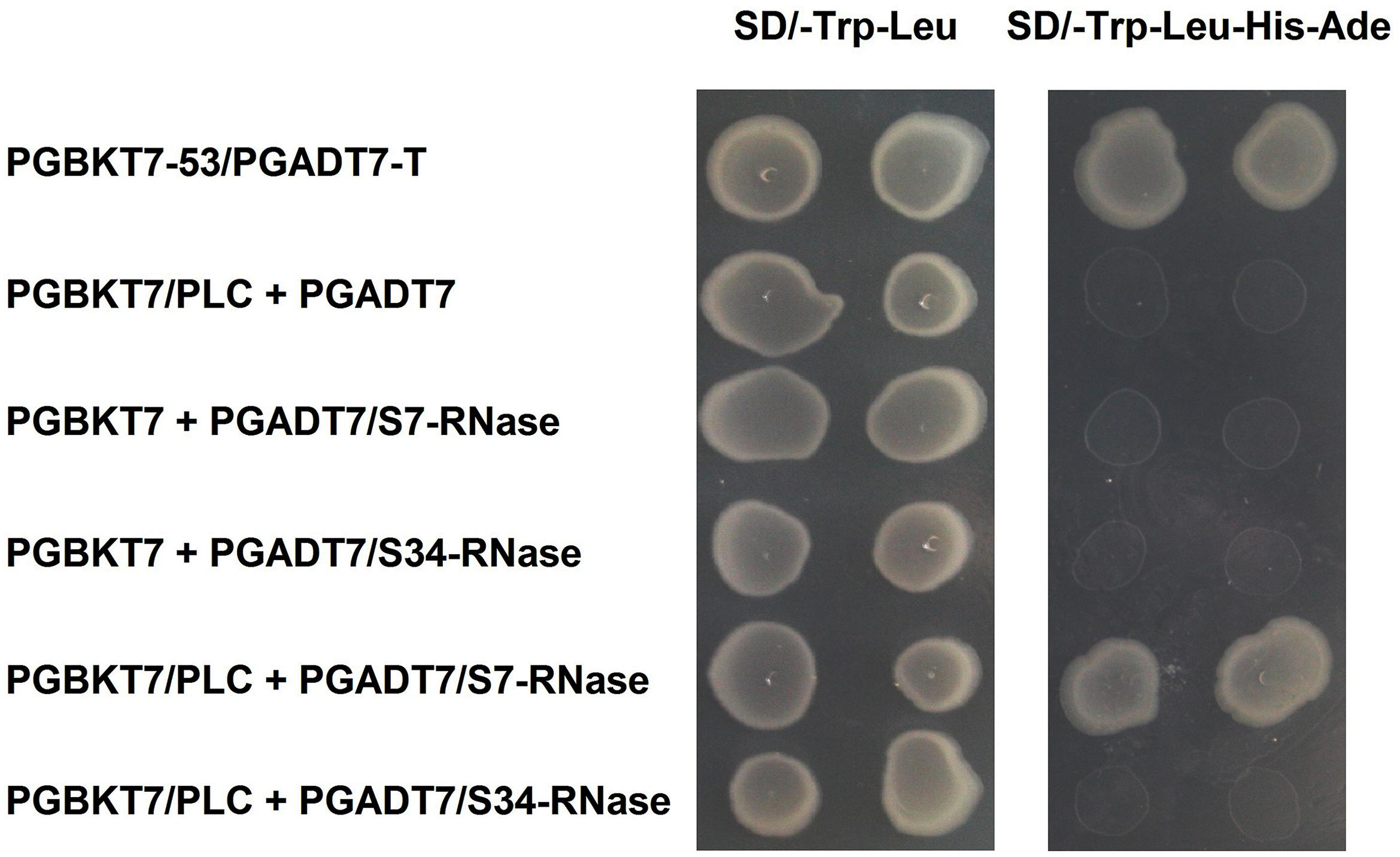
FIGURE 5. Interaction of PLC in the pollen tube with S-RNase in the style of Pyrus x bretschneideri Rehd in yeast two-hybrid assays. Positive control (PGBKT7-53/PGADT7-T); Negative control (PGBKT7/PLC + PGADT7; PGBKT7 + PGADT7/S7-RNase; PGBKT7 + PGADT7/S34-RNase); Experimental group (PGBKT7/PLC + PGADT7/S7-RNase; PGBKT7/PLC + PGADT7/S34-RNase). The SD/-Trp/-Leu/X-α-Gal (DDO/X, Right) and SD/-Trp/-Leu/-His/-Ade/X-α-Gal (QDO/X, Left) plates were incubated at 30°C for 3 days and then visualized. Co-transformed yeast containing pGBKT7/PLC and pGADT7/S34-RNase plasmids grew on the SD medium without Trp and Leu, but it could not grow on SD medium lacking Trp, Leu, His, and Ade. Yeast cells with pGBKT7/PLC and pGADT7/S7-RNase grew on SD medium lacking additional nutrients.
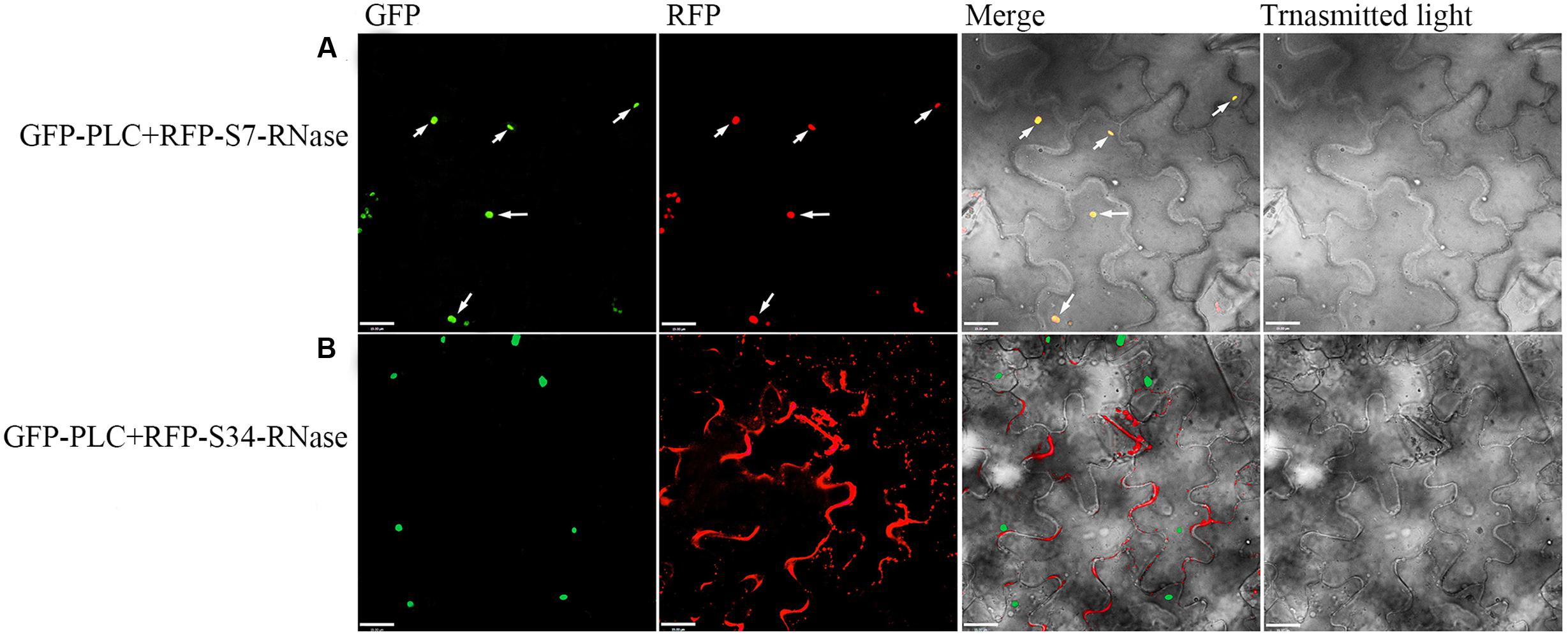
FIGURE 6. Colocalization of the PLC with S-RNase. Dual-view analyses of cells expressing both GFP-PLC (removing the C2 region bound to the membrane) and RFP-S-RNase. (A) Co-transformed with GFP-PLC and RFP-S7-RNase. (B) Co-transformed with GFP-PLC and RFP-S34-RNase. The images of GFP fluorescence, DesRed fluorescence, brightfield, and the merged were obtained through fluorescence microscopy, and were displayed from left to right respectively. The merged image (yellow) shows the high degree of colocalization.
PLC Polymorphisms in Pollen Tubes
Self-specific direct interactions between PLC and self-S-RNase require PLC polymorphisms at a level matching the S-RNase polymorphisms. To clone the PLC cDNA from Pyrus pyrifolia pollen tubes, one degenerate oligonucleotide, designed according to a conserved amino acid sequence, was used for PCR analyses with cDNA from ‘Housui’ pollen tubes as templates. Two DNA fragments (approximately 750 bp) were amplified. A BLAST analysis indicated that these two fragments were homologous to plant PI-PLCs. Sequence alignments of the two DNA fragments revealed the encoded amino acid sequences (according to Open Reading Frame Finder1) with a polymorphism frequency of 22% (Figure 7A). The two partial PLC genes were determined to be PI-PLC 2-like (LOC103936790) according to Protein Blast (P. x bretschneideri), with identities of 96 and 83%.
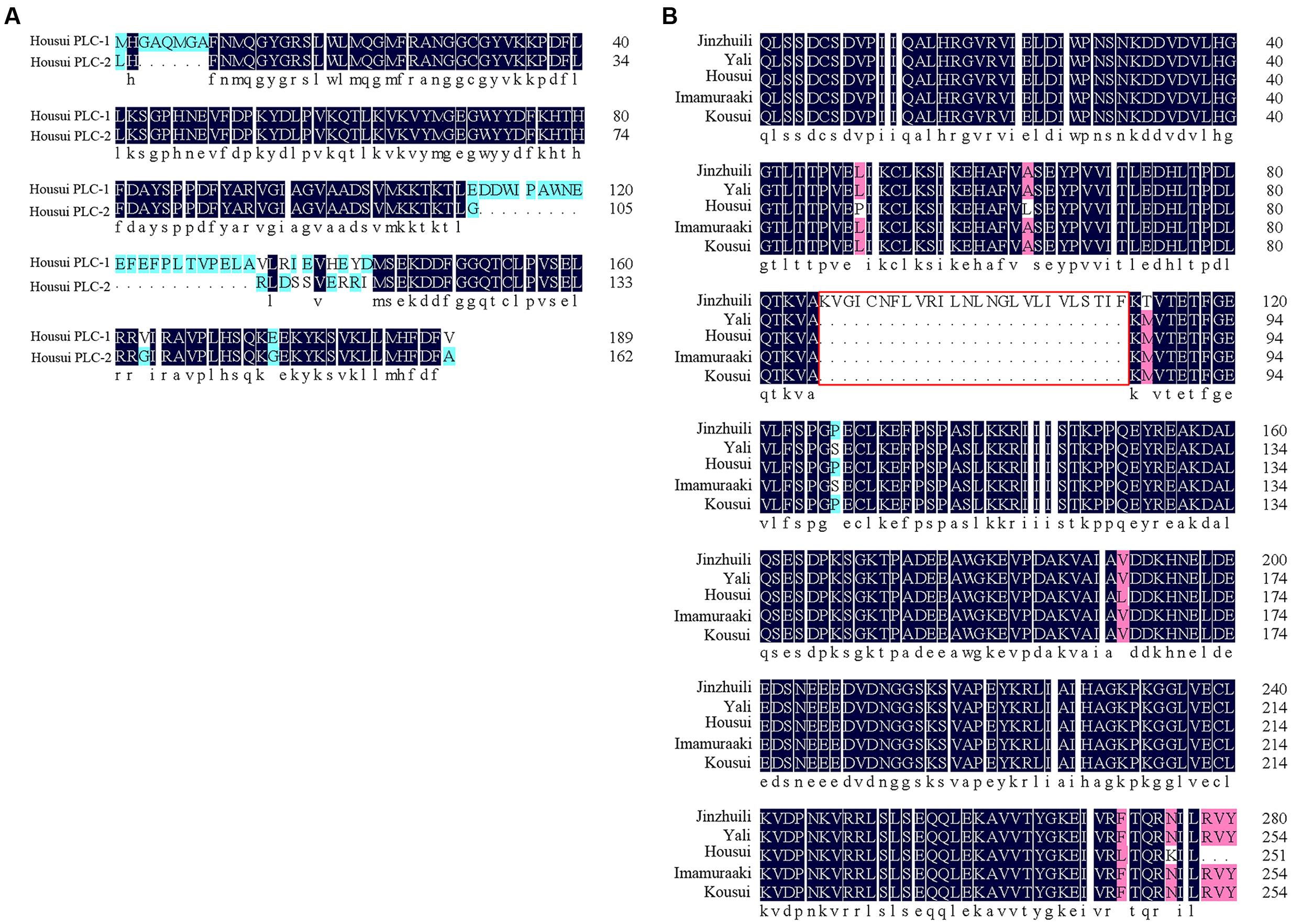
FIGURE 7. Partial PLC amino acid sequence alignment. (A) Amino acid sequence of the ‘Housui’ partial Y-domain and subsequent amino acids. (B) Partial C2- and Y-domains of PLC. The amino acid sequence of self-compatible ‘Jinzhuili’ was compared with that of SI varieties (‘Housui,’ GenBank: EU295486.1).
For fruit trees, including pear trees, it is very difficult to construct a stable transgenic system. However, we obtained ‘Jinzhuili’ as a naturally occurring self-compatible mutant of ‘Yali.’ Field pollination tests showed that >76% of flowers set fruits, and many pollen tubes grew down to the base of the style during ‘Jinzhuili’ self-pollination and cross-pollination of ‘Yali’ (female) × ‘Jinzhuili’ (male). In contrast, during ‘Yali’ self-pollination and cross-pollination of ‘Jinzhuili’ (female) × ‘Yali’ (male), fruit set did not occur, and most pollen tubes were arrested at the upper part of the style. The results indicate that ‘Yali’ and ‘Jinzhuili’ had a normal SI response in the style, which rejected competent ‘Yali’ pollen, but accepted ‘Jinzhuili’ pollen. Further molecular analyses revealed that both ‘Yali’ – and ‘Jinzhuili’ – labeled S21-RNase and S34-RNase genes were transcribed normally with identical mRNA sequences. Therefore, we suggest that it was most likely ‘Jinzhuili’ had lost pollen SI activity, resulting in a style that behaved normally during the SI response, similar to that of the wild-type ‘Yali’ (Supplementary Figure S2).
We used a previously described method to clone PLC genes from ‘Jinzhuili’ and ‘Yali.’ The amino acid sequence alignments (Figure 7B) showed that the PLC of ‘Jinzhuili’ had a 26-amino acid insertion. We inserted this nucleotide fragment to encode the 26-amino acid peptide segment into the PLC gene of ‘Dangshansuli’ (initially used in the yeast-two-hybrid experiment as shown in Figure 5). This mutated PLC protein did not interact with either the S7-RNAse or the S34-RNAse in our yeast-two-hybrid experiment (Figure 8 and Supplementary Figure S1). The self-S-RNase could not identify mutant PLC, and was unable to inhibit PLC activity. Therefore, ‘Jinzhuili’ exhibited self-compatibility. These results suggest that the PLC in the pollen tube is one of the male determinants in the self-incompatibility reaction.
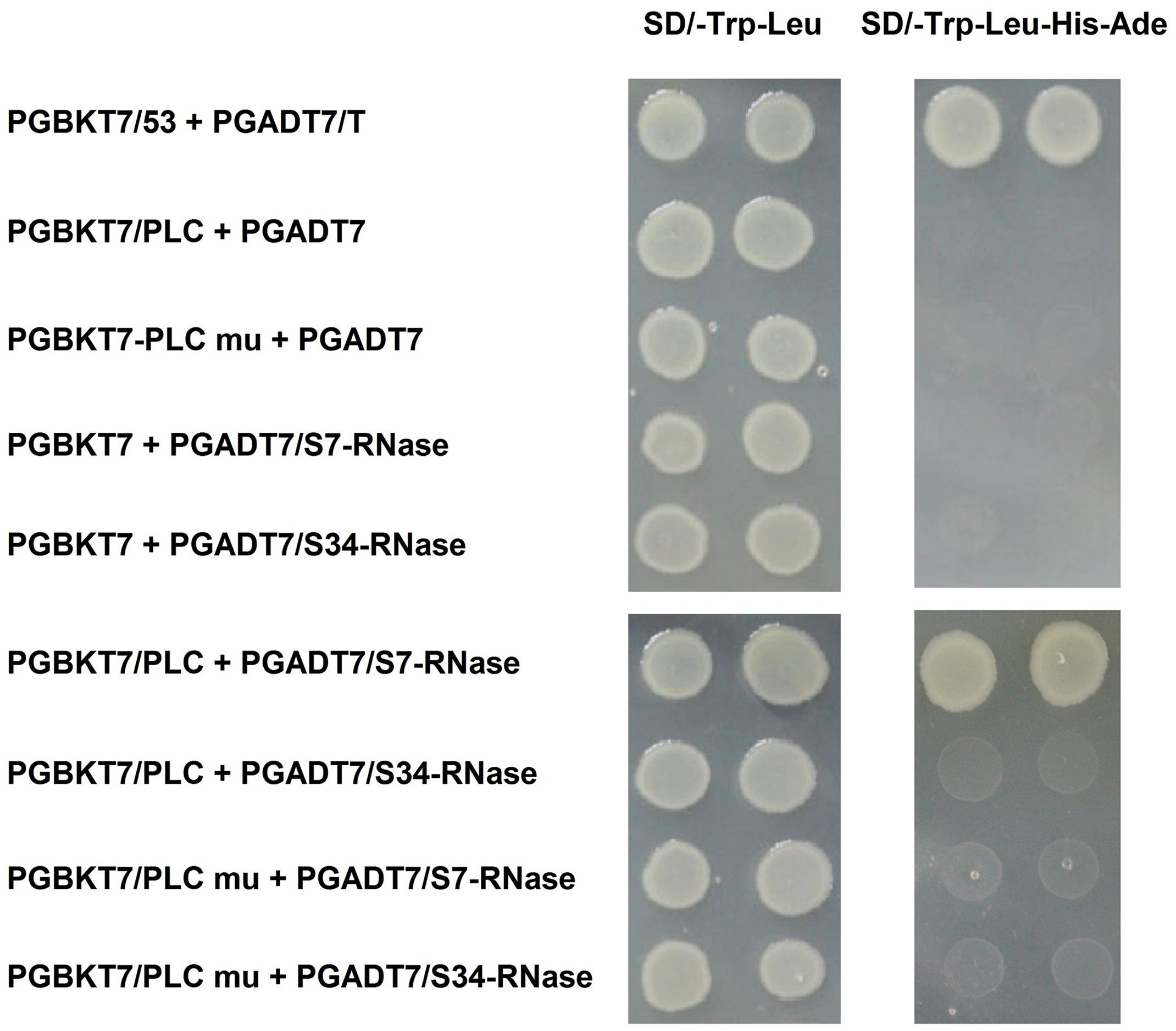
FIGURE 8. Interaction of inserting the mutated PLC (PLC mu) into the pollen tube with S-RNase in the style of P. x bretschneideri Rehd in yeast two-hybrid assays. Positive control (PGBKT7-53/PGADT7-T); Negative control (PGBKT7/PLC + PGADT7; PGBKT7/PLC mu + PGADT7; PGBKT7 + PGADT7/S7-RNase; PGBKT7 + PGADT7/S34-RNase); Experimental group (PGBKT7/PLC + PGADT7/S7-RNase; PGBKT7/PLC + PGADT7/S34-RNase; PGBKT7/PLC mu + PGADT7/S7-RNase; PGBKT7/PLC mu + PGADT7/S34-RNase). The SD/-Trp/-Leu/X-α-Gal (DDO/X, Left) and SD/-Trp/-Leu/-His/-Ade/X-α-Gal (QDO/X, Right) plates were incubated at 30°C for 3 days, and then visualized. Co-transformed yeast containing pGBKT7/PLC and pGADT7/S34-RNase plasmids grew on the SD medium without Trp and Leu, but it could not grow on SD medium lacking Trp, Leu, His, and Ade. Yeast cells with pGBKT7/PLC and pGADT7/S7-RNase grew on SD medium without Trp and Leu, also can grow without Trp, Leu, His, and Ade. The results indicate that there is interaction between PLC and S7-RNase proteins, but there is no interaction between PLC mu and S7-RNase protein.
Analysis of Gene Expression and Proteome Difference in PLC Gene in Pollen Tube
We explored the difference in gene expression at the early stage after pollination, and compared gene expression levels in the styles of ‘Yali’ and ‘Jinzhuili’ 0.5 h after self-pollination. Although the differences in gene expression were compared after pollination, the GO terms associated with “pollen tube growth,” “cell tip growth,” “pollen tube development,” and “pollination” identified cell activities consistent with pollen growth activity (data unpublished). We searched for Pyrus PLC genes in the GenBank database (Supplementary Table S1). The differences in gene expression levels were determined based on high-throughput sequencing results (Supplementary Table S1). Importantly, 0.5 h after pollination, the pollen grain had just germinated on the stigma, and there was no difference in pollen tube growth between plants that underwent compatible and incompatible pollinations (Supplementary Figure S3). Therefore, at this time point, differences in gene expression between stigmas derived from compatible and incompatible pollinations likely reflect differences related to the compatibility of the interaction rather than differences in pollen tube size or viability.
In theory, if the PLC gene is expressed specifically in pollen tubes, its expression during compatible pollination should be higher than that of the control or during incompatible pollination. This is because pollen tubes can grow during compatible pollination. The values of Log2(Jinzhuili 0.5 h/ Jinzhuili CK) and Log2(Jinzhuili 0.5 h/ Yali 0.5 h) were positive for only LOC103958605, LOC103936790, and LOC103966250, although the p-value for LOC103966250 was higher than 0.05 (i.e., 0.252186). Therefore, we selected these three genes for quantitative PCR analysis, and detected the expression levels at 0.5 h after pollination. The expression levels in the pollen tube after compatible pollination were significantly higher (p < 0.01) than those after incompatible pollination and the expression level of non-pollinated styles. During compatible pollination, the expression levels were 7, 10, and 14 times higher than those during incompatible pollination (Figures 9A–C). Moreover, based on Supplementary Table S1, we selected LOC103932386, which exhibited no differential expression, for further RT-PCR analysis. We confirmed that the expression level did not change following pollination (Figure 9D). These results suggest that the three genes of LOC103958605, LOC103936790, and LOC103966250, were expressed specifically in the pollen tube, and they were related to pollen tube growth in the style. The expression levels of these three genes in the styles were higher (p < 0.01) after incompatible pollination than in non-pollinated styles.
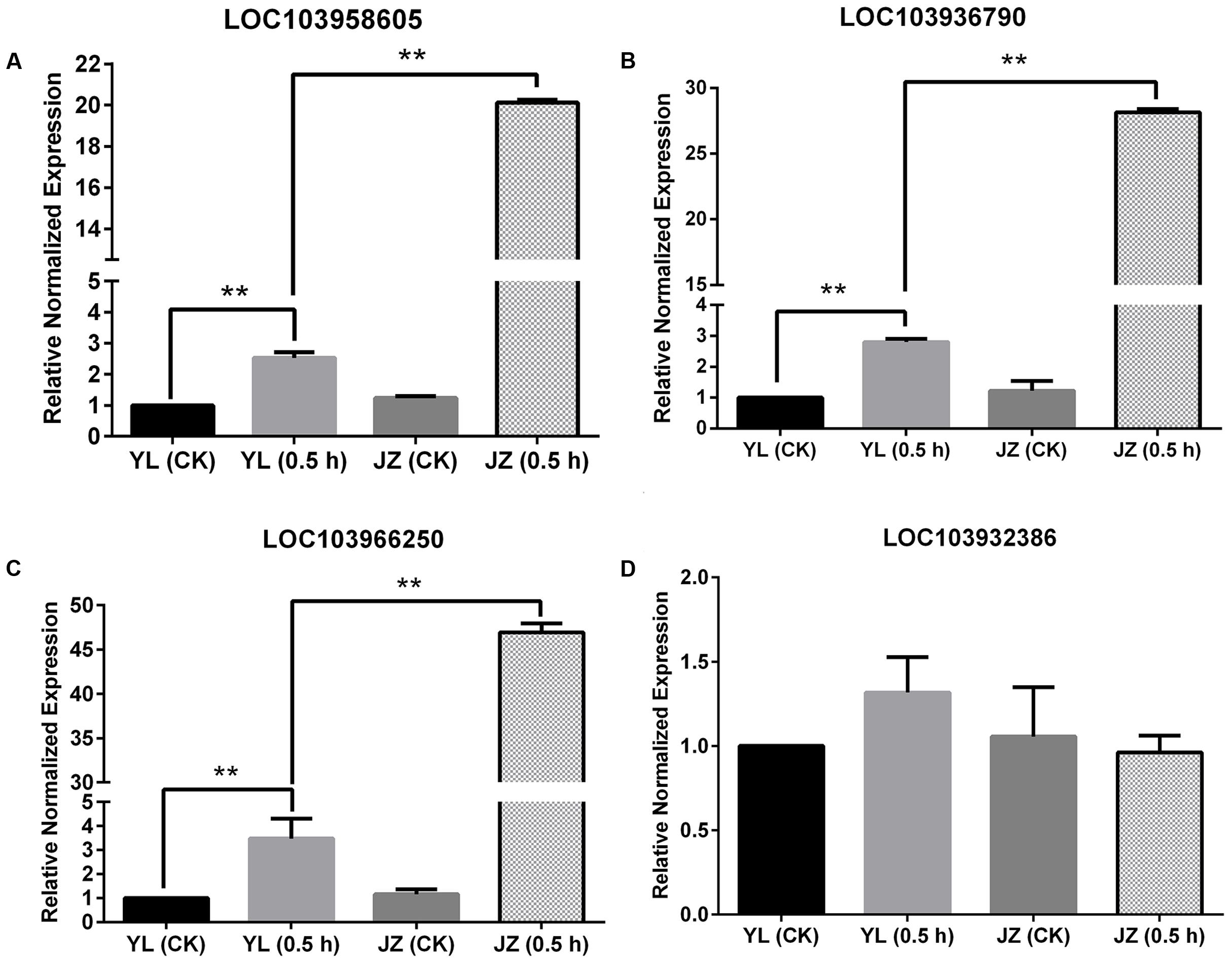
FIGURE 9. Expression of four PI-PLC genes in styles based on quantitative PCR. Comparison of the expression of four PLC genes during compatible and incompatible pollination. ‘Yali’ styles were collected 0.5 h after self-pollination [YL (0.5 h)]. ‘Jinzhuili’ styles were collected 0.5 h after self-pollination [JZ (0.5 h)]. Non-pollinated ‘Jinzhuili’ [JZ (CK)] and ‘Yali’ [YL (CK)] styles were collected 0.5 h without pollination. The GeneID of the four PLC genes are as follows: (A) LOC103958605. (B) LOC103936790. (C) LOC103966250. (D) LOC103932386. ∗∗p < 0.01. Primer sequences are provided in electronic Supplementary Table S2.
By using the iTRAQ technique, we measured the style proteome of the ‘Jinzhuili’ and ‘Yali’ after 0.5 h of self-pollination respectively. The expression of PI-PLC related proteins was different under ‘Jinzhuili’ (0.5 h) – vs. – ‘Yali’ (0.5 h), ‘Yali’ (0.5 h) – vs. – ‘Jinzhuili’ (CK), ‘Jinzhuili’ (0.5 h) – vs. – ‘Jinzhuili’ (CK) (Table 1). In the case of self-compatibility comparing to self-incompatibility, PI-PLC-related protein up-regulated expression was significant (Protein ID: XP_009373787.1, XP_009343292.1, XP_009344939.1). The proteins, XP_009343292.1 and XP_009344939.1, were up-regulated only in the case of self-compatibility. In particular, the changes in XP-009344939.1 protein expression were consistent with the results of high-throughput sequencing and RT-PCR. Although expression of the XP-009346269.1 protein was down-regulated in styles derived from compatible pollination, and up-regulated in styles derived from incompatible pollination, the differences were not significant. These results indicate that by using the ITRAQ technology to analyze PLC protein expression differences, it could be useful to distinguish between compatibility and incompatibility.
S-RNase inhibited PLC activity and arrested pollen tube growth. Furthermore, we cloned a gene fragment from PI-PLC 2-like gene (LOC103936790), and observed that the expression levels of the other two PLC genes (LOC103958605 and LOC103966250) were relatively higher after compatible pollination. According to proteomic analysis, there are other PLC genes involved in self-incompatibility. Therefore, it requires further research on the role of these PLC genes.
Discussion
In Solanaceae, Plantaginaceae, and Rosaceae species, SI is genetically controlled by a single S-locus with multiple haplotypes. The pistil-S gene encodes a T2 family ribonuclease, namely S-RNase (Xue et al., 2014), but the identity of the pollen-S gene has not been confirmed. Current research suggests that the SLF (S-locus F-box) gene determines male specificity in SI. The SLF gene product forms SCF (SKP1/Cullin1/F-box) complexes that serve as putative E3 ubiquitin ligases, which interact with S-RNases and cause non-self-S-RNase degradation during compatible pollination (McClure, 2006). However, S-RNases from the style continuously enter pollen tubes. Immunolocalization studies showed an abundance of S-RNase inside compatible and incompatible Solanum chacoense pollen tubes (Luu et al., 2000). Furthermore, there was no evidence of S-RNase degradation by using 3H-labeled S2-RNase and in vitro grown Nicotiana alata pollen tubes (Gray et al., 1991). We were unable to determine the differences of S-RNase between pollen tubes treated with self-S-RNase and non-self-S-RNase by using a colloidal gold technique during in vitro experiments (Qu et al., 2016b). McClure (2006) proposed that S-RNase compartmentalization into vacuoles could restrict S-RNase cytotoxicity, which enabled compatible pollination. If the vacuoles ruptured and S-RNase was released into the cytoplasm, the resulting cytotoxicity would inhibit pollen tube growth during incompatible pollination. During the SI response in Nicotiana species, when the incompatible pollen tube growth was fully inhibited 6 days after pollination, the vacuolar membrane system was still intact in 80% of pollen tubes in vivo (Roldán et al., 2012). Therefore, the S-RNase degradation and S-RNase compartmentalization models cannot be used to fully explain the physiological function of the Pollen-S gene during GSI responses. It is noteworthy that even though Rosaceae species also employ S-RNase and SLF as the female and male determinants respectively, the role of SLF in SI has not been established. The Japanese pear has a ‘non-self-recognition by multiple factors’ SI system, while a ‘self-recognition by a single factor’ system exists in Prunus species (Kakui et al., 2011).
The quantities of S-proteins differ among cultivars. However, they do not correlate with the growth of selfed pollen tubes. For example, there are much more S4-proteins in ‘Nijisseiki’ and ‘Kikusui’ than in ‘Yakumo,’ even though ‘Nijisseiki’ and ‘Kikusui’ exhibit intermediate levels of SI, and ‘Yakumo’ experiences strong SI (Zhang and Hiratsuka, 1999). Qin et al. (2006) observed that in accordance with the average values of S11- and S12-RNase for a single Solanum chacoense style, the levels of the S11- and S12-RNases can differ by up to 10-fold within a genotype. Additionally, the S12-RNase levels can differ by over threefold when different genotypes are compared. Surprisingly, the abundance of S12-RNase in different styles of the same plant can differ by over 20-fold. Therefore, S-RNase is not only responsible for just degrading RNA during the self-incompatible response. Early grafting experiments conducted by Lush and Clarke (1997) showed that growth inhibition caused by incompatible styles could be reversed in some pollen tubes, suggesting that the cytotoxic effect of S-RNase was not only caused by its role in RNA degradation.
The S-RNase protein structure is consistent with a dual role as both a cytotoxin and a recognition protein (McClure, 2006). Recently, several new functions of S-like RNases unrelated to ribonuclease activity have been uncovered, indicating that their cytotoxicity may be a result of their effects on proteins rather than RNA (Levava et al., 2006; Zhang et al., 2009; Luhtala and Parker, 2010). S-RNases modify the actins in the cytoskeleton of self-pollen tubes in vitro. These modifications occur prior to the arrest of pollen tube growth. The S-RNases may initiate programmed cell death during self-pollen tube growth inhibition in vitro in P. pyrifolia GSI responses (Liu et al., 2007; Wang et al., 2010).
Phosphoinositides are most likely present in every plant cell, but their most prominent function is pollen development and pollen tube polarity (Heilmann and Ischebeck, 2016). Phosphatidylinositol-specific phospholipase C controls pollen tube tip growth by regulating the levels of its substrate PIP2 and its cleavage product IP3 (Franklin-Tong et al., 1996; Zonia and Munnik, 2004; Monteiro et al., 2005). The maintenance of a Ca2+ gradient and vesicle secretion in pollen tube apices is essential for growth. Phosphatidic acid, PIP2, and IP3 play vital roles in the regulation of these processes (Monteiro et al., 2005; Krinke et al., 2007). Helling et al. (2006) did not study the relationship between Ca2+ and PI-PLC in pollen tube apices, but they proposed that IP3 generated at the flanks of the pollen tube tips during hydrolysis of PIP2 by PI-PLC may help establish the tip-focused cytoplasmic Ca2+ gradient essential for the growth of these cells. Moreover, Kost et al. (1999) also hypothesized that PLC-mediated hydrolysis of tip-localized PIP2 and IP3-induced Ca2+ influx into the cytoplasm may be involved in the establishment of the tip-focused Ca2+ gradient. Additionally, the putative IP3-regulated Ca2+ channels, which allow Ca2+ to enter from the extracellular matrix, may be present in the pollen tube plasma membrane. However, IP3-sensitive Ca2+ channels have not been identified in plants (Munnik et al., 1998; Meijer and Munnik, 2003). Dellis et al. (2006) believed that the IP3 receptors (IP3R) contribute more directly to Ca2+ entry across the plasma membrane. In the present study, we demonstrated that hyperpolarization-activated Ca2+ channels in the plasma membrane of P. pyrifolia pollen tube apices were sensitive to IP3. When IP3 was added to pipette solutions, the Ca2+ currents presented significant increases, or they were recovered if they had been suppressed by U-73122. Thus, we suggest that active PLC located at the pollen tube tips cleaves PIP2 to IP3, which then increases the extracellular Ca2+ influx through hyperpolarization-activated Ca2+ channels. Ca2+ influx at the apices of pollen tubes not only maintains the Ca2+ gradient from tip to base, but also stimulates pollen tube elongation.
Although it is difficult to assess direct correlations among ion flux, ion gradients, delivery and turnover of vesicles, and pollen growth (Messerli et al., 1999), there is no doubt they are closely linked (Blakeslee et al., 2004). The growing pollen tube tip is filled with secretory vesicles that fuse with the membrane to drive pollen tube elongation (Hepler, 2015). Campanoni and Blatt (2007) suggested that exocytosis from the Golgi apparatus enriched the ion channels at the tip of growing pollen tubes through site-directed exocytosis or lateral diffusion. We speculated that Ca2+ channel on the membrane maintains the sensitivity of IP3. Moreover, based on transmission electron microscopy, we observed that a large number of vesicles were fusing with the cell membrane at the tip of pollen tube (data unpublished). According to patch clamp technique, IP3 regulates the extracellular calcium influx through the Ca2+ channel within the cytoplasmic membrane (Vaca and Kunze, 1995; Dellis et al., 2006). These results are consistent with our findings that IP3 can increase extracellular calcium influx at apical pollen tube.
The PI-PLC activity assay showed that self-S-RNase (‘Housui’) decreased the activity of PLC extracted from ‘Housui’ pollen tubes. In the pollen tube, the PLC gene was inserted into the mutant, which resulted in the conversion from the self-incompatible ‘Yali’ cultivar to the self-compatible ‘Jinzhuili’ cultivar. The PLC only interacted with one S-RNase according to yeast two hybrid technology in ‘Dangshansuli,’ but if the PLC insertion mutation is artificially performed, the mutated PLC could not interact with any S-RNase. These results were consistent with the inhibition model proposed by Wu et al. (2012a) in their research. The amino acid sequences of pollen PLC have polymorphisms that may match the S-RNase polymorphisms in the style. Moreover, IP3 can enable full recovery of Ca2+ currents following suppression by self-S-RNase or U-73122. Thus, self-S-RNase selectively blocks the activity of a pollen-specific PLC located in the apical plasma membrane of pollen tubes, and decreases Ca2+ influx through hyperpolarization-activated Ca2+ channels. This ultimately leads to a decrease or elimination of the Ca2+ gradient.
The transcriptomic analyses of pollen–pistil interactions have provided a valuable approach and a wealth of information on which to base future functional analyses (Dresselhaus and Franklin-Tong, 2013). According to the pathway enrichment analysis of differentially expressed proteins based on KEGG database, the pathway of plant–pathogen interaction was significant after pollination (Supplementary Figure S4). Therefore, we suggested that the PLC activity in pollen tube was selectively blocked by self-S-RNase during GSI in P. pyrifolia, but the process was very slow due to abundance of S-RNase (about 20 ng) in P. pyrifolia styles (Zhang and Hiratsuka, 2005). The Ca2+ gradient was gradually eliminated, and the growth rate of the incompatible pollen tube decreased, but it was not completely arrested (Herrero and Dickinson, 1980; Lush and Clarke, 1997). Roldán et al. (2012) also suggested that the inhibition of pollen tube growth was a progressive process rather than a rapid and uncontrolled collapse of cellular structures. As the incompatible pollen tube growth slowed down, S-RNase accumulated and began to degrade RNA when it exceeded the threshold concentration in the pollen tube, which caused further arrest. This is consistent with the dependence of S-haplotype-specific pollen rejection on S-RNase concentration (Hiratsuka et al., 2001; Takayama and Isogai, 2005).
Many studies on the GSI of Rosaceae focused the fate of pollen tubes within the style, while ignoring the viability of pollen in the stigma (Romain and Dirk, 2014). Furthermore, no studies have so far examined the changes in gene expression that accompany mutual interactions between pollen and stigma (Kao and Huang, 1994). In this study, we investigated the changes in gene expression that occur in the gametophyte during early stage after self-compatible and self-incompatible pollinations in Pyrus. At the early stage of pollination, under incompatible pollination, the expression of PLC gene in pollen tube was lower than that under compatible pollination. The low expression of PLC gene may be related to the slow growth of pollen tubes in the style, but what factors induce the low expression of PLC gene need further exploration. Vossen et al. (2010) believed that PLC isoforms in plant were required for the hypersensitive response (HR) and disease resistance. However, there is a differential requirement of PLC isoforms for the tomato [Solanum lycopersicum (SI)] immune response, and SIPLC4 is specifically required for Cf-4 function, while SIPLC6 may be a more general component of resistance protein signaling. We found that the expression of PLC isoforms was significantly increased under compatible pollination according to the results of iTRAQ. Perhaps these PLCs are involved in self-incompatibility, but the specific role may not be the same.
In summary, it was found that self-S-RNase inhibited the activity of PLC enzyme, and there was a positive correlation between the growth of pollen tube and the expression of PLC gene during the process of mutual recognition between pollen and stigma. Moreover, self-S-RNase decreased the activity of Ca2+ channels and disrupted the Ca2+ gradient at the tip of the growing pollen tube during the GSI response through PLC-IP3 single pathway. ‘Jinzhuili’ is a naturally occurring SC mutant from SI of ‘Yali,’ because the S-RNase cannot recognize the insertion of mutated PLC in the pollen tube. Thus, we suggested that PLC in the pollen tube is one of the determinant male specificity factors during SI interactions. We proposed a new model to explain these results (Figure 10).
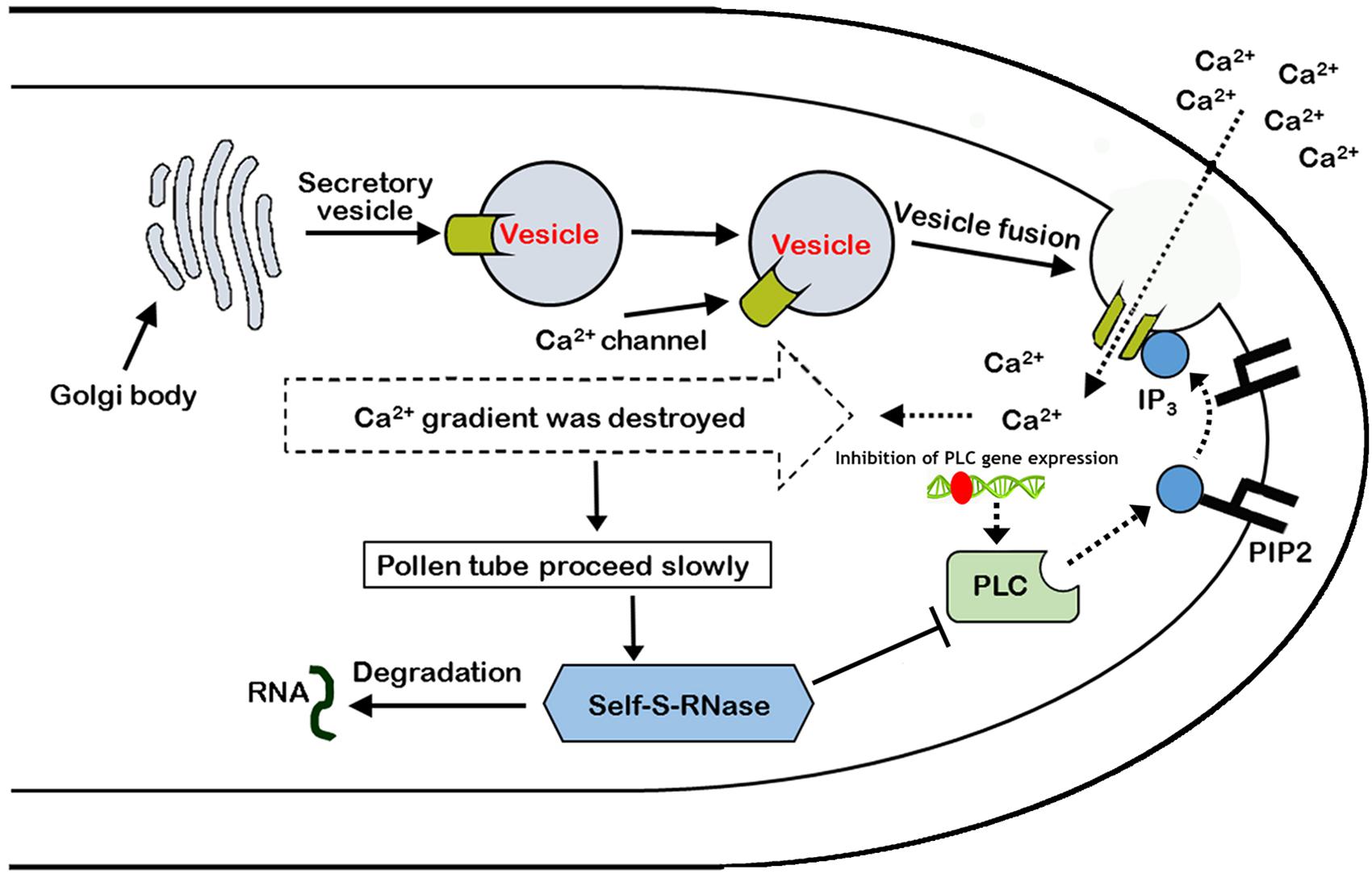
FIGURE 10. Model of the arrest of pollen-tube growth by self-S-RNase in Pyrus pyrifolia. PLC in pollen tube, located at pollen tube apices, cleaves PIP2 to IP3 and DAG (1,2-Diacylglycerol). Inositol 1,4,5-trisphosphate stimulates extracellular free-Ca2+ influx through Ca2+ channels, maintaining the Ca2+ gradient. If the activity of the special apical tube PLC is blocked by self-S-RNase, the concentration of IP3 decreases at the apex. In addition to the inhibition of PLC activity, the expression of PLC gene was also inhibited by unknown factors. As a result, the activity of Ca2+ channels is suppressed, and extracellular free-Ca2+ influx decreases. Additionally, the high Ca2+ gradient at the tip cannot be generated and maintained (Campanoni and Blatt, 2007). Therefore, the pollen tube eventually stops growing. Roldán et al. (2012) concluded that the style rejected incompatible pollen through S-RNase degrading the RNA, which was shown to be essential for the SI response, but the procession is at the late stage after major cellular dismantling of incompatible pollen tubes. In this self-incompatible model, the PLC activity and expression in the non-compatible pollen tube coincide with the behavior of the PLC between plant and pathogen. Type T symbol represents inhibition of PLC enzyme activity. Broken and solid lines represent negative and positive regulatory activities, respectively. Red ellipse represents an unknown factor that inhibits PLC gene expression.
Materials and Methods
Samples
We used two pear varieties of ‘Housui’ and ‘Imamuraaki’ to investigate GSI in P. pyrifolia. The S-genotypes of ‘Housui’ and ‘Imamuraaki’ are S3S5 and S1S6, respectively (Ishimizu et al., 1998). The two varieties are completely self-incompatible after self-pollination, but show strong compatibility when cross-pollinated.
Pollen Collection, S-RNase Extraction, and Isolation of Protoplasts from Pollen Tubes
We collected ‘Housui’ (P. pyrifolia Nakai) pollen from blooming flowers in a fruit experimental garden in Jiangsu, China. The collected pollen was stored at -20°C. S-RNase was extracted from ‘Housui’ and ‘Imamuraaki’ styles according to previously described methods (Hiratsuka et al., 2001). The final S-RNase concentration was 1.0 μg μl-1 with 0.15 units of activity in the medium. The medium composed of 0.55 mM Ca(NO3)2, 1.6 mM H3BO3, 1.6 mM MgSO4, 1 mM KNO3, 440 mM sucrose and 5 mM 5, 2-(N-Morpholino) ethane sulfonic acid hydrate (MES), and pH was adjusted to pH 6.0 with Tris. Protoplasts were isolated from pollen tube apices according to our established method (Qu et al., 2007).
Pollen Tube Staining with fluo-4/AM
Pollen tubes were cultured for 2.0 h, and then stained with 1 μM fluo-4/AM dye (Dôjindo Laboratories) for 15 min at room temperature in the dark according to our methods (Qu et al., 2016a). We used a TCS SP-2 MP AOBS Confocal System (Leica) to observe the tubes. The fluorescence intensity was measured with Image Pro Plus software (Qu et al., 2016a).
Electrophysiology and Data Analysis
Pipettes were pulled from borosilicate glass blanks and coated with Sylgard (184 Silicone Elastomer kit; Dow Corning). Pipettes with solution had resistance values ranging from 15 to 35 MΩ in 10 mM CaCl2. Whole-cell currents across the plasma membrane of protoplasts isolated from pollen tubes were measured with an Axon 200B amplifier (Axon Instruments). Whole-cell preparations were obtained by forming a giga seal in the cell-attached mode, and the membrane was ruptured with a short burst of extra suction. A substantial increase in capacitance indicated that the whole-cell configuration was achieved, and the series resistance and capacitance were adjusted accordingly. Voltage-pulses of 2.5 s were used to elicit voltage-dependent currents. Data were sampled at 2 kHz and filtered at 0.5 kHz. Records were stored and analyzed by using pClamp 9.0 (Axon Instruments). The junction potential was corrected according to a published report (Amtmann and Sanders, 1997). All experiments were conducted at room temperature (20–22°C). The current-voltage curves were constructed by using the current values measured at the final voltage.
Experimental Solutions
The basal solution consisted of 10 mM CaCl2, 0.2 mM glucose, and 0.05 mM MES. The bath solutions were adjusted to 800 mOsM and pH 5.8 with mannitol and Tris. The internal basal pipette solution contained 1 mM MgCl2, 0.1 mM CaCl2, 4 mM Ca(OH)2, 10 mM EGTA, 2 mM MgATP, 10 mM HEPES, 100 mM CsCl, and 0.1 mM GTP, with pH 7.3. The solution was adjusted to 1,100 mOsM with mannitol and Tris. The free Ca2+ concentration of the pipette solution was approximately 10 nM. Details regarding the bath and pipette solutions are provided in the figure legends.
Preparation of Plasma Membranes
Mature pollen grains (0.1 g, approximately 1 × 107 cells ml-1) were suspended in 10 ml medium and cultured for 3 h at 25°C. Subsequently, pollen tubes were washed with 0.8 M sorbitol and quickly frozen with liquid nitrogen. The tubes were then homogenized in 20 mM Hepes-Tris (pH 7.8), 1 mM EGTA (pH 8.0), 0.25 mM sorbitol, 0.5% (w/v) BSA, 1 mM PMSF, 5 mM DTT, and 0.7% PVPP for membrane isolation. Tissue debris was removed by centrifugation at 10,000 × g for 20 min at 4°C. The supernatant was further centrifuged at 100,000 × g for 60 min at 4°C for microsomal membrane preparation. Highly purified plasma membranes were prepared from the pellet through two-phase partitioning (Tate and Gruner, 1989). All steps were performed at 4°C. Protein concentrations were measured by using a bicinchoninic acid protein assay reagent with BSA as a standard (Smith et al., 1985). The cell membrane extracted from the experimental procedure can be used to evaluate the activity of phospholipase C (Liu et al., 2006).
PI–PLC Activity Assay
Phospholipase C activity was assayed according to a published method (Helling et al., 2006). The standard reaction mixture contained 50 mM Tris-maleate (pH 6.0), 0.8 mM Ca2+/EGTA, 0.08% (w/v) sodium deoxycholate, 10 μl self-S-RNase (‘Housui’) or non-self-S-RNase (‘Imamuraaki’), 0.74 kBq [3H]PIP2 (251.6 GBq mmol-1, head group labeled; Perkin-Elmer), and 10 μg membrane protein in a final volume of 50 μl. The reaction was initiated by adding micellar substrate solutions, and stopped after 20 min at 25°C by adding 1 ml chloroform: methanol (2: 1, v/v) and 250 μl 1 M HC1. After vortexing and a brief centrifugation, the radioactive reaction products were recovered in the upper phase, and quantified with a liquid scintillation counter (LS 6500 Multi-Purpose Scintillation Counter; Beckman Coulter). Background radioactivity detected in control samples incubated without protein was subtracted from all data.
Yeast Two-Hybrid Interaction Assays
The diploid P. x bretschneideri Rehd. is the first pear species with its genome comprehensively sequenced (Wu et al., 2012b). We compared the sequence of our cloned PLC cDNA fragments isolated from pollen tubes with that of the ‘DangshanSuli’ (P. x bretshneideri Rehd.) genome to determine the full-length PLC sequence. The full-length cDNA sequences of S7-RNase and S34-RNase (DQ414813) were based on the P. x bretschneideri Rehd. genome and GenBank sequences, respectively. The S7-RNase and S34-RNase cDNA sequences were separately cloned into the pGADT7 vector (Clontech). The PLC cDNA sequence was cloned into the pGBKT7 vector (Clontech). Comparing with the PLC gene mutation of the ‘Jinzhuili,’ we inserted 78 nucleotide residues, which encoded 26 amino acid residues, into the same mutant position in the ‘Dangshansuli’ PLC gene. The mutation of the ‘Dangshansuli’ PLC gene was also cloned into the pGBKT7 vector (Clontech). Each bait/prey pair was introduced to the AH109 yeast strain (Clontech). As a control for auto-activation false-positives, each bait was also co-transformed into the yeast strain with an empty AD vector, and each prey was co-transformed with an empty BD vector. The bait/prey pair colonies that grew on all selective media (Trp-Leu and Trp-Leu-Ade-His) were considered positive for interaction.
Microscopy and Live-Cell Imaging
For Tobacco leaves expressing PLC-GFP and S-RNase-RFP, fluorescence localization and image processing were performed as described in Francinallami et al. (2011) and Zhong et al. (2012).
Preparation of RNA Samples
Total RNA was extracted from ‘Housui’ and ‘Imamuraaki’ pollen grains as previously described (Tao et al., 1999), and treated with DNase I (Invitrogen). The integrity of the RNA was checked by electrophoresis, and concentrations were determined by spectrophotometry. Total RNA (1 μg) was used for first-strand cDNA synthesis with the Micro-FastTrack 2.0 mRNA Isolation Kit (Invitrogen) by following the manufacturer’s instructions. The cDNA then served as templates for subsequent polymerase chain reaction (PCR) amplifications.
Reverse Transcription-PCR Analysis of PI-PLC in ‘Housui’ and ‘Imamuraaki’ Cultivars
We reverse-transcribed mRNA to synthesize first-stand cDNA by using the adapter primer NotI-(dT)18. The 3′-region of cDNA encoding PI-PLC was amplified with NotI-(dT)18 and a forward primer (5′-CAGCTKAGYAGYGAYTGYAGTG-3′) designed based on the X-domain region. The 5′-ends of the cDNA were amplified by using the 5′-RACE System 2.0 (Invitrogen) with reverse primers (5′-TGAACATWCCDTGCATRAACCAAAG-3′ and 5′-TNAGDATYTTCCGCTGAGTYAACCTG-3′) specific for PI-PLC. The partial-length cDNA encoding PI-PLC was obtained through 3′ and 5′ RACE cloning.
High Throughput Analysis of Differential Gene Expression
‘Jinzhuili’ is a spontaneous self-compatible mutant of ‘Yali’ (P. x bretschneideri Rehd., S21S34). ‘Yali’ is a major Chinese pear cultivar, which displays a typical S-RNase-based GSI (Wu et al., 2013). We collected the styles from 100 flowers of each cultivar 0.5 h after self-pollination. We used non-pollinated ‘Jinzhuili’ styles as controls. Total RNA was extracted from each sample with Takara reagent (Takara Bio) by following the manufacturer’s protocol. RNA quantity, purity, and quality were analyzed with a spectrophotometer (SMA4000 UV-VIS spectrophotometer; Varian, Inc.). Double-stranded cDNA libraries were then constructed by using the TruSeq RNA Sample Preparation kit v2 (Illumina). Each sample pool was sequenced on the MiSeq (Illumina) platform with sequence runs of 2 × 250 paired-end reads at The Central Facility of the Institute for Molecular Biology and Biotechnology, McMaster University, Canada.
The P. pyrifolia reference genome and annotation data were obtained from GenBank2. The transcriptome genome coverage was deduced based on our transcriptome data (4.0 Gb) and the P. x bretschneideri Rehd. genome data (432 Mb) from the Centre of Pear Engineering Technology Research, State Key Laboratory of Crop Genetics and Germplasm Enhancement (Wu et al., 2012b).
In our project, we sequenced three samples on the Illumina HiSeq platform. On average, we generated about 4.46 Gb bases from each sample. After mapping sequenced reads to the reference genome and reconstructing transcripts, we obtained 7,251 novel transcripts from all samples. Among them, 4,997 were previously unknown splicing variants from known genes, 999 were novel coding transcripts without any known features, and the remaining 1,255 were long non-coding RNA. The sequencing reads containing low quality, adaptor polluted and high content of unknown bases (N) were removed before downstream analyses. After filtering, the read-quality metrics were generated (Supplementary Table S3). The filtered reads were mapped to the reference genome by using HISAT (Kim et al., 2015). On average, 59.79% of the reads were mapped, and the uniformity of the mapping results between samples suggested that the results of these analyses were comparable. The mapping details are summarized in Supplementary Table S4.
GO Analysis
Gene ontology (GO) analysis was performed to determine the biological implications of the expression of unique genes in significant or representative profiles of genes that were differentially expressed. We downloaded GO annotations from NCBI3, UniProt4, and the Gene Ontology Consortium5. Fisher’s exact test was conducted to identify significant GO categories, and false discovery rate (FDR) was used to correct the p-values (Dupuy et al., 2007).
Pathway Analysis
Pathway analysis was used to determine the significant pathway(s) of the DEGs based on the KEGG database. Fisher’s exact test was conducted to select significant pathways, and the threshold of significance was defined according to the p-value and FDR (Kanehisa et al., 2004; Yi et al., 2006; Draghici et al., 2007).
Quantitative Real-Time RT-PCR to Determine Gene Expression in Style
To estimate mRNA expression levels of PLC genes in style after compatible and incompatible pollination, qRT-PCR analysis was performed. After 0.5 h of artificial pollination, we collected the styles and immediately placed them into liquid nitrogen for preservation. Total RNA was extracted with TRIzol reagent (Invitrogen), and the first-strand cDNA was synthesized from 2 μg of total RNA with an Omniscript RT kit (Qiagen). The primers used for RT-PCR are shown in Supplementary Table S2. Actin gene was used as the reference gene. All quantitative RT–PCR experiments were performed in biological triplicate and technical duplicate. Relative quantification was normalized to the housekeeping control gene ubiquitin, and FC (fold-change) in gene expression was calculated with the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Proteome Analysis
The three samples used in high-throughput sequencing were used to perform the proteome analysis. The total protein of each corresponding group was blocked, digested and labeled according to the iTRAQ protocol (BGI, Co., Ltd, China). The labeling was as follows: ‘Yali’ 0.5 h (113), ‘Jinzhuili’ Control (114) and ‘Jinzhuili’ 0.5 h (115). All LC-ESI-MSMS analysis was based on the Triple TOF 5600 system (SCIEX, Framingham, MA, United States) fitted with a Nanospray III source (SCIEX, Framingham, MA, United States), and a pulled quartz tip as the emitter (New Objectives, Woburn, MA, United States). The IQuant software was used to conduct protein identification, tag impurity correction, data normalization, missing value imputation, protein ratio calculation, statistical analysis, and results presentation for iTRAQ (Wen et al., 2014). The three samples went through a batch of machine tests. No difference in the number of proteins was identified between samples, and no abnormal sample was found. The labeling efficiency and label quality after mass spectrometry were subject to quality-control tests. The ratios of the medians of tag intensities were close to 1. The iTRAQ labeling efficiency was 96.44%. Protein identification was based on unique peptide segments with its threshold set at a minimum of one unique peptide segment. The peptide segments retained for quantification were the segments that uniquely identified individual proteins. The search was performed against P. x bretschneideri Rhd. genome data.
Author Contributions
HQ wrote paper and designed and analyzed the experiments. YG performed the experiments. YW designed the experiment and collected the pollen and done the physiology experiments and revised the manuscript. SZ designed the part of experiments. All of the authors analyzed the results and approved the final version of the manuscript.
Funding
The work was supported by Natural Science Foundation of Shandong (ZR2015CM033), experts working in the fruit system of Shandong Province and Taishan Scholars Research Group, and National Pear Industry System.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We would like to thank all scientists for helping us to improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01164/full#supplementary-material
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/gorf/gorf.html
- ^ http://www.ncbi.nlm.nih.gov/nuccore/
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://www.uniprot.org/
- ^ http://www.geneontology.org/
References
Akagi, T., Henry, I. M., Morimoto, T., and Tao, R. (2016). Insights into the Prunus-specific S-RNase-based self-incompatibility system from a genome-wide analysis of the evolutionary radiation of S locus-related f-box genes. Plant Cell Physiol. 57, 1281–1294. doi: 10.1093/pcp/pcw077
Amtmann, A., and Sanders, D. (1997). A unified procedure for the correction of liquid junction potentials in patch clamp experiments on endo-and plasma membranes. J. Exp. Bot. 48, 361–364. doi: 10.1093/jxb/48.Special_Issue.361
Ashkani, J., and Rees, D. J. G. (2016). A comprehensive study of molecular evolution at the self-incompatibility locus of Rosaceae. J. Mol. Evol. 82, 128–145. doi: 10.1007/s00239-015-9726-4
Blakeslee, J. J., Bandyopadhyay, A., Peer, W. A., Makam, S. N., and Murphy, A. S. (2004). Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 134, 28–31. doi: 10.1104/pp.103.031690
Campanoni, P., and Blatt, M. R. (2007). Membrane trafficking and polar growth in root hairs and pollen tubes. J. Exp. Bot. 58, 65–74. doi: 10.1093/jxb/erl059
Cruz-Garcia, F., Hancock, C. N., and McClure, B. (2003). S-RNase complexes and pollen rejection. J. Exp. Bot. 54, 123–130. doi: 10.1093/jxb/erg045
De Franceschi, P., Dondini, L., and Sanzol, J. (2012). Molecular bases and evolutionary dynamics of self-incompatibility in the Pyrinae (Rosaceae). J. Exp. Bot. 63, 4015–4032. doi: 10.1093/jxb/ers108
Dellis, O., Dedos, S. G., Tovey, S. C., Taufiq-Ur-Rahman, Dubel, S. J., and Taylor, C. W. (2006). Ca2+ entry through plasma membrane IP3 receptors. Science 313, 229–233. doi: 10.1126/science.1125203
Dowd, P. E., Coursol, S., Skirpan, A. L., Kao, T.-H., and Gilroy, S. (2006). Petunia phospholipase C1 is involved in pollen tube growth. Plant Cell 18, 1438–1453. doi: 10.1105/tpc.106.041582
Draghici, S., Khatri, P., Tarca, A. L., Amin, K., Done, A., Voichita, C., et al. (2007). A systems biology approach for pathway level analysis. Genome Res. 17, 1537–1545. doi: 10.1101/gr.6202607
Dresselhaus, T., and Franklin-Tong, N. (2013). Male-female cross-talk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6, 1018–1036. doi: 10.1093/mp/sst061
Dupuy, D., Bertin, N., Hidalgo, C., Venkatesan, K., Tu, D., Lee, D., et al. (2007). Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 25, 663–668. doi: 10.1038/nbt1305
Evdonin, A. L., Guzhova, I. V., Margulis, B. A., and Medvedeva, N. D. (2004). Phospholipase C inhibitor, u73122, stimulates release of hsp-70 stress protein from A431 human carcinoma cells. Cancer Cell Int. 4:2. doi: 10.1186/1475-2867-4-2
Francinallami, M., Saumonneau, A., Lavenant, L., Bouder, A., Sparkes, I., Hawes, C., et al. (2011). Dynamic trafficking of wheat γ-gliadin and of its structural domains in tobacco cells, studied with fluorescent protein fusions. J. Exp. Bot. 62, 4507–4520. doi: 10.1093/jxb/err159
Franklin-Tong, V. E., Drobak, B. K., Allan, A. C., Watkins, P. A., and Trewavas, A. J. (1996). Growth of pollen tubes of Papaver rhoeas is regulated by a slow-moving calcium wave propagated by inositol 1,4, 5-trisphosphate. Plant Cell 8, 1305–1321. doi: 10.1105/tpc.8.8.1305
Franklin-Tong, V. E., and Franklin, F. C. H. (2003). Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. Trends Plant Sci. 8, 598–605. doi: 10.1016/j.tplants.2003.10.008
Franklin-Tong, V. E., Hackett, G., and Hepler, P. K. (1997). Ratio-imaging of Ca2+i in the self-incompatibility response in pollen tubes of Papaver rhoeas. Plant J. 12, 1375–1386. doi: 10.1046/j.1365-313x.1997.12061375.x
Franklin-Tong, V. E., Holdaway-Clarke, T. L., Straatman, K. R., Kunkel, J. G., and Hepler, P. K. (2002). Involvement of extracellular calcium influx in the self-incompatibility response of Papaver rhoeas. Plant J. 29, 333–345. doi: 10.1046/j.1365-313X.2002.01219.x
Fujii, S., Kubo, K., and Takayama, S. (2016). Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants 2:16130. doi: 10.1038/nplants.2016.130
Gray, J. E., Mcclure, B. A., Bonig, I., Anderson, M. A., and Clarke, A. E. (1991). Action of the style product of the self-incompatibility gene of Nicotiana alata (S-RNase) on in vitro-grown pollen tubes. Plant Cell 3, 271–283. doi: 10.1105/tpc.3.3.271
Hegedűs, A., Lénárt, J., and Halász, J. (2012). Sexual incompatibility in Rosaceae fruit tree species: molecular interactions and evolutionary dynamics. Biol. Plant. 56, 201–209. doi: 10.1007/s10535-012-0077-3
Heilmann, I., and Ischebeck, T. (2016). Male functions and malfunctions: the impact of phosphoinositides on pollen development and pollen tube growth. Plant Reprod. 29, 3–20. doi: 10.1007/s00497-015-0270-6
Helling, D., Possart, A., Cottier, S., Klahre, U., and Kost, B. (2006). Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 18, 3519–3534. doi: 10.1105/tpc.106.047373
Hepler, P. K. (2015). The pollen tube clear zone: clues to the mechanism of polarized growth. J. Integr. Plant Biol. 57, 79–92. doi: 10.1111/jipb.12315
Herrero, M., and Dickinson, H. (1980). Pollen tube growth following compatible and incompatible intraspecific pollinations in Petunia hybrida. Planta 148, 217–221. doi: 10.1007/BF00380030
Hiratsuka, S., Zhang, S.-L., Nakagawa, E., and Kawai, Y. (2001). Selective inhibition of the growth of incompatible pollen tubes by S-protein in the Japanese pear. Sex. Plant Reprod. 13, 209–215. doi: 10.1007/s004970000058
Hodgkin, T., Lyon, G. D., and Dickinson, H. G. (1988). Recognition in flowering plants: a comparison of the brassica self-incompatibility system and plant pathogen interactions. New Phytol. 110, 557–569. doi: 10.1111/j.1469-8137.1988.tb00296.x
Ishimizu, T., Norioka, S., Nakanishi, T., and Sakiyama, F. (1998). S-genotype of Japanese pear ’Hosui’. J. Jpn. Soc. Hortic. Sci. 67, 35–38. doi: 10.2503/jjshs.67.35
Kakui, H., Kato, M., Ushijima, K., Kitaguchi, M., Kato, S., and Sassa, H. (2011). Sequence divergence and loss-of-function phenotypes of S locus F-box brothers genes are consistent with non-self recognition by multiple pollen determinants in self-incompatibility of Japanese pear (Pyrus pyrifolia). Plant J. 68, 1028–1038. doi: 10.1111/j.1365-313X.2011.04752.x
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280. doi: 10.1093/nar/gkh063
Kao, T. H., and Huang, S. (1994). Gametophytic self-incompatibility: a mechanism for self/nonself discrimination during sexual reproduction. Plant Physiol. 105, 461–466. doi: 10.1104/pp.105.2.461
Kao, T. H., and McCubbin, A. G. (1996). How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proc. Natl. Acad. Sci. U.S.A. 93, 12059–12065. doi: 10.1073/pnas.93.22.12059
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., et al. (1999). Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145, 317–330. doi: 10.1083/jcb.145.2.317
Kovaleva, L., and Zakharova, E. (2003). Hormonal status of the pollen-pistil system at the progamic phase of fertilization after compatible and incompatible pollination in Petunia hybrida L. Sex. Plant Reprod. 16, 191–196. doi: 10.1007/s00497-003-0189-1
Kovaleva, L. V., Zakharova, E. V., Skorobogatova, I. V., and Karsunkina, N. P. (2002). Gametophyte sporophyte interactions in the pollen pistil system: 3. hormonal status at the progamic phase of fertilization. Russ. J. Plant Physiol. 49, 492–495.
Krinke, O., Novotná, Z., Valentová, O., and Martinec, J. (2007). Inositol trisphosphate receptor in higher plants: is it real? J. Exp. Bot. 58, 361–376. doi: 10.1093/jxb/erl220
Levava, R., Patricia, S., Menashe, B. E., Betty, S., and Oded, S. (2006). ACTIBIND, an actin-binding fungal T2-RNase with antiangiogenic and anticarcinogenic characteristics. Cancer 106, 2295–2308. doi: 10.1002/cncr.21878
Li, J., Zhang, Y., Song, Y., Zhang, H., Fan, J., Li, Q., et al. (2017). Electrostatic potentials of the S-locus f-box proteins contribute to the pollen S specificity in self-incompatibility in Petunia hybrida. Plant J. 89, 45–57. doi: 10.1111/tpj.13318
Liu, H. T., Huang, W. D., Pan, Q. H., Weng, F. H., Zhan, J. C., Liu, Y., et al. (2006). Contributions of PIP(2)-specific-phospholipase C and free salicylic acid to heat acclimation-induced thermotolerance in pea leaves. J. Plant Physiol. 163, 405–416. doi: 10.1016/j.jplph.2005.04.027
Liu, Z. Q., Xu, G. H., and Zhang, S. L. (2007). Pyrus pyrifolia stylar S-RNase induces alterations in the actin cytoskeleton in self-pollen and tubes in vitro. Protoplasma 232, 61–67. doi: 10.1007/s00709-007-0269-4
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luhtala, N., and Parker, R. (2010). T2 Family ribonucleases: ancient enzymes with diverse roles. Trends Biochem. Sci. 35, 253–259. doi: 10.1016/j.tibs.2010.02.002
Lush, W., and Clarke, A. E. (1997). Observations of pollen tube growth in Nicotiana alata and their implications for the mechanism of self-incompatibility. Sex. Plant Reprod. 10, 27–35. doi: 10.1007/s004970050064
Luu, D. T., Qin, X., Morse, D., and Cappadocia, M. (2000). S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407, 649–651. doi: 10.1038/35036623
Mak, D. O., McBride, S., Raghuram, V., Yue, Y., Joseph, S. K., and Foskett, J. K. (2000). Single-channel properties in endoplasmic reticulum membrane of recombinant type 3 inositol trisphosphate receptor. J. Gen. Physiol. 115, 241–256. doi: 10.1085/jgp.115.3.241
Malhó, R., Read, N. D., Pais, M. S., and Trewavas, A. J. (1994). Role of cytosolic free calcium in the reorientation of pollen tube growth. Plant J. 5, 331–341. doi: 10.1111/j.1365-313X.1994.00331.x
McClure, B. (2006). New views of S-RNase-based self-incompatibility. Curr. Opin. Plant Biol. 9, 639–646. doi: 10.1016/j.pbi.2006.09.004
Meijer, H. J., and Munnik, T. (2003). Phospholipid-based signaling in plants. Annu. Rev. Plant Biol. 54, 265–306. doi: 10.1146/annurev.arplant.54.031902.134748
Meng, D., Gu, Z., Li, W., Wang, A., Yuan, H., Yang, Q., et al. (2014). Apple MdABCF assists in the transportation of S-RNase into pollen tubes. Plant J. Cell Mol. Biol. 78, 990–1002. doi: 10.1111/tpj.12524
Messerli, M. A., Danuser, G., and Robinson, K. R. (1999). Pulsatile influxes of H+, K+ and Ca2+ lag growth pulses of Lilium longiflorum pollen tubes. J. Cell Sci. 112, 1497–1509.
Mogami, H., Lloyd Mills, C., and Gallacher, D. (1997). Phospholipase C inhibitor, U73122, releases intracellular Ca2+, potentiates Ins(1,4,5)P3-mediated Ca2+ release and directly activates ion channels in mouse pancreatic acinar cells. Biochem. J. 324, 645–651. doi: 10.1042/bj3240645
Monteiro, D., Liu, Q., Lisboa, S., Scherer, G., Quader, H., and Malhó, R. (2005). Phosphoinositides and phosphatidic acid regulate pollen tube growth and reorientation through modulation of [Ca2+]c and membrane secretion. J. Exp. Bot. 56, 1665–1674. doi: 10.1093/jxb/eri163
Munnik, T., Irvine, R., and Musgrave, A. (1998). Phospholipid signalling in plants. Biochim. Biophys. Acta 1389, 222–272. doi: 10.1016/S0005-2760(97)00158-6
Pierson, E., Miller, D., Callaham, D., Van Aken, J., Hackett, G., and Hepler, P. (1996). Tip-localized calcium entry fluctuates during pollen tube growth. Dev. Biol. 174, 160–173. doi: 10.1006/dbio.1996.0060
Qin, X., Liu, B., Soulard, J., Morse, D., and Cappadocia, M. (2006). Style-by-style analysis of two sporadic self-compatible Solanum chacoense lines supports a primary role for S-RNases in determining pollen rejection thresholds. J. Exp. Bot. 57, 2001–2013. doi: 10.1093/jxb/erj147
Qu, H., Shang, Z., Zhang, S., Liu, L., and Wu, J. (2007). Identification of hyperpolarization-activated calcium channels in apical pollen tubes of Pyrus pyrifolia. New Phytol. 174, 524–536. doi: 10.1111/j.1469-8137.2007.02069.x
Qu, H., Xing, W., Wu, F., and Wang, Y. (2016a). Rapid and inexpensive method of loading fluorescent dye into pollen tubes and root hairs. PLoS ONE 11:e0152320. doi: 10.1371/journal.pone.0152320
Qu, H., Zhang, Z., Wu, F., and Wang, Y. (2016b). The role of Ca2+ and Ca2+ channels in the gametophytic self-incompatibility of Pyrus pyrifolia. Cell Calcium 60, 299–308. doi: 10.1016/j.ceca.2016.06.006
Quinet, M., Kelecom, S., Raspé, O., and Jacquemart, A. L. (2014). S-genotype characterization of 13 North Western European pear (Pyrus communis) cultivars. Sci. Hortic. 165, 1–4. doi: 10.1016/j.scienta.2013.10.023
Roldán, J. A., Rojas, H. J., and Goldraij, A. (2012). Disorganization of F-actin cytoskeleton precedes vacuolar disruption in pollen tubes during the in vivo self-incompatibility response in Nicotiana alata. Ann. Bot. 110, 787–795. doi: 10.1093/aob/mcs153
Romain, S., and Dirk, A. (2014). Cytological evidence for gametophytic self-incompatibility in the genus Veronica. Turk. J. Bot. 38, 197–201. doi: 10.3906/bot-1302-2
Silva, N. F., and Goring, D. R. (2001). Mechanisms of self-incompatibility in flowering plants. Cell. Mol. Life Sci. 58, 1988–2007. doi: 10.1007/PL00000832
Smith, P., Krohn, R. I., Hermanson, G., Mallia, A., Gartner, F., Provenzano, M., et al. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85. doi: 10.1016/0003-2697(85)90442-7
Steinhorst, L., and Kudla, J. (2013). Calcium - a central regulator of pollen germination and tube growth. Biochim. Biophys. Acta 1833, 1573–1581. doi: 10.1016/j.bbamcr.2012.10.009
Takayama, S., and Isogai, A. (2005). Self-incompatibility in plants. Annu. Rev. Plant Biol. 56, 467–489. doi: 10.1146/annurev.arplant.56.032604.144249
Tao, R., Yamane, H., Sugiura, A., Murayama, H., Sassa, H., and Mori, H. (1999). Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J. Am. Soc. Hortic. Sci. 124, 224–233.
Tate, M. W., and Gruner, S. M. (1989). Temperature dependence of the structural dimensions of the inverted hexagonal (HII) phase of phosphatidylethanolamine-containing membranes. Biochemistry 28, 4245–4253. doi: 10.1021/bi00436a019
Vaca, L., and Kunze, D. L. (1995). IP3-activated Ca2+ channels in the plasma membrane of cultured vascular endothelial cells. Am. J. Physiol. 269, 733–738.
Vossen, J. H., Fradin, E. F., Van, D. B., Grardy, C. M., Ekengren, S. K., Meijer, H. J. G., et al. (2010). Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J. Cell Mol. Biol. 62, 224–239. doi: 10.1111/j.1365-313X.2010.04136.x
Wang, C. L., Wu, J., Xu, G.-H., Gao, Y.-B., Chen, G., Wu, J. Y., et al. (2010). S-RNase disrupts tip-localized reactive oxygen species and induces nuclear DNA degradation in incompatible pollen tubes of Pyrus pyrifolia. J. Cell Sci. 123, 4301–4309. doi: 10.1242/jcs.075077
Wen, B., Zhou, R., Feng, Q., Wang, Q., Wang, J., and Liu, S. (2014). IQuant: an automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 14, 2280–2285. doi: 10.1002/pmic.201300361
Wheeler, M. J., Graaf, B. H. J. D., Hadjiosif, N., Perry, R. M., Poulter, N. S., Osman, K., et al. (2009). Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature 459, 992–995. doi: 10.1038/nature08027
Wilkins, K. A., Poulter, N. S., and Franklin-Tong, V. E. (2014). Taking one for the team: self-recognition and cell suicide in pollen. J. Exp. Bot. 65, 1331–1342. doi: 10.1093/jxb/ert468
Wu, J., Gu, C., Khan, M. A., Wu, J., Gao, Y., Wang, C., et al. (2012a). Molecular determinants and mechanisms of gametophytic self-incompatibility in fruit trees of Rosaceae. Crit. Rev. Plant Sci. 32, 53–68. doi: 10.1270/jsbbs.66.116
Wu, J., Li, M., and Li, T. (2013). Genetic features of the spontaneous self-compatible mutant, ‘Jin Zhui’ (Pyrus bretschneideri Rehd.). PLoS ONE 8:e76509. doi: 10.1371/journal.pone.0076509
Wu, J., Wang, Z., Shi, Z., Zhang, S., Ming, R., Zhu, S., et al. (2012b). The genome of pear (Pyrus bretschneideri Rehd.). Genome Res. 23, 396–408. doi: 10.1101/gr.144311.112
Wu, J. Z., Lin, Y., Zhang, X. L., Pang, D. W., and Zhao, J. (2008). IAA stimulates pollen tube growth and mediates the modification of its wall composition and structure in Torenia fournieri. J. Exp. Bot. 59, 2529–2543. doi: 10.1093/jxb/ern119
Xue, Y., Liu, W., Fan, J., Li, J., Song, Y., Li, Q., et al. (2014). SCFSLF-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petunia hybrida. Front. Genet. 5, 558. doi: 10.3389/fgene.2014.00228
Yang, Q., Zhang, D., Li, Q., Cheng, Z., and Xue, Y. (2007). Heterochromatic and genetic features are consistent with recombination suppression of the self-incompatibility locus in Antirrhinum. Plant J. 51, 140–151. doi: 10.1111/j.1365-313X.2007.03127.x
Yi, M., Horton, J. D., Cohen, J. C., Hobbs, H. H., and Stephens, R. M. (2006). WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics 7:30. doi: 10.1186/1471-2105-7-30
Zhang, S. L., and Hiratsuka, S. (1999). Variations in s-protein levels in styles of Japanese pears and the expression of self-incompatibility. J. Jpn. Soc. Hortic. Sci. 68, 911–918. doi: 10.2503/jjshs.68.911
Zhang, S. L., and Hiratsuka, S. (2005). Analyses of pollen-tube growth and biological action of S-RNase in the style of self-compatible Japanese pear. Sci. Hortic. 104, 169–178. doi: 10.1016/j.scienta.2004.08.017
Zhang, Y. J., Zhao, Z.-H., and Xue, Y.-B. (2009). Roles of proteolysis in plant self-incompatibility. Annu. Rev. Plant Biol. 60, 21–42. doi: 10.1146/annurev.arplant.043008.092108
Zhong, G., Wei, W., Guan, Q., Ma, Z., Wei, H., Xu, X., et al. (2012). Phosphoribosyl pyrophosphate synthetase, as a suppressor of the sepH mutation in Aspergillus nidulans, is required for the proper timing of septation. Mol. Microbiol. 86, 894–907. doi: 10.1111/mmi.12026
Keywords: phospholipase C, S-RNase, gametophytic self-incompatibility, IP3, pollen tube
Citation: Qu H, Guan Y, Wang Y and Zhang S (2017) PLC-Mediated Signaling Pathway in Pollen Tubes Regulates the Gametophytic Self-incompatibility of Pyrus Species. Front. Plant Sci. 8:1164. doi: 10.3389/fpls.2017.01164
Received: 19 November 2016; Accepted: 16 June 2017;
Published: 06 July 2017.
Edited by:
Patrick H. Masson, University of Wisconsin-Madison, United StatesReviewed by:
Wei-Hua Tang, Shanghai Institute of Plant Physiology and Ecology (CAS), ChinaImara Yasmin Perera, North Carolina State University, United States
Copyright © 2017 Qu, Guan, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongzhang Wang, wangyzh304@163.com
 Haiyong Qu
Haiyong Qu Yaqin Guan1
Yaqin Guan1