- 1Department of Plant Sciences, University of Hyderabad, Hyderabad, India
- 2Department of Biotechnology, Indian Institute of Rice Research, Hyderabad, India
Ribosomal proteins (RPs) are indispensable in ribosome biogenesis and protein synthesis, and play a crucial role in diverse developmental processes. Our previous studies on Ribosomal Protein Large subunit (RPL) genes provided insights into their stress responsive roles in rice. In the present study, we have explored the developmental and stress regulated expression patterns of Ribosomal Protein Small (RPS) subunit genes for their differential expression in a spatiotemporal and stress dependent manner. We have also performed an in silico analysis of gene structure, cis-elements in upstream regulatory regions, protein properties and phylogeny. Expression studies of the 34 RPS genes in 13 different tissues of rice covering major growth and developmental stages revealed that their expression was substantially elevated, mostly in shoots and leaves indicating their possible involvement in the development of vegetative organs. The majority of the RPS genes have manifested significant expression under all abiotic stress treatments with ABA, PEG, NaCl, and H2O2. Infection with important rice pathogens, Xanthomonas oryzae pv. oryzae (Xoo) and Rhizoctonia solani also induced the up-regulation of several of the RPS genes. RPS4, 13a, 18a, and 4a have shown higher transcript levels under all the abiotic stresses, whereas, RPS4 is up-regulated in both the biotic stress treatments. The information obtained from the present investigation would be useful in appreciating the possible stress-regulatory attributes of the genes coding for rice ribosomal small subunit proteins apart from their functions as house-keeping proteins. A detailed functional analysis of independent genes is required to study their roles in stress tolerance and generating stress- tolerant crops.
Introduction
Ribosomal proteins (RPs) constitute the protein part of the ribosomes and have a significant role in ribosome biogenesis, protein synthesis, cell growth, development, and apoptosis (Ramakrishnan and White, 1998; Naora and Naora, 1999; Maguire and Zimmermann, 2001; Wang Z. et al., 2013). RPs have long been known primarily for their housekeeping functions. But, there are several reports on their extra-ribosomal functions in animal systems (Warner and McIntosh, 2009). The knockdown of individual RPs leads to P53 accumulation, cell death and certain developmental abnormalities in Zebra fish (Uechi et al., 2006; Danilova et al., 2008). Mutations in certain RPs were associated with Diamond-Blackfan Anemia (DBA) and increased risk of cancer (Gazda et al., 2008). On the contrary, specific RPs have been used as therapeutic targets in cancer treatment. Overexpression of RPS2 has been linked with the survival of prostate tumors, whereas suppression of RPS2 or RPS3a resulted in apoptosis (Wang et al., 2009).
The number of RPs varies within eukaryotes. In yeast 78 RPs (32 of the small subunit and 46 of large subunit) are encoded by 137 genes (Planta, 1997). Two hundred and forty nine genes encode the 80 RPs of Arabidopsis; 32 are small subunit, and 48 are large subunit proteins (Barakat et al., 2001; Wang J. et al., 2013). In rice, 34 large subunit proteins are encoded by 123 genes (Moin et al., 2016b) showing that multiple copies of genes encode an individual ribosomal protein in eukaryotes (McIntosh and Bonham-Smith, 2006). This can be correlated with the requirement of ample supply of RPs during rapid growth phase (Filipowicz and Hohn, 2012). Most of these genes are specifically expressed during a certain developmental stage or in a particular tissue, or they may be functionally redundant. The subsequent lack of sufficient quantity of individual RPs can lead to non-lethal phenotypic abnormalities and aberrant translational efficiency (Hulm et al., 2005; Komili et al., 2007; Degenhardt and Bonham-Smith, 2008).
Several studies have demonstrated important roles of RPs in development and physiology of organisms as their reduced level in the organisms resulted in detrimental effects. Semi-dominant mutation of cytoplasmic ribosomal protein RPL27aC resulted in multiple developmental defects such as changes in leaf patterning, inflorescence, floral meristem function and seed set (Szakonyi and Byrne, 2011). In Arabidopsis, PFL codes for the ribosomal protein S18 and a mutation at PFL locus resulted in developmental abnormalities involving reduced fresh weight, pointed first leaves and stunted growth indicating the importance of the corresponding protein in meristem activity (Van Lijsebettens et al., 1994). RPL18aB is presumed to have a critical role in male gametophyte development and embryo pattern formation in Arabidopsis with the rpl18aB mutants exhibiting defects in pollen growth (Yan et al., 2016). Genes coding for cytoplasmic ribosomal proteins, L10a, L9, and L5 play an important role in determining the adaxial and abaxial fate of leaf during development via their interaction with adaxial HD-ZIPIII gene, REVOLUTA, and abaxial KANADI gene (Pinon et al., 2008). Aberrant expression of ribosomal protein S6 in Arabidopsis leads to a deleterious effect on survival of cells indicating their role in a complex translational regulation. Insertion of ribosomal protein S6 gene in antisense orientation resulted in over-expression of antisense ribosomal protein S6 RNA, which led to a reduced level of rps6 expression. This might be the reason for subdued expression of some other specific proteins, which is manifested in the form of complex phenotypic disorders including multiple leaves and flower formation in close vicinity (Morimoto et al., 2002). In Saccharomyces cerevisiae, RPs have been predicted to participate in several extra-ribosomal functions, which are related to ribosome assembly, replicative life span, DNA repair, adhesive growth and filament formation (Lu et al., 2015).
T-DNA insertional mutagenesis in one of the RPS27 genes (AtRS27A) led to strong growth inhibition and the formation of tumor-like structure in place of auxiliary roots in the presence of methyl methanesulfonate (MMS) in growth medium (Revenkova et al., 1999). In Arabidopsis, delayed cell division and complete developmental arrest was observed in RPS5 hetero- and homozygous mutants, respectively (Weijers et al., 2001). The mutation in a key translation initiation factor, RPL24 in Arabidopsis resulted in perturbed gynoecium development with diminished ovary and lengthened gynophore (Nishimura et al., 2004).
In addition to their crucial roles in developmental processes in plants, several reports also focused on their expression pattern in response to several external stimuli. The expression of several RP genes is also regulated by phytohormones, ABA and cytokinin. ABA negatively regulates the expression of RPS14, RPS16, RPS13a, and RPL30, whereas cytokinin induced the expression of these genes at both the transcript and protein levels (Cherepneva et al., 2003). In rice, RPS4, 7, 8, 9, 10, 19, 26, and RPL2, 5, 18, 44 were among the early responsive genes that were up-regulated under salt stress (Kawasaki et al., 2001). Nutrition deficiency also resulted in the differential expression pattern of RP genes in Arabidopsis. Comparison of differentially expressed RP genes under Fe and Pi deficiency in Arabidopsis roots revealed that both the RPS and Ribosomal Protein Large subunit (RPL) genes were differentially expressed under Fe deficiency, whereas only RPL genes were differentially expressed under Pi deficiency (Wang J. et al., 2013). Induction of soybean RPs, RPS13, RPS6, and RPL37 in response to cold treatment gave a clue to their probable role in secondary signaling under low-temperature conditions (Kim et al., 2004). These reports point toward the differential expression of RP genes in response to stress treatments leading to differential accumulation of RPs in the ribosomal apparatus, which might help ribosome remodeling and selective translation to cope up with nutrition deficiency or other unfavorable conditions (Wang J. et al., 2013).
Screening of a large-pool of activation tagged mutant population of rice for high water-use efficiency revealed the significant upregulation of RPL genes (6 and 23a) in our previous studies (Moin et al., 2016a). This subsequently persuaded us to study the regulatory role of all the RPL genes under different external stimuli (Moin et al., 2016b). This study led another group to identify the insect resistance roles of NlRPL5 in rice as an important responsive gene in resistance to the brown plant hopper, Nilaparvata lugens (Zhu et al., 2017). The objective of the present study is to ascertain whether RPS genes are stress responsive like the RPL genes (Moin et al., 2016b).
Hence, we have attempted to analyse the differential expression patterns of ribosomal protein small subunit genes (RPS genes, one from each family) for their spatiotemporal and stress induced regulation along with a comparative analysis with the large subunit counterparts and in silico studies on their promoter sequences, gene structures, protein properties and phylogeny. These observations are reported here.
Materials and Methods
RPS Gene Sequence Retrieval
A list of all ribosomal protein family (both large and small subunit) was obtained using a keyword search, “ribosomal” in the RGAP-DB1 and Phytozome v112 databases. From the list, a total of 56 ribosomal small subunit genes (40S) were shortlisted by their names starting with the letter “S” as a prefix (S for Small subunit). Nucleotide sequences of all the 56 genes were retrieved from RGAP-DB. These 56 sequences were further confirmed for their ribosomal origin through nucleotide and protein BLAST3 search in the NCBI4 and Hidden Markov Model (HMM) of Pfam5 databases, respectively. The availability of only 34 ribosomal small subunit proteins, which are coded by 56 genes indicated the existence of paralogs of the genes coding for more than one RPS protein. For the gene expression analysis, we selected 34 RPS genes, one from each group representing their corresponding paralogous groups. Identical gene copies were, thus, excluded from further analyses. We have listed the 34 genes along with their paralogs in the Supplementary Table 9. The sequences of RPS genes were retrieved from RGAP-DB (a Nipponbare database). Our analysis of gene expression is performed in an indica rice variety (Samba Mahsuri). Hence, we cross-checked the sequence similarity of the coding as well as the 5′- upstream regions of the genes selected for analysis in the present investigation in both indica and japonica sequence databases by doing a detailed BLAST analysis of the japonica sequences on indica genome databases, OryGeneDB6 and EnsemblPlants7. Further, we have looked for the similarity of rice (40S) RPS genes with RPS genes of Arabidopsis and performed a BLAST search of the Rice sequences in TAIR8 database and listed the similarity percentage in Table 1.

Table 1. Similarity of Ribosomal protein small subunit (RPS) genes in rice with that in Arabidopsis.
Chromosomal Location of RPS Genes
To obtain the genome-wide chromosomal distribution of all RPS genes on 12 chromosomes of rice, we had submitted the locus numbers (obtained from RGAP-DB) of 56 RPS genes as an input to the OrygenesDB. Based on the OrygenesDB outputs, all RPS gene locations were indicated on the chromosome maps, and the number of genes on each chromosome has also been shown. Genes, which were selected for expression analysis, have been shown in bold in Figure 1.
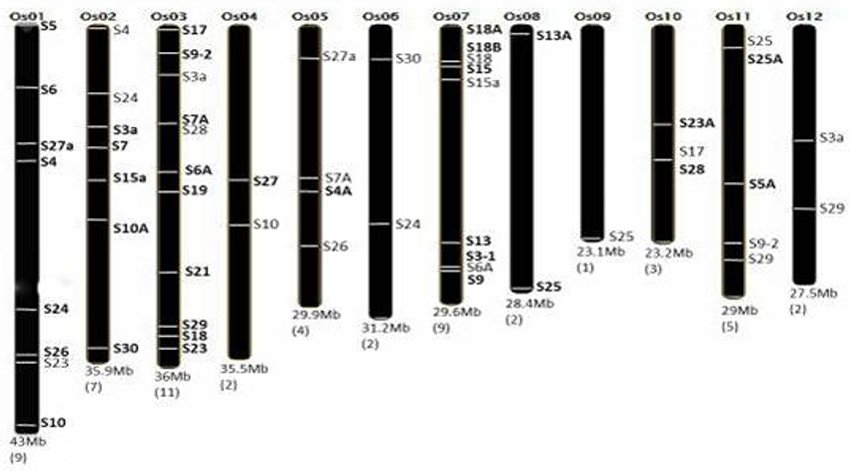
Figure 1. Distribution of RPS genes in rice genome. Number of RPS genes corresponding to the particular chromosome is mentioned in bracket below. The RPS genes and their location on the particular chromosome are marked accordingly. The chromosome number and size are also mentioned. This figure is adapted and modified from Moin et al. (2016b), under CC BY 4.0 license.
Structural Features of RPS Genes
To predict the gene structure features, such as intron and exon number, size, etc., we had submitted the genomic and corresponding cDNA sequences of the selected 34 genes to the Gene Structure Display Server (GSDSv2)9.
Putative Promoter Survey of 34 RPS Genes
Since all the 34 genes exhibited differential expression under multiple stress treatments, we checked for the presence of putative cis-elements in the putative promoter regions of the corresponding genes. To verify this, we have used an in silico approach, in which the nucleotide sequence of about ≤ 1 kb upstream of each of the 34 RPS genes was retrieved from RGAP-DB, and the sequences were cross-checked for their identity/ deviation with the indica genome in OrygeneDB and EnsemblPlants databases. Further, the sequences that exhibited the highest similarity with indica genome in the databases were submitted to PlantCARE (Cis-Acting Regulatory Elements) Database10 (Rombauts et al., 1999; Dhadi et al., 2009; Ding et al., 2011) for identifying the regulatory elements. We have compared the putative promoter regions of some of the significant genes in two other databases, such as PlantPAN 2.011 and the JASPAR12 and found the functional similarity of the elements with respect to PlantCare database confirming the authenticity of our observations.
Ribosomal Protein Structural and Phylogenic Properties
Predicted protein sequences available from the RGAP-DB were used for predicting the protein structure, molecular weight, isoelectric point (pI) prediction in silico by the online tool PSORT13 and ExPASyProtParam14. By submitting the protein sequences to SMART15 (Simple Modular Architecture Research Tool), we have identified various ribosomal domains and motifs. Sequence submission to ExPASyProtParam helped us in obtaining the GRAVY indices of RPSs, which indicated the probable hydrophobicity of the proteins by their negative values for the indices that are relevant for the hydrophobic proteins.
To predict the three-dimensional structure and to check for the presence of ligand binding sites on each RPS, we have used the Phyre216 (Protein Homology/AnalogY Recognition Engine v2; Kelley et al., 2015) and 3DLigandSite17 programs (Wass et al., 2010), respectively. Results from these tools predicted the protein properties like percentage of α-helix, β-sheet, disordered protein fraction and ligands of each protein, ligand binding residues on the proteins and 3D model of the ligand bound protein. Multiple sequence alignment using ClustalW18 also deciphered the phylogenetic relationships among the 34 RPS proteins in MEGA619 (Molecular Evolutionary Genetic Analysis) platform, which constructed an unrooted phylogenetic tree showing evolutionary sequence similarity among the proteins with bootstrap values of 100.
Plant Material, Growth Conditions and Stress Treatments
Oryza sativa L. ssp. indica var. Samba Mahsuri (BPT-5204) seeds were surface sterilized using 70% ethanol for 50 s followed by washing with 4% sodium hypochlorite for 15–20 min and 5–6 washes with sterile double-distilled water. After this, the seeds were dried on sterile blotting paper and were placed on solid MS media at 28 ± 2°C and 16 h light/8 h dark photoperiod cycle (Moin et al., 2016b).
For native expression studies, the surface sterilized seeds were allowed to germinate on solid MS medium. Some of the seeds were soaked for 16 h to collect the embryo and endosperm tissues using a sterile surgical blade. Shoot and root apical meristems of 3 d old seedlings and roots and shoots of 7 d old seedlings were collected for the expression studies. After transferring the seedlings from MS medium to soil in the pots; roots, shoots, leaves, root-shoot transition region, flowers, grains and panicles were collected (Moin et al., 2016b).
For abiotic stress treatments, 7 d old seedlings were exposed to ABA (100 μM), PEG (10%), NaCl (250 μM), and H2O2 (10 μM) by dipping them in the respective solutions. Around five seedlings in replicates were collected after 15 min, 3, 6, 12, 24, 48, and 60 h duration. Roots and shoots were collected separately for RNA isolation and cDNA synthesis. The untreated seedlings in water were used as control samples for normalization in qRT-PCR (Moin et al., 2016b).
To analyse the expression of the RPS genes under biotic stress, rice samples infected with Xanthomonas oryzae pv. oryzae (Xoo) and Rhizoctonia solani were collected 20 and 25 d post infection, respectively. For the biotic stress experiments, 1-month-old rice plants were used for treatments. The untreated samples of the same age were used as corresponding controls. The suspension of Xoo bacterium was applied at the edge of the leaves using a sterile blade (Moin et al., 2016b) and agar blocks of Rhizoctonia solani were placed on the leaf sheaths (Park et al., 2008) as per the standard protocols (and maintained under controlled culture room conditions.
All the samples were collected as three biological and technical replicates, and qRT-PCR was performed with their respective control samples. Rice specific act1 and β-tub genes were used as internal reference genes for normalization and the mean values of relative fold change were calculated as per ΔΔCT method (Livak and Schmittgen, 2001). For native expression studies, the normalization was performed with only rice specific act1 and β-tub genes (ΔCT). Bar diagrams and heat maps were generated by Sigmaplotv11 and Morpheus20 programs, respectively using the means of the fold change that were calculated from the qRT-PCR data.
RNA Isolation, cDNA Synthesis and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from the treated and untreated samples using Tri-Reagent (Takara Bio, UK) according to the manufacturer's protocol. The concentration of the isolated RNA was checked using a Nanodrop spectrophotometer, and 2 ug of RNA was used to synthesize first strand cDNA using reverse transcriptase (Takara Bio, UK). The synthesized cDNA was diluted ten times, and 2 μl of it was used to perform qRT-PCR and 10 μM of the primers for each gene was used per reaction. The primers specific to RPS genes were designed by using the on line tool Primer-321 the sequences of forward and reverse primers are listed in Supplementary Table 5. The amplification protocol included an initial denaturation at 94°C for 2 min, with 40 cycles of 94°C for 30 s, primer specific annealing temperature for 25 s, an extension of 72°C for 30 s. This was followed by constructing a melt curve at the end to estimate the amplification efficiency of each gene. The fold change was calculated as per ΔΔCT method, and the mean of these values was considered as the final fold change of the transcript levels (Moin et al., 2016b). Means of relative fold change were plotted in Sigmaplot v11 with standard error to produce the bar diagrams for all the genes in a specific stress.
Results
Chromosomal Distribution
Rice genome has 12 chromosomes carrying 56 RPS genes. Based upon the OrygeneDB output, the RPS genes were found to be distributed throughout the rice genome covering all 12 chromosomes. Both the arms of chromosome randomly carried the RPS genes. Each chromosome carried at least one member of the RPS gene family. Most of the genes are located on chromosomes 3, 1, 7, and 11, with chromosome 3 bearing the highest number of eleven RPS genes. Both the chromosomes 1 and 7 carried a total of eight and nine genes, respectively. Chromosome 9 is the only example with only one RPS gene present near the telomere region of its long arm. The chromosome 10 and 1 carried three and eight RPS genes, respectively (Figure 1).
Gene Structure Prediction
RPS genes, like other eukaryotic genes, showed the organization of introns, exons and UTRs. On submitting the nucleotide sequences of the 34 RPS genes in the Gene Structure Display Server (GSDS), we have obtained the predicted gene structure showing all exons, introns, UTRs for each gene and the number and relative position of each of the components. The BLAST analysis of RGAP-DB sequences with indica variety database showed that the coding regions of the 34 genes in both the varieties were similar. The percent similarity of the coding and the 5′-upstream regions of the genes found from the BLAST analysis has been listed in Supplementary Table 1. The similarity of RPS genes in rice with the corresponding RPS gene in Arabidopsis has also been searched with the help of BLAST analysis in TAIR database. All the 34 RPS genes studied in this report exhibited very high similarity with their counterparts in Arabidopsis also. The minimum similarity percentage is 78 and the maximum is 95.
Results from the GSDS suggested that most of the RPS genes consisted of both introns and exons (Supplementary Figure 1). RPS25a has a long stretch of intronic sequence covering most of its length. It is also the longest gene among the selected genes with a total length of approximately 6 kb. RPS17 and RPS30 lacked introns and possessed a single and small CDS flanked by UTRs. RPS6, RPS19, RPS23a, and RPS7 lacked UTRs and are only composed of intronic and exonic sequences. RPS23a is the smallest gene with a total size of 0.5 kb. The number of exons also varied from a minimum of two in RPS23a and RPS29 to a maximum of three exons in RPS3a. The details of properties related to the gene structure have been provided in Supplementary Table 2.
Putative Promoter Analysis
In accordance with the observations regarding the stress responsive differential expression and tissue specific expression of several RPS genes, we tried to correlate the transcript levels with the presence of stress-responsive elements in the 5′ upstream regions of the individual genes. Among the 5′ regions of the RPS genes, we have shortlisted some of the common stress-responsive elements that were present in single or multiple copies, and we have listed the elements that might have significance in tissue specificity of expression in Supplementary Table 7. Most of the genes have elements for meristem and endosperm specificity in their promoter regions.
All the RPS genes have at least two stress responsive elements in their immediate upstream regions (up to 1 kb). RPS30 has one copy of ABRE and TGACG, and RPS18b has one copy of LTR and TC rich repeat as their cis-acting regulatory elements (CAREs). Other than these two genes, putative promoter regions of the rest of the genes have more than two elements. SARE, EIRE, motifIIb, DRE and ERE, which are responsive to SA, elicitor, ABA, dehydration and ethylene, respectively and are present in single copies in the putative promoter regions of some of the genes. RPS23 showed five repeats of ABREs, three copies of TGACG motif, three copies of CGTCA motif and one copy each of TGA, Box W1, W box, and GARE elements. RPS4a exhibited three copies of ABREs, two repeats each of TGA, TGACG, and CGTCA elements. RPS15 has two repeats of MBS. Similarly, the genes (RPS4, 10a, 6a, and 5) that have exhibited remarkable up-regulation under biotic stress conditions have multiple MeJa responsive elements and some of them contain box W1 and TCA elements that are related to responsiveness under pathogen attack. All the stress responsive cis-acting elements were pictorially represented by their physical mapping in Figure 2 and Table 2 mentioning the function of the corresponding motifs. The elements having roles in tissue specificity for each of the genes have been listed in Supplementary Table 7. The BLAST analysis for the 5′-upstream regions of the genes of both the varieties showed that 8 out of 34 genes had shown a slight variation in the sequences of upstream regions (97%) in the two varieties. The differences in regulatory elements due to the dissimilarity in their sequences have been compared and shown in the Supplementary Figure 7. The upstream regions could not be identified for three genes (S15a, S18a, and S18b) for which the promoter analysis has been performed using the japonica sequences only.
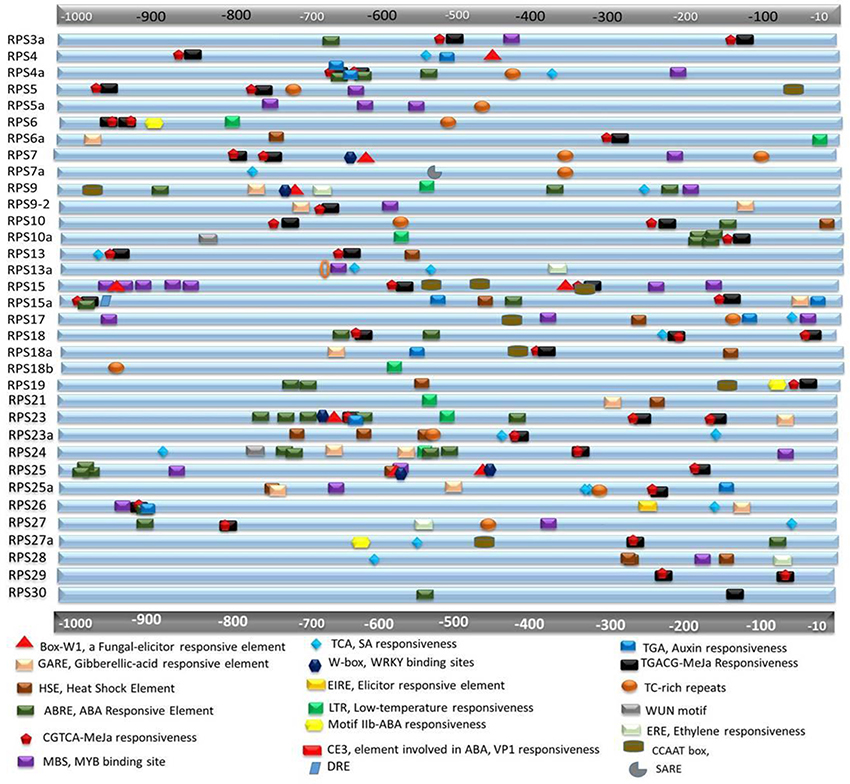
Figure 2. In silico analysis for the presence of cis-regulatory elements in the putative promoter regions of RPS genes. About≤1 kb upstream region of each gene is retrieved and searched for cis-regulatory elements that might be responsible for stress responsiveness. Different elements have been demonstrated in different shapes and colors and they are approximately placed according to their actual location in the genome. The functions of each of the elements are mentioned below the figure.
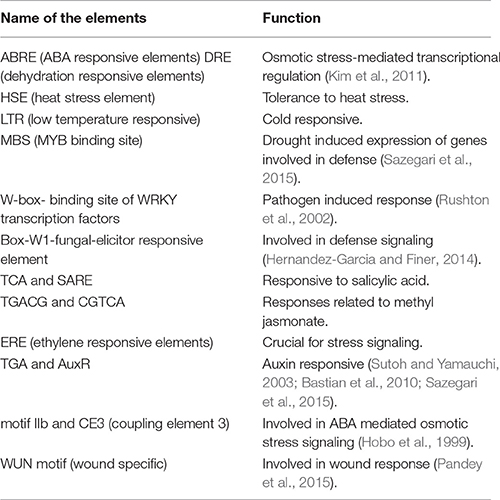
Table 2. Functions of different stress responsive elements found in the promoter regions of the RPS genes.
Protein Properties and Phylogeny
The submission of the protein sequences from RGAP-DB to the ExPasyProtParam server revealed many putative properties. Their molecular weights (MW) varied from approximately 7 to 30 kDa with RPS4 having the highest MW of 29.8 kDa and RPS30 with the lowest MW of 6.9 kDa. In accordance with the MW, the RPS4 is the largest protein with a sequence of 265 amino acids, while RPS30 is the smallest with 62 amino acids. In silico analysis using ExPasyProtParam server helped us to get the theoretical pI-values of each of the RPS. Most of them were in the range of 9 to 12 with very few exceptions out of this range. For example, RPS6 has a very low pI of 4.86. Most of the RPS proteins are rich in positively charged amino acids like Arg and Lys compared to negatively charged amino acids like Asp or Glu. The only exceptions are the RPS21, which has an equal number of both the types and RPS6 having more of acidic amino acids. Percentage composition of secondary protein structures (disordered protein, alpha helix and beta sheet) has variations within the RPS family. RPS4 has the minimum value of 14% while the RPS30 has the maximum value of 76%, respectively; S10a exhibited 10%, and S29 and S13 have 64% α- helical content, respectively. The range on the percentage of β-sheet content also varied with RPS13, 13a having 0% and S27 showing 49% of the β-sheet content.
The Grand Average of Hydrophobicity (GRAVY) indices were below the value of zero for all 34 RPS proteins suggesting the hydrophobic nature of RPS proteins. We have also identified some of the interacting ligands of these RPS proteins from the 3DLigandSite. The metal and non-metal ligands include IF-1, FMN, FAD, Mg2+, Zn2+ etc. and the specific residues involved in the ligand binding have been listed in the Supplementary Table 3. Most of the RPS proteins have Low Complexity Region (LCR) motifs, which are the stretches of biased amino acid sequences having a long repetition of single or few amino acids and they have been connected to many protein functions and interactions. LCRs that are located terminally in a protein have been considered to be translationally enriched and have stress responsive roles (Coletta et al., 2010). Three dimension protein structures from 3D ligand site have been included in Supplementary Figure 2. The ribosomal protein sequences were submitted to SMART database, and it was found that all of them exhibited ribosomal domains. Some of the proteins also contain KOW domain (for e.g., RPS4, RPS4a). RPS6 has a SPT2 domain; RPS5a has a HLH domain, a CARD domain and an ANTAR domain. RPS18a has a SANT domain, which is a MYB- related domain that binds to DNA and helps in transcriptional regulation (Feller et al., 2011) and it also has a homeobox domain HALZ. All the domains associated with each of the RPS subunits are listed in Supplementary Table 8.
Protein sequence data was exploited to construct an unrooted phylogenetic tree of 34 RPS proteins in the MEGA6 software. Alignment with ClustalW employing a neighbor joining algorithm resulted in the generation of a tree that showed the evolutionary relationship between the RPS proteins. The tree suggested that a considerable number of cladogenesis events occurred during the course of evolution in the RPS gene family that was reflected in their protein sequence similarity and diversity. The earliest divergence separated the proteins into two major clades, of which one contains five proteins (RPS13, 13a, 3a, 19, and 21) and the other major clade carried all the other proteins, which has further diverged several times. These five RPS genes were checked for similarity with their paralogs and depicted as a phylogenetic tree in Supplementary Figure 3B. While RPS 19 and 21 have no paralogous members, RPS13 and RPS3a have two paralogous members, and they have shown similarity with their corresponding group. Nodes, from where some pairs of genes were diversified showed the bootstrap value of 100. Such high value indicates the high significance of the clade. The phylogenetic tree showing the evolutionary relationships among the RPSs is shown in Supplementary Figure 3.
Native Expression Profiling of the RPS Genes in Different Tissues
To gain insights into the expression patterns of RPS genes in rice development and also to provide a comparative analysis between RPs of both the large and small subunits, we performed qRT PCR of 34 RPS genes in 13 different tissues as described in Moin et al. (2016a) for the RPL genes. The mean values were utilized to construct heat maps to demonstrate clearly the native expression pattern of the RPS genes (Figure 3). The tissues used to study the native expression pattern included embryo and endosperm from 16 h old germinating seed, plumule and radicle from 3 d old seedlings; shoot and root of 7 d old seedling; shoot, root, flag leaves, panicle, flower, grains and root-shoot transition of mature 20 d old plants (after transfer to green house).
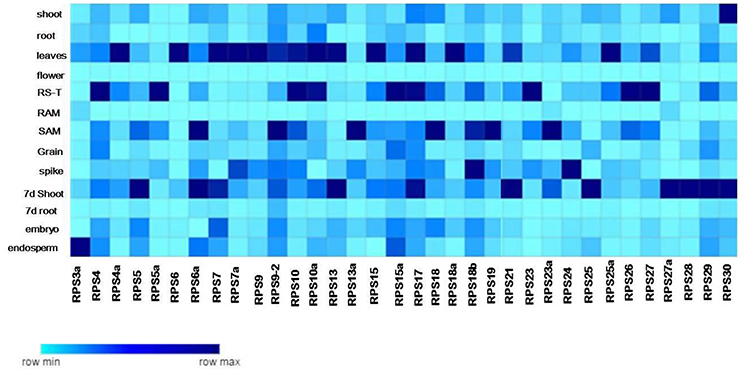
Figure 3. Tissue specific expression of the RPS genes. The native expression of 34 RPS genes in 13 tissues is determined by qRT-PCR with single normalization with rice actin. The mean values of the fold change of the biological and technical replicates are represented in the form of a heat map.
The complete tissue-specific up-regulation pattern of the RPS genes in native conditions has been summarized in Supplementary Table 4 and also in the form of heat maps (Figure 3). The up-regulation of RPS genes was significant in mature leaves (25 genes), shoots (21 genes), 7 d shoots (26 genes), plumules (21 genes), and root-shoot transition (23 genes). Apart from these four, seven and 15 genes were up-regulated in seedling radicles, 7 d old root and mature roots, respectively. An analogous pattern of gene expression was also observed under different stress conditions as well, where more number of RPS genes was found to be up-regulated in shoot than in the root tissues showing that the majority of the RPS genes were up-regulated in shoots, leaves and plumules compared to the other organs.
About 17 RPS genes showed expression in rice grains and panicles indicating their involvement in inflorescence development and grain filling. None of the genes was expressed in florets implying that RPS proteins have probably no significant roles in the development of this meiotically active tissue. About 18 and 15 RPS genes were up-regulated in embryo and endosperm, respectively showing that these genes might have a role in seed development and maturity. RPS23a, RPS27a, RPS10a, and RPS7 have exhibited no significant up-regulation in any of the tissues and RPS23 is up-regulated only in root-shoot transition. A considerable up-regulation of RPS28 and RPS29 was observed in roots under different stresses. RPS4, RPS5a, RPS7a, RPS9, RPS9-2, RPS13, RPS6, RPS17, RPS18b, RPS24, and RPS29 were up-regulated in most of the tissues, while RPS13a, RPS18a, RPS4 were up-regulated in shoots throughout the duration of all the stress conditions. Similarly, these genes were also expressive in shoots, leaves, plumules and in native conditions as well.
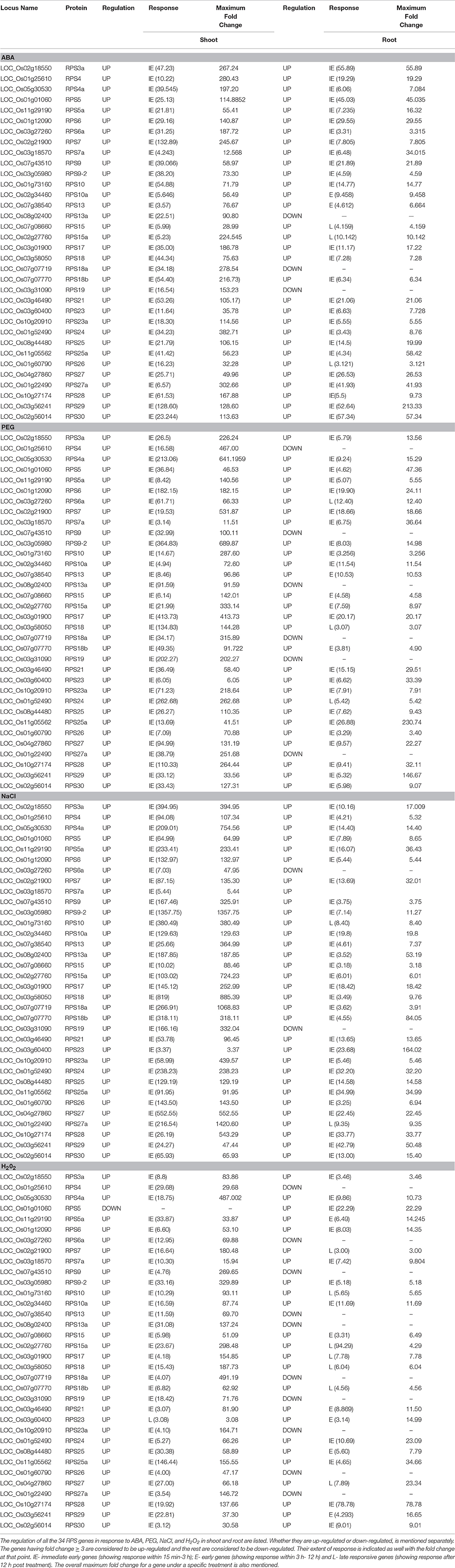
Differential Expression Analysis of the RPS Genes in Response to Abiotic Stress Treatments
Our findings on the screening of a large-pool of activation tagged mutant population for high water-use efficiency revealed the significant up-regulation of RPL genes (RPL6 and RPL23A) by the integrated enhancers in two of the mutant plants with sustained productivity under limited water conditions (Moin et al., 2016a). In the follow-up study, an extensive tissue specific and differential expression study of the entire RPL gene family was carried out, which showed significant up-regulation of certain RPL genes under abiotic and biotic stresses (Moin et al., 2016b). Results from these two studies prompted us to further investigate the expression of 40S Ribosomal Protein genes in response to abiotic and biotic stress conditions and to arrive at proper combinations of RPS and RPL genes for subsequent genetic manipulation. For studying their differential expression, we have selected 34 RPS candidate genes from the respective paralogous groups in response to four abiotic stresses (ABA, PEG, NaCl, and H2O2) at seven time points and two biotic stresses (BLB and SB). ABA, PEG, NaCl, and H2O2 induce a stress similar to osmotic, drought, salinity and oxidative stress, respectively (Gao et al., 2007; Hamayun et al., 2010; Hossain et al., 2015; Sah et al., 2016). Treatment of the plants with these stress-inducing agents would mimic the corresponding stress situations and consequently activate numerous stress responsive pathways. Also, we also analyzed their native expression pattern in different tissues.
The genes that exhibited ≥ 3 fold transcript level on the log2 scale were considered as up-regulated. The pattern of expression of the genes depended mostly on the type of stress signal and also the tissue in which the expression was studied. We have categorized the genes in terms of their timing and the intensity of response. Some of the genes were up-regulated as early as 15 min-3 h after the onset of the stress, while a few of them responded between 3 and 12 h after exposure while others were up-regulated 12 h after the application of the stress. We have differentiated them as Immediate Early (IE), Early (E), and Late (L) expressing genes, respectively.
Since the level of expression varied from 3 to several 100 folds, we have further categorized the up-regulated genes as highly expressive genes (≥30 fold), moderately expressive (between 10 and 30 fold) and low-expressive (between 3 and 10 fold). The expression of the RPS genes in the shoot samples has been observed to be higher compared to the corresponding root samples under almost all the abiotic stresses at different time intervals. Under some conditions, the up-regulation of the genes in the shoot samples has been detected to be several 100 folds, which is not the case for the corresponding root samples. In the case of shoot samples, all the genes showed significant up-regulation for all the stress conditions for at least one time point, and they were predominantly IE type in their response, except under H2O2 treatment.
Although all the genes were up-regulated under all the stress conditions in shoots, their mode of response varied with the nature of the stress. For example, ABA induced the up-regulation of 26% of the genes (RPS7, 15a, 18a, 25a, 4, 7a, 10, 10a, and 13a) and these maintained a significant expression (≥3-fold) until the last time point used (60 h). Of these, 50% were highly expressive (RPS7, 15a, S8a, and 25a) with >30-fold transcript level throughout the stress treatment up to 60 h time point. RPS7 and RPS18a showed up-regulation up to several 100 folds. PEG and NaCl also induced the up-regulation of a majority of genes in shoots until 60 h time point, except RPS7a and RPS23 and RPS5, RPS10a, and RPS23a under PEG and NaCl treatments, respectively, which became down-regulated.
Some of the RPS genes maintained remarkably high transcript levels (≥100 fold). These include RPS4a, RPS9, RPS9-2, RPS13, RPS17, RPS19, RPS27, and RPS30 under PEG treatment and RPS4a, RPS7, RPS9-2, RPS18a, and RPS23a under NaCl treatment. The up-regulation of RPS4, RPS7, RPS9, RPS10, RPS19, and RPS26 under salt treatment is in agreement with the earlier studies (Kawasaki et al., 2001). Unlike in ABA, PEG, and NaCl treatments, several RPS genes were late responding in shoots under H2O2 treatment. Moreover, the level of up-regulation was also not as high as in other treatments. About 60% of the genes were up-regulated 15 min post treatment. About 30 genes have been found to maintain a considerable level of expression, and among these, RPS4a, RPS9-2, RPS15a, RPS18, and RPS28 have exhibited ≥100 fold up-regulation. In roots, RPS19 was down-regulated persistently in response to all the abiotic stresses at all the time intervals. RPS10a, RPS18, RPS18a, RPS18b, RPS24, RPS27a, and RPS6a were also observed to maintain a very low transcript level. Some of the genes that were initially up-regulated, subsequently, became down-regulated with the progression of the stress treatment. The expression pattern of all the genes under different stress treatments in the shoot and root tissues has been represented in the form of heat maps (Figures 4A,B).
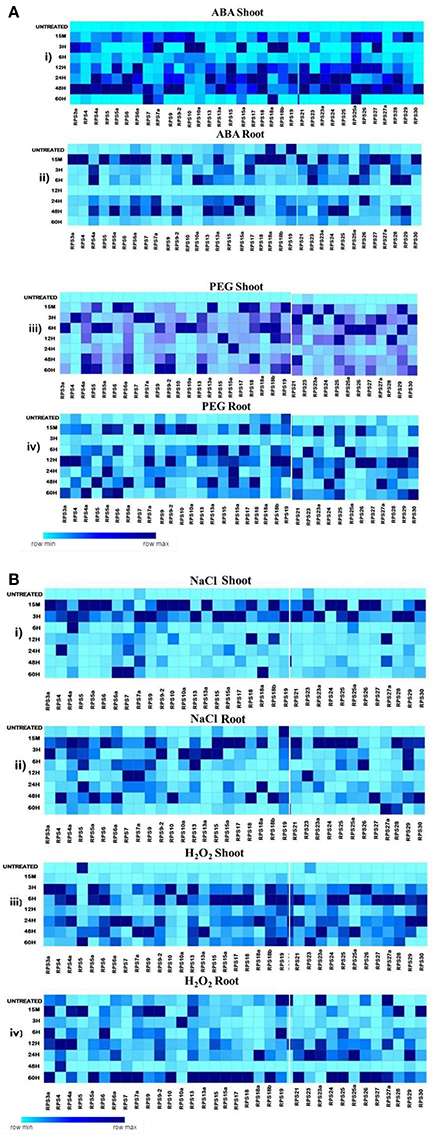
Figure 4. Heat map representation of RPS genes in response to ABA, PEG, NaCl and H2O2 treatments. The 7d old seedlings were treated with ABA(100 μM), PEG (10%), NaCl (250 μM), and H2O2(10 μM) and qRT-PCR is used to determine the level of expression of 34 RPS genes in the shoot and the root tissues at the time points mentioned on the left side. The fold change has been normalized by ΔΔCT method with respect to the unstressed seedlings maintained in water. Rice specific act1 and β-tub genes were used as endogenous reference genes. The mean values of the fold changes of the biological and technical replicates were considered for preparing the heat maps. A-(i), (ii), (iii), and (iv) represent the expression of the genes in shoot and root tissues under ABA and PEG respectively and B- (i), (ii), (iii), and (iv) represent the expression of the genes in shoot and root tissues under NaCl and H2O2 respectively. The darkest color depicts the highly up-regulated genes whereas the lightest color represents the less up-regulated and down-regulated genes.
There is an overlap in the expression of the genes that are up or down-regulated in response to the stresses at several time points, and this has been demonstrated in the form of Venn diagrams (Supplementary Figures 4, 5). Table 3 presents a detailed list of genes that were up/down-regulated in different stress conditions in the shoot and root tissues separately. The percentage of genes over-expressed at certain time point under the stress conditions in shoot and root has been represented in the form of Pie charts (Figures 5A,B), and the genes are tabulated in the tables that accompanied the figures. The genes that were commonly up-regulated under all the treatments, particularly during the early response may have significance in stress related signaling (Kilian et al., 2007) as the early responsive genes play a crucial role in defense against the environmental stresses (Mahajan and Tuteja, 2005; Ouyang et al., 2007).
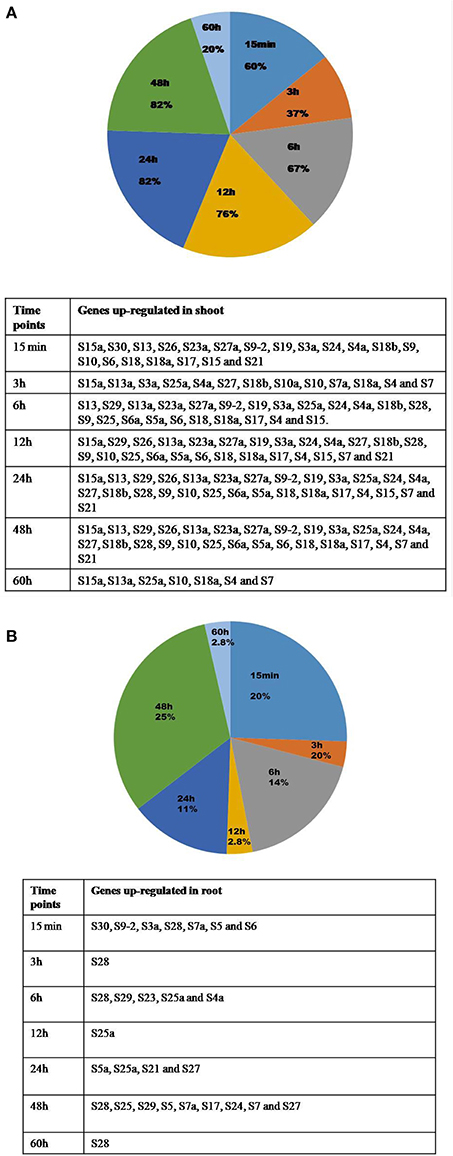
Figure 5. Percentages of genes up-regulated in shoot and root under all the stresses at different time points. The percentage of genes that are upregulated in shoot (A) and root (B) at different time intervals after the application of the stress is represented in form of pie charts with the genes enlisted in the respective tables.
Differential Expression of RPS Genes upon Infection with Xanthomonas oryzae pv. oryzae (Xoo) and Rhizoctonia solani
Apart from the four abiotic stresses, the expression level of 34 RPS genes was also studied in response to the treatments with Xoo and R. solani pathogens that cause very serious Bacterial Leaf Blight (BLB) and Sheath Blight (SB) diseases, respectively in rice. In the BLB infected samples, 14 (40%) genes were down-regulated, and the rest of them were up-regulated (60%). RPS6a (26 fold), RPS9 (18 fold), RPS10a (29 fold), and RPS4 (13 fold) showed a high level of transcript upregulation when compared with the other up-regulated genes.
The qRT-PCR analysis of 34 RPS genes was performed from both the shoot and the leaf samples infected with R. solani. The up-regulation of genes was observed particularly in the shoot tissues for a majority of the RPS genes. Thirteen (37%) genes were down-regulated in response to this pathogen in the shoot region whereas the 22 RPS (63%) genes were up-regulated. Among the genes that were expressive, RPS4 and RPS5 have exhibited 22 and 14 fold up-regulation, respectively, which is the highest compared with the other genes. The expression pattern of all the genes under two biotic stresses has been depicted graphically in Figures 6A,B.
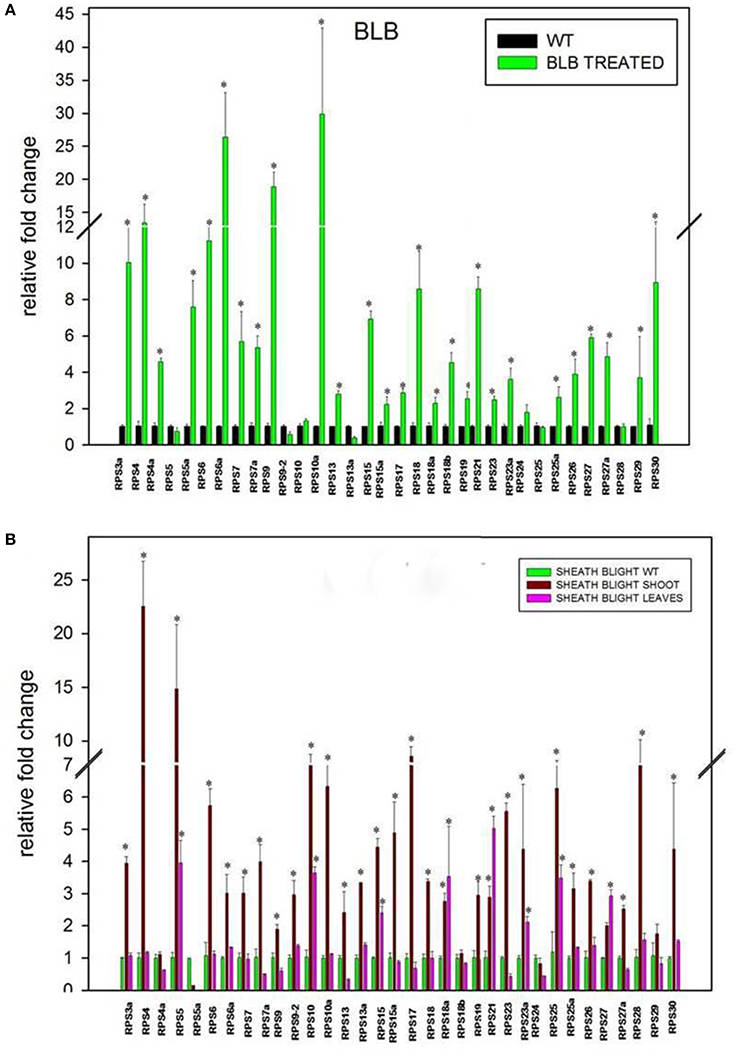
Figure 6. Differential expression of RPS genes in response to Xanthomonas oryzae pv. oryzae (Xoo) (A) and Rhizoctonia solani (B), which cause Bacterial Leaf Blight (BLB) and Sheath Blight (SB) respectively. The expression of 34 RPS genes was determined in BLB and SB infected leaf samples after 20 and 25 d respectively after infection. The expression was normalized with corresponding untreated samples in the identical conditions. The statistical significance of expression analysis has been represented with asterisks *P < 0.05.
Comparative Expression Analysis of RPL and RPS Genes
Previously available information about the RPL genes (Moin et al., 2016b) has been utilized and subsequently combined with the information obtained from the current study on the expression patterns of RPS genes. The details regarding their stress responsiveness and tissue-specific expression pattern under native condition have been summarized in tabular form to select suitable combinations of RPS and RPL genes for stress alleviation in transgenic plants. All the RPL and RPS genes up-regulated in response to four individual stress treatments were depicted in Supplementary Table 6. Highly up-regulated RPL and RPS genes in each of the 13 different tissues under native conditions have been represented in Figure 7. Four heat maps combining the expression of RPL and RPS genes under four abiotic stresses have been represented separately (Figures 8 A–D) and Supplementary Figure 6 shows the comparative expression analysis of both RPL and RPS genes under 13 different tissues in the form of a heat map.
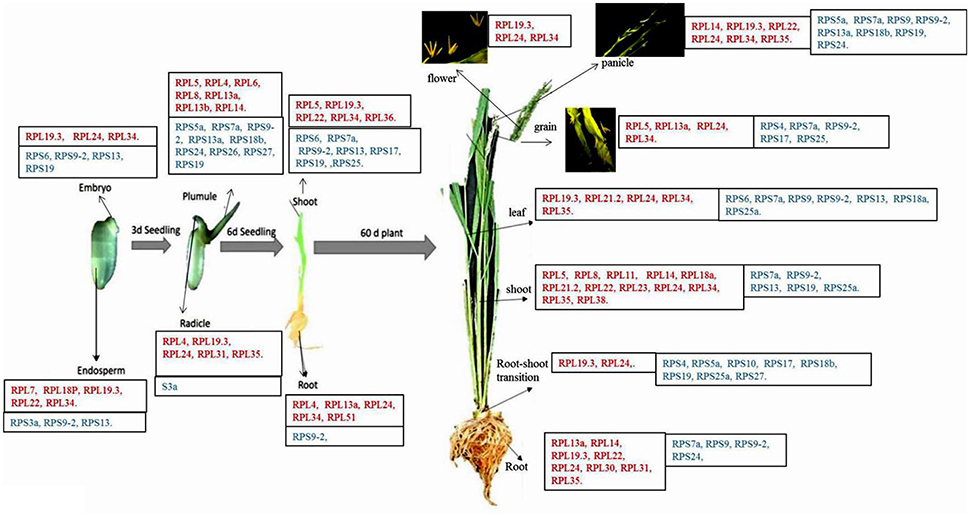
Figure 7. Native expression of RPL and RPS genes in different tissues. Some of the RPL and RPS genes showing considerably higher expression than the other genes in each of the 13 different tissues under native conditions have been listed in the boxes (Moin et al., 2016b). In each box, the names of the genes mentioned in red color belong to RPL gene family and the names of the genes mentioned in blue color belong to RPS gene family. The figure has been adapted and updated from Figure 3B of Moin et al. (2016b) under CC BY 4.0 license.
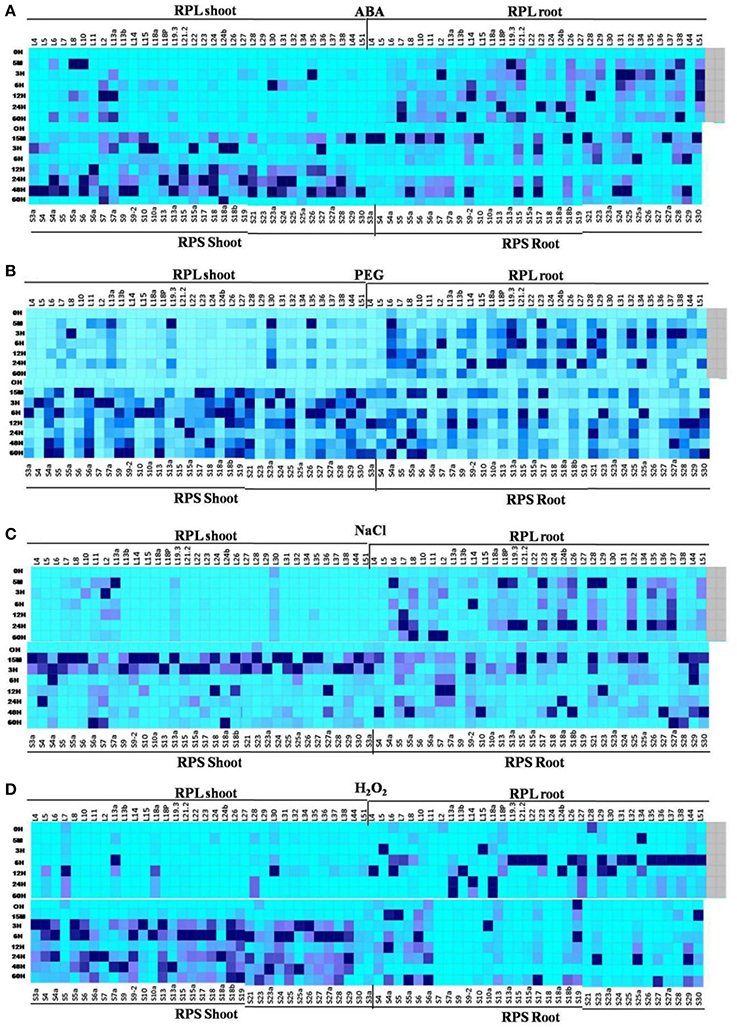
Figure 8. combined heat maps depicting differential expression of RPL and RPS genes under 4 abiotic stresses in shoot and root tissues. Differential expression of all the RPL and RPS genes under 4 abiotic stress is depicted together in 4 heat maps (A- ABA, B- PEG, C- NaCl, and D- H2O2). The mean of the fold change values of the biological and technical replicates are represented in the form of heat maps.
One of the intriguing observations from this comparative analysis is that majority of the RPL genes were up-regulated in both shoots and roots of the plant under native conditions. But under all abiotic stress conditions, the maximum up-regulation was observed primarily in roots than in shoots. On the contrary, under the native conditions, the expression of RPS genes was mostly confined to shoots and the leaves compared to roots. Taken together, the up-regulation of the RPS genes and RPL genes was predominantly noticed in shoots and roots, respectively at all the time intervals under all the stress conditions.
According to the previous studies, RPL23A, L19.3, and L38 have shown to exhibit a high level of expression under ABA, PEG and NaCl treatments. RPL6, 12, 23A, 18A, and 13A have maintained high up-regulation under H2O2 treatment (Moin et al., 2016b). In the present study, RPS13a, 18a, 4, and 4a have shown significant up-regulation in almost all the stresses in shoots, while RPS29 and RPS28 were highly expressive in roots. In the case of RPL genes, NaCl caused down-regulation of a majority of genes when compared to ABA and PEG. On the contrary, almost all the RPS genes showed up-regulation under salt stress in shoots. The H2O2 treatment caused down-regulation of 60 and 80% of RPL genes in shoots and roots, respectively when compared with MeJA, SA and cold treatments, which induced up-regulation of many of them. Most of the RPS genes were up-regulated under all the stresses in shoots, whereas in roots, 8, 22, 5, and 28% of the RPS genes were down-regulated under ABA, PEG, NaCl, and H2O2 treatments, respectively. The H2O2 treatment induced the down-regulation of many of them. RPL30, 44, 22, 14, 29, 13A, and 15 were down-regulated under ABA, PEG, and NaCl treatments and RPS27a, 10, 20, 6, and 9 were down-regulated in response to all the treatments (ABA, PEG, NaCl, and H2O2). RPL10 was significantly activated upon infection with Xoo pathogen, whereas among the RPS genes, RPS4 became commonly up-regulated under both Xoo and R. solani treatments.
Discussion
Abiotic and nutrient stresses affect global agriculture by drastically reducing crop yield and productivity. Keeping the expanding world population in mind, stress management is one of the growing concerns for researchers worldwide. Rice is one of the most important staple food crops feeding a large portion of the world's population and is highly affected by some abiotic stresses, which include salinity, extreme temperatures, drought (Gao et al., 2007) and assailants like pests, fungi, virus and bacteria, etc. Being sessile, plants are highly vulnerable to these detrimental factors that drastically affect their growth and productivity. Hence, they have developed highly organized signaling responses and various other complex events to withstand and curtail the deleterious effects of these stress factors.
The role of RPs in ribosome assembly and house-keeping functions was very well recognized in several earlier studies on animal and plant systems. In addition to that, mutations in these RPs have led to physiological defects in several organisms. Effect on global protein synthesis due to these reduced RP functions could be one of the reasons behind developmental abnormalities. However, these physiological defects imply the potential extra-ribosomal and regulatory roles apart from the regular housekeeping functions for the RPs (Lu et al., 2015).
In this study, we have made comprehensive expression analyses of Ribosomal Protein Small subunit genes (RPS) in rice under multiple abiotic and biotic stresses and compared their expression with large subunit genes (RPL) genes (Moin et al., 2016b). Based on the differential and native expression data, we have short-listed the promising RPS and RPL combinations of candidate genes that might be useful in manipulating stress tolerance in rice. Also, we have presented an in silico analysis of the RPS protein properties and the putative promoter elements of the studied genes. With our previous study, which dealt with the extensive investigation on rice RPL genes (Moin et al., 2016b), we have tried to provide comprehensive information on the stress induced expression of all the ribosomal protein (RPS and RPL) genes in rice as a whole.
The Bacterial Leaf Blight (BLB) and Sheath Blight (SB) diseases are two of the major constraints in rice production with devastating influence on rice yield and its productivity (Zheng et al., 2013; Khan et al., 2014).
Availability of high-quality rice genome database helped us retrieve the genomic, CDS and protein sequences along with the 5′ upstream regions of the selected genes. A total of 56 RPS gene sequences were retrieved from the search in the databases, and finally, 34 genes were short listed excluding the identical members. The distribution of RPS genes appeared to be throughout the rice genome with each of the 12 chromosomes carrying at least one RPS gene family member with chromosome 3 exhibiting a maximum of 11 RPS genes. The RPs are mostly rich in positively charged amino acids, which can be correlated with the hypothesis that positively charged amino acids might play a significant role in electrostatic protein-RNA interactions (Law et al., 2006) within the ribosomal complex. Since the RPs are involved mostly in interactions that make up the ribosome complex along with rRNAs, the RPs are expected to have similar isoelectric pH range to facilitate their molecular interactions. Proteins with high isoelectric points tend to interact with DNA, RNA and other biomolecules that are negatively charged whereas the proteins with low isoelectric points prevent non-specific interaction with nucleic acids (Takakura et al., 2016). In a recent study, RPS6 has been reported to interact with a plant specific histone-deacetylase-2B and also with nucleosome assembly protein-1 thereby regulating the transcription of r-DNA in Arabidopsis (Son et al., 2015; Xiong and Sheen, 2015). Having a low isoelectric point might, therefore, be beneficial for RPS6 to minimize non-specific interactions. The RPs also have several domains associated with them like KOW, SPT2, HLH, CARD, ANTAR, HALZ, SANT domains etc. These domains have different functions including interaction with other proteins, transcription regulation, protein dimerization, apoptosis, binding to RNA and so on (Kyrpides et al., 1996; BouchierHayes and Martin, 2002; Shu and Zhulin, 2002; Jones, 2004; Novoseler et al., 2005; Feller et al., 2011).
Under native conditions in 13 different tissues, the RPS genes are prevalently up-regulated in 45 d old mature leaves, shoots and root-shoot transition and 7 d shoots and plumules and some of them were up-regulated in roots, panicles, embryo, endosperm and grains. But, none of them was found to be expressive in flowers. The role of RPS genes might be presumed to be involved in modulating developmental stages in vegetative tissues, mainly shoots and not to a greater extent in reproductive tissues. The activity of RPS18A promoter of Arabidopsis was seen in meristematic tissues and embryonic stage and mutations in these loci resulted in the formation of pointed leaves (Van Lijsebettens et al., 1994). This can be correlated with the transcriptional up-regulation of RPS18, RPS18a, and RPS18b in embryo, plumules, shoots, leaves, and root-shoot transition in the present study.
Around 60 and 20% of the genes were up-regulated immediately after treatment in shoots and roots, respectively under all the stress conditions. Up-regulation of these genes might fall under immediate response to the stress, and these genes are presumed to encode non-specific master regulators needed for plant core environmental stress response proteins. The immediate-early response to a certain stress might sometimes be non-specific, and responses with continuous up-regulation are considered to be specific ones (Kilian et al., 2007). And, the genes that are up-regulated at around 1 h time points can be assumed to have a pivotal role in early defense signaling and might have a function in reconstructing the proteins synthesis apparatus (Ouyang et al., 2007). This has led us to look for continuity in the up-regulation of immediate-early responsive RPS genes.
There are a considerable number of genes for which the up-regulation is maintained throughout the duration of the stress for example, RPS13a, 18a, 4 and 4a. Subsequently, RPS29 has shown a remarkably high transcript level (≥100 fold) in roots under ABA and PEG treatments and also showed a moderate level of expression (11-fold) under H2O2 treatment. RPS28 has consistently displayed a moderately high level of expression in all the stress conditions in root tissues.
On the other hand, RPL23A, 6, 8, L19.3, and L38 among the members of large subunit genes have consistently shown up-regulation under ABA, PEG, NaCl treatments. RPL18a, L24a, L24b, L30, and L34 were significantly overexpressed under oxidative stress. Moreover, RPL23A, one of the candidate genes that were identified using an activation tagging approach in water use efficiency (Moin et al., 2016a) might have a potential role to be utilized in modifying abiotic stress tolerance in rice orchestrating the responses of other candidate genes from both the subunits. Among the RPS genes, RPS3a, RPS6, S15a, S17, S18a, S23a, S24, S25, S27a, and S28 are substantially up-regulated in shoots under a minimum of two abiotic stress conditions. RPL10 and RPS4 are significantly up-regulated under pathogen treatments. RPS18a has few significant protein domains associated with it and RPS18a has shown upregulation throughout all the stress treatments. Hence, the candidate genes overexpressed in small and large subunit categories listed in this study might be crucial in shortlisting genes to manipulate stress tolerance in rice by generating transgenic plants overexpressing the genes of both the subunits either individually or in combination.
It is crucial to look into the response of plants in the presence of multiple stress factors in order to completely evaluate the resistance responses of the candidate genes as the plant responses to a combination of stresses differ from individual stress responses (Atkinson and Urwin, 2012). In this context, the genes that are mostly up-regulated under all the stress conditions are significant. On the contrary, RPS23 and RPS7a have maintained a low level of expression at all the time points under almost all the abiotic stresses in shoots, while RPS19 has consistently exhibited down-regulation in roots under all the abiotic stress conditions. Except at certain time points under ABA treatment, RPS27a, 10, 6, and 9 have shown either down-regulation or no change in expression under rest of the stress conditions. RPS18, 18a, 15, and 13 have consistently displayed a low level of expression irrespective of the abiotic stress condition.
Apart from the genes that have been up-regulated in response to stress, there are also certain genes (mentioned above) that did not respond to the stress factors effectively in the respective tissues, probably indicating the limitations of their roles in stress tolerance. Under biotic stress, RPS4, 10a, and 6a were up-regulated in BLB infected leaf samples, and RPS4 and 5 were up-regulated in sheath blight infected leaf samples. Since RPS4 has been up-regulated remarkably under all the abiotic and biotic stress conditions; it can be considered as one of the important RPS genes having both abiotic and biotic stress responsive roles.
The cis-regulatory elements (CREs) in the 5' UTR regions play significant roles in activation and suppression of gene expression that can confer tolerance to stresses (Ibraheem et al., 2010; Hernandez-Garcia and Finer, 2014). Throughout the putative promoter regions of almost all the RPS genes, regulatory elements that respond to abiotic and biotic stress treatments are distributed in multiple copies justifying their enhanced expression under various stress treatments.
The alarming change in climatic conditions will have a drastic consequence on global agriculture providing impetus to the already existing abiotic and biotic stresses. Temperature increase is more likely to be accompanied by recurrent flood, drought, enhanced salinity and prevalent occurrence of pathogen attacks (Atkinson and Urwin, 2012). This propels the researchers to produce transgenic plants with better yielding traits, which can withstand the altered environmental conditions. Ribosomal protein genes are important in this context because ribosome constitutes the protein synthesis machinery in a cell and under unfavorable conditions, the composition of the ribosome has been thought to change in order to assist biased protein translation (Wang J. et al., 2013). Moreover, some of the Genome-wide RNA sequencing technology and microarray analyses of other crop plants like Sorghum and alfalafa also revealed ribosomal protein genes to be important in response to abiotic stress (Fracasso et al., 2016; Liu et al., 2017).
Peptide mass spectrometry provides information that RPs of both prokaryotes and eukaryotes undergo a high degree of post-translational modifications (Arragain et al., 2010). Six RPs of Escherichia coli are methylated, three are acetylated, and one of them is methyl-thiolated. Although these modifications might have specific regulatory functions, the actual significance of these modifications is unclear (Nesterchuk et al., 2011). In plants, RPs are mostly methylated and methylation appears to be important for ribosome biogenesis and perhaps, its function (Friso and van Wijk, 2015). Apart from methylation, phosphorylation and glycosylation also play critical roles in post-translational modification of ribosomal proteins. Modulation of diurnal protein synthesis in plants has been predicted to have some link with differential phosphorylation of some ribosomal proteins. In higher plants, phosphorylation status of ribosomal protein S6 contributes to rapid adjustments of growth under environmental changes. Auxin enhances S6 protein phosphorylation in maize, which selectively increases ribosomal protein synthesis (Turkina et al., 2011). In terms of glycosylation, N glycosylation of Asn165 residue of Rps3, which is secreted in cancer cell lines, regulates the migration and invasive phenotype of the cancer cells (Kim et al., 2016). Overall the post-translational modification of RPs is important for the accuracy of the decoding machinery (Arragain et al., 2010) and it might also establish an additional RNA-protein contact which might be beneficial (Polikanov et al., 2015).
Differential expression of RPL and RPS genes under abiotic and biotic stresses, as we have described in the present study, indicates their role in stress responsiveness and might also modulate stress tolerance. The RPL and RPS genes that are remarkably up-regulated in response to almost all the stresses in comparison to the other genes can be considered as the cardinal RP genes that are essential to maintain the integrity of ribosomal units as well as ensuring sustained and undisturbed protein synthesis even under stress. This study provides an insight into the changes in the expression of RP genes in response to the environmental condition. Thus, the stress responsive increase in expression of several ribosomal protein genes can be considered for further exploitation as a resource in crop improvement programs.
Author Contributions
AS and SD performed the experiments. AS, MM, and PK designed the experiments and prepared the manuscript. AB and MD helped in qRT-PCR experiments and analysis. MSM performed the Xanthomonas oryzae pv. oryzae and Rhizoctonia solani infection on rice plants. PK supervised the work. AS, MM, and PK edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The funding for the current work has been obtained through a grant sponsored by DBT, GOI to PK with grant number BT/PR13105/AGR/02/684/2009. AS, MM are grateful to DBT for proving Research Fellowships. AB is also thankful to DBT for Project Fellowships.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01553/full#supplementary-material
Abbreviations
RPL, Ribosomal Protein Large subunit; RPS, Ribosomal Protein Small subunit; RP, Ribosomal proteins; ABA, Abscisic Acid; PEG, Polyethylene Glycol; NaCl, Sodium chloride; H2O2, Hydrogen peroxide; BLB, Bacterial Leaf Blight; SB, Sheath Blight.
Footnotes
1. ^http://rice.plantbiology.msu.edu/index.shtml
2. ^https://phytozome.jgi.doe.gov/pz/portal.html
3. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
4. ^https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
6. ^http://orygenesdb.cirad.fr/tools.html
7. ^http://plants.ensembl.org/Oryza_indica/Tools/Blast?db=core;tl=dxXz9p4PlES2NtPR-17828772
8. ^https://www.arabidopsis.org/Blast/
9. ^http://gsds.cbi.pku.edu.cn/
10. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
11. ^http://plantpan2.itps.ncku.edu.tw/promoter.php
12. ^http://jaspar.genereg.net/cgi-bin/jaspar_db.pl?rm=browse&db=core&tax_group=plants
13. ^https://psort.hgc.jp/form.html
14. ^http://web.expasy.org/cgi-bin/protparam/protparam
15. ^http://smart.embl-heidelberg.de/
16. ^http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index
17. ^http://www.sbg.bio.ic.ac.uk/3dligandsite/
18. ^http://www.genome.jp/tools/clustalw/
19. ^http://www.megasoftware.net/
References
Arragain, S., Garcia-Serres, R., Blondin, G., Douki, T., Clemancey, M., Latour, J. M., et al. (2010). Post-translational modification of ribosomal proteins structural and functional characterization of rimo from Thermotoga maritima, a radical s-adenosylmethionine methylthiotransferase. J. Biol. Chem. 285, 5792–5801. doi: 10.1074/jbc.M109.065516
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Barakat, A., Szick-Miranda, K., Chang, I.-F., Guyot, R., Blanc, G., Cooke, R., et al. (2001). The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 127, 398–415. doi: 10.1104/pp.010265
Bastian, R., Dawe, A., Meier, S., Ludidi, N., Bajic, V. B., and Gehring, C. (2010). Gibberellic acid and cGMP-dependent transcriptional regulation in Arabidopsis thaliana. Plant Signal. Behav. 5, 224–232. doi: 10.4161/psb.5.3.10718
BouchierHayes, L., and Martin, S. J. (2002). CARD games in apoptosis and immunity. EMBO Rep, 3, 616–621. doi: 10.1093/embo-reports/kvf139
Cherepneva, G. N., Schmidt, K. H., Kulaeva, O. N., Oelmüller, R., and Kusnetsov, V. V. (2003). Expression of the ribosomal proteins S14, S16, L13a and L30 is regulated by cytokinin and abscisic acid: implication of the involvement of phytohormones in translational processes. Plant Sci. 165, 925–932. doi: 10.1016/S0168-9452(03)00204-8
Coletta, A., Pinney, J. W., Solís, D. Y. W., Marsh, J., Pettifer, S. R., and Attwood, T. K. (2010). Low-complexity regions within protein sequences have position-dependent roles. BMC Systems Biol. 4:43. doi: 10.1186/1752-0509-4-43
Danilova, N., Sakamoto, K. M., and Lin, S. (2008). Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood 112, 5228–5237. doi: 10.1182/blood-2008-01-132290
Degenhardt, R. F., and Bonham-Smith, P. C. (2008). Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 147, 128–142. doi: 10.1104/pp.107.111799
Dhadi, S. R., Krom, N., and Ramakrishna, W. (2009). Genome-wide comparative analysis of putative bidirectional promoters from rice, Arabidopsis and Populus. Gene 429, 65–73. doi: 10.1016/j.gene.2008.09.034
Ding, Y., Chen, Z., and Zhu, C. (2011). Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J. Exp. Bot. 62, 3563–3573. doi: 10.1093/jxb/err046
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Filipowicz, W., and Hohn, T. (eds.). (2012). Post-Transcriptional Control of Gene Expression in Plants. Springer Science & Business Media. doi: 10.1007/978-94-009-0353-1
Fracasso, A., Trindade, L. M., and Amaducci, S. (2016). Drought stress tolerance strategies revealed by RNA-Seq in two sorghum genotypes with contrasting WUE. BMC Plant Biol. 16:115. doi: 10.1186/s12870-016-0800-x
Friso, G., and van Wijk, K. J. (2015). Post-translational protein modifications in plant metabolism. Plant Physiol. 169, 1469–1487. doi: 10.1104/pp.15.01378
Gao, J. P., Chao, D. Y., and Lin, H. X. (2007). Understanding abiotic stress tolerance mechanisms: recent studies on stress response in rice. J. Integr. Plant Biol. 49, 742–750. doi: 10.1111/j.1744-7909.2007.00495.x
Gazda, H. T., Sheen, M. R., Vlachos, A., Choesmel, V., O'Donohue, M. F., Schneider, H., et al. (2008). Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 83, 769–780. doi: 10.1016/j.ajhg.2008.11.004
Hamayun, M., Khan, S. A., Shinwari, Z. K., Khan, A. L., Ahmad, N., and Lee, I. J. (2010). Effect of polyethylene glycol induced drought stress on physio-hormonal attributes of soybean. Pak. J. Bot. 42, 977–986.
Hernandez-Garcia, C. M., and Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Hobo, T., Asada, M., Kowyama, Y., and Hattori, T. (1999). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19, 679–689. doi: 10.1046/j.1365-313x.1999.00565.x
Hossain, M. A., Bhattacharjee, S., Armin, S.-M., Qian, P., Xin, W., Li, H.-Y., et al. (2015). Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front. Plant Sci. 6:420. doi: 10.3389/fpls.2015.00420
Hulm, J. L., McIntosh, K. B., and Bonham-Smith, P. C. (2005). Variation in transcript abundance among the four members of the Arabidopsis thaliana RIBOSOMAL PROTEIN S15a gene family. Plant Sci. 169, 267–278. doi: 10.1016/j.plantsci.2005.04.001
Ibraheem, O., Botha, C. E., and Bradley, G. (2010). In silico analysis of cis-acting regulatory elements in 5′ regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput. Biol. and Chem. 34, 268–283. doi: 10.1016/j.compbiolchem.2010.09.003
Jones, S. (2004). An overview of the basic helix-loop-helix proteins. Genome Biol. 5:226. doi: 10.1186/gb-2004-5-6-226
Kawasaki, S., Borchert, C., Deyholos, M., Wang, H., Brazille, S., Kawai, K., et al. (2001). Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13, 889–905. doi: 10.1105/tpc.13.4.889
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Khan, M. A., Naeem, M., and Iqbal, M. (2014). Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur. J. Plant Pathol. 139, 27–37. doi: 10.1007/s10658-014-0377-x
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Kim, J. S., Mizoi, J., Yoshida, T., Fujita, Y., Nakajima, J., Ohori, T., et al. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52, 2136–2146. doi: 10.1093/pcp/pcr143
Kim, K.-Y., Park, S.-W., Chung, Y.-S., Chung, C.-H., Kim, J.-I., and Lee, J. H. (2004). Molecular cloning of low-temperature-inducible ribosomal proteins from soybean. J. Exp. Bot. 55, 1153–1155. doi: 10.1093/jxb/erh125
Kim, Y., Lee, M. S., Kim, H. D., and Kim, J. (2016). Ribosomal protein S3 (rpS3) secreted from various cancer cells is N-linked glycosylated. Oncotarget 7:80350. doi: 10.18632/oncotarget.10180
Komili, S., Farny, N. G., Roth, F. P., and Silver, P. A. (2007). Functional specificity among ribosomal proteins regulates gene expression. Cell 131, 557–571. doi: 10.1016/j.cell.2007.08.037
Kyrpides, N. C., Woese, C. R., and Ouzounis, C. A. (1996). KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 21, 425–426. doi: 10.1016/S0968-0004(96)30036-4
Law, M. J., Linde, M. E., Chambers, E. J., Oubridge, C., Katsamba, P. S., et al. (2006). The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res. 34, 275–285. doi: 10.1093/nar/gkj436
Liu, W., Xiong, C., Yan, L., Zhang, Z., Ma, L., Wang, Y., et al (2017). Transcriptome analyses reveal candidate genes potentially involved in al stress response in alfalfa. Front. Plant Sci. 8:26. doi: 10.3389/fpls.2017.00026
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, H., Zhu, Y. F., Xiong, J., Wang, R., and Jia, Z. (2015). Potential extra-ribosomal functions of ribosomal proteins in Saccharomyces cerevisiae. Microbiol. Res. 177, 28–33. doi: 10.1016/j.micres.2015.05.004
Maguire, B. A., and Zimmermann, R. A. (2001). The ribosome in focus. Cell 104, 813–816. doi: 10.1016/S0092-8674(01)00278-1
Mahajan, S., and Tuteja, N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. doi: 10.1016/j.abb.2005.10.018
McIntosh, K. B., and Bonham-Smith, P. C. (2006). Ribosomal protein gene regulation: what about plants? Botany 84, 342–362. doi: 10.1139/b06-014
Moin, M., Bakshi, A., Saha, A., Udaya Kumar, M., Reddy, A. R., Rao, K. V., et al. (2016a). Activation tagging in indica rice identifies ribosomal proteins as potential targets for manipulation of water-use efficiency and abiotic stress tolerance in plants. Plant Cell Environ. 39, 2440–2459. doi: 10.1111/pce.12796
Moin, M., Bakshi, A., Saha, A., Dutta, M., Madhav, S. M., and Kirti, P. B. (2016b). Rice ribosomal protein large subunit genes and their spatio-temporal and stress regulation. Front. Plant Sci. 7:1284. doi: 10.3389/fpls.2016.01284
Morimoto, T., Suzuki, Y., and Yamaguchi, I. (2002). Effects of partial suppression of ribosomal protein S6 on organ formation in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 66, 2437–2443. doi: 10.1271/bbb.66.2437
Naora, H., and Naora, H. (1999). Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol. Cell Biol. 77, 197–205. doi: 10.1046/j.1440-1711.1999.00816.x
Nesterchuk, M. V., Sergiev, P. V., and Dontsova, O. A. (2011). Post-translational modifications of ribosomal proteins in Escherichia coli. Acta Naturae 3, 22–33.
Nishimura, T., Wada, T., and Okada, K. (2004). A key factor of translation reinitiation, ribosomal protein L24, is involved in gynoecium development in Arabidopsis. Biochem. Soc. Trans. 32, 611–613. doi: 10.1042/BST0320611
Novoseler, M., Hershkovits, G., and Katcoff, D. J. (2005). Functional domains of the yeast chromatin protein Sin1p/Spt2p can bind four-way junction and crossing DNA structures. J. Biol. Chem. 280, 5169–5177. doi: 10.1074/jbc.M406249200
Ouyang, B., Yang, T., Li, H., Zhang, L., Zhang, Y., Zhang, J., et al. (2007). Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J. Exp. Bot. 58, 507–520. doi: 10.1093/jxb/erl258
Pandey, S., Subramanaym Reddy, C., Yaqoob, U., Negi, Y. K., and Arora, S. (2015). In silico analysis of cis acting regulatory elements CAREs in upstream regions of ascorbate glutathione pathway genes from Oryza sativa. Biochem. Physiol. 4:2. doi: 10.4172/2168-9652.1000159
Park, D. S., Sayler, R. J., Hong, Y. G., Nam, M. H., and Yang, Y. (2008). A method for inoculation and evaluation of rice sheath blight disease. Plant Dis. 92, 25–29. doi: 10.1094/PDIS-92-1-0025
Pinon, V., Etchells, J. P., Rossignol, P., Collier, S. A., Arroyo, J. M., Martienssen, R. A., et al. (2008). Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135, 1315–1324. doi: 10.1242/dev.016469
Planta, R. J. (1997). Regulation of ribosome synthesis in yeast. Yeast 13, 1505–1518. doi: 10.1002/(SICI)1097-0061(199712)13:16<1505::AID-YEA229>3.0.CO;2-I
Polikanov, Y. S., Melnikov, S. V., Söll, D., and Steitz, T. A. (2015). Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344. doi: 10.1038/nsmb.2992
Ramakrishnan, V., and White, S. W. (1998). Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends Biochem. Sci. 23, 208–212. doi: 10.1016/S0968-0004(98)01214-6
Revenkova, E., Masson, J., Koncz, C., Afsar, K., Jakovleva, L., and Paszkowski, J. (1999). Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J. 18, 490–499. doi: 10.1093/emboj/18.2.490
Rombauts, S., Déhais, P., Rouzé, P., and Van Montagu, M. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Rushton, P. J., Reinstädler, A., Lipka, V., Lippok, B., and Somssich, I. E. (2002). Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen-and wound-induced signaling. Plant Cell 14, 749–762. doi: 10.1105/tpc.010412
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. doi: 10.3389/fpls.2016.00571
Sazegari, S., Niazi, A., and Ahmadi, F. S. (2015). A study on the regulatory network with promoter analysis for Arabidopsis DREB-genes. Bioinformation 11:101. doi: 10.6026/97320630011101
Shu, C. J., and Zhulin, I. B. (2002). ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 27, 3–5. doi: 10.1016/S0968-0004(01)02036-9
Son, O., Kim, S., Shin, Y. J., Kim, W. Y., Koh, H. J., and Cheon, C. I. (2015). Identification of nucleosome assembly protein 1 (NAP1) as an interacting partner of plant ribosomal protein S6 (RPS6) and a positive regulator of rDNA transcription. Biochem. Biophys. Res. Commun. 465, 200–205. doi: 10.1016/j.bbrc.2015.07.150
Sutoh, K., and Yamauchi, D. (2003). Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 34, 635–645. doi: 10.1046/j.1365-313X.2003.01753.x
Szakonyi, D., and Byrne, M. E. (2011). Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 65, 269–281. doi: 10.1111/j.1365-313X.2010.04422.x
Takakura, Y., Sofuku, K., Tsunashima, M., and Kuwata, S. (2016). Lentiavidins: Novel avidin-like proteins with low isoelectric points from shiitake mushroom (Lentinulaedodes). J. Biosci. Bioeng. 121, 420–423. doi: 10.1016/j.jbiosc.2015.09.003
Turkina, M. V., Årstrand, H. K., and Vener, A. V. (2011). Differential phosphorylation of ribosomal proteins in Arabidopsis thaliana plants during day and night. PLoS ONE 6:e29307. doi: 10.1371/journal.pone.0029307
Uechi, T., Nakajima, Y., Nakao, A., Torihara, H., Chakraborty, A., Inoue, K., et al. (2006). Ribosomal protein gene knockdown causes developmental defects in zebrafish. PLoS ONE 1:e37. doi: 10.1371/journal.pone.0000037
Van Lijsebettens, M., Vanderhaeghen, R., De Block, M., Bauw, G., Villarroel, R., and Van Montagu, M. (1994). An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 13:3378.
Wang, J., Lan, P., Gao, H., Zheng, L., Li, W., and Schmidt, W. (2013). Expression changes of ribosomal proteins in phosphate-and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics 14:783. doi: 10.1186/1471-2164-14-783
Wang, M., Hu, Y., and Stearns, M. E. (2009). RPS2: a novel therapeutic target in prostate cancer. J. Exp. Clin. Cancer Res. 28:6. doi: 10.1186/1756-9966-28-6
Wang, Z., Hou, J., Lu, L., Qi, Z., Sun, J., Gao, W., et al. (2013). Small ribosomal protein subunit S7 suppresses ovarian tumorigenesis through regulation of the PI3K/AKT and MAPK pathways. PLoS ONE 8:e79117. doi: 10.1371/journal.pone.0079117
Warner, J. R., and McIntosh, K. B. (2009). How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11. doi: 10.1016/j.molcel.2009.03.006
Wass, M. N., Kelley, L. A., and Sternberg, M. J. (2010). 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 38, W469–W473. doi: 10.1093/nar/gkq406
Weijers, D., Franke-van Dijk, M., Vencken, R. J., Quint, A., Hooykaas, P., and Offringa, R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–4299.
Xiong, Y., and Sheen, J. (2015). Novel links in the plant TOR kinase signaling network. Curr. Opin. Plant Biol. 28, 83–91. doi: 10.1016/j.pbi.2015.09.006
Yan, H., Chen, D., Wang, Y., Sun, Y., Zhao, J., Sun, M., et al. (2016). Ribosomal protein L18aB is required for both male gametophyte function and embryo development in Arabidopsis. Sci. Rep. 6:31195. doi: 10.1038/srep31195
Zheng, A., Lin, R., Zhang, D., Qin, P., Xu, L., Ai, P., et al. (2013). The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat. Commun. 4:1424. doi: 10.1038/ncomms2427
Keywords: rice, ribosomal protein small subunit (RPS) genes, ribosomal proteins, stress responses, gene expression
Citation: Saha A, Das S, Moin M, Dutta M, Bakshi A, Madhav MS and Kirti PB (2017) Genome-Wide Identification and Comprehensive Expression Profiling of Ribosomal Protein Small Subunit (RPS) Genes and their Comparative Analysis with the Large Subunit (RPL) Genes in Rice. Front. Plant Sci. 8:1553. doi: 10.3389/fpls.2017.01553
Received: 24 April 2017; Accepted: 25 August 2017;
Published: 15 September 2017.
Edited by:
Andy Pereira, University of Arkansas, United StatesReviewed by:
Ing-Feng Chang, National Taiwan University, TaiwanEls J. M. Van Damme, Ghent University, Belgium
Copyright © 2017 Saha, Das, Moin, Dutta, Bakshi, Madhav and Kirti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: P. B. Kirti, pbkirti@uohyd.ac.in
 Anusree Saha
Anusree Saha Shubhajit Das1
Shubhajit Das1 Mazahar Moin
Mazahar Moin M. S. Madhav
M. S. Madhav P. B. Kirti
P. B. Kirti