Description of the viral community in fecal content of common carp (Cyprinus carpio) living in a naturally preserved wetland on Mexican plateau
- 1Department of Medicine and Zootechnics of Birds, Faculty of Veterinary Medicine and Zootechnics, National Autonomous University of Mexico, Mexico City, Mexico
- 2Department of Bioinformatics in Infectious Diseases, Center for Research on Infectious Diseases, National Institute of Public Health, Morelos, Mexico
- 3Department of Ethology, Wildlife and Laboratory Animals Faculty of Veterinary Medicine and Zootechnics, National Autonomous University of Mexico, Mexico City, Mexico
- 4Department of Biotechnology in Animal Health, National Center for Disciplinary Research in Animal Health and Safety, National Institute of Agricultural and Livestock Forestry Research, Mexico City, Mexico
Introduction: The common carp (Cyprinus carpio) is a species that has been used worldwide as a source of protein for humans because of its adaptive capacity to the environment; however, it is also an invasive species. In Mexico, it was introduced to provide an easily accessible source of protein to people in rural areas until its extensive nationwide distribution. The community of viruses in these fish is not completely known.
Methods: Massive sequencing, i.e., whole metagenome shotgun (WMS) sequencing, and bioinformatic analysis were used to describe the viral community of common carp (Cyprinus carpio) living in a natural preserved wetland.
Results: Through sequencer Illumina NextSeq 500/550 we got 484,953,684 sequences from ten samples of fecal content of carp (5 samples to DNA and 5 samples to RNA). The virome comprised 30 viral families (13 DNA families and 17 RNA families), among which viruses related to vertebrates stood out, for example Orthoherpesviridae, Parvoviridae, Astroviridae, Poxviridae, Hantaviridae and Picornaviridae.
Discussion: The most abundant viral taxa corresponded to bacteriophages, most of which are of polyphyletic origin. The results of this work contribute to the knowledge of the different viral taxa found in the common carp in the wild and indicate that these viral taxa may play a role in health and productivity of fish farms.
1 Introduction
The virome formed by viral communities has been studied since the beginning of this millennium. Initially, non-cultivable viral taxa were identified in seas and oceans (Breitbart et al., 2002). Later, viral families were characterized in the intestinal content of humans (Breitbart et al., 2003) and animals that due to their economic importance, proximity to humans, and host to or vector of known viruses, could transmit a virus that affects public health or animal health (Cann et al., 2005; Chen et al., 2017; Kwok et al., 2020; Tan et al., 2021).
The viral community of fish has been little studied; currently, there are seven articles that describe the virome in more than 30 species from various organs, including the spleen, liver, gills and skin, notably, only one of them analyzed stool samples (Xi et al., 2023). Three research used metagenomics for their descriptions (Filipa-Silva et al., 2020; Gadoin et al., 2021; Xi et al., 2023) and the remaining four performed metatranscriptomics (Costa et al., 2021; Geoghegan et al., 2021; Perry et al., 2022; Grimwood et al., 2023).The aforementioned studies have mainly studied abundance and viral diversity, which help to understand the viral ecology of the most diverse group of vertebrates on the planet.
For the present, the only research related to common carp (Cyprinus carpio) was carried out in Basin, Australia (Costa et al., 2021). In contrast, the viromes for freshwater have been widely studied, and most have reported viral families of bacteriophages and viruses related to the flora or fauna of the study site (Aguirre de Cárcer et al., 2016; Cai et al., 2016; Chopyk et al., 2018; Gu et al., 2018; Moon et al., 2020; Rusiñol et al., 2020; Palermo et al., 2021).
Carp is a species that has been food for humans since ancient Roman times, and between the 12th and 14th centuries AD, unintentional selection occurred that represented one of the first steps toward carp domestication (FAO, 2005). To date, carp has been introduced in practically all countries where there is an opportunity for successful reproduction and is one of the most cultivated species worldwide (FAO, 2023a). The common carp (Cyprinus carpio) was introduced to Mexico from Europe at the end of the 19th century. Subsequently, the Mexican government imported more carp to improve the quality of the diet, providing another protein option for Mexican families as well as sources of work for rural populations (National Institute of Fisheries, 2018). Currently, common carp is cultivated in different states of the Mexican Republic, with Tamaulipas, Jalisco and Michoacán being the main producers in 2021 (National Aquaculture and Fisheries Commission, 2021).
The common carp is an omnivorous animal that can eat a wide variety of natural foods, with a diet consisting of zooplankton, worms, mollusks, crustaceans, insects (which can be eggs and pupae), stems, leaves and soft parts of plants, seeds of aquatic plants, fish eggs, small fish and larval fish. Additionally, it has an adaptive capacity that allows for changes in food preferences based on the availability of resources (Hoole et al., 2001; FAO, 2005; FAO, 2023b). Feeding by carp occurs mainly in the deepest parts of the body of water that they inhabit (benthic behavior); the optimal temperature of the feeding environment is 20 to 25°C, with feeding stopping at 8°C (FAO, 2023b). These feeding characteristics can modify ecosystem conditions if carp are released to the wild, which is why the National Commission for the Knowledge and Use of Biodiversity (Comisión Nacional para el Conocimiento y Uso de la Biodiversidad - CONABIO) and the Invasive Species Specialist Group (ISSG) consider carp a species with invasive potential (CONABIO (2011); GISD, 2013).
Considering the feeding behavior, adaptability and broad diet of carp, this fish can provide and expand the information on the identification and presence of viral communities in freshwater natural habitats that could be important for health fishes. Therefore, the objective of this study was to describe, through massive DNA and RNA sequencing, the viral community in the fecal content of wild Cyprinus carpio.
2 Materials and methods
2.1 Study site
The wetland “Laguna de Chignahuapan” is located in the municipality of Almoloya del Río (19°09’15.9”N 99°29’41.7”W) within Cienegas de Lerma. It is a naturally preserved wetland where there is a carp population and is listed on “The List of Wetlands of International Importance” by Ramsar (Ramsar, 2023).This is the only wetland of “Cienegas de Lerma” best preserved and no outbreak of illness due to viral infection has been reported.
2.2 Carp
The fish were caught by the person in charge of the wetland. Ten wild carp were obtained. The carp were collected from January 30 to February 20, 2022, and were transported to the Department of Bird Medicine and Animal Husbandry (Departamento de Medicina y Zootecnia de Aves - DMZA) at the School of Veterinary Medicine and Animal Husbandry (FMVZ-UNAM, for its acronym in Spanish) under refrigeration conditions inside a cooler with ice for human consumption (4°C). Once at the DMZA, the carp were euthanized using a piercing pin in accordance with the “Aquatic Animal Health Code” of the World Organization for Animal Health (WOAH) (OMSA, 2019). Subsequently, to avoid contamination of the sample with the microbiome present in carp skin, the fish were sprayed with 70% ethanol (Givens et al., 2015; Li et al., 2015; Cui et al., 2017). Biometrics as maximum high, partial high, length, body weight and sex were measured (Supplementary Material 1). The collection of and procedures involving carp were approved by the Institutional Subcommittee for the Care and Use of Experimental Animals (Subcomité Institucional para el Cuidado y uso de Animales Experimentales - SICUAE) of FMVZ-UNAM, protocol number SICUA. DC-2021/4-8.
2.3 Collection of samples
Anatomical dissection was carried out in the necropsy room of the DMZA under aseptic conditions using sterile surgical tools to dissect the final part of the lower esophagus (10 cm). The fecal content of each fish (average 1.13 grams) was collected individually in DNAse-free and RNAse-free sterile plastic tubes with a screw cap, and once collected, the samples were stored immediately at -70°C. In addition, we performed histopathologic analysis of gills, spleen, liver and kidney, there was no evidence of tissue injuries. Results not shown.
2.4 Obtaining viral particles
The collected intestinal content samples were allowed to thaw at room temperature inside a NUAIRE laminar flow hood (model NO. NU-440-400), after which the samples were resuspended in Viral Transport Media (GIBCO, Thermo Fisher Scientific, A48750DK) at a ratio of 1:1 and homogenized by gentle manual shaking. The samples were centrifuged at 3150 x g for 15 minutes to precipitate the organic material at the bottom of the tube, and the supernatant was collected. To remove residual content, the supernatant was centrifuged at 15,000 x g for 15 minutes at 4°C. To remove eukaryotic and prokaryotic cells, the supernatant was passed through 0.45-µm and 0.22-µm filters, 0.4-1 ml were recovered. Finally, each sample was treated with DNase TURBO (Thermo Fisher, Invitrogen, Catalog number: AM2238) and RNase Cocktail (Thermo Fisher, Invitrogen, Product code 10036284), following the instructions of the manufacturer using 200 µl of sample for each reaction. The reaction was carried out at 37°C for 1 hour and stopped at -20°C.
2.5 Obtaining nucleic acids from viral particles
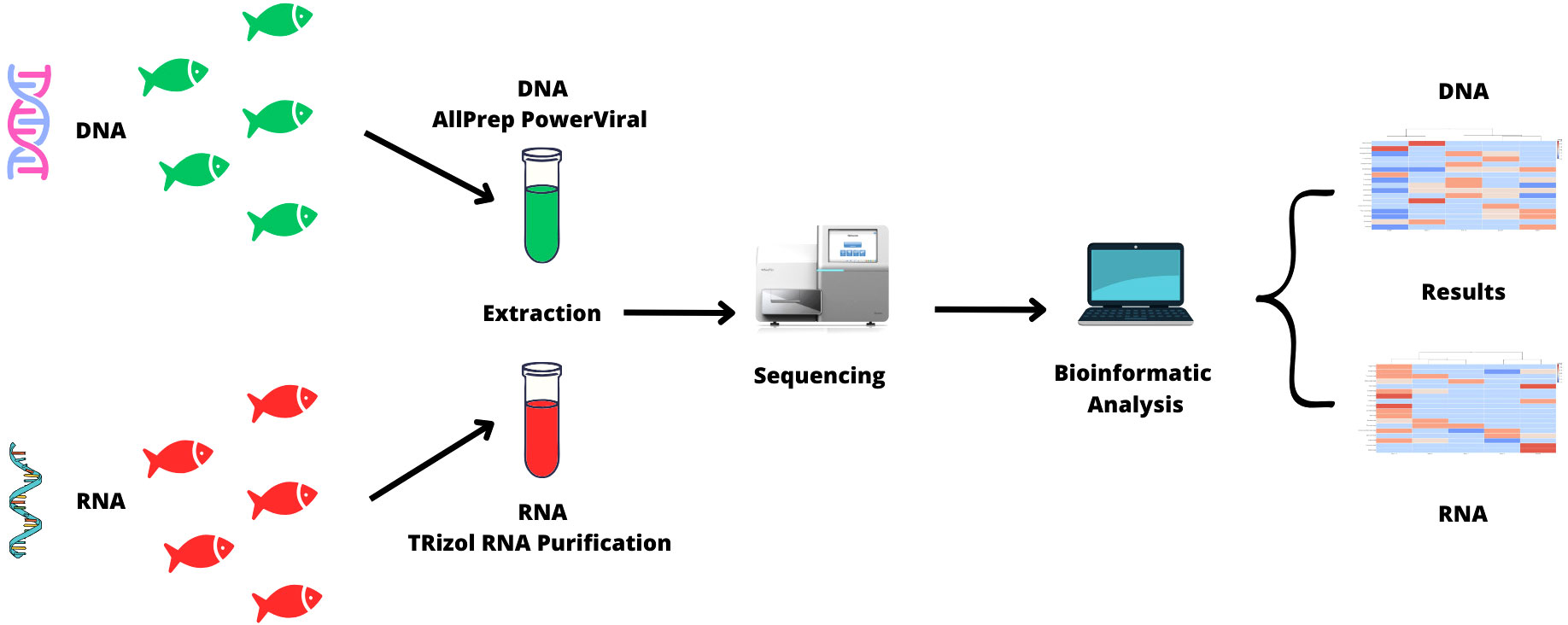
DNA was obtained from 5 samples using a commercial kit (AllPrep PowerViral DNA/RNA Kit, Qiagen, Hiden Germany, Cat. No. 28000-50) following the protocol recommended by the manufacturer. RNA was obtained from other 5 samples using a TRIzol RNA Purification kit and Phasemaker Tubes Complete System (Invitrogen, Cat No. A33254), for both material genetic 200 µl of sample were used for each reaction. The purified DNA and RNA samples were used for massive whole metagenome shotgun (WMS) sequencing at the University Unit for Massive Sequencing (Unidad Universitaria de Secuenciación Masiva - UUSM) of the Institute of Biotechnology (IBT-UNAM). A summary of the processing of the 10 samples is shown in Figure 1.
2.6 Preparation of WMS libraries and massive sequencing
Library preparation and sequencing was carried out by UUSM. Genetic material was quantified using the Qubit™ dsDNA HS Assay Kit and the Qubit ™ RNA HS Assay Kit, none of the samples showed a concentration of 1 mg/µl. The DNA libraries were constructed using the Illumina DNA Prep Kit following the manufacturer’s protocol, and the RNA libraries were constructed using the Illumina TruSeq Stranded mRNA Sample Prep Kit following the manufacturer’s protocol, where DNA libraries had average 600 bp and final loading concentration 1.2-1.3 pM. By the other hand, RNA libraries had average 260 bp and final loading concentration 4 nM. Both libraries were sequenced using the Illumina NextSeq 500/550 High Output Kit v2.5. For the DNA libraries, sequencing was carried out using 2x150 cycles, and for the RNA samples, sequencing was carried out using 2x75 cycles (Supplementary Material 2A).
2.7 Bioinformatic analysis
The raw sequences were filtered by quality Phred Q> 30 threshold using Trimmomatic V0.40 (Bolger et al., 2014). Bowtie2 was used to remove the host genome (Langmead and Salzberg, 2012) using the reference genome for Cyprinus carpio (GenBank LN590701.1) and the mitochondrial genome of Cyprinus carpio (GenBank NC-001606). Sequences belonging to bacteria and archaea were identified using Kraken2 with a kmers of size 50 (Wood et al., 2019) for later removal. The filtered sequences were searched in BLASTn using a value of e = 1x10-5. For DNA samples, the virus database in GenBank (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?VirusLineage_ss=DNA%20viruses&SeqType_s=Genome&SourceDB_s=RefSeq) (download date 19/4/2023) was used, and for RNA samples, NCBI’s RNA RefSeq viral database was searched https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?VirusLineage_ss=RNA%20viruses&SeqType_s=Genome&SourceDB_s=RefSeq (download date 4/19/2023). BLASTn results were viewed and analyzed in MEGAN Community Edition (v6.12.3) (Huson et al., 2016) and R version 4.2.2. with the ggplot2 library.
Using MEGAN, the Shannon-Wiener index and rarefaction curves were calculated at the viral family level for DNA and RNA samples (respectively) using normalized counts (sequence counts were normalized to the smallest sample size). Likewise, to visualize the composition of viral communities at the DNA and RNA levels by means of beta diversity, the Bray−Curtis dissimilarity was calculated. The relative abundance of the most representative viral taxa (abundance> 1%) in the samples was visualized by bar graphs generated in ggplot2, and the abundance by mean normalized counts was visualized by heatmaps generated in MEGAN.
Finally, the sequences classified as viral by BLASTn were aligned with sequences in NCBI RefSeq viral protein database https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?SeqType_s=Protein (download date 04/22/2023) using BLASTx with e = 1x10-5 as the threshold for positive matches. Similarly, the MEGAN program was used to generate heatmaps of the results for the DNA and RNA samples (Supplementary Material 2B).
3 Results
3.1 Cleaning and taxonomic assignment
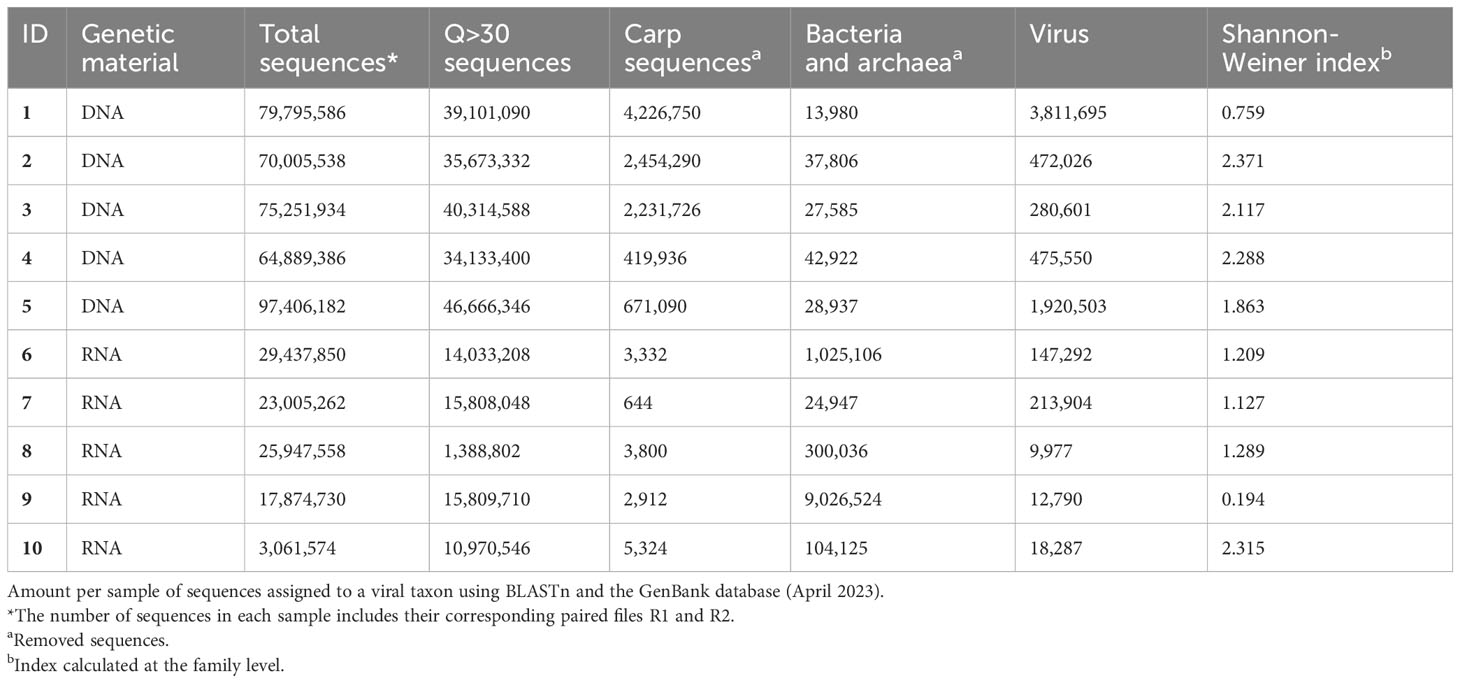
Ten fecal content samples from Cyprinus carpio were processed: five samples were used to extract DNA, and five samples were used to extract RNA (NCBI BioProject accession PRJNA1011357, publicly available 02/29/2024). A total of 386,988,626 DNA sequences were obtained, of which 195,888,756 (50.61%) were with Phred Q quality > 30. Approximately 5% of sequences with Phred Q quality > 30 were removed because they were sequences from carp, bacteria or archaea genomes. A total of 6,960,375 viral sequences were identified by means of BLASTn (Table 1) and classified into 13 viral DNA families. A total of 97,965,058 RNA sequences were obtained, of which 58,010,314 sequences (59.21%) sequences with Phred Q quality> 30 were preserved. Approximately ~18.09% of the sequences with Phred Q quality > 30 were removed because they were sequences in the carp, bacteria or archaea genomes. BLASTn alignment identified 402,250 viral sequences classified into 17 viral families, a group of unclassified Picornaviral and a group of unclassified RNA viruses (Table 1).
3.2 Abundance and viral diversity
The most abundant group of viruses were bacteriophages, approximately 90% of viral sequences corresponded to viral families related to bacteria; these sequences were classified into polyphyletic viruses (before known as Caudovirales) and 7 families: all of them detected in DNA samples (Ackermannviridae, Autographiviridae, Chaseviridae, Herelleviridae, Schitoviridae, Zobellviridae and Demerecviridae). Vertebrate-related viruses were the second most abundant group of viruses, approximately 7% of viral sequences corresponded were classified to viral families related to vertebrates (rodents, fish, reptiles, birds and humans); these sequences were classified into 10 families: 4 detected in DNA samples (Iridoviridae, Poxviridae, Orthoherpesviridae (before known as Herpesviridae (Benkő et al., 2021)) and Parvoviridae) and 6 detected in RNA samples (Astroviridae, Coronaviridae, Hantaviridae, Nodaviridae, Picornaviridae and Retroviridae). Viral families related to invertebrates had at less 1.4% of sequences classified into 4 families: 1 in DNA samples (Mimiviridae) and 3 classified in RNA samples (Chuviridae, Dicistroviridae and Iflaviridae). Approximately 0.7% of viral sequences corresponded to viral families related to plants these sequences were classified into 7 families, with all 7 detected in RNA samples (Botourmiaviridae, Caulimoviridae, Partitiviridae, Solemoviridae, Tombusviridae, Tospoviridae and Virgaviridae). While viral sequences related to algae and phytoplankton were the least group of viruses with less than 1% of sequences classified into 2 families: 1 detected in DNA (Phycodnaviridae) and 1 detected in RNA (Marnaviridae). Ultimately, unsorted RNA virus (including unclassified Picornavirales) sequences represented less than 1%.
Three of five DNA samples (60%) and 1 of 5 RNA sample (20%) showed a Shannon-Weiner index between 2-3, representing homogeneous diversity, and the rest of the samples, 40% to DNA samples and 80% to RNA samples, had low diversity (<2), representing heterogeneous diversity (Table 1). For both the DNA and RNA samples, diversity was analyzed through rarefaction curves at the level of viral families. The rarefaction curve for DNA samples indicated that each sample reached its respective plateau between 10,000 and 20,000 sequences (Supplementary Material 3), and the rarefaction curve for RNA indicated that each sample reached an asymptotic level between 12,000 and 12,500 sequences (Supplementary Material 4).
For the DNA samples, bacteriophages were the most representative families (90.14%), among which poliphyletic viruses were identified in all carp. The viral family related to vertebrate that was detected in all carp was Poxviridae, with an abundance of 0.3 to 5.4%. Nevertheless, it was not the only viral family related to vertebrate; Iridoviridae was also detected in carp 1 and 5 (20% of carps) with abundances <4%, Orthoherpesviridae and Parvoviridae in carp 5 (10% of carps) with abundances of 0.4% and 0.2%, respectively. Viral taxa related to invertebrates represented 0.8% of viral sequences among which Mimiviridae had abundances of 0.11 to 6.12%. Algal-related viral families represented 0.55% of viral sequences, among which Phycodnaviridae had an abundance of 0.23 to 2.9% in all samples (Figure 2).
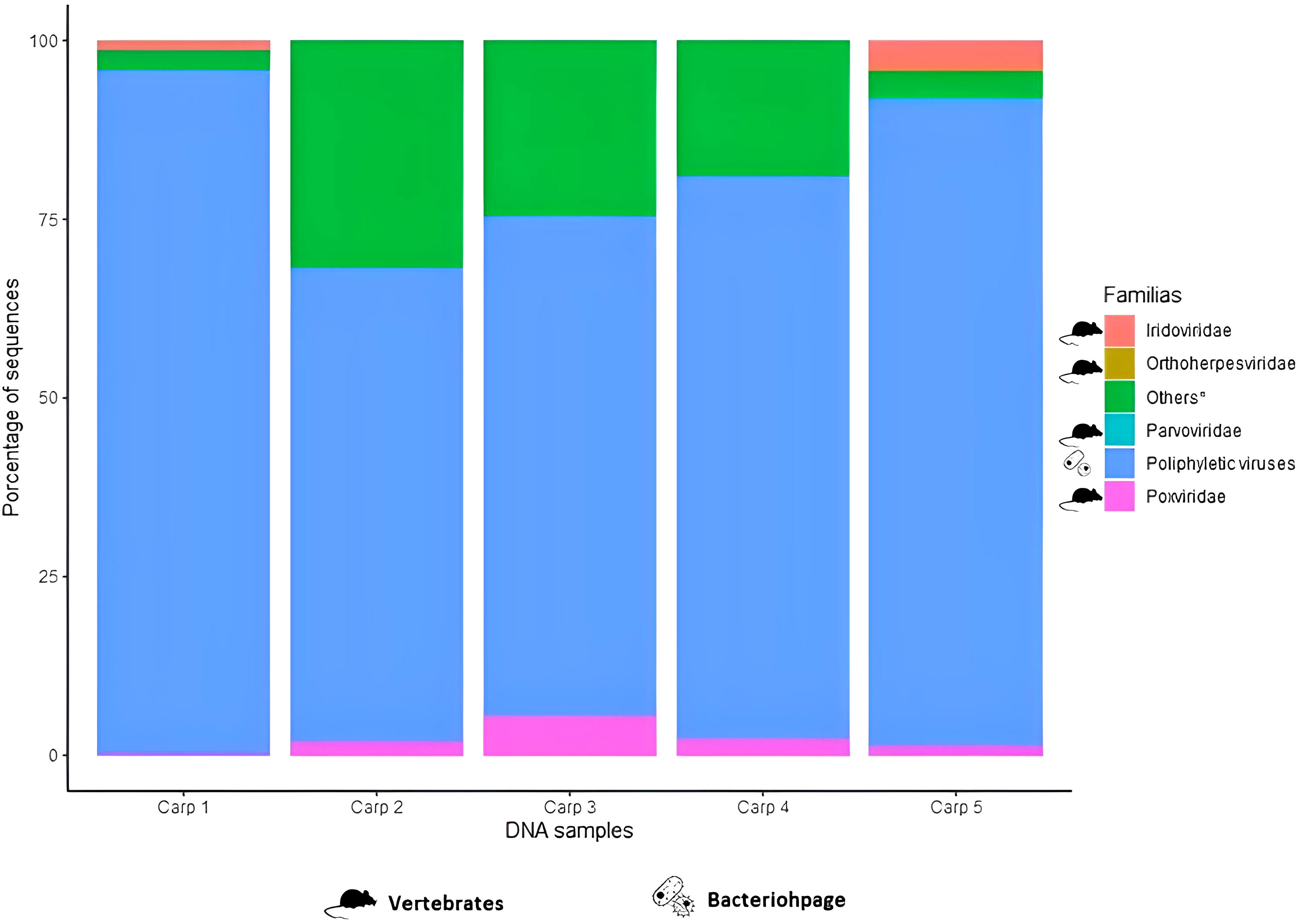
Figure 2 Relative frequency of the most representative viral families in DNA samples extracted from the fecal content of common carp (Cyprinus carpio).* Viral families with an abundance <1% or present in only a few samples were included in “Other”.
For RNA samples, viral families related to vertebrates were most representative sequences with 75%. For all RNA samples, there were sequences belong to Picornaviridae with abundances of 1 to 80% and Solemoviridae with 0.4 to 97%. Four carps (80%) had viral families related to vertebrates: Astroviridae (0.1 to 1.8% of abundance), Hantaviridae (0.2 to ~8.1%), Nodaviridae (0.6 to 1.1%) and Retroviridae (0.005%). Viral families related to plants were also present in 4 carps: Virgaviridae (0.89 to 7.2%) y Partitiviridae (0.35 to 66%). Viral families related to invertebrates represented 9.57% of RNA viral sequences, which Dicistroviridae were detected in four carps (80%) with abundance of 2.7 a 49.63% (Figure 3).
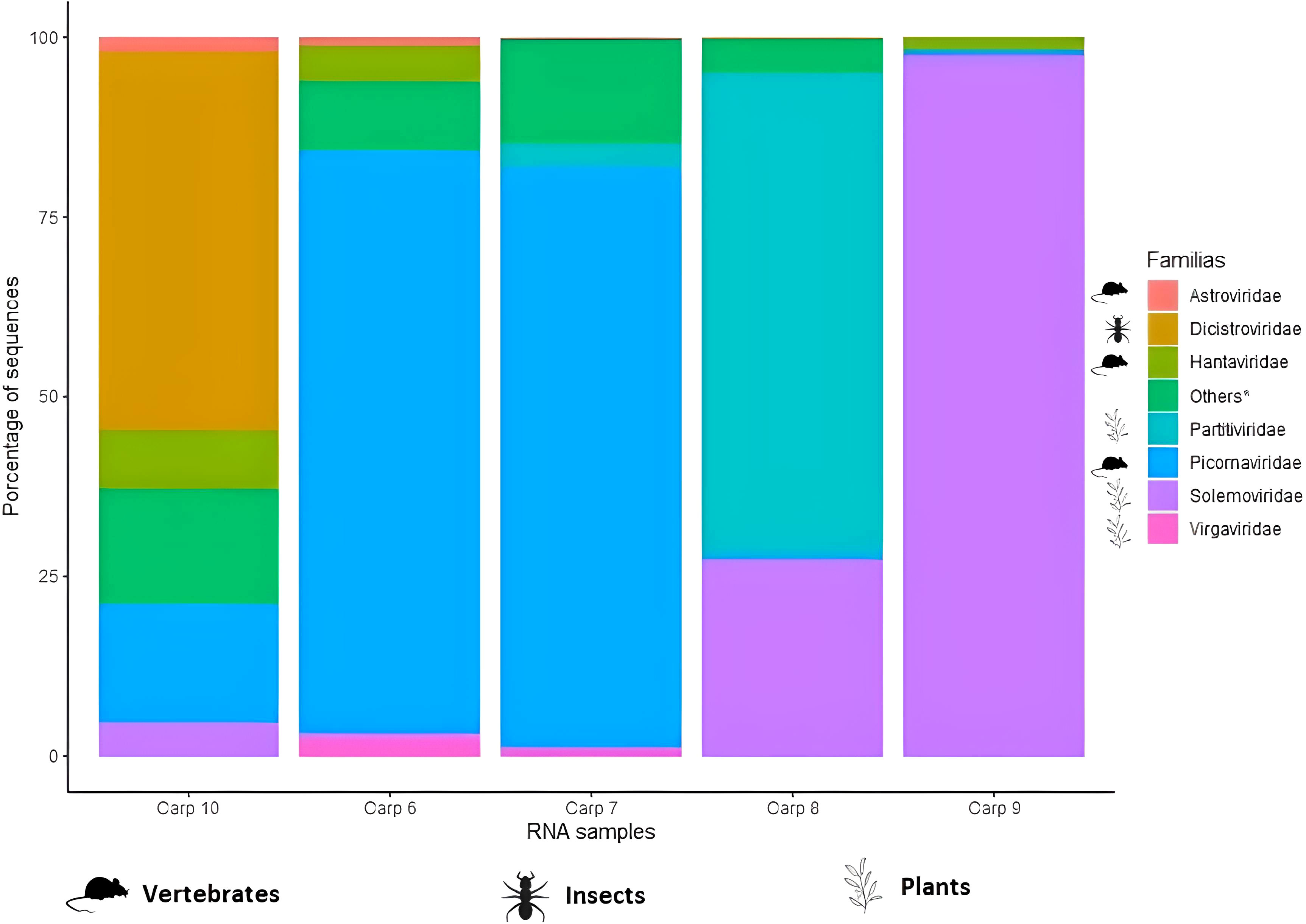
Figure 3 Relative frequency of the most representative viral families in RNA samples extracted from the fecal content of common carp (Cyprinus carpio). * Viral families with an abundance <1% or present in only a few samples were included in “Other”.
3.3 Beta diversity
As seen in Figure 4, unsupervised hierarchical grouping (dendrogram) carp 2 and 5 were grouped in same clade as they contained viral families that were only detected in these samples, such as Ackermannviridae in carp 1 and the aforementioned Orthoherpesviridae and Parvoviridae in carp 5, with carp 1 being the sample with the greatest dissimilarity in relation to the others (Bray-Curts index 0.622-0.725). While, carps 2 and 4 were grouped together in the dendogram as they were similar in their viral community, as well as in proportion of reads assigned to each viral family (Bray-Curtis index 0.188), on the other hand, carp 2 showed a dissimilarity of 1.179 and 0.156 with carps 3 and 4, respectively, however; by showing a higher abundance of counts assigned to viral families related to bacteria, it was grouped close to these carps (Figure 4). Similarly, the heat map of RNA samples had a similar architecture to DNA in terms of the dendogram, as it clustered carps 8 and 9 together due to their high abundance in viral families such as Chuviridae, Caulimoviridae and Retroviridae in tent 8 and Solemoviridae and unqualified Picornavirales in carp 9. The carp 6 and 7 samples were highly similar to each other (Bray-Curtis index 0.05) and were pooled together, being carp 7 the RNA sample with the highest dissimilarity in relation to the others (Bray-Curtis index 0.853-0.980). Carp 10 was not pooled with carp 6 and 7 due to high abundance in 12 viral families (Figure 5).
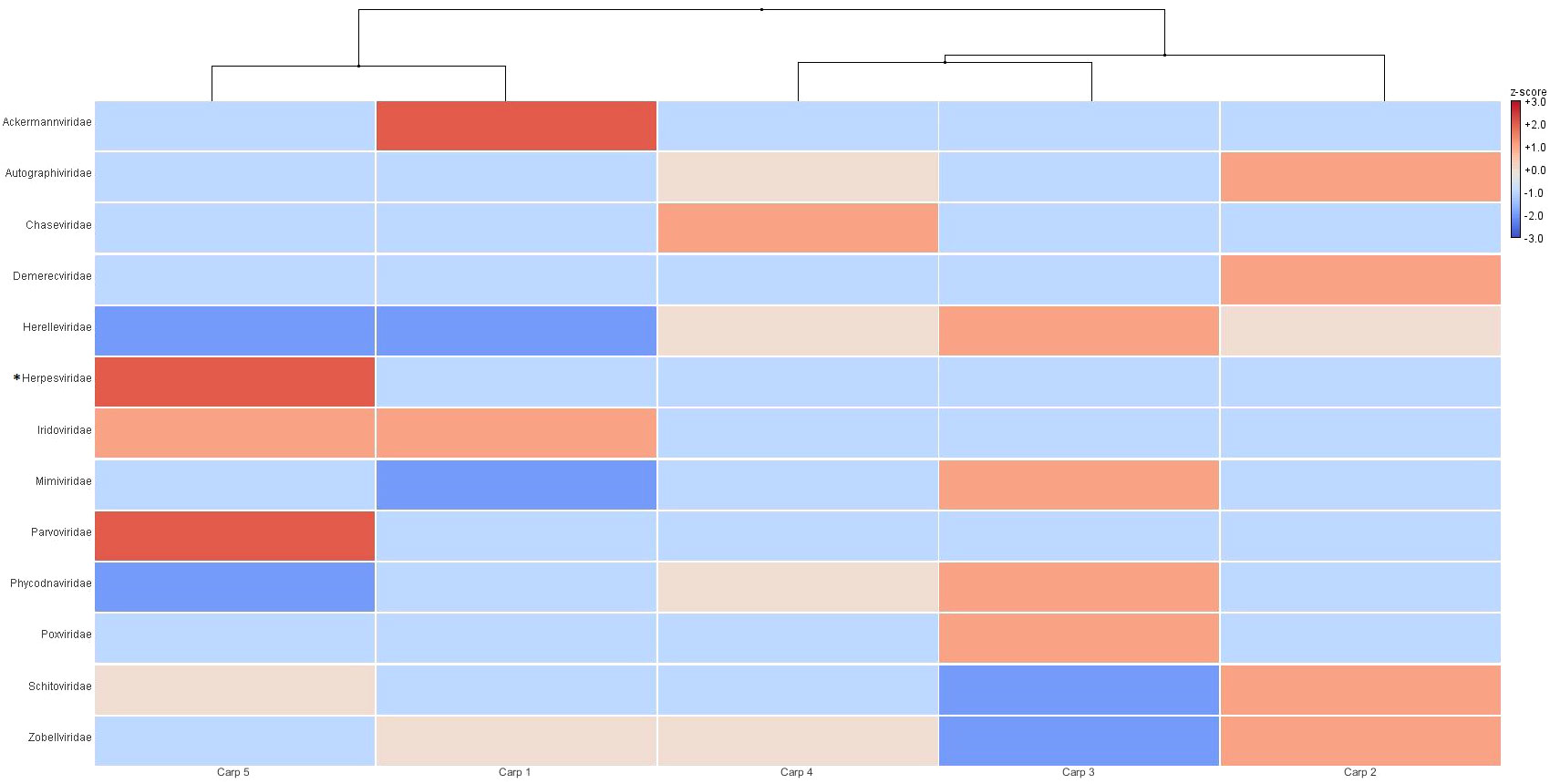
Figure 4 Unsupervised hierarchical grouping of DNA samples at the family level. The relative abundance of viral families was obtained from the BLASTn analysis, and the assigned sequence counts were normalized, analyzed and visualized in MEGAN Community Edition. The color code indicates the relative abundance in relation to the mean (red indicates greater abundance, and blue indicates lower abundance). *Sequences in this taxon now are classified in Orthoherpesviridae family by ICTV (International Committe on Taxonomy of Viruses).
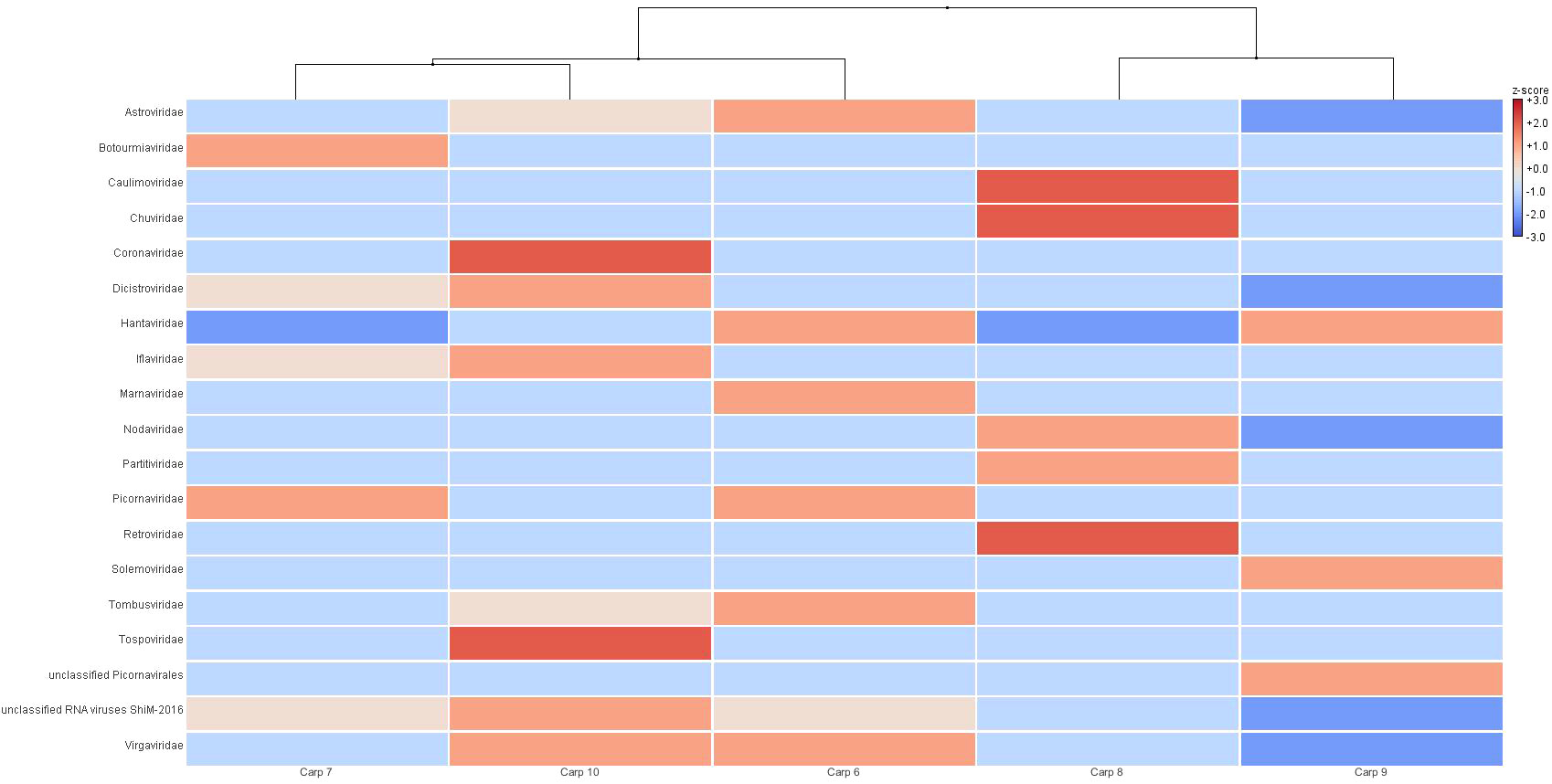
Figure 5 Unsupervised hierarchical grouping of RNA samples at the family level. The relative abundance of viral families was obtained from the BLASTn analysis, and the assigned sequence counts were normalized, analyzed and visualized in MEGAN Community Edition. The color code indicates the relative abundance in relation to the mean (red indicates greater abundance, and blue indicates lower abundance).
4 Discussion
Our analysis of fecal contents of wild carp revealed a viral community composed of 30 viral families, which 17 correspond to RNA viral families and 13 to DNA viral families. Among these 30 viral taxa, 21 have been detected in other studies of viral populations in different fishes, the most remarkable similarity being presence of all the vertebrate-related viral families reported in the present work. The Poxviridae family has been detected in 5 different studies conducted in Australia (Costa et al., 2021), China (Xi et al., 2023), Portugal (Filipa-Silva et al., 2020) and New Zealand (Perry et al., 2022; Grimwood et al., 2023), followed by Hantaviridae and Picornaviridae, which have been reported in 4 studies conducted in Australia (both) (Costa et al., 2021; Geoghegan et al., 2021) and New Zealand (Perry et al., 2022; Grimwood et al., 2023) for Hantaviridae, and, China (Xi et al., 2023) and Portugal (Filipa-Silva et al., 2020) for Picornaviridae. For its part, Astroviridae and Parvoviridae families have been detected in three different researches, wich were caried out in Australia (Costa et al., 2021; Geoghegan et al., 2021) and New Zealand (Grimwood et al., 2023), while the sites reporting Parvoviridae were conducted in China (Xi et al., 2023), Portugal (Filipa-Silva et al., 2020), and Gulf of Guinea (Gadoin et al., 2021). Likewise, Orthoherpesviridae was reported as Herpesviridae in three different investigations performed in China (Xi et al., 2023), Portugal (Filipa-Silva et al., 2020), and New Zealand (Grimwood et al., 2023). It is noteworthy that the present research shows the largest number of viral families related to vertebrates detected so far in fish, the study performed by Xi et al (Xi et al., 2023). is the second with 5 viral families, which was the only one that used samples of fecal contents for its research. Differences in the presence or absence of some viral families between the common carp viral community and the viromes described in other marine and freshwater fish species could be due to the study site, study subject, type of sample and abiotic factors, such as; temperature, ultraviolet radiation and pH (Sahu et al., 2023), that could affect the composition of viral communities.
It is important to emphasize that in the seven previous studies on viroma in fishes living in natural aquatic environments, viral families that affect fish have been detected, for example; Alloherpesviridae, Poxviridae, Iridoviridae, Picornaviridae, Caliciviridae, Rhabdoviridae, among others (Filipa-Silva et al., 2020; Costa et al., 2021; Gadoin et al., 2021; Geoghegan et al., 2021; Perry et al., 2022; Grimwood et al., 2023; Xi et al., 2023), but in absence of disease. Likewise, in the present description of viroma in carp fecal contents, viral families that might affect fishes including carp, such as Iridoviridae, Picornaviridae, Poxviridae and Nodaviridae, were detected without any outbreak of diseases.
Other authors have reported illness in common carp by Koi ranavirus (unclassified Iridovirus/Ranavirus), carp picornavirus (unclassified Picornavirus) (Lange et al., 2014; George et al., 2015; Kaviarasu et al., 2020) and carp poxvirus on fish farms (Ono et al., 1986). This scenario shows how limited our knowledge is in terms of viral pathogenesis and spread of these viruses in fishes living in a natural aquatic environment versus fish farms.
The detection of these viral families in common carp could represent the proximity they have with the diverse hosts where the viruses replicate, even without determining whether fish could participate in the transmission of these viral taxa, so further studies of molecular biology, isolates and epidemiology would be needed to corroborate or refute the participation of fish living in natural aquatic environment in the transmission of viruses to farm fishes.
In viromes described for freshwater environments, viral families related to aquatic or terrestrial vertebrates have been detected (with the exception of Hantaviridae) and were identified in the present work (Rooks et al., 2010; Fancello et al., 2012; Ge et al., 2013; Wan et al., 2013; Green et al., 2015; Green et al., 2018; Kim et al., 2015; Mohiuddin and Schellhorn, 2015; Cai et al., 2016; Alexyuk et al., 2017; Gu et al., 2018; Potapov et al., 2019; Chopyk et al., 2020; Rusiñol et al., 2020; Palermo et al., 2021) which point out, if these environments also participate in spreading viruses to other animals taxonomically different from fish.
Although there are no fish viruses that cause zoonoses or diseases to another taxonomic group of animals other than fish, the detection of these viral families in the fecal content of common carp in the wild could represent a public or animal health risk similar to that with fecal matter of domestic animals through viral families such as Astroviridae and Parvoviridae, which have been identified in the virome of the fecal content of dogs, cats, cattle, small ruminants, pigs and productive birds, as well as Retroviridae detected in cattle, small rumiants, pigs, productive birds and dogs. Coronaviridae reported in cattle, pigs, dogs and cats. Orthoherpesviridae (before known as Herpesviridae) detected in cattle, pigs, productive birds, dogs and cats. Picornaviridae and Poxviridae has not been detected in cats, Iridoviridae only reported in cattle and productive birds, Hantaviridae have been only detected in production birds (Zhang et al., 2014; Kwok et al., 2020; Shi et al., 2021). The viromes of the wild common carp and domestic animals differ mainly because of a greater diversity of vertebrate-related viruses and bacteriophages in domestic animals. However, viruses detected in the feces could only pass-through digestive tract. In order to know if those viruses replicated in common carp and participate as mechanical carriers or reservoirs in the natural aquatic environment, experimental studies are required in the future.
The confirmation of viral families related to vertebrates by BLASTx (except Coronaviridae, Hantaviridae and Retroviridae) indicates that common carp is a species to consider for the presence of viral families that could represent a risk to public health and animal health; however, more detailed studies (molecular, isolates and epidemiological) are need to be able to deny or confirm the presence of viral species in common carp that cause diseases to animals or humans. Importantly, the virome described in the present work represents a part of the viral community present in the “Laguna de Chignahuapan” wetland and habitats in the region.
The viral families related to plants accounted for a small percentage of sequences (0.70%) but comprised the second most diverse group after viruses related to vertebrates; 7 viral families were identified: Botourmiaviridae, Caulimoviridae, Partitiviridae, Solemoviridae, Tombusviridae, Tospoviridae and Virgaviridae. These may have been present in fecal samples due to the feeding behavior of carp when eating various types of plants or their debris (García-Berthou, 2001). Only Caulimoviridae and Tombusviridae have been reported in fishes viromas (Gadoin et al., 2021; Perry et al., 2022); for the viromes in freshwater fish, only Partitiviridae, Tombusviridae and Virgaviridae have been detected (Djikeng et al., 2009; Rusiñol et al., 2020).
The unsupervised hierarchical clustering shows that carps even sharing the same environment might present differences in their viral populations, with viral taxa existing only in some carps, such as Ackermannviridae family in carp 1, Orthoherpesviridae and Parvoviridae in carp 5 and Coronaviridae in carp 10. These viral communities could be affected by biotic (microbial community and extracellular enzymes) and abiotic factors (sedimentation, oxygen, pH, ultraviolet radiation and salinity) in presence, absence and frequency of virus taxa. However, those factors have not yet been assessed for their influence on the fish viroma, but have been tested in free DNA from aquatic environments (Sahu et al., 2023).
The results for the common carp suggest variability within the dynamic ecosystem of Laguna de Chignahuapan, unlike a closed ecosystem animal farm, such as commercial poultry or pigs for human consumption, where these ecosystems have biosecurity measures that focus on reducing the entry of microorganisms that could cause disease in animals. Consequently, the virome in these farm animals comprises fewer viral families than did the virome of carp in this study (Shan et al., 2011; Lima et al., 2017). In pigs, a constant presence of some viral families (Picornaviridae, and Caudovirales before known as Myoviridae and Podoviridae) can be observed in all individuals (Yang et al., 2022).
In virome studies of other fish, viral diversity has been explored from different perspectives; for example, in tuna, a “core” virome comprising bacteriophages, Microviridae, Circoviridae and Geminiviridae was identified (Gadoin et al., 2021). In gilthead seabream and horse mackerel, the differences in the virome between the liver and skin of the same species are explained by the sampling regions (Filipa-Silva et al., 2020). For marine fish studied in Australia, initially, the authors considered the study of diversity based on behavioral differences of the different species, and later, the virome was analyzed by grouping the fish by taxonomic order (Geoghegan et al., 2021). These analyses of diversities have been conducted based on the particular objective of each study.
5 Conclusion
The fecal content virome of the common carp (Cyprinus carpio) from a natural preserved wetland contained an abundant community of phages and secondly other viral taxa related to terrestrial and aquatic vertebrates, with some of these viral families having been detected in different marine and freshwater fish viromes studies.
This information provides a first overview of the viral taxa that may be present in a natural and still preserved aquatic environment. However, although some viral families affecting farms fishes have been reported to cause disease, there is not sufficient evidence of viral fish illness in naturally aquatic environment. Therefore, the role of common carp in natural environments as potential reservoirs of different viruses needs further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject, PRJNA1011357.
Ethics statement
The animal study was approved by Institutional Subcommittee for the Care and Use of Animals Experimental. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
OT-M: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Software, Writing – original draft, Investigation, Visualization. EG-L: Methodology, Software, Supervision, Writing – review & editing. OR-C: Methodology, Supervision, Writing – review & editing. EL-R: Methodology, Supervision, Writing – review & editing. GG-E: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The funding of this project was part of UNAM-DGAPA-PAPIIT IN218021 program. Conahcyt supplied scholarship to T-MO with CVU 893995.
Acknowledgments
We would like to express our sincere gratitude to PhD. Chávez-Maya F for providing technical assistance during sample processing, as well as National Institute of Public Health for providing access to its server to expedite data analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/faquc.2024.1289423/full#supplementary-material
References
Aguirre de Cárcer D., López-Bueno A., Alonso-Lobo J. M., Quesada A., Alcamí A. (2016). Metagenomic analysis of lacustrine viral diversity along a latitudinal transect of the Antarctic Peninsula. FEMS Microbiol. Ecol. 92, 1–10. doi: 10.1093/femsec/fiw074
Alexyuk M. S., Turmagambetova A. S., Alexyuk P. G., Bogoyavlenskiy A. P., Berezin V. E. (2017). Comparative study of viromes from freshwater samples of the Ile-Balkhash region of Kazakhstan captured through metagenomic analysis. Virusdisease 28, 18. doi: 10.1007/s13337-016-0353-5
Benkő M., Brandt C. R., Bryant N. A., Dastjerdi A., Davison A. J., Depledge D. P., et al. (2021). Abolish 6 species and rename 1 family, 4 genera and 124 species in the order Herpesvirales. Int. Committee Taxonomy Viruses: ICTV.
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Breitbart M., Hewson I., Felts B., Mahaffy J. M., Nulton J., Salamon P., et al. (2003). Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol 185, 6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003
Breitbart M., Salamon P., Andresen B., Mahaffy J. M., Segall A. M., Mead D., et al. (2002). Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U.S.A. 99, 14250. doi: 10.1073/pnas.202488399
Cai L., Zhang R., He Y., Feng X., Jiao N. (2016). Metagenomic analysis of virioplankton of the subtropical jiulong river estuary, China. Viruses 8, 35. doi: 10.3390/v8020035
Cann A. J., Fandrich S. E., Heaphy S. (2005). Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30, 151–156. doi: 10.1007/s11262-004-5624-3
Chen L., Liu B., Wu Z., Jin Q., Yang J. (2017). DRodVir: A resource for exploring the virome diversity in rodents. J. Genet. Genomics 44, 259. doi: 10.1016/j.jgg.2017.04.004
Chopyk J., Allard S., Nasko D. J., Bui A., Mongodin E. F., Sapkota A. R. (2018). Agricultural freshwater pond supports diverse and dynamic bacterial and viral populations. Front. Microbiol. 0. doi: 10.3389/fmicb.2018.00792
Chopyk J., Nasko D. J., Allard S., Bui A., Pop M., Mongodin E. F., et al. (2020). Seasonal dynamics in taxonomy and function within bacterial and viral metagenomic assemblages recovered from a freshwater agricultural pond. Environ. Microbiome 15, 1–16. doi: 10.1186/s40793-020-00365-8
CONABIO. (2011). Cyprinus carpio carpio Linnaeus, 1758 Common carp. Consideration of Invasiveness of Exotic Species, 4-6. Available online at: http://www.conabio.gob.mx/institucion/proyectos/resultados/LI007_Anexo_9_Ficha_Cyprinus%20_carpio_carpio.pdf (Accessed July 30, 2020).
Costa V. A., Mifsud J. C. O., Gilligan D., Williamson J. E., Holmes E. C., Geoghegan J. L. (2021). Metagenomic sequencing reveals a lack of virus exchange between native and invasive freshwater fish across the Murray–Darling Basin, Australia. Virus Evol. 7, 1-15. doi: 10.1093/ve/veab034
Cui J., Xiao M., Liu M., Wang Z., Liu F., Guo L., et al. (2017). Coupling metagenomics with cultivation to select host-specific probiotic micro-organisms for subtropical aquaculture. J. Appl. Microbiol. 123, 1274–1285. doi: 10.1111/jam.2017.123.issue-5
Djikeng A., Kuzmickas R., Anderson N. G., Spiro D. J. (2009). Metagenomic analysis of RNA viruses in a fresh water lake. PloS One 4, 1-14. doi: 10.1371/journal.pone.0007264
Fancello L., Trape S., Robert C., Boyer M., Popgeorgiev N., Raoult D., et al. (2012). Viruses in the desert: a metagenomic survey of viral communities in four perennial ponds of the Mauritanian Sahara. ISME J. 7, 359–369. doi: 10.1038/ismej.2012.101
FAO (2005)Cyprinus carpio. Cultured Aquatic Species Information Programme. In: Fisheries and Aquaculture Division. Available online at: https://www.fao.org/fishery/en/culturedspecies/cyprinus_carpio/en (Accessed May 31, 2023).
FAO (2023a)Common carp - Cyprinus carpio. In: Aquaculture Feed and Fertilizer Resources Information System. Available online at: https://www.fao.org/fishery/affris/species-profiles/common-carp/common-carp-home/en/ (Accessed June 4, 2023).
FAO (2023b). “Common carp - Natural food and feeding habits,” in Aquaculture Feed and Fertilizer Resources Information System. Available at: https://www.fao.org/fishery/affris/species-profiles/mandarin-fish/natural-food-and-feeding-habits/en/. (Accessed February 28, 2024).
Filipa-Silva A., Parreira R., Martínez-Puchol S., Bofill-Mas S., Barreto Crespo M. T., Nunes M. (2020). The unexplored virome of two atlantic coast fish: contribution of next-generation sequencing to fish virology. Foods 2020 9, 1634. doi: 10.3390/foods9111634
Gadoin E., Desnues C., Monteil-Bouchard S., Bouvier T., Auguet J. C., Roque D’Orbcastel E., et al. (2021). Fishing for the virome of tropical tuna. Viruses 13, 1–8. doi: 10.3390/v13071291
García-Berthou E. (2001). Size-and depth-dependent variation in habitat and diet of the common carp (Cyprinus carpio). Aquat Sci. 63, 466–476. doi: 10.1007/S00027-001-8045-6/METRICS
Ge X., Wu Y., Wang M., Wang J., Wu L., Yang X., et al. (2013). Viral metagenomics analysis of planktonic viruses in East Lake, Wuhan, China. Virol. Sin. 28, 280–290. doi: 10.1007/s12250-013-3365-y
Geoghegan J. L., Di Giallonardo F., Wille M., Ortiz-Baez A. S., Costa V. A., Ghaly T., et al. (2021). Virome composition in marine fish revealed by meta-transcriptomics. Virus Evol. 7, 1–11. doi: 10.1093/ve/veab005
George M. R., John K. R., Mansoor M. M., Saravanakumar R., Sundar P., Pradeep V. (2015). Isolation and characterization of a ranavirus from koi, Cyprinus carpio L., experiencing mass mortalities in India. J. Fish Dis. 38, 389–403. doi: 10.1111/jfd.12246
GISD (2013). Cyprinus carpio. In: Invasive Species Specialist Group. Available online at: https://www.invasivespecies.net/web/20090116213211/http://www.issg.org/ (Accessed July 30, 2020).
Givens C. E., Ransom B., Bano N., Hollibaugh J. T. (2015). Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar. Ecol. Prog. Ser. 518, 209–223. doi: 10.3354/meps11034
Green J. C., Rahman F., Saxton M. A., Williamson K. E. (2015). Metagenomic assessment of viral diversity in Lake Matoaka, a temperate, eutrophic freshwater lake in southeastern Virginia, USA. Aquat. Microbial Ecol. 75, 117–128. doi: 10.3354/ame01752
Green J. C., Rahman F., Saxton M. A., Williamson K. E. (2018). Quantifying aquatic viral community change associated with stormwater runoff in a wet retention pond using metagenomic time series data. Aquat. Microbial Ecol. 81, 19–35. doi: 10.3354/ame01856
Grimwood R. M., Fortune-Kelly G., Holmes E. C., Ingram T., Geoghegan J. L. (2023). Host specificity shapes fish viromes across lakes on an isolated remote island. Virology 587, 1-14. doi: 10.1016/j.virol.2023.109884
Gu X., Tay Q. X. M., Te S. H., Saeidi N., Goh S. G., Kushmaro A., et al. (2018). Geospatial distribution of viromes in tropical freshwater ecosystems. Water Res. 137, 220–232. doi: 10.1016/j.watres.2018.03.017
Hoole D., Bucke D., Burgess P., Wellby I. (2001). Diseases of carp and other cyprinid fish. (England: Fishing News Books). David. doi: 10.1002/9780470999752
Huson D. H., Beier S., Flade I., Górska A., El-Hadidi M., Mitra S. (2016). MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PloS Comput. Biol. 12, 1004957. doi: 10.1371/journal.pcbi.1004957
Kaviarasu D., John K. R., George M. R., Ahilan B., Padmavathy P. R., Petchimuthu M. (2020). Experimental Infection with Koi Ranavirus (KIRV) of Common Carp (Cyprinus carpio L.) and Rohu (Labeo rohita H.). Int. J. Curr. Microbiol. Appl. Sci. 9, 3234–3243. doi: 10.20546/ijcmas.2020.911.388
Kim Y., Aw T. G., Teal T. K., Rose J. B. (2015). Metagenomic investigation of viral communities in ballast water. Environ. Sci. Technol. 49, 8396–8407. doi: 10.1021/acs.est.5b01633
Kwok K. T. T., Nieuwenhuijse D. F., Phan M. V. T., Koopmans M. P. G. (2020). Virus metagenomics in farm animals: A systematic review. Viruses 12, 1–22. doi: 10.3390/v12010107
Lange J., Groth M., Fichtner D., Granzow H., Keller B., Walther M., et al. (2014). Virus isolate from carp: Genetic characterization reveals a novel picornavirus with two aphthovirus 2A-like sequences. J. Gen. Virol. 95, 80–90. doi: 10.1099/vir.0.058172-0
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li T., Long M., Gatesoupe F. J., Zhang Q., Li A., Gong X. (2015). Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing. Microb. Ecol. 69, 25–36. doi: 10.1007/s00248-014-0480-8
Lima D. A., Cibulski S. P., Finkler F., Teixeira T. F., Varela A. P. M., Cerva C., et al. (2017). Faecal virome of healthy chickens reveals a large diversity of the eukaryote viral community, including novel circular ssDNA viruses. J. Gen. Virol. 98, 690–703. doi: 10.1099/jgv.0.000711
Mohiuddin M., Schellhorn H. (2015). Spatial and temporal dynamics of virus occurrence in two freshwater lakes captured through metagenomic analysis. Front. Microbiol., 960, 6. doi: 10.3389/fmicb.2015.00960
Moon K., Jeon J. H., Kang I., Park K. S., Lee K., Cha C.-J., et al. (2020). Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 8, 1–15. doi: 10.1186/s40168-020-00863-4
National Aquaculture and Fisheries Commission (2021). Statistical Yearbook Of Aquaculture And Fisheries Of The National Commission Of Aquaculture And Fisheries (Mexico: Secretariat of Agriculture and Social Development).
National Institute of Fisheries. (2018) Aquaculture - Common carp. Commercial aquaculture. Available online at: https://www.gob.mx/imipas/acciones-y-programas/acuacultura-carpa-comun (Accessed June 11, 2023).
OMSA (2019). Aspectos relativos al bienestar en el aturdimiento y la matanza de peces de cultivo para consumo humano 9–12.
Ono S.-I., Nagai A., Sugai N. (1986). A histopathological study on juvenile colorcarp, cyprinus carpio, showing edema. Fish Pathol. 21, 167–175. doi: 10.3147/jsfp.21.167
Palermo C. N., Shea D. W., Short S. M. (2021). Analysis of different size fractions provides a more complete perspective of viral diversity in a freshwater embayment. Appl. Environ. Microbiol. 87, 1–15. doi: 10.1128/AEM.00197-21
Perry B. J., Darestani M. M., Ara M. G., Hoste A., Jandt J. M., Dutoit L., et al. (2022). Viromes of freshwater fish with lacustrine and diadromous life histories differ in composition. Viruses 14, 1-15. doi: 10.3390/v14020257
Potapov S. A., Tikhonova I. V., Krasnopeev A. Y., Kabilov M. R., Tupikin A. E., Chebunina N. S., et al. (2019). Metagenomic analysis of virioplankton from the pelagic zone of lake baikal. Viruses 11, 1-15. doi: 10.3390/v11110991
Ramsar (2023) The List of Wetlands of International Importance. Available online at: https://www.ramsar.org.
Rooks D. J., Smith D. L., Mcdonald J. E., Woodward M. J., Mccarthy A. J., Allison H. E. (2010). 454-pyrosequencing: A molecular battiscope for freshwater viral ecology. Genes (Basel) 1, 210–226. doi: 10.3390/genes1020210
Rusiñol M., Martínez-Puchol S., Timoneda N., Fernández-Cassi X., Pérez-Cataluña A., Fernández-Bravo A., et al. (2020). Metagenomic analysis of viruses, bacteria and protozoa in irrigation water. Int. J. Hyg Environ. Health 224, 113440. doi: 10.1016/j.ijheh.2019.113440
Sahu A., Kumar N., Pal Singh C., Singh M. (2023). Environmental DNA (eDNA): Powerful technique for biodiversity conservation. J. Nat. Conserv. 71, 1-15. doi: 10.1016/j.jnc.2022.126325
Shan T., Li L., Simmonds P., Wang C., Moeser A., Delwart E. (2011). The fecal virome of pigs on a high-density farm. J. Virol. 85, 11697–11708. doi: 10.1128/JVI.05217-11
Shi Y., Tao J., Li B., Shen X., Cheng J., Liu H. (2021). The gut viral metagenome analysis of domestic dogs captures snapshot of viral diversity and potential risk of coronavirus. Front. Vet. Sci. 8. doi: 10.3389/fvets.2021.695088
Tan C. W., Yang X., Anderson D. E., Wang L. F. (2021). Bat virome research: the past, the present and the future. Curr. Opin. Virol. 49, 68–80. doi: 10.1016/j.coviro.2021.04.013
Wan X.-F., Barnett J. L., Cunningham F., Chen S., Yang G., Nash S., et al. (2013). Detection of African swine fever virus-like sequences in ponds in the Mississippi Delta through metagenomic sequencing. Virus Genes 46, 441–446. doi: 10.1007/s11262-013-0878-2
Wood D. E., Lu J., Langmead B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 1–13. doi: 10.1186/s13059-019-1891-0
Xi Y., Jiang X., Xie X., Zhao M., Zhang H., Qin K., et al. (2023). Viromics reveals the high diversity of viruses from fishes of the tibet highland. Microbiol. Spectr. 11, 1-17. doi: 10.1128/spectrum.00946-23
Yang S., Zhang D., Ji Z., Zhang Y., Wang Y., Chen X., et al. (2022). Viral metagenomics reveals diverse viruses in tissue samples of diseased pigs. Viruses 14, 1-16. doi: 10.3390/v14092048
Keywords: fish, virome, mass sequencing, whole metagenome shotgun, viral diversity, vertebrate viruses
Citation: Torres-Meza OA, Godoy-Lozano EE, Rico-Chávez O, Loza-Rubio E and García-Espinosa G (2024) Description of the viral community in fecal content of common carp (Cyprinus carpio) living in a naturally preserved wetland on Mexican plateau. Front. Aquac. 3:1289423. doi: 10.3389/faquc.2024.1289423
Received: 05 September 2023; Accepted: 21 February 2024;
Published: 18 March 2024.
Edited by:
Prapansak Srisapoome, Kasetsart University, ThailandReviewed by:
Neelesh Kumar, Rani Lakshmi Bai Central Agricultural University, IndiaPrabhugouda Siriyappagouder, Nord University, Norway
Copyright © 2024 Torres-Meza, Godoy-Lozano, Rico-Chávez, Loza-Rubio and García-Espinosa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gary García-Espinosa, gary@unam.mx
 Oscar Abel Torres-Meza
Oscar Abel Torres-Meza Elizabeth Ernestina Godoy-Lozano
Elizabeth Ernestina Godoy-Lozano Oscar Rico-Chávez
Oscar Rico-Chávez Elizabeth Loza-Rubio
Elizabeth Loza-Rubio Gary García-Espinosa
Gary García-Espinosa