Editorial: Self-organizing and excitable signaling networks in cell biology
- 1Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, CA, United States
- 2Department of Molecular and Cellular Biology, University of California, Davis, Davis, CA, United States
- 3Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, CA, United States
Editorial on the Research Topic
Self-organizing and excitable signaling networks in cell biology
Introduction
In the past 2 decades, advances in live cell imaging using fluorescent microscopy have revealed that certain signaling networks display intriguing spatiotemporal patterns characteristic of excitable systems. Classic examples of excitable systems include electrical waves in neuromuscular cells, calcium waves in fertilized eggs, and the Belousov-Zhabotinsky reaction. Excitable systems have been defined in various ways but generally possess the following features: 1) the existence of a resting, an excited, and a refractory state, and 2) the capacity to propagate the excited state as waves. Upon receiving a suprathreshold stimulus, the system transitions from the resting state into an excited state, followed by a refractory, or unresponsive state before returning to the resting state. Waves in excitable systems mutually annihilate due to the system’s inability to reactivate during the refractory period. These properties are also evident in various signaling networks, including the actin cytoskeleton and its regulators and the Ras/PI3K network that operates downstream of numerous receptors. The high temporal resolution of live-cell imaging has revealed that the activities of these networks spontaneously organize into dynamic spatial patterns across a range of scales, spanning from individual cells to populations of cells. While some stimuli trigger excitable activity, others can modulate the threshold for excitation, providing a means to integrate multiple inputs to mediate crucial cellular functions in both prokaryotic and eukaryotic cells (Loose et al., 2011; Huang et al., 2013; Tang et al., 2014; Yang et al., 2018; Di Talia and Vergassola, 2022; Gallagher et al., 2022) (Figure 1).
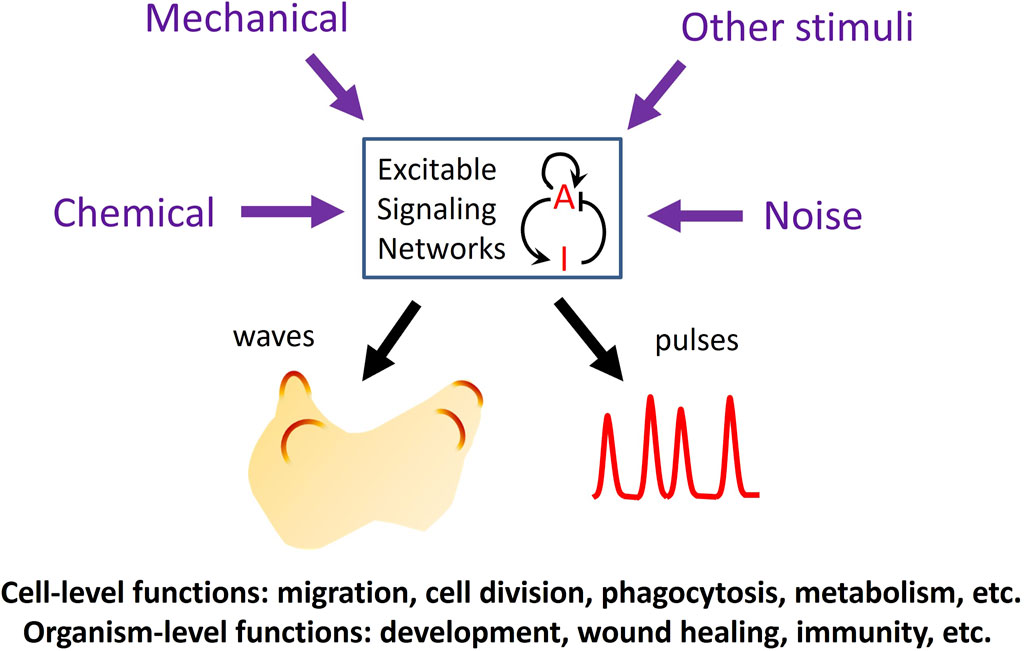
Figure 1. Functions of excitable signaling networks. The activity of an excitable signaling network, represented by a prototypical activator (A)-inhibitor (I) system, can be tuned by various types of external stimuli, which can enter the network through different pathways to modulate the frequency of the waves or pulses of activation. Excitability plays crucial roles in processes at both the cellular and organismal levels.
Excitability of cortical actin network
Many cellular processes involve changes in cell morphology driven by the polymerization of actin. Live imaging of actin and its regulators revealed that the polymerization activity of the cortical network propagates as mutually annihilating waves (Vicker, 2002; Gerisch et al., 2004; Weiner et al., 2007). These cortical waves have been observed in cells undergoing migration, mitosis, embryonic development, and immune activation. Functionally, cortical excitability coordinates the cellular morphological changes in response to environmental stimuli.
In mast cells, traveling waves and oscillations of F-actin are driven by feedback interactions between signaling molecules such as Cdc42 and FBP17 (Wu et al., 2013). A research article in this issue by Tong et al. provides molecular insights into the nucleation mechanism of these actin waves through antagonism between microtubule and actin polymerization. Specifically, they find that the microtubule-binding formin FHDC1 (FH2 domain-containing protein 1), when released from depolymerizing microtubules, mediates actin polymerization to initiate actin waves. Disruption of FHDC1 causes defective cell polarity and division. These findings add to our understanding of the molecular mechanisms and functions of cortical wave initiation.
Waves of F-actin and Rho activation have also been observed in the eggs and embryonic cells of frogs and starfish. In these cells, cortical waves are regulated by mitotic spindles and are implicated in cytokinesis (Bement et al., 2024). Sepaniac et al. studied how actin waves respond to wounding and change over the course of embryonic development in the Xenopus embryo. They found that wounding causes waves to converge and stop, pulling the cell-cell junction toward the wound, possibly contributing to the healing process. Interestingly, over the developmental time course, cortical excitability is replaced by epithelial excitability in which waves of actin and calcium propagate across the cell population rather than individual cell cortices. This observation hints at possible roles of excitability in both individual cells and across the cell population during embryonic development, as has been demonstrated in other processes discussed below.
From individual cells to cell populations in receptor signaling networks
Ras/PI3K signaling in Dictyostelium discoideum has been a particularly revealing model for signaling network excitability operating at multiple scales (Devreotes et al., 2017). Matsuoka et al. review recent progress in Dictyostelium cell migration directed by the excitable network involving Ras GTPases and their downstream effectors including PI3K, PTEN, and phosphoinositides PI(3,4,5)P3 and PI(4,5)P2. Within an individual cell, these signaling molecules can undergo spontaneous symmetry breaking to generate polarity and coordinate remodeling of the actin cytoskeleton, driving cell migration. When coupled to upstream gradient-sensing receptors, excitation of the Ras/PI3K signaling network can be spatially biased by chemical gradients to mediate chemotaxis. This excitable behavior of Ras signaling can be simulated mathematically using reaction-diffusion equations.
As a population, Dictyostelium cells respond to starvation by initiating a developmental program that culminates in the formation of fruiting bodies. In this process, periodic waves of cAMP, a chemoattractant for Dictyostelium, propagate across the cell population to coordinate the collective migration of cells toward each other. Drawing on biochemical and genetic analyses, Jaiswal et al. present an integrated model for the generation of outwardly propagating multicellular waves of cAMP in developing Dictyostelium. In their model, multiple positive and negative feedback loops between MEK/ERK and cAMP/PKA signaling pathways underlie the activation, adaptation, and re-activation responses to cAMP. This model underscores the complexity of molecular interactions involved in the generation of oscillatory cAMP waves.
Findings in Dictyostelium have led to understanding excitable Ras and actin signaling in higher eukaryotes. For example, waves of Ras-PI3K activation and F-actin can generate protrusions in mammalian epithelial cells, and induce pulsatile activation of ERK, an important regulator of cell proliferation (Albeck et al., 2013; Aoki et al., 2013; Regot et al., 2014; Yang et al., 2018; Goglia et al., 2020; Zhan et al., 2020). Propagation of ERK activity waves across the population can coordinate the collective migration and survival of epithelial cells (Aoki et al., 2017; Hino et al., 2020; 2022; Gagliardi et al., 2021), connecting excitable networks to the physiological function of tissues.
Excitability in biological vs. chemical systems
Excitable biochemical systems in eukaryotic cells, including the actin and Ras signaling networks, generally involve complex interactions between a large number of molecular species. Such systems are very challenging to analyze in their intact state, due to the difficulty of experimentally manipulating them in the sophisticated ways required to dissect their molecular functions. In contrast, simple excitable systems composed of a handful of components have been found in prokaryotes and chemical reactions. Riedl and Sixt compared the excitable dynamics of polymerizing actin with that of the Belousov-Zhabotinsky reaction, a well studied excitable chemical system composed of five reagents. Despite the vastly different magnitude of their complexities, remarkable similarities are shared between the two systems such as the responses of the propagating waves to geometric constraints or external fields as well as emergent collective behaviors.
Conclusion
The papers in this Research Topic capture various aspects of the fascinating phenomenon of self-organized and excitable dynamics of signaling networks in different organisms, highlighting their roles in cell migration, embryonic development, and cell proliferation. Since these processes are involved in the pathogenesis of many diseases, future research into the molecular basis and the functions of signaling excitability may lead to new targets for intervention.
Author contributions
C-HH: Writing–original draft, Writing–review and editing. JA: Writing–review and editing. PD: Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH grants R01GM136711 (to C-HH), the Sol Goldman Pancreatic Cancer Research Center (to C-HH), a Johns Hopkins Discovery Award (to C-HH), 5R35GM139621 (to JA), R01HL151983 (to JA), NIH R35GM118177 (to PD), and DARPA HR0011-16-C-0139 (to PD).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albeck, J. G., Mills, G. B., and Brugge, J. S. (2013). Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell 49, 249–261. doi:10.1016/j.molcel.2012.11.002
Aoki, K., Kondo, Y., Naoki, H., Hiratsuka, T., Itoh, R. E., and Matsuda, M. (2017). Propagating wave of ERK activation orients collective cell migration. Dev. Cell 43, 305–317. doi:10.1016/j.devcel.2017.10.016
Aoki, K., Kumagai, Y., Sakurai, A., Komatsu, N., Fujita, Y., Shionyu, C., et al. (2013). Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell 52, 529–540. doi:10.1016/j.molcel.2013.09.015
Bement, W. M., Goryachev, A. B., Miller, A. L., and von Dassow, G. (2024). Patterning of the cell cortex by Rho GTPases. Nat. Rev. Mol. Cell Biol. 25, 290–308. doi:10.1038/s41580-023-00682-z
Devreotes, P. N., Bhattacharya, S., Edwards, M., Iglesias, P. A., Lampert, T., and Miao, Y. (2017). Excitable signal transduction networks in directed cell migration. Annu. Rev. Cell Dev. Biol. 33, 103–125. doi:10.1146/annurev-cellbio-100616-060739
Di Talia, S., and Vergassola, M. (2022). Waves in embryonic development. Annu. Rev. Biophys. 51, 327–353. doi:10.1146/annurev-biophys-111521-102500
Gagliardi, P. A., Dobrzyński, M., Jacques, M.-A., Dessauges, C., Ender, P., Blum, Y., et al. (2021). Collective ERK/Akt activity waves orchestrate epithelial homeostasis by driving apoptosis-induced survival. Dev. Cell 56, 1712–1726.e6. doi:10.1016/j.devcel.2021.05.007
Gallagher, K. D., Mani, M., and Carthew, R. W. (2022). Emergence of a geometric pattern of cell fates from tissue-scale mechanics in the Drosophila eye. Elife 11, e72806. doi:10.7554/eLife.72806
Gerisch, G., Bretschneider, T., Müller-Taubenberger, A., Simmeth, E., Ecke, M., Diez, S., et al. (2004). Mobile actin clusters and traveling waves in cells recovering from actin depolymerization. Biophys. J. 87, 3493–3503. doi:10.1529/biophysj.104.047589
Goglia, A. G., Wilson, M. Z., Jena, S. G., Silbert, J., Basta, L. P., Devenport, D., et al. (2020). A live-cell screen for altered erk dynamics reveals principles of proliferative control. Cell Syst. 10, 240–253. doi:10.1016/j.cels.2020.02.005
Hino, N., Matsuda, K., Jikko, Y., Maryu, G., Sakai, K., Imamura, R., et al. (2022). A feedback loop between lamellipodial extension and HGF-ERK signaling specifies leader cells during collective cell migration. Dev. Cell. 57, 2290–2304.e7. doi:10.1016/j.devcel.2022.09.003
Hino, N., Rossetti, L., Marín-Llauradó, A., Aoki, K., Trepat, X., Matsuda, M., et al. (2020). ERK-mediated mechanochemical waves direct collective cell polarization. Dev. Cell 53, 646–660. doi:10.1016/j.devcel.2020.05.011
Huang, C.-H., Tang, M., Shi, C., Iglesias, P. A., and Devreotes, P. N. (2013). An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat. Cell Biol. 15, 1307–1316. doi:10.1038/ncb2859
Loose, M., Kruse, K., and Schwille, P. (2011). Protein self-organization: lessons from the min system. Annu. Rev. Biophys. 40, 315–336. doi:10.1146/annurev-biophys-042910-155332
Regot, S., Hughey, J. J., Bajar, B. T., Carrasco, S., and Covert, M. W. (2014). High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734. doi:10.1016/j.cell.2014.04.039
Tang, M., Wang, M., Shi, C., Iglesias, P. A., Devreotes, P. N., and Huang, C.-H. (2014). Evolutionarily conserved coupling of adaptive and excitable networks mediates eukaryotic chemotaxis. Nat. Commun. 5, 5175. doi:10.1038/ncomms6175
Vicker, M. G. (2002). Eukaryotic cell locomotion depends on the propagation of self-organized reaction-diffusion waves and oscillations of actin filament assembly. Exp. Cell Res. 275, 54–66. doi:10.1006/excr.2001.5466
Weiner, O. D., Marganski, W. A., Wu, L. F., Altschuler, S. J., and Kirschner, M. W. (2007). An actin-based wave generator organizes cell motility. PLoS Biol. 5, e221. doi:10.1371/journal.pbio.0050221
Wu, M., Wu, X., and De Camilli, P. (2013). Calcium oscillations-coupled conversion of actin travelling waves to standing oscillations. Proc. Natl. Acad. Sci. U. S. A. 110, 1339–1344. doi:10.1073/pnas.1221538110
Yang, J.-M., Bhattacharya, S., West-Foyle, H., Hung, C.-F., Wu, T.-C., Iglesias, P. A., et al. (2018). Integrating chemical and mechanical signals through dynamic coupling between cellular protrusions and pulsed ERK activation. Nat. Commun. 9, 4673. doi:10.1038/s41467-018-07150-9
Keywords: signaling network, Belousov-Zhabotinsky oscillatory reaction, excitable system, cell migration and proliferation, mathematical modeling, live cell imaging, actin cytoskeletal dynamics
Citation: Huang C-H, Albeck JG and Devreotes PN (2024) Editorial: Self-organizing and excitable signaling networks in cell biology. Front. Cell Dev. Biol. 12:1430911. doi: 10.3389/fcell.2024.1430911
Received: 10 May 2024; Accepted: 13 May 2024;
Published: 04 June 2024.
Edited and reviewed by:
Claudia Tanja Mierke, Leipzig University, GermanyCopyright © 2024 Huang, Albeck and Devreotes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan-Hsiang Huang, chuang29@jhmi.edu; John G. Albeck, jgalbeck@ucdavis.edu; Peter N. Devreotes, pnd@jhmi.edu
 Chuan-Hsiang Huang
Chuan-Hsiang Huang John G. Albeck
John G. Albeck Peter N. Devreotes
Peter N. Devreotes