Comparison of the enzymatic depolymerization of polyethylene terephthalate and AkestraTM using Humicola insolens cutinase
- 1Division of Biotechnology, Department of Chemistry, Faculty of Engineering (LTH), Lund University, Lund, Sweden
- 2Facultad de Ciencias, Universidad Nacional de Colombia, Sede Medellín, Medellín, Colombia
- 3Centre for Analysis and Synthesis, Department of Chemistry, Faculty of Engineering (LTH), Lund University, Lund, Sweden
The enzymatic depolymerization of synthetic polyesters has become of great interest in recycling plastics. Most of the research in this area focuses on the depolymerization of polyethylene terephthalate (PET) due to its widespread use in various applications. However, the enzymatic activity on other commercial polyesters is less frequently investigated. Therefore, AkestraTM attracted our attention, which is a copolymer derived from PET with a partially biobased spirocyclic acetal structure. In this study, the activity of Humicola insolens cutinase (HiCut) on PET and AkestraTM films and powder was investigated. HiCut showed higher depolymerization activity on amorphous PET films than on Akestra™ films. However, an outstanding performance was achieved on AkestraTM powder, reaching 38% depolymerization in 235h, while only 12% for PET powder. These results are consistent with the dependence of the enzymes on the crystallinity of the polymer since Akestra™ is amorphous while the PET powder has 14% crystallinity. On the other hand, HiCut docking studies and molecular dynamic simulations (MD) suggested that the PET-derived mono (hydroxyethyl)terephthalate dimer (MHET)2 is a hydrolyzable ligand, producing terephthalic acid (TPA), while the Akestra™-derived TPA-spiroglycol ester is not, which is consistent with the depolymerization products determined experimentally. MD studies also suggest ligand-induced local conformational changes in the active site.
Introduction
According to the latest report Global Plastics Outlook (OECD, 2022), plastic waste is to triplicate by 2060 unless serious policies are implemented. In the report, it is also clear that low recycling percentages and mismanagement of plastic wastes, significantly contribute to the plastic contamination in terrestrial and aquatic environments. Therefore, the development of sustainable and high-quality recycling methods becomes fundamental. As such, enzymatic degradation provides a powerful tool that works under mild conditions with good specificity towards the ester bond (Zimmermann and Billig, 2010), resulting in high quality chemicals and monomers for closed-loop recycling applications (Wei and Zimmermann, 2017).
With enzymatic degradation technology, polyethylene terephthalate (PET) is the most intensively studied polymer: more than 15 PET hydrolases, and many more variants, have been described so far (Qi et al., 2021). Yet, novel plastics using biobased building blocks that retain comparable or even better properties became popular (Hatti-Kaul et al., 2020), and their enzymatic depolymerization also needs to be assessed (Arza et al., 2019; Wang et al., 2020; Li et al., 2021). Among the newly developed biobased polyesters, a partially biobased polymer AkestraTM (Perstorp AB) presents a high Tg (90–110°C) and transparency, which shows high potential in various applications, such as hot-filling food packaging, bottles, teapots, and others. Previously, enzymatic treatments of AkestraTM using Ideonella sakaiensis PETase (IsPETase) and a marine PET hydrolase (PET2) showed only 0.13% of depolymerization (Wagner-Egea et al., 2021) and no detectable depolymerization products (Wagner-Egea et al., 2022), respectively. Both enzymatic processes were performed at 37°C (IsPETase) and 55°C (PET2), temperatures significantly lower than the glass transition temperature of AkestraTM. Thus, this factor is important for an efficient depolymerization.
Humicola insolens cutinase (HiCut), with an optimal temperature of 70°C (Fabbri et al., 2021), can perform more closely to polymer’s glass transition temperature (Tg∼72°C for PET, 90–110°C for AkestraTM), where amorphous regions are more accessible to enzyme hydrolysis (Wei et al., 2019). Several degradation studies have been conducted using HiCut yielding a high degradation rate in PET (Ronkvist et al., 2009; Machado de Castro et al., 2017; Machado de Castro et al., 2018; Machado de Castro et al., 2019; de Queiros Eugenio et al., 2021) and polyester-polyurethane copolymer (Di Bisceglie et al., 2022). Compared to other cutinases, HiCut showed a good performance in degrading polyesters with other biobased aromatic units such as furan or thiophene units (Weinberger et al., 2017; Gigli et al., 2019). Herein, biocatalytic degradation of commercial polyester AkestraTM is demonstrated for the first time. Moreover, molecular docking of HiCut with PET and AkestraTM, together with molecular dynamic simulations provided insights into the enzyme/ligand interactions including the ligand effect on the conformational stabilization of a flexible predicted loop.
Materials and methods
Chemicals, enzyme and polymers
All chemicals and reagents were purchased, at the highest purity, from Sigma-Aldrich. The commercial liquid-preparation of Humicola insolens cutinase (HiCut) was purchased from Novozymes (product Novozym® 51032).
Amorphous PET film was purchased from Goodfellow (product number ES30-FM-000145), RamaPET N180 from Indorama, and AkestraTM courtesy of Perstorp AB. The called powder polymers were prepared through a dissolution-precipitation pretreatment as previously described (Wagner-Egea et al., 2021). The physical properties of the polymers used in this study are summarized in Table 1.
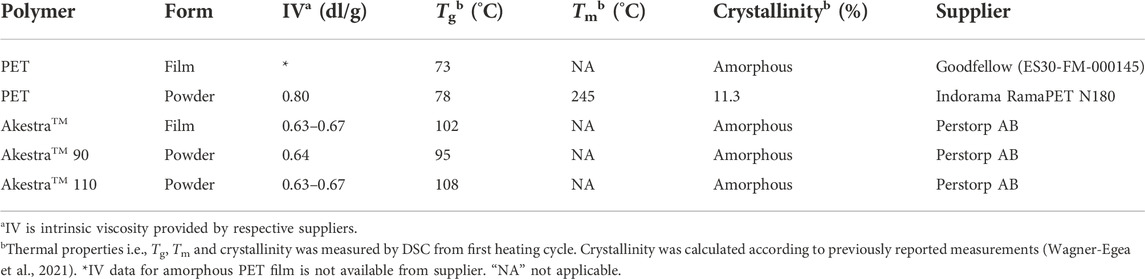
TABLE 1. Properties of the films according to provider specifications, and properties of the powder before dissolution-precipitation (crystallinity was determined after dissolution-precipitation).
Moreover, according to the manufacturer, AkestraTM 90 has 20 mol% content in spiroacetal units, while AkestraTM 110 has 45 mol% (with respect to terephthalate units). Figure 1 shows the structure of both terephthalate-based polyesters evaluated.
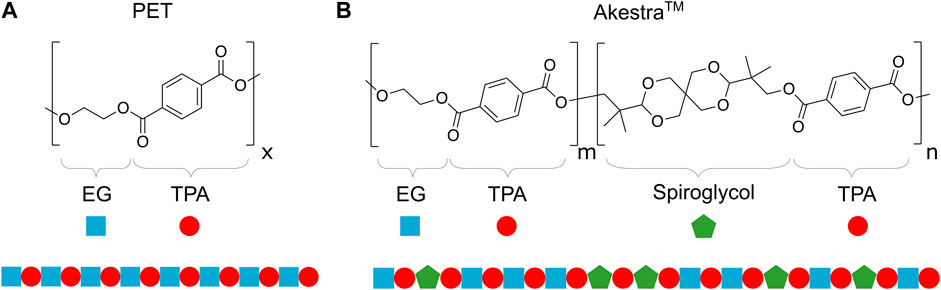
FIGURE 1. Chemical structures of the polyesters with schematic representation. (A) PET. (B) AkestraTM.
Protein characterization: Purity, concentration, and thermal stability
The concentration of the commercial HiCut was determined by the bicinchoninic acid (BCA) method following the kit manufacturer protocol (BCA1 from Sigma Aldrich), obtaining a stock concentration of 53.5 g/L. Moreover, SDS-PAGE analysis using pre-casted 12% acrylamide gels (Bio-Rad) shows no signs of degradation, and the predominant protein of the preparation has a molar mass of around 20 kDa, consistent with the theoretically calculated molecular weight. The activity of the batch used was determined in 344 ± 31 U/mL, by using pNP-butyrate as a substrate (Supplementary information, Supplementary Figure S1).
The thermal stability was evaluated by Differential Scanning Fluorometry (DSF) using the Prometheus NT 48 nano DSF (NanoTemper Technologies GmbH, Munich, Germany). Triplicate samples of 1 mg/ml HiCut, with and without 1 mM CaCl2, were prepared in 50 µL of a 1 M Tris-HCl Buffer. The samples were analyzed by heating from 20 to 90°C at a rate of 1°C per minute. Protein unfolding was monitored by measuring the intrinsic fluorescence at emission wavelengths of 330 and 350 nm.
Enzymatic reactions
Reactions were performed by soaking 20 mg of the polymer in 2 ml of a 1 M Tris-HCl buffer with a pH of 7.5, 10% (v/v) dimethyl sulfoxide (DMSO), and 0.085 mg/ml of the enzyme (Ronkvist et al., 2009; Wagner-Egea et al., 2021). Triplicates, including negative control (reaction mixture without the enzyme), were incubated at 70°C and 200 rpm for 235 h. Samples at various times were retrieved by diluting 200 µL of sample in 500 µL of DMSO, filtered with a 0.2 µm PES sterile membrane, and transferred to glass vials for further HPLC analyses. For reactions with powder polymers, a second dilution in DMSO was needed.
HPLC and LC-MS analysis
Reaction products of the enzymatic degradation were analyzed by High-Performance Liquid Chromatography (HPLC) and Liquid Chromatography-Mass Spectrometry (LC-MS) with methods described previously (Wagner-Egea et al., 2021). The HPLC used is an Ultimate 3000 RS (Dionex) equipped with a UV/Vis detector (SPD-20A) and the LC-MS is an Ultimate 3000 RS HPLC-system (ThermoFisher Scientific) coupled with an LTQ Velos Pro Ion trap mass spectrom (ThermoFisher Scientific) using a heated electrospray ionization source (HESI-II). The wavelength of the UV was set to 260 nm, and the MS was operated in negative mode, over a mass range of m/z 50–700. The HESI-source temperature was set to 300°C, and spray voltage was 3.0 kV. Helium was used as collision gas, and nitrogen gas was used as nebulizing gas. LC-MS/MS was conducted using data dependent acquisition (DDA) with CID fragmentation energy setting of 35%. A hydrophobic C18 column (Kinetex®1.7 μm XB-C18 100 Å, LC Column50 × 2.1 mm) was used, and a mobile phase consisting of 20 and 80% mixture of acetonitrile and formic acid (0.1%), respectively. The flow rate was fixed at 0.4 ml/min for 3 min per sample. For certain trials the program was extended to 20 min, to verify the wash-out of the column and to search for additional degradation products. The same flow was kept, and the standard conditions were held for 8 min, whereafter mobile phase A (acetonitrile) increased to 50% (phase B was formic acid 0.1%) over 1 min, was held on this level for 4 min, then decreased back to 20% over 1 min and finally, the starting conditions were held for another 6 min.
Fourier-transform infrared spectroscopy
The residual PET and AkestraTM, both films and powders, from 192 h of enzymatic reactions, were washed with detergent, rinsed with ultrapure water, and left drying under airflow. Then, infrared spectra of the samples were recorded at a resolution of 4 cm−1 on a NicoletTMiSTM5 FTIR Spectrometer (Thermo Fisher Scientific). The data analysis was performed with OMINIC software.
Surface electron microscopy imaging
The surface morphology of PET and AkestraTM films after 192 h of enzymatic reactions was examined by Surface Electron Microscopy (SEM) on a JSM-6700F (JEOL) model at an accelerating voltage of 10 kV. Samples, including negative controls, were sputter coated with a 15 nm layer of Au/Pd to provide sufficient electrical conductivity.
Nuclear magnetic resonance spectroscopy
AkestraTM samples were dissolved in CDCl3 and subjected to nuclear magnetic resonance (NMR) spectroscopy analysis, performed at 100.61 MHz carbon frequency on a Bruker DR X400 spectrometer.
Molecular docking and molecular dynamics simulation
To investigate the interactions between enzyme (receptor) and substrate (ligand), a molecular docking of HiCut was performed with PET and AkestraTM. The ligands used were mono (hydroxyethyl)terephthalate dimer (MHET)2 (Wagner-Egea et al., 2022) and terephthalate-spiroglycol ester (TPA-spiroglycol) (Figure 4B). The crystallographic structure of HiCut was previously resolved (Kold et al., 2014) and 3D structure of 3.00 Å resolution deposited in the Protein Data Bank, accession code 4OYY. Then, ligands and enzyme were docked using Autodock Vina (Trott and Olson, 2010) implemented in YASARA (Krieger et al., 2002), with visual analysis in USCF Chimera v 1.15 (Pettersen et al., 2004).
After choosing the best-docked complexes, molecular dynamic simulations (MD) were performed in YASARA (Krieger and Vriend, 2015). The MD simulation conditions included a pH 7.4, 0.9% NaCl, 0.97774 g/ml water density, and 343 K for 50 ns. Simulation snapshots were saved every 250 ps. The simulation cell was a cube 10 Å around all atoms with periodic boundaries. The forcefield used was AMBER14 (Case et al., 2014), with long-range coulomb forces. Detailed trajectory analysis and ligand-receptor binding energy calculations were performed by using Poisson-Boltzmann and Surface Area Solvation (MM/PBSA) methods (Genheden and Ryde, 2015).
Differential scanning calorimetry
A TA Instruments DSC Q2000 was used for analysis of thermal properties and crystallinity. The PET and Akestra (both films and powders) were analyzed before enzymatic degradation. The amorphous samples were heated from room temperature to 200°C at the rate of 10°C/min and PET powder was heated from room temperature to 300°C at the rate of 10°C/min under nitrogen with a purge rate of 50 ml min−1. The crystallinity of PET powder was calculated according to previously reported method (Wagner-Egea et al., 2021).
Results and discussion
HiCut thermal stability
The melting temperature (Tm) of HiCut, in buffer Tris-HCl pH 7.5, was 80°C free of DMSO and 77°C in presence of 10% DMSO (Figure 2). Previously, Baker et al. (2012) reported a Tm at 62.7°C (pH 8) and 64.3°C (pH 5) with sodium phosphate and sodium acetate buffers, respectively. This highly suggests that the buffer composition and pH used in this work are more suitable for HiCut thermostability. The addition of 10% DMSO decreases the HiCut Tm only in 3°C, indicating that 70°C is a suitable temperature for performing reactions with 10% DMSO. On the other hand, additional CaCl2 1 mM did not show any significant effect on the thermal stability of HiCut.
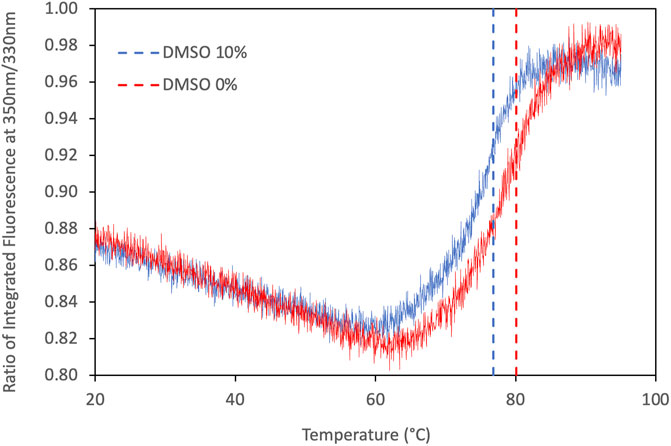
FIGURE 2. Thermograms of HiCut DSF analyses. Dotted lines corresponds to melting temperature in presence of 10% DMSO (77°C) and free of DMSO (80°C).
Depolymerization products and reaction profile
HiCut depolymerization products of PET, determined by HPLC, were terephthalic acid (TPA) at ∼0.61 min, mono (hydroxyethyl)terephthalate (MHET) at ∼0.76 min, and bis(hydroxyethyl)terephthalate (BHET) at ∼0.96 min of retention time (Figure 3A). These products were confirmed by LC-MS (Supplementary information, Supplementary Figure S6). Moreover, a peak eluting between DMSO and TPA was identified as Tris, a component of the buffer used for the reaction mix. In the first 9 h of reaction, the main product consisted of MHET with small amounts of BHET and TPA, while by later (192 h), the main product was TPA. This result indicates that HiCut can further degrade MHET into TPA, as reported by others (Ronkvist et al., 2009; Machado de Castro et al., 2019; de Queiros Eugenio et al., 2021; Kaabel et al., 2021). Having relatively pure TPA as the main product is highly desirable because it is a monomer that can be directly used for a re-polymerization of PET (Wierckx et al., 2015) or synthesis of other polymers such as AkestraTM or other value-added products (Roy et al., 2013).
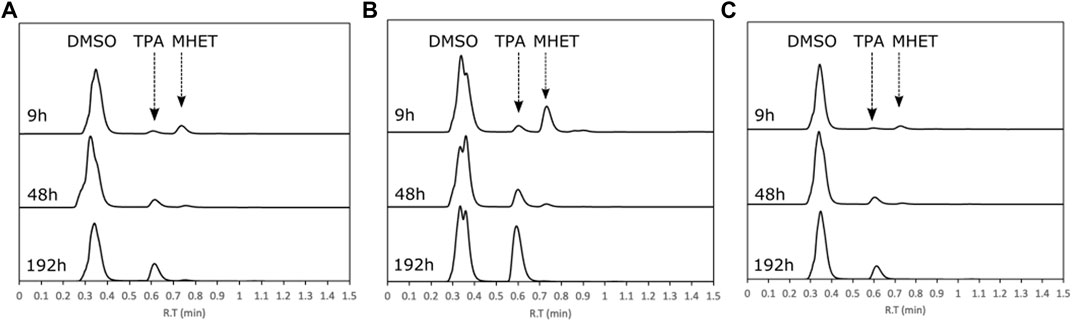
FIGURE 3. Analysis of HiCut depolymerization products by HPLC. Chromatograms of samples taken after 9, 48, and 192 h of reaction from powder of (A) PET, (B) AkestraTM90, and (C) AkestraTM110.
HiCut depolymerization of the two grades of AkestraTM, both 90 and 110, produced TPA and MHET in the first hours of the reaction, but later, after 48 h, the dominant product was TPA (Figures 3B,C). LC-MS product analysis at the end of the reaction (192 h), with a longer analytical time, showed the TPA peak (Figure 4A) and a second peak at 14.3 min of retention time, identified as a TPA-spiroglycol ester (Figure 4B), indicating that HiCut is ineffective in hydrolyzing the TPA-spiroglycol ester bond (Figure 4C). Further hydrolysis product (spiroglycol) was not detected.
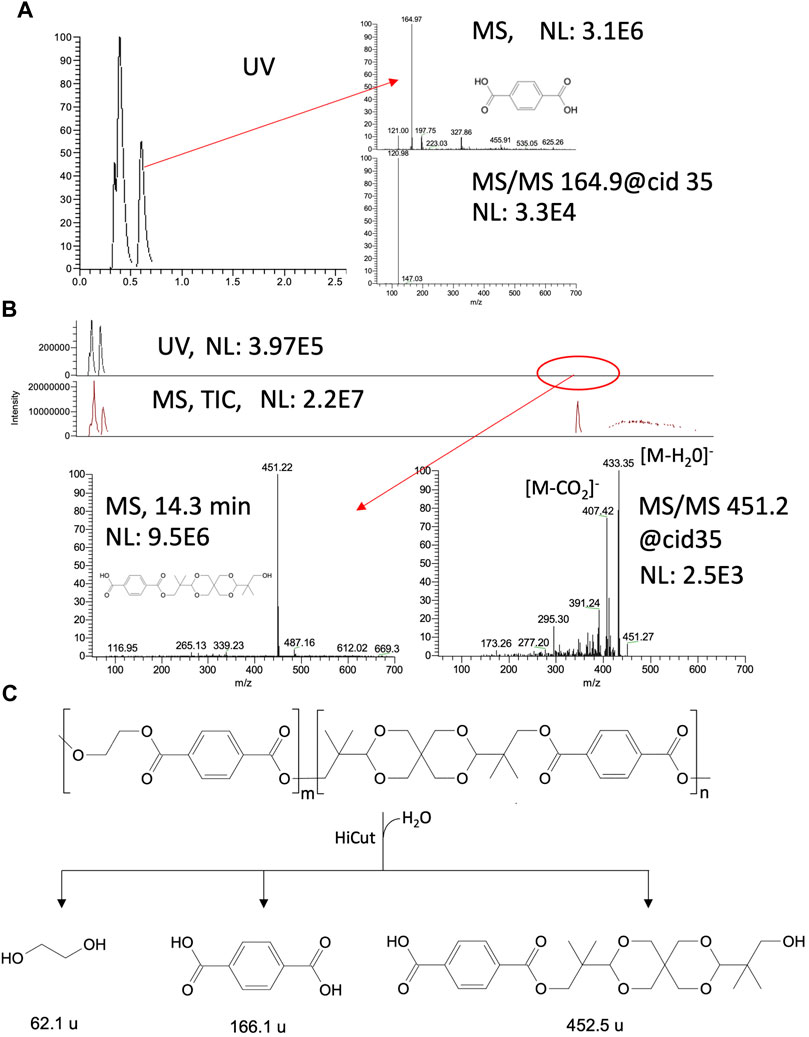
FIGURE 4. HiCut-depolymerization of AkestraTM powders, 90 or 110 after 192 h of reaction. (A) LC-MS analysis of TPA, (B) LC-MS analysis of TPA-spiroglycol. (C) Enzymatic depolymerization of AkestraTM proposed.
HiCut activity on PET and AkestraTM
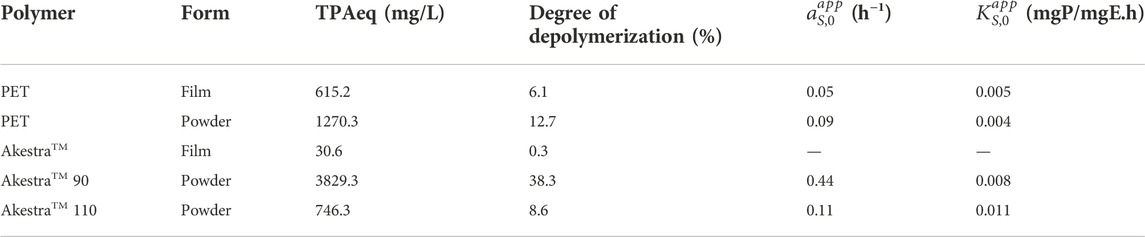
The HiCut activity on PET and AkestraTM was assessed here as the concentration (mg/L) of terephthalic acid equivalents (TPAeq) produced over time. For the HPLC quantification (Supplementary information, Supplementary Figure S2 and Supplementary Table S1), the products MHET and BHET were regarded as TPAeq, assuming the same molar extinction coefficient ε = 17,000M−1cm−1 conferred by the aromatic ring. Thus, the degradation of amorphous PET films reached a product concentration of 615.2 mg/L, while AkestraTM film degradation was only 30.6 mg/L (Figure 5A). In case of amorphous powders (Figure 5B), HiCut yielded 1270.3, 3829.3 and 746.3 mg/L TPAeq for PET, AkestraTM 90 and AkestraTM 110, respectively (Table 2). It should be noted that the rate of enzymatic depolymerization of polyesters can be influenced by many intrinsic factors of the polymers (e.g., chemical structures, Tg, crystallinity, molecular weight, physical forms of the samples) as well as external factors (e.g., choice of enzymes, temperature, pH, etc.). It is therefore rather challenging to isolate the impact of one particular factor and draw a meaningful conclusion based on limited experiments. In this work, we specifically compared the results of the seemingly similar samples, aiming to provide a first insight. When the two powders of different grades of AkestraTM 90 and 110 were compared (Figure 5B), it is interesting to observe that AkestraTM 90 showed a significantly higher depolymerization rate than AkestraTM 110. These two samples have the same physical form (powders), crystallinity (fully amorphous) and similar molecular weight (as reflected by their similar IV values, ∼0.64 and ∼0.63–0.67 dl/g for AkestraTM 90 and 110, respectively, Table 1), so the large difference in their depolymerization rate could be attributed to their difference in Tg and chemical structures. AkestraTM 90 has a lower Tg than AkestraTM 110, which is a factor that has been reported to enhance other enzymatic depolymerization (Carniel et al., 2021). Furthermore, AkestraTM 110 also has higher content of spiroglycol units (∼45 mol%) than that of AkestraTM 90 (∼20 mol%, according to supplier), which could influence the rate of enzymatic degradation according to literature (Hu et al., 2021). In addition, the two AkestraTM (90 and 110) powders also differ in their distributions of the structural dyads and monomer sequence lengths (Supplementary information, Supplementary Figure S3), as shown by the presence of multiple aromatic or carbonyl 13C signals (8 (Ar-CH), 9 (Ar-C) and 10 (C=O) (Supplementary information, 13C NMR of AkestraTM 90 and 110, Supplementary Figures S4A–C). The possible impact of this factor on the rate of enzymatic degradation of the two AkestraTM polyesters remained to be unraveled (Japu et al., 2012; Lavilla et al., 2012; Kupczak et al., 2021).
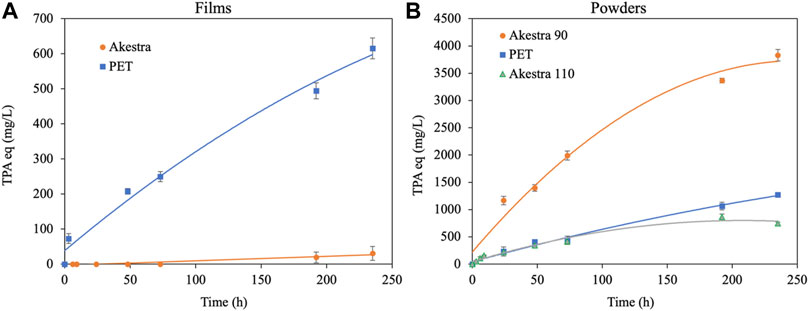
FIGURE 5. Enzymatic degradation products in TPAeq as a function of time. (A) PET (blue squares) and AkestraTM (orange dots) films. (B) PET (blue squares), AkestraTM 90 (orange dots) and AkestraTM 110 (green triangles) powders.
When PET powder was compared to the two AkestraTM powders, it is more complicated because PET not only differ from AkestraTM in their chemical structures and Tg, but also crystallinity. In principle, the low degradation products obtained with PET powder, compared with AkestraTM 90, could be related to its 11% crystallinity that limits the access to polymer chains for depolymerization (Webb et al., 2012). Previously, other authors also reported a decreased performance of HiCut with semi-crystalline polymers (Ronkvist et al., 2009; Weinberger et al., 2017; Machado de Castro et al., 2019), making it the major drawback to implementing this enzyme. In the meantime, the lower Tg of PET compared to AkestraTM 110 might be the dominating factor that resulted in the observed slightly faster depolymerization rate of PET powder compared to that of AkestraTM 110 powder (Figure 5B).
When the degradation rate of the two films were compared (PET and Akestra, Figure 5A), apparently PET film showed much faster degradation rate. These two samples were both fully amorphous, but differ largely in their Tg and chemical structures. The lower Tg of PET (73°C) compared to AkestraTM film (102°C) could be an important factor that caused the significantly higher rate of PET degradation. The possible impacts of other structural factors, such as the presence of spiroacetal units and molecular weight, remained to be unraveled.
Except for AkestraTM films, the experimental data was fitted according to first order kinetics with the coexistence of a sufficient substrate (20 mg of polymer in 2 ml), to the next equation (Miyawaki et al., 2017) to find the apparent inactivation rate constant
Where
From the kinetic constants and the saturation curves, it is possible to conclude that Humicola insolens cutinase has higher initial activity (
Finally, it should be noted that commercial polyesters like PET commonly contain a few percent of cyclic oligomers. In addition, enzymatic degradation (and hydrolysis) of polyesters may also lead to the formation of different oligomers or even micro- or nano-sized particles before they finally turned into TPA. The exact degradation process might be complicated and diverse, depending on the conditions and nature of polymers. This paper only aims to gain a preliminary insight into the production rate of the final TPA product, in correlation with the polymer and enzyme structures. Although we have not observed any oligomers in our experimental results, it could not be ruled out that oligomers (either those originally present in the materials or those intermediate degradation products) were not detected due to their low aqueous solubility. In the future, deeper insight into the degradation mechanism including the possible impacts of oligomers might be obtained by extraction of the reaction medium using organic solvents (Tsochatzis et al., 2020; Diamantidou et al., 2022). In addition, the possible formation of micro-or nano-sized particles during the degradation would also be an interesting subject to study.
Scanning electron microscopy analysis
AkestraTM and amorphous PET films treated with HiCut for 192 h were analyzed by SEM (Figure 6). Untreated films (control) presented a smooth surface in both polymers, while the enzyme treated PET showed a granulated and irregular surface, and the enzyme treated AkestraTM films showed big cracks. These results suggest that the enzyme acts mainly on the surface of the films and confirms a surface erosion process (Mueller, 2006).
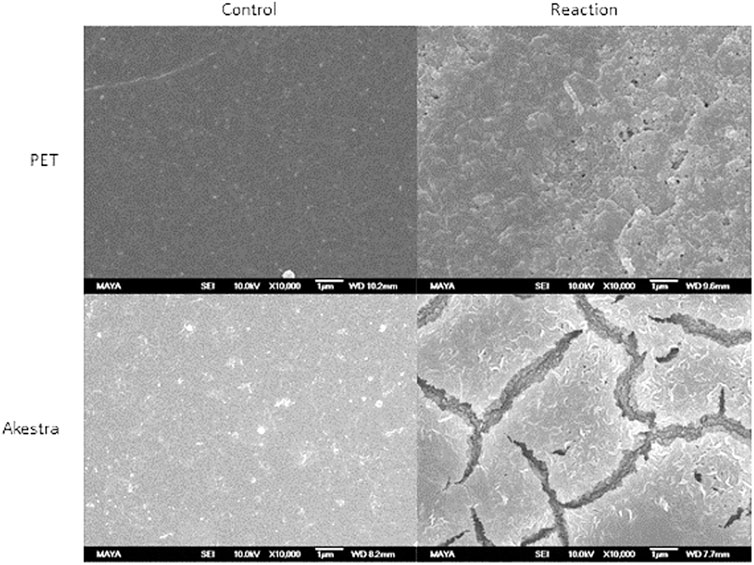
FIGURE 6. SEM images of PET and AkestraTM films after 192 h of reaction with HiCut (X10000). Controls on the left and enzymatically treated samples on the right.
FTIR analysis
FTIR spectroscopy was used to characterize the polymer films or powders after the enzymatic treatment. All the spectra (Supplementary information, Supplementary Figure S5) were normalized by using the small peak at 1410 cm−1 (benzene ring vibrations) as the reference (Drobota et al., 2013). In general, all the characteristic peaks for PET (e.g., 1714, 1240, 1093, 1016, 870 and 720 cm−1) and AkestraTM (e.g., 1201 and 1163 cm−1) remained visible after the HiCut treatment (Figure 7A–E). For the powders of PET, AkestraTM 90 and 110, and AkestraTM110 film, reduced intensity of the FTIR peaks at 720, 1093 and 1240 cm−1 (Figure 7) was observed after enzymatic treatment, compared to the negative control samples, which was consistent with the occurrence of surface degradation (Ioakeimidis et al., 2016). For PET film (Figure 7D), no significant difference was observed in the FTIR spectra after the enzymatic treatment compared to the control sample, indicating its relatively slow degradation rate.
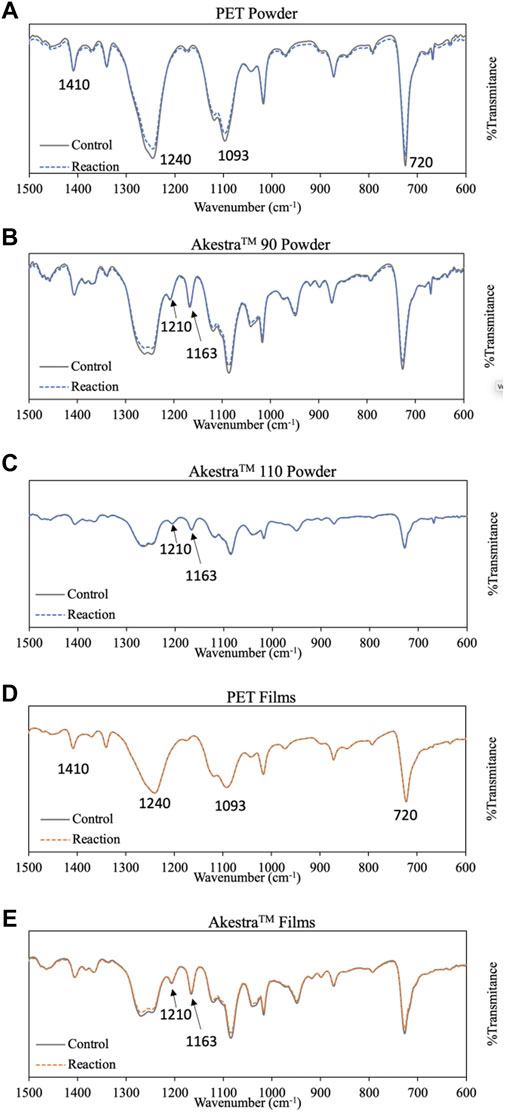
FIGURE 7. Zoomed-in FTIR spectra of polyester samples after HiCut treatment compared to the negative control samples. (A) PET Powder, (B) AkestraTM 90 Powder, (C) AkestraTM 110 powder, (D) PET film and (E) AkestraTM film. The data was normalized at the peak at 1410 cm−1 as a reference (Drobota et al., 2013).
Molecular docking and molecular dynamics
Interactions between HiCut and the hydrolysable (MHET)2 and non-hydrolysable TPA-spiroglycol ligands were studied by molecular docking. Compared to the molecular structure of Fusarium solani cutinase (56% sequence identity) (Martinez et al., 1992), HiCut shares a well-conserved catalytic triad, Ser105, His173, and Asp160, and the oxyanion hole, that stabilizes the negative charge on the carbonyl oxygen in the transition state.
Initially, the docking showed that both (MHET)2 and TPA-spiroglycol interact with HiCut with the carbonyl group oriented to the catalytic amino acids and oxyanion hole (Figures 8A,B). The ligands are surrounded by hydrophobic amino acids, Leu66, Phe70, Leu167, and Ile169, mainly in the subsite corresponding to the terminal terephthalate moiety. The presence of hydrophobic amino acids in the catalytic site of other PET active enzymes (Roth et al., 2014; Sulaiman et al., 2014; Kitadokoro et al., 2019) has also highlighted important features of the interaction with polyesters. The subsites corresponding to the ethylene glycol and second TPA in the case of (MHET)2 (Figure 8A), and spiroglycol for TPA-spiroglycol (Figure 8B), are surrounded by Leu174, Tyr104, and polar amino acids Thr29, Glu30, and Thr37. The interaction with the ligand (MHET)2 seems favored for the two T-stacking interactions, Phe70 and Tyr104, with both aromatic TPA rings. While the ligand TPA-spiroglycol showed only one with Phe70. In addition, the molecular dynamic simulations showed conformational changes that could not favor the HiCut/TPA-spiroglycol interactions.
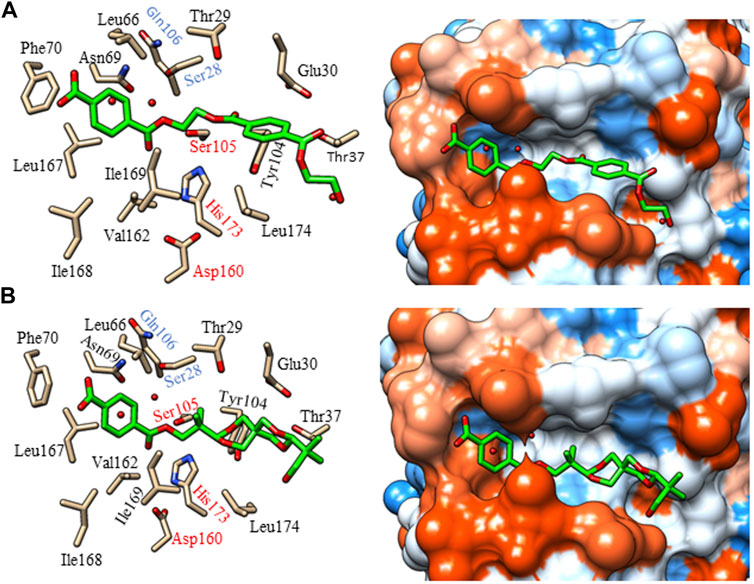
FIGURE 8. Molecular docking of HiCut with hydrolyzable and non-hydrolyzable ligands (in green). (A) Complexes HiCut/(MHET)2 and (B) HiCut/TPA-spiroglycol. On the right, amino acids surround the ligands. Catalytic amino acids are labeled in red, and the suggested amino acids for the oxyanion hole are in blue. On the left, the representation of the hydrophobicity surfaces is according to the Kyte-Doolittle scale, which is from dodger blue for the most hydrophilic to white and orange-red for the most hydrophobic (Kyte and Doolittle, 1982).
Both complexes, HiCut/(MHET)2 and HiCut/ TPA-spiroglycol, and HiCut free of ligand were subjected to molecular dynamics simulations. The simulated conditions included 70°C, pH 7.4, and 0.9% NaCl ions for 50 ns. The analysis of the trajectories showed an important variation of the root-mean-square (RMSD) of alpha carbons positions, up to 1.6Å in the free enzyme (Figure 9A). However, the complexes HiCut/(MHET)2 and HiCut/TPA-spiroglycol, showed significantly lower RMSD values, with averages of 0.8 and 0.9Å, respectively (Figure 9A). These results indicate that both ligands stabilize conformational changes in the enzyme. Indeed, the root-mean-square fluctuations (RMSF) analysis of alpha carbons revealed that the loop α8- α9 located in the active site suffers conformational changes (Figure 9B), mainly in Asp160, His173, Ile169, and Leu174. Both ligands stabilized this region in form of an alpha helix (Figures 9B,C). It is also remarkable the conformation changes in the ligand TPA-spiroglycol, in which the carbonyl oxygen turns towards the side opposite to the oxyanion hole and catalytic triad (Figure 9B), indicating an unfavorable conformation for the reaction. Distance fluctuations between the catalytic Ser105 and carbonyl carbon of both ligands (Figure 8B) indicate the esterase activity occurs in the ester bond of the second unit of (MHET)2, which corresponds to the spiroglycol moiety in the TPA-spiroglycol ligand, explaining its impossibility for hydrolysis. These results are consistent with the experimental evidence discussed above regarding the LC-MS analysis.
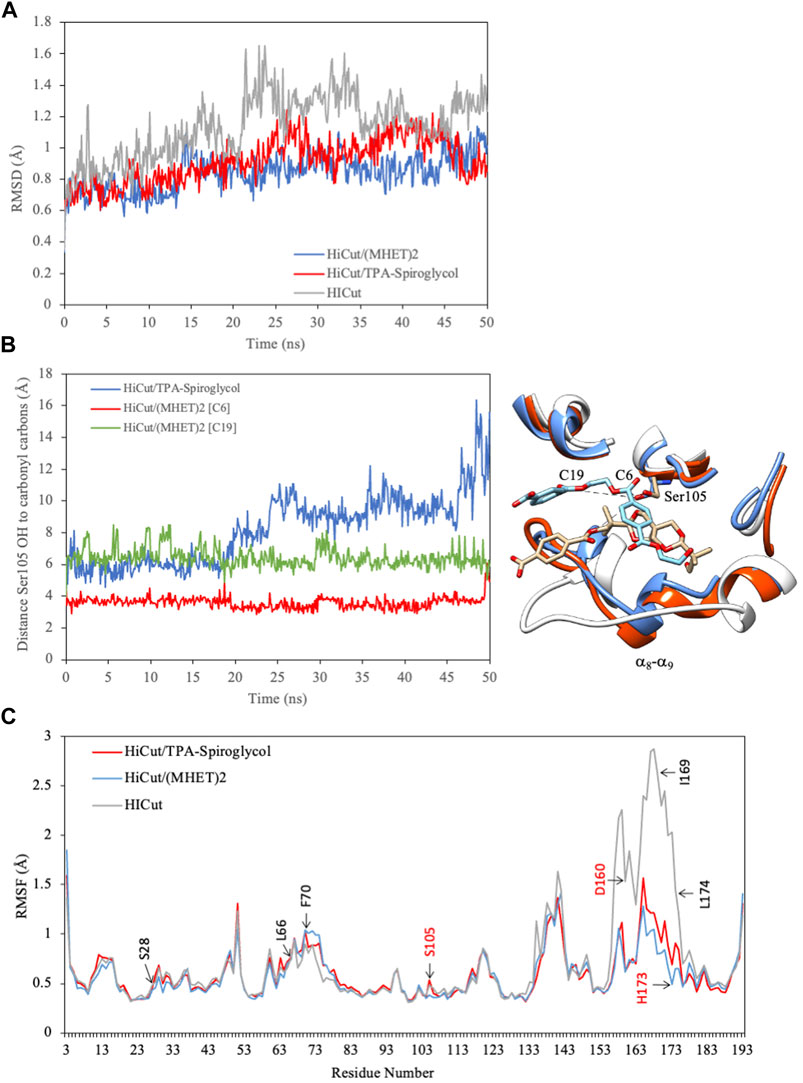
FIGURE 9. Molecular dynamics simulation of free HiCut and in complexes with ligands. (A) RMSD of alfa carbons. In grey free enzyme, with (MHET)2 in blue and with TPA-spiroglycol ester in red. (B) Distances fluctuation between the oxygen of the Ser105 (nucleophile) to the carbonyl carbons of the ester bonds in TPA-spiroglycol (blue) and (MHET)2 (green for C19 and red for C6). In the right the conformational changes in the active site loop (α8- α9) and ligands. (MHET)2 is with carbon atoms in blue and oxygens in red, while TPA-spiroglycol is with carbon atoms in brown and oxygens in red. (C) RMSF of alfa carbons labeling the amino acids surrounding the active site.
Conclusion
Humicola insolens cutinase (HiCut) has so far been the only enzyme capable of efficient degradation of the novel polymer AkestraTM, and its performance, as reported for other polyester-degrading enzymes, is highly dependent on Tg and crystallinity of the polymer. This characteristic is the major drawback to enzyme depolymerization, and since many polyesters (e.g., PET) used in consumer products have high crystalline percentages, exploring suitable pretreatment methods could improve the enzymatic action. The docking and molecular dynamics simulations, together with the experimental data, have shown the capability of HiCut to depolymerize AkestraTM yielding terephthalic acid and an ester of terephthalic acid and spiroglycol. Both building blocks are of high value for re-polymerization for the same or different polyesters or synthesis of other products. Finally, these results could set the basis for future process development and optimization research.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
JL-P and BZ designed the study. LA-L performed most of the experiments and wrote the draft of the manuscript. SM and CT performed experiments. All authors analyzed the data, participated in the revision of the manuscript, and approved the article. JL-P and BZ acquired the research funds. JL-P administrated the project.
Funding
This research was financed by the Crafoord Foundation (grant number 2020-0774), The Royal Physiographic Society in Lund, Swedish Research Council for Sustainable Development (Formas, No. 2021-01107) and Mistra Foundation (the “STEPS” project, No. 2016/1489).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fceng.2022.1048744/full#supplementary-material
References
Arza, C. R., Wang, P., Linares-Pastén, J., and Zhang, B. (2019). Synthesis, thermal, rheological characteristics, and enzymatic degradation of aliphatic polyesters with lignin-based aromatic pendant groups. J. Polym. Sci. Part A Polym. Chem. 57, 2314–2323. doi:10.1002/pola.29534
Baker, P. J., Poultney, C., Liu, Z., Gross, R., and Montclare, J. K. (2012). Identification and comparison of cutinases for synthetic polyester degradation. Appl. Microbiol. Biotechnol. 93, 229–240. doi:10.1007/s00253-011-3402-4
Carniel, A., De Abreu Waldow, V., and De Castro, A. M. (2021). A comprehensive and critical review on key elements to implement enzymatic PET depolymerization for recycling purposes. Biotechnol. Adv. 52, 107811. doi:10.1016/j.biotechadv.2021.107811
Case, D. A., Babin, V., Berryman, J., Betz, R., Cai, Q., Cerutti, D., et al. (2014). Amber 14. San Francisco: University of California.
De Queiros Eugenio, E., Pereira Campisano, I. S., Machado De Castro, A., Zarur Coelho, M. A., and Pereira Langone, M. A. (2021). Experimental and mathematical modeling approaches for biocatalytic post-consumer poly (ethylene terephthalate) hydrolysis. J. Biotechnol. 341, 76–85. doi:10.1016/j.jbiotec.2021.09.007
Di Bisceglie, F., Quartinello, F., Vielnascher, R., Guebitz, G. M., and Pellis, A. (2022). Cutinase-Catalyzed polyester-polyurethane degradation: Elucidation of the hydrolysis mechanism. Polymers 14, 411. doi:10.3390/polym14030411
Diamantidou, D., Mastrogianni, O., Tsochatzis, E., Theodoridis, G., Raikos, N., Gika, H., et al. (2022). Liquid chromatography-mass spectrometry method for the determination of polyethylene terephthalate and polybutylene terephthalate cyclic oligomers in blood samples. Anal. Bioanal. Chem. 414, 1503–1512. doi:10.1007/s00216-021-03741-6
Fabbri, F., Bertolini, F. A., Guebitz, G. M., and Pellis, A. (2021). Biocatalyzed synthesis of flavor esters and polyesters: A design of experiments (DoE) approach. Int. J. Mol. Sci. 22, 8493. doi:10.3390/ijms22168493
Genheden, S., and Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. drug Discov. 10, 449–461. doi:10.1517/17460441.2015.1032936
Gigli, M., Quartinello, F., Soccio, M., Pellis, A., Lotti, N., Guebitz, G. M., et al. (2019). Enzymatic hydrolysis of poly (1, 4-butylene 2, 5-thiophenedicarboxylate)(PBTF) and poly (1, 4-butylene 2, 5-furandicarboxylate)(PBF) films: A comparison of mechanisms. Environ. Int. 130, 104852. doi:10.1016/j.envint.2019.05.046
Hatti-Kaul, R., Nilsson, L. J., Zhang, B., Rehnberg, N., and Lundmark, S. (2020). Designing biobased recyclable polymers for plastics. Trends Biotechnol. 38, 50–67. doi:10.1016/j.tibtech.2019.04.011
Hu, H., Tian, Y., Kong, Z., Ying, W. B., Chen, C., Li, F., et al. (2021). A high performance copolyester with “locked” biodegradability: Solid stability and controlled degradation enabled by acid-labile acetal. ACS Sustain. Chem. Eng. 9, 2280–2290. doi:10.1021/acssuschemeng.0c08274
Ioakeimidis, C., Fotopoulou, K., Karapanagioti, H., Geraga, M., Zeri, C., Papathanassiou, E., et al. (2016). The degradation potential of PET bottles in the marine environment: An ATR-FTIR based approach. Sci. Rep. 6, 23501–23508. doi:10.1038/srep23501
Japu, C., Alla, A., De Ilarduya, A. M., García-Martín, M. G., Benito, E., Galbis, J. A., et al. (2012). Bio-based aromatic copolyesters made from 1, 6-hexanediol and bicyclic diacetalized D-glucitol. Polym. Chem. 3, 2092–2101. doi:10.1039/c2py20145c
Kaabel, S., Therien, J. D., Deschênes, C. E., Duncan, D., Friščić, T., and Auclair, K. (2021). Enzymatic depolymerization of highly crystalline polyethylene terephthalate enabled in moist-solid reaction mixtures. Proc. Natl. Acad. Sci. U. S. A. 118, e2026452118. doi:10.1073/pnas.2026452118
Kitadokoro, K., Kakara, M., Matsui, S., Osokoshi, R., Thumarat, U., Kawai, F., et al. (2019). Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase. FEBS J. 286, 2087–2098. doi:10.1111/febs.14781
Kold, D., Dauter, Z., Laustsen, A. K., Brzozowski, A. M., Turkenburg, J. P., Nielsen, A. D., et al. (2014). Thermodynamic and structural investigation of the specific SDS binding of Humicola insolens cutinase. Protein Sci. 23, 1023–1035. doi:10.1002/pro.2489
Krieger, E., Koraimann, G., and Vriend, G. (2002). Increasing the precision of comparative models with YASARA nova—A self-parameterizing force field. Proteins. 47, 393–402. doi:10.1002/prot.10104
Krieger, E., and Vriend, G. (2015). New ways to boost molecular dynamics simulations. J. Comput. Chem. 36, 996–1007. doi:10.1002/jcc.23899
Kupczak, M., Mielańczyk, A., and Neugebauer, D. (2021). The influence of polymer composition on the hydrolytic and enzymatic degradation of polyesters and their block copolymers with PDMAEMA. Materials 14, 3636. doi:10.3390/ma14133636
Kyte, J., and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. doi:10.1016/0022-2836(82)90515-0
Lavilla, C., De Ilarduya, A. M., Alla, A., García-MartíN, M. D. G., Galbis, J., and MuñOz-Guerra, S. (2012). Bio-based aromatic polyesters from a novel bicyclic diol derived from D-mannitol. Macromolecules 45, 8257–8266. doi:10.1021/ma3013288
Li, X., Ilk, S., Linares-Pastén, J. A., Liu, Y., Raina, D. B., Demircan, D., et al. (2021). Synthesis, enzymatic degradation, and polymer-miscibility evaluation of nonionic antimicrobial hyperbranched polyesters with indole or isatin functionalities. Biomacromolecules 22, 2256–2271. doi:10.1021/acs.biomac.1c00343
Machado De Castro, A., Carniel, A., Nicomedes Junior, J., Da Conceição Gomes, A., and Valoni, É. (2017). Screening of commercial enzymes for poly (ethylene terephthalate)(PET) hydrolysis and synergy studies on different substrate sources. J. Industrial Microbiol. Biotechnol. 44, 835–844. doi:10.1007/s10295-017-1942-z
Machado De Castro, A., Carniel, A., Sirelli, L., Lopes Dias, M., Cabral De Menezes, S. M., Chinelatto Junior, L. S., et al. (2018). Enzyme-catalyzed simultaneous hydrolysis-glycolysis reactions reveals tunability on PET depolymerization products. Biochem. Eng. J. 137, 239–246. doi:10.1016/j.bej.2018.06.007
Machado De Castro, A., Carniel, A., Stahelin, D., Chinelatto Junior, L. S., De Angeli Honorato, H., and Cabral De Menezes, S. M. (2019). High-fold improvement of assorted post-consumer poly (ethylene terephthalate)(PET) packages hydrolysis using Humicola insolens cutinase as a single biocatalyst. Process Biochem. 81, 85–91. doi:10.1016/j.procbio.2019.03.006
Martinez, C., De Geus, P., Lauwereys, M., Matthyssens, G., and Cambillau, C. (1992). Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature 356, 615–618. doi:10.1038/356615a0
Miyawaki, O., Kanazawa, T., Maruyama, C., and Dozen, M. (2017). Static and dynamic half-life and lifetime molecular turnover of enzymes. J. Biosci. Bioeng. 123, 28–32. doi:10.1016/j.jbiosc.2016.07.016
Mueller, R.-J. (2006). Biological degradation of synthetic polyesters—enzymes as potential catalysts for polyester recycling. Process Biochem. 41, 2124–2128. doi:10.1016/j.procbio.2006.05.018
OECD (2022). Global Plastics Outlook: Policy Scenarios to 2060. Paris: OECD Publishing. doi:10.1787/aa1edf33-en
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi:10.1002/jcc.20084
Qi, X., Yan, W., Cao, Z., Ding, M., and Yuan, Y. (2021). Current advances in the biodegradation and bioconversion of polyethylene terephthalate. Microorganisms 10, 39. doi:10.3390/microorganisms10010039
Ronkvist, Å. M., Xie, W., Lu, W., and Gross, R. A. (2009). Cutinase-catalyzed hydrolysis of poly (ethylene terephthalate). Macromolecules 42, 5128–5138. doi:10.1021/ma9005318
Roth, C., Wei, R., Oeser, T., Then, J., Föllner, C., Zimmermann, W., et al. (2014). Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 98, 7815–7823. doi:10.1007/s00253-014-5672-0
Roy, P. K., Ramanan, A., and Rajagopal, C. (2013). Post consumer PET waste as potential feedstock for metal organic frameworks. Mater. Lett. 106, 390–392. doi:10.1016/j.matlet.2013.05.058
Sulaiman, S., You, D.-J., Kanaya, E., Koga, Y., and Kanaya, S. (2014). Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase. Biochemistry 53, 1858–1869. doi:10.1021/bi401561p
Trott, O., and Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Tsochatzis, E. D., Lopes, J. A., Kappenstein, O., Tietz, T., and Hoekstra, E. J. (2020). Quantification of PET cyclic and linear oligomers in teabags by a validated LC-MS method–in silico toxicity assessment and consumer’s exposure. Food Chem. 317, 126427. doi:10.1016/j.foodchem.2020.126427
Wagner-Egea, P., Aristizábal-Lanza, L., Tullberg, C., Wang, P., Bernfur, K., Grey, C., et al. (2022). Marine PET hydrolase (PET2): Assesment of terephthalate- and -indole-based polyesters depolymerization. Submited for publication.
Wagner-Egea, P., Tosi, V., Wang, P., Grey, C., Zhang, B., and Linares-Pastén, J. A. (2021). Assessment of is PETase-assisted depolymerization of terephthalate aromatic polyesters and the effect of the thioredoxin fusion domain. Appl. Sci. 11, 8315. doi:10.3390/app11188315
Wang, P., Linares-Pastén, J. A., and Zhang, B. (2020). Synthesis, molecular docking simulation, and enzymatic degradation of AB-type indole-based polyesters with improved thermal properties. Biomacromolecules 21, 1078–1090. doi:10.1021/acs.biomac.9b01399
Webb, H. K., Arnott, J., Crawford, R. J., and Ivanova, E. P. (2012). Plastic degradation and its environmental implications with special reference to poly (ethylene terephthalate). Polymers 5, 1–18. doi:10.3390/polym5010001
Wei, R., Breite, D., Song, C., Gräsing, D., Ploss, T., Hille, P., et al. (2019). Biocatalytic degradation efficiency of postconsumer polyethylene terephthalate packaging determined by their polymer microstructures. Adv. Sci. 6, 1900491. doi:10.1002/advs.201900491
Wei, R., and Zimmermann, W. (2017). Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb. Biotechnol. 10, 1302–1307. doi:10.1111/1751-7915.12714
Weinberger, S., Canadell, J., Quartinello, F., Yeniad, B., Arias, A., Pellis, A., et al. (2017). Enzymatic degradation of poly (ethylene 2, 5-furanoate) powders and amorphous films. Catalysts 7, 318. doi:10.3390/catal7110318
Wierckx, N., Prieto, M. A., Pomposiello, P., De Lorenzo, V., O'connor, K., and Blank, L. M. (2015). Plastic waste as a novel substrate for industrial biotechnology. Microb. Biotechnol. 8, 900–903. doi:10.1111/1751-7915.12312
Keywords: plastic degradation, biocatalysts, PET–poly(terephtalate ethylene), Akestra, cutinases, Humicola insolens cutinase (HiC)
Citation: Aristizábal-Lanza L, Mankar SV, Tullberg C, Zhang B and Linares-Pastén JA (2022) Comparison of the enzymatic depolymerization of polyethylene terephthalate and AkestraTM using Humicola insolens cutinase. Front. Chem. Eng. 4:1048744. doi: 10.3389/fceng.2022.1048744
Received: 19 September 2022; Accepted: 24 November 2022;
Published: 13 December 2022.
Edited by:
Antoni Sánchez, Universitat Autònoma de Barcelona, SpainCopyright © 2022 Aristizábal-Lanza, Mankar, Tullberg, Zhang and Linares-Pastén. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Javier A. Linares-Pastén, javier.linares_pasten@biotek.lu.se
 Lucía Aristizábal-Lanza
Lucía Aristizábal-Lanza Smita V. Mankar
Smita V. Mankar Cecilia Tullberg
Cecilia Tullberg Baozhong Zhang
Baozhong Zhang Javier A. Linares-Pastén
Javier A. Linares-Pastén