Fermentative Production of N-Alkylated Glycine Derivatives by Recombinant Corynebacterium glutamicum Using a Mutant of Imine Reductase DpkA From Pseudomonas putida
- 1Genetics of Prokaryotes, Faculty of Biology and CeBiTec, Bielefeld University, Bielefeld, Germany
- 2Fermentation Technology, Technical Faculty and CeBiTec, Bielefeld University, Bielefeld, Germany
- 3Microbial Processes and Technology Division, National Institute for Interdisciplinary Science and Technology, Council of Scientific & Industrial Research, Trivandrum, India
Sarcosine, an N-methylated amino acid, shows potential as antipsychotic, and serves as building block for peptide-based drugs, and acts as detergent when acetylated. N-methylated amino acids are mainly produced chemically or by biocatalysis, with either low yields or high costs for co-factor regeneration. Corynebacterium glutamicum, which is used for the industrial production of amino acids for decades, has recently been engineered for production of N-methyl-L-alanine and sarcosine. Heterologous expression of dpkA in a C. glutamicum strain engineered for glyoxylate overproduction enabled fermentative production of sarcosine from sugars and monomethylamine. Here, mutation of an amino acyl residue in the substrate binding site of DpkA (DpkAF117L) led to an increased specific activity for reductive alkylamination of glyoxylate using monomethylamine and monoethylamine as substrates. Introduction of DpkAF117L into the production strain accelerated the production of sarcosine and a volumetric productivity of 0.16 g L−1 h−1 could be attained. Using monoethylamine as substrate, we demonstrated N-ethylglycine production with a volumetric productivity of 0.11 g L−1 h−1, which to the best of our knowledge is the first report of its fermentative production. Subsequently, the feasibility of using rice straw hydrolysate as alternative carbon source was tested and production of N-ethylglycine to a titer of 1.6 g L−1 after 60 h of fed-batch bioreactor cultivation could be attained.
Introduction
N-methylated amino acids are non-proteinogenic amino acids found in plants, mammals, and microorganisms. Substitution of proteinogenic amino acids in a peptide with an N-methylated derivative mimics the native peptide, yet, often leads to increased stability against proteases and better membrane permeability (Chatterjee et al., 2013; Di Gioia et al., 2016). Further, N-methylated amino acids are known to stabilize—similar to L-proline—discrete conformations as shown for the ATPase inhibitor efrapeptin C (Dutt Konar et al., 2013). Natural peptides containing N-methylated amino acids such as the potential anti-cancer drugs enniatins, which disrupt membrane potential by forming stacked sandwich complexes with cations in the cell membrane (Kamyar et al., 2004), and bouvardin, which blocks protein synthesis by interacting with eukaryotic 80S ribosomes (Zalacaín et al., 1982), serve as a model for synthetic peptides (peptidomimetics). Several studies showed enhanced characteristics of peptidomimetics with incorporated N-methylated amino acids: for example Clingitide, a synthetic pentapeptide, showed increased affinity for integrin receptors (Mas-Moruno et al., 2010), while N-methyltubulysin showed enhanced antimitotic activity (Patterson et al., 2008). In addition to the incorporated amino acid derivatives, monomeric N-alkylated amino acids occur in nature as intermediates of metabolic pathways such as sarcosine (N-methylglycine) in degradation of creatine and choline, or N-methylglutamate in the C1 assimilation from monomethylamine by methylotrophic bacteria like Methyloversatilis universalis (Latypova et al., 2010). Free N-alkylated amino acids also occur in green tea leaves, e.g., the flavor enhancing N5-ethylated glutamine derivative L-theanine (Sakato, 1950). Studies on the effect of the anesthetic lidocaine showed pain reducing properties (Eipe et al., 2016). Further studies on this mechanism showed that the lidocaine metabolite N-ethylglycine (NEtGly) acts as an inhibitor of glycine uptake and inhibits pain signaling. Hence, NEtGly is a promising candidate for chronical pain treatment (Werdehausen et al., 2015).
Chemical synthesis of N-methylated amino acids has been studied extensively and main routes involve reductive amination, ring opening of 5-oxazolidinones and direct methylation (Aurelio et al., 2004; Gentilucci et al., 2010). Recently, the biocatalytic synthesis of N-alkylated amino acids has been developed (Hyslop et al., 2019). Notably, sustainable fermentative production of N-methylated amino acids was established in 2018 using two independent pathways. Pathway interception of the C1 assimilation pathway of Methylobacterium extorquens and implementation of the first two genes coding for γ-glutamylmethylamide synthetase and N-methylglutamate synthase enabled glycerol-based production of N-methylglutamate in Pseudomonas putida KT2440 (Mindt et al., 2018b). The second pathway for production of N-methylated amino acids exploited the side activity of the imine reductase DpkA from Pseudomonas putida. Natively, DpkA catalyzes the reduction of piperidein-2-carboxylate, but in addition DpkA catalyzes the reductive methylamination of 2-oxo acids. Expression of dpkA in recombinant Corynebacterium glutamicum strains overproducing the relevant 2-oxo acid precursor enabled fermentative production of N-methylalanine (NMeAla) (Mindt et al., 2018a) and sarcosine (Mindt et al., 2019), respectively, when monomethylamine (MMA) was added to the growth medium.
The industrially relevant production host C. glutamicum is used for decades for safe production of amino acids in the food and feed industries. Since the genetic tool box for C. glutamicum is developed very well, it was metabolically engineered for production of many value-added compounds, in particular proteinogenic amino acids, but also for non-proteinogenic amino acids like trans-hydroxyproline (Yi et al., 2014; Falcioni et al., 2015), pipecolic acid (Pérez-García et al., 2016, 2017a), 7-chloro- or 7-bromo-tryptophan (Veldmann et al., 2019a,b) and the N-methylated amino acids NMeAla (Mindt et al., 2018a) and sarcosine (Mindt et al., 2019). The immediate precursors of several amino acids, 2-oxo acids such as 2-oxovalerate (Krause et al., 2010), pyruvate (Wieschalka et al., 2012), and 2-oxoisocaproate (Bückle-Vallant et al., 2014) are also efficiently produced by recombinant C. glutamicum strains. The 2-oxo acid glyoxylate is not an immediate precursor of glycine biosynthesis in C. glutamicum. Nevertheless, a glyoxylate overproducing strain could be generated by interrupting the glyoxylate bypass of the TCA cycle (Zahoor et al., 2014) as basis for production of sarcosine (N-methylglycine) (Mindt et al., 2019).
By site-directed mutagenesis of the substrate binding site of the imine reductase DpkA, we could change the specific activity for methyl- and ethylamination of glyoxylate and this mutant led to faster fermentative production of sarcosine (N-methylglycine) and N-ethylglycine when expressed in the production strain. Potential of using biomass derived sugars for value added products such as N-ethylglycine was also demonstrated in a fed-batch bioreactor process.
Materials and Methods
Bacterial Strains and Culture Conditions
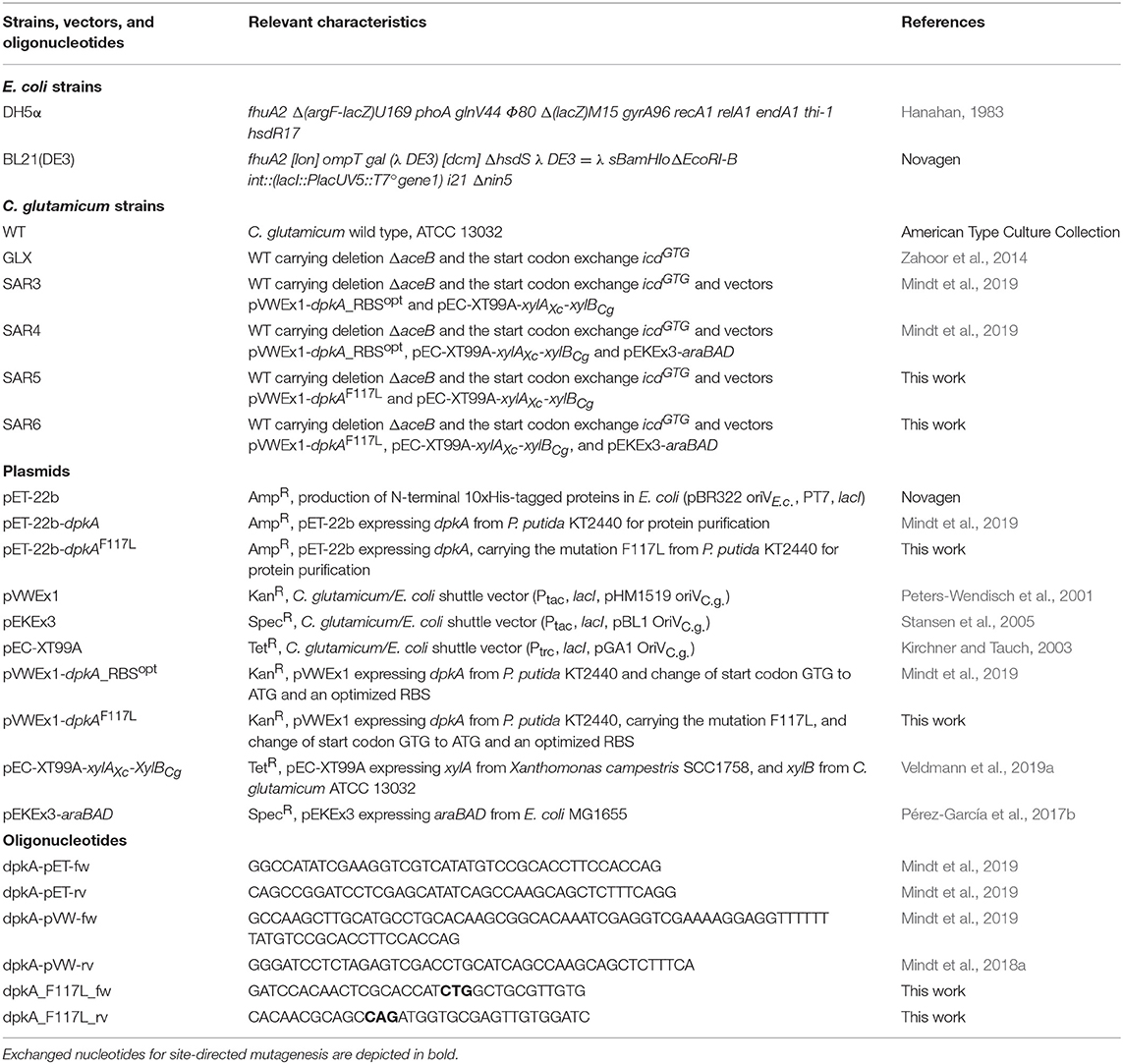
All bacterial strains, plasmids, and primers used were listed in Table 1. Escherichia coli DH5α (Hanahan, 1983) was cultivated in lysogeny broth (LB) at 37°C and 180 rpm. Pre-cultures of C. glutamicum were cultivated in LB, and main cultures for growth and production experiments where grown in standard CGXII medium (Eggeling and Bott, 2005) with reduced nitrogen content (2 g L−1 ammonium sulfate and 0.5 g L−1 urea) and given concentrations of carbon source and/or monomethylamine (MMA) at 30°C on a rotary shaker (120 rpm) in baffled shake flasks. All media were inoculated from a fresh LB or BHI agar plate, when necessary, ampicillin (100 μg mL−1), kanamycin (25 μg mL−1), spectinomycin (100 μg mL−1), and/or tetracycline (5 μg mL−1) were added to the medium. For induction of gene expression from the vectors pVWEx1, pEC-XT99A, and pEKEx3, medium was supplemented with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG). For growth and production experiments, C. glutamicum cells were harvested (3,200 × g, 7 min), washed once with TN buffer pH 6.3 (50 mM TrisHCl, 50 mM NaCl) and the minimal medium was inoculated to an optical density at 600 nm (OD600) of 1. Determination of OD600 was performed using V-1200 spectrophotometer (VWR, Radnor, PA, USA). Tolerance test of C. glutamicum for monoethylamine (MEA) was performed in the Biolector microfermentation system (m2p-labs, Aachen, Germany) in 48-well flower plates or in 100 mL flasks. Cultivation in 48-flower plates was performed in a volume of 1 mL at a shaking frequency of 1,200 rpm. Growth of cultures was followed by backlight scatter at 620 nm and at a signal gain factor of 20.
Fed-Batch Bioreactor Process
For production of N-ethylglycine a stirred tank reactor cultivation of C. glutamicum was performed with an initial working volume of 2 L (3.7 L KLF, Bioengineering AG, Switzerland) at pH 7.0, 30°C, 0.2 bar overpressure, and an aeration rate of 1.25 NL min−1 enriched with 20% (v/v) CO2 in order to enhance carboxylating reactions in C. glutamicum. The pH value was controlled by the automatic addition of 10% (w/w) H3PO4 and 4 M KOH. Struktol® J647 serves as antifoam agent and was added manually when necessary. Two set-points for the relative dissolved oxygen saturation (rDOS) of 30% (stirrer speed) and 60% (feeding pump), respectively, serves for preventing first, oxygen limitations by enhancing stirrer speed gradually in steps of 2%, and second, substrate limitations by feeding of rice straw hydrolysate and potassium acetate (see section Preparation of Lignocellulosic Hydrolysates) until rDOS fell again under 60%. The initial stirrer speed was set to 200 min−1. Samples were taken automatically every 2 h within the first 34 h and every 4 h afterwards and kept to 4°C unil analysis. For fermentation, a BHI medium overnight culture of C. glutamicum strain SAR6 was used to inoculate a shake flask preculture with CGXII medium that contained reduced nitrogen content (2 g L−1 ammonium sulfate and 0.5 g L−1 urea) and rice straw hydrolysate as sole carbon source (at a concentration equivalent to 10 g L−1 sugars). The fermenter medium (2 L of CGXII minimal medium lacking MOPS buffer, containing reduced nitrogen content and rice straw hydrolysate as sole carbon source (at a concentration equivalent to 4 g L−1 sugars) was inoculated with 350 mL of this preculture. After reaching an optical density of about 5, 40 mL of 5 M monoethylamine solution were added. After 24 h of cultivation, 100 mL of 60 g L−1 potassium acetate solution were added.
Preparation of Lignocellulosic Hydrolysates
Rice straw procured from a local market in Trivandrum, Kerala (India), was used as the raw material for the hydrolysis. Dried rice straw was powdered in a knife mill to a particle size ≤1 mm and sealed in polythene bags till further use. The milled and processed biomass was treated using diluted acid (H2SO4) with slight modifications on the method described by Gopinath et al. (2011). A solid loading of 10% (dry w/v) and an acid loading of 4% (v/v) were used for the hydrolysis at 134°C for 35 min. The hydrolysed biomass obtained was cooled, neutralized with 10 M KOH and wet sieved. The liquid fraction was centrifuged at 4,000 rpm for 20 min to remove remaining particles and filter-sterilized (Millipore, Massachusetts, USA) before fermentation.
Molecular Genetic Techniques and Strain Construction
Transformation of competent Escherichia coli DH5α (prepared by RbCl method) was performed by heat shock (Green and Sambrook, 2012) and transformation of C. glutamicum by electroporation (Eggeling and Bott, 2005). PCR-fragments were amplified using the respective primer (see Table 1) with ALLin™ HiFi DNA Polymerase (highQu GmbH, Kraichtal, Germany). The mutation F117L was introduced into dpkA using the respective primers (see Table 1) followed by cross-over PCR for alignment of the PCR products. PCR fragments were assembled to linearized vectors (pET-22b with NdeI, pVWEx1 with BamHI) by Gibson assembly (Gibson et al., 2009). The expression host E. coli BL21(DE3) was transformed with plasmids for protein purification (pET-22b), C. glutamicum was transformed with expression vectors pVWEx1, pEKEx3, and pEC-XT99A.
Enzyme Preparation and Kinetic Characterization
For enzyme characterization, fresh cultures of E. coli BL21(DE3) derived strains were inoculated from an overnight culture to an OD600 of 0.05 in 500 mL LB in 2 L baffled flask. When cultures reached OD600 of 0.5–0.6, 1 mM IPTG was added for induction of gene expression and cultures were transferred to 20°C and 180 rpm for further 3.5 h. Subsequently, cells were harvested at 4°C and cell pellets were stored at −20°C for further analysis. Starting with crude extract preparation, cells, and enzyme solutions were kept on ice or at 4°C. Crude extract preparations, protein purification (Mindt et al., 2019), and reductive activity assays (Muramatsu et al., 2005; Mindt et al., 2018a) were performed as described previously. For determination of Michaelis constants (Km), various 2-oxo acid concentrations from 0.5 to 30 mM were added. The Km values were determined using Origin with the add-on “Enzyme kinetics” and calculations for kcat were performed with respect to Michaelis and Menten (translation: Johnson and Goody, 2011) (MDpkA = 35.14 kDa). Specific activity was given in units, where one unit (U) was defined as the amount of enzyme required to convert 1 μmol substrate within 1 min.
Quantification of Amines, Organic Acids, and Sugars in Fermentation Broth
Amines, oxo acids, and carbohydrates were determined using high-performance liquid chromatography (HPLC) (1200 series, Agilent Technologies Deutschland GmbH, Böblingen, Germany). For quantification, supernatants of the cultures were harvested regularly by centrifugation (20,200 × g, 15 min at 4°C) and stored at −20°C for further analysis. Derivatization of amines with 9-fluorenylmethyl chlorocarbonate was performed as described elsewhere (Mindt et al., 2018b). Separation was performed on a reversed phase HPLC using a column system consisting of a pre-column (LiChrospher 100 RP18 EC-5μ (40 × 4 mm), CS-Chromatographie Service GmbH, Langerwehe, Germany) and a main column (LiChrospher 100 RP18 EC-5μ (125 × 4 mm), CS Chromatographie Service GmbH). Fluorescent derivatives were detected by a fluorescence detector (FLD G1321A, 1200 series, Agilent Technologies) with excitation and emission wavelength of 263 and 310 nm, respectively. L-proline was used as internal standard.
Concentrations of organic acids and sugars were determined using an amino exchange column (Aminex, 300 × 8 mm, 10 μm particle size, 25 Å pore diameter, CS Chromatographie Service GmbH) with 5 mM sulfuric acid as mobile phase and a refractive index detector (RID G1362A, 1200 series, Agilent Technologies) at 210 nm.
Results and Discussion
Site-Directed Mutagenesis of the Active Site of the N-Methylamino Acid Dehydrogenase DpkA
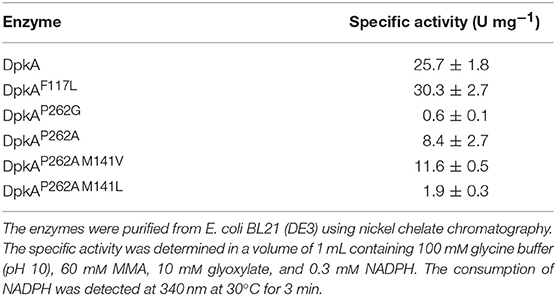
In a previous study, we established fermentative production of sarcosine by methylamination of glyoxylate using DpkA and showed that xylose was superior as source of carbon and energy than glucose (Mindt et al., 2019). To improve the low volumetric productivities observed for this sarcosine production process, we focused on changing an amino acyl residue of the binding site of the enzyme DpkA here.
Our approach was based on the protein structure of a DpkA homolog from Pseudomonas syringae (PsDpkA) for which the residues of the active site could be identified by co-crystallization of PsDpkA with the native substrate pyrroline-2-carboxylate (Pyr2C) and redox cofactor NADPH (Figure 1; PDB: 2CWH; Goto et al., 2005). The catalytic site comprises S43, H54, R58, and T166 and substrate binding involves the three amino acid residues F127, M151, and P272 (Figure 1A; Goto et al., 2005). These residues are conserved in DpkA from P. putida KT2440 that shares amino acid similarity of 79.6% (catalytic site: S33, H44, R48, and T156; substrate binding site: F117, M141, and P262; Figure 1B). Due to structural similarities of the native substrate Pyr2C and the imines formed by 2-oxo acids and alkylamines, we assumed that the non-native imine substrates are recognized in a similar manner. The wild-type DpkA and several mutants (Table 2) were purified from recombinant E. coli and their specific activity for the reductive amination of glyoxylate using MMA was determined. Mutagenesis of the three residues F117, M141, and P262 revealed altered specific activities for the reductive amination of glyoxylate using MMA (Table 2). The mutations P262I, P262L (data not shown), and P262G led to almost complete loss of activity, while the mutation P262A reduced the activity to 30% of the wildtype enzyme. Combined mutagenesis of positions P262 and M141 showed only minor improvement of the specific reductive amination of glyoxylate. Additionally, mutation F117L was considered under the assumption that it favors aliphatic interaction with imines formed by glyoxylate and either MMA or MEA (Figure 1). Although being aware that residues are often changed to Ala, our intention was to keep the aliphatic character of the binding pocket due to potential interactions with the alkyl-moiety of the N-alkylated product. We did not choose Ala since alanine is much smaller compared to phenylalanine (in the wildtype, which showed high activity for methylation) and our aim was to increase the activity toward ethylation. Since the size of the desired product increased by one carbon (from methylation to ethylation), we chose the aliphatic leucine as substitution to increase the binding pocked slightly and intended a potential aliphatic interaction of the leucine residue and the N-alkyl moiety. This mutant (DpkAF117L) was the only one in the chosen library which showed improved activity for the reductive amination of glyoxylate.
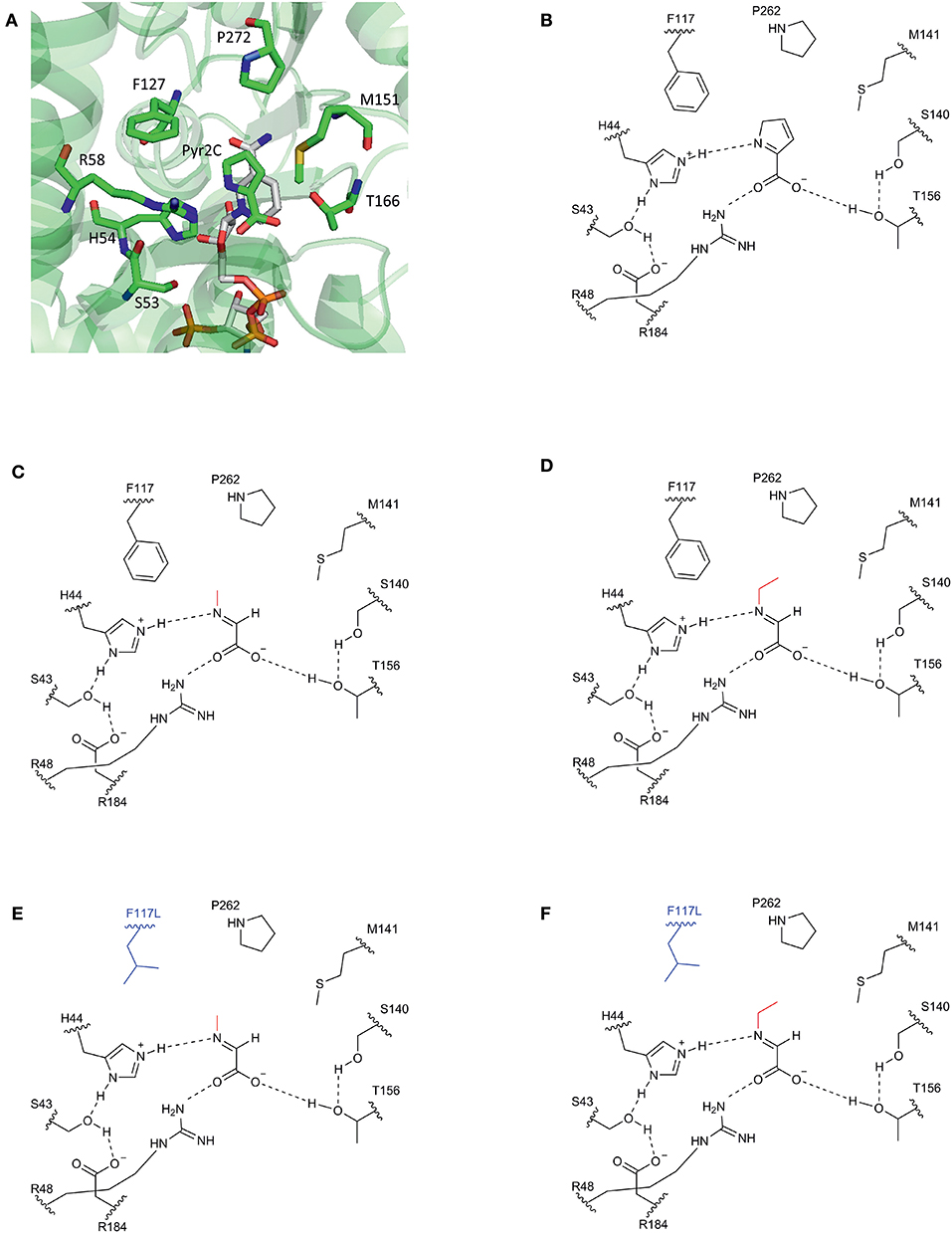
Figure 1. Schematic view of the substrate binding site of DpkA. (A) Active site of DpkA from Pseudomonas syringae (PDB: 2CWH). The native substrate pyrroline-2-carboxylate (Pyr2C; carbon atoms in green) and the cofactor NADPH (carbon atoms in light gray) are bound to the active site. The pyrrole ring of Pyr2C is recognized by the three amino acid residues Phe117, Pro262, and Met141 (carbon atoms in green). Schematic view of the active site of DpkA from Pseudomonas putida with the native substrate Pyr2C (B), the imine formed by glyoxylate and MMA (C), and the imine formed by glyoxylate and MEA (D). Potential substrate binding site of the mutant DpkAF117L with the imine formed by glyoxylate and MMA (E) and the imine formed by glyoxylate and MEA (F). The alkyl moiety from the alkylamine is shown in red. The mutated moiety Phe117Leu is depicted in blue (Based on Goto et al., 2005).
The mutant DpkAF117L was the most promising one in the prior analysis, therefore the substrate affinities (pyruvate and glyoxylate as 2-oxo acid substrates, MMA and MEA as alkylamine substrates) and catalytic efficiencies of wildtype DpkA and DpkAF117L were determined (see Supplementary Figures S1, S2). Interestingly, characterization of the wild-type enzyme DpkA with non-native substrates revealed substrate preference for glyoxylate and MEA over glyoxylate and MMA. Methylamination of pyruvate showed the highest catalytic efficiency (Table 3). Mutation of phenylalanine at position 117 by leucine enabled an increased specific methylamination activity toward glyoxylate (30.3 ± 2.7 U mg−1; wild-type enzyme 25.7 ± 1.8 U mg−1; Mindt et al., 2019), while the catalytic efficiency was similar. Yet, the catalytic efficiency for reductive methylamination of pyruvate (7.95 s−1 mM−1, wt: 6.66 s−1 mM−1) and for reductive ethylamination of glyoxylate (7.61 s−1 mM−1, wt: 6.44 s−1 mM−1) was enhanced by this mutant (Table 3; Figure 2). Further biochemical characterization of the mutant DpkAF117L may unravel the influence of this phenylalanine residue on substrate binding of other non-native aliphatic substrates as well as of the native substrates (Pip2C and Pyr2C). Thus, the mutant DpkAF117L is the most promising candidate to enhance volumetric productivity for fermentative production of sarcosine as well as NEtGly. While a thorough biochemical analysis of DpkA and its mutants is required to unravel the underlying molecular mechanism, our aim to improve reductive ethylamination of glyoxylate was met.
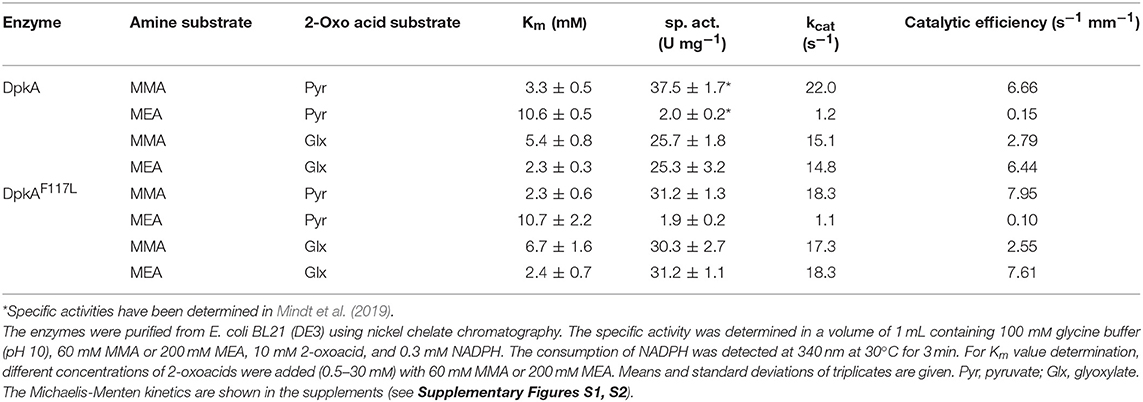
Table 3. Parameters of 10xHis-DpkA and the mutant 10xHis-DpkAF117L with various 2-oxo acid substrates and MMA and MEA as amine substrates.
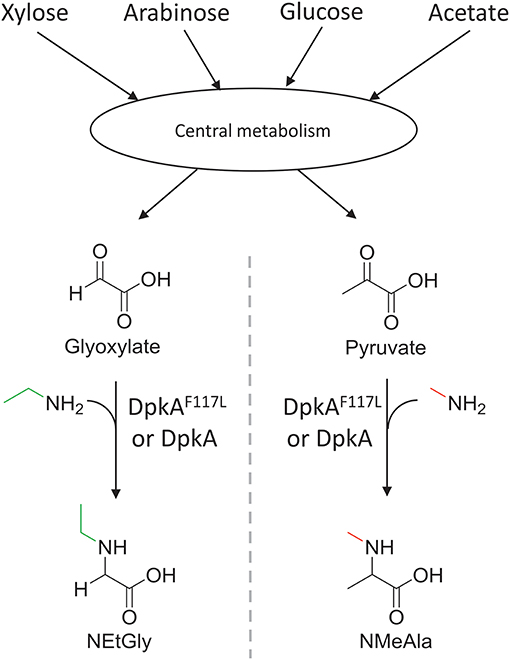
Figure 2. Schematic view of the sugar-based production of N-alkylated amino acids by C. glutamicum. Sugars (C5 and C6) and acetate of lignocellulosic hydrolysates are catabolized in the central carbon metabolism and converted to 2-oxo acids. Subsequently, the imine reductase DpkA from P. putida catalyzes the reductive amination of the 2-oxoacids yielding N-alkylated amino acids. The mutant DpkAF117L catalyzes the faster formation of NEtGly than the wild-type enzyme.
The kinetic studies of Mihara et al. on DpkA revealed a sequential Ter Bi mechanism: NADPH, pyruvate, and methylamine sequentially bind to the active site, followed by sequential release of NMeAla and NADP+ (Mihara et al., 2005). Due to this finding, DpkA may catalyze imine formation in addition to the reduction of the imine to its corresponding amine. Additional to catalysis by N-methylamino acid dehydrogenases, imine formation is catalyzed by other enzyme classes like amino acid dehydrogenases (Brunhuber et al., 2000) and reductive aminases (Aleku et al., 2017; Sharma et al., 2018). Surprisingly, several studies of classical imine reductases demonstrated that IREDs catalyze imine reduction, but not their formation from (alkyl)amines and ketones (Huber et al., 2014; Scheller et al., 2015; Wetzl et al., 2016). For the reductive alkylamination of 2-oxo acids by DpkA via a sequential Ter Bi mechanism, the presence of different alkylamines should not change Km values for the same 2-oxo acid since the 2-oxo acid is expected to bind prior to the alkylamine. However, the substrate affinity of the wild type enzyme for glyoxylate in the presence of MMA (Km 5.4 mM) was more than two times lower than in the presence of MEA (Km 2.3 mM; Table 3). Due to the contradictory findings to the study of Mihara et al. (2005), further investigation of the reaction mechanism for different non-native substrate combinations and a detailed analysis of the coordination of non-native substrates in the active site are needed to rationally engineer DpkA. Depending on the knowledge of the structure and the reaction mechanism, there are three different approaches for protein engineering: random, rational and semi-rational (Arnold, 2001; Reetz and Carballeira, 2007; Gupta and Tawfik, 2008). Alternatively, semi-rational enzyme design, in which the structural information is taken into account to generate mutant libraries (Lutz, 2010), may be used. This strategy was successfully applied e.g., to change an leucine dehydrogenase from Bacillus stereothermophilus to an enantioselective amine dehydrogenase (Abrahamson et al., 2012) or to change the cofactor specificity of an IRED from NADPH to NADH (Borlinghaus and Nestl, 2018).
Expression of DpkAF117L in C. glutamicum Led to Enhanced Sarcosine and N-Ethylglycine Production
Corynebacterium glutamicum was shown to withstand high concentrations of MMA (Mindt et al., 2018a) and sarcosine (Mindt et al., 2019). In order to determine possible effects of MEA on growth of C. glutamicum, cells were grown in minimal medium supplemented with increasing concentrations of MEA (0.05[ENTX]X02013[/ENTX]1.5 M). Extrapolation of the graph revealed a diminished growth rate to about half maximal at MEA concentrations of 0.7 M (Figure 3). Albeit the C2-compound showed a stronger negative impact on growth of C. glutamicum compared to methylamine (1.8 M; Mindt et al., 2018a), high concentrations of both alkylamines are tolerated by C. glutamicum, and hence, this bacterium is a suitable host for fermentative production of N-alkylated amino acids.
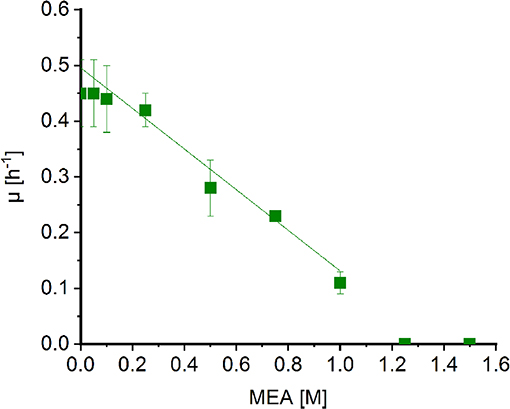
Figure 3. Effect of MEA on growth of Corynebacterium glutamicum wild type. Growth rates of C. glutamicum cultivated in minimal medium supplemented with increasing concentrations of MEA (green boxes) in Biolector microfermentation system in 48-well flower plates. Effect of MEA for half maximal growth rates was determined by extrapolation and linear fitting (OriginLab, Northampton, MA).
To compare the influence of the substitution of phenylalanine residue at position 117 to leucine in DpkA on sarcosine production, vector pVWEx1-dpkAF117L was constructed. This vector differs from vector pVWEx1-dpkA_RBS°pt (Mindt et al., 2019) only by the amino acid exchange F117L. Transformation of the glyoxylate production strain with this plasmid yielded strain SAR5 [i.e., strain GLX(pVWEx1-dpkAF117L) (pEC-XT99A-xylAXc-xylBCg); Table 1]. Strain SAR5 and isogenic strain SAR3 [i.e., strain GLX(pVWEx1-dpkA_RBS°pt) (pEC-XT99A-xylAXc-xylBCg); Table 1] were cultivated in minimal medium supplemented with 5 g L−1 xylose, 30 g L−1 potassium acetate and 6.2 g L−1 MMA (i.e., 200 mM). Production of sarcosine from both strains started 24 h after inoculation and yielded similar titers after 72 h cultivation (SAR3 10.4 ± 0.5 g L−1; SAR5 10.5 ± 0.3 g L−1). Notably, the mutated imine reductase DpkAF117L allowed faster fermentative production of sarcosine (vol. prod. for 56 h: 0.16 g L−1 h−1) compared to the wild type enzyme (vol. prod. for 56 h: 0.12 g L−1 h−1) (Figures 4A,B). Carbon source utilization by SAR5 was faster than by SAR3. SAR5 stopped NEtGly production earlier (at around 48 h), which we believe is due to the fact that the carbon sources were depleted. Lactate was not detected as byproduct. Although DpkA and DpkAF117L showed higher catalytic efficiency for methylamination of pyruvate as compared to glyoxylate, the finding that neither strain produced N-methylalanine as side product indicates that the glyoxylate overproduction base strain provided sufficient glyoxylate while maintaining sub-threshold pyruvate concentrations.
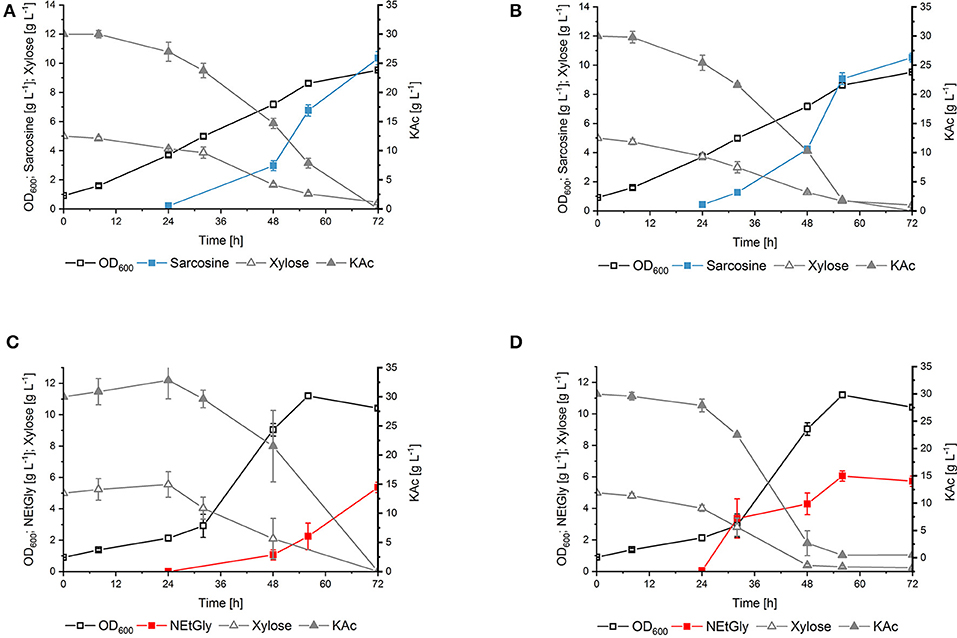
Figure 4. Production sarcosine and NEtGly by C. glutamicum strains in shake flask fermentation. The growth, production, and consumption of carbon source of SAR3 (A,C) and SAR5 (B,D) in minimal medium supplemented with 5 g L−1 xylose, 30 g L−1 potassium acetate, and 6.2 g L−1 MMA (A,B) or 9.0 g L−1 MEA (C,D) was followed. Biomass formation (open black boxes), sarcosine production (filled turquoise boxes), NEtGly production (filled red boxes), xylose (open gray triangles), and potassium acetate (filled gray triangles) are given as means of triplicates with standard deviations.
DpkA favors reductive ethylamination of glyoxylate over reductive ethylamination of pyruvate by one order of magnitude (6.44 s−1 mM−1 as compared to 0.15 s−1 mM−1; Table 3). This preference is even larger for the mutant DpkAF117L (7.61 s−1 mM−1as compared to 0.10 s−1 mM−1; Table 3). Since the Km values for glyoxylate were in the lower millimolar range for DpkA (around 2–7 mM; Table 3), we expected that reductive ethylamination of glyoxylate should proceed in vivo. Thus, both strains expressing the mutant or the wild type enzyme (SAR5 and SAR3, respectively) were cultivated with 9.0 g L−1 MEA (or 200 mM) instead of MMA. Indeed, both strains produced NEtGly to comparable product titers (5.7 ± 0.3 g L−1 for SAR5 and 5.4 ± 0.3 g L−1for SAR3 accumulated within 72 h; Figures 4C,D), while NEtAla was not formed as by-product. As expected from the higher catalytic efficiency of DpkAF117L, strain SAR5 showed faster consumption of potassium acetate and xylose as well as faster NEtGly production (volumetric productivity within 56 h: SAR5 0.11 g L−1 h−1, SAR3 0.04 g L−1 h−1). To the best of our knowledge, this is the first demonstration of fermentative production of NEtGly.
Interestingly, the strains showed different growth behavior during production of sarcosine und NEtGly. Growth of the engineered C. glutamicum strains (both SAR3 and SAR5) in NEtGly producing experiments is improved compared to sarcosine production (Figure 4). This may be due to the lower kcat value of DpkA and DpkAF117L for the reductive amination of glyoxylate with MEA (Table 3). Accumulation of glyoxylate intracellularly inhibits the growth of C. glutamicum (Zahoor et al., 2014), thus, faster consumption of the aldehyde in presence of MEA may allow for faster growth.
Production of NEtGly From Lignocellulosic Hydrolysates
In order to test the efficacy of lignocellulosic hydrolysate for fermentative production of NEtGly, strain SAR5 was transformed with plasmid pEKEx3-araBAD to yield strain SAR6, which is able to also consume arabinose. The hydrolysis of rice straw with 4% sulfuric acid (see section Preparation of Lignocellulosic Hydrolysates) resulted in 14.7 g L−1 of xylose, 7.2 g L−1 of glucose, 4.3 g L−1 of arabinose, and 4.3 g L−1 of acetate. Strains SAR4 and SAR6 are able to utilize the four carbon sources xylose, glucose, arabinose, and acetate present in the hydrolysis liquor. The hydrolysis liquor was mixed with 18 g L−1 potassium acetate (1:1.85 ratio) to obtain the feed used during fermentation. During the first growth phase of the fermentation process (up to 8 h of cultivation, see Figure 5), glucose was depleted completely and lactate accumulated transiently up to a concentration of 0.1 g L−1. At that time, i.e., after reaching an optical density of about 5, 40 mL of 5 M ethylamine solution was added. From 8 to 24 h of cultivation, the feed was increased. While xylose and arabinose concentrations in the bioreactor medium varied between 0.5 and 2 g L−1, glucose was not detected and only little acetate was observed. During this period, growth continued and NEtGly production started. After 24 h of cultivation, 100 mL of 60 g L−1 potassium acetate solution were added and after 28 h the feed was stopped. After 30 h of cultivation, growth reached the maximal OD600 of 23.3 (Figure 5). In the last phase (from 30 to 66 h), acetate and the pentoses were utilized slowly. NEtGly was produced steadily and the concentration more than doubled from 30 to 66 h. A final concentration of 1.6 g L−1 of NEtGly was obtained.
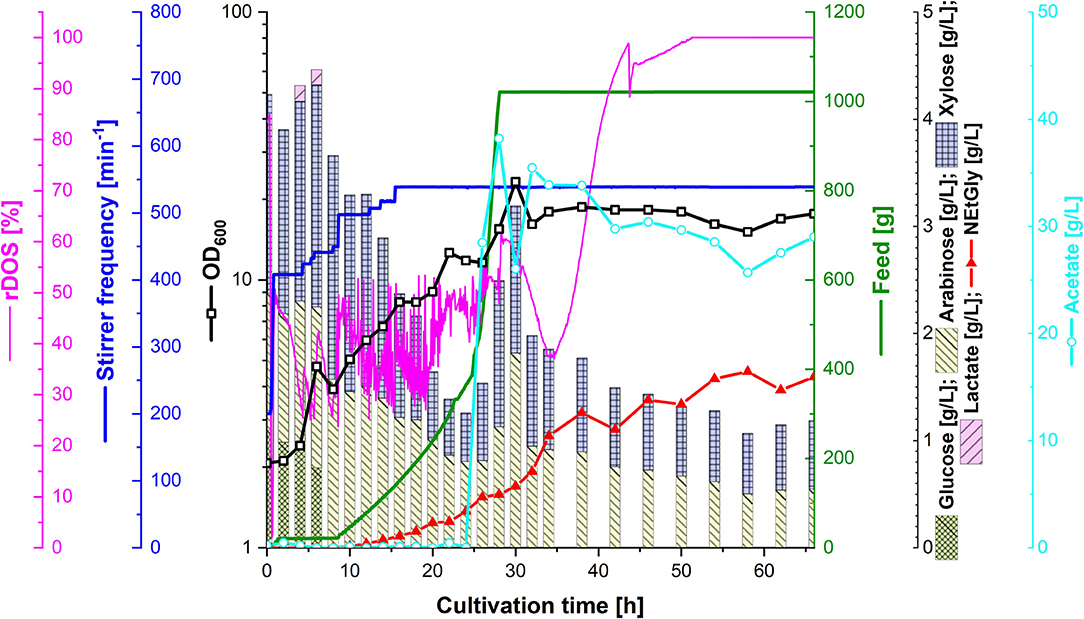
Figure 5. Production of NEtGly from lignocellulosic hydrolysate by C. glutamicum SAR6 in a 2 L scale fed-batch bioreactor process. The fermentation took place in low nitrogen minimal medium supplemented with feed (green line) consisting of rice straw hydrolysate and 18 g L−1 potassium acetate (1:1.85 ratio). After cells reached an OD600 (open black boxes) of 5, 40 mL of 5 M ethylamine solution were added; after 24 h of cultivation, 100 mL of 60 g L−1 potassium acetate solution were added. The relative dissolved oxygen saturation (rDOS; pink line) was measured to control the stirrer frequency (dark blue line) and the feed supplementation as described in section Fed-Batch Bioreactor Process. Samples for HPLC analysis of sugar content (cumulative stacked columns), acetate content (light blue open circles) and NEtGly production (red triangles) were taken automatically.
Bioreactor cultivation showed that the production of NEtGly is scalable and can be based on lignocellulosic hydrolysate. However, titer, yield, and productivity were lower than in shake flasks experiments using pure chemicals as carbon sources. To some extent, C. glutamicum shows recalcitrance against typical inhibitors present in lignocellulosic hydrolysates such as hydroxymethylfurfural and methylfurfural (Gopinath et al., 2011), however, it cannot be excluded that other inhibitors impaired hydrolysate-based NEtGly production. Unlike E. coli and yeasts, C. glutamicum can simultaneously utilize acetate present in hydrolysates with other carbon sources (Wendisch et al., 2000). Pentose utilization that is slower than glucose utilization was accelerated by using alternative donors for enzymes of xylose catabolism and/or by pentose uptake engineering (Sasaki et al., 2009; Meiswinkel et al., 2013; Brüsseler et al., 2018). Uptake of acetate by MctC (Jolkver et al., 2009) or methylammonium (and likely also ethylammonium) uptake by Amt (Meier-Wagner et al., 2001) have not yet been targeted to improve production based on these substrates. Currently, it is unknown how NEtGly is exported from the C. glutamicum cell. While some export due to diffusion is possible, it is likely that an export system accepting NEtGly is encoded in the C. glutamicum genome. While transport engineering is expected to affect volumetric productivity, improvement of yield and titer requires other approaches of strain and process development.
In this study, we established fermentative production of the N-alkylated amino acid NEtGly. Expression of the N-methylamino acid dehydrogenase/IRED gene dpkA from P. putida in a recombinant C. glutamicum strain allowed reductive amination of glyoxylate forming NEtGly, when MEA was added. However, the xylose-based process had a low volumetric productivity (0.04 g L−1 h−1). By site directed mutagenesis of the substrate binding site, we could identify a mutant showing increased catalytic efficiency toward ethylamination of glyoxylate. Implementation of the mutant DpkAF117L enabled faster production of NEtGly with a final titer of 6.1 ± 0.3 g L−1 and a volumetric productivity of 0.11 g L−1 h−1.
Data Availability Statement
All datasets generated for this study are included in the manuscript/Supplementary Files.
Author Contributions
MM and VW planned and designed the experiments. MM and MH performed the enzymatic characterization. MM constructed production strains and performed shake flask cultivations. SH and KS prepared lignocellulosic hydrolysates and SH and JR performed bioreactor cultivations. MM, MH, SH, JR, KN, and VW analyzed the data. MM drafted the manuscript. VW and KN finalized the manuscript. All authors agreed to the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Thomas Schäffer from the Department of Fermentation Technology, Bielefeld University for help during bioreactor cultivations. We acknowledge support for the Article Processing Charge by the Deutsche Forschungsgemeinschaft and the Open Access Publication Fund of Bielefeld University. VW and KN acknowledge funding within the Indo-German BIOCON project by DBT and BMBF (01DQ17009).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2019.00232/full#supplementary-material
References
Abrahamson, M. J., Vázquez-Figueroa, E., Woodall, N. B., Moore, J. C., and Bommarius, A. S. (2012). Development of an amine dehydrogenase for synthesis of chiral amines. Angew. Chem. Int. Ed. 51, 3969–3972. doi: 10.1002/anie.201107813
Aleku, G. A., France, S. P., Man, H., Mangas-Sanchez, J., Montgomery, S. L., Sharma, M., et al. (2017). A reductive aminase from Aspergillus oryzae. Nat. Chem. 9, 961–969. doi: 10.1038/nchem.2782
Arnold, F. H. (2001). Combinatorial and computational challenges for biocatalyst design. Nature 409, 253–257. doi: 10.1038/35051731
Aurelio, L., Brownlee, R. T. C., and Hughes, A. B. (2004). Synthetic preparation of N-methyl-α-amino acids. Chem. Rev. 104, 5823–5846. doi: 10.1021/cr030024z
Borlinghaus, N., and Nestl, B. M. (2018). Switching the cofactor specificity of an imine reductase. ChemCatChem 10, 183–187. doi: 10.1002/cctc.201701194
Brunhuber, N. M. W., Thoden, J. B., Blanchard, J. S., and Vanhooke, J. L. (2000). Rhodococcus L-phenylalanine dehydrogenase: Kinetics, mechanism, and structural basis for catalytic specifity. Biochemistry 39, 9174–9187. doi: 10.1021/bi000494c
Brüsseler, C., Radek, A., Tenhaef, N., Krumbach, K., Noack, S., and Marienhagen, J. (2018). The myo-inositol/proton symporter IolT1 contributes to D-xylose uptake in Corynebacterium glutamicum. Bioresour. Technol. 249, 953–961. doi: 10.1016/j.biortech.2017.10.098
Bückle-Vallant, V., Krause, F. S., Messerschmidt, S., and Eikmanns, B. J. (2014). Metabolic engineering of Corynebacterium glutamicum for 2-ketoisocaproate production. Appl. Microbiol. Biotechnol. 98, 297–311. doi: 10.1007/s00253-013-5310-2
Chatterjee, J., Rechenmacher, F., and Kessler, H. (2013). N-methylation of peptides and proteins: an important element for modulating biological functions. Angew. Chem. Int. Ed. 52, 254–269. doi: 10.1002/anie.201205674
Di Gioia, M. L., Leggio, A., Malagrin,ò, F., Romio, E., Siciliano, C., and Liguori, A. (2016). N-methylated α-amino acids and peptides: synthesis and biological activity. Mini Rev. Med. Chem. 16, 683–690. doi: 10.2174/1389557516666160322152457
Dutt Konar, A., Vass, E., Hollósi, M., Majer, Z., Grüber, G., Frese, K., et al. (2013). Conformational properties of secondary amino acids: replacement of pipecolic acid by N-methyl-L-alanine in efrapeptin C. Chem. Biodivers. 10, 942–951. doi: 10.1002/cbdv.201300086
Eggeling, L., and Bott, M. (2005). Handbook of Corynebacterium glutamicum. Boca Raton, FL: CRC Press.
Eipe, N., Gupta, S., and Penning, J. (2016). Intravenous lidocaine for acute pain: an evidence-based clinical update. BJA Educ. 16, 292–298. doi: 10.1093/bjaed/mkw008
Falcioni, F., Bühler, B., and Schmid, A. (2015). Efficient hydroxyproline production from glucose in minimal media by Corynebacterium glutamicum: hydroxyproline production by C. glutamicum. Biotechnol. Bioeng. 112, 322–330. doi: 10.1002/bit.25442
Gentilucci, L., De Marco, R., and Cerisoli, L. (2010). Chemical modifications designed to improve peptide stability: incorporation of non-natural amino acids, pseudo-peptide bonds, and cyclization. Curr. Pharm. Des. 16, 3185–3203. doi: 10.2174/138161210793292555
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Gopinath, V., Meiswinkel, T. M., Wendisch, V. F., and Nampoothiri, K. M. (2011). Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose-utilizing Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 92, 985–996. doi: 10.1007/s00253-011-3478-x
Goto, M., Muramatsu, H., Mihara, H., Kurihara, T., Esaki, N., Omi, R., et al. (2005). Crystal structures of Δ1-piperideine-2-carboxylate/Δ1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition and stereochemistry of the reaction. J. Biol. Chem. 280, 40875–40884. doi: 10.1074/jbc.M507399200
Green, M. R., and Sambrook, J. (2012). Molecular Cloning: A Laboratory Manual, 4th Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Gupta, R. D., and Tawfik, D. S. (2008). Directed enzyme evolution via small and effective neutral drift libraries. Nat. Methods 5, 939–942. doi: 10.1038/nmeth.1262
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Huber, T., Schneider, L., Präg, A., Gerhardt, S., Einsle, O., and Müller, M. (2014). Direct reductive amination of ketones: structure and activity of S-selective imine reductases from Streptomyces. ChemCatChem 6, 2248–2252. doi: 10.1002/cctc.201402218
Hyslop, J. F., Lovelock, S. L., Watson, A. J. B., Sutton, P. W., and Roiban, G.-D. (2019). N-Alkyl-α-amino acids in nature and their biocatalytic preparation. J. Biotechnol. 293, 56–65. doi: 10.1016/j.jbiotec.2019.01.006
Johnson, K. A., and Goody, R. S. (2011). The original Michaelis constant: translation of the 1913 Michaelis–Menten paper. Biochemistry 50, 8264–8269. doi: 10.1021/bi201284u
Jolkver, E., Emer, D., Ballan, S., Krämer, R., Eikmanns, B. J., and Marin, K. (2009). Identification and characterization of a bacterial transport system for the uptake of pyruvate, propionate, and acetate in Corynebacterium glutamicum. J. Bacteriol. 191, 940–948. doi: 10.1128/JB.01155-08
Kamyar, M., Rawnduzi, P., Studenik, C. R., Kouri, K., and Lemmens-Gruber, R. (2004). Investigation of the electrophysiological properties of enniatins. Arch. Biochem. Biophys. 429, 215–223. doi: 10.1016/j.abb.2004.06.013
Kirchner, O., and Tauch, A. (2003). Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 104, 287–299. doi: 10.1016/S0168-1656(03)00148-2
Krause, F. S., Blombach, B., and Eikmanns, B. J. (2010). Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl. Environ. Microbiol. 76, 8053–8061. doi: 10.1128/AEM.01710-10
Latypova, E., Yang, S., Wang, Y.-S., Wang, T., Chavkin, T. A., Hackett, M., et al. (2010). Genetics of the glutamate-mediated methylamine utilization pathway in the facultative methylotrophic beta-proteobacterium Methyloversatilis universalis FAM5. Mol. Microbiol. 75, 426–439. doi: 10.1111/j.1365-2958.2009.06989.x
Lutz, S. (2010). Beyond directed evolution—semi-rational protein engineering and design. Curr. Opin. Biotechnol. 21, 734–743. doi: 10.1016/j.copbio.2010.08.011
Mas-Moruno, C., Rechenmacher, F., and Kessler, H. (2010). Cilengitide: the first anti-angiogenic small molecule drug candidate. Design, synthesis and clinical evaluation. Anticancer Agents Med. Chem. 10, 753–768. doi: 10.2174/187152010794728639
Meier-Wagner, J., Nolden, L., Jakoby, M., Siewe, R., Krämer, R., and Burkovski, A. (2001). Multiplicity of ammonium uptake systems in Corynebacterium glutamicum: role of Amt and AmtB. Microbiol. Read. Engl. 147, 135–143. doi: 10.1099/00221287-147-1-135
Meiswinkel, T. M., Gopinath, V., Lindner, S. N., Nampoothiri, K. M., and Wendisch, V. F. (2013). Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb. Biotechnol. 6, 131–140. doi: 10.1111/1751-7915.12001
Mihara, H., Muramatsu, H., Kakutani, R., Yasuda, M., Ueda, M., Kurihara, T., et al. (2005). N-methyl-L-amino acid dehydrogenase from Pseudomonas putida. A novel member of an unusual NAD(P)-dependent oxidoreductase superfamily. FEBS J. 272, 1117–1123. doi: 10.1111/j.1742-4658.2004.04541.x
Mindt, M., Heuser, M., and Wendisch, V. F. (2019). Xylose as preferred substrate for sarcosine production by recombinant Corynebacterium glutamicum. Bioresour. Technol. 281, 135–142. doi: 10.1016/j.biortech.2019.02.084
Mindt, M., Risse, J. M., Gruß, H., Sewald, N., Eikmanns, B. J., and Wendisch, V. F. (2018a). One-step process for production of N-methylated amino acids from sugars and methylamine using recombinant Corynebacterium glutamicum as biocatalyst. Sci. Rep. 8:12895. doi: 10.1038/s41598-018-31309-5
Mindt, M., Walter, T., Risse, J. M., and Wendisch, V. F. (2018b). Fermentative production of N-methylglutamate from glycerol by recombinant Pseudomonas putida. Front. Bioeng. Biotechnol. 6:159. doi: 10.3389/fbioe.2018.00159
Muramatsu, H., Mihara, H., Kakutani, R., Yasuda, M., Ueda, M., Kurihara, T., et al. (2005). The putative malate/lactate dehydrogenase from Pseudomonas putida is an NADPH-dependent Δ1-piperideine-2-carboxylate/Δ1-pyrroline-2-carboxylate reductase involved in the catabolism of D-lysine and D-proline. J. Biol. Chem. 280, 5329–5335. doi: 10.1074/jbc.M411918200
Patterson, A. W., Peltier, H. M., and Ellman, J. A. (2008). Expedient synthesis of N-methyl tubulysin analogues with high cytotoxicity. J. Org. Chem. 73, 4362–4369. doi: 10.1021/jo800384x
Pérez-García, F., Max Risse, J., Friehs, K., and Wendisch, V. F. (2017a). Fermentative production of L-pipecolic acid from glucose and alternative carbon sources. Biotechnol. J. 12:1600646. doi: 10.1002/biot.201600646
Pérez-García, F., Peters-Wendisch, P., and Wendisch, V. F. (2016). Engineering Corynebacterium glutamicum for fast production of L-lysine and L-pipecolic acid. Appl. Microbiol. Biotechnol. 100, 8075–8090. doi: 10.1007/s00253-016-7682-6
Pérez-García, F., Ziert, C., Risse, J. M., and Wendisch, V. F. (2017b). Improved fermentative production of the compatible solute ectoine by Corynebacterium glutamicum from glucose and alternative carbon sources. J. Biotechnol. 258, 59–68. doi: 10.1016/j.jbiotec.2017.04.039
Peters-Wendisch, P. G., Schiel, B., Wendisch, V. F., Katsoulidis, E., Möckel, B., Sahm, H., et al. (2001). Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3, 295–300.
Reetz, M. T., and Carballeira, J. D. (2007). Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat. Protoc. 2, 891–903. doi: 10.1038/nprot.2007.72
Sakato, Y. (1950). Studies on the chemical constituents of tea. J. Agric. Food Chem. 23, 262–267. doi: 10.1271/nogeikagaku1924.23.262
Sasaki, M., Jojima, T., Kawaguchi, H., Inui, M., and Yukawa, H. (2009). Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl. Microbiol. Biotechnol. 85, 105–115. doi: 10.1007/s00253-009-2065-x
Scheller, P. N., Lenz, M., Hammer, S. C., Hauer, B., and Nestl, B. M. (2015). Imine reductase-catalyzed intermolecular reductive amination of aldehydes and ketones. ChemCatChem 7, 3239–3242. doi: 10.1002/cctc.201500764
Sharma, M., Mangas-Sanchez, J., France, S. P., Aleku, G. A., Montgomery, S. L., Ramsden, J. I., et al. (2018). A mechanism for reductive amination catalyzed by fungal reductive aminases. ACS Catal. 8, 11534–11541. doi: 10.1021/acscatal.8b03491
Stansen, C., Uy, D., Delaunay, S., Eggeling, L., Goergen, J.-L., and Wendisch, V. F. (2005). Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 71, 5920–5928. doi: 10.1128/AEM.71.10.5920-5928.2005
Veldmann, K. H., Dachwitz, S., Risse, J. M., Lee, J.-H, Sewald, N., and Wendisch, V. F. (2019b) Bromination of L-tryptophan in a fermentative process with Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 7:219.
Veldmann, K. H., Minges, H., Sewald, N., Lee, J.-H., and Wendisch, V. F. (2019a). Metabolic engineering of Corynebacterium glutamicum for the fermentative production of halogenated tryptophan. J. Biotechnol. 291, 7–16. doi: 10.1016/j.jbiotec.2018.12.008
Wendisch, V. F., de Graaf, A. A., Sahm, H., and Eikmanns, B. J. (2000). Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 182, 3088–3096. doi: 10.1128/JB.182.11.3088-3096.2000
Werdehausen, R., Mittnacht, S., Bee, L. A., Minett, M. S., Armbruster, A., Bauer, I., et al. (2015). The lidocaine metabolite N-ethylglycine has antinociceptive effects in experimental inflammatory and neuropathic pain. Pain 156, 1647–1659. doi: 10.1097/j.pain.0000000000000206
Wetzl, D., Gand, M., Ross, A., Müller, H., Matzel, P., Hanlon, S. P., et al. (2016). Asymmetric reductive amination of ketones catalyzed by imine reductases. ChemCatChem 8, 2023–2026. doi: 10.1002/cctc.201600384
Wieschalka, S., Blombach, B., and Eikmanns, B. J. (2012). Engineering Corynebacterium glutamicum for the production of pyruvate. Appl. Microbiol. Biotechnol. 94, 449–459. doi: 10.1007/s00253-011-3843-9
Yi, Y., Sheng, H., Li, Z., and Ye, Q. (2014). Biosynthesis of trans-4-hydroxyproline by recombinant strains of Corynebacterium glutamicum and Escherichia coli. BMC Biotechnol. 14:44. doi: 10.1186/1472-6750-14-44
Zahoor, A., Otten, A., and Wendisch, V. F. (2014). Metabolic engineering of Corynebacterium glutamicum for glycolate production. J. Biotechnol. 192, 366–375. doi: 10.1016/j.jbiotec.2013.12.020
Keywords: Corynebacterium glutamicum, imine reductase, metabolic engineering, enzyme engineering, N-alkylated amino acids, N-methylamino acids, N-ethylglycine, sarcosine
Citation: Mindt M, Hannibal S, Heuser M, Risse JM, Sasikumar K, Nampoothiri KM and Wendisch VF (2019) Fermentative Production of N-Alkylated Glycine Derivatives by Recombinant Corynebacterium glutamicum Using a Mutant of Imine Reductase DpkA From Pseudomonas putida. Front. Bioeng. Biotechnol. 7:232. doi: 10.3389/fbioe.2019.00232
Received: 17 June 2019; Accepted: 09 September 2019;
Published: 26 September 2019.
Edited by:
Zhi-Qiang Liu, Zhejiang University of Technology, ChinaReviewed by:
Ki Jun Jeong, Korea Advanced Institute of Science and Technology (KAIST), South KoreaXiaowei Li, Chalmers University of Technology, Sweden
Copyright © 2019 Mindt, Hannibal, Heuser, Risse, Sasikumar, Nampoothiri and Wendisch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Volker F. Wendisch, volker.wendisch@uni-bielefeld.de
 Melanie Mindt
Melanie Mindt Silvin Hannibal
Silvin Hannibal Maria Heuser
Maria Heuser Joe Max Risse
Joe Max Risse Keerthi Sasikumar
Keerthi Sasikumar K. Madhavan Nampoothiri3
K. Madhavan Nampoothiri3  Volker F. Wendisch
Volker F. Wendisch