Prevalence and Serotype Diversity of Salmonella in Apparently Healthy Cattle: Systematic Review and Meta-Analysis of Published Studies, 2000–2017
- 1Department of Microbiology, Immunology and Veterinary Public Health, College of Veterinary Medicine and Agriculture, Addis Ababa University, Bishoftu, Ethiopia
- 2Department of Veterinary Public Health and Food Safety, Faculty of Veterinary Medicine, Ghent University, Merelbeke, Belgium
- 3Food Animal Environmental Systems Research Unit, United States Department of Agriculture, Agricultural Research Service, Bowling Green, KY, United States
- 4Department of Veterinary Biomedical Sciences, College of Veterinary Medicine, Long Island University, Greenvale, NY, United States
- 5Department of Nutrition, Genetics and Ethology, Faculty of Veterinary Medicine, Ghent University, Merelbeke, Belgium
Salmonellosis is a leading cause of foodborne illnesses in humans with cattle being one of the reservoirs for Salmonella. We estimated a pooled prevalence of Salmonella in apparently healthy cattle and examined serotype diversity through systematic review and meta-analysis of studies published between 2000 and 2017. Peer reviewed publications reporting the prevalence of Salmonella in cattle were searched through five electronic databases (PubMed, Google scholar, Agricola, Scopus, CAB direct) and through manual search. We obtained 71 publications with 75 datasets consisting a total of 52,766 animals examined and 5,010 Salmonella positive cattle from 29 countries in six continents (except from Antarctica). Pooled prevalence of Salmonella in cattle was 9% (95% confidence interval: 7–11%). Significantly high heterogeneity (I2 = 98.7%, P < 0.01) was observed among all studies as well as within continents. Prevalence varied from 2% (Europe) to 16% (North America). Overall, 143 different serotypes were reported with the most diverse serotypes being reported from Africa (76 different serotypes) followed by North America (49 serotypes). The 10 most frequently reported serotypes (Montevideo, Typhimurium, Kentucky, Meleagridis, Anatum, Cerro, Mbandaka, Muenster, Newport, and Senftenberg) accounted for 65% of the isolates for which specific serotype information was reported. Salmonella Montevideo and S. Dublin are the most frequently reported serotypes in North America and Europe, respectively, while S. Typhimurium was the most frequent in Africa, Asia and Australasia. Our results indicated variability both in the prevalence and serotype diversity of Salmonella in cattle across continents. Although all Salmonella serotypes are potentially pathogenic to humans, five (Montevideo, Typhimurium, Anatum, Mbandaka, and Newport) of the top 10 serotypes identified in this study are among the serotypes most commonly associated with clinical illnesses in humans.
Background
Foodborne illnesses pose public health and economic burdens both in developed and developing countries (1, 2). Annually, foodborne illnesses are responsible for an estimated 600 million cases, 420,000 deaths, and 33 million disability adjusted life years lost worldwide. Salmonella is a major cause of foodborne illnesses in humans (3–5). Salmonella are Gram-negative, non-spore forming, mostly motile, facultative anaerobic bacilli within the family Enterobacteriaceae. The species Salmonella enterica consists of six subspecies and more than 2,579 serovars (6, 7). Based on the clinical profiles of infections caused in humans S. enterica can be divided into typhoidal—which are human specific—and non-typhoidal Salmonella (NTS)—having a broad host range (6). The NTS serotypes are leading causes of bacterial diarrhea and invasive bacterial infections in young children, the elderly and the immune-compromised individuals throughout the world. Salmonella Typhimurium and S. Enteritidis together account for approximately 50% of all isolates globally reported from human clinical cases (8–10). The global incidence of diarrheal disease due to the NTS accounts for about 94 million enteric infections each year, of which 80.3 million cases are considered foodborne and resulting in 155,000 human deaths annually (11). Human salmonellosis is also recognized as an important socioeconomic disease posing considerable economic burden in the world (12, 13).
Salmonella colonizes mainly the intestinal tracts of humans and animals including cattle. Foods of animal origin are important sources of Salmonella infections in humans (13–18). Humans acquire the infection mainly through consumption of contaminated products including beef and beef products (19), by direct contact with infected animals or their environment (20) and by direct human-to-human transmission (21). Carcass contamination with Salmonella during slaughter, particularly under unsatisfactory hygienic operations, poses a significant public health risk (22–25). The transfer of NTS to food processing plants and equipment used for food preparation also plays an important role, ultimately leading to the risk of salmonellosis after the consumption of contaminated foods (21). Knowledge about the overall occurrence of Salmonella and the diversity of serotypes in cattle provides important information for decision making and to promote reliable efforts toward prevention and control of foodborne salmonellosis associated with cattle. Therefore, the objectives of this study were to determine the prevalence of Salmonella in apparently healthy cattle, and to assess the diversity of Salmonella serotypes associated with cattle production systems through a systematic review and meta-analysis of peer-reviewed publications between 2000 and 2017.
Methods
Systematic Review of the Literature
Preferred reporting items for systematic reviews and meta-analysis protocols (PRISMA-P) 2015 checklist was followed for the systematic review and meta-analysis of studies reporting Salmonella serotypes and prevalence in cattle (26). Five electronic databases were searched: PubMed (http://www.ncbi.nlm.nih.gov/pubmed), Google scholars (https://scholar.google.com/), Agricola (http://agricola.nal.usda.gov/), Scopus (http://www.scopus.com/), and CAB direct (http://www.cabdirect.org/). Additional publications were obtained by the manual scanning of the reference list from the retrieved publications. Salmonella, cattle, and prevalence were the main key words used for the search. The search was conducted with alternative terms for each key term using the general protocol [Salmonella AND (cattle OR bovine OR heifer OR bull OR bullock OR ruminant OR steer OR cow OR cull OR calf OR calves OR yearling OR beef OR dairy OR feedlot) AND (prevalence OR isolation OR identification OR “antimicrobial resistance” OR “antimicrobial susceptibility”)], that was modified and tailored to search strategies of each database when needed.
Relevance Screening
The retrieved articles were imported to Refworks to manage and exclude duplicated studies (27). The duplicated records were excluded manually after making the bibliography list and prior to the eligibility assessment. The eligibility criteria were: (i) articles published in English between January 1, 2000 (since full articles could not be available online, publications prior to 2000 were not considered) and January 4, 2017 (the last date of literature search); (ii) reported on apparently healthy cattle (no statement is given about the inclusion of sick/diseased animals) from different production categories (dairy, beef, mixed) and sample sources (slaughter plant/abattoir/slaughter house, dairy farm, beef farm, ranch, feedlot, grazing point, market place, mixed cattle farm); (iii) samples collected from the intestinal content (feces from the rectum and other intestinal contents); (iv) prevalence report from any part of the world; and (v) cross-sectional study in which animal level prevalence was reported or could be calculated from the information provided in the publication during data extraction. The exclusion criteria were: (i) irrelevant records to the objective of the review; (ii) articles on sick or diseased cattle; (iii) non-cross-sectional study design; (iv) report on inappropriate samples such as ground or pen fecal or pooled fecal samples from which animal level prevalence was unknown, lymph nodes, rumen contents or other body parts of cattle; (v) when only citations or abstracts were available.
Data Extraction
A peer-reviewed publication that describes prevalence of Salmonella in cattle was considered as a study unit. Cattle were considered positive for Salmonella when samples from the intestinal contents were tested and confirmed positive. When different prevalence reports in the content from various sites of intestinal tract were observed in a single study, we considered this one with the highest proportion for better precision to minimize under estimation. From each eligible publication, we extracted the following information: author, year of publication, year of study, study location (country and continent), detection method, production type (beef, dairy, and mixed), sampling location (abattoir, farm, market, ranch, grazing points, feedlot), age (calves and adults), amount of tested samples, sample size, number of Salmonella positive samples and serotypes identified, and number within each serotype. The extracted information was entered to a Microsoft excel spread sheet for quality assessment and data preparation for analysis.
Data Analysis
Frequency distributions were used to describe the characteristics of the eligible publications and the diversity and proportion of Salmonella serotypes. Meta-analysis was conducted using the metaprop-one package (28), a Stata based program specifically designed for binominal data, that allows the computation of studies with 0 or 100% prevalence. Analysis was done in STATA version 14 (29). The prevalence of Salmonella in cattle was defined as the proportion of Salmonella positives based on the intestinal content samples. The pooled prevalence of Salmonella was computed by meta-analysis from the prevalence values of the individual publications by accounting for potential heterogeneity between studies and weighted on sample size (30). A logistic-normal random-effects model was used to model the within-study variability. The 95% confidence intervals (CIs) for the proportion of cattle Salmonella positive for the separate publications and their pooled prevalence was computed with the exact binomial method with the Freeman-Tukey double arcsine transformation which gives the CIs within admissible values. Further analysis of sub-groups of the overall estimate was performed according to age, production type, detection method, and continent categories. Heterogeneity of the effect sizes among the publications was assessed by Cochrane Q test and inverse variance index (I2) test and quantified as recommended by Higgins and Thompson (31). A P < 0.01 was set as an indication of a statistically significant heterogeneity. The basic results from the meta-analysis were visually presented using forest plots. Frequency distributions were used to describe the characteristics of the eligible studies and the diversity and proportion of Salmonella serotypes.
Results
Systematic Review of the Literature
A flow chart showing the systematic literature search procedure is shown in Figure 1. A total of 2,655 records were retrieved from the five search engines (PubMed, Google scholar, Agricola, Scopus, and CAB direct) and by manual search. After de-duplicating the references, 1,753 publications were retained for further screening. After relevance screening of the titles and abstracts, 1,625 articles were excluded resulting in 128 potentially eligible full articles. Further in-depth eligibility assessment of the full articles resulted in 71 eligible publications for data extraction and analysis. The references of all the eligible articles are listed in Supplementary Table 1.
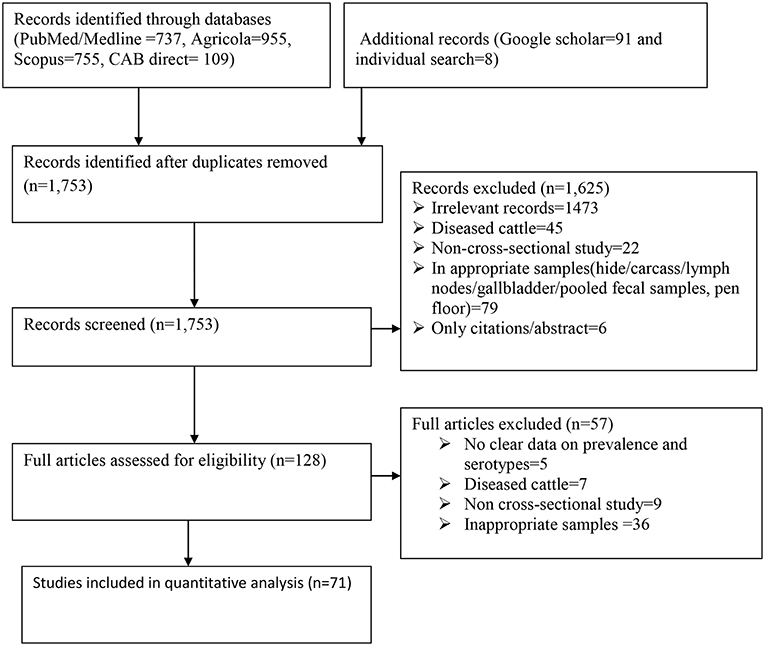
Figure 1. Flow diagram for the selection of studies included in the meta-analysis for the prevalence of Salmonella in apparently healthy cattle.
Data Extraction and Meta-Analysis
Data were extracted from the 71 peer-reviewed publications comprising 75 data sets. Two separate datasets were extracted from three publications (32–34) based on age and from one study (35) based on sampling points. Therefore, 75 data sets (hereafter referred to as studies) comprising fecal samples or swabs from 52,766 animals were included in the meta-analysis. Salmonella was detected in 5,010 of the animals. Over two-thirds (68%) of the studies used ≤ 10 g of feces, and 91% of the studies used traditional culture methods for the detection of Salmonella. The publications represented 29 countries across six continents except Antarctica. While 80% of the countries were represented by one or two publications, the United States was the most represented with 25 publications. Forty percent of the studies were conducted on samples collected at processing plants. Characteristics of the publications are shown in Table 1.
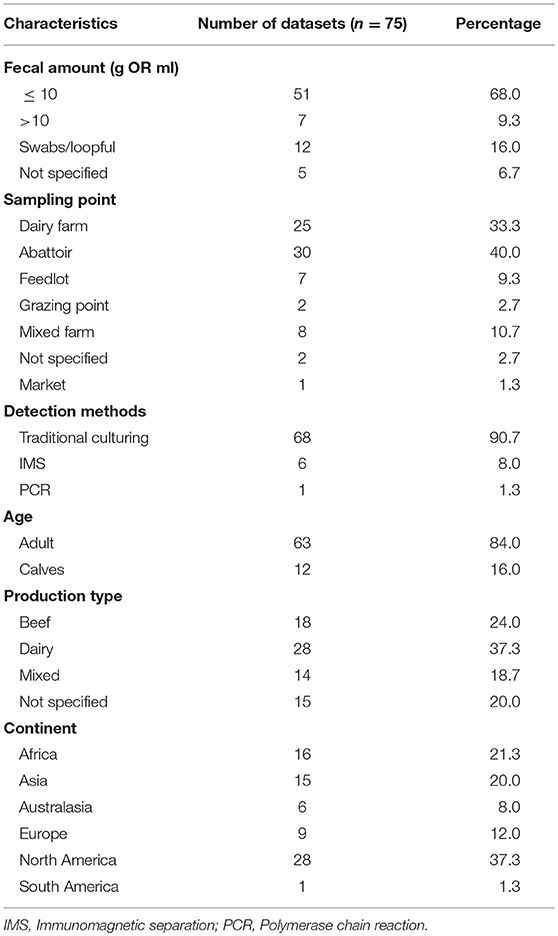
Table 1. Description of the eligible publications included in the systematic review and meta-analysis of Salmonella in apparently healthy cattle.
Overall pooled prevalence of Salmonella in cattle was 9% (95% CI: 7–11%). Results of individual studies along with the effect of sizes are shown in Figure 2. Study prevalence values ranged from 0 to 95%. Test of heterogeneity demonstrated the presence of a high degree of heterogeneity (I2 = 98.7%, P < 0.01) among the studies. To account for some of the variability separate stratified meta-analyses were performed by age, production type, detection method, and continent (Table 2). The pooled prevalence of Salmonella is higher in the adult cattle [9% (95% CI: 7–12%)] than in the calves [6% (95% CI: 2–11%)], in beef cattle [14% (95% CI: 7–23%)] than in other production types, and in North America [16% (95% CI:12–20%)] than in other continents. Studies within each category of the strata defined by detection method and continent, showed significantly high degrees of heterogeneities (P < 0.01). However, no significant heterogeneity was observed between the age groups, among production types and when comparing only between immunomagnetic separation (IMS) and non-IMS detection methods (P > 0.01).
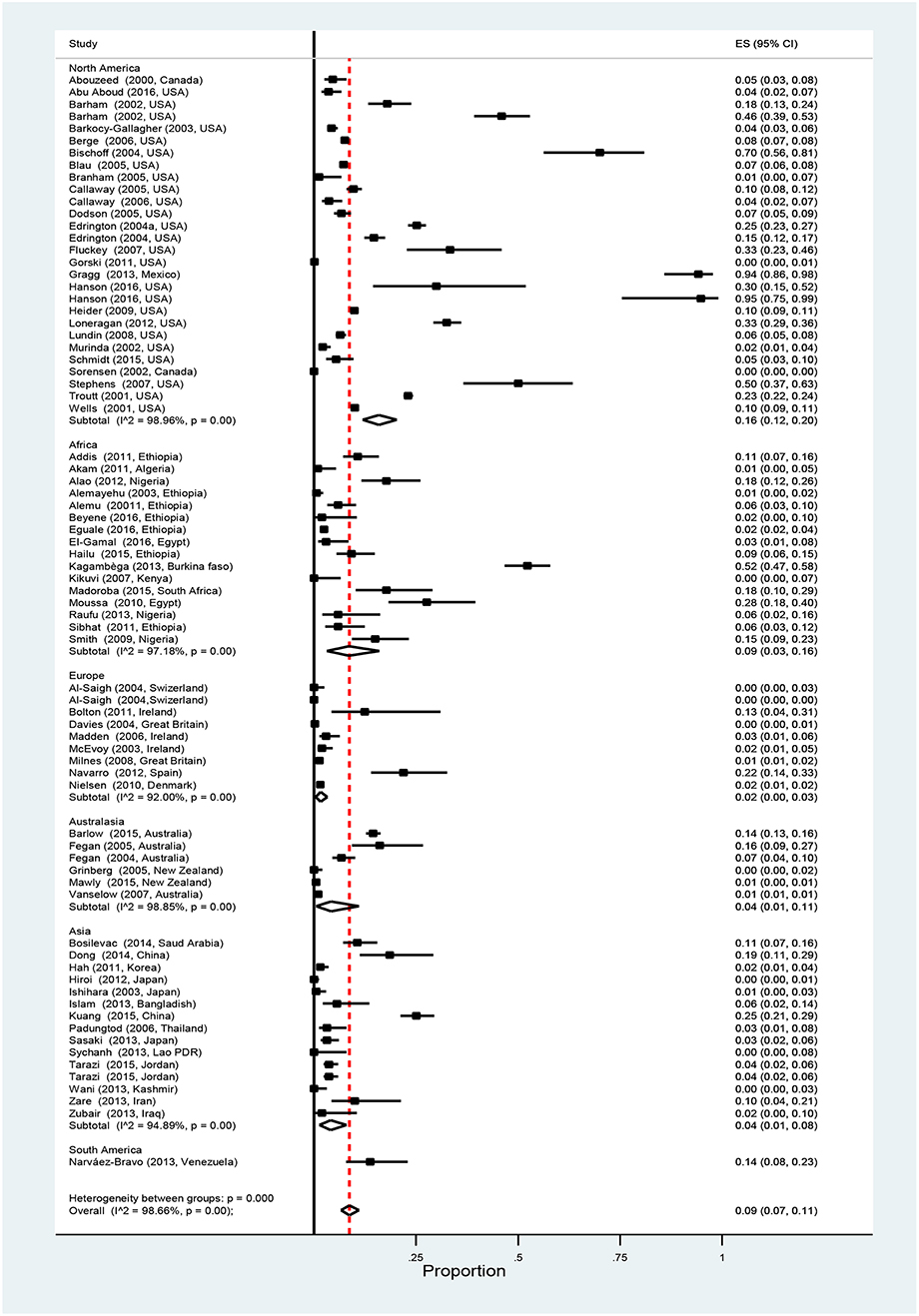
Figure 2. Forest plot showing estimated individual and overall Salmonella prevalence in apparently healthy cattle (ES, effect size; CI, confidence interval; I2, Inverse variance index).
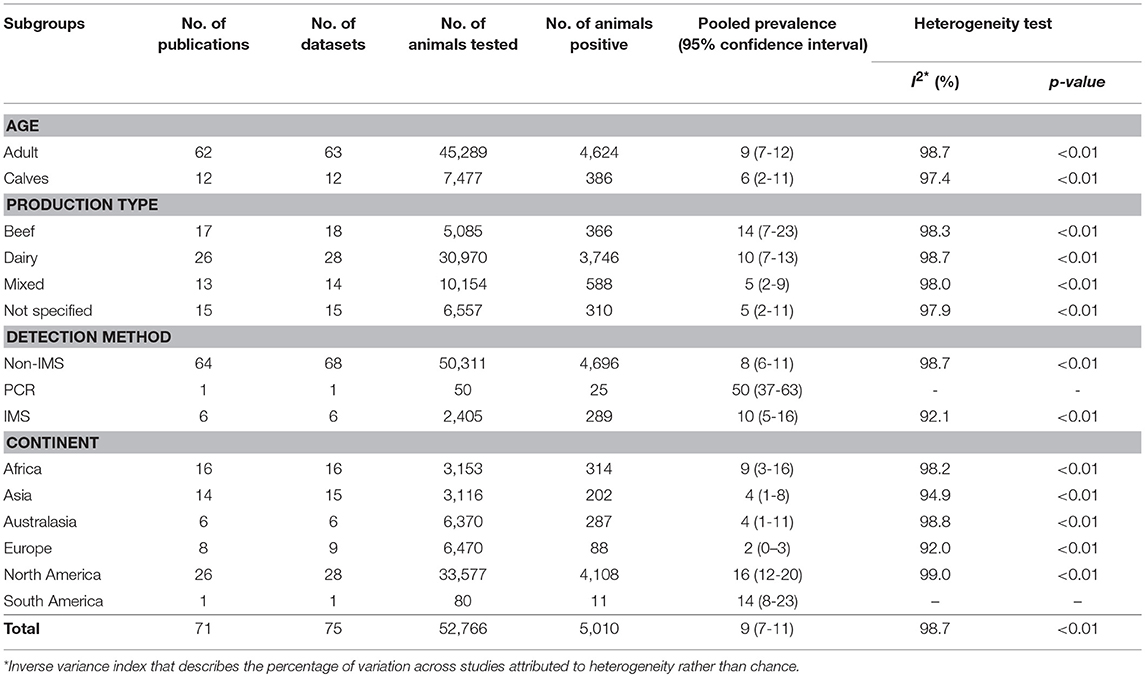
Table 2. Pooled prevalence of Salmonella in apparently healthy cattle determined by meta-analysis of 75 datasets studies by age, production type, detection method, and continent.
Diversity of Serotypes
Serotype information was not reported for 1,926 Salmonella positive cattle from a total of 16,175 cattle examined in 27 publications representing 29 data sets. In the remaining 44 publications representing 46 datasets for which serotype information was available, 3,191 Salmonella isolates were reported from 3,084 Salmonella positive cattle from a total of 36,591 cattle examined. Among the 3,191 isolates with serotyping information, specific serotypes were reported in 91.6% (2,923/3,191) of the isolates while 2.8% of the isolates were untypable, and the remaining 5.6% were reported as “other serotypes” where the list of which was not stated in the publication.
Overall, 143 different serotypes were reported among the 2,923 Salmonella isolates listed in the data sets included in the meta-analysis. The most frequently (with ≥1%) reported serotypes are shown in Table 3 and the list of serotypes (<1%) categorized as “others” in the latter table is presented in the Supplementary Table 2. The 10 most frequently reported cattle associated serotypes across all studies were S. Montevideo, Typhimurium, Kentucky, Meleagridis, Anatum, Cerro, Mbandaka, Muenster, Newport, and Senftenberg. These 10 most frequently isolated serotypes comprised 69.5% (2,032/2,923) of total isolates for which specific serotypes were reported. There were variations in the frequency and diversity of Salmonella serotypes in the six continents for which publications were retrieved (Table 4). S. Montevideo was the most frequent reported serotype from North America, while this serotype did not belong to the five most frequently reported serotypes in most other continents. Salmonella Typhimurium was the most frequently reported serotype in Africa, Asia, and Australasia, while S. Dublin was the most frequently reported serotype in Europe. The most diverse serotypes were reported from Africa (76 different serotypes) followed by North America (49 different serotypes), Australasia (39 serotypes), Asia (23 serotypes), Europe (12 serotypes), and South America (2 serotypes).
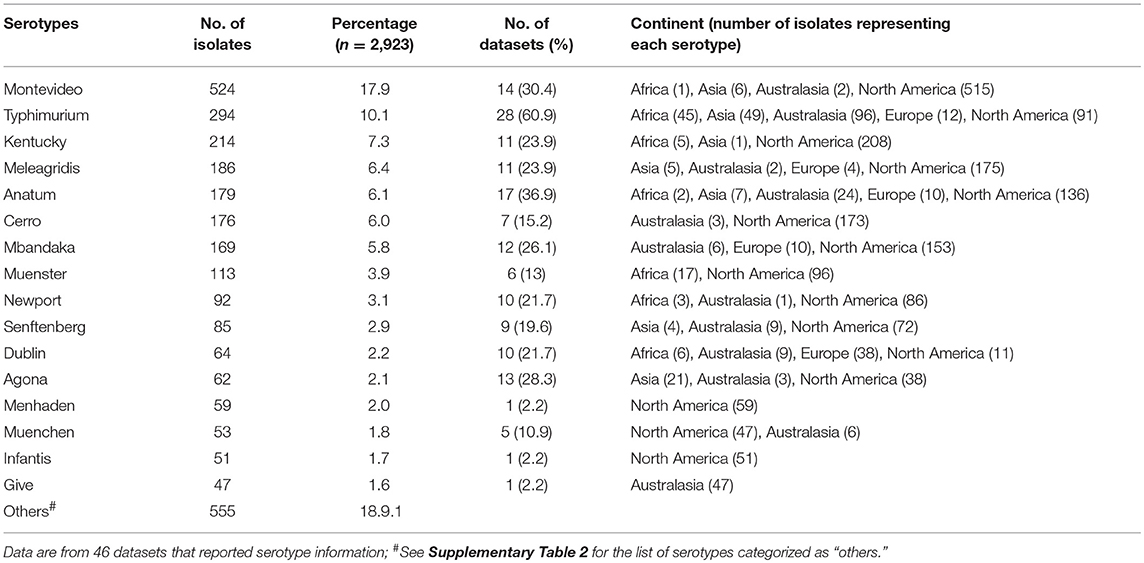
Table 3. Salmonella isolates by serotype in descending order of frequency across studies reporting specific serotypes.
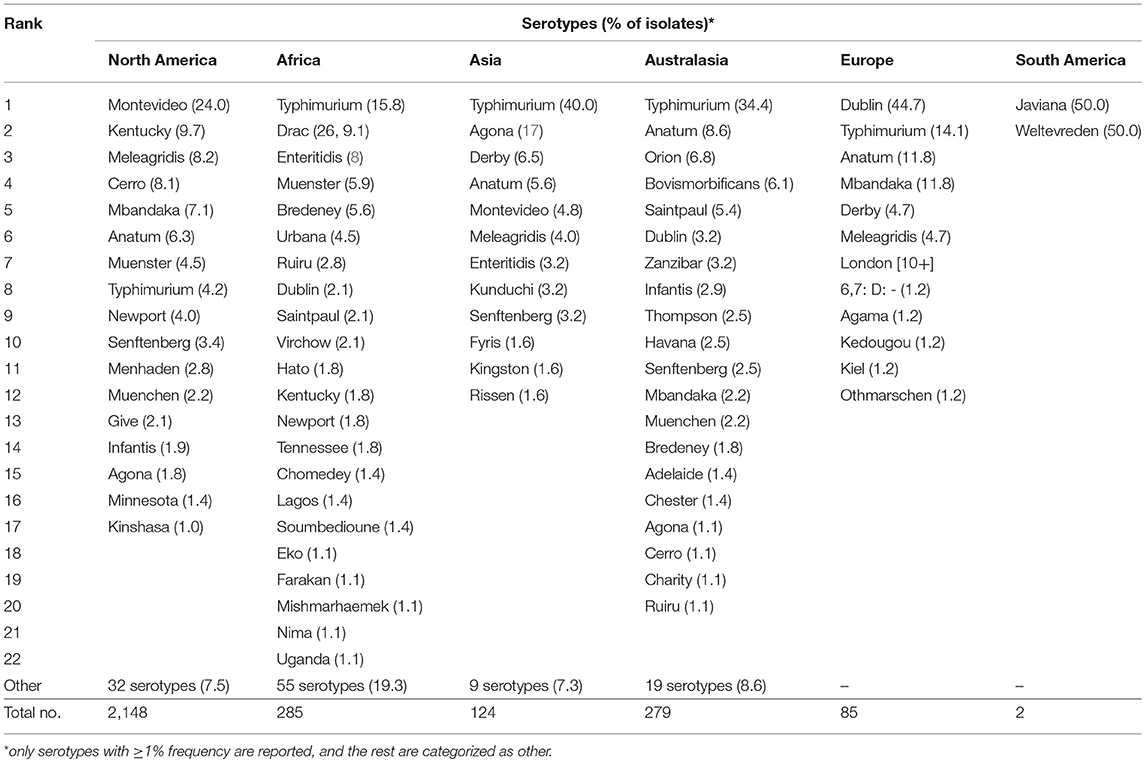
Table 4. Salmonella isolates by serotype within six continents in descending order of frequency in studies reporting specific serotypes.
Discussion
To the best of our knowledge, this is the first estimate of the overall Salmonella prevalence and the diversity of serotypes in apparently healthy cattle. We used a systematic method to identify articles reporting the prevalence of Salmonella and the serotypes in such cattle, followed by a quantitative meta-analysis to estimate the overall prevalence of Salmonella at the global level.
Salmonella colonizes the gastrointestinal tract of food animals (7) and is shed via feces (36–39). Cattle are asymptomatic carriers or reservoirs for Salmonella and may function as a source of foodborne infection (8, 23, 24). A number of serotypes frequently isolated from humans have been isolated from sick or asymptomatic cattle and some human cases have also been linked to direct exposure to cattle (20). Knowing the prevalence and diversity of Salmonella serotypes in cattle can provide important information necessary to develop preventive measures and strategies at different stages of the food chain such as application hazard analysis and critical control point (HACCP) programs in beef and milk production industries to ensure food safety (40).
There was high heterogeneity in the estimated Salmonella prevalence among the studies included in the analysis. The Salmonella prevalence can vary depending on the detection method used, the amount of sample processed, production type, and geographical variation in the distribution of the Salmonella (32, 41). The overall pooled prevalence of 9% is higher compared to other reported national level prevalence values ranging from 0.2 to 7.1% (42–46). This is not surprising since our meta-analysis provides a precise estimate (with narrow confidence interval) as it includes a higher amount of samples and total number of positive cattle for Salmonella by pooling 75 datasets from 71 publications.
The prevalence was higher in the adult cattle [9% (95%CI: 7–12%)] than in the young age group [6%(95%CI: 2–11%)]. Although the effect of age needs further investigation, this variation can presumably be in part due to variation in the number of studies included in the meta-analysis in each age group. In the young age group there were 12 publications representing only 14.2% (n = 7,477) of total cattle examined compared to 63 publications in the adult cattle with 86% of the total cattle examined. Over 70% of the publications were conducted at processing plants and in culled dairy cows destined for slaughter perhaps because of the higher public health significance at the final stage of production chain that is close to consumers (47). Even though Salmonella colonizes the intestinal tracts of cattle, there is no difference in the colonization and shedding of Salmonella between healthy calves and adult cattle (7). However, a higher prevalence of Salmonella shedding animals occurs when asymptomatic chronically infected carrier cattle are present on the farm and stay on the farm for long periods (45), which may contribute to transmission and persistence of Salmonella on the farm.
Although not statistically significant, the prevalence was higher in beef cattle compared to dairy cattle. This apparent difference can be attributed to how the animals were sampled. In most of the studies culled dairy cows were sampled at farms before shipment as opposed to beef cattle which were commonly sampled at the processing plants. Temporary restriction or complete feed withdrawal (48) and exposure to stress such as transport (42, 49, 50) can result in increased fecal shedding of Salmonella in feedlot cattle prior to slaughter.
Variations in prevalence that ranged from 2% (Europe) to 16% (North America) in various continents of the world could partly be attributed to the differences in the number of publications and the number of cattle samples included in the analysis. For North America, 26 publications (28 data sets) were retrieved consisting of 33,577 cattle samples, being the majority of the articles. In contrast, the very low prevalence estimate (2%) observed in Europe, was estimated only from 8 publications (9 data sets) in which 6,470 cattle sampled. The prevalence in South America was 14%, however this does not represent the pooled estimate as only one article was included in the analysis. The differences might also be associated with the differences in the monitoring and surveillance mechanisms among the continents (51).
Difference in the prevalence was also observed among categories of detection methods. In the majority (91%) of the studies, Salmonella was detected using traditional culturing methods which are in general considered less sensitive methods. Limited number of studies used immunomagnetic separation beads or PCR. Moreover, variation in the sensitivity of culture detection methods can influence the prevalence and consequently the observed heterogeneity (52).
In this systematic review, S. Montevideo and S. Typhimurium were the two most frequent and dominant serotypes reported where S. Montevideo was majorly reported from North America. Salmonella Typhimurium is one of the major serotypes that accounted for human clinical cases globally (10). Human infections and outbreaks due to S. Montevideo is also increasing around the globe (53)and reported in the USA, Europe, Australia, and Asia (54–56). There were differences in the most commonly reported serotypes and their proportions among different continents. Salmonella Typhimurium which is historically associated with cattle ranked number one in Africa, Asia, and Australasia. In North America and Europe, however, S. Montevideo and S. Dublin ranked number one, respectively. The implication of the shift in serotype with respect to public health requires further study. Interestingly, among the top 10 Salmonella serotypes identified in this study, S. Montevideo, S. Typhimurium, S. Anatum, S. Mbandaka, and S. Newport are among the World Health Organization's top 20 serotypes associated with human salmonellosis across the world (52). Spatial and temporal effects on the distribution and diversity of Salmonella have been reported (57, 58), which may explain the observed differences in the serotype diversity among the studies reporting Salmonella. Some of the serotypes reported in the present review were identified as the dominant serotypes elsewhere in cattle at varying proportions. For instance, in the USA, S. Newport (48.7%) and S. Typhimurium (7.1%) (59); in Ethiopia, S. Typhimurium (17.4%), S. Newport (13%) and S. Anatum (5.8%) (42), and in Europe, S. Typhimurium (38.6%) were reported to be the most frequent and dominant serotypes (60). On the contrary, none of these serotypes were reported from the national survey of Salmonella serotypes in cattle carried out in Japan (41).
All non-typhoidal Salmonella serotypes except a few serotypes which are host-specific, can potentially cause disease in humans and reside in one or more animal species (61). Salmonella serotypes were reported to be linked to several outbreaks following the consumption of contaminated beef, milk, and products thereof (62). S. Enteritidis and S. Typhimurium are the two most important serotypes transmitted from animals to humans in most parts of the world (51, 60, 63, 64). In the USA, 29 cases of diarrheal illness caused by S. Typhimurium were associated with the consumption of raw milk or raw-milk products from dairy cattle (65). During the period 1973–2011, of the 1,965 Salmonella outbreaks where a food vehicle was implicated, 96 were attributed to beef, accounting for 3,684 illnesses in USA. S. Newport and S. Typhimurium accounted for 18 and 17% of illnesses, and 29 and 18% of hospitalizations, respectively (19). The multidrug-resistant S. Typhimurium DT104 has also been associated with outbreaks related to beef contamination and resulted in hospitalization rates twice as that of other foodborne salmonellosis cases (65). From a total of 1,168 foodborne outbreaks of human salmonellosis in 2013 reported by the European member states, 1.6% of the cases were attributed to beef and beef products (60). This systematic review showed that S. Typhimurium was the most frequently reported serotype from cattle in Africa, Asia, and Australasia. Cattle could also contribute to the invasive non-typhoidal Salmonella disease in people who have contact with cattle feces. This is particularly important in regions like Africa where invasive non-typhoidal Salmonella infections are endemic as reviewed by Marks et al. (66). All the above evidence supports the importance of cattle and cattle associated serotypes for human salmonellosis.
Besides the datasets from the publications included in this review and meta-analysis, other relevant information was available in new articles that were published in the years 2017 and 2018 while the manuscript was under preparation by the authors. During this period, 6 full articles and three published abstracts representing 11 datasets were retrieved using the search engines (67–75). The majority of these studies were reported from Africa (67, 68, 70–74) except for two studies from Europe (69) and South America (75). Among the total of 5,868 cattle examined, 9.2% (554/6018), which is nearly equal to the pooled prevalence estimate, were reported to be positive for Salmonella species with different serotypes. The global level pooled prevalence of Salmonella in cattle was higher (9%) as compared to the pooled prevalence estimates of E. coli O157 (5.68%), which is also excreted by cattle showing the relative public health importance of Salmonella (76).
Conclusions
This study based on systematic reviews and meta-analysis provides an overall prevalence of Salmonella and serotype diversity in apparently healthy cattle at a global level. The results indicated variations in the level of Salmonella carriage in cattle across the world, and the presence of a diverse number of Salmonella serotypes. The estimated Salmonella prevalence was higher in North America. The predominant detection method is traditional culturing. Because of the possibility of Salmonella contamination of carcasses during slaughter and milk during milking, cattle can be a potential source of Salmonella and can lead to public health risk and economic loss if the necessary hygienic measures are not properly followed.
Author Contributions
FG, GA, RA, LD, and LDZ designed the study and identified the search engines and key words for literature search. FG, GA, and LD analyzed the data. FG wrote the manuscript. GA, RA, LD, LDZ, and SG revised the manuscript.
Funding
FG is supported by Ghent University through its doctoral grants for candidates from developing countries and Addis Ababa University. Mention of trade names or commercial products by a USDA author (GA) in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2019.00102/full#supplementary-material
References
1. Glavin MO. A single microbial sea: food safety as a global concern. SAIS Rev. (2003) 23:203–20. doi: 10.1353/sais.2003.0012
2. Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. (2015) 12:e1001940. doi: 10.1371/journal.pmed.1001940
3. D'Aoust JY. Salmonella. In: Lund BM, Baire-Parker AC, Gould GW, editors. The Microbiology Safety and Quality of Food. Gaithersburg, MD: Aspen Publishers (1999). p. 1233–99.
4. Schlundt J, Toyofuku H, Jansen J, Herbst SA. Emerging food-borne zoonoses. Rev Sci Tech. (2004) 23:513–34. doi: 10.20506/rst.23.2.1506
5. Rosel K, Delia G. Can Participation Improve Food Safety?. Food Safety and Informal Markets: Animal Products In Sub-Saharan Africa. (2014). Available online at: https://cgspace.cgiar.org/handle/10568/42438
6. Tindall BJ, Grimont PA, Garrity GM, Euzeby JP. Nomenclature and taxonomy of the genus Salmonella. Int J Syst Evol Microbiol. (2005) 55:521–4. doi: 10.1099/ijs.0.63580-0
7. Andino A, Hanning I. Salmonella enterica.: survival, colonization, and virulence differences among serovars. Sci World J. (2015) 2015:520179. doi: 10.1155/2015/520179
8. Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. (2012) 379:2489–9. doi: 10.1016/S0140-6736(11)61752-2
9. de Freitas Neto OC, Penha Filho RA, Barrow P, Berchieri JrA. Sources of human non-typhoid salmonellosis: a review. Rev Bras Ciência Avícola. (2010) 12:01–11. doi: 10.1590/S1516-635X2010000100001
10. Tennant SM, MacLennan CA, Simon R, Martin LB, Khan MI. Non-typhoidal Salmonella disease: current status of vaccine research and development. Vaccine. (2016) 34:2907–10. doi: 10.1016/j.vaccine.2016.03.072
11. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'brien SJ, et al. The global burden of non-typhoidal Salmonella gastroenteritis. Clin Infect Dis. (2010) 50:882–9. doi: 10.1086/650733
12. Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. Global burden of invasive non-typhoidal Salmonella disease, 2010. Emerg Infect Dis. (2015) 21:941–9. doi: 10.3201/eid2106.140999
13. Tauxe RV, Doyle MP, Kuchenmüller T, Schlundt J, Stein CE. Evolving public health approaches to the global challenge of foodborne infections. Int J Food Microbiol. (2010) 139:S16–28. doi: 10.1016/j.ijfoodmicro.2009.10.014
14. Buncic S, Sofos J. Interventions to control Salmonella contamination during poultry, cattle and pig slaughter. Food Res Int. (2012) 45:641–55. doi: 10.1016/j.foodres.2011.10.018
15. EFSA. Analysis of the baseline survey on the prevalence of Salmonella in holdings with breeding pigs, in the EU, 2008, part A: Salmonella. prevalence estimates. EFSA J. (2009) 7:1377. doi: 10.2903/j.efsa.2009.1377
16. Ejeta G, Molla B, Alemayehu D, Muckle CA. Salmonella serotypes isolated from minced meat beef, mutton and pork in Addis Ababa, Ethiopia. Rev Med Vet. (2004) 155:547–51. Available online at: https://www.revmedvet.com/2004/RMV155_547_551.pdf
17. Pires SM, Evers EG, van Pelt W, Ayers T, Scallan E, Angulo FJ, et al. Attributing the human disease burden of foodborne infections to specific sources. Foodborne Pathog Dis. (2009) 6:417–24. doi: 10.1089/fpd.2008.0208
18. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. (2011) 17:7–15. doi: 10.3201/eid1701.P11101
19. Laufer AS, Grass J, Holt K, Whichard JM, Griffin PM, Gould LH. Outbreaks of Salmonella infections attributed to beef–United States, 1973–2011. Epidemiol Infect. (2015) 143:2003–13. doi: 10.1017/S0950268814003112
20. Hoelzer K, Switt AI, Wiedmann M. Animal contact as a source of human non-typhoidal salmonellosis. Vet Res. (2011) 42:34. doi: 10.1186/1297-9716-42-34
21. Pui CF, Wong WC, Chai LC, Tunung R, Jeyaletchumi P, Hidayah N, et al. Salmonella: a foodborne pathogen. Int Food Res J. (2011) 18:465–73. Available online at: http://www.ifrj.upm.edu.my/18%20(02)%202011/(1)%20IFRJ-2010-306.pdf
22. Abdunaser D, Almabrouk F, Ashraf W, Yves M, Olivier C, Moez S. Quantitative risk assessment of human salmonellosis linked to the consumption of ground beef. Iraqi J Vet Sci. (2009) 23: 263–73. Available online at: https://pastel.archives-ouvertes.fr/pastel-00003795/document
23. Agga GE, Arthur TM, Schmidt JW, Wang R, Brichta-Harhay DM. Diagnostic accuracy of rectoanal mucosal swab of feedlot cattle for detection and enumeration of Salmonella enterica. J Food Prot. (2016) 79:531–7. doi: 10.4315/0362-028X.JFP-15-409
24. Chaney WE, Agga GE, Nguyen SV, Arthur TM, Bosilevac JM, Dreyling E, et al. Rapid detection and classification of Salmonella enterica. shedding in feedlot cattle utilizing the roka bioscience atlas salmonella detection assay for the analysis of rectoanal mucosal swabs. J Food Prot. (2017) 80:1760–7. doi: 10.4315/0362-028X.JFP-17-124
25. Mannion C, Fanning J, McLernon J, Lendrum L, Gutierrez M, Duggan S, et al. The role of transport, lairage and slaughter processes in the dissemination of Salmonella spp. in pigs in Ireland. Food Res Int. (2012) 45:871–9. doi: 10.1016/j.foodres.2011.02.001
26. Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. (2015)4:1. doi: 10.1186/2046-4053-4-1
27. ProQuest. RefWorks. (2016). Available online at: https://www.refworks.com/
28. Nyaga VN, Arbyn M, Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Arch Public Heal. (2014) 72:39. doi: 10.1186/2049-3258-72-39
30. Borenstein M, Hedges LV, Higgins JP, Rothstein HR. Introduction to Meta-Analysis. Hoboken, NJ: John Wiley & Sons (2009).
31. Higgins J, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. (2002) 21:1539–58. doi: 10.1002/sim.1186
32. Al-Saigh H, Zweifel C, Blanco J, Blanco JE, Blanco M, Usera MA, et al. Fecal shedding of Escherichia coli O157, Salmonella, and Campylobacter. in Swiss cattle at slaughter. J Food Prot. (2004) 67:679–84. doi: 10.4315/0362-028X-67.4.679
33. Hanson DL, Loneragan GH, Brown TR, Nisbet DJ, Hume ME, Edrington TS. Evidence supporting vertical transmission of Salmonella in dairy cattle. Epidemiol Infect. (2016) 144:962–7. doi: 10.1017/S0950268815002241
34. Tarazi YH, Abo-Shehada MN. Herd-and individual-level prevalence of and risk factors for Salmonella spp. fecal shedding in dairy farms in Al-Dhulail Valley, Jordan. Trop Anim Health Prod. (2015) 47:1241–8. doi: 10.1007/s11250-015-0854-z
35. Barham AR, Barham BL, Johnson AK, Allen DM, Blanton JR Jr, Miller MF. Effects of the transportation of beef cattle from the feed yard to the packing plant on prevalence levels of Escherichia coli. O157 and Salmonella spp. J Food Prot. (2002) 65:280–3. doi: 10.4315/0362-028X-65.2.280
36. Abouzeed YM, Hariharan H, Poppe C, Kibenge FS. Characterization of Salmonella. isolates from beef cattle, broiler chickens and human sources on Prince Edward Island. Compar Immun Microbiol Infect Dis. (2000) 23:253–66. doi: 10.1016/S0147-9571(99)00079-X
37. Callaway TR, Keen JE, Edrington TS, Baumgard LH, Spicer L, Fonda ES, et al. Fecal prevalence and diversity of Salmonella species in lactating dairy cattle in four states. J Dairy Sci. (2005) 88:3603–8. doi: 10.3168/jds.S0022-0302(05)73045-9
38. Loneragan GH, Thomson DU, McCarthy RM, Webb HE, Daniels AE, Edrington TS, et al. Salmonella. diversity and burden in cows on and culled from dairy farms in the Texas high plains. Foodborne Pathog Dis. (2012) 9:549–55. doi: 10.1089/fpd.2011.1069
39. Narváez-Bravo C, Rodas-González A, Fuenmayor Y, Flores-Rondon C, Carruyo G, Moreno M, et al. Salmonella. on feces, hides and carcasses in beef slaughter facilities in Venezuela. Int J Food Microbiol. (2013) 166:226–30. doi: 10.1016/j.ijfoodmicro.2013.07.009
40. Tietjen M, Fung DYC. Salmonellae and food safety. Crit Rev Microbiol. (1995) 21:53–83. doi: 10.3109/10408419509113534
41. Ishihara K, Takahashi T, Morioka A, Kojima A, Kijima M, Asai T, et al. National surveillance of Salmonella enterica in food-producing animals in Japan. Acta Vet Scand. (2009) 51:35. doi: 10.1186/1751-0147-51-35
42. Tadesse G, Tessema TS. A meta-analysis of the prevalence of Salmonella in food animals in Ethiopia. BMC Microbiol. (2014) 14:270. doi: 10.1186/s12866-014-0270-y
43. Dargatz DA, Fedorka-Cray PJ, Ladely SR, Ferris KE. Survey of Salmonella serotypes shed in feces of beef cows and their antimicrobial susceptibility patterns. J Food Prot. (2000) 63:1648–53. doi: 10.4315/0362-028X-63.12.1648
44. Davies RH, Dalziel R, Gibbens JC, Wilesmith JW, Ryan JM, Evans SJ, et al. National survey for Salmonella in pigs, cattle and sheep at slaughter in Great Britain (1999–2000). J Appl Microbiol. (2004) 96:750–60. doi: 10.1111/j.1365-2672.2004.02192.x
45. Fegan N, Vanderlinde P, Higgs G, Desmarchelier P. Quantification and prevalence of Salmonella in beef cattle presenting at slaughter. J Appl Microbiol. (2004) 97:892–8. doi: 10.1111/j.1365-2672.2004.02380.x
46. Al Mawly J, Grinberg A, Prattley D, Moffat J, French N. Prevalence of endemic enteropathogens of calves in New Zealand dairy farms. N Z Vet J. (2015) 63:147–52. doi: 10.1080/00480169.2014.966168
47. Sofos JN, Geornaras I. Overview of current meat hygiene and safety risks and summary of recent studies on biofilms, and control of Escherichia coli O157:H7 in nonintact, and Listeriamonocytogenes in ready-to-eat, meat products. Meat Sci. (2010)86:2–14. doi: 10.1016/j.meatsci.2010.04.015
48. Beach JC, Murano EA, Acuff GR. Prevalence of Salmonella and Campylobacter in beef cattle from transport to slaughter. J Food Prot. (2002) 65:1687–93. doi: 10.4315/0362-028X-65.11.1687
49. Corrier DE, Purdy CW, DeLoach JR. Effects of marketing stress on fecal excretion of Salmonella spp. in feeder calves. Am J Vet Res. (1990) 51:866–9.
50. Millemann Y, Gaubert S, Remy D, Colmin C. Evaluation of IS200-PCR and comparison with other molecular markers to trace Salmonella enterica subsp. enterica serotype Typhimurium bovine isolates from farm to meat. J Clin Microbiol. (2000) 38:2204–9.
51. Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, et al. Global monitoring of Salmonellaserovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis. (2011) 8:887–900. doi: 10.1089/fpd.2010.0787
52. Eriksson E, Aspan A. Comparison of culture, ELISA and PCR techniques for Salmonella detection in faecal samples for cattle, pig and poultry. BMC Vet Res. (2007) 3:21. doi: 10.1186/1746-6148-3-21
53. Lalsiamthara J, Lee JH. Pathogenic traits of Salmonella Montevideo in experimental infections in vivo and in vitro. Sci Rep. (2017) 7:46232. doi: 10.1038/srep46232
54. Kim J, Park Y, Lee S, Song W, Jeong SH, Yoo YA, et al. Bacteremia due to Salmonella enterica serotype montevideo producing plasmid-mediated AmpC β-Lactamase (DHA-1). Case Rep. (2004) 34:214–7. Available online at: http://www.annclinlabsci.org/content/34/2/214.full.pdf
55. Harada T, Sakata J, Kanki M, Seto K, Taguchi M, Kumeda Y. Molecular epidemiological investigation of a diffuse outbreak caused by Salmonella enterica serotype montevideo isolates in Osaka Prefecture, Japan. Foodborne Pathog Dis. (2011) 8:1083–8. doi: 10.1089/fpd.2011.0862
56. CDC. Centers for Diseases Control and Prevention. Salmonella Typhimurium infection associated with raw milk and cheese consumption-Pennsylvania. Morb Mortal Wkly Rep. (2007) 56:1161–4. Available online at: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5644a3.htm
57. Besser TE, Goldoft M, Pritchett LC, Khakhria R, Hancock DD, Rice DH, et al. Multiresistant Salmonella Typhimurium DT104 infections of humans and domestic animals in the Pacific Northwest of the United States. Epidemiol Infect. (2000) 124:193–200. doi: 10.1017/S0950268899003283
58. Galland JC, Troutt HF, Brewer RL, Osburn BI, Braun RK, Sears P, et al. Diversity of Salmonella serotypes in cull (market) dairy cows at slaughter. J Am V Medical Assoc. (2001) 219:1216–20. doi: 10.2460/javma.2001.219.1216
59. Jackson CR, Fedorka-Cray PJ, Wineland N, Tankson JD, Barrett JB, Douris A, et al. Introduction to United States Department of Agriculture VetNet: status of Salmonella and Campylobacter databases from 2004 through 2005. Foodborne Pathog Dis. (2007) 4:241–8. doi: 10.1089/fpd.2006.0067
60. EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. (2015) 13:3991. doi: 10.2903/j.efsa.2015.3991
61. Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, et al. Host adapted serotypes of Salmonella enterica. Epidemiol Infect. (2000) 125:229–55.
62. Schneider JL, White PL, Weiss J, Norton D, Lidgard J, Gould LH, et al. Multistate outbreak of multidrug-resistant Salmonella Newport infections associated with ground beef, October to December 2007. J Food Prot. (2011) 74:1315–9. doi: 10.4315/0362-028X.JFP-11-046
63. Eng SK, Pusparajah P, AbMutalib NS, Ser HL, Chan KG, Lee LH. Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci. (2015) 8:284–93. doi: 10.1080/21553769.2015.1051243
64. Greig JD, Ravel A. Analysis of foodborne outbreak data reported internationally for source attribution. Int J Food Microbiol. (2009) 130:77–87. doi: 10.1016/j.ijfoodmicro.2008.12.031
65. Yousef AE, Carlstrom C. Salmonella. In: Yousef AE, Carlstrom C, editors. Food Microbiology: A Laboratory Manual. John Wiley and Sons Inc (2003). p. 165–205.
66. Marks F, von Kalckreuth V, Aaby P, Adu-Sarkodie Y, El Tayeb MA, Ali M, et al. Incidence of invasive Salmonella disease in sub-Saharan Africa: a multicentre population-based surveillance study. Lancet Global Health. (2017) 5:10–23. doi: 10.1016/S2214-109X(17)30022-0
67. Aziz SA, Abdel-Latef GK, Shany SA, Rouby SR. Molecular detection of Integron and antimicrobial resistance genes in multidrug resistant Salmonella isolated from poultry, calves and human in Beni-Suef governorate, Egypt. Beni-Suef University. J Basic App Sci. (2018)7: 535–42. doi: 10.1016/j.bjbas.2018.06.005
68. Ball TA, Fedorka-Cray PJ, Horovitz J, Thakur S. Molecular characterization of Salmonella spp. from cattle and chicken farms in Uganda. Online J Public Health Inform. (2018) 10:e148. doi: 10.5210/ojphi.v10i1.8934
69. Cetin E, Serbetcioglu T, Temelli S, Eyigor A. Non-typhoid Salmonella carriage, serovar profile and antimicrobial resistance phenotypes in slaughter cattle. J Food Safety. (2018) e12603. doi: 10.1111/jfs.12603
70. Fashae K, Leekitcharoenphon P, Hendriksen RS. Phenotypic and genotypic comparison of salmonellae from diarrhoeic and healthy humans and cattle, Nigeria. Zoonoses Public Health. (2018) 65:185–95. doi: 10.1111/zph.12427
71. Ketema L, Ketema Z, Kiflu B, Alemayehu H, Terefe Y, Ibrahim M, et al. Prevalence and antimicrobial susceptibility profile of Salmonella serovars isolated from slaughtered cattle in Addis Ababa, Ethiopia. BioMed Res Int. (2018)2018:9794869. doi: 10.1155/2018/9794869
72. Kore K, Asrade B, Demissie K, Aragaw K. Characterization of Salmonella isolated from apparently healthy slaughtered cattle and retail beef in Hawassa, southern Ethiopia. Prev Vet Med. (2017) 147:11–6. doi: 10.1016/j.prevetmed.2017.08.018
73. Nouichi S, Ouatouat R, Can HY, Mezali L, Belkader C, Ouar-Korichi M, et al. Prevalence and antimicrobial resistance of Salmonella isolated from bovine and ovine samples in slaughterhouses of Algiers, Algeria. J Hellenic Vet Med Soc. (2018) 69:863–72. doi: 10.12681/jhvms.16441
74. Takele S, Woldemichael K, Gashaw M, Tassew H, Yohannes M, Abdissa A. Prevalence and drug susceptibility pattern of Salmonella isolates from apparently healthy slaughter cattle and personnel working at the Jimma municipal abattoir, south-West Ethiopia. Trop Dis Travel Med Vaccines. (2018) 4:13. doi: 10.1186/s40794-018-0072-6
75. Fuenmayor Y, Rodas-González A, Carruyo G, Hoet AE, Wittum T, Narváez-Bravo C. Salmonella prevalence and antimicrobial drug resistance in dual-purpose cattle operations in the eastern region of Zulia State, Venezuela. Foodborne Pathog Dis. (2018). 16:205–13. doi: 10.1089/fpd.2018.2515
Keywords: Salmonella, cattle, prevalence, serotypes, systematic review, meta-analysis
Citation: Gutema FD, Agga GE, Abdi RD, De Zutter L, Duchateau L and Gabriël S (2019) Prevalence and Serotype Diversity of Salmonella in Apparently Healthy Cattle: Systematic Review and Meta-Analysis of Published Studies, 2000–2017. Front. Vet. Sci. 6:102. doi: 10.3389/fvets.2019.00102
Received: 26 January 2019; Accepted: 19 March 2019;
Published: 09 April 2019.
Edited by:
Armanda Bastos, University of Pretoria, South AfricaReviewed by:
Becky Gosling, Animal and Plant Health Agency, United KingdomSmita Gopinath, Yale University, United States
Copyright © 2019 Gutema, Agga, Abdi, De Zutter, Duchateau and Gabriël. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fanta D. Gutema, fantadesissa@aau.edu.et
 Fanta D. Gutema
Fanta D. Gutema Getahun E. Agga
Getahun E. Agga Reta D. Abdi
Reta D. Abdi Lieven De Zutter
Lieven De Zutter Luc Duchateau
Luc Duchateau Sarah Gabriël
Sarah Gabriël