Role and regulation of heme iron acquisition in gram-negative pathogens
- Department of Biology, University of Richmond, Richmond, VA, USA
Bacteria that reside in animal tissues and/or cells must acquire iron from their host. However, almost all of the host iron is sequestered in iron-containing compounds and proteins, the majority of which is found within heme molecules. Thus, likely iron sources for bacterial pathogens (and non-pathogenic symbionts) are free heme and heme-containing proteins. Furthermore, the cellular location of the bacterial within the host (intra or extracellular) influences the amount and nature of the iron containing compounds available for transport. The low level of free iron in the host, coupled with the presence of numerous different heme sources, has resulted in a wide range of high-affinity iron acquisition strategies within bacteria. However, since excess iron and heme are toxic to bacteria, expression of these acquisition systems is highly regulated. Precise expression in the correct host environment at the appropriate times enables heme iron acquisitions systems to contribute to the growth of bacterial pathogens within the host. This mini-review will highlight some of the recent findings in these areas for gram-negative pathogens.
Introduction
Almost all living organisms require iron for growth. One notable exception is the Lyme disease pathogen, Borrelia burgdorferi, which uses manganese in place of iron (Posey and Gherardini, 2000). Iron is critical for a wide range of cellular functions; however, high levels of iron are toxic because iron catalyzes the formation of reactive oxygen species, and iron acquisition by cells is highly regulated as a result. In the complex interaction between human host and bacterium, iron plays a critical role. Free ferric (Fe3+) iron is poorly soluble in aerobic conditions at neutral pHs; however, ferrous (Fe2+) iron is much more soluble. Additionally, the host sequesters free iron in iron binding proteins (such as ferritin, transferrin, lactoferrin) and in heme and hemoproteins to prevent iron toxicity and to withhold nutrients from pathogens, thereby limiting pathogen growth. Thus, free iron is not readily available to the bacterial pathogen inside the host. Pathogens have evolved numerous mechanisms to capture this limited supply of free iron and iron from host iron proteins. Since the type of iron available varies depending on the location of the pathogen within the human host and since pathogens occupy a wide variety of host niches, there is a diversity of iron acquisition mechanisms employed by both intracellular and extracellular pathogens. This mini-review focuses on acquisition of iron in gram-negative pathogens from one of the most abundant sources—host heme.
Availability of Heme and Heme-Containing Molecules in the Human Host
Approximately 70% of the iron in the human body is within heme, a heterocyclic organic ring called porphryin covalently bound to one ferrous iron atom (Bridges and Seligman, 1995). Heme is critical for functions including oxygen transport, enzymatic reactions, and cellular respiration. Heme is synthesized in almost all human cell types (the majority in erythroid cells, and to a lesser extent in hepatocytes) and can be obtained from the diet (reviewed in Hamza and Dailey, 2012).
Heme is an essential biomolecule; however, excess free heme is toxic to cells due to its lipophilic nature, lipid peroxidation capacity, and ability to catalyze the production of reactive oxygen species (reviewed in Anzaldi and Skaar, 2010). Thus, over 95% of the heme is bound to proteins (hemoproteins), the majority of which are intracellular (Bridges and Seligman, 1995). The intracellular free heme pool is approximately 0.1 μM, which is less than 0.1% of total cellular heme (Granick et al., 1975). The majority of heme in the human body (~67%) is in hemoglobin, which is primarily found in erythrocytes (Bridges and Seligman, 1995). Other major hemoproteins include myoglobin and cytochromes. Recently, additional hemoproteins have been described, including cytoglobin and neuroglobin, which appear to play a role in oxygen homeostasis/oxygen stress (Liu et al., 2012b; Watanabe et al., 2012; Storz et al., 2013). Additional heme binding proteins exist that are most likely important in scaffolds for synthesis and scavenging heme. The existence of heme chaperones for incorporating heme into apo-hemoproteins has been proposed, but such proteins have yet to been identified in humans (Severance and Hamza, 2009). All of these proteins represent potential heme sources for intracellular pathogens.
Although the majority is intracellular, limited amounts of heme can be found extracellular and thus available to extracellular pathogens. One of the major locations for extracellular heme is in blood hemoglobin (estimated to be 80–800 nM in serum) (Schryvers and Stojiljkovic, 1999). Hemoglobin from lysed erythrocytes is bound by haptoglobin for eventual recycling by macrophage and hepatocytes (Tolosano et al., 2010). Free heme, from damaged hemoglobin, is bound by serum hemopexin and, to a lesser extent, serum albumin. In the gut, dietary heme may be bioavailable to bacteria, either free or complexed with hemopexin. Heme levels are thought to be low in the respiratory track; however, since the heme auxotroph Haemophilus influenzae can live in this environment, there must be enough heme to support bacterial growth (Fournier et al., 2011). The urogenital track has varying amounts of heme: the bladder, urethra, and male genital track likely have low heme levels; however, there may be high heme levels in the female urogenital track during menses (Schryvers and Stojiljkovic, 1999). Finally, even in environments where heme is typically low, heme and hemoproteins are released by cells damaged during infection.
Bacterial Heme Transporters and Liberation of Iron from Heme
Host microenvironments that have potential heme sources have selected for bacteria with high-affinity heme transport systems which locate and transport heme into the bacterial cell. Heme auxotrophs can use the intact heme for insertion into bacterial hemoproteins. Additionally for both heme prototrophs and autotrophs alike, the iron can be extracted from the heme for other uses (e.g., building Fe-S cluster proteins). Most commonly, there is direct uptake of heme by a cell surface receptor which binds heme or host hemoproteins. A variation of this method includes bipartite systems in which a lipoprotein facilitates heme or hemoproteins binding to the cell surface receptor (Lewis et al., 1998, 2006). Alternatively, some pathogens produce hemophores, small secreted proteins that capture free heme or heme bound to host hemoproteins and then deliver this heme to bacterial surface receptors (Cescau et al., 2007).
There are over 30 well-characterized outer membrane heme receptors that transport heme in gram-negative pathogens, although there are many more putative receptors in genomic databases (Table 1). The overall structure of these proteins includes a membrane spanning beta-barrel with extracellular loops that bind to free heme, host hemoproteins, or bacterial hemophores (reviewed in Wilks and Burkhard, 2007). Most are characterized by the presence of FRAP/NPNL domains with a conserved histidine residue that coordinates that heme (Stojiljkovic et al., 1995), although there are reports of heme transporters lacking some of these elements (e.g., PhuR from Pseudomonas aeruginosa) suggesting that there are other motifs for heme coordination in outer membrane heme transporters (Tong and Guo, 2009). The energy for heme transport is transduced from the inner to the outer membrane using the TonB/ExbB/ExbD system (reviewed in Krewulak and Vogel, 2011). Thus, all heme outer membrane transporters have a characteristic “TonB box” motif, through which the receptor interacts with TonB. Given the presence of multiple hemoproteins as potential iron sources, there are at least two strategies for bacteria to optimize access to heme iron (Figure 1). Some species have multiple receptors, presumably for different hemoproteins or for expression in different host environments (e.g., Haemophilus influenza). Other species have one outer membrane receptor capable of binding to multiple hemoproteins (e.g., Yersinia enterocolitica HemR), suggesting the recognition is at the level of the heme molecule (Stojiljkovic and Hantke, 1992; Bracken et al., 1999).
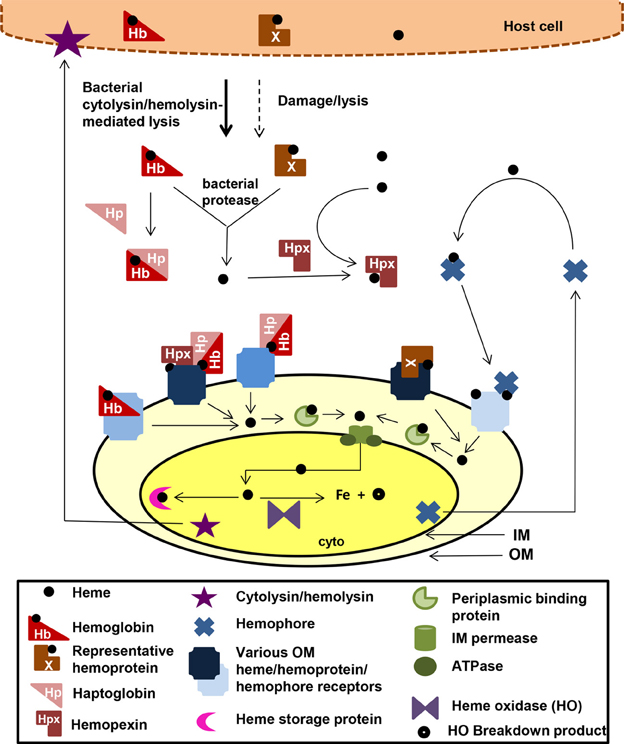
Figure 1. Mechanisms for heme iron acquisition from the host by gram-negative bacteria. Bacteria factors damage host cells releasing heme, Hb, and other hemoproteins. Additionally, secreted bacterial hemophores capture host heme. Extracellular host Hb and heme may be bound by host Hp and Hpx, respectively. A bacterium could acquire iron from these host heme sources using one or more TonB-dependent outer membrane (OM) receptors for these heme compounds, which transport the heme through the outer membrane into the periplasm. Some OM receptors are specific for one molecule, whereas others have a broad specificity for multiple hemoproteins. Transport though the periplasmic and across the inner membrane is facilitated by ABC transport systems (green). Inside the bacterium, the heme is degraded using heme oxidases or stored in heme storage protein. Intracellular pathogens would have access to host heme and hemoproteins via similar mechanisms. cyto, bacterial cytoplasm; IM, bacterial inner/cytoplasmic membrane; OM, bacterial outer membrane. Although all the OMR are TonB-dependent, TonB is not shown in the figure.
Once the heme molecule has been transported through the outer membrane receptor, ABC transport systems then transport heme though the periplasm, across the inner membrane, and into the cytoplasm (Table 1 and Figure 1). Each ABC transport system consists of a high-affinity periplasmic ligand-binding protein which shuttles heme through the periplasm, two subunits of a cytoplasmic membrane permease, and a peripheral membrane ATPase that supplies the energy for transport. Although there is low sequence homology among the approximately 50 identified periplasmic heme binding proteins, all but one has a conserved tyrosine which is believed to coordinate heme (Tong and Guo, 2009). Frequently, these ABC transporter genes are located in the same operon as or near outer membrane receptor genes; however, orphan ABC transporters that can transport heme exist (e.g., the E. coli DppABCD system, which also transports dipeptides) (Letoffe et al., 2006).
Upon entry into the bacterial cell, heme storage, transfer and degradation proteins sequester heme and facilitate extraction of the iron from heme (Table 1 and Figure 1). Bacterial proteins that sequester heme likely prevent heme from catalyzing the formation of reactive oxygen species [e.g., Shigella dysenteriae ShuS Wyckoff et al. (2005)]. Other cytoplasmic heme-binding proteins transfer heme to heme degradation proteins [e.g., Pseudomonas aeruginosa PhuS Lansky et al. (2006)]. Many pathogens contain homologues of mammalian heme oxygenases (HO), enzymes that cleave the heme to release the iron, generating biliverdin and CO as end products (e.g., Pseudomonas aeruginosa HO and Neisseria meningitidis HemO). Recently, new structural classes of HOs have been identified such as the “split-barrel fold class” in Helicobacter pylori (HugZ) and Campylobacter jejuni (ChuZ) (Guo et al., 2008; Zhang et al., 2011). Additional bacterial enzymes that degrade heme to liberate iron, but release different end products than those released by classical HOs, have been identified. For example, MhuD in Mycobacterium tuberculosis cleaves heme to release the iron, generating a novel tetrapyrrole product of called mycobilin, but not CO (Nambu et al., 2013).
For pathogens that can transport heme, the ability to increase the local concentration of heme and/or hemoproteins would be advantageous for growth in the host. Production of cytolysins/hemolysins that lyse cells releasing hemoproteins is common in almost all extracellular and facultative intracellular pathogens that use heme as an iron source. Additionally, some pathogens secrete proteases that degrade hemoproteins to release heme. For example, Porphyromonas gingivalis produces hemolysins to lyse cells and proteases called gingipans that have hemaglutin domains and degrade hemoproteins (Chu et al., 1991; Sroka et al., 2001). Alternatively, some bacteria secrete hemophores, small, secreted proteins that capture free heme or heme bound to host hemoproteins and that deliver the heme to bacterial cells. There are several distinct families of hemophores, which share little to no sequence similarity, suggesting convergent evolution of this strategy for increasing local heme concentration (Table 1; Figure 1). The first class of hemophores identified was the HasA group, initially characterized in Serratia marcescens (Letoffe et al., 1994). HasA captures heme, using conserved His32 and Tyr75 residues, and relays it to the outer membrane receptor HasR for transport. Homologues of the HasA/HasR system have only been found in gram-negative bacteria including Yersinia pestis (Rossi et al., 2001) and Pseudomonas aeruginosa (Ochsner et al., 2000). A second type of hemophore, only found in Haemophilus influenza, is HxuA, which captures heme from hemopexin, and the released heme is transported into the cell by outer membrane heme transporters (Fournier et al., 2011).
Regulation of Expression of Heme Iron Acquisition Genes by Iron, Heme and Other Stimuli
Most genes encoding components of heme iron acquisition systems are not transcribed in iron-replete condition because (1) high-affinity heme iron acquisition is generally not needed and, thus, would be energetically wasteful and (2) excess iron is cytotoxic. One of the most common mechanisms of regulation of heme iron acquisition system expression by iron levels utilizes iron-responsive transcriptional regulators that repress transcription of high-affinity iron acquisition systems when iron is plentiful. The prototypical example is Fur (ferric uptake regulation). In the classical model of iron-repression, Fe-Fur binds to a DNA sequence called the Fur-box in promoters of many high-affinity iron acquisition genes. Fe-Fur occupation of the promoter prevents RNA polymerase binding, thereby repressing transcription. When iron levels decrease, the Fe-Fur equilibrium shifts, Apo-Fur cannot bind to the Fur-box, and transcription occurs [for review Carpenter et al. (2009)]. DtxR and IdeR are iron responsive regulators with similar functions to Fur, and most heme acquisition genes are regulated by repressor proteins from the Fur or DtxR families (Table 1).
Not only is excess iron toxic to bacteria, but heme can also be cytotoxic due to its ability to catalyze the formation of reactive oxygen species, its peroxidase activity, and its lipophilic nature which disrupts cell membranes. Thus, for these reasons and for energetic reasons similar to those for iron regulation, expression of a subset of heme iron acquisition systems is regulated by heme levels in some pathogens. In Bartonella quintana, transcription of the hut operon increases when heme concentrations are lower than required for optimal growth, but decreases at very high heme concentrations. The decrease in expression is predicted to be mediated by the heme-responsive Irr transcriptional regulator, which is only found in some alpha-proteobacteria (Parrow et al., 2009). Bordetella pertussis employs an extracytoplasmic function σ factor (ECF) called HurI and its cognate anti-sigma factor HurR to modulate transcription of the bhuRSTUV heme uptake operon by heme though a mechanisms in which iron regulation and heme regulation converge. In low iron, Fur repression of hurIR is relieved; however, HurI is inactive because it is bound by HurR when heme is absent. Heme binding by BhuB alleviates HurR repression of HurI activity, and HurI can activate transcription of the bhuRSTUV operon. (Vanderpool and Armstrong, 2003, 2004). In the presence of heme, the Vibrio vulnificus LysR-family transcriptional regulator HupR increases transcription of the Fur-regulated outer membrane heme receptor gene hupA (Litwin and Quackenbush, 2001). In Pseudomonas aeruginosa, transcription of the phu operon is up-regulated via an uncharacterized, but Fur-independent, mechanism (Kaur et al., 2009). Regulatory patterns like these enable expression of heme iron acquisition systems when some heme is available for transport and/or prevent expression of the systems when heme levels are too high. It is unclear why more heme iron acquisition systems are not under such control; however, most expression studies have not formally tested this possibility and, thus, this mode of regulation may be more widespread than reported.
In addition to heme/iron levels, other host-related environmental stimuli may fine-tune expression of heme iron acquisition genes, allowing integration of the iron/heme conditions with other physiological and environmental signals. The cyclic AMP receptor protein, which actives transcription when glucose levels are low, activates expression of Vibrio vulnificus hupA (Oh et al., 2009). In Shigella dysenteriae and pathogenic E. coli, expression of the Fur-regulated outer membrane heme receptor genes shuA and chuA increases at 37°C due to post-transcriptional regulation by the 5' untranslated region of these genes (Kouse et al., 2013). The Fur-regulated Yersinia pestis hasRADEB and Vibrio vulnificus hupA genes have increased expression at 37°C and 40°C, respectively, as compared to lower temperatures (Rossi et al., 2001; Oh et al., 2009). phuR and hasA expression in Pseudomonas aeruginosa and hmuRY expression in Porphyromonas gingivalis are quorum/cell density-regulated (Arevalo-Ferro et al., 2003; Wu et al., 2009). Haemophilus influenzae and Neisseria meningitidis overlay phase variation on expression of heme acquisition systems, perhaps to counteract the host response to immunogenic OMPs (Ren et al., 1999; Richardson and Stojiljkovic, 1999). Finally, the pathogen's niche may change during the course of infection due to the interaction between host and pathogen and the movement of the pathogen through the host, and available iron sources may change as a result. Tissue specific expression of heme receptors has been show in several pathogens including Yersinia enterocolitica, where hemR expression is higher in spleen and peritoneum, as compared to liver and intestinal lumen. Furthermore, peritoneum expression of hemR is higher than in in vitro iron-limited media suggesting there are additional host specific signals besides low iron that allow for maximal hemR expression (Jacobi et al., 2001). Finally, there are examples of transcriptional regulation by other regulators suggesting there are more regulatory signals and integration with other regulatory pathways to be discovered.
In summary, each pathogen fine-tunes expression of heme iron acquisition genes to generate the appropriate physiological response for each environmental niche. This response is characterized by particular host heme iron sources/levels, total iron levels, other environmental inputs, and the phylogenetic history of the pathogen. Thus, there are varying patterns of regulation of heme iron acquisition system and regulation of the expression of these systems sometimes overlaps with other global regulatory circuits, creating intricate regulatory pathways in some pathogens. Alternatively, regulation of heme acquisition systems in other pathogens may be relatively simple (e.g., only regulated by an iron-responsive transcriptional regulator) because the pathogen is in a stable environment with low free iron and access to heme.
Conclusions and Future Outlook
Although much is known about heme transport mechanisms and their regulation in many of well-studied pathogens, these topics have not been investigated as extensively in less-common and emerging pathogens, leaving the potential for novel discoveries. Furthermore, the possible fates of the transported heme molecule within the bacterial cell are just beginning to be clarified fully. Additional families of heme iron acquisition and utilization proteins may be waiting to be identified using biochemical (e.g., heme binding assays), genetic (e.g., complementation of E. coli heme mutants), and bioinformatic (e.g., mining expression databases for Fur- or iron-regulated genes and searching for heme binding motifs in proteins databases) approaches. Defining the role of each particular heme iron acquisition system in virulence is ongoing for many pathogens, but has been complicated by the presence of redundant systems in some pathogens and/or the use of certain systems in just one niche in the host. Thus, deletions of particular heme iron acquisition genes do not always show an effect in all animal models. It is clear, however that in many pathogens there is a role for some heme iron acquisition proteins, demonstrating the importance of heme for pathogenesis (Henderson and Payne, 1994; Morton et al., 2004, 2007, 2009a; Palyada et al., 2004; Domenech et al., 2005; Brickman et al., 2006; Hagan and Mobley, 2009). A more complete description of heme acquisition and utilization in human pathogens may serve as a reference point for understanding iron acquisition in non-pathogenic symbiotic bacteria that reside in humans and other animals, an area that is currently under-investigated. With respect to gene regulation, expression of the genes encoding most heme iron acquisition systems increases when iron is low due to alleviation of transcriptional repression by iron-responsive transcriptional repressors. However, whether heme levels and/or other regulatory RNAs or proteins modulate this expression further has not been examined for many of these genes.
Pathogens and their human hosts have evolved together, and as a consequence, there is a complex interplay between sequestration of iron from the pathogen by the host and elaboration of mechanism to capture that iron by the pathogen. From the host side, human hemoglobin is quite variable in amino acid sequence; thus, individuals may have differing susceptibility to pathogens due to differences in the ability of the pathogen to bind hemoglobin to access the heme (Pishchany and Skaar, 2012). Thus, bacteria pathogen acquisition of heme iron could have been a driving force for hemoglobin evolution. From the pathogen side, the fact that most heme is intracellular and bound to hemoproteins may have been a selective pressure for intracellular growth and protease/hemolysin production in pathogen evolution. Furthermore, heme acquisition genes have been found associated with mobile genetic elements in some pathogens (e.g., Neisseria meningitidis and Shigella dysenteriae), suggesting potential for rapid spread of these genes via horizontal gene transfer (Wyckoff et al., 1998; Kahler et al., 2001).
Author Contributions
Laura Runyen-Janecky conceived and wrote the entire manuscript.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aduse-Opoku, J., Slaney, J. M., Rangarajan, M., Muir, J., Young, K. A., and Curtis, M. A. (1997). The Tla protein of Porphyromonas gingivalis W50: a homolog of the RI protease precursor (PrpRI) is an outer membrane receptor required for growth on low levels of hemin. J. Bacteriol. 179, 4778–4788.
Agiato, L. A., and Dyer, D. W. (1992). Siderophore production and membrane alterations by Bordetella pertussis in response to iron starvation. Infect. Immun. 60, 117–123.
Antunes, L. C., Imperi, F., Towner, K. J., and Visca, P. (2011). Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 162, 279–284. doi: 10.1016/j.resmic.2010.10.010
Anzaldi, L. L., and Skaar, E. P. (2010). Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78, 4977–4989. doi: 10.1128/IAI.00613-10
Arevalo-Ferro, C., Hentzer, M., Reil, G., Gorg, A., Kjelleberg, S., Givskov, M., et al. (2003). Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5, 1350–1369. doi: 10.1046/j.1462-2920.2003.00532.x
Asuthkar, S., Velineni, S., Stadlmann, J., Altmann, F., and Sritharan, M. (2007). Expression and characterization of an iron-regulated hemin-binding protein, HbpA, from Leptospira interrogans serovar Lai. Infect. Immun. 75, 4582–4591. doi: 10.1128/IAI.00324-07
Bracken, C. S., Baer, M. T., Abdur-Rashid, A., Helms, W., and Stojiljkovic, I. (1999). Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J. Bacteriol. 181, 6063–6072.
Brickman, T. J., Anderson, M. T., and Armstrong, S. K. (2007). Bordetella iron transport and virulence. Biometals 20, 303–322. doi: 10.1007/s10534-006-9031-1
Brickman, T. J., Vanderpool, C. K., and Armstrong, S. K. (2006). Heme transport contributes to in vivo fitness of Bordetella pertussis during primary infection in mice. Infect. Immun. 74, 1741–1744. doi: 10.1128/IAI.74.3.1741-1744.2006
Bridges, K. R., and Seligman, P. A. (1995). “Disorders of iron metabolism,” in Blood: Principles and Practice of Hematology, eds R. I. Handin, S. E. Lux, and T. P. Stossel (Philadelphia, PA: JB Lippincott Company), 1433–1472.
Carpenter, B. M., Whitmire, J. M., and Merrell, D. S. (2009). This is not your mother's repressor: the complex role of fur in pathogenesis. Infect. Immun. 77, 2590–2601. doi: 10.1128/IAI.00116-09
Carrizo-Chavez, M. A., Cruz-Castaneda, A., and Olivares-Trejo Jde, J. (2012). The frpB1 gene of Helicobacter pylori is regulated by iron and encodes a membrane protein capable of binding haem and haemoglobin. FEBS Lett. 586, 875–879. doi: 10.1016/j.febslet.2012.02.015
Cescau, S., Cwerman, H., Letoffe, S., Delepelaire, P., Wandersman, C., and Biville, F. (2007). Heme acquisition by hemophores. Biometals 20, 603–613. doi: 10.1007/s10534-006-9050-y
Chen, C. J., Elkins, C., and Sparling, P. F. (1998). Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect. Immun. 66, 987–993.
Chen, C. J., Sparling, P. F., Lewis, L. A., Dyer, D. W., and Elkins, C. (1996). Identification and purification of a hemoglobin-binding outer membrane protein from Neisseria gonorrhoeae. Infect. Immun. 64, 5008–5014.
Chu, L., Bramanti, T. E., Ebersole, J. L., and Holt, S. C. (1991). Hemolytic activity in the periodontopathogen Porphyromonas gingivalis: kinetics of enzyme release and localization. Infect. Immun. 59, 1932–1940.
Cope, L. D., Thomas, S. E., Latimer, J. L., Slaughter, C. A., Muller-Eberhard, U., and Hansen, E. J. (1994). The 100 kDa haem:haemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol. Microbiol. 13, 863–873. doi: 10.1111/j.1365-2958.1994.tb00478.x
Cope, L. D., Yogev, R., Muller-Eberhard, U., and Hansen, E. J. (1995). A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177, 2644–2653.
Cornelis, P., Matthijs, S., and Van Oeffelen, L. (2009). Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22, 15–22. doi: 10.1007/s10534-008-9193-0
Dashper, S. G., Hendtlass, A., Slakeski, N., Jackson, C., Cross, K. J., Brownfield, L., et al. (2000). Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J. Bacteriol. 182, 6456–6462. doi: 10.1128/JB.182.22.6456-6462.2000
Datta, S., and Crosa, J. H. (2012). Identification and characterization of a novel outer membrane protein receptor required for hemin utilization in Vibrio vulnificus. Biometals 25, 275–283. doi: 10.1007/s10534-011-9501-y
Davies, B. W., Bogard, R. W., and Mekalanos, J. J. (2011). Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc. Natl. Acad. Sci. U.S.A. 108, 12467–12472. doi: 10.1073/pnas.1107894108
Domenech, P., Reed, M. B., and Barry, C. E. 3rd. (2005). Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73, 3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005
Dyer, D. W., West, E. P., and Sparling, P. F. (1987). Effects of serum carrier proteins on the growth of pathogenic neisseriae with heme-bound iron. Infect. Immun. 55, 2171–2175.
Fournier, C., Smith, A., and Delepelaire, P. (2011). Haem release from haemopexin by HxuA allows Haemophilus influenzae to escape host nutritional immunity. Mol. Microbiol. 80, 133–148. doi: 10.1111/j.1365-2958.2011.07562.x
Gao, J. L., Nguyen, K. A., and Hunter, N. (2010). Characterization of a hemophore-like protein from Porphyromonas gingivalis. J. Biol. Chem. 285, 40028–40038. doi: 10.1074/jbc.M110.163535
Gonzalez-Lopez, M. A., and Olivares-Trejo, J. J. (2009). The gene frpB2 of Helicobacter pylori encodes an hemoglobin-binding protein involved in iron acquisition. Biometals 22, 889–894. doi: 10.1007/s10534-009-9240-5
Graber, K. R., Smoot, L. M., and Actis, L. A. (1998). Expression of iron binding proteins and hemin binding activity in the dental pathogen Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 163, 135–142. doi: 10.1111/j.1574-6968.1998.tb13037.x
Granick, S., Sinclair, P., Sassa, S., and Grieninger, G. (1975). Effects by heme, insulin, and serum albumin on heme and protein synthesis in chick embryo liver cells cultured in a chemically defined medium, and a spectrofluorometric assay for porphyrin composition. J. Biol. Chem. 250, 9215–9225.
Grenier, D., Leduc, A., and Mayrand, D. (1997). Interaction between Actinobacillus actinomycetemcomitans lipopolysaccharides and human hemoglobin. FEMS Microbiol. Lett. 151, 77–81. doi: 10.1111/j.1574-6968.1997.tb10397.x
Guegan, R., Camadro, J. M., Saint Girons, I., and Picardeau, M. (2003). Leptospira spp. possess a complete haem biosynthetic pathway and are able to use exogenous haem sources. Mol. Microbiol. 49, 745–754. doi: 10.1046/j.1365-2958.2003.03589.x
Guo, Y., Guo, G., Mao, X., Zhang, W., Xiao, J., Tong, W., et al. (2008). Functional identification of HugZ, a heme oxygenase from Helicobacter pylori. BMC Microbiol. 8:226. doi: 10.1186/1471-2180-8-226
Hagan, E. C., and Mobley, H. L. (2009). Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 71, 79–91. doi: 10.1111/j.1365-2958.2008.06509.x
Hamza, I., and Dailey, H. A. (2012). One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim. Biophys. Acta 1823, 1617–1632. doi: 10.1016/j.bbamcr.2012.04.009
Harland, D. N., Dassa, E., Titball, R. W., Brown, K. A., and Atkins, H. S. (2007). ATP-binding cassette systems in Burkholderia pseudomallei and Burkholderia mallei. BMC Genomics 8:83. doi: 10.1186/1471-2164-8-83
Harrison, A., Santana, E. A., Szelestey, B. R., Newsom, D. E., White, P., and Mason, K. M. (2013). Ferric uptake regulator and its role in the pathogenesis of nontypeable Haemophilus influenzae. Infect. Immun. 81, 1221–1233. doi: 10.1128/IAI.01227-12
Hayashida, H., Poulsen, K., and Kilian, M. (2002). Differences in iron acquisition from human haemoglobin among strains of Actinobacillus actinomycetemcomitans. Microbiology 148, 3993–4001.
Helms, S. D., Oliver, J. D., and Travis, J. C. (1984). Role of heme compounds and haptoglobin in Vibrio vulnificus pathogenicity. Infect. Immun. 45, 345–349.
Henderson, D. P., and Payne, S. M. (1993). Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol. Microbiol. 7, 461–469. doi: 10.1111/j.1365-2958.1993.tb01137.x
Henderson, D. P., and Payne, S. M. (1994). Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun. 62, 5120–5125.
Ho, W. W., Li, H., Eakanunkul, S., Tong, Y., Wilks, A., Guo, M., et al. (2007). Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins. J. Biol. Chem. 282, 35796–35802. doi: 10.1074/jbc.M706761200
Hornung, J. M., Jones, H. A., and Perry, R. D. (1996). The hmu locus of Yersinia pestis is essential for utilization of free haemin and haem–protein complexes as iron sources. Mol. Microbiol. 20, 725–739. doi: 10.1111/j.1365-2958.1996.tb02512.x
Jackson, L. A., Ducey, T. F., Day, M. W., Zaitshik, J. B., Orvis, J., and Dyer, D. W. (2010). Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon. J. Bacteriol. 192, 77–85. doi: 10.1128/JB.00741-09
Jacobi, C. A., Gregor, S., Rakin, A., and Heesemann, J. (2001). Expression analysis of the yersiniabactin receptor gene fyuA and the heme receptor hemR of Yersinia enterocolitica in vitro and in vivo using the reporter genes for green fluorescent protein and luciferase. Infect. Immun. 69, 7772–7782. doi: 10.1128/IAI.69.12.7772-7782.2001
Jones, C. M., and Niederweis, M. (2011). Mycobacterium tuberculosis can utilize heme as an iron source. J. Bacteriol. 193, 1767–1770. doi: 10.1128/JB.01312-10
Kahler, C. M., Blum, E., Miller, Y. K., Ryan, D., Popovic, T., and Stephens, D. S. (2001). exl, an exchangeable genetic island in Neisseria meningitidis. Infect. Immun. 69, 1687–1696. doi: 10.1128/IAI.69.3.1687-1696.2001
Kaur, A. P., Lansky, I. B., and Wilks, A. (2009). The role of the cytoplasmic heme-binding protein (PhuS) of Pseudomonas aeruginosa in intracellular heme trafficking and iron homeostasis. J. Biol. Chem. 284, 56–66. doi: 10.1074/jbc.M806068200
Kouse, A. B., Righetti, F., Kortmann, J., Narberhaus, F., and Murphy, E. R. (2013). RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS ONE 8:e63781. doi: 10.1371/journal.pone.0063781
Krewulak, K. D., and Vogel, H. J. (2011). TonB or not TonB: is that the question. Biochem. Cell Biol. 89, 87–97. doi: 10.1139/O10-141
Kumar, P., Sannigrahi, S., and Tzeng, Y. L. (2012). The Neisseria meningitidis ZnuD zinc receptor contributes to interactions with epithelial cells and supports heme utilization when expressed in Escherichia coli. Infect. Immun. 80, 657–667. doi: 10.1128/IAI.05208-11
Kvitko, B. H., Goodyear, A., Propst, K. L., Dow, S. W., and Schweizer, H. P. (2012). Burkholderia pseudomallei known siderophores and hemin uptake are dispensable for lethal murine melioidosis. PLoS Negl. Trop. Dis. 6:e1715. doi: 10.1371/journal.pntd.0001715
Lansky, I. B., Lukat-Rodgers, G. S., Block, D., Rodgers, K. R., Ratliff, M., and Wilks, A. (2006). The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the delta-regioselective heme oxygenase. J. Biol. Chem. 281, 13652–13662. doi: 10.1074/jbc.M600824200
Law, D., and Kelly, J. (1995). Use of heme and hemoglobin by Escherichia coli O157 and other Shiga-like-toxin-producing E. coli serogroups. Infect. Immun. 63, 700–702.
Letoffe, S., Delepelaire, P., and Wandersman, C. (2006). The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 12891–12896. doi: 10.1073/pnas.0605440103
Letoffe, S., Ghigo, J. M., and Wandersman, C. (1994). Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc. Natl. Acad. Sci. U.S.A. 91, 9876–9880. doi: 10.1073/pnas.91.21.9876
Lewis, J. P. (2010). Metal uptake in host-pathogen interactions: role of iron in Porphyromonas gingivalis interactions with host organisms. Periodontol. 2000 52, 94–116. doi: 10.1111/j.1600-0757.2009.00329.x
Lewis, J. P., Plata, K., Yu, F., Rosato, A., and Anaya, C. (2006). Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology 152, 3367–3382. doi: 10.1099/mic.0.29011-0
Lewis, L. A., Sung, M. H., Gipson, M., Hartman, K., and Dyer, D. W. (1998). Transport of intact porphyrin by HpuAB, the hemoglobin-haptoglobin utilization system of Neisseria meningitidis. J. Bacteriol. 180, 6043–6047.
Lima, A., Zunino, P., D'Alessandro, B., and Piccini, C. (2007). An iron-regulated outer-membrane protein of Proteus mirabilis is a haem receptor that plays an important role in urinary tract infection and in in vivo growth. J. Med. Microbiol. 56, 1600–1607. doi: 10.1099/jmm.0.47320-0
Litwin, C. M., and Byrne, B. L. (1998). Cloning and characterization of an outer membrane protein of Vibrio vulnificus required for heme utilization: regulation of expression and determination of the gene sequence. Infect. Immun. 66, 3134–3141.
Litwin, C. M., and Quackenbush, J. (2001). Characterization of a Vibrio vulnificus LysR homologue, HupR, which regulates expression of the haem uptake outer membrane protein, HupA. Microb. Pathog. 31, 295–307. doi: 10.1006/mpat.2001.0472
Liu, M., Boulouis, H. J., and Biville, F. (2012a). Heme degrading protein HemS is involved in oxidative stress response of Bartonella henselae. PLoS ONE 7:e37630. doi: 10.1371/journal.pone.0037630
Liu, X., Follmer, D., Zweier, J. R., Huang, X., Hemann, C., Liu, K., et al. (2012b). Characterization of the function of cytoglobin as an oxygen-dependent regulator of nitric oxide concentration. Biochemistry 51, 5072–5082. doi: 10.1021/bi300291h
Liu, X., Olczak, T., Guo, H. C., Dixon, D. W., and Genco, C. A. (2006). Identification of amino acid residues involved in heme binding and hemoprotein utilization in the Porphyromonas gingivalis heme receptor HmuR. Infect. Immun. 74, 1222–1232. doi: 10.1128/IAI.74.2.1222-1232.2006
Mason, K. M., Raffel, F. K., Ray, W. C., and Bakaletz, L. O. (2011). Heme utilization by nontypeable Haemophilus influenzae is essential and dependent on Sap transporter function. J. Bacteriol. 193, 2527–2535. doi: 10.1128/JB.01313-10
Mey, A. R., and Payne, S. M. (2001). Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42, 835–849. doi: 10.1046/j.1365-2958.2001.02683.x
Mills, M., and Payne, S. M. (1995). Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J. Bacteriol. 177, 3004–3009.
Mills, M., and Payne, S. M. (1997). Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect. Immun. 65, 5358–5363.
Morton, D. J., Seale, T. W., Bakaletz, L. O., Jurcisek, J. A., Smith, A., Vanwagoner, T. M., et al. (2009a). The heme-binding protein (HbpA) of Haemophilus influenzae as a virulence determinant. Int. J. Med. Microbiol. 299, 479–488. doi: 10.1016/j.ijmm.2009.03.004
Morton, D. J., Seale, T. W., Vanwagoner, T. M., Whitby, P. W., and Stull, T. L. (2009b). The dppBCDF gene cluster of Haemophilus influenzae: role in heme utilization. BMC Res. Notes 2:166. doi: 10.1186/1756-0500-2-166
Morton, D. J., Seale, T. W., Madore, L. L., Vanwagoner, T. M., Whitby, P. W., and Stull, T. L. (2007). The haem-haemopexin utilization gene cluster (hxuCBA) as a virulence factor of Haemophilus influenzae. Microbiology 153, 215–224. doi: 10.1099/mic.0.2006/000190-0
Morton, D. J., Smith, A., Ren, Z., Madore, L. L., Vanwagoner, T. M., Seale, T. W., et al. (2004). Identification of a haem-utilization protein (Hup) in Haemophilus influenzae. Microbiology 150, 3923–3933. doi: 10.1099/mic.0.27238-0
Morton, D. J., Whitby, P. W., Jin, H., Ren, Z., and Stull, T. L. (1999). Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect. Immun. 67, 2729–2739.
Murray, G. L., Ellis, K. M., Lo, M., and Adler, B. (2008). Leptospira interrogans requires a functional heme oxygenase to scavenge iron from hemoglobin. Microbes Infect. 10, 791–797. doi: 10.1016/j.micinf.2008.04.010
Nambu, S., Matsui, T., Goulding, C. W., Takahashi, S., and Ikeda-Saito, M. (2013). A new way to degrade heme: the Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. J. Biol. Chem. 288, 10101–10109. doi: 10.1074/jbc.M112.448399
Occhino, D. A., Wyckoff, E. E., Henderson, D. P., Wrona, T. J., and Payne, S. M. (1998). Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29, 1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x
Ochsner, U. A., Johnson, Z., and Vasil, M. L. (2000). Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146 (Pt 1), 185–198.
Ochsner, U. A., and Vasil, M. L. (1996). Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. U.S.A. 93, 4409–4414. doi: 10.1073/pnas.93.9.4409
O'Connell, W. A., Hickey, E. K., and Cianciotto, N. P. (1996). A Legionella pneumophila gene that promotes hemin binding. Infect. Immun. 64, 842–848.
Oh, M. H., Lee, S. M., Lee, D. H., and Choi, S. H. (2009). Regulation of the Vibrio vulnificus hupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect. Immun. 77, 1208–1215. doi: 10.1128/IAI.01006-08
Olczak, T., Simpson, W., Liu, X., and Genco, C. A. (2005). Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol. Rev. 29, 119–144. doi: 10.1016/j.femsre.2004.09.001
Palyada, K., Threadgill, D., and Stintzi, A. (2004). Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186, 4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004
Parrow, N. L., Abbott, J., Lockwood, A. R., Battisti, J. M., and Minnick, M. F. (2009). Function, regulation, and transcriptional organization of the hemin utilization locus of Bartonella quintana. Infect. Immun. 77, 307–316. doi: 10.1128/IAI.01194-08
Perkins-Balding, D., Ratliff-Griffin, M., and Stojiljkovic, I. (2004). Iron transport systems in Neisseria meningitidis. Microbiol. Mol. Biol. Rev. 68, 154–171. doi: 10.1128/MMBR.68.1.154-171.2004
Perry, R. D. (2004). “Yersinia,” in Iron Transport in Bacteria, eds J. H. Crosa, A. R. Mey, and S. M. Payne (Washington, DC: ASM Press), 219–240.
Perry, R. D., and Brubaker, R. R. (1979). Accumulation of iron by yersiniae. J. Bacteriol. 137, 1290–1298.
Pickett, C. L., Auffenberg, T., Pesci, E. C., Sheen, V. L., and Jusuf, S. S. (1992). Iron acquisition and hemolysin production by Campylobacter jejuni. Infect. Immun. 60, 3872–3877.
Pishchany, G., and Skaar, E. P. (2012). Taste for blood: hemoglobin as a nutrient source for pathogens. PLoS Pathog. 8:e1002535. doi: 10.1371/journal.ppat.1002535
Posey, J. E., and Gherardini, F. C. (2000). Lack of a role for iron in the Lyme disease pathogen. Science 288, 1651–1653. doi: 10.1126/science.288.5471.1651
Raghu, B., Sarma, C. R., and Venkatesan, P. (1993). Effect of hemoglobin on the growth of mycobacteria and the production of siderophores. Indian J. Pathol. Microbiol. 36, 376–382.
Ratliff, M., Zhu, W., Deshmukh, R., Wilks, A., and Stojiljkovic, I. (2001). Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403. doi: 10.1128/JB.183.21.6394-6403.2001
Ren, Z., Jin, H., Whitby, P. W., Morton, D. J., and Stull, T. L. (1999). Role of CCAA nucleotide repeats in regulation of hemoglobin and hemoglobin-haptoglobin binding protein genes of Haemophilus influenzae. J. Bacteriol. 181, 5865–5870.
Rhodes, E. R., Menke, S., Shoemaker, C., Tomaras, A. P., McGillivary, G., and Actis, L. A. (2007). Iron acquisition in the dental pathogen Actinobacillus actinomycetemcomitans: what does it use as a source and how does it get this essential metal. Biometals 20, 365–377. doi: 10.1007/s10534-006-9058-3
Richardson, A. R., and Stojiljkovic, I. (1999). HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J. Bacteriol. 181, 2067–2074.
Ridley, K. A., Rock, J. D., Li, Y., and Ketley, J. M. (2006). Heme utilization in Campylobacter jejuni. J. Bacteriol. 188, 7862–7875. doi: 10.1128/JB.00994-06
Rossi, M. S., Fetherston, J. D., Letoffe, S., Carniel, E., Perry, R. D., and Ghigo, J. M. (2001). Identification and characterization of the hemophore-dependent heme acquisition system of Yersinia pestis. Infect. Immun. 69, 6707–6717. doi: 10.1128/IAI.69.11.6707-6717.2001
Sander, A., Kretzer, S., Bredt, W., Oberle, K., and Bereswill, S. (2000). Hemin-dependent growth and hemin binding of Bartonella henselae. FEMS Microbiol. Lett. 189, 55–59. doi: 10.1111/j.1574-6968.2000.tb09205.x
Schneider, S., Sharp, K. H., Barker, P. D., and Paoli, M. (2006). An induced fit conformational change underlies the binding mechanism of the heme transport proteobacteria-protein HemS. J. Biol. Chem. 281, 32606–32610. doi: 10.1074/jbc.M607516200
Schryvers, A. B., and Stojiljkovic, I. (1999). Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32, 1117–1123. doi: 10.1046/j.1365-2958.1999.01411.x
Senkovich, O., Ceaser, S., McGee, D. J., and Testerman, T. L. (2010). Unique host iron utilization mechanisms of Helicobacter pylori revealed with iron-deficient chemically defined media. Infect. Immun. 78, 1841–1849. doi: 10.1128/IAI.01258-09
Severance, S., and Hamza, I. (2009). Trafficking of heme and porphyrins in metazoa. Chem. Rev. 109, 4596–4616. doi: 10.1021/cr9001116
Shalom, G., Shaw, J. G., and Thomas, M. S. (2007). In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699. doi: 10.1099/mic.0.2007/006585-0
Shizukuishi, S., Tazaki, K., Inoshita, E., Kataoka, K., Hanioka, T., and Amano, A. (1995). Effect of concentration of compounds containing iron on the growth of Porphyromonas gingivalis. FEMS Microbiol. Lett. 131, 313–317. doi: 10.1111/j.1574-6968.1995.tb07793.x
Shoji, M., Shibata, Y., Shiroza, T., Yukitake, H., Peng, B., Chen, Y. Y., et al. (2010). Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis: its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 10:152. doi: 10.1186/1471-2180-10-152
Simpson, W., Olczak, T., and Genco, C. A. (2000). Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 182, 5737–5748. doi: 10.1128/JB.182.20.5737-5748.2000
Sroka, A., Sztukowska, M., Potempa, J., Travis, J., and Genco, C. A. (2001). Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J. Bacteriol. 183, 5609–5616. doi: 10.1128/JB.183.19.5609-5616.2001
Stoebner, J. A., and Payne, S. M. (1988). Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect. Immun. 56, 2891–2895.
Stojiljkovic, I., and Hantke, K. (1992). Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 11, 4359–4367.
Stojiljkovic, I., Hwa, V., de Saint Martin, L., O'Gaora, P., Nassif, X., Heffron, F., et al. (1995). The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol 15, 531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x
Storz, J. F., Opazo, J. C., and Hoffmann, F. G. (2013). Gene duplication, genome duplication, and the functional diversification of vertebrate globins. Mol. Phylogenet. Evol. 66, 469–478. doi: 10.1016/j.ympev.2012.07.013
Stull, T. L. (1987). Protein sources of heme for Haemophilus influenzae. Infect. Immun. 55, 148–153.
Suits, M. D., Pal, G. P., Nakatsu, K., Matte, A., Cygler, M., and Jia, Z. (2005). Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proc. Natl. Acad. Sci. U.S.A. 102, 16955–16960. doi: 10.1073/pnas.0504289102
Thomas, M. S. (2007). Iron acquisition mechanisms of the Burkholderia cepacia complex. Biometals 20, 431–452. doi: 10.1007/s10534-006-9065-4
Thompson, J. M., Jones, H. A., and Perry, R. D. (1999). Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect. Immun. 67, 3879–3892.
Tolosano, E., Fagoonee, S., Morello, N., Vinchi, F., and Fiorito, V. (2010). Heme scavenging and the other facets of hemopexin. Antioxid. Redox Signal. 12, 305–320. doi: 10.1089/ars.2009.2787
Tong, Y., and Guo, M. (2007). Cloning and characterization of a novel periplasmic heme-transport protein from the human pathogen Pseudomonas aeruginosa. J. Biol. Inorg. Chem. 12, 735–750. doi: 10.1007/s00775-007-0226-x
Tong, Y., and Guo, M. (2009). Bacterial heme-transport proteins and their heme-coordination modes. Arch Biochem Biophys 481, 1–15. doi: 10.1016/j.abb.2008.10.013
Torres, A. G., and Payne, S. M. (1997). Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23, 825–833. doi: 10.1046/j.1365-2958.1997.2641628.x
Tuanyok, A., Kim, H. S., Nierman, W. C., Yu, Y., Dunbar, J., Moore, R. A., et al. (2005). Genome-wide expression analysis of iron regulation in Burkholderia pseudomallei and Burkholderia mallei using DNA microarrays. FEMS Microbiol. Lett. 252, 327–335. doi: 10.1016/j.femsle.2005.09.043
Tullius, M. V., Harmston, C. A., Owens, C. P., Chim, N., Morse, R. P., McMath, L. M., et al. (2011). Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U.S.A. 108, 5051–5056. doi: 10.1073/pnas.1009516108
Vanderpool, C. K., and Armstrong, S. K. (2001). The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183, 4278–4287. doi: 10.1128/JB.183.14.4278-4287.2001
Vanderpool, C. K., and Armstrong, S. K. (2003). Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185, 909–917. doi: 10.1128/JB.185.3.909-917.2003
Vanderpool, C. K., and Armstrong, S. K. (2004). Integration of environmental signals controls expression of Bordetella heme utilization genes. J. Bacteriol. 186, 938–948. doi: 10.1128/JB.186.4.938-948.2004
Watanabe, S., Takahashi, N., Uchida, H., and Wakasugi, K. (2012). Human neuroglobin functions as an oxidative stress-responsive sensor for neuroprotection. J. Biol. Chem. 287, 30128–30138. doi: 10.1074/jbc.M112.373381
Wegele, R., Tasler, R., Zeng, Y., Rivera, M., and Frankenberg-Dinkel, N. (2004). The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279, 45791–45802. doi: 10.1074/jbc.M408303200
Whitby, P. W., Vanwagoner, T. M., Springer, J. M., Morton, D. J., Seale, T. W., and Stull, T. L. (2006). Burkholderia cenocepacia utilizes ferritin as an iron source. J. Med. Microbiol. 55, 661–668. doi: 10.1099/jmm.0.46199-0
Wilks, A., and Burkhard, K. A. (2007). Heme and virulence: how bacterial pathogens regulate, transport and utilize heme. Nat. Prod. Rep. 24, 511–522. doi: 10.1039/b604193k
Worst, D. J., Otto, B. R., and De Graaff, J. (1995). Iron-repressible outer membrane proteins of Helicobacter pylori involved in heme uptake. Infect. Immun. 63, 4161–4165.
Wu, J., Lin, X., and Xie, H. (2009). Regulation of hemin binding proteins by a novel transcriptional activator in Porphyromonas gingivalis. J. Bacteriol. 191, 115–122. doi: 10.1128/JB.00841-08
Wyckoff, E. E., Duncan, D., Torres, A. G., Mills, M., Maase, K., and Payne, S. M. (1998). Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28, 1139–1152. doi: 10.1046/j.1365-2958.1998.00873.x
Wyckoff, E. E., Lopreato, G. F., Tipton, K. A., and Payne, S. M. (2005). Shigella dysenteriae ShuS promotes utilization of heme as an iron source and protects against heme toxicity. J. Bacteriol. 187, 5658–5664. doi: 10.1128/JB.187.16.5658-5664.2005
Wyckoff, E. E., Schmitt, M., Wilks, A., and Payne, S. M. (2004). HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186, 4142–4151. doi: 10.1128/JB.186.13.4142-4151.2004
Zhang, R., Zhang, J., Ding, H., Lu, D., Hu, Y., Wang Da, C., et al. (2011). Crystallization and preliminary crystallographic studies of Campylobacter jejuni ChuZ, a member of a novel haem oxygenase family. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1228–1230. doi: 10.1107/S1744309111026194
Zhou, D., Qin, L., Han, Y., Qiu, J., Chen, Z., Li, B., et al. (2006). Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol. Lett. 258, 9–17. doi: 10.1111/j.1574-6968.2006.00208.x
Zhu, W., Hunt, D. J., Richardson, A. R., and Stojiljkovic, I. (2000). Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 182, 439–447. doi: 10.1128/JB.182.2.439-447.2000
Keywords: heme, hemin, hem, hemoglobin, iron, pathogens, regulation, Fur
Citation: Runyen-Janecky LJ (2013) Role and regulation of heme iron acquisition in gram-negative pathogens. Front. Cell. Infect. Microbiol. 3:55. doi: 10.3389/fcimb.2013.00055
Received: 02 August 2013; Accepted: 10 September 2013;
Published online: 08 October 2013.
Edited by:
Frédéric J. Veyrier, Institut Pasteur, FranceReviewed by:
Erin R. Murphy, Ohio University Heritage College of Osteopathic Medicine, USAZehava Eichenbaum, Georgia State University, USA
Copyright © 2013 Runyen-Janecky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura J. Runyen-Janecky, Department of Biology, Gottwald Science Center, University of Richmond, Richmond, VA 23173, USA e-mail: lrunyenj@richmond.edu
 Laura J. Runyen-Janecky
Laura J. Runyen-Janecky