Identification and mechanism prediction of mulberroside A metabolites in vivo and in vitro of rats using an integrated strategy of UHPLC-Q-Exactive Plus Orbitrap MS and network pharmacology
- 1Shandong University of Traditional Chinese Medicine, Jinan, China
- 2Shandong Academy of Chinese Medicine, Jinan, China
- 3State Key Laboratory for Quality Research of Chinese Medicines, Macau University of Science and Technology, Taipa, Macao SAR, China
Mulberroside A is a polyhydroxylated stilbene active component of Morus alba L. Studies have shown that it has antitussive, antiasthmatic, tyrosinase and antioxidation activities. However, little is known about the metabolism of it in vitro and in vivo. In our study, an integrated strategy on the basis of UHPLC-Q-Exactive Plus Orbitrap MS and network pharmacology was established to comprehensively research the metabolic characteristic of mulberroside A for the first time. Plasma, urine, feces and liver tissues of rats in the blank group and drug group were collected after intragastric administration of mulberroside A at a dose of 150 mg/kg, and rat liver microsomes were cultured for in vitro metabolism experiment. The biological samples were processed by different methods and analyzed in positive and negative ion modes using UHPLC-Q-Exactive Plus Orbitrap MS. A total of 72 metabolites were finally identified based on the accurate molecular mass, retention time, MS/MS spectra and related literatures combined with the Compound Discoverer 3.1. The metabolic pathways were mainly hydrolysis, glucuronidation, hydrogenation, sulfation, hydroxylation, methylation and their composite reactions. In addition, a network pharmacology method was used to predict the mechanism of action of mulberroside A and its metabolites. In the end, 7 metabolites with high gastrointestinal absorption and drug-likeness and 167 targets were screened by Swiss ADME and Swiss Target Prediction. 1702 items of GO analysis and 158 related signaling pathways of KEGG were enriched using Metascape. This study established a novel integrated strategy based on UHPLC-Q-Exactive Plus Orbitrap MS and network pharmacology, which could systematically analyze the metabolism behavior of mulberroside A in vivo and in vitro of rats and provide basis for the further research of mulberroside A.
1 Introduction
Mulberroside A, presenting at relatively high abundance in the roots and twigs of Morus alba L. (Zhou et al., 2013), is a natural polyhydroxylated stilbene compound (Yu et al., 2021). Furthermore, it possesses various therapeutic effects through pharmacological studies, including hypoglycemic (Li et al., 2014), antidiabetic (Xu et al., 2021), anti-inflammatory (Chung et al., 2003), analgesic (Zhang and Shi, 2010), hepatic protective function (Zhang et al., 2008), nephroprotective (Wang et al., 2011), the inhibitory effect of melanogenesis (Park et al., 2011) and neuroprotective (Wang C. P. et al., 2014) effects, etc.
Drug metabolism, also known as drug biotransformation, refers to the process of chemical structure change of drugs under the action of drug enzymes (Jiang et al., 2014). Once a drug enters the body, it is usually converted into various forms through different metabolic pathways (Prasad et al., 2011). Drug metabolism research can elucidate the scientific connotation of drugs, clarify the pharmacodynamic active ingredients, guide the rational use of clinical drugs, and promote the improvement of dosage forms and quality control, which has great significance in the drug discovery process. However, as far as we know, the in vivo and in vitro metabolism study of mulberroside A has been poorly understood. A pharmacokinetic study on mulberroside A revealed that its oral absolute bioavailability was poor in rats (Qiu et al., 1996b). In Qiu F’s previous metabolic research on mulberroside A, four metabolites were isolated from the urine and bile of rats after oral administration of mulberroside A (Qiu et al., 1996a). After that, they reported the identification of 3 metabolites of mulberroside A in the gastrointestinal contents and feces of rats (Zhaxi et al., 2010). However, whether mulberroside A is metabolized prior to be absorbed into the blood has not been reported so far. Therefore, it is important to carry out a comprehensive metabolism study of mulberroside A. Then, we could better understand its metabolic mechanism and promote research for the further potential therapeutic applications.
Contemporarily, ultra-high performance liquid chromatography (UHPLC) coupled with mass spectrometry (MS), especially high-resolution mass spectrometry (HRMS), has been widely used in drug metabolism analysis (Dong et al., 2021; Yuan et al., 2021). UHPLC has a powerful separation capability, which is currently one of the most effective tools for the separation of complex components. In recent years, network pharmacology has been widely used in the field of traditional Chinese medicine with its unique advantages. The organic combination of metabolite identification and network pharmacology could effectively reveal the mechanism of drug action in the body. In our study, an integrated strategy on the basis of UHPLC-Q-Exactive Plus Orbitrap MS and network pharmacology was developed. This strategy was cost-effective to characterize chemical transformations and further gain comprehensive knowledge about mulberroside A.
2 Materials and methods
2.1 Chemicals and reagents
The reference standard, mulberroside A, was purchased from Chengdu Must Biotechnology Co., Ltd. (Sichuan, China), with purity greater than 98% determined using HPLC-UV analysis. Rat liver microsomes were purchased from Wuxi Xinrun Biotechnology Co., Ltd. (Jiangsu, China). HPLC grade methanol, formic acid, and acetonitrile were provided by Thermo Fisher Scientific (Fair Lawn, NJ, United States). Water was obtained from Watsons (Guangdong, China). Grace Pure SPE C18-Low solid-phase extraction cartridges (200 mg/3 ml, 59 μm, 70 Å) were purchased from Grace Davison Discovery Science (Deerfield, IL, United States).
2.2 Animals and drug administration
Six male SD rats (200 ± 10 g) were provided by Jinan Pengyue Experimental Animal Breeding Co., Ltd. (Shandong, China). The animals were housed in a controlled environment with the temperature held at 24 ± 2 °C, relative humidity held at 60±5%, and kept on a 12 h light/12 h dark regime. After 7 days of acclimation, the rats were randomly divided into drug group (n=3) and control group (n=3). Before the experiment, all rats were fasted for 12 h with free access to water. Mulberroside A was dissolved in normal saline. The rats in the drug group were given with a dose of 150 mg/kg/d orally for 3 days. A same dose of normal saline was administered to the rats in the control group.
2.3 Sample collection
2.3.1 Plasma and liver tissue sample collection
0.5 ml of blood samples were taken from the suborbital venous plexus of rats at 0.5, 1, 1.5, 2, 4, 6, 12, and 24 h after the last administration. The blood samples collected at different times were mixed and placed in a centrifuge tube coated with sodium heparin. After that, they were centrifuged for 10 min at 3,500 rpm to separate plasma sample. Then, it was frozen in a refrigerator at -80°C. The centrifugation was performed on Centrifuge 5430R (Eppendorf, Germany).
24 h after the last administration, liver tissue was obtained and quenched in liquid nitrogen and then stored at -80°C. 1 g of liver tissue was added to 10 ml of normal saline to homogenize at 4°C and then centrifuged at 3,500 rpm for 10 min to obtain the supernatant.
2.3.2 Urine and feces sample collection
Two groups of rats were housed in the metabolic cage. The urine and feces were collected within 24 h after the last administration. The urine samples were frozen in the refrigerator at -80°C after centrifuged at 12,000 rpm for 15 min, and the feces samples were freeze-dried and ground into powder.
2.3.3 Liver microsomes incubation
MgCl2 and liver microsomes were dissolved in PBS (pH=7.4), formulated into a solution with a MgCl2 concentration of 3 mM and a protein concentration of 1 mg/ml. The solution was used to dissolve mulberroside A with a final drug concentration of 0.1 mg/ml. Drug group and blank group were set up. 900 µl of the above mixture was added to each well of the 6-well plate in the drug group while the blank group was given an equal volume of drug-negative solution. After preheating at 37°C for 5 min, 100 µl NADPH (final concentration of 25 mg/ml) was added to start the reaction and continue to incubate at 37°C. At 5, 10, 15, 30, 45, 60, 120, 240 min after the start of the reaction, 100 µl of the system solution was mixed with 200 µl of cold acetonitrile to terminate the reaction. Then, the supernatant was obtained after centrifuged at 20,000 rpm for 10 min.
2.4 Sample preparation
The SPE method for precipitation and concentration of protein and solid residues was applied to pretreat all the biological samples. Before loading and elution, 2 g feces was added to 10 ml pure water, sonicated for 0.5 h, and centrifuged to obtain the supernatant. Then, plasma, urine, feces, and liver tissues samples (1 ml) were added to SPE cartridges pretreated with methanol (3 ml) and deionized water (3 ml), respectively. After that, the SPE cartridges were successively washed with deionized water (3 ml) and methanol (3 ml). The methanol eluent was collected and dried under N2 at 4°C. The residue was then redissolved in 300 µl methanol and centrifuged for 15 min at 20,000 rpm (4°C).
In addition, plasma samples were treated with methanol (3 ml) and acetonitrile (3 ml), respectively. Then, they were centrifuged at 3,500 rpm. The obtained supernatant was dried under N2 at room temperature. The residue was then redissolved in 300 µl methanol and centrifuged for 15 min at 20,000 rpm (4°C).
All the biological samples from the same group were merged into a collective sample for the further instrumental analysis.
2.5 Instruments and analytical conditions
2.5.1 UHPLC parameters
The separation was performed on Vanquish UHPLC (Thermo Scientific, Germany) with a ACQUITY UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm). The flow rate was 0.3 ml/min and the injection volume was 5 µl. The column temperature was maintained at 35°C. The mobile phase consists of acetonitrile(A) and 0.1% formic acid-water (B). The gradient elution condition was set as: 0–5.0 min, 95% B; 5.0–10.0 min, 95%–70% B; 10.0–15.0 min, 70%–50% B; 15.0–32.0 min, 50%–10% B; 32.0–32.1 min, 10%–95% B; 32.1–35.0 min, 95% B.
2.5.2 HRMS parameters
The MS analysis was performed on Q-Exactive Plus Orbitrap MS (Thermo Scientific, Germany) with a heated electrospray ionization (HESI) source. A high-resolution mass spectrum was acquired at full scan in a mass range of m/z 80–m/z 1,200 at a resolution of 70,000 with AGC target at 3e6. The dd-MS2 data were obtained at a resolution of 17,500 with AGC target at 1e6. Mass spectrometric detection was performed in positive and negative ion modes. The ion source parameters were set as follows: sheath gas and auxiliary gas of nitrogen (purity ≥99.99%) with the flow rate of 45 arb and 10 arb; capillary temperature of 320 °C; spray voltage of 3,800/3,500 V (+/-); probe heater temperature of 320°C. The stepped normalized collision energy (NCE) was set at 15, 30 and 45. S-Lens RF Level was 50.00.
2.6 Data processing and statistical analyses
Thermo Scientific Xcalibur 3.1 workstation and Compound Discoverer 3.1 software were used to process and analyze the collected mass spectrometry data. The chemical formulas attributed to the selected peaks were calculated using a formula predictor by setting the parameters as follows: C [5–40], H [5–60], O [2–30], S [0–2], N [0–3] and the ring double bond (RDB) equivalent value [3–20]. The HRMS data obtained from UHPLC-Q-Exactive Plus Orbitrap MS was imported into the Compound Discoverer, and the structure information of mulberroside A was added to Compound Discoverer at the same time. Then, Compound Discoverer gave the information of possible metabolites according to the structural characteristics of mulberroside A, HRMS information and the corresponding databases. In addition, the metabolites were selected by Swiss ADME (http://www.swissadme.ch/) and the targets were predicted using Swiss Target Prediction (http://www.swisstargetprediction.ch/). All genes were submitted to Metascape (Zhou et al., 2019) (http://metascape.org) for GO and KEGG analysis and Cytoscape 3.8.2 was used for the construction of metabolite-target network and metabolite-target-pathway network. The visual analysis of GO and KEGG enrichment results was performed using Bioinformatics online platform (http://www.bioinformatics.com.cn/).
3 Results and discussion
3.1 The establishment of an analytical strategy
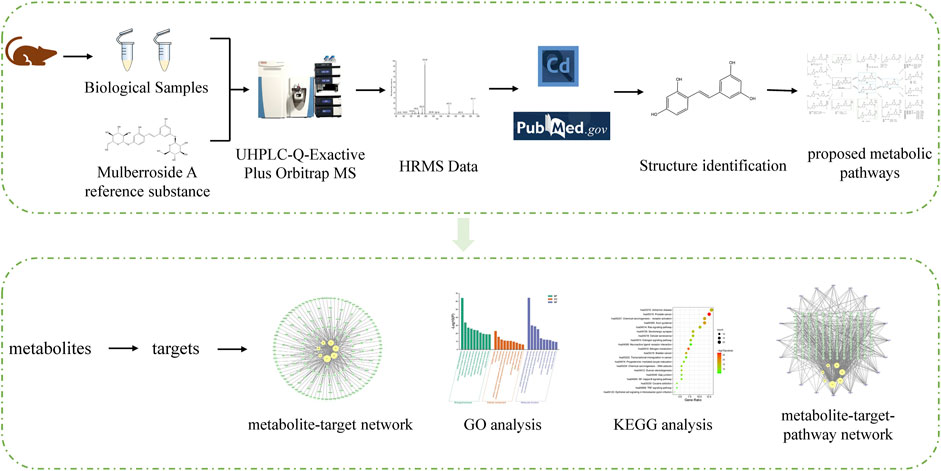
In order to understand the metabolism and mechanism of mulberroside A comprehensively, an efficient and integrated strategy was established based on the UHPLC-Q-Exactive Plus Orbitrap MS and network pharmacology, as shown in Figure 1. (1) The rats were given the mulberroside A solution by intragastric administration and biological samples were collected. Then the treated biological samples and standard mulberroside A samples were injected into UHPLC-Q-Exactive Plus Orbitrap MS to obtain HRMS data. (2) The information such as accurate molecular mass, retention time and MS/MS spectra of compounds was obtained by Thermo Scientific Xcalibur 3.1 and then the compounds were identified based on the information and related literatures. After that, the metabolic pathway of mulberroside A could be predicted on the basis of metabolites and metabolic reactions. (3) Swiss ADME and Swiss Target Prediction were applied to screen the metabolites with high gastrointestinal absorption and drug-likeness and predict the targets of these selected metabolites, which were used to map the metabolite-target network by Cytoscape 3.8.2. Then, Metascape was used for GO and KEGG analysis. In the end, the metabolite-target-pathway network was also constructed by Cytoscape 3.8.2.
3.2 Analysis of the mass spectrometry fragmentation behaviors of mulberroside A
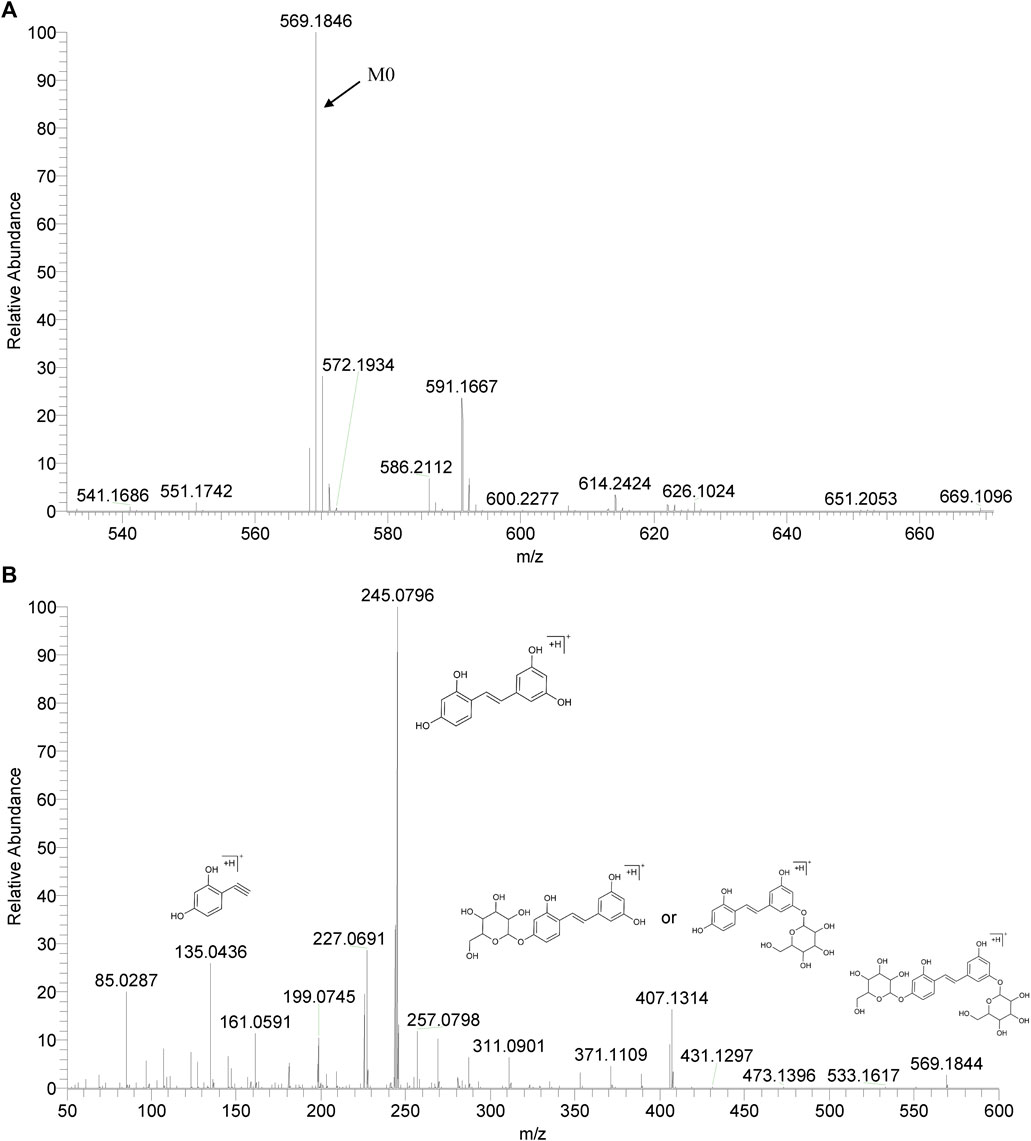
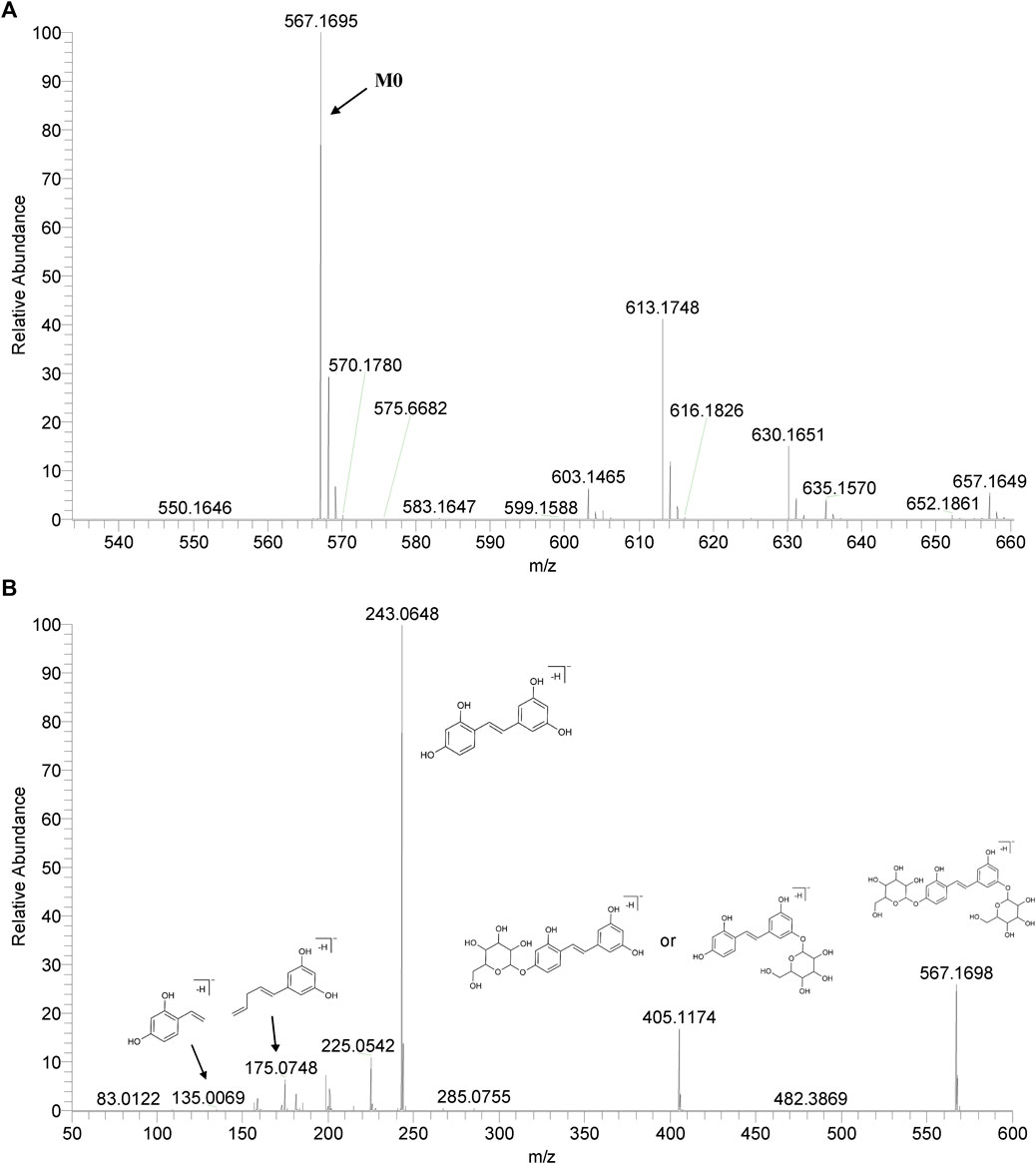
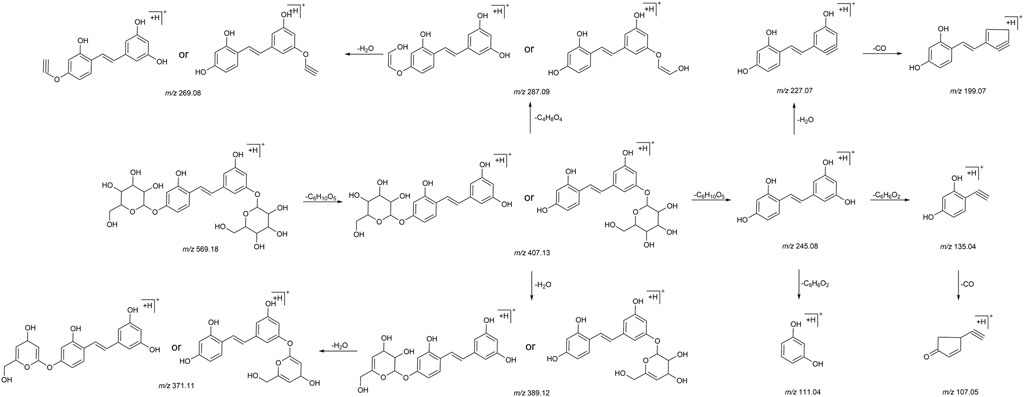
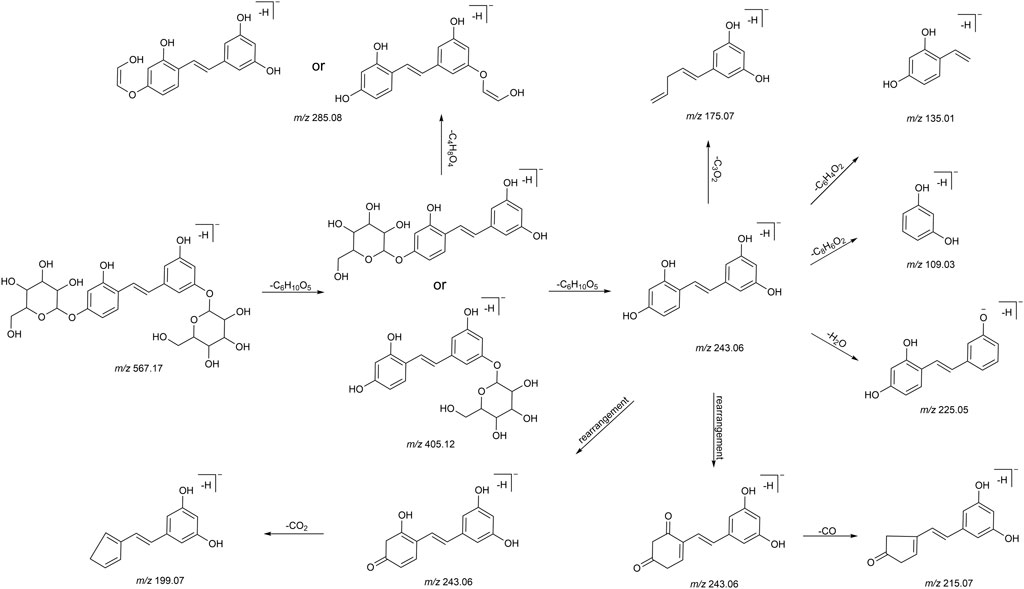
We summarized the mass spectrometry fragmentation behaviors of mulberroside A according to related literatures reported and the MS2 information of the reference substances (Sun, 2004; Pan et al., 2015; Xu, 2015; Guo et al., 2018; Zhang, 2018; Liang et al., 2021). As shown in Figure 2 and Figure 3, the [M+H]+ and [M-H]- was generated in positive and negative ion modes at m/z 569.1846 and m/z 567.1695, respectively, in the ESI-MS spectra of mulberroside A. Characteristic fragments were detected in positive ion mode: m/z 407 [M+H-glucose]+ ([M+H-Glu]+), m/z 245 [M+H-2Glu]+, m/z 227 [M+H-2Glu-H2O]+, m/z 199 [M+H-2Glu-H2O-CO]+, m/z 135 [M+H-2Glu-C6H6O2]+. Representative ions were detected in negative ion mode as follows: m/z 405 [M-H-Glu]-, m/z 243 [M-H-2Glu]-, m/z 225 [M-H-2Glu-H2O]-, m/z 215 [M-H-2Glu-CO]-, m/z 199 [M-H-2Glu-CO2]-, m/z 181 [M-H-2Glu-CO2-H2O]-, m/z 175 [M-H-2Glu-C3O2]−, m/z 157 [M-H-2Glu-C3O2-H2O]−. The mass fragmentation behavior of mulberroside A in positive and negative ion modes are shown in Figure 4 and Figure 5 respectively.
3.3 Detection and structural elucidation of mulberroside A metabolites
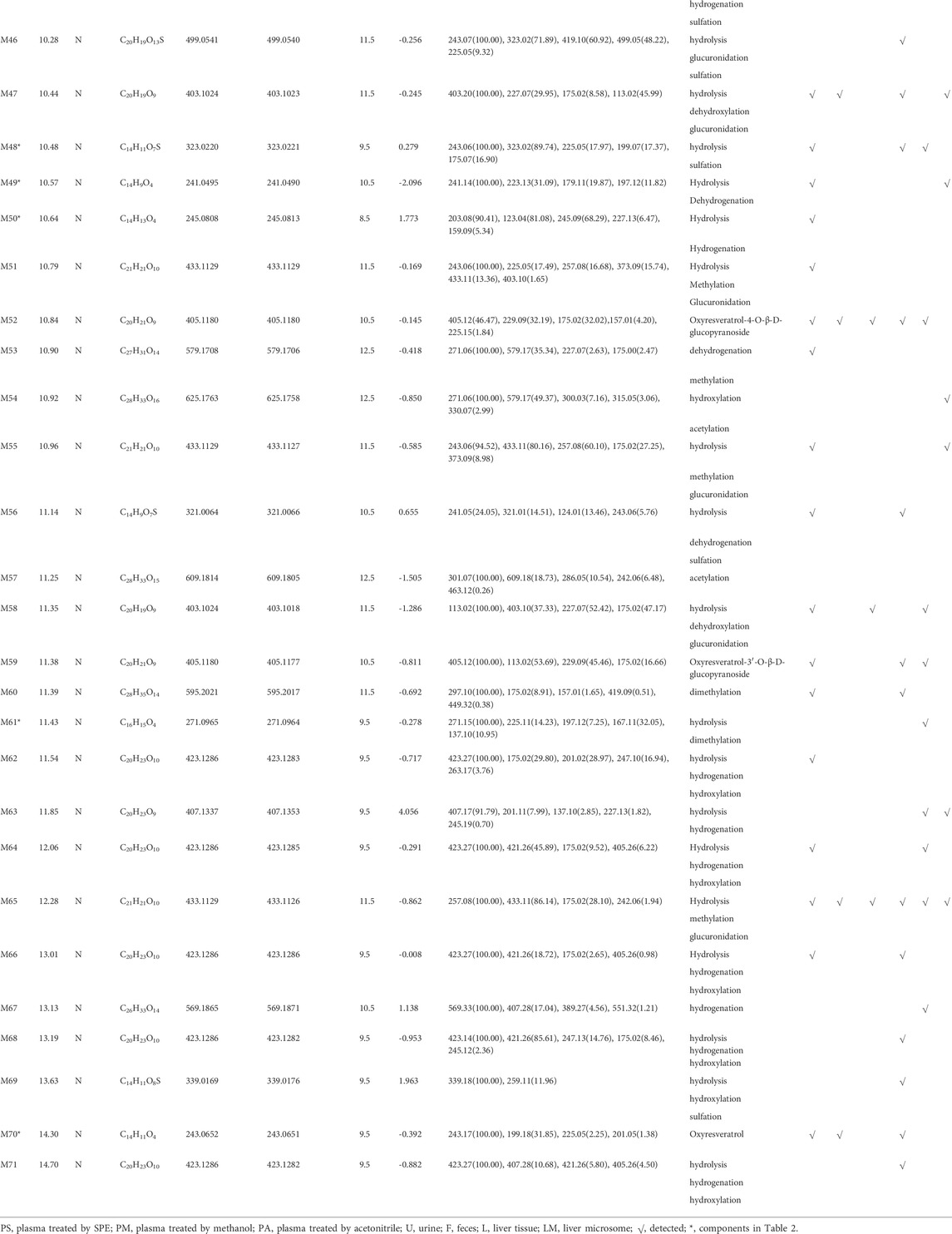
A total of 76 metabolites were finally identified based on the accurate molecular mass, retention time, MS/MS spectra and related literature reports in the plasma, urine, feces, liver tissues and liver microsomes. The chromatographic and MS data of these detected metabolites were summarized in Table 1.
3.3.1 Identification of mulberroside A metabolites
M0 was identified as the prototype of mulberroside A with the [M-H]- ions at m/z 567.1707 (C26H31O14, -0.197 ppm) in negative ion modes. The fragment ions at m/z 243, m/z 567, m/z 405, m/z 225 were detected, which were consistent with the mass fragmentation behavior of the reference substance of mulberroside A. Therefore, M0 was characterized as the prototype of mulberroside A. M27 possessed the same theoretical [M-H]- and product ions with M0, but compared with M0, it was eluted later than M0. So, M27 was deduced as isomer of mulberroside A.
M1 and M21 generated [M-H]- ions at m/z 285.0607 (C12H13O8, 0.618 ppm) and m/z 311.0767 (C14H15O8, 1.659 ppm) respectively. The neutral loss of 176 Da (m/z 285→m/z 109 and m/z 311→m/z 135) was generated in their ESI-MS/MS spectra, indicating that their structures contain glucuronic acid (Glu A). In addition, the product ion at m/z 175 ([Glu A-H]-) was detected, which also proved the existence of Glu A. According to pubchem database (https://pubchem.ncbi.nlm.nih.gov/) and molecular formulas, M1 was identified as β-D-Glucopyranosiduronic Acid-3-Hydroxyphenyl and M21 was characterized as isomer of 6-(4-Ethenyl-2-hydroxyphenoxy)-3,4,5-trihydroxyoxane-2-carboxylic acid. They were the glucuronidation metabolites of compounds formed by the rupture of C1-α bond or C1′-β bond in mulberroside A.
M2 and M6 possessed the same theoretical [M-H]- ions at m/z 583.1652 (C26H31O15, mass error within 2 ppm) with the retention time of 7.72 and 8.20 min, respectively. They were 16 Da more massive than M0, which suggested that they might be the hydroxylation products of mulberroside A. The fragment ion at m/z 259 ([M-H-2Glu]-) was 16 Da more massive than m/z 243, providing evidence for hydroxylation reaction. The product ions at m/z 421 ([M-H-Glu]-) was formed by losing Glu. Therefore, it was speculated that M2 and M6 were hydroxylation products of mulberroside A.
M7 was eluted at 8.20 min with the [M-H]- at m/z 599.1602 (C26H31O16, -0.736 ppm). It was 32 Da (2O) more massive than M0, indicating that it might be the dihydroxylation product of mulberroside A. The fragment ions involved m/z 137, m/z 599, m/z 125, m/z 437, and m/z 275. The [M-H]- ion lost Glu to form m/z 437 ([M-H-Glu]-), and lost another Glu to form m/z 275 ([M-H-2Glu]-). We first assumed that two benzene rings were each bound to a hydroxyl group. Then it was split by Chem Draw, and we found that the α-β bond of the product ion at m/z 275 ([M-H-2Glu]-) broke to form C7H5O32-, which molecular weight was exactly m/z 137. From these, it was speculated that M7 was dihydroxylation product of mulberroside A and two hydroxyl groups were bonded to the benzene rings on both sides.
M5 and M29, with the molecular formula of C27H33O16, showed the same theoretical [M-H]- ions at m/z 613.1763 and they were 46 Da (CH2O2) more massive than M0. The loss of 30 Da (m/z 435→m/z 405) suggested the existence of methoxy. In addition, the product ions at m/z 243, m/z 405, m/z 567 were representative ions of mulberroside A. As a consequence, it was deemed that M5 and M29 were dihydroxylation and methylation products of mulberroside A.
M9 eluted at 8.24 min and generated the [M-H]- at m/z 601.1769 (C26H33O16, 1.046 ppm). It was 34 Da (H2O2) more massive than M0, which indicated that it was hydrogenation and hydroxylation product of mulberroside A. The fragment ions at m/z 137, m/z 243, m/z 601, m/z 567, m/z 439 ([M-H-Glu]-) were observed in its ESI-MS/MS spectrum. The α-β bond of M9 was broken and then lost Glu to form m/z 137 (C7H5O3−), while m/z 243 and m/z 567 were characteristic ions of mulberroside A. Therefore, M9 was identified as vicinal diols formed by dihydroxylation at α-β bond of mulberroside A.
M54, with the retention time of 10.92 min, was 58 Da (C2H2O2) more massive than M0. The fragment ion at m/z 579 was fragmented by losing of CO and H2O from [M-H]- ion, while m/z 271 was identified as C12H15O7− formed by the fracture of the bonds C-1 and α. The neutral loss of 30 Da (m/z 330→m/z 300) indicated that methoxy was contained in the structure. Hence, M54 was finally characterized as hydroxylation and acetylation product of mulberroside A.
M57 was 42 Da (C2H2O) more massive than M0 with the molecular formula of C28H33O15. It was speculated that M57 was acetylation product of mulberroside A. In the ESI-MS/MS spectrum of M57, the fragment ion at m/z 463 removed Glu to form m/z 301, then lose -CH3 to form the ion m/z 286. The product ion at m/z 297 was 30 Da more massive than m/z 267 ([M-H-2Glu]-), which proved the existence of methoxy.
M60 owned the [M-H]- ion at m/z 595.2017 (C28H35O14, -0.692 ppm) with the retention time of 11.39 min and was 28 Da (C2H4) more massive than M0. We speculated that M60 was dimethylation product of mulberroside A. In its ESI-MS/MS spectrum, the loss of 30 Da (m/z 449→m/z 419) was detected, which confirmed our prediction. In addition, the fragment ion at m/z 297 was identified as C14H18O7− formed by the rupture of α-β bond.
M67 was 2 Da more massive than M0 and generated the [M-H]- at m/z 569.1871 (C26H33O14, 1.138 ppm) with the retention time of 13.13 min, which indicated that it might be hydrogenation product of mulberroside A. The product ion at m/z 407 ([M-H-Glu]-) was 2 Da more massive than that of m/z 405. Thus, M67 was ultimately characterized as hydrogenation product of mulberroside A.
Metabolites M4 and M28, with the [M-H]- ions at m/z 665.1384 (0.239 ppm) and m/z 665.1390 (1.156 ppm) respectively, was 98 Da more massive than M0, suggesting that they might be the hydrogenation, hydroxylation and sulfation metabolite of mulberroside A. The fragment ions at m/z 96.96 ([SO4H]-) had the highest abundance, which was a base peak, indicating that S may be contained. In addition, m/z 243, m/z 567, m/z 405 were characteristic fragments of mulberroside A. Therefore, it was speculated that M4 and M28 were hydrogenation, hydroxylation and sulfation products of mulberroside A.
M52, M59 and M70 generated the [M-H]- ions at m/z 405.1180 (-0.145 ppm), m/z 405.1177 (-0.811 ppm), and m/z 243.0651 (-0.392 ppm), respectively. M52 and M59 were 162 Da less massive than M0 while M70 was 324 Da less massive than M0. Moreover, the product ion at m/z 225 was included in their ESI-MS/MS spectra and m/z 199 was detected in the ESI-MS/MS spectra of M70. It was speculated that they were hydrolysates of mulberroside A. M52 and M59 were formed by losing a molecule of Glu while M70 was formed by losing two molecules of Glu. In the experiment of Zhaxi et al., they isolated three metabolites of mulberroside A from rat intestinal contents and feces. The three metabolites were identified as oxyresveratrol-2-O-β-D-glucopyranoside, oxyresveratrol-3′-O-β-D-glucopyranoside and oxyresveratrol according to ESI-MS spectrum, 1H and 13C-NMR spectroscopy (Zhaxi et al., 2010). In their study, oxyresveratrol-3′-O-β-D-glucopyranoside (31.3 min) was eluted later than oxyresveratrol-2-O-β-D-glucopyranoside (27.8 min). However, the hydrolysis products of mulberroside A that formed by losing a Glu should be oxyresveratrol-4-O-β-D-glucopyranoside and oxyresveratrol-3′-O-β-D-glucopyranoside. Hence, it was speculated that oxyresveratrol-2-O-β-D-glucopyranoside was formed by oxyresveratrol combined with a Glu. Therefore, in our experiment, M52, M59 were deduced as oxyresveratrol-4-O-β-D-glucopyranoside and oxyresveratrol-3′-O-β-D-glucopyranoside based on the retention time. Furthermore, M70 was identified as oxyresveratrol according to the literature (Xu et al., 2014).
M53 was deemed as the dehydrogenation and methylation product of mulberroside A, with the [M-H]- ion at m/z 579.1706 (C27H31O14, -0.418 ppm), which was 12 Da more massive than M0. The DPI at m/z 271 was deduced as C12H15O7− formed by the rupture of α-β bond.
3.3.2 Identification of oxyresveratrol-4-O-β-D-glucopyranoside or oxyresveratrol-3′-O-β-D-glucopyranoside metabolites
M3, M16 and M25, with the retention time of 7.98, 8.77 and 9.29 min, possessed the same theoretical [M-H]- ion at m/z 581.1501 (C26H29O15, mass error within 1.00 ppm). They were 176 Da more massive than M52 or M59, which suggested that they may be the glucuronidation products of M52 or M59. In the ESI-MS/MS spectrum, the product ions at m/z 243 ([M-H-Glu A-Glu]-), m/z 405 ([M-H-Glu A]-), m/z 225 ([M-H-Glu A-Glu-H2O]-) were detected. Therefore, M3, M16 and M25 were identified as glucuronidation metabolites of oxyresveratrol-4-O-β-D-glucopyranoside or oxyresveratrol-3′-O-β-D-glucopyranoside.
M8, detected at 8.20 min, generated the [M-H]- ion at m/z 437.1079 (C20H21O11, 0.042 ppm) and was 32 Da more massive than M52 or M59, indicating that it may be the dihydroxylation metabolite of oxyresveratrol-4-O-β-D-glucopyranoside or oxyresveratrol-3′-O-β-D-glucopyranoside. The fragment ions at m/z 275 ([M-H-Glu]-) and m/z 257 ([M-H-Glu-H2O]-) were detected in its ESI-MS/MS spectrum. In addition, the fragment of m/z 137 was also detected, which was presumed to be C7H6O3− formed by α-β bond breakage.
M13 eluted at 8.57 min, was deduced as hydrogenation and glucuronidation product of M52 or M59. The successive loss of 162 and 176 Da (m/z 583→m/z 421→m/z 245) indicated that Glu and Glu A may be contained. Besides, the DPI at m/z 245 ([M-H-Glu-Glu A]-) was 2 Da more massive than m/z 243 (characteristic ion of M52 and M59). Eventually, M13 was identified as hydrogenation and glucuronidation metabolite of oxyresveratrol-4-O-β-D-glucopyranoside or oxyresveratrol-3′-O-β-D-glucopyranoside. In the ESI-MS/MS spectrum of M14, the successive loss of Glu A, Glu and Glu A (m/z 759→m/z 583→m/z 421→m/z 245) was detected, which indicated that two molecules of Glu A and a molecule of Glu may be contained. So, M14 was characterized as hydrogenation and bis-glucuronidation product of oxyresveratrol-4-O-β-D-glucopyranoside or oxyresveratrol-3′-O-β-D-glucopyranoside.
M31, with the [M-H]- ion at m/z 485.0748 (C20H21O12S, -0.048ppm), was 80 Da more massive than M52 or M59, which implied that it might be the sulfation product. In its ESI-MS/MS spectrum, the product ions at m/z 405 ([M-H-SO3]-) and m/z 243 ([M-H-SO3-Glu]-) confirmed the metabolite identification. Hence, M31 was tentatively characterized as sulfation product of oxyresveratrol-4-O-β-D-glucopyranoside or oxyresveratrol-3′-O-β-D-glucopyranoside.
M63 was 2 Da more massive than M52 or M59, indicating that they were hydrogenation metabolites of M52 or M59. The fragment ion at m/z 201 were detected in its ESI-MS/MS spectrums, which was formed by the successive loss of Glu and CO2. In addition, the product ion at m/z 227 ([M-H-Glu-H2O]-) was also detected.
M41 possessed the [M-H]- ion at m/z 435.1271 (C21H23O10, -3.363ppm), with the retention time of 10.00 min. The DPIs at m/z 417 [M-H-H2O]-, m/z 403 and m/z 193 were generated in its ESI-MS/MS spectrum. Therefore, it was speculated that M41 was hydroxylation and methylation metabolite of oxyresveratrol-4-O-β-D-glucopyranoside or oxyresveratrol-3′-O-β-D-glucopyranoside. The fragment of m/z 403 was presumed to be formed by the loss of H2O from m/z 421 (m/z 405+16) and m/z 193 may be C11H14O3− formed by benzene ring opening.
M62, M64, M66, M68 and M71, with the same molecular formula of C20H23O10, had the same theoretical [M-H]- ion at m/z 423.1286. They were 18 Da more massive than M52 or M59, suggesting that they may be hydrogenation and hydroxylation products of M52 or M59. The DPIs at m/z 405, m/z 201, m/z 263 and m/z 247 were detected, which provided evidence for the metabolite identification.
3.3.3 Identification of oxyresveratrol metabolites
M17, M24, M33, M36 and M46 were 256 Da more massive than M70 with the same theoretical [M-H]- ions at m/z 499.0541 (C20H19O13S, error<1.00 ppm). In the ESI-MS/MS spectra, the successive loss of Glu A and SO3 (m/z 499 [M-H]-→m/z 419 [M-H-SO3]-→m/z 243 [M-H-SO3-Glu A]-, m/z 499 [M-H]-→m/z 323 [M-H-Glu A]-→m/z 243 [M-H-Glu A-SO3]-) was observed. Therefore, they were eventually characterized as glucuronidation and sulfation products of oxyresveratrol.
M34, M43 and M44 were tentatively identified as glucuronidation products of M70 according to the neutral loss of 176 Da (m/z 419→m/z 243). In addition, the fragment ion at m/z 175 ([Glu A-H]-) provided evidence for the existence of glucuronic acid group. M48 was 80 Da more massive than M70, with the [M-H]- ion at m/z 323.02209 (C14H11O7S, 0.279 ppm), implying that it may be sulfation product of M70. The neutral loss of 80 Da (m/z 323→m/z 243) proved the existence of SO3 in its ESI-MS/MS spectrum.
M50 was 2 Da more massive than M70 and it was preliminarily deduced as the hydrogenation metabolite of oxyresveratrol. It was produced by hydrogenation reduction reaction occur at the α-β bond according to the product ion at m/z 123, which was formed by the rupture of α-β bond. The fragment ions at m/z 203 ([[M-H-CO-CH2]−]-) and m/z 159 ([M-H-C3O2-H2O]−) were generated in its ESI-MS/MS spectra. Besides, the DPI at m/z 227 ([M-H-H2O]-) was also detected. M49 was 2 Da less massive than M70, indicating that it might be the dehydrogenation product of oxyresveratrol. The DPIs at m/z 197 ([M-H-CO2]-), m/z 179 ([M-H-CO2-H2O]-) and m/z 223 ([M-H-H2O]-) provided evidence for the metabolite identification. Finally, M50 was identified as hydrogenation metabolite of oxyresveratrol and M49 was characterized as dehydrogenation product of oxyresveratrol.
M10, M51, M55 and M65, with the same theoretical [M-H]- ion at m/z 433.1129 (C21H21O10, error ≤1.00 ppm), were 190 Da more massive than M70. The DPI at m/z 257 ([M-H-Glu A]-) and the continuous neutral loss of 30 Da (m/z 433→m/z 403→m/z 373) were detected, suggesting that Glu A and -OCH3 might be contained. Thus, they were eventually deduced as methylation and glucuronidation metabolites of oxyresveratrol.
M11, M19 and M20, eluted at 8.43 min, 8.84 min and 8.94 min respectively, were deduced as hydrogenation and bis-glucuronidation products of M70. In the ESI-MS/MS spectra, the successive loss of 176 Da (m/z 597→m/z 421→m/z 245) indicated that two molecules of Glu A might be contained. Furthermore, the DPI at m/z 175 confirmed the existence of Glu A and the DPI at m/z 245 ([M-H-2Glu A]-) was 2 Da more massive than m/z 243 ([M-H]- ion of M70). Therefore, they were eventually identified as hydrogenation and glucuronidation products of oxyresveratrol.
M12, eluted at 8.52 min, was 174 Da more massive than M70. The neutral loss of 176 Da (m/z 421→m/z 245 and m/z 417→m/z 241) was detected in both ESI-MS/MS spectra, which was suggested that Glu A might be contained in its structure. What’s more, the DPI at m/z 241 of M12 was 2 Da less massive than m/z 243 ([M-H]- ion of M70). Thus, M12 was characterized as dehydrogenation and glucuronidation metabolite of oxyresveratrol. M22 and M37, with the retention time of 9.19 and 9.82 min, were 178 Da more massive than M70. The neutral loss of 176 Da (m/z 421→m/z 245 and m/z 417→m/z 241) provided evidence for the existence of Glu A. In addition, the DPI at m/z 245 was 2 Da more massive than m/z 243. Finally, they were deduced as hydrogenation and glucuronidation product of oxyresveratrol.
M15, M23 and M26 were identified as bis-glucuronidation products of M70 according to the successive loss of Glu A (m/z 595→m/z 419→m/z 243) with the retention time of 8.76 min, 9.25 min and 9.31 min, respectively. M18, M30, M32 and M35, with the molecular formula of C20H21O13S, generated the same theoretical [M-H]- ions at m/z 501.0697. The successive loss of 176 Da (m/z 421→m/z 245) and 80 Da (m/z 501→m/z 421, m/z 325→m/z 245) was detected in their ESI-MS/MS spectra, indicating that they might be the glucuronidation and sulfation products of M70. In addition, the DPI at m/z 245 was 2 Da more massive than the [M-H]- ion of M70 (m/z 243). So, they were deduced as the hydrogenation, glucuronidation and sulfation products of oxyresveratrol.
M38, M47 and M58 were deduced as the dehydroxylation and glucuronidation metabolites of oxyresveratrol, with the molecular formula of C20H19O9. The DPIs at m/z 227 [M-H-Glu A]- (m/z 403–176 Da, m/z 243–16 Da) provided the evidence for the metabolite identification. The successive loss of 176 and 16 Da (m/z 435→m/z 259→m/z 243) was observed in the ESI-MS/MS spectrum of M39, indicating that Glu A and -OH may be contained. Therefore, M39 was identified as hydroxylation and glucuronidation product of oxyresveratrol. M40 was characterized as hydration and glucuronidation metabolite of oxyresveratrol based on the successive loss of Glu A and H2O (m/z 437→m/z 261→m/z 243).
M42 and M45 were deemed to be the hydrogenation and sulfation products of oxyresveratrol while M56 was identified as dehydrogenation and sulfation metabolite of oxyresveratrol. The DPIs at m/z 245 (m/z 325–80 Da, m/z 243+2 Da) and m/z 241 (m/z 321–80 Da, m/z 243–2 Da) provided the evidence for the metabolite identification.
M61 possessed the [M-H]- ion at m/z 271.0964 (C16H15O4, -0.278 ppm), which was 28 Da (C2H4) more massive than M70. It was presumed to be the dimethylation product of oxyresveratrol. The successive loss of 30 Da (m/z 197→m/z 167→m/z 137) provided evidence for the existence of methoxy.
M69, with the [M-H]- ion at m/z 339.0176 (C14H11O8S, 1.963 ppm), was eluted at 13.63 min. The neutral loss of 80 Da (m/z 339→m/z 259) was observed in the ESI-MS/MS spectrum, indicating that SO3 might be contained in its structure. Besides, the fragment ion at m/z 259 was 16 Da more massive than m/z 243, which suggested that it may be hydroxylation product of M70. Hence, it was speculated that M69 was hydroxylation and sulfation product of oxyresveratrol.
3.4 Analysis of metabolic reaction of mulberroside A
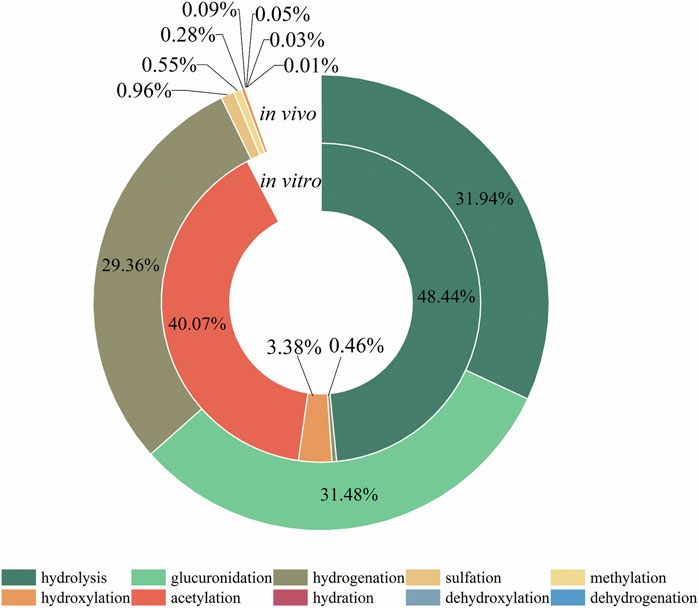
The metabolic reactions of mulberroside A in vivo and in vitro of rats were hydrolysis, hydrogenation, hydroxylation, methylation, acetylation, sulfation, glucuronidation and so on. In this paper, the product peak areas of each type of metabolic reaction were summarized, as shown in Figure 6. The results showed that the metabolic reactions of mulberroside A in vivo were mainly hydrolysis, glucuronidation and hydrogenation. The total peak area of hydrolysis metabolites accounted for the largest proportion. Therefore, it was preliminarily speculated that after entering the body, mulberroside A was hydrolyzed into aglycone firstly, and then a series of metabolic reactions occur. While in vitro, the most abundant compounds were hydrolysate and acetylation products of mulberroside A.
3.5 Analysis of proposed metabolic characteristics of mulberroside A
In this experiment, a total of 72 metabolites including mulberroside A were detected, 40 in blood, 37 in urine, 12 in feces, 9 in liver, and 10 in liver microsomes. The prototype component was only detected in rat plasma samples, which indicated that mulberroside A could be absorbed into the blood, and then a series of metabolic reactions occurred. There was no significant difference on the type of metabolites in plasma and urine, and the metabolites in feces were mainly glucuronidation, sulfation and methylation products of oxyresveratrol-4-O-β-D-glucopyranoside or oxyresveratrol-3′-O-β-D-glucopyranoside. It was speculated that mulberroside A was metabolized in 3 pathways in vivo and in vitro of rats. (1) The metabolic reactions such as sulfation, hydroxylation and methylation were occurred on the basis of mulberroside A (M2, M4, M5, M6, M7, M9, M28, M29, etc.). In this pathway, glucuronidation reaction was undetected. (2) Mulberroside A was hydrolyzed to form oxyresveratrol-4-O-β-D-glucopyranoside (M52) or oxyresveratrol-3′-O-β-D-glucopyranoside (M59), on which metabolic reactions took place, such as M3, M8, M13, M14, M16, M25, M31 and so on. (3) Mulberroside A continued to be hydrolyzed and lose another Glu to produce oxyresveratrol (M70). Then, groups such as glucuronic acid, sulfate and methyl were bound to oxyresveratrol (M10, M11, M15, M17, M18, M19, etc.). Besides, β-D-Glucopyranosiduronic Acid 3-Hydroxyphenyl (M1) and isomer of 6-(4-Ethenyl-2-hydroxyphenoxy)-3,4,5-trihydroxyoxane-2-carboxylic acid (M21) were formed by the rupture of C1′-β bond. The proposed metabolic pathways of mulberroside A are shown in Figure 7.
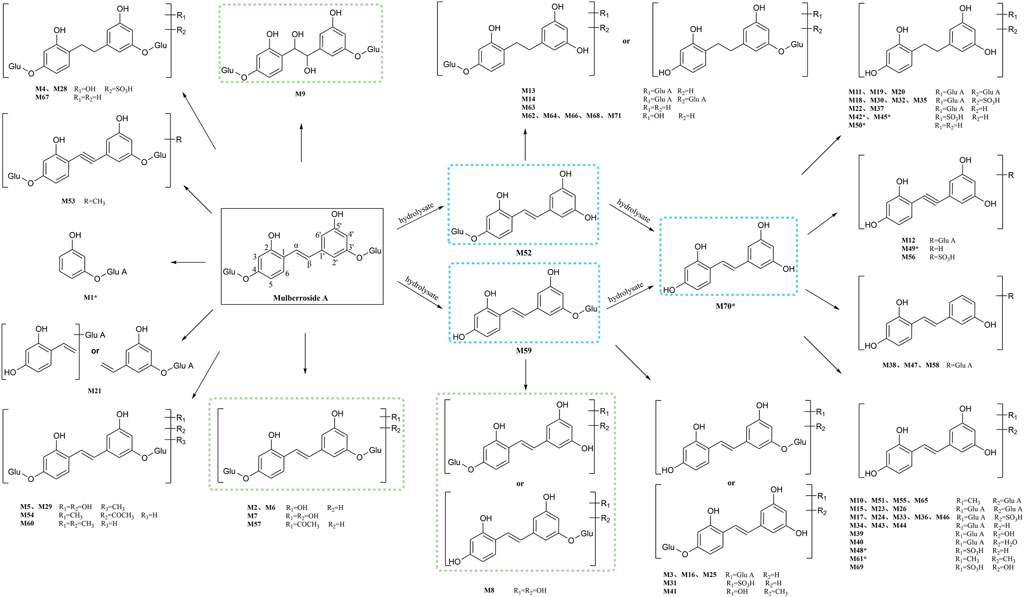
FIGURE 7. The proposed metabolic pathways of mulberroside A in vivo and in vitro of rats. Note: Glu: glucose; Glu A: glucuronic acid; component with blue flame: metabolite in vivo and in vitro; component with green flame: metabolite in vitro; *: components in Table 2.
Furthermore, the in vivo and in vitro metabolites of mulberroside A were mostly based on its hydrolyzed products oxyresveratrol-4-O-β-D-glucopyranoside or oxyresveratrol-3′-O-β-D-glucopyranoside and oxyresveratrol. The main metabolic reactions were hydrolysis, hydroxylation, methylation, sulfation, glucuronidation and their compound reactions. The metabolic result showed that the main forms of mulberroside A in vivo were the glucuronidation and sulfation products of its aglycone, while in vitro they were mainly the hydroxylation and hydrolysis products of its prototype. The in vitro pharmacokinetic study of mulberroside A also showed that the glucose group on mulberroside A was rapidly hydrolyzed to generate monoglycosides and its aglycone oxyresveratrol when incubated anaerobically with intestinal bacteria (Mei et al., 2012), which was consisted with our findings.
3.6 Comparison of three different sample preparation methods
In the study of drug metabolism, deproteinization of biological samples is essential on account of a large number of proteins were contained in plasma, which interferes with the identification of metabolites. At present, the main methods of protein removal are organic solvent precipitation (methanol, acetonitrile, methanol-ethanol, methanol-acetonitrile-acetone, etc.) and solid phase extraction method (Xu et al., 2011). In our experiment, the plasma samples of rats were treated by solid phase extraction, methanol precipitation and acetonitrile precipitation respectively. In the end, 36 metabolites were detected in the plasma samples treated by SPE while 17 metabolites were detected in the plasma samples treated by methanol precipitation and 15 metabolites in the plasma samples treated by acetonitrile precipitation. It was obvious that solid phase extraction method was significantly better than the other two methods, which was consistence with the study results of Liu et al. (Liu et al., 2020). It was speculated that solid phase extraction plays the role of separation, enrichment and concentration in the process of sample pretreatment. On the contrary, biological samples may be diluted by methanol precipitation and acetonitrile precipitation. In addition, the type of organic solvents and the proportion of different solvents also play important roles in the pretreatment of biological samples.
3.7 Network pharmacology analysis of mulberroside A and its metabolites
3.7.1 Construction and analysis of metabolite-target network
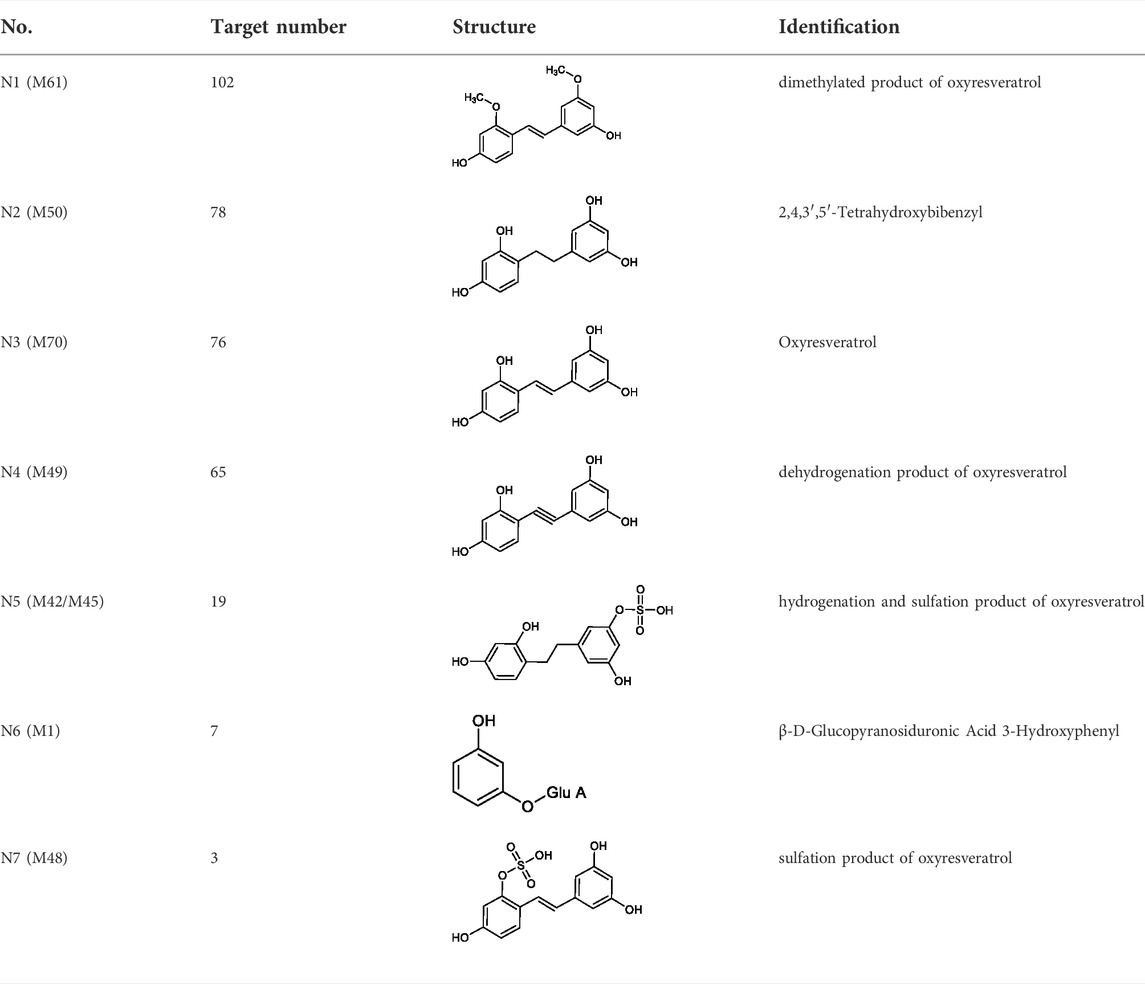
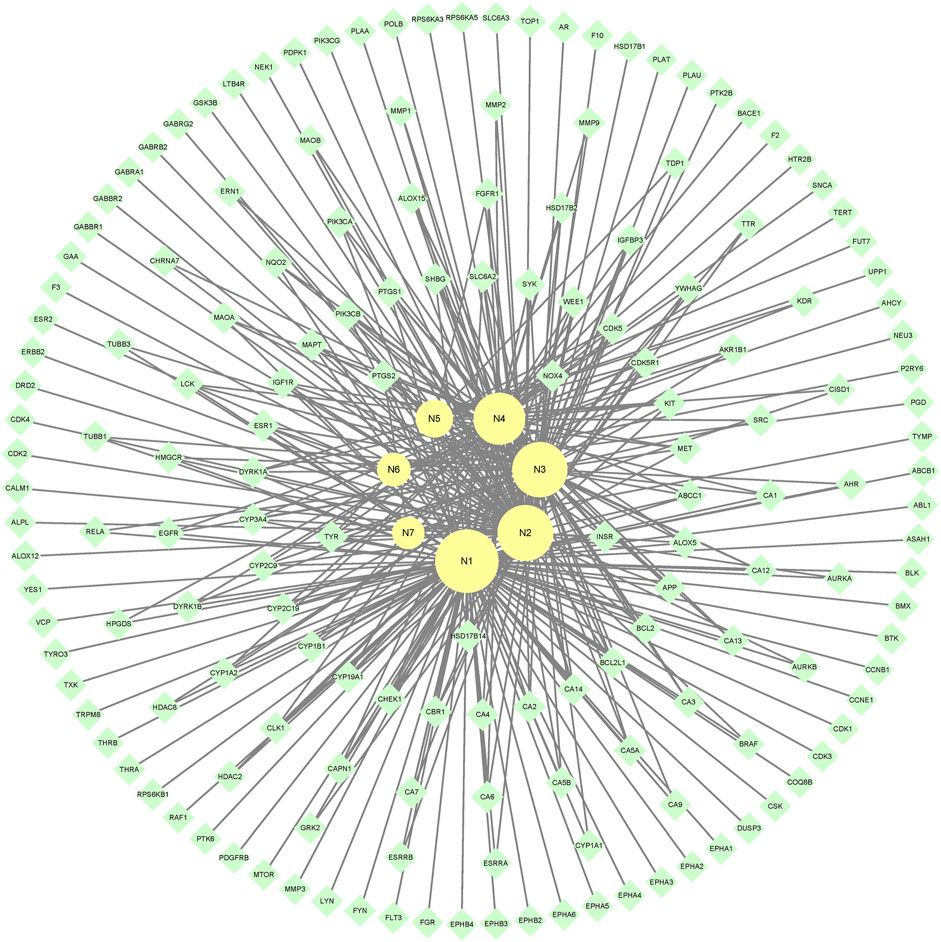
7 metabolites (N1–N7) with high gastrointestinal absorption and drug-likeness were selected by Swiss ADME. The information of the 7 metabolites is shown in Table 2. However, the Swiss ADME results showed that mulberroside A had no drug-likeness and no target of mulberroside A was obtained by Swiss Target Prediction. So, mulberroside A was not included. As a result, a total of 167 targets were obtained using Swiss Target Prediction. Then, the metabolite-target network with 174 nodes and 350 edges was constructed based on the information by Cytoscape 3.8.2, as shown in Figure 8. The area of the component graph in the figure is proportional to the degree value, so it can be seen intuitively that N1 has the largest effect, followed by N2 and N3.
3.7.2 GO enrichment analysis of target proteins
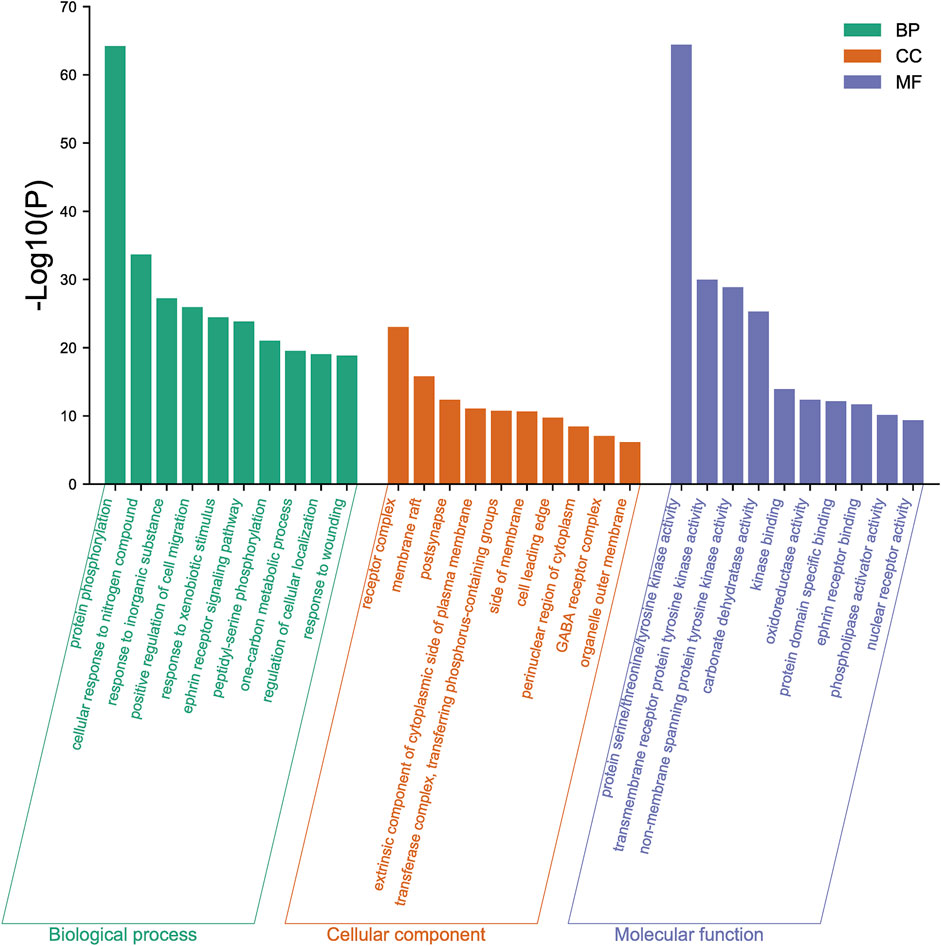
The 167 targets were submitted to the Metascape for GO enrichment analysis, including molecular functions (MF), biological processes (BP) and cellular components (CC). A total of 1702 GO items were obtained finally (164 of MF, 1427 of BP and 111 of CC) (p < 0.01). The top 10 clusters with their representative enriched terms (one per cluster) were selected and they are shown in Figure 9. The serine/threonine/tyrosine kinase activity, transmembrane receptor protein tyrosine kinase activity, non-membrane spanning protein tyrosine kinase, carbonate dehydratase activity and kinase binding were involved in MF. In the aspect of BP, there were mainly protein phosphorylation, cellular response to nitrogen compound, response to inorganic substance, positive regulation of cell migration and response to xenobiotic stimulus. The receptor complex, membrane raft, postsynapse, extrinsic component of cytoplasmic side of plasma membrane and transferase complex, transferring phosphorus-containing groups were mainly included in the category of CC.
3.7.3 KEGG analysis of target proteins
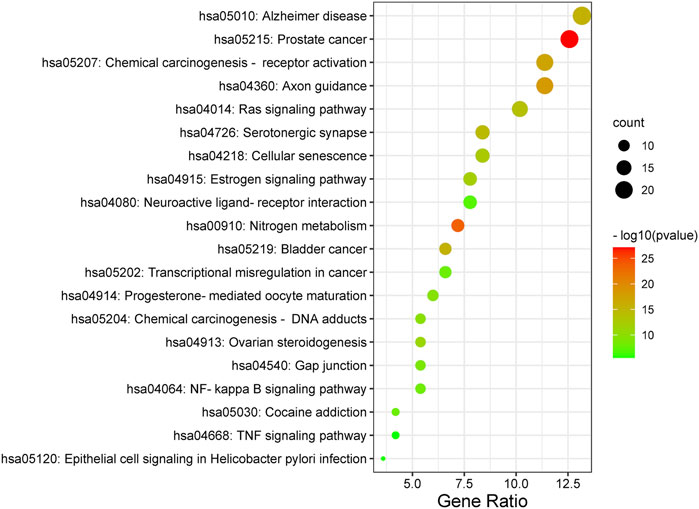
Furthermore, the Metascape was also applied in KEGG analysis. In the end, 158 signal pathways were enriched (p < 0.01). The result suggested that the pathways of mulberroside A metabolites were mainly involved Axon guidance, Alzheimer disease, Serotonergic synapse, NF-kappa B signaling pathway, Neuroactive ligand-receptor interaction, Epithelial cell signaling in Helicobacter pylori infection, TNF signaling pathway and so on (Figure 10, Supplementary Table S1).
3.7.4 Construction and analysis of metabolite-target-pathway network
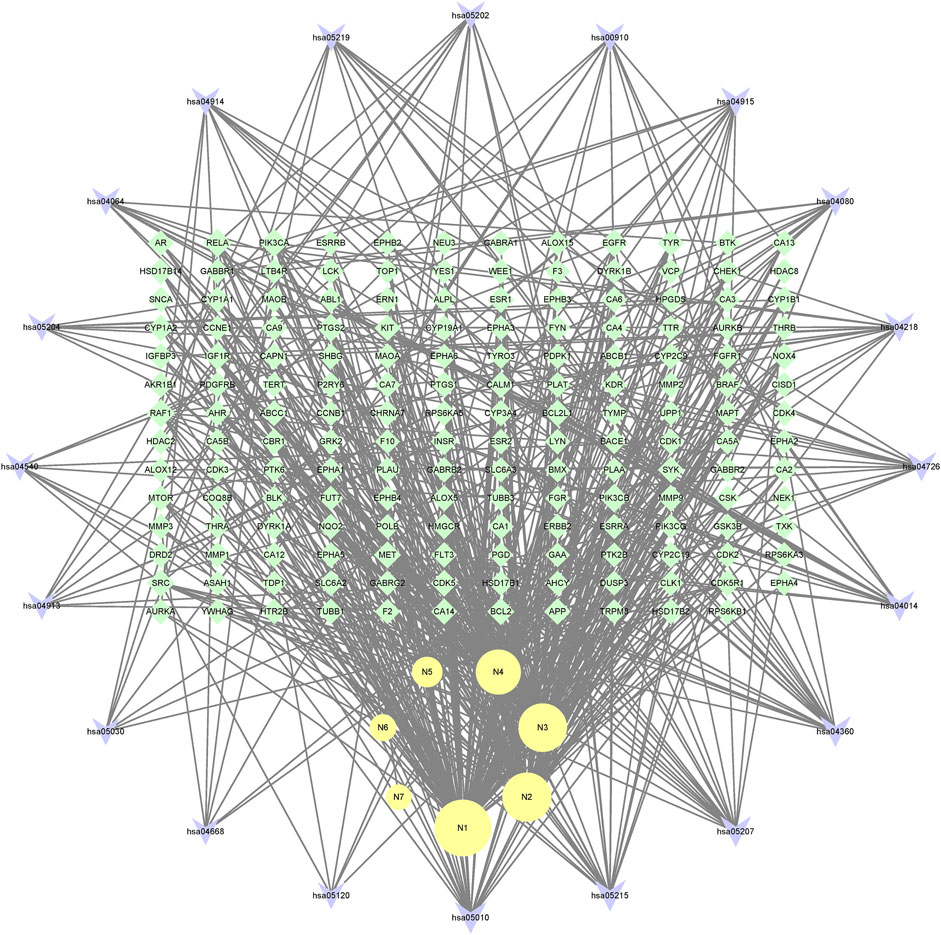
In the end, the 7 metabolites (N1–N7), 167 targets and the top 20 pathways were imported to Cytoscape 3.8.2 for metabolite-target-pathway network. As a result, the metabolite-target-pathway network, with 194 nodes and 602 edges, was constructed, as shown in Figure 11.
Studies have shown that mulberroside A has the effect of neuroprotection (Wang C. P. et al., 2014). From the metabolite-target-pathway network, it can be seen intuitively that the metabolites N1, N2, N3 and N4 were involved in pathways of Axon guidance, Alzheimer disease and Serotonergic synapse. Axon guidance and serotonin (5-HT) play key roles in neuronal network (Negishi et al., 2005; Migliarini et al., 2013). Therefore, it was speculated that the metabolites N1, N2, N3 and N4 exert the neuroprotective effect by Axon guidance and Serotonergic synapse. Besides, oxyresveratrol (N3) has the effect of neuroprotection (Lee et al., 2019) and can be used to treat alzheimer disease (Lakshmi et al., 2021), which proved the results. From the metabolite-target-pathway network (Figure 11), it could be seen intuitively that EGFR may play an important role. EGFR is a tyrosine kinase which can regulate cellular homeostasis. Duan’s study suggested that mulberroside A could suppress the migration and invasion of renal cancer A498 cells through inhibit the expression of p-EGFR (Duan et al., 2019). So, it was indicated that mulberroside A could exert nephroprotective effect by regulating the expression of EGFR. In the pathway of Melanogenesis, tyrosinase (TYR) and tyrosinase-related protein 1 (TYRP1) are key proteins in melanin production. The study of Wang et al. showed that mulberroside A can suppress the activity of tyrosinase (Wang S. et al., 2014). Thus, it can be concluded that mulberroside A could inhibit the production of melanin by inhibiting the activity of tyrosinase.
Furthermore, metabolites N1, N2, N3 and N4 were also associated with NF-kappa B signaling pathway, TNF signaling pathway and Epithelial cell signaling in Helicobacter pylori infection. RPS6KA5 was the key target of N2, which exerts anti-inflammatory effect by decreasing the expression of TNF-α and inhibiting the activation of NF-κB (Wang C. P. et al., 2014). It should be noted that NF-kappa B signaling pathway plays a crucial role in mulberroside A-induced suppression of P-Glycoprotein (Li et al., 2014) and research has shown that insulin regulates P-glycoprotein via PKC/NF-κB pathway (Liu et al., 2009). Thus, it was speculated that mulberroside A has the inflammatory and antidiabetic effects after being metabolized to N1, N2, N3 and N4.
4 Conclusion
In the present study, an integrated strategy based on the UHPLC-Q-Exactive Plus Orbitrap MS and network pharmacology was firstly established to elucidate the metabolic mechanism of mulberroside A. In order to detect as many metabolites as possible, not only biological samples (plasma, urine, feces, liver tissues) were prepared in vivo of rats, but also the liver microsomes of rats were incubated in vitro. For the treatment of plasma samples, three methods (solid phase extraction, methanol precipitation and acetonitrile precipitation) were applied. We concluded that solid phase extraction was the most suitable method for plasma sample of mulberroside A. As a result, a total of 72 metabolites including mulberroside A were detected by UHPLC-Q-Exactive Plus Orbitrap MS. The metabolites and metabolic pathways of mulberroside A in vivo and in vitro of rats were also elaborated. The main metabolite reactions of mulberroside A were hydrolysis, glucuronidation and hydrogenation. Furthermore, the results of network pharmacology study showed that the component that exerts pharmacological effects in the body is not mulberroside A but its metabolites, which mainly include oxyresveratrol (aglycon of mulberroside A), dehydrogenation, hydrogenation, and demethylated products of oxyresveratrol.
The integrated strategy established in our study could elucidate the metabolic pathways and mechanism of mulberroside A in vivo and in vitro of rats more comprehensively, which would provide more theoretical basis for the study of mulberroside A. It would be conducive to better understand the metabolic action and promote research for the further potential therapeutic application of mulberroside A.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by BIN ZHOU Medical University.
Author contributions
JS and HZ conceived and designed the experiment. XZ and PD performed the experiment. XZ, FW, YY and LL analyzed data and wrote the manuscript. PD and HZ revised the manuscript. YL provided technical support. All authors contributed to manuscript read and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (no. 82004037), the Key Research and Development program of Shandong Province (no. 2018GSF119018) and Major Science and technology innovation engineering projects of Shandong Province (no. 2018CXGC1301).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.981173/full#supplementary-material
References
Chung, K. O., Kim, B. Y., Lee, M. H., Kim, Y. R., Chung, H. Y., Park, J. H., et al. (2003). In-vitro and in-vivo anti-inflammatory effect of oxyresveratrol from Morus alba L. J. Pharm. Pharmacol. 55 (12), 1695–1700. doi:10.1211/0022357022313
Dong, P. P., Shi, L., Wang, S. P., Jiang, S., Li, H. R., Dong, F., et al. (2021). Rapid profiling and identification of vitexin metabolites in rat urine, plasma and faeces after oral administration using a UHPLC-Q-exactive Orbitrap mass spectrometer coupled with multiple data-mining methods. Curr. Drug Metab. 22 (3), 185–197. doi:10.2174/1389200221999210101232841
Duan, C., Han, J., Zhang, C., Wu, K., and Lin, Y. (2019). Inhibition of kidney cancer cell growth by Mulberroside-A is mediated via mitochondrial mediated apoptosis, inhibition of cell migration and invasion and targeting EGFR signalling pathway. J. buon 24 (1), 296–300.
Guo, Z. H., Jia, Z. X., Chen, K. K., Liu, J., Xu, W. J., Duan, F. P., et al. (2018). Studies on constituents of Polygonum multiflorum extract in vivo and in vitro based on UPLC-Q-TOF-MS. China J. Chin. Mat. Med. 43 (13), 2796–2805. doi:10.19540/j.cnki.cjcmm.20180419.002
Jiang, Y. B., Shan, J. H., Wang, Y., Cai, Y. H., and Tian, Y. P. (2014). Application progress of LC-MS/MS technology in drug metabolism research. Chin. J. Pharm. Anal. 34 (03), 385–391. doi:10.16155/j.0254-1793.2014.03.022
Lakshmi, S., Varija Raghu, S., Elumalai, P., and Sivan, S. (2021). Alkoxy glycerol enhanced activity of Oxyresveratrol in Alzheimer's disease by rescuing Tau protein. Neurosci. Lett. 759, 135981. doi:10.1016/j.neulet.2021.135981
Lee, H. J., Feng, J. H., Sim, S. M., Lim, S. S., Lee, J. Y., and Suh, H. W. (2019). Effects of resveratrol and oxyresveratrol on hippocampal cell death induced by kainic acid. Anim. Cells Syst. Seoul. 23 (4), 246–252. doi:10.1080/19768354.2019.1620853
Li, Y. H., Huang, L., Zeng, X. Z., Zhong, G. P., Ying, M. J., Huang, M., et al. (2014). Down-regulation of P-gp expression and function after mulberroside A treatment: Potential role of protein kinase C and NF-kappa B. Chem. Biol. Interact. 213, 44–50. doi:10.1016/j.cbi.2014.02.004
Liang, Y. L., Shi, X. H., Bai, J. Q., Huang, Z. H., Xu, W., Huang, J., et al. (2021). Analysis of metabolites and metabolism pathway of tetrahydroxystilbene glucoside in rats with UHPLC-MS and mass defect filter method. China Pharm. 32 (06), 675–681.
Liu, H. Y., Yang, H. W., Wang, D. L., Liu, Y. C., Liu, X. D., Li, Y., et al. (2009). Insulin regulates P-glycoprotein in rat brain microvessel endothelial cells via an insulin receptor-mediated PKC/NF-kappaB pathway but not a PI3K/Akt pathway. Eur. J. Pharmacol. 602 (2–3), 277–282. doi:10.1016/j.ejphar.2008.11.026
Liu, Z. H., Wang, S. P., Dong, F., Lin, Y., Li, H. R., Shi, L., et al. (2020). Comprehensive analysis of resveratrol metabolites in rats using ultra high performance liquid chromatography coupled with high resolution mass spectrometry. Arab. J. Chem. 13 (9), 7055–7065. doi:10.1016/j.arabjc.2020.07.011
Mei, M., Ruan, J. Q., Wu, W. J., Zhou, R. N., Lei, J. P., Zhao, H. Y., et al. (2012). In vitro pharmacokinetic characterization of mulberroside A, the main polyhydroxylated stilbene in mulberry (Morus alba L.), and its bacterial metabolite oxyresveratrol in traditional oral use. J. Agric. Food Chem. 60 (9), 2299–2308. doi:10.1021/jf204495t
Migliarini, S., Pacini, G., Pelosi, B., Lunardi, G., and Pasqualetti, M. (2013). Lack of brain serotonin affects postnatal development and serotonergic neuronal circuitry formation. Mol. Psychiatry 18 (10), 1106–1118. doi:10.1038/mp.2012.128
Negishi, M., Oinuma, I., and Katoh, H. (2005). Plexins: Axon guidance and signal transduction. Cell Mol. Life Sci. 62 (12), 1363–1371. doi:10.1007/s00018-005-5018-2
Pan, Z. R., Liang, H. L., Liang, Z. H., and Xu, W. (2015). An ultra-high-pressure liquid chromatography/linear ion trap-Orbitrap mass spectrometry method coupled with a diagnostic fragment ions-searching-based strategy for rapid identification and characterization of chemical components in Polygonum cuspidatum. Chin. J. Chromatogr. 33 (01), 22–28.
Park, K. T., Kim, J. K., Hwang, D., Yoo, Y., and Lim, Y. H. (2011). Inhibitory effect of mulberroside A and its derivatives on melanogenesis induced by ultraviolet B irradiation. Food Chem. Toxicol. 49 (12), 3038–3045. doi:10.1016/j.fct.2011.09.008
Prasad, B., Garg, A., Takwani, H., and Singh, S. (2011). Metabolite identification by liquid chromatography-mass spectrometry. Trac-Trends Anal. Chem. 30 (2), 360–387. doi:10.1016/j.trac.2010.10.014
Qiu, F., Komatsu, K., Saito, K., Kawasaki, K., Yao, X., and Kano, Y. (1996a). Pharmacological properties of traditional medicines (XXI): Analysis of plasma, urine and bile of rats after oral administration of water extract of Mori Cortex. Nat. Med. 50 (2), 103–108.
Qiu, F., Komatsu, K., Saito, K., Kawasaki, K., Yao, X., and Kano, Y. (1996b). Pharmacological properties of traditional medicines. XXII. Pharmacokinetic study of mulberroside A and its metabolites in rat. Biol. Pharm. Bull. 19 (11), 1463–1467. doi:10.1248/bpb.19.1463
Sun, J. H. (2004). Studies on pharmacokinetics and metabolites of stilbene glycoside. Shijiazhuang: Hebei Medical University. [master’s thesis]. [Shijiazhuang (Hebei province)].
Wang, C. P., Wang, Y., Wang, X., Zhang, X., Ye, J. F., Hu, L. S., et al. (2011). Mulberroside A possesses potent uricosuric and nephroprotective effects in hyperuricemic mice. Planta Med. 77 (8), 786–794. doi:10.1055/s-0030-1250599
Wang, C. P., Zhang, L. Z., Li, G. C., Shi, Y. W., Li, J. L., Zhang, X. C., et al. (2014a). Mulberroside A protects against ischemic impairment in primary culture of rat cortical neurons after oxygen-glucose deprivation followed by reperfusion. J. Neurosci. Res. 92 (7), 944–954. doi:10.1002/jnr.23374
Wang, S., Liu, X. M., Zhang, J., and Zhang, Y. Q. (2014b). An efficient preparation of mulberroside A from the branch bark of mulberry and its effect on the inhibition of tyrosinase activity. PLoS One 9 (10), e109396. doi:10.1371/journal.pone.0109396
Xu, J., Tian, Y. P., Chen, Y. H., Zhang, R. P., Yang, F., Song, Y. M., et al. (2011). Study on pretreatment of metabonomic plasma samples and its rapid high resolution liquid chromatography-mass spectrometry. Chin. J. Anal. Chem. 39 (12), 1793–1797.
Xu, W., Fu, Z. Q., Lin, J., Huang, X. C., Chen, D., Yu, H. M., et al. (2014). Qualitative and quantitative analysis of major constituents in Tetrastigma hemsleyanum by HPLC-Q-TOF-MS and UPLC-QqQ-MS. China J. Chin. Mat. Med. 39 (22), 4365–4372.
Xu, W. (2015). The applicaiton of liqud chromatography coupled with high resolution mass spectrometry technology on chemical study of two Chinese herbs. Guangzhou: Guangzhou University of Chinese Medicine. [doctor’s thesis]. [Guangzhou (Guangdong province)].
Xu, Y. Y., Guo, H. L., Zhao, T. T., Fu, J., and Xu, Y. H. (2021). Mulberroside A from cortex mori enhanced gut integrity in diabetes. Evid. Based Complement. Altern. Med. 2021, 6655555. doi:10.1155/2021/6655555
Yu, R., Wen, S. Y., Wang, Q. N., Wang, C. Y., Zhang, L. P., Wu, X. X., et al. (2021). Mulberroside A repairs high fructose diet-induced damage of intestinal epithelial and blood-brain barriers in mice: A potential for preventing hippocampal neuroinflammatory injury. J. Neurochem. 157 (6), 1979–1991. doi:10.1111/jnc.15242
Yuan, J. Q., Wang, H., Wang, Y. T., Wang, Z. J., Huo, Q., Dai, X. L., et al. (2021). Rapid identification of 3,6'-disinapoyl sucrose metabolites in alzheimer's disease model mice using UHPLC-orbitrap mass spectrometry. Molecules 27 (1). doi:10.3390/molecules27010114
Zhang, J. P. (2018). The study of isolation and identification of metabolites of Polygonum multiflorum in rats. Guangzhou: Guangzhou University of Chinese Medicine. [master’s thesis]. [Guangzhou (Guangdong province)].
Zhang, Z. F., Jin, J., and Shi, L. G. (2008). Protective function of cis-mulberroside A and oxyresveratrol from Ramulus mori against ethanol-induced hepatic damage. Environ. Toxicol. Pharmacol. 26 (3), 325–330. doi:10.1016/j.etap.2008.06.008
Zhang, Z. F., and Shi, L. G. (2010). Anti-inflammatory and analgesic properties of cis-mulberroside A from Ramulus mori. Fitoterapia 81 (3), 214–218. doi:10.1016/j.fitote.2009.09.005
Zhaxi, M., Chen, L. X., Li, X. G., Komatsu, K., Yao, X. S., and Qiu, F. (2010). Three major metabolites of mulberroside A in rat intestinal contents and feces. Planta Med. 76 (4), 362–364. doi:10.1055/s-0029-1186160
Zhou, J., Li, S. X., Wang, W., Guo, X. Y., Lu, X. Y., Yan, X. P., et al. (2013). Variations in the levels of mulberroside A, oxyresveratrol, and resveratrol in mulberries in different seasons and during growth. Sci. World J. 2013, 380692. doi:10.1155/2013/380692
Keywords: mulberroside A, UHPLC-Q-Exactive Plus Orbitrap MS, network pharmacology, metabolites, metabolic pathways
Citation: Zhang X, Dong P, Song J, Zhang H, Wang F, Liu Y, Yan Y and Li L (2022) Identification and mechanism prediction of mulberroside A metabolites in vivo and in vitro of rats using an integrated strategy of UHPLC-Q-Exactive Plus Orbitrap MS and network pharmacology. Front. Chem. 10:981173. doi: 10.3389/fchem.2022.981173
Received: 29 June 2022; Accepted: 28 July 2022;
Published: 27 September 2022.
Edited by:
Alberto Salomone, University of Turin, ItalyReviewed by:
Svante Vikingsson, RTI International, United StatesWei Cai, Hunan University of Medicine, China
Copyright © 2022 Zhang, Dong, Song, Zhang, Wang, Liu, Yan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huimin Zhang, huiminzhang@163.com
†These authors have contributed equally to this work and share first authorship
 Xiao Zhang
Xiao Zhang Pingping Dong
Pingping Dong Jian Song1
Jian Song1  Huimin Zhang
Huimin Zhang Feiran Wang
Feiran Wang Linlin Li
Linlin Li