miRNAs in NK Cell-Based Immune Responses and Cancer Immunotherapy
- 1Department of Experimental Medicine, University of Genoa, Genoa, Italy
- 2Center of Excellence for Biomedical Research, University of Genoa, Genoa, Italy
- 3Histological and Anatomical Pathology Unit, Department of Laboratory and Service, E.O. Galliera Hospital, Genova, Italy
- 4Tumor Immunology Laboratory (TIMM) Laboratory at Sahlgrenska Cancer Center, Department of Infectious Diseases, Institute of Biomedicine, University of Gothenburg, Gothenburg, Sweden
- 5IRCCS Ospedale Policlinico San Martino, UO Bioterapie, Genoa, Italy
- 6Department of Earth Science, Environment and Life (DISTAV), University of Genoa, Genoa, Italy
The incidence of certain forms of tumors has increased progressively in recent years and is expected to continue growing as life expectancy continues to increase. Tumor-infiltrating NK cells may contribute to develop an anti-tumor response. Optimized combinations of different cancer therapies, including NK cell-based approaches for targeting tumor cells, have the potential to open new avenues in cancer immunotherapy. Functional inhibitory receptors on NK cells are needed to prevent their attack on healthy cells. Nevertheless, disruption of inhibitory receptors function on NK cells increases the cytotoxic capacity of NK cells against cancer cells. MicroRNAs (miRNAs) are small non-coding RNA molecules that target mRNA and thus regulate the expression of genes involved in the development, maturation, and effector functions of NK cells. Therapeutic strategies that target the regulatory effects of miRNAs have the potential to improve the efficiency of cancer immunotherapy. Interestingly, emerging evidence points out that some miRNAs can, directly and indirectly, control the surface expression of immune checkpoints on NK cells or that of their ligands on tumor cells. This suggests a possible use of miRNAs in the context of anti-tumor therapy. This review provides the current overview of the connections between miRNAs and regulation of NK cell functions and discusses the potential of these miRNAs as innovative biomarkers/targets for cancer immunotherapy.
miRNAs as Key Regulators of Gene Expression
About 2,000 human miRNAs are currently recognized. These are small RNAs which originate from longer precursors (Pri-miRNAs) mainly transcribed by the RNA polymerase II (Macfarlane and Murphy, 2010). These Pri-miRNAs undergo a precisely coordinated maturation process involving several steps. In the nucleus, the RNAse III Drosha, supported by the DiGeorge critical region 8 (DGCR8), converts them in short hairpin intermediates of 70–120 nucleotides-long (Pre-miRNAs) (Gregory et al., 2004). After transport to the cytoplasm by exportin 5 (Yi et al., 2003), Pre-miRNAs are then processed by the RNAse III Dycer into mature miRNAs which are duplexes of approximately 22 nucleotides. One strand of the duplex is incorporated along with the argonaute protein in the miRNA-induced silencing complex RISC (Diederichs and Haber, 2007). These complexes promote the pairing of miRNA nucleotide sequences to their target sequences on 3′ UTR sequences of mRNAs and RISC cofactors mediate site-specific cleavage, degradation of the target mRNA, or inhibition of its translation in protein (Gu and Kay, 2010). It is well-known that miRNA can repress the expression of hundreds of different mRNAs. Furthermore, as several different miRNA target sequences may be present on the 3′ UTR of a single mRNA, complex networks of cooperative regulation by several miRNAs may affect the stability or the translation of a multitude of mRNAs (Filipowicz et al., 2008; Liu et al., 2009, 2014). As miRNA recognition sequences appear to be present on most of the protein-coding human mRNAs, the role of miRNAs as regulators of gene expression is quite relevant in mammalian development physiology and pathology (Dallaire and Simard, 2016; Peng and Croce, 2016; Ivanova et al., 2018; Johnson, 2019; Horsburgh et al., 2017). Therefore, miRNAs and in particular those present in body fluids and blood, either as free molecules or included in extracellular vesicles, are receiving increasing attention as possible disease biomarkers (Mori et al., 2019).
NK Cells as Innate Immune Cells With a Key Role in Fighting Viral Infections and in the Surveillance Against Malignant Transformation
Natural Killer (NK) cells represent cytotoxic, innate lymphoid cells (ILCs) (Cortez and Colonna, 2016), and their main function is to provide the organism with a rapid immune response against infections, autologous transformed cells, and allogeneic cells (Vivier et al., 2011; Del Zotto et al., 2017; Freud et al., 2017). In fact, NK cells do not need to be primed with antigens to become fully functional and the mechanisms of non-self recognition do not rely on genomic recombination and subsequent cell clone expansion events.
Nowadays it is recognized that these cells mediate immune-surveillance also via regulatory functions by secreting cytokines, primarily interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α), and by interacting with other immune or adaptive immune cells (Marcenaro et al., 2006; Vivier et al., 2011; Riise et al., 2015; Pesce et al., 2017b; Bernson et al., 2019). In turn, NK cells can respond to different types of chemokines and cytokines produced by other immune cells (Marcenaro et al., 2005a,b, 2006; Moretta et al., 2006; Parodi et al., 2015; Pesce et al., 2016).
NK cells are not a homogeneous population, but there are different NK subsets that differ in phenotype, maturational step, and functions. Among circulating mature NK cells, two main subsets can be identified: regulatory NKs (CD56bright/CD16−), which are the most abundant in secondary lymphoid organs (SLO) and display the ability to secrete high amounts of pro-inflammatory cytokines, and cytotoxic NKs (CD56dim/CD16+), which represent about 90% of circulating NK cells (Farag and Caligiuri, 2006; Carrega and Ferlazzo, 2012; Del Zotto et al., 2017).
SLO have been suggested being the anatomical sites where NK cells complete their maturation process that is associated with the transition from a CD56bright to a CD56dim phenotype, acquisition of self-tolerance and lytic activity (Romagnani et al., 2007; Yu et al., 2010).
Both the killing and immune-regulative functions of NK cells depend on a balance of activating or inhibiting signals that originate from NK receptors (NKRs) (activating NKR-aNKR and inhibitory NKR -iNKR-, respectively). The iNKRs include the human leucocyte antigens (HLA) class I-specific Killer Ig-like receptors (KIRs) recognizing allotypic determinants shared by groups of classical HLA-ABC alleles (Moretta et al., 1996), the leukocyte immunoglobulin-like receptor subfamily B member 1 (LILRB1) that is specific for different HLA-class I molecules (Cosman et al., 1997), and the CD94/NKG2A heterodimer specific for HLA-E (Braud et al., 1998) ad well as additional non-HLA-I specific inhibitory receptors, including programmed cell death protein 1 (PD-1), T cell immunoreceptor with Ig and ITIM domains (TIGIT), T-cell immunoglobulin domain and mucin domain 3 (TIM-3), lymphocyte-activation gene 3 (LAG-3), and CD96 (Di Vito et al., 2019). The activating NKRs (aKIRs) include non-HLA-specific receptors such as NCRs (NKp30, NKp44 and NKp46), NKG2D, DNAM-1, NKp80, CD59, NTB-A, and 2B4 (Moretta et al., 2001) as well as the activating HLA class I-specific Killer Ig-like receptors and the HLA-E specific CD94/NKG2C heterodimer. NK cells can also express different Toll-like receptors (TLR), including TLR2, TLR3, TLR5, TLR7, TLR8, and TLR9 (Sivori et al., 2004; Hart et al., 2005; Tsujimoto et al., 2005; Marcenaro et al., 2008; Voo et al., 2014). These receptors, by recognizing conserved pathogen structures, induce NK cell activation (Della Chiesa et al., 2014).
During NK cell development/differentiation, CD94/NKG2A is the first HLA-I specific receptor to be expressed. It appears on the most immature CD56bright CD16neg/dim NK cell subset. After several maturation steps, CD56bright cells become CD56dim CD16+, lose NKG2A and acquire KIR and LILRB1 receptors (Di Santo, 2006; Freud and Caligiuri, 2006; Romagnani et al., 2007). The most mature NK cells are KIR+ (and/or LILRB1+), NKG2A− CD16bright and express the marker of terminal differentiation, CD57 (Moretta et al., 2004; Bjorkstrom et al., 2010; Marcenaro et al., 2017).
Under homeostatic conditions, NK cells continuously receive inhibitory signals mainly originating from the interaction between iNKRs and a large spectrum of classical and non-classical HLA-I molecules expressed on the surface of autologous cells (self-cells). Allogeneic or viral-infected or tumor cells often downregulate or lack altogether the expression of these antigens and therefore fail to be recognized as self-cells by the iNKRs. Under these conditions, the signaling from the aNKRs, engaged with ligands displayed on target cells, prevails, and the NK-mediated killing of these non-self cells is unleashed. Notably, in tumors that maintain the expression of HLA-I molecules, the iNKRs function as immune checkpoints and block the cytotoxic activity of NK cells (Romagne et al., 2009; Vey et al., 2012; Kohrt et al., 2014).
Several strategies have been forwarded to strenghten NK cell activity against HLA-I-expressing cancer cells. For example, IL-2-based immunotherapy allows NK cells to override inhibitory signals from acute myeloid leukemia (AML) blasts (Hallner et al., 2019), and recently, immunotherapies based on the use of therapeutic monoclonal antibodies specific for iNKRs, in particular anti-pan-KIR2D (lirilumab) (Romagne et al., 2009; Kohrt et al., 2014; Vey et al., 2018) and anti-NKG2A (monalizumab), have been developed (André et al., 2018; Tinker et al., 2019; Zaghi et al., 2019). These agents efficiently disrupt the interaction between these NK cell immune checkpoints and their ligands, and will in this way enable NK cells to efficiently kill also HLA-I+ tumor cells (Chiossone et al., 2017; Di Vito et al., 2019).
In addition, the microenvironment of chronic infections and tumors may lead to NK cell phenotypic changes and impairment of NK cell functions (Bi and Tian, 2017). The most frequently NK cell phenotypic changes are represented by downregulation of the aNKRs expression (Costello et al., 2002; Romero et al., 2006; Carlsten et al., 2009; Pesce et al., 2015; Han et al., 2018; Poznanski and Ashkar, 2019) and/or upregulation/de novo expression of iNKRs (Carlsten et al., 2009; Di Vito et al., 2019; Sanchez-Correa et al., 2019). In fact, it has been unveiled that besides T lymphocytes also NK cells can express PD-1, an immune checkpoint specific for the PD-L1/2 molecules often displayed on the surface of tumor cells (Pesce et al., 2019b).
PD-1 is expressed on a subset of fully mature (KIR+CD57+NKG2A−) NK cells from one-fourth of human cytomegalovirus (HCMV) seropositive individuals (Della Chiesa et al., 2016; Pesce et al., 2017a; Mariotti et al., 2019). Increased proportions of PD-1+ NK cells can be observed in patients affected by different types of tumors (Beldi-Ferchiou et al., 2016; Pesce et al., 2017a, 2019a,b; André et al., 2018). Accordingly, studies suggest a role for NK cells in immunotherapy targeting the PD-1/PD-L1 axis (Hsu et al., 2018) and this is clinically relevant for patients with tumors characterized by a T cell resistant (HLA-Ineg) phenotype.
Apart from the wide-spread use of checkpoint inhibitors in melanoma, lung cancer etc., agents blocking the PD-1/PD-L1 axis are currently being evaluated in clinical trials on both hematologic and solid tumors as monotherapy or in combination with other agents, including other forms of immune checkpoint blockade, such as anti-panKIR2D and anti-NKG2A antibodies in the case of HLA-I+ tumor cells (Moretta et al., 1996, 2001; Cosman et al., 1997; Braud et al., 1998; Sivori et al., 2004; Marcenaro et al., 2008; Di Vito et al., 2019).
In summary, NK cell activation depends on the nature of interactions between inhibitory/activating receptors on their surface and the relative ligands on target cells, and thus receptor/ligand pairs could represent key checkpoints in the regulation of anti-tumor NK cell activity and in the planning of innovative NK cell-based immunotherapy.
miRNAs as Regulators of NK Cells Survival, Development/Maturation, and Functions
Numerous studies showed that miRNAs play a relevant role in the regulation of NK cell survival, development/maturation, activation, proliferation, cytotoxicity, and cytokine production both in healthy and pathological conditions (i.e., tumors/viral infections) by targeting receptors or factors involved in transcriptional expression (Table 1).
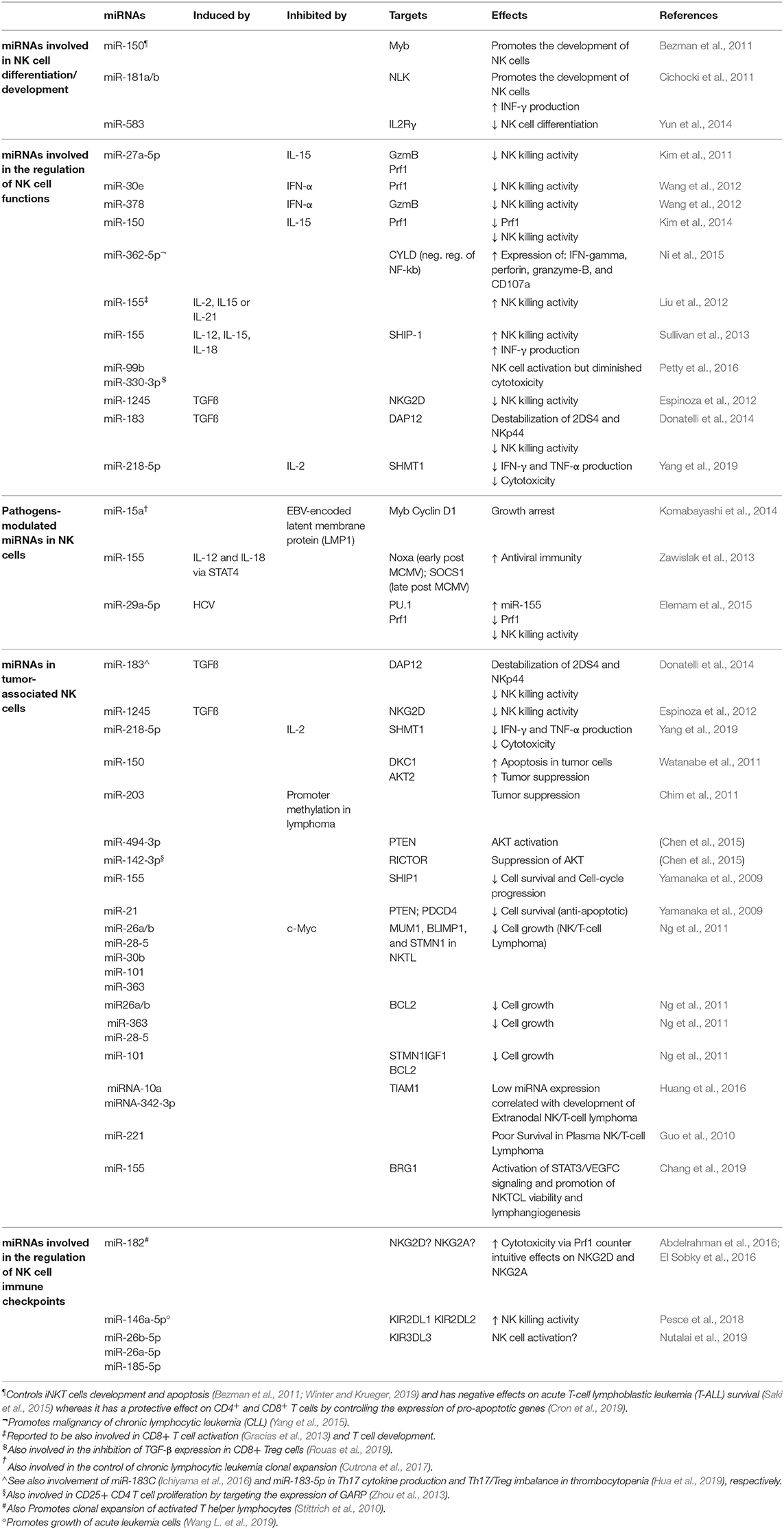
Table 1. Examples of miRNAs expressed in NK cells and involved in the modulation of several aspects of NK cell development and functions.
miRNAs Involved in NK Cell Differentiation/Development
The first evidence of the important role played by miRNAs within the immune system was provided by genetic studies showing a critical requirement for Dicer in vivo. Conditional deletion of Dicer in various hematopoietic lineages in mice produced defects, such as impaired cell differentiation, proliferation, and survival (Muljo et al., 2005; Cobb et al., 2006; Koralov et al., 2008; Liston et al., 2008; Fedeli et al., 2009).
Bezman et al. (2010) investigated the role of miRNAs by ablation of the miRNA biogenesis pathway, through deletion of Dicer or Dgcr8 in the mature murine peripheral NK cells. Dicer- and Dgcr8- deficient NK cells showed an increased cell death supporting the important role of miRNAs in controlling cell survival. Moreover, Dicer- and Dgcr8-deficient NK cells are able to respond efficiently through their cytokine receptors; however, function of their immunoreceptor tyrosine-based activation motif (ITAM)-containing aKIRs is impaired.
By using another molecular approach, Sullivan et al. (2012) eliminated Dicer during the earliest stages of murine NK cell development in the bone marrow to better characterize the phenotypic features derived from global loss of mature miRNA expression. These studies confirmed that the absence of miRNAs led to reduced numbers and percentages of NK cells, and a decreased in vitro survival/proliferation.
Studies utilizing next-generation sequencing in mouse and human reported some information about the miRNA repertoire in resting CD56+ CD3− human or NK1.1+CD3− murine NK cells and upon cytokine activation (Fehniger et al., 2010; Liu et al., 2012; Wang et al., 2012). Furthermore, Ni et al. (2015) identified the miRNA profiles of human NK cells from different compartments (peripheral blood, cord blood, and uterine decidua). Very recently, our group, by analyzing peripheral blood NK cells from 10 different human healthy donors, identified a 108 miRNA signature able to discriminate CD56bright from CD56dim NK cell subsets independently from their surface phenotype (Pesce et al., 2018). Interestingly, we found some miRNAs (miR-146a-5p, miR-92a-3p, miR-223-3p, miR-873-5p, miR-31a-5p, hsa-miR-130a-5p, miR-181a-2-3p) with consistent differential expression in the two NK cell subsets, and with an intermediate expression in the CD56bright/CD16dim NK cell subset, which represents a transition phase in the NK cell maturation process of NK cells.
A key miRNA for NK cell development is miR-150. A gain of function miR-150 transgene in mouse was demonstrated to drive the development and maturation of NK cells. In line with this, mice with a targeted deletion of miR-150 instead display cell lineage–intrinsic defect in their ability to generate mature NK cells (Bezman et al., 2011).
Additional miRNAs relevant for NK cell development and maturation are miR-181 and miR-583. Cichocki and coworkers found that miR-181 promotes NK cell development via inhibition of the Nemo like kinase (NLK) (Cichocki et al., 2011) while Yun and collaborators showed that the miR-583 targets and downregulates IL2Rγ in NK cells acting as a negative regulator of their differentiation process (Yun et al., 2014).
miRNAs Involved in the Regulation of NK Cell Functions
Accumulating evidence suggests that distinct miRNAs may play regulative roles on NK cell functions both in terms of cytotoxicity and cytokine production. In this context, miR-27a-5p (Kim et al., 2011), miR-378, miR-30e (Wang et al., 2012), and miR-150 (Kim et al., 2014), were proposed as negative regulators of NK cell killing ability. In particular, miR-378 was found to target granzyme b (Gzmb) (Wang et al., 2012), miR-30e and miR-150 have as target perforin (Prf1) (Wang et al., 2012; Kim et al., 2014) while miR-27a-5p targets both (Kim et al., 2011). By contrast, Prf1, Gzmb, IFN-γ, and CD107a in human NK cells were all upregulated after miR-362-5p overexpression. Ni and collaborators indeed found that this miRNA targets the mRNA coding for the cylindromatosis lysine 63 deubiquitase (CYLD) and suggested that miR-362-5p promotes NK cell effector functions (Ni et al., 2015).
miR-155 should also be included among the miRNAs that enhance NK cell functions. In particular, IL-2, IL-15, and IL-21 upregulate this miRNA, which in turn, enhances NK cell cytotoxicity (Liu et al., 2012). Moreover, miR-155 extensively regulates the NK cell activation threshold regulating molecules involved in NK cell activation and their IFN-γ release by modulating the expression of the phosphatase SHIP-1, T-bet/Tim-3, or the activation of several signaling pathways, including those involving PI3K, NF-kB, and calcineurin (Trotta et al., 2012; Sullivan et al., 2013; Cheng et al., 2015). Similarly, miR-181 was also found to promote IFN-γ production in primary NK cells in response to cytokine stimulation through regulation of the Notch pathway (Cichocki et al., 2011).
In a study aimed to identify miRNAs potentially involved in the pathogenesis of chronic fatigue syndrome or myalgia encephalomyelitis in peripheral blood mononucleate cells (PBMC), 34 miRNAs were found upregulated compared to healthy controls and 2 of these (miR-99b, miR-330-3p) were confirmed having the most important deregulation in NK cells in terms of cytotoxic activity (Petty et al., 2016).
Pathogens-Modulated miRNAs in NK Cells
The host's immune responses must be strictly regulated by an sophisticated balance between positive and negative signals during the fight against pathogens.
One of the mechanisms by which pathogens can break this balance is that of interfering with the regulatory role of miRNAs.
TLRs are receptors of the innate immune system that directly recognize conserved structures of both viral and bacterial origin that are present and functional on NK cells (Della Chiesa et al., 2014). It has been recently demonstrated that several miRNAs, including miR-21, miR-146, miR-155, and let-7 family can bind to TLRs (acting also as physiological ligands for these receptors) or proteins in TLR signaling pathways. These interactions can regulate the expression and the transcriptional responses of TLRs (Bayraktar et al., 2019). In addition, some miRNAs and miRNA-containing exosomes can selectively activate innate immune effector cells, including NK cells, via the TLR1–NF-kB signaling pathway (He et al., 2013).
Enhancement of NK cell cytotoxicity with upregulation of Prf1 was described as associated with miR-182 overexpression in NK cells derived from hepatocellular carcinoma (HCC) patients (Abdelrahman et al., 2016). However, a subsequent study from the same group reported contradicting roles of this miRNA in both NK cells and in hepatocytes infected by hepatitis C virus (HCV) (El Sobky et al., 2016).
Komabayashi and collaborators demonstrated that the Epstein-Barr virus (EBV)-encoded latent membrane protein 1 (LMP1) is able to downregulate the expression of the miR-15a and increase MYB and cyclin D1 in cell lines with an NK cell phenotype (Komabayashi et al., 2014), thus suggesting that miR-15a may have a role in the repression of NK cell proliferation. In line with this, Cheng and collaborators found that the down-regulation of miR-155 suppressed IFN-γ production through Tim-3 signaling and lead to HCV evading immune clearance (Cheng et al., 2015). However, the specific target of miR-155 in the context of these two studies remains unknown.
miR-155 was found to be induced by IL-2 and IL-18 via STAT4 and able to reduce the expression of NOXA and SOCS1 at distinct stages of homeostasis and activation. As NK cells of mice with a targeted miR-155 deletion displayed dramatically diminished effector activities and reduce memory cell numbers in both lymphoid and non-lymphoid tissues afterwards murine cytomegalovirus (MCMV) infection, these findings suggest that miR-155 promotes antiviral immunity (Zawislak et al., 2013). However, a study by Elemam et al. (2015) reached contrasting conclusions as they observed that HCV infection might abolish NK cell cytotoxicity via modification of PU.1 (a key transcription factor in the NK cell development), and Prf1/NKG2D expression by miR-29a-5p and miR-155 overexpression, respectively.
miRNAs in Tumor-Associated NK Cells
Several studies demonstrated that miRNAs might also act as oncogenes or tumor suppressor genes in different human cancer histotypes. Most of the endogenous miRNAs that have been characterized so far modulate NK cell antitumor activity in the tumor microenvironment (TME). TGFß released by tumor cells in the TME is a powerful inhibitor of the NK cell killing activity, and Donatelli and collaborators have shown that a specific miRNA, miR183, is induced by TGFß (Donatelli et al., 2014). They also formally proved that this miRNA downregulates the expression of the DNAX activating protein 12 kDa (DAP12) that was found critically involved in the stabilization of KIR2DS4 and NKp44 receptors on the plasma membrane and required for their signaling activities. Interestingly, loss of DAP12 was also identified as a common trait in tumor-infiltrating lymphocytes in lung cancer (Donatelli et al., 2014). TGFß has also been reported to increase post-transcriptionally the levels of mature miR-1245 which suppresses NKG2D expression, thus spoiling NKG2D-mediated immune responses and enhancing the tumor supporting properties of the TME (Espinoza et al., 2012).
Yang and collaborators reported that miR-218-5p suppresses the NK-mediated killing of lung adenocarcinoma by targeting Serine Hydroxymethyltransferase 1 (SHMT1) (Yang et al., 2019).
Several studies identified miRNAs (miR-203, miR-494-3p, miR-142-3p, miR-155, miR-21) that affect NK cell lymphoma survival and apoptosis modulating different pathways including the PTEN-AKT-mTOR pathway (Yamanaka et al., 2009; Chim et al., 2010, 2011; Ichimura et al., 2010; Chen et al., 2015).
Recent findings provided evidence on the role of some miRNAs as tumor suppressors, such as miR-150 that is involved in the pathogenesis of malignant lymphoma, by increasing the incidence of apoptosis and reducing cancer cell proliferation (Watanabe et al., 2011).
Several studies were performed using as model the NK/T cell lymphoma (NKTL), a progressive malignancy with unfavorable prognosis without a specific treatment, and most of them were pursued to identify dysregulated miRNAs that can affect targets involved in the oncogenesis of NKTL. In this context, Ng and collaborators found that miR-26a, miR-26b, miR-28-5, miR-30b, miR-101, and miR-363 were downregulated, possibly via MYC, in NKTL and NK cell lines compared to normal NK cells and that the suppressed miRNA expression allowed increased expression of genes implicated in oncogenesis (Ng et al., 2011). Furthermore, in a recent study focused on NKTL, Huang and collaborators reported data suggesting that miR-10a and miR-342-3p may be implicated in the development of NKTL through the T-lymphoma invasion and metastasis inducing factor 1 (TIAM1) pathway, which has a crucial role in the development of several types of human cancer (Huang et al., 2016). Other miRNAs such as miR-221 and miR-155, associated with promotion of NKTL viability, have been proposed as potential molecular markers of NKTL (Guo et al., 2010; Chang et al., 2019).
Recently, growing evidence has shown that extracellular vesicles (EVs) released by NK cells transport miRNAs capable of exerting a strong anti-tumor effect in immunosuppressive TME (Fabbri, 2020). Previous studies have also demonstrated that NK cell-derived exosomes have tumor-specific accumulation with no cytotoxic activity against normal tissues (Lugini et al., 2012). Meanwhile, microenvironment acidic pH promotes the traffic of this EVs in tumor cells (Parolini et al., 2009). In addition, NK cell-derived exosomes also exhibit the benefits of being stable vesicles and maintain their biological activities. Thus, NK cell-derived exosomes can both facilitate tumor targeting and act as direct antitumor agent. These properties make them more suitable for clinical applications, thus suggesting a possible use of NK-cell derived EVs as anticancer agents as a new avenue for tumor therapy (Wang H. et al., 2019).
miRNAs Involved in the Regulation of NK Cell Immune Checkpoints
Immune checkpoints have a key role in regulating the intensity of immune responses of lymphocytes by performing inhibitory functions. The use of immune checkpoint inhibitors in immunotherapy has driven anti-cancer treatment on a novel level. Emerging evidence suggests that some miRNAs can control the expression of immune checkpoints on the surface of NK cells or that of their ligands on tumor cells. This suggests a possible use of miRNAs in the context of anti-tumor therapy.
In a recent study we proved that the miR-146a-5p is able to downregulate both KIR2DL1 and KIR2DL2, two HLA-specific inhibitory receptors belonging to KIR family (Pesce et al., 2018) (Figure 1A). Furthermore, in silico functional characterization of miR-146a-5p gene targets, identified CD94, HLA-C, HLA-E, Prf1, and several other KIRs genes as additional targets. These results are in line with the higher levels of miR-146a-5p found in CD56bright NK cells and with other studies suggesting that this miRNA is engaged in the regulation of NK cell maturation via the STAT1 (Xu et al., 2017) and NF-kappaB (Wang et al., 2018) signaling pathways. A different research group identified very recently three miRNAs, miR-26a-5p, miR-26b-5p, and miR-185-5p, as inhibitors of the expression of an additional KIR, the KIR3DL3, which is included in the iNKRs group but it is still poorly characterized (Nutalai et al., 2019) (Figure 1A). Therefore, the role of these miRNAs in NK cells development or function remains to be defined.
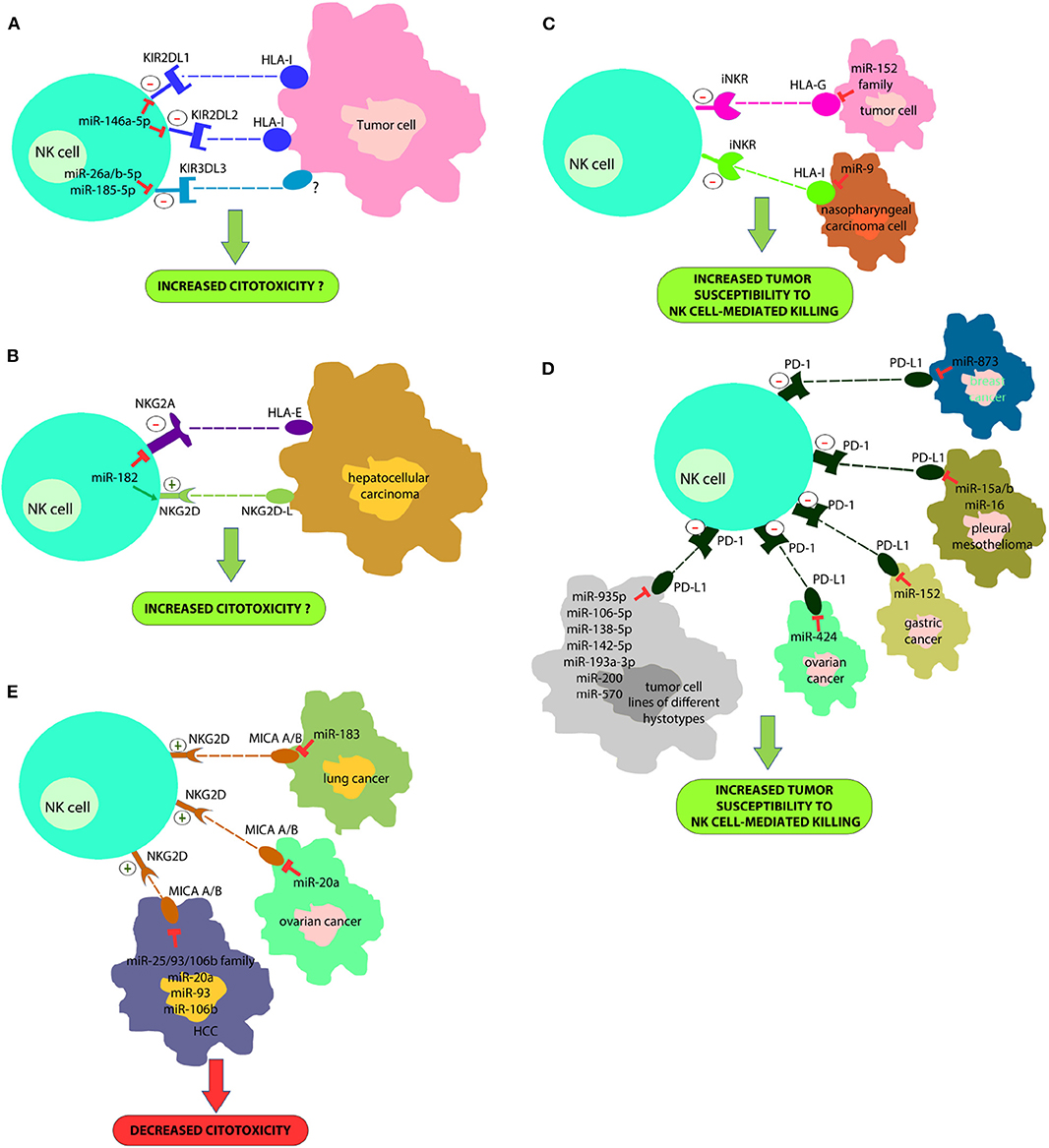
Figure 1. A new strategy for improving NK cell-based immunotherapy: miRNAs can directly regulate the expression of different NK cell immune checkpoints (including KIRs, PD-1, NKG2A, and other iNKR) (A,B) or their ligands (HLA-I, PD-L1, HLA-E/G) (C,D). In addition, some miRNAs can also regulate the expression of activating NK cell receptors (i.e., NKG2D) or their ligands (e.g., MIC A/B) (E). This effect can deeply impact on NK cell ability to recognize and kill cancer cells. In particular, a downregulation of immune-checkpoints or immune-checkpoints' ligands can restore an appropriate NK cell anti-tumor activity; on the contrary, a downregulation of activating receptor expression or their ligands can affect their anti-tumor potential. This suggests innovative miRNA-based therapeutic approaches to unleash NK cell effector functions in the cancer treatment.
The study of Abdelrahman and collaborators showed that enhancement of NK cell cytotoxicity by miR-182 in human hepatocellular carcinoma and increased Prf1 expression were indirect effects likely mediated by a complex modulation of NKG2D and NKG2A levels in these cells at different stages of the disease (Abdelrahman et al., 2016) (Figure 1B).
Regarding miRNAs regulating PD-1 expression, miR-28 (Li et al., 2016), miR138 (Wei et al., 2016), and miR-4717 (Zhang et al., 2015) have been found to target this immune checkpoint, and to induce T cell exhaustion. It has been demonstrated that miR-4717 play a role in chronic Hepatitis B Virus (HBV) infection, where this miRNA is significantly reduced (Zhang et al., 2015). Since also NK cells may express PD-1, it cannot be excluded that these miRNAs may play an important regulatory role also in these innate cells.
Notably, some mi-RNAs target additional immune checkpoints playing a critical role for cytotoxic immune cell functions, such as miR-28 targeting TIM-3 (Li et al., 2016), expressed by both T and NK cells (Di Vito et al., 2019), and miR-16, miR-138, and miR-195 targeting cytotoxic T-lymphocyte antigen 4 (CTLA-4), mainly expressed by T cells (Wei et al., 2016; Tao et al., 2018).
miRNAs Involved in the Regulation of the Expression of Ligands for NK Cell Receptors
Tumor immune evasion is not restricted to the upregulation of immune checkpoint proteins, but also to the dysregulation in the expression of immune checkpoint ligands, including classical and non-classical HLA-I molecules or ligands for activating NK cell receptors. In this context, it has been found that miR-9 is involved in the downmodulation of the expression of HLA-I molecules in human cancer cells, preventing the detection of cancer cells by the immune system (Gao et al., 2013) (Figure 1C). This suggests that tumors overexpressing this miRNA might become resistant to CD8+ T-cell mediated killing but susceptible to NK cell-mediated attack. In addition, the tumor-suppressive miR-148 family has been found to regulate the expression of HLA-G, a ligand for different NK cell inhibitory receptors (Mandelboim et al., 1997; Seliger, 2016) (Figure 1D).
Recently, it has been demonstrated that some miRNAs directly target the 3′-UTR of PD-L1 mRNA and others the PD-1/PD-L1 indirectly by targeting the related signaling pathways (Wang et al., 2017; Gao et al., 2019; Omar et al., 2019). miR-15a, miR-15b and miR-16 were discovered to downregulate the PD-L1 expression in malignant pleural mesothelioma cell line (Kao et al., 2017) (Figure 1D). miR-34a was found to be inversely correlated with PD-L1 expression in 44 AML samples (Wang et al., 2015) (Figure 1D). miR-935p, miR-106b-5p, miR-138-5p, miR-142-5p, miR-193a-3p, miR-200, and miR-570 overexpression downregulate PD-L1 in tumor cell lines of different hystotypes (Chen et al., 2014; Guo et al., 2015; Cioffi et al., 2017; Jia et al., 2017; Kao et al., 2017) (Figure 1D). miR-152 was found to regulate PD-L1 in gastric cancer tissues (Guo et al., 2015), while miR-424 regulates the PD-L1 expression in chemo-resistant ovarian cancer patients (Xu et al., 2016) (Figure 1D). Notably, miR-873 decreased the stemness and resistance to chemotherapy of breast cancer cells, depending on PD-L1 and the downstream PI3K/Akt and ERK1/2 signaling, by directly inhibiting PD-L1 expression. This suggests that miR-873/PD-L1 regulatory axis may represent a new therapeutic target in breast cancer. Such data are interesting for employing miRNAs as useful diagnostic targets and valuable biomarkers for prognosis in the PD-1/PD-L1 blockade therapy. miR-155, a key component of inflammatory responses, is dysregulated in different cancer cell types. In this context, it has been recently demonstrated that the induction of miRNA-155 expression, suppresses the expression of PD-L1 in both primary lymphatic endothelial cells and fibroblasts, by exposing these cells to the TNF-a and IFN-γ proinflammatory cytokines (Yee et al., 2017).
Moreover, different miRNAs downregulate MHC class I chain-related protein A/B (MICA/B) expression (NKG2D NK-cell receptor ligands) and this represents another mechanism of immune suppression targeting NK cells cytotoxicity. These miRNAs include miR-183 that targets MIC A/B in lung cancer (Trinh et al., 2019), miR-20a that induces the same effect in ovarian cancer (Xie et al., 2014) and miR-25/93/106b family, miR-20a, miR-93 and miR-106b that act in HCC (Kishikawa et al., 2013) (Figure 1E).
Concluding Remarks
Recently, there has been a substantial evolution in cancer therapy, mainly oriented toward immunotherapy approaches, in substitution or in combination with classical therapy. Cancer immunotherapy represents a promising new era in cancer management due to the relatively high safety margins and selectivity, compared to the classical cancer chemotherapeutic agents.
miRNAs have come to light over the last years as key actors in epigenetic regulation and for their capacity to modulate tumor immunity by directly regulating the expression of genes involved in the activation or suppression of the immune response. In this review, we focused our attention on the current state of knowledge concerning the involvement of miRNAs in various physiologic processes of NK cells. In particular, we discussed their abilty to regulate NK immune responses and their potential implications in resistance to cancer immunotherapy, with main focus on immune checkpoints. In this context, several miRNAs have been found to modulate different immune checkpoints/ligands interaction, including the PD-1/PD-L1 axis or their upstream genes. Future studies comparing miRNAs' expression profiles in patients who respond to immune checkpoint blockade immunotherapies as compared to non-responders will help to disclose the potential role of miRNAs as non-invasive predictive biomarkers for monitoring the response and clinical outcomes to immunotherapy with immune checkpoint inhibitors.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by Fondazione Associazione Italiana per la Ricerca sul Cancro (IG 2017-20312 and 5x1000-21147); Fondazione Roche (Progetto Roche per la Ricerca 2017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdelrahman, M. M., Fawzy, I. O., Bassiouni, A. A., Gomaa, A. I., Esmat, G., Waked, I., et al. (2016). Enhancing NK cell cytotoxicity by miR-182 in hepatocellular carcinoma. Hum. Immunol. 77, 667–673. doi: 10.1016/j.humimm.2016.04.020
André, P., Denis, C., Soulas, C., Bourbon-Caillet, C., Lopez, J., Arnoux, T., et al. (2018). Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell 175, 1731–1743.e13. doi: 10.1016/j.cell.2018.10.014
Bayraktar, R., Bertilaccio, M. T. S., and Calin, G. A. (2019). The interaction between two worlds: microRNAs and toll-like receptors. Front. Immunol. 10:1053. doi: 10.3389/fimmu.2019.01053
Beldi-Ferchiou, A., Lambert, M., Dogniaux, S., Vély, F., Vivier, E., Olive, D., et al. (2016). PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget 7, 72961–72977. doi: 10.18632/oncotarget.12150
Bernson, E., Christenson, K., Pesce, S., Pasanen, M., Marcenaro, E., Sivori, S., et al. (2019). Downregulation of HLA class I renders inflammatory neutrophils more susceptible to NK cell-induced apoptosis. Front. Immunol. 10:2444. doi: 10.3389/fimmu.2019.02444
Bezman, N. A., Cedars, E., Steiner, D. F., Blelloch, R., Hesslein, D. G. T., and Lanier, L. L. (2010). Distinct requirements of microRNAs in NK cell activation, survival, and function. J. Immunol. 185, 3835–3846. doi: 10.4049/jimmunol.1000980
Bezman, N. A., Chakraborty, T., Bender, T., and Lanier, L. L. (2011). miR-150 regulates the development of NK and iNKT cells. J. Exp. Med. 208, 2717–2731. doi: 10.1084/jem.20111386
Bi, J., and Tian, Z. (2017). NK cell exhaustion. Front. Immunol. 8:760. doi: 10.3389/fimmu.2017.00760
Bjorkstrom, N. K., Riese, P., Heuts, F., Andersson, S., Fauriat, C., Ivarsson, M. A., et al. (2010). Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 116, 3853–3864. doi: 10.1182/blood-2010-04-281675
Braud, V. M., Allan, D. S. J., O'Callaghan, C. A., Söderström, K., D'Andrea, A., Ogg, G. S., et al. (1998). HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799. doi: 10.1038/35869
Carlsten, M., Norell, H., Bryceson, Y. T., Poschke, I., Schedvins, K., Ljunggren, H.-G., et al. (2009). Primary human tumor cells expressing CD155 impair tumor targeting by down-regulating DNAM-1 on NK cells. J. Immunol. 183, 4921–4930. doi: 10.4049/jimmunol.0901226
Carrega, P., and Ferlazzo, G. (2012). Natural killer cell distribution and trafficking in human tissues. Front. Immunol. 3:347. doi: 10.3389/fimmu.2012.00347
Chang, Y., Cui, M., Fu, X., Zhang, L., Li, X., Li, L., et al. (2019). MiRNA-155 regulates lymphangiogenesis in natural killer/T-cell lymphoma by targeting BRG1. Cancer Biol. Ther. 20, 31–41. doi: 10.1080/15384047.2018.1504721
Chen, H.-H., Huang, W.-T., Yang, L.-W., and Lin, C.-W. (2015). The PTEN-AKT-mTOR/RICTOR pathway in nasal natural killer cell lymphoma is activated by miR-494-3p via PTEN but inhibited by miR-142-3p via RICTOR. Am. J. Pathol. 185, 1487–1499. doi: 10.1016/j.ajpath.2015.01.025
Chen, L., Gibbons, D. L., Goswami, S., Cortez, M. A., Ahn, Y.-H., Byers, L. A., et al. (2014). Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 5:5241. doi: 10.1038/ncomms6241
Cheng, Y. Q., Ren, J. P., Zhao, J., Wang, J. M., Zhou, Y., Li, G. Y., et al. (2015). MicroRNA-155 regulates interferon-γ production in natural killer cells via Tim-3 signalling in chronic hepatitis C virus infection. Immunology 145, 485–497. doi: 10.1111/imm.12463
Chim, C. S., Wong, K. Y., Leung, C. Y., Chung, L. P., Hui, P. K., Chan, S. Y., et al. (2011). Epigenetic inactivation of the hsa-miR-203 in haematological malignancies. J. Cell. Mol. Med. 15, 2760–2767. doi: 10.1111/j.1582-4934.2011.01274.x
Chim, C. S., Wong, K. Y., Qi, Y., Loong, F., Lam, W. L., Wong, L. G., et al. (2010). Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis 31, 745–750. doi: 10.1093/carcin/bgq033
Chiossone, L., Vienne, M., Kerdiles, Y. M., and Vivier, E. (2017). Natural killer cell immunotherapies against cancer: checkpoint inhibitors and more. Semin. Immunol. 31, 55–63. doi: 10.1016/j.smim.2017.08.003
Cichocki, F., Felices, M., McCullar, V., Presnell, S. R., Al-Attar, A., Lutz, C. T., et al. (2011). Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J. Immunol. 187, 6171–6175. doi: 10.4049/jimmunol.1100835
Cioffi, M., Trabulo, S. M., Vallespinos, M., Raj, D., Kheir, T. B., Lin, M.-L., et al. (2017). The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget 8:21609–25. doi: 10.18632/oncotarget.15450
Cobb, B. S., Hertweck, A., Smith, J., O'Connor, E., Graf, D., Cook, T., et al. (2006). A role for Dicer in immune regulation. J. Exp. Med. 203, 2519–2527. doi: 10.1084/jem.20061692
Cortez, V. S., and Colonna, M. (2016). Diversity and function of group 1 innate lymphoid cells. Immunol. Lett. 179, 19–24. doi: 10.1016/j.imlet.2016.07.005
Cosman, D., Fanger, N., Borges, L., Kubin, M., Chin, W., Peterson, L., et al. (1997). A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 7, 273–282. doi: 10.1016/S1074-7613(00)80529-4
Costello, R. T., Sivori, S., Marcenaro, E., Lafage-Pochitaloff, M., Mozziconacci, M.-J., Reviron, D., et al. (2002). Defective expression and function of natural killer cell-triggering receptors in patients with acute myeloid leukemia. Blood 99, 3661–3667. doi: 10.1182/blood.V99.10.3661
Cron, M. A., Maillard, S., Truffault, F., Gualeni, A. V., Gloghini, A., Fadel, E., et al. (2019). Causes and consequences of miR-150-5p dysregulation in myasthenia gravis. Front. Immunol. 10:539. doi: 10.3389/fimmu.2019.00539
Cutrona, G., Matis, S., Colombo, M., Massucco, C., Baio, G., Valdora, F., et al. (2017). Effects of miRNA-15 and miRNA-16 expression replacement in chronic lymphocytic leukemia: implication for therapy. Leukemia 31, 1894–1904. doi: 10.1038/leu.2016.394
Dallaire, A., and Simard, M. J. (2016). The implication of microRNAs and endo-siRNAs in animal germline and early development. Dev. Biol. 416, 18–25. doi: 10.1016/j.ydbio.2016.06.007
Del Zotto, G., Marcenaro, E., Vacca, P., Sivori, S., Pende, D., Della Chiesa, M., et al. (2017). Markers and function of human NK cells in normal and pathological conditions. Cytometry B Clin. Cytom. 92, 100–114. doi: 10.1002/cyto.b.21508
Della Chiesa, M., Marcenaro, E., Sivori, S., Carlomagno, S., Pesce, S., and Moretta, A. (2014). Human NK cell response to pathogens. Semin. Immunol. 26, 152–160. doi: 10.1016/j.smim.2014.02.001
Della Chiesa, M., Pesce, S., Muccio, L., Carlomagno, S., Sivori, S., Moretta, A., et al. (2016). Features of memory-like and PD-1+ human NK cell subsets. Front. Immunol. 7:351. doi: 10.3389/fimmu.2016.00351
Di Santo, J. P. (2006). Natural killer cell developmental pathways: a question of Balance. Annu. Rev. Immunol. 24, 257–286. doi: 10.1146/annurev.immunol.24.021605.090700
Di Vito, C., Mikulak, J., Zaghi, E., Pesce, S., Marcenaro, E., and Mavilio, D. (2019). NK cells to cure cancer. Semin. Immunol. 41:101272. doi: 10.1016/j.smim.2019.03.004
Diederichs, S., and Haber, D. A. (2007). Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131, 1097–1108. doi: 10.1016/j.cell.2007.10.032
Donatelli, S. S., Zhou, J.-M., Gilvary, D. L., Eksioglu, E. A., Chen, X., Cress, W. D., et al. (2014). TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc. Natl. Acad. Sci. U.S.A. 111, 4203–4208. doi: 10.1073/pnas.1319269111
El Sobky, S. A., El-Ekiaby, N. M., Mekky, R. Y., Elemam, N. M., Mohey Eldin, M. A., El-sayed, M., et al. (2016). Contradicting roles of miR-182 in both NK cells and their host target hepatocytes in HCV. Immunol. Lett. 169, 52–60. doi: 10.1016/j.imlet.2015.10.013
Elemam, N. M., Mekky, R. Y., El-Ekiaby, N. M., El Sobky, S. A., El Din, M. A. M., Esmat, G., et al. (2015). Repressing PU.1 by miR-29a* in NK cells of HCV patients, diminishes its cytolytic effect on HCV infected cell models. Hum. Immunol. 76, 687–694. doi: 10.1016/j.humimm.2015.09.021
Espinoza, J. L., Takami, A., Yoshioka, K., Nakata, K., Sato, T., Kasahara, Y., et al. (2012). Human microRNA-1245 down-regulates the NKG2D receptor in natural killer cells and impairs NKG2D-mediated functions. Haematologica 97, 1295–1303. doi: 10.3324/haematol.2011.058529
Fabbri, M. (2020). Natural killer cell-derived vesicular miRNAs: a new anticancer approach? Cancer Res. 80, 17–22. doi: 10.1158/0008-5472.CAN-19-1450
Farag, S. S., and Caligiuri, M. A. (2006). Human natural killer cell development and biology. Blood Rev. 20, 123–137. doi: 10.1016/j.blre.2005.10.001
Fedeli, M., Napolitano, A., Wong, M. P. M., Marcais, A., de Lalla, C., Colucci, F., et al. (2009). Dicer-dependent microRNA pathway controls invariant NKT cell development. J. Immunol. 183, 2506–2512. doi: 10.4049/jimmunol.0901361
Fehniger, T. A., Wylie, T., Germino, E., Leong, J. W., Magrini, V. J., Koul, S., et al. (2010). Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 20, 1590–1604. doi: 10.1101/gr.107995.110
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114. doi: 10.1038/nrg2290
Freud, A. G., and Caligiuri, M. A. (2006). Human natural killer cell development. Immunol. Rev. 214, 56–72. doi: 10.1111/j.1600-065X.2006.00451.x
Freud, A. G., Mundy-Bosse, B. L., Yu, J., and Caligiuri, M. A. (2017). The broad spectrum of human natural killer cell diversity. Immunity 47, 820–833. doi: 10.1016/j.immuni.2017.10.008
Gao, F., Zhao, Z.-L., Zhao, W.-T., Fan, Q.-R., Wang, S.-C., Li, J., et al. (2013). miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem. Biophys. Res. Commun. 431, 610–616. doi: 10.1016/j.bbrc.2012.12.097
Gao, L., Guo, Q., Li, X., Yang, X., Ni, H., Wang, T., et al. (2019). MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine 41, 395–407. doi: 10.1016/j.ebiom.2019.02.034
Gracias, D. T., Stelekati, E., Hope, J. L., Boesteanu, A. C., Doering, T. A., Norton, J., et al. (2013). The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat. Immunol. 14, 593–602. doi: 10.1038/ni.2576
Gregory, R. I., Yan, K.-P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., et al. (2004). The microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240. doi: 10.1038/nature03120
Gu, S., and Kay, M. A. (2010). How do miRNAs mediate translational repression? Silence 1:11. doi: 10.1186/1758-907X-1-11
Guo, H.-Q., Huang, G.-L., Guo, C.-C., Pu, X.-X., and Lin, T.-Y. (2010). Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis. Markers 29, 251–258. doi: 10.1155/2010/474692
Guo, W., Tan, W., Liu, S., Huang, X., Lin, J., Liang, R., et al. (2015). MiR-570 inhibited the cell proliferation and invasion through directly targeting B7-H1 in hepatocellular carcinoma. Tumor Biol. 36, 9049–9057. doi: 10.1007/s13277-015-3644-3
Hallner, A., Bernson, E., Hussein, B. A., Ewald Sander, F., Brune, M., Aurelius, J., et al. (2019). The HLA-B−21 dimorphism impacts on NK cell education and clinical outcome of immunotherapy in acute myeloid leukemia. Blood 133, 1479–1488. doi: 10.1182/blood-2018-09-874990
Han, B., Mao, F.-Y., Zhao, Y.-L., Lv, Y.-P., Teng, Y.-S., Duan, M., et al. (2018). Altered NKp30, NKp46, NKG2D, and DNAM-1 expression on circulating NK cells is associated with tumor progression in human gastric cancer. J. Immunol. Res. 2018:6248590. doi: 10.1155/2018/6248590
Hart, O. M., Athie-Morales, V., O'Connor, G. M., and Gardiner, C. M. (2005). TLR7/8-mediated activation of human NK cells results in accessory cell-dependent IFN-γ production. J. Immunol. 175, 1636–1642. doi: 10.4049/jimmunol.175.3.1636
He, S., Chu, J., Wu, L.-C., Mao, H., Peng, Y., Alvarez-Breckenridge, C. A., et al. (2013). MicroRNAs activate natural killer cells through Toll-like receptor signaling. Blood 121, 4663–4671. doi: 10.1182/blood-2012-07-441360
Horsburgh, S., Fullard, N., Roger, M., Degnan, A., Todryk, S., Przyborski, S. et al. (2017). MicroRNAs in the skin: role in development, homoeostasis and regeneration. Clin. Sci. 131, 1923–1940. doi: 10.1042/CS20170039.
Hsu, J., Hodgins, J. J., Marathe, M., Nicolai, C. J., Bourgeois-Daigneault, M.-C., Trevino, T. N., et al. (2018). Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J. Clin. Invest. 128, 4654–4668. doi: 10.1172/JCI99317
Hua, M., Li, J., Wang, C., Shao, L., Hou, M., Peng, J., et al. (2019). Aberrant expression of microRNA in CD4+ cells contributes to Th17/Treg imbalance in primary immune thrombocytopenia. Thromb. Res. 177, 70–78. doi: 10.1016/j.thromres.2019.03.005
Huang, H., Fan, L., Zhan, R., Wu, S., and Niu, W. (2016). Expression of microRNA-10a, microRNA-342-3p and their predicted target gene TIAM1 in extranodal NK/T-cell lymphoma, nasal type. Oncol. Lett. 11, 345–351. doi: 10.3892/ol.2015.3831
Ichimura, A., Ruike, Y., Terasawa, K., Shimizu, K., and Tsujimoto, G. (2010). MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Mol. Pharmacol. 77, 1016–1024. doi: 10.1124/mol.109.063321
Ichiyama, K., Gonzalez-Martin, A., Kim, B.-S., Jin, H. Y., Jin, W., Xu, W., et al. (2016). The microRNA-183-96-182 cluster promotes T helper 17 cell pathogenicity by negatively regulating transcription factor Foxo1 expression. Immunity 44, 1284–1298. doi: 10.1016/j.immuni.2016.05.015
Ivanova, E., Bozhilova, R., Kaneva, R., and Milanova, V. (2018). The dysregulation of microRNAs and the role of stress in the pathogenesis of mental disorders. Curr. Top. Med. Chem. 18, 1893–1907. doi: 10.2174/1568026619666181130135253
Jia, L., Xi, Q., Wang, H., Zhang, Z., Liu, H., Cheng, Y., et al. (2017). miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem. Biophys. Res. Commun. 488, 425–431. doi: 10.1016/j.bbrc.2017.05.074
Johnson, J. L. (2019). Elucidating the contributory role of microRNA to cardiovascular diseases (a review). Vascul. Pharmacol. 114, 31–48. doi: 10.1016/j.vph.2018.10.010
Kao, S. C., Cheng, Y. Y., Williams, M., Kirschner, M. B., Madore, J., Lum, T., et al. (2017). Tumor suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma. J. Thoracic Oncol. 12, 1421–1433. doi: 10.1016/j.jtho.2017.05.024
Kim, N., Kim, M., Yun, S., Doh, J., Greenberg, P. D., Kim, T.-D., et al. (2014). MicroRNA-150 regulates the cytotoxicity of natural killers by targeting perforin-1. J. Allergy Clin. Immunol. 134, 195–203. doi: 10.1016/j.jaci.2014.02.018
Kim, T.-D., Lee, S. U., Yun, S., Sun, H.-N., Lee, S. H., Kim, J. W., et al. (2011). Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood 118, 5476–5486. doi: 10.1182/blood-2011-04-347526
Kishikawa, T., Otsuka, M., Yoshikawa, T., Ohno, M., Takata, A., Shibata, C., et al. (2013). Regulation of the expression of the liver cancer susceptibility gene MICA by microRNAs. Sci. Rep. 3:2739. doi: 10.1038/srep02739
Kohrt, H. E., Thielens, A., Marabelle, A., Sagiv-Barfi, I., Sola, C., Chanuc, F., et al. (2014). Anti-KIR antibody enhancement of anti-lymphoma activity of natural killer cells as monotherapy and in combination with anti-CD20 antibodies. Blood 123, 678–686. doi: 10.1182/blood-2013-08-519199
Komabayashi, Y., Kishibe, K., Nagato, T., Ueda, S., Takahara, M., and Harabuchi, Y. (2014). Downregulation of miR-15a due to LMP1 promotes cell proliferation and predicts poor prognosis in nasal NK/T-cell lymphoma. Am. J. Hematol. 89, 25–33. doi: 10.1002/ajh.23570
Koralov, S. B., Muljo, S. A., Galler, G. R., Krek, A., Chakraborty, T., Kanellopoulou, C., et al. (2008). Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 132, 860–874. doi: 10.1016/j.cell.2008.02.020
Li, Q., Johnston, N., Zheng, X., Wang, H., Zhang, X., Gao, D., et al. (2016). miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget 7, 53735–50. doi: 10.18632/oncotarget.10731
Liston, A., Lu, L.-F., O'Carroll, D., Tarakhovsky, A., and Rudensky, A. Y. (2008). Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 205, 1993–2004. doi: 10.1084/jem.20081062
Liu, B., Li, J., and Cairns, M. J. (2014). Identifying miRNAs, targets and functions. Brief Bioinformatics 15, 1–19. doi: 10.1093/bib/bbs075
Liu, B., Li, J., Tsykin, A., Liu, L., Gaur, A. B., and Goodall, G. J. (2009). Exploring complex miRNA-mRNA interactions with Bayesian networks by splitting-averaging strategy. BMC Bioinformatics 10:408. doi: 10.1186/1471-2105-10-408
Liu, X., Wang, Y., Sun, Q., Yan, J., Huang, J., Zhu, S., et al. (2012). Identification of microRNA transcriptome involved in human natural killer cell activation. Immunol. Lett. 143, 208–217. doi: 10.1016/j.imlet.2012.02.014
Lugini, L., Cecchetti, S., Huber, V., Luciani, F., Macchia, G., Spadaro, F., et al. (2012). Immune surveillance properties of human NK cell-derived exosomes. J. Immunol. 189, 2833–2842. doi: 10.4049/jimmunol.1101988
Macfarlane, L.-A., and Murphy, P. R. (2010). MicroRNA: biogenesis, function and role in cancer. Curr. Genomics 11, 537–561. doi: 10.2174/138920210793175895
Mandelboim, O., Pazmany, L., Davis, D. M., Valés-Gómez, M., Reyburn, H. T., Rybalov, B., et al. (1997). Multiple receptors for HLA-G on human natural killer cells. Proc. Natl. Acad. Sci. U.S.A. 94, 14666–14670. doi: 10.1073/pnas.94.26.14666
Marcenaro, E., Della Chiesa, M., Bellora, F., Parolini, S., Millo, R., Moretta, L., et al. (2005a). IL-12 or IL-4 prime human NK cells to mediate functionally divergent interactions with dendritic cells or tumors. J. Immunol. 174, 3992–3998. doi: 10.4049/jimmunol.174.7.3992
Marcenaro, E., Dondero, A., and Moretta, A. (2006). Multi-directional cross-regulation of NK cell function during innate immune responses. Transplant. Immunol. 17, 16–19. doi: 10.1016/j.trim.2006.09.019
Marcenaro, E., Ferranti, B., Falco, M., Moretta, L., and Moretta, A. (2008). Human NK cells directly recognize Mycobacterium bovis via TLR2 and acquire the ability to kill monocyte-derived DC. Int. Immunol. 20, 1155–1167. doi: 10.1093/intimm/dxn073
Marcenaro, E., Ferranti, B., and Moretta, A. (2005b). NK-DC interaction: on the usefulness of auto-aggression. Autoimmun. Rev. 4, 520–525. doi: 10.1016/j.autrev.2005.04.015
Marcenaro, E., Notarangelo, L. D., Orange, J. S., and Vivier, E. (2017). Editorial: NK cell subsets in health and disease: new developments. Front. Immunol. 8:1363. doi: 10.3389/978-2-88945-350-4
Mariotti, F. R., Petrini, S., Ingegnere, T., Tumino, N., Besi, F., Scordamaglia, F., et al. (2019). PD-1 in human NK cells: evidence of cytoplasmic mRNA and protein expression. Oncoimmunology 8:1557030. doi: 10.1080/2162402X.2018.1557030
Moretta, A., Bottino, C., Vitale, M., Pende, D., Biassoni, R., Mingari, M. C., et al. (1996). Receptors for HLA class-i molecules in human natural killer cells. Annu. Rev. Immunol. 14, 619–648. doi: 10.1146/annurev.immunol.14.1.619
Moretta, A., Bottino, C., Vitale, M., Pende, D., Cantoni, C., Mingari, M. C., et al. (2001). Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 19, 197–223. doi: 10.1146/annurev.immunol.19.1.197
Moretta, L., Bottino, C., Pende, D., Vitale, M., Mingari, M., and Moretta, A. (2004). Different checkpoints in human NK-cell activation. Trends Immunol. 25, 670–676. doi: 10.1016/j.it.2004.09.008
Moretta, L., Ferlazzo, G., Bottino, C., Vitale, M., Pende, D., Mingari, M. C., et al. (2006). Effector and regulatory events during natural killer-dendritic cell interactions. Immunol. Rev. 214, 219–228. doi: 10.1111/j.1600-065X.2006.00450.x
Mori, M. A., Ludwig, R. G., Garcia-Martin, R., Brandão, B. B., and Kahn, C. R. (2019). Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 30, 656–673 doi: 10.1016/j.cmet.2019.07.011
Muljo, S. A., Ansel, K. M., Kanellopoulou, C., Livingston, D. M., Rao, A., and Rajewsky, K. (2005). Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 202, 261–269. doi: 10.1084/jem.20050678
Ng, S.-B., Yan, J., Huang, G., Selvarajan, V., Tay, J. L.-S., Lin, B., et al. (2011). Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood 118, 4919–4929. doi: 10.1182/blood-2011-07-364224
Ni, F., Guo, C., Sun, R., Fu, B., Yang, Y., Wu, L., et al. (2015). MicroRNA transcriptomes of distinct human NK cell populations identify miR-362-5p as an essential regulator of NK cell function. Sci. Rep. 5:9993. doi: 10.1038/srep09993
Nutalai, R., Gaudieri, S., Jumnainsong, A., and Leelayuwat, C. (2019). Regulation of KIR3DL3 expression via mirna. Genes 10:603. doi: 10.3390/genes10080603
Omar, H. A., El-Serafi, A. T., Hersi, F., Arafa, E. A., Zaher, D. M., Madkour, M., et al. (2019). Immunomodulatory MicroRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. 286, 3540–3557. doi: 10.1111/febs.15000
Parodi, M., Pedrazzi, M., Cantoni, C., Averna, M., Patrone, M., Cavaletto, M., et al. (2015). Natural killer (NK)/melanoma cell interaction induces NK-mediated release of chemotactic High Mobility Group Box-1 (HMGB1) capable of amplifying NK cell recruitment. Oncoimmunology 4:e1052353. doi: 10.1080/2162402X.2015.1052353
Parolini, I., Federici, C., Raggi, C., Lugini, L., Palleschi, S., De Milito, A., et al. (2009). Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 284, 34211–34222. doi: 10.1074/jbc.M109.041152
Peng, Y., and Croce, C. M. (2016). The role of MicroRNAs in human cancer. Signal. Transduct. Target Ther. 1:15004. doi: 10.1038/sigtrans.2015.4
Pesce, S., Belgrano, V., Greppi, M., Carlomagno, S., Squillario, M., Barla, A., et al. (2019a). Different features of tumor-associated NK cells in patients with low-grade or high-grade peritoneal carcinomatosis. Front. Immunol. 10:1963. doi: 10.3389/fimmu.2019.01963
Pesce, S., Greppi, M., Grossi, F., Del Zotto, G., Moretta, L., Sivori, S., et al. (2019b). PD/1-PD-Ls checkpoint: insight on the potential role of NK cells. Front. Immunol. 10:1242. doi: 10.3389/fimmu.2019.01242
Pesce, S., Greppi, M., Tabellini, G., Rampinelli, F., Parolini, S., Olive, D., et al. (2017a). Identification of a subset of human natural killer cells expressing high levels of programmed death 1: A phenotypic and functional characterization. J. Allergy Clin. Immunol. 139, 335–346.e3. doi: 10.1016/j.jaci.2016.04.025
Pesce, S., Moretta, L., Moretta, A., and Marcenaro, E. (2016). Human NK cell subsets redistribution in pathological conditions: a Role for CCR7 receptor. Front. Immunol. 7:414. doi: 10.3389/fimmu.2016.00414
Pesce, S., Squillario, M., Greppi, M., Loiacono, F., Moretta, L., Moretta, A., et al. (2018). New miRNA signature heralds human NK cell subsets at different maturation steps: involvement of miR-146a-5p in the regulation of KIR expression. Front. Immunol. 9:2360. doi: 10.3389/fimmu.2018.02360
Pesce, S., Tabellini, G., Cantoni, C., Patrizi, O., Coltrini, D., Rampinelli, F., et al. (2015). B7-H6-mediated downregulation of NKp30 in NK cells contributes to ovarian carcinoma immune escape. OncoImmunology 4:e1001224. doi: 10.1080/2162402X.2014.1001224
Pesce, S., Thoren, F. B., Cantoni, C., Prato, C., Moretta, L., Moretta, A., et al. (2017b). The innate immune cross talk between NK cells and eosinophils is regulated by the interaction of natural cytotoxicity receptors with eosinophil surface ligands. Front. Immunol. 8:510. doi: 10.3389/fimmu.2017.00510
Petty, R. D., McCarthy, N. E., Le Dieu, R., and Kerr, J. R. (2016). MicroRNAs hsa-miR-99b, hsa-miR-330, hsa-miR-126 and hsa-miR-30c: potential diagnostic biomarkers in natural killer (NK) cells of patients with chronic fatigue syndrome (CFS)/myalgic encephalomyelitis (ME). PLoS ONE 11:e0150904. doi: 10.1371/journal.pone.0150904
Poznanski, S. M., and Ashkar, A. A. (2019). What defines NK cell functional fate: phenotype or metabolism? Front. Immunol. 10:1414. doi: 10.3389/fimmu.2019.01414
Riise, R. E., Bernson, E., Aurelius, J., Martner, A., Pesce, S., Della Chiesa, M., et al. (2015). TLR-stimulated neutrophils instruct NK cells to trigger dendritic cell maturation and promote adaptive T cell responses. J. Immunol. 195, 1121–1128. doi: 10.4049/jimmunol.1500709
Romagnani, C., Juelke, K., Falco, M., Morandi, B., D'Agostino, A., Costa, R., et al. (2007). CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J. Immunol. 178, 4947–4955. doi: 10.4049/jimmunol.178.8.4947
Romagne, F., Andre, P., Spee, P., Zahn, S., Anfossi, N., Gauthier, L., et al. (2009). Preclinical characterization of 1-7F9, a novel human anti-KIR receptor therapeutic antibody that augments natural killer-mediated killing of tumor cells. Blood 114, 2667–2677. doi: 10.1182/blood-2009-02-206532
Romero, A. I., Thorén, F. B., Brune, M., and Hellstrand, K. (2006). NKp46 and NKG2D receptor expression in NK cells with CD56dim and CD56bright phenotype: regulation by histamine and reactive oxygen species. Br. J. Haematol. 132, 91–98. doi: 10.1111/j.1365-2141.2005.05842.x
Rouas, R., Merimi, M., Najar, M., El Zein, N., Fayyad-Kazan, M., Berehab, M., et al. (2019). Human CD8+ CD25 + CD127 low regulatory T cells: microRNA signature and impact on TGF-β and IL-10 expression. J. Cell. Physiol. 234, 17459–17472. doi: 10.1002/jcp.28367
Saki, N., Abroun, S., Soleimani, M., Hajizamani, S., Shahjahani, M., Kast, R. E., et al. (2015). Involvement of microRNA in T-cell differentiation and malignancy. Int. J. Hematol. Oncol. Stem Cell Res. 9, 33–49.
Sanchez-Correa, B., Valhondo, I., Hassouneh, F., Lopez-Sejas, N., Pera, A., Bergua, J. M., et al. (2019). DNAM-1 and the TIGIT/PVRIG/TACTILE axis: novel immune checkpoints for natural killer cell-based cancer immunotherapy. Cancers 11:E877. doi: 10.3390/cancers11060877
Seliger, B. (2016). Role of microRNAs on HLA-G expression in human tumors. Human Immunol. 77, 760–763. doi: 10.1016/j.humimm.2016.04.006
Sivori, S., Falco, M., Della Chiesa, M., Carlomagno, S., Vitale, M., Moretta, L., et al. (2004). CpG and double-stranded RNA trigger human NK cells by Toll-like receptors: induction of cytokine release and cytotoxicity against tumors and dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 101, 10116–10121. doi: 10.1073/pnas.0403744101
Stittrich, A.-B., Haftmann, C., Sgouroudis, E., Kühl, A. A., Hegazy, A. N., Panse, I., et al. (2010). The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat. Immunol. 11, 1057–1062. doi: 10.1038/ni.1945
Sullivan, R. P., Fogel, L. A., Leong, J. W., Schneider, S. E., Wong, R., Romee, R., et al. (2013). MicroRNA-155 tunes both the threshold and extent of NK cell activation via targeting of multiple signaling pathways. J. Immunol. 191, 5904–5913. doi: 10.4049/jimmunol.1301950
Sullivan, R. P., Leong, J. W., Schneider, S. E., Keppel, C. R., Germino, E., French, A. R., et al. (2012). MicroRNA-deficient NK cells exhibit decreased survival but enhanced function. J. Immunol. 188, 3019–3030. doi: 10.4049/jimmunol.1102294
Tao, Z., Xu, S., Ruan, H., Wang, T., Song, W., Qian, L., et al. (2018). MiR-195/-16 family enhances radiotherapy via T cell activation in the tumor microenvironment by blocking the PD-L1 immune checkpoint. Cell. Physiol. Biochem. 48, 801–814. doi: 10.1159/000491909
Tinker, A. V., Hirte, H. W., Provencher, D., Butler, M., Ritter, H., Tu, D., et al. (2019). Dose-ranging and cohort-expansion study of monalizumab (IPH2201) in patients with advanced gynecologic malignancies: a trial of the canadian cancer trials group (CCTG): IND221. Clin. Cancer Res. 25, 6052–6060 doi: 10.1158/1078-0432.CCR-19-0298
Trinh, T. L., Kandell, W. M., Donatelli, S. S., Tu, N., Tejera, M. M., Gilvary, D. L., et al. (2019). Immune evasion by TGFβ-induced miR-183 repression of MICA/B expression in human lung tumor cells. OncoImmunology 8:e1557372. doi: 10.1080/2162402X.2018.1557372
Trotta, R., Chen, L., Ciarlariello, D., Josyula, S., Mao, C., Costinean, S., et al. (2012). miR-155 regulates IFN-γ production in natural killer cells. Blood 119, 3478–3485. doi: 10.1182/blood-2011-12-398099
Tsujimoto, H., Uchida, T., Efron, P. A., Scumpia, P. O., Verma, A., Matsumoto, T., et al. (2005). Flagellin enhances NK cell proliferation and activation directly and through dendritic cell-NK cell interactions. J. Leukoc. Biol. 78, 888–897. doi: 10.1189/jlb.0105051
Vey, N., Bourhis, J.-H., Boissel, N., Bordessoule, D., Prebet, T., Charbonnier, A., et al. (2012). A phase 1 trial of the anti-inhibitory KIR mAb IPH2101 for AML in complete remission. Blood 120, 4317–4323. doi: 10.1182/blood-2012-06-437558
Vey, N., Karlin, L., Sadot-Lebouvier, S., Broussais, F., Berton-Rigaud, D., Rey, J., et al. (2018). A phase 1 study of lirilumab (antibody against killer immunoglobulin-like receptor antibody KIR2D; IPH2102) in patients with solid tumors and hematologic malignancies. Oncotarget 9, 17675–17688. doi: 10.18632/oncotarget.24832
Vivier, E., Raulet, D. H., Moretta, A., Caligiuri, M. A., Zitvogel, L., Lanier, L. L., et al. (2011). Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–49. doi: 10.1126/science.1198687
Voo, K. S., Bover, L., Harline, M. L., Weng, J., Sugimoto, N., and Liu, Y.-J. (2014). Targeting of TLRs inhibits CD4 + regulatory T cell function and activates lymphocytes in human peripheral blood mononuclear cells. J. Immunol. 193, 627–634. doi: 10.4049/jimmunol.1203334
Wang, H., Chen, S., and Ye, X. (2019). Cocktail strategy based on NK cell-derived exosomes and their biomimetic nanoparticles for dual tumor therapy. Cancers 11:1560. doi: 10.3390/cancers11101560
Wang, H., Zhang, Y., Wu, X., Wang, Y., Cui, H., Li, X., et al. (2018). Regulation of human natural killer cell IFN-γ production by microRNA-146a via targeting the NF-κB signaling pathway. Front. Immunol. 9:293. doi: 10.3389/fimmu.2018.00293
Wang, L., Zhang, H., and Lei, D. (2019). microRNA-146a promotes growth of acute leukemia cells by downregulating ciliary neurotrophic factor receptor and activating JAK2/STAT3 signaling. Yonsei Med. J. 60, 924–934. doi: 10.3349/ymj.2019.60.10.924
Wang, P., Gu, Y., Zhang, Q., Han, Y., Hou, J., Lin, L., et al. (2012). Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J. Immunol. 189, 211–221. doi: 10.4049/jimmunol.1200609
Wang, Q., Lin, W., Tang, X., Li, S., Guo, L., Lin, Y., et al. (2017). The roles of microRNAs in regulating the expression of PD-1/PD-L1 immune checkpoint. Int. J. Mol. Sci. 18:2540. doi: 10.3390/ijms18122540
Wang, X., Li, J., Dong, K., Lin, F., Long, M., Ouyang, Y., et al. (2015). Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal. 27, 443–452. doi: 10.1016/j.cellsig.2014.12.003
Watanabe, A., Tagawa, H., Yamashita, J., Teshima, K., Nara, M., Iwamoto, K., et al. (2011). The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia 25, 1324–1334. doi: 10.1038/leu.2011.81
Wei, J., Nduom, E. K., Kong, L. -Y., Hashimoto, Y., Xu, S., Gabrusiewicz, K., et al. (2016). MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro. Oncol. 18, 639–648. doi: 10.1093/neuonc/nov292
Winter, S. J., and Krueger, A. (2019). Development of unconventional T cells controlled by MicroRNA. Front. Immunol. 10:2520. doi: 10.3389/fimmu.2019.02520
Xie, J., Liu, M., Li, Y., Nie, Y., Mi, Q., and Zhao, S. (2014). Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell. Mol. Immunol. 11, 495–502. doi: 10.1038/cmi.2014.30
Xu, D., Han, Q., Hou, Z., Zhang, C., and Zhang, J. (2017). miR-146a negatively regulates NK cell functions via STAT1 signaling. Cell. Mol. Immunol. 14, 712–720. doi: 10.1038/cmi.2015.113
Xu, S., Tao, Z., Hai, B., Liang, H., Shi, Y., Wang, T., et al. (2016). miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 7:11406. doi: 10.1038/ncomms11406
Yamanaka, Y., Tagawa, H., Takahashi, N., Watanabe, A., Guo, Y.-M., Iwamoto, K., et al. (2009). Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood 114, 3265–3275. doi: 10.1182/blood-2009-06-222794
Yang, P., Ni, F., Deng, R.-Q., Qiang, G., Zhao, H., Yang, M.-Z., et al. (2015). MiR-362-5p promotes the malignancy of chronic myelocytic leukaemia via down-regulation of GADD45α. Mol. Cancer 14:190. doi: 10.1186/s12943-015-0465-3
Yang, Q., Li, J., Hu, Y., Tang, X., Yu, L., Dong, L., et al. (2019). MiR-218-5p suppresses the killing effect of natural killer cell to lung adenocarcinoma by targeting SHMT1. Yonsei Med. J. 60, 500–508. doi: 10.3349/ymj.2019.60.6.500
Yee, D., Shah, K. M., Coles, M. C., Sharp, T. V., and Lagos, D. (2017). MicroRNA-155 induction via TNF-α and IFN-γ suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J. Biol. Chem. 292, 20683–20693. doi: 10.1074/jbc.M117.809053
Yi, R., Qin, Y., Macara, I. G., and Cullen, B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016. doi: 10.1101/gad.1158803
Yu, J., Mao, H. C., Wei, M., Hughes, T., Zhang, J., Park, I., et al. (2010). CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK-cell subsets. Blood 115, 274–281. doi: 10.1182/blood-2009-04-215491
Yun, S., Lee, S. U., Kim, J. M., Lee, H.-J., Song, H. Y., Kim, Y. K., et al. (2014). Integrated mRNA-microRNA profiling of human NK cell differentiation identifies MiR-583 as a negative regulator of IL2Rγ expression. PLoS ONE 9:e108913. doi: 10.1371/journal.pone.0108913
Zaghi, E., Calvi, M., Marcenaro, E., Mavilio, D., and Di Vito, C. (2019). Targeting NKG2A to elucidate natural killer cell ontogenesis and to develop novel immune-therapeutic strategies in cancer therapy. J. Leukoc. Biol. 105, 1243–1251. doi: 10.1002/JLB.MR0718-300R
Zawislak, C. L., Beaulieu, A. M., Loeb, G. B., Karo, J., Canner, D., Bezman, N. A., et al. (2013). Stage-specific regulation of natural killer cell homeostasis and response against viral infection by microRNA-155. Proc. Natl. Acad. Sci. U.S.A. 110, 6967–6972. doi: 10.1073/pnas.1304410110
Zhang, G., Li, N., Li, Z., Zhu, Q., Li, F., Yang, C., et al. (2015). microRNA-4717 differentially interacts with its polymorphic target in the PD1 3′ untranslated region: a mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget 6, 18933–18944. doi: 10.18632/oncotarget.3662
Keywords: human NK cells, NK cell receptors, microRNA, immune checkpoint, immunotherapy, gene expression
Citation: Pesce S, Greppi M, Ferretti E, Obino V, Carlomagno S, Rutigliani M, Thoren FB, Sivori S, Castagnola P, Candiani S and Marcenaro E (2020) miRNAs in NK Cell-Based Immune Responses and Cancer Immunotherapy. Front. Cell Dev. Biol. 8:119. doi: 10.3389/fcell.2020.00119
Received: 30 November 2019; Accepted: 11 February 2020;
Published: 25 February 2020.
Edited by:
Francesco Fazi, Sapienza University of Rome, ItalyReviewed by:
Enrico Maggi, University of Florence, ItalySaula Checquolo, Sapienza University of Rome, Italy
Copyright © 2020 Pesce, Greppi, Ferretti, Obino, Carlomagno, Rutigliani, Thoren, Sivori, Castagnola, Candiani and Marcenaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simona Sivori, simona.sivori@unige.it; Emanuela Marcenaro, emanuela.marcenaro@unige.it
†These authors have contributed equally to this work
‡These authors share senior authorship
 Silvia Pesce
Silvia Pesce Marco Greppi
Marco Greppi Elisa Ferretti
Elisa Ferretti Valentina Obino
Valentina Obino Simona Carlomagno
Simona Carlomagno Mariangela Rutigliani
Mariangela Rutigliani Fredrik B. Thoren
Fredrik B. Thoren Simona Sivori
Simona Sivori Patrizio Castagnola
Patrizio Castagnola Simona Candiani
Simona Candiani Emanuela Marcenaro
Emanuela Marcenaro