Genomic Insight into Mechanisms of Reversion of Antibiotic Resistance in Multidrug Resistant Mycobacterium tuberculosis Induced by a Nanomolecular Iodine-Containing Complex FS-1
- 1Scientific Center for Anti-Infectious Drugs, Almaty, Kazakhstan
- 2Department of Biochemistry, Centre for Bioinformatics and Computational Biology, University of Pretoria, Pretoria, South Africa
Drug induced reversion of antibiotic resistance is a promising way to combat multidrug resistant infections. However, lacking knowledge of mechanisms of drug resistance reversion impedes employing this approach in medicinal therapies. Induction of antibiotic resistance reversion by a new anti-tuberculosis drug FS-1 has been reported. FS-1 was used in this work in combination with standard anti-tuberculosis antibiotics in an experiment on laboratory guinea pigs infected with an extensively drug resistant (XDR) strain Mycobacterium tuberculosis SCAID 187.0. During the experimental trial, genetic changes in the population were analyzed by sequencing of M. tuberculosis isolates followed by variant calling. In total 11 isolates obtained from different groups of infected animals at different stages of disease development and treatment were sequenced. It was found that despite the selective pressure of antibiotics, FS-1 caused a counter-selection of drug resistant variants that speeded up the recovery of the infected animals from XDR tuberculosis. Drug resistance mutations reported in the genome of the initial strain remained intact in more sensitive isolates obtained in this experiment. Variant calling in the sequenced genomes revealed that the drug resistance reversion could be associated with a general increase in genetic heterogeneity of the population of M. tuberculosis. Accumulation of mutations in PpsA and PpsE subunits of phenolpthiocerol polyketide synthase was observed in the isolates treated with FS-1 that may indicate an increase of persisting variants in the population. It was hypothesized that FS-1 caused an active counter-selection of drug resistant variants from the population by aggravating the cumulated fitness cost of the drug resistance mutations. Action of FS-1 on drug resistant bacteria exemplified the theoretically predicted induced synergy mechanism of drug resistance reversion. An experimental model to study the drug resistance reversion phenomenon is hereby introduced.
Introduction
The dilemma of acquired drug resistance by pathogenic bacteria is generally recognized (Tenover, 2001; Morens et al., 2004). This problem is complicated by a common crisis in development of new antibiotics (Spellberg et al., 2008; van den Boogaard et al., 2009; Piddock, 2012). Drug induced reversion of antibiotic resistance is a prospective approach to combat drug resistant infections. However, the mechanisms of this phenomenon are not understood and even the terminology has not been well defined. In early publications, this term referred to a suppression of antibiotic resistance proteins such as drug efflux pumps and/or beta-lactamases (Reading and Cole, 1977; Rodrigues et al., 2013). Another definition was given in a recent review by Baym et al. (2016), where this phenomenon was considered as an active drug induced counter-selection of resistant variants from populations of pathogens. Several theoretic clauses were discussed in the mentioned paper to explain the resistance reversion that occurred despite the presence of selective antibiotics. Drug induced over-burdening of the resistant strains by an aggravated fitness cost was considered as a possible mechanism. The fitness cost relates to adverse side effects of drug resistance mutations reducing the viability, adaptability, and growth rate of the mutants (Cohen et al., 2003; Gagneux et al., 2006). A mathematic model by Cohen and Murray (2004) has predicted that multidrug resistant pathogens would be rather unlikely to appear due to the cumulated fitness cost. Unfortunately, this prediction was rather over-optimistic. Subsequent compensatory mutations reduce the fitness cost to such a level that the population could retain the mutants for an extended period of time even after the withdrawal of the selective pressure of antibiotics (Pym et al., 2002; Luciani et al., 2009). However, the idea to aggravate the cumulated fitness cost by supplementary drugs in an effort to combat drug resistance still seems attractive.
Tuberculosis remains one of the most dangerous threats to public health. This problem has been complicated by a wide distribution of drug resistant infections that basically brought about a return to the pre-antibiotic era (Cohn et al., 1997; Nouvel et al., 2006; Zager and McNerney, 2008). Multidrug resistant (MDR) tuberculosis pathogens were defined by the WHO as those resistant to isoniazid and rifampicin; and extensively drug resistant (XDR) variants being additionally resistant to fluoroquinalones and at least one second-line injectable antibiotics. Treatment of MDR and XDR tuberculosis requires an application of complex formulations of antibiotics (Iseman, 1993) that often cannot be administered due to life threatening side effects (Awofeso, 2008).
Drug resistance in Mycobacterium tuberculosis is associated with spontaneous mutations in functional genomic loci. More than 1,000 putative drug resistance mutations have been predicted (Sandgren et al., 2009). Over the last 40 years, no new antibiotics against tuberculosis have been developed and only recently several new drugs have been proposed: Bedaquiline (Mahajan, 2013), Delamanid (Gupta et al., 2015) and FS-1 (Ilin and Kulmanov, 2014; Kalykova et al., 2016). However, resistance to Bedaquiline and Delamanid has already been reported (Hoffmann et al., 2016). FS-1 is an iodine-containing nanomolecular complex showing an antimicrobial effect (Kalykova et al., 2016). Active units of FS-1 are aggregated micelles containing complexes of triiodide molecules coordinated by metal ions and integrated into a dextrin-polypeptide moiety. The basic formula of the micelle is:
where L–dextrin-polypeptide ligand; Me–Li/Mg ions; n, m, x, y, and k–variable integers ≥1; molecular mass of the micelles is in the range of 30–300 kD.
In the blood plasma, the micelles bind to blood albumins. The mean residence time (MRT) of FS-1 estimated as a geometric average of the time of elimination of the drug from an organism (Cawello, 1999), was 24.6 h. Disintegration of the micelles causes a dissociation of triiodides into iodine molecules, which are the active antimicrobial agents of FS-1.
FS-1 passed preclinical and clinical trials and in 2015 it was accepted as a new anti-MDR/XDR drug in Kazakhstan (Ilin and Kulmanov, 2014). FS-1 is of great interest for this study because of the reported antibiotic resistance reversion induced by this drug. It was hypothesized that FS-1 could influence the composition of bacterial populations by removal of the most resistant variants of M. tuberculosis, probably due to higher sensitivity to oxidative stresses (Cohen and Murray, 2004; Ilin et al., 2017).
The aim of this work was to perform a laboratory experiment on animals infected with a XDR-TB strain to identify genetic changes in bacterial populations induced by FS-1. The strain M. tuberculosis SCAID 187.0 was used in this study. It was isolated from a patient with tuberculosis showing extensive drug resistance (Ilin et al., 2015). The present study proved the efficacy of the combinatorial treatment of infected animals by conventional anti-tuberculosis antibiotics supplemented with FS-1. An increased susceptibility to antibiotics was observed in M. tuberculosis isolates from the FS-1 treated animals. Sequencing of the isolates demonstrated that FS-1 caused recognizable changes in the M. tuberculosis populations by removal of the most resistant clonal lines, which were dominant in the untreated animals and in the animals treated solely by antibiotics. Reduction of the antibiotic resistance correlated with an increased genetic heterogeneity and accumulation of mutated variants of PpsA and truncated PpsE subunits of the phenolpthiocerol polyketide synthase.
Materials and Methods
Isolation and Identification of Clinical Cultures of M. tuberculosis
M. tuberculosis strains were isolated on a regular basis from patients' sputum samples during the combinatorial therapy of antibiotics and FS-1 (Ilin et al., 2017). Sputum samples were inoculated into liquid Löwenstein-Jensen medium (HiMedia Laboratories, India) and cultivated at 37°C for 8 weeks. Culture growth was controlled visually, by microscopy of Ziehl–Neelsen stained smears and by standard diagnostic biochemical tests including the positive catalase, niacin and nicotinamidase activities, negative Tween-80 hydrolysis and susceptibility to sodium salicylate (Segal and Bloch, 1956).
Minimal Inhibitory Concentration of FS-1
Minimal inhibitory concentration (MIC) of FS-1 was tested on the type strain M. tuberculosis H37Rv and several clinical XDR isolates by serial dilution of FS-1 in liquid Löwenstein-Jensen medium with 1.75, 0.35, 0.175, 0.0875, 0.0437, 0.0218, and 0.0109 mg/ml of FS-1. Culture suspensions of 5 × 108 cells/ml were inoculated in aliquots of 0.2 ml into test tubes with 5 ml of the medium with FS-1 and without the compound for the positive growth control. Each concentration of FS-1 was triple replicated. Tubes were incubated for 10 days at 37°C.
Laboratory Animals
In total, 105 healthy male and female 6–8 weeks old Hartley guinea pigs weighing 350–450 g were obtained from the Kazakh Scientific Research Veterinary Institute, Almaty, Kazakhstan. Animals were held under barrier conditions in a biosafety level III animal laboratory at 23 ± 2°C, humidity 50 ± 10%, on a 12 h light/dark cycle. All the animals received the standard forage (Ltd. Assortiment Agro, Russia) and deionized water consumed ad libitum.
For blood collection and necropsy, the animals were sacrificed by an overdose of sodium pentobarbital (JSC KievMedPreparat, Ukraine).
The study was approved by the Ethics Committee of the Scientific Center for Anti-Infectious Drugs (no 29, 05.06.2014). Animal care complied with the Guide for the Care and Use of Laboratory Animals (National Research Council, 2010) and with the Kazakhstan governmental guidelines.
Infection and Treatment of the Animals by Anti-Tuberculosis Antibiotics and FS-1
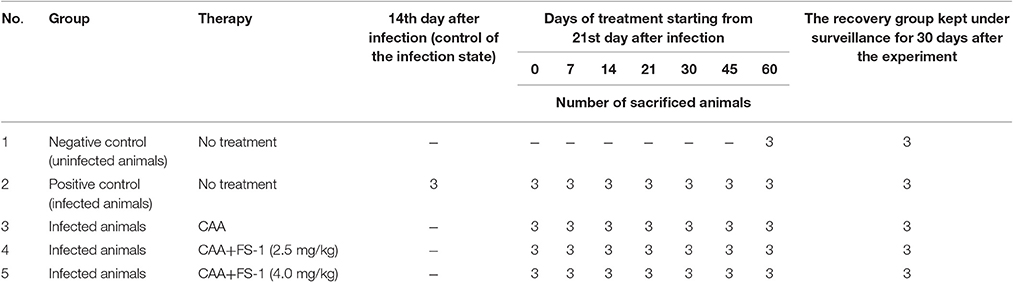
Guinea pigs were infected subcutaneously with 1 × 106 CFU/ml of the virulent strain M. tuberculosis SCAID 187.0. Bacterial suspensions were prepared ex tempore 2 h before infection from the bacterial culture cultivated for 10 days at 37°C on solid Löwenstein-Jensen medium. Development of tuberculosis in the animals was monitored weekly by changes in behavior, general appearance and body weight loss. On the 14th day after infection, three infected animals from the positive control group were sacrificed to confirm the infection state by necropsy and histopathological parameters.
Anti-tuberculosis antibiotics were administrated starting on the 21st day after infection. Antibiotic dosage corresponded to the XDR-TB treatment regimen approved in Kazakhstan by the Ministry of Health. A combined formulation of 4–5 antibiotics is usually applied for XDR-TB treatment (Iseman, 1993). The complex of anti-tuberculosis antibiotics (CAA) included pyrazinamide (per os 100 mg/kg), cycloserine (per os 20.0 mg/kg), prothionamide (per os 20.0 mg/kg), capreomycin (intramuscular 20.0 mg/kg), and amikacin (intramuscular 50.0 mg/kg). FS-1 was administrated per os 30 min before CAA in doses of 2.5 and 4.0 mg/kg, which were respectively the therapeutic and maximal tolerable doses of the drug determined during the clinical trials (Kalykova et al., 2016). The treatment course lasted for 60 days. The animals were weighed daily and the dosage of the antibiotics and FS-1 were adjusted individually for every animal. The experimental design is shown in Table 1.
Blood Sampling and Histological Studies
Blood and tissue collections were performed for hematological, biochemical, microbiological and histological studies on the 14th and 21st days after the infection and on the 14th, 30th, 45th and 60th days of the treatment. Whole blood collection was performed by a cardiac puncture of the narcotized animals. Blood samples were placed in 1 ml Vacumed test tubes with K3-EDTA anticoagulant (FL Medical, Italy) and then analyzed by the Analyzer Human Humacount (Human GmbH, Germany). The remaining volumes of the blood samples were placed in 1.5 ml vials without anticoagulants and allowed to clot at room temperature for 60 min. The samples were then centrifuged at 1800 × g for 5 min. The serum was decanted and stored at −80°C prior to the analysis by Analyzer A25 (BioSystems Diagnostics Pvt. Ltd., India) to identify concentrations of the chemical compounds. The statistical reliability of observed differences was confirmed by Student's t-test. P-values smaller or equal to 0.05 were considered as significant.
Necropsy
Pieces of 10 × 20 mm were sampled in several repeats from the areas in-between necrolysis and intact tissues from lungs, livers, spleens, kidneys and thyroid glands of the sacrificed animals. The samples were fixed in 10% paraformaldehyde (Sigma, USA) buffered with PBS (Sigma, USA) at pH 7.0. Paraffin embedding and sectioning were performed by using the conventional methodology (Ross and Pawlina, 2006). Then the tissue samples were trimmed and processed by microscopic examination after staining with hematoxylin and eosin (Sigma, USA), Masson's trichrome and Van Gieson's stains (Bio Optica, Italy).
Isolation and Storage of M. tuberculosis Cultures from the Infected Animals
Tissue samples of approximately the same size (10 × 20 mm) were homogenized in sterilized laboratory mortars and re-suspended in 4 volumes of 6% sulphuric acid by shaking for 10 min at room temperature. The obtained suspension was left to precipitate and the supernatant fluid was removed. Deposited materials were washed out 3 times with 4 volumes of sterile saline (0.9% NaCl in distilled water) and the aliquots of 1 ml were inoculated onto solid Löwenstein-Jensen medium for further cultivation at 37°C for 8 weeks. Culture growth was controlled visually. Selected colonies (from 10 to 25 per group of isolates) were checked individually for susceptibility to antibiotics. The isolated cultures were frozen at −80°C in a protective medium (3% gelatin and 10% sucrose in water) and then lyophilized by ALPHA 1-2 LD-plus (Martin Christ Gefriertrocknungsanlagen GmbH, Germany) for extended storage at −30°C.
Susceptibility to Antibiotics
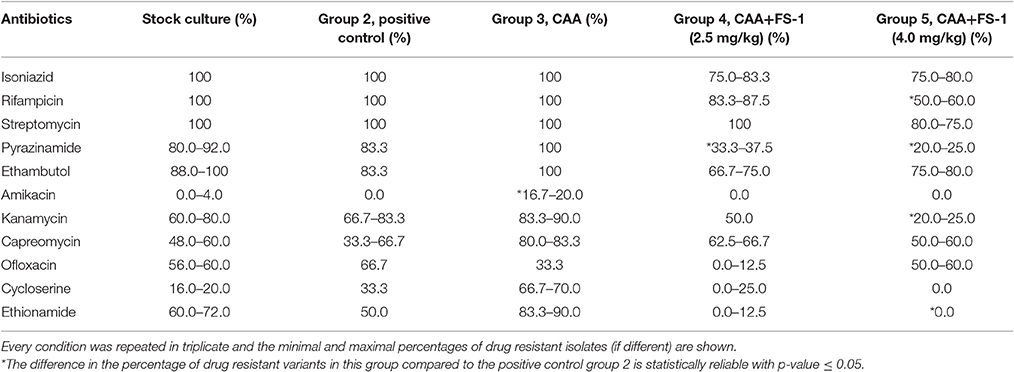
Susceptibility to all antibiotics but pyrazinamide was tested by growing the cultures for 4 weeks at 37°C in test-tubes on solid Löwenstein-Jensen medium supplemented with antibiotics in recommended concentrations (Krüüner et al., 2006). Medium pH was adjusted to 6.8. Susceptibility to pyrazinamide was tested on solid medium FAST-3L (BIOK, Russia) with pH adjusted to 5.0. The following antibiotics in μg/ml were used: isoniazid (0.2), rifampicin (40.0), streptomycin (4.0), pyrazinamide (200.0), ethambutol (2.0), amikacin (30.0), kanamycin (30.0), capreomycin (40.0), ofloxacin (2.0), cycloserine (40.0), and ethionamide (40.0). Percentages of drug resistant variants among M. tuberculosis isolates from different groups of infected animals were calculated. To compare the percentages of drug resistance in different groups, the standard error was calculated as the square root of [p × (1–p)] / n, where p, an average percentage of drug resistant isolates in a group; n, number of repeats of the experiment.
Minimal inhibition concentrations (MIC) of the antibiotics were determined by the serial dilution method in a synthetic nutrient medium containing: KH2PO4—1.5 g/l, Na2HPO4—2.5 g/l, MgSO4× 7H2O—0.5 g/l, sodium citrate—1.5 g/l, ammonium iron (III) hydrocitrate—0.05 g/l, L-asparagine—0.1 g/l, glycerol—30 ml, sucrose—220 g/l, bovine serum (Sigma Aldrich)—100 ml/l and bacteriological agar (HiMedia Laboratories, India)—0.5 g/l. Antibiotics were dissolved in the medium to obtain the required serial dilutions. Then the diluted solutions of the antibiotics were dispensed by 700 μl per well into 48-well plates (BD Falcon, NU, USA). Each well was inoculated with 100 μl of ex tempore prepared suspensions of bacterial cells, 108 CFU/ml. Plates were incubated for 8 days at 37°C. Then aliquots of 100 μl of 0.05% (w/v) resazurin (Sigma-Aldrich, St. Louis, MO) were added to each well and the plates were incubated for a further 24 h. Bacterial growth was detected by changes in the medium color from violet to pink. MIC values were identified in 3 repeats. Medium contamination with and without antibiotics and the growth of M. tuberculosis on the medium without antibiotics were controlled. The following serial dilutions of the antibiotics were used: isoniazid—0.35, 0.3, 0.25, 0.2, 0.15, 0.1, and 0.05 μg/ml; rifampicin—6.0, 5.0, 4.0, 3.0, 2.0, 1.0, and 0.5 μg/ml; streptomycin—6.0, 5.0, 4.0, 3.0, 2.0, 1.0, and 0.5 μg/ml; pyrazinamide—350, 300, 250, 200, 150, 100, and 50 μg/ml; and ethionamide—17.5, 15.0, 12.5, 10.0, 7.5, 5.0, and 2.5 μg/ml.
DNA Extraction, Sequencing and Comparison
DNA samples were extracted from the isolates by the Cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). The eluted DNA was quantified using the Qubit dsDNA BR Assay Kit (Life Technologies, USA). In total, twelve DNA samples were prepared (Table 2) and sequenced by Macrogen (South Korea) using Illumina Hiseq 2000 paired-end technology.
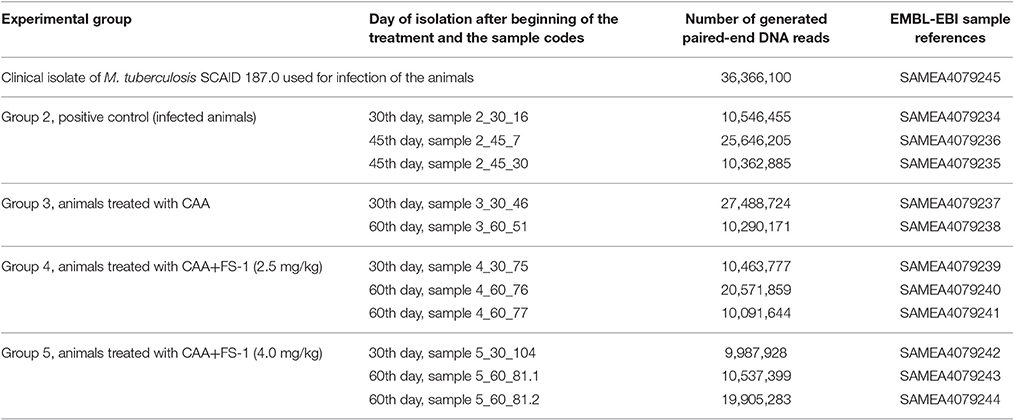
Table 2. Sequencing of M. tuberculosis isolates from the infected animals on different days of the experiment.
Variant calling was performed by aligning the DNA reads to the reference sequence SCAID 187.0 [CP012506] using CLC Genomics Workbench 7.0.3. Statistical validation of the identified variants was performed by the Quality based variant detection algorithm with the PHRED-type quality cut-off set to 20 for the central nucleotide residue and to 15 for five neighboring residues. Maximum of two gaps or mismatches were allowed per read.
Results
The experiment on laboratory animals was designed to simulate the clinical trial of FS-1 and induction of an antibiotic resistance reversion under the selective pressure of antibiotics. MIC of FS-1 was identified for the clinical isolate M. tuberculosis SCAID 187 during the first isolation and after 60 days of the combinatorial treatment by CAA+FS-1. Prior to the treatment, the determined MIC was 27.7 ± 2.4 μg/ml and after the treatment course the isolates from the same patients became even more sensitive to FS-1 with the MIC values 21.1 ± 2.0 μg/ml. This indicates that the application of FS-1 did not cause any resistance selection for the drug. An interesting observation was that the clinical XDR-TB isolate was even more sensitive to FS-1 than the drug sensitive type strain M. tuberculosis H37Rv (MIC was around 48 μg/ml). However, the hypothesis of a higher susceptibility of multidrug resistant variants has yet to be confirmed in an additional study with a greater number of isolates.
Drug Resistance Reversion Induced In vitro by FS-1
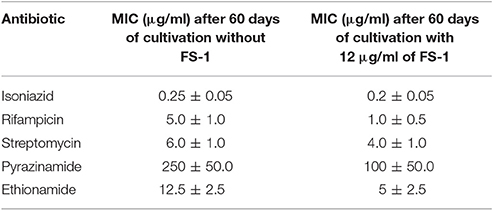
It was found that the cultivation of the XDR strain SCAID 187.0 for 60 days in six passages on the medium with a sub-lethal dose of 12 μg/ml of FS-1 reversed the susceptibility of the bacteria to the tested antibiotics. MIC values determined for the cultures cultivated with FS-1 compared to the control condition (cultivated on the same medium without FS-1) are shown in Table 3 (see also Supplementary Figure 1). The values in Table 3 were confirmed in three replications of the experiment. More than two folds MIC reduction observed for rifampicin, ethionamide and pyrazinamide was considered as statistically reliable. These results cannot be attributed to any direct interaction between FS-1 and the antibiotics as the susceptibility was tested on the medium without FS-1. The increased susceptibility to the antibiotics may be explained by a counter-selection of the most resistant variants from the population.
Combinatorial Treatment of XDR-TB Infected Laboratory Animals with Antibiotics and FS-1
Miliary tuberculosis was diagnosed by necropsy of the sacrificed animals and by microscopy of histological sections. Granulomatous lungs, spleens and livers with multiple disseminated tubercles and significantly enlarged lymph nodes were observed (Supplementary Figure 2). Untreated animals died during the 60 days of the experiment. Contrary, the infected animals subjected to combinatorial treatment with FS-1 in doses of 2.5 and 4 mg/kg recovered from the disease. Fast recovery of these animals could even be seen by the positive body weight dynamics compared to the other groups of animals (Supplementary Table 1). The antibiotic therapy without FS-1 showed only a transient effect followed by a subsequent recurrence of the disease during the 30 days of the recovery period (Figure 1). Necropsy of the sacrificed animals and the results of the biochemical blood tests also confirmed the positive effect of FS-1 (Supplementary Figure 3 and Supplementary Table 2). In general, the biochemical blood test parameters in the groups treated by CAA+FS-1 corresponded to those in the intact animals of the negative control group (Supplementary Table 2). It was concluded that the hepatotoxicity of the anti-tuberculosis antibiotics was alleviated by FS-1 either by a direct action on hepatocytes, or by a faster removal of the tuberculosis infection. Differences revealed by these tests between the animals administrated with 2.5 and 4.0 mg/kg of FS-1 were statistically insignificant.
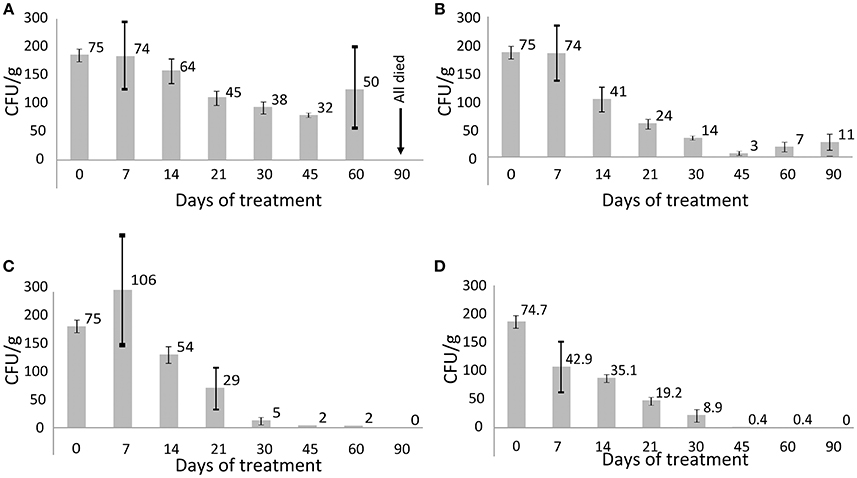
Figure 1. CFU titres in different groups of animals infected with the XDR M. tuberculosis. Average values are indicated and the bars depict standard deviations. (A) Group 2, positive control (untreated animals); (B) group 3 treated with CAA; (C) group 4 treated with CAA+FS-1 (2.5 mg/kg); (D) group 5 treated with CAA+FS-1 (4.0 mg/kg).
M. tuberculosis isolates from the animals treated for 45 days by CAA+FS-1 showed an increased susceptibility to the antibiotics compared to the isolates from the positive control group and especially to those treated solely by CAA (Table 4). There was a clear trend of a dose dependent drug resistance reversion induced by FS-1 despite some variations between the isolates from different animals. Contrary, the treatment of the animals infected with XDR-TB solely by CAA caused a selection of more resistant variants.
Comparison of Genome Sequences of M. tuberculosis Isolates
Eight drug resistance mutations listed in Table 5 were found in the reference genome M. tuberculosis SCAID 187.0 (CP012506). All of them remained unchanged in the isolates showing reduced drug resistance (Table 4). This finding was consistent with the results of sequencing of a series of clinical isolates obtained during the clinical trial of FS-1 (Ilin et al., 2017). Variant calling in the experimental isolates revealed 30 polymorphic sites, which were found in at least two isolates per group with the frequency of minor alleles above 1% and a read coverage above 10 (Table 6). Only two polymorphic loci were present in the isolates from the animals treated solely by CAA. This indicates a purifying selection caused by the antibiotics. Fifteen polymorphic loci were discovered in the isolates from the untreated animals. M. tuberculosis isolates from the animals treated by CAA+FS-1 showed the highest genetic heterogeneity with 20 and 26 identified polymorphic loci in groups 4 and 5, respectively. Almost all polymorphic loci were in hyper-variable PPE-PGRS genes. However, the treatment of the animals with FS-1 caused an accumulation of mutations in subunits A and E of the phenolpthiocerol synthase. One mutation causing a P-to-H substitution in a proline rich linker region between the acyltransferase and dehydratase domains was characteristic for 37–38% of the DNA reads generated from the isolates of groups 4 and 5. A frameshift mutation caused by an insertion of 2 nucleotides in the mid part of the ppsE subunit was found in 95% of the reads generated from the isolates on the 30th day of the treatment by CAA + 4.0 mg/kg FS-1. In the M. tuberculosis cultures isolated from animals in this group on the 60th day of the treatment the percentage of this mutation lowered to 50%. These mutations in ppsA and ppsE were not found in the initial strain SCAID 187.0 or in the isolates from the other groups of animals.
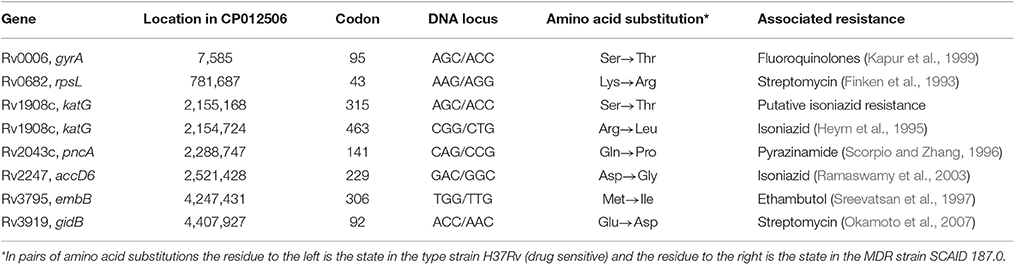
Table 5. Canonical drug resistance mutations found in the genome of the strain M. tuberculosis SCAID 187.0 [CP012506].
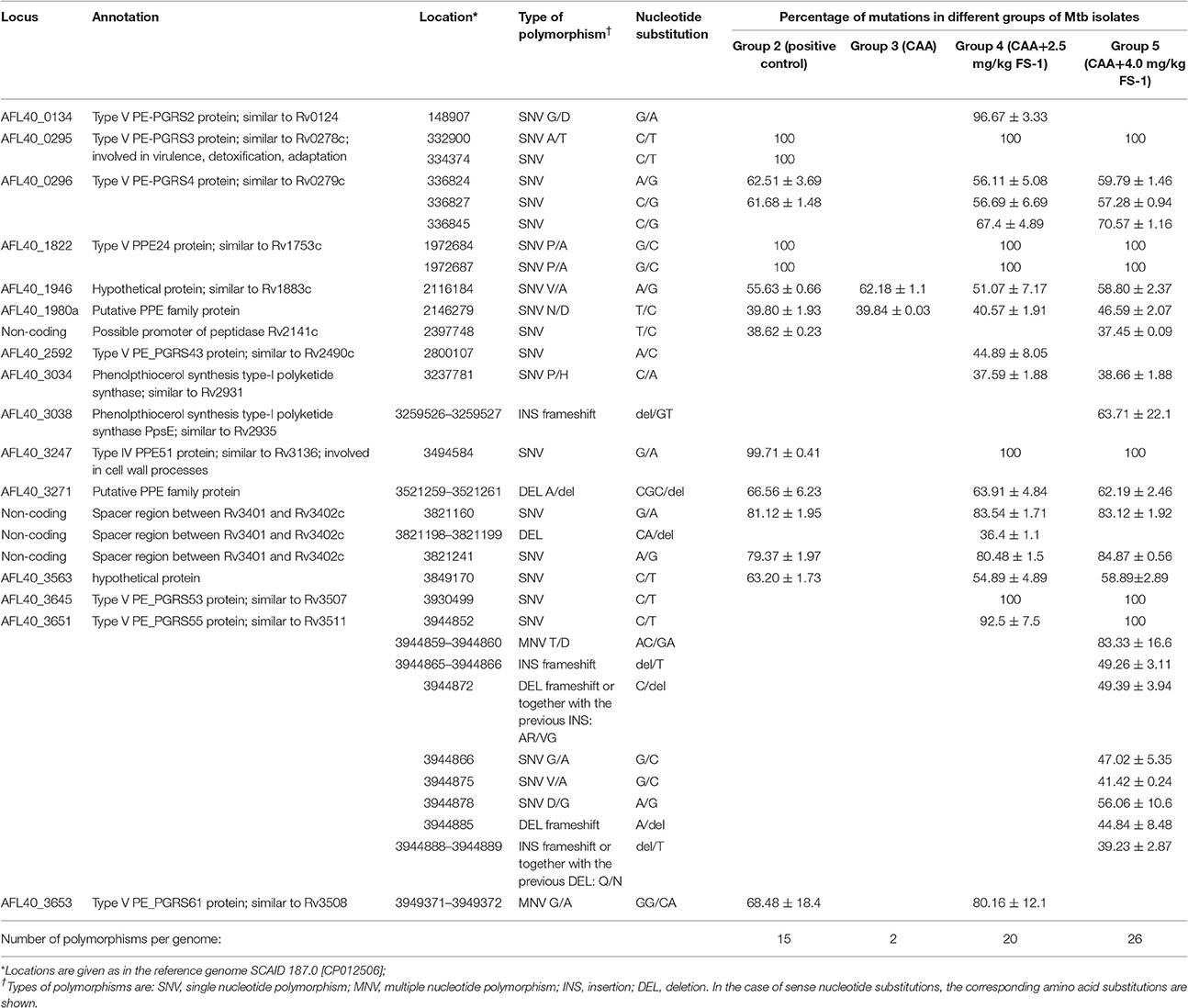
Table 6. Genomic polymorphisms in M. tuberculosis isolates from different groups of experimental animals identified by the quality based variant calling against the reference genome SCAID 187.0 [CP012506].
Discussion
The experiment on laboratory animals confirmed the therapeutic effect of FS-1 administrated in combination with the antibiotics of the standard regiment. It was hypothesized that the induced drug resistance reversion presumably contributed to the therapeutic effect by FS-1. Remarkably, the drug resistance reversion was demonstrated also in vitro where no direct interaction between FS-1 and the antibiotics was possible. There are evidences that FS-1 can cause a counter-selection of the drug resistant variants of M. tuberculosis (Ilin et al., 2017).
Sequencing of the isolates obtained from different groups of animals revealed dissimilar frequencies and allelic states of genomic polymorphisms. Selection of the resistant clonal lines was caused by the CAA treatment. Contrary, no resistance selection was observed in the M. tuberculosis isolates from the animals threated by CAA+FS-1. The combinatorial therapy led to a complete sanitation of the infected animals from Mycobacterial bacilli in 45–60 days of the treatment (Figure 1) which was also confirmed by the blood test results and necropsy (Supplementary Figures 2–3, Supplementary Tables 1–2).
M. tuberculosis isolates obtained during the combinatorial treatment showed an increased susceptibility to antibiotics (Table 4) associated with an increased level of genetic heterogeneity (Table 6). However, all the “canonical” drug resistance mutations identified in the initial strain M. tuberculosis SCAID 187.0 (Table 5) remained intact in the isolates despite the increased susceptibility. This suggests that the known antibiotic resistance mutations might not be sufficient to produce sustainable drug resistance which requires the presence of a specific genetic context rendered by other polymorphic genes.
Majority of the polymorphic loci were located in hyper-variable PPE-PGRS genes. These genes are three fold more mutable than the average. It was hypothesized that their major role was an antigenic redress of bacterial cells (Banu et al., 2002). Several additional functions of PPE-PGRS proteins were proposed in a recent publication by Fishbein et al. (2015). Selection of PPE-PGRS mutants was also observed in M. tuberculosis cultures grown in human macrophages (Guerrini et al., 2016). However, it should also be considered that these mutations could just be genetic markers of different competing clonal lines in the population, which differ in gene transcription or DNA methylation patterns (Heusipp et al., 2007).
CAA+FS-1 treatment of the infected animals caused an accumulation of disruptive mutations in ppsE and ppsA. A similar result was obtained recently by sequencing a series of TB isolates from patients treated with FS-1 during clinical trials (Ilin et al., 2017). This polyketide synthase operon encodes synthesis of a methyl-branched fatty acid phthiocerol dimycocerosate (PDIM) that is a characteristic cell wall compound of Mycobacteria (Azad et al., 1997; Camacho et al., 2001). PDIM has been proven to be an important virulence factor (Yu et al., 2012). However, other authors noted that impairing of the PDIM biosynthesis improved the persistence ability of the pathogens (Torrey et al., 2016). Variations in the PDIM operon may allow finding a compromise between pathogenicity, viability and ability to persist. An interesting observation was that the frequency of the frameshift mutations in ppsE reached the 95% maximum after 30 days of the treatment by CAA + 4 mg/kg FS-1, but then decreased to 50% implying that the persistent variants did not overcome the continuous treatment by FS-1.
The observed increase in the genetic heterogeneity of the isolates under influence of FS-1 may indicate an accumulation of random mutations causing a general metabolic retardation. Indeed, on average it took longer to mature the isolates on the late stages of the treatment with FS-1; however, the growth rate results were inconsistent. Such strains could persist for longer in the treated animals without being able to maintain the infection, while in the animals treated solely by CAA the selected drug resistant variants caused recurrences of the disease (Figure 1). Counter-selection of the drug resistant variants from the population may be associated with the fitness cost aggravation by FS-1 (Ilin et al., 2017). It was reported that the drug resistant mutants were more sensitive to oxidative stress (Cohen and Murray, 2004). Under the combinatorial action of the antibiotics and FS-1, the XDR-TB population failed to find genetic variants fitting both selective pressures. Late isolates appeared to be more susceptible to the antibiotics and to FS-1. To the best of our knowledge, this work was the first experimental model to study the drug induced antibiotic resistance reversion by the induced synergy mechanism that had been predicted theoretically in a recent publication (Baym et al., 2016). Application of drug resistance reversion induced in clinics may allow reducing the frequency of side effect complications by shortening the treatment course of the standard antibiotics and it will improve the efficacy of curing of XDR-TB infection that currently is below a 20% success rate (Pietersen et al., 2014).
Author Contributions
AI and MK planning of experiments and general managements; RI, GA, and ML work with M. tuberculosis isolates, biochemical experiments; RI work with laboratory animals; IK extraction of DNA and sequencing; OR bioinformatics support; AI, RI, ML, and OR preparation and edition of the manuscript.
Funding
The research was funded by the grant 0115PK00389 provided by the Ministry for investments and development of Kazakhstan. Bioinformatic analysis was funded by the grant #93664 provided for ONR by the National Research Foundation (NRF) of South Africa.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to Dr. Rian Pierneef (Centre for Bioinformatics and Computational Biology, Department of Biochemistry, University of Pretoria, Pretoria, South Africa) for reviewing and correcting the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fcimb.2017.00151/full#supplementary-material
Abbreviations
MDR-TB, multidrug resistant tuberculosis; CAA, complex of anti-tuberculosis antibiotics; PDIM, phthiocerol dimycocerosate; SCAID, Scientific Center for Anti-Infectious Drugs; XDR-TB, extensively drug resistant tuberculosis.
References
Awofeso, N. (2008). Anti-tuberculosis medication side-effects constitute major factor for poor adherence to tuberculosis treatment. Bull. World Health Organ. 86, B–D. doi: 10.2471/blt.07.043802
Azad, A. K., Sirakova, T. D., Fernandes, N. D., and Kolattukudy, P. E. (1997). Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J. Biol. Chem. 272, 16741–16745. doi: 10.1074/jbc.272.27.16741
Banu, S., Honore, N., Saint-Joanis, B., Philpott, D., Prevost, M. C., and Cole, S. T. (2002). Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol. Microbiol. 44, 9–19. doi: 10.1046/j.1365-2958.2002.02813.x
Baym, M., Stone, L. K., and Kishony, R. (2016). Multidrug evolutionary strategy to reverse antibiotic resistance. Science 351:aad3292. doi: 10.1126/science.aad3292
Camacho, L. R., Constant, P., Raynaud, C., Laneelle, M. A., Triccas, J. A., Gicquel, B., et al. (2001). Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276, 19845–19854. doi: 10.1074/jbc.M100662200
Cawello, W. (1999). Parameters for Compartment Free Pharmacokinetics: Standardisation of Study Design, Data Analysis and Reporting. Aachen: Shaker Verlag.
Cohen, T., and Murray, M. (2004). Modeling epidemics of multidrug-resistant M. tuberculosis of heterogeneous fitness. Nat. Med. 10, 1117–1121. doi: 10.1038/nm1110
Cohen, T., Sommers, B., and Murray, M. (2003). The effect of drug resistance on the fitness of Mycobacterium tuberculosis. Lancet Infect. Dis. 3, 13–21. doi: 10.1016/S1473-3099(03)00483-3
Cohn, D. L., Bustreo, F., and Raviglione, M. C. (1997). Drug-resistant tuberculosis: review of the worldwide situation and the WHO/IUATLD Global Surveillance Project. Clin. Infect. Dis. 24, S121–S130. doi: 10.1093/clinids/24.Supplement_1.S121
Finken, M., Kirschner, P., Meier, A., Wrede, A., and Böttger, E. C. (1993). Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 9, 1239–1246. doi: 10.1111/j.1365-2958.1993.tb01253.x
Fishbein, S., van Wyk, N., Warren, R. M., and Sampson, S. L. (2015). Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol. Microbiol. 96, 901–916. doi: 10.1111/mmi.12981
Gagneux, S., Long, C. D., Small, P. M., Van, T., Schoolnik, G. K., and Bohannan, B. J. (2006). The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312, 1944–1946. doi: 10.1126/science.1124410
Guerrini, V., Subbian, S., Santucci, P., Canaan, S., Gennaro, M. L., and Pozzi, G. (2016). Experimental evolution of Mycobacterium tuberculosis in human macrophages results in low-frequency mutations not associated with selective advantage. PLoS ONE 11:e0167989. doi: 10.1371/journal.pone.0167989
Gupta, R., Geiter, L. J., Wells, C. D., Gao, M., Cirule, A., and Xiao, H. (2015). Delamanid for extensively drug-resistant tuberculosis. N. Engl. J. Med. 373, 291–292. doi: 10.1056/NEJMc1415332
Heusipp, G., Fälker, S., and Schmidt, M. A. (2007). DNA adenine methylation and bacterial pathogenesis. Int. J. Med. Microbiol. 297, 1–7. doi: 10.1016/j.ijmm.2006.10.002
Heym, B., Alzari, P. M., Honoré, N., and Cole, S. T. (1995). Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 15, 235–245. doi: 10.1111/j.1365-2958.1995.tb02238.x
Hoffmann, H., Kohl, T. A., Hofmann-Thiel, S., Merker, M., Beckert, P., Jaton, K., et al. (2016). Delamanid and Bedaquiline resistance in Mycobacterium tuberculosis ancestral Beijing genotype causing extensively drug-resistant tuberculosis in a Tibetan Refugee. Am. J. Respir. Crit. Care Med. 193, 337–340. doi: 10.1164/rccm.201502-0372LE
Ilin, A. I., and Kulmanov, M. E. (2014). Antibacterial Agent for Treating Infectious Diseases of Bacterial Origin. Patent US20140010782 A1—2014-09-03. Available online at: https://www.google.com/patents/US20140010782
Ilin, A. I., Kulmanov, M. E., Korotetskiy, I. S., Akhmetova, G. K., Lankina, M. V., Shvidko, S. V., et al. (2015). Complete genome sequence of the multidrug- resistant clinical isolate Mycobacterium tuberculosis 187.0, used to study an effect of drug susceptibility reversion by a new medicinal drug FS-1. Genome Announc. 3:e01272-15. doi: 10.1128/genomeA.01272-15
Ilin, A. I., Kulmanov, M. E., Korotetskiy, I. S., Lankina, M. V., Akhmetova, G. K., Shvidko, S. V., et al. (2017). Constraints of drug resistance in Mycobacterium tuberculosis–prospects for pharmacological reversion of susceptibility to antibiotics. Open Conf. Proc. J. 8, 33–43. doi: 10.2174/2210289201708010033
Iseman, M. D. (1993). Treatment of multidrug-resistant tuberculosis. N. Engl. J. Med. 329, 784–791. doi: 10.1056/NEJM199309093291108
Kalykova, A., Kustova, T., Sakipova, Z., Ibragimova, N., Islamov, R., Vetchý, D., et al. (2016). Acute and subchronic toxicity studies of the original drug FS-1. Acta Vet. Brno 85, 9–16. doi: 10.2754/avb201685010009
Kapur, V., Li, L. L., Hamrick, M. R., Plikaytis, B. B., Shinnick, T. M., Telenti, A., et al. (1999). Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch. Pathol. Lab. Med. 119, 131–138.
Krüüner, A., Yates, M. D., and Drobniewski, F. A. (2006). Evaluation of MGIT 960-based antimicrobial testing and determination of critical concentrations of first- and second-line antimicrobial drugs with drug-resistant clinical strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 44, 811–818. doi: 10.1128/JCM.44.3.811-818.2006
Luciani, F., Sisson, S. A., Jiang, H., Francis, A. R., and Tanaka, M. M. (2009). The epidemiological fitness cost of drug resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 106, 14711–14715. doi: 10.1073/pnas.0902437106
Mahajan, R. (2013). Bedaquiline: first FDA-approved tuberculosis drug in 40 years. Int. J. Appl. Basic Med. Res. 3, 1–2. doi: 10.4103/2229-516X.112228
Morens, D. M., Folkers, G. K., and Fauci, A. S. (2004). The challenge of emerging and re-emerging infectious diseases. Nature 430, 242–249. doi: 10.1038/nature02759
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
National Research Council (2010). Guide for the Care and Use of Laboratory Animals, 8th Edn. Washington, DC: The National Academy Press.
Nouvel, L. X., Kassa-Kelembho, E., Vultos, T. D., Zandanga, G., Rauzier, J., Lafoz, C., et al. (2006). Multidrug-resistant Mycobacterium tuberculosis, Bangui, Central African Republic. Emerg. Infect. Dis. 12, 1454–1456. doi: 10.3201/eid1209.060361
Okamoto, S., Tamaru, A., Nakajima, C., Nishimura, K., Tanaka, Y., Tokuyama, S., et al. (2007). Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 63, 1096–1106. doi: 10.1111/j.1365-2958.2006.05585.x
Piddock, L. J. V. (2012). The crisis of no new antibiotics – what is the way forward? Lancet Infect. Dis. 12, 249–253. doi: 10.1016/S1473-3099(11)70316-4
Pietersen, E., Ignatius, E., Streicher, E. M., Mastrapa, B., Padanilam, X., Pooran, A., et al. (2014). Long-term outcomes of patients with extensively drug-resistant tuberculosis in South Africa: a cohort study. Lancet 383, 1230–1239. doi: 10.1016/S0140-6736(13)62675-6
Pym, A. S., Saint-Joanis, B., and Cole, S. T. (2002). Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70, 4955–4960. doi: 10.1128/IAI.70.9.4955-4960.2002
Ramaswamy, S. V., Reich, R., Dou, S. J., Jasperse, L., Pan, X., Wanger, A., et al. (2003). Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47, 1241–1250. doi: 10.1128/AAC.47.4.1241-1250.2003
Reading, C., and Cole, M. (1977). Clavulanic acid: a beta-lactamase-inhiting beta-lactam from Streptomyces clavuligerus. Antimicrob. Agents Chemother. 11, 852–857. doi: 10.1128/AAC.11.5.852
Rodrigues, L., Villellas, C., Bailo, R., Viveiros, M., and Aínsa, J. A. (2013). Role of the Mmr efflux pump in drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 751–757. doi: 10.1128/AAC.01482-12
Sandgren, A., Strong, M., Muthukrishnan, P., Weiner, B. K., Church, G. M., and Murray, M. B. (2009). Tuberculosis drug resistance mutation database. PLoS Med. 6:e1000002. doi: 10.1371/journal.pmed.1000002
Scorpio, A., and Zhang, Y. (1996). Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2, 662–667. doi: 10.1038/nm0696-662
Segal, W., and Bloch, H. (1956). Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 72, 132–141.
Spellberg, B., Guidos, R., Gilbert, D., Bradley, J., Boucher, H. W., Scheld, W. M. Jr., et al. (2008). Infectious diseases society of America. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the infectious diseases society of America. Clin. Infect. Dis. 46, 155–164. doi: 10.1086/524891
Sreevatsan, S., Stockbauer, K. E., Pan, X., Kreiswirth, B. N., Moghazeh, S. L., Jacobs, W. R. Jr., et al. (1997). Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob. Agents Chemother. 41, 1677–1681.
Tenover, F. C. (2001). Development and spread of bacterial resistance to antimicrobial agents: an overview. Clin. Infect. Dis. 33, S108–S115. doi: 10.1086/321834
Torrey, H. L., Keren, I., Via, L. E., Lee, J. S., and Lewis, K. (2016). High persister mutants in Mycobacterium tuberculosis. PLoS ONE 11:e0155127. doi: 10.1371/journal.pone.0155127
van den Boogaard, J., Kibiki, G. S., Kisanga, E. R., Boeree, M. J., and Aarnoutse, R. E. (2009). New drugs against tuberculosis: problems, progress, and evaluation of agents in clinical development. Antimicrob. Agents Chemother. 53, 849–862. doi: 10.1128/AAC.00749-08
Yu, J., Tran, V., Li, M., Huang, X., Niu, C., Wang, D., et al. (2012). Both phthiocerol dimycocerosates and phenolic glycolipids are required for virulence of Mycobacterium marinum. Infect. Immun. 80, 1381–1389. doi: 10.1128/IAI.06370-11
Keywords: Mycobacterium tuberculosis, drug resistance reversion, new drug, next generation sequencing, genome polymorphism
Citation: Ilin AI, Kulmanov ME, Korotetskiy IS, Islamov RA, Akhmetova GK, Lankina MV and Reva ON (2017) Genomic Insight into Mechanisms of Reversion of Antibiotic Resistance in Multidrug Resistant Mycobacterium tuberculosis Induced by a Nanomolecular Iodine-Containing Complex FS-1. Front. Cell. Infect. Microbiol. 7:151. doi: 10.3389/fcimb.2017.00151
Received: 29 January 2017; Accepted: 11 April 2017;
Published: 08 May 2017.
Edited by:
Fabiana BIGI, Instituto Nacional de Tecnología Agropecuaria (INTA), ArgentinaReviewed by:
Paul M. Coussens, Michigan State University, USAMax Maurin, Université Grenoble Alpes, France
Copyright © 2017 Ilin, Kulmanov, Korotetskiy, Islamov, Akhmetova, Lankina and Reva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleksandr I. Ilin, ilin_ai@mail.ru
Oleg N. Reva, oleg.reva@up.ac.za
 Aleksandr I. Ilin
Aleksandr I. Ilin Murat E. Kulmanov1
Murat E. Kulmanov1  Ilya S. Korotetskiy
Ilya S. Korotetskiy Oleg N. Reva
Oleg N. Reva