PBX1 as Pioneer Factor: A Case Still Open
- Institute of Neurology (Edinger Institute), University Hospital Frankfurt, J. W. Goethe University, Frankfurt, Germany
Pioneer factors are proteins that can recognize their target sites in barely accessible chromatin and initiate a cascade of events that allows for later transcriptional activation of the respective genes. Pioneer factors are therefore particularly well-suited to initiate cell fate changes. To date, only a small number of pioneer factors have been identified and studied in depth, such as FOXD3/FOXA1, OCT4, or SOX2. Interestingly, several recent studies reported that the PBC transcription factor PBX1 can access transcriptionally inactive genomic loci. Here, we summarize the evidence linking PBX1 with transcriptional pioneer functions, suggest potential mechanisms involved and discuss open questions to be resolved.
Transcriptional Pioneer Factors
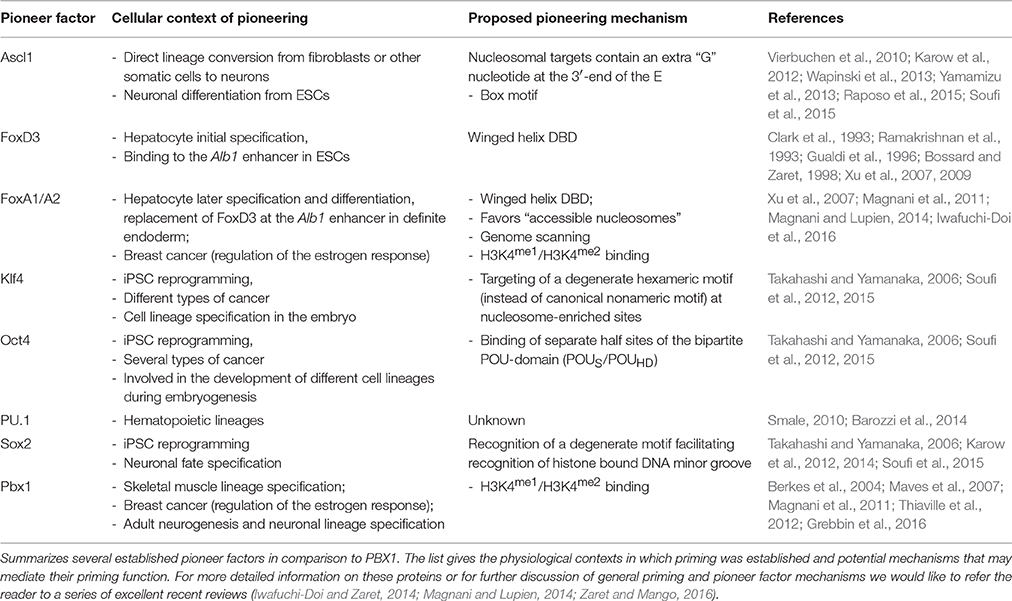
Transcriptional activation or repression of tissue specific genes is typically controlled by a set of sequence specific transcription factors (TFs), together with additional co-regulatory proteins. In addition, there is accumulating evidence that efficient de novo activation of previously silent genes often also depends on DNA–protein interactions and chromatin modifications that occur long before mRNA transcripts of these genes can be detected. These processes are carried out by a special type of DNA binding proteins, termed pioneer TFs (Smale, 2010; Iwafuchi-Doi and Zaret, 2014). Two of the first identified and best studied pioneer factors are the FOX family members FOXD3 and FOXA1. FOXD3 is bound to an enhancer of the liver specific albumin (Alb1) gene already in embryonic stem cells (ESCs), although the gene is not transcriptionally activated until hepatocyte differentiation (Gualdi et al., 1996; Bossard and Zaret, 1998; Xu et al., 2007). During endodermal differentiation, FOXD3 gets downregulated and FOXA1 becomes upregulated and subsequently binds this Alb1 enhancer together with GATA-4 (Gualdi et al., 1996; Bossard and Zaret, 1998). FOXD3 thus serves as a placeholder for FOXA1 and both proteins provide an early molecular anchor for other TFs at the Alb1 enhancer, facilitating later Alb1 gene activation upon differentiation into hepatocytes (Smale, 2010).
Since the 1990s, a number of additional pioneer TFs were discovered. The emerging definition of a pioneer factor comprises the ability to (1) engage its target site(s) in closed chromatin prior to gene activation, (2) increase chromatin accessibility for other proteins at this site, and (3) establish competence for cell fate changes and thus play a key role in cellular (re-) programming (Iwafuchi-Doi and Zaret, 2014). For about two decades, the pressing question of how pioneer factors can access genomic sites in silent chromatin remained a largely unresolved issue, yet recent structural investigations from the Zaret laboratory provide novel advances in the understanding of possible binding modes of pioneer factors to nucleosomal DNA (Soufi et al., 2015). Previously, it was postulated that pioneer factors may access their binding sites in compacted chromatin because of a local destabilization of the nucleosome-DNA contact. Suggested mechanisms of destabilization were the presence of poly(dA-dT) sequences or of histone variants such as H3.3 or H2A.Z (Sekinger et al., 2005; Zhang et al., 2005; Jin et al., 2009). However, genome-wide approaches found that many regulatory regions do not coincide with DNA sequences that assemble into unstable nucleosomes (Zaret and Carroll, 2011). Instead, there is accumulating evidence that pioneer TFs can recognize their target DNA binding sites even in compacted chromatin. The prototype pioneer factor FOXA1, for instance, associates with DNA through a “winged helix”-type DNA binding domain, which structurally resembles the linker histones H1 and H5 (Clark et al., 1993; Ramakrishnan et al., 1993). The FOXA1 C-terminus, on the other hand, can bind to core histones independently of the protein's DNA-binding domain (Cirillo et al., 2002; Sekiya et al., 2009). These structural characteristics enable FOXA1 to penetrate nucleosomal chromatin and, by competition, to displace linker histones. FOXA1 thereby paves the way for other TFs to bind. Consequently, pioneer factors, like linker histones, are retained on mitotic chromosomes and thus might assume a “bookmarking” function during mitosis (Yan et al., 2006; Taube et al., 2010; Zaret et al., 2010; Iwafuchi-Doi et al., 2016). However, not all known pioneer factors possess such linker histone-like properties or winged helix motifs. For example, the reprogramming factors OCT4, SOX2, and KLF4 share the ability to target partial recognition motives on nucleosomes where only one face of the DNA is accessible, yet possess very different DNA binding domains. In addition, recent studies suggest that occupancy of enhancer regions by nucleosomes may actually favor pioneer factor binding and thereby mediate cooperativity between factors that would not necessarily interact on naked DNA (Iwafuchi-Doi and Zaret, 2014; Iwafuchi-Doi et al., 2016).
But what if the nucleosome units are inaccessible, hidden in tight heterochromatin? For a number of pioneer TFs special chromatin binding properties have been reported. An example is the sequence-independent affinity of some bona-fide pioneer TFs to histone modifications, such as mono- or di-methylation of lysine 4 on histone 3 (H3K4me1 or H3K4me2), epigenetic modifications that occur on active enhancers (Cirillo et al., 2002; Sekiya et al., 2009; Magnani et al., 2011). The current view is that this initial, global recruitment to chromatin enables the pioneer TFs to scan the surrounding sequences for their recognition motives (Soufi et al., 2015). Other heterochromatin regions, like those where H3K9me2 or H3K9me3 marks are deposited, remain inaccessible even to pioneer factors (Soufi et al., 2012; Iwafuchi-Doi and Zaret, 2014). Collectively, these studies have begun to shed light onto the versatile mechanisms used by pioneer factors to engage their target sites in closed chromatin (Table 1). However, the sequence of events during initial heterochromatin opening, involving chromatin modifications, pioneer factors, and possibly additional components, are still subject of debate (Choukrallah and Matthias, 2014). In addition, mechanistic details are only known for a small number of pioneer factors at present. Given the enormous complexity of cell lineage decisions during embryonic development and the recent advancements to revert these decisions by cellular reprogramming strategies, many more TFs with pioneering activity may wait to be discovered.
PBC Proteins in Development, Adult Stem Cells, and Cancer
Pre-B cell leukemia (PBC) TFs are evolutionarily conserved, atypical homeodomain proteins. Phylogenetically, they constitute one class of the Three Amino acid Loop Extension-homeodomain (TALE-HD) superclass, a separate branch of the homeodomain proteins characterized by the name-giving Three Amino acid Loop Extension (“TALE”) between the first and second alpha-helix of the homeodomain (Gehring et al., 1994; Bürglin and Affolter, 2016). In animals, TALE-HD proteins can be subdivided into five classes (PBC, MEIS, IRO, MKX, TGIF), of which the PBC-class, consisting of pre-B cell leukemic homeobox (PBX) 1–4 proteins in mammals, will be addressed in more detail here.
PBC proteins were originally identified as HOX-cofactors in D. melanogaster, because mutants of the fly PBC protein extradenticle displayed homeotic transformations similar to those seen in Hox-mutant animals, without altering the expression of the respective Hox genes themselves (Peifer and Wieschaus, 1990; Rauskolb et al., 1993, 1995). Subsequent studies revealed that this protein class contributes to the correct patterning of the anterior–posterior and proximal–distal body axes, confers regional identity in the embryo and is involved in the regulation of proliferation, apoptosis, and differentiation during embryogenesis (Berkes et al., 2004; Ferretti et al., 2011; Gordon et al., 2011; Koss et al., 2012; Yao et al., 2013). For instance, knockout of Pbx1 in mouse embryos leads to embryonic lethality at E15/E16 with hypoplasia or aplasia of several organs, including impaired hematopoiesis, incomplete development of the thymus, spleen agenesis, pancreas hypoplasia, second branchial arch transformation, malformations of cervical vertebrae, ribs, and proximal limbs, and failure of septation of the cardiac outflow tract (DiMartino et al., 2001; Selleri et al., 2001; Manley et al., 2004; Brendolan et al., 2005; Stankunas et al., 2008). The unifying concept emerging from these studies is that PBX proteins act at or near the top of multiple cell fate hierarchies.
Corroborating their multifaceted roles during embryogenesis, dysregulation of PBC proteins is also a frequent phenomenon in cancer. In fact, the mammalian PBX1 protein was first identified in a chromosomal translocation [t(1;19) (q23;p13.3)] in pre-B cell acute lymphoblastic leukemia (ALL) that resulted in the expression of an oncogenic E2A-PBX1 fusion protein (Carroll et al., 1984; Williams et al., 1984; Kamps et al., 1990, 1991). Oncogenic roles of HOX/PBX dimers have also been reported in many other cancers and can be blocked by peptide-based inhibition (Morgan et al., 2007, 2010; Ando et al., 2014; Kelly et al., 2016). In addition, PBX1 plays an important role in estrogen receptor alpha (ERα)-positive breast carcinogenesis (Magnani et al., 2011). In contrast to their rather well-studied roles in embryonic development and cancerous malignancies, the contribution of PBC proteins to adult stem cell niches is still largely unexplored. In the hematopoietic system, PBX1 regulates long term hematopoietic stem cell quiescence, limits myeloid maturation and preserves a lymphoid potential in multipotent progenitor (MPP) and common myeloid progenitor (CMP) pools, thereby regulating and maintaining progenitor reservoirs (Ficara et al., 2013).
The subventricular zone (SVZ) in rodents is an adult stem cell niche that provides new neurons and glia to the brain. In brief, adult stem cells residing in the SVZ produce young neurons, termed neuroblasts, via an intermediate population of transient amplifying progenitor cells. Neuroblasts leave the SVZ and migrate into the olfactory bulb where they terminally differentiate to distinct types of interneurons that are continuously replaced in the existing circuitry as part of a life-long remodeling of the olfactory system. A number of TFs, including DLX2 and PAX6, bias progenitor cells toward a general neuronal fate and promote their subsequent maturation to defined types of interneurons (Hack et al., 2005; Brill et al., 2008). In cooperation with PAX6 and DLX2, the TALE-HD protein MEIS2 regulates neuronal cell fate acquisition, as well as the terminal differentiation of neuroblasts into dopaminergic periglomerular neurons (Agoston et al., 2014). Recently, we characterized the contribution of PBX1 to adult SVZ neurogenesis (Grebbin et al., 2016). We observed high PBX1 expression in rapidly proliferating SVZ progenitors and neuroblasts, as well as in subsets of their progenies in the olfactory bulb, including dopaminergic neurons. Targeted deletion of Pbx1 in transient amplifying progenitor cells (in a Pbx2-deficient background to prevent functional compensation by this structurally related gene) significantly reduced the production of neurons and increased the generation of oligodendrocytes in vitro and in vivo, establishing Pbx1 as an early lineage regulator of SVZ neurogenesis. Loss of Pbx1 expression in neuronally committed neuroblasts, by contrast, severely compromised cell survival. By chromatin immunoprecipitation from endogenous tissues or isolated cells, we identified the neuron-specific gene doublecortin (Dcx) and the dopaminergic neuron marker gene tyrosine hydroxylase (Th), as direct PBX1 target genes.
Notably, PBX1 binds to its target sites in promoter/enhancer regions of these genes already in undifferentiated progenitor cells and hence at times that significantly precede the transcriptional activation of both genes. The Dcx gene encodes a microtubule-associated protein, which is expressed by all migrating neuroblasts and therefore a frequently used marker for young neurons (Francis et al., 1999; Gleeson et al., 1999). Primary cultures of proliferating adult SVZ neural stem- and progenitor cells (neurospheres) are DCX negative, but the protein becomes quickly upregulated once differentiation is induced. Although undifferentiated neurospheres do not yet express DCX and the Dcx locus exhibits very low levels of the activating epigenetic histone modification H3K4me3, a significant enrichment of PBX1 at the Dcx promoter was observed, indicating that PBX chromatin binding preceded Dcx gene activation. In contrast to the relatively fast upregulation of pan-neuronal markers like Dcx, terminal differentiation and integration of adult generated neurons into the existing circuitry of the olfactory bulb represent the last steps of neuronal turnover during adult neurogenesis. Full maturation of dopaminergic neurons is a particularly slow process and ~2 months pass before an adult SVZ-generated neuron reaches the olfactory bulb and achieves a level of cellular maturation at which Th expression is initiated (Brill et al., 2008). Unexpectedly, PBX1 occupies the Th promoter/proximal enhancer already in progenitor cells and newborn neuroblasts in the SVZ, suggesting that priming by PBX1 can precede transcriptional activation of the Th gene by several months.
PBX Pioneering?
Our observation that PBX1 may bind its targets in silent gene loci does not stand alone. In fact, the first evidence for a pioneering role of PBX1 was provided by Berkes et al. (2004). The authors observed that during skeletal muscle differentiation PBX1 is constitutively bound to the promoter of the Myogenin gene. Differentiation and activation of Myogenin expression are subsequently initiated by the pro-myogenic transcription factor MYOD through interaction with pre-bound PBX1. At the onset of myogenic differentiation, PBX1 thus seems to serve as a platform for MYOD binding in inactive chromatin, thereby preparing genes of the skeletal muscle lineage for activation (Berkes et al., 2004; Maves et al., 2007). This finding was supported in 2011 by a report describing PBX1 as a pioneer factor in ERα-signaling in breast cancer (Magnani et al., 2011). Using the cell line MCF7 as model to investigate the activation of oncogenic ERα target genes during breast cancer progression, the study identified PBX1 as “partner” pioneer factor to FOXA1. PBX1 is pre-bound to shared PBX1-ERα binding sites proximal to genes involved in cancer cell proliferation prior to estrogen application and its binding to these sites remains following estrogen treatment. Although FOXA1 and PBX1 pioneering functions are independent from each other, the presence of both factors has a synergistic effect on chromatin openness on shared binding sites (Magnani et al., 2011). Together, PBX1 pre-loading to chromatin before estrogen treatment and its ability to induce chromatin opening argue for a pioneering function in this context.
In contrast to FOXA1, little is known about the mechanisms that may allow PBX1 to access silent chromatin. However, the detailed structural analysis of the iPSC reprogramming factors Oct4, Sox2, Klf4, and c-Myc may provide a hint. Specifically, the length of the basic helix-loop-helix domain of bHLH proteins appears to inversely correlate with pioneer activity, as short bHLH basic regions facilitate nucleosomal DNA binding, whereas proteins with longer bHLH basic regions depend on the cooperation with other pioneer factors (Nair and Burley, 2003; Sauvé et al., 2004; Soufi et al., 2015). The ability of a bHLH protein to access closed chromatin thereby appears to be either defined by the way helix 1 contacts DNA or by cooperation with additional factors, leading to recognition of target sequences which may contain partial, degenerate or altered motifs (Soufi et al., 2015). In this context it is intriguing to consider that the TALE-homeodomain, owing to the insertion of three amino acids between helix 1 and helix 2, is structurally distinct from other homeodomains (Gehring et al., 1994; Bürglin and Affolter, 2016). This raises the possibility that the TALE-HD might be especially suited to recognize its consensus motif on nucleosomal DNA, but to date mechanistic details remain unknown.
An alternative scenario was described by Magnani et al. who reported that PBX1, as had been previously shown for FOXA1, preferably associates with nucleosomes carrying H3K4me1 and H3K4me2 modifications (Magnani et al., 2011; Sérandour et al., 2011; Jozwik et al., 2016). Mono- and di-methylation of H3K4 mark distal enhancers that are linked to active or poised genes and can be found as biochemical intermediates preceding the triple-methylation of H3K4 at transcriptionally active promoters. As H3K4me1 deposition is catalyzed by the Set/MLL family proteins MLL3 and MLL4, this would paradoxically suggest that FOXA1 and PBX1 binding to DNA requires prior histone modification by MLL3/4, a notion that is difficult to reconcile with the proven or proposed pioneering activity of FOXA1 and PBX1, respectively (Hu et al., 2013). A possible explanation comes from the observation that the relationship between pioneer factor binding and H3K4 epigenetic modification seems to be bidirectional: The pioneer factors FOXA1 and PU.1 not only preferentially bind to chromatin carrying H3K4me1/H3K4me2 modifications, but their association in turn promotes H3K4me1/2 deposition (Heinz et al., 2010; Sérandour et al., 2011). The essential question of which is present on the chromatin first, the nucleosomal modification or the pioneer factor, cannot be conclusively answered. At present, examples for both cases exist and quite possibly, pioneer factor binding and epigenetic histone modifications stabilize each other. Moreover, unknown additional players might be involved, such as long or short non-coding RNAs. In any respect, further efforts are needed to decipher the whole spectrum of mechanisms used by pioneer factors to access closed chromatin.
In conclusion, multiple lines of evidence, obtained from in vivo and in vitro studies and made in the context of embryonic development, adult stem cell differentiation and cancer, suggest that PBX1 may act as pioneer factor. The three hallmarks of pioneer factor function—target site binding in closed chromatin, the ability to increase DNA access for other proteins and active involvement in cell fate specification or cellular (re-)programming—have all been demonstrated for PBX1. Yet, each of these features has been investigated in a different physiological setting and with different tools. A unifying model is therefore still missing. In addition, little is known about whether the other three members of the mammalian PBC family, PBX2-4, also possess the ability to recognize their target sites in silent chromatin or even pioneer factor activity. A coherent assessment of PBX pioneering function by standardized approaches is therefore an important next step in research on this protein family.
Author Contributions
DS and BG jointly wrote the manuscript and approved it for publication.
Funding
Research related to this Review was funded by the Deutsche Forschungsgemeinschaft, grant SCHU1218/3-1 to DS and a Ludwig Edinger Fellowship to BG.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CC and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
References
Agoston, Z., Heine, P., Brill, M. S., Grebbin, B. M., Hau, A.-C., Kallenborn-Gerhardt, W., et al. (2014). Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development 141, 28–38. doi: 10.1242/dev.097295
Ando, H., Natsume, A., Senga, T., Watanabe, R., Ito, I., Ohno, M., et al. (2014). Peptide-based inhibition of the HOXA9/PBX interaction retards the growth of human meningioma. Cancer Chemother. Pharmacol. 73, 53–60. doi: 10.1007/s00280-013-2316-5
Barozzi, I., Simonatto, M., Bonifacio, S., Yang, L., Rohs, R., Ghisletti, S., et al. (2014). Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol. Cell 54, 844–857. doi: 10.1016/j.molcel.2014.04.006
Berkes, C. A., Bergstrom, D. A., Penn, B. H., Seaver, K. J., Knoepfler, P. S., and Tapscott, S. J. (2004). Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14, 465–477. doi: 10.1016/S1097-2765(04)00260-6
Bossard, P., and Zaret, K. S. (1998). GATA transcription factors as potentiators of gut endoderm differentiation. Development 125, 4909–4917.
Brendolan, A., Ferretti, E., Salsi, V., Moses, K., Quaggin, S., Blasi, F., et al. (2005). A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development 132, 3113–3126. doi: 10.1242/dev.01884
Brill, M. S., Snapyan, M., Wohlfrom, H., Ninkovic, J., Jawerka, M., Mastick, G. S., et al. (2008). A dlx2- and pax6-dependent transcriptional code for periglomerular neuron specification in the adult olfactory bulb. J. Neurosci. 28, 6439–6452. doi: 10.1523/JNEUROSCI.0700-08.2008
Bürglin, T. R., and Affolter, M. (2016). Homeodomain proteins: an update. Chromosoma 125, 497–521. doi: 10.1007/s00412-015-0543-8
Carroll, A. J., Crist, W. M., Parmley, R. T., Roper, M., Cooper, M. D., and Finley, W. H. (1984). Pre-B cell leukemia associated with chromosome translocation 1;19. Blood 63, 721–724.
Choukrallah, M. A., and Matthias, P. (2014). The interplay between chromatin and transcription factor networks during b cell development: who pulls the trigger first? Front. Immunol. 5:156. doi: 10.3389/fimmu.2014.00156
Cirillo, L. A., Lin, F. R., Cuesta, I., Friedman, D., Jarnik, M., and Zaret, K. S. (2002). Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289. doi: 10.1016/S1097-2765(02)00459-8
Clark, K. L., Halay, E. D., Lai, E., and Burley, S. K. (1993). Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420. doi: 10.1038/364412a0
DiMartino, J. F., Selleri, L., Traver, D., Firpo, M. T., Rhee, J., Warnke, R., et al. (2001). The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood 98, 618–626. doi: 10.1182/blood.V98.3.618
Ferretti, E., Li, B., Zewdu, R., Wells, V., Hebert, J. M., Karner, C., et al. (2011). A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev. Cell 21, 627–641. doi: 10.1016/j.devcel.2011.08.005
Ficara, F., Crisafulli, L., Lin, C., Iwasaki, M., Smith, K. S., Zammataro, L., et al. (2013). Pbx1 restrains myeloid maturation while preserving lymphoid potential in hematopoietic progenitors. J. Cell Sci. 126, 3181–3191. doi: 10.1242/jcs.125435
Francis, F., Koulakoff, A., Boucher, D., Chafey, P., Schaar, B., Vinet, M. C., et al. (1999). Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron 23, 247–256.
Gehring, W. J., Affolter, M., and Bürglin, T. (1994). Homeodomain proteins. Annu. Rev. Biochem. 63, 487–526. doi: 10.1146/annurev.bi.63.070194.002415
Gleeson, J. G., Lin, P. T., Flanagan, L. A., and Walsh, C. A. (1999). Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron 23, 257–271.
Gordon, J. A. R., Hassan, M. Q., Koss, M., Montecino, M., and Selleri, L., van Wijnen, A. J., et al. (2011). Epigenetic regulation of early osteogenesis and mineralized tissue formation by a HOXA10-PBX1-associated complex. Cells Tissues Organs 194, 146–150. doi: 10.1159/000324790
Grebbin, B. M., Hau, A.-C., Groß, A., Anders-Maurer, M., Schramm, J., Koss, M., et al. (2016). Pbx1 is required for adult subventricular zone neurogenesis. Development 143, 2281–2291. doi: 10.1242/dev.128033
Gualdi, R., Bossard, P., Zheng, M., Hamada, Y., Coleman, J. R., and Zaret, K. S. (1996). Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 10, 1670–1682. doi: 10.1101/gad.10.13.1670
Hack, M. A., Saghatelyan, A., de Chevigny, A., Pfeifer, A., Ashery-Padan, R., Lledo, P.-M., et al. (2005). Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat. Neurosci. 8, 865–872. doi: 10.1038/nn1479
Heinz, S., Benner, C., Spann, N., Bertolino, E., Lin, Y. C., Laslo, P., et al. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. doi: 10.1016/j.molcel.2010.05.004
Hu, D., Gao, X., Morgan, M. A., Herz, H. M., Smith, E. R., and Shilatifard, A. (2013). The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 33, 4745–4754. doi: 10.1128/MCB.01181-13
Iwafuchi-Doi, M., Donahue, G., Kakumanu, A., Watts, J. A., Mahony, S., Pugh, B. F., et al. (2016). The pioneer transcription factor foxa maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62, 79–91. doi: 10.1016/j.molcel.2016.03.001
Iwafuchi-Doi, M., and Zaret, K. S. (2014). Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692. doi: 10.1101/gad.253443.114
Jin, C., Zang, C., Wei, G., Cui, K., Peng, W., Zhao, K., et al. (2009). H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat. Genet. 41, 941–945. doi: 10.1038/ng.409
Jozwik, K. M., Chernukhin, I., Serandour, A. A., Nagarajan, S., and Carroll, J. S. (2016). FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Rep. 17, 2715–2723. doi: 10.1016/j.celrep.2016.11.028
Kamps, M. P., Look, A. T., and Baltimore, D. (1991). The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Genes Dev. 5, 358–368.
Kamps, M. P., Murre, C., Sun, X. H., and Baltimore, D. (1990). A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell 60, 547–555. doi: 10.1016/0092-8674(90)90658-2
Karow, M., Sánchez, R., Schichor, C., Masserdotti, G., Ortega, F., Heinrich, C., et al. (2012). Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell 11, 471–476. doi: 10.1016/j.stem.2012.07.007
Karow, M., Schichor, C., Beckervordersandforth, R., and Berninger, B. (2014). Lineage-reprogramming of pericyte-derived cells of the adult human brain into induced neurons. J. Vis. Exp. e51433. doi: 10.3791/51433
Kelly, Z., Moller-Levet, C., McGrath, S., Butler-Manuel, S., Kavitha Madhuri, T., Kierzek, A. M., et al. (2016). The prognostic significance of specific HOX gene expression patterns in ovarian cancer. Int. J. Cancer 139, 1608–1617. doi: 10.1002/ijc.30204
Koss, M., Bolze, A., Brendolan, A., Saggese, M., Capellini, T. D., Bojilova, E., et al. (2012). Congenital asplenia in mice and humans with mutations in a Pbx/Nkx2-5/p15 module. Dev. Cell 22, 913–926. doi: 10.1016/j.devcel.2012.02.009
Magnani, L., Ballantyne, E. B., Zhang, X., and Lupien, M. (2011). PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer. PLoS Genet. 7:e1002368. doi: 10.1371/journal.pgen.1002368
Magnani, L., and Lupien, M. (2014). Chromatin and epigenetic determinants of estrogen receptor alpha (ESR1) signaling. Mol. Cell. Endocrinol. 382, 633–641. doi: 10.1016/j.mce.2013.04.026
Manley, N. R., Selleri, L., Brendolan, A., Gordon, J., and Cleary, M. L. (2004). Abnormalities of caudal pharyngeal pouch development in Pbx1 knockout mice mimic loss of Hox3 paralogs. Dev. Biol. 276, 301–312. doi: 10.1016/j.ydbio.2004.08.030
Maves, L., Waskiewicz, A. J., Paul, B., Cao, Y., Tyler, A., Moens, C. B., et al. (2007). Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development 134, 3371–3382. doi: 10.1242/dev.003905
Morgan, R., Pirard, P. M., Shears, L., Sohal, J., Pettengell, R., and Pandha, H. S. (2007). Antagonism of HOX/PBX dimer formation blocks the in vivo proliferation of melanoma. Cancer Res. 67, 5806–5813. doi: 10.1158/0008-5472.CAN-06-4231
Morgan, R., Plowright, L., Harrington, K. J., Michael, A., and Pandha, H. S. (2010). Targeting, HOX and PBX transcription factors in ovarian cancer. BMC Cancer 10:89. doi: 10.1186/1471-2407-10-89
Nair, S. K., and Burley, S. K. (2003). X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205. doi: 10.1016/S0092-8674(02)01284-9
Peifer, M., and Wieschaus, E. (1990). Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 4, 1209–1223. doi: 10.1101/gad.4.7.1209
Ramakrishnan, V., Finch, J. T., Graziano, V., Lee, P. L., and Sweet, R. M. (1993). Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362, 219–223. doi: 10.1038/362219a0
Raposo, A. A. S. F., Vasconcelos, F. F., Drechsel, D., Marie, C., Johnston, C., Dolle, D., et al. (2015). Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. 10, 1544–1556. doi: 10.1016/j.celrep.2015.02.025
Rauskolb, C., Peifer, M., and Wieschaus, E. (1993). Extradenticle, a regulator of homeotic gene activity, is a homolog of the homeobox-containing human proto-oncogene pbx1. Cell 74, 1101–1112. doi: 10.1016/0092-8674(93)90731-5
Rauskolb, C., Smith, K. M., Peifer, M., and Wieschaus, E. (1995). Extradenticle determines segmental identities throughout Drosophila development. Development 121, 3663–3673.
Sauvé, S., Tremblay, L., and Lavigne, P. (2004). The NMR solution structure of a mutant of the Max b/HLH/LZ free of DNA: insights into the specific and reversible DNA binding mechanism of dimeric transcription factors. J. Mol. Biol. 342, 813–832. doi: 10.1016/j.jmb.2004.07.058
Sekinger, E. A., Moqtaderi, Z., and Struhl, K. (2005). Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18, 735–748. doi: 10.1016/j.molcel.2005.05.003
Sekiya, T., Muthurajan, U. M., Luger, K., Tulin, A. V., and Zaret, K. S. (2009). Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 23, 804–809. doi: 10.1101/gad.1775509
Selleri, L., Depew, M. J., Jacobs, Y., Chanda, S. K., Tsang, K. Y., Cheah, K. S., et al. (2001). Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128, 3543–3557.
Sérandour, A. A., Avner, S., Percevault, F., Demay, F., Bizot, M., Lucchetti-Miganeh, C., et al. (2011). Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 21, 555–565. doi: 10.1101/gr.111534.110
Smale, S. T. (2010). Pioneer factors in embryonic stem cells and differentiation. Curr. Opin. Genet. Dev. 20, 519–526. doi: 10.1016/j.gde.2010.06.010
Soufi, A., Donahue, G., and Zaret, K. S. (2012). Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell 151, 994–1004. doi: 10.1016/j.cell.2012.09.045
Soufi, A., Garcia, M. F., Jaroszewicz, A., Osman, N., Pellegrini, M., and Zaret, K. S. (2015). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568. doi: 10.1016/j.cell.2015.03.017
Stankunas, K., Shang, C., Twu, K. Y., Kao, S.-C., Jenkins, N. A., Copeland, N. G., et al. (2008). Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ. Res. 103, 702–709. doi: 10.1161/CIRCRESAHA.108.175489
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Taube, J. H., Allton, K., Duncan, S. A., Shen, L., and Barton, M. C. (2010). Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J. Biol. Chem. 285, 16135–16144. doi: 10.1074/jbc.M109.088096
Thiaville, M. M., Stoeck, A., Chen, L., Wu, R.-C., Magnani, L., Oidtman, J., et al. (2012). Identification of PBX1 target genes in cancer cells by global mapping of PBX1 binding sites. PLoS ONE 7:e36054. doi: 10.1371/journal.pone.0036054
Vierbuchen, T., Ostermeier, A., Pang, Z. P., Kokubu, Y., Südhof, T. C., and Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. doi: 10.1038/nature08797
Wapinski, O. L., Vierbuchen, T., Qu, K., Lee, Q. Y., Chanda, S., Fuentes, D. R., et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635. doi: 10.1016/j.cell.2013.09.028
Williams, D. L., Look, A. T., Melvin, S. L., Roberson, P. K., Dahl, G., Flake, T., et al. (1984). New chromosomal translocations correlate with specific immunophenotypes of childhood acute lymphoblastic leukemia. Cell 36, 101–109. doi: 10.1016/0092-8674(84)90078-3
Xu, J., Pope, S. D., Jazirehi, A. R., Attema, J. L., Papathanasiou, P., Watts, J. A., et al. (2007). Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 12377–12382. doi: 10.1073/pnas.0704579104
Xu, J., Watts, J. A., Pope, S. D., Gadue, P., Kamps, M., Plath, K., et al. (2009). Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 23, 2824–2838. doi: 10.1101/gad.1861209
Yamamizu, K., Piao, Y., Sharov, A. A., Zsiros, V., Yu, H., Nakazawa, K., et al. (2013). Identification of transcription factors for lineage-specific ESC differentiation. Stem cell Rep. 1, 545–559. doi: 10.1016/j.stemcr.2013.10.006
Yan, J., Xu, L., Crawford, G., Wang, Z., and Burgess, S. M. (2006). The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol. Cell. Biol. 26, 155–168. doi: 10.1128/MCB.26.1.155-168.2006
Yao, Z., Farr, G. H. III, Tapscott, S. J., and Maves, L. (2013). Pbx and Prdm1a transcription factors differentially regulate subsets of the fast skeletal muscle program in zebrafish. Biol. Open 2, 546–555. doi: 10.1242/bio.20133921
Zaret, K. S., Caravaca, J. M., Tulin, A., and Sekiya, T. (2010). Nuclear mobility and mitotic chromosome binding similarities between pioneer transcription factor FoxA and linker histone H1. Cold Spring Harb. Symp. Quant. Biol. 75, 219–226. doi: 10.1101/sqb.2010.75.061
Zaret, K. S., and Carroll, J. S. (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241. doi: 10.1101/gad.176826.111
Zaret, K. S., and Mango, S. E. (2016). Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 37, 76–81. doi: 10.1016/j.gde.2015.12.003
Keywords: TALE homeodomain protein, PBX, pioneer factor, adult stem cell, cell fate specification, chromatin remodeling
Citation: Grebbin BM and Schulte D (2017) PBX1 as Pioneer Factor: A Case Still Open. Front. Cell Dev. Biol. 5:9. doi: 10.3389/fcell.2017.00009
Received: 30 November 2016; Accepted: 31 January 2017;
Published: 14 February 2017.
Edited by:
Thimios Mitsiadis, University of Zurich, SwitzerlandReviewed by:
Eumorphia Remboutsika, BSRC ≪Alexander Fleming≫, GreeceStavros Malas, The Cyprus Institute of Neurology and Genetics, Cyprus
Claudio Cantù, University of Zurich, Switzerland
Copyright © 2017 Grebbin and Schulte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dorothea Schulte, dorothea.schulte@kgu.de
†Present Address: Britta M. Grebbin, Institute for Tumorbiology and Experimental Therapy, Frankfurt, Germany
 Britta M. Grebbin†
Britta M. Grebbin†  Dorothea Schulte
Dorothea Schulte