Novel insights into the ontogeny of basophils
- Institute of Research, Tokyo Medical and Dental University (TMDU), Tokyo, Japan
Basophils are the least common granulocytes, accounting for <1% of peripheral blood leukocytes. In the last 20 years, analytical tools for mouse basophils have been developed, and we now recognize that basophils play critical roles in various immune reactions, including the development of allergic inflammation and protective immunity against parasites. Moreover, the combined use of flow cytometric analyses and knockout mice has uncovered several progenitor cells committed to basophils in mice. Recently, advancements in single-cell RNA sequencing (scRNA-seq) technologies have challenged the classical view of the differentiation of various hematopoietic cell lineages. This is also true for basophil differentiation, and studies using scRNA-seq analysis have provided novel insights into basophil differentiation, including the association of basophil differentiation with that of erythrocyte/megakaryocyte and the discovery of novel basophil progenitor cells in the mouse bone marrow. In this review, we summarize the recent findings of basophil ontogeny in both mice and humans, mainly focusing on studies using scRNA-seq analyses.
1 Introduction
Basophils are the rarest granulocytes, accounting for ∼1% of peripheral blood leukocytes in both mice and humans. Since their discovery more than 140 years ago by Paul Ehrlich, the functional significance of basophils in health and diseases remained largely unnoticed, partly due to their rarity and functional similarities with tissue-resident mast cells, including basophilic granules, surface expression of high-affinity IgE receptors (FcεRI), and release of histamines upon activation. Hence, basophils had erroneously been considered as blood-circulating mast cells that exert only redundant functions to mast cells. However, recent studies have revealed that basophils and mast cells display distinct transcriptomic signatures (1), indicating the non-redundant role of basophils. Intriguingly, basophils are found in the peripheral blood of some snapping turtles and teleost fishes (2, 3), indicating the evolutional conservation of basophils and the unique role of basophils in animals.
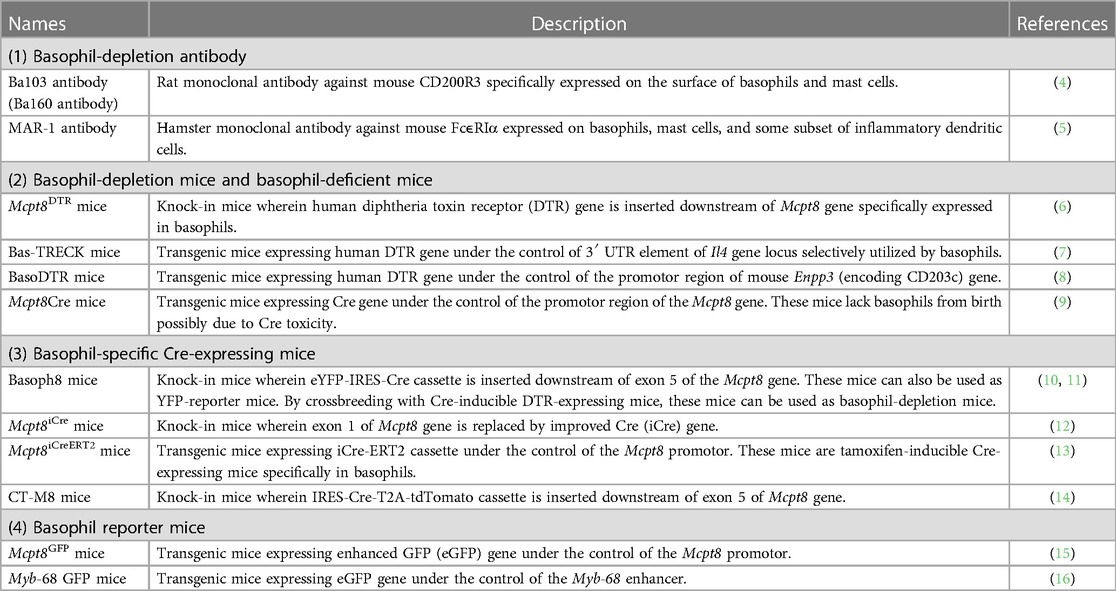
In these 10–20 years, researchers have developed an array of analytical tools for mouse basophils, such as basophil-depleting antibodies, engineered mice that can specifically deplete basophils, basophil reporter mice, and basophil-specific Cre-expressing mice (Table 1) (4–16). Under such tools, basophils are now found to play a critical role in various immune reactions, including chronic allergic inflammation, protective immunity against parasites, autoimmune reactions, and tumor immunity (17–20). Recent advancements in single-cell transcriptomic techniques have shed new light on the understanding of basophil biology. Studies using single-cell RNA sequencing (scRNA-seq) analysis have elucidated the critical roles of basophils in alveolar macrophage maturation, repair from myocardial infarction, kidney fibrosis, and induction and resolution of skin allergic inflammation (21–26). Moreover, single-cell transcriptomics have also revealed the differentiation trajectory of basophils and mast cells. Fate tracking experiments and single-cell transcriptomic analyses have identified the developmental origins of mast cells which is discussed in detail by other reviews (27–29). Therefore, in this review, we will focus on novel insights into the ontogeny of basophils, mainly identified by using single-cell transcriptomic techniques.
2 Progenitor populations committed to basophils and mast cells
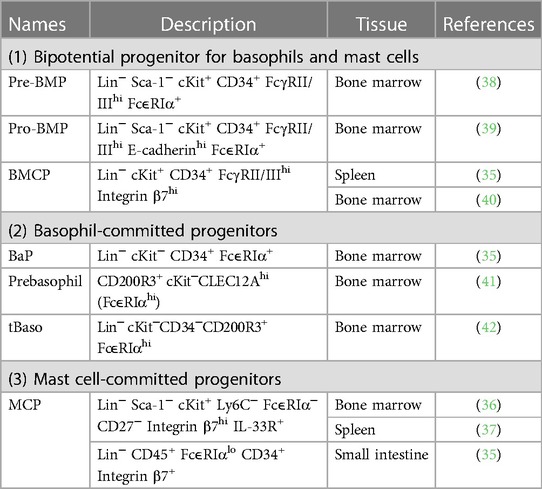
Differentiation and maturation of basophils occur in the bone marrow, and mature basophils circulate in the bloodstream under homeostatic conditions (30, 31). In contrast, mucosal-type mast cells initially differentiate within the bone marrow, and immature mast cells migrate to the tissues where they undergo further maturation (27–29). Moreover, recent fate tracking studies identified that connective tissue-type mast cells arise from embryonic origin and appear to be independent of bone marrow progenitor under homeostatic conditions (32–34). Several progenitor populations which possess potentials to differentiate into basophils or mast cells are identified in mouse bone marrow (Table 2) (35–42). The unipotent basophil progenitors (BaPs) and progenitors for mast cells (mast cell progenitors; MCPs) are identified in the mouse bone marrow (35) and in the intestine, spleen and bone marrow (35–37), respectively.
In mice, the differentiation trajectory of basophils and mast cells are closely overlapped. Consistent with this assumption, ex vivo culture of bone marrow-derived progenitor cells can generate both basophils and mast cells, indicating the presence of common basophil and mast cell precursors in the bone marrow (43). Indeed, bipotential progenitor cells that can produce both basophils and mast cells have been identified in the mouse bone marrow and spleen. In 2013, Qi et al. identified that FcεRIαhi granulocyte–macrophage progenitors (GMPs) in the bone marrow possess a highly enriched capacity for differentiating into basophils and mast cells, and FcεRIαhi GMPs were considered pre-basophil/mast cell progenitors (pre-BMPs) (38). Single-cell colony-forming assay identified that ∼40% of pre-BMPs can produce both basophil and mast cell lineages, indicating the presence of basophil–mast cell common progenitors in pre-BMP populations. A later study has identified E-cadherin+FcεRIα− GMPs named as pro-BMPs in the bone marrow, which are the immediate precursor of pre-BMPs and can differentiate into basophils and mast cells (39).
Besides pre- and pro-BMPs in the bone marrow, bipotential basophil/mast cell progenitor cells defined as Lin−Sca-1−c-Kit+ Integrin β7hi FcγRII/IIIhi basophil/mast cell progenitors (BMCPs) are also found in the mouse spleen (35). Colony-forming experiments identified that BMCPs can differentiate into both basophils and mast cells. However, the differentiation potential of spleen BMCPs is controversial because a later study indicates that BMCPs in the spleen preferentially differentiate into mast cells and have little capacity for differentiating into basophils (44). Although the first report only identified BMCPs in the spleen and not in the bone marrow (35), a recent study has identified the presence of BMCP in the mouse bone marrow (40). Single-cell colony-forming assay revealed that ∼10% of BMCPs in the bone marrow can differentiate into both basophils and mast cells. Collectively, in mice, bipotential basophil and mast cell progenitor cells, including pre-BMPs and BMCPs, produce unipotent progenitor cells of each cell type, such as BaPs and MCPs, which further differentiate into mature basophils and mast cells, respectively. However, whether BMCPs and pre-BMPs are the different cell types is unclear. Comprehensive single-cell transcriptomic analysis that can characterize both BMCPs and pre-BMPs is required to resolve this problem.
Human basophils have been believed to differentiate from common basophil/eosinophil progenitors (45). In the 1980s, the methylcellulose-based culture of human bone marrow or peripheral blood cells was reported to promote the generation of hybrid cells containing both basophilic and eosinophilic granules (45, 46). Moreover, hybrid basophilic/eosinophilic cells can also be detected in the peripheral blood or bone marrow in patients with myeloid leukemia (47). Another study showed that hybrid basophilic/eosinophilic cells can be generated in vitro from CD133lo/−CD34+ cells derived from umbilical cord blood (48). Therefore, basophils may differentiate from bipotential basophil/eosinophil progenitors in humans, although a later study identified that hybrid basophilic/eosinophilic cells mainly differentiate into eosinophils but not into basophils (49). However, whether the differentiation of human basophils is associated with that of human mast cells is unclear, in contrast to the case of mouse basophils.
3 Basophil differentiation trajectory uncovered with scRNA-seq analyses
Classically, hematopoietic stem cells initially segregate into common myeloid progenitors (CMPs) and common lymphoid progenitors (CLPs), and CMPs further differentiate into megakaryocyte/erythrocyte progenitors (MEPs) and granulocyte/macrophage progenitors (GMPs) (Figure 1A) (50–52). In this model, all members of granulocytes, namely basophils, eosinophils, and neutrophils, are derived from the same precursor cells. Providing that basophils and mast cells differentiate from bipotential common progenitor cells in mice, basophils and mast cells may be derived from GMPs. In line with this assumption, both pre-BMPs and BMCPs can be induced ex vivo from GMPs (35, 38). However, several studies have challenged the classical model of hematopoietic cell differentiation. Franco et al. identified that single-cell culture of Sca-1loFlt3− CMPs but not Sca-1lo GMPs can produce mast cells, indicating the differentiation trajectory of mast cells and granulocytes can be segregated at the CMP stage (53). Moreover, recent studies using scRNA-seq have identified that the differentiation trajectory of basophil/eosinophil/mast cell lineages is coupled with that of erythrocyte/megakaryocyte rather than that of neutrophil/monocyte in both mice and humans (54–58), leading to the generation of the revised model of hematopoietic cell differentiation (Figure 1B).
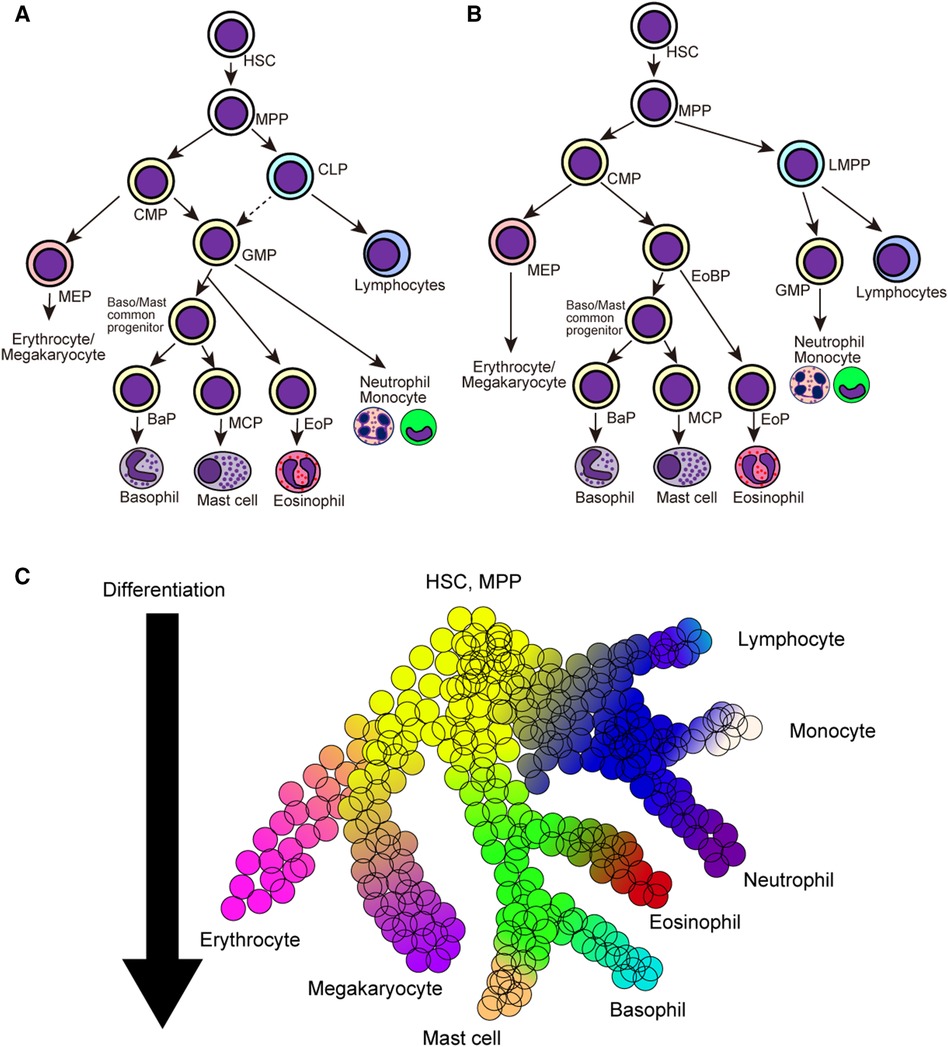
Figure 1. Classical and revised model of hematopoietic cell differentiation. (A) Classical model of hematopoietic cell differentiation tree. Hematopoietic stem cells (HSCs) and multi-potential progenitors (MPPs) firstly differentiate into common myeloid precursors (CMPs) and common lymphoid precursors (CLPs), and CMPs further differentiate into megakaryocyte/erythrocyte precursors (MEPs) and granulocyte/macrophage precursors (GMPs). (B) The revised model of hematopoietic cell differentiation. HSCs and MPPs differentiate into common myeloid precursors (CMPs) and lympho-myeloid primed progenitors (LMPPs). CMPs differentiate into MEPs and eosinophil-basophil progenitor (EoBP), while LMPPs differentiate into GMPs and lymphoid progenitor cells. (C) Continuous hematopoietic differentiation landscape model emerged after the advancements in the scRNA-seq analysis. Hematopoietic cell lineages continuously change their cellular states during the differentiation. HSC, hematopoietic stem cell; MPP, multi-potential progenitor; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; LMPP, lympho-myeloid primed progenitor; MEP, megakaryocyte/erythrocyte progenitor; GMP, granulocyte/macrophage progenitor; BaP, basophil progenitor; MCP, mast cell progenitor; EoP, eosinophil progenitor; EoBP, eosinophil/basophil progenitor.
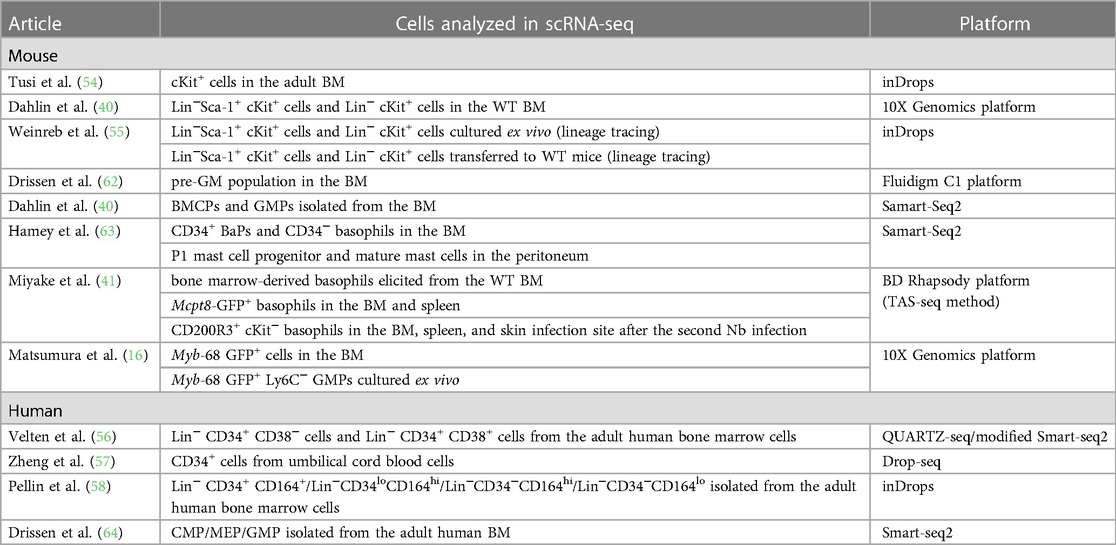
Advancements in single-cell transcriptomic techniques have exposed novel and comprehensive insights into the differentiation trajectories of various hematopoietic cell lineages. A series of single-cell transcriptomic datasets have challenged the classical stepwise hematopoietic cell differentiation model wherein cells are transitioned to the discrete progenitor states during their differentiation. Instead, multiple studies have proposed the continuous differentiation landscape model, which indicates a continuous change in cellular states during the differentiation of various hematopoietic cell lineages (Figure 1C) (59–61). This is also true for basophil/mast cell differentiation, and recent studies with scRNA-seq analysis have elucidated continuous gene expression changes during basophil differentiation (Table 3) (16, 40, 41, 54–58, 62–64). In the following sections, we summarize the recent findings in basophil differentiation mainly focusing on the following points: (1) the association of the basophil differentiation trajectory to the erythrocyte/megakaryocyte differentiation trajectory in mice and humans; (2) the association of basophil differentiation with the differentiation of mast cells or eosinophils; (3) the characterization of basophil progenitor cells located upstream of mature basophils.
3.1 Association of basophil differentiation trajectory to erythrocyte/megakaryocyte differentiation trajectory
Drissen et al. conducted a scRNA-seq analysis of mouse pre-granulocyte–macrophage progenitors (pre-GMs) and identified the presence of distinct pre-GM populations, namely GATA1+ and GATA1− pre-GMs (62). In vitro culture experiments identified that GATA1− pre-GMs preferentially differentiate into neutrophils and monocytes, whereas GATA1+ pre-GMs can differentiate into both eosinophil/mast cell and erythrocyte/megakaryocyte lineages. Tusi et al. conducted a scRNA-seq analysis of c-Kit+ cell populations isolated from adult mouse bone marrow by using the inDrops technique (54). Population balance analysis predicted that the differentiation trajectory of basophils/mast cells was coupled with erythrocyte differentiation rather than neutrophil or monocyte differentiation. Supporting this notion, ∼25% of single-cell cultured c-Kit+CD55+CD105+CD71−CD150+ progenitor cells produced both TER-119+ erythroid cells and FcεRI+ basophils. Weinreb et al. traced hematopoietic lineage differentiation by combining DNA barcoding with scRNA-seq technologies (55). They conducted scRNA-seq analysis of DNA-barcoded mouse hematopoietic stem cells cultured ex vivo for 2, 4, and 6 days. Lineage tracing identified the presence of progenitor cells that differentiate into basophil/mast cell/eosinophil and erythrocyte/megakaryocyte lineages. Lineage hierarchy tree constructed by clonal couplings inferred that the differentiation trajectory of basophil/mast cell/eosinophil is coupled with that of erythrocyte/megakaryocyte in vitro. However, the conclusion was different when the authors used scRNA-seq datasets obtained from in vivo differentiated DNA-barcoded hematopoietic stem cells, indicating further experimental evidence to verify the coupling of differentiation pathways between basophil/mast cell and erythrocyte lineages.
The association of basophil and erythrocyte differentiation is also reported in humans. Görgens et al. identified that CD133lo/−CD34+ hematopoietic progenitor cells produce both basophils/eosinophils and erythrocytes/megakaryocytes, although these progenitor populations have limited potential to differentiate into neutrophils (48). Drissen et al. conducted scRNA-seq analysis of Lin−CD34+CD38+CD123+CD45RA− CMPs in the human bone marrow and identified the heterogeneity within the CMP population (64). Thus, they showed that CMP populations can be subdivided by surface expressions of two markers: CD114 (G-CSFR) and CD131 (IL-3Rβ). Single-cell culture experiments showed that CD114+ CMPs produce neutrophil and monocyte lineages, whereas CD131+ CMPs produce both eosinophil/mast cell/basophil and megakaryocyte/erythrocyte lineages.
A series of scRNA-seq analyses further support the association of basophil/eosinophil differentiation and erythrocyte/megakaryocyte differentiation. Velten et al. combined flow cytometry and scRNA-seq analyses of human Lin−CD34+ adult human bone marrow cells and identified that hematopoietic stem cells gradually acquire lineage-committed gene expression profiles during their differentiation (56). Based on the gene expression profiles, early progenitors committed to eosinophils/basophils/mast cells expressing CLC, HDC, and PRG2 are identified in the Lin−CD34+CD38+ population. Progenitors of eosinophils/basophils/mast cells display CD34+CD38+CD10−CD45RA−CD135mid MEP-like surface expression phenotype, indicating the close relationship between eosinophil/basophil/mast cell and erythrocyte/megakaryocyte differentiation. Similarly, scRNA-seq analysis of CD34+ human cord blood-derived progenitor cells revealed the coupling of eosinophil/basophil/mast cell differentiation to erythrocyte differentiation by using a minimum spanning tree algorithm (57). Pellin et al. conducted scRNA-seq analysis of Lin− adult human bone marrow cells and showed the potential association between the differentiation trajectory of basophils and erythrocytes/megakaryocytes by using population balance analysis (58). Ex vivo culture experiments identified that Lin−CD34+CD135 (Flt3)+ bone marrow precursor cells preferentially differentiate into monocytes, whereas Lin−CD34+CD135− cells can produce both basophils and megakaryocytes. A recent study identified that FcεRIα+Integrin-β7+CD203c− CMPs can produce basophils, mast cells, and erythrocytes, whereas FcεRIα+CD203c+ CMPs produce only basophils and mast cells but not erythrocytes (65). These results support the notion that the differentiation trajectory of basophils/eosinophils/mast cells is associated with that of erythrocytes/megakaryocytes but not that of neutrophils/monocytes.
3.2 Association of the differentiation trajectory of basophils with that of mast cells and eosinophils
Besides the association of the differentiation trajectory of basophils with that of the erythrocytes/megakaryocytes, scRNA-seq analysis revealed the coupled differentiation pathways of basophils and mast cells in mice. Dahlin et al. confirmed the association between basophil and mast cell differentiation pathway by using scRNA-seq analysis of mouse hematopoietic progenitor populations in the bone marrow (40). Force-directed graph visualization identified that entry points into basophil and mast cell differentiation are close, indicating the coupled differentiation trajectory of basophils and mast cells. By contrast, the association of the basophil differentiation trajectory with that of eosinophils is less clarified in mice. The scRNA-seq datasets by Dahlin et al. mapped the eosinophil differentiation trajectory close to that of neutrophils (40). By contrast, scRNA-seq datasets of c-Kit+ bone marrow progenitor cells mapped basophils and eosinophils in the same cluster (54, 58), indicating the closely coupled differentiation trajectory. Assessment of the transcriptome of eosinophils by using single-cell RNA-seq is technically challenging, possibly because of the abundant amounts of RNase in their granules, their sensitivity to the shear stress, and degranulation. Recently, Gurtner et al. reported the scRNA-seq data of eosinophils isolated from various tissues in mice by preventing shear stress and resulting mRNA degradation (66). By using such approaches, the relationship among the ontogeny of eosinophil, mast cell, and basophil lineages would be clarified.
Contrary to the case of mice, the association of the basophil-differentiation trajectory with that of mast cells or eosinophils is less clear in humans. Because studies using scRNA-seq have positioned basophils close to mast cells or eosinophils (56, 57), the differentiation of basophils, mast cells, and eosinophils may be closely coupled to each other in humans.
3.3 Identification of basophil lineage-committed progenitors by using scRNA-seq
In addition to the characterization of the basophil differentiation trajectory, single-cell transcriptomic analyses were also used to identify basophil-committed progenitor cell populations. By combining highly sensitive scRNA-seq (67) and flow cytometric analyses, Miyake et al. identified CLEC12Ahic-Kitlo pre-basophils within the mouse bone marrow and bone marrow-derived basophils (BMBAs) (Figure 2) (41). CLEC12Ahi pre-basophils and CLEC12Alo mature basophils showed distinct cell morphology and cell surface markers. CLEC12Ahi pre-basophils showed kidney-shaped indented nuclei with large cell bodies, whereas CLEC12Alo mature basophils showed ring-shaped nuclei with smaller cell bodies. CLEC12Ahi pre-basophils and CLEC12Alo mature basophils in the bone marrow showed FcεRIαhiCD49bint and FcεRIαloCD49bhi surface expression profiles, respectively. RNA velocity and pseudotime trajectory analyses inferred the sequential differentiation trajectory from Fcer1ahiCd34hiKithi pre-BMP-like populations into Clec12ahiCd9intKitlo pre-basophils that further differentiate into Clec12aloCd9hiKitlo mature basophils. Consistently, ex vivo culture of CLEC12AhiCD9int pre-basophils for 24 h promoted the differentiation into CLEC12AloCD9hi mature basophils even without IL-3. Notably, CD34 + FcεRIα BaPs also showed CLEC12AhiCD9int phenotype, and only 10% of CLEC12Ahi populations showed positive but low expression of CD34, indicating the possibility that CD34+ BaPs are included in the CLEC12Ahi pre-basophil populations. Supporting this notion, scRNA-seq analysis revealed that a small fraction (10%–20%) of Clec12ahi cells in the pre-basophil cluster showed Cd34hiKitlo phenotype. Transcriptomic analysis revealed that pre-basophils and mature basophils highly expressed genes associated with cell proliferation and effector functions, respectively. Consistently, pre-basophils showed higher proliferative capacity than mature basophils. By contrast, mature basophils showed greater capacity for degranulation and de novo cytokine production in response to antigen/IgE stimulation. Notably, when basophils are stimulated with non-IgE stimulation, such as IL-3, IL-3+ IL-18, IL-3 + IL-33, and IL-3 + LPS, pre-basophils produced a greater amount of IL-4 than mature basophils, indicating that the responsiveness to stimulants is different between pre-basophils and mature basophils. Similarly, CD34+ basophil progenitors produced a greater amount of type 2 and proinflammatory cytokines than mature basophils in response to IL-33 stimulation (68). Thus, the bulk transcriptomic analysis identified that pre-basophils showed limited gene expression changes in response to antigen/IgE stimulation although FcεRIα expression on pre-basophils was higher than mature basophils. Bulk transcriptomic analysis further showed that IL-3-stimulated pre-basophils and mature basophils showed distinct gene expression profiles.
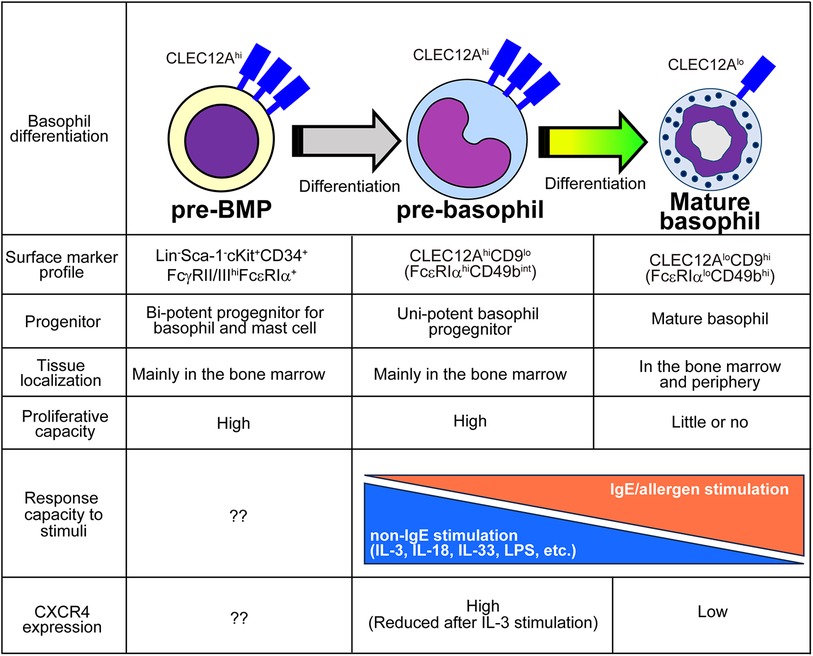
Figure 2. Characteristics of pre-BMPs, pre-basophils, and mature basophils. The distinct properties among pre-BMPs, pre-basophils and mature basophils are summarized.
Reanalysis of previously published scRNA-seq analysis dataset obtained by Weinreb et al. identified Clec12ahi pre-basophils among Mcpt8+ basophil populations. Likewise, pre-basophil-like precursor populations were also reported by other groups. The scRNA-seq analysis of basophil lineage populations by using Myb-68 GFP+ cells identified two heterogeneous basophil populations, namely Fcer1ahi basophil1 and Fcer1aint basophil2 populations (16), which correspond to pre-basophils and mature basophils, respectively. Park et al. identified CD34−CD200R3+FcεRIαhi transitional basophils (tBasos) in the bone marrow with high proliferative capacity (42).
Under homeostatic conditions, pre-basophils reside in the bone marrow and are rarely detected outside the bone marrow. By contrast, 7 days after infection with intestinal helminth Nippostrongylus brasiliensis (Nb), CLEC12Ahi pre-basophils egress the bone marrow and can be detected in the spleen, peripheral blood, and lungs. Notably, CLEC12Ahi pre-basophils detected in the infected lungs retained their proliferative capacity and expressed IL-4 comparable to mature basophils. Moreover, the scRNA-seq analysis of basophils accumulating in the skin infection site after the second Nb infection demonstrated that pre-basophils detected in the skin infection site show transcriptional profiles similar to those of pre-basophils in the bone marrow of noninfected mice.
IL-3 promotes blood basophilia during Nb infections (69, 70). Notably, mice treated with IL-3 complexes also showed the emergence of CLEC12Ahi pre-basophils in the peripheral blood. Therefore, IL-3 upregulation caused by helminth infection may promote the egress of pre-basophils from the bone marrow. Further mechanistic study identified that IL-3 promotes the downregulation of chemokine receptor CXCR4 on basophils in the bone marrow, which prevents the retention of pre-basophils in the bone marrow and promotes the appearance of pre-basophils in the periphery.
Contrary to the case of mouse basophils, the committed progenitors for human basophils remain less identified. Providing that human basophil-like cells can be induced by culturing CD34+ progenitor cells isolated from the adult human bone marrow in the presence of IL-3, progenitors possessing basophil-differentiation potential may be found in the CD34+ cells in the bone marrow (71). Similarly, human mast cells can be generated ex vivo by culturing CD34+c-Kit+ cells isolated from the peripheral blood (72, 73), indicating the presence of mast cell-committed progenitors in the peripheral blood CD34+c-Kit+ cells. In line with this, recent studies have discovered Lin−CD34hic-Kitint/hiFcεRIα+ mast cell-committed precursor cells in the peripheral blood (74, 75).
4 Transcription factors regulating basophil/mast cell fates
Several transcription factors (TFs) regulate the differentiation into basophils and mast cells (Figure 3 and Table 4) (16, 37–39, 42, 44, 76–83). These TFs are divided into two groups: (1) TFs important for both basophil and mast cell lineages; (2) TFs important for basophils but not for mast cells, or vice versa.
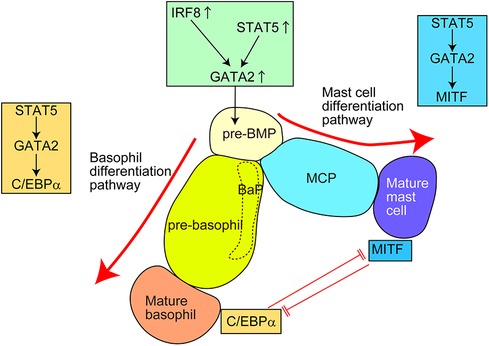
Figure 3. Transcription factors (TFs) regulating the differentiation of basophils and mast cells in mice. For the generation of common basophil/mast cell progenitors (pre-BMPs), the upregulation of GATA2, possibly caused by upregulation of STAT5 and/or IRF8, is required. For the differentiation into basophils, the STAT5–GATA2–C/EBPα signaling pathway is required. For the differentiation into mast cells, the STAT5–GATA2–MITF signaling pathway is required. Pre-BMP, pre-basophil/mast cell progenitor; BaP, basophil progenitor; MCP, mast cell progenitor.
4.1 TFs regulating both basophil and mast cell fates
STAT5 and GATA2 regulate basophil and mast cell differentiation. Bone marrow chimeric mice reconstituted with STAT5-deficient cells showed a markedly reduced number of basophils and tissue mast cells (76, 77). Induced GATA2-deficiency significantly reduced the surface expression of FcεRIα and E-cadherin on basophils and c-Kit on mast cells (39, 78), which almost abolished the generation of basophils and tissue mast cells (38, 78). Considering that STAT5 can directly bind to the promotor region of GATA2 and GATA2 overexpression restores the phenotype of induced STAT5-deficiency, the STAT5–GATA2 signaling may be critical for basophil and mast cell differentiation. Moreover, the STAT5–GATA2 signaling is critical for the survival of mature basophils and mast cells. Notably, haploinsufficiency of Gata2 gene affects the generation of mast cells but not basophils, indicating that mast cells are more heavily dependent on GATA2 TF than basophils, especially regarding their generation. A recent study further identified that GATA2 directly promoted chromatin remodeling at super-enhancer regions thus robustly regulating the expression of mast cell-associated genes and responsiveness to antigen/IgE stimuli (84). Thus, STAT5–GATA2 signaling is critical for both the differentiation and maintenance of basophils and mast cells in mice. However, the factor(s) inducing STAT5 activity for basophil/mast cell differentiation under homeostatic conditions remains unclear.
IL-3 is a growth factor promoting basophil differentiation and proliferation. The ex vivo culture of bone marrow cells in the presence of IL-3 induces the generation of BMBAs. Moreover, systemic administration of IL-3 complexes in mice increases the number of mature basophils and basophil precursor cells in the bone marrow and spleen (77, 85). Thymic stromal lymphopoietin (TSLP) is another growth factor that can promote the differentiation from basophil precursor cells into mature basophils. Systemic administration of TSLP increases the number of mature basophils in the spleen in an IL-3–IL-3R-independent manner (86). Notably, IL-3-elicited basophils and TSLP-elicited basophils show transcriptionally and functionally distinct properties. IL-3-elicited basophils are superior to TSLP-elicited basophils in the degranulation capacity in response to IgE-dependent stimuli. By contrast, TSLP-elicited basophils are more responsive to stimulation by IL-3, IL-18, and IL-33, and produce a greater amount of IL-4 compared with IL-3-elicited basophils. However, neither IL-3 nor TSLP is essential for basophil differentiation, which was evidenced by both IL-3 receptor-deficient mice and TSLP-receptor mice showing a normal number of basophils in the bone marrow (86). Moreover, basophil number was also intact even in the deficiency in the IL-3 and TSLP receptors (86), indicating that factor(s) other than IL-3 or TSLP played critical roles for the STAT5-mediated basophil differentiation.
IRF8 plays critical roles in the development of basophils and mast cells. Mechanistically, IRF8 regulates GATA2 expression in granulocyte progenitors; therefore, IRF8 deficiency impairs the generation of bipotent pre-BMPs in the bone marrow, leading to impaired development of basophils and mast cells in the bone marrow (79). IRF8-deficient mice showed significantly reduced pre-BMPs, BaPs, mature basophils, and MCPs in the bone marrow. However, IRF8-deficient mice showed a normal number of MCPs and mature mast cells in the tissues (e.g., intestine, skin, peritoneal cavity). Considering that the number of BMCPs in the spleen was unaffected by IRF8 deficiency, multiple pathways might have existed to retain tissue mast cell numbers.
The role of GATA1 is implicated in basophil and mast cell differentiation, although the contribution of GATA1 in mast cell differentiation remains controversial (87–89). Among hematopoietic cell lineages, GATA1 plays critical roles in the differentiation of erythrocytes, megakaryocytes, and eosinophils. GATA1-deficient mice result in embryonic lethality due to severe anemia, whereas ΔdblGATA mice lacking the double-GATA enhancer site in the Gata1 promotor region show selective deficiency in eosinophil lineages with only mild anemia (90). The number of mature basophils and basophil progenitor cells in the bone marrow is partially reduced in ΔdblGATA mice, possibly due to the decreased Gata1 expression in basophils (80). Moreover, basophils from ΔdblGATA mice show impaired degranulation and cytokine production in response to antigen/IgE stimulation. By contrast, GATA1-low mutant mice (91) that lack the upstream enhancer and promotor sequences of Gata1 gene displayed an increased frequency of basophils in the bone marrow. Interestingly, basophils from GATA1-low mutant mice show reduced CD49b expression, indicating the possibility that GATA1 directly regulates CD49b expression or GATA1 promotes the differentiation from CD49blo pre-basophils into CD49bhi mature basophils. Importantly, GATA1-low mutant mice showed increased Gata2 expression in basophils, whereas ΔdblGATA mice showed normal Gata2 expression in basophils (39, 80). Therefore, the discrepancy in the basophil numbers between ΔdblGATA mice and GATA1-low mutant mice might stem from the extent of the GATA2 expression in basophils.
A recent report identified that the use of the Myb-68kb enhancer region is largely restricted to basophil and mast cell lineages (16). CRISPR-mediated depletion of Myb-68 enhancer results in the reduced Myb expression and impaired generation of basophils and mast cells in vitro, indicating the role of Myb-68 enhancer in basophil and mast cell differentiation.
4.2 TFs regulating the fate of basophils but not mast cells
C/EBPα and MITF are critical TFs regulating basophil and mast cell differentiation, respectively (38). For differentiation into basophils, the STAT5–GATA2 signaling induces C/EBPα activity to promote basophil-associated gene expression. By contrast, for differentiation into mast cells, the STAT5 signaling induces MITF activity to promote mast cell-associated gene expression. In line with this, MITF-deficient mice lack mature mast cells in various tissues (81, 82). C/EBPα and MITF repress the expression of other genes by directly binding to the promotor region of MITF and CEBP/α gene, respectively. Consistently, inducible knockdown of the C/EBPα gene promotes the differentiation from pre-BMPs to mast cells. Interestingly, inducible loss of the C/EBPα gene in mature basophils induces the generation of c-Kit+ mast cell-like cells. Conversely, approximately half of the bone marrow cells from MITF-mutant mice failed to generate c-Kit+CD49b+ mast cells but generated c-Kit−CD49b+ basophil-like cells when cultured in the presence of IL-3 for 28 days. A similar phenomenon was observed in Ikaros-deficient mice (37). When bone marrow cells from Ikaros-deficient mice were cultured with IL-3 and stem cell factor (SCF) for 6 weeks, some cells showed c-Kit−FcεRIα+ basophil-like surface expression phenotype. Mechanistically, Ikaros directly binds to the promotor region of Cebpa and represses the C/EBPα expression possibly through histone modification.
RUNX1, a critical regulator of hematopoietic stem cells, is involved in basophil differentiation. Distal (P1) and proximal (P2) promotors regulate RUNX1 expression (92). Deficiency in P1 promotor results in significantly reduced mature basophils and basophil progenitor cells in the bone marrow (44). However, P1-RUNX1 deficiency has a limited impact on the number of mast cells and eosinophils, indicating the critical roles of P1-RUNX1 in basophil lineage differentiation. The zinc-finger TF promyelocytic leukemia zinc finger (PLZF encoded by the Zbtb16 gene) is essential for the development of several innate lymphoid cells, including NKT cells, mucosal-associated invariant T (MAIT) cells, and group 2 innate lymphoid cells (ILC2s) (93). A recent study showed that Zbtb16 was highly expressed in basophil progenitor cells (83). PLZF deficiency partially reduced the number of basophil progenitors and mature basophils in the bone marrow. PLZF deficiency also influenced IL-4 production of basophils in response to IgE-dependent stimulation. Therefore, PLZF plays key roles in the development and function of basophils. Although PLZF is expressed in mature mast cells, the effect of PLZF deficiency on mast cells remains unclear.
A recent report has identified that the TF NFIL3 is upregulated during basophil maturation. However, basophil-specific Nifl3-deficient mice show a normal number of mature basophil and basophil precursor (tBaso) populations, indicating the possible roles of NFIL3 in the effector functions of mature basophils. Indeed, basophil-specific Nifl3-deficiency partly impairs IgE-mediated activation of mature basophils (42). Moreover, basophil-specific Nifl3-deficient mice show reduced ear thickening in the hapten oxazolone-induced atopic dermatitis model (42), possibly due to the reduced IL-4 expression in skin-infiltrating basophils.
4.3 TFs involved in human basophil differentiation
A genetic association study using whole genome sequencing data from Estonian Biobank identified the strong association of basophil counts with the SNPs in CEBPA and GATA2 gene loci (94), indicating that GATA2 and CEBP/α possibly regulate basophil differentiation in humans. The mutation in the +39-kb enhancer region of CEBPA (rs787444187) influences the basophil count but not the number of other myeloid lineages. Interestingly, ATAC-seq analysis identified that the open chromatin region containing CEBPA +39-kb enhancer is only present in CMPs but not in GMPs or MEPs, indicating that basophils possibly differentiate from CMPs but not from GMPs or MEPs in humans, which is consistent with the previous findings. When the CEBPA +39-kb enhancer region is mutated in human hematopoietic progenitor cells, the differentiation into basophils is partially blocked, but the differentiation into mast cells is rather enhanced (94), possibly through reduced CEBPA expression.
GATA2 deficiency syndrome is caused by heterozygous and loss-of-function mutation in GATA2 and displays severe abnormalities in multiple myeloid and lymphoid lineages, including monocytopenia, neutropenia, and dendritic cell deficiency (95). Moreover, patients with GATA2 deficiency syndrome develop myeloid neoplasms, including myelodysplastic syndrome and acute myeloid leukemia, with a median age of onset at 17 years. Consistent with the findings in Gata2-deficient mice, mast cells generated from hematopoietic progenitor cells isolated from GATA2 deficiency syndrome showed reduced FcεRIα and c-Kit expressions, resulting in the defective degranulation capacity in response to antigen/IgE and SCF stimulation (96). Moreover, patients with GATA2 deficiency syndrome have decreased surface expression of FcεRIα on peripheral blood basophils. Interestingly, patients with GATA2-deficiency syndrome had a reduced prevalence of IgE-medicated hypersensitivity syndrome possibly due to the reduced FcεRI expression on basophils and mast cells. However, whether GATA2 deficiency in humans affects the generation of basophils and mast cells in the bone marrow remains unclear, although the frequency of basophils in the peripheral blood is unaffected by GATA2 deficiency in humans.
5 Conclusion and perspectives
Recent advancements in the development of scRNA-seq techniques have brought us novel insights into the developmental pathways of hematopoietic cell lineages. This is also the case in the differentiation of basophils and mast cells, and recent studies using scRNA-seq analysis have elucidated the differentiation trajectory of basophils and mast cells in both mice and humans. Indeed, scRNA-seq analyses have confirmed the associated differentiation trajectory between basophils and mast cells in mice. Moreover, a series of scRNA-seq analyses have shown the potential coupling of basophil-differentiation pathway with the erythrocyte/megakaryocyte differentiation pathway in mice and humans. In addition, scRNA-seq studies have identified a novel progenitor population of basophils in the mouse bone marrow.
Knockout mice studies have elucidated several TFs regulating the fate of basophils and mast cells in mice. Further studies using both knockout mice and scRNA-seq analysis would further provide valuable insights into the mechanisms of basophil differentiation in mice. Compared with the case of mouse basophils, TFs regulating human basophils remain unclear. The combined use of scRNA-seq analysis with CRISPR screening or other perturbations (97, 98) would further accelerate our understanding of human basophil differentiation, which might connect to the identification of novel drug targets in basophil-related disorders, including allergic diseases and parasitic infections.
Author contributions
KM: Writing – original draft, Writing – review & editing. JI: Writing – original draft, Writing – review & editing. HK: Writing – original draft, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article.
This work was supported by research grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology [22K007115 (KM)], SENSHIN Medical Research Foundation (KM), Takeda Science Foundation (KM), The Naito Foundation (KM), Astellas Foundation for Research on Metabolic Disorders (KM), Daiichi Sankyo Foundation of Life Science (KM), The Chemo-SeroTherapeutic Research Institute (KM), Grant-in-Aid for JSPS Fellows 23KJ0837 (JI), and JST ACT-X, Grant Number JPMJAX232I (KM).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declared that they were an editorial board member of Frontiers, at the time of submission.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dwyer DF, Barrett NA, Austen KF, Immunological Genome Project C. Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol. (2016) 17(7):878–87. doi: 10.1038/ni.3445
2. Mead KF, Borysenko M, Findlay SR. Naturally abundant basophils in the snapping turtle, Chelydra serpentina, possess cytophilic surface antibody with reaginic function. J Immunol. (1983) 130(1):334–40 doi: 10.4049/jimmunol.130.1.334
3. Odaka T, Suetake H, Maeda T, Miyadai T. Teleost basophils have IgM-dependent and dual Ig-independent degranulation systems. J Immunol. (2018) 200(8):2767–76. doi: 10.4049/jimmunol.1701051
4. Obata K, Mukai K, Tsujimura Y, Ishiwata K, Kawano Y, Minegishi Y, et al. Basophils are essential initiators of a novel type of chronic allergic inflammation. Blood. (2007) 110(3):913–20. doi: 10.1182/blood-2007-01-068718
5. Denzel A, Maus UA, Rodriguez Gomez M, Moll C, Niedermeier M, Winter C, et al. Basophils enhance immunological memory responses. Nat Immunol. (2008) 9(7):733–42. doi: 10.1038/ni.1621
6. Wada T, Ishiwata K, Koseki H, Ishikura T, Ugajin T, Ohnuma N, et al. Selective ablation of basophils in mice reveals their nonredundant role in acquired immunity against ticks. J Clin Invest. (2010) 120(8):2867–75. doi: 10.1172/JCI42680
7. Sawaguchi M, Tanaka S, Nakatani Y, Harada Y, Mukai K, Matsunaga Y, et al. Role of mast cells and basophils in IgE responses and in allergic airway hyperresponsiveness. J Immunol. (2012) 188(4):1809–18. doi: 10.4049/jimmunol.1101746
8. Matsuoka K, Shitara H, Taya C, Kohno K, Kikkawa Y, Yonekawa H. Novel basophil- or eosinophil-depleted mouse models for functional analyses of allergic inflammation. PLoS One. (2013) 8(4):e60958. doi: 10.1371/journal.pone.0060958
9. Ohnmacht C, Schwartz C, Panzer M, Schiedewitz I, Naumann R, Voehringer D. Basophils orchestrate chronic allergic dermatitis and protective immunity against helminths. Immunity. (2010) 33(3):364–74. doi: 10.1016/j.immuni.2010.08.011
10. Sullivan BM, Liang HE, Bando JK, Wu D, Cheng LE, McKerrow JK, et al. Genetic analysis of basophil function in vivo. Nat Immunol. (2011) 12(6):527–35. doi: 10.1038/ni.2036
11. Pellefigues C, Mehta P, Prout MS, Naidoo K, Yumnam B, Chandler J, et al. The Basoph8 mice enable an unbiased detection and a conditional depletion of basophils. Front Immunol. (2019) 10:2143. doi: 10.3389/fimmu.2019.02143
12. Shibata S, Miyake K, Tateishi T, Yoshikawa S, Yamanishi Y, Miyazaki Y, et al. Basophils trigger emphysema development in a murine model of COPD through IL-4-mediated generation of MMP-12-producing macrophages. Proc Natl Acad Sci U S A. (2018) 115(51):13057–62. doi: 10.1073/pnas.1813927115
13. Yoshikawa S, Oh-Hora M, Hashimoto R, Nagao T, Peters L, Egawa M, et al. Pivotal role of STIM2, but not STIM1, in IL-4 production by IL-3-stimulated murine basophils. Sci Signal. (2019) 12(576):eaav2060. doi: 10.1126/scisignal.aav2060
14. Tchen J, Simon Q, Chapart L, Pellefigues C, Karasuyama H, Miyake K, et al. CT-M8 mice: a new mouse model demonstrates that basophils have a nonredundant role in lupus-like disease development. Front Immunol. (2022) 13:900532. doi: 10.3389/fimmu.2022.900532
15. Miyake K, Shiozawa N, Nagao T, Yoshikawa S, Yamanishi Y, Karasuyama H. Trogocytosis of peptide-MHC class II complexes from dendritic cells confers antigen-presenting ability on basophils. Proc Natl Acad Sci U S A. (2017) 114(5):1111–6. doi: 10.1073/pnas.1615973114
16. Matsumura T, Totani H, Gunji Y, Fukuda M, Yokomori R, Deng J, et al. A Myb enhancer-guided analysis of basophil and mast cell differentiation. Nat Commun. (2022) 13(1):7064. doi: 10.1038/s41467-022-34906-1
17. Charles N. Autoimmunity, IgE and FcepsilonRI-bearing cells. Curr Opin Immunol. (2021) 72:43–50. doi: 10.1016/j.coi.2021.03.003
18. Peng J, Siracusa MC. Basophils in antihelminth immunity. Semin Immunol. (2021) 53:101529. doi: 10.1016/j.smim.2021.101529
19. Miyake K, Ito J, Karasuyama H. Role of basophils in a broad spectrum of disorders. Front Immunol. (2022) 13:902494. doi: 10.3389/fimmu.2022.902494
20. Poto R, Loffredo S, Marone G, Di Salvatore A, de Paulis A, Schroeder JT, et al. Basophils beyond allergic and parasitic diseases. Front Immunol. (2023) 14:1190034. doi: 10.3389/fimmu.2023.1190034
21. Cohen M, Giladi A, Gorki AD, Solodkin DG, Zada M, Hladik A, et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell. (2018) 175(4):1031–44.e18. doi: 10.1016/j.cell.2018.09.009
22. Sicklinger F, Meyer IS, Li X, Radtke D, Dicks S, Kornadt MP, et al. Basophils balance healing after myocardial infarction via IL-4/IL-13. J Clin Invest. (2021) 131(13):e136778. doi: 10.1172/JCI136778
23. Doke T, Abedini A, Aldridge DL, Yang YW, Park J, Hernandez CM, et al. Single-cell analysis identifies the interaction of altered renal tubules with basophils orchestrating kidney fibrosis. Nat Immunol. (2022) 23(6):947–59. doi: 10.1038/s41590-022-01200-7
24. Leyva-Castillo JM, Sun L, Wu SY, Rockowitz S, Sliz P, Geha RS. Single-cell transcriptome profile of mouse skin undergoing antigen-driven allergic inflammation recapitulates findings in atopic dermatitis skin lesions. J Allergy Clin Immunol. (2022) 150(2):373–84. doi: 10.1016/j.jaci.2022.03.002
25. Miyake K, Ito J, Takahashi K, Nakabayashi J, Brombacher F, Shichino S, et al. Single-cell transcriptomics identifies the differentiation trajectory from inflammatory monocytes to pro-resolving macrophages in a mouse skin allergy model. Nat Commun. (2024) 15(1):1666. doi: 10.1038/s41467-024-46148-4
26. Strakosha M, Vega-Mendoza D, Kane J, Jain A, Sun L, Rockowitz S, et al. Basophils play a protective role in the recovery of skin barrier function from mechanical injury in mice. J Invest Dermatol. (2024). doi: 10.1016/j.jid.2023.12.024
27. Grootens J, Ungerstedt JS, Nilsson G, Dahlin JS. Deciphering the differentiation trajectory from hematopoietic stem cells to mast cells. Blood Adv. (2018) 2(17):2273–81. doi: 10.1182/bloodadvances.2018019539
28. Derakhshan T, Boyce JA, Dwyer DF. Defining mast cell differentiation and heterogeneity through single-cell transcriptomics analysis. J Allergy Clin Immunol. (2022) 150(4):739–47. doi: 10.1016/j.jaci.2022.08.011
29. St. John AL, Rathore APS, Ginhoux F. New perspectives on the origins and heterogeneity of mast cells. Nat Rev Immunol. (2023) 23(1):55–68. doi: 10.1038/s41577-022-00731-2
30. Huang H, Li Y, Liu B. Transcriptional regulation of mast cell and basophil lineage commitment. Semin Immunopathol. (2016) 38(5):539–48. doi: 10.1007/s00281-016-0562-4
31. Sasaki H, Kurotaki D, Tamura T. Regulation of basophil and mast cell development by transcription factors. Allergol Int. (2016) 65(2):127–34. doi: 10.1016/j.alit.2016.01.006
32. Gentek R, Ghigo C, Hoeffel G, Bulle MJ, Msallam R, Gautier G, et al. Hemogenic endothelial fate mapping reveals dual developmental origin of mast cells. Immunity. (2018) 48(6):1160–71.e5. doi: 10.1016/j.immuni.2018.04.025
33. Li Z, Liu S, Xu J, Zhang X, Han D, Liu J, et al. Adult connective tissue-resident mast cells originate from late erythro-myeloid progenitors. Immunity. (2018) 49(4):640–53.e5. doi: 10.1016/j.immuni.2018.09.023
34. Tauber M, Basso L, Martin J, Bostan L, Pinto MM, Thierry GR, et al. Landscape of mast cell populations across organs in mice and humans. J Exp Med. (2023) 220(10):e20230570. doi: 10.1084/jem.20230570
35. Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, et al. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci U S A. (2005) 102(50):18105–10. doi: 10.1073/pnas.0509148102
36. Chen CC, Grimbaldeston MA, Tsai M, Weissman IL, Galli SJ. Identification of mast cell progenitors in adult mice. Proc Natl Acad Sci U S A. (2005) 102(32):11408–13. doi: 10.1073/pnas.0504197102
37. Rao KN, Smuda C, Gregory GD, Min B, Brown MA. Ikaros limits basophil development by suppressing C/EBP-alpha expression. Blood. (2013) 122(15):2572–81. doi: 10.1182/blood-2013-04-494625
38. Qi X, Hong J, Chaves L, Zhuang Y, Chen Y, Wang D, et al. Antagonistic regulation by the transcription factors C/EBPα and MITF specifies basophil and mast cell fates. Immunity. (2013) 39(1):97–110. doi: 10.1016/j.immuni.2013.06.012
39. Wanet A, Bassal MA, Patel SB, Marchi F, Mariani SA, Ahmed N, et al. E-cadherin is regulated by GATA-2 and marks the early commitment of mouse hematopoietic progenitors to the basophil and mast cell fates. Sci Immunol. (2021) 6(56):eaba0178. doi: 10.1126/sciimmunol.aba0178
40. Dahlin JS, Hamey FK, Pijuan-Sala B, Shepherd M, Lau WWY, Nestorowa S, et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in kit mutant mice. Blood. (2018) 131(21):e1–11. doi: 10.1182/blood-2017-12-821413
41. Miyake K, Ito J, Nakabayashi J, Shichino S, Ishiwata K, Karasuyama H. Single cell transcriptomics clarifies the basophil differentiation trajectory and identifies pre-basophils upstream of mature basophils. Nat Commun. (2023) 14(1):2694. doi: 10.1038/s41467-023-38356-1
42. Park J, Cho Y, Yang D, Yang H, Lee D, Kubo M, et al. The transcription factor NFIL3/E4BP4 regulates the developmental stage-specific acquisition of basophil function. J Allergy Clin Immunol. (2024) 153(1):132–45. doi: 10.1016/j.jaci.2023.09.029
43. Metcalf D, Ng AP, Baldwin TM, Di Rago L, Mifsud S. Concordant mast cell and basophil production by individual hematopoietic blast colony-forming cells. Proc Natl Acad Sci U S A. (2013) 110(22):9031–5. doi: 10.1073/pnas.1307711110
44. Mukai K, BenBarak MJ, Tachibana M, Nishida K, Karasuyama H, Taniuchi I, et al. Critical role of P1-Runx1 in mouse basophil development. Blood. (2012) 120(1):76–85. doi: 10.1182/blood-2011-12-399113
45. Denburg JA, Telizyn S, Messner H, Lim B, Jamal N, Ackerman SJ, et al. Heterogeneity of human peripheral blood eosinophil-type colonies: evidence for a common basophil-eosinophil progenitor. Blood. (1985) 66(2):312–8. doi: 10.1182/blood.V66.2.312.312
46. Leary AG, Ogawa M. Identification of pure and mixed basophil colonies in culture of human peripheral blood and marrow cells. Blood. (1984) 64(1):78–83. doi: 10.1182/blood.V64.1.78.78
47. Weil SC, Hrisinko MA. A hybrid eosinophilic-basophilic granulocyte in chronic granulocytic leukemia. Am J Clin Pathol. (1987) 87(1):66–70. doi: 10.1093/ajcp/87.1.66
48. Görgens A, Radtke S, Mollmann M, Cross M, Durig J, Horn PA, et al. Revision of the human hematopoietic tree: granulocyte subtypes derive from distinct hematopoietic lineages. Cell Rep. (2013) 3(5):1539–52. doi: 10.1016/j.celrep.2013.04.025
49. Boyce JA, Friend D, Matsumoto R, Austen KF, Owen WF. Differentiation in vitro of hybrid eosinophil/basophil granulocytes: autocrine function of an eosinophil developmental intermediate. J Exp Med. (1995) 182(1):49–57. doi: 10.1084/jem.182.1.49
50. Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. (2007) 26(6):726–40. doi: 10.1016/j.immuni.2007.06.004
51. Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. (2008) 132(4):631–44. doi: 10.1016/j.cell.2008.01.025
52. Luc S, Buza-Vidas N, Jacobsen SE. Delineating the cellular pathways of hematopoietic lineage commitment. Semin Immunol. (2008) 20(4):213–20. doi: 10.1016/j.smim.2008.07.005
53. Franco CB, Chen CC, Drukker M, Weissman IL, Galli SJ. Distinguishing mast cell and granulocyte differentiation at the single-cell level. Cell Stem Cell. (2010) 6(4):361–8. doi: 10.1016/j.stem.2010.02.013
54. Tusi BK, Wolock SL, Weinreb C, Hwang Y, Hidalgo D, Zilionis R, et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature. (2018) 555(7694):54–60. doi: 10.1038/nature25741
55. Weinreb C, Rodriguez-Fraticelli A, Camargo FD, Klein AM. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science. (2020) 367(6479):eaaw3381. doi: 10.1126/science.aaw3381
56. Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol. (2017) 19(4):271–81. doi: 10.1038/ncb3493
57. Zheng S, Papalexi E, Butler A, Stephenson W, Satija R. Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol Syst Biol. (2018) 14(3):e8041. doi: 10.15252/msb.20178041
58. Pellin D, Loperfido M, Baricordi C, Wolock SL, Montepeloso A, Weinberg OK, et al. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun. (2019) 10(1):2395. doi: 10.1038/s41467-019-10291-0
59. Laurenti E, Gottgens B. From haematopoietic stem cells to complex differentiation landscapes. Nature. (2018) 553(7689):418–26. doi: 10.1038/nature25022
60. Zhang Y, Gao S, Xia J, Liu F. Hematopoietic hierarchy—an updated roadmap. Trends Cell Biol. (2018) 28(12):976–86. doi: 10.1016/j.tcb.2018.06.001
61. Liggett LA, Sankaran VG. Unraveling hematopoiesis through the lens of genomics. Cell. (2020) 182(6):1384–400. doi: 10.1016/j.cell.2020.08.030
62. Drissen R, Buza-Vidas N, Woll P, Thongjuea S, Gambardella A, Giustacchini A, et al. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat Immunol. (2016) 17(6):666–76. doi: 10.1038/ni.3412
63. Hamey FK, Lau WWY, Kucinski I, Wang X, Diamanti E, Wilson NK, et al. Single-cell molecular profiling provides a high-resolution map of basophil and mast cell development. Allergy. (2021) 76(6):1731–42. doi: 10.1111/all.14633
64. Drissen R, Thongjuea S, Theilgaard-Monch K, Nerlov C. Identification of two distinct pathways of human myelopoiesis. Sci Immunol. (2019) 4(35):eaau7148. doi: 10.1126/sciimmunol.aau7148
65. Grootens J, Ungerstedt JS, Wu C, Hamberg Levedahl K, Nilsson G, Dahlin JS. CD203c distinguishes the erythroid and mast cell-basophil differentiation trajectories among human FcεRI+ bone marrow progenitors. Allergy. (2020) 75(1):211–4. doi: 10.1111/all.13981
66. Gurtner A, Borrelli C, Gonzalez-Perez I, Bach K, Acar IE, Nunez NG, et al. Active eosinophils regulate host defence and immune responses in colitis. Nature. (2023) 615(7950):151–7. doi: 10.1038/s41586-022-05628-7
67. Shichino S, Ueha S, Hashimoto S, Ogawa T, Aoki H, Wu B, et al. TAS-seq is a robust and sensitive amplification method for bead-based scRNA-seq. Commun Biol. (2022) 5(1):602. doi: 10.1038/s42003-022-03536-0
68. Tsuzuki H, Arinobu Y, Miyawaki K, Takaki A, Ota SI, Ota Y, et al. Functional interleukin-33 receptors are expressed in early progenitor stages of allergy-related granulocytes. Immunology. (2017) 150(1):64–73. doi: 10.1111/imm.12667
69. Lantz CS, Min B, Tsai M, Chatterjea D, Dranoff G, Galli SJ. IL-3 is required for increases in blood basophils in nematode infection in mice and can enhance IgE-dependent IL-4 production by basophils in vitro. Lab Invest. (2008) 88(11):1134–42. doi: 10.1038/labinvest.2008.88
70. Shen T, Kim S, Do JS, Wang L, Lantz C, Urban JF, et al. T cell-derived IL-3 plays key role in parasite infection-induced basophil production but is dispensable for in vivo basophil survival. Int Immunol. (2008) 20(9):1201–9. doi: 10.1093/intimm/dxn077
71. MacGlashan D Jr. Modulating the human basophil phenotype during its development and maturation: basophils derived from in vitro cultures of CD34(+) progenitor cells. Methods Mol Biol. (2020) 2163:69–83. doi: 10.1007/978-1-0716-0696-4_6
72. Rottem M, Okada T, Goff JP, Metcalfe DD. Mast cells cultured from the peripheral blood of normal donors and patients with mastocytosis originate from a CD34+/Fc epsilon RI- cell population. Blood. (1994) 84(8):2489–96. doi: 10.1182/blood.V84.8.2489.2489
73. Maaninka K, Lappalainen J, Kovanen PT. Human mast cells arise from a common circulating progenitor. J Allergy Clin Immunol. (2013) 132(2):463–9.e3. doi: 10.1016/j.jaci.2013.02.011
74. Dahlin JS, Malinovschi A, Ohrvik H, Sandelin M, Janson C, Alving K, et al. Lin- CD34hi CD117int/hi FcepsilonRI+ cells in human blood constitute a rare population of mast cell progenitors. Blood. (2016) 127(4):383–91. doi: 10.1182/blood-2015-06-650648
75. Wu C, Boey D, Bril O, Grootens J, Vijayabaskar MS, Sorini C, et al. Single-cell transcriptomics reveals the identity and regulators of human mast cell progenitors. Blood Adv. (2022) 6(15):4439–49. doi: 10.1182/bloodadvances.2022006969
76. Shelburne CP, McCoy ME, Piekorz R, Sexl V, Roh KH, Jacobs-Helber SM, et al. Stat5 expression is critical for mast cell development and survival. Blood. (2003) 102(4):1290–7. doi: 10.1182/blood-2002-11-3490
77. Ohmori K, Luo Y, Jia Y, Nishida J, Wang Z, Bunting KD, et al. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J Immunol. (2009) 182(5):2835–41. doi: 10.4049/jimmunol.0802870
78. Li Y, Qi X, Liu B, Huang H. The STAT5-GATA2 pathway is critical in basophil and mast cell differentiation and maintenance. J Immunol. (2015) 194(9):4328–38. doi: 10.4049/jimmunol.1500018
79. Sasaki H, Kurotaki D, Osato N, Sato H, Sasaki I, Koizumi S, et al. Transcription factor IRF8 plays a critical role in the development of murine basophils and mast cells. Blood. (2015) 125(2):358–69. doi: 10.1182/blood-2014-02-557983
80. Nei Y, Obata-Ninomiya K, Tsutsui H, Ishiwata K, Miyasaka M, Matsumoto K, et al. GATA-1 regulates the generation and function of basophils. Proc Natl Acad Sci U S A. (2013) 110(46):18620–5. doi: 10.1073/pnas.1311668110
81. Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. (1993) 74(2):395–404. doi: 10.1016/0092-8674(93)90429-t
82. Morii E, Oboki K, Ishihara K, Jippo T, Hirano T, Kitamura Y. Roles of MITF for development of mast cells in mice: effects on both precursors and tissue environments. Blood. (2004) 104(6):1656–61. doi: 10.1182/blood-2004-01-0247
83. Zhang S, Vieth JA, Krzyzanowska A, Henry EK, Denzin LK, Siracusa MC, et al. The transcription factor PLZF is necessary for the development and function of mouse basophils. J Immunol. (2019) 203(5):1230–41. doi: 10.4049/jimmunol.1900068
84. Li Y, Gao J, Kamran M, Harmacek L, Danhorn T, Leach SM, et al. GATA2 regulates mast cell identity and responsiveness to antigenic stimulation by promoting chromatin remodeling at super-enhancers. Nat Commun. (2021) 12(1):494. doi: 10.1038/s41467-020-20766-0
85. Li Y, Qi X, Zhao D, Urban JF, Huang H. IL-3 expands pre-basophil and mast cell progenitors by upregulating the IL-3 receptor expression. Cell Immunol. (2022) 374:104498. doi: 10.1016/j.cellimm.2022.104498
86. Siracusa MC, Saenz SA, Hill DA, Kim BS, Headley MB, Doering TA, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. (2011) 477(7363):229–33. doi: 10.1038/nature10329
87. Ohneda K, Moriguchi T, Ohmori S, Ishijima Y, Satoh H, Philipsen S, et al. Transcription factor GATA1 is dispensable for mast cell differentiation in adult mice. Mol Cell Biol. (2014) 34(10):1812–26. doi: 10.1128/MCB.01524-13
88. Ghinassi B, Sanchez M, Martelli F, Amabile G, Vannucchi AM, Migliaccio G, et al. The hypomorphic Gata1low mutation alters the proliferation/differentiation potential of the common megakaryocytic-erythroid progenitor. Blood. (2007) 109(4):1460–71. doi: 10.1182/blood-2006-07-030726
89. Migliaccio AR, Rana RA, Sanchez M, Lorenzini R, Centurione L, Bianchi L, et al. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J Exp Med. (2003) 197(3):281–96. doi: 10.1084/jem.20021149
90. Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. (2002) 195(11):1387–95. doi: 10.1084/jem.20020656
91. Vannucchi AM, Bianchi L, Cellai C, Paoletti F, Carrai V, Calzolari A, et al. Accentuated response to phenylhydrazine and erythropoietin in mice genetically impaired for their GATA-1 expression [GATA-1(low) mice]. Blood. (2001) 97(10):3040–50. doi: 10.1182/blood.v97.10.3040
92. de Bruijn M, Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood. (2017) 129(15):2061–9. doi: 10.1182/blood-2016-12-689109
93. Maeda T. Regulation of hematopoietic development by ZBTB transcription factors. Int J Hematol. (2016) 104(3):310–23. doi: 10.1007/s12185-016-2035-x
94. Guo MH, Nandakumar SK, Ulirsch JC, Zekavat SM, Buenrostro JD, Natarajan P, et al. Comprehensive population-based genome sequencing provides insight into hematopoietic regulatory mechanisms. Proc Natl Acad Sci U S A. (2017) 114(3):E327–36. doi: 10.1073/pnas.1619052114
95. Peters IJA, de Pater E, Zhang W. The role of GATA2 in adult hematopoiesis and cell fate determination. Front Cell Dev Biol. (2023) 11:1250827. doi: 10.3389/fcell.2023.1250827
96. Desai A, Sowerwine K, Liu Y, Lawrence MG, Chovanec J, Hsu AP, et al. GATA-2-deficient mast cells limit IgE-mediated immediate hypersensitivity reactions in human subjects. J Allergy Clin Immunol. (2019) 144(2):613–7.e14. doi: 10.1016/j.jaci.2019.05.007
97. Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell. (2016) 167(7):1883–96.e15. doi: 10.1016/j.cell.2016.11.039
Keywords: single-cell transcriptome, basophil, eosinophil, erythrocyte, megakaryocyte, progenitor, pre-basophil
Citation: Miyake K, Ito J and Karasuyama H (2024) Novel insights into the ontogeny of basophils. Front. Allergy 5:1402841. doi: 10.3389/falgy.2024.1402841
Received: 18 March 2024; Accepted: 1 May 2024;
Published: 13 May 2024.
Edited by:
Masao Yamaguchi, Teikyo University Chiba Medical Center, JapanReviewed by:
Nicolas Gaudenzio, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceDaniel P. Potaczek, University of Marburg, Germany
© 2024 Miyake, Ito and Karasuyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kensuke Miyake, miyake.mbch@tmd.ac.jp
 Kensuke Miyake
Kensuke Miyake Junya Ito
Junya Ito Hajime Karasuyama
Hajime Karasuyama