Draft genome analysis for Enterobacter kobei, a promising lead bioremediation bacterium
- 1Agricultural Biotechnology Department, College of Agriculture and Food Sciences, King Faisal University, Al-Ahsa, Saudi Arabi
- 2Biochemistry Department, Faculty of Agriculture, Cairo University, Giza, Egypt
- 3Genetics Department, Faculty of Agriculture, Cairo University, Giza, Egypt
- 4Biology Department, College of Science and Humanities in Al-Kharj, Prince Sattam Bin Abdulaziz University, Al-Kharj, Saudi Arabia
- 5Department of Chemistry, College of Science, King Faisal University, Al-Ahsa, Saudi Arabia
- 6Microbiology Department, Faculty of Agriculture, Cairo University, Giza, Egypt
- 7College of Biotechnology, Misr University for Science and Technology (MUST), 6th October City, Egypt
Lead pollution of the environment poses a major global threat to the ecosystem. Bacterial bioremediation offers a promising alternative to traditional methods for removing these pollutants, that are often hindered by various limitations. Our research focused on isolating lead-resistant bacteria from industrial wastewater generated by heavily lead-containing industries. Eight lead-resistant strains were successfully isolated, and subsequently identified through molecular analysis. Among these, Enterobacter kobei FACU6 emerged as a particularly promising candidate, demonstrating an efficient lead removal rate of 83.4% and a remarkable lead absorption capacity of 571.9 mg/g dry weight. Furthermore, E. kobei FACU6 displayed a remarkable a maximum tolerance concentration (MTC) for lead reaching 3,000 mg/L. To further investigate the morphological changes in E. kobei FACU6 in response to lead exposure, scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were employed. These analyses revealed significant lead adsorption and intracellular accumulation in treated bacteria in contrast to the control bacterium. Whole-genome sequencing was performed to gain deeper insights into E. kobei’s lead resistance mechanisms. Structural annotation revealed a genome size of 4,856,454 bp, with a G + C content of 55.06%. The genome encodes 4,655 coding sequences (CDS), 75 tRNA genes, and 4 rRNA genes. Notably, genes associated with heavy metal resistance and their corresponding regulatory elements were identified within the genome. Furthermore, the expression levels of four specific heavy metal resistance genes were evaluated. Our findings revealed a statistically significant upregulation in gene expression under specific environmental conditions, including pH 7, temperature of 30°C, and high concentrations of heavy metals. The outstanding potential of E. kobei FACU6 as a source of diverse genes related to heavy metal resistance and plant growth promotion makes it a valuable candidate for developing safe and effective strategies for heavy metal disposal.
1 Introduction
While rapid technological advancements and industrialization contribute to fulfilling human needs and increasing their national income, they are considered the main source of hazardous waste material pollution, particularly heavy metals and other toxic compounds (Hubeny et al., 2021). Heavy metals, either essential (e.g., nickel (Ni), iron (Fe), manganese (Mn) and copper (Cu) or toxic [e.g., lead (Pb), mercury (Hg), arsenic (As), silver (Ag), cadmium (Cd) and chromium (Cr)], are generally harmful to living organisms even at low concentrations (Lemire et al., 2013). Likewise, it was reported that Pb, Hg, Cd, Cr, and As were considered the most toxic and carcinogenic metals (Mohamed et al., 2009; Masindi and Muedi, 2018). The toxicity of Pb leads to a variety of symptoms in the neurological system as well as the hematologic, hepatic, and renal systems (El-Beltagi and Mohamed, 2010; Flora et al., 2012). Pb also results in carcinogenesis, altered enzyme specificity, and disruption of cell membranes (Olaniran et al., 2013). Pb interacts with several proteins in renal cells, among them those that have been related to lead toxicity (Briffa et al., 2020).
Due to extensive industrial usage of metals, hazardous heavy metals have accumulated in the environment in significant amounts through effluents. These effluents are non-degradable, posing challenges for environmental disposal (Herawati et al., 2000). So, heavy metal accumulation negatively impacts the environment, especially when it enters the food chain (Tchounwou et al., 2012). The major sources of Pb in the environment include industrial processes, including the manufacturing of batteries, pigments, and metals, as well as the production of goods like lead arsenate pesticides or lead water pipes (Gadd, 2010). Therefore, it is crucial to investigate and identify an appropriate approach for treating this industrial wastewater before its discharge into the ecosystem, especially with the defects and problems that direct conventional treatment methods such as electrochemical removal, ion exchange and chemical precipitation (Gunatilake, 2015). Additionally, there is a need for effective and cheap technology to remove heavy metals using eco-friendly methods.
The use of biological agents as alternatives to chemical approaches for heavy metal removal has gained increasing attention. In this context, bioremediation can be regarded as a key, and effective method for ecological restoration and environmental cleanup (Abdelhaleem et al., 2019). Heavy metal-resistant bacteria have been isolated from different contaminated sites and studies on the interactions between these bacteria and heavy metals and other environmental pollutants to remove and reduce pollution hazards have been reported, such as lead (Mitra et al., 2021; Elarabi et al., 2023), chromium (Mohamed et al., 2020), cadmium (Saini and Dhania, 2020), arsenic (Balali-Mood et al., 2021), mercury (Kumari et al., 2020), nickel (Jan et al., 2019), herbicides (Elarabi et al., 2020) and polycyclic aromatic hydrocarbons (Abdelhaleem et al., 2019). There are five major mechanisms of heavy metal resistance (HMR) in the bacterial cell, including 1) extracellular barriers where the plasma membrane, cell wall, or capsule can prohibit the entry of metal ions into the cell; 2) metal ions active transport (efflux), which considers a method to export metal ions from the cytoplasm and includes three families of proteins [Cation Diffusion Facilitator (CDF), P-type ATPases and Resistance and Nodulation, Cell Division (RND)]; 3) extracellular sequestration, which contains insoluble chemicals or metal ions collected by biological components in the outer membrane; 4) intracellular sequestration, which prevents exposure to essential cellular components by accumulating metal ions in the cytoplasm in forms that are not bioavailable; and 5) reoxidation of metal ions (Choudhury and Srivastava, 2001).
Bacterial resistance mechanisms to heavy metals can be categorized into two main types: biochemical and molecular (Ma et al., 2016). The genetic determinants of resistance, encoded by genes located on bacterial chromosomes or plasmids, constitute the molecular mechanisms (von Rozycki and Nies, 2009). On the other hand, the bacterial resistance to heavy metals can be assessed by utilizing the bacteria’s Minimum Inhibitory Concentration (MIC) (Parameswari et al., 2010). Enterobacter plays great role in the bioremediation of multiple heavy metals (González-Henao and Ghneim-Herrera, 2021). It was reported that Enterobacter cloacae have high resistance levels to lead, cadmium, nickel and chromium (Pattnaik et al., 2020). Enterobacter mori and Enterobacter aerogenes were considered cadmium-resistant bacteria (Pramanik et al., 2018a). Otherwise, E. kobei was used as a lead-resistant bacterium (Abdollahi et al., 2020). Several recent advancements address the critical need to prevent the migration of heavy metals such as lead (Pb) and copper (Cu) into surrounding environments. These advancements include: Immobilization using loess and nanoscale zerovalent iron (nZVI). Wen et al. (2023) demonstrated the effectiveness of this method in immobilizing lead and reducing its mobility. Microbial Induced Carbonate Precipitation (MICP), this approach uses microbes to induce the precipitation of carbonates, which trap and immobilize heavy metals. Xue et al. (2023) and Xie et al. (2023a) further explored its application for remediating Pb-contaminated soil and water bodies. Biopolymer-assisted Enzyme-Induced Carbonate Precipitation (EICP), this technique utilizes enzymes and biopolymers to precipitate carbonates, effectively immobilizing heavy metals and preventing their migration. Xie et al. (2023b) highlighted its wide applicability for heavy metal remediation. Electrokinetic technology coupled with a biological permeable reactive barrier, this method uses electrical currents to drive the movement of contaminants towards a reactive barrier composed of biological materials, where heavy metals are removed and immobilized. Wang et al. (2023) studied its effectiveness in remediating heavy metal-contaminated loess. Combining these recent applications with the use of heavy metal-resistant bacteria has the potential to significantly improve the effectiveness of heavy metal removal.
The dramatic reduction in sequencing costs has made new genome sequencing technology increasingly available and affordable (Shendure and Ji, 2008). This has significantly enhanced our understanding of the biology of any organism, facilitated the identification of genome rearrangements, and simplified the investigation of novel genes during bacterial exposure to stress (Land et al., 2015). Also, genome analysis offers the opportunity to confirm the bacterial ability to resist unknown heavy metals (Huo et al., 2014). Additionally, this technique reflects the evolutionary dynamics of heavy metals and their relationship to environment (Ridge et al., 2008). Our investigation focused on isolating and characterizing a lead-resistant bacterium for bioremediation applications. By sequencing and analyzing its draft genome, we gained insights into the genetic mechanisms underlying its lead resistance. Moreover, the expression of four crucial genes associated with heavy metal resistance was studied.
2 Materials and methods
2.1 Sampling and evaluation of the samples’ physicochemical characteristics
Untreated industrial wastewater samples for bacterial isolation were collected from two sites in Sadat City, Menoufia Governorate, Egypt (30°26′30.6″N 30°38′24.7″E) in October 2019 (Supplementary Figure S1). The pH and electrical conductivity were measured following the method described by Elarabi et al. (2023) in triplicate. Using an atomic absorption spectrophotometer (Buck Model 210 VGP), the concentrations of As, Cd, Cr, Cu, Fe, Mn, Ni, Pb, and Zn ions were determined in the samples’ final solutions according to Page et al. (1983).
2.2 Pb-resistance bacterial isolation
The C4H6O4Pb, Na2HAsO4. 7H2O and K2Cr2O7 were purchased from Sigma and CdCo3 was purchased from Oxford laboratory. Pb-resistant bacteria were isolated and counted using the serial dilution method (Harley and Prescott, 2002). 0.1 mL of the diluted suspension was inoculated onto Luria Bertani (LB) media (10 g/L peptone, 5 g/L yeast extract, 10 g/L NaCl and 20 g/L agar: pH 7.00) supplemented with six different C4H6O4Pb concentrations (50, 100, 250, 500, 1,000, and 1,200 mg/L). Each isolate was assigned a lead resistance strain (LRS) name code.
2.3 Molecular identification
According to the manufacturer’s instructions, genomic DNA was purified from the bacterial isolates using the Simple™ Bacterial DNA Isolation Kit (Gene Direx, Inc., Cat. No. SN023-0100, Taiwan). Two universal primers targeting the 16S rRNA gene, 27F, and 1492R, were used for molecular identification (Supplementary Table S1). The PCR reaction was carried out in a 50 μL total volume containing the following components: 50 ng/μL DNA, 2X One PCR™ Master Mix (Gene Direx, Cat. No. MB203-0100, Taiwan), 10 pmol of each primer, and the remainder of the volume to 50 μL with nuclease-free water. The PCR conditions were as follows: an initial denaturation step at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 1 min, and extension at 72°C for 2 min. A final extension step at 72°C for 5 min was then performed. PCR products were purified using ExoSAP-IT™ PCR Product Cleanup Reagent (Applied Biosystems, United States, Cat. No. 78201). Purified DNA was sequenced by Sangon Biotech Co., Ltd, Macrogen, Korea. The 16S rRNA gene sequences of the isolates and their closely related strains were aligned using ClustalOmega version 1.2.4 (Madeira et al., 2019). The 16S rRNA gene sequence of each bacterial isolate was submitted to the National Center for Biotechnology Information (NCBI) GenBank database for comparison with published sequences. To further verify the identification, the sequences were also analyzed using the EzBioCloud Database (https://www.ezbiocloud.net/). Prior to phylogenetic analysis, the sequence alignment was trimmed using trimAl version 1.4.rev22 (Capella-Gutiérrez et al., 2009). Highly homologous sequences were selected and aligned using Clustal Omega. A phylogenetic tree was constructed using MEGA 11 software (Tamura et al., 2021) employing the Maximum Likelihood method under the Kimura 2-parameter model. Bootstrap analysis with 1,000 replications was performed to assess the confidence of the tree branches.
2.4 Estimation of the effect of Pb on cell survival, Pb biosorption efficiency and Pb uptake
The lead (Pb) resistance of bacterial isolates was measured using both the Minimum Inhibition Concentration (MIC) and the Maximum Tolerance Concentration (MTC) (Malik and Jaiswal, 2000). Bacterial isolates were cultured on LB agar plates supplemented with 10 different concentrations of lead acetate (ranging from 1,400 to 2,800 mg/L). Plates were incubated at 30°C for 15 days (Elarabi et al., 2023). After incubation, colony-forming units (CFUs) were counted to determine the survival of bacterial isolates. To assess the Pb removal ability of the isolates, LB broth supplemented with 2,600 mg/L lead acetate was inoculated with each isolate (this concentration was chosen as it is suitable and not stressful for all bacterial isolates). Pb absorption levels (mg/L), residual Pb ion concentration in supernatants, and Pb biosorption percentage were determined using inductively coupled plasma atomic emission spectroscopy (ICP-AES), following the method described by Shetty and Rajkumar (2009). Additionally, the survival and suppression percentages were calculated as described by Elarabi et al. (2023).
2.5 Microscopic analysis
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analyses were conducted at the Cairo University Research Park (CURP), Faculty of Agriculture, Cairo University, Giza, Egypt, to determine potential Pb stress cells exhibit any structural changes and where metal accumulated. For SEM, a drop of the suspension from the bacterial culture, both with and without Pb, was placed on an aluminum stub and subjected to the procedures outlined by Khan et al. (2016). The gold film was then applied to samples using a sputter coater (Denton, Desk V HP) and viewed under a SEM (Nova NanoSEM 450). The TEM for the Pb-treated and untreated bacteria was carried out using a Tecnai G 20 transmission electron microscope (FEI, Limeil-Brevannes, France) SA 9900 at 200 kV (Burghardt and Droleskey, 2006). Images were captured using a high-resolution digital CCD camera and Olympus Soft Imaging System’s (Germany) iTEM image processing software.
2.6 Draft genome analysis
Following the manufacturer’s instructions, total DNA was extracted from E. kobei FACU6 using the QIAamp® DNA Minikit (QIAGEN, Cat. No. 51304, Germany). The bacterium’s genome was then sequenced using the Illumina MiSeq™ platform (Illumina, United States) at the Genomics Research Program at the Children’s Cancer Hospital-Egypt 57357, Cairo, Egypt, employing NextGen High Throughput Sequencing technology. A conventional Illumina shotgun library, Nextera XT DNA Library Prep, was constructed and sequenced using the MiSeq platform. Paired-end sequencing generated 363,533 reads with an average length of 2 × 300 bp. The genome data was analyzed following the method described by Elarabi et al. (2023).
2.7 Determination of E. kobei FACU6 ability to resist other heavy metals
To confirm the ability of the E. kobei FACU6 strain to resist three different heavy metals (As, Cr, Cd), E. kobei FACU6 strain was grown on LB media supplemented with 1,500 mg/L Na2HAsO4. 7H2O (Pramanik et al., 2018a), 200 mg/L K2Cr 2O7 (Iyer et al., 2004) or 1,000 mg/L CdCO3 (Pramanik et al., 2018b) to resist As, Cr and Cd respectively, in separate experiments and together in the same media. All cultures were incubated for 15 days at 30°C.
2.8 RNA extraction quantitative polymerase chain reaction (qPCR)
The E. kobei FACU6 strain, exhibiting the highest lead resistance, was used to evaluate the gene expression of various heavy metal (As, Cd, and Pb) resistance genes. The changes in mRNA levels of the arsB, arsC, cadA, and pbrC genes were assessed under different conditions of pH (5, 7, and 9), temperature (25, 30, and 42°C), and heavy metal concentrations (low, medium, and high) for each of Cd, As, and Pb. E. kobei FACU6 was cultured in LB medium supplemented with the three elements and incubated at 28°C for 3–15 days. The “low” concentration was 50 mg/L for all heavy metals. The “medium” concentrations were 500 mg/L for Cd, 1,250 mg/L for Pb, and 500 mg/L for As. The “high” concentrations were 1,000 mg/L for Cd, 2,500 mg/L for Pb, and 1,000 mg/L for As. In the combined three heavy metals experiment, 500 mg/L of each heavy metal was added to the medium. Total RNA was isolated from both metal-treated and untreated cells using the Simply™ Gene Direx Total RNA Isolation Kit (Cat. No. NA017-0100, Taiwan) according to the manufacturer’s instructions. Reverse transcription was performed using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific™, Cat. No. K1621, United States) with 250 ng RNA, following the manufacturer’s instructions. qPCR reactions were carried out in a 10 µL reaction volume containing 5 µL of 2X Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific™, Cat. No. K0221, United States), 2 µL of 10X diluted cDNA, and 1 µL of each forward and reverse primer (Supplementary Table S1). The qPCR temperature program was as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The 16S rRNA and DNA gyrase genes were used as reference genes.
2.9 Statistics analysis
GraphPad Prism 8 and R software were used to conduct one-way analysis of variance (ANOVA) and least significant difference (LSD) tests, respectively. The relative expression fold changes were calculated using the equation 2−ΔΔCT, as described by Livak and Schmittgen (2001). In addition, a two-way ANOVA test of multiple comparisons was performed in the gene expression experiment to measure whether the two datasets were significantly different (p > 0.005) using GraphPad Prism 8 software.
3 Results
3.1 Physicochemical analysis of collected samples
Electric conductivity (EC) and other physicochemical parameters were measured. As well as heavy metal contents were determined, as shown in Supplementary Table S2. The obtained samples had a slightly acidic, while the EC was low. In addition, the main hazardous metal concentrations in collected samples were measured. The findings showed that sample no. 1’s As, Cr, Cd, and Mn concentrations were found to be greater than United States Environmental Protection Agency (US EPA) screening limits, Conversely, Pb and Fe concentrations were higher in both samples (Supplementary Table S2) (US EPA, 2022). While the Pb concentrations in the two samples were similar, Cu and Zn were lower concentrations than the permissible concentration limit.
3.2 Selection of Pb resistant isolates
Twenty bacterial isolates were chosen due to their ability to survive on LB media supplemented with 1,200 mg/L of lead acetate. The MIC and MTC for these isolates were then determined by gradually increasing the Pb concentrations. The results showed that eight isolates named (LRS1, LRS2, LRS6, LRS9, LRS10, LRS15, LRS18, and LRS19) had a high MIC that reached 2,900 mg/L and a MTC of up to 2,800 mg/L (Supplementary Table S3). LRS19 exhibited the highest MIC and MTC (3,100 and 3,000 mg/L), respectively.
3.3 Molecular identification of the selected isolates
The 16S rRNA genes of eight bacterial isolates were amplified, generating a 1,500 bp band that were subsequently purified and sequenced. The partial sequences of the 16S rRNA gene were aligned and deposited to NCBI under accession numbers MT912742, MT912763, MW599729, MT912790, MT912758, MT912751, MW599731, and MW599732 as K. quasipneumoniae subsp. ouasipneumoniae strain FACU3 (98.71% similarity), Klebsiella quasivariicola strain FACU (99.22% similarity), Citrobacter freundiistrain FACU 2 (99.52% similarity), K. quasipneumoniae subsp. quasipneumoniae strain FACU4 (98.55% similarity), K. quasipneumoniae subsp. quasipneumoniae strain FACU2 (99.66% similarity), K. variicola subsp. variicola strain FACU (99.73% similarity), Enterobacter sichuanensis strain FACU (99.9% similarity) and Enterobacter kobei strain FACU6 with (98.37% similarity), respectively. The phylogenetic tree was constructed as shown in Supplementary Figure S2. Also, the bacteria were deposited in the Culture Collection at Ain Shams University (CCASU WDCM1186, Cairo, Egypt) under designations CCASU-2022-26 to CCASU-2022-33 for Klebsiella quasipneumoniae subsp. quasipneumoniae FACU3, K.quasivariicola FACU, C. freundii FACU 2, K. quasipneumoniae subsp. quasipneumoniae FACU4, K. quasipneumoniae subsp. Quasipneumoniae FACU2, Klebsiella variicola subsp. variicola FACU, E. sichuanensis FACU and E. kobei FACU6, respectively.
3.4 Determination of the Pb effect on bacterial survival, biosorption efficiency and Pb uptake
The bacterial strain’s ability to resist Pb was evaluated through measurements of bacterial survival (%), biosorption efficiency (%) and lead uptake (mg/g dry weight). Significant differences were observed between the strains using a one-way ANOVA test and LSD tests. Additionally, the metal concentration in biomass (mg/g) was evaluated. The bacterial strains under investigation in this study have demonstrated a high efficiency in the uptake of heavy metals from the culture medium, with efficiencies ranging from 52.0% to 83.79%. This significant uptake has led to a substantial reduction in the residual concentration of heavy metals, as detailed in Supplementary Figure S3; Table 1. It was found that strain LRS19 (E. kobei), which can absorb 571.9 mg/g dry weight on strongly supplemented LB with 2,600 mg/L lead acetate had a highly significant percentage of bacterial survival under Pb stress (86.69%) and the best efficiency of biosorption (83.4%). The metal concentration in biomass was measured to be 1,024 mg/g in E. kobei while LRS15 (K. variicola) demonstrated the lowest significant percentage of bacterial survival under Pb stress (3.2%), with a biosorption efficiency of roughly 52.1 percent, capable of absorbing 87.6 mg/g dry weight and accumulate around 68 mg/g.
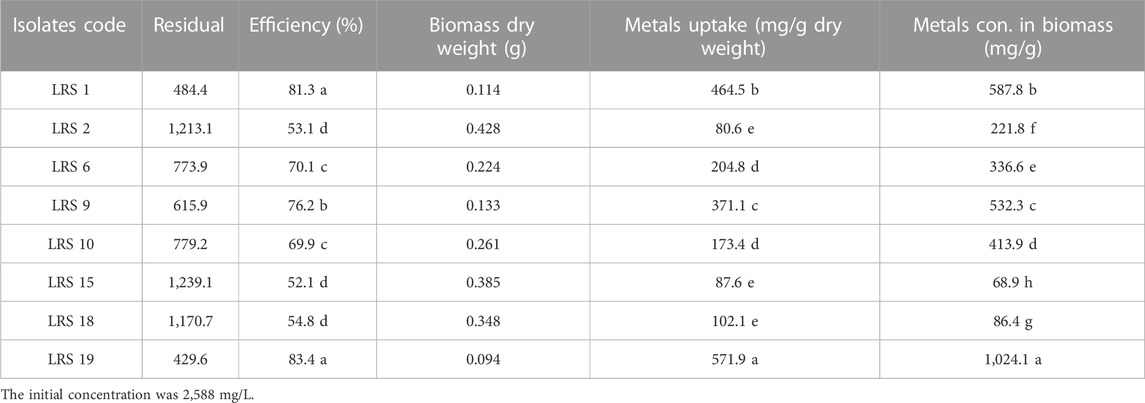
TABLE 1. Determination of the Pb effect on bacterial survival, biosorption efficiency, Pb uptake and metal concentration in the biomass.
3.5 Cellular morphological response to Pb stress by SEM and TEM
The adsorption of Pb onto the cell surface of E. kobei was observed using SEM. Comparing Pb-treated cells to the control, the SEM images revealed changes in cell morphology. In this study, The E. kobei cells in the control showed smooth exterior surfaces, were transparent, and had a uniform shape. All untreated bacteria displayed smooth, crystal-clear cell walls (Figure 1A), whereas the treated E. kobei cells were clumped or aggregated and had some shining particles on their surface (Figure 1B). Cells with clumped granular materials, deformed cells, protrusions that resembled configurations, and irregularly sized cells were all observed. After the biosorption of Pb, the bacterial cell wall was deformed. The SEM images demonstrated that Pb caused significant alterations in the shape of the E. kobei cell. By using TEM examination, it was possible to compare treated and untreated conditions’ Pb adsorption and intracellular accumulation. The biosorption ability of E. kobei was observed in the treated condition as numerous black dots, which were electron-dense materials or granules, were deposited mostly inside the cell or attached to the cell surface. In the images taken with TEM, an accumulation of Pb can be seen within the cell cytoplasm of E. kobei (Figures 2B, D) compared to untreated cells (Figures 2A, C).
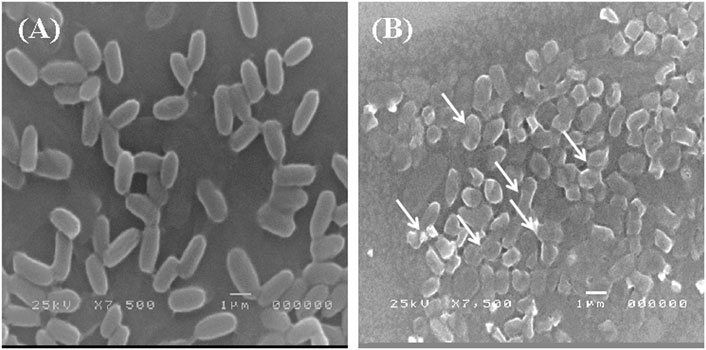
FIGURE 1. Scanning electron microscopic view of lead resistant E. kobei: (A) showing SEM pattern for absence of lead (control) and (B) for presence of lead. Arrows indicate the deformation in the bacterial cell compared with the untreated cells.
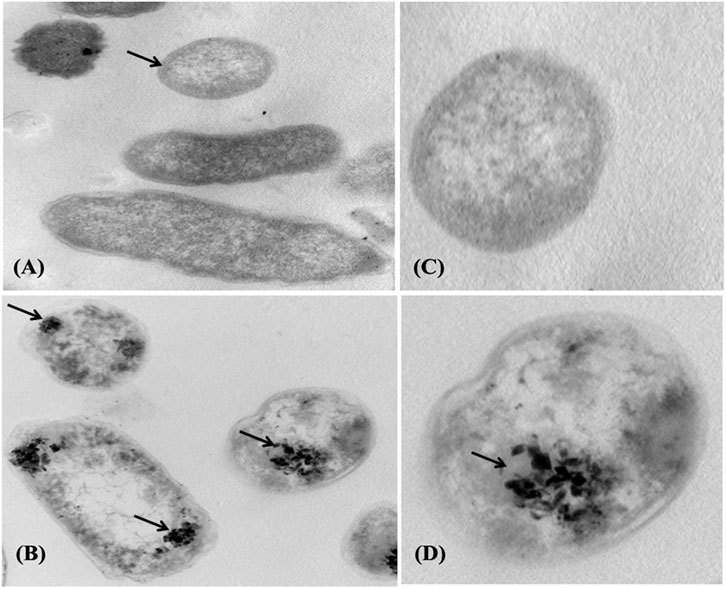
FIGURE 2. Transmission electron micrograph of the E. kobei strain cultured in liquid LB medium supplemented with 3000 mg/L lead acetate. (A) and (C) control - absence of metals; (B) and (D) with Pb. Arrows indicate locations of clearest contrasts of metal accumulation.
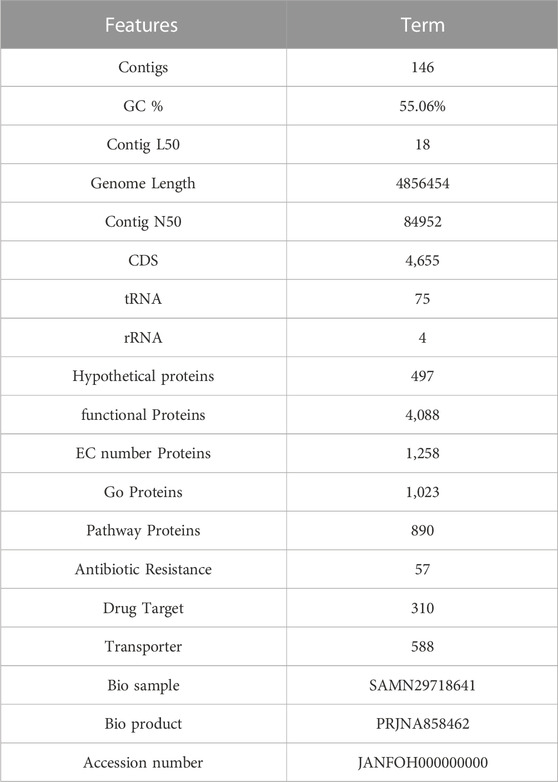
3.6 Genome analysis
The genome analysis revealed 146 contigs with a total size of 4,856,454 bp and a G + C content of 55.06%, as shown in Table 2. The contigs’ N50 and L50 values were 84,952 bp and 18, respectively. Then 4655 CDS, 75 tRNAs and 4 rRNAs were identified using genome annotation. 497 putative proteins and 4,088 proteins with functional annotations were found among the protein-coding genes. The proteins with functional annotations included 1,258 proteins with Enzyme Commission (EC) numbers, 1,023 proteins with Gene Ontology (GO) annotations, and 890 proteins that were linked to Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways. According to the transporter categorization database (TCDB), 588 of the identified genes have similarities to recognized transporters, and 57 antibiotic-resistant genes according to CARD. DrugBank listed 310 gene targets for drugs.
Detailed properties and statistics of genome, in addition to the phylogeny relationship of E. kobei FACU6 between Enterobacter genus, are summarized in Table 3; Figures 3A, B. Together with the top 10 gene ontologies and gene count for biological processes (BP), cellular components (CC), and molecular functions, as depicted in Figure 3C. In addition, three plasmids having IncFII, Col and IncFIB replicons with identities of about 96.92%, 96.12%, and 98.93%, respectively, when comparing to the databases in PlasmidFinder, on three different contigs, were identified in this genome. The three plasmids were also confirmed using the Abricate database. Furthermore, Figure 3D shows the subsystem coverage and category distribution of the entire E. kobei FACU6 genome. The results demonstrated that this genome contains approximately 27 subsystems. One of these subsystems was the virulence, disease, and defense subsystem, which contained approximately 57 genes. Twenty-nine heavy metal genes were annotated in the E. kobei FACU 6 genome.
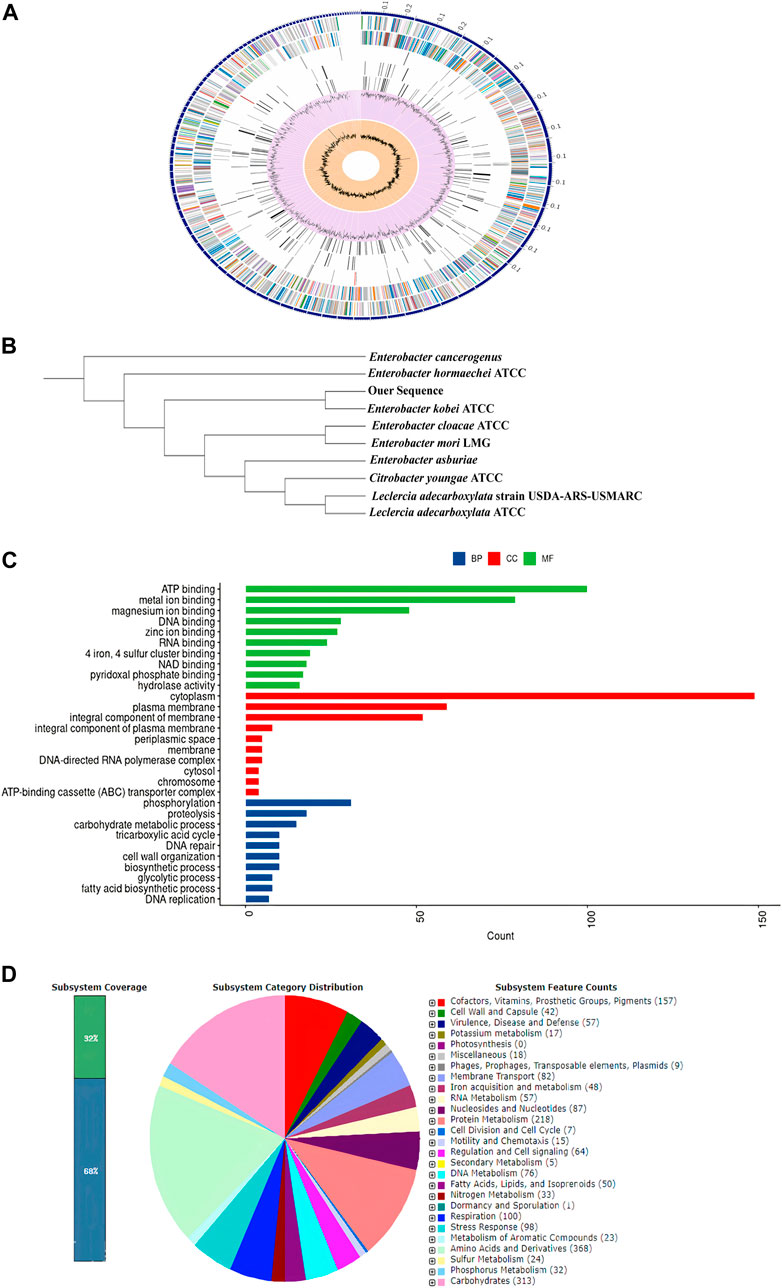
FIGURE 3. (A): Using the PATRIC annotation tool, circular graphical representation of the E. kobei FACU6 genome’s distribution was created. The contigs are shown on the outer ring, followed by CDS on the forward and reverse strands, RNA genes, and CDS with homology to known antimicrobial resistance genes and known virulence factors. The GC content and GC skew are shown on the inner ring. (B): Phylogenetic tree generated by IQ-TREE using the GTR1F1I1G4 model and visualized by iTol. (C): Top 10 gene ontologies produced by UniprotR for biological process (BP), cellular component (CC), and molecular function. (D): RAST was used to analyze subsystem coverage and category distribution of the entire E. kobei FACU 6 genome. The counts for each subsystem feature and the subsystem coverage are shown in the pie chart. Genes were displayed in brackets for each subsystem group.
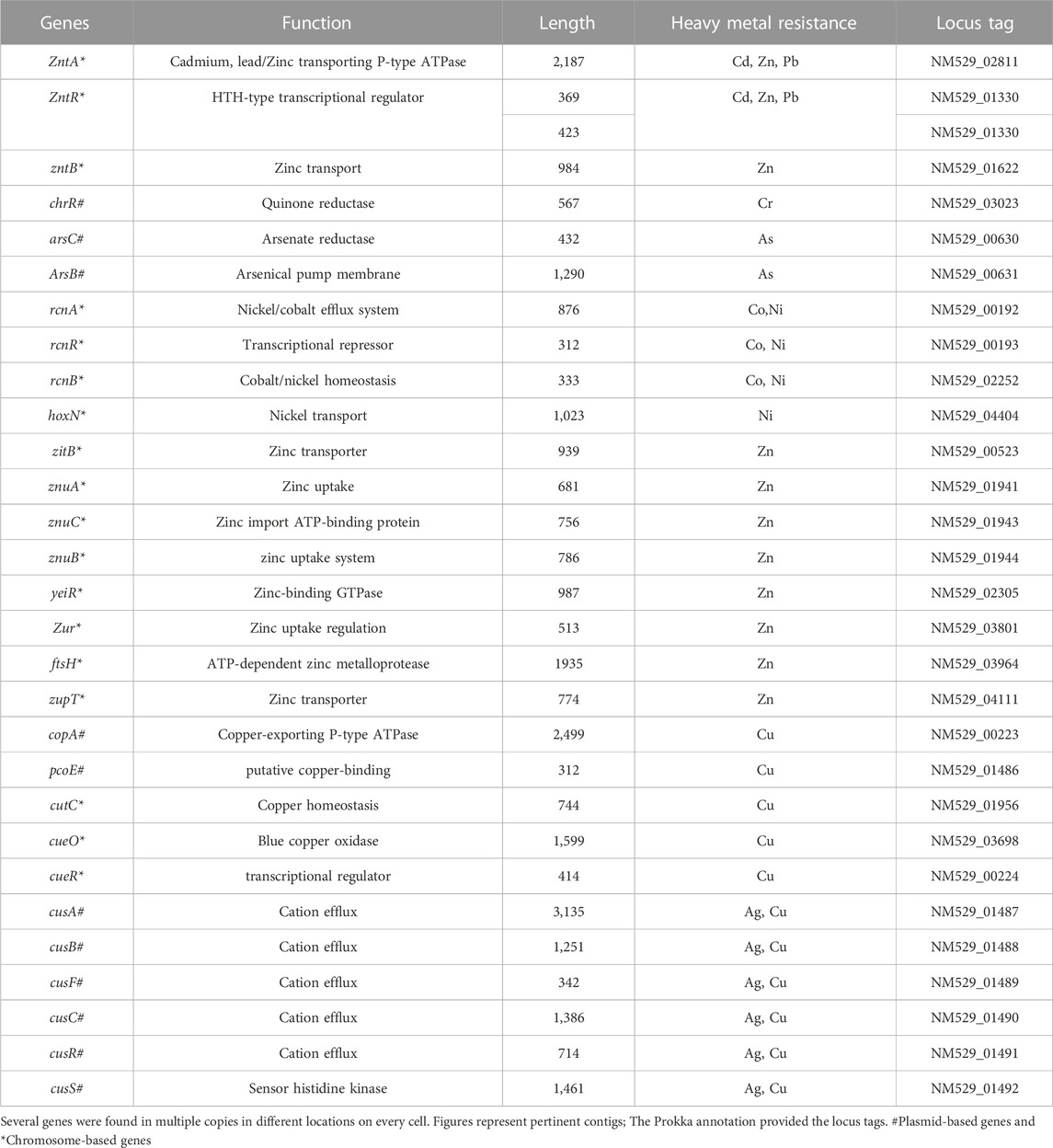
Some of those genes, including Pb, Cd, Cr, As, Ni, Co, Cu, and silver (Ag), the genes identified were discovered to be implicated in heavy metal resistance genes. In addition, resistance, including those associated with lead (Pb), cadmium (Cd), chromium (Cr), arsenic (As), nickel (Ni), cobalt (Co), copper (Cu), and silver (Ag).
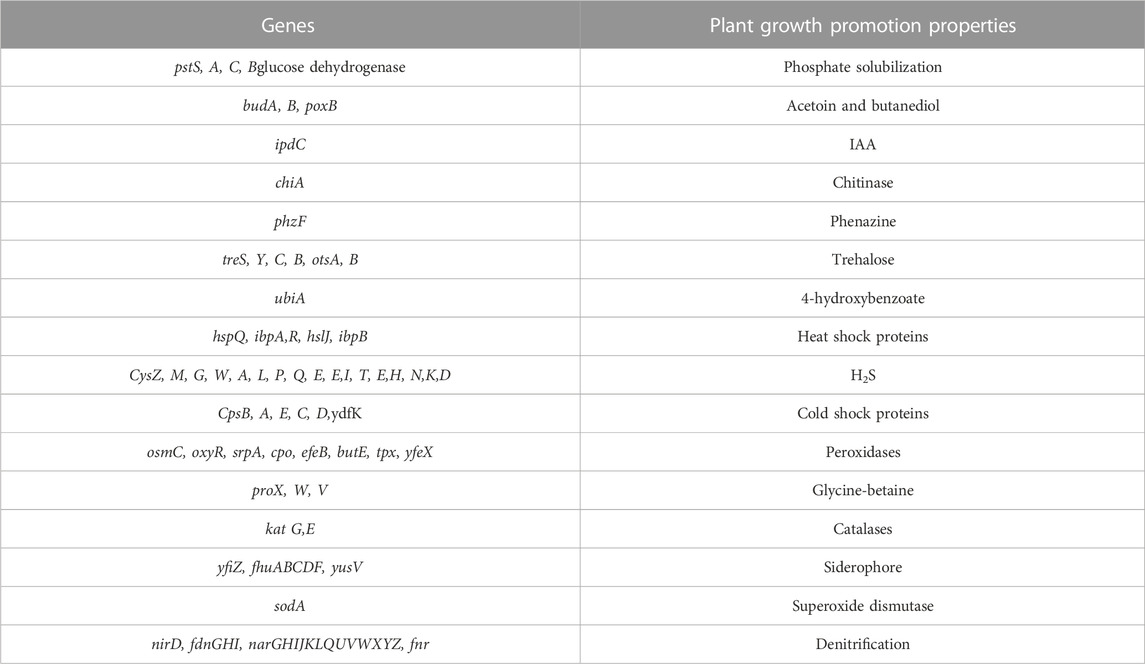
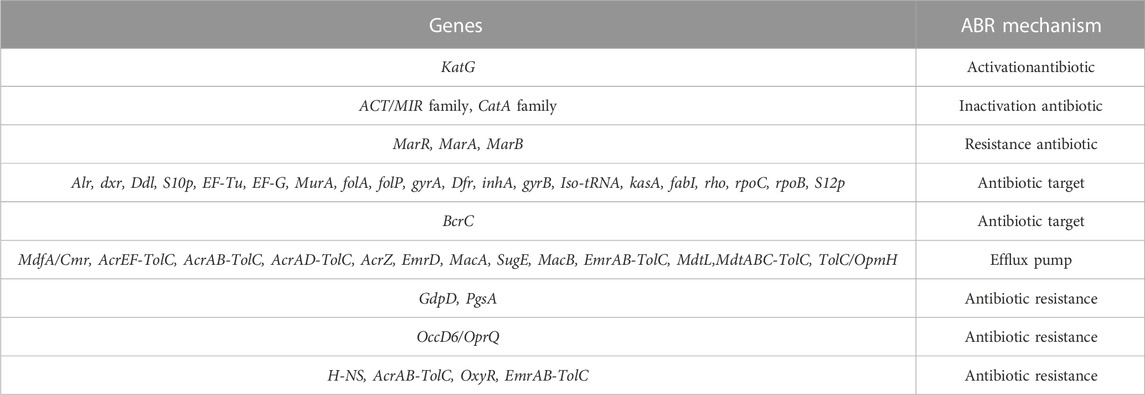
Some of these genes, those related to resistance against heavy metals such asPb, Cd, Cr, As, Ni, Co, Cu, and silver (Ag), to be heavy metal resistance. In addition, some genes were found in complete clusters, such as the zinc resistance gene cluster (Figure 4). The E. kobei FACU 6 zinc resistance gene cluster’s chromosomal region was compared to four other bacteria (E. cloacae subsp. cloacae ATCC 13047, Enterobacter sp 838, E. hormaechei ATCC 49162 and E. mori LMG 2506). The results displayed complete similarity in the zinc resistance gene cluster between these species. These results indicated the ability of E. kobei FACU 6 to resist various heavy metals, including Zn, Cr and Cd. Moreover, this genome harbored about 65 plant growth-promoting genes (PGPGs) like genes involved in indole acetic acid (IAA) production, acetoin and butanediol synthesis, trehalose metabolism, phosphate solubilization, chitinase, phenazine, 4-hydroxybenzoate, cold shock proteins, heat shock proteins, glycine-betaine, H2S, peroxidases, catalases, superoxide and siderophore production (Table 4). The majority of the AMR genes were engaged in conferring resistance via efflux pumps or changing antibiotic targets, as shown by Table 5, which summarizes the identified AMR genes in this genome and their corresponding mechanisms. Supplementary Table S4 shows the results of the RGI tool’s analysis of the E. kobei FACU6 resistome, which included 1 perfect hit and 19 strict hits. The strain is expected to be resistant to carbapenem, cephalosporin, cephamycin, and penam due to the presence of several resistant genes.
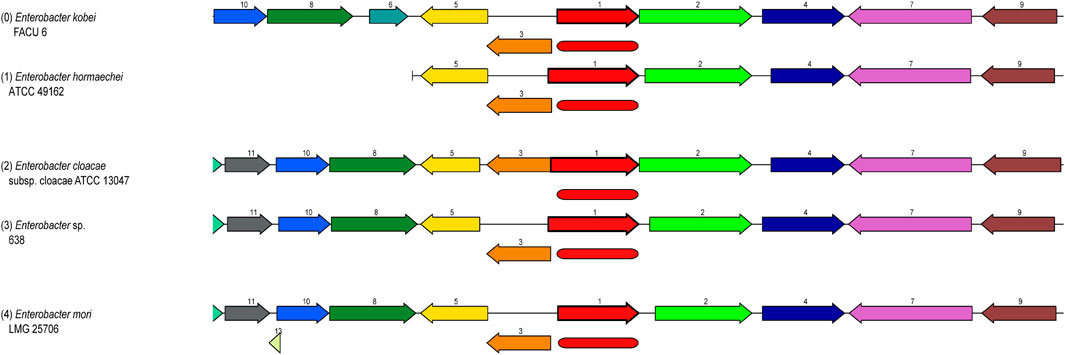
FIGURE 4. Zinc resistance gene cluster: Four closely related organisms’ chromosome regions were compared with the region of the focal gene (top). The focus gene, which is red and has the number 1, is shown in the graphic. The same number and color are used to identify groups of genes with identical sequences (1-Zinc ABC transporter, substrate-binding protein ZnuA, 2-Murein DD-endopeptidase MepM, 3-Zinc ABC transporter, ATP-binding protein ZnuC, etc.). Myristoyltransferase for 4-Lipid A biosynthesis, ZnuB, a putative protein, a 5-Zinc ABC transporter and kinase 7-pyruvate, RuvB, an ATP-dependent DNA helicase for the Holiday junction, HexR, the RpiR family, and 11-Crossover junction endodeoxy ribonuclease RuvCand 10-Holliday junction ATP-dependent DNA helicase RuvA).
3.7 Evaluation of E. kobei FACU6 ability to resist other heavy metals
The ability of the E. kobei FACU6 strain to resist additional heavy metals was assessed. It was found that E. kobei FACU6 could grow in media supplemented with As, Cr, or Cd in addition to Pb. The results showed that E. kobei FACU6 could tolerate 1,500 mg/L, 200 mg/L and 1,000 mg/L of As, Cr, and Cd, respectively. In addition, E. kobei FACU6 was able to grow in LB media supplemented with the four heavy metals. To confirm the ability of E. kobei FACU6 to resist different heavy metals, qPCR was utilized to determine messenger RNA expression levels of two As resistance genes (arsB, arsC), one Cd resistance gene (cadA) and the pbrC gene for Pb resistance under different conditions. There was a highly significant difference between pH 7, 5, and 9. The expression of arsB, arsC, cadA, and pbrC genes in E. kobei FACU6 increased significantly (p 0.05) under pH 7, with a fold change of 2–3 times (Figure 5A). For the different temperatures tested, all genes were downregulated at 42°C, while their expression at 25°C and 30°C was upregulated with fold changes ranging from 2 to 3.5 times (Figure 5B). Furthermore, the effect of heavy metal concentration on gene expression of these genes was determined individually and combined in LB media. Gene expression has a positive relationship with the increase in heavy metal concentrations, with highly significant differences between them (Figures 5C, D). The previous results concluded that E. kobei FACU6 is considered a multi-heavy metal resistant isolate due to its ability to resist Pb, Ar, Cr, and Cd with high tolerance concentrations.
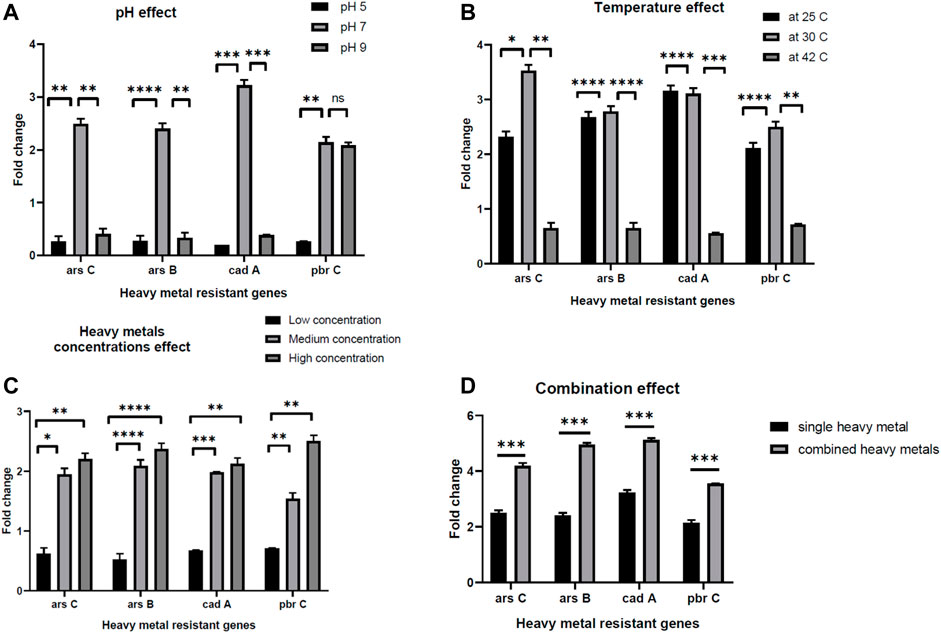
FIGURE 5. The effect of pH, temperature, heavy metal concentrations and combination effect (A–D) on gene expression of arsC, arsB, cadA, pbrC genes in E. kobei FACU6.
4 Discussion
Isolating bacteria resistant to heavy metals from contaminated sites is a critical step in the bioremediation process. In this study, the industrial zone of Sadat City was chosen for bacterial isolation since it is one of the biggest industrial zones in Egypt. In this region, it is common to find industries in batteries, plumbing, painting, wiring, electric cables, raw material mining, and ceramic glazing. The physicochemical properties of both soil samples were analyzed. The pH was slightly acidic, which was reflected in the EC, which had low conductivity. The lead industries’ byproducts may cause low pH and EC (Nirbhavane et al., 2017). Also, it was observed that sample no. 1 was more contaminated than sample no. 2, with high levels of several heavy metals that exceeded the allowed concentration limit (US EPA, 2022). According to their MICs, eight bacterial isolates were selected. These isolates were molecularly identified and submitted to GenBank. Five strains belonged to the genus Klebsiella, two to the genus Enterobacter, and one to the genus Citrobacter. According to reports, the Enterobacter and Klebsiella species are useful bacteria in the treatment of antibiotic and heavy metal resistance (Wang et al., 2018). Citrobacter freundii has also been applied to obtain biosurfactants and remove heavy metals (Gomaa and El-Meihy, 2019).
In addition, significant variations in bacterial survival, biosorption efficiency, and lead uptake were observed among the strains. The results indicated that the significantly low percentages of bacterial survival, biosorption efficiency, and lead uptake may be attributed to the strains reaching the level of toxicity, and thus lead pressure influenced cell survival. If toxic pressure is insufficient to kill the organisms, it can increase cell membrane permeability, subsequently increasing toxic ions’ uptake (Odokuma and Akponah, 2010). The results showed that the E. kobei (LRS 19) strain displayed the highest level of Pb resistance, whereas the K. variicola (LRS 15) strain exhibited the lowest level. While the MIC of the promising strain E. kobei FACU6 reaches 3,000 mg/L with efficiency (83.4%), Abdollahi et al. (2020) tested the ability of E. kobei isolated from soil collected from sites near a lead-zinc factory in Iran to resist lead and found that it had an accumulation capacity of 25% in addition to its ability to resist cadmium. Moreover, the binding of lead ions on bacterial cell surface was also observed using SEM. The cellular surface of a bacterial cell has been considered the most potent sorbent that adsorbs metal ions and minimizes their harmful effects (Kang et al., 2020). In Gram-negative bacteria, the cells have negatively charged lipopolysaccharides in their cell walls provide the ability to bind to cationic metal ions and give them a metal binding feature (Khan et al., 2015). Since metals are not biodegradable and cannot be broken down. Organisms can detoxify metal ions by enclosing the active component in a protein or storing them insoluble in intracellular granules. Furthermore, in E. kobei FACU6, the TEM data showed a clear distinction between control and Pb-treated cells in terms of the size and thickness of the cell wall. On the cell membrane and in the cytoplasm, black granules were observed. Generally, heavy metals inevitably enter the cells, accompanied by nutrient uptake by microorganisms (Cuypers et al., 2010). Consequently, accumulated heavy metals may harm bacterial cells (Cuypers et al., 2010). To ensure their survival, normal growth, and metabolism, bacterial strains employ various defense mechanisms, including extracellular sequestration, biosorption, precipitation, efflux, intracellular bioaccumulation, and changes in cell morphology (Naik and Dubey, 2013). The first structure in a cell to interact with metal ions is the cell wall, which has carboxyl, phosphate, sulfhydryl carbonyl or hydroxyl groups as functional groups. These groups play a significant and fundamental role in the biosorption of metal ions (Khan et al., 2022). They negatively charge the cell wall and produce insoluble compounds when they bind to lead ions. The cell wall, composed of organic macromolecules including polypeptides, polysaccharides, and proteins, could also adsorb Pb by electrostatic forces like Van der Waal’s forces, covalent or ionic bonds (Qiao et al., 2019). Thus, this could explain the molecular mechanisms of lead resistance in E. kobei FACU6.
Bacteria possess high surface-to-volume ratios and active sites in their cell walls, which serve as potential metal accumulators (Sah et al., 2022). This characteristic likely contributes to the observed high efficiency of heavy metal uptake. However, an interesting observation was made regarding the metal concentration within the bacterial biomasses. Despite the high uptake, the concentration within the biomasses was found to be significantly lower or moderated for all strains investigated. For instance, the metal concentration within the LRS1 strain biomass was recorded at 9.23 mg/g. This discrepancy could be attributed to this discrepancy, including adsorption of metals onto the bacterial cell surface or extracellular precipitation (Maity et al., 2023). SEM result provided visual evidence supporting these mechanisms. Furthermore, it was found that LRS19 bacterial strain has ability to reduce heavy metal concentrations in their environment through intracellular accumulation, which clarifies the high metal concentration in its biomass (1,024 mg/g). However, it is important to note that the relative contribution of each mechanism can vary depending on specific environmental conditions and types of heavy metals present (Ayele and Godeto, 2021). To further investigate the Pb-resistant genes and removal mechanism and metabolic pathways, whole-genome sequencing (WGS) was performed for E. kobei FACU6. The genome was larger than previously reported E. kobei M4-VN that was previously disclosed (Thanh et al., 2020). E. kobei FACU6 contained a high tRNA gene content. The significant tRNA gene content may be a result of the cell’s ability to regulate gene expression in microorganisms in habitats with a wide range of conditions (Rodríguez-Rojas et al., 2016). In addition, three plasmids, IncFII, Col and IncFIB replicons, were found in the E. kobei genome. Referring to the minimal genome sequence information, revealed that the genome possesses an extensive number of heavy metal resistance genes and gene clusters, as shown in Table 4. Compared to metal-resistant bacterium E. kobei CPE isolated from wastewater in the UK, which carried arsenic, mercury, and tellurium resistance genes (Ludden et al., 2017), the draft genome of E. kobei FACU6 revealed the presence of arsBC pair genes encoding for the proteins arsA and arsB, which participate in arsenic resistance but didn’t have any specific resistance genes for tellurium and mercury. The genome also contains clusters and system genes involved in the transport and resistance to metals like cadmium, zinc, nickel, and cobalt. E. kobei FACU6 lacks the Pbr, Czc, and Cad operons, although earlier studies suggested that heavy metals (Zn, Pb, Co, and Cd) translocating P-type ATPase genes can also function in lead resistance. The E. kobei FACU6 genome contained the zntA gene, which encodes a zinc/cadmium/lead-transporting P-type ATPase (Hui et al., 2022). Not only zinc but also lead and cadmium greatly increased the expression of zntA, in a process uncontrolled and mediated through zntR (Binet and Poole, 2000). zntR and zntA genes were located at different locations on the chromosome (Baya et al., 2021). This confirms our findings about the lead resistance of E. kobei FACU6. Lead-resistant bacteria can use a variety of methods, including biosorption, efflux mechanisms, induced precipitation, extracellular sequestration, and intracellular lead bioaccumulation, in Pb bioremediation process (Jarosławiecka and Piotrowska-Seget, 2014). When Pb is present, an influx transporter, such as P-type ATPases, can move and deposit Pb into the periplasmic region of the bacterial cell (Huang et al., 2018). The resistance gene Pbr functions as efflux pump to transport and immobilize Pb on cell surface in insoluble forms as Pb concentration rises, minimizing the toxicity caused by Pb (Jarosławiecka and Piotrowska-Seget, 2014). Microorganisms can transform harmful metal ions into insoluble complexes like phosphate, carbonate and sulfate during precipitation, lowering their concentration and, as a result, the toxicity of contaminated areas (Naik and Dubey, 2013). ZntA stimulates metal translocation across inner membrane, and molecule is transported to periplasm from the cytoplasm (Klein and Lewinson, 2011). While González Henao and Ghneim-Herrera (2021) examined the absence of pbr genes in several genera, including Bacillus, Pseudomonas, Klebsiella, Microbacterium, Agrobacterium, Rhodococcus, and Enterobacter, Hynninen et al. (2009) suggested that lead detoxification by ATPases and phosphatases was a common mechanism for lead tolerance in these microorganisms. Therefore, more research is needed to understand these metal resistance genes and systems in many bacterial species. ZntB, which serves as a zinc efflux method, was present in E. kobei FACU6 (Worlock and Smith, 2002). ZitB is crucial for zinc homeostasis at low zinc concentrations, but znt A is crucial at high zinc concentrations (Grass et al., 2002). Zur gene and znuABC operon were identified in E. kobei FACU6, and it was reported that the zur gene product reportedly responds to intracellular zinc concentration to regulate the zinc transporter expressed by znuABC gene cluster (Patzer and Hantke, 2000). In addition to transporting cadmium and copper ions, ZupT also mediates the absorption of zinc (Grass et al., 2002). YieR participates in metal hemeostasis (Blaby-Haas et al., 2012). Previous research has shown that E. kobei possesses several zinc resistance gene systems and gene clusters.
The nickel-cobalt efflux system’s rcn-operon (rcnA, R, and B) was found in E. kobei (Blaha et al., 2011). Hox N gene, which is characterized as a high-affinity nickel transport protein facilitating nickel transport, was also present in E. kobei (Wolfram et al., 1995). Our results showed that E. kobei FACU6 additionally possessed various arsenic and chromium resistance genes. The arsenate reductase (arsC) protein is necessary for arsenic resistance. Arsenate reductase, which is encoded by the existing operon (arsRBC), was able to convert arsenate into arsenite, and the integral membrane protein arsB encoded the remaining steps of the process. This prevented arsenic buildup by expelling it out of the cytoplasm (Yang et al., 2012). Notably, the quinone reductase gene (chrR) contributes to chromate bioremediation by decreasing chromate (Paul et al., 2020). In addition to the cus ABCFRS operon, which is involved in the silver efflux system, gene systems or gene clusters were encoded for copper hemeostasis, such as copA, pcoE, cutC, cueO, and R (Gudipaty et al., 2012). Moreover, E. kobei was considered a multi-heavy metal resistant bacteria and it harbors antibiotic resistant genes. Their AMR mechanisms revealed that most genes contributed to resistance through efflux pumps and altered antibiotic targets. Multiple resistant genes were expected to confer resistance to carbapenem, cephalosporin, cephamycin, and penam. This has been verified by numerous studies showing that heavy metal induction can affect both antibiotic-resistance genes and heavy metal-resistance genes in bacteria. These studies also demonstrated that heavy metal contamination of ecosystem can affect antibiotic resistance genes (Chen et al., 2019). It was important to note that genes that support plant growth in E. kobei can help increase nutrient availability, resistance to oxidative stress, and suppression of biotic and abiotic stress. E. kobei genome contains the indole pyruvate decarboxylase (ipd) gene, which converts tryptophan to IAA (Straub et al., 2013). Moreover, the tryptophan biosynthesis-related trp cluster (trpA, B, C, R, and S) genes were identified. These genes may contribute to the production of tryptophan, a building block for the biosynthesis of the IAA hormones that promote plant development (Duca et al., 2014). Genes for glucose dehydrogenase activity and the phosphate-specific transport system (pst) operon, which are involved in the solubilization of mineral phosphates in soil, were found in the genome of E. kobei (Brito et al., 2020). Moreover, E. kobei had genes involved in hydrogen sulfide (H2S) biosynthesis, which contribute to seed germination and accelerate plant growth (Dooley et al., 2013). Genes involved in the production of 2, 3-butanediol and acetoin, which are known to promote plant growth, were also found in E. kobei, including budA, B, C, and poxB (Ryu et al., 2003). Furthermore, many genes encoding catalases, peroxidases, and superoxide dismutase, all of which reduce oxidative stress in plants, were also found in the E. kobei genome (Rai et al., 2013). Additionally, the E. kobei genome contained the genes phzF and ubiA, which were responsible for the synthesis of phenazine and 4-hydroxybenzoate, respectively. These genes also supported plant growth by reducing osmotic stress and promoting plant defense and communication, respectively (Yuan et al., 2020). In addition, genes that allow bacteria to resist abiotic stress such as heat shock, cold shock, trehalose synthesis, and glycine-betaine production were discovered. This bacterium can be employed in biocontrol because the chiA gene, which encodes chitinase synthesis and is important for the nutritional cycling of chitin, was discovered (Veliz et al., 2017). E. kobei genome contained yfiZ, yusV, and fhu clusters, which participate in iron acquisition (Veliz et al., 2017). According to Papaspyrou et al. (2014), denitrification had ecological importance, and the findings revealed the genes in the control of nitrate reduction to ammonia. Taking the aforementioned results into consideration, it can be concluded that E. kobei FACU6 possesses not only multi-level heavy metal resistance but also the potential to promote plant growth.
To confirm the E. Kobei FACU6 genome annotation results, the ability of the bacteria to grow in LB media containing a mixture of heavy metals was assessed, along with the gene expression of some genes involved in heavy metal resistance (arsB, arsC, cadA, and pbrC). In the gene expression experiment, several factors were used to assess the heavy metal resistance genes, including pH, temperature, heavy metal concentration, and combined heavy metals in LB media. A highly significant difference was observed between pH 7, 5, and 9. This may be because optimal heavy metal removal occurs at pH 7, which is consistent with the findings of Gudipaty et al. (2012). Alkaline and acidic pH values decrease heavy metal removal due to the ease of heavy metal reactions with H+ atthese pH levels, creating competition between H+ and the active site on the biosorbent (Vásquez et al., 2007). Changing in temperature will affect numerous factors that are critical to heavy metal removal (Ashokkumar et al., 2017). Temperature plays a significant role in the adsorption of heavy metals. Increasing temperature increases the rate of adsorbate diffusion across the external boundary layer, leading to enhanced adsorption. Additionally, the solubility of heavy metals increases with temperature, making them more bioavailable (Bandowe et al., 2014). However, within a suitable range, rising temperature also stimulates the activity of microorganisms, enhancing their metabolism and enzyme activity, thereby accelerating bioremediation (Igiri et al., 2018). Consistent with our study, our study, Babai and Ron (1998) found that fusing an Escherichia coli gene responsive to heavy metals with β-galactosidase did not increase β-galactosidase expression following a temperature shift to 42°C for 30 min.
Highly significant positive correlations were observed between gene expressions and increasing in heavy metal concentrations. The results suggest that gene expression was induced by the increase in heavy metal concentrations. The fold change reached two-fold with the high concentrations in contrast to the low concentrations. Additionally, when the three heavy metals were combined, the fold change increased by 1.5–2 times for each gene compared to its expression in individual experiments. In conclusion, E. kobei strain FACU6 exhibited significant levels of heavy metal tolerance that could be used in bioremediation applications. The ability to survive in many ecological niches was demonstrated by the existence of distinct metal transport/resistant genes and plant growth-promoting genes. However, further research is needed to fully understand these processes and optimize them for practical applications.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA858462.
Author contributions
HE-B: Conceptualization, Investigation, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. AH: Data curation, Formal Analysis, Methodology, Writing–original draft, Writing–review and editing. ZA: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. HA: Conceptualization, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. NE: Conceptualization, Data curation, Formal Analysis, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. AA: Conceptualization, Investigation, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. AH: Data curation, Formal Analysis, Methodology, Writing–original draft, Writing–review and editing. HR: Conceptualization, Data curation, Formal Analysis, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported under Project No. PSAU/2023/01/9075, from Prince Sattam bin Abdulaziz University.
Acknowledgments
Authors gratefully acknowledge the Prince Sattam bin Abdulaziz University (Project No. PSAU/2023/01/9075), for supporting this research work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1335854/full#supplementary-material
References
Abdelhaleem, H. A. R., Zein, H. S., Azeiz, A., Sharaf, A. N., and Abdelhadi, A. A. (2019). Identification and characterization of novel bacterial polyaromatic hydrocarbon-degrading enzymes as potential tools for cleaning up hydrocarbon pollutants from different environmental sources. Environ. Toxicol. Pharmacol. 67, 108–116. doi:10.1016/j.etap.2019.02.009
Abdollahi, S., Golchin, A., and Shahryari, F. (2020). Lead and cadmium-resistant bacterial species isolated from heavy metal-contaminated soils show plant growth-promoting traits. Int. Microbiol. 23, 625–640. doi:10.1007/s10123-020-00133-1
Ashokkumar, P., Loashini, V. M., and Bhavya, V. (2017). Effect of pH, temperature and biomass on biosorption of heavy metals by Sphaerotilus natans 33. Int. J. Microbiol. Mycol. 6 (1), 32–38.
Ayele, A., and Godeto, Y. G. (2021). Bioremediation of chromium by microorganisms and its mechanisms related to functional groups. J. Chem. 2021 (2), 1–21. doi:10.1155/2021/7694157
Babai, R., and Ron, E. Z. (1998). An Escherichia coli gene responsive to heavy metals. FEMS Microbiol. Lett. 167 (2), 107–111. doi:10.1111/j.1574-6968.1998.tb13215.x
Balali-Mood, M., Naseri, K., Tahergorabi, Z., Khazdair, M. R., and Sadeghi, M. (2021). Toxic mechanisms of five heavy metals: mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 12, 643972–644019. doi:10.3389/fphar.2021.643972
Bandowe, B. A. M., Bigalke, M., Boamah, L., Nyarko, E., Saalia, F. K., and Wilcke, W. (2014). Polycyclic aromatic compounds (PAHs and oxygenated PAHs) and trace metals in fish species from Ghana (West Africa): bioaccumulation and health risk assessment. Environ. Int. 65, 135–146. doi:10.1016/j.envint.2013.12.018
Baya, G., Muhindi, S., Ngendahimana, V., and Caguiat, J. (2021). Potential whole-cell biosensors for detection of metal using MerR family proteins from Enterobacter sp. YSU and Stenotrophomonas maltophilia OR02. Micromachines 12 (2), 142–217. doi:10.3390/mi120.20142
Binet, M. R. B., and Poole, R. K. (2000). Cd(II), Pb(II) and Zn(II) ions regulate expression of the metal-transporting P-type ATPase zntA in Escherichia coli. FEBS Lett. 473 (1), 67–70. doi:10.1016/S0014-5793(00).01509-X
Blaby-Haas, C. E., Flood, J. A., Crécy-Lagard, V. D., and Zamble, D. B. (2012). YeiR: a metal-binding GTPase from Escherichia coli involved in metal homeostasis. Metallomics 4, 488–497. doi:10.1039/c2mt20012k
Blaha, D., Arous, S., Blériot, C., Dorel, C., Mandrand-Berthelot, M. A., and Rodrigue, A. (2011). The Escherichia coli metallo-regulator RcnR represses rcnA and rcnR transcription through binding on a shared operator site: insights into regulatory specificity towards nickel and cobalt. Biochimie 93, 434–439. doi:10.1016/j.biochi.2010.10.016
Briffa, J., Sinagra, E., and Blundell, R. (2020). Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 6, e04691. doi:10.1016/j.heliyon.2020.e04691
Brito, L. F., López, M. G., Straube, L., Passaglia, L. M. P., and Wendisch, V. F. (2020). Inorganic phosphate solubilization by rhizosphere bacterium Paenibacillus sonchi: gene expression and physiological functions. Front. Microbiol. 11, 1–21. doi:10.3389/fmicb.2020.588605
Burghardt, R. C., and Droleskey, R. (2006). Transmission electron microscopy. Curr. Protoc. Microbiol. 3, doi:10.1002/9780471729259.mc02b01s03
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 (15), 1972–1973. doi:10.1093/bioinforma.tics/btp348
Chen, J., Li, J., Zhang, H., Shi, W., and Liu, Y. (2019). Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in northern China. Front. Microbiol. 10, 1916–2012. doi:10.3389/fmicb.2019.01916
Choudhury, R., and Srivastava, S. (2001). Zinc resistance mechanisms in bacteria. Curr. Sci. 81 (7), 768–775. https://www.jstor.org/stable/24106396.
Cuypers, A., Plusquin, M., Remans, T., Jozefczak, M., Keunen, E., Gielen, H., et al. (2013). Increased growth and germination success in plants following hydrogen sulfide administration. PLoS One 8, 620488–e62055. doi:10.1371/journal.pone.0062048
Duca, D., Lorv, J., Patten, C. L., Rose, D., and Glick, B. R. (2014). Indole-3-acetic acid in plant-microbe interactions. Antonie Leeuwenhoek 106 (1), 85–125. doi:10.1007/s10482-013-0095-y
Elarabi, N. I., Abdelhadi, A. A., Ahmed, R. H., Saleh, I., Arif, I. A., Osman, G., et al. (2020). Bacillus aryabhattai FACU: a promising bacterial strain capable of manipulate the glyphosate herbicide residues. Saudi J. Biol. Sci. 27, 2207–2214. doi:10.1016/j.sjbs.2020.06.050
Elarabi, N. I., Halema, A. A., Abdelhadi, A. A., Henawy, A. R., Samir, O., and Abdelhaleem, H. (2023). Draft genome of Raoultella planticola, a high lead resistance bacterium from industrial wastewater. Amb. Express 13, 14. doi:10.1186/s13568-023-01519-w
El-Beltagi, H. S., and Mohamed, A. A. (2010). Changes in non-protein thiols, some antioxidant enzymes activity and ultrastructural alteration in radish plant (Raphanus sativus L.) grown under lead toxicity. Not. Bot. HortiAgrobotanici Cluj-Napoca 38, 76–85. doi:10.15835/nbha3834631
Flora, G., Gupta, D., and Tiwari, A. (2012). Toxicity of lead: a review with recent updates. Interdiscip. Toxicol. 5, 47–58. doi:10.2478/v10102-012-0009-2
Gadd, G. M. (2010). Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156, 609–643. doi:10.1099/mic.0.037143-0
Gomaa, E. Z., and El-Meihy, R. M. (2019). Bacterial biosurfactant from Citrobacter freundii MG812314.1 as a bioremoval tool of heavy metals from wastewater. Bull. Natl. Res. Cent. 43, 69. doi:10.1186/s42269-019-0088-8
González Henao, S., and Ghneim-Herrera, T. (2021). Heavy metals in soils and the remediation potential of bacteria associated with the plant microbiome. Front. Environ. Sci. 9, 1–17. doi:10.3389/fenvs.2021.604216
Grass, G., Wong, M. D., Rosen, B. P., Smith, R. L., and Rensing, C. (2002). Zupt is a Zn(II) uptake system in Escherichia coli. J. Bacteriol. 184, 864–866. doi:10.1128/JB.184.3.864-866.2002
Gudipaty, S. A., Larsen, A. S., Rensing, C., and Mcevoy, M. M. (2012). Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol. Lett. 330, 30–37. doi:10.1111/j.1574-6968.2012.02529.x
Gunatilake, S. K. (2015). Methods of removing heavy metals from Industrial Wastewater. J. Multidiscip. Eng. Sci. Stud. 1, 12–18.
Harley, J. P., and Prescott, L. M. (2002). Laboratory exercises in microbiology. Fifth Edition. Boston, MA, USA: McGraw-Hill Companies, 466.
Herawati, N., Suzuki, S., Hayashi, K., Rivai, I. F., and Koyama, H. (2000). Cadmium, copper, and zinc levels in rice and soil of Japan, Indonesia, and China by soil type. Bull. Environ. Contam. Toxicol. 64, 33–39. doi:10.1007/s001289910006
Huang, F., Wang, Z. H., Cai, Y. X., Chen, S. H., Tian, J. H., and Cai, and K. Z. (2018). Heavy metal bioaccumulation and cation release by growing Bacillus cereus RC-1 under culture conditions. Ecotoxicol. Environ. Saf. 157, 216–226. doi:10.1016/j.ecoenv.2018.03.077
Hubeny, J., Harnisz, M., Korzeniewska, E., Buta, M., Zieliński, W., Rolbiecki, D., et al. (2021). Industrialization as a source of heavy metals and antibiotics which can enhance the antibiotic resistance in wastewater, sewage sludge and river water. PLoS One 16, doi:10.1371/journal.pone.0252691
Hui, C. Y., Guo, Y., Liu, L., and Yi, J. (2022). Recent advances in bacterial biosensing and bioremediation of cadmium pollution: a mini-review. World J. Microbiol. Biotechnol. 38, 9. doi:10.1007/s11274-021-03198-w
Huo, Y. Y., Li, Z. Y., Cheng, H., Wang, C. S., and Xu, X. W. (2014). High quality draft genome sequence of the heavy metal resistant bacterium Halomonas zincidurans type strain B6T. Stand. Genomic Sci. 9, 30. doi:10.1186/1944-3277-9-30
Hynninen, A., Touze, T., Pitkanen, L., Mengin-Lecreulx, D., and Virta, M. (2009). An efflux transporter PbrA and a phosphatase PbrB cooperate in a lead-resistance mechanism in bacteria. Mol. Microbiol. 74, 384–394. doi:10.1111/j.1365-2958.2009.06868.x
Igiri, B. E., Okoduwa, S. I. R., Idoko, G. O., Akabuogu, E. P., Adeyi, A. O., and Ejiogu, I. K. (2018). Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a Review. J. Toxicol. 27, 1–16. doi:10.1155/2018/2568038
Iyer, A., Mody, K., and Jha, B. (2004). Accumulation of hexavalent chromium by an exopolysaccharide producing marine Enterobacter cloaceae. Mar. Pollut. Bull. 49, 974–977. doi:10.1016/j.marpolbul.2004.06.023
Jan, R., Khan, M. A., Asaf, S., Lubna, L. I. J., and Kim, K. M. (2019). Metal resistant endophytic bacteria reduces cadmium, nickel toxicity, and enhances expression of metal stress related genes with improved growth of Oryza sativa, via regulating its antioxidant machinery and endogenous hormones. Plants 8 (10), 363. doi:10.3390/PLANTS8100363
Jarosławiecka, A., and Piotrowska-Seget, Z. (2014). Lead resistance in micro-organisms. Microbiology 160, 12–25. doi:10.1099/mic.0.070284-0
Kang, W., Zheng, J., Bao, J., Wang, Z., Zheng, Y., He, J. Z., et al. (2020). Characterization of the copper resistance mechanism and bioremediation potential of an Acinetobacter calcoaceticus strain isolated from copper mine sludge. Environ. Sci. Pollut. Res. 27, 7922–7933. doi:10.1007/s11356-019-07303-3
Khan, S. A., Khan, S. B., and Asiri, A. M. (2016). Electro-catalyst based on cerium doped cobalt oxide for oxygen evolution reaction in electrochemical water splitting. J. Mater. Sci. Mater. Electron. 27, 5294–5302. doi:10.1007/s10854-016-4427-3
Khan, Z., Elahi, A., Bukhari, D. A., and Rehman, A. (2022). Cadmium sources, toxicity, resistance and removal by microorganisms-A potential strategy for cadmium eradication. J. Saudi Chem. Soc. 26 (6), 101569. doi:10.1016/j.jscs.2022.101569
Khan, Z., Hussain, S. Z., Rehman, A., Zulfiqar, S., and Shakoori, A. (2015). Evaluation of cadmium resistant bacterium, Klebsiella pneumoniae, isolated from industrial wastewater for its potential use to bioremediate environmental cadmium. Pak. J. Zool. 47, 1533–1543.
Klein, J. S., and Lewinson, O. (2011). Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence. Metallomics 3, 1098–1108. doi:10.1039/c1mt00073j
Kumari, S., Amit, J. R., Mishra, N., and aand Singh, D. K. (2020). Recent developments in environmental mercury bioremediation and its toxicity: a review. Environ. Nanotechnol. Monit. Manag. 13, 100283. doi:10.1016/j.enmm.2020.100283
Land, M., Hauser, L., Jun, S. R., Nookaew, I., Leuze, M. R., Ahn, T. H., et al. (2015). Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics 15, 141–161. doi:10.1007/s10142-015-0433-4
Lemire, J. A., Harrison, J. J., and Turner, R. J. (2013). Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 11, 371–384. doi:10.1038/nrmicro3028
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Ludden, C., Reuter, S., Judge, K., Gouliouris, T., Blane, B., Coll, F., et al. (2017). Sharing of carbapenemase-encoding plasmids between Enterobacteriaceae in UK sewage uncovered by MinION sequencing. Microb. genomics 3, e000114. doi:10.1099/mgen.0.000114
Ma, Y., Oliveira, R. S., Freitas, H., and Zhang, C. (2016). Biochemical and molecular mechanisms of plant-microbe-metal interactions: relevance for phytoremediation. Front. Plant. Sci. 7, 918–919. doi:10.3389/fpls.2016.00918
Madeira, F., Park, Y. M., Lee, J. B. N., Gur, T., Madhusoodanan, N., Basutkar, P., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47 (W1), W636–W641. doi:10.1093/nar/gkz268
Maity, S., Sarkar, D., Poddar, K., Patil, P., and Sarkar, A. (2023). Biofilm-mediated heavy metal removal from aqueous system by multi-metal-resistant bacterial strain Bacillus sp. GH-s29. Appl. Biochem. Biotech. 195, 4832–4850. doi:10.1007/s12010-022-04288-7
Malik, A., and Jaiswal, R. (2000). Metal resistance in Pseudomonas strains isolated from soil treated with industrial wastewater. World J. Microbiol. Biotechnol. 16, 177–182. doi:10.1023/A:1008905902282
Masindi, V., and Muedi, K. L. (2018). “Environmental contamination by heavy metals,” in Heavy metals (Nappanee, IN, United States: InTech), 412. doi:10.5772/intechopen.76082
Mitra, A., Chatterjee, S., Kataki, S., Rastogi, R. P., and Gupta, D. K. (2021). Bacterial tolerance strategies against lead toxicity and their relevance in bioremediation application. Environ. Sci. Pollut. Res. 28, 14271–14284. doi:10.1007/s11356-021-12583-9
Mohamed, A. A., El-Beltagi, H. S., and Rashed, M. M. (2009). Cadmium stress induced change in some hydrolytic enzymes, free radical formation and ultrastructural disorders in radish plant. Electron. J. Environ. Agric. Food Chem. 8, 969–983.
Mohamed, M. S. M., El-Arabi, N. I., El-Hussein, A., El-Maaty, S. A., and Abdelhadi, A. A. (2020). Reduction of chromium-VI by chromium-resistant Escherichia coli FACU: a prospective bacterium for bioremediation. Folia Microbiol. 65, 687–696. doi:10.1007/s12223-020-00771-y
Naik, M. M., and Dubey, S. K. (2013). Lead resistant bacteria: lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicol. Environ. Saf. 98, 1–7. doi:10.1016/j.ecoenv.2013.09.039
Nirbhavane, H. M., Bagde, U. S., Nirbhavane, H. M., Bagde, U. S., and Nirbhavane, H. M. (2017). Resistance by Enterobacter spp. towards several antimicrobial drugs and heavy metals: a review. Afr. J. Biotechnol. 16, 826–841. doi:10.5897/ajb2017.15875
Odokuma, L. O., and Akponah, E. (2010). Effect of concentration and contact time on heavy metal uptake by three bacterial isolates. J. Environ. Chem. Toxicol. 2, 84–97.
Olaniran, A. O., Balgobind, A., and Pillay, B. (2013). Bioavailability of heavy metals in soil: impact on microbial biodegradation of organic compounds and possible improvement strategies. Int. J. Mol. Sci. 14, 10197–10228. doi:10.3390/ijms140510197
Page, A. L., Miller, R. H., and Keeney, D. R. (1983). “Methods of soil analysis. Part 2. Chemical and microbiological properties,” in Soil science society of America (Madison, Wisconsin, USA: American Society of Agronomy), 1159. doi:10.2134/agronmonogr9.2.2ed
Papaspyrou, S., Smith, C. J., Dong, L. F., Whitby, C., Dumbrell, A. J., and Nedwell, D. B. (2014). Nitrate reduction functional genes and nitrate reduction potentials persist in deeper estuarine sediments why? PLoS One 9, e94111. doi:10.1371/journal.pone.0094111
Parameswari, E., Lakshmanan, A., and Thilagavathi, T. (2010). Biosorption and metal tolerance potential of filamentous fungi isolated from metal polluted ecosystem. Electron J. Environ. Agric. Food Chem. 9, 664–671.
Pattnaik, S., Dash, D., Mohapatra, S., Pattnaik, M., Marandi, A. K., Das, S., et al. (2020). Improvement of rice plant productivity by native Cr(VI) reducing and plant growth promoting soil bacteria Enterobacter cloacae. Chemosphere 240, 124895. doi:10.1016/j.chemosphere.2019.124895
Patzer, S. I., and Hantke, K. (2000). The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275, 24321–24332. doi:10.1074/jbc.M001775200
Paul, M., Pranjaya, P. P., and Thatoi, H. (2020). In silico studies on structural, functional, and evolutionary analysis of bacterial chromate reductase family responsible for high chromate bioremediation efficiency. SN Appl. Sci. 2, 1997–2021. doi:10.1007/s42452-020-03830-8
Pramanik, K., Mitra, S., Sarkar, A., and Maiti, T. K. (2018a). Alleviation of phytotoxic effects of cadmium on rice seedlings by cadmium resistant PGPR strain Enterobacter aerogenes MCC 3092. J. Hazard Mater. 351, 317–329. doi:10.1016/j.jhazmat.2018.03.009
Pramanik, K., Mitra, S., Sarkar, A., Soren, T., and Maiti, T. K. (2018b). Characterization of a Cd2-resistant plant growth promoting rhizobacterium (Enterobacter sp.) and its effects on rice seedling growth promotion under Cd2-stress in vitro. Agri. Nal. Res. 52, 215e221. doi:10.1016/j.anres.2018.09.007
Qiao, W., Zhang, Y., Xia, H., Luo, Y., Liu, S., Wang, S., et al. (2019). Bioimmobilization of lead by Bacillus subtilis X3 biomass isolated from lead mine soil under promotion of multiple adsorption mechanisms. R. Soc. open Sci. 6, 181701. doi:10.1098/rsos.181701
Rai, G. K., Rai, N. P., Rathaur, S., Kumar, S., and Singh, M. (2013). Expression of rd29A: AtDREB1A/CBF3 in tomato alleviates drought-induced oxidative stress by regulating key enzymatic and non-enzymatic antioxidants. Plant Physiol. biochem. 69, 90–100. doi:10.1016/j.plaphy.2013.05.002
Ridge, P. G., Zhang, Y., and Gladyshev, V. N. (2008). Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One 3, e1378. doi:10.1371/journal.pone.0001378
Rodríguez-Rojas, F., Tapia, P., Castro-Nallar, E., Undabarrena, A., Muñoz-Díaz, P., Arenas-Salinas, M., et al. (2016). Draft genome sequence of a multi-metal resistant bacterium Pseudomonas putida ATH-43 isolated from Greenwich Island, Antarctica. Front. Microbiol. 7, 1777–1785. doi:10.3389/fmicb.2016.01777
Ryu, C. M., Faragt, M. A., Hu, C. H., Reddy, M. S., Wei, H. X., Paré, P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. 100, 4927–4932. doi:10.1073/pnas.0730845100
Sah, J., Sarkar, M., Mandal, P., and Pal, A. (2022). Comparative study of heavy metal uptake and analysis of plant growth promotion potential of multiple heavy metal-resistant bacteria isolated from arable land. Curr. Microbiol. 79, 1–13. doi:10.1007/s00284-021-02704-5
Saini, S., and Dhania, G. (2020). “Cadmium as an environmental pollutant: ecotoxicological effects, health hazards, and bioremediation approaches for its detoxification from contaminated sites,” in Bioremediation of industrial waste for environmental safety (Singapore: Springer), 357–387. doi:10.1007/978-981-13-3426-9_15
Sanders, E. R. (2012). Aseptic laboratory techniques: plating methods. J. Vis. Exp. 11 (63), e3064. doi:10.3791/3064
Shendure, J., and Ji, H. (2008). Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145. doi:10.1038/nbt1486
Shetty, R., and Rajkumar, S. (2009). Biosorption of Cu (II) by metal resistant Pseudomonas sp. Int. J. Environ. Res. 3 (1), 121–128. doi:10.22059/ijer.2009.40
Straub, D., Yang, H., Liu, Y., Tsap, T., and Ludewig, U. (2013). Root ethylene signalling is involved in Miscanthus sinensis growth promotion by the bacterial endophyte Herbaspirillum frisingense GSF30 T. J. Exp. Bot. 64, 4603–4615. doi:10.1093/jxb/ert276
Tchounwou, P. B., Yedjou, C. G., Patlolla, A. K., and Sutton, D. J. (2012). Molecular, clinical and environmental toxicicology volume 3: environmental toxicology. Mol. Clin. Environ. Toxicol. 101, 133–164. doi:10.1007/978-3-7643-8340-4_6
Thanh, N. C., Fujino, Y., Hiromasa, Y., and Doi, K. (2020). Draft genome sequence of Enterobacter kobei M4-VN, isolated from potatoes with Soft rot disease. Microbiol. Resour. Announc. 9, e00908. doi:10.1128/mra.00908-20
US, EPA (2022). Regional screening levels (RSLs)—user ’ s guide regional screening levels (RSLs). https://www.epa.gov/risk/regional-screeninglevels-rslsusers-guide (Accessed May 29, 2022).
Vásquez, T. G. P., Botero, A. E. C., de Mesquita, L. M. S., and Torem, M. L. (2007). Biosorptive removal of Cd and Zn from liquid streams with a Rhodococcus opacus strain. Min. Eng. 20, 939–944. doi:10.1016/j.mineng.2007.03.014
Veliz, E. A., Martínez-Hidalgo, P., and Hirsch, A. M. (2017). Chitinase-producing bacteria and their role in biocontrol. AIMS Microbiol. 3, 689–705. doi:10.3934/microbiol.2017.3.689
von Rozycki, T., and Nies, D. H. (2009). Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Int. J. Gen. Mol. Microbiol. 96, 115–139. doi:10.1007/s10482-008-9284-5
Wang, L., Cheng, W. C., Xue, Z. F., Xie, Y. X., and Lv, X. J. (2023). Study on Cu-and Pb-contaminated loess remediation using electrokinetic technology coupled with biological permeable reactive barrier. J. Environ. Manage. 348, 119348. doi:10.1016/j.jenvman.2023.119348
Wang, X., Huang, N., Shao, J., Hu, M., Zhao, Y., and Huo, M. (2018). Coupling heavy metal resistance and oxygen flexibility for bioremoval of copper ions by newly isolated Citrobacter freundii JPG1. J. Environ. Manage. 226, 194–200. doi:10.1016/j.jenvman.2018.08.042
Wen, S., Cheng, W. C., Li, D., and Hu, W. (2023). Immobilizing lead using loess and nanoscale zerovalent iron (nZVI)-amended loess: insights from macroscopic and microscopic tests. Environ.Tech. Innov. 103228, 103228. doi:10.1016/j.eti.2023.103228
Wolfram, L., Friedrich, B., and Eitinger, T. (1995). The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. J. Bacteriol, J. Bacteriol. 177, 1840–1843. doi:10.1128/jb.177.7.1840-1843.1995
Worlock, A. J., and Smith, R. L. (2002). ZntB is a novel Zn2+ transporter in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184, 4369–4373. doi:10.1128/JB.184.16.4369-4373.2002
Xie, Y. X., Cheng, W. C., Wang, L., Xue, Z. F., Rahman, M. M., and Hu, W. (2023). Immobilizing copper in loess soil using microbial-induced carbonate precipitation: insights from test tube experiments and one-dimensional soil columns. J. Hazard. Mater. 444, 130417. doi:10.1016/j.jhazmat.2022.130417
Xie, Y. X., Cheng, W. C., Wang, L., Xue, Z. F., and Xu, Y. L. (2023). Biopolymer-assisted enzyme-induced carbonate precipitation for immobilizing Cu ions in aqueous solution and loess. Environ. Sci. Pollut. Res. 1-13, 116134–116146. doi:10.1007/s11356-023-30665-8
Xue, Z. F., Cheng, W. C., Wang, L., Xie, Y. X., and Qin, P. (2023). Effect of a harsh circular environment on self-healing microbial-induced calcium carbonate materials for preventing Pb2+ migration. Environ.Tech. Innov. 32, 103380. doi:10.1016/j.eti.2023.103380
Yang, H. C., Fu, H. L., Lin, Y. F., and Rosen, B. P. (2012). Pathways of arsenic uptake and efflux. Curr. Top. Membr. 69, 325–358. doi:10.1016/B978-0-12-394390-3.00012-4
Keywords: bioremediation, genome analysis, heavy metal resistant bacteria, lead, qPCR, TEM
Citation: El-Beltagi HS, Halema AA, Almutairi ZM, Almutairi HH, Elarabi NI, Abdelhadi AA, Henawy AR and Abdelhaleem HAR (2024) Draft genome analysis for Enterobacter kobei, a promising lead bioremediation bacterium. Front. Bioeng. Biotechnol. 11:1335854. doi: 10.3389/fbioe.2023.1335854
Received: 09 November 2023; Accepted: 18 December 2023;
Published: 08 January 2024.
Edited by:
Wen-Chieh Cheng, Xi’an University of Architecture and Technology, ChinaReviewed by:
Yi-Xin Xie, Xi’an University of Architecture and Technology, ChinaN. F. Islam, N.N. Saikia College, India
Copyright © 2024 El-Beltagi, Halema, Almutairi, Almutairi, Elarabi, Abdelhadi, Henawy and Abdelhaleem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hossam S. El-Beltagi, helbeltagi@kfu.edu.sa; Asmaa A. Halema, assma.ahmed@agr.cu.edu.eg; Zainab M. Almutairi, z.almutairi@psau.edu.sa
†These authors have contributed equally to this work and share first authorship
 Hossam S. El-Beltagi
Hossam S. El-Beltagi Asmaa A. Halema
Asmaa A. Halema Zainab M. Almutairi
Zainab M. Almutairi Hayfa Habes Almutairi5
Hayfa Habes Almutairi5  Abdelhadi A. Abdelhadi
Abdelhadi A. Abdelhadi Ahmed R. Henawy
Ahmed R. Henawy Heba A. R. Abdelhaleem
Heba A. R. Abdelhaleem