Tackling mosaicism in gene edited livestock
- 1Independent researcher, Ashburton, New Zealand
- 2Sydney School of Veterinary Science, Faculty of Science, The University of Sydney, Camden, NSW, Australia
- 3NSW Department of Primary Industries, Elizabeth Macarthur Agricultural Institute, Menangle, NSW, Australia
The farming of livestock has a critical role to play in global nutritional security and poverty alleviation. To meet these goals through more efficient, environmentally sustainable and animal welfare focused means, gene editing technologies could be integrated into current breeding programs. A common issue with gene editing in livestock zygotes is the high incidence of genetic mosaicism. Genetic mosaicism, characterised by a single individual carrying distinct genotypes in different cell lineages, can lead to inconsistent presentation of a desired trait phenotypically, or the absence of the intended genotype in the animal’s germline. This review explores the present status of genetic mosaicism associated with CRISPR-Cas9 gene editing in cattle, sheep, and pigs, and identifies four areas for refinement; (1) the type of CRISPR-Cas9 genome editor used; (2) the CRISPR-Cas9 formats and timing of gene editing during embryonic development; (3) the method used to deliver the genome editor and (4) the genetic screening strategies applied. We also discuss alternatives to direct zygote gene editing, including surrogate sire technology and blastomere separation, which circumvent the production of mosaic offspring. By exploring these avenues for reducing mosaicism, gene editing protocols in livestock could become more efficient and effective, which will ultimately pave the way for traits to be introduced that improve animal welfare standards and help address gaps in the security of global nutrition access.
1 Mosaicism in gene edited livestock
Since early agriculturalists first domesticated livestock from wild species, the genetic composition of these animals has been altered through the selection of specific animals with desirable traits for breeding (Wiener and Wilkinson, 2011; Frantz et al., 2020). Gene editing has the potential to be a molecular tool in modern breeding systems for introducing desirable traits or removing deleterious traits from livestock, through the precise alteration of an animal’s genome (Bishop and Van Eenennaam, 2020). However, the relatively high frequency of mosaicism observed in gene edited founders has limited the efficient application of the technology (Hennig et al., 2020; Lee et al., 2020; Lin and Van Eenennaam, 2021). This review focuses on CRISPR-Cas9 from Streptococcus pyogenes (Box 1), the most commonly used CRISPR genome editor in livestock, and its association with mosaicism when generating gene edited livestock. To fully realise the benefits of gene editing in livestock, it is vital that breeders have access to gene editing technologies that result in low frequencies of mosaic offspring and can be practically integrated into on-farm livestock embryo transfer programs.
Mosaicism is defined by the presence of two or more cell lineages carrying different genotypes in an individual. It can arise naturally during the early stages of embryonic development through errors introduced during DNA replication (Biesecker and Spinner, 2013). The prevalence of mosaicism at CRISPR-Cas9 target alleles can be increased through the introduction of genome editors to an embryo (Hennig et al., 2020). If editing of DNA takes place after the initial embryonic division following fertilisation, variable editing across cells of the multicellular embryo leads to the production of genetically mosaic offspring, as depicted in Figure 1A. The potential for founding animals to have heterozygous germlines for the desired edits, or not carry the genetic alteration in their germline at all is problematic in livestock breeding due to their relatively long generation intervals (Lin and Van Eenennaam, 2021). Mosaicism is therefore a key technical impediment to the efficient and widespread application of gene editing in zygotes to improve livestock genetics.
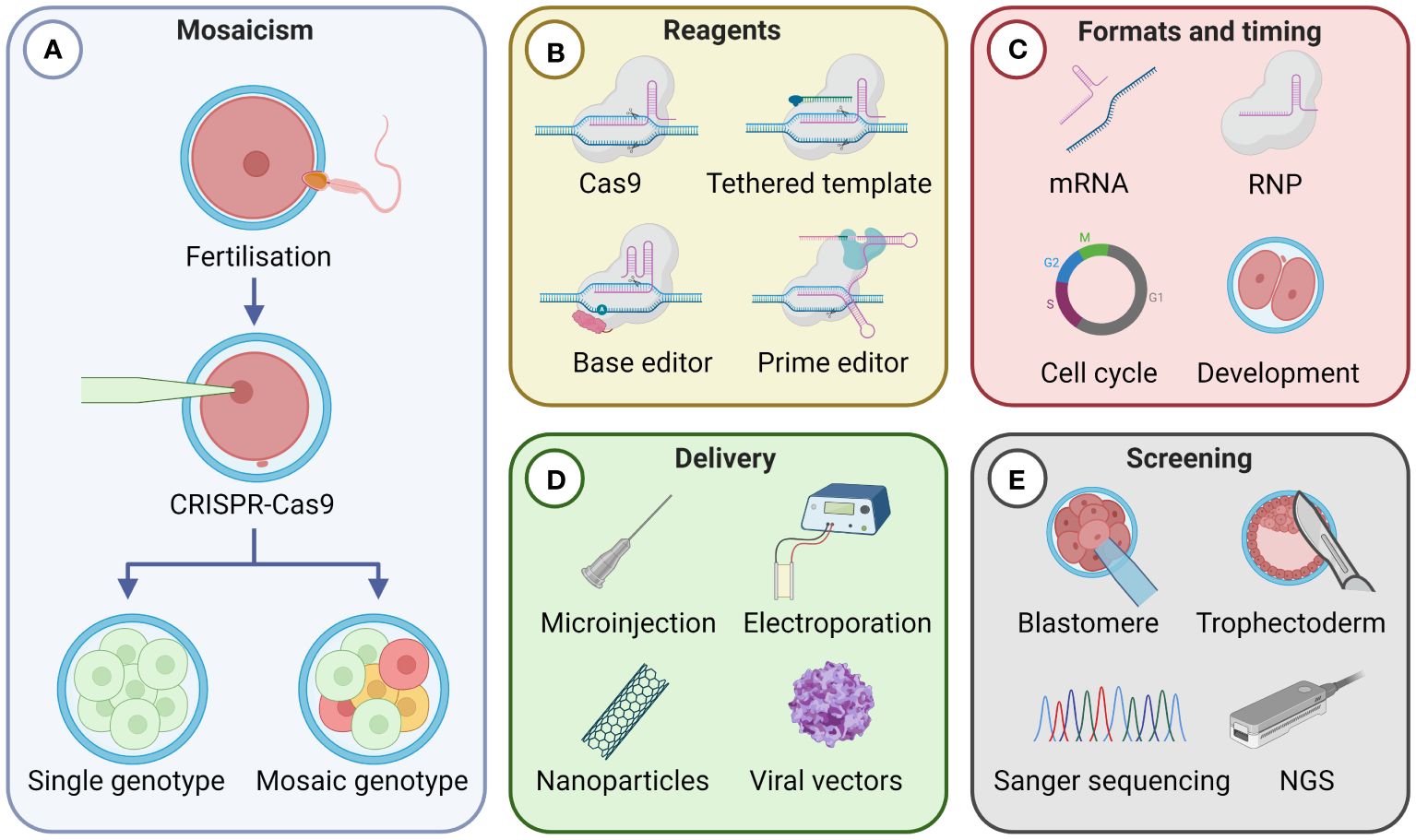
Figure 1 Factors influencing the frequency of mosaicism resulting from CRISPR-Cas9 gene editing in livestock zygotes. (A) Mosaicism arises when CRISPR-Cas9 introduces edits asymmetrically in a multicellular embryo, resulting in multiple genotypes. A single genotype occurs when editing takes place at the single-cell stage or symmetrically at a multicellular stage. Four key factors that can impact the frequency of mosaicism are: (B) the type of CRISPR-Cas9 reagents used; (C) the CRISPR-Cas9 format used and timing of gene editing activity during embryonic development; (D) the delivery method of the genome editor; and (E) the genetic screening strategies applied. dsDNA, double-stranded DNA; mRNA, messenger RNA; RNP, Cas9 ribonucleoprotein; NGS, next generation sequencing. Created with BioRender.com.
For mosaicism to not be present in progeny, gene editing must either occur during the single-cell zygote stage, when a single genome is present, or symmetrically throughout a multicellular embryo. Currently, when performing direct zygotic gene editing in livestock using conventional CRISPR-Cas9 reagents, mosaicism is the expected outcome rather than the exception (Crispo et al., 2015; Wang et al., 2015; Petersen et al., 2016; Tao et al., 2016; Vilarino et al., 2017; Zhang et al., 2017; Daigneault et al., 2018; Lamas-Toranzo et al., 2019; Namula et al., 2019). An alternative to performing genome editing in zygotes that does not generate mosaic progeny involves somatic cell nuclear transfer (SCNT), also known as cloning. With SCNT, cultured cells are edited and screened in vitro. The nucleus of a correctly gene edited cell is then transferred into an enucleated oocyte (Bondioli, 2018). Since the resulting animal develops from a single genome, the offspring will not be mosaic. However, SCNT is technically demanding, costly, and relatively inefficient, making it impractical for widespread commercial application as a breeding technology in livestock (Vajta, 2007; Walton, 2018). In contrast, the simplicity of direct zygotic gene editing procedures, using microinjection, electroporation or viral and non-viral strategies, makes the prospect of these methods being integrated into current livestock breeding systems more appealing.
This review examines four key areas regarding mosaicism in gene edited livestock: (1) the type of CRISPR-Cas9 genome editor used; (2) the delivered CRISPR-Cas9 format and timing of gene editing during embryonic development; (3) the method used to deliver the genome editor; and (4) the genetic screening strategies applied (Figures 1B-E). Finally, we will consider alternative breeding technologies that could contribute to reducing the presence of mosaicism in gene edited livestock.
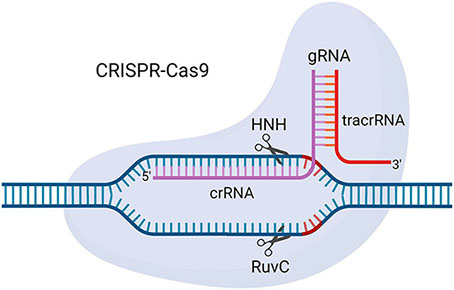
Box 1. CRISPR-Cas9 from Streptococcus pyogenes.
CRISPR-Cas9 is a genome editing tool developed by repurposing a bacterial antiviral defence system. It consists of two components: a CRISPR-associated protein 9 (Cas9) enzyme with endonuclease activity via HNH and RuvC domains, and a guide RNA (gRNA) that guides Cas9 to the target site in the genome through Watson-Crick base pairing with the target sequence. The gRNA is made up of a user-defined CRISPR RNA (crRNA) that base pairs with the target sequence and a trans-activating crRNA (tracrRNA) that facilitates interaction between the Cas9 enzyme and crRNA (Doudna & Charpentier, 2014; Wang et al., 2016). Optimization of the CRISPR-Cas9 system has led to the development of variants such as the single guide RNA (sgRNA) system, which combines the crRNA and tracrRNA into a single RNA molecule, nickase Cas9 (nCas9), which has single-strand DNA nicking capacity, and catalytically dead Cas9 (dCas9), which does not cut DNA but can be fused to other enzymes to deliver them to target DNA sequences of interest (Qi et al., 2013; Shen et al., 2014; Doench et al., 2016). Adapted from CRISPR/Cas9, by BioRender.com.
2 CRISPR-Cas9 genome editors and enhancers
Since the adaptation of CRISPR-Cas9 into a genome editing tool in 2012, the system has been modified to expand its capabilities beyond its native function of generating double-strand DNA breaks (DSBs) (Haifeng Wang et al., 2016). The three genome-editing Cas9 variants with predominant use presently are: (1) conventional Cas9 (Cas9) when used for cut and repair editing, (2) catalytically inactive, dead Cas9 (dCas9), fused with secondary proteins to confer specific enzymatic activity at the target site, and (3) nickase Cas9 (nCas9), which introduce single-strand breaks and are utilised in prime editing and base editing (Figure 1B) (Qi et al., 2013; Shen et al., 2014; Doench et al., 2016).
Conventional Cas9 gene editing exploits host DNA repair pathways to introduce changes in the nucleotide sequence. This can be an insertion or deletion (indel) of non-specific nucleotides at the DSB site through the non-homologous end-joining (NHEJ) pathway, or precise alterations through either the microhomology-mediated end-joining (MMEJ) pathway or through homology-directed repair (HDR) when a nucleotide template is provided with homologous arms around the DSB site (Liu et al., 2019; Sansbury et al., 2019). The preferential instigation of host DNA repair pathways is dynamic across developmental stages and cell lineages, although generally, HDR occurs at a lower efficiency than NHEJ in mammalian embryos (Miyaoka et al., 2016; Yang et al., 2020; Meyenberg et al., 2021; Zhang et al., 2022). Once an NHEJ event has occurred that disrupts the gRNA recognition sequence through the introduction of an indel, the potential for the editing by Cas9-mediated HDR is lost (Paquet et al., 2016). Therefore, methods to improve the efficiency of HDR repair and reduce the prevalence of NHEJ could decrease the frequency of mosaic progeny.
However, it should be noted that reductions in NHEJ will not intrinsically decrease mosaicism rates unless alternative precise repair pathways, such as HDR, are successful in the zygote. If NHEJ is inhibited but alternative precise editing pathways aren’t initiated until later stages of development, where CRISPR-Cas9 reagents are unevenly distributed, asymmetrically active or have different rates of editing success across the cells of a multicellular embryo, there may be no beneficial decrease in mosaicism.
2.1 Small molecule enhancers
Understanding the recruitment of native proteins that instigate particular DNA repair pathways and how they interact through the progression of the cell cycle has led to the identification of molecular regulators for promoting HDR or reducing NHEJ. The potential use of small molecules to enhance HDR in zygotes and reduce the occurrence of mosaicism in gene edited animals is an area of active research (Yang et al., 2020). For example, introducing key molecules such as RS-1, which stimulates Rad51—a protein essential for HDR—has been shown to increase Cas9-mediated HDR efficiency, resulting in an 8.5-fold increase in rabbit embryos and a 1.6-fold increase in zebrafish embryos (Song et al., 2016; Zhang et al., 2018). Alternatively, inhibiting gene expression of proteins associated with NHEJ, such as KU70 or KU80 with small interfering RNA (siRNA), has been found to increase the probability of the desired HDR genome edit occurring by 1.6- to 3-fold in pig foetal fibroblasts (Li et al., 2018). The use of small molecule cell cycle regulators, such as nocodazole and ABT, to extend the time cells spend in cell cycle phases in which HDR is most active (S and G2 phases) has been demonstrated to bias host DNA repair pathways and achieved 3.4 and 6.8-fold increase in HDR in human pluripotent stem cells, respectively (Yang et al., 2016). The application of these molecules when gene editing livestock zygotes could improve HDR efficiency, and thereby reduce the frequency of mosaicism by minimising the incident rate of indel introduction.
2.2 Cas9 fused to geminin
Engineered variants of Cas nucleases have also been developed which assist in further reducing mosaicism through cell cycle manipulation. These modified variants restrict endonuclease activity of Cas9 to specific phases of the cell cycle. One such example is a fusion of Cas9 to the N-terminal region of human geminin (hGeminin), which resulted in a cell-cycle-tailored activity with low levels in G1 but high expression in S/G2/M. The presence of hGeminin targets this fusion for ubiquitin-mediated degradation in the M and G1 phases of the cell cycle (Richardson et al., 2023). The enhanced activity of Cas9-hGeminin in cell cycle phases characterised by elevated rates of HDR has been demonstrated to augment the HDR rate by as much as 87% in HEK cells (Gutschner et al., 2016). The use of Cas9-hGeminin to reduce the incidence of mosaicism warrants testing in livestock zygotes.
2.3 Cas9 tethering of DNA repair template
Alternative Cas9 fusion proteins, such as PCV-Cas9 and Cas9-mSA, can covalently tether to a oligonucleotide repair template, thereby increasing HDR efficiency through proximity. Enhancements of HDR efficiency, compared to conventional Cas9 with a non-tethered oligonucleotide template, were demonstrated by Aird et al. (2018), who achieved up to a 30-fold increase in HDR efficiency using PCV-Cas9 in HEK cells, and Gu et al. (2020) achieved up to a 95% increase when editing two-cell mouse embryos with a Cas9-mSA based approach. These increases are attributed to the spatial and temporal co-localisation of the DSB and the oligonucleotide template. Although the efficacy of this method in livestock zygotes is yet to be determined, the notable improvements in HDR efficiency indicates its potential as a promising avenue for further research.
2.4 Prime editing
Another engineered Cas9 variant which could enhance the precision of gene editing in livestock while reducing the rate of unintended alterations is prime editing (PE). In the PE system, one of the endonuclease domains of Cas9 is catalytically inactivated, resulting in a Cas9 with only single-strand nicking capability (nCas9). In the PE system, nCas9 is fused with an engineered reverse transcriptase enzyme and complexed with a prime-editing guide RNA (pegRNA). The pegRNA is a ssRNA encoding the guide RNA, a repair template, and a primer sequence to promote initiation of reverse transcriptase activity. PE can mediate insertions, deletions, and base-to-base conversions. It offers advantages over conventional Cas9 based HDR strategies for specific alterations because the single-strand nicking approach prevents the initiation of the NHEJ pathway, reducing the potential for unwanted indels (Anzalone et al., 2019; Scholefield and Harrison, 2021).
In a study by Gao et al. (2021) aimed at producing gene edited mice, separate cohorts of mouse zygotes were microinjected with either PE or Cas9/HDR reagents. Targeted sequencing analysis of genomic DNA from the mice was performed to assess the accuracy of on-target editing. The results showed an average of 56% correct on-target editing in the HDR founder mice, in contrast to 21% in PE founder mice. However, all HDR founders had undesired indels, whereas none of the PE founders had any indels. In vitro data using porcine embryonic fibroblasts from Qi et al. (2023) identified that using standard PE protocols has limited efficiency in pigs. To improve efficiency in the agriculturally important species of pigs, they optimised the reverse transcriptase process to improve PE efficiency up to 29-fold (4-fold average across 12 target gene edits) which offers promise of further optimisation in efficiency for the accurate PE system. The lack of DSB and the unique ‘copy-and-paste’ mechanism of PE suggest it has potential in reducing mosaicism when generating gene edited livestock zygotes.
2.5 Base editing
Similar to PE, another Cas9-based gene editing strategy that circumvents DSBs while enabling single base changes is base editing (BE). The first-generation BE, termed BE1, utilises dCas9 complexed with a deaminase enzyme (Komor et al., 2016). This ensemble mediates the transformation of C-G to T-A editing through an intermediary U-G base pair, that is subsequently read as T-A during DNA replication. The most commonly used cytosine base editors (CBE), BE3 and BE4, consist of a Cas9 nickase fused to APOBEC1, a cytidine deaminase, and one or two uracil glycosylase inhibitors (UGI). Directed by a gRNA, nCas9 exposes the target DNA strand through homologous pairing, allowing APOBEC1 to deaminate cytosine specifically within the window of base pair interaction. UGI inhibits repair of the resulting mismatch, and a subsequent nick in the target strand leads to the incorporation of adenine opposite the uracil, culminating in a T-A pairing post-replication (Rees and Liu, 2018; Caso and Davies, 2022).
Addressing the limitations of CBEs in correcting A-to-G or T-to-C mutations, led to the development of adenine base editors (ABEs). These editors, evolved from an Escherichia coli tRNA adenosine deaminase (TadA), include a mutated TadA variant for enhanced efficiency, paired with a natural TadA for structural support, and a Cas9 nickase. Like the CBE systems, the ABE systems function by exposing the non-target DNA strand through homologous pairing for the mutated TadA-mediated adenine deamination to occur at base pairs within a defined window. This editing results in an inosine-cytosine pairing, which is converted to a C-G pair during DNA replication (Gaudelli et al., 2017; Caso and Davies, 2022).
Luo et al. (2022) used a BE3 to knockout the SOX2 gene in bovine embryos. Genotyping showed that a premature stop codon was successfully introduced in SOX2 in 87.1% (101 out of 116) bovine blastocysts and only 9 (out of 116) embryos display mosaicism. In contrast, Zhou et al. (2020) used a ABE to introduce a point mutation in the sheep BMPR1B by microinjection in sheep zygotes. Six out of 8 lambs born were heterozygous carriers for the desired substitution in the BMPR1B gene, but all edited lambs were mosaic. For single base changes and gene knockouts, base editors, particular CBEs, warrant further investigation in livestock embryos as a potential tool to decrease the rates of mosaicism.
A further strength of base editors in relation to zygotic gene editing is that that they are not reliant on being active during a cycle cell phase in which the HDR repair pathway is active, and therefore activity can occur directly after fertilisation. The strategic avoidance of DSBs across these engineered gene editing platforms demonstrates the desire to avoid the NHEJ pathway from being activated. Due to its inability to direct specific base pair modifications, NHEJ is a key contributor to the production of mosaic offspring from genome edited zygotes, and progress in avoiding mosaicism in gene edited livestock will likely emerge from these strategies.
3 Timing of gene editing activity and formats of CRISPR-Cas9
The timing of gene editing in embryos is crucial for reducing the frequency of mosaicism. The level of mosaicism can be affected by two aspects of gene editing timing: the developmental stage at which gene editing reagents are introduced, and the timing of CRISPR activity as influenced by reagent format and accessibility to the genome target. Considering when the CRISPR reagents are introduced, with each embryonic cell division, the number of alleles to edit doubles, meaning the potential for distinct edits to occur in different cells of blastomeres increases. Non-mosaic genotypes can therefore be achieved with the least likelihood of multiple allelic variants arising when gene editing occurs in a single cell zygote, prior to DNA replication, with only two target alleles present.
One study reported that electroporation of Cas9 protein and sgRNA into mouse pre-mitotic zygotes (5 hours (h) post fertilisation) using both NHEJ (n=10) and HDR (n=4; with DNA repair template) strategies, no mosaicism for gene edited alleles was detected. In mice, genome replication begins 5.5 h post fertilisation, which suggests that introduction of genome editing reagents before then reduces the incidence of mosaicism (Hashimoto et al., 2016); (Yamauchi et al., 2009). In pigs, sheep, and cattle zygotes, the first round of DNA replication occurs 12-15, 10-12, and 18 h after fertilisation, respectively (Lin and Van Eenennaam, 2021). This provides a greater window for the delivery of genome editing components in single cell livestock zygotes compared to mice. It must be acknowledged that while early introduction of gene-editing reagents into a zygote is likely to assist in reducing mosaicism, nuclear import, catalytic activation and genome accessibility are further factors that will influence genome editing success.
Refining in vitro fertilisation (IVF) protocols in mammalian livestock species to allow for CRISPR-Cas9 introduction into zygotes can provide opportunities to reduce gene editing associated mosaicism. Conventional cattle and sheep IVF protocols typically incubate oocytes with sperm overnight (Ferré et al., 2020), even though most of the oocytes are penetrated by sperm within 6 h (Ward et al., 2002; Berland et al., 2011; Anzalone et al., 2021). Therefore, introducing the CRISPR-Cas9 components after the usual IVF incubation period could result in gene editing occurring around the time of DNA replication during the cell cycle S-phase of the zygote (Coticchio et al., 2023). Lamas-Toranzo et al. (2019) found that in cattle, reducing the gamete co-incubation time from 20 to 10 h did not affect blastocyst development rates and reduced Cas9-mediated mosaicism in progeny from 70% in the 20 h control group to 10-30% in the 10 h group.
An alternative to gene editing at the zygotic stage involves the delivery of CRISPR-Cas9 prior to fertilisation. Introducing CRISPR-Cas9 ribonucleoprotein (RNP) complexes into mature second metaphase (MII) bovine oocytes can significantly decrease mosaicism rates. Specifically, Lamas-Toranzo et al. (2019) observed a reduction in mosaicism from 100% in embryos where Cas9 RNP was microinjected 20 h after gamete co-incubation, to 30% in those injected at the MII oocyte stage. However, the advantages of oocyte delivery were less marked in sheep. Marcela Vilarino et al. (2017) reported that while microinjecting MII sheep oocytes or zygotes with Cas9 mRNA resulted in similar mutation rates (~59%), there was no significant difference in mosaicism rates between groups (37.5% (3/8) versus 66.7% (4/6) for MII-injected and zygote-injected, respectively). This less pronounced difference might be attributable to the use of Cas9 mRNA in the sheep study, which exhibits extended activity compared to the Cas9 RNP which was used the bovine study.
In experimental trials in human embryos, Ma et al. (2017) demonstrated that microinjecting CRISPR-Cas9 RNP into M-phase oocytes eliminated mosaicism. Furthermore, they observed no reduction in HDR editing efficiency, even when CRISPR-Cas9 was introduced into MII oocytes at the time of intracytoplasmic sperm injection (ICSI). However, as noted by Lin and Van Eenennaam (2021) in their review, the early timing of CRISPR-Cas9 delivery into oocytes can reduce fertilisation rates, particularly when oocytes are co-incubated with cumulus cells and spermatozoa for a reduced duration (Ward et al., 2002). Collectively, these findings suggest that the introduction of CRISPR-Cas9 RNP into oocytes may offer a viable alternative to zygotic gene editing, potentially lowering mosaicism rates. These examples of delivering CRISPR-Cas9 both before and after fertilisation demonstrate the impact that developmental timing of gene editor delivery has on mosaicism rates. Optimisation of IVF protocols for the reduction of mosaicism will be a critical element for tackling mosaicism in gene edited livestock.
In addition to the timing of CRISPR-Cas9 reagent introduction, it is also important when the reagents are enzymatically active and the subsequent persistence of their activity. Ideally, CRISPR-Cas9 reagents will have immediate activity in the zygote, successfully edit the target alleles in the desired manner, and then be rapidly degraded or inactivated (Wang et al., 2020).The molecular format of Cas9 and gRNA is a determinant of when genome editing activity is initiated. CRISPR-Cas9 reagents are most commonly delivered as double-stranded DNA, mRNA, or as a pre-complexed RNP (Menchaca et al., 2020).
The flow of molecular information from DNA to RNA to protein is a key consideration in the timing of Cas9’s activity and persistence. DNA based vectors require transcription and translation, complexing of Cas9 with the sgRNA and transport into the nucleus before gene editing can occur, whilst mRNA does not require transcription (Liu et al., 2017). Transcription and translation are under regulation from zygotic factors, with any delays further postponing the activation of Cas9-mediated editing (Mehravar et al., 2019). Direct introduction of pre-complexed RNP allows the Cas9 enzyme and its associated gRNA to exhibit immediate catalytic activity, provided they are efficiently transported into the nucleus (Hennig et al., 2020). This efficiency hinges on the presence of appropriate nuclear localisation signals (NLS) on the Cas9 enzyme. NLS sequences may vary between species, influencing the efficiency with which Cas9 is transported into the nucleus (Lu et al., 2021).
The initial approach to CRISPR-based gene editing in livestock involved the delivery of Cas9 and gRNA as double-stranded DNA plasmid (Han et al., 2014; Ni et al., 2014; Whitworth et al., 2014). This method was predominantly chosen for its feasibility of reagent development and modification in conventional molecular biology laboratories. However, as discussed above, this approach has associated delays in gene editing activity. For example, Cas9 delivered by plasmid electroporation into human K562 cells was not readily expressed until 12 h and expression persisted at high levels until at least 72 h when the time course concluded (Kim et al., 2014). The delayed onset of the intended activity and the persistence of expression from dsDNA plasmids is highly conducive to the generation of mosaic animals. An additional concern with this approach is that dsDNA has the potential to integrate into the host genome, as highlighted when in the process of creating hornless cattle using TALENs genome editing, progeny were identified to be carrying a partial plasmid integration by regulators (Norris et al., 2020). Furthermore, Lillico et al. (2016) identified two piglets from their porcine zygote ZFN genome editing pipeline which carried integrations of the HDR plasmid. As off-target edits should be identified when commercial products are being produced, using dsDNA requires screening for plasmid integration and presents an additional regulatory hurdle if identified.
To mitigate these issues, Cas9 and gRNA can also be encoded and delivered as RNA molecules. In this instance the reagents will not randomly integrate into the host genome and can be directly translated and translocated to the nucleus for more immediate endonuclease activity (Yip, 2020). However, for most zygotic applications, the positives of mRNA use are superseded by those of Cas9 RNP complexes, which are now the preferred method and have been associated with reducing the incidence of mosaicism, due to their capacity to be delivered in a precise molecular quantity, immediate activity and rapid degradation (Hashimoto et al., 2016; Tu et al., 2017). When introduced into cells, Cas9-induced indels are measurable very shortly after electroporation, with robust editing observed within 3 h (Bloomer et al., 2022). In an in vitro context of cultured cells, Cas9 RNP complexes are degraded to the extent that they are barely detectable after 24 h and not detectable at 48 h - the shorter time frame of activity limits the potential for off-target gene editing activity (Kim et al., 2014). Furthermore, Han et al. (2017) found that Cas9 RNP delivery induced off-target mutations at a lower frequency (0.8%) than plasmid transfection (4.7%) with on-target mutations being of similar frequency. Taken together, with Cas9 RNP now being widely commercially available, it is generally regarded as the preferred format for reducing mosaicism in gene edited livestock zygotes.
4 Delivery methods
The choice of delivery method for CRISPR-Cas9 reagent introduction into zygotes influences editing efficiency, embryo viability, and the level of mosaicism. Currently, the most prevalent methods for introducing CRISPR-Cas9 reagents into zygotes are microinjection, electroporation or viral or non-viral strategies (Figure 1D). Each method requires the fine-tuning of parameters and concentrations tailored to achieve optimal editing conditions in each species. For instance, Tanihara et al. (2019) demonstrated that increasing the concentration of Cas9 RNP delivered through microinjection from 20 to 100 ng/μl increased the percentage of biallelic gene edited embryos from 0 to 16.7%. Notably, the increase in Cas9 RNP concentration did not adversely affect blastocyst development rates or the degree of mosaicism. Selection of the delivery method depends on factors such as the developmental stage of the embryos, the molecular format’s compatibility with the delivery method (for example, viral vectors are restricted to nucleic acids, while other methods provide greater flexibility), the technical expertise required and equipment availability.
Initially, microinjection was the predominant technique for introducing CRISPR-Cas9 reagents into zygotes. However, as microinjection requires specialised equipment and considerable expertise, its application beyond research environments with low throughput is limited. Utilised in various livestock including cattle, sheep, and pigs, microinjection can show inconsistency in the delivery site of reagents (nucleus, cytoplasm, or both) and high variability in the rates of mosaic offspring (Hai et al., 2014; Bevacqua et al., 2016; Zhang et al., 2017). Moreover, the mechanical process of injecting zygotes involves the physical piercing of the plasma membrane, with or without the use of a Piezo drill, which can result in a proportion of the zygotes lysing. To minimise losses due to lysis, zygotes are commonly treated with reagents, such as cytochalasin B, that increase the fluidity of the lipid bilayer during the procedure (Scott and Hu, 2019). Another difficulty with microinjection in livestock zygotes is their high lipid content (Sturmey et al., 2009), which makes the visualisation of pronuclei impossible without performing an additional manipulation to polarise the cytoplasmic lipid droplets. In a comparison of microinjection and electroporation delivery methods, subsequent development to the blastocyst stage was superior following electroporation (Le et al., 2020). Unless there are considerable advancements in robotic microinjection technology, its widespread adoption in agricultural settings remains improbable.
In recent years there has been a rise in the use of electroporation and viral vectors due to their higher throughput, improved embryo survival rates, and reduced dependency on technical expertise and specialised equipment. Among these, electroporation is emerging as a preferred CRISPR-Cas9 delivery method in livestock zygotes (Lin and Van Eenennaam, 2021; Mahdi et al., 2022; Gim et al., 2023). This technique involves applying electrical pulses to zygotes in a medium containing CRISPR-Cas9 reagents. High voltage pulses create temporary pores in the zona pellucida and plasma membrane, and low voltage pulses then draw polarised molecules through the medium and into the zygote’s cytoplasm. operators. However, with benefits such as the minimal equipment required, including a stereomicroscope, an electroporator, an electroporation cuvette or slide, and standard molecular biological tools, its ease of setup and portability enhance its appeal for working with large animals (Takemoto, 2020). Compared to microinjection, electroporation inflicts less physical damage on the zygotes, once the optimal parameters have been established, it significantly improves throughput, allowing simultaneous treatment of about 100 zygotes (Alghadban et al., 2020; Lin and Van Eenennaam, 2021). Regarding mosaicism, variations observed between the microinjection and electroporation methods are likely due to the timing of delivery and the format of the CRISPR-Cas9 reagents used, rather than the delivery method itself.
Recombinant adeno-associated viruses (rAAVs) have effectively been used to deliver Cas9 and a repair template into mammalian embryos (Wang et al., 2019). As small episomal viruses capable of transducing zygotes, rAAVs rarely integrate into the host genome but necessitate transcription and translation after transduction. Therefore, gene editing activity is delayed until after the embryo initiates cellular transcription and translation, which has the potential to lead to increased rates of mosaicism. A study by Lin and Van Eenennaam (2021) involving the transduction of cattle and sheep embryos with rAAV and subsequent mosaicism analysis of the blastocysts revealed that genetic mosaicism is a frequent outcome in embryos gene edited with Cas9-expressing rAAV. In cattle, 50% of the blastocysts showed more than two distinct alleles, while in sheep, all blastocysts presented mosaic patterns Furthermore, introducing rAAV gene editing agents later than 6-8 h post-insemination was linked to higher mosaicism rates, with 100% of embryos treated 20 h post-insemination being mosaic (Lin and Van Eenennaam, 2021). While rAAVs offer practical advantages in gene editing, such as requiring less equipment for the delivery of reagents, they are unlikely to play a significant role in reducing mosaicism in gene edited livestock.
Non-viral delivery strategies, particularly those involving transfection agents like cationic liposome-based reagents (for example, jetCRISPR by Polyplus-transfection and CRISPRMAX by Invitrogen), have proven effective for the delivery of DNA, RNA, and proteins into mammalian oocytes and zygotes (Lin et al., 2021; Hirata et al., 2021a, 2021b). Hirata et al. (2021b) successfully employed lipofection (using jetCRISPR) to introduce Cas9-RNP into porcine zygotes devoid of zona pellucida (ZP), 10 hours post-insemination. This approach yielded 7 of 9 piglets with the desired gene edit, though NGS read frequency analysis suggested that at least 3 piglets were likely mosaic. A significant limitation of transfection agents is their reduced efficacy in the presence of the ZP, a formidable physical barrier. Consequently, ZP-free oocytes and zygotes, which are more challenging to manipulate and exhibit lower viability, are often used. Addressing this, Piñeiro-Silva et al. (2023) sought to optimise lipofection in ZP-intact oocytes. They generated gene edited porcine embryos via lipofection of Cas9-RNP into ZP-intact oocytes during IVF. This was achieved using CRISPRMAX reagent, hypothesising that the smaller liposomes generated with this reagent could penetrate the ZP. Their modified protocol resulted in a relatively low on-target editing rate (8.7 to 20%) but notably, no mosaicism in the 6 embryos evaluated. Further investigation is needed to better understand whether dsDNA or ssDNA templates could be introduced using this method for an HDR gene editing approach. However, given that in vitro small liposomes have the capacity to translocate ssODNs across a somatic cell membrane it is likely to be a feasible strategy for small templates. Thus, the application of non-viral transfection reagents during IVF presents a promising strategy to mitigate mosaicism with minimal expertise and equipment required.
An alternative approach that has similar practical benefits over microinjection and electroporation are inorganic nanoparticles, notably carbon nanotubes (CNTs). CNTs are emerging as a promising strategy for CRISPR-Cas9 delivery, as their pronounced aspect ratio and rigidity enables them to passively traverse the cell membrane and be internalised. Munk et al (2016) incubated bovine embryos in a medium containing CNTs complexed with DNA encoding Green Fluorescent Protein (GFP). and observed GFP expression by day three of embryo culture (Munk et al., 2016). The recent advancements in using CNTs for Cas9 RNP delivery to plant cells (Cunningham, 2022), together with prior experiments in bovine embryos, indicate a potential for similar applications in livestock zygotes. Further research into using CNTs for Cas9 RNP delivery in livestock zygotes is called for, particularly due to its straightforward application. Introducing CNTs complexed with Cas9 RNP into the medium during IVF could facilitate early delivery of Cas9 RNP into zygotes, which may reduce the incidence of mosaicism.
5 Genetic screening
Preimplantation genetic screening (PGS) is a technique commonly used in IVF but is not widely used when gene editing livestock embryos (Weissman et al., 2017). PGS can help determine if gene editing has occurred, whether the editing is heterozygous or homozygous, and help identify mosaicism in embryos. PGS involves extracting a biopsy from an embryo, isolating DNA from the sample, and conducting analyses on the DNA. These analyses typically include PCR amplification and DNA sequencing. Biopsies can be obtained through several methods, these include the blade biopsy, where a portion of a morula or the polar trophectoderm from a blastocyst is sliced off, or through needle biopsy, where blastomere cells are aspirated from a morula-stage embryo (Figure 1E) (Cenariu et al., 2012; Yang et al., 2022). Taking a biopsy from a developing embryo requires balancing the need for enough cells to enable accurate DNA screening with the need to maintain the developmental competency of the embryo (Aoyama & Kato, 2020). When screening for mosaicism, it is especially important to take enough cells to increase the likelihood of identifying mosaic embryos.
One challenge in applying PGS to livestock gene editing programs is adapting human PGS protocols in on-farm laboratory environments. Many human PGS protocols rely on cryopreserving embryos while DNA sequencing is performed (Coates et al., 2017). For livestock PGS, it is desirable to biopsy and screen embryos without an interim period of cryopreservation to allow for high throughput and better viability. Portable real-time DNA sequencers, such as Oxford Nanopore, coupled with isothermal PCR reactions can enable rapid sequencing of embryo biopsies with minimal equipment in on-farm settings. However, there is a risk that the relatively small number cells biopsied do not represent the entire embryo and may not identify mosaic embryos (Vilarino et al., 2018).
After birth, founder gene edited livestock can be screened for mosaicism by DNA sequencing of various tissues, such as ear, hair follicles, and germ cells. It is important to never assess founder animals based on phenotype alone, as their genetic value may be hidden within their germ cells (Bunton-Stasyshyn et al., 2022). In male progeny, it is relatively straightforward to sequence sperm or testicular tissue to determine the germline genotype, but in females, oocyte screening is less practical due to the challenges of accessing these cells and their finite nature. For this reason, it is recommended to use Y-chromosome bearing sperm to generate male founder animals whenever possible to facilitate postnatal genetic screening. If a founder animal is mosaic with the correct germline genotype, the founder can be bred, and the phenotype and genotype should be confirmed in the offspring. As the generation intervals are relatively long in livestock species, obtaining single genotype founder animals at a high frequency is required to realise widespread on-farm application of gene editing as a breeding technology.
6 Emerging strategies
Direct zygote editing is currently the most practical application of CRISPR-Cas9 gene editing in livestock, as it can be easily integrated into existing IVF programs (McFarlane et al., 2019). However, emerging technologies such as gene editing of spermatogonial stem cells (SSCs) and blastomere separation may offer solutions to the issue of mosaicism in gene edited founder animals. Although these techniques may not be immediately available at the farm level due to the technical expertise and laboratory equipment required, their potential is significant and is worth highlighting.
SSCs can be collected from the testes of male animals through testicular biopsies and enzymatic digestion of testicular tissue. Mouse and rat SSCs have been successfully differentiated to functional spermatids and there are several groups actively developing SSC differentiation protocols for livestock species (Sato et al., 2011; Matsumura et al., 2023). Once collected, SSCs can be cultured in vitro, gene edited, and screened to confirm the presence of desired alterations (Tang et al., 2018). The edited SSCs could then be differentiated into spermatids and used for ICSI to fertilize embryos (Figure 2A) or transplanted into the testes of male animals that lack germ cells (Chapman et al., 2015). The transplanted SSCs can colonise the testes and produce gene edited sperm that carry the genotype of interest. Ciccarelli et al. (2020) demonstrated the process of SSC transplantation into germline ablated adult pigs and goats, where donor stem cells led to sustained donor-derived spermatogenesis. This technique could allow hardy and virile breeds to carry sperm that are homozygous for a specific desirable trait from other breeds, generating heterozygous offspring in an extensive environment.
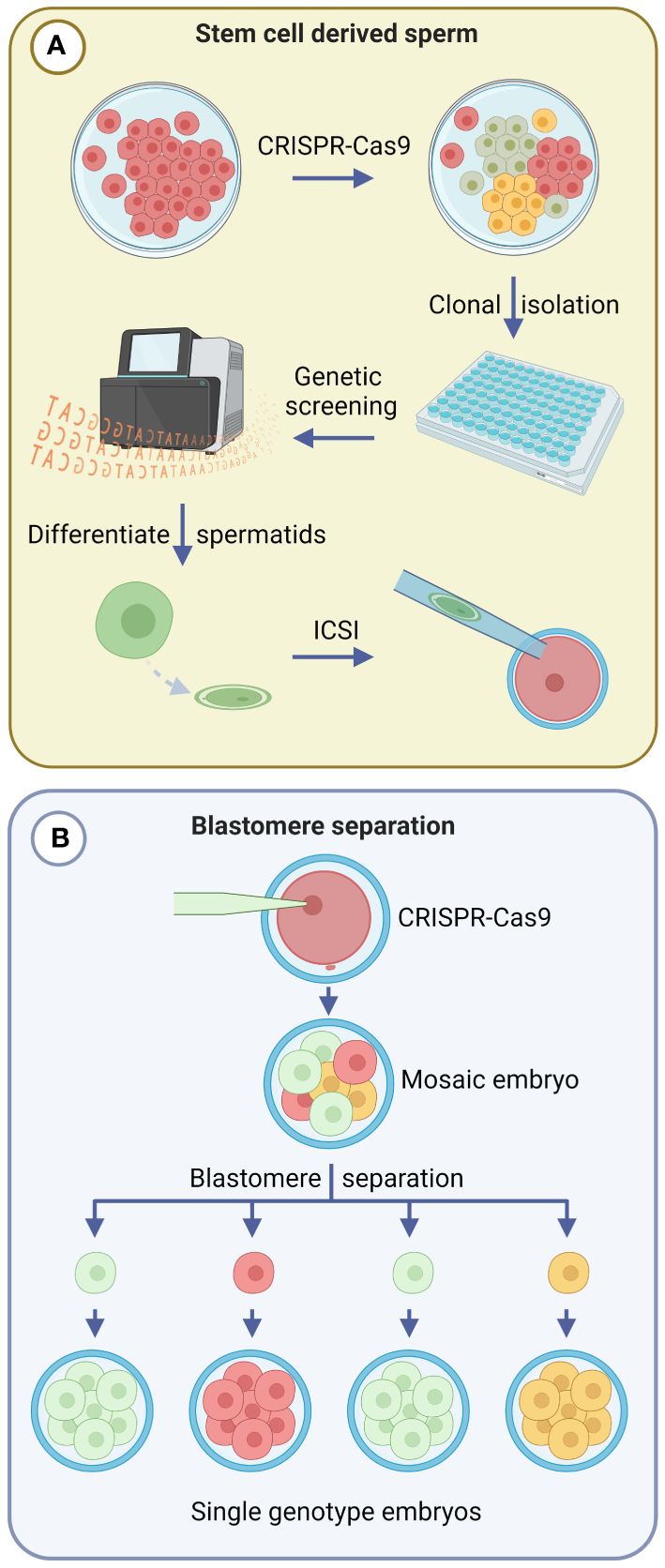
Figure 2 Emerging gene editing strategies that prevent mosaicism in gene edited offspring. Green, yellow and red cell colours indicate different genotypes. (A) Spermatogonial stem cells (SSCs) can be gene edited in vitro and clonal populations screened to confirm gene editing outcomes. Confirmed SSCs can then be differentiated into spermatozoa and undergo intracytoplasmic sperm injection (ICSI) to fertilize embryos. (B) Blastomere separation involves mechanical or chemical methods to split 2-, 4-, or 8-cell stage gene edited embryos, and facilitates the production of single genotype embryos from individual blastomeres. Created with BioRender.com.
In the 1980s, microblade splitting of morula and blastocyst stage embryos was developed as a strategy to increase the number of offspring obtained from embryo transfer programs in cattle, sheep and goats (Willadsen, 1980; Ozil et al., 1982; Willadsen and Godke, 1984; Udy, 1987; Seike et al., 1989; Velásquez et al., 2017). This method involved the bisection of each collected in vivo-derived embryo to form two equivalent demi-embryos, thereby doubling the number of embryos transferred to recipient females. Studies in which individual blastomeres of 2-, 4-, and 8-cell sheep embryos were separated, had previously demonstrated that totipotency is maintained for several cleavage divisions, such that each blastomere can form a blastocyst and give rise to offspring (Willadsen, 1979, 1981) (Figure 2B). From 14 originating sheep embryos, SWilladsen (1981) obtained 56 embryos after blastomere separation that resulted in the birth of 18 lambs. Methods of blastomere separation include mechanical separation using a micromanipulator and chemical disaggregation after removing the ZP (Casser et al., 2019). Due to the technical challenges involved, blastomere separation has been restricted to research studies, and has not yet been integrated into breeding programs.
The integration of blastomere separation with gene editing remains largely unexplored, although the foundations of such an approach have been employed in cynomolgus monkeys to evaluate the editing efficacy of Cas9 reagents in individual blastomeres (Tu et al., 2017). To integrate gene editing with blastomere separation, a zygote would be gene edited using Cas9-based reagents, and cultured up to the morula stage, which would then undergo bisection into single blastomeres. Each blastomere could be cultured further after being placed into an evacuated ZP, or by using the well-of-the-well system (Tagawa et al., 2008). Each resulting blastocyst could then be screened by biopsy to confirm the desired genotype prior to transfer into a recipient female. This strategy could eliminate the risk of Cas9-mediated mosaicism, facilitating the creation of a gene edited animal from a single blastomere, in a manner analogous to SCNT. In addition, serial separation of blastomeres following subsequent cleavage divisions may be a useful strategy to multiply embryos of a single genotype.
While gene editing using SSCs or blastomere separation can avoid the production of mosaic animals, these techniques may not meet the high throughput breeding requirements of livestock farming systems in their current forms due to the technical expertise and laboratory equipment required. Advanced robotics and microfluidics may overcome some of these challenges in the future. Further research is necessary to address these concerns and evaluate the benefits and limitations of these techniques for livestock breeding.
7 Conclusion
Applications of gene editing have the potential to revolutionise livestock breeding and contribute to addressing production, welfare, and environmental challenges whilst aiming to achieve global nutritional security. Cas9 has supplanted previous genome editing technologies, and CRISPR continues to be an innovative field in which new discoveries will offer improved prospects for efficient genome editing in livestock. However, to make Cas9-based zygote gene editing a practical and cost-effective breeding technology that can be reliably used on-farm, it is important to reduce the frequency of mosaicism currently seen in gene edited founder animals. This review has outlined potential tools and techniques that could reduce mosaicism in mammalian livestock species in the hope of stimulating further research in this space. Our goal was to highlight the many opportunities available for improvements, and that a multifaceted approach that combines genome editor selection, format and timing, delivery, and genetic screening, is likely needed to see significant reductions in mosaic founders. While further innovations are likely necessary, we are optimistic that the combination of the techniques presented will help reduce mosaicism to levels that enable the on-farm application of livestock gene editing in the near future. The potential benefits of gene editing in livestock are vast and this technology will play an increasingly important role in meeting the growing demand for safe, sustainable, and nutritious animal protein.
Author contributions
HS: Writing – original draft, Writing – review & editing. CG: Writing – original draft, Writing – review & editing. GM: Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the McGarvie Smith Institute.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aird E. J., Lovendahl K. N., St Martin A., Harris R. S., Gordon W. R. (2018). Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun. Biol. 1, 54. doi: 10.1038/s42003-018-0054-2
Alghadban S., Bouchareb A., Hinch R., Hernandez-Pliego P., Biggs D., Preece C., et al. (2020). Electroporation and genetic supply of Cas9 increase the generation efficiency of CRISPR/Cas9 knock-in alleles in C57BL/6J mouse zygotes. Sci. Rep. 10, 17912. doi: 10.1038/s41598-020-74960-7
Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi: 10.1038/s41586-019-1711-4
Anzalone D. A., Palazzese L., Czernik M., Sabatucci A., Valbonetti L., Capra E., et al. (2021). Controlled spermatozoa–oocyte interaction improves embryo quality in sheep. Sci. Rep. 11, 22629. doi: 10.1038/s41598-021-02000-z
Aoyama N., Kato K. (2020). Trophectoderm biopsy for preimplantation genetic test and technical tips: A review. Reprod. Med. Biol. 19, 222–231. doi: 10.1002/rmb2.12318
Berland M., Frei M., Peralta O., Ratto M. (2011). Time exposure period of bovine oocytes to sperm in relation to embryo development rate and quality. ISRN Vet. Sci 2011 257627. doi: 10.5402/2011/257627
Bevacqua R. J., Fernandez-Martín R., Savy V., Canel N. G., Gismondi M. I., Kues W. A., et al. (2016). Efficient edition of the bovine PRNP prion gene in somatic cells and IVF embryos using the CRISPR/Cas9 system. Theriogenology 86, 1886–1896. e1881. doi: 10.1016/j.theriogenology.2016.06.010
Biesecker L. G., Spinner N. B. (2013). A genomic view of mosaicism and human disease. Nat. Rev. Genet. 14, 307–320. doi: 10.1038/nrg3424
Bishop T. F., Van Eenennaam A. L. (2020). Genome editing approaches to augment livestock breeding programs. J. Exp. Biol. 223, jeb207159. doi: 10.1242/jeb.207159
Bloomer H., Khirallah J., Li Y., Xu Q. (2022). CRISPR/Cas9 ribonucleoprotein-mediated genome and epigenome editing in mammalian cells. Adv. Drug Delivery Rev. 181, 114087. doi: 10.1016/j.addr.2021.114087
Bondioli K. R. (2018). Cloning of livestock by somatic cell nuclear transfer. Anim. Biotechnol. 2: Emerg. Breed. Technol. 2, 1–20. doi: 10.1007/978-3-319-92348-2_1
Bunton-Stasyshyn R. K., Codner G. F., Teboul L. (2022). Screening and validation of genome-edited animals. Lab. Anim. 56, 69–82. doi: 10.1177/00236772211016922
Caso F., Davies B. (2022). Base editing and prime editing in laboratory animals. Lab. Anim. 56, 35–49. doi: 10.1177/0023677221993895
Casser E., Israel S., Boiani M. (2019). Multiplying embryos: experimental monozygotic polyembryony in mammals and its uses. Int. J. Dev. Biol. 63, 143–155. doi: 10.1387/ijdb.190016mb
Cenariu M., Pall E., Cernea C., Groza I. (2012). Evaluation of bovine embryo biopsy techniques according to their ability to preserve embryo viability. J. BioMed. Biotechnol. 2012, 541384. doi: 10.1155/2012/541384
Chapman K. M., Medrano G. A., Jaichander P., Chaudhary J., Waits A. E., Nobrega M. A., et al. (2015). Targeted germline modifications in rats using CRISPR/Cas9 and spermatogonial stem cells. Cell Rep. 10, 1828–1835. doi: 10.1016/j.celrep.2015.02.040
Ciccarelli M., Giassetti M. I., Miao D., Oatley M. J., Robbins C., Lopez-Biladeau B., et al. (2020). Donor-derived spermatogenesis following stem cell transplantation in sterile NANOS2 knockout males. Proc. Natl. Acad. Sci. U.S.A. 117, 24195–24204. doi: 10.1073/pnas.2010102117
Coates A., Bankowski B. J., Kung A., Griffin D. K., Munne S. (2017). Differences in pregnancy outcomes in donor egg frozen embryo transfer (FET) cycles following preimplantation genetic screening (PGS): a single center retrospective study. J. Assisted Reprod. Genet. 34, 71–78. doi: 10.1007/s10815-016-0832-z
Coticchio G., Cimadomo D., Cermisoni G. C., Rienzi L., Papaleo E., Ubaldi F. M., et al. (2023). The first mitotic division: a perilous bridge connecting the zygote and the early embryo. Hum. Reprod. 38, 1019–1027. doi: 10.1093/humrep/dead067
Crispo M., Mulet A., Tesson L., Barrera N., Cuadro F., dos Santos-Neto P., et al. (2015). Efficient generation of myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes. PloS One 10, e0136690. doi: 10.1371/journal.pone.0136690
Cunningham F. J. (2022). Conjugating CRISPR-cas9 machinery to single-walled carbon nanotubes for plant cellular delivery (University of California, Berkeley).
Daigneault B. W., Rajput S., Smith G. W., Ross P. J. (2018). Embryonic POU5F1 is required for expanded bovine blastocyst formation. Sci. Rep. 8, 7753. doi: 10.1038/s41598-018-25964-x
Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191. doi: 10.1038/nbt.3437
Doudna J. A., Charpentier E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. doi: 10.1126/science.1258096
Ferré L. B., Kjelland M. E., Strøbech L. B., Hyttel P., Mermillod P., Ross P. J. (2020). Review: Recent advances in bovine in vitro embryo production: reproductive biotechnology history and methods. Animal 14, 991–1004. doi: 10.1017/S1751731119002775
Frantz L. A., Bradley D. G., Larson G., Orlando L. (2020). Animal domestication in the era of ancient genomics. Nat. Rev. Genet. 21, 449–460. doi: 10.1038/s41576-020-0225-0
Gao P., Lyu Q., Ghanam A. R., Lazzarotto C. R., Newby G. A., Zhang W., et al. (2021). Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome Biol. 22, 83. doi: 10.1186/s13059-021-02304-3
Gaudelli N. M., Komor A. C., Rees H. A., Packer M. S., Badran A. H., Bryson D. I., et al. (2017). Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi: 10.1038/nature24644
Gim G.-M., Eom K.-H., Kwon D.-H., Jung D.-J., Kim D.-H., Yi J.-K., et al. (2023). Generation of double knockout cattle via CRISPR-Cas9 ribonucleoprotein (RNP) electroporation. J. Anim. Sci. Biotechnol. 14, 103. doi: 10.1186/s40104-023-00902-8
Gu B., Posfai E., Gertsenstein M., Rossant J. (2020). Efficient generation of large-fragment knock-in mouse models using 2-cell (2C)-homologous recombination (HR)-CRISPR. Curr. Protoc. Mouse Biol. 10, e67. doi: 10.1002/cpmo.67
Gutschner T., Haemmerle M., Genovese G., Draetta G. F., Chin L. (2016). Post-translational regulation of cas9 during G1 enhances homology-directed repair. Cell Rep. 14, 1555–1566. doi: 10.1016/j.celrep.2016.01.019
Hai T., Teng F., Guo R., Li W., Zhou Q. (2014). One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 24, 372–375. doi: 10.1038/cr.2014.11
Han X., Liu Z., Ma Y., Zhang K., Qin L. (2017). Cas9 ribonucleoprotein delivery via microfluidic cell-deformation chip for human T-cell genome editing and immunotherapy. Adv. Biosyst. 1, 1600007. doi: 10.1002/adbi.201600007
Han H., Ma Y., Wan T., Lian L., Tian X., R H., et al. (2014). One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front. Agr. Sci. Eng. 1, 2–5. doi: 10.15302/J-FASE-2014007
Hashimoto M., Yamashita Y., Takemoto T. (2016). Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev. Biol. 418, 1–9. doi: 10.1016/j.ydbio.2016.07.017
Hennig S. L., Owen J. R., Lin J. C., Young A. E., Ross P. J., Van Eenennaam A. L., et al. (2020). Evaluation of mutation rates, mosaicism and off target mutations when injecting Cas9 mRNA or protein for genome editing of bovine embryos. Sci. Rep. 10, 22309. doi: 10.1038/s41598-020-78264-8
Hirata M., Wittayarat M., Namula Z., Anh Le Q., Lin Q., Takebayashi K., et al. (2021a). Lipofection-mediated introduction of CRISPR/Cas9 system into porcine oocytes and embryos. Animals 11, 578. doi: 10.3390/ani11020578
Hirata M., Wittayarat M., Namula Z., Le Q. A., Lin Q., Takebayashi K., et al. (2021b). Generation of mutant pigs by lipofection-mediated genome editing in embryos. Sci. Rep. 11, 23806. doi: 10.1038/s41598-021-03325-5
Kim S., Kim D., Cho S. W., Kim J., Kim J.-S. (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019. doi: 10.1101/gr.171322.113
Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi: 10.1038/nature17946
Lamas-Toranzo I., Galiano-Cogolludo B., Cornudella-Ardiaca F., Cobos-Figueroa J., Ousinde O., Bermejo-Álvarez P. (2019). Strategies to reduce genetic mosaicism following CRISPR-mediated genome edition in bovine embryos. Sci. Rep. 9, 14900. doi: 10.1038/s41598-019-51366-8
Le Q. A., Hirata M., Nguyen N. T., Takebayashi K., Wittayarat M., Sato Y., et al. (2020). Effects of electroporation treatment using different concentrations of Cas9 protein with gRNA targeting Myostatin (MSTN) genes on the development and gene editing of porcine zygotes. Anim. Sci. J. 91, e13386. doi: 10.1111/asj.13386
Lee K., Uh K., Farrell K. (2020). Current progress of genome editing in livestock. Theriogenology 150, 229–235. doi: 10.1016/j.theriogenology.2020.01.036
Li G., Liu D., Zhang X., Quan R., Zhong C., Mo J., et al. (2018). Suppressing Ku70/Ku80 expression elevates homology-directed repair efficiency in primary fibroblasts. Int. J. Biochem. Cell Biol. 99, 154–160. doi: 10.1016/j.biocel.2018.04.011
Lillico S. G., Proudfoot C., King T. J., Tan W., Zhang L., Mardjuki R., et al. (2016). Mammalian interspecies substitution of immune modulatory alleles by genome editing. Sci. Rep. 6, 21645. doi: 10.1038/srep21645
Lin Q., Le Q. A., Takebayashi K., Thongkittidilok C., Wittayarat M., Hirata M., et al. (2021). Timing and duration of lipofection-mediated CRISPR/Cas9 delivery into porcine zygotes affect gene-editing events. BMC Res. Notes 14, 1–6. doi: 10.1186/s13104-021-05800-8
Lin J. C., Van Eenennaam A. L. (2021). Electroporation-mediated genome editing of livestock zygotes. Front. Genet. 12, 648482. doi: 10.3389/fgene.2021.648482
Liu M., Rehman S., Tang X., Gu K., Fan Q., Chen D., et al. (2019). Methodologies for improving HDR efficiency. Front. Genet. 9, 691. doi: 10.3389/fgene.2018.00691
Liu C., Zhang L., Liu H., Cheng K. (2017). Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control Release 266, 17–26. doi: 10.1016/j.jconrel.2017.09.012
Lu J., Wu T., Zhang B., Liu S., Song W., Qiao J., et al. (2021). Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Communicat. Signaling 19, 60. doi: 10.1186/s12964-021-00741-y
Luo L., Shi Y., Wang H., Wang Z., Dang Y., Li S., et al. (2022). Base editing in bovine embryos reveals a species-specific role of SOX2 in regulation of pluripotency. PloS Genet. 18, e1010307. doi: 10.1371/journal.pgen.1010307
Ma H., Marti-Gutierrez N., Park S. W., Wu J., Lee Y., Suzuki K., et al. (2017). Correction of a pathogenic gene mutation in human embryos. Nature 548, 413–419. doi: 10.1038/nature23305
Mahdi A. K., Medrano J. F., Ross P. J. (2022). Single-step genome editing of small ruminant embryos by electroporation. Int. J. Mol. Sci. 23. doi: 10.3390/ijms231810218
Matsumura T., Katagiri K., Yao T., Ishikawa-Yamauchi Y., Nagata S., Hashimoto K., et al. (2023). Generation of rat offspring using spermatids produced through in vitro spermatogenesis. Sci. Rep. 13, 12105. doi: 10.1038/s41598-023-39304-1
McFarlane G. R., Salvesen H. A., Sternberg A., Lillico S. G. (2019). On-farm livestock genome editing using cutting edge reproductive technologies. Front. Sustain. Food Syst. 3. doi: 10.3389/fsufs.2019.00106
Mehravar M., Shirazi A., Nazari M., Banan M. (2019). Mosaicism in CRISPR/Cas9-mediated genome editing. Dev. Biol. 445, 156–162. doi: 10.1016/j.ydbio.2018.10.008
Menchaca A., Dos Santos-Neto P. C., Mulet A. P., Crispo M. (2020). CRISPR in livestock: From editing to printing. Theriogenology 150, 247–254. doi: 10.1016/j.theriogenology.2020.01.063
Meyenberg M., Ferreira da Silva J., Loizou J. I. (2021). Tissue specific DNA repair outcomes shape the landscape of genome editing. Front. Genet. 12. doi: 10.3389/fgene.2021.728520
Miyaoka Y., Berman J. R., Cooper S. B., Mayerl S. J., Chan A. H., Zhang B., et al. (2016). Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci. Rep. 6, 23549. doi: 10.1038/srep23549
Munk M., Ladeira L. O., Carvalho B. C., Camargo L. S. A., Raposo N. R. B., Serapião R. V., et al. (2016). Efficient delivery of DNA into bovine preimplantation embryos by multiwall carbon nanotubes. Sci. Rep. 6, 33588. doi: 10.1038/srep33588
Namula Z., Wittayarat M., Hirata M., Hirano T., Nguyen N. T., Le Q. A., et al. (2019). Genome mutation after the introduction of the gene editing by electroporation of Cas9 protein (GEEP) system into bovine putative zygotes. In Vitro Cell. Dev. Biology-Animal 55, 598–603. doi: 10.1007/s11626-019-00385-w
Ni W., Qiao J., Hu S., Zhao X., Regouski M., Yang M., et al. (2014). Efficient gene knockout in goats using CRISPR/cas9 system. PloS One 9, e106718. doi: 10.1371/journal.pone.0106718
Norris A. L., Lee S. S., Greenlees K. J., Tadesse D. A., Miller M. F., Lombardi H. A. (2020). Template plasmid integration in germline genome-edited cattle. Nat. Biotechnol. 38, 163–164. doi: 10.1038/s41587-019-0394-6
Ozil J. P., Heyman Y., Renard J. P. (1982). Production of monozygotic twins by micromanipulation and cervical transfer in the cow. Vet. Rec 110, 126–127. doi: 10.1136/vr.110.6.126
Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., et al. (2016). Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129. doi: 10.1038/nature17664
Petersen B., Frenzel A., Lucas-Hahn A., Herrmann D., Hassel P., Klein S., et al. (2016). Efficient production of biallelic GGTA 1 knockout pigs by cytoplasmic microinjection of CRISPR/Cas9 into zygotes. Xenotransplantation 23, 338–346. doi: 10.1111/xen.12258
Piñeiro-Silva C., Navarro-Serna S., Belda-Pérez R., Gadea J. (2023). Production of genetically modified porcine embryos via lipofection of zona-pellucida-intact oocytes using the CRISPR/cas9 system. Animals 13, 342. doi: 10.3390/ani13030342
Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Qi Y., Zhang Y., Tian S., Zong R., Yan X., Wang Y., et al. (2023). An optimized prime editing system for efficient modification of the pig genome. Sci. China Life Sci. 66, 2851–2861. doi: 10.1007/s11427-022-2334-y
Rees H. A., Liu D. R. (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. doi: 10.1038/s41576-018-0059-1
Richardson C., Kelsh R. N., R J. R. (2023). New advances in CRISPR/Cas-mediated precise gene-editing techniques. Dis. Model. Mech. 16. doi: 10.1242/dmm.049874
Sansbury B. M., Hewes A. M., Kmiec E. B. (2019). Understanding the diversity of genetic outcomes from CRISPR-Cas generated homology-directed repair. Commun. Biol. 2, 458. doi: 10.1038/s42003-019-0705-y
Sato T., Katagiri K., Yokonishi T., Kubota Y., Inoue K., Ogonuki N., et al. (2011). In vitro production of fertile sperm from murine spermatogonial stem cell lines. Nat. Commun. 2, 472. doi: 10.1038/ncomms1478
Scholefield J., Harrison P. T. (2021). Prime editing – an update on the field. Gene Ther. 28, 396–401. doi: 10.1038/s41434-021-00263-9
Scott M. A., Hu Y. C. (2019). Generation of CRISPR-edited rodents using a piezo-driven zygote injection technique. Methods Mol. Biol. 1874, 169–178. doi: 10.1007/978-1-4939-8831-0_9
Seike N., Teranishi M., Yamada S., Takakura R., Nagao Y., Kanagawa H. (1989). Increase in calf production by the transfer of bisected bovine embryos. Nihon Juigaku Zasshi 51, 1193–1199. doi: 10.1292/jvms1939.51.1193
Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., et al. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402. doi: 10.1038/nmeth.2857
Song J., Yang D., Xu J., Zhu T., Chen Y. E., Zhang J. (2016). RS-1 enhances CRISPR/Cas9-and TALEN-mediated knock-in efficiency. Nat. Commun. 7, 10548. doi: 10.1038/ncomms10548
Sturmey R., Reis A., Leese H., McEvoy T. (2009). Role of fatty acids in energy provision during oocyte maturation and early embryo development. Reprod. Domest. Anim. 44, 50–58. doi: 10.1111/j.1439-0531.2009.01402.x
Tagawa M., Matoba S., Narita M., Saito N., Nagai T., Imai K. (2008). Production of monozygotic twin calves using the blastomere separation technique and Well of the Well culture system. Theriogenology 69, 574–582. doi: 10.1016/j.theriogenology.2007.11.003
Takemoto T. (2020). Zygote electroporation for CRISPR/Cas9 delivery to generate genetically modified mice. Electroporat. Protocols: Microorganism Mamm. System Nanodevice 2050, 121–126. doi: 10.1007/978-1-4939-9740-4_13
Tang L., Bondareva A., González R., Rodriguez-Sosa J. R., Carlson D. F., Webster D., et al. (2018). TALEN-mediated gene targeting in porcine spermatogonia. Mol. Reprod. Dev. 85, 250–261. doi: 10.1002/mrd.22961
Tanihara F., Hirata M., Nguyen N. T., Le Q. A., Hirano T., Otoi T. (2019). Effects of concentration of CRISPR/Cas9 components on genetic mosaicism in cytoplasmic microinjected porcine embryos. J. Reprod. Dev. 65, 209–214. doi: 10.1262/jrd.2018-116
Tao L., Yang M., Wang X., Zhang Z., Wu Z., Tian J., et al. (2016). Efficient biallelic mutation in porcine parthenotes using a CRISPR-Cas9 system. Biochem. Biophys. Res. Commun. 476, 225–229. doi: 10.1016/j.bbrc.2016.05.100
Tu Z., Yang W., Yan S., Yin A., Gao J., Liu X., et al. (2017). Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci. Rep. 7, 42081. doi: 10.1038/srep42081
Udy G. B. (1987). Commercial splitting of goat embryos. Theriogenology 28, 837–847. doi: 10.1016/0093-691X(87)90035-5
Vajta G. (2007). Somatic cell nuclear transfer in its first and second decades: successes, setbacks, paradoxes and perspectives. Reprod. biomed. Online 15, 582–590. doi: 10.1016/S1472-6483(10)60391-4
Velásquez A. E., Manríquez J., Castro F. O., Cox J. F., Rodriguez-Alvarez L. (2017). Embryo splitting affects the transcriptome during elongation stage of in vitro–produced bovine blastocysts. Theriogenology 87, 124–134. doi: 10.1016/j.theriogenology.2016.08.014
Vilarino M., Rashid S. T., Suchy F. P., McNabb B. R., van der Meulen T., Fine E. J., et al. (2017). CRISPR/Cas9 microinjection in oocytes disables pancreas development in sheep. Sci. Rep. 7, 17472. doi: 10.1038/s41598-017-17805-0
Vilarino M., Suchy F. P., Rashid S. T., Lindsay H., Reyes J., McNabb B. R., et al. (2018). Mosaicism diminishes the value of pre-implantation embryo biopsies for detecting CRISPR/Cas9 induced mutations in sheep. Transgenic Res. 27, 525–537. doi: 10.1007/s11248-018-0094-x
Walton M. (2018). Commercial applications of SCNT in livestock. Anim. Biotechnol. 2: Emerg. Breed. Technol. 2, 21–35. doi: 10.1007/978-3-319-92348-2_2
Wang Y., Du Y., Shen B., Zhou X., Li J., Liu Y., et al. (2015). Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci. Rep. 5, 8256. doi: 10.1038/srep08256
Wang H., La Russa M., Qi L. S. (2016). CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264. doi: 10.1146/annurev-biochem-060815-014607
Wang H., Lu H., Lei Y. S., Gong C. Y., Chen Z., Luan Y. Q., et al. (2020). Development of a self-Restricting CRISPR-Cas9 system to reduce off-Target effects. Mol. Ther. Methods Clin. Dev. 18, 390–401. doi: 10.1016/j.omtm.2020.06.012
Wang D., Niu Y., Ren L., Kang Y., Tai P. W. L., Si C., et al. (2019). Gene delivery to nonhuman primate preimplantation embryos using recombinant adeno-associated virus. Adv. Sci. (Weinh) 6, 1900440. doi: 10.1002/advs.201900440
Ward F., Enright B., Rizos D., Boland M., Lonergan P. (2002). Optimization of in vitro bovine embryo production: effect of duration of maturation, length of gamete co-incubation, sperm concentration and sire. Theriogenology 57, 2105–2117. doi: 10.1016/S0093-691X(02)00696-9
Weissman A., Shoham G., Shoham Z., Fishel S., Leong M., Yaron Y. (2017). Preimplantation genetic screening: results of a worldwide web-based survey. Reprod. biomed. Online 35, 693–700. doi: 10.1016/j.rbmo.2017.09.001
Whitworth K. M., Lee K., Benne J. A., Beaton B. P., Spate L. D., Murphy S. L., et al. (2014). Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol. Reprod. 91, 78. doi: 10.1095/biolreprod.114.121723
Wiener P., Wilkinson S. (2011). Deciphering the genetic basis of animal domestication. Proc. R. Soc. B: Biol. Sci. 278, 3161–3170. doi: 10.1098/rspb.2011.1376
Willadsen S. M. (1979). A method for culture of micromanipulated sheep embryos and its use to produce monozygotic twins. Nature 277, 298–300. doi: 10.1038/277298a0
Willadsen S. (1980). The viability of early cleavage stages containing half the normal number of blastomeres in the sheep. Reproduction 59, 357–362. doi: 10.1530/jrf.0.0590357
Willadsen S. M. (1981). The development capacity of blastomeres from 4- and 8-cell sheep embryos. J. Embryol Exp. Morphol 65, 165–172
Willadsen S. M., Godke R. A. (1984). A simple procedure for the production of identical sheep twins. Vet. Rec 114, 240–243. doi: 10.1136/vr.114.10.240
Yamauchi Y., Ward M. A., Ward W. S. (2009). Asynchronous DNA replication and origin licensing in the mouse one-cell embryo. J. Cell Biochem. 107, 214–223. doi: 10.1002/jcb.22117
Yang H., Ren S., Yu S., Pan H., Li T., Ge S., et al. (2020). Methods favoring homology-directed repair choice in response to CRISPR/cas9 induced-double strand breaks. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21186461
Yang D., Scavuzzo M. A., Chmielowiec J., Sharp R., Bajic A., Borowiak M. (2016). Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 6, 21264. doi: 10.1038/srep21264
Yang H., Yang D., Zhu Q., Wang K., Zhang C., Chen B., et al. (2022). Application of two blastocyst biopsy strategies in preimplantation genetic testing treatment and assessment of their effects. Front. Endocrinol. 13. doi: 10.3389/fendo.2022.852620
Yip B. H. (2020). Recent advances in CRISPR/cas9 delivery strategies. Biomolecules 10. doi: 10.3390/biom10060839
Zhang X., Li T., Ou J., Huang J., Liang P. (2022). Homology-based repair induced by CRISPR-Cas nucleases in mammalian embryo genome editing. Protein Cell 13, 316–335. doi: 10.1007/s13238-021-00838-7
Zhang X., Li W., Wu Y., Peng X., Lou B., Wang L., et al. (2017). Disruption of the sheep BMPR-IB gene by CRISPR/Cas9 in in vitro-produced embryos. Theriogenology 91, 163–172.e162. doi: 10.1016/j.theriogenology.2016.10.025
Zhang Y., Zhang Z., Ge W. (2018). An efficient platform for generating somatic point mutations with germline transmission in the zebrafish by CRISPR/Cas9-mediated gene editing. J. Biol. Chem. 293, 6611–6622. doi: 10.1074/jbc.RA117.001080
Keywords: mosaicism, CRISPR-Cas9, livestock, gene editing, agriculture, genetic engineering
Citation: Salvesen HA, Grupen CG and McFarlane GR (2024) Tackling mosaicism in gene edited livestock. Front. Anim. Sci. 5:1368155. doi: 10.3389/fanim.2024.1368155
Received: 10 January 2024; Accepted: 16 May 2024;
Published: 03 June 2024.
Edited by:
Francisco Javier Navas González, University of Cordoba, SpainReviewed by:
Simon Geoffrey Lillico, University of Edinburgh, United KingdomDongshan Yang, University of Michigan, United States
Copyright © 2024 Salvesen, Grupen and McFarlane. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gus R. McFarlane, gus.mcfarlane@dpi.nsw.gov.au
 Hamish A. Salvesen
Hamish A. Salvesen Christopher G. Grupen
Christopher G. Grupen Gus R. McFarlane
Gus R. McFarlane