A murine model of peanut-allergic asthma
- 1Department of Dermatology, University of Zurich, Zurich, Switzerland
- 2Department of Dermatology, University Hospital Zurich, Zurich, Switzerland
Objectives: Peanut allergy is an IgE-mediated food allergy that is associated with asthma in certain patients. With increasing prevalence, its great impact on the quality of life, and a lack of treatment options, the need for new therapy options is a given. Hence, models for research and development are required. This study aimed to establish a murine model of allergic airway inflammation induced by peanut allergens.
Methods: C3H mice were sensitised by intraperitoneal injections of peanut allergen extract and challenged by an intranasal application of the same extract. The assessment of airway inflammation involved the analysis of immune cells in the bronchoalveolar lavage fluid as measured by flow cytometry. Inflammatory reactions in the lung tissue were also studied by histology and quantitative PCR. Moreover, peanut-specific immune responses were studied after re-stimulation of spleen cells in vitro.
Results: Sensitisation led to allergen-specific IgE, IgA, and IgG1 seroconversion. Subsequent nasal exposure led to allergic airway inflammation as manifested by structural changes such as bronchial smooth muscle hypertrophy, mucus cell hyperplasia, infiltration of eosinophil cells and T cells, as well as an upregulation of genes expressing IL-4, IL-5, IL-13, and IFN-γ. Upon re-stimulation of splenocytes with peanut allergen, increased secretion of both T-helper type 2 (Th2) and Th1 cytokines was observed.
Conclusion: We successfully established a peanut-associated asthma model that exhibited many features characteristic of airway inflammation in human patients with allergic asthma. The model holds potential as a tool for investigating novel therapeutic approaches aimed at preventing the development of allergic asthma.
Introduction
Food allergy is described as an excessive response of the immune system to otherwise harmless food proteins (1), occasionally resulting in respiratory symptoms upon allergen exposure (2). Individuals with coexisting food allergy and asthma often experience more severe asthmatic reactions (3). Among food allergens, peanuts are known to induce the most severe allergic manifestations, often leading to systemic responses with a high risk of anaphylaxis. The dominant allergen trigger in peanut allergy is the storage protein Ara h 2 (4). Unlike other allergies that may be outgrown with age, peanut allergy tends to persist, significantly impacting patients throughout their lives (5–7).
Patients with peanut allergy typically experience intermittent airway obstruction with chest tightness, wheezing, and dyspnoea (8). Food allergy-associated asthma varies in severity, ranging from mild to severe, and may potentially end up in fatal anaphylactic shock with respiratory arrest (2). Asthma is mediated by allergen-specific IgE antibodies and attributed to an inflammation of the lower respiratory tract with increased mucus production, swelling, bronchial constriction, and infiltration of immune cells such as lymphocytes, eosinophils, and mast cells in lung tissue (8, 9). Chronic food-allergic asthma results in structural changes in the lung with airway wall remodelling and the thickening of smooth muscle cells and of the basement membrane (10).
The current treatments for food allergy symptoms are limited to allergen avoidance, bronchodilators inhalation, as well as an epinephrine autoinjector in case of anaphylaxis (5). These measures therefore only provide a short-term symptomatic relief and do not serve as a curative treatment. As the prevalence of food allergy and asthma rises globally, these conditions pose a growing health concern (11, 12).
The pathophysiological process of peanut-allergic asthma involves a type I hypersensitivity reaction (13). Following an initial peanut allergen exposure, dendritic cells (DCs) present the allergen to T-helper type 2 (Th2) lymphocytes. This induces the release of cytokines such as IL-4 and IL-13 that drive B cells to switch their production of immunoglobulin to the IgE isotype (13, 14). The formation of allergen-specific IgE antibodies results in sensitisation and disrupted tolerance (4, 15). Newly produced allergen-specific IgE antibodies then bind high affinity FcεRI on mast cells (16). Upon subsequent allergen exposure, the allergen crosslinks bound IgE antibodies, induces mast cells to de-granulate (15), releasing into the bloodstream vasoactive substances such as histamine, leukotrienes, and tryptase that contribute to early- and late-phase allergic reactions (13, 14, 17). The Th2-like cell responses and cytokines, including IL-4, IL-5, IL-13, and TNF-α, play a central role in the immunological pathway in patients with allergic asthma (18). IL-4, in addition to its role in the IgE isotype switch in B cells, contributes to the differentiation of the Th2 cells, while supressing a Th1 cell response (14, 19). An essential feature of allergic asthma is the increased migration of eosinophils into the sputum, stimulated and recruited by IL-5 (14, 19–21). Eosinophilia has demonstrated to be responsible for bronchial hyper-responsiveness, and for increased mucus production in allergy-induced asthmatic airways (14, 22). IL-13 acts similarly to IL-4 but also plays an additional role in the regulation of mucus production (14). TNF-α stimulates airway epithelial cells and increases the number of cell adhesion molecules, facilitating migration of lymphocytes to the site of inflammation (14, 19). The characteristic Th2 secretion pattern ultimately induces chronic tissue inflammation in the lungs, attracting more inflammatory effector cells (14, 15), thus establishing a positive feedback (9).
Although multiple murine models of food allergies exist, they do not assimilate the complete human allergic pathology and clinical manifestations (23). Our study aims to establish a comprehensive mouse model of peanut allergy-induced asthma. Because mice do not spontaneously develop allergies, this has to be artificially induced, considering factors such as sensitisation protocols, allergen form, adjuvants, and mouse strains (23). Building upon a previously published peanut allergy mouse model (24) characterised by elevated allergen-specific IgE titres and a Th2-like cytokine profile, our goal is to create a robust model. The successful development of such a model could serve as a standardised tool for evaluating novel therapeutic approaches against peanut allergy. Furthermore, this groundwork may be adaptable to other allergic disorders, including conditions like asthma triggered by respiratory allergens such as tree pollen or house dust mites.
Material and methods
Mice and ethics
Female C3H mice were purchased from Envigo (Horst, the Netherlands). The mice were kept in a pathogen-free and animal-friendly environment at ca. 21°C and with a 12–12 h light–dark cycle at the Laboratory Animal Services Centre, University of Zurich. They had open access to water and chow, and were kept in unit cages of five. The mice were not further randomised. Housing, care, and treatment of the mice were carried out according to good animal practices, following the Swiss guidelines. The mice entered the experiments at ca. 6 weeks of age. The experiments were approved by the ethical review board and Cantonal Veterinary Office of Zurich (license ZH 147/2021).
Peanut allergen sensitisation and induction of allergic airway inflammation
The mice were sensitised by intraperitoneal (IP) injections of 4.2 μg of a peanut allergen extract skin-prick-test solution (Allergopharma, Reinbek, Germany) mixed with 150 μg aluminium hydroxide (alum) adjuvant from InvivoGen (San Diego, CA, USA) and phosphate buffered saline (PBS) constituting a total of 50 μl as previously described in a peanut allergen model of anaphylaxis (24). The allergen–alum mixture was left at room temperature one hour before injection. The IP sensitisation was performed once or twice with a seven days interval. One week after the last sensitisation, the mice were challenged for five consecutive days by intranasal (IN) application of 42 μg of a purified peanut allergen extract (Inbio, Cardiff, UK) mixed with PBS in a total volume of 50 μl. The control groups of mice were kept non-sensitised and non-challenged as baseline control or kept non-sensitised but challenged. Before each challenge dose, the mice were transiently anesthetised with 2% isoflurane. Forty-eight hours after the last challenge, the mice were euthanised by IP injection of a xylazine (90 μg, Rompun-2%, Bayer Health Care, Leverkusen, Germany) and ketamine (1,800 μg, Ketasol-100, Graeub AG, Bern, Switzerland) mixture. Spleen, lung, blood, and bronchoalveolar lavage fluid (BALF) were collected for further experiments and analysis. An experimental scheme is illustrated in Figure 1A.
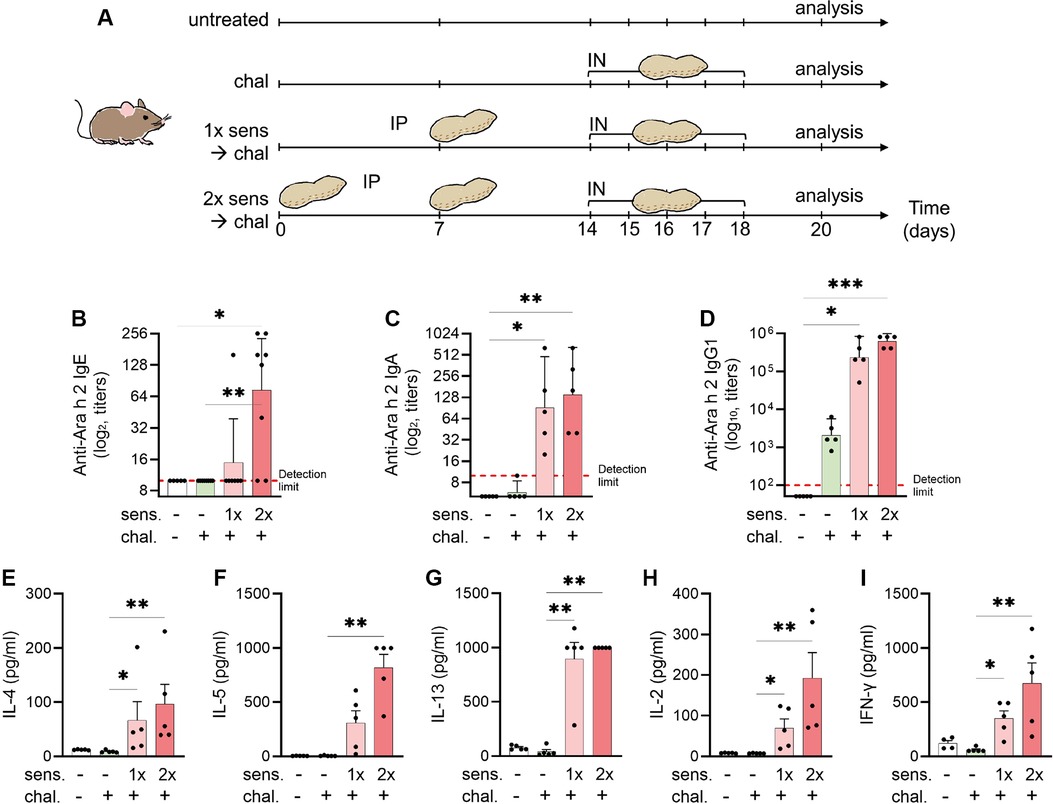
Figure 1. Experimental setup for a murine model of peanut-allergic asthma and detection of allergen-specific serum antibodies and cytokines. (A) C3H mice were sensitised (sens) IP and challenged (chal) IN for analysis of allergic airway inflammation (n = 5 per group). Sensitisation with peanut extract was done on days 0 and 7 (sens 2×) or day 7 only (sens 1×). The challenge with peanut extract was performed on days 14–18. Euthanasia and analysis followed on day 20. Blood sera were analysed by ELISA for the presence of Ara-h-2-specific IgE (B), IgA (C), and IgG1 (D) antibodies. Antibodies titres are shown as geometric mean with 95% CI. (E–I) Splenocytes were re-stimulated with peanut allergen extract and the supernatants were analysed for the content of IL-4 (E), IL-5 (F), IL-13 (G), IL-2 (H), and IFN-γ (I) by ELISA and illustrated as mean ± SEM. Statistical analyses were performed using the Kruskal–Wallis with Dunn test. *p < 0.05; **p < 0.01. All data are representative of the two experiments.
Splenocytes isolation for in vitro stimulation
Excised spleens were aseptically processed into a single cell suspension, passed through a cell strainer and washed with PBS and foetal calf serum (FCS, 2%). The samples were centrifuged and the pellet treated with red blood cell lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) and washed twice with PBS. The cells were then resuspended in RPMI 1640 cell culture medium supplemented with 10% FCS and 1% L-glutamine. The cells were counted with a MoxiFlow (Orflo, Idaho, USA) and plated on round-bottom 96-well plates at 0.5 × 106 cells in a volume of 100 μl and in triplicates. Peanut allergen extract was added at 50 μg/ml and the plates incubated at 37°C with 5% CO2 for 24 or 96 h. The supernatants were frozen at −20°C for a later analysis of cytokines by enzyme-linked immunosorbent assay (ELISA).
Enzyme-linked immunosorbent assay
We applied the ELISA for detection of cytokines in the supernatant of cultured splenocytes and for the detection of Ara-h-2-specific antibodies in murine blood serum. The ELISAs were performed on 96-well Maxisorb plates. The absorbance was read at 450 nm with a Spark microplate reader (Tecan, Männedorf, Switzerland).
For the detection of peanut-specific IgE, the plates were coated with anti-mouse-IgE (BioRad, California, USA) in carbonate buffer at pH 9.6. After blocking for 1 h with 5% skimmed milk in PBS with 0.05% polysorbate 20, serial dilutions of the mouse serum were added. Next, biotin natural Ara h 2 (Inbio) was added, and the ELISA was further developed by incubation of streptavidin-conjugated horse-radish peroxidase (BioLegend, San Diego, CA, USA) followed by tetramethylbenzidine substrate (eBioscience, as purchased from Thermo Fisher Scientific, Basel, Switzerland). The colorimetric enzyme reaction was stopped with 2N sulphuric acid. Each incubation step was followed by washing the plates thrice with 0.05% polysorbate 20 in PBS.
To measure Ara-h-2-specific IgA and IgG1 levels, the plates were coated with 1 µg/ml of purified natural Ara h 2 (Inbio), and serially diluted mouse serum was added. Detection was carried out using biotinylated biotin rat anti-mouse IgA (BD Pharmingen, Franklin Lakes, USA) and rat goat-mouse IgG1 antibody (Abcam, Cambridge, UK), and the assay was developed with HRP-conjugated streptavidin and TMB, following the same procedure as described previously.
To determine the antibody titre, we measured the optical density in sera from naïve mice. The seroconversion threshold was set by calculating the mean plus 3 standard deviations above the mean using the naive mouse serum data. The anti-Ara h 2 IgE, IgA, and IgG1 titres were defined as the reciprocal final dilution at which OD were higher than the threshold.
For cytokine detection in a supernatant of splenocytes cultures, kits from eBioscience were applied. Briefly, the plates were coated with anti-cytokine capture antibodies in carbonate buffer. The culture samples and controls were diluted 1:1 in test diluent following the manufacturer's protocol. The tested cytokines included IL-2 (24-h cultures) and IL-4, IL-5, IL-13, and IFN-γ (96-h cultures). Plotting of standard curves and equations for the best linear fit were then applied to calculate the concentration levels of cytokines.
Flow cytometry of bronchoalveolar lavage fluid
To collect the BALF from mice, a 22-gauge needle was used as a catheter and gently inserted into the trachea of a supine mouse. A cold solution of PBS (1 ml) was injected and aspirated to harvest cellular contents from the lung lumen of mice for subsequent ex vivo analysis. The process was repeated twice. The processing of BALF for flow cytometry was performed on ice. The total BALF volume was measured, filtered through sterile gauze, and then centrifuged at 4°C and 1,500 rpm for 5 min before being resuspended in cold PBS. After cell counting with MoxiFlow, the samples were once again centrifuged and the pellet resuspended in PBS with 2% FCS (FACS buffer) on a 96-well plate. The Fc receptors on cells were blocked by incubating the cells with anti-CD16/32 (eBioscience) on ice for 15 min. The cell staining was performed by incubating cells with a cocktail of fluorescently labelled antibodies (1:100) in the dark for 30 min for identification of T cells, B cells, macrophages, dendritic cells, neutrophils, and eosinophil cells. The following fluorescent antibodies were used: CD3e-FITC, CD19-FITC, CD11c-PE-Cy7, CD11b-PE, LY-6G-PerCP-Cy5.5, MHC II-APC, and SiglecF-BV421. All antibodies were purchased from BD Biosciences (Allschwil, Switzerland) or eBioscience. The cells were fixed in 1% paraformaldehyde and on ice for 30 min, washed in FACS buffer, and kept in the dark before acquisition on flow cytometer LSR II Fortessa 4l (BD, Heidelberg, Germany) operating on BD FACS DIVA software. The obtained data were analysed with Flow Jo 10 software (FlowJo LLC, Ashland, OR).
Histology
Mice lung tissue was harvested, fixed in 4% formalin in PBS overnight, dehydrated, and embedded in paraffin. Histological sections of 5 μm thickness were cut using an HM 325 Rotary Microtome (Thermo Fisher Scientific). The sections were further stained with haematoxylin and eosin (H&E) and Periodic acid–Schiff (PAS) using a standardised in-house procedure. The stained sections were scanned with Zeiss Axio Scan.Z1 Slidescanner (Zeiss, Oberkochen, Germany) and Akoya Vectra Polaris (Akoya Biosciences, Marlborough, MA, USA). QuPath (version 0.5.1) was used for the H&E staining quantification.
Quantitative polymerase chain reaction
Cytokine gene expression in the lungs was measured using a two-step quantitative polymerase chain reaction (qPCR). Tissue samples were lysed and homogenised with TissueLyser II (Qiagen, Venlo, the Netherlands) and TRIzol Reagent (Invitrogen). Following the manufacturer's protocol, total RNA was then extracted in a first step. RNA purity and concentration were assessed with a Nanodrop (Witec AG, Sursee, Switzerland). The RNA was stored at −70°C until reverse transcription into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). For qPCR experiments, cDNA samples were diluted to 1:10 and measured on a LightCycler480 (Roche, Basel, Switzerland) using specific PCR Primers (Table 1) and FastStart Essential DNA Green Master (Roche Diagnostics, Rotkreuz, Switzerland) for amplification and detection. Peptidylprolyl isomerase A (PPIA) was selected as the housekeeping gene as described (25) and as confirmed in pre-tests with NormFinder (MOMA, Aarhus, Denmark), having the lowest inter and intra group differences in gene expression (25, 26). The expression levels of genes of interest were then normalised to PPIA. Up to 50 PCR cycles were conducted, and the cycle threshold value (Ct) was determined when the fluorescent signal of amplified DNA was first measured above the background fluorescence. The test samples were measured as triplicates for every gene of interest. The mean Ct value of each set of triplicate values was inserted in the double delta Ct formula (2-ΔΔCt) and used for the calculation of the fold change of gene expression as compared with PPIA expression.
Statistical analysis
The acquired data from all experiments were statistically analysed using GraphPad Prism software (version 9, GraphPad, La Jolla, CA, USA). The descriptive statistics were calculated and expressed as means, and standard error of the means were calculated. Antibody titres are presented as geometric means with 95% confidence intervals. All data were defined as non-parametric and analysed using Kruskal–Wallis tests, followed by Dunn's post-hoc tests for multiple comparisons. The significance level was set at 95%, with p < 0.05.
Results
Allergen-specific IgE seroconversion in mice after sensitisation with peanut extract
Because peanut allergy and allergic asthma are considered IgE mediated, we measured peanut-allergen-specific (Ara h 2) IgE antibodies in mice sera by ELISA. The mice that were not sensitised had no detectable Ara-h-2-specific IgE titre (Figure 1B). A single IP sensitisation with peanut allergen extract did not trigger IgE seroconversion (geometric mean 9.4), but two injections of the allergen extract caused a statistically significant (p < 0.05) induction of allergen-specific IgE antibodies (geometric mean 73.6), and seroconversion was detected in all mice. Similarly, in the assessment of IgA levels (Figure 1C), the primary isotype associated with mucosal immunity, there was a notable increase in anti-Ara h 2 titres among mice sensitised once (geometric mean 91.90) and twice (geometric mean 139.3) to peanut, compared with non-sensitised controls (geometric mean 5). In addition, we evaluated IgG1 titres (Figure 1D) targeting the key peanut allergen Ara h 2. The mice sensitised either once (geometric mean 235,253) or twice (geometric mean 620,838) prior to the challenge exhibited markedly elevated titres compared with the non-sensitised mice (geometric mean 50).
Increased secretion of Th2- and Th1-like cytokines in mice with peanut-allergen sensitisation
After the peanut sensitisation and intranasal challenge of mice, the animals were euthanised and the harvested spleen cells were cultured in vitro with peanut protein extract. When measuring the cytokine secretion from non-sensitised mice, we observed that IL-4, IL-5, IL-13, IL-2, and IFN-γ were hardly detectable, independent of the mice receiving an intranasal challenge with peanut allergen or not (Figures 1E–I). By contrast, when the mice were sensitised prior to the intranasal challenge, a clear increase in secretion of all tested cytokines was observed. While there was a tendency of more cytokine secretion in cells from mice sensitised by two IP injections of peanut allergen extract, the differences between mice with single or double sensitisation were not statistically significant. The increased cytokine levels in cell cultures from mice that were sensitised twice compared with challenged-only were significant for all cytokines (p < 0.01). The Th2-like cytokines, IL-4 (Figure 1E), IL-5 (Figure 1F), and IL-13 (Figure 1G), in sensitised mice showed a 10–100-fold increase as compared with non-sensitised but challenged mice. The measurement of IL-2 (Figure 1H), which is a facultative Th2/Th1-like cytokine, was also significantly increased in sensitised mice as compared with challenged-only mice and the Th1-like cytokine. Interestingly, also the Th1-like cytokine IFN-γ (Figure 1I) was increased in sensitised mice.
Infiltration of eosinophils and T cells in bronchoalveolar lavage fluid of peanut-allergen-sensitised mice
To further analyse the phenotype of mice sensitised and intranasally challenged with peanut allergen extract, a BAL was performed and the BALF samples were analysed by flow cytometry for the content of leukocytes. Different cell populations were distinguished based on their expressed surface antigens (Figure 2A). The percentage of CD3+ T cells in the BALF was significantly higher (p < 0.01) in the double-sensitised mice than in the non-sensitised challenged mice (Figure 2B). A similar trend was also noted for CD11c+ dendritic cells and especially for CD11b+ SiglecF+ eosinophils (p < 0.05). When comparing mice sensitised once and twice for the same cell types, the double-sensitised mice showed a tendency for higher percentages of positive cells, but the differences were not statically significant. No differences in the frequency of macrophages, Ly6G+ neutrophils, or CD19+ B cells were determined when comparing naïve, non-sensitised, and sensitised mice.
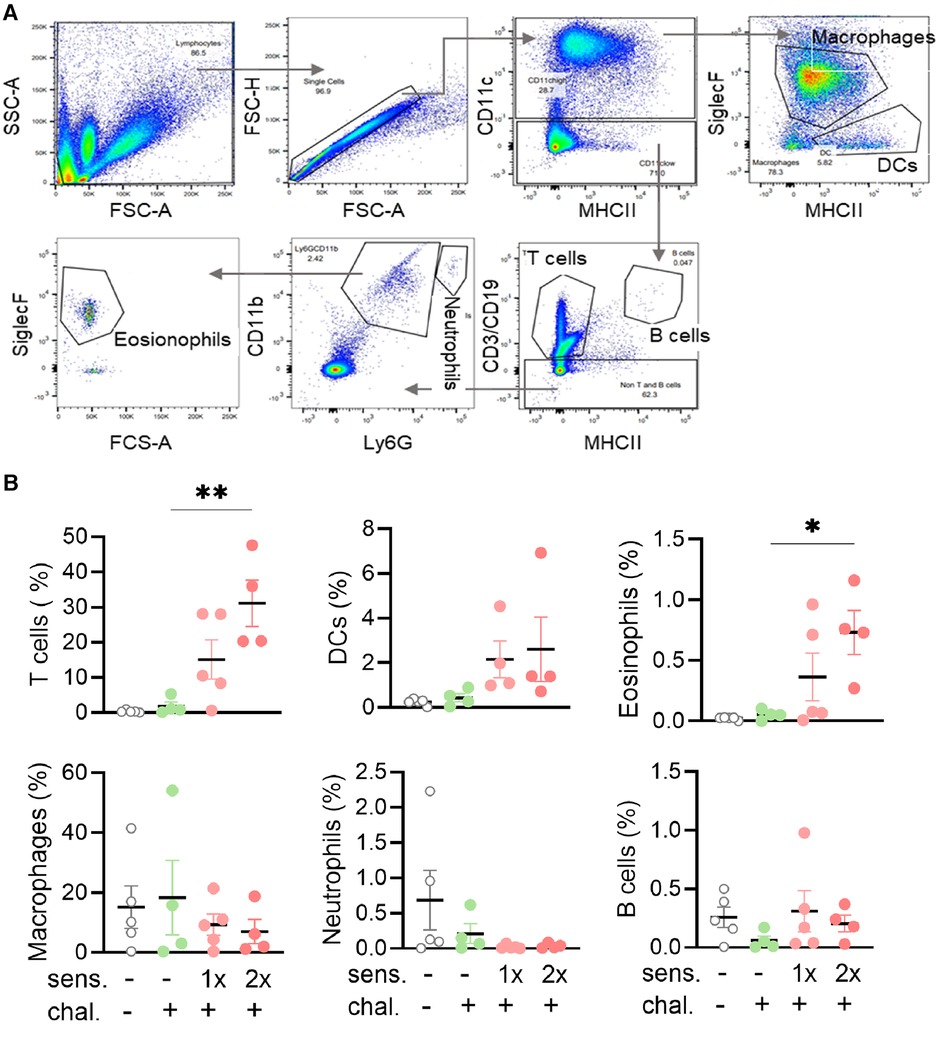
Figure 2. Flow cytometry gating strategy with percentages and absolute cell numbers of various immune cells found in BALF. (A) BALF was isolated and the leukocytes and single cells thereof identified with side scatter (SSC-A) and forward scatter area (FSC-A/FSC-H) and height (FSC-H). Sequential gating for MHC II, CD11b, CD11c, Ly6G, and SiglecF led to the identification of six leukocyte subpopulations. (B) BALF samples were analysed for leukocyte phenotypes. The percentages of T cells, B cells, DCs, macrophages, neutrophils, and eosinophils groups are shown as mean ± SEM. Data were analysed using the Kruskal–Wallis with Dunn's test. *: p < 0.05, **: p < 0.01. sens, sensitised; chal, challenged.
Upregulated expression of genes encoding for Th2- and Th1-like cytokines in mice after sensitisation with peanut allergen
To complement the immune analysis of lungs from peanut-allergen-sensitised mice challenged intranasally, we conducted qPCR analyses on lung tissue samples for quantification of the expression of mRNA encoding for cytokines (Figure 3). The samples from the sensitised and challenged mice were compared with those from challenged-only mice. For all the genes of interest, few to no changes were observed in the challenged-only mice. The sensitised mice exhibited an increased expression of the Th2-like and asthma-associated cytokines, IL-4 (Figure 3A), IL-5 (Figure 3B), and IL-13 (Figure 3C), as well as mRNA encoding for pro-inflammatory TNF-α (Figure 3D) and the Th1 cytokine IFN-γ (Figure 3E).
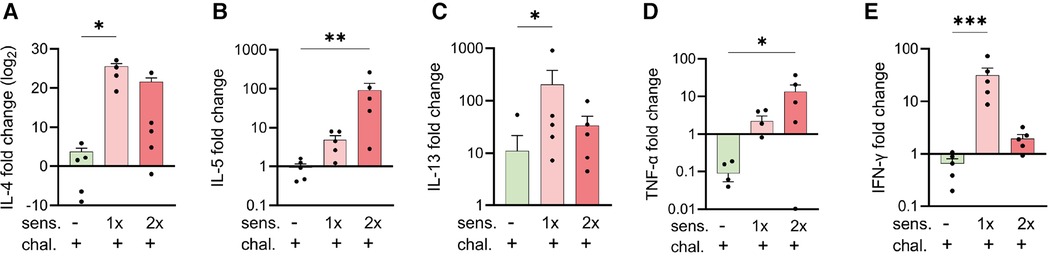
Figure 3. Expression of cytokine genes in peanut-allergen-treated mice via qPCR analysis. Lung tissue samples from mice described in Figure 1 were isolated, and gene expression patterns of cytokines were analysed by qPCR. Fold changes of IL-4 (A), IL-5 (B) IL-13 (C), TNF-α (D), and IFN-γ (E) over untreated mice were normalised to the housekeeping gene PPIA. Data are shown as mean ± SEM and analysed with the Kruskal–Wallis with Dunn multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001. sens, sensitised.
Histological signs of inflammation in lung tissue of peanut-allergen-sensitised mice
One notable characteristic of asthma is the modification of lung tissue morphology caused by inflammation and the infiltration of immune cells, resulting in increased mucus production. Consequently, we assessed lung tissue sections from sensitised and challenged mice using H&E staining to examine cell infiltration, and PAS staining to identify carbohydrate macromolecules commonly present in the mucus. No signs of inflammation were observed in the lungs of non-sensitised untreated mice (Figure 4A) or in the lungs of non-sensitised mice that received an intranasal challenge with peanut allergen extract (Figure 4B). By contrast, lung tissue from mice sensitised once prior to the peanut allergen challenge exhibited inflammatory infiltrate (Figure 4C). However, when mice were sensitised with two intraperitoneal injections prior to the peanut allergen challenge, a much more severe allergic airway inflammation was observed (Figure 4D). In Figure 4A inset, the relative cell counts from lung tissue sections are presented. Statistical analysis revealed a significant difference in cell infiltration between non-sensitised mice with or without challenge and mice sensitised twice and challenged (p < 0.05). Further examination of the lung tissue (Figures 4E–H) revealed notable structural changes, including bronchial smooth muscle hypertrophy and mucus cells hyperplasia in addition to cellular infiltration in double-sensitised and challenged mice (Figure 4H). In addition, PAS staining (Figures 4I–N) showed increased numbers of PAS-positive areas in the airways of double-sensitised and challenged mice (Figure 4N), suggestive of increased mucus production. The immune cell infiltration was mostly localised around bronchioles and composed of macrophages with foamy multinucleated, giant foreign body cells. An accumulation of many eosinophilic granulocytes, few neutrophils, and very few lymphocytes was also noted. In addition, de-granulated mast cells appear to be present in sensitised lung tissue.
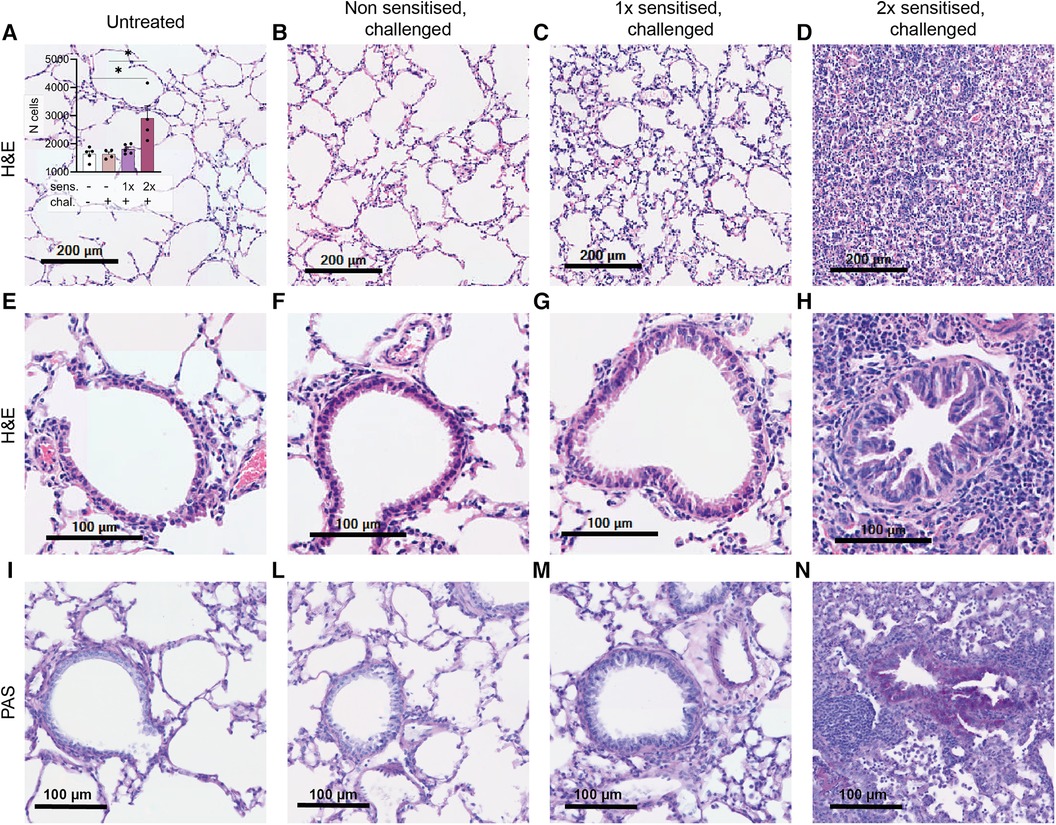
Figure 4. H&E and PAS staining of lung tissue sections. Lung tissue was obtained from mice that were treated as described in Figure 1. Representative H&E staining of alveoli (upper panel; A–D) with the relative cell counts (inset in A) and H&E and PAS staining of bronchioles (lower panels, E–H and I–N) are shown from untreated, challenged-only (non-sensitised, challenged), single-sensitised and challenged (1× sensitised, challenged), and double-sensitised and challenged mice (2× sensitised, challenged). Scale bar, 200 µm (upper panel) and 100 µm (lower panels). Data are shown as mean ± SEM and analysed with the Kruskal–Wallis with Dunn multiple comparison test. *p < 0.05.
Discussion
The classical acute model of allergic asthma in mice is based on ovalbumin (27, 28). While the ovalbumin model can reveal mechanisms involved in the pathogenesis of asthma and be useful for treatment targets of allergic inflammation (29), it is important also to include allergens that go beyond ovalbumin and that cause asthma in humans. Therefore, acute and chronic animal models have been developed with aeroallergens. However, patients with allergic sensitisation to foods such as peanut allergens are at risk for allergic inflammation of the airways (30, 31), and robust animal models of food allergy-associated asthma are needed. Therefore, this study describes an acute model of peanut-allergic airway inflammation in mice induced by IP sensitisation followed by intranasal allergen using a peanut allergen extract. This experimental setting of a systemic sensitisation phase and a pulmonary challenge phase is standard in IgE-mediated allergen-specific asthma in animals (32, 33). Our model demonstrated consistency with other murine models of allergy, in terms of allergen-specific IgE seroconversion, elevated Th2 cytokines, and remodelling of the airways, as manifested by structural changes such as bronchial smooth muscle hypertrophy, mucus cell hyperplasia, and infiltration of eosinophil cells. The latter is an important feature of asthma in humans (34–37).
While intranasal sensitisation would more resemble sensitisation in humans (38), we chose the IP route of administration as this has proven to require fewer doses, hence is fast, and since we have successfully used this route for the study of peanut-allergic anaphylaxis with the same allergen extract and for the same strain of mice (24, 39). Previous studies with other allergens often use two IP injections for sensitisation. Interestingly, we could show a single sensitisation was sufficient to trigger allergen-specific Th2 cytokine production, but that a second sensitisation was required to induce the remodelling of the lung tissue that is observed in asthma. A second sensitisation dose was also required to significantly raise the Ara-h-2-specific IgE levels, a further prerequisite of a mouse model of allergic asthma. When compared with the acute ovalbumin model of asthma (40), similar patterns of eosinophilic granulocyte accumulation in sensitised mice exposed to nasal allergen challenge were observed, both in BALF and in the lung tissue, further emphasising the role of eosinophils in driving allergic airway inflammation also in murine models of asthma (21, 41).
Our described model was also characterised by the involvement of T cells. The T-cell number in the BALF was increased in peanut-allergic mice with lung pathology. T-cell-derived cytokine production was determined in lung tissues as well as in lymphocytes after in vitro stimulation with peanut allergens, for instance by IL-2 release supportive of T-cell proliferation. Of interest, increased secretion of IFN-γ in mice subjected to nasal sensitisation and challenge was observed. However, this result aligns with other reports in murine models of peanut allergy (24, 42). Hence, the data suggest that the inflammatory response in this murine model might consist of an interplay of Th1- and Th2-associated processes (43, 44). To understand this underlying role of the Th1–Th2 interplay, further investigations would be required as the current study does not provide a definite explanation to that question. Although the standard Th1–Th2 paradigm remains a useful framework for understanding T-cell heterogeneity, different subsets of T cells may play different important roles in allergic asthma suggesting that asthma is not simply a Th2-type disease. Indeed, next to Th1 subsets, also Th17 cells, Th9 cells, and Tregs have been ascribed various roles in the pathology of asthma (45). Although the quantity of pulmonary B cells remained unchanged after nasal exposure to peanut allergens in our model, allergic B-cell responses may be ascribed a pivotal function in the production of allergen-specific IgE antibodies, rather than the proliferation and infiltration of B cells in lung tissue (46, 47).
Boyaka et al. were the first to describe lung reactivity in a mouse model of peanut allergy (42). In the work from 2005, the authors used a combined oral and nasal sensitisation regimen. Our model provides a cost-effective, simple, and fast method to establish peanut-allergic asthma in mice using a combined systemic and nasal sensitisation regimen, and the model may offer a valuable tool to study allergic asthma and potential novel therapeutic application for the treatment of peanut allergy. However, it is essential to acknowledge that our proposed model does not fully replicate the physiological sensitisation process in human peanut allergy. Therefore, recent efforts have been made to apply humanised mouse models in preclinical allergy (48) and asthma (49). Also the choice of the IP route for allergen exposure deviates from the typical gastrointestinal or epicutaneous routes for natural sensitisation (15, 50, 51). Furthermore, our study was limited to monitoring humoral and cellular immune responses and changes in gene expression. No conclusions can be drawn about the expected clinical effects like wheezing, dyspnoea, and reduced lung function (52, 53). Consequently, our murine model not only provides a partial understanding of the pathomechanisms underlying allergic asthma in humans, but also a starting point for development of a peanut-allergen-associated mouse model.
Given the lack of curative treatment options, the increasing prevalence, and a high socioeconomic burden associated with frequent emergency admissions, novel therapy options are urgently needed (54). Recently, oral immunotherapy (OIT) has been recommended as a potential treatment option for peanut allergy (55, 56). However, effectiveness was only seen in children and permanent desensitisation was not achieved (55). Patients already suffering from severe allergic airway inflammation undergoing OIT are at an increased risk of systemic allergic reactions, leaving an uncertainty regarding the safety of conventional immunotherapy (23, 55, 57, 58). Ongoing research is exploring novel therapeutic avenues. Passive allergen immunotherapy with monoclonal antibodies (mAbs) has demonstrated promising outlooks on reducing disease symptoms for birch- and cat- allergic patients (59, 60) and in mouse models of peanut allergy (39, 50, 61, 62). The development of additional mouse models that recapitulate different pathological aspects of peanut allergy, including complications linked to allergic asthma and airways inflammation, are therefore needed for therapeutic assessment. Our model, which effectively recapitulates airways inflammation, can be a valuable tool for researching and assessing the potential effects of novel therapeutic approaches. This model holds promise for making significant advancements in the pursuit of a cure for peanut-allergic asthma.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by the Ethical Review Board and Cantonal Veterinary Office of Zurich (license ZH 147/2021). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MP: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. NA: Data curation, Investigation, Writing – original draft, Formal Analysis, Writing – review & editing, Methodology. VH: Formal Analysis, Investigation, Writing – review & editing, Data curation, Methodology. IK: Formal Analysis, Writing – review & editing. TK: Funding acquisition, Writing – review & editing. PJ: Data curation, Formal Analysis, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article.
The project received funds from the University of Zurich (to TK).
Acknowledgements
The authors are grateful to Agathe Duda for support with ELISA measurement and Zuzanna Kotkowska and Gabriele Fenini for support with qPCR measurements.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer-review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Anvari S, Miller J, Yeh CY, Davis MC. IgE-mediated food allergy. Clin Rev Allergy Immunol. (2019) 57(2):244–60. doi: 10.1007/s12016-018-8710-3
2. Chong KW, Ruiz-Garcia M, Patel N, Boyle RJ, Turner PJ. Reaction phenotypes in IgE-mediated food allergy and anaphylaxis. Ann Allergy Asthma Immunol. (2020) 124(5):473–8. doi: 10.1016/j.anai.2019.12.023
3. Wang J, Liu AH. Food allergies and asthma. Curr Opin Allergy Clin Immunol. (2011) 11(3):249–54. doi: 10.1097/ACI.0b013e3283464c8e
4. Hemmings O, Du Toit G, Radulovic S, Lack G, Santos AF. Ara h 2 is the dominant peanut allergen despite similarities with Ara h 6. J Allergy Clin Immunol. (2020) 146(3):621–630.e5. doi: 10.1016/j.jaci.2020.03.026
5. Sampson HA, Aceves S, Bock SA, James J, Jones S, Lang D, et al. Food allergy: a practice parameter update—2014. J Allergy Clin Immunol. (2014) 134(5):1016–25.e43. doi: 10.1016/j.jaci.2014.05.013
6. Iweala OI, Choudhary SK, Commins SP. Food allergy. Curr Gastroenterol Rep. (2018) 20(5):17. doi: 10.1007/s11894-018-0624-y
7. Bégin P, Paradis L, Paradis J, Picard M, Des Roches A. Natural resolution of peanut allergy: a 12-year longitudinal follow-up study. J Allergy Clin Immunol Pract. (2013) 1(5):528–30.e1–4. doi: 10.1016/j.jaip.2013.05.008
8. Mims JW. Asthma: definitions and pathophysiology. Int Forum Allergy Rhinol. (2015) 5(Suppl 1):S2–6. doi: 10.1002/alr.21609
9. Hansen I, Klimek L, Mosges R, Hormann K. Mediators of inflammation in the early and the late phase of allergic rhinitis. Curr Opin Allergy Clin Immunol. (2004) 4(3):159–63. doi: 10.1097/00130832-200406000-00004
10. Fang L, Sun Q, Roth M. Immunologic and non-immunologic mechanisms leading to airway remodeling in asthma. Int J Mol Sci. (2020) 21(3):757. doi: 10.3390/ijms21030757
11. Cosme-Blanco W, Arroyo-Flores E, Ale H. Food allergies. Pediatr Rev. (2020) 41(8):403–15. doi: 10.1542/pir.2019-0037
12. Sicherer SH, Sampson HA. Food allergy: a review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J Allergy Clin Immunol. (2018) 141(1):41–58. doi: 10.1016/j.jaci.2017.11.003
13. Dispenza MC. Classification of hypersensitivity reactions. Allergy Asthma Proc. (2019) 40(6):470–3. doi: 10.2500/aap.2019.40.4274
14. Chung KF, Barnes PJ. Cytokines in asthma. Thorax. (1999) 54(9):825–57. doi: 10.1136/thx.54.9.825
15. De Martinis M, Sirufo MM, Suppa M, Ginaldi L. New perspectives in food allergy. Int J Mol Sci. (2020) 21(4):1474. doi: 10.3390/ijms21041474
16. Rios EJ, Kalesnikoff J. FcεRI expression and dynamics on mast cells. Methods Mol Biol. (2015) 1220:239–55. doi: 10.1007/978-1-4939-1568-2_15
17. Poowuttikul P, Seth D. Anaphylaxis in children and adolescents. Pediatr Clin North Am. (2019) 66(5):995–1005. doi: 10.1016/j.pcl.2019.06.005
18. Levine SJ, Wenzel SE. Narrative review: the role of Th2 immune pathway modulation in the treatment of severe asthma and its phenotypes. Ann Intern Med. (2010) 152(4):232–7. doi: 10.7326/0003-4819-152-4-201002160-00008
19. Borish LC, Steinke JW. 2. Cytokines and chemokines. J Allergy Clin Immunol. (2003) 111(2 Suppl):S460–75. doi: 10.1067/mai.2003.108
20. Peebles RS Jr, Aronica MA. Proinflammatory pathways in the pathogenesis of asthma. Clin Chest Med. (2019) 40(1):29–50. doi: 10.1016/j.ccm.2018.10.014
21. Bossley CJ, Fleming L, Gupta A, Regamey N, Frith J, Oates T, et al. Pediatric severe asthma is characterized by eosinophilia and remodeling without T(H)2 cytokines. J Allergy Clin Immunol. (2012) 129(4):974–82.e13. doi: 10.1016/j.jaci.2012.01.059
22. Lee JJ, Dimina D, Macias MP, Ochkur SI, McGarry MP, O'Neill KR, et al. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. (2004) 305(5691):1773–6. doi: 10.1126/science.1099472
23. Schülke S, Albrecht M. Mouse models for food allergies: where do we stand? Cells. (2019) 8(6):546. doi: 10.3390/cells8060546
24. Paolucci M, Homère V, Waeckerle-Men Y, Wuillemin N, Bieli D, Pengo N, et al. Strain matters in mouse models of peanut-allergic anaphylaxis: systemic IgE-dependent and Ara h 2-dominant sensitization in C3H mice. Clin Exp Allergy. (2023) 53(5):550–60. doi: 10.1111/cea.14279
25. He JQ, Sandford AJ, Wang IM, Stepaniants S, Knight DA, Kicic A, et al. Selection of housekeeping genes for real-time PCR in atopic human bronchial epithelial cells. Eur Respir J. (2008) 32(3):755–62. doi: 10.1183/09031936.00129107
26. Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. (2004) 64(15):5245–50. doi: 10.1158/0008-5472.Can-04-0496
27. Fuchs B, Braun A. Improved mouse models of allergy and allergic asthma—chances beyond ovalbumin. Curr Drug Targets. (2008) 9(6):495–502. doi: 10.2174/138945008784533589
28. Kumar RK, Herbert C, Foster PS. The “classical” ovalbumin challenge model of asthma in mice. Curr Drug Targets. (2008) 9(6):485–94. doi: 10.2174/138945008784533561
29. Nials AT, Uddin S. Mouse models of allergic asthma: acute and chronic allergen challenge. Dis Model Mech. (2008) 1(4–5):213–20. doi: 10.1242/dmm.000323
30. Simpson AB, Yousef E, Hossain J. Association between peanut allergy and asthma morbidity. J Pediatr. (2010) 156(5):777–81.e1. doi: 10.1016/j.jpeds.2009.11.080
31. di Palmo E, Gallucci M, Cipriani F, Bertelli L, Giannetti A, Ricci G. Asthma and food allergy: which risks? Medicina (Kaunas). (2019) 55(9):509. doi: 10.3390/medicina55090509
32. Aun MV, Bonamichi-Santos R, Arantes-Costa FM, Kalil J, Giavina-Bianchi P. Animal models of asthma: utility and limitations. J Asthma Allergy. (2017) 10:293–301. doi: 10.2147/JAA.S121092
34. Prieto J, Van Der Ploeg I, Roquet A, Gigliotti D, Dahlén B, Eklund A, et al. Cytokine mRNA expression in patients with mild allergic asthma following low dose or cumulative dose allergen provocation. Clin Exp Allergy. (2001) 31(5):791–800. doi: 10.1046/j.1365-2222.2001.01078.x
35. Breiteneder H, Peng YQ, Agache I, Diamant Z, Eiwegger T, Fokkens WJ, et al. Biomarkers for diagnosis and prediction of therapy responses in allergic diseases and asthma. Allergy. (2020) 75(12):3039–68. doi: 10.1111/all.14582
36. Ogulur I, Pat Y, Ardicli O, Barletta E, Cevhertas L, Fernandez-Santamaria R, et al. Advances and highlights in biomarkers of allergic diseases. Allergy. (2021) 76(12):3659–86. doi: 10.1111/all.15089
37. Li XM, Serebrisky D, Lee SY, Huang CK, Bardina L, Schofield BH, et al. A murine model of peanut anaphylaxis: T- and B-cell responses to a major peanut allergen mimic human responses. J Allergy Clin Immunol. (2000) 106(1 Pt 1):150–8. doi: 10.1067/mai.2000.107395
38. Dolence JJ. Induction of peanut allergy through inhalation of peanut in mice. Methods Mol Biol. (2021) 2223:19–35. doi: 10.1007/978-1-0716-1001-5_2
39. Paolucci M, Wuillemin N, Homère V, Bieli D, Köhli A, Ballmer-Weber B, et al. Targeting Ara h 2 with human-derived monoclonal antibodies prevents peanut-induced anaphylaxis in mice. Allergy. (2023) 78(6):1605–14. doi: 10.1111/all.15659
40. Kim DI, Song MK, Lee K. Comparison of asthma phenotypes in OVA-induced mice challenged via inhaled and intranasal routes. BMC Pulm Med. (2019) 19(1):241. doi: 10.1186/s12890-019-1001-9
41. Tran TN, Khatry DB, Ke X, Ward CK, Gossage D. High blood eosinophil count is associated with more frequent asthma attacks in asthma patients. Ann Allergy Asthma Immunol. (2014) 113(1):19–24. doi: 10.1016/j.anai.2014.04.011
42. Fischer R, McGhee JR, Vu HL, Atkinson TP, Jackson RJ, Tomé D, et al. Oral and nasal sensitization promote distinct immune responses and lung reactivity in a mouse model of peanut allergy. Am J Pathol. (2005) 167(6):1621–30. doi: 10.1016/s0002-9440(10)61246-1
43. Gans MD, Gavrilova T. Understanding the immunology of asthma: pathophysiology, biomarkers, and treatments for asthma endotypes. Paediatr Respir Rev. (2020) 36:118–27. doi: 10.1016/j.prrv.2019.08.002
44. Cui J, Pazdziorko S, Miyashiro JS, Thakker P, Pelker JW, Declercq C, et al. TH1-mediated airway hyperresponsiveness independent of neutrophilic inflammation. J Allergy Clin Immunol. (2005) 115(2):309–15. doi: 10.1016/j.jaci.2004.10.046
45. Lloyd CM, Hessel EM. Functions of T cells in asthma: more than just T(H)2 cells. Nat Rev Immunol. (2010) 10(12):838–48. doi: 10.1038/nri2870
46. Hoh RA, Joshi SA, Lee JY, Martin BA, Varma S, Kwok S, et al. Origins and clonal convergence of gastrointestinal IgE(+) B cells in human peanut allergy. Sci Immunol. (2020) 5(45):eaay4209. doi: 10.1126/sciimmunol.aay4209
47. Meng W, Zhang B, Schwartz GW, Rosenfeld AM, Ren D, Thome JJC, et al. An atlas of B-cell clonal distribution in the human body. Nat Biotechnol. (2017) 35(9):879–84. doi: 10.1038/nbt.3942
48. Burton OT, Stranks AJ, Tamayo JM, Koleoglou KJ, Schwartz LB, Oettgen HC. a humanized mouse model of anaphylactic peanut allergy. J Allergy Clin Immunol. (2017) 139(1):314–22.e9. doi: 10.1016/j.jaci.2016.04.034
49. Wang B, Hu J, Liu Y, Liu Q, Li D. Food allergy promotes a Th2/Th17 response that drives house dust mite-induced allergic airway inflammation in humanized mice. Clin Exp Immunol. (2020) 202(3):300–7. doi: 10.1111/cei.13504
50. Storni F, Cabral-Miranda G, Roesti E, Zha L, Engeroff P, Zeltins A, et al. A single monoclonal antibody against the peanut allergen Ara h 2 protects against systemic and local peanut allergy. Int Arch Allergy Immunol. (2020) 181(5):334–41. doi: 10.1159/000505917
51. van Splunter M, Liu L, van Neerven RJJ, Wichers HJ, Hettinga KA, de Jong NW. Mechanisms underlying the skin-gut cross talk in the development of IgE-mediated food allergy. Nutrients. (2020) 12(12):3830. doi: 10.3390/nu12123830
52. Chheang C, Guinand S, von Garnier C, Sartori C. New perspectives of biological therapy for severe asthma in adults and adolescents. Swiss Med Wkly. (2022) 152:w30176. doi: 10.4414/smw.2022.w30176
53. Pagovich OE, Wang B, Chiuchiolo MJ, Kaminsky SM, Sondhi D, Jose CL, et al. Anti-hIgE gene therapy of peanut-induced anaphylaxis in a humanized murine model of peanut allergy. J Allergy Clin Immunol. (2016) 138(6):1652–62.e7. doi: 10.1016/j.jaci.2016.03.053
54. Lange L, Klimek L, Beyer K, Blümchen K, Novak N, Hamelmann E, et al. White paper on peanut allergy—part 1: epidemiology, burden of disease, health economic aspects. Allergo J Int. (2021) 30(8):261–9. doi: 10.1007/s40629-021-00189-z
55. Pajno GB, Fernandez-Rivas M, Arasi S, Roberts G, Akdis CA, Alvaro-Lozano M, et al. EAACI Guidelines on allergen immunotherapy: IgE-mediated food allergy. Allergy. (2018) 73(4):799–815. doi: 10.1111/all.13319
56. Vickery BP, Vereda A, Casale TB, Beyer K, du Toit G, Hourihane JO, et al. AR101 oral immunotherapy for peanut allergy. N Engl J Med. (2018) 379(21):1991–2001. doi: 10.1056/NEJMoa1812856
57. Kim EH, Patel C, W A. Burks: immunotherapy approaches for peanut allergy. Expert Rev Clin Immunol. (2020) 16(2):167–74. doi: 10.1080/1744666x.2019.1708192
58. Nurmatov U, Dhami S, Arasi S, Pajno GB, Fernandez-Rivas M, Muraro A, et al. Allergen immunotherapy for IgE-mediated food allergy: a systematic review and meta-analysis. Allergy. (2017) 72(8):1133–47. doi: 10.1111/all.13124
59. Gevaert P, De Craemer J, De Ruyck N, Rottey S, de Hoon J, Hellings PW, et al. Novel antibody cocktail targeting Bet v 1 rapidly and sustainably treats birch allergy symptoms in a phase 1 study. J Allergy Clin Immunol. (2022) 149(1):189–99. doi: 10.1016/j.jaci.2021.05.039
60. Shamji MH, Singh I, Layhadi JA, Ito C, Karamani A, Kouser L, et al. Passive prophylactic administration with a single dose of anti-Fel d 1 monoclonal antibodies REGN1908-1909 in cat allergen-induced allergic rhinitis: a randomized, double-blind, placebo-controlled clinical trial. Am J Respir Crit Care Med. (2021) 204(1):23–33. doi: 10.1164/rccm.202011-4107OC
61. Durham SR, Shamji MH. Allergen immunotherapy: past, present and future. Nat Rev Immunol. (2023) 23(5):317–28. doi: 10.1038/s41577-022-00786-1
Keywords: mouse model, peanut allergy, asthma, histology, cytokines
Citation: Paolucci M, Antz N, Homère V, Kolm I, Kündig TM and Johansen P (2024) A murine model of peanut-allergic asthma. Front. Allergy 5:1378877. doi: 10.3389/falgy.2024.1378877
Received: 30 January 2024; Accepted: 11 April 2024;
Published: 25 April 2024.
Edited by:
Ana Rebane, University of Tartu, EstoniaReviewed by:
Kapil Sirohi, National Jewish Health, United StatesBrandi T. Johnson-Weaver, Duke University, United States
© 2024 Paolucci, Antz, Homère, Kolm, Kündig and Johansen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pål Johansen pal.johansen@usz.ch
 Marta Paolucci
Marta Paolucci Nathalie Antz1
Nathalie Antz1  Thomas M. Kündig
Thomas M. Kündig Pål Johansen
Pål Johansen