SapF-mediated heme-iron utilization enhances persistence and coordinates biofilm architecture of Haemophilus
- Center for Microbial Pathogenesis, The Research Institute at Nationwide Children's Hospital, The Ohio State University School of Medicine, Columbus, OH, USA
Non-typeable Haemophilus influenzae (NTHI) is a common commensal bacterium that resides in the human upper respiratory tract of healthy individuals. NTHI is also a known causative agent of multiple diseases including sinusitis, otitis media, as well as exacerbates disease severity of patients with cystic fibrosis and chronic obstructive pulmonary disease. We have previously shown that the Sap transporter mediates resistance to host antimicrobial peptides (AMPs) and import of the iron-containing compound heme. Here, we analyzed the contribution of the Sap structural ATPase protein, SapF, in these essential functions. In contrast to SapD, SapF was dispensable for NTHI survival when exposed to AMPs in vitro. SapF was responsible for heme utilization and recovery of depleted internal heme-iron stores. Further, a loss of SapF resulted in morphological plasticity and enhanced community development and biofilm architecture, suggesting the potential role of heme-iron availability in coordinating the complexity of NTHI biofilm architecture. SapF was required for colonization of the nasopharynx and acute infection of the middle ear, as SapF deficiency correlated with a statistically significant decrease in NTHI persistence in vivo. These data suggest that SapF is required for proper heme utilization which directly impacts NTHI survival. Thus, these studies further support a role for the Sap complex in the transport of multiple substrates and further defines substrate specificity for the two ATPase subunits. Given the multiple essential functions provided by the Sap transporter, this complex could prove to be an effective therapeutic target for the treatment of NTHI diseases.
Introduction
Non-typeable Haemophilus influenzae (NTHI) is a Gram-negative nasopharyngeal commensal microorganism, and opportunistic pathogen that can mediate human airway diseases such as otitis media (OM), acute sinusitis, chronic bronchitis, pneumonia, and exacerbations in patients with cystic fibrosis and chronic obstructive pulmonary disease (Klein, 1997; St Geme, 2000; Sethi and Murphy, 2001; Murphy, 2003; Roman et al., 2004). Commensal microorganisms must adapt to various micro-environmental cues for long-term colonization of the host. Bacterial acquisition of nutrients and resistance to host bactericidal mechanisms are vital for survival as a commensal microorganism. Disruption of commensal-host homeostasis, however, can potentiate disease development. Pathogenesis is a multifactorial and dynamic process that begins with bacterial migration to a privileged site and culminates with unrestricted bacterial growth. The unrestricted growth is dependent upon multiple complex and dynamic interactions between the microbes, the varied microenvironments encountered in the host, and interactions with host immune effectors. Acquisition of essential nutrients in this hostile environment and bacterial strategies to thwart innate immune clearance mechanisms are critical for NTHI pathogenesis.
The propensity for NTHI to build and reside within a biofilm, as observed in samples taken from patients with OM as well as animals with experimentally induced OM, further increases the biological complexity of NTHI lifestyles (Ehrlich et al., 2002; Hall-Stoodley et al., 2006; Jurcisek and Bakaletz, 2007; Jurcisek et al., 2007; Post et al., 2007). The organized biofilm community contributes tremendously to pathogenesis and, importantly, to the prolonged, recurrent and difficult-to-treat nature of these diseases. The development of structured biofilm formation in other pathogens (e.g., Pseudomonas aeruginosa) is conditional upon availability of nutrients (Banin et al., 2005; Moreau-Marquis et al., 2009; Wu and Outten, 2009). NTHI, similarly with other microbes that reside on and within the mucosal epithelium, requires iron for survival and for key bacterial processes (Markel et al., 2007). Microbes adapt to dynamic-host environments with specific mechanisms to compete with their host for essential iron. Host iron sequestration (“nutritional immunity”) influences microbial growth and biofilm formation (Ong et al., 2006). Haemophilus, a strictly obligate human commensal, lacks the ability to synthesize the protoporphyrin ring required for iron sequestration and, therefore, must possess multiple mechanisms to acquire both heme and iron for use in cytoplasmic enzymes (Stojiljkovic and Perkins-Balding, 2002). Proteins that function in heme acquisition include homologues of hemoglobin and hemoglobin–haptoglobin binding proteins (Maciver et al., 1996; Ren et al., 1998; Morton et al., 2004a), a heme-hemopexin utilization system (Cope et al., 1994), the Hup heme utilization protein (Morton et al., 2004b), and the HbpA heme-binding lipoprotein (Hanson et al., 1992; Morton et al., 2005). In addition, the Sap transporter proteins were recently shown to bind and transport heme across the bacterial inner membrane into the cytoplasm (Mason et al., 2011).
The sap (sensitivity to antimicrobial peptides) operon (sapABCDFZ) encodes a periplasmic substrate binding protein, SapA, two transmembrane permease proteins (SapB and SapC), two ATPase subunits (SapD and SapF) which drive substrate transport across the inner membrane, and SapZ of unknown function (Mason et al., 2003, 2005, 2006, 2011). Importantly, we demonstrated that the NTHI Sap ATP-binding-cassette (ABC) transporter is multifunctional, which not only mediates heme transport, but uptake of host innate antimicrobial peptides (AMPs), both functions that are critical for survival of NTHI in vivo (Mason et al., 2011; Shelton et al., 2011). The Sap system is wide spread in Gram-negative bacteria and is known to mediate innate immune resistance in strains of Salmonella, Proteus, Pasteurella, Escherichia coli (E. coli), Vibrio, Erwinia, Actinobacillus, and Haemophilus ducreyi (Groisman et al., 1992; Parra-Lopez et al., 1993, 1994; Groisman, 1994; Lopez-Solanilla et al., 1998; Nakamura et al., 1998; Chen et al., 2000; Harms et al., 2001; McCoy et al., 2001; Paustian et al., 2002; Mount et al., 2007, 2010; Lone et al., 2009). AMPs target bacterial membranes to disrupt the electrochemical gradient, cause leakage of cytoplasmic contents, and ultimately kill the microorganism (Sitaram and Nagaraj, 2002; Brogden, 2005). To survive these host insults, NTHI must effectively neutralize these pathogen-targeted bactericidal mechanisms. Modification of lipooligosaccharide (LOS) and Lipid A acylation serve as a first line of defense against these peptides (Lysenko et al., 2000; Starner et al., 2002). We extended these investigations and show that SapA directly binds AMPs, signals increased sap gene expression, and subsequently enhances a bacterial AMP-resistant phenotype (Mason et al., 2003, 2006, 2005). Further, SapA was essential for survival of NTHI in an experimental mammalian model of human airway disease (Mason et al., 2005, 2006). Importantly, SapA is required for neutralizing β-defensin activity in the mammalian host via transport and subsequent degradation of these peptides (Shelton et al., 2011).
The contribution of the Sap system to NTHI virulence is dependent upon transporter ATPase activity. We have previously shown that an NTHI strain deficient in one of the ATPase subunits, SapD, is susceptible to killing by recombinant chinchilla β-defensin-1, an ortholog of human β-defensin-3, (Mason et al., 2005, 2006) and is unable to maintain potassium homeostasis when the outer membrane is compromised. Moreover, SapD is required for persistence in a mammalian host (Mason et al., 2005, 2006). Although SapD and SapF are homologous proteins sharing ATPase function, the contribution of SapF for energizing transport of specific substrates through the Sap permease has not been described.
Here, we evaluated the transport of known Sap-dependent substrates in the absence of the SapF ATPase. We constructed an unmarked, nonpolar sapF deletion mutant strain and investigated the biological consequence of SapF deficiency on Sap-mediated AMP resistance, heme utilization, community architecture, and NTHI persistence in a mammalian animal model. We demonstrated that AMP resistance was independent of SapF function when NTHI was exposed to physiological levels of AMPs, concentrations previously shown to be lethal for the sapA and sapD mutants (Mason et al., 2006, 2011). Further, SapF was required for heme-iron utilization and influences the extent of biofilm architecture and community development. Importantly, SapF was essential for persistence of NTHI in the middle ear and nasopharynx. Together, these data further support a critical multifunctional role for the Sap complex in transport of substrates that are essential for robust growth and subversion of host innate immune effectors. Moreover, this study demonstrates that two homologous ATPase subunits encoded within the same operon can serve independent functions.
Materials and Methods
Generation of a sapF Deletion Mutant Strain
NTHI strain 86-028NP is a minimally passaged clinical isolate obtained from Nationwide Children's Hospital which has been extensively characterized in the chinchilla model of OM and has been sequenced and annotated (Harrison et al., 2005, 2007). The parental NTHI strain 86-028NP::rpsLA128G is a streptomycin resistant strain constructed as previously described (Carruthers et al., 2012). Construction of an unmarked, non-polar deletion mutant of the sapF gene was performed by the recombineering strategy as previously described (Tracy et al., 2008; Shelton et al., 2011). Briefly, primers 5′ TTCACTAAAAATCCACCAACCA 3′ and 5′ TGGACGTA TAAAAATTGTAGATGG 3′ were used to amplify sapF and 1 kb of the flanking DNA both 5′ and 3′ to sapF. The subsequent amplicon was ligated into the pGEM-T Easy vector (pAV001) and transformed into E. coli strain DY380. In parallel, primers, 5′ AAAACAACCGCCACCCCTTTTATACTTAATTGCAAAGG AAATGAATAATGATTCCGGGGATCCGTCGACC 3′ and 5′ AAAGGGACGAGTGTTCATTTTTCCTTTCC TTAGTGAGTGTTTTTGTCTTTTGTAGGCTGGAGCTGCTTCG 3′, each containing 50 base pairs of DNA homologous to the 5′ and 3′ ends of the sapF gene were used to amplify the spec-rpsL cassette from pRSM2832. This amplicon was then electroporated into strain E. coli DY380/pAV001 to form strain DY380/pAV002, in which the sapF gene in pAV001, has been replaced by the cassette. The plasmid pAV002 was then used to transform NTHI 86-028NP::rpsLA128G and transformants were selected by growth on spectinomycin-containing chocolate II agar plates. To generate a nonpolar deletion mutant, the sapF mutant was transformed with plasmid pRSM2947, grown at 32°C, and FLP expression was induced using anhydrotetracycline. The cells were cured of the plasmid by growth at 37°C.
Construction of sapF Complement
Complementation of the sapF mutant was constructed as previously described (Mason et al., 2006). Briefly sapF was amplified from NTHI 86-028NP and ligated downstream the sap promoter into pSpec1 and transformed into Haemophilus strain Rd. A spectinomycin resistant clone was isolated and the plasmid stored as pSWS001. This plasmid was then transformed into NTHI 86-028NP::rpsLA128GΔsapF and a spectinomycin resistant complemented strain was stored as NTHI 86-028NP::rpsLA128GΔsapF/pSWS001.
Construction of GFP Reporter Strains
Green fluorescent protein (GFP) reporter strains were engineered to monitor morphological plasticity phenotypes in the sapF deletion mutant strain and the parental strain NTHI 86-028NP::rpsLA128G. An EcoRI/SphI restriction fragment was removed from pRSM2211 (Mason et al., 2003). The fragment contains the NTHI OMP P2 promoter cloned upstream the gfpmut3 gene. The P2:gfp fusion was cloned into the EcoR1/SphI site of pSpec1, constructed as previously described (Mason et al., 2006) and the plasmid saved as pGM1.1. This plasmid was then transformed into NTHI 86-028NP generating strain RSM2713. The plasmid (pGM1.1) was isolated from RSM2713 and used to transform parental strain NTHI 86-028NP::rpsLA128G and the ΔsapF strain, generating strain NTHI 86-028NP::rpsLA128G/pGM1.1 and NTHI 86-028NP::rpsLA128GΔsapF/pGM1.1, respectively.
Growth Conditions
NTHI was grown on chocolate II agar (Becton Dickinson, Sparks, MD), in brain heart infusion broth supplemented with 2 μg heme/mL and 1 μg NAD/mL (supplemented brain-heart infusion; sBHI) or in defined iron-source (DIS) medium supplemented with 0, 2, or 20 μg heme/mL at 37°C in 5% CO2, prepared as previously described (Mason et al., 2006). To monitor the requirement of the SapF ATPase on heme import and growth after heme starvation, the bacterial strains were grown overnight on chocolate agar at 37°C and 5% CO2, sub-cultured into nitric acid-washed 15 mL round bottom glass tubes containing pre-warmed DIS medium with either 0 or 2 μg heme/mL and grown for 16 h to deplete internal stores of heme for use in subsequent experiments, as previously described (Mason et al., 2011). Heme-iron utilization was monitored in the iron-starved or non-starved cultures by measuring bacterial growth every hour for 16 h using the Synergy HT kinetic plate reader (BioTek, Bath, UK) as previously described (Mason et al., 2011). Number of viable bacteria was confirmed by serial dilution and plating to determine the colony forming units (CFU) per mL of culture.
Bactericidal Assays
Bactericidal assays were performed to evaluate AMP resistance as previously described (Mason et al., 2005). Briefly, bacteria were grown overnight on chocolate II agar plates, inoculated into 5 mL of pre-warmed sBHI medium, grown for 3 h at 37°C in 5% CO2, and then diluted with sBHI to 108 CFU/mL. The bacteria were further diluted 1:100 in 10 mM sodium phosphate (pH 7.4) containing 1% sBHI to 106 CFU/mL. In a 96-well polypropylene plate (Falcon), 90 μL of 10 mM sodium phosphate containing 1% sBHI was added to each well. Human β-defensin (hBD-3, PeproTech) or human cathelicidin LL-37 (Phoenix Pharmaceuticals) was serially diluted in the wells so that each well retained 90 μL of the appropriate concentration of peptide. 10 μL bacteria (104 CFU) were added to each well, and the plate was incubated for 1 hour at 37°C in 5% CO2. Following incubation, bacterial concentrations were determined by serial dilution and growth on chocolate II agar, and data were expressed as CFU/mL of viable bacteria.
Biofilm Growth on Abiotic Surface
Starved and non-starved cultures were used to determine the influence of heme starvation and SapF deficiency on biofilm structure and organization. Starved and non-starved cultures were matched for CFU/mL in DIS medium and 2.5 × 105 bacteria were added to each well of a Nunc Lab-Tek8-well chamber slide containing DIS medium supplemented with or without 2 μg heme/mL. The chamber slide was then incubated at 37°C with 5% CO2 and biofilms were grown for 48 hours with a media change at 24 hours. The biofilms were stained with BacLite™ LIVE/DEAD stain (Molecular Probes, Grand Island, NY) for the strains not containing the pGM1.1 GFP reporter plasmid. Biofilm structure and organization were imaged with an Axiovert 200M inverted epifluorescence microscope equipped with the Apotome attachment for improved fluorescence resolution and an Axiocam MRM CCD camera (Carl Zeiss Inc., Thornwood, NY). Experiments were repeated in triplicate and representative samples are shown.
Animal Modeling
Healthy adult chinchillas (Chinchilla lanigera) purchased from Rauscher's Chinchilla ranch (LaRue, OH) with no evidence of middle ear infection were used to assess the biological impact of SapF function. Chinchillas were anesthetized with 2 mg xylazine/kg (Fort Dodge Animal Health, Fort Dodge, IA) and 10 mg ketamine/kg (Phoenix Scientific Inc., St. Joseph, MO) and 86-028NP::rpsLA128G or 86-028NP::rpsLA128GΔsapF were inoculated transbullarly into the middle ear (86-028NP::rpsLA128G= 4.19 × 103; 86-028NP::rpsLA128G ΔsapF = 8.34 × 103) of a chinchilla or via passive inhalation for nasopharyngeal colonization (86-028NP::rpsLA128G = 1.34 × 108; 86-028NP::rpsLA128GΔsapF = 2.05 × 108). At the indicated days post-inoculation, middle ear effusions were collected by epitympanic taps through the superior bullae, and directly obtained from the inferior bullae. Nasopharyngeal lavage fluids were obtained by passive inhalation of 500 μL of pyrogen free sterile saline into one naris and collection of the lavage through the other naris as the liquid was exhaled. Middle ear infection was monitored daily by video otoscopy for signs of inflammation and middle ear fluid accumulation. All animal experiments were performed using accredited conditions for animal welfare approved by the Institutional Animal Care and Use Committee (Welfare Assurance Number A3544-01) at The Research Institute at Nationwide Children's Hospital, AR08-00027.
Results
SapF is not Required for AMP Resistance
We have previously shown that the SapA and SapD Sap transporter subunits are required for resistance to the cationic peptide (r)cBD-1 (the chinchilla homologue of human β-defensin-3, hBD-3; Mason et al., 2005, 2006). We further demonstrated that AMP transport to the bacterial cytoplasm, as a mechanism to decrease periplasmic accumulation of these toxic peptides, was dependent upon SapBC permease production (Shelton et al., 2011). We hypothesized that the two ATPase subunits, SapD and SapF, would energize transport of AMP molecules through the SapBC permease. To this end, we exposed the unmarked, nonpolar sapF mutant or the marked, non-polar sapD mutant (Mason et al., 2006) to increasing concentrations of the AMPs hBD-3 or LL-37 and monitored bacterial survival compared to that of the parental (NTHI 86-028NP::rpsLA128G) or wild type (NTHI 86-028NP) strains, respectively. In marked contrast to the SapD ATPase, whereby the sapD mutant was susceptible to both hBD-3 and LL-37 (Figures 1B,D, respectively), we observed that the SapF ATPase was dispensable for conferring the resistant phenotype at all concentrations of LL-37 and at hBD-3 concentrations less than 2 μg/ml (microgram/ml) (Figures 1C,A, respectively). These data suggest that each ATPase may mediate unique functions with the Sap ABC transporter.
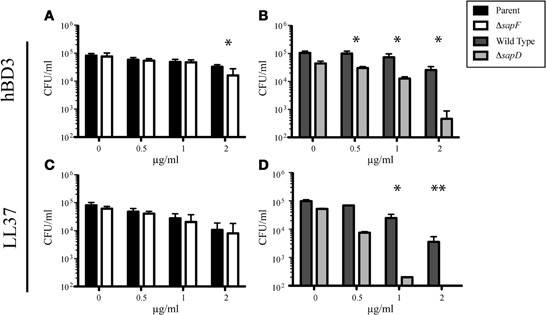
Figure 1. The SapF ATPase is dispensable for resistance to the AMPs hBD-3 and LL-37. The parent strain and the SapF ATPase-deficient strain (A and C) or the Wild Type and SapD-ATPase deficient strain (B and D) were incubated with increasing concentrations of hBD-3 (A and B) or LL-37 (C and D) for 1 h and then plated on chocolate agar for enumeration of viable bacteria. Data represents the average number of surviving bacteria from at least three independent experiments. Error bars represent the standard deviation of the mean of cultures assayed in triplicate from six (n = 6; A and C) or three (n = 3; B and D) independent experiments. *Denotes p < 0.05 by a two-tailed Student's t-test; **Denotes p < 0.01.
The sapF ATPase is Required for Heme-Iron Utilization
We previously demonstrated that transport of heme across the inner membrane requires the Sap transporter permease, SapBC, and the SapA periplasmic binding protein (Mason et al., 2011). For technical reasons associated with the requirement of SapD for potassium homeostasis, we have not been able to determine the contribution of SapD to heme-iron transport in NTHI. However, as SapF is not required for potassium homeostasis (data not shown), we determined whether SapF was required for the energy-dependent transport of heme. Thus, we predicted that loss of SapF would impair heme-iron utilization, measured by changes in bacterial growth, particularly under conditions of heme-iron starvation. We first monitored whether loss of SapF altered growth kinetics of NTHI compared to that of the parental strain when grown in DIS medium. We observed a delay in entrance into logarithmic growth in the sapF mutant compared to that of the parent strain (Figure 2A). However, the SapF-deficient strain maintained a similar growth rate compared to that of the parent strain throughout the remainder of the culture period, suggesting a compensatory mechanism for heme acquisition in the absence of SapF protein or a requirement of SapF for heme acquisition during the transition from stationary phase to logarithmic growth.
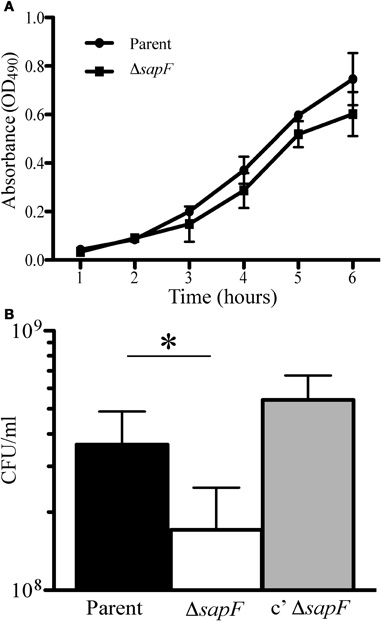
Figure 2. Growth characteristics in the absence of the SapF ATPase. The parent strain (circle) and SapF-deficient strain (square) were grown overnight on chocolate agar, diluted in DIS medium, matched for optical density (OD490) and inoculated into DIS medium containing 2 μg heme/mL (A). Growth was assessed by measuring the OD490 every hour for 5 h. Error bars represent the standard deviation of the mean of cultures assayed in triplicate from three independent assays (n = 2). (B) The parental strain (black bar), the SapF-deficient strain (white bar), and the complemented SapF-deficient strain (gray bar) were incubated in DIS medium in the absence of heme-iron for 16 hours. The resultant cultures were matched for optical density (OD490) and plated for number of CFU. *denotes p < 0.05 by a two-tailed Student's t-test (N = 5).
In order to monitor whether environmental heme-iron starvation would provoke more severe growth defects, the parent and sapF mutant strains were depleted of internal stores of heme-iron by incubation of each strain in DIS medium without heme for 16 h. Bacteria were matched for optical density and viable bacteria were enumerated by plating and overnight growth on agar (See “Material and Methods”; Figure 2B). We observed a statistically significant decrease (∼2-fold) in the number of colonies observed in the absence of SapF when compared to that of the parental strain (Figure 2B). The addition of sapF in trans restored the number of colonies produced by the sapF mutant (Figure 2B).
We next investigated whether SapF was required for recovery from heme-iron depletion of internal stores during planktonic growth (Figure 3). To this end, the parent strain, a SapF-deficient strain, or a SapF-complemented mutant strain was heme-iron starved for 16 h in DIS medium depleted of iron sources. The bacteria were then cultured in the presence of one of two concentrations of heme for 16 h (Figure 3). The heme-iron-starved parent strain failed to grow when cultured in heme-iron-depleted medium, which indicated that the microorganism was sufficiently starved of all internal iron stores (Figure 3A). Growth was restored to the parental strain when cultured in the presence of either concentration of heme-iron (Figures 3B,C). Although no growth defects were observed with the SapF-deficient strain during heme-iron starvation (data not shown), the bacteria demonstrated some modest growth but failed to regain logarithmic growth in the presence of heme-iron (Figures 3B,C). Complementation of the sapF mutation fully restored the growth of the heme-iron-starved microorganism compared to that of the parent strain (Figures 3B,C). These data indicated that SapF is critical for heme acquisition and utilization in NTHI.
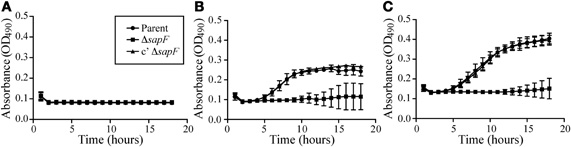
Figure 3. Role of SapF ATPase in heme utilization. The parent strain (circle), the SapF-deficient strain (square), and the complemented SapF-deficient strain (triangle) were iron-starved for 16 h in DIS medium in the absence of heme, matched for CFU, and subcultured in DIS medium containing 0 (A), 2 (B), or 20 μg heme/mL (C). Growth was assessed by measuring the increase in OD490 every 30 min for 16 h. Error bars represent the standard deviation of the mean of cultures assayed in triplicate from three independent experiments (n = 3).
SapF Deficiency Promotes Robust Biofilm Architecture
A biofilm is a highly organized, multicellular network of bacteria encased in a matrix which results in a robust architecture. Iron availability has been shown to influence biofilm formation (Johnson et al., 2005, 2008; Hindre et al., 2008), and iron availability is limited in the human host. Given the observation that SapF contributes to the uptake and utilization of heme-iron, we investigated whether the loss of SapF ATPase likewise resulted in changes to NTHI biofilm architecture. Biofilm development was monitored for the sapF mutant strain or the parental strain when cultured in a heme-iron-replete medium (Figure 4). The parental strain forms a biofilm that primarily consists of a mat-like architecture with sporadic spiked short towers (Figures 4A,C). In marked contrast, loss of SapF resulted in more robust, thicker tower formation (Figures 4B,D). The differences in biofilm architecture were more readily observed when comparing the three-dimensional surface plot produced from a series of fluorescent optical sections (Figures 4C,D). The altered architecture in the absence of SapF suggests that environmental stresses (i.e., heme-iron depletion) may enhance the overall complexity of NTHI biofilms.
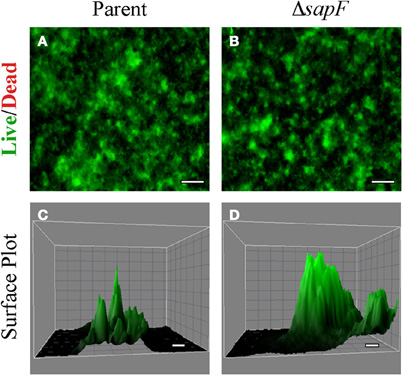
Figure 4. Biofilm density and architecture in the absence of SapF. The parental strain (A, C) or the SapF-deficient strain (B, D) were grown in DIS medium without heme on a glass chamber slide for 48 h. Biofilm structure was assessed by fluorescence microscopy following staining with the Live/Dead viability stain (see “Materials and Methods”). The surface area of a representative portion of the biofilm was generated from a series of optical sections using ImageJ software for the parental strain (C), and the SapF-deficient strain (D). Scale bar = 100 μm (A, B) or 10 μm (C, D).
Since we demonstrated that SapF activity was essential for heme acquisition and utilization by NTHI, we monitored NTHI biofilm formation and architecture following heme-iron starvation in the presence or absence of heme for 24 h. Biofilms that develop following restoration of heme-iron to depleted cultures revealed phenotypic changes in the sapF mutant strain (Figure 5B) compared to the parent strain (Figure 5A). The biofilms were more punctate in appearance, reflective of bacterial aggregates that appeared to be primed for bacterial growth in the presence of heme. Complementation of the sapF mutation restored a smoother biofilm phenotype similar to that of the parental strain (compare Figures 5A,C). These alterations in biofilm architecture and bacterial morphology dependent upon SapF function were exaggerated under heme-iron starvation conditions (Figure 5E). Changes in bacterial morphology were also evident in the parental and complemented mutant strains when subjected to heme-iron starvation conditions. These data suggest that heme-iron starvation influences changes in NTHI biofilm architecture.
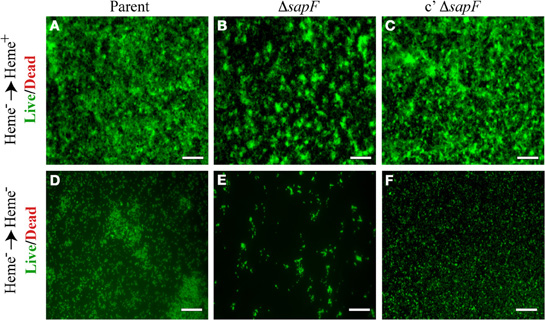
Figure 5. Biofilm formation in the presence and absence of heme-iron. The parental strain (A, D), SapF-deficient strain (B, E), and complemented Sap-deficient strain (C, F) were cultured in DIS medium without heme for 16 hours, matched for OD490 units, and then sub-cultured into glass chamber slides with DIS medium containing 0 or 2 μg heme/mL for 24 h. Biofilms were stained with the Live/Dead viability stain and the structure was visualized by fluorescence microscopy (all images 20 × magnification). Scale bar = 100 μm.
SapF Deficiency Provokes Morphological Plasticity of NTHI
We became interested in investigating the punctate structures (Figure 5E) and overall architecture of the more robust tower structures (Figure 4D) formed in the absence of SapF. In contrast to the mat-like architecture of the parental strain (Figure 6A), microscopic evaluation at higher magnification revealed that the towers were formed from individuals that appear in an open lacework architecture (Figure 6B). In many cases the live dead stain revealed a chain of nucleoids that appear to be floating in the culture medium but securely attached to the biofilm proper. The apparently suspended nucleoid chain suggested that the NTHI may be forming bacterial chains or filaments. To better characterize the interbacterial interactions of the NTHI individuals within the biofilms, we transitioned to the use of an NTHI strain that constitutively produces GFP to visualize the intact cytoplasm for better differentiation of overall cell length. We thus sought to examine alterations in biofilm growth and architecture when both the parent and mutant strain were depleted of internal heme-iron stores. The environmentally depleted bacteria were placed into a biofilm chamber slide for 24 h of culture in the absence of environmental heme-iron (Figures 6C,D). Depletion of heme-iron resulted in the inhibition of cell division leading to the formation of elongated NTHI with a continuous cytoplasm as indicated by the uniform distribution of GFP (Figures 6C,D). These filamentous NTHI formed patches or clusters that were readily observed even at lower magnification (Figure 5E). This phenotypic change in morphology was accentuated in the sapF mutant strain as a majority of viable bacteria were filamentous compared to the parent strain (compare Figures 6C,D). These data demonstrate morphological plasticity occurs under conditions of heme-iron limitation. The morphological plasticity observed here can account for the decreases in CFU observed above (Figure 2B). In addition to the filamentous form being enriched at early stages in biofilm development in response to heme-iron limitation, filamentous NTHI were also observed throughout the towers formed in the absence of SapF (Figure 6B). Bacterial filamentation in response to environmental stressors has been shown to provide selective survival advantages to the bacterium (Justice et al., 2008; Horvath et al., 2011). Since the host microenvironment is initially iron deplete, the ability to filament may provide NTHI a survival advantage during colonization and biofilm formation.
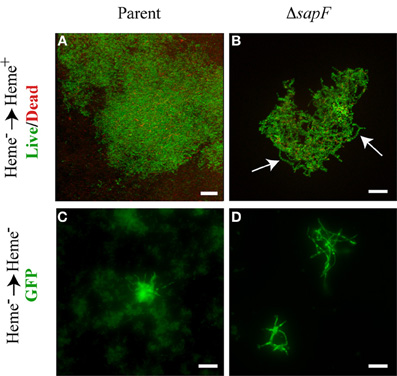
Figure 6. Morphological plasticity in the absence of SapF. The parental strain (A, C) and the SapF-deficient strain (B, D) were cultured in DIS medium without heme for 16 h, matched for OD490 units, and then sub cultured into chamber slides (1 × 106 CFU/mL starved) with DIS medium with (A, B) or without heme (C, D) for 48 h. (A, B) Biofilms were stained with the Live/Dead viability stain and colony morphology was analyzed by fluorescence microscopy. 3D rendering of sequential optical sections of the SapF-deficient strain demonstrating lace-like architecture composed of chaining nucleoids (B) as compared to mat-like architecture of the parent (A). The parent strain (86-028NP rpsLA128G/pGM1.1; C) forms fewer filamentous individuals while the SapF-deficient strain (86-028NP rpsLA128GΔsapF/pGM1.1; D) readily filaments under heme-starved conditions. All images were taken under 63 × magnification. Scale bar = 10 μm.
Loss of SapF-Mediated Heme-Iron Acquisition Attenuates NTHI Persistence in vivo
Recently, multiple functions of the Sap transporter have been revealed through the use of strains deficient in the periplasmic binding protein, SapA (Mason et al., 2006, 2011; Shelton et al., 2011). We have demonstrated here that SapF was required for heme utilization, and coordination of NTHI biofilm architecture, traits that are important to establish acute OM. We have also previously shown that resistance to AMPs is essential for persistence of NTHI in vivo (Shelton et al., 2011). Therefore, we took advantage of our observation that SapF is dispensable for antimicrobial resistance to elucidate the contribution of heme-iron utilization toward colonization of the chinchilla nasopharynx and middle ear (Figure 7). We hypothesized that heme-iron utilization is a key trait required for survival in a host environment. In order to test this hypothesis we infected four chinchillas each with the parental NTHI strain or the sapF mutant strain and monitored bacterial survival in the nasopharynx and middle ear over a 14-day period. The absence of SapF dramatically attenuates persistence in the nasopharynx (Figures 7A,C) and middle ear (Figures 7B,D) as evidenced by the more rapid clearance of NTHI by day 14 (Chi square p = 0.04 and p = 0.05, respectively). The sapF mutant was cleared from the nasopharynx in three of the four animals on day 14 (Figures 7A,C). Moreover, the sapF mutant was cleared from the middle ear in five of eight ears on day 14 (Figures 7B,D). In contrast, the majority of the ears infected with the parental isolate had ongoing infections two weeks following bacterial inoculation. It is clear that a loss of SapF alters NTHI persistence and that SapF is required for normal long-term infection of the middle ear space, suggesting that heme-iron utilization is required for persistence in the host.
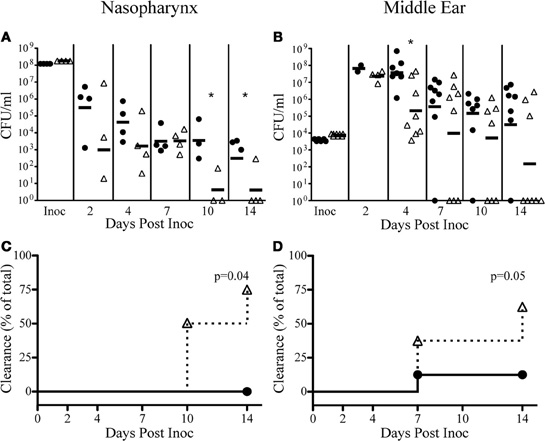
Figure 7. SapF is required for colonization of a mammalian host and infection of the middle ear. The parent strain (black circles) or SapF-deficient strain (white triangles) were inoculated via passive inhalation for nasopharyngeal colonization (A) and transbullarly into the middle ear of a chinchilla (B). At the indicated days post-inoculation, nasopharyngeal lavage or middle ear fluids were obtained and bacterial persistence was enumerated. The SapF-deficient strain was significantly attenuated in the nasopharynx (days 10 and 14) and in the middle ear (day 4) and was cleared more rapidly from the nasopharynx (3 of 4 animals; A, C) and the middle ear (4 of 7 ears; B, D), compared to the parent strain (Chi square p = 0.04 and p = 0.05, respectively). Each cohort contained four animals [nasopharynx: n = 4; middle ear: n = 8 (left and right ears)]. *Denotes p < 0.05 by a two-tailed Mann–Whitney U-test.
Discussion
We previously described distinct functions for the Sap transporter in Haemophilus growth and survival, yet we currently do not understand how individual transporter protein components coordinate and cooperate to confer multiple biological functions. In this study, we evaluated the biological relevance for SapF ATPase function in NTHI heme-iron utilization, AMP susceptibility, community architecture, and virulence. We demonstrated that SapF functions to acquire heme but is dispensable for AMP resistance. Interestingly, although the sap operon encodes SapD and SapF ATPase proteins that share homology and are thus predicted to provide energy dependence for similar biological functions, previous results indicate that SapD functions to impart unique and separate functions of the transporter complex (Mason et al., 2006). SapD-deficient NTHI is sensitive to physiological concentrations of AMPs [0.5–2.0 μg/mL in nasal secretions from healthy donors; (Groisman, 1994)] whereas SapF activity was dispensable. Further, SapD-deficient NTHI was unable to support critical uptake of potassium upon compromise of the outer membrane (Mason et al., 2006). We propose that SapD energizes potassium uptake via the TrkA/H potassium importer (Mason et al., 2006). The inability of SapF to compensate and restore AMP resistance and potassium uptake indicated unique functional roles for this protein. In addition, we demonstrated that the SapF ATPase was critical for acquisition and utilization of the essential iron-containing compound heme and for establishment of community architecture. Given that the two ATPase subunits facilitate transport of unique substrates, we were able to independently elucidate the contribution of AMP resistance and heme-iron utilization in the acute stages of NTHI disease. The SapD and SapF ATPase components confer unique functions that may contribute in a complementary manner to sustain bacterial infection in the host. The ability of each subunit to energize transport of independent substrates, in part, explains the presence of two highly homologous proteins encoded within the same operon. Given these differences, we are currently investigating the substrate specificity for each of the two Sap permease components.
The evidence presented here supports a critical role for SapF in heme utilization by NTHI. In heme-iron replete medium, the SapF-deficient strain was delayed in entry to logarithmic growth from stationary phase, yet the rate of growth was comparable between both strains during exponential growth. Following heme-iron starvation of both strains, however, SapF was required for recovery from depletion of heme-iron from internal stores. These data suggest that SapF functions to increase internal stores of heme-iron within the cells which can be utilized under starvation conditions, such as those of host micro-environments.
We demonstrated that SapF contributes to biofilm community architecture. The altered architecture observed in the absence of SapF suggests that environmental stresses (i.e., heme-iron depletion) may enhance the overall complexity of NTHI biofilms. Indeed, we observed accentuated morphological plasticity of NTHI due to heme-iron limitation. Morphological plasticity has been observed in a number of pathogens and, where tested, is essential for persistence in a host environment (Justice et al., 2006, 2008). We hypothesize that morphological plasticity of NTHI will result in formation of communities that contain large bacterial filaments that likely contribute to biofilm architecture and stability in the host. This morphological plasticity is particularly relevant considering that in vivo NTHI must sense and respond to host iron sequestration (iron-deplete middle ear) and colonize the mucosal epithelium. It is well accepted that the human host is an iron-restricted environment (Ong et al., 2006), in fact, host “nutritional immunity” serves to “starve” the microorganism of essential iron sources and thus inhibit bacterial growth. Subsequent host inflammation and disease progression should increase iron availability for the microorganism, to thus augment community development. Using scanning electron microscopy, we have observed NTHI filamentation of the parental isolate and a Sap transporter mutant strain that is genetically starved of heme-iron (unpublished). We hypothesize that iron starvation at the host cell interface may trigger morphological plasticity (i.e., filamentation) of Haemophilus. These data are consistent with a recent observation of Haemophilus filaments adhered to middle ear mucosa epithelium of chinchillas (Leroy et al., 2007). We hypothesize that morphological plasticity associated with heme-iron restriction (genetically induced in the sap mutant strain) dictates alterations in NTHI communities that alter host response efforts. Further elucidation of the contribution of morphological plasticity to community architecture and virulence are currently under investigation.
We have previously demonstrated that SapA, the Sap transporter substrate binding protein, directly counters host AMP lethality in vivo (Shelton et al., 2011). This finding is significant in that it establishes a direct link between Sap transporter function and NTHI survival in the host. Our data clearly support multiple functions for the Sap transporter, and we have begun to identify Sap associated proteins required for each specific function. Here, we determined that SapF does not mediate AMP resistance, yet functions to maintain heme homeostasis between NTHI and the microenvironment. Use of these mutant strains in vivo will begin to define the impact of AMP exposure and iron availability on NTHI pathogenesis. There are multiple gaps in our knowledge of this dynamic interplay, particularly how nutritional limitation dictates microbial virulence factors that equip survival of bacteria in the host, both as benign nasopharyngeal commensal or opportunistic trespasser of privileged sterile sites. Elucidation of NTHI responses to nutrient limitation has great likelihood to serve as a prototype for other mucosal opportunists. The Sap transporter appears to contribute a vital role in these survival methods and understanding the role of each of its component proteins could result in targeted neutralization of the transporter to prevent disease.
In summary, we have elucidated the roles of specific Sap transporter components by mutational analysis of independent constituents of the transporter complex. Through this approach we have also observed changes at the single cell level that contribute to the ability of NTHI to persist in the host. Further investigations are needed to better understand how each of the subunits cooperates for transport of multiple substrates.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by an R21 A1070825 from NIAID/NIH to Kevin M. Mason.
References
Banin, E., Vasil, M. L., and Greenberg, E. P. (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 102, 11076–11081.
Brogden, K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250.
Carruthers, M. D., Tracy, E. N., Dickson, A. C., Ganser, K. B., Munson, R. S. Jr., and Bakaletz, L. O. (2012). Nontypeable Haemophilus influenzae type IV pili: biological roles of the products encoded by the pil and com operons. J. Bacteriol. 194. [Epub ahead of print].
Chen, H. Y., Weng, S. F., and Lin, J. W. (2000). Identification and analysis of the sap genes from Vibrio fischeri belonging to the ATP-binding cassette gene family required for peptide transport and resistance to antimicrobial peptides. Biochem. Biophys. Res. Commun. 269, 743–748.
Cope, L. D., Thomas, S. E., Latimer, J. L., Slaughter, C. A., Muller-Eberhard, U., and Hansen, E. J. (1994). The 100 kDa haem:haemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol. Microbiol. 13, 863–873.
Ehrlich, G. D., Veeh, R., Wang, X., Costerton, J. W., Hayes, J. D., Hu, F. Z., Daigle, B. J., Ehrlich, M. D., and Post, J. C. (2002). Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 287, 1710–1715.
Groisman, E. A. (1994). How bacteria resist killing by host-defense peptides. Trends Microbiol. 2, 444–449.
Groisman, E. A., Parra-Lopez, C., Salcedo, M., Lipps, C. J., and Heffron, F. (1992). Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. U.S.A. 89, 11939–11943.
Hall-Stoodley, L., Hu, F. Z., Gieseke, A., Nistico, L., Nguyen, D., Hayes, J., Forbes, M., Greenberg, D. P., Dice, B., Burrows, A., Wackym, P. A., Stoodley, P., Post, J. C., Ehrlich, G. D., and Kerschner, J. E. (2006). Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296, 202–211.
Hanson, M. S., Slaughter, C., and Hansen, E. J. (1992). The hbpA gene of Haemophilus influenzae type b encodes a heme-binding lipoprotein conserved among heme-dependent Haemophilus species. Infect. Immun. 60, 2257–2266.
Harms, C., Domoto, Y., Celik, C., Rahe, E., Stumpe, S., Schmid, R., Nakamura, T., and Bakker, E. P. (2001). Identification of the ABC protein SapD as the subunit that confers ATP dependence to the K+-uptake systems TrkH and TrkG from Escherichia coli K-12. Microbiology 147, 2991–3003.
Harrison, A., Dyer, D. W., Gillaspy, A., Ray, W. C., Mungur, R., Carson, M. B., Zhong, H., Gipson, J., Gipson, M., Johnson, L. S., Lewis, L., Bakaletz, L. O., and Munson, R. S. Jr. (2005). Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 187, 4627–4636.
Harrison, A., Ray, W. C., Baker, B. D., Armbruster, D. W., Bakaletz, L. O., and Munson, R. S. Jr. (2007). The OxyR regulon in nontypeable Haemophilus influenzae. J. Bacteriol. 189, 1004–1012.
Hindre, T., Bruggemann, H., Buchrieser, C., and Hechard, Y. (2008). Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology 154, 30–41.
Horvath, D. J. Jr., Li, B., Casper, T., Partida-Sanchez, S., Hunstad, D. A., Hultgren, S. J., and Justice, S. S. (2011). Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli. Microbes Infect. 13, 426–437.
Johnson, M., Cockayne, A., and Morrissey, J. A. (2008). Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect. Immun. 76, 1756–1765.
Johnson, M., Cockayne, A., Williams, P. H., and Morrissey, J. A. (2005). Iron-responsive regulation of biofilm formation in staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 187, 8211–8215.
Jurcisek, J. A., and Bakaletz, L. O. (2007). Biofilms formed by nontypeable Haemophilus influenzaein vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. 189, 3868–3875.
Jurcisek, J. A., Bookwalter, J. E., Baker, B. D., Fernandez, S., Novotny, L. A., Munson, R. S. Jr., and Bakaletz, L. O. (2007). The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol. Microbiol. 65, 1288–1299.
Justice, S. S., Hunstad, D. A., Cegelski, L., and Hultgren, S. J. (2008). Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6, 162–168.
Justice, S. S., Hunstad, D. A., Seed, P. C., and Hultgren, S. J. (2006). Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc. Natl. Acad. Sci. U.S.A. 103, 19884–19889.
Klein, J. O. (1997). Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr. Infect. Dis. J. 16, S5–S8.
Leroy, M., Cabral, H., Figueira, M., Bouchet, V., Huot, H., Ram, S., Pelton, S. I., and Goldstein, R. (2007). Multiple consecutive lavage samplings reveal greater burden of disease and provide direct access to the nontypeable Haemophilus influenzae biofilm in experimental otitis media. Infect. Immun. 75, 4158–4172.
Lone, A. G., Deslandes, V., Nash, J. H., Jacques, M., and Macinnes, J. I. (2009). Modulation of gene expression in Actinobacillus pleuropneumoniae exposed to bronchoalveolar fluid. PLoS One 4:e6139. doi: 10.1371/journal.pone.0006139
Lopez-Solanilla, E., Garcia-Olmedo, F., and Rodriguez-Palenzuela, P. (1998). Inactivation of the sapA to sapF locus of Erwinia chrysanthemi reveals common features in plant and animal bacterial pathogenesis. Plant Cell 10, 917–924.
Lysenko, E. S., Gould, J., Bals, R., Wilson, J. M., and Weiser, J. N. (2000). Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect. Immun. 68, 1664–1671.
Maciver, I., Latimer, J. L., Liem, H. H., Muller-Eberhard, U., Hrkal, Z., and Hansen, E. J. (1996). Identification of an outer membrane protein involved in utilization of hemoglobin-haptoglobin complexes by nontypeable Haemophilus influenzae. Infect. Immun. 64, 3703–3712.
Markel, T. A., Crisostomo, P. R., Wang, M., Herring, C. M., Meldrum, K. K., Lillemoe, K. D., and Meldrum, D. R. (2007). The struggle for iron: gastrointestinal microbes modulate the host immune response during infection. J. Leukoc. Biol. 81, 393–400.
Mason, K. M., Bruggeman, M. E., Munson, R. S., and Bakaletz, L. O. (2006). The non-typeable Haemophilus influenzae Sap transporter provides a mechanism of antimicrobial peptide resistance and SapD-dependent potassium acquisition. Mol. Microbiol. 62, 1357–1372.
Mason, K. M., Munson, R. S. Jr., and Bakaletz, L. O. (2003). Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 71, 3454–3462.
Mason, K. M., Munson, R. S. Jr., and Bakaletz, L. O. (2005). A mutation in the sap operon attenuates survival of nontypeable Haemophilus influenzae in a chinchilla model of otitis media. Infect. Immun. 73, 599–608.
Mason, K. M., Raffel, F. K., Ray, W. C., and Bakaletz, L. O. (2011). Heme utilization by nontypeable Haemophilus influenzae is essential and dependent on Sap transporter function. J. Bacteriol. 193, 2527–2535.
McCoy, A. J., Liu, H., Falla, T. J., and Gunn, J. S. (2001). Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 45, 2030–2037.
Moreau-Marquis, S., O'Toole, G. A., and Stanton, B. A. (2009). Tobramycin and FDA-approved iron chelators eliminate Pseudomonas aeruginosa biofilms on cystic fibrosis cells. Am. J. Respir. Cell Mol. Biol. 41, 305–313.
Morton, D. J., Bakaletz, L. O., Jurcisek, J. A., VanWagoner, T. M., Seale, T. W., Whitby, P. W., and Stull, T. L. (2004). Reduced severity of middle ear infection caused by nontypeable Haemophilus influenzae lacking the hemoglobin/hemoglobin-haptoglobin binding proteins (Hgp) in a chinchilla model of otitis media. Microb. Pathog. 36, 25–33.
Morton, D. J., Madore, L. L., Smith, A., Vanwagoner, T. M., Seale, T. W., Whitby, P. W., and Stull, T. L. (2005). The heme-binding lipoprotein (HbpA) of Haemophilus influenzae: role in heme utilization. FEMS Microbiol. Lett. 253, 193–199.
Morton, D. J., Smith, A., Ren, Z., Madore, L. L., VanWagoner, T. M., Seale, T. W., Whitby, P. W., and Stull, T. L. (2004). Identification of a haem-utilization protein (Hup) in Haemophilus influenzae. Microbiology 150, 3923–3933.
Mount, K. L., Townsend, C. A., and Bauer, M. E. (2007). Haemophilus ducreyi is resistant to human antimicrobial peptides. Antimicrob. Agents Chemother. 51, 3391–3393.
Mount, K. L., Townsend, C. A., Rinker, S. D., Gu, X., Fortney, K. R., Zwickl, B. W., Janowicz, D. M., Spinola, S. M., Katz, B. P., and Bauer, M. E. (2010). Haemophilus ducreyi SapA contributes to cathelicidin resistance and virulence in humans. Infect. Immun. 78, 1176–1184.
Murphy, T. F. (2003). Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 16, 129–134.
Nakamura, T., Yamamuro, N., Stumpe, S., Unemoto, T., and Bakker, E. P. (1998). Cloning of the trkAH gene cluster and characterization of the Trk K+-uptake system of Vibrio alginolyticus. Microbiology 144, 2281–2289.
Ong, S. T., Ho, J. Z., Ho, B., and Ding, J. L. (2006). Iron-withholding strategy in innate immunity. Immunobiology 211, 295–314.
Parra-Lopez, C., Baer, M. T., and Groisman, E. A. (1993). Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12, 4053–4062.
Parra-Lopez, C., Lin, R., Aspedon, A., and Groisman, E. A. (1994). A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 13, 3964–3972.
Paustian, M. L., May, B. J., Cao, D., Boley, D., and Kapur, V. (2002). Transcriptional response of Pasteurella multocida to defined iron sources. J. Bacteriol. 184, 6714–6720.
Post, J. C., Hiller, N. L., Nistico, L., Stoodley, P., and Ehrlich, G. D. (2007). The role of biofilms in otolaryngologic infections: update 2007. Curr. Opin. Otolaryngol. Head Neck Surg. 15, 347–351.
Ren, Z., Jin, H., Morton, D. J., and Stull, T. L. (1998). hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect. Immun. 66, 4733–4741.
Roman, F., Canton, R., Perez-Vazquez, M., Baquero, F., and Campos, J. (2004). Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 42, 1450–1459.
Sethi, S., and Murphy, T. F. (2001). Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14, 336–363.
Shelton, C. L., Raffel, F. K., Beatty, W. L., Johnson, S. M., and Mason, K. M. (2011). Sap transporter mediated import and subsequent degradation of antimicrobial peptides in Haemophilus. PLoS Pathog. 7:e1002360. doi: 10.1371/journal.ppat.1002360
Sitaram, N., and Nagaraj, R. (2002). Host-defense antimicrobial peptides: importance of structure for activity. Curr. Pharm. Des. 8, 727–742.
St Geme, J. W. 3rd. (2000). The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 19, S41–S50.
Starner, T. D., Swords, W. E., Apicella, M. A., and McCray, P. B. Jr. (2002). Susceptibility of nontypeable Haemophilus influenzae to human beta-defensins is influenced by lipooligosaccharide acylation. Infect. Immun. 70, 5287–5289.
Stojiljkovic, I., and Perkins-Balding, D. (2002). Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21, 281–295.
Tracy, E., Ye, F., Baker, B. D., and Munson, R. S. Jr. (2008). Construction of non-polar mutants in Haemophilus influenzae using FLP recombinase technology. BMC Mol. Biol. 9, 101.
Keywords: otitis media, biofilm, NTHI, SapF, heme, iron
Citation: Vogel AR, Szelestey BR, Raffel FK, Sharpe SW, Gearinger RL, Justice SS and Mason KM (2012) SapF-mediated heme-iron utilization enhances persistence and coordinates biofilm architecture of Haemophilus. Front. Cell. Inf. Microbio. 2:42. doi: 10.3389/fcimb.2012.00042
Received: 15 December 2011; Paper pending published: 12 January 2012;
Accepted: 14 March 2012; Published online: 03 April 2012.
Edited by:
W. Edward Swords, Wake Forest University Health Sciences, USAReviewed by:
Eric Krukonis, University of Michigan, USAVincent J. Starai, The University of Georgia, USA
Copyright: © 2012 Vogel, Szelestey, Raffel, Sharpe, Gearinger, Justice and Mason. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Kevin M. Mason, Center for Microbial Pathogenesis, The Research Institute at Nationwide Children's Hospital, Rm. W531, The Ohio State University College of Medicine, 700 Children's Dr, Columbus, OH 43205-2696, USA. e-mail: kevin.mason@nationwidechildrens.org