Application of artificial scaffold systems in microbial metabolic engineering
- 1College of Pharmaceutical Science, Zhejiang University of Technology, Hangzhou, China
- 2HIM-BGI Omics Center, Zhejiang Cancer Hospital, Hangzhou Institute of Medicine (HIM), Chinese Academy of Sciences (CAS), Hangzhou, China
In nature, metabolic pathways are often organized into complex structures such as multienzyme complexes, enzyme molecular scaffolds, or reaction microcompartments. These structures help facilitate multi-step metabolic reactions. However, engineered metabolic pathways in microbial cell factories do not possess inherent metabolic regulatory mechanisms, which can result in metabolic imbalance. Taking inspiration from nature, scientists have successfully developed synthetic scaffolds to enhance the performance of engineered metabolic pathways in microbial cell factories. By recruiting enzymes, synthetic scaffolds facilitate the formation of multi-enzyme complexes, leading to the modulation of enzyme spatial distribution, increased enzyme activity, and a reduction in the loss of intermediate products and the toxicity associated with harmful intermediates within cells. In recent years, scaffolds based on proteins, nucleic acids, and various organelles have been developed and employed to facilitate multiple metabolic pathways. Despite varying degrees of success, synthetic scaffolds still encounter numerous challenges. The objective of this review is to provide a comprehensive introduction to these synthetic scaffolds and discuss their latest research advancements and challenges.
1 Introduction
Recently, the successful synthesis of diverse natural products has been achieved through the introduction of heterologous metabolic pathways into microbial cell factories (Srinivasan and Smolke, 2020; Yuan et al., 2022; Zhang et al., 2022). These synthetic pathways are collectively built with heterologous enzymes selected from various sources and are not accompanied by their regulatory partners in the new host (Tran et al., 2023). Thus, these unregulated enzymes may not be able to channel intermediates from the input reactions to the formation of end products properly (Dueber et al., 2009; Liu et al., 2023). In contrast, metabolic enzymes of a native pathway can be formed multi-enzyme complexes (Tittes et al., 2022), enzyme molecular scaffolds (Artzi et al., 2017), reaction microchambers [e.g., arom multienzyme complexes (Lumsden and Coggins, 1977), or caveolae, etc. (Polka et al., 2016)] to mediate the catalytic cascades coordinately. The structural entities play a pivotal role in facilitating efficient substrate transfer between adjacent enzyme active sites (Castellana et al., 2014). Thus, adopting a synthetic scaffold is one of the strategies to co-ordinate the none-native enzymes in microbial cell factories. To address the challenges several synthetic scaffolds have been devised for the precise modulation of enzyme activity. For instance, enzymes can be assembled on scaffolds made of DNA or protein, where protein-protein or DNA interactions are employed to facilitate the formation of cascading complexes among enzymes (Gad and Ayakar, 2021). These DNA or protein scaffolds are thought to form channels conducive to continuous metabolism, directing metabolic intermediates from one enzyme to another to regulate the spatial distribution of enzymes and increase their local concentrations (Tippmann et al., 2017). The utilization of synthetic scaffolds serves to significantly decrease interenzyme distances, thereby effectively restricting the diffusion of intermediate metabolitesand concurrently attenuates cellular cytotoxicity (Conrado et al., 2008). Though, the synthetic scaffolds had been successfully applied to metabolic engineering, trial and error are still the only way we may learn.
This review provides a detailed account of the applications of artificially synthesized scaffolds through specific case studies and comprehensively summarizes the latest advancements in various scaffold assembly methods. Additionally, it explores the potential challenges faced by artificially synthesized scaffolds. At the same time, the possible role of the current hot artificial intelligence (AI) technology in the application of artificial stent systems is also discussed.
2 Protein scaffold
Paired protein scaffolders fall into three main categories, including protein-peptide, peptide-peptide, and protein-protein pairs. These scaffolds can be fused directly with target enzymes to induce assembly, and they achieve enzyme assembly through non-covalent or covalent interactions between ligands and receptors in the scaffolds, with little effect on enzyme properties (Price et al., 2016; Chen et al., 2023).
2.1 Protein–peptide pair
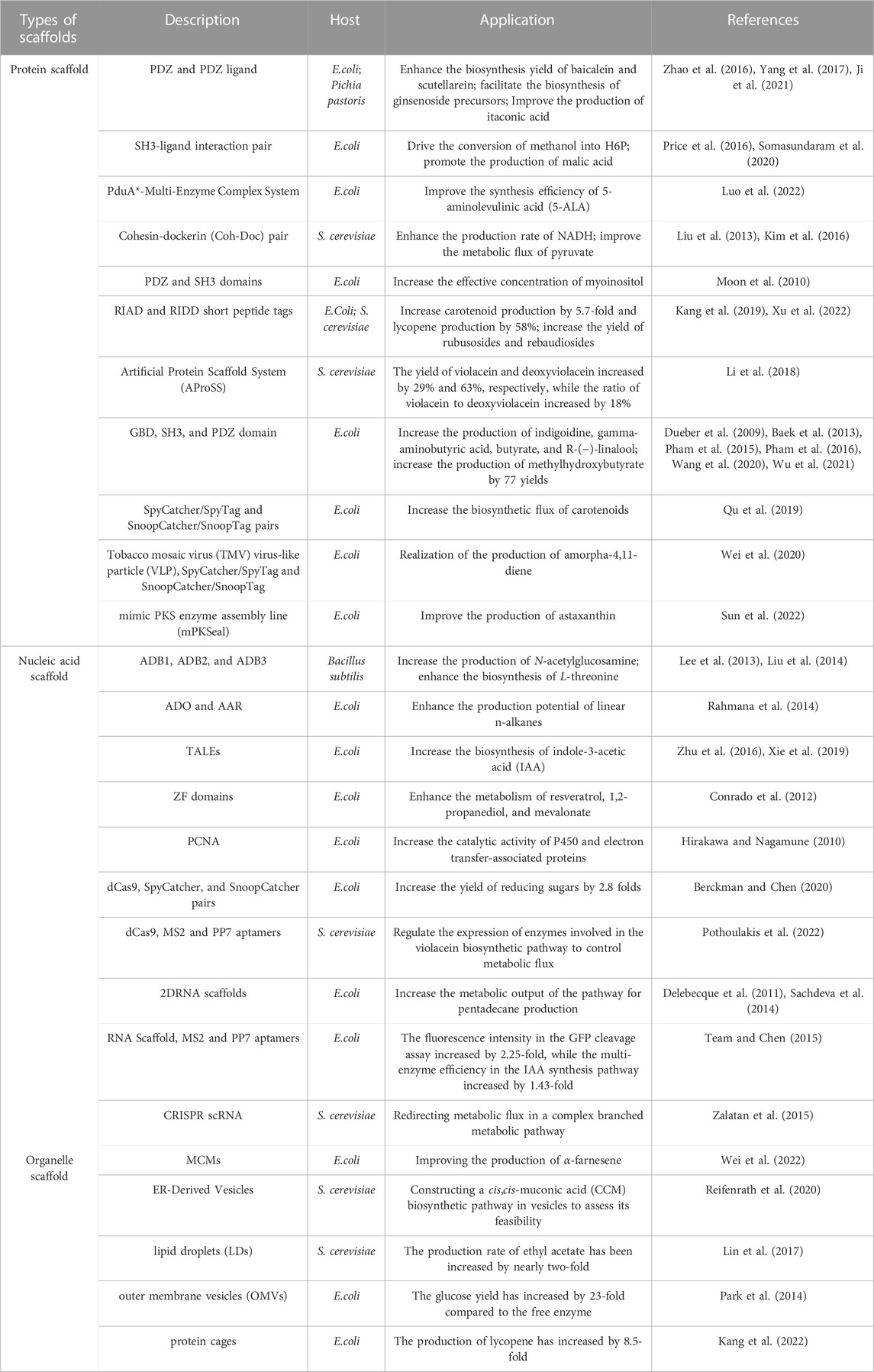
Protein-peptide interaction recognition domains are widely present in various cells, where they participate in the assembly of intracellular complexes and play diverse cellular functions. Currently, several modular protein domains [e.g., PDZ domain, SH3 domains, GTPase binding domain (GBD), GBD1SH31PDZ2, GBD1SH32PDZ4, and GBD1SH34PDZ4] and their corresponding partners have been identified (Pawson, 2007). The PDZ domain (also known as GLGF repeats or DHR domains) is typically an essential component of multi-domain scaffold proteins involved in cell polarity and intercellular interactions (Fanning and Anderson, 1996). It can selectively recognize the C-terminal peptide sequences on its partner protein and then assemble them into a complex and target specific subcellular localization sites (Tonikian et al., 2008). Based on this protein-peptide interaction, Gao et al. (2014) proposed a scaffold-free self-assembly strategy. This strategy was successfully demonstrated using the NAD (H) cycle system with L-tert-leucine as a model, achieving scaffold-free self-assembly technology. They fused the PDZ (PSD95/Dlg1/zo-1) domain and corresponding ligands (PDZlig) from metazoan cells separately with the octameric leucine dehydrogenase (LDH, derived from Bacillus subtilis BEST7613) and the dimeric formate dehydrogenase (FDH, derived from Lodderomyces elongisporus NRRL YB4239) (Figure 1A). The fusion proteins self-assembled into extended supramolecular interaction networks, significantly enhancing the efficiency and structural stability of the coenzyme cycling system involving NAD (H). Compared to their non-assembled counterparts, they exhibited better performance (Gao et al., 2014).
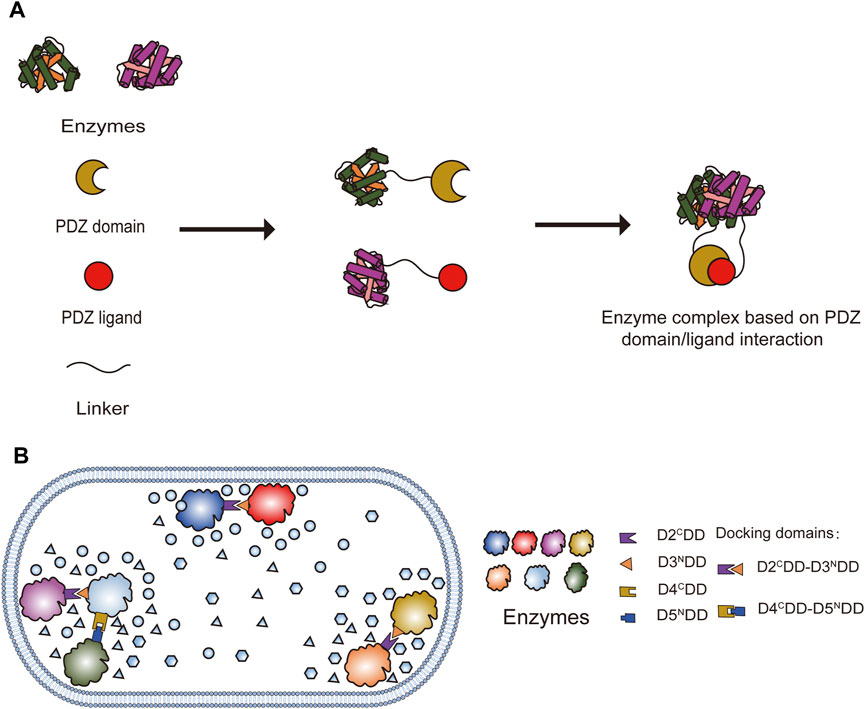
FIGURE 1. Protein scaffold; (A) Enzyme complex assembly based on PDZ domain and ligand interaction, adapted from (Gao et al., 2014); (B) Enzyme complex assembly based on mPKSeal strategy, adapted from (Sun et al., 2022).
The SRC Homology 3 Domain (or SH3 domain) is a small protein domain containing 60 amino acid residues that are folded into beta-barrels with five or six β-strands arranged as two tightly packed anti-parallel β sheets (Schlessinger, 1994). It typically binds to proline-rich peptides in its respective binding partner. A SH3-ligand interaction strategy was used to successfully assemble methanol dehydrogenase (Mdh), 3-hexulose-6-phosphate synthase (Hps), and 6-phospho-3-hexuloseisomerase (Phi) into highly efficient enzyme complexes, significantly improving the conversion efficiency of methanol to fructose-6-phosphate (F6P) (Price et al., 2016). Meanwhile, in Escherichia coli, lactate dehydrogenase was utilized as an NADH scavenger to establish an “NADH sink.” By combining these two strategies, a 97-fold increase in extracellular F6P production and a 9-fold improvement in intracellular methanol consumption were successfully achieved (Price et al., 2016).
The GTPase binding domain (GBD) from the actin polymerization switch N-WASP could be recognized by the GTP-bound Cdc42. Dueber et al. (2009), used the GTPase binding domain (GBD), the SH3 domain, and the PDZ domain to build a synthetic scaffold to provide modular control over metabolic pathway flux. By varying the numbers of these three domains (GBDxSH3yPDZz; x, y, z = number of domain repeats) to control the co-localization ratio of the interacting catalytic enzymes (e.g., atoB, HMGS, and HMGR), the optimal scaffold quantity is GBD1SH32PDZ2, and this engineering strategy led to a 77-fold increase in malic acid production (Dueber et al., 2009). By constructing a self-assembly enzyme reactor in E. coli, the stoichiometric ratio of two enzymes in the baicalein synthesis pathway was regulated to form an enzyme complex. This strategy significantly increased the titers of baicalein and scutellarein by 6.6 and 1.4 folds, respectively (Ji et al., 2021). Wei et al. utilized tobacco mosaic virus (TMV) virus-like particle (VLP) as a protein scaffold and orthogonal reactive protein pairs (SpyCatcher/SpyTag and SnoopCatcher/SnoopTag) as a linking module to assemble terpene biosynthesis in E. coli, enabling the production of amorpha-4,11-diene (Wei et al., 2020).
2.2 Peptide–peptide pair
In nature, there are numerous examples of optimizing metabolic pathway performance by forming multienzyme complexes. A prominent example is polyketide synthases (PKSs), which are considered among the most intricate proteins in nature. PKSs are classified into types I, II, and III and are involved in the synthesis of numerous compounds (Nivina et al., 2019). Through the sequential action of multiple catalytic modules, type I modular polyketide synthases are capable of extending, modifying, and terminating polyketide peptide chains. These interrelated modules interact with each other through docking domains (DDs) mediated by folding regions at the C- and N-termini (Weissman, 2016). Sun et al. (2022) utilized the DDs of type I cis-AT-PKS as mediators to develop a multi-enzyme assembly strategy named mimic PKS enzyme assembly line (mPKSeal), which mimics the assembly line of PKS enzymes (Figure 1B). This strategy was applied in engineered E. coli to enhance astaxanthin production and possesses the ability to co-locate enzymes within the cell, enabling the assembly of two or three enzyme units in different cellular environments (Sun et al., 2022). Their research also found that DDs from different PKSs but located on the same molecular evolutionary tree also possess enzyme assembly activity. The mPKSeal enzyme assembly strategy has tremendous potential for enhancing the efficiency of biocatalytic reactions by regulating the spatial positioning of enzymes without altering their abundance. These short-chain DDs have little significant impact on the catalytic activity of most enzyme assemblies, but they have a more pronounced effect on certain specific membrane proteins. Thus, this is an issue that needs to be noted when dealing with membrane proteins (Sun et al., 2022).
Kang et al. (2019) developed a scaffold-free modular enzyme assembly, which incorporated short peptide tags RIDD and RIAD derived from cAMP-dependent protein kinase (PKA) (Wong and Scott, 2004) and the A kinase-anchoring proteins (AKAPs) (Sarma et al., 2010), respectively. In E. coli, researchers successfully assembled enzyme complexes by combining the interaction peptides of RIAD and RIDD with the isopentenyl diphosphate isomerase (IDI) and Geranylgeranyl diphosphate synthase (CrtE), involved in the carotenoid biosynthesis pathway. This led to a significant increase in the production of carotenoids. Furthermore, in S. cerevisiae, the assembly of these two short peptides with the IDI and CrtE for the biosynthesis of lycopene resulted in a 58% increase in lycopene production (Kang et al., 2019). Xu et al. (2022) assembled two cytochrome P450 enzymes, ent-kaurene oxidase (KO) and kaurenoic acid 13α-hydroxylase (KAH), using RIAD and RIDD, successfully increasing the production of rubusoside and rebaudiosides in yeast. Fink et al. (2020), through orthogonally designed coiled-coil interaction domains, cluster resveratrol biosynthetic pathway enzymes, thereby increasing the yield of resveratrol in E. coli. The yield of resveratrol produced by this method is higher than that of direct enzyme fusion and internal protein-mediated fusion. At the same time, the biosynthesis of mevalonate in yeast was improved by this clustering method.
2.3 Protein-protein pair
Protein-protein pairs of special peptides in TatB/TatC can spontaneously interact to form aggregates. Henriques de Jesus et al. (2017) realized the co-localization of enzymes by exchanging the membrane anchors of the dhurrin biosynthesis pathway enzymes into TatB and TatC components of the twin-arginine translocation pathway with self-assembly properties. This method achieved a 4-fold increase in dhurrin titer and reduced the amount of intermediates and side products. CipA and CipB are two small proteins that form protein crystalline inclusions (PCIs) in the cytoplasm of Photorhabdus luminescens. Wang Y. et al. (2017) used CipA as a protein scaffold to bring together multiple enzymes (Vio enzymes) of the violacein biosynthetic pathway to explore its application in vivo. They found that the violacein production in the complex was significantly increased with fewer side-products (Wang Y. et al., 2017). More recently, Park et al. (2022) applied CipB scaffold proteins to bring P450s and reductase in close proximity, facilitating electron transfer between them. The development of strains producing lutein, apigenin, (+)-nootkatone, and L-3, 4-dihydroxyphenylalanine (l-DOPA) in E. coli has demonstrated the universal applicability of this electronic channel strategy. By using an implicit negative design, Sahtoe et al. (2022) generated beta sheet-mediated heterodimers capable of assembling into a variety of complexes. Their implicit negative design principle makes it possible to design higher-order asymmetric polyprotein complexes by rigid fusion of components through structured helical linkers. Moreover, due to the small size of the unfused protomers, the complex can be functionalized by easily fusing with the protein of interest by subunits (Sahtoe et al., 2022).
The scaffold assembly can realize the orderly arrangement of multiple enzymes, shorten the spatial distance of enzymes, accelerate sequential catalysis, and achieve a high yield. The composite formed by the scaffold assembly strategy can significantly improve the efficiency of enzyme catalysis in the biosynthesis of natural products and has broad application prospects in the fields of metabolic engineering and synthetic biology. However, until now, only four enzymes could be assembled sequentially. The reason is that the assembly of the enzyme requires the fusion expression of the scaffold and the enzyme through the joint, and misfolding is easy to occur during the fusion process, which will affect the assembly performance. On the other hand, the assembly of multiple enzymes is affected by steric hindrance, making it difficult to achieve sequential arrangement (Chen et al., 2023).
3 Nucleic acid scaffold
3.1 DNA scaffold
In addition to using protein scaffold approaches, the DNA double helix can serve as an alternative scaffold system. Compared with protein scaffolds, nucleic acid scaffolds have higher flexibility and maneuverability. With the advancement of gene editing technology, several molecular tools are available for efficient and specific DNA targeting, such as zinc finger proteins (ZFPs), transcription activator-like effector (TALE) proteins, and CRISPR-Cas (Kim and Kim, 2014). Moreover, the plasmid DNA as a configurable, stable, and robust scaffold for arranging biosynthetic enzymes in the cytoplasm is proposed.
3.1.1 DNA scaffold based on zinc finger protein
In E. coli, the plasmid DNAs equipped with corresponding zinc finger protein binding sites were designed to assemble three different biosynthetic pathways to produce resveratrol, 1,2-propanediol, or mevalonate (Conrado et al., 2012). By varying the enzymatic ratios and the base pairs between each enzyme, the catalytic efficiency is improved, which leads to better production of final products. This similar approach had been applied to using zinc finger proteins (ZFPs) as adaptors to anchor the L-threonine biosynthetic genes. By using DNA scaffold assembly, the accumulation of the intermediate homoserine is reduced and significantly increases the efficiency of L-threonine biosynthesis due to the shortening of the distance between enzymes and the enhancement of the local concentration of metabolic products (Lee et al., 2013). In addition, Rahmana et al. (2014) utilized fusion proteins of chimeric acyl-ACP reductase (AAR) and aldehyde decarbonylase (ADO), or zinc finger proteins, as guides to assemble ADO/AAR with DNA scaffolds. The strain containing the fusion protein ADO-AAR showed a 4.8-fold increase in the production of branched alkanes. On the DNA scaffold, when the stoichiometric ratio of ADO to AAR was 3:1, the strain exhibited an 8.8-fold increase in production, reaching the optimal level of branched alkane synthesis (Rahmana et al., 2014). Liu et al. (2014) constructed a B. subtilis strain capable of producing N-acetylglucosamine (GlcNAc) and, for the first time, utilized DNA scaffolds to regulate the activities of glucosamine-6-phosphate synthase and GlcNAc-6-phosphate N-acetyltransferase, resulting in a GlcNAc titer of 4.55 g/L.
3.1.2 DNA scaffold based on TALEs
TALEs (transcription activator-like effectors) are effectors from the family III in Xanthomonas bacteria (Boch and Bonas, 2010), and different TALEs share similar structural domains. These domains are capable of binding to the host cell genome and act as transcription factors to recognize specific DNA repeat sequences (Boch et al., 2009). Based on the binding characteristics of TALEs to DNA, Zhu et al. (2016) developed a TALE-based DNA scaffold system and applied it to the biosynthesis of indole-3-acetic acid (IAA). Furthermore, in a modified TALE-DNA scaffold system, three fusion enzymes were successfully assembled in E. coli and significantly increased the production of a mevalonate-producing tri-enzymatic pathway (Xie et al., 2019) (Figure 2).
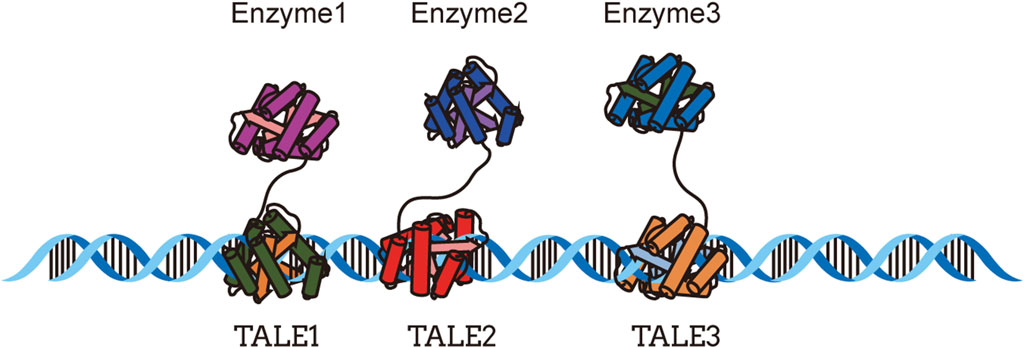
FIGURE 2. DNA scaffold system based on TALE, adapted from (Xie et al., 2019).
3.1.3 DNA scaffold based on CRISPR-Cas
CRISPR-associated (Cas) nucleases are a class of DNA-binding proteins distinct from zinc finger enzymes and transcriptional activator-like effector proteins. Through the guidance of RNA molecules complementary to DNA sequences, the CRISPR-Cas DNA scaffold achieves Cas-specific customization (Lim et al., 2020). The Cas9 protein is ideally suited for modular enzyme assembly, with a high affinity for DNA and the ability to bind to specific DNA sequences (Berckman and Chen, 2019). By combining the dCas9 nuclease (from Streptococcus pyogenes) with the Spycatcher-Spytag chemical binding system, successful modular assembly of five enzymatic pathways involved in violacein biosynthesis was achieved, resulting in a significant increase in violacein production (Lim et al., 2020). In another study, two orthogonal SpyCatcher and SnoopCatcher pairs were bioconjugated onto two different dCas9 proteins, enabling them to guide the enzyme assembly to the DNA scaffold, resulting in a 2.8-fold increase in reducing sugar production compared to the unassembled enzyme (Berckman and Chen, 2020).
3.1.4 DNA scaffold based on PCNA
Proliferating cell nuclear antigen (PCNA) is a trimeric ring-shaped protein (Moldovan et al., 2007) that binds to DNA as a scaffold for DNA-related enzymes. The fusion protein between the PCNA and the functional protein can act as a nanoscale part and self-assemble to form a functional nanohybrid complex. Fusion of three PCNA proteins with bacterial cytochrome P450 or one of the two electron transfer-related proteins can form a stable heterotrimer complex, resulting in increased local ferridoxin concentrations of P450 and ferredoxin reductase and high catalytic activity of electron transfer within the complex (Hirakawa and Nagamune, 2010).
3.2 RNA scaffold
By binding to the adapter, the RNA scaffold achieves highly specific binding to the target enzyme. Delebecque et al. (2011) designed and assembled multidimensional RNA structures to spatially organize proteins in cells and utilized this RNA scaffold to optimize a biosynthetic pathway for hydrogen production. A fluorescent protein library containing 8 aptamers and corresponding RNA domains was successfully assembled by fusing the active viral scaffold made of RNA with engineered proteins and specific RNA domains. The scaffold enables the co-localization of fragmented green fluorescent proteins to achieve precise measurement of cellular activity (Sachdeva et al., 2014). The application of this RNA scaffold to the synthesis pathway enzymes of pentadecane and succinic acid demonstrated the assembly of 0D, 1D, and 2D scaffolds. In the biosynthesis pathway of pentadecane, the 2D-assembled scaffold resulted in a 2.4-fold increase in pentadecane production (Sachdeva et al., 2014).
CRISPR-associated RNA scaffolds provide a powerful approach to the construction of synthetic gene programs. By inducing the expression of the dCas9 protein, we can achieve gene activation and inhibition, thereby enabling the directed expression of complex branching metabolic pathways (Zalatan et al., 2015). Pothoulakis et al. (2022) developed an RNA design approach for RNA origami scaffolds (termed sgRNAO) by recruiting activation domains from fused single-guide RNAs and RNA origami scaffolds to control gene expression in yeast. They successfully applied sgRNAOs to regulate the expression of enzymes involved in the violacein biosynthetic pathway (Pothoulakis et al., 2022).
Compared to DNA or protein-based scaffolds, RNA scaffolds, as non-coding synthetic scaffolds, offer greater flexibility. They can control protein spatial organization, such as distances and orientations between bound proteins, chemical dosage, and complex sizes, among others (Delebecque et al., 2012). However, although RNA scaffolds have certain advantages, they also have obvious disadvantages, including high synthesis costs, easy hydrolysis by nucleases, large environmental factors, and structural instability, which limit their application. Therefore, it requires further research and development to resolve these drawbacks (Geraldi et al., 2021).
4 Cellular scaffolds
4.1 Natural cellular scaffolds
As subcellular structures within cells, the integrity and autonomy of organelles have sparked scientists interest in using them as scaffolds for enzyme assembly (Liu et al., 2023). Bacterial microcompartments (BMCs) (Kerfeld et al., 2018) are self-assembling organelles composed of enzymatic cores that participate in the metabolism of various organic compounds such as 1,2-propanediol (Petit et al., 2013), ethanolamine (Petit et al., 2013), fucose, and rhamnose (Parsons et al., 2008), playing a crucial role in carbon fixation processes (Lawrence et al., 2014). In E. coli, reconstitution of recombinant microcompartments can be achieved by translocating the entire propanediol utilization (Pdu) operon from Citrobacter freundii (Parsons et al., 2008). The Zymomonas mobilis enzymes pyruvate decarboxylase (Pdc) and alcohol dehydrogenase (Adh) can be targeted to PduP of C. freundii to form a simple ethanol bioreactor inside the Pdu microcompartment shell (Lawrence et al., 2014). The enzymes Pdc and Adh, necessary for ethanol production, are expressed heterologously using a foreign host and targeted to the protein shell. In strains containing target enzymes, the ethanol yield significantly increases when the protein shell content is highest, including the strains producing shell proteins P18-Pdc and D18-Adh (Lawrence et al., 2014). In another study, the known Pdu (D18 and P18) targeting peptides were fused with four different 1,2-propanediol synthetic enzymes to create fusion proteins that target the empty Pdu BMC system. The fusion strategy of targeting peptides with all proteins involved in 1,2-propanediol synthesis significantly increased the product yield (Lee et al., 2016). Studies have shown that BMCs have great potential for constructing organelle scaffolds and are relatively easy to design, especially when it comes to the metabolism of toxic intermediates (Lawrence et al., 2014). Nielsen et al. (2013) transferred the biosynthetic pathway of the aromatic defense compound dhurrin [D-glucopyranosyloxy-(S)-p-hydroxymandelonitrile, a cyanogenic glucoside] to plant chloroplasts, utilizing photo-induced water splitting as the electron source to drive product synthesis in a light-dependent manner. The biosynthetic pathway of dhurrin involves three ER-localized enzymes, including two P450 enzymes, CYP79A1 and CYP71E1, as well as an NADPH cytochrome P450 oxidoreductase, POR. The chloroplast stroma provides a reducing environment for P450 enzymes, thereby enhancing their stability (Nielsen et al., 2013). Gram-negative bacteria release spherical nanoscale particles called outer membrane vesicles (OMVs) during their growth process. These vesicles have a composition similar to the bacterial outer membrane, containing lipopolysaccharides (LPSs), outer membrane proteins (OMPs), and phospholipids (Beveridge, 1999). In order to achieve the goal of hijacking the bacterial cell export pathway to simultaneously produce, package, and release an active enzyme, phosphotriesterase (PTE), Alves et al. (2015) attempted to establish synthetic linkages between enzymes and proteins known to exist in the outer membrane. They used the SpyCatcher/SpyTag (SC/ST) bioconjugated system to connect OmpA proteins present in OMVs to phosphotriesterase from Brevundimonas diminuta. A PTE-SpyCatcher (PTE-SC) fusion protein and a SpyTag transmembrane porin protein (OmpA-ST) were constructed. The coexpression of OmpA-ST with PTE-SC not only reduced the toxicity of PTE and improved the overall PTE production level, but also enhanced the stability of packaging enzymes against repeated freeze-thaw cycles (Alves et al., 2015). By employing a truncated ice nucleation protein anchoring motif (INP) on OMVs, a trivalent protein scaffold containing three divergent cohesin domains was utilized for site-specific expression of a three-enzyme cascade, resulting in a 23-fold increase in glucose production (Park et al., 2014) (Figure 3A). Yang et al. (2021) developed a metabolically engineered strain of E. coli to produce seven natural colorants, and they significantly increased the yield of seven natural colorants through cell morphological engineering, IMV and OMV formation, and fermentation optimization strategies. S. cerevisiae possesses various subcellular compartments, making it an ideal host for building heterologous natural product biosynthesis. Shi et al. (2021) used the PLN1 protein to target endoplasmic reticulum-localized cytochrome P450 enzymes and protopanaxadiol (PPD) synthase (PPDs) towards lipid droplets [DDs, the storage organelle for dammarenediol-II (DD)], resulting in a 394% increase in the conversion of DD to PPD and an elevated conversion rate of DD to 86.0%. Peroxisomes are organelles involved in fatty acid degradation. In yeast, significant improvements in the production of fatty acid derivatives, such as fatty alcohols, alkanes, and olefins, can be achieved by engineering peroxisomes. For example, increasing the number of peroxisomes can triple the production of fatty acid derivatives (Zhou et al., 2016).
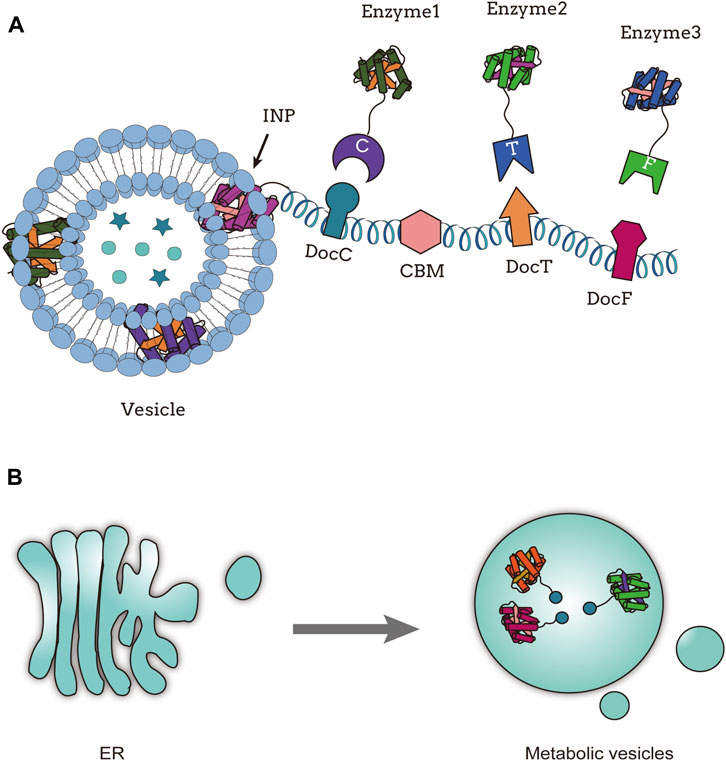
FIGURE 3. Organelle scaffold; (A) Functional assembly of multienzyme systems on outer membrane vesicles, adapted from (Park et al., 2014); (B) Compartmental assembly of endoplasmic reticulum derived metabolic vesicles, adapted from (Reifenrath et al., 2020).
4.2 Synthetic organelle scaffold
Cellular organelle scaffolds can be artificially designed to meet various metabolic pathway requirements. Lipids are widely present in cells and can form cell membranes, with many proteins anchored to these membrane structures. Inspired by this, researchers have attempted to use lipids as synthetic scaffolds to achieve co-assembly of lipids and target proteins. They discovered that, unlike most bacteriophages, bacteriophage ϕ6 contains a protein nucleocapsid surrounded by a lipid envelope and several membrane proteins (Sinclair et al., 1975). During infection of the natural host (Stitt and Mindich, 1983) and in strains of E. coli expressing genes encoding bacteriophages ϕ6 viral proteins (Johnson and Mindich, 1994), assembly intermediates of bacteriophage ϕ6 with lipid-like structures were found. When only genes encoding the three viral proteins P8, P9, and P12 are expressed in E. coli, circular particles composed of a mixture of lipids and proteins can be observed by cryo-electron microscopy (Sarin et al., 2012). Myhrvold et al. (2016) engineered synthetic lipid-containing scaffolds (SLSs) in E. coli. The scaffold consists of the membrane protein P9 and the non-structural protein P12, which are required for the formation of the particle structure. The target proteins are fused to the C-terminus of P9 to position them on the scaffold. TnaA and FMO enzymes involved in indigo biosynthesis were fused to the C-terminus of P9 to construct lipid scaffolds for increased indigo production (Myhrvold et al., 2016). By utilizing a protein scaffold based on the plant lipid droplet protein oleosin and cohesin-dockerin interaction pairs recruited upstream enzymes, the final three reaction steps of yeast ester biosynthesis were assembled on lipid droplets (LDs) within yeast cells. This resulted in a doubling of the synthesis rate of ethyl acetate (Lin et al., 2017). There are derived vesicles containing metabolic pathway enzymes in the endoplasmic reticulum (“metabolic vesicles”). Reifenrath et al. (2020) integrate the three enzymes involved in the production of cis,cis-muconic acid into yeast vesicles, construct an ER-derived synthetic cell envelope containing the metabolic pathway, and map the yeast metabolism (Figure 3B). Wei et al. (2022) used β-Cav1 caveolar vesicles as an enzyme assembly scaffold to immobilize enzymes involved in the biosynthesis of α-farnesene from isopentenyl diphosphate and dimethylallyl pyrophosphate through non-covalent interactions or covalent protein reactions on the β-Cav1 caveolar vesicles. They successfully constructed a multienzyme complex called multi-enzyme caveolar membranes (MCMs), which increased the catalytic efficiency of α-farnesene by 10-fold compared to the non-assembled enzymes (Wei et al., 2022).
5 Discussion and summary
With the advancement of biotechnology, building synthetic pathways into heterologous microbial hosts became possible (Jiang et al., 2021). However, this none-native expression of catalytic enzymes still faces numerous issues and challenges, including low productivity and yield, rapid diffusion and degradation of key intermediates, and the accumulation of toxic metabolites (Jiang et al., 2021). To address these issues, scientists have been organizing these enzymes into molecular complexes in space to enhance the local concentration of enzymes and metabolites, thereby improving reaction flux (Wang S. Z. et al., 2017). Currently, various assembly strategies have been derived based on the interactions of biomacromolecules such as nucleic acids, peptides, and scaffold proteins (Table 1). In comparison, the spatial organization of biosynthetic pathway enzymes through synthetic scaffolds has proven to be an effective method for enhancing reaction rates and biosynthetic yields while also improving host viability. Despite achieving a certain level of success, the precise prediction of artificial scaffold assembly remains challenging due to the complexity of enzyme structures. Furthermore, the catalytic efficiency of multi-enzyme complexes is influenced by numerous parameters, and the impact mechanism of linker sequences on both the multi-enzyme complex and substrate channeling effects remains undisclosed. Therefore, further research is needed to address the factors in the design-built-test-learn (DBTL) cycles of establishing artificial scaffold systems.
The computational inference of biological systems has emerged as a transformative field, driven by the confluence of advanced computational methods, machine learning techniques, and ever-expanding biological data. The best-known case is protein structure prediction, and it has been a grand challenge for decades. Since the advancement of the computational implication of a protein’s spatial arrangement of atoms and computing power, several artificial intelligence (AI) systems have been successfully applied to predict the structure of protein complexes (e.g., the AlphaFold-Multimer, RoseTTAFolds, and trRosetta) (Baek et al., 2021; Du et al., 2021; Brems et al., 2022; Ivanov et al., 2022). In addition, the use of artificial intelligence computational methods can effectively find scaffold proteins from protein interactors and fully reveal their functions (Oh and Yi, 2016). Currently, we may de novo design protein structures for specific purposes (Watson et al., 2023).
In contrast to protein-based scaffold systems, using nucleic acid as a scaffold provides its unique advantages. The DNA scaffold systems are more stable, easier to design, and have higher stability (Chen et al., 2014; Siu et al., 2015). However, the DNA scaffolds also serve several drawbacks, such as high cost and high error rates in self-assembly, making the folding process in living cells susceptible to environmental factors such as temperature and ions and difficult to manipulate. In addition, the DNA topology is single in dimension and has the problem of supercoiling (Polka et al., 2016). DNA origami is a technology for designing complex three-dimensional DNA structures. This has been broadly applied in nanotechnology, creating nanoscale structures for various purposes, including drug delivery and molecular computing (Zhou et al., 2023). Thus, DNA origami could greatly assist in resolving the issues and unlocking the great potential of using DNA scaffolds in metabolic engineering. Alternatively, the RNA can also be used as a scaffold for the self-assembly of multi-enzyme complexes. However, its discrete structure prevents the formation of complex geometric shapes in living organisms, limiting its application compared to other types of scaffolds (Lv et al., 2020). AI models also open up new opportunities for predicting the secondary and tertiary structures of RNA molecules, providing insights into designing new functions and interactions and improving stability (Sato and Hamada, 2023).
Cellular scaffolds constructed based on different organelles or membranes have achieved varying degrees of success. However, due to the levels of complexity of biological parts (e.g., membranes, proteins, and protein interactions), the working mechanism of such scaffold systems still needs to be further explored. Synthetic biology aims to combine multidisciplinary disciplines that pursue the development new biological parts or systems. The promise of using AI technology to resolve complex biological questions is now is gradually revealed. Combining these two technologies, we can better understand the mechanisms of different types of artificial scaffolds and design streamlined artificial scaffolds that provide a broader range of engineering applications.
Author contributions
NL: Writing–review and editing, Writing–original draft. WD: Writing–review and editing, Supervision. HY: Funding acquisition, Writing–review and editing, Supervision. J-HL: Supervision, Writing–review and editing. T-YC: Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by HIM-BGI Omics Center, Zhejiang Cancer Hospital, Hangzhou Institute of Medicine (HIM), Chinese Academy of Sciences (CAS), Hangzhou, China 310022. The funder did not participate in the designing, performing, or reporting of the current study.
Acknowledgments
Sincerely thanks to teacher T-YC for revising and polishing the whole article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alves, N. J., Turner, K. B., Daniele, M. A., Oh, E., Medintz, I. L., and Walper, S. A. (2015). Bacterial nanobioreactors--directing enzyme packaging into bacterial outer membrane vesicles. ACS Appl. Mater Interfaces 7 (44), 24963–24972. doi:10.1021/acsami.5b08811
Artzi, L., Bayer, E. A., and Moraïs, S. (2017). Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 15 (2), 83–95. doi:10.1038/nrmicro.2016.164
Baek, J. M., Mazumdar, S., Lee, S. W., Jung, M. Y., Lim, J. H., Seo, S. W., et al. (2013). Butyrate production in engineered Escherichia coli with synthetic scaffolds. Biotechnol. Bioeng. 110 (10), 2790–2794. doi:10.1002/bit.24925
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373 (6557), 871–876. doi:10.1126/science.abj8754
Berckman, E. A., and Chen, W. (2019). Exploiting dCas9 fusion proteins for dynamic assembly of synthetic metabolons. Chem. Commun. (Camb) 55 (57), 8219–8222. doi:10.1039/c9cc04002a
Berckman, E. A., and Chen, W. (2020). A modular approach for dCas9-mediated enzyme cascading via orthogonal bioconjugation. Chem. Commun. (Camb) 56 (77), 11426–11428. doi:10.1039/d0cc04196c
Beveridge, T. J. (1999). Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181 (16), 4725–4733. doi:10.1128/jb.181.16.4725-4733.1999
Boch, J., and Bonas, U. (2010). Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. doi:10.1146/annurev-phyto-080508-081936
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326 (5959), 1509–1512. doi:10.1126/science.1178811
Brems, M. A., Runkel, R., Yeates, T. O., and Virnau, P. (2022). AlphaFold predicts the most complex protein knot and composite protein knots. Protein Sci. 31 (8), e4380. doi:10.1002/pro.4380
Castellana, M., Wilson, M. Z., Xu, Y., Joshi, P., Cristea, I. M., Rabinowitz, J. D., et al. (2014). Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat. Biotechnol. 32 (10), 1011–1018. doi:10.1038/nbt.3018
Chen, R., Chen, Q., Kim, H., Siu, K. H., Sun, Q., Tsai, S. L., et al. (2014). Biomolecular scaffolds for enhanced signaling and catalytic efficiency. Curr. Opin. Biotechnol. 28, 59–68. doi:10.1016/j.copbio.2013.11.007
Chen, Z., Wu, T., Yu, S., Li, M., Fan, X., and Huo, Y. X. (2023). Self-assembly systems to troubleshoot metabolic engineering challenges. Trends Biotechnol. doi:10.1016/j.tibtech.2023.06.009
Conrado, R. J., Varner, J. D., and DeLisa, M. P. (2008). Engineering the spatial organization of metabolic enzymes: mimicking nature's synergy. Curr. Opin. Biotechnol. 19 (5), 492–499. doi:10.1016/j.copbio.2008.07.006
Conrado, R. J., Wu, G. C., Boock, J. T., Xu, H., Chen, S. Y., Lebar, T., et al. (2012). DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res. 40 (4), 1879–1889. doi:10.1093/nar/gkr888
Delebecque, C. J., Lindner, A. B., Silver, P. A., and Aldaye, F. A. (2011). Organization of intracellular reactions with rationally designed RNA assemblies. Science 333 (6041), 470–474. doi:10.1126/science.1206938
Delebecque, C. J., Silver, P. A., and Lindner, A. B. (2012). Designing and using RNA scaffolds to assemble proteins in vivo. Nat. Protoc. 7 (10), 1797–1807. doi:10.1038/nprot.2012.102
Du, Z., Su, H., Wang, W., Ye, L., Wei, H., Peng, Z., et al. (2021). The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 16 (12), 5634–5651. doi:10.1038/s41596-021-00628-9
Dueber, J. E., Wu, G. C., Malmirchegini, G. R., Moon, T. S., Petzold, C. J., Ullal, A. V., et al. (2009). Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27 (8), 753–759. doi:10.1038/nbt.1557
Fanning, A. S., and Anderson, J. M. (1996). Protein-protein interactions: PDZ domain networks. Curr. Biol. 6 (11), 1385–1388. doi:10.1016/s0960-9822(96)00737-3
Fink, T., Stevovic, B., Verwaal, R., Roubos, J. A., Gaber, R., Bencina, M., et al. (2020). Metabolic enzyme clustering by coiled coils improves the biosynthesis of resveratrol and mevalonate. Amb. Express 10 (1), 97. doi:10.1186/s13568-020-01031-5
Gad, S., and Ayakar, S. (2021). Protein scaffolds: a tool for multi-enzyme assembly. Biotechnol. Rep. (Amst) 32, e00670. doi:10.1016/j.btre.2021.e00670
Gao, X., Yang, S., Zhao, C., Ren, Y., and Wei, D. (2014). Artificial multienzyme supramolecular device: highly ordered self-assembly of oligomeric enzymes in vitro and in vivo. Angew. Chem. Int. Ed. Engl. 53 (51), 14027–14030. doi:10.1002/anie.201405016
Geraldi, A., Khairunnisa, F., Farah, N., Bui, L. M., and Rahman, Z. (2021). Synthetic scaffold systems for increasing the efficiency of metabolic pathways in microorganisms. Biol. (Basel) 10 (3), 216. doi:10.3390/biology10030216
Henriques de Jesus, M. P. R., Zygadlo Nielsen, A., Busck Mellor, S., Matthes, A., Burow, M., Robinson, C., et al. (2017). Tat proteins as novel thylakoid membrane anchors organize a biosynthetic pathway in chloroplasts and increase product yield 5-fold. Metab. Eng. 44, 108–116. doi:10.1016/j.ymben.2017.09.014
Hirakawa, H., and Nagamune, T. (2010). Molecular assembly of P450 with ferredoxin and ferredoxin reductase by fusion to PCNA. Chembiochem 11 (11), 1517–1520. doi:10.1002/cbic.201000226
Ivanov, Y. D., Taldaev, A., Lisitsa, A. V., Ponomarenko, E. A., and Archakov, A. I. (2022). Prediction of monomeric and dimeric structures of CYP102A1 using AlphaFold2 and AlphaFold multimer and assessment of point mutation effect on the efficiency of intra- and interprotein electron transfer. Molecules 27 (4), 1386. doi:10.3390/molecules27041386
Ji, D., Li, J., Xu, F., Ren, Y., and Wang, Y. (2021). Improve the biosynthesis of baicalein and scutellarein via manufacturing self-assembly enzyme reactor in vivo. ACS Synth. Biol. 10 (5), 1087–1094. doi:10.1021/acssynbio.0c00606
Jiang, Y., Zhang, X., Yuan, H., Huang, D., Wang, R., Liu, H., et al. (2021). Research progress and the biotechnological applications of multienzyme complex. Appl. Microbiol. Biotechnol. 105 (5), 1759–1777. doi:10.1007/s00253-021-11121-4
Johnson, M. D., and Mindich, L. (1994). Plasmid-directed assembly of the lipid-containing membrane of bacteriophage phi 6. J. Bacteriol. 176 (13), 4124–4132. doi:10.1128/jb.176.13.4124-4132.1994
Kang, W., Ma, T., Liu, M., Qu, J., Liu, Z., Zhang, H., et al. (2019). Modular enzyme assembly for enhanced cascade biocatalysis and metabolic flux. Nat. Commun. 10 (1), 4248. doi:10.1038/s41467-019-12247-w
Kang, W., Ma, X., Kakarla, D., Zhang, H., Fang, Y., Chen, B., et al. (2022). Organizing enzymes on self-assembled protein cages for cascade reactions. Angew. Chem. Int. Ed. Engl. 61 (52), e202214001. doi:10.1002/anie.202214001
Kerfeld, C. A., Aussignargues, C., Zarzycki, J., Cai, F., and Sutter, M. (2018). Bacterial microcompartments. Nat. Rev. Microbiol. 16 (5), 277–290. doi:10.1038/nrmicro.2018.10
Kim, H., and Kim, J. S. (2014). A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15 (5), 321–334. doi:10.1038/nrg3686
Kim, S., Bae, S. J., and Hahn, J. S. (2016). Redirection of pyruvate flux toward desired metabolic pathways through substrate channeling between pyruvate kinase and pyruvate-converting enzymes in Saccharomyces cerevisiae. Sci. Rep. 6, 24145. doi:10.1038/srep24145
Lawrence, A. D., Frank, S., Newnham, S., Lee, M. J., Brown, I. R., Xue, W. F., et al. (2014). Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth. Biol. 3 (7), 454–465. doi:10.1021/sb4001118
Lee, J. H., Jung, S. C., Bui le, M., Kang, K. H., Song, J. J., and Kim, S. C. (2013). Improved production of L-threonine in Escherichia coli by use of a DNA scaffold system. Appl. Environ. Microbiol. 79 (3), 774–782. doi:10.1128/AEM.02578-12
Lee, M. J., Brown, I. R., Juodeikis, R., Frank, S., and Warren, M. J. (2016). Employing bacterial microcompartment technology to engineer a shell-free enzyme-aggregate for enhanced 1,2-propanediol production in Escherichia coli. Metab. Eng. 36, 48–56. doi:10.1016/j.ymben.2016.02.007
Li, T., Chen, X., Cai, Y., and Dai, J. (2018). Artificial Protein Scaffold System (AProSS): an efficient method to optimize exogenous metabolic pathways in Saccharomyces cerevisiae. Metab. Eng. 49, 13–20. doi:10.1016/j.ymben.2018.07.006
Lim, S., Kim, J., Kim, Y., Xu, D., and Clark, D. S. (2020). CRISPR/Cas-directed programmable assembly of multi-enzyme complexes. Chem. Commun. (Camb) 56 (36), 4950–4953. doi:10.1039/d0cc01174f
Lin, J. L., Zhu, J., and Wheeldon, I. (2017). Synthetic protein scaffolds for biosynthetic pathway colocalization on lipid droplet membranes. ACS Synth. Biol. 6 (8), 1534–1544. doi:10.1021/acssynbio.7b00041
Liu, F., Banta, S., and Chen, W. (2013). Functional assembly of a multi-enzyme methanol oxidation cascade on a surface-displayed trifunctional scaffold for enhanced NADH production. Chem. Commun. (Camb) 49 (36), 3766–3768. doi:10.1039/c3cc40454d
Liu, M., Wang, Y., Jiang, H., Han, Y., and Xia, J. (2023). Synthetic multienzyme assemblies for natural product biosynthesis. Chembiochem 24 (6), e202200518. doi:10.1002/cbic.202200518
Liu, Y., Zhu, Y., Ma, W., Shin, H. D., Li, J., Liu, L., et al. (2014). Spatial modulation of key pathway enzymes by DNA-guided scaffold system and respiration chain engineering for improved N-acetylglucosamine production by Bacillus subtilis. Metab. Eng. 24, 61–69. doi:10.1016/j.ymben.2014.04.004
Lumsden, J., and Coggins, J. R. (1977). The subunit structure of the arom multienzyme complex of Neurospora crassa. A possible pentafunctional polypeptide chain. Biochem. J. 161 (3), 599–607. doi:10.1042/bj1610599
Luo, Z., Pan, F., Zhu, Y., Du, S., Yan, Y., Wang, R., et al. (2022). Synergistic improvement of 5-aminolevulinic acid production with synthetic scaffolds and system pathway engineering. ACS Synth. Biol. 11 (8), 2766–2778. doi:10.1021/acssynbio.2c00157
Lv, X., Cui, S., Gu, Y., Li, J., Du, G., and Liu, L. (2020). Enzyme assembly for compartmentalized metabolic flux control. Metabolites 10 (4), 125. doi:10.3390/metabo10040125
Moldovan, G. L., Pfander, B., and Jentsch, S. (2007). PCNA, the maestro of the replication fork. Cell 129 (4), 665–679. doi:10.1016/j.cell.2007.05.003
Moon, T. S., Dueber, J. E., Shiue, E., and Prather, K. L. (2010). Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab. Eng. 12 (3), 298–305. doi:10.1016/j.ymben.2010.01.003
Myhrvold, C., Polka, J. K., and Silver, P. A. (2016). Synthetic lipid-containing scaffolds enhance production by colocalizing enzymes. ACS Synth. Biol. 5 (12), 1396–1403. doi:10.1021/acssynbio.6b00141
Nielsen, A. Z., Ziersen, B., Jensen, K., Lassen, L. M., Olsen, C. E., Møller, B. L., et al. (2013). Redirecting photosynthetic reducing power toward bioactive natural product synthesis. ACS Synth. Biol. 2 (6), 308–315. doi:10.1021/sb300128r
Nivina, A., Yuet, K. P., Hsu, J., and Khosla, C. (2019). Evolution and diversity of assembly-line polyketide synthases. Chem. Rev. 119 (24), 12524–12547. doi:10.1021/acs.chemrev.9b00525
Oh, K., and Yi, G. S. (2016). Prediction of scaffold proteins based on protein interaction and domain architectures. BMC Bioinforma., 17 (Suppl. 6), 220. doi:10.1186/s12859-016-1079-5
Park, M., Sun, Q., Liu, F., DeLisa, M. P., and Chen, W. (2014). Positional assembly of enzymes on bacterial outer membrane vesicles for cascade reactions. PLoS One 9 (5), e97103. doi:10.1371/journal.pone.0097103
Park, S. Y., Eun, H., Lee, M. H., and Lee, S. Y. (2022). Metabolic engineering of Escherichia coli with electron channelling for the production of natural products. Nat. Catal. 5 (8), 726–737. doi:10.1038/s41929-022-00820-4
Parsons, J. B., Dinesh, S. D., Deery, E., Leech, H. K., Brindley, A. A., Heldt, D., et al. (2008). Biochemical and structural insights into bacterial organelle form and biogenesis. J. Biol. Chem. 283 (21), 14366–14375. doi:10.1074/jbc.M709214200
Pawson, T. (2007). Dynamic control of signaling by modular adaptor proteins. Curr. Opin. Cell Biol. 19 (2), 112–116. doi:10.1016/j.ceb.2007.02.013
Petit, E., LaTouf, W. G., Coppi, M. V., Warnick, T. A., Currie, D., Romashko, I., et al. (2013). Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8 (1), e54337. doi:10.1371/journal.pone.0054337
Pham, V. D., Lee, S. H., Park, S. J., and Hong, S. H. (2015). Production of gamma-aminobutyric acid from glucose by introduction of synthetic scaffolds between isocitrate dehydrogenase, glutamate synthase and glutamate decarboxylase in recombinant Escherichia coli. J. Biotechnol. 207, 52–57. doi:10.1016/j.jbiotec.2015.04.028
Pham, V. D., Somasundaram, S., Lee, S. H., Park, S. J., and Hong, S. H. (2016). Redirection of metabolic flux into novel gamma-aminobutyric acid production pathway by introduction of synthetic scaffolds strategy in Escherichia coli. Appl. Biochem. Biotechnol. 178 (7), 1315–1324. doi:10.1007/s12010-015-1948-9
Polka, J. K., Hays, S. G., and Silver, P. A. (2016). Building spatial synthetic biology with compartments, scaffolds, and communities. Cold Spring Harb. Perspect. Biol. 8 (8), a024018. doi:10.1101/cshperspect.a024018
Pothoulakis, G., Nguyen, M. T. A., and Andersen, E. S. (2022). Utilizing RNA origami scaffolds in Saccharomyces cerevisiae for dCas9-mediated transcriptional control. Nucleic Acids Res. 50 (12), 7176–7187. doi:10.1093/nar/gkac470
Price, J. V., Chen, L., Whitaker, W. B., Papoutsakis, E., and Chen, W. (2016). Scaffoldless engineered enzyme assembly for enhanced methanol utilization. Proc. Natl. Acad. Sci. U. S. A. 113 (45), 12691–12696. doi:10.1073/pnas.1601797113
Qu, J., Cao, S., Wei, Q., Zhang, H., Wang, R., Kang, W., et al. (2019). Synthetic multienzyme complexes, catalytic nanomachineries for cascade biosynthesis in vivo. ACS Nano 13 (9), 9895–9906. doi:10.1021/acsnano.9b03631
Rahmana, Z., Sung, B. H., Yi, J. Y., Bui le, M., Lee, J. H., and Kim, S. C. (2014). Enhanced production of n-alkanes in Escherichia coli by spatial organization of biosynthetic pathway enzymes. J. Biotechnol. 192 Pt A, 187–191. doi:10.1016/j.jbiotec.2014.10.014
Reifenrath, M., Oreb, M., Boles, E., and Tripp, J. (2020). Artificial ER-derived vesicles as synthetic organelles for in vivo compartmentalization of biochemical pathways. ACS Synth. Biol. 9 (11), 2909–2916. doi:10.1021/acssynbio.0c00241
Sachdeva, G., Garg, A., Godding, D., Way, J. C., and Silver, P. A. (2014). In vivo co-localization of enzymes on RNA scaffolds increases metabolic production in a geometrically dependent manner. Nucleic Acids Res. 42 (14), 9493–9503. doi:10.1093/nar/gku617
Sahtoe, D. D., Praetorius, F., Courbet, A., Hsia, Y., Wicky, B. I. M., Edman, N. I., et al. (2022). Reconfigurable asymmetric protein assemblies through implicit negative design. Science 375 (6578), eabj7662. doi:10.1126/science.abj7662
Sarin, L. P., Hirvonen, J. J., Laurinmäki, P., Butcher, S. J., Bamford, D. H., and Poranen, M. M. (2012). Bacteriophage ϕ6 nucleocapsid surface protein 8 interacts with virus-specific membrane vesicles containing major envelope protein 9. J. Virol. 86 (9), 5376–5379. doi:10.1128/jvi.00172-12
Sarma, G. N., Kinderman, F. S., Kim, C., von Daake, S., Chen, L., Wang, B. C., et al. (2010). Structure of D-AKAP2:PKA RI complex: insights into AKAP specificity and selectivity. Structure 18 (2), 155–166. doi:10.1016/j.str.2009.12.012
Sato, K., and Hamada, M. (2023). Recent trends in RNA informatics: a review of machine learning and deep learning for RNA secondary structure prediction and RNA drug discovery. Brief. Bioinform 24 (4), bbad186. doi:10.1093/bib/bbad186
Schlessinger, J. (1994). SH2/SH3 signaling proteins. Curr. Opin. Genet. Dev. 4 (1), 25–30. doi:10.1016/0959-437x(94)90087-6
Shi, Y., Wang, D., Li, R., Huang, L., Dai, Z., and Zhang, X. (2021). Engineering yeast subcellular compartments for increased production of the lipophilic natural products ginsenosides. Metab. Eng. 67, 104–111. doi:10.1016/j.ymben.2021.06.002
Sinclair, J. F., Tzagoloff, A., Levine, D., and Mindich, L. (1975). Proteins of bacteriophage phi6. J. Virol. 16 (3), 685–695. doi:10.1128/jvi.16.3.685-695.1975
Siu, K. H., Chen, R. P., Sun, Q., Chen, L., Tsai, S. L., and Chen, W. (2015). Synthetic scaffolds for pathway enhancement. Curr. Opin. Biotechnol. 36, 98–106. doi:10.1016/j.copbio.2015.08.009
Somasundaram, S., Jeong, J., Irisappan, G., Kim, T. W., and Hong, S. H. (2020). Enhanced production of malic acid by Co-localization of phosphoenolpyruvate carboxylase and malate dehydrogenase using synthetic protein scaffold in Escherichia coli. Biotechnol. Bioprocess Eng. 25 (1), 39–44. doi:10.1007/s12257-019-0269-1
Srinivasan, P., and Smolke, C. D. (2020). Biosynthesis of medicinal tropane alkaloids in yeast. Nature 585 (7826), 614–619. doi:10.1038/s41586-020-2650-9
Stitt, B. L., and Mindich, L. (1983). Morphogenesis of bacteriophage phi 6: a presumptive viral membrane precursor. Virology 127 (2), 446–458. doi:10.1016/0042-6822(83)90157-5
Sun, X., Yuan, Y., Chen, Q., Nie, S., Guo, J., Ou, Z., et al. (2022). Metabolic pathway assembly using docking domains from type I cis-AT polyketide synthases. Nat. Commun. 13 (1), 5541. doi:10.1038/s41467-022-33272-2
Team, Z. J. C., and Chen, M. (2015). RNA scaffold: designed to Co-localize enzymes. Methods Mol. Biol. 1316, 105–112. doi:10.1007/978-1-4939-2730-2_9
Tippmann, S., Anfelt, J., David, F., Rand, J. M., Siewers, V., Uhlen, M., et al. (2017). Affibody scaffolds improve sesquiterpene production in Saccharomyces cerevisiae. ACS Synth. Biol. 6 (1), 19–28. doi:10.1021/acssynbio.6b00109
Tittes, Y. U., Herbst, D. A., Martin, S. F. X., Munoz-Hernandez, H., Jakob, R. P., and Maier, T. (2022). The structure of a polyketide synthase bimodule core. Sci. Adv. 8 (38), eabo6918. doi:10.1126/sciadv.abo6918
Tonikian, R., Zhang, Y., Sazinsky, S. L., Currell, B., Yeh, J. H., Reva, B., et al. (2008). A specificity map for the PDZ domain family. PLoS Biol. 6 (9), e239. doi:10.1371/journal.pbio.0060239
Tran, K.-N. T., Kumaravel, A., and Hong, S. H. (2023). Impact of the synthetic scaffold strategy on the metabolic pathway engineering. Biotechnol. Bioprocess Eng. 28 (3), 379–385. doi:10.1007/s12257-022-0350-z
Wang, L., Sun, Y., Lv, D., Liu, B., Guan, Y., and Yu, D. (2020). Protein scaffold optimizes arrangement of constituent enzymes in indigoidine synthetic pathway to improve the pigment production. Appl. Microbiol. Biotechnol. 104 (24), 10493–10502. doi:10.1007/s00253-020-10990-5
Wang, S. Z., Zhang, Y. H., Ren, H., Wang, Y. L., Jiang, W., and Fang, B. S. (2017a). Strategies and perspectives of assembling multi-enzyme systems. Crit. Rev. Biotechnol. 37 (8), 1024–1037. doi:10.1080/07388551.2017.1303803
Wang, Y., Heermann, R., and Jung, K. (2017b). CipA and CipB as scaffolds to organize proteins into crystalline inclusions. ACS Synth. Biol. 6 (5), 826–836. doi:10.1021/acssynbio.6b00323
Watson, J. L., Juergens, D., Bennett, N. R., Trippe, B. L., Yim, J., Eisenach, H. E., et al. (2023). De novo design of protein structure and function with RFdiffusion. Nature 620 (7976), 1089–1100. doi:10.1038/s41586-023-06415-8
Wei, Q., He, S., Qu, J., and Xia, J. (2020). Synthetic multienzyme complexes assembled on virus-like particles for cascade biosynthesis in cellulo. Bioconjug Chem. 31 (10), 2413–2420. doi:10.1021/acs.bioconjchem.0c00476
Wei, Q., Wang, Y., Liu, Z., Liu, M., Cao, S., Jiang, H., et al. (2022). Multienzyme assembly on caveolar membranes in cellulo. ACS Catal. 12 (14), 8372–8379. doi:10.1021/acscatal.2c01906
Weissman, K. J. (2016). Genetic engineering of modular PKSs: from combinatorial biosynthesis to synthetic biology. Nat. Prod. Rep. 33 (2), 203–230. doi:10.1039/c5np00109a
Wong, W., and Scott, J. D. (2004). AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5 (12), 959–970. doi:10.1038/nrm1527
Wu, J., Wang, X., Xiao, L., Wang, F., Zhang, Y., and Li, X. (2021). Synthetic protein scaffolds for improving R-(−)-Linalool production in Escherichia coli. J. Agric. Food Chem. 69 (20), 5663–5670. doi:10.1021/acs.jafc.1c01101
Xie, S. S., Qiu, X. Y., Zhu, L. Y., Zhu, C. S., Liu, C. Y., Wu, X. M., et al. (2019). Assembly of TALE-based DNA scaffold for the enhancement of exogenous multi-enzymatic pathway. J. Biotechnol. 296, 69–74. doi:10.1016/j.jbiotec.2019.03.008
Xu, Y., Wang, X., Zhang, C., Zhou, X., Xu, X., Han, L., et al. (2022). De novo biosynthesis of rubusoside and rebaudiosides in engineered yeasts. Nat. Commun. 13 (1), 3040. doi:10.1038/s41467-022-30826-2
Yang, D., Park, S. Y., and Lee, S. Y. (2021). Production of rainbow colorants by metabolically engineered Escherichia coli. Adv. Sci. (Weinh) 8 (13), e2100743. doi:10.1002/advs.202100743
Yang, Z., Gao, X., Xie, H., Wang, F., Ren, Y., and Wei, D. (2017). Enhanced itaconic acid production by self-assembly of two biosynthetic enzymes in Escherichia coli. Biotechnol. Bioeng. 114 (2), 457–462. doi:10.1002/bit.26081
Yuan, W., Jiang, C., Wang, Q., Fang, Y., Wang, J., Wang, M., et al. (2022). Biosynthesis of mushroom-derived type II ganoderic acids by engineered yeast. Nat. Commun. 13 (1), 7740. doi:10.1038/s41467-022-35500-1
Zalatan, J. G., Lee, M. E., Almeida, R., Gilbert, L. A., Whitehead, E. H., La Russa, M., et al. (2015). Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160 (1-2), 339–350. doi:10.1016/j.cell.2014.11.052
Zhang, J., Hansen, L. G., Gudich, O., Viehrig, K., Lassen, L. M. M., Schrubbers, L., et al. (2022). A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 609 (7926), 341–347. doi:10.1038/s41586-022-05157-3
Zhao, C., Gao, X., Liu, X., Wang, Y., Yang, S., Wang, F., et al. (2016). Enhancing biosynthesis of a ginsenoside precursor by self-assembly of two key enzymes in Pichia pastoris. J. Agric. Food Chem. 64 (17), 3380–3385. doi:10.1021/acs.jafc.6b00650
Zhou, Y., Dong, J., and Wang, Q. (2023). Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales. NPG Asia Mater. 15 (1), 25. doi:10.1038/s41427-023-00470-3
Zhou, Y. J., Buijs, N. A., Zhu, Z., Gómez, D. O., Boonsombuti, A., Siewers, V., et al. (2016). Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition. J. Am. Chem. Soc. 138 (47), 15368–15377. doi:10.1021/jacs.6b07394
Keywords: multienzyme complexes, enzyme molecular scaffolds, reaction microcompartments, microbial cell factory, synthetic scaffold
Citation: Liu N, Dong W, Yang H, Li J-H and Chiu T-Y (2023) Application of artificial scaffold systems in microbial metabolic engineering. Front. Bioeng. Biotechnol. 11:1328141. doi: 10.3389/fbioe.2023.1328141
Received: 26 October 2023; Accepted: 12 December 2023;
Published: 22 December 2023.
Edited by:
Luan Luong Chu, Phenikaa University, VietnamReviewed by:
Jin-Ho Lee, Kyungsung University, Republic of KoreaRichard Kelwick, Imperial College London, United Kingdom
Tianwen Wang, Anhui Polytechnic University, China
Copyright © 2023 Liu, Dong, Yang, Li and Chiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsan-Yu Chiu, qiucanyu@genomics.cn
 Nana Liu
Nana Liu Wei Dong2
Wei Dong2  Tsan-Yu Chiu
Tsan-Yu Chiu