Ureaplasma Species Multiple Banded Antigen (MBA) Variation Is Associated with the Severity of Inflammation In vivo and In vitro in Human Placentae
- 1Faculty of Health, School of Biomedical Sciences, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia
- 2Division of Neonatology, Cincinnati Children's Hospital Medical Centre, Cincinnati, OH, USA
- 3Division of Neonatology, Department of Paediatrics, University of Minnesota, Minneapolis, MN, USA
Background: The multiple banded antigen (MBA), a surface-exposed lipoprotein, is a proposed virulence factor of Ureaplasma spp. We previously demonstrated that the number of Ureaplasma parvum MBA size variants in amniotic fluid was inversely proportional to the severity of chorioamnionitis in experimentally infected pregnant sheep. However, the effect of ureaplasma MBA size variation on inflammation in human pregnancies has not been reported.
Methods: Ureaplasmas isolated from the chorioamnion of pregnant women from a previous study (n = 42) were speciated/serotyped and MBA size variation was demonstrated by PCR and western blot. Results were correlated with the severity of chorioamnionitis and cord blood cytokines. In vitro, THP-1-derived macrophages were exposed to recombinant-MBA proteins of differing sizes and NF-κB activation and cytokine responses were determined.
Results: MBA size variation was identified in 21/32 (65.6%) clinical isolates (in 10 clinical isolates MBA size variation was unable to be determined). Any size variation (increase/decrease) of the MBA (regardless of Ureaplasma species or serovar) was associated with mild or absent chorioamnionitis (P = 0.023) and lower concentrations of cord blood cytokines IL-8 (P = 0.04) and G-CSF (P = 0.008). In vitro, recombinant-MBA variants elicited different cytokine responses and altered expression of NF-κB p65.
Conclusion: This study demonstrates that size variation of the ureaplasma MBA protein modulates the host immune response in vivo and in vitro.
Introduction
The human Ureaplasma species (U. parvum and U. urealyticum) are prevalent colonizers of the lower genital tract and are known to colonize up to 80% of women and 50% of men (Cassell et al., 1993; Volgmann et al., 2005). Although these microorganisms were traditionally considered to be of low virulence (Volgmann et al., 2005), more recent evidence suggests that Ureaplasma spp. may be virulent pathogens of the female upper genital tract. The Ureaplasma spp. are the microorganisms isolated most frequently from the amniotic fluid and placentae of women (Hillier et al., 1988; Knox et al., 1997; Namba et al., 2010; Sweeney et al., 2016), either in the presence or absence of histological chorioamnionitis (Sweeney et al., 2016). These microorganisms have been associated with spontaneous abortion and miscarriage (Naessens et al., 1987, 1988), preterm birth (Hillier et al., 1988; DiGiulio et al., 2010), chorioamnionitis (Hillier et al., 1988; Namba et al., 2010; Sweeney et al., 2016), and preterm premature rupture of membranes (Jacobsson et al., 2009; DiGiulio et al., 2010). Despite the fact that Ureaplasma spp. have been isolated from up to 42% of pregnancies that end prematurely (Namba et al., 2010), the pathogenesis of ureaplasmas is not always clear; particularly as not all women who are infected with Ureaplasma spp. experience preterm birth or adverse pregnancy outcomes (Gerber et al., 2003). Several hypotheses have been proposed to explain this variation in pathology/outcomes of women infected with Ureaplasma spp. Researchers have suggested that there may be “virulent” Ureaplasma species or serovars (Naessens et al., 1988; Knox and Timms, 1998; De Francesco et al., 2009; Eun et al., 2013); however, links to a particular species/serovar with a disease state or adverse outcomes have not been consistent (Zheng et al., 1992). Others have suggested that virulence may not be limited to a single species or serovar of ureaplasmas, but instead may be associated with antigenic variation of the pathogen itself (Zheng et al., 1992, 1995, 1996; Dando et al., 2012).
The multiple banded antigen (MBA) is a major surface-exposed (Zheng et al., 1995; Shimizu et al., 2008), immunodominant antigen of Ureaplasma spp., which activates NF-κB and the production of cytokines by signaling via Toll-like receptors (TLRs) 1, 2, and 6 (Shimizu et al., 2008). More recently, Knox et al. (2010) demonstrated that the number of U. parvum MBA size variants present within the amniotic fluid of pregnant sheep (an ovine model of Ureaplasma spp. infection) was inversely proportional to the severity of inflammation within the chorioamnion. Sheep infected intraamniotically with the same strain and dose of U. parvum had divergent inflammatory responses within their chorioamnion: when >9 MBA size variants were present, there was little or no inflammation within the chorioamnion; by contrast, when there were five or fewer MBA size variants, severe inflammation of the chorioamnion was observed (Knox et al., 2010). Antigenic variation is a common feature of a wide range of pathogens; and to date the mechanisms known to govern Ureaplasma spp. antigenic variation include: (i) slipped strand mispairing and/or nucleotide insertions or deletions of simple repeating sequences and (ii) DNA rearrangements via site-specific recombination (Citti et al., 2010). While these studies have demonstrated a potential role for MBA size variation and variability in chorioamnionitis or pregnancy outcomes; it is currently unclear if MBA size variation can occur during infection of human placentae and if this variation results in similar outcomes to what we have previously reported in an ovine model.
Recently, our research group demonstrated that the Ureaplasma spp. were the most prevalent microorganisms (42/535; 7.8%) isolated from late preterm or term placentae and that their presence (but not the presence of other bacteria) was associated with chorioamnionitis. However, not all women whose placentae were infected/colonized with ureaplasmas had chorioamnionitis: in 15/42 (35.7%), no histological chorioamnionitis was observed (Sweeney et al., 2016). Using these same Ureaplasma spp. clinical isolates, we further investigated the role of MBA virulence in human ureaplasmas. We hypothesized size variation of the MBA protein (and gene) would occur in human Ureaplasma spp. clinical isolates, and that differences in the host immune response (in vivo and in vitro) would contribute to the severity of inflammation.
Materials and Methods
Ethics Statement
The use of human blood and tissues was approved by the review boards of the Good Samaritan Hospital (approval 09105-09-067) and Cincinnati Children's Hospital Medical Center (approval 2009-0236). All patients gave permission for the collection of their placentae upon delivery and for their medical records (demographic data and pregnancy/neonatal outcomes) to be recorded in a de-identified database, as described in our previous study (Shepard and Lunceford, 1976; Sweeney et al., 2016). Chorioamnion inflammation (chorioamnionitis) scores were performed by a pathologist, blinded to the microbiological findings, and the severity of chorioamnionitis was graded according to the guidelines set by Redline et al. (2003). All subjects gave written informed consent in accordance with the Declaration of Helsinki for their medical records to be recorded within a de-identified database. The work within this study was also submitted to the Human Research Ethics Committee (HREC) of the Queensland University of Technology and considered exempt from approval, as patient samples and data were de-identified prior to shipment/accessing of patient medical records. The QUT ethics committee also approved the production of recombinant proteins for use in in vitro experiments.
Ureaplasma spp. Clinical Isolates
Tissue samples were excised from placentae and snap frozen prior to transport to Queensland University of Technology (QUT). At QUT, placental tissue samples were homogenized and ureaplasmas were cultured and ureaplasma DNA extracted as previously described (Sweeney et al., 2016). Clinical ureaplasma isolate cultures were also utilized for extraction of Ureaplasma proteins as previously described (Dando et al., 2012).
Extraction of DNA from Cultured Ureaplasmas
Positive ureaplasma cultures from low passage (≤ 2 passages) isolates were centrifuged at 4,500 × g for 20 min (Allegra XR-15, Beckman Coulter, Australia) and nucleic acid was extracted from the resulting bacterial pellet using the QIAamp mini DNA extraction kit (Qiagen, Australia). All extracted DNA was stored at −20°C until required.
Speciation and Serotyping of Ureaplasma spp. Clinical Isolates
The upstream conserved region of the Ureaplasma spp. multiple banded antigen (mba) gene was performed as previously presented (Sweeney et al., 2016; Accession numbers: KY796009, KY796010, KY796011, KY796012, KY796013, KY796014, KY796015, KY796016, KY796017, KY796018, KY796019, KY796020, KY796021, KY796022, KY796023, KY796024, KY796025, KY796026, KY796027, KY796028, KY796029, KY796030, KY796031, KY796032, KY796033, KY796034, KY796035, KY796036, KY796037, KY796038, KY796039, KY796040, KY796041, KY796042, KY796043, KY796044, KY796045, KY796046, KY796047, KY796048, and KY796049) and the mba gene was used to serotype the U. parvum and U. urealyticum clinical isolates.
PCR Assays Targeting the Downstream Repetitive Region of the mba Gene
The downstream repetitive region of U. parvum clinical isolates was amplified using previously published assays (Knox et al., 2010; Dando et al., 2012; Robinson et al., 2013). These primers amplified U. parvum serovars 1 and 6; or serovars 3 and 14 and revealed size variation within the mba gene.
Western Blotting of Ureaplasma spp. Multiple Banded Antigen (MBA) Protein
Cultures of each clinical isolate and Ureaplasma spp. ATCC strain were centrifuged at 4,500 × g for 20 min. The supernatant was then discarded and the pellet resuspended in 100 μL of sterile PBS. The suspensions were stored at −20°C prior to use. Extracted proteins (30 μg) were then used for western blot analysis, as previously described, to identify size variation in the MBA protein (Dando et al., 2012).
Cord Blood Cytokine Analysis
Cord blood was collected at the time of delivery from the umbilical vein using a sterile Viacord collection kit containing an anticoagulant, and the blood components separated by centrifugation. Concentrations of cytokines/chemokines within cord blood plasma were then determined using MILLIPLEX® MAP Human Cytokine/Chemokine magnetic bead panel (Millipore, USA). Concentrations of cytokines/chemokines were calculated from standard curves using recombinant proteins and the results were expressed in pg/mL.
Production of Recombinant MBA Proteins from Ureaplasmas
DNA from ATCC U. parvum serovar 6 and a selection of U. parvum serovar 6 clinical isolates (#27, #50, and #122 and #334B) were utilized to amplify the mba gene. The mba gene was selected from these clinical isolates (as they are representative the large variety of sizes observed within our U. parvum serovar 6 clinical isolates) in order to determine if the size of the mba gene (and the expressed MBA protein) alters the host immune response. All mba genes were selected from U. parvum serovar 6 isolates that were isolated from preterm placentae (<37 weeks of gestation). PCR assays were performed in 50 μL reactions, containing: 4 μL of DNA template, 50 μM dNTPs (ThermoFisher Scientific, Australia), 1.5 mM MgCl2 (ThermoFisher Scientific), 1 μM forward and reverse PCR primers (forward primer: ACATTAGGAGTTACC; reverse primer: TTATTTTCTAGCAGC; Sigma Aldrich) 2 units of pfu polymerase (Promega, Australia) and sterile DNAse/RNAse-free dH20 (Gibco, Australia). PCR cycling was performed using the PTC-2000 thermocycler (Bio-Rad, Australia) and cycling consisted of: denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, primer annealing at 55°C for 30 s and extension at 72°C for 2.5 min. PCR fragments were then purified using the PureLink PCR amplicon purification kit (ThermoFisher Scientific) and sequencing was performed (Life Technologies) to confirm the correct mba gene sequence.
The purified mba genes were ligated into the pRSET A plasmid (kindly provided by Professor Ken Beagley) and transformed into chemically competent DH5α Escherichia coli. Positive transformants for each mba gene/protein were then selected (a minimum of ten per gene/protein) for further analysis, to confirm the presence of the mba gene within the plasmid. Plasmids containing the correct insert were then transfected into BL21 E. coli and stored at −80°C for future use.
BL21 E. coli strains containing the mba genes were then used for large-scale protein production. This involved culturing E. coli strains until logarithmic growth was achieved, followed by induction of protein expression using isopropyl β-D-thiogalactoside (IPTG; Bioline, Australia). E. coli were then allowed to produce the protein of interest for 3 h, before being collected by centrifugation. The resulting cell pellet was resuspended in sterile phosphate-buffered saline (PBS). E. coli were then sonicated prior to protein purification.
MBA proteins were produced with a 6xHis-tag for use in ion metal chromatography using Talon® resin (ClonTech, Australia). MBA proteins were bound to the Talon® resin for 1 h before a series of washing steps with low concentrations (10 mM) of imidazole to remove any non-specific proteins. After washing, the protein of interest was eluted from the resin using 500 mM imidazole. Purified rMBA proteins were evaluated by western blot, using MBA-specific primary antibodies (kindly provided by Emeritus Dr Patricia Quinn, The Hospital for Sick Children, Toronto) to demonstrate the recombinant proteins were of the correct size. All proteins were confirmed to be free of contaminating LPS; any LPS detected was removed using a high capacity endotoxin removal kit (Life Technologies, Australia).
In vitro Stimulation of THP-1 Macrophages Using Recombinant MBA Proteins
THP-1 monocyte cells were grown in Roswell Park Memorial Institute (RPMI; ThermoFisher Scientific) media containing 10% fetal bovine serum, 1,000 U/mL benzylpenicillin and 0.05 mM β-mercaptoethanol. Cells were seeded into 48-well plates (Corning Life Sciences, Australia) at 1 × 105 cells/well and supplemented with 10 ng/mL phorbol 12-myristate 13-acetate (PMA; Sigma Aldrich) to induce the differentiation of THP-1 monocytes to macrophages for 72 h (Park et al., 2007). After differentiation, the adherent cell monolayer was washed using sterile PBS and then exposed to 10 μg/mL of: ATCC U. parvum serovar 6 recombinant protein (~75 kDa), and U. parvum serovar 6 clinical isolate recombinant MBA proteins #27 (~37 kDa), #50 (~60 kDa), #122 (~70 kDa), and #334B (~75 kDa), suspended in RPMI culture media. Controls included no recombinant proteins (negative control), macrophages stimulated with 100 ng/mL Escherichia coli lipopolysaccharide (E. coli LPS; Sigma Aldrich) and macrophages exposed to 2 × 107 colony forming units (CFU) of live or UV-inactivated (killed) U. parvum serovar 6. After 24 h, cell culture supernatant was collected for enzyme-linked immunosorbent assays (ELISA) for TNF-α, IL-1β IL-6, IL-8, IL-10, and G-CSF (ELISAkit.com, Australia). THP-1 cells exposed to each treatment group were also prepared for western blot analysis. Cells from each experimental group were scraped from the culture vessel (Sarstedt Pty. Ltd., Australia) and incubated in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, protease inhibitor cocktail, 50 mM Tris pH 8.0) at 4°C for 1 h, to extract the total cell protein. For SDS-PAGE electrophoresis, 60 μg of cell lysate from each sample was loaded into a gel for subsequent western blotting and transfer to a nitrocellulose membrane. The membrane was then stained with Ponceau S dye (Sigma Aldrich), in order to visualize the protein bands of the correct sizes, before the membrane was cut and blocked in 5% skim milk solution for 1 h. The membranes were then probed with human phosphorylated NF-κB p65 primary antibody (Abcam, Australia) or human β-actin primary antibody (Abcam) overnight at 4°C. The membranes were washed and probed with a rabbit anti-human IgG secondary antibody (Sigma Aldrich) for 1 h. Protein bands were visualized using 3′3′-diaminobenzidine (DAB) with metal enhancer (Sigma Aldrich). The developed membranes were then imaged using ChemiDoc MP imaging system (Bio-Rad) and densitometry analysis was performed using ImageJ software (NIH).
Statistical Analysis
All data are presented as the mean value, plus or minus the standard error of the mean (SEM). Data were analyzed using analysis of variance (ANOVA) and included adjustments for multiple comparisons. Statistical significance was accepted as P < 0.05.
Results
Speciation and Serotyping of Ureaplasma Clinical Isolates
The upstream conserved portion of the mba gene of 42 Ureaplasma spp. isolates were sequenced (Sweeney et al., 2016). The majority of these isolates were confirmed to be U. parvum (36/42; 85.7%) and only 6 (14.3%) U. urealyticum isolates were identified.
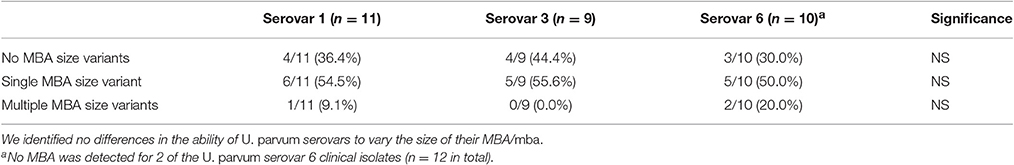
Of the 36 U. parvum clinical isolates, four were unable to be serotyped by sequencing; however, the remaining isolates were serotyped as U. parvum serovar 1 (11/32; 34.4%), serovar 3 (9/32; 28.1%), and serovar 6 (12/32; 37.5%). No U. parvum serovar 14 clinical isolates were detected. Of the U. urealyticum clinical isolates, only two of the six clinical isolates were able to be serotyped, and these were identified as U. urealyticum serovar 8 and serovar 10.
Ureaplasma Species and Serovars: Association with Adverse Pregnancy or Neonatal Outcomes
There were no differences in outcomes for neonates exposed to U. parvum or U. urealyticum (Table 1).
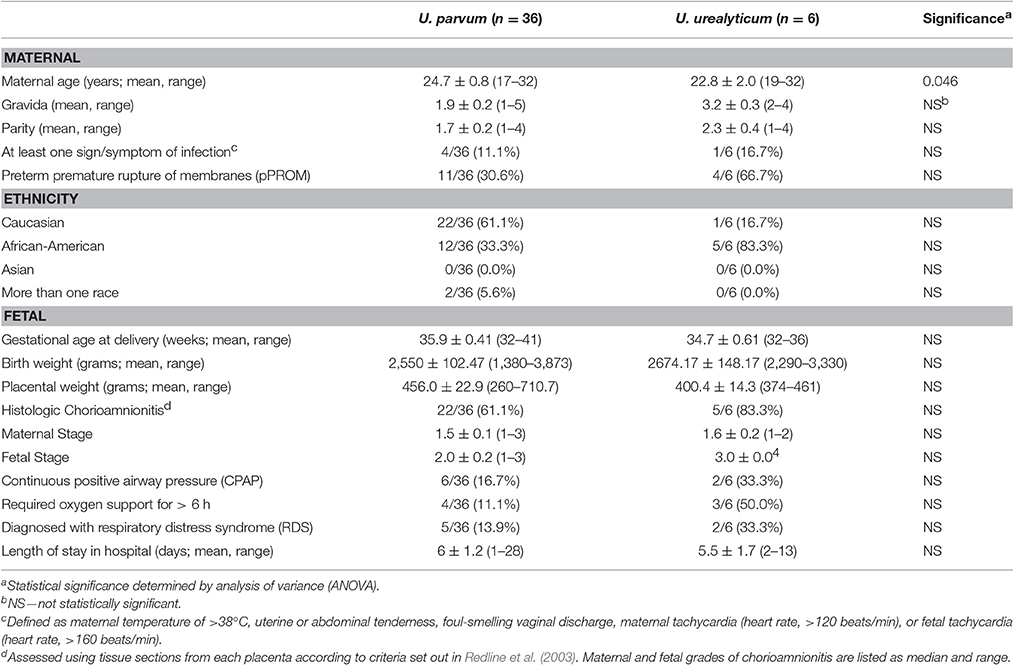
Table 1. Maternal and neonatal demographic and outcome data for women whose placentae were infected with U. parvum or U. urealyticum.
We also compared the outcomes of mothers and their infants exposed to the most common serovars within our study: U. parvum serovars 1, 3, and 6. Women whose placentae were infected with U. parvum serovar 3 (n = 9) were younger (21.4 ± 0.9 years) than women in whom U. parvum serovars 1 (n = 11) or 6 (n = 12) were identified (25.6 ± 1.3 and 26.0 ± 1.4 years, respectively; P = 0.024) (Table 2). There were no other differences between these three groups of women, nor were there any differences in the incidence of adverse neonatal outcomes, including the prevalence of histological chorioamnionitis.
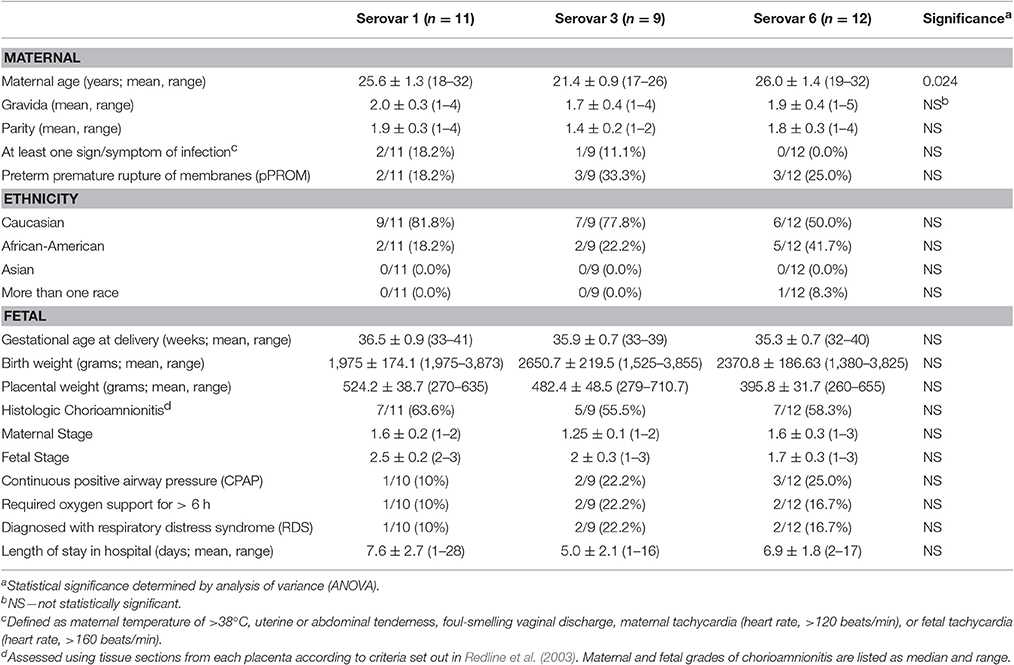
Table 2. Maternal and neonatal demographic/outcome data for women whose placentae were infected with U. parvum serovars 1, 3, or 6.
PCR and Western Blot Targeting the MBA Protein and mba Gene
The MBA protein and mba gene of ureaplasma clinical isolates were compared to American Type Culture Collection (ATCC) strain controls (serovar 1, 3, 6, 8, and 10, which served as positive controls for the size/expression of the MBA/mba). Numerous MBA/mba size variants were detected within low-passage (≤ passage 2) clinical isolates (Figure 1). For some ureaplasma clinical isolates, there was no identifiable MBA/mba size variation, i.e., the MBA protein and mba gene were the same size as the antigens/genes of the ATCC strain serovars (Serovar 1 isolates: 1A, 1B, 262T, 507; Serovar 3 isolates: 33A, 33B, 322T, 325; and Serovar 6 isolates: 334A, 334B, 364A); for other clinical isolates, variation in the size of their MBA protein and mba gene band(s) were observed. These clinical isolates demonstrated either a “single MBA/mba size variant,” which was considered to be an individual protein/gene band that differed in size when compared to the corresponding ATCC strain controls (Serovar 1 isolates: 43, 301, 473T, 483T, 498A, 498B; Serovar 3 isolates: 44A, 44B, 314T, 365, 435; Serovar 6 isolates: 27, 50, 55B, 122, and 310T; Serovar 8 isolate: 8; and Serovar 10 isolate 300); or in some cases, “multiple MBA/mba variant” bands were seen, where more than one MBA/mba band was visualized by PCR or western blot (Serovar 1 isolates: 290T; Serovar 6 isolates: 182, 429) (Figure 1). We did not see any difference in the propensity for these Ureaplasma isolates to vary the size of their MBA/mba, according to the species or serovar identified (Table 3).
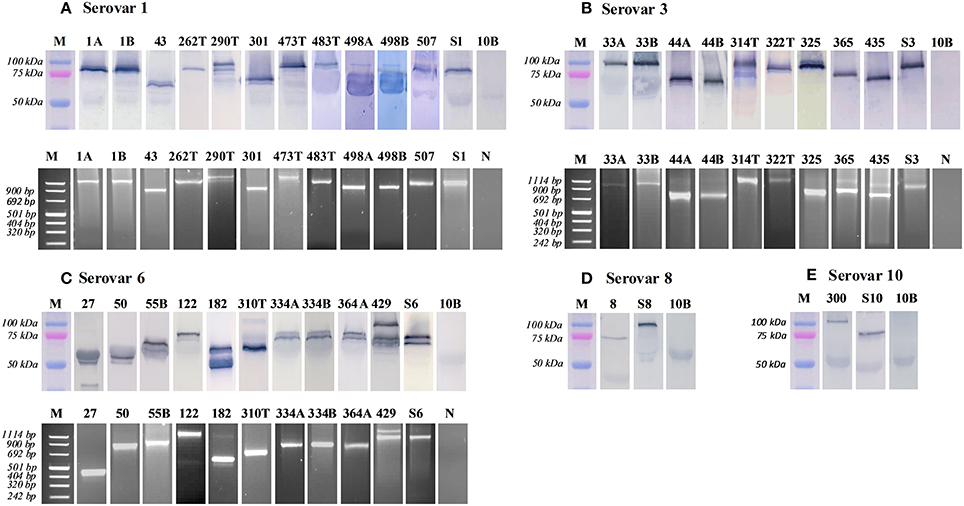
Figure 1. Variation of the MBA protein and mba gene was detected by western blot and PCR. MBA/mba size variation was characterized by comparing the protein and PCR amplicons to the ATCC strain serovar control and a molecular weight marker bands. Images were cropped and displayed at their correct molecular weight and sorted according to their serovar. M, protein marker; 10B, 10B media control; N, negative control; S, serovar 1, 3, 6, 8, 10—ATCC positive controls; A/B, twin pregnancies; T, term pregnancy (where all others are late preterm: 32–36 weeks of gestation). (A) serovar 1. (B) serovar 3. (C) serovar 6. (D) serovar 8. (E) serovar 10.
For some clinical isolates, n = 2 U. parvum serovar 6 isolates and n = 8 U. urealyticum isolates the MBA protein and mba gene were not detected or visualized. Therefore, we were unable to determine if MBA/mba size variation occurred in these clinical isolates.
MBA/mba Size Variation was Associated with Altered Immune Responses In vivo
The maternal demographic data of ureaplasma-infected women in which MBA/mba variation was identified was compared to those women in whom no MBA/mba size variation was seen. No differences were observed (data not shown). However, a major finding of this study was that Ureaplasma spp. MBA/mba size variation was associated with differences in the incidence of histological chorioamnionitis as graded by a US pathologist (Figure 2A). Cord blood cytokines from women in which no microorganisms were detected (by culture and PCR) were compared to the cord blood cytokine profiles from women whose placentae were found to be infected with Ureaplasma spp. Placentae which harbored ureaplasmas with no MBA/mba size variation demonstrated a higher prevalence of histological chorioamnionitis (9/11; 81.8%), when compared to placentae in which the ureaplasmas present expressed either a single or multiple MBA/mba size variant(s) (10/21; 47.6% p = 0.03). No differences in the severity of inflammation within the maternal and fetal sides of these membranes were observed (Figure 2A). There were no other differences in pregnancy or neonatal outcomes associated with ureaplasma MBA/mba size variation.
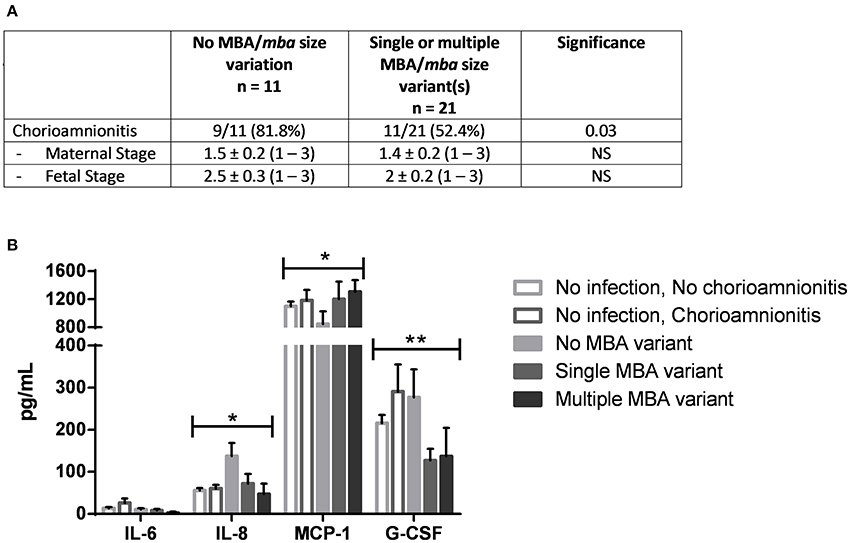
Figure 2. MBA/mba size variation was associated with the incidence and severity of histological chorioamnionitis. (A) Histological chorioamnionitis was more prevalent in placentae infected with Ureaplasma spp. that did not vary their MBA/mba, when compared to placentae infected with Ureaplasma spp. that expressed a single or multiple MBA/mba size variants (B) MBA/mba size variation was also associated with altered levels of cord blood cytokines in vivo. *P < 0.05, **P < 0.01.
Cord blood samples collected at the time of delivery were also tested for inflammatory markers (cytokines, chemokines, and growth factors including IL-1β, IL-6, IL-8, MCP-1, and G-CSF). In vivo, when ureaplasma MBA/mba size variation occurred within infected placentae, lower concentrations of IL-8 (67.7 pg/mL) and G-CSF (128.7 pg/mL) were detected, when compared to the concentrations of these same cytokines in placentae which were infected with Ureaplasma spp. but demonstrated no MBA/mba size variation (IL-8: 137.7 pg/mL, G-CSF: 277.0 pg/mL; P = 0.044, P = 0.008, respectively; Figure 2B).
In contrast, levels of MCP-1 in cord blood were significantly elevated when ureaplasmas which demonstrated MBA/mba size variation were isolated from placentae, when compared to the cord blood collected from those placentae which were infected with ureaplasmas that showed no variation in the size of their MBA/mba (P = 0.048).
MBA Size Variation was Also Associated with Altered Immune Responses In vitro
To further investigate the role of MBA size variation and the host response, we challenged differentiated human THP-1 (macrophages) with recombinant MBA proteins of differing sizes, using Escherichia coli lipopolysaccharide (LPS) as a positive control. These recombinant MBA proteins were produced from U. parvum serovar 6 ATCC strain (control), and from U. parvum serovar 6 clinical isolates (isolates: #27, #50, #122, and #334B; see Figure 1).
When THP-1 macrophages were exposed to the E. coli LPS control, robust immune responses were detected for the cytokines TNF-α, IL-1β, IL-8, and G-CSF. In vitro, the ATCC U. parvum recombinant MBA (rMBA; ~75 kDa) protein elicited the strongest immune response of all rMBA proteins tested; and stimulated the production of TNF-α, IL-1β, IL-8, and G-CSF (Figure 3). Similarly, the equivalent size rMBA protein of isolate #334B (~75 kDa) elicited similar immune responses. In contrast, recombinant proteins that were smaller in size (compared to ATCC U. parvum serovar 6 and the rMBA #334B) elicited diminished cytokine responses. The rMBA #50 protein (~60 kDa) elicited lower concentrations of TNF-α (36.6 ± 25.9 pg/mL; P = 0.024) and G-CSF (5.3 ± 1.3 pg/mL; P = 0.044), when compared to the concentrations seen in response to the rMBA of ATCC U. parvum serovar 6.; while the rMBA #122 (~70 kDa) elicited lower levels of TNF-α (35.2 ± 1.1 pg/mL; P = 0.020), IL-1β (14.9 ± 2.1 pg/mL; P = 0.045), and G-CSF (5.3 ± 1.7 pg/mL; P = 0.039). The smallest rMBA #27 (~37 kDa) elicited the production of cytokines at similar levels to that of our negative (no treatment) controls (TNF-α: P = 0.002, IL-1β: P = 0.033, G-CSF: P < 0.001; Figure 3), with the exception of IL-8. Interestingly, regardless of the challenge (LPS, U. parvum controls, no treatment and rMBA proteins) there were no differences in the concentrations of IL-8. Similarly, no significant differences were seen in IL-6 and IL-10 cytokine levels in cell culture supernatants (data not shown).
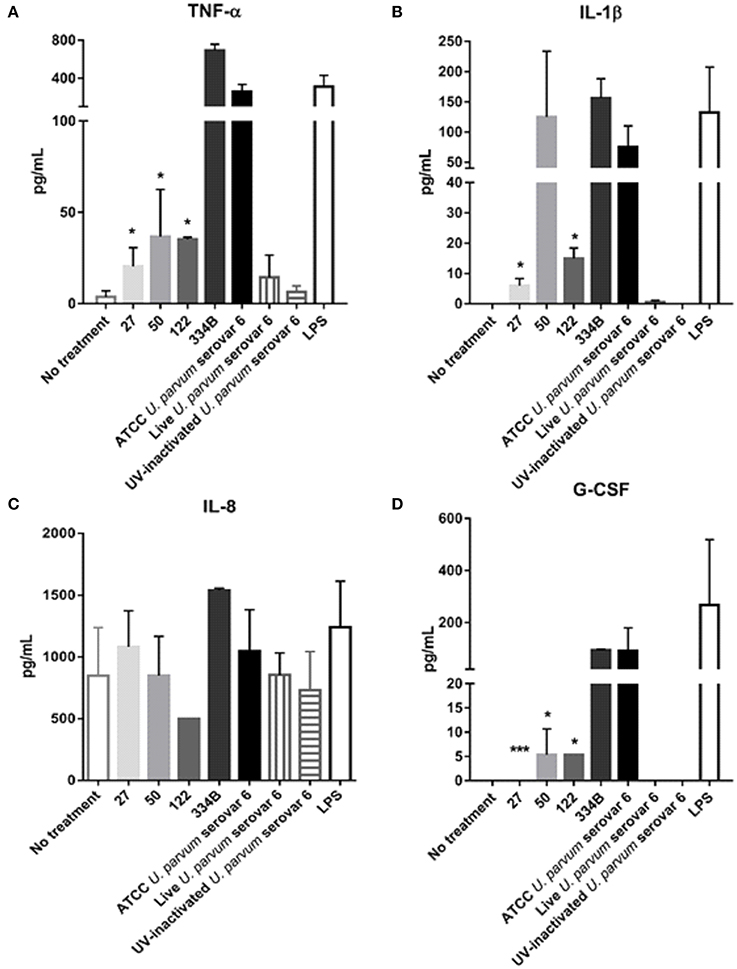
Figure 3. Recombinant MBA proteins of differing sizes elicit varying immune responses in THP-1 macrophages in vitro. Assays were conducted on cell culture supernatant taken from differentiated THP-1 (macrophages) exposed to recombinant proteins, or controls, for 24 h. ELISA assays for (A) TNF-a, (B) IL-lB, (C) IL-8, and (D) G-CSF expressed in picograms per milliliter of culture supernatant. *P < 0.05, **P < 0.01, ***P < 0.001.
Since altered cytokine responses were observed when macrophages were exposed to different rMBA protein size variants, we further investigated the activation/expression of NF-κB p65, the protein complex that controls transcription of DNA and cytokine production, in cells exposed to the different sized rMBA proteins. Western blot and densitometry analyses revealed that the expression of NF-κB was highest for cell lysates exposed to LPS, ATCC U. parvum serovar 6 and rMBA #334B (Figures 4A,B). By contrast, diminished expression of NF-κB p65 was observed in cell lysates after exposure to rMBAs #27 and #122, relative to host (β-actin) controls (Figure 4).
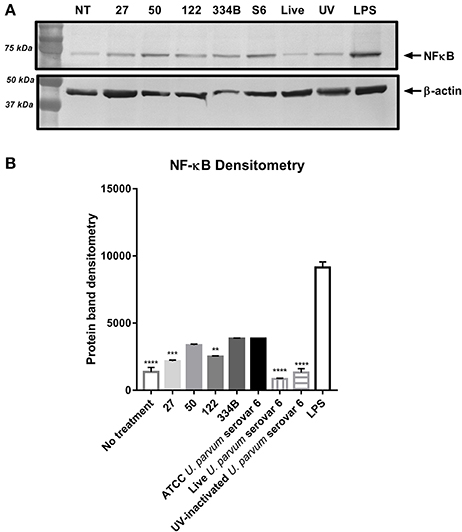
Figure 4. NF -KB p65 expression alters upon exposure to different size recombinant MBA proteins in vitro. (A) NF-KB p 65 expression is altered upon exposure to recombinant MBA proteins of differing sizes, when normalized to host (β-actin) controls. (B) Densitometry analyses from n = 3 experiments, performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
The pathogenic role of Ureaplasma spp. remains controversial, as ureaplasma infection within placentae is not always associated with inflammation and adverse pregnancy outcomes (Gerber et al., 2003). Furthermore, the host immune response which influences the development of pathology/disease has not been well-studied during Ureaplasma spp. infections. A previous study by our group demonstrated that Ureaplasma spp. were the most prevalent bacteria detected within the chorioamnion of women (7.8%) who delivered late preterm or at term, but not all women infected with ureaplasmas developed histological chorioamnionitis (Sweeney et al., 2016). In this current study, we investigated the factors previously implicated in adverse pregnancy outcomes and inflammation during Ureaplasma spp. infection.
Similar to our previous study, in which U. parvum was the most frequently isolated bacterial species (86%) (Sweeney et al., 2016), in this current study we identified that U. parvum serovars 1, 3, and 6 were the most common ureaplasma serovars detected within infected placentae. However, we found that regardless of the Ureaplasma species or serovar isolated within the chorioamnion, the incidence of chorioamnionitis and other adverse pregnancy/neonatal outcomes did not differ (Tables 1, 2). Previous studies have suggested the existence of “virulent” Ureaplasma species or serovars (Naessens et al., 1988; Knox and Timms, 1998; Knox et al., 2003; Dando et al., 2012; Sweeney et al., 2016); however, ureaplasmas may not be intrinsically “virulent” or “avirulent,” rather, that other factors may instead contribute to adverse outcomes during ureaplasma infections. Furthermore, our findings are consistent with previous studies that have shown Ureaplasma spp. to be associated with chorioamnionitis (Hillier et al., 1988; Cassell et al., 1993; Namba et al., 2010), but ureaplasmas may also be detected within the placentae of women with no evidence of chorioamnionitis and the pregnancy may continue until term delivery, as was also reported by Gerber et al. (2003).
The most significant finding of this study was that Ureaplasma spp. isolated from the placentae of human pregnancies demonstrated differences in the size of their MBA protein and mba gene. Some of the clinical ureaplasma isolates obtained from chorioamnion tissue expressed no MBA size variation (and their proteins were the same size as our ATCC strain controls), while other clinical isolates expressed a single or multiple MBA size variants (as evidenced by single or multiple MBA bands which differed in size, when compared to the control serovar ATCC strains). While we saw no differences in the propensity of the different Ureaplasma species or serovars to vary the size of their MBA/mba (Table 3), our study demonstrated that a lack of MBA/mba size variation in vivo was associated with a significantly higher incidence of histological chorioamnionitis (Figure 2A) and elevated levels of the cord blood cytokines IL-8 and G-CSF (Figure 2B). By contrast, when MBA/mba size variation occurred, this was associated with a significant (~30%) reduction in the incidence of histological chorioamnionitis and significantly lower levels of the cord blood cytokines IL-8 and G-CSF (P = 0.04 and P = 0.008; Figure 2B).
Previous studies have indicated that the severity of chorioamnionitis varied, depending on the numbers of Ureaplasma spp. present within placental tissue (Jacobsson et al., 2009; Kasper et al., 2010; Kacerovsky et al., 2011); however, within our previous study of n = 535 placentae we did not find an association between the numbers of ureaplasmas present within the chorioamnion and the severity of chorioamnionitis (Sweeney et al., 2016). Instead, the results of this current study support the proposal that the severity of inflammation is associated with the degree of MBA/mba size variation.
We further investigated the role of MBA/mba size variation using an in vitro cell culture model: differentiated THP-1 (macrophage) cells were stimulated with rMBA U. parvum serovar 6 proteins of differing sizes. Of the rMBA proteins tested, the ATCC U. parvum serovar 6 protein elicited the greatest cytokine response, followed closely by the rMBA protein #334B, which was equivalent in size to that of the ATCC strain. In contrast, rMBA serovar 6 proteins that differed in size to the ATCC U. parvum serovar 6 strain and #334B proteins elicited lower concentrations of cytokines TNF-α, IL-1β, and G-CSF. For the rMBAs #27 and #122 significantly lower concentrations of IL-1β were also elicited when these proteins were exposed to THP-1 cells (Figures 3A–D). However, no differences were observed in the concentrations of IL-6 or IL-10 in the cell culture supernatant (data not shown). Of great interest, the smallest recombinant MBA protein #27 elicited minimal cytokine production and these concentrations of cytokines were similar to those in response to the negative (no treatment) control, indicating that the macrophages within this study may be unable to recognize this protein. These findings were confirmed by the results of densitometry analyses of NF-κB p65, the protein complex that controls transcription of DNA and cytokine production (Rahman and McFadden, 2011). Altered expression of NF-κB p65 was demonstrated in cell lysates exposed to rMBA proteins that were different in size, when compared to ATCC U. parvum serovar 6 and #334B recombinant proteins. NF-κB p65 expression was decreased for recombinant proteins #27 and #122 (Figures 4A,B), and this corresponded directly to the diminished cytokine levels seen in vitro (Figure 3).
It was first reported in 2008 that the Ureaplasma spp. surface-exposed lipoproteins, predominantly the MBA, induced an inflammatory response that resulted in the induction of NF-κB though TLRs 1, 2, and 6; and this study further proposed that size variation of the MBA may affect the stimulatory activity of the ureaplasma MBA and its ability to interact with TLRs (Shimizu et al., 2008). Others have reported the cytokine responses elicited when: (i) adult human monocytes and term and preterm neonatal cord blood were exposed to low (103 color changing units [CCU]) or high doses (106 CCU) of U. parvum serovars 3 (Manimtim et al., 2001), (ii) human monocytes (THP-1 cells) were treated with heat-killed U. urealyticum (Li et al., 2000), (iii) THP-1 macrophages were exposed to surface lipoproteins of Ureaplasma urealyticum (Peltier et al., 2007), and (iv) human amniotic epithelial cells were exposed to Ureaplasma spp. serovars 2, 3, and 14 (1 × 108 bacteria/mL) (Triantafilou et al., 2013). In each of these in vitro experiments, the production of TNF-α was elevated in response to the ureaplasmas/antigens presented. However, the production of other cytokines (IL-1β, IL-8, IL-6, and IL-10) varied depending on the cell type, the bacterial or antigenic load, and the presence of LPS or steroids. A major strength of our current study was the correlation of immune responses in differentiated THP-1 (macrophages) to the same ureaplasma serovar (live or UV-inactivated U. parvum serovar 6) and rMBA proteins of differing sizes that were synthesized from low-passage U. parvum serovar 6 clinical isolates originally isolated from human placentae. These experiments confirm that rMBA proteins of differing sizes elicit varying concentrations of cytokines (IL-1β, IL-8, MCP-1, and G-CSF), depending on the size of the MBA antigens expressed by the ureaplasmas in vitro or in vivo.
Antigenic variation is an important mechanism used by microorganisms to mediate their interaction with the host and/or environment and this variation is thought to be an essential strategy employed by microorganisms to assist in pathogen survival, particularly in the presence of a host immune response (Citti et al., 2010). Antigen variation is not a unique trait of Ureaplasma spp., many microbes possess the ability to vary their surface exposed antigens (Citti et al., 2010; Foley, 2015); however, MBA/mba size variation has been reported previously in a sheep model of intraamniotic ureaplasma infection (Knox et al., 2010; Dando et al., 2012; Robinson et al., 2013). To the best of our knowledge, this current study is the first to identify variation of the Ureaplasma spp. MBA/mba in human placentae, and to demonstrate that this variation is associated with the incidence and severity of chorioamnionitis during pregnancy. Furthermore, this study is also the first to demonstrate that levels of cord blood cytokines differ when the placenta of pregnant women infected with ureaplasmas express different MBA/mba size variants.
Previous studies have investigated the role of size variation of the MBA/mba. Initial studies demonstrated the ability of Ureaplasma spp. isolated from neonates to vary their surface-exposed MBA/mba (Zheng et al., 1992) and that variation of the MBA/mba correlated with the number of repeating units in the downstream (surface-exposed) portion of the protein and gene (Zheng et al., 1995). These authors suggested that further knowledge of MBA size variation “would be requisite to understanding the role that these antigens and their associated size variation may play in the success or failure of these organisms as pathogens” (Zheng et al., 1995). Further studies by this same group utilized antibody-reactive peptide scanning and showed that differences in the numbers of repeating units within the MBA were associated with altered recognition of the MBA by sera which contained anti-ureaplasma antibodies (Zheng et al., 1996). In this previous study an interesting trend was observed; the ability of the monoclonal antibodies to bind to the MBA increased with the number of repeating units present (Zheng et al., 1996). This finding correlates with our data, which demonstrated that MBA size variation resulted in altered immune responses, both in vivo and in vitro and suggests that MBA size variation may be a mechanism by which ureaplasmas can evade either innate or adaptive immune responses.
Zimmerman et al. (2009, 2013) in in vitro experiments demonstrated inversion events that occurred between the mba and other regions with the genome of U. parvum serovars 3. These events were confirmed on both the genomic and protein level. They concluded that these DNA inversion events are dynamic and result in the high-frequency, broad spectrum, antigenic variation of these pathogens. While in these studies, DNA inversion events were not characterized in vivo, studies by Dando et al. (2012) identified ovine anti-ureaplasma IgG antibodies within maternal and fetal serum that were collected after sheep were infected intraamniotically with the same U. parvum low passage clinical isolate. This study revealed that the anti-ureaplasmal antibodies from these animals reacted with more than one ureaplasma MBA size variant, and that the IgG reactivity differed between animals (Dando et al., 2012). This study confirmed that MBA/mba size variation occurred in vivo and we and others (Zheng et al., 1992, 1995, 1996; Monecke et al., 2003; Zimmerman et al., 2009, 2011, 2013) propose that this variation is an important immune evasion mechanism for Ureaplasma spp. Similar antigenic variation leading to immune evasion has been noted in several other well-studied organisms, including Plasmodium falciparum, Mycoplasma pulmonis, and the human immunodeficiency virus (Lipsitch and O'Hagan, 2007; Citti et al., 2010). Further studies are required to understand the immune pressures which may trigger MBA/mba size variation in vivo and in vitro and to determine how these changes of the organism facilitates immune evasion.
In summary, this is the first study to demonstrate MBA/mba size variation within ureaplasmas isolated from the human chorioamnion. Our data suggest that Ureaplasma species or serovars are not intrinsically “virulent” or “avirulent,” but instead that variation of the ureaplasma MBA protein may play an important role in modulating the immune response: when MBA/mba size variation occurred in vivo there was a decrease in the incidence of chorioamnionitis and lower levels of the cord blood cytokines IL-8 and G-CSF. This hypothesis was also supported by our in vitro findings that recombinant MBA proteins of different sizes elicited altered cytokine responses and augmented expression of NF-κB p65 in macrophages. While variation in the size of the MBA/mba did not always result in an abolished immune response in vitro and in vivo, the responses to these variants were often diminished and this is consistent with an immune evasion event. The ability of these microorganisms to alter/modulate the host immune response may be a contributing factor to the virulence of the Ureaplasma spp. in establishing chronic, asymptomatic infections in utero, and highlights the need for future studies of these microorganisms as underestimated pathogens of pregnancy.
Author Contributions
Conceived and designed the experiments: ES, SK, TG, SS, AJ, and CK. Performed the experiments/generated data within the manuscript: ES, SK, SM, TG, AJ, and CK. Analyzed/Interpreted the data within the manuscript: ES, SK, SM, TG, SS, AJ, and CK. Contributed to the writing/revising of the manuscript: ES, SK, SM, TG, SS, AJ, and CK.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the nurses at the Good Samaritan Hospital, in particular, Dr. Donna Lambers MD, Peggy Walsh RN, Rita Doeger RN, and Laurie Bambrick RN for their assistance in consenting research subjects and collection of the high quality placental samples at the Good Samaritan Hospital, Cincinnati. We gratefully acknowledge the assistance of Karen Henderson, and Thomas Panke MD for placenta specimen collection and processing for pathology evaluation at the Good Samaritan Hospital, Cincinnati. We thank Manuel Alvarez Jr. for assisting in specimen handling. We also thank the Research Flow Cytometry Core facility of CCHMC, in particular, Dr. Claire Chougnet and Casey Wells for performing and analyzing the cord blood cytokine data. We are very grateful to Dr. Charles Armitage and Professor Ken Beagley for their assistance in the production of recombinant MBA proteins and provision of the pRSET. A plasmid and also to Dr. Willa Huston for supplying the THP-1 cell line for use in this project. The authors are also indebted to Emeritus Dr. Patricia Quinn, for the provision of the serovar-specific rabbit antisera used within this study.
References
Cassell, G. H., Waites, K. B., Watson, H. L., Crouse, D. T., and Harasawa, R. (1993). Ureaplasma urealyticum intrauterine infection: role in prematurity and disease in newborns. Clin. Microbiol. Rev. 6, 69–87. doi: 10.1128/CMR.6.1.69
Citti, C., Nouvel, L. X., and Baranowski, E. (2010). Phase and antigenic variation in mycoplasmas. Future Microbiol. 5, 1073–1085. doi: 10.2217/fmb.10.71
Dando, S. J., Nitsos, I., Kallapur, S. G., Newnham, J. P., Polglase, G. R., Pillow, J. J., et al. (2012). The role of the multiple banded antigen of Ureaplasma parvum in intra-amniotic infection: major virulence factor or decoy? PLoS ONE 7:e29856. doi: 10.1371/journal.pone.0029856
De Francesco, M. A., Negrini, R., Pinsi, G., Peroni, L., and Manca, N. (2009). Detection of Ureaplasma biovars and polymerase chain reaction-based subtyping of Ureaplasma parvum in women with or without symptoms of genital infections. Eur. J. Clin. Microbiol. Infect. Dis. 28, 641–646. doi: 10.1007/s10096-008-0687-z
DiGiulio, D. B., Romero, R., Kusanovic, J. P., Gómez, R., Kim, C. J., Seok, K. S., et al. (2010). Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am. J. Reprod. Immunol. 64, 38–57. doi: 10.1111/j.1600-0897.2010.00830.x
Eun, H. S., Lee, S. M., Park, M. S., Park, K. I., Namgung, R., and Lee, C. (2013). Serological investigation of Ureaplasma urealyticum in Korean preterm infants. Korean J. Pediatr. 56, 477–481. doi: 10.3345/kjp.2013.56.11.477
Foley, J. (2015). Mini-review: strategies for variation and evolution of bacterial antigens. Comput. Struct. Biotechnol. J. 13, 407–416. doi: 10.1016/j.csbj.2015.07.002
Gerber, S., Vial, Y., Hohlfeld, P., and Witkin, S. S. (2003). Detection of Ureaplasma urealyticum in second-trimester amniotic fluid by polymerase chain reaction correlates with subsequent preterm labor and delivery. J. Infect. Dis. 187, 518–521. doi: 10.1086/368205
Hillier, S. L., Martius, J., Krohn, M., Kiviat, N., Holmes, K. K., and Eschenbach, D. A. (1988). A case-control study of chorioamnionic infection and histologic chorioamnionitis in prematurity. N. Engl. J. Med. 319, 972–978. doi: 10.1056/NEJM198810133191503
Jacobsson, B., Aaltonen, R., Rantakokko-Jalava, K., Morken, N. H., and Alanen, A. (2009). Quantification of Ureaplasma urealyticum DNA in the amniotic fluid from patients in PTL and pPROM and its relation to inflammatory cytokine levels. Acta Obstet. Gynecol. Scand. 88, 63–70. doi: 10.1080/00016340802572646
Kacerovsky, M., Pliskova, L., Bolehovska, R., Musilova, I., Hornychova, H., Tambor, V., et al. (2011). The microbial load with genital mycoplasmas correlates with the degree of histologic chorioamnionitis in preterm PROM. Am. J. Obstet. Gynecol. 205, 213.e211–217. doi: 10.1016/j.ajog.2011.04.028
Kasper, D. C., Mechtler, T. P., Reischer, G. H., Witt, A., Langgartner, M., Pollak, A., et al. (2010). The bacterial load of Ureaplasma parvum in amniotic fluid is correlated with an increased intrauterine inflammatory response. Diagn. Microbiol. Infect. Dis. 67, 117–121. doi: 10.1016/j.diagmicrobio.2009.12.023
Knox, C. L., Allan, J. A., Allan, J. M., Edirisinghe, W. R., Stenzel, D., Lawrence, F. A., et al. (2003). Ureaplasma parvum and Ureaplasma urealyticum are detected in semen after washing before assisted reproductive technology procedures. Fertil. Steril. 80, 921–929. doi: 10.1016/S0015-0282(03)01125-7
Knox, C. L., Cave, D. G., Farrell, D. J., Eastment, H. T., and Timms, P. (1997). The role of Ureaplasma urealyticum in adverse pregnancy outcome. Aust. N. Z. J. Obstet. Gynaecol. 37, 45–51. doi: 10.1111/j.1479-828X.1997.tb02216.x
Knox, C. L., Dando, S. J., Nitsos, I., Kallapur, S. G., Jobe, A. H., Payton, D., et al. (2010). The severity of chorioamnionitis in pregnant sheep is associated with in vivo variation of the surface-exposed multiple-banded antigen/gene of Ureaplasma parvum. Biol. Reprod. 83, 415–426. doi: 10.1095/biolreprod.109.083121
Knox, C. L., and Timms, P. (1998). Comparison of PCR, nested PCR, and random amplified polymorphic DNA PCR for detection and typing of Ureaplasma urealyticum in specimens from pregnant women. J. Clin. Microbiol. 36, 3032–3039.
Li, Y. H., Brauner, A., Jonsson, B., van der Ploeg, I., Söder, O., Holst, M., et al. (2000). Ureaplasma urealyticum-induced production of proinflammatory cytokines by macrophages. Pediatr. Res. 48, 114–119. doi: 10.1203/00006450-200007000-00020
Lipsitch, M., and O'Hagan, J. J. (2007). Patterns of antigenic diversity and the mechanisms that maintain them. J. R. Soc. Interface 4, 787–802. doi: 10.1098/rsif.2007.0229
Manimtim, W. M., Hasday, J. D., Hester, L., Fairchild, K. D., Lovchik, J. C., and Viscardi, R. M. (2001). Ureaplasma urealyticum modulates endotoxin-induced cytokine release by human monocytes derived from preterm and term newborns and adults. Infect. Immun. 69, 3906–3915. doi: 10.1128/IAI.69.6.3906-3915.2001
Monecke, S., Helbig, J. H., and Jacobs, E. (2003). Phase variation of the multiple banded protein in Ureaplasma urealyticum and Ureaplasma parvum. Int. J. Med. Microbiol. 293, 203–211. doi: 10.1078/1438-4221-00239
Naessens, A., Foulon, W., Breynaert, J., and Lauwers, S. (1988). Serotypes of Ureaplasma urealyticum isolated from normal pregnant women and patients with pregnancy complications. J. Clin. Microbiol. 26, 319–322.
Naessens, A., Foulon, W., Cammu, H., Goossens, A., and Lauwers, S. (1987). Epidemiology and pathogenesis of Ureaplasma urealyticum in spontaneous abortion and early preterm labor. Acta Obstet. Gynecol. Scand. 66, 513–516. doi: 10.3109/00016348709015726
Namba, F., Hasegawa, T., Nakayama, M., Hamanaka, T., Yamashita, T., Nakahira, K., et al. (2010). Placental features of chorioamnionitis colonized with Ureaplasma species in preterm delivery. Pediatr. Res. 67, 166–172. doi: 10.1203/PDR.0b013e3181c6e58e
Park, E. K., Jung, H. S., Yang, H. I., Yoo, M. C., Kim, C., and Kim, K. S. (2007). Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 56, 45–50. doi: 10.1007/s00011-007-6115-5
Peltier, M. R., Freeman, A. J., Mu, H. H., and Cole, B. C. (2007). Characterization of the macrophage-stimulating activity from Ureaplasma urealyticum. Am. J. Reprod. Immunol. 57, 186–192. doi: 10.1111/j.1600-0897.2006.00460.x
Rahman, M. M., and McFadden, G. (2011). Modulation of NF-kappaB signalling by microbial pathogens. Nat. Rev. Microbiol. 9, 291–306. doi: 10.1038/nrmicro2539
Redline, R. W., Faye-Petersen, O., Heller, D., Qureshi, F., Savell, V., and Vogler, C. (2003). Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns. Pediatr. Dev. Pathol. 6, 435–448. doi: 10.1007/s10024-003-7070-y
Robinson, J. W., Dando, S. J., Nitsos, I., Newnham, J., Polglase, G. R., Kallapur, S. G., et al. (2013). Ureaplasma parvum serovar 3 multiple banded antigen size variation after chronic intra-amniotic infection/colonization. PLoS ONE 8:e62746. doi: 10.1371/journal.pone.0062746
Shepard, M. C., and Lunceford, C. D. (1976). Differential agar medium (A7) for identification of Ureaplasma urealyticum (human T mycoplasmas) in primary cultures of clinical material. J. Clin. Microbiol. 3, 613–625.
Shimizu, T., Kida, Y., and Kuwano, K. (2008). Ureaplasma parvum lipoproteins, including MB antigen, activate NF-{kappa}B through TLR1, TLR2 and TLR6. Microbiology 154(Pt 5), 1318–1325. doi: 10.1099/mic.0.2007/016212-0
Sweeney, E. L., Kallapur, S. G., Gisslen, T., Lambers, D. S., Chougnet, C. A., Stephenson, S. A., et al. (2016). Placental infection with ureaplasma species is associated with histologic chorioamnionitis and adverse outcomes in moderately preterm and late-preterm infants. J. Infect. Dis. 213, 1340–1347. doi: 10.1093/infdis/jiv587
Triantafilou, M., De Glanville, B., Aboklaish, A. F., Spiller, O. B., Kotecha, S., and Triantafilou, K. (2013). Synergic activation of toll-like receptor (TLR) 2/6 and 9 in response to Ureaplasma parvum & urealyticum in human amniotic epithelial cells. PLoS ONE 8:e61199. doi: 10.1371/annotation/1dc00176-e096-4621-9494-2d848dac8262
Volgmann, T., Ohlinger, R., and Panzig, B. (2005). Ureaplasma urealyticum-harmless commensal or underestimated enemy of human reproduction? A review. Arch. Gynecol. Obstet. 273, 133–139. doi: 10.1007/s00404-005-0030-1
Zheng, X., Lau, K., Frazier, M., Cassell, G. H., and Watson, H. L. (1996). Epitope mapping of the variable repetitive region with the MB antigen of Ureaplasma urealyticum. Clin. Diagn. Lab. Immunol. 3, 774–778.
Zheng, X., Teng, L. J., Watson, H. L., Glass, J. I., Blanchard, A., and Cassell, G. H. (1995). Small repeating units within the Ureaplasma urealyticum MB antigen gene encode serovar specificity and are associated with antigen size variation. Infect. Immun. 63, 891–898.
Zheng, X., Watson, H. L., Waites, K. B., and Cassell, G. H. (1992). Serotype diversity and antigen variation among invasive isolates of Ureaplasma urealyticum from neonates. Infect. Immun. 60, 3472–3474.
Zimmerman, C. U., Rosengarten, R., and Spergser, J. (2011). Ureaplasma antigenic variation beyond MBA phase variation: DNA inversions generating chimeric structures and switching in expression of the MBA N-terminal paralogue UU172. Mol. Microbiol. 79, 663–676. doi: 10.1111/j.1365-2958.2010.07474.x
Zimmerman, C. U., Rosengarten, R., and Spergser, J. (2013). Interaction of the putative tyrosine recombinases RipX (UU145), XerC (UU222), and CodV (UU529) of Ureaplasma parvum serovar 3 with specific DNA. FEMS Microbiol. Lett. 340, 55–64. doi: 10.1111/1574-6968.12077
Keywords: Ureaplasma species, preterm birth, chorioamnionitis, multiple banded antigen (MBA), virulence, host-microbe Interactions
Citation: Sweeney EL, Kallapur SG, Meawad S, Gisslen T, Stephenson S-A, Jobe AH and Knox CL (2017) Ureaplasma Species Multiple Banded Antigen (MBA) Variation Is Associated with the Severity of Inflammation In vivo and In vitro in Human Placentae. Front. Cell. Infect. Microbiol. 7:123. doi: 10.3389/fcimb.2017.00123
Received: 05 January 2017; Accepted: 27 March 2017;
Published: 13 April 2017.
Edited by:
Margaret E. Bauer, Indiana University School of Medicine, USAReviewed by:
Azadeh Farzin, Johns Hopkins University, USABryan Troxell, North Carolina State University, USA
Copyright © 2017 Sweeney, Kallapur, Meawad, Gisslen, Stephenson, Jobe and Knox. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emma L. Sweeney, el.sweeney@qut.edu.au
 Emma L. Sweeney
Emma L. Sweeney Suhas G. Kallapur
Suhas G. Kallapur Simone Meawad1
Simone Meawad1  Sally-Anne Stephenson
Sally-Anne Stephenson Alan H. Jobe
Alan H. Jobe